Abstract
While CD4 cell counts are widely used to predict disease progression in human immunodeficiency virus (HIV)-infected patients, they are poorly explanatory of the progression to AIDS or death after the introduction of chemotherapy. Changes in HIV load (as measured by RNA PCR) have been shown to be a much better predictor of the risk of disease progression. Since the interrelationship of these markers is of great clinical interest, we modeled the time-averaged return of CD4 cell count and change in viral load subsequent to therapy with the HIV protease inhibitor indinavir. We found that CD4 cell return was significantly related to both the baseline CD4 count (r2 = 0.86, P < 0.001) and the decline in HIV RNA PCR-determined viral load (also referred to in this work as the HIV RNA PCR decline) (r2 = 0.60, P < 0.01). Simultaneously modeling both influences in a linked nonlinear model (r2 = 0.93, P < 0.001) demonstrated that (i) the starting number of CD4 cells accounted for the majority of the change in CD4 cell return and (ii) the return of CD4 cells attributable to viral load decrease was 50% of maximal with only a decrease of approximately 0.2 log of HIV RNA as modeled from the first 12 weeks of therapy. Much greater viral inhibition beyond that necessary for maximal CD4 cell return is possible. Given that HIV RNA PCR decline is more strongly linked to ultimate clinical course in HIV disease, our findings indicate that CD4 return is potentially misleading as an indicator of antiviral effect, since it is determined more by the starting CD4 value than by viral load decline and since near-maximal changes occur with minimal antiviral effect.
While CD4 cell counts are widely used to predict disease progression in human immunodeficiency virus (HIV)-infected patients, they are variable and poorly explanatory of the progression to AIDS or death after the introduction of chemotherapy (17). Despite these limitations, CD4 cell counts have been employed by the Food and Drug Administration as a surrogate marker to provide evidence of therapeutic agent effectiveness.
Recently, a number of investigations have shown that HIV RNA PCR determination is an excellent predictor of prognosis for patients infected with the HIV (7, 10). Perhaps even more importantly, O’Brien and colleagues (13) demonstrated that the change in HIV load as measured by RNA PCR after antiretroviral chemotherapy was significantly linked to the risk of subsequent progression and/or death in subjects who did or did not receive zidovudine.
As HIV RNA PCR-determined viral load at baseline and its change with antiretroviral intervention have been shown to be a much better surrogate marker, the following questions arise: what is its relationship to CD4 cell count changes induced by therapy and how much antiviral effect is needed to induce these effects? In order to answer these questions, we examined the change in the number of HIV RNA PCR copies/ml and the change in CD4 cell count subsequent to initiation of protease inhibitor therapy to determine if there was a relationship between viral load change and CD4 cell return.
MATERIALS AND METHODS
For the interrelationship between viral load and changes in CD4 cell counts, we examined the viral load data available for 14 of the 15 patients we had previously investigated for CD4 cell changes, turnover, and half-life determinations after treatment with the HIV protease inhibitor indinavir (15). Neither virologic data nor its interrelationship with CD4 cell count changes was analyzed in that report. Clinical data from five of these patients have been previously reported (16). For the subjects in this analysis, the average baseline CD4 cell count ranged from 14 to 345 cells/μl and the baseline number of copies of log10 HIV RNA determined by PCR ranged from 4.45 to 5.35. The doses of indinavir used all had similar antiviral activity and ranged from 600 to 800 mg every 6 h (q6h) and 800 to 1,000 mg q8h (14, 16). As previously described (15), CD4 cell counts were obtained every 2 weeks for 12 weeks and then either every 2 or 4 weeks for 24 weeks. The average number of CD4 cells over the 24-week interval was calculated by determining the area under the CD4-time curve to week 24, without extrapolation, by employing the LAGRAN program of Rocci and Jusko (13a). This value was then divided by 24, providing the time-averaged CD4 cell count over 24 weeks. The baseline value was the mean of two independent determinations. Screening values for CD4 and viral load were not included because of a potential regression to the mean effect. The baseline value served as the independent variable in a sigmoid-Emax effect model analysis, where the 24-week average CD4 cell count was the dependent variable. Sigmoidal relationships are the classical relationships seen in pharmacologic interventions. This fits the biology of the model processes, which are at steady state until the changes induced by the protease inhibitor, and there is a maximal-effect limit to the relationship (e.g., CD4 cell counts cannot exceed normal range and HIV RNA cannot be detected below some value). As an example, the general form of a sigmoid-Emax equation adapted for evaluation of CD4 return is Return = Emax ∗ StartH/(StartH + Start50H) where the Emax is the maximal effect, Start is the baseline or starting CD4 lymphocyte count, Start50 is the starting CD4 lymphocyte count at which 50% of the maximal effect occurs, and H is the sigmoidicity. The modeling process was performed by employing the ADAPT II package of programs of D’Argenio and Schumitzky (3a), a package of nonlinear least-squares regression programs (Biomedical Simulations Resource, University of Southern California, Los Angeles, Calif.). The amount of the variance explained by the regression (r2) is as calculated by ADAPT II. The P values, adjusted for the appropriate degrees of freedom, are determined from the correlation coefficient (r) and are two sided.
In order to obtain an estimate of the effect of antiviral chemotherapy upon viral load, we modeled all the viral load data from baseline through week 12. HIV RNA PCR was determined at the baseline and on a biweekly basis through week 12. The determination was performed by the Roche AMPLICOR assay, which has a lower limit of sensitivity of 200 copies/ml. Data after week 12 were not examined because the potential emergence of resistance of virus to indinavir could confound the analysis. CD4 data were included to week 24 because of the longer half-life of CD4 cells compared to that of HIV RNA and the persistence of CD4 changes even after HIV RNA returned to the baseline (14, 16). The model system employed recognized two separate compartments of viral replication: a lymph node compartment, which was not sampled, and the sampled blood compartment. The two compartments were linked by first-order transfer rate constants, and a first-order clearance term removed virus from the lymph node compartment. Two viral generation rates are employed, one in the absence and one in the presence of protease inhibitor. The generation rate in the absence of protease inhibitor is arbitrarily fixed to 1, so that the generation rate in the presence of inhibitor represents the relative downturn in generation rate induced by the protease inhibitor. The rates are turned off and on by piecewise input functions based on the time of treatment initiation. The differential equations employed for the modeling process have been previously published (16). The log10 of the generation rate in the presence of inhibitor represents the log drop in HIV RNA PCR determination from one steady state to the next (baseline to the new steady state induced by the protease inhibitor). This value was used as the viral load change induced by the protease inhibitor in further analyses. With this estimate of viral load decline, we again employed a sigmoid-Emax model to link the HIV RNA PCR-determined viral load change (also referred to in this work as HIV RNA PCR change) and the time-averaged return of CD4 cells, with viral load decline being the independent variable. We also evaluated a two-independent-variable model, with baseline CD4 cell count and modeled HIV RNA PCR decline serving as the independent variables. The model employed was two linked sigmoid-Emax models. This was parameterized so that the fraction of the maximal CD4 cell return attributable to each variable could be determined.
RESULTS
As expected from the analysis with 15 patients, in these 14 subjects the initial CD4 cell count exerted a major effect upon the numbers of CD4 cells returning with therapy (r2 = 0.86, P < 0.001) up to a maximal value. The viral load decline seen in our patients, as well as the calculated (from the model parameters—intercompartmental transfer rate constants and clearance constant) viral generation half time, is presented in Table 1. As can be seen, the viral generation half time averages 3.0 days, with a median of 2.5 days and a range out to almost 7 days. The viral load drop averaged 2.56 log10, with a median of 2.71 log10. The range varied from a net increase in viral load to a 4.70 log10 decline.
TABLE 1.
Results of two-compartment modeling of the effect of indinavir on viral productiona
| Statistical parameter | Indinavir-induced downturn in viral generation (log10) | Viral generation half-life (days) |
|---|---|---|
| Mean | 2.56 | 3.0 |
| SD | 1.34 | 1.7 |
| Median | 2.71 | 2.5 |
| Range | −0.42–4.7 | 1.4–6.4 |
Based on two-compartment modeling of 14 subjects performed by using simultaneous inhomogenous differential equations. Half-lifes were calculated by using the rate constants generated from the solution of the equations and the standard formula for half-life.
To examine the issue of whether HIV RNA PCR change also influenced CD4 cell count, we performed another sigmoid-Emax analysis, this time employing the modeled HIV RNA PCR decline as the independent variable and time-averaged CD4 cell count as the dependent variable by using nonlinear regression in the ADAPT II program package. HIV RNA PCR change with protease inhibitor administration was significantly correlated with the time-averaged CD4 cell count (r2 = 0.60, P < 0.01). The modeled HIV RNA decline using 12 weeks of data associated with 50% of the maximal amount of time-averaged CD4 cell return was only 0.2 log. The sigmoidicity (steepness of the curve) identified was very large (>21). This indicates that essentially all CD4 cell return attributable to a drop in HIV RNA PCR has occurred by 0.3 log unit of decline.
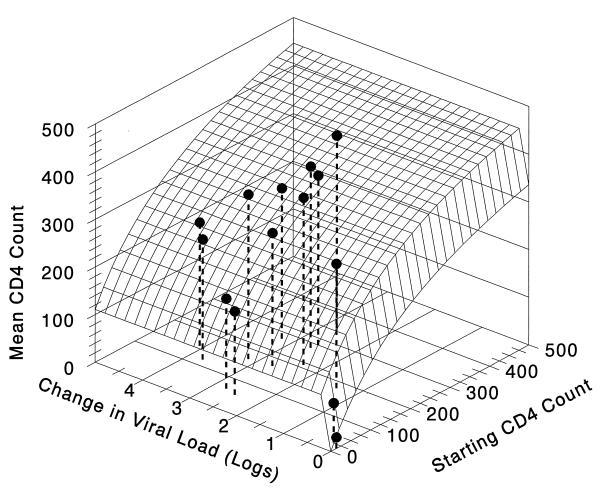
Since the two different independent variables were shown to affect time-averaged CD4 cell count, with baseline CD4 cell count being the stronger of the two influences, we then examined the interrelationship of these variables in two linked sigmoid-Emax models. The models used baseline CD4 cell count and HIV RNA PCR decline as the independent variables, with time-averaged CD4 cell count as the dependent variable. The model fit the data quite well, with an r2 of 0.93 (P < 0.001). The results of this three-dimensional relationship are displayed in Fig. 1. The surface in Fig. 1 demonstrates that the time-averaged CD4 cell count return attributable to viral load suppression rapidly achieves a near-maximal effect with little HIV RNA change. In the two-independent-variable model, the HIV RNA PCR decline that produces 50% of the maximal change in CD4 return (attributable to RNA decline) is 0.1 log10 copies/ml, with a sigmoidicity of 26 and a maximal CD4 cell return of 110 cells. On the other hand, the CD4 cell return attributable to the baseline CD4 cell count was 50% maximal at a baseline CD4 cell count of 291, with a maximal CD4 return of 535 cells/μl. Therefore, the baseline CD4 cell count still accounted for the majority of the variance explained by the relationship and the majority of the returning cells.
FIG. 1.
CD4 return with indinavir therapy and effect of RNA drop and starting CD4 count. The three-dimensional plot of CD4 cell return as a function of both baseline CD4 cell count and the modeled HIV RNA PCR decline induced by indinavir therapy is shown. The lines defining the edges of the surface are the direct two-dimensional relationships, with the response surface showing the interactive effect. It is clear that the amount of CD4 cell return attributable to viral load decline is maximal at very low values of HIV RNA PCR change. The equation for the surface is Average number of CD4 cells with therapy (cells/milliliter) = 110.3*{(log RNA decline)26.1/[(log RNA decline)26.1 + 0.09826.1]} + 534.6*{(starting number of CD4 cells)0.98/[(starting number of CD4 cells)0.98 + 291.30.98]}. The model fits the data quite well (r2 = 0.93, P < 0.001).
DISCUSSION
We have previously examined the influence of the starting CD4 count on the return of these cells induced by protease inhibitor therapy (15). In this analysis, we have been able to incorporate the effect of the decrease in HIV-1 RNA PCR-determined copy number on CD4 cell return and also to build a combined model of both RNA PCR-determined copy number change along with baseline CD4 cell count. Our analysis indicates that only small changes in viral load account for maximal changes in CD4 cell count return after initiation of a protease inhibitor. The starting CD4 cell count explained the majority of the change in CD4 cell count induced by the antiviral effect of the protease inhibitor. We feel that this is likely to reflect a cell reserve problem, with later-stage patients demonstrating a smaller number of cells with which to repopulate. Previous investigations of ours (15) as well as others (9) demonstrate that CD4 cell replication is quite active. CD4 cell numbers are a balance between rapid turnover and rapid, unchecked virally mediated destruction. When the virally mediated destruction is checked by protease inhibitor administration in late-stage patients (e.g., <50 cells/μl), it is likely that, although the CD4 cell turnover is rapid, there is an insufficient number of them to allow large changes in total CD4 cell numbers.
The relationships demonstrated in these analyses between CD4 cell return and changes in viral load are consistent with clinical trial data. Meng et al. (11), examining the effects of zidovudine dosages of 50 mg q8h, demonstrated a decreased CD4 cell return, relative to concurrent treatments, but also relative to other similar historical groups receiving larger doses of zidovudine. However, once the dose of zidovudine increases past 300 mg/day up to 1,500 mg/day (3, 5, 12, 18), no further dose dependence is seen with regard to CD4 cell return. Clearly, the decline in HIV RNA would be expected to be quite low with a dose of 50 mg of zidovudine q8h. Data from O’Brien et al. (13) indicated that the average HIV RNA PCR drop was 0.6 log for a 1,500-mg/day dose of zidovudine. Further, with protease inhibitor (indinavir)-nucleoside combination trials, patients receiving combination therapy over the first 24 weeks did not have CD4 cell counts which were different from those of patients receiving indinavir alone, even though changes in viral load were greater and more prolonged (package inset for Crixivan; Merck, Inc.). Why protease inhibitors as a class appear to give greater CD4 cell returns for the degree of antiviral effect is unknown. One could speculate that this is due to the effect on viral load in the lymphoid compartment by the protease inhibitors which is not seen with nucleosides (1, 2, 8, 20).
The values reported in this work for viral half-life are slightly longer and somewhat more variable than those reported initially by Wei et al. (19) and Ho et al. (9), who used a one-compartment model. Our two-compartment model is closer to the physiologic realities than a log-linear or linear one-compartment model. In a two-compartment model, the unmeasured noncirculating compartment is considered in the analysis and a steady state-to-steady state change is modeled, while prior one-compartment models require the changes observed to continue unchanged to zero viral load and assume complete blockage of new virion production. In addition, we examined more patients demonstrating broader arrays of antiviral effect from the protease inhibitor than in earlier studies. Despite these differences in methodology, the implications of each model are similar, even with the disparities in calculated viral generation half-lives.
The data from our analyses also provide insight into why HIV RNA PCR change is a better surrogate marker for HIV disease, particularly in cases with antiretroviral intervention. Clearly, two patients could have starting CD4 counts of 100, but one could have a viral load decline of 1.0 log which took 12 weeks to return to the baseline and the other could have a viral load decline of 3 log units which took 48 weeks to return to baseline. In both instances, the same amplitude of CD4 cell return would result. However, in one patient, viral replication is under much better control relative to the other patient (viral generation for the second patient at maximal effect is 1/100 that of the first patient). Indeed, if one subscribes to the models of Frost and McLean (6) or De Jong et al. (4), even though two patients had a return of CD4 cell numbers to the same level, the patient with the increase in CD4 cells would be much more at risk to be attacked by the virus under less-tight control, leading to a more-rapid decline in the cellular gain, ultimately returning to the CD4 baseline more rapidly and placing the patient at increased risk of opportunistic infection and death. If these models are correct, the deeper and longer the HIV RNA PCR drop, the longer the CD4 cell return will last and the relative risk of the patient for opportunistic infection and/or death will be less in any defined time frame.
In summary, modeling of time-averaged CD4 cell return after initiation of protease inhibitor therapy demonstrated that the cellular numbers over the first 24 weeks of drug administration are related to both baseline CD4 cell count and the size of the change in HIV RNA PCR induced by the drug. The CD4 cell return attributable to viral load decrease maximizes quickly, allowing patients with very different viral load changes to have essentially the same number of CD4 cells return in the short term. However, these may merely represent new targets for the virus under less stringent control by the protease inhibitor. Consequently, less ultimate good for the patient in terms of progression, survivorship benefit, or outgrowth of resistant virus may occur. Therefore, in terms of explaining the benefit accruing to patients with antiretroviral chemotherapy, it is not surprising that viral load change explains more of the benefit of antiretroviral chemotherapy (is a better surrogate marker) than does CD4 cell count. Our results indicate that CD4 cell return as an indicator of the clinical activity of an antiviral therapy is misleading, since it is determined more by the starting CD4 value than viral load change and large increases can occur with minimal antiviral effect. Another implication of our results is that the use of CD4 cell return by the Food and Drug Administration in approval of antivirals may be suboptimal. Whether the same degree of CD4 return compared to that of decline in HIV RNA will occur with other antiviral agents remains to be investigated.
ACKNOWLEDGMENT
This study was supported in part by NIH grant NO1-AI-15104-015.
REFERENCES
- 1.Cavert W, Staskus K, Zupanic M, Wietgrefe S, Notermans D, Danner S, Henry K, Mills R, Haase A T. Abstracts of the 4th Conference on Retroviruses and Opportunistic Infections, Washington, D.C. 1997. Quantitative in situ hybridization measurement of HIV-1 RNA clearance kinetics from lymphoid tissue cellular compartments during triple drug therapy, abstr. LB9. [Google Scholar]
- 2.Cohen O J, Pantaleo G, Holodniy M, Fox C H, Orenstein J M, Schnittman S, Niu M, Graziosi C, Pavlakis G N, Lalezari J, Bartlett J A, Steigbigel R T, Cohn J, Novak R, McMahon D, Bilello J, Fauci A S. Antiretroviral monotherapy in early stage human immunodeficiency virus disease has no detectable effect on virus load in peripheral blood and lymph nodes. J Infect Dis. 1996;173:849–856. doi: 10.1093/infdis/173.4.849. [DOI] [PubMed] [Google Scholar]
- 3.Collier A C, Bozzette S, Coombs R W, Causey D M, Schoenfeld D A, Spector S A, Petinelli C B, Daivies G, Richman D D, Leedom J M, Kidd P, Corey L. A pilot study of low-dose zidovudine in human immunodeficiency virus infection. N Engl J Med. 1990;323:1015–1021. doi: 10.1056/NEJM199010113231502. [DOI] [PubMed] [Google Scholar]
- 3a.D’Argenio D Z, Schumitzky A. ADAPT II user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Los Angeles, Calif: Biomedical Simulations Resource; 1997. [Google Scholar]
- 4.De Jong M D, Veenstra J, Stilianakis N I, Schuurman R, Lange J M A, DeBoer R J, Boucher C A B. Host-parasite dynamics and outgrowth of virus containing a single K70R amino acid change in reverse transcriptase are responsible for the loss of human immunodeficiency virus type 1 RNA load suppression by zidovudine. Proc Natl Acad Sci USA. 1996;93:5501–5506. doi: 10.1073/pnas.93.11.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischl M A, Parker C B, Pettinelli C, Wulfsohn M, Hirsch M S, Collier A C, Antoniskis D, Ho M, Richman D D, Fuchs E, Merigan T C, Reichman R C, Gold J, Steigbigel N, Leoung G S, Rasheed S, Tsiatis A the AIDS Clinical Trial Group. A randomized conrolled trial of a reduced daily dose of zidovudine in patients with the acquired immunodeficiency syndrome. N Engl J Med. 1990;323:1008–1014. doi: 10.1056/NEJM199010113231501. [DOI] [PubMed] [Google Scholar]
- 6.Frost S D, McLean A R. Quasispecies dynamics and the emergence of drug resistance during zidovudine therapy of HIV infection. AIDS. 1994;8:323–332. doi: 10.1097/00002030-199403000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Galetto-Lacour A, Yerly S, Perneger T V, Baumberger C, Hirschel B, Perrin L the Swiss HIV Cohort Study Group. Prognostic value of viremia in patients with long standing human immunodeficiency virus infection. J Infect Dis. 1996;173:1388–1393. doi: 10.1093/infdis/173.6.1388. [DOI] [PubMed] [Google Scholar]
- 8.Haase A T, Henry K, Zupancic M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z-Q, Dailey P J, Balfour H H, Jr, Erice A, Perelson A S. Quantitative analysis of HIV-1 infection in lymphoid tissue. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 9.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 10.Mellors J W, Rinaldo C R, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 11.Meng T C, Fischl M A, Boota A M, Spector S A, Bennett D, Bassiakos Y, Lai S, Wright B, Richman D D. Combination therapy with zidovudine and dideoxycytidine in patients with advanced human immunodeficiency virus infection. Ann Intern Med. 1992;116:13–20. doi: 10.7326/0003-4819-116-1-13. [DOI] [PubMed] [Google Scholar]
- 12.Nordic Medical Research Councils’ HIV Therapy Group. Double blind dose-response study of zidovudine in AIDS and advanced HIV infection. Br Med J. 1992;304:13–17. doi: 10.1136/bmj.304.6818.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Brien W A, Hartigan P M, Martin D, Esenhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D the Veterans Affairs Cooperative Study Group on AIDS. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 13a.Rocci M L, Jr, Jusko W J. LAGRAN program for area and moments in pharmacokinetic analysis. Computer Programs in Biomedicine. 1983;16:203–216. doi: 10.1016/0010-468x(83)90082-x. [DOI] [PubMed] [Google Scholar]
- 14.Stein, D., G. Drusano, R. Steigbigel, P. Berry, J. Mellors, D. McMahon, H. Teppler, C. Hildebrand, M. Nessly, and J. Chodakewitz. Two year followup of patients treated with indinavir 800 mg q8h, abstr. 195. In Abstracts of the 4th Conference on Retroviruses and Opportunistic Infections, Washington, D.C.
- 15.Stein D S, Drusano G L. Modeling of the change in CD4 lymphocyte cell counts in patients before and after administration of the human immunodeficiency virus protease inhibitor indinavir. Antimicrob Agents Chemother. 1997;41:449–453. doi: 10.1128/aac.41.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein D S, Fish D G, Bilello J A, Preston S L, Martineau G L, Drusano G L. A 24 week open label phase I/II evaluation of the HIV protease inhibitor MK-639 (indinavir) AIDS. 1996;10:485–492. doi: 10.1097/00002030-199605000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Stein D S, Korvick J A, Vermund S H. CD4 lymphocyte cell enumeration for prediction of clinical course of HIV disease: a review. J Infect Dis. 1992;165:352–363. doi: 10.1093/infdis/165.2.352. [DOI] [PubMed] [Google Scholar]
- 18.Volberding P A, Lagakos S W, Koch M A, Pettinelli C, Myers M W, Booth D K, Balfour H H, Jr, Reichman R C, Bartlett J A, Hirsch M S, Murphy R L, Hardy W D, Soeiro R, Fischl M A, Bartlett J G, Merigan T C, Hyslop N E, Richman D D, Valentine F T, Corey L the AIDS Clinical Trials Group of the National Institutes of Health. Zidovudine in asymptomatic human immunodeficiency virus infection. N Engl J Med. 1990;322:941–949. doi: 10.1056/NEJM199004053221401. [DOI] [PubMed] [Google Scholar]
- 19.Wei X, Ghosh S K, Taylor M E, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 20.Wong J K, Gunthard H F, Havlir D V, et al. Abstracts of the 4th Conference on Retroviruses and Opportunistic Infections, Washington, D.C. 1997. Reduction of HIV in blood and lymph nodes after potent antiretroviral therapy, abstr. LB10. [Google Scholar]