Abstract
The gut microbiome is recognized as a key modulator of sepsis development. However, the contribution of the gut mycobiome to sepsis development is still not fully understood. Here, we demonstrated that the level of Candida albicans was markedly decreased in patients with bacterial sepsis, and the supernatant of Candida albicans culture significantly decreased the bacterial load and improved sepsis symptoms in both cecum ligation and puncture (CLP)-challenged mice and Escherichia coli-challenged pigs. Integrative metabolomics and the genetic engineering of fungi revealed that Candida albicans-derived phenylpyruvate (PPA) enhanced the bactericidal activity of macrophages and reduced organ damage during sepsis. Mechanistically, PPA directly binds to sirtuin 2 (SIRT2) and increases reactive oxygen species (ROS) production for eventual bacterial clearance. Importantly, PPA enhanced the bacterial clearance capacity of macrophages in sepsis patients and was inversely correlated with the severity of sepsis in patients. Our findings highlight the crucial contribution of commensal fungi to bacterial disease modulation and expand our understanding of the host-mycobiome interaction during sepsis development.
Keywords: Candida albicans, Phenylpyruvate, Sepsis, Macrophage, Bacterial clearance
Subject terms: Infection, Antimicrobial responses
Introduction
Sepsis, characterized by a dysregulated inflammatory response to a bacterial infection and clearance, is defined as a life-threatening organ dysfunction syndrome [1]. Despite the significantly increased awareness of the pathological and molecular basis of sepsis, as well as many advances in its diagnosis and treatment, the mortality rate of sepsis remains approximately 25–30% and may even increase to 40–50% when including the consequences of septic shock [2–4]. To combat sepsis progression, novel treatment strategies are being designed to enhance host defenses for the timely clearance of bacteria.
Macrophages, a significant contributor to the host defense against microbial infections, recognize microbial pathogens and secrete inflammatory cytokines and chemokines to facilitate the engulfment of pathogens in phagosomes [5]. In addition, the production of reactive oxygen species (ROS) begins abruptly after phagocytosis. A high level of ROS production directly contributes to the rapid and efficient killing of phagocytized pathogens [6, 7]. In macrophages, nicotinamide adenine dinucleotide phosphate (NADPH), which is regulated by sirtuin 2 (SIRT2), is a key modulator of ROS generation [8]. SIRT2 mediates deacetylation and activation of glucose-6-phosphate dehydrogenase (G6PD) to supply cytosolic NADPH to counteract the damage caused by excessive ROS [9]. Loss of SIRT2 increases bacterial phagocytosis by macrophages [10]. Hence, controlling SIRT2-mediated ROS production in macrophages is a potential therapeutic strategy for sepsis.
Accumulating evidence suggests that gut microorganisms regulate pathophysiological functions in humans [11–14]. However, the roles of gut fungi in health and disease have been underappreciated compared to those of commensal gut bacteria, and the study of resident fungal communities is still in its infancy. Intestinal fungi, including Saccharomyces cerevisiae [15], Saccharomyces boulardii [16], Malassezia [17], and Candida albicans (C. albicans) [18], may influence gut microbiota community structure, alter the generation and consumption of key metabolites, and influence host health and disease development in the gut and at peripheral sites. Our previous findings showed that C. albicans can directly promote the progression of liver diseases [19, 20], indicating its role in pathogenesis. Recently, the importance of commensal fungi-derived metabolites [21] in relation to the functions of several organs was highlighted [22–25]. However, interactions between the metabolites from gut commensal fungi and the host in the context of sepsis remain poorly understood. In this study, we systematically evaluated the impact of phenylpyruvate (PPA) derived from commensal C. albicans on sepsis development, which may provide a novel therapeutic approach to limit the severity of sepsis.
Results
A C. albicans metabolite, but not C. albicans alone, ameliorated bacterial sepsis
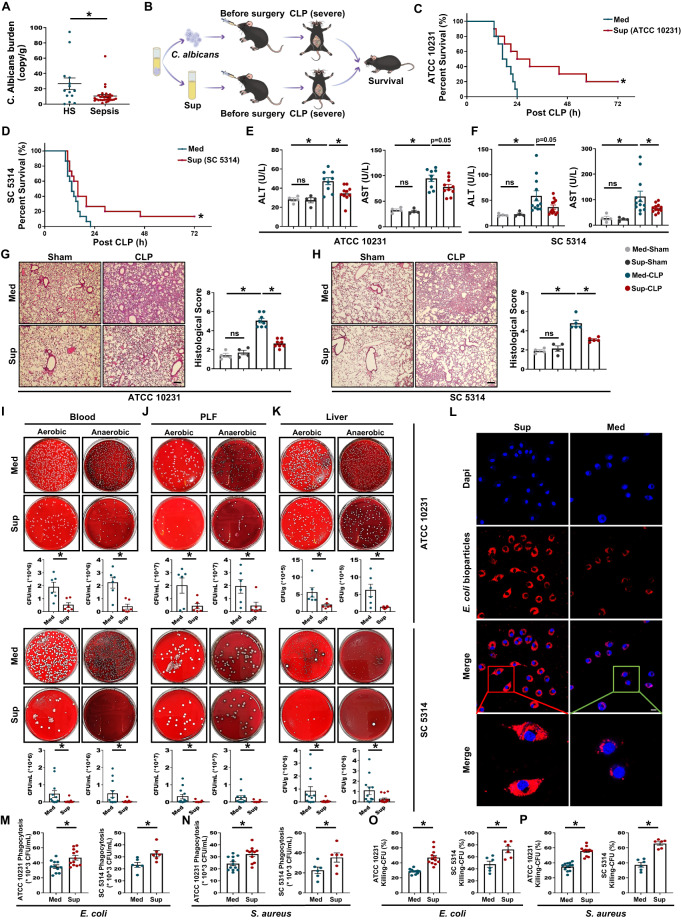
Clinicians primarily view fungi negatively as they focus on invasive infections caused by C. albicans and other fungal species. Recent studies have shown that fungal commensals provide multiple benefits in protecting against invasive infection [18]. To understand whether there was a potential correlation between C. albicans and bacterial sepsis, we first assessed the abundance of C. albicans in the feces of healthy subjects and patients with bacterial sepsis (no antifungal drugs were taken as indicated by clinical diagnosis). Unexpectedly, C. albicans was significantly decreased in bacterial sepsis patients compared to healthy subjects (Fig. 1A). C. albicans is the most common fungus in the human intestine, but not in mice [26]. To generate a murine sepsis model that more closely resembles human sepsis and to further evaluate the effect of C. albicans on bacterial sepsis, we employed the following two widely used strains of commensal C. albicans: ATCC 10231 and ATCC MYA-2876 (SC 5314). Live or heat-killed C. albicans was orally administered, and both strains significantly increased sepsis-related mortality after CLP compared to that after CLP without C. albicans administration (Figs. 1B and S1A, B), indicating the harmful effects of C. albicans. Surprisingly, administration of the supernatant (Sup) from cultures of either C. albicans strain prolonged the survival time of CLP mice compared to that of CLP mice administered blank medium (Med, Fig. 1B–D). In addition, improvement of sepsis patients was confirmed by the serum levels of ALT, AST, and cytokines (TNF-α and IL-6) at 12 h after CLP (Figs. 1E, F and S1C, D) and the diminished pathological changes in the lungs (Fig. 1G, H). Moreover, supernatant-treated mice had significantly reduced mRNA levels of TNFα, IL-1B, CXCL1, CXCL2, and CCL4 in the peritoneal lavage fluid (PLF, Fig. S1E), lung (Fig. S1F), and liver (Fig. S1G) samples compared to the medium-treated controls. These results suggest the presence of molecules that protect against sepsis in the supernatant of C. albicans culture.
Fig. 1.
Effects of gut commensal C. albicans on bacterial sepsis. A C. albicans burden in bacterial sepsis patients (n = 30) and healthy subjects (HS, n = 14) was determined using a C. albicans SYBR qPCR Kit. B Experimental design of the CLP model and pretreatments. C, D Mice received an oral gavage of 300 μL blank medium (Med), supernatant of C. albicans ATCC 10231 [Sup (ATCC 10231)] or SC 5314 [Sup (SC 5314)] 3 h before CLP. The survival rate (n = 10–15) was determined up to 72 h after CLP using the Kaplan‒Meier method with the log-rank test. E, F Serum ALT and AST levels (n = 4–12). G, H HE staining and histological score of the lungs (n = 4–8). Scale bar, 50 μm. I–K Blood, peritoneal lavage fluid (PLF), and liver samples were collected from the mice at 12 h after CLP. After the samples were serially diluted, 100 μL of each dilution was plated on Columbia blood agar plates and incubated at 37 °C for 14–16 h under aerobic or anaerobic conditions. Representative images and quantified results of bacterial load in blood (I), PLF (J) and liver (K) samples (n = 6–13). L Representative fluorescence images of medium- or supernatant (ATCC 10231)-pretreated BMDMs with pHrodo red E. coli bioparticles and blue nuclear staining (DAPI) over the course of 45 min. Scale bar, 10 μm. M, N Phagocytosis of E. coli or S. aureus by BMDMs after 3 h of pretreatment with medium or supernatant (n = 6–12). O, P Bacterial killing by BMDMs based on the gentamicin protection assay at 60 min post E. coli or S. aureus infection after 3 h of pretreatment with medium or supernatant (n = 6–12). Data are presented as the mean ± SEM. *p < 0.05 by two-tailed Student’s t test
C. albicans culture supernatant promoted bacterial clearance by macrophages
As effective and rapid bacterial clearance is a fundamental determinant of sepsis outcomes [27], we treated mice with supernatant from C. albicans culture and assessed the bacterial counts (12 h after CLP) in blood, PLF, and liver samples. Treatment with the supernatant significantly reduced the bacterial counts (Fig. 1I–K). We therefore investigated whether the supernatant modulates the intrinsic antibacterial functions of phagocytes (including neutrophils and macrophages). Bone marrow-derived macrophages (BMDMs) and neutrophils were isolated and treated with supernatant from C. albicans culture or blank medium. In neutrophils, preincubation with the supernatant did not influence bacterial phagocytosis and killing upon live Escherichia coli (E. coli, gram-negative) or Staphylococcus aureus (S. aureus, gram-positive) infection (Fig S2A, B). Next, fluorescent E. coli bioparticles, E. coli, or S. aureus were added to assess the phagocytosis capacity of the BMDMs, and there was increased phagocytosis of all three by the supernatant-treated BMDMs compared to that of the medium-treated BMDMs (Fig. 1L–N). In addition, to determine the bactericidal activity, we infected BMDMs with E. coli or S. aureus. The supernatant-treated BMDMs had significantly increased bactericidal activity after bacterial infection compared to the medium-treated BMDMs (Fig. 1O, P). These results suggest that the beneficial effect of the C. albicans culture supernatant was associated with increased macrophage bactericidal activity.
The protective effect of C. albicans culture supernatant was dependent on phenylpyruvate (PPA)
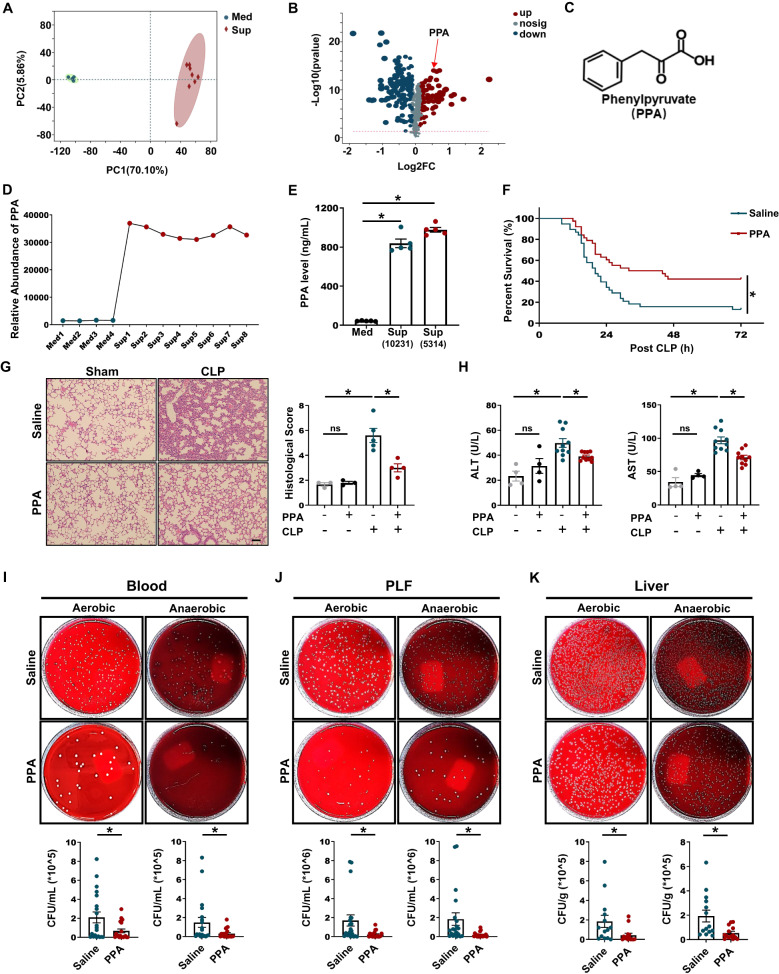
To determine which C. albicans metabolite contributes to the protective effect in CLP-induced septic mice, we performed a nontargeted metabolomics analysis of both the supernatant and blank medium. Principal component analysis (PCA) showed a prominent shift in the metabolic profile of the supernatant (Fig. 2A) compared to that of the medium. A volcano plot showed that 57 compounds were significantly enriched in the supernatant (fold change > 1.2, P < 0.001, Fig. 2B). We focused on PPA because (1) its abundance was significantly higher in the supernatant than in the medium (Fig. 2C, D), as confirmed by quantitative gas chromatography–mass spectrometry (GC/MS, Figs. 2E and S3A); (2) PPA-treated mice had a significantly longer survival time than untreated CLP mice (Fig. 2F), and multiple organ damage at 12 h after CLP (involving pathological changes in the lungs, ALT, AST, and in the expression of inflammatory factors) was also markedly decreased by PPA (Figs. 2G, H and S4A–C); and (3) the total bacterial load in blood, PLF, and liver samples was significantly reduced by PPA (Fig. 2I–K).
Fig. 2.
The C. albicans metabolite PPA protected against polymicrobial sepsis. A, B PCA scatter plots and volcano plots of metabolomics data based on blank medium and supernatant from C. albicans culture (n = 4–8). C Molecular structure of PPA. D Relative abundance of PPA detected by nontargeted metabolomics. E PPA levels in the medium and supernatant detected by GC/MS (n = 5). F–K Mice were treated with saline or 40 mg/kg PPA 3 h before CLP. F Survival rates of saline- or PPA-pretreated CLP mice were analyzed using the Kaplan‒Meier method with the log-rank test (n = 38). G HE staining and histological scores (n = 3–5). H Serum ALT and AST levels (n = 4–10) and bacterial load in the I blood, J PLF and K liver (n = 14–20) samples. Scale bars, 50 μm. Data are presented as the mean ± SEM. *p < 0.05 by two-tailed Student’s t test. ns not significant
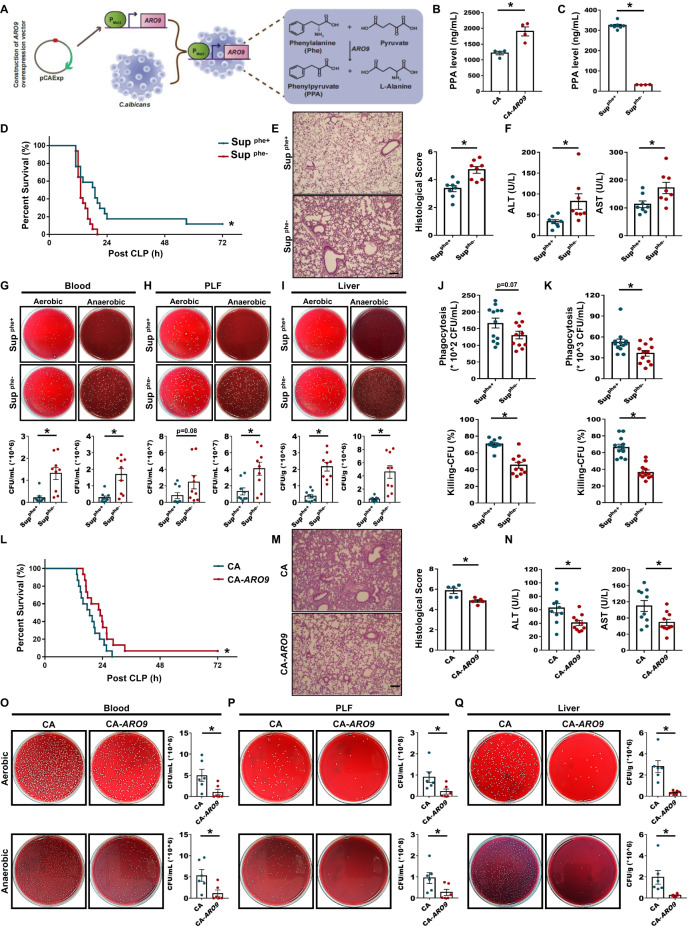
To the best of our knowledge, in C. albicans, PPA is mainly produced by conversion of the aromatic amino acid phenylalanine (Phe) by aromatic-amino-acid:2-oxoglutarate transaminase (ARO9, Fig. 3A). To verify the ability of C. albicans to produce PPA and determine the mechanism of PPA production, the supernatants of wild-type or ARO9-overexpressing C. albicans and the supernatant of wild-type C. albicans cultured in synthetic low-dextrose (SLD) medium with or without 300 μg/mL Phe (Supphe+ and Supphe−, respectively) were used. GC/MS analysis of ARO9-overexpressing vs. wild-type C. albicans supernatant revealed that ARO9 overexpression enhanced PPA production (Fig. 3B). GC/MS analysis also showed that wild-type C. albicans cultured in SLD medium without Phe (Supphe−) produced negligible PPA compared to wild-type C. albicans cultured in SLD medium with Phe (Supphe+) (Fig. 3C).
Fig. 3.
Decreased PPA production by C. albicans impaired protection against sepsis. A Schematic diagram illustrating the workflow for aromatic-amino-acid:2-oxoglutarate transaminase-overexpressing C. albicans and the metabolic process of C. albicans. PPA is mainly produced by the conversion of phenylalanine (Phe) by aromatic-amino-acid:2-oxoglutarate transaminase. B PPA levels in the supernatant of wild-type and ARO9-overexpressing C. albicans detected by GC/MS (n = 4). C PPA levels in Supphe+ and Supphe− (supernatant of C. albicans cultured in SLD medium containing 300 or 0 μg/mL Phe, respectively) detected by GC/MS (n = 4–7). D Survival rate of CLP mice (n = 17) treated with Supphe+ or Supphe− 3 h before CLP. HE staining and histological scores (E, n = 8), serum ALT and AST levels (F, n = 8), and bacterial load in blood (G), PLF (H), and liver (I, n = 9) samples. Scale bars, 50 μm. Phagocytosis and bacterial killing by BMDMs with Supphe+ or Supphe- pretreatment prior to E. coli (J) or S. aureus infection (K, n = 11–12). L Survival rate of CLP mice treated with wild-type or ARO9-overexpressing C. albicans 3 h before CLP (n = 15). HE staining and histological scores (M, n = 5), serum ALT and AST levels (N, n = 10), and bacterial load in blood (O, n = 6), PLF (P, n = 6), and liver (Q, n = 6) samples. Scale bars, 50 μm. Data are presented as the mean ± SEM. *p < 0.05 by two-tailed Student’s t test. The survival rates of septic mice were analyzed using the Kaplan‒Meier method with the log-rank test
To study whether the substrate Phe and downstream derivative phenyllactic acid (PLA) of PPA influence the pathological progression of sepsis, we pretreated mice with Phe or PLA and then performed CLP. There was no significant difference in the survival rate between the two groups (Fig. S5A, B), demonstrating that neither Phe nor PLA affected sepsis progression. Next, we investigated whether the lack of Phe affects C. albicans growth. As shown in Fig. S5A, D, no significant difference between the two groups was observed, indicating that the absence of Phe does not affect the growth of C. albicans. However, compared to Supphe+ mice, Supphe- mice exhibited significantly shorter survival time after CLP (Fig. 3D), had more pathological changes in the lungs, such as alveolar septal thickening (Fig. 3E), and had marked increases in ALT, AST (Fig. 3F), and inflammatory mediator expression (Fig. S5E–G). In addition, we explored whether the lack of C. albicans-derived PPA affected the bacterial load in blood, PLF, and liver samples after CLP. Compared to Supphe+ mice, Supphe− mice had a greater number of bacterial colonies, indicating that a lack of PPA indeed significantly increased the bacterial load (Fig. 3G–I). Furthermore, Supphe−-treated BMDMs exhibited decreased phagocytosis and bacterial killing compared to Supphe+-treated BMDMs (Fig. 3J, K).
Next, we pretreated mice with live wild-type or ARO9-overexpressing C. albicans (CA or CA-ARO9, respectively) before CLP and found that the latter had improved survival rates (Fig. 3L), decreased lung injury (Fig. 3M) and serum ALT and AST (Fig. 3N), and reduced bacterial colony-forming units (CFUs) in blood, PLF, and liver samples (Fig. 3O–Q). These data clearly demonstrate the key function of C. albicans-derived PPA in protecting against sepsis.
Macrophages were required for the PPA-mediated protection against sepsis
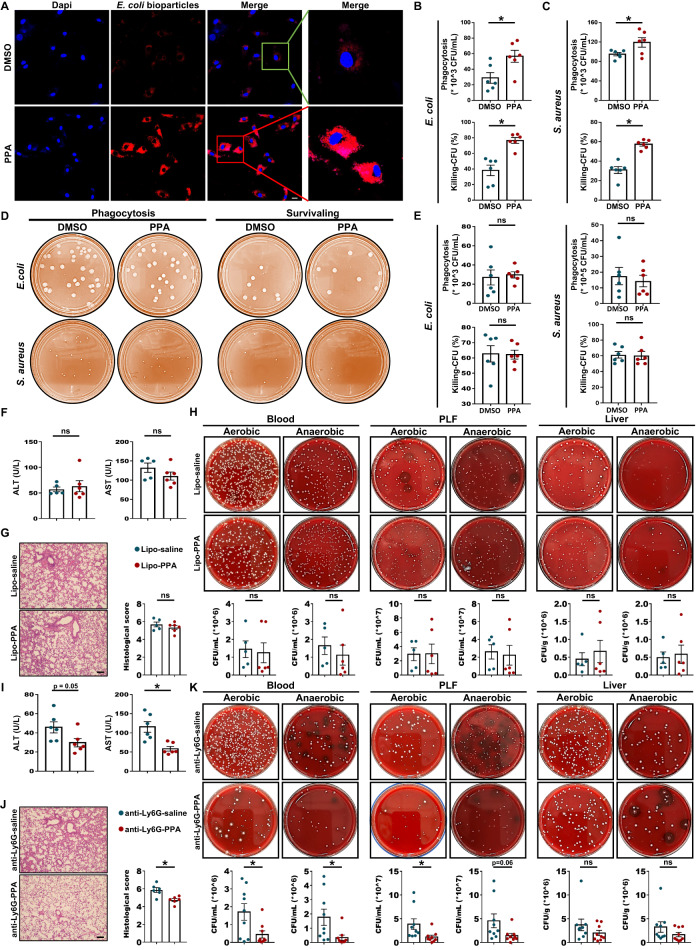
To further determine the role of PPA in sepsis protection, we then assessed the effect of PPA on bacterial growth, and no significant differences in bacterial growth (E. coli and S. aureus) were found between the PPA-treated and untreated groups (Fig. S6A, B), confirming that PPA may affect bacterial clearance by facilitating the bactericidal activity of host immune cells. To verify this hypothesis, we carried out in vitro studies. Initially, we assessed the cytotoxicity of 1–500 μM PPA by using Cell Counting Kit-8 (CCK-8) assays in BMDMs infected with heat-killed E. coli or S. aureus and found that PPA had no toxic effects on cell viability at the concentrations tested (Fig. S6C, D). Hence, a concentration of 100 μM PPA was used in the following experiments. As expected, PPA significantly increased the phagocytotic capacity of BMDMs (Fig. 4A–C). In addition, it also increased bacterial killing, as evidenced by the number of CFUs (Fig. 4B, C). In contrast, there was no significant change in the phagocytosis and bacterial killing of neutrophils when comparing the PPA-treated and negative control groups (Fig. 4D, E).
Fig. 4.
Macrophages were required for the PPA-mediated beneficial effects in CLP-induced sepsis. A Confocal micrographs showing the phagocytosis of pHrodo red E. coli bioparticles by BMDMs with or without pretreatment with PPA (100 μM). Scale bars, 10 μm. B, C Phagocytosis by BMDMs with or without pretreatment with PPA (100 μM) for 18 h prior to E. coli or S. aureus infection (MOI, 20) for 45 min (n = 6). Separate samples were incubated for another hour to assess the bacterial killing activity (n = 6). D, E Neutrophils were pretreated with or without PPA for 18 h and then infected with E. coli or S. aureus (MOI, 100) for 45 min. Representative images and quantification results of phagocytosis and bacterial killing are presented (n = 6). Serum ALT and AST levels (F, n = 5-6), HE staining and histological scores (G, n = 5-6) and bacterial load (H) in blood, PLF, and liver (H, n = 5–6) samples from macrophage-depleted mice with or without PPA treatment. Scale bars, 50 μm. Serum ALT and AST levels (I, n = 6), HE staining and histological scores (J, n = 6) and bacterial load (H) in blood, PLF, and liver (K, n = 9) samples from neutrophil-depleted mice with or without PPA treatment. Scale bars, 50 μm. Data are presented as the mean ± SEM. *p < 0.05 by two-tailed Student’s t test. ns not significant
Next, we further investigated the role of macrophages and neutrophils in PPA-mediated protection against sepsis in vivo. Mice were treated with clodronate liposomes (Lipo) or Ly6G antibody to deplete macrophages and neutrophils prior to CLP, respectively. Depleting macrophages dramatically impaired the organ protection (Fig. 4F, G) and bacterial clearance (Fig. 4H) of septic mice treated with PPA. Consistently, the mRNA expression of cytokines in PLF from septic mice after pretreatment with Lipo was not alleviated by PPA administration (Fig. S7A). In contrast, enhanced organ protection (Fig. 4I, J) and bacterial clearance (except liver, Fig. 4K), as well as markedly decreased expression of inflammatory factors (Fig. S7B), were still observed in PPA-treated septic mice after the depletion of neutrophils. These results suggest that macrophages may be associated with the beneficial effect of PPA in sepsis.
PPA directly binds to SIRT2 in macrophages
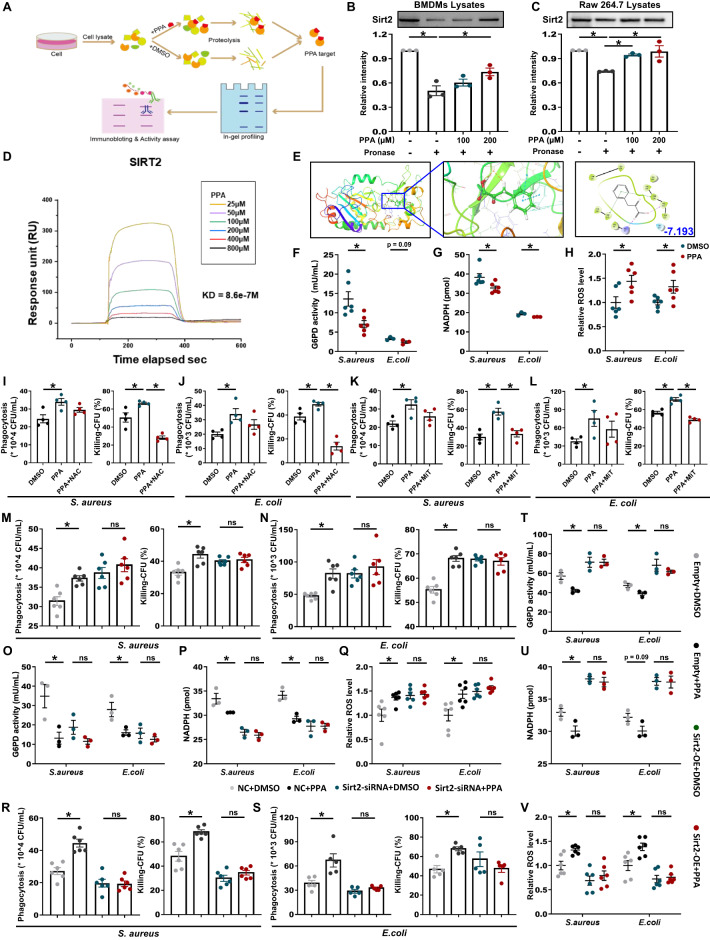
Next, we explored the molecular mechanisms of PPA in the regulation of bactericidal activity. We conducted a drug affinity responsive target stabilization (DARTS) assay to identify the potential molecular target of PPA. DARTS detects ligand-bound targets in lysates based on the principle of small-molecule binding proteins being protected and thereby enriched during proteolytic digestion (Fig. 5A). To conduct the DARTS assay, we incubated PPA or DMSO with BMDMs lysate or Raw 264.7 lysate and then conducted pronase degradation. We found that a Coomassie blue-stained band at 40–55 kD was enriched by PPA treatment, likely due to PPA binding leading to resistance to pronase degradation (Fig. S8A, B). Next, virtual screening involving a molecular docking/scoring strategy was carried out to identify the most likely PPA target, and SIRT2 was the top-ranked protein among the proteins that fell within the expected molecular weight range (40–55 kD) and were associated with bacterial clearance.
Fig. 5.
PPA inhibited SIRT2 activity and increased ROS production in BMDMs. A Schematic diagram showing target identification of PPA in macrophage lysates. Immunoblot analysis of SIRT2 in pronase-digested (B) BMDMs and (C) Raw 264.7 cell lysates (n = 3). D Surface-plasmon resonance (SPR) analysis of PPA binding to SIRT2. E Docking analysis of the interactions between PPA and SIRT2. The SIRT2 residues that are likely to participate in the interactions with PPA are labeled. BMDMs were treated with or without PPA (100 μM) for 18 h. G6PD activity (F), NADPH levels (G), and ROS levels (H) in E. coli- or S. aureus-infected BMDM lysates (n = 3–6). I–L Phagocytosis and bacterial killing by BMDMs with or without N-acetylcysteine (NAC) or mitoTEMPOL (MiT) pretreatment (n = 4). M, N Phagocytosis and bacterial killing by BMDMs transfected with SIRT2 siRNA or negative control siRNA for 48 h in the presence or absence of PPA (n = 6). O G6PD activity, P NADPH levels, and Q ROS levels in E. coli- or S. aureus-infected BMDM lysates with the indicated treatments (n = 3–6). R, S Phagocytosis and bacterial killing by BMDMs with SIRT2 overexpression or empty plasmid transfection for 48 h in the presence or absence of PPA (n = 5–6). T G6PD activity, U NADPH levels, and V ROS levels in E. coli- or S. aureus-infected BMDM lysates with the indicated treatments (n = 3–6). Data are presented as the mean ± SEM. *p < 0.05 by two-tailed Student’s t test. ns not significant
The PPA–SIRT2 interactions were confirmed by SIRT2 immunoblotting using DARTS-processed BMDMs and Raw 264.7 lysates (Fig. 5B, C). Furthermore, the PPA–SIRT2 interactions were evaluated by cellular thermal shift assay (CETSA). As expected, PPA incubation stabilized SIRT2 in the heat-denatured BMDMs and Raw 264.7 lysates (Fig. S8C, D). Moreover, surface-plasmon resonance (SPR) analysis showed a direct interaction between PPA and SIRT2 that was dose-dependent, suggesting a high binding affinity (Fig. 5D). Molecular docking analysis results identified the four key residues present in the active pocket (ARG97, PHE119, VAL233, and PHE234) of the SIRT2 protein that may interact with the PPA molecule (Fig. 5E). We further examined whether the binding between PPA and SIRT2 influences SIRT2 biochemical activities by testing the acetylation level of α-tubulin, as SIRT2 inhibition is correlated with tubulin hyperacetylation [28]. As expected, PPA was found to increase the acetylation level of α-tubulin after bacterial infection (Fig. S8E). Collectively, these data suggest a direct interaction between PPA and SIRT2 in macrophages and inhibition of the deacetylation activity of SIRT2 after bacterial infection.
PPA-mediated bacterial killing relies on SIRT2-dependent ROS production
ROS generation by the NADPH oxidase complex is critical for the host’s defense against pathogens [29–31]. SIRT2 regulates NADPH homeostasis and ROS production by controlling G6PD acetylation and activities [9]. In this context, we investigated whether PPA could enhance ROS production by macrophages after bacterial infection by regulating the SIRT2/G6PD/NADPH axis. Interestingly, PPA treatment significantly reduced G6PD activity and NADPH levels in BMDMs after E. coli or S. aureus infection (Fig. 5F, G). As a result, PPA significantly increased the ROS level in macrophages (Fig. 5H), indicating that the key mechanisms behind the PPA-mediated bacterial killing by macrophages were related to an increase in ROS levels.
Next, we tested the effects of N-acetylcysteine (NAC), a ROS inhibitor, on PPA-mediated bacterial killing by BMDMs. Bacterial killing was significantly decreased by NAC pretreatment followed by PPA treatment compared to that following PPA-only treatment (Fig. 5I, J). Similarly, pretreatment with mitoTEMPOL (MiT), a mtROS scavenger, also decreased bacterial killing by macrophages (Fig. 5K, L), indicating that the effect of PPA on bacterial killing by macrophages was dependent on ROS generation.
To further validate the SIRT2-dependent effects of PPA on bacterial clearance, we next knocked down SIRT2 in BMDMs using SIRT2 siRNA (Fig. S9A). SIRT2 knockdown (relative to the negative control siRNA group) significantly diminished PPA’s promotion of bacterial clearance by BMDMs after S. aureus or E. coli infection (Fig. 5M, N) and significantly abolished PPA’s inhibition of G6PD activity in BMDMs (Fig. 5O) and intracellular NADPH production (Fig. 5P), thereby abolishing PPA’s promotion of ROS production in BMDMs (Fig. 5Q). In addition, SIRT2 overexpression (relative to the empty plasmid transfection group) significantly reversed PPA’s promotion of bacterial clearance by BMDMs after S. aureus or E. coli infection (Figs. 5R, S and S9B) and significantly reversed PPA’s inhibition of G6PD activity and NADPH production (Fig. 5T, U), which resulted in insufficient ROS production and thus limited clearance of bacteria after PPA treatment (Fig. 5V). Collectively, these results indicate the indispensable nature of SIRT2-dependent ROS production in the PPA-induced increase in bacterial killing by BMDMs.
Evaluation of C. albicans culture supernatant and PPA treatment in a porcine model of E. coli-induced sepsis
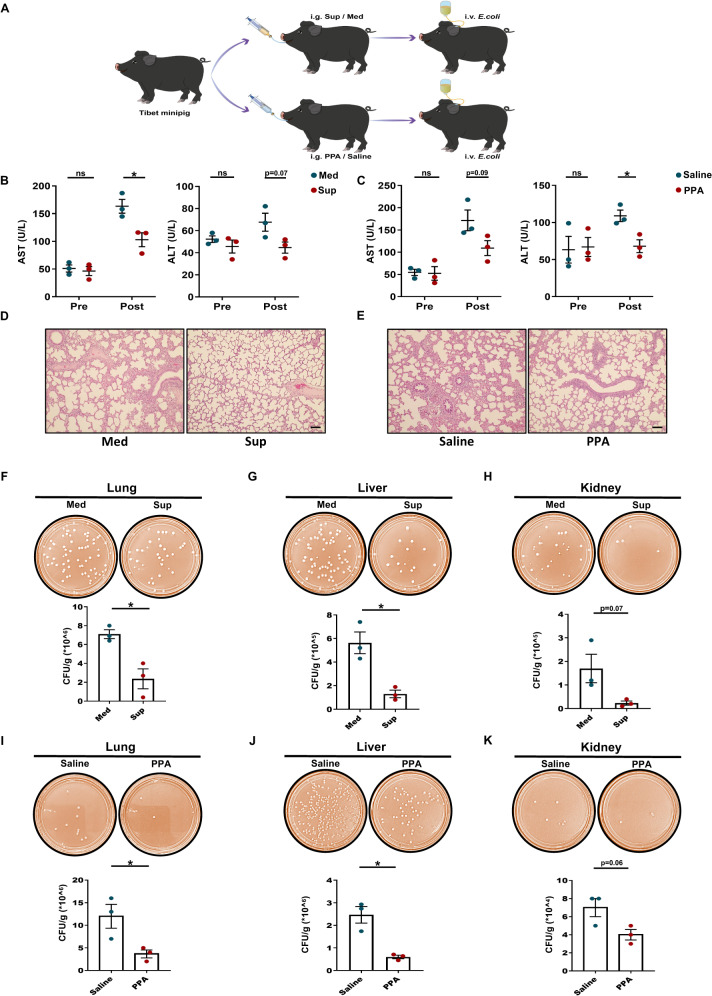
To further verify the protective effect of C. albicans culture supernatant (Sup) and PPA in vivo in a clinically relevant large-animal model of sepsis, 12 pigs were prepared as described in the Methods section and Fig. 6A. No obvious differences in health or behavior were observed before the experiment. Serum concentrations of ALT and AST were recorded after euthanasia of the pigs, and the values before E. coli administration were also calculated. Although no significant differences were observed before E. coli administration, serum biomarkers indicated that liver damage was substantially reduced in the Sup- or PPA-treated septic pigs compared to the corresponding controls (Fig. 6B, C). Moreover, compared with vehicle pretreatment, histological analysis revealed that lungs from Sup- or PPA-treated pigs exhibited less pulmonary vascular congestion and edema (Fig. 6D, E). We then determined whether Sup or PPA could enhance bacterial clearance in septic pigs. Organ samples (lung, liver, and kidney) were obtained after euthanasia and kept on ice before tissue homogenization. Bacterial load analysis revealed that Sup administration dramatically reduced the bacterial load in all analyzed organs (Fig. 6F–H). Similarly, pretreatment with PPA also significantly decreased bacterial counts in the lung, liver, and kidney (Fig. 6I–K). Taken together, in agreement with the mouse data, these data indicate that administration of Sup or PPA promotes bacterial clearance and has a protective effect on the organs of septic pigs.
Fig. 6.
C. albicans culture supernatant and PPA protect against E. coli-induced sepsis in pigs. A Basic scheme of the experimental protocol. Serum ALT and AST before (B) Sup or (C) PPA pretreatment and after euthanasia of the pigs (n = 3). D Representative HE staining image of the lungs of septic pigs with or without Sup treatment. Scale bar, 50 μm. E Representative HE staining image of the lungs of septic pigs with or without PPA treatment. Scale bar, 50 μm. Effect of Sup (F–H) or PPA (I–K) on bacterial load in the lungs, liver, and kidneys of septic pigs. Bacterial colonies were cultured on plates, and CFUs were counted (n = 3). Data are presented as the mean ± SEM. *p < 0.05 by two-tailed Student’s t test. ns not significant
PPA enhanced bacterial killing by human phagocytes and was negatively correlated with a marker of sepsis severity in patients
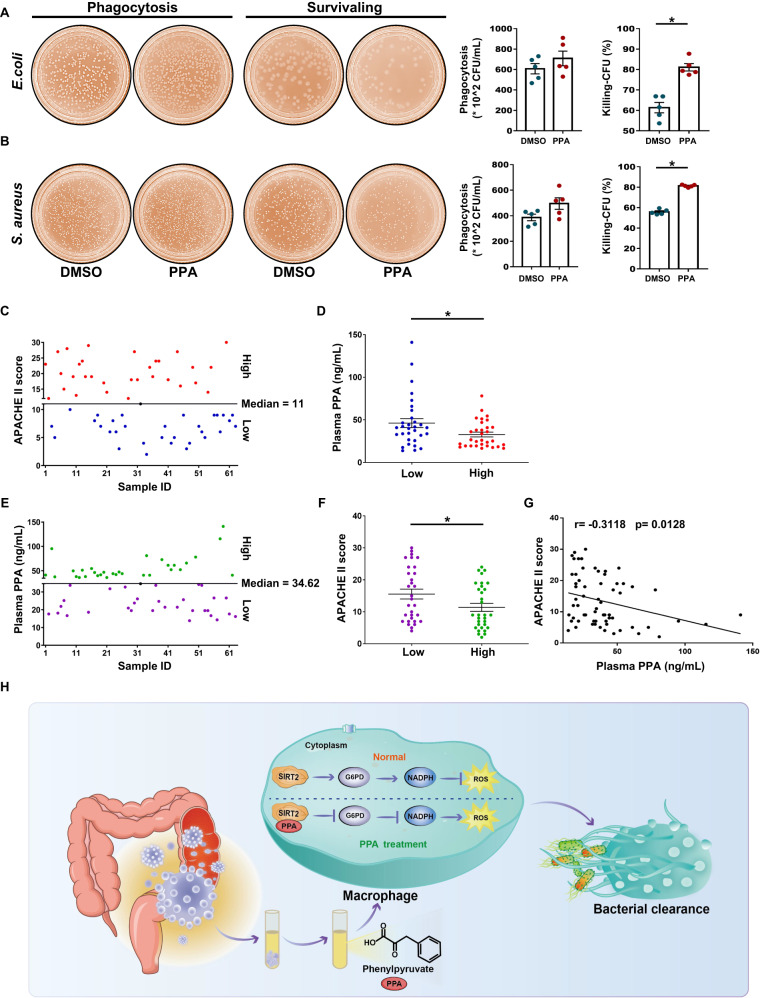
We finally investigated whether these basic findings could be translated to clinical settings by analyzing the role of PPA in the function of human macrophages. Human blood monocyte-derived macrophages (MDMs) from sepsis patients were isolated and incubated with PPA. PPA treatment increased E. coli and S. aureus killing by MDMs (Fig. 7A, B), suggesting that PPA may be used as a novel compound to enhance the bacterial clearance capacity of macrophages.
Fig. 7.
PPA modulated bacterial killing by human phagocytes and was associated with the disease outcome of sepsis patients. A, B Representative images and quantitative results of phagocytosis and killing of E. coli and S. aureus by human blood monocyte-derived macrophages (MDMs) from sepsis patients in the dimethyl sulfoxide (DMSO) and PPA groups (n = 5). Sepsis patients were divided into two groups based on APACHE II scores as follows: high group (>median) and low group (<median) (C, n = 63). PPA levels were compared between the two groups (D, n = 31). E, F Sepsis patients were divided into two groups based on the circulating levels of PPA as follows: a high group (>median) and a low group (<median) (E, n = 63). The APACHE II score was compared between the two groups (F, n = 31). G Correlation analysis of plasma PPA levels with APACHE II scores (n = 63). H Schematic diagram depicting enhanced bacterial clearance by C. albicans-derived PPA via regulation of SIRT2-dependent ROS production in macrophages. Data are presented as the mean ± SEM. *p < 0.05 by two-tailed Student’s t test. ns not significant
To determine whether poor disease outcomes in humans are associated with decreased PPA, we evaluated the circulatory level of PPA in sepsis patients with higher Acute Physiology and Chronic Health Evaluation (APACHE) II scores (>median value) and found a clear reduction in the PPA levels in these patients (Fig. 7C, D). We further divided the sepsis patients into two groups according to the circulating level of PPA. Interestingly, in the patients with a higher plasma level of PPA (>median value), the APACHE II score was significantly reduced compared to that in the patients with a lower plasma PPA level (<median value) (Fig. 7E, F). More strikingly, the plasma PPA level had a significant negative correlation with the APACHE II score in sepsis patients (r = −0.3118, p = 0.0128, Fig. 7G). Collectively, these findings demonstrate that PPA could have a key role in bacterial sepsis in humans.
Discussion
At present, how the gut mycobiota and associated metabolites affect human health and disease is largely unknown. To the best of our knowledge, we report for the first time that a metabolite derived from C. albicans, PPA, has a protective effect against bacterial sepsis. Herein, we provide a systematic analysis of the role of PPA, which promotes the bactericidal activity of macrophages, reduces bacterial load and organ damage, and prolongs the survival time of septic mice. Mechanistically, PPA inhibited SIRT2-mediated activation of G6PD after bacterial challenge and reduced intracellular NADPH levels, thereby producing more ROS to clear bacteria (Fig. 7H). Collectively, our findings highlight the diverse roles played by C. albicans during sepsis and support the importance of PPA in clearing bacteria from patients with bacterial sepsis.
Although all healthy humans and laboratory animals carry numerous fungal species, thus far, researchers have mostly focused on the role of the bacterial microbiota. However, commensal fungi have a profound effect on the nutritional status, metabolism, and immunity of the host [32–34]. The beneficial effects of fungal colonization on hosts include resistance to pathogens and fine tuning of the immune system [35, 36]. A recent review highlighted the beneficial effects of fungal transfer during fecal microbiota transplantation [37]. Among the commensal fungal species colonizing the human intestine, C. albicans is the most dominant. However, C. albicans is an opportunistic fungus; under certain predisposing conditions, such as immunosuppression, antibiotic treatment, or prolonged stay in intensive care units, it can cause superficial infections and even enter the bloodstream, resulting in systemic candidiasis. Conversely, recent studies have explored the beneficial effects of C. albicans on the host. For example, intestinal colonization by C. albicans protected against subsequent challenge with other virulent fungi [38]. In addition to these direct host–microbe interactions, mouse gut colonization by C. albicans improved host immunity, resulting in a systemic increase in fungal-specific Th17 CD4+ T cells and IL-17-responsive circulating neutrophils, which protected the mice against more invasive bacterial and fungal species [39]. Moreover, the effect of C. albicans on homeostasis in humans is not limited to direct fungi–host interactions because various metabolites derived from this yeast, including toxins and antibacterial agents, may exert biological effects on humans. For example, exposure of monocytes to the C. albicans-derived polysaccharide β-1,3-glucan causes epigenetic changes such as histone methylation and induces a stronger immune response against other fungi during subsequent colonization [36]. Another example involves farnesol produced by C. albicans, which may alter the regulation of quorum sensing in P. aeruginosa [40]. These observations improved our understanding of the roles of commensal C. albicans in numerous physiological and disease processes. However, the specific interactions between C. albicans-derived metabolites and the host immune system, especially regarding their contribution to the pathogenesis of sepsis, are still largely unknown. Here, we uncovered the crucial role of an important C. albicans-derived functional metabolite, PPA, in the development of bacterial sepsis. By using metabolomics analysis and blocking PPA generation, we provide compelling evidence of the protective effect of C. albicans-derived PPA against sepsis progression. Of note, interactions among microbes can be very complex, ranging from cooperation to predation [41]. Previous studies have already shown that colonization with specific bacterial species can reduce Candida load in vivo [42], and several E. coli strains have been found to inhibit C. albicans growth in vitro [43]. We also found that PPA production by C. albicans cocultured with E. coli was significantly reduced compared to that of C. albicans cultured alone (Fig. S10A, B). Therefore, it can be speculated that in a patient with bacterial sepsis, the overgrowth of pathogenic bacteria, such as E. coli, may lead to restricted growth of C. albicans in the intestines, which in turn leads to decreased PPA production.
In the pharmaceutical industry, PPA has been used to produce d-phenylalanine, the raw material for antidiabetic, antineoplastic, and anti-AIDS compounds, and peptide antibiotics. In addition, PPA is used to synthesize PLA, which is used as an antimicrobial agent, food preservative, and flavor compound [44]. However, the potential beneficial effects of PPA against diseases in both humans and animals are not well known, although PPA administration via a chow diet effectively alleviated the anxiety and depression behavior of mice and improved their exercise capacity [45]. In the current study, PPA promoted the bactericidal activity of macrophages, reduced the bacterial load and organ damage, and decreased mortality in CLP-induced septic mice. These findings demonstrated the protective effect of PPA against sepsis. Of note, PPA accumulates in patients with a phenylalanine-4-hydroxylase deficiency, which causes phenylketonuria [46]. Nevertheless, an appropriate pharmacological dose of PPA may have therapeutic effects on conditions such as sepsis in humans and animals. However, we cannot rule out the possibility that other key metabolites derived from C. albicans may also mediate sepsis progression. With the development of metabolomics analysis, more functional metabolites may be discovered in the future.
Regulating ROS is vital for mounting an appropriate immune response and for avoiding any excessive oxidative damage to resident host cells/tissues. We have now provided solid evidence demonstrating that PPA is a natural SIRT2 antagonist, and it can increase ROS production for bacterial killing by macrophages by inhibiting SIRT2 activity and downstream signaling. SIRT2 is one of the seven members of the family of NAD+-dependent histone deacetylases, which are most highly expressed in myeloid cells [47, 48]. Increasing evidence implicates SIRT2 in the regulation of metabolism and cellular energy responses [49, 50]. To date, many substrates of SIRT2 have been identified, including α-tubulin [48], histone H3/H4 [8, 51], phosphoenolpyruvate carboxykinase 1 (PEPCK1) [52], and G6PD [9]. Among these, G6PD is linked to ROS levels; SIRT2-dependent deacetylation of G6PD stimulates NADPH production, thereby reducing the cellular ROS level and decreasing the level of cell death. In addition, SIRT2 deficiency promotes bacterial phagocytosis by macrophages but not cytokine production [10]. These findings suggest a significant role for SIRT2 in ROS regulation and bacterial clearance by macrophages. Moreover, PPA impairs the oxidative phase of the pentose phosphate pathway by reducing G6PD activity, which may compromise NADPH production and eventually lead to increased ROS production [53]. Our current results link PPA-mediated bacterial clearance to SIRT2-dependent ROS production. Based on SIRT2 knockdown and overexpression in BMDMs, we confirmed that PPA inhibits SIRT2, which in turn reduces G6PD activity, decreases intracellular NADPH, and increases ROS, thereby accelerating bacterial clearance by BMDMs. Thus, inhibitors targeting SIRT2, such as PPA, may be used as novel molecules for bacterial clearance and sepsis treatment.
In summary, we clearly demonstrated that PPA derived from C. albicans enhances the bactericidal activity of macrophages and reduces the mortality of septic mice. Therefore, effective strategies such as treatment with PPA and similar SIRT2 inhibitors should be explored, which may provide a new approach for the clinical management of sepsis.
Materials and methods
Study approval
All animal experiments were performed in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals, with the approval of the Institutional Animal Care and Use Committee (IACUC) of Southern Medical University (approval number: SMUL2022308 & SMUL2021094). The Medical Ethics Committee of the local institution approved all human studies (approval numbers: NFEC-2021-139 and LAEC-2020-055).
Fungal strains and growth conditions
C. albicans (ATCC 10231, ATCC MYA-2876 (also known as SC 5314)) was cultured under aerobic conditions in liquid or solid Sabouraud dextrose broth (SDB) medium at 30 °C. For preparation of the supernatant, C. albicans was washed and resuspended in different media as follows: SDB containing 10% fetal bovine serum (FBS), complete RPMI 1640 medium or synthetic low-dextrose (SLD) medium (0.17% yeast nitrogen base (YNB, Y1251, Sigma) with 0.1% glucose and 5 mg/mL ammonium sulfate) with or without phenylalanine (Phe) and grown for 2–5 days. The ARO9 open reading frame (ORF) under the control of the MET3 promoter was inserted into the pCaExp plasmid to construct pCaEXP-ARO9 plasmids that can overexpress the ARO9 gene. Then, we used the lithium acetate method to transform the linearized pCaEXP-ARO9 into C. albicans (SC 5314) and select positive colonies in SDB-selective solid culture medium. PCR was used to investigate the integration.
Cells and cell lines
Bone marrow-derived macrophages (BMDMs) were isolated and differentiated into macrophages as described previously [54]. Briefly, BMDMs were generated by culturing bone marrow cells obtained from male mice in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 1% penicillin–streptomycin (15140-122, Gibco) and 20 ng/mL macrophage colony-stimulating factor (M-CSF, 416-ML, R&D Systems). Neutrophils were isolated from mouse bone marrow as described previously [55]. The bone marrow cells were flushed with complete culture medium (RPMI 1640 medium with 10% FBS, 1% penicillin‒streptomycin and 2 mM EDTA) and subsequently collected by centrifugation at 427 × g for 7 min at 4 °C. Then, Histopaque 1119 (3 mL, 11191, Sigma) and Histopaque 1077 (3 mL, 10771, Sigma) were added layer by layer to the centrifuge tube, and marrow cells were carefully laid on the uppermost layer. The tube was centrifuged at 850 × g for 30 min at RT without braking. The cells suspended between Histopaque 1119 and Histopaque 1077 were murine neutrophils. Ficoll density-gradient centrifugation was used to isolate peripheral blood mononuclear cells (PBMCs) from human blood samples. Human monocyte-derived macrophages (MDMs) were cultured for 7 days in RPMI 1640 medium containing 10% FBS and 1% penicillin‒streptomycin-glutamine with 20 ng/mL human macrophage colony-stimulating factor (hMCSF) from Novoprotein (Suzhou, China). The macrophage cell line Raw 264.7 was cultured in DMEM supplemented with 10% FBS and 1% penicillin‒streptomycin at 37 °C in a humidified atmosphere containing 5% CO2.
Animal models
Mouse
All mice (male, C57BL/6J, 6–8 weeks old) were housed with free access to food and water. Polymicrobial sepsis was induced by cecal ligation and puncture (CLP) as described previously [56]. Briefly, after giving anesthesia to the mice, the cecum was ligated and punctured twice with an 18 G needle to establish the severe sepsis model. The survival rate was monitored every 1 h for the first 3 days. Before CLP surgery, the mice received the indicated culture supernatant of C. albicans (300 µL) or vehicle medium (300 µL) through oral gavage administration 3 h before CLP surgery. For PPA (156-06-9, Sigma–Aldrich) treatment, mice were administered 40 mg/kg PPA dissolved in saline and sonicated by oral gavage 3 h before CLP surgery.
Pig
Male Tibetan minipigs weighing 10 kg (range 8.8–11 kg) were used and allocated into one of two arms. The C. albicans culture supernatant arm consisted of subgroups receiving E. coli and the supernatant of C. albicans culture or E. coli and vehicle medium. The PPA arm consisted of subgroups receiving E. coli and PPA or E. coli and saline. Live E. coli was infused intravenously at an increasing rate as follows: 3 × 107 bacteria/30 min from 0 to 30 min, 3 × 108 bacteria/30 min from 30 to 60 min, and 3 × 109 bacteria/30 min from 60 to 90 min. Injection of bacteria started 3 h after the supernatant of C. albicans culture or PPA treatment was given.
Human samples
A group of 94 individuals, including 14 healthy controls, were recruited for this study. Sepsis was diagnosed according to the sepsis 3.0 criteria [1]. Detailed information on the patients is given in Supplementary Tables 1 and 2. Plasma and stool samples were collected and stored at −80 °C until further use.
Detection of C. albicans load
Feces collected from humans were suspended in 600 μL of a buffer (SA and SC) mix containing 15 μL of proteinase K. The mixture was incubated at 70 °C for 15 min, and the fungal genomic DNA was isolated from the feces with the use of a TIANamp Stool DNA Kit (DP328, Tiangen). To detect the C. albicans burden in feces, a C. albicans SYBR qPCR Kit (14-60210, TIANDZ) was used according to the manufacturers’ instructions.
Serum biochemistry
Blood was collected in tubes containing EDTA after venipuncture and centrifuged at 12,000 rpm for 10 min at 4 °C. Alanine transaminase (ALT) and aspartate transaminase (AST) levels were determined with commercially available kits according to the manufacturers’ instructions.
Measurement of cytokines
Blood was collected 12 h after CLP surgery, and serum samples were prepared by centrifugation at 12,000 rpm for 10 min at 4 °C. The levels of TNF-α and IL-6 in the serum were tested using commercially available kits [EMC102a.96 (TNF-α), EMC004.96 (IL-6), NeoBioscience, Beijing, China].
Histological procedures
Lung tissue was fixed in 4% paraformaldehyde, embedded in paraffin, and then sliced into 5-μm-thick sections. Tissue sections were stained with H&E and examined with a light microscope (Olympus, Tokyo). Pulmonary injury score parameters included the thickness of alveolar walls, alveolar fusion, hemorrhage in the air compartment, leukocyte infiltration and tissue exudation.
Analysis of bacterial load
Mouse
For bacterial load analysis, blood, peritoneal lavage fluid (PLF) and homogenized liver samples were collected 12 h after CLP surgery. After preparing serial dilutions, 100 μL of each sample was plated on Columbia blood agar plates and incubated under aerobic or anaerobic conditions at 37 °C for 14–16 h. CFUs were counted, and the results were expressed as CFU/mL of fluid (blood and PLF) or as CFU/g tissue (liver).
Pig
Lung, liver and kidney tissue were homogenized in PBS (10 mg/500 μL) using a tissue homogenizer. The solid tissue was removed by centrifugation at 1000 rpm for 1 min, and the supernatants were serially diluted, plated on LB agar plates, and then incubated at 37 °C overnight. The number of colonies was counted after 16–18 h.
RNA extraction and real-time PCR assay
Total RNA was extracted from PLF, liver and lung using the FastPure Cell/Tissue Total RNA isolation kit (RC112-01, Vazyme). The cDNA was synthesized using total RNA by ReverTra Ace® qPCR RT Master Mix (FSQ-101, Toyobo). The expression levels of mRNAs were determined by an ABI 7500 Real-Time PCR System using TB Green® Premix Ex Taq™ II (RR820B, Takara). All the primer sequences used for each targeted gene are listed in Supplementary Table 3.
Coculture experiments
C. albicans cells were cultured overnight, diluted to an OD600 of 0.1, and allowed to grow to an OD600 of 0.6. Overnight cultures of E. coli were centrifuged and resuspended in fresh LB. Subsequently, C. albicans was cocultured with E. coli or an equal volume of LB for 16–20 h. Supernatants were centrifuged and collected for GC/MS analysis.
Metabolomics analysis
Sample preparation
The supernatant of C. albicans was prepared as described above followed by a freeze-drying procedure. In the experiment, 20 mg of sample was mixed with 400 μL of methanol-water (4:1, v/v) containing 0.02 mg/mL of 2-chloro-L-phenylalanine and then homogenized for 6 min at −10 °C. This solution was ultrasonically extracted on ice for 30 min and centrifuged for 15 min at 15,000 × g at 4 °C. Twenty microliters of the supernatant was used for LC‒MS/MS analysis.
LC‒MS/MS analysis
LC‒MS/MS was performed on a UHPLC-Q Exactive system (Thermo Scientific, USA). The LC was run with the following conditions: Column: Acquity UPLC HSS T3 (100 mm × 2.1 mm i.d., 1.8 µm; Waters, Milford); Solvent A: 95:5 water/acetonitrile mixture (0.1% (v/v) formic acid); and Solvent B: acetonitrile: isopropanol: water 47.5: 47.5: 5 (0.1% (v/v) formic acid). The column was maintained at 40 °C, and separation was conducted as follows: 0% B–5% B over 0–0.1 min, 5% B–25% B over 0.1–2 min, 25% B–100% B over 2–9 min, 100% B–100% B over 9–13 min; 0% A–100% A over 13–13.1 min, and 13.1–16 min holding at 100% A at a flow rate of 0.4 mL/min. The mass spectrometer was operated with spray voltages of 3.5 kV and −2.8 kV in positive or negative mode. Centroid data were collected from 70 to 1050 m/z with a 70,000 resolution. Progenesis QI (Waters Corporation, Milford, USA) was used for peak picking and alignment to screen the metabolic biomarkers that displayed significant changes between the medium and supernatant groups. Several standard metabolome databases, including the human metabolome database (http://www.hmdb.ca/), MassBank (http://massbank.jp/Index), and METLIN (http://metlin.scripps.edu/index.php), were used to identify the assigned biomarkers by matching the acquired precursor and fragment ions.
Quantitative analysis of PPA
For detection of PPA content, 100 μL of serum sample or supernatant of C. albicans culture was aspirated, and 900 μL of acetonitrile containing 0.1% formic acid was added to precipitate the proteins. The solution was ultrasonically extracted on ice for 20 min and then centrifuged at 15,000 rpm for 10 min at 4 °C. The supernatant was transferred into a new tube and dried by a nitrogen-blowing instrument. Then, the compounds in the dried residue were extracted using a water-acetic ether mixture (v/v: 2:7) and centrifuged at 15,000 rpm for 10 min at 4 °C. The supernatant was collected and dried under nitrogen, and the compounds were converted to TMS derivatives with 100 μL of BSTFA/TMCS (100:1) and 20 μL of pyridine followed by GC/MS analysis.
CCK-8 assay
Cell proliferation was tested by the Cell Counting Kit-8 (CCK-8, Dojindo) assay according to the manufacturer’s instructions. Briefly, 104 cells/well were evenly distributed and grown in 96-well plates and then treated with the indicated concentrations of PPA for 18 h, followed by incubation with heat-killed E. coli or S. aureus for 45 min. Finally, CCK-8 reagent was added to each well, and the plates were incubated at 37 °C for 3 h. The optical density at 450 nm was assessed using a microplate reader (SpectraMax M5, Molecular Devices).
Phagocytosis assay
To analyze the phagocytic capacity of BMDMs, cells were treated with the supernatant or PPA (100 µM) for 3 or 18 h, followed by incubation with pHrodo red E. coli BioParticles (P35361, Thermo Fisher Scientific) for 45 min at 37 °C. After washing three times with PBS, cell nuclei were stained with antifading mounting medium (DAPI, S2110, Solarbio), followed by visualization using a confocal LSM 880 microscope (Carl Zeiss Microimaging, Jena).
A suspension of 106 neutrophils was seeded in 24-well plates in RPMI medium with the supernatant or PPA (100 µM) for 3 or 18 h, followed by incubation with E. coli or S. aureus (multiplicity of infection, MOI = 1:100) at 37 °C for 45 min. Neutrophils were separated from bacteria in the suspension by centrifugation (400 × g) for 3 min at 4 °C and then suspended in RPMI medium containing 50 μg/mL gentamicin for 30 min to kill any adherent extracellular bacteria. Finally, neutrophils were washed twice to remove gentamicin and lysed with 0.1% Triton X-100 on ice for 20 min to release intracellular bacteria. All samples were serially diluted and plated on LB agar to determine the CFU counts.
Bacterial killing assay
BMDMs with the indicated treatments were infected with E. coli or S. aureus at 37 °C for 45 min, and then the cells were washed two times with PBS containing gentamycin (0.5 µg/mL). Fresh DMEM containing 50 µg/mL gentamicin was added and cells were incubated for 5 min to remove the extracellular bacteria. To determine the amount of internalized bacteria (t = 0 h), macrophages were washed two times with cold PBS and lysed in PBS containing 0.5% Triton X-100. Other cells were incubated for another 60 min (t = 1 h). Bacterial killing was calculated from the percentage of colonies present at t = 1 h compared to that at t = 0 h as follows: [(CFU count at 0 h—CFU count at 1 h)/(CFU count at 0 h)] × 100% [57].
In neutrophils, cells were infected with E. coli or S. aureus at 37 °C for 45 min at an MOI of 1:100, resuspended in medium containing 50 μg/mL gentamycin for 30 min to remove extracellular bacteria, and then lysed in PBS containing 0.1% Triton 100 for assessment of uptake (t = 0 h). Additional samples were incubated for 1 additional hour (t = 1 h) to assess bacterial killing as described above.
In vivo neutrophil and macrophage depletion
For neutrophil depletion, each mouse was injected intravenously with 200 μg of anti-mouse Ly6G antibodies (BE0075-1, BioXCell) 24 h before CLP. For macrophage depletion, mice were intraperitoneally administered 150 μL clodronate-loaded liposomes (Liposoma, Amsterdam) 24 h before CLP according to the manufacturer’s instructions.
Western blotting analysis
Cell protein was extracted by a commercial kit (KGP10100, KeyGEN Biotech), and the protein concentrations were determined by using the Pierce™ BCA protein assay kit (23225, Thermo). Then, protein samples were separated by 10% or 12% SDS‒PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (IPVH00010, Millipore). PVDF membranes were blocked with 5% skim milk for 1 h and then incubated with corresponding primary antibodies at 4 °C overnight. Rabbit anti-SIRT2 (19655-1-AP, 1:2000 dilution; Proteintech), mouse anti-α-tubulin (3873, 1:2000 dilution; CST) and rabbit anti-acetyl-α-tubulin (5335, 1:2000 dilution; CST) antibodies were used. Subsequently, after washing with TBS-T, membranes were incubated at 37 °C for 1 h with the following secondary antibodies: horse-anti-mouse IgG-HRP (7076, 1:2000 dilution; CST) and goat-anti-rabbit IgG-HRP (7074, 1:2000 dilution; CST). The bands were analyzed using the ChemiDoc MP imaging system (Bio-Rad, USA) with Image Lab 5.2.1 software.
Transient transfection
The siRNA and NC oligonucleotide sequences were synthesized by RiboBio (Guangzhou, China). The SIRT2-overexpression plasmids and empty controls were synthesized by GeneChem (Shanghai, China). Transfection of SIRT2 siRNA or overexpression plasmid was performed with a riboFECT CP Transfection Kit (C10511-05, RiboBio) or INVI DNA RNA Transfection Reagent™ (IV1216075, Invigentech), respectively. Cells were harvested for assay 48 h after transfection.
ROS detection assay
BMDMs with the indicated treatments were incubated with or without PPA for 18 h and then infected with E. coli or S. aureus for 45 min. After incubation, the cells were washed with PBS three times and then incubated for an additional 1 h. Subsequently, BMDMs were treated with 5 μM of the fluorogenic probe H2DCFDA (HY-D0940, MCE) for 30 min at 37 °C. The emitted fluorescence was detected using a fluorescence microplate reader with 488/525 nm excitation/emission filters.
Determination of NADPH levels and G6PD activity in BMDMs
NADPH levels and G6PD activity were assessed in the lysates of BMDMs with the indicated treatments by using an NADP/NADPH assay kit (S0179, Beyotime) and a G6PD assay kit (S0189, Beyotime) according to the manufacturer’s instructions.
Drug affinity responsive target stability assay
The drug affinity responsive target stability (DARTS) assay was conducted as described previously [58], with minor modifications using BMDMs and Raw 264.7 cells. Briefly, untreated cells were lysed with a mammalian protein extraction reagent (78501, Thermo Scientific) containing freshly added protease inhibitors on ice for 15–30 min. After centrifugation (18,000 × g for 15 min) and the addition of 1× TNC buffer [500 mM Tris·HCl (pH 8.0), 500 mM NaCl, 100 mM CaCl2], lysates were equally divided into 4 tubes (400 μg of total protein, 4 μg/μL) and gently mixed with the indicated concentrations of PPA or an equal volume of DMSO, followed by incubation for 2 h at RT. Incubated samples were digested by pronase (4693116001, Roche) at a 1:400 ratio (w/w) for 30 min. Then, a loading buffer was added into the lysates and boiled for 10 min, and the samples were separated by SDS‒PAGE and stained with Coomassie Blue or for western blotting analysis.
Cellular thermal shift assay
A cellular thermal shift assay (CETSA) was performed as previously reported with minor modifications [59]. Briefly, BMDMs or Raw 264.7 cells were collected, pelleted, washed with PBS, and lysed by freeze‒thawing using liquid nitrogen. The lysate was equally divided into two tubes and incubated with PPA (100 μM) or an equal volume of DMSO for 2 h at RT. Subsequently, the lysates were heat-treated at 37, 41, 44, 47, 50, 53, 56, 59, 63 and 67 °C for 3 min and immediately cooled for 3 min at RT, followed by centrifugation at 15,000 rpm for 15 min at 4 °C. Then, the supernatant was boiled after adding SDS‒PAGE loading buffer and analyzed by immunoblotting.
Surface-plasmon resonance
The interaction between PPA and SIRT2 was quantitatively measured using the PlexArray® HT System (PlexArray, USA) with a 3D MES/HEMA sensor chip. Purified recombinant SIRT2 protein (P1226, Finetest) was immobilized on the protein chip and activated with 10 mM NiSO4. Gradient concentrations of PPA (25, 50, 100, 200, 400, 800 μmol/L) were injected as analytes. Data were analyzed using PlexArray HT specialized software (Plexera Bioscience).
Molecular docking analysis
The SIRT2 protein crystal structure was downloaded from the protein data bank (PDB ID 5YQM), and the structure of PPA was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Schrodinger-Maestro software (version 11.1) was used to analyze the binding sites between PPA and SIRT2. After removing the crystal water and FBP and adding the hydrogen atoms, PPA was then docked into SIRT2 based on evolutionary optimization of the ligand pose. The binding site of PCA was defined as proteinaceous residues located within a distance of 1.74 Å radius from PPA.
Statistical analysis
Statistical analysis was performed using Prism 9.5.1 (GraphPad Software). Survival studies were analyzed using log-rank (Mantel‒Cox) tests. Statistical significance was evaluated by two-tailed Student’s t test or one-way analysis of variance followed by the Bonferroni post hoc test. Correlation significance was determined by using linear regression analysis. Unless otherwise stated, data are presented as the mean ± SEM. P values < 0.05 were considered statistically significant. N represents the number of biological replicates.
Supplementary information
Author contributions
PG, RL, QY, LX, RW, FM and TC performed the experiments and analyzed the data; JL, ZZ, YH and HZ collected all the clinical data; HP, HC and YJ provided technical support; KSN revised the manuscript; WG, YC, BS, and PC designed the study, interpreted the data, drafted and edited the manuscript, and supervised the study.
Funding
This study was supported by the National Natural Science Foundation of China (32271230 and 32071124) to PC; the NIH Grant (P30DK120515) to BS; the National Natural Science Foundation of China (82270581) to YC; the National Key R&D Project of China (2018YFC0115301), the National Natural Science Foundation of China (81974070), the Shenzhen Science and Technology Program (JCYJ20210324131010027) and the Research Foundation of Shenzhen Hospital of Southern Medical University (PT2018GZR10) to WG.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Peng Gu, Ruofan Liu, Qin Yang, Li Xie, Rongjuan Wei.
Contributor Information
Wei Gong, Email: gongwei@smu.edu.cn.
Ye Chen, Email: yechen@smu.edu.cn.
Peng Chen, Email: perchen@smu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-023-01070-5.
References
- 1.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315:801–10. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavaillon JM, Singer M, Skirecki T. Sepsis therapies: learning from 30 years of failure of translational research to propose new leads. EMBO Mol Med. 2020;12:e10128. doi: 10.15252/emmm.201810128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busani S, Serafini G, Mantovani E, Venturelli C, Giannella M, Viale P, et al. Mortality in patients with septic shock by multidrug resistant bacteria: risk factors and impact of sepsis treatments. J Intensive Care Med. 2019;34:48–54. doi: 10.1177/0885066616688165. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J, Vincent JL, Adhikari NK, Machado FR, Angus DC, Calandra T, et al. Sepsis: a roadmap for future research. Lancet Infect Dis. 2015;15:581–614. doi: 10.1016/S1473-3099(15)70112-X. [DOI] [PubMed] [Google Scholar]
- 5.Liu YC, Zou XB, Chai YF, Yao YM. Macrophage polarization in inflammatory diseases. Int J Biol Sci. 2014;10:520–29. doi: 10.7150/ijbs.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabani R, Volchuk A, Jerkic M, Ormesher L, Garces-Ramirez L, Canton J, et al. Mesenchymal stem cells enhance Nox2-dependent reactive oxygen species production and bacterial killing in macrophages during sepsis. Eur Respir J. 2018;51:1702021. doi: 10.1183/13993003.02021-2017. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Wu H, Chen L, Wen Y, Kong X, Gao WQ. Park7 interacts with p47(phox) to direct NADPH oxidase-dependent ROS production and protect against sepsis. Cell Res. 2015;25:691–706. doi: 10.1038/cr.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eskandarian HA, Impens F, Nahori MA, Soubigou G, Coppee JY, Cossart P, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 9.Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen LL, Liu LX, et al. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. 2014;33:1304–20. doi: 10.1002/embj.201387224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciarlo E, Heinonen T, Theroude C, Herderschee J, Mombelli M, Lugrin J, et al. Sirtuin 2 deficiency increases bacterial phagocytosis by macrophages and protects from chronic staphylococcal infection. Front Immunol. 2017;8:1037. doi: 10.3389/fimmu.2017.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correale J, Hohlfeld R, Baranzini SE. The role of the gut microbiota in multiple sclerosis. Nat Rev Neurol. 2022;18:544–58.. doi: 10.1038/s41582-022-00697-8. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan V, Kanwar SS. Lipopeptide(s) associated with human microbiome as potent cancer drug. Semin Cancer Biol. 2021;70:128–33.. doi: 10.1016/j.semcancer.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Malka O, Kalson D, Yaniv K, Shafir R, Rajendran M, Ben-David O, et al. Cross-kingdom inhibition of bacterial virulence and communication by probiotic yeast metabolites. Microbiome. 2021;9:70. doi: 10.1186/s40168-021-01027-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R. Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: a door to the body. Front Immunol. 2021;12:578386. doi: 10.3389/fimmu.2021.578386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong S, Lan T, Zeng L, Luo H, Yang X, Li N, et al. Gut microbiota mediates diurnal variation of acetaminophen induced acute liver injury in mice. J Hepatol. 2018;69:51–59. doi: 10.1016/j.jhep.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terciolo C, Dapoigny M, Andre F. Beneficial effects of Saccharomyces boulardii CNCM i-745 on clinical disorders associated with intestinal barrier disruption. Clin Exp Gastroenterol. 2019;12:67–82. doi: 10.2147/CEG.S181590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, et al. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 2015;11:e1005614. doi: 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao TY, Haslam DB, Bennett RJ, Way SS. Friendly fungi: symbiosis with commensal Candida albicans. Trends Immunol. 2022;43:706–17.. doi: 10.1016/j.it.2022.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu H, Duan Y, Lang S, Jiang L, Wang Y, Llorente C, et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J Hepatol. 2020;72:391–400. doi: 10.1016/j.jhep.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Investig. 2017;127:2829–41.. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mogilnicka I, Ufnal M. Gut mycobiota and fungal metabolites in human homeostasis. Curr Drug Targets. 2019;20:232–40.. doi: 10.2174/1389450119666180724125020. [DOI] [PubMed] [Google Scholar]
- 22.Mendoza SR, Zamith-Miranda D, Takacs T, Gacser A, Nosanchuk JD, Guimaraes AJ. Complex and controversial roles of eicosanoids in fungal pathogenesis. J Fungi. 2021;7:254. doi: 10.3390/jof7040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panpetch W, Somboonna N, Bulan DE, Issara-Amphorn J, Worasilchai N, Finkelman M, et al. Gastrointestinal colonization of Candida albicans increases serum (1–>3)-beta-d-glucan, without candidemia, and worsens cecal ligation and puncture sepsis in murine model. Shock. 2018;49:62–70. doi: 10.1097/SHK.0000000000000896. [DOI] [PubMed] [Google Scholar]
- 24.Wagener J, MacCallum DM, Brown GD, Gow NA. Candida albicans chitin increases arginase-1 activity in human macrophages, with an impact on macrophage antimicrobial functions. mBio. 2017;8:e01820–16.. doi: 10.1128/mBio.01820-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. Pathogenic yeasts cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun. 2001;69:2957–63. doi: 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amatullah H, Shan Y, Beauchamp BL, Gali PL, Gupta S, Maron-Gutierrez T, et al. DJ-1/park7 impairs bacterial clearance in sepsis. Am J Respir Crit Care Med. 2017;195:889–905. doi: 10.1164/rccm.201604-0730OC. [DOI] [PubMed] [Google Scholar]
- 28.Rumpf T, Schiedel M, Karaman B, Roessler C, North BJ, Lehotzky A, et al. Selective SIRT2 inhibition by ligand-induced rearrangement of the active site. Nat Commun. 2015;6:6263. doi: 10.1038/ncomms7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou J, Chen Q, Wu X, Zhao D, Reuveni H, Licht T, et al. S1pr3 signaling drives bacterial killing and is required for survival in bacterial sepsis. Am J Respir Crit Care Med. 2017;196:1559–70.. doi: 10.1164/rccm.201701-0241OC. [DOI] [PubMed] [Google Scholar]
- 30.Pizzolla A, Hultqvist M, Nilson B, Grimm MJ, Eneljung T, Jonsson IM, et al. Reactive oxygen species produced by the nadph oxidase 2 complex in monocytes protect mice from bacterial infections. J Immunol. 2012;188:5003–11. doi: 10.4049/jimmunol.1103430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong X, Thimmulappa R, Craciun F, Harvey C, Singh A, Kombairaju P, et al. Enhancing Nrf2 pathway by disruption of keap1 in myeloid leukocytes protects against sepsis. Am J Respir Crit Care Med. 2011;184:928–38. doi: 10.1164/rccm.201102-0271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang S, Duan Y, Liu J, Torralba MG, Kuelbs C, Ventura-Cots M, et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology. 2020;71:522–38.. doi: 10.1002/hep.30832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–16. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider SM, Girard-Pipau F, Filippi J, Hebuterne X, Moyse D, Hinojosa GC, et al. Effects of Saccharomyces boulardii on fecal short-chain fatty acids and microflora in patients on long-term total enteral nutrition. World J Gastroenterol. 2005;11:6165–69. doi: 10.3748/wjg.v11.i39.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang TT, Shao TY, Ang W, Kinder JM, Turner LH, Pham G, et al. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe. 2017;22:809–16.. doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quintin J, Saeed S, Martens J, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–32. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam S, Bai X, Shkoporov AN, Park H, Wu X, Lan P, et al. Roles of the gut virome and mycobiome in faecal microbiota transplantation. Lancet Gastroenterol Hepatol. 2022;7:472–84.. doi: 10.1016/S2468-1253(21)00303-4. [DOI] [PubMed] [Google Scholar]
- 38.Tso G, Reales-Calderon JA, Tan A, Sem X, Le GTT, Tan TG, et al. Experimental evolution of a fungal pathogen into a gut symbiont. Science. 2018;362:589–95.. doi: 10.1126/science.aat0537. [DOI] [PubMed] [Google Scholar]
- 39.Shao TY, Ang W, Jiang TT, Huang FS, Andersen H, Kinder JM, et al. Commensal Candida albicans positively calibrates systemic th17 immunological responses. Cell Host Microbe. 2019;25:404–17.. doi: 10.1016/j.chom.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cugini C, Calfee MW, Farrow JR, Morales DK, Pesci EC, Hogan DA. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- 41.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–66. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 42.Tachedjian G, Aldunate M, Bradshaw CS, Cone RA. The role of lactic acid production by probiotic lactobacillus species in vaginal health. Res Microbiol. 2017;168:782–92.. doi: 10.1016/j.resmic.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Cabral DJ, Penumutchu S, Norris C, Morones-Ramirez JR, Belenky P. Microbial competition between escherichia coli and Candida albicans reveals a soluble fungicidal factor. Micro Cell. 2018;5:249–55.. doi: 10.15698/mic2018.05.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo Z, Yu S, Zeng W, Zhou J. Comparative analysis of the chemical and biochemical synthesis of keto acids. Biotechnol Adv. 2021;47:107706. doi: 10.1016/j.biotechadv.2021.107706. [DOI] [PubMed] [Google Scholar]
- 45.Wang T, Liao Z, Man D, Yang J, He Z, Zeng Y, et al. Effects of phenylpyruvate on anxiety, depression and exercise capacity in C57BL/6J male mice. Acta Agric Univ Jiangxiensis. 2019;41:551–57. [Google Scholar]
- 46.van Spronsen FJ, Blau N, Harding C, Burlina A, Longo N, Bosch AM. Phenylketonuria. Nat Rev Dis Prim. 2021;7:36. doi: 10.1038/s41572-021-00267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sidorova-Darmos E, Wither RG, Shulyakova N, Fisher C, Ratnam M, Aarts M, et al. Differential expression of sirtuin family members in the developing, adult, and aged rat brain. Front Aging Neurosci. 2014;6:333. doi: 10.3389/fnagi.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human sir2 ortholog, sirt2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–44. doi: 10.1016/S1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 49.Gomes P, Fleming OT, Cavadas C. Emerging role of sirtuin 2 in the regulation of mammalian metabolism. Trends Pharmacol Sci. 2015;36:756–68.. doi: 10.1016/j.tips.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Jing E, Gesta S, Kahn CR. Sirt2 regulates adipocyte differentiation through foxo1 acetylation/deacetylation. Cell Metab. 2007;6:105–14. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, et al. Sirt2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–61. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, et al. Acetylation regulates gluconeogenesis by promoting pepck1 degradation via recruiting the ubr5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosa AP, Jacques CE, Moraes TB, Wannmacher CM, Dutra AM, Dutra-Filho CS. Phenylpyruvic acid decreases glucose-6-phosphate dehydrogenase activity in rat brain. Cell Mol Neurobiol. 2012;32:1113–18. doi: 10.1007/s10571-012-9834-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Assouvie A, Daley-Bauer LP, Rousselet G. Growing murine bone marrow-derived macrophages. Methods Mol Biol. 2018;1784:29–33. doi: 10.1007/978-1-4939-7837-3_3. [DOI] [PubMed] [Google Scholar]
- 55.Swamydas M, Luo Y, Dorf ME, Lionakis MS. Isolation of mouse neutrophils. Curr Protoc Immunol. 2015;110:3–20. doi: 10.1002/0471142735.im0320s110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang P, Mu X, Zhao H, Li Y, Wang L, Wolfe V, et al. Administration of GDF3 into septic mice improves survival via enhancing LXRα-mediated macrophage phagocytosis. Front Immunol. 2021;12:647070. doi: 10.3389/fimmu.2021.647070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pai MY, Lomenick B, Hwang H, Schiestl R, McBride W, Loo JA, et al. Drug affinity responsive target stability (darts) for small-molecule target identification. Methods Mol Biol. 2015;1263:287–98. doi: 10.1007/978-1-4939-2269-7_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100–22. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.