Abstract
SEL1L-mediated endoplasmic reticulum-associated degradation (ERAD) plays critical roles in controlling protein homeostasis by degrading misfolded or terminal unfolded proteins. However, it remains unclear how SEL1L regulates peripheral T-cell survival and homeostasis. Herein, we found that SEL1L deficiency led to a greatly reduced frequency and number of mature T cells, which was further validated by adoptive transfer experiments or bone marrow chimera experiments, accompanied by the induction of multiple forms of cell death. Furthermore, SEL1L deficiency selectively disrupted naïve CD8+ T-cell homeostasis, as indicated by the severe loss of the naïve T-cell subset but an increase in the memory T-cell subset. We also found that SEL1L deficiency fueled mTORC1/c-MYC activation and induced a metabolic shift, which was largely attributable to enhanced expression of the IL-15 receptor α and β chains. Mechanistically, single-cell transcriptomic profiling and biochemical analyses further revealed that Sel1l−/− CD8+ T cells harbored excessive ER stress, particularly aberrant activation of the PERK-ATF4-CHOP-Bim pathway, which was alleviated by supplementing IL-7 or IL-15. Importantly, PERK inhibition greatly resolved the survival defects of Sel1l−/− CD8+ T cells. In addition, IRE1α deficiency decreased mTORC1 signaling in Sel1l−/− naïve CD8+ T cells by downregulating the IL-15 receptor α chain. Altogether, these observations suggest that the ERAD adaptor molecule SEL1L acts as an important checkpoint for preserving the survival and homeostasis of peripheral T cells by regulating the PERK signaling cascade and IL-15 receptor-mediated mTORC1 axis.
Keywords: T-cell homeostasis, Endoplasmic reticulum-associated degradation, ER stress response, PERK, IRE1α
Subject terms: T cells, Cellular immunity
Introduction
The stringent proteostasis system ensures the delicate regulation of a balanced and functional proteome within the cell [1]. Misfolded or aggregated proteins in the endoplasmic reticulum are selectively recognized and degraded via the endoplasmic reticulum-associated degradation (ERAD) pathway [2, 3]. Mammalian cells have evolved remarkably elaborate ERAD systems, and accumulating evidence indicates that there are at least 10 ERAD branches with their respective ubiquitin ligases and substrates [4]. The complex containing suppressor/enhancer of Lin-12-like (SEL1L) and HRD1 is the most conserved ERAD branch across eukaryotes and harbors a substantial amount of substrates [2]. SEL1L is an important adaptor molecule that maintains HRD1 stability and recruits substrates [5–7]. Previous studies have indicated that the SEL1L/HRD1 ERAD complex has indispensable physiological significance for regulating food uptake, water balance, systemic energy homeostasis and lipid metabolism [5, 8–10]. Notably, SEL1L deletion in brown adipocytes leads to the formation of megamitochondria, which are morphologically enlarged and abnormally shaped with perforating ER tubules. Accordingly, mitochondrial function is greatly impaired due to dysregulated ER-mitochondria contacts and mitochondrial dynamics [11]. In addition, loss of SEL1L in hematopoietic stem cells (HSCs) drives hyperproliferation and impairs long-term self-renewal potential [12, 13]. Moreover, SEL1L has been shown to preserve quiescence and the self-renewal capacity of HSCs via restricted mTOR activity and MPL, the master regulator of HSC identity. Similarly, a recent study suggested that SEL1L acts as a critical protein quality-control checkpoint governing thymic β-selection and the DN3 to DN4 transition. Interestingly, that study demonstrated that the Notch intracellular domain is critically required for the transcription of core ERAD genes, including SEL1L and HRD1 [14].
Naïve T-cell homeostasis is maintained via tonic T-cell receptor signaling as well as cytokine stimulation [15]. Given the critical role of SEL1L in regulating HSC quiescence and thymocyte development, we thus aimed to investigate whether and how the SEL1L-mediated ERAD pathway regulates peripheral naïve T-cell homeostasis under steady-state conditions. Herein, by using a T-cell-specific Sel1l-knockout mouse model, we demonstrated that SEL1L deficiency led to significantly decreased numbers of mature T cells in both the thymus and periphery. Consistently, Sel1l−/− T cells were largely outcompeted by WT T cells in adoptive transfer and competitive bone marrow transplantation experiments. Importantly, naïve CD8+ rather than CD4+ T-cell maintenance was heavily disrupted in the absence of SEL1L, as indicated by marked loss of the CD62L+CD44− naïve T-cell population and increases in both the CD62L+CD44+ and CD62L−CD44+ T-cell populations. Mechanistically, we demonstrated that SEL1L deficiency resulted in PERK-ATF4-CHOP-Bim hyperactivation and that inhibition of PERK largely rescued Sel1l−/− CD8+ T-cell survival. Furthermore, SEL1L deficiency selectively impaired tonic TCR signaling in mature CD8+ rather than CD4+ T cells by decreasing surface TCRβ and CD3 expression. Additionally, SEL1L deficiency resulted in elevated IL-15 receptor expression and signaling, which enhanced mTORC1/c-MYC activation. In contrast, IRE1α deletion attenuated mTORC1 signaling in Sel1l−/− naïve CD8+ T cells by reducing the IL-15 receptor α chain. Taken together, these findings provide important insights into the role of the SEL1L-mediated ERAD pathway in preserving the survival and homeostasis of mature CD8+ T cells.
Results
SEL1L deficiency leads to significantly decreased numbers of T cells in the thymus and periphery
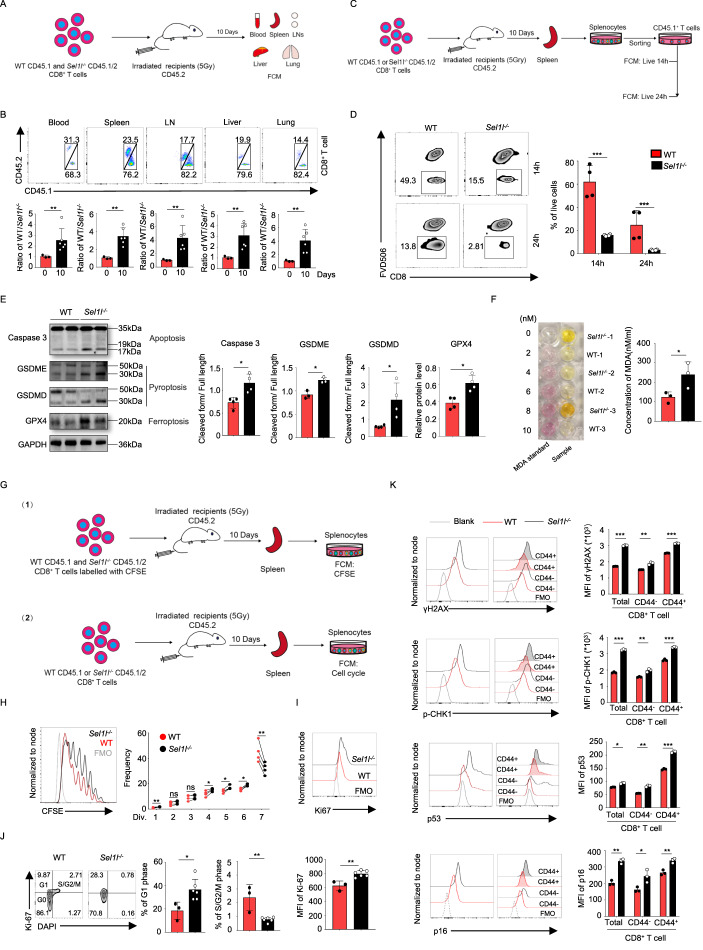
To probe the role of SEL1L in regulating T-cell development and homeostasis, we generated T-cell-specific Sel1l-knockout mice by crossing Sel1lflox/flox mice with the CD4-Cre transgenic mouse line to bypass the potential block of SEL1L deficiency on early T-cell development. We first confirmed the specific deletion of SEL1L in T cells by both genomic PCR analysis and Western blot analyses (Fig. S1A). Next, we sought to determine the impacts of SEL1L deficiency on T-cell development. Notably, the frequencies and numbers of SP4+ and SP8+ thymocytes were strikingly decreased in the absence of SEL1L, whereas the percentage of DP thymocytes was slightly increased (Fig. 1A, B), and the total number of DP thymocytes remained comparable.
Fig. 1.
SEL1L deletion decreases the number of T cells in a cell-intrinsic manner. Representative flow cytometry plots and quantification of the frequencies (A) and absolute numbers (B) of single-positive CD4+ (SP4+) cells, CD8+ (SP8+) cells, double-positive CD4+CD8+ cells (DP cells), and double-negative cells (DN cells) in thymus tissues from 8-week-old Sel1lflox/flox (WT) and Sel1lflox/flox:CD4-Cre (Sel1l−/−) mice (WT, n = 5; Sel1l−/−, n = 5). Representative flow cytometry plots (C) and quantification of the frequencies and absolute numbers (D) of T cells separately in spleens and lymph nodes from 8-week-old WT and Sel1l−/− mice (WT, n = 5; Sel1l−/−, n = 5). Representative flow cytometry plots and quantification of the frequencies (E) and absolute numbers (F) of CD8+ and CD4+ T cells separately in spleens and lymph nodes from 8-week-old WT and Sel1l−/− mice (WT, n = 5; Sel1l−/−, n = 5). G Schematic of the competitive BMT experiment using whole bone marrow cells from WT and Sel1l−/− mice as donors. H, I Ratio of WT to Sel1l−/− donor-derived CD45+ cells (H) and B cells, CD4+T cells and CD8+ cells in the peripheral blood of the recipient mice at the indicated time points in the competitive BMT experiment (n = 10). Ratios of WT to Sel1l−/− donor-derived DN, DP, SP4+, SP8+ cells in thymus tissues (J) and B cells, CD4+ T cells and CD8+ cells in the spleens and lymph nodes (K) of the recipient mice at the indicated time points in the competitive BMT experiment (n = 10). The data are shown as the mean ± SD, and statistical significance was determined by unpaired two-tailed Student’s t test (A–F). The data are representative of at least three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001)
Consistent with the fact that the frequencies of both SP4+ and SP8+ thymocytes were decreased in the thymus, we observed a significant reduction in total CD3+ T cells in both Sel1l−/− spleens and peripheral LNs (pLNs) (Fig. 1C, D). Among these T cells, Sel1l−/− splenic CD8+ T cells were particularly decreased with a 5-fold reduction, whereas CD4+ T cells were ~2-fold reduced (Fig. 1E, F). Consistently, Sel1l−/− CD8+ T cells were particularly decreased with a 3-fold reduction, whereas CD4+ T cells were ~1.6-fold reduced in peripheral blood (Fig. S1F). In addition, the frequencies and numbers of CD4+CD25+Foxp3+ regulatory T cells and NK T cells were also reduced in Sel1l−/− thymus, spleen and pLNs compared to their WT counterparts (Fig. S1B, D). In contrast, the percentages and numbers of γδ T cells, NK cells and B cells were significantly increased in both the spleen and pLNs (Fig. S1C–E).
To further confirm that the decreased levels of thymic or peripheral CD8+ T cells were attributed to SEL1L deficiency in a cell-intrinsic manner, we performed adoptive transfer of mixed bone marrow (BM). To this end, hosts were lethally irradiated and reconstituted with equal amounts of WT or Sel1l−/− BM cells, and the development of diverse subsets of CD8+ or CD4+ T cells was evaluated (Fig. 1G). As expected, the frequencies of WT or Sel1l−/− BM-derived total CD45+ cells or B cells remained comparable at 8 or 12 weeks post reconstitution (Fig. 1H, I). Notably, the percentages of both CD8+ and CD4+ T cells were drastically reduced in the peripheral blood, spleen and pLNs (Fig. 1I, K), suggesting that loss of SEL1L led to incompetence of T cells in the competitive reconstitution setting. Consistently, the percentages of SP4+ and SP8+ thymocytes were also reduced in the absence of SEL1L, although the frequencies of DNs and DPs remained comparable (Fig. 1J). Given all the evidence, we conclude that SEL1L is indispensable for the development and maintenance of T cells, in particular CD8+ T cells, under steady-state conditions in an intrinsic manner.
SEL1L is required for the survival and homeostatic proliferation of CD8+ T cells
The size of the T-cell pool is tightly regulated and contingent on T-cell survival and homeostatic proliferation [16]. To further investigate the mechanisms underlying the decreased number of T cells, we evaluated the survival and proliferative capacity of WT and Sel1l−/− T cells. Since the CD8+ T-cell number was mostly impacted by SEL1L deficiency, we focused primarily on this population in subsequent studies. To this end, WT (CD45.1) and Sel1l−/− (CD45.1/CD45.2) CD8+ T cells were sorted and cotransferred into sublethally irradiated (5 Gy) recipient (CD45.2) mice, and their proportions were evaluated under homeostatic conditions (Fig. 2A). Indeed, the ratio of WT to Sel1l−/− CD8+ T cells was significantly increased in the peripheral blood, spleen, lymph nodes, liver and lung tissues, indicating that Sel1l−/− CD8+ T cells were outcompeted under competitive circumstances (Fig. 2B).
Fig. 2.
SEL1L deletion impairs CD8+ T-cell survival and homeostatic proliferation in a cell-intrinsic manner. A, B Experimental scheme of the adoptive cotransferred CD8+ T-cell population in vivo. CD45.2 recipient mice were irradiated with 5 Gy and administered equal proportions of sorted splenic CD8+ T cells from WT (CD45.1) and Sel1l−/− (CD45.1/CD45.2) mice the next day. After 10 days, blood, spleen, lymph node, liver and lung samples were harvested to detect the frequency of transferred WT and Sel1l−/− CD8+ T-cell populations by flow cytometry (A). Representative flow cytometry plots showing the populations of transferred WT and Sel1l−/− CD8+ T cells from blood, spleen, lymph node, liver and lung samples. The ratio of WT to Sel1l−/− CD8+ T cells was also quantified (n = 3–6) (B). C, D Experimental scheme for analysis of adoptively transferred CD8+ T-cell survival in vivo. CD45.2 recipient mice were irradiated with 5 Gy and separately administered equal numbers of sorted CD8+ T cells from WT (CD45.1) or Sel1l−/− (CD45.1/CD45.2) mice the next day (C). After 10 days, the spleen was harvested to sort CD45.1 CD8+ T cells, and the sorted CD45.1 splenic T cells were cultured in complete medium ex vivo for 14 and 24 h. Then, the survival of the sorted T cells was detected by flow cytometry (D) (WT, n = 4; Sel1l−/−, n = 4). E Western blot of the expression of cleaved Caspase3, GSDME, GSDMD, GPX4 and GAPDH with the relative band intensity (cleaved length normalized to full length) in unprimed CD8+ T cells from 8-week-old WT and Sel1l−/− mice. Each lane shows the results of pooled CD8+ T cells from two mice. F The lipid peroxidation marker MDA was detected with a microplate reader in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 3). G–J Experimental scheme of the homeostatic proliferation assay in vivo. CD45.2 recipient mice irradiated with 5 Gy were cotransferred with an equal number of sorted CD8+ T cells labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) to detect the transferred homeostatic proliferation of CD8+ T cells. CD45.2 recipient mice irradiated with 5 Gy were separately administered the same amounts of sorted CD8+ T cells from WT (CD45.1) or Sel1l−/− (CD45.1/CD45.2) mice, and the cell cycle phase was detected (G). Representative flow cytometry analysis of the proliferation of the transferred WT and Sel1l−/− CD8+ T cells (n = 4) (H). Representative flow cytometry plots showing Ki-67 expression (I) and the proportions of WT and Sel1l−/− CD8+ T cells in different phases of the cell cycle in the spleen (WT, n = 3; Sel1l−/−, n = 6) (J). K Representative flow cytometry analysis of γH2AX, p-CHK1, p53 and p16 expression in splenic CD8+ T cells from 6-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 3). The data are shown as the mean ± SD, and statistical significance was determined by unpaired two-tailed Student’s t test (B, D–F, I–K) and paired two-tailed Student’s t test (H). The data are representative of two or three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001)
Next, we further investigated whether the reduced number of Sel1l−/− CD8+ T cells is attributed to increased cell death or impaired proliferation under homeostatic conditions. In this regard, the transferred WT and Sel1l−/− CD8+ T cells from sublethally irradiated recipient mice were sorted and cultured in complete medium for 14 or 24 h, and their survival was detected (Fig. 2C). Indeed, the ex vivo viability of transferred Sel1l−/− CD8+ T cells was severely reduced compared to that of WT cells (Fig. 2D). Moreover, the survival of both unprimed and activated Sel1l−/− CD8+ T cells was impaired (Fig. S2A, B). Cell death can occur in multiple forms in response to distinct stresses; thus, we sought to characterize the key cell death modalities of Sel1l−/− CD8+ T cells [17]. As shown in Fig. 2E, the expression of cleaved caspase 3 (an apoptosis marker) and cleaved GSDMD and GSDME (a pyroptosis marker) was increased in Sel1l−/− CD8+ T cells, while the expression of LC-3 and p62 (an autophagic cell death marker) and pMLKL (a necroptosis marker) remained comparable between WT and Sel1l−/− CD8+ T cells (Fig. S2D). Ferroptosis is a newly identified form of cell death and is largely iron dependent [18]. We also measured the lipid peroxidation level by determining the levels of malondialdehyde (MDA), which is a typical product of ferroptosis. Indeed, we observed increased levels of MDA in Sel1l−/− CD8+ T cells (Fig. 2F), and this was also further confirmed by BODIPY C11 staining (Fig. S2C). However, the expression of glutathione peroxidase 4 (GPX4) was upregulated (Figs. 2E and S2E), which may have been related to a compensatory mechanism for coping with ferroptosis in Sel1l−/− CD8+ T cells. Altogether, these observations suggest that Sel1l−/− CD8+ T cells displayed evident survival defects. Furthermore, as shown in Fig. 2G, H, we found that Sel1l−/− CD8+ T cells exhibited markedly decreased homeostatic proliferation compared with that of WT CD8+ T cells. Next, we investigated whether SEL1L deficiency controls the cell cycling of CD8+ T cells under steady-state conditions. Surprisingly, we found upregulated Ki67 expression and an increased frequency of cells in G1 phase but a decreased percentage of cells in S/G2/M phase among the transferred Sel1l−/− CD8+ T cells (Fig. 2I, J), indicating that Sel1l−/− CD8+ T cells held proliferative capacity but were arrested at G1 phase under homeostatic conditions. It is well known that DNA damage is one of the leading causes of G1/S arrest. We next sought to determine whether Sel1l−/− CD8+ T cells display increased DNA damage responses. To this end, we measured several key regulators of the DNA damage response, such as γ-H2AX, p-CHK1, p53 and p16. Notably, among all tested regulators, γ-H2AX, p-CHK1, p53 and p16 exhibited markedly increased expression in Sel1l−/− CD8+ T cells (Figs. 2K and S3A). These data suggest that SEL1L is required for the survival and homeostatic proliferation of CD8+ T cells via control of the DNA damage response.
SEL1L deficiency induces intense ER stress in CD8+ T cells
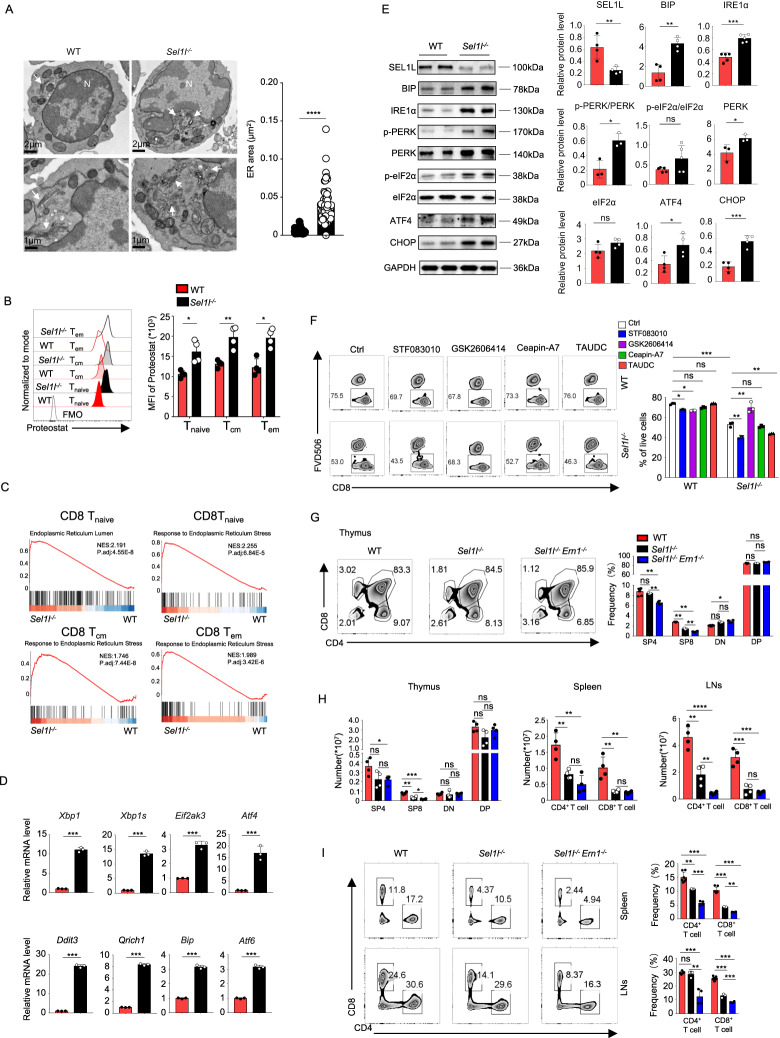
Previous studies have indicated that SEL1L deficiency leads to abnormal ER morphology in acinar cells and macrophages [7, 19]. Next, we sought to investigate the morphological alterations of the ER within CD8+ T cells by transmission electron microscopy. In line with previous studies, swollen, fragmented and dilated balloon-like ERs were also observed in the Sel1l−/− CD8+ T cells, whereas the WT counterparts displayed long, thin, sheet-like, densely packed cisternae (Fig. 3A). Additionally, we noticed increased side scatter area (SSC-A) within Sel1l−/− CD8+ T cells, indicating increased granule content (Fig. S3B). Given the critical role of ERAD in controlling protein homeostasis by degrading misfolded or aggregated proteins [2], we further explored whether SEL1L deficiency triggers dysregulated protein quality control and leads to a severe ER stress response. In this regard, we measured protein aggregation with a PROTEOSTAT protein aggregation assay, and our observations indeed revealed that SEL1L deficiency resulted in significant accumulation of protein aggregates (Fig. 3B).
Fig. 3.
SEL1L deficiency induces intense ER stress in CD8+ T cells. A Representative TEM images showing the ultrastructure of unprimed CD8+ T cells. The quantitation of the ER area is shown on the right (n = 60 ER areas from 12 WT CD8+ T cells and 70 ER areas from 13 Sel1l−/− CD8+ T cells, pooled from three mice per genotype). The arrows point to the ER. N nucleus. B Representative flow cytometry plots showing the MFI from the PROTEOSTAT assay in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 4). C GSEA plots depicting the enriched gene sets between WT and Sel1l−/− CD8+ T cells. Tnaive naïve T-cell, Tcm central memory T-cell, Tem effector memory T-cell. D RT‒qPCR analysis of ER stress signature gene expression in splenic WT and Sel1l−/− CD8+ T cells (Xbp1, Xbp1s, Eif2ak3, ATF4, Ddit3, Qrich1, Bip, Atf6), normalized to the expression of β-actin (WT, n = 3; Sel1l−/−, n = 3). E Lysates from isolated CD8+ T cells in 8-week-old WT and Sel1l−/− mice were subjected to immunoblotting with SEL1L, BIP, IRE1α, p-PERK, PERK, p-eFI2α, eFI2α, ATF4, CHOP and GAPDH antibodies. Each lane shows the results of pooled CD8+ T cells from two mice. F Representative flow cytometry plots showing the percentage of live CD8+ T cells in WT and Sel1l−/− splenic CD8+ T cells after treatment with the IRE1α inhibitor STF083010 (50 μM), the PERK inhibitor GSK2606414 (1 μM), the ATF6 inhibitor ceapin-A7 (1.2 μM) and the pan-ER stress inhibitor TAUDC (0.5 mM) for 24 h. This is representative of three independent biological replicates. G Representative flow cytometry plots showing the frequencies of SP4+ T cells, SP8+ T cells, DN T cells, and DP T cells in thymus tissues from 6-week-old WT, Sel1l−/− and DKO mice (WT, n = 7; Sel1l−/−, n = 3; DKO, n = 4). H Absolute numbers of CD8+ and CD4+ T cells in the thymus, spleen and lymph nodes from 6-week-old WT, Sel1l−/− and DKO mice (WT, n = 4; Sel1l−/−, n = 4; DKO, n = 4). I Representative flow cytometry plots showing the frequency of CD8+ and CD4+ T cells separately in spleens and lymph nodes from 6-week-old WT, Sel1l−/− and DKO mice (WT, n = 7; Sel1l−/−, n = 3; DKO n = 4). The data are shown as the mean ± SD, and statistical significance was determined by unpaired two-tailed Student’s t test (A, B, D, E), Dunnett’s t test (G) and one-way ANOVA followed by the SNK-q post hoc test (G–I). The data are representative of two or three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ****p < 0.0001)
Next, to further explore the ER stress response in Sel1l−/− CD8+ T cells [20], we performed single-cell transcriptomic sequencing to investigate endoplasmic reticulum stress and the unfolded protein response (UPR) in distinct Sel1l−/− CD8+ T-cell subsets. The specific, detailed annotations of each subset are given in Fig. S5H. GSEA confirmed that the ER stress response as well as the UPR were enriched signaling pathways in the absence of SEL1L, as indicated by our single-cell transcriptomic analysis (Fig. 3C). In addition, we carried out qRT‒PCR assays to measure the mRNA expression of key ER stress response genes. Notably, we noticed markedly increased expression of Xbp1, Xbp1s, Eif2ak3, ATF4, Ddit3, Qrich1, Bip and Atf6 in Sel1l−/− CD8+ T cells compared with their WT counterparts (Fig. 3D). Along the same line, several key executors of distinct branches of the UPR were analyzed at the protein level in unprimed WT and Sel1l−/− CD8+ T cells. Consistently, BIP, phosphorylation of PERK and total PERK as well as IRE1α was drastically increased in Sel1l−/− CD8+ T cells (Fig. 3E). Furthermore, p-eIF2α, ATF4 and CHOP, several key downstream indicators of PERK activation, were also upregulated in the absence of SEL1L (Fig. 3E).
Persistent and intense ER stress and protein misfolding initiate intracellular ROS accumulation, which leads to cell death [21]. Therefore, we further investigated the role of ROS generation in the survival defects of Sel1l−/− CD8+ T cells. Unprimed CD8+ T cells produced more ROS in the absence of SEL1L (Fig. S4A). N-acetylcysteine (NAC) is a widely used ROS scavenger, but we found that NAC slightly decreased intracellular ROS levels (Fig. S4B) and marginally improved the survival of Sel1l−/− CD8+ T cells (Fig. S4C). Mitochondria are key organelles that produce large amounts of ROS [22]. Next, we specifically measured mitochondrial-derived ROS production with the probe MitoSox and found comparable levels of MitoSox between Sel1l−/− and WT CD8+ T cells (Fig. S4D). The ER also produces ROS during the oxidative protein folding process through ERO1 [23]. We thus hypothesized that the increased levels of ROS may have been derived from ER stress. ER ROS scavenger treatment decreased intracellular ROS levels (Fig. S4E) but did not rescue the survival of Sel1l−/− CD8+ T cells (Fig. S4F). These findings indicate that a high level of intracellular ROS is likely not the primary reason for impaired survival in Sel1l−/− CD8+ T cells.
To explore the role of ER stress in regulating Sel1l−/− CD8+ T-cell survival, an IRE1α inhibitor (STF083010), a PERK inhibitor (GSK2606414), an ATF6 inhibitor (ceapin-A7), and a pan-ER stress inhibitor (TAUDC) were utilized to treat WT and Sel1l−/− CD8+ T cells. Our data showed that blockade of PERK signaling significantly improved cell survival, whereas IRE1α inhibition slightly impaired cell viability (Fig. 3F). Given the significantly increased IRE1α expression in Sel1l−/− CD8+ T cells, we wondered whether deficiency of Ern1, the gene encoding IRE1α, could resolve the survival defects of Sel1l−/− CD8+ T cells. To answer this question, we generated T-cell-specific Sel1l−/−Ern1−/− DKO mice by crossing Sel1lflox/flox:CD4-Cre mice with Ern1flox/flox mice and confirmed the deletion of SEL1L and IRE1α in the thymus and activated CD8+ T cells by Western blot analyses (Fig. S8B). We therefore measured the percentages of CD8+ T cells in the thymus, spleen, and pLNs and found that deletion of Ern1 further exacerbated the decrease in T-cell number caused by SEL1L deficiency, which was in accordance with the observations with in vitro IRE1a inhibitor treatment (Fig. 3F–I). These data demonstrate that upregulation of IRE1α might represent a cellular defense mechanism and play protective roles to relieve ER stress in the context of SEL1L deletion and that PERK activation is the critical trigger of survival defects.
Inhibition of PERK signaling rescues Sel1l−/− CD8+ T-cell survival
Given that PERK inhibition rescued the survival of Sel1l−/− CD8+ T cells, we sought to further explore the potential effects of PERK inhibition on the survival of Sel1l−/− CD8+ T cells in vivo. We noticed that CD8+ T-cell numbers began to decrease in 1-week-old Sel1l−/− mice (Fig. S9A–C). Therefore, ~3-day-old Sel1l−/− mice were administered the PERK inhibitor GSK2606414 i.p. every four days (Fig. 4A). After 44 days of treatment, we demonstrated that GSK2606414 administration, compared with vehicle treatment, significantly increased both the frequency and absolute numbers of cells in the SP8+ subset (Fig. 4B). Notably, the percentages and absolute numbers of CD8+ T and CD4+ T cells in the spleen and pLNs were evidently rescued by administration of GSK2606414 (Fig. 4C). Taken together, our data indicate that loss of SEL1L leads to aberrant PERK activation and the ER stress response and that inhibition of PERK signaling greatly attenuates the reductions in percentages and absolute numbers of CD8+ T cells.
Fig. 4.
Inhibition of PERK signaling rescues the survival of Sel1l−/− T cells. A Experimental scheme to assess the percentage and absolute number of T cells in Sel1−/− mice treated with or without GSK2606414 every four days for 44 days by intraperitoneal injection starting from 3 days after birth. After 44 days, the thymus, spleen and LNs were harvested (WT, n = 4; Sel1l−/− vehicle, n = 4; Sel1l−/− vehicle, n = 4). B Representative flow cytometry plots showing the frequencies and absolute numbers of SP4+ T cells, SP8+ T cells, DN T cells, and DP T cells in thymus tissues from 6-week-old WT and Sel1l−/− mice treated with or without GSK2606414. PERKi: PERK inhibitor. C Representative flow cytometry plots and quantification of the frequencies and absolute numbers of CD8+ and CD4+ T cells in spleens and lymph nodes from 6-week-old WT and Sel1l−/− mice treated with or without GSK2606414 every other day. D Experimental scheme of the ratio of WT to Sel1l−/− CD8+ T cells and proliferation in recipients treated with or without GSK2606414 (vehicle, n = 4; PERKi, n = 4). E Representative flow cytometry analysis showing the ratio of cotransferred WT to Sel1l−/− naïve CD8+ and CD4+ T-cell populations. F Representative flow cytometry analysis of CFSE intensity in splenic WT and Sel1l−/− naïve CD8+ T cells. G, H Quantification of the percentages and absolute numbers of T cells in Sel1l−/− mice treated with or without GSK2606414 (G). After 40 days, splenic CD8+ T cells were sorted and cultured with or without GSK2606414 for 16 h for the survival assay (WT, n = 3; Sel1l−/− vehicle, n = 3; Sel1l−/− vehicle, n = 3) (H). I Lysates from isolated CD8+ T cells treated with GSK2606414 for 24 h from 8-week-old WT and Sel1l−/− mice were subjected to immunoblotting with p-PERK, PERK, p-eFI2α, ATF4, CHOP, Bim and GAPDH antibodies, and the relative intensity was calculated (ER stress effector molecules normalized to GAPDH). The WT and Sel1l−/− lanes show pooled CD8+ T cells from three or four mice. J Immunoblot in unprimed CD8+ T cells treated with GSK2606414 for 20 h from 8-week-old WT and Sel1l−/− mice, with the relative intensity (cleaved length normalized to full length) being representative of three independent biological replicates. Each lane shows the results of pooled CD8+ T cells from three mice. The data are shown as the mean ± SD, and statistical significance was determined by one-way ANOVA followed by the SNK-q post hoc test (B, C, E, F, H) and paired two-tailed Student’s t test (I). The data are representative of three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ****p < 0.0001)
Next, we sought to further determine whether the rescue effects were due to increased survival or proliferation upon GSK2606414 treatment. To this end, equal amounts of WT and Sel1l−/− naïve CD8+ T cells were labeled with CFSE and cotransferred into recipients. The recipients were than intraperitoneally injected with GSK2606414 every other day for 10 days (Fig. 4D). Notably, without GSK2606414 treatment, Sel1l−/− naïve CD8+ T cells were largely outcompeted by WT cells, and the ratio of WT to Sel1l−/− CD8+ T cells was ~8–10 on day 10 post transfer (Fig. 4E). Intriguingly, upon GSK2606414 treatment, the ratio of WT to Sel1l−/− CD8+ T cells was close to 1 on day 10 post transfer (Fig. 4E). Moreover, we observed that GSK2606414 treatment impaired the proliferation of CD8+ T cells, as suggested by the CFSE dilution assay (Fig. 4F). In this regard, we further confirmed that administration of GSK2606414 greatly improved the survival of Sel1l−/− CD8+ T cells (Fig. 4G, H). Previous studies have indicated that several members of the Bcl-2 family, such as Bcl-2, Bcl-xL, Bax and Bim, are downstream molecules of the PERK-ATF4-CHOP axis [24, 25], and we found that Bim expression was indeed increased in Sel1l−/− CD8+ T cells (Fig. S10E). To reveal the underlying mechanisms by which the PERK inhibitor rescued Sel1l−/− CD8+ T-cell survival, we investigated the expression of Bim in Sel1l−/− CD8+ T cells treated with GSK2606414 for 24 h. Western blot analysis showed that GSK2606414 treatment significantly reduced p-PERK, PERK, p-eIF2α, ATF4, CHOP and Bim expression in Sel1l−/− CD8+ T cells (Fig. 4I). In this regard, our results indicated that SEL1L deficiency led to PERK-p-eIF2α-ATF4-CHOP-Bim hyperactivation and that inhibition of PERK activation improved cell survival. In addition, PERK inhibitor treatment led to reduced levels of cleaved caspase3 and GSDMD in Sel1l−/− CD8+ T cells (Fig. 4J), indicating that PERK inhibition may decrease apoptosis and pyroptosis of Sel1l−/− CD8+ T cells. Taken together, the findings suggest that PERK inhibition attenuates the reduction in CD8+ T-cell number in Sel1l−/− mice by preserving cell survival.
SEL1L is indispensable for the maintenance of peripheral naïve CD8+ T cells
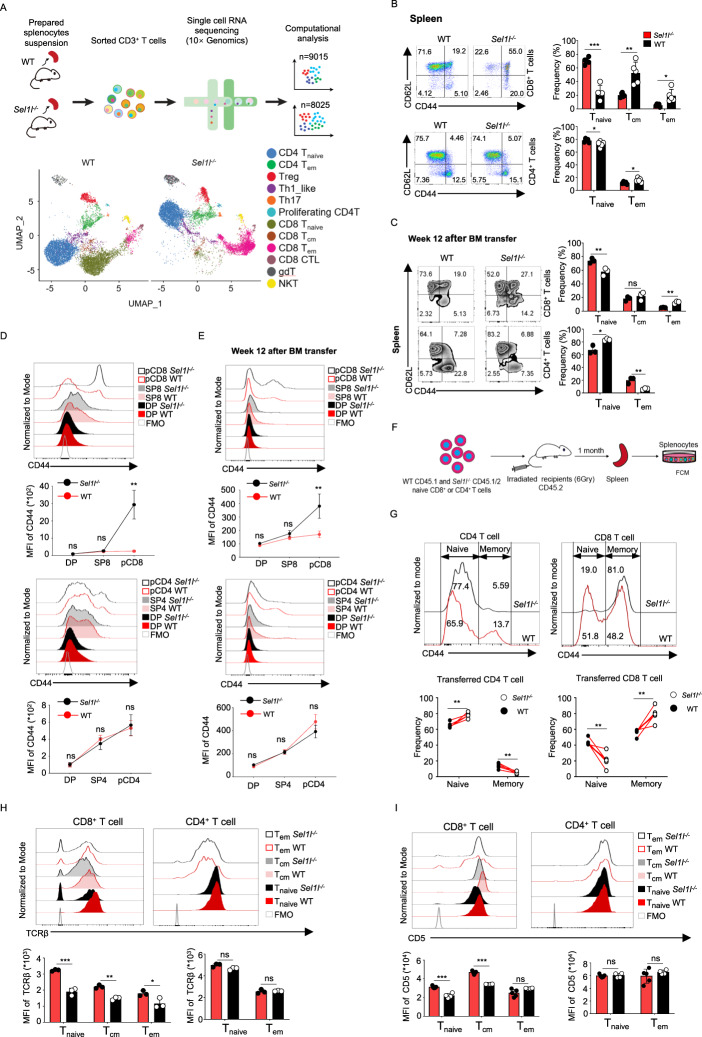
Given the greatly reduced number of T cells in the periphery, we sought to determine whether naïve T-cell homeostasis was impacted in the absence of SEL1L. We carried out single-cell transcriptome sequencing (scRNA-seq) of CD3+ T cells sorted from 8-week-old WT and Sel1l−/− mice (pooled from 3 mice for each genotype) using the 10x Genomics platform (Fig. 5A). After quality control and filtering, we obtained single-cell transcriptomes of 9015 T cells from WT mice and 8025 T cells from Sel1l−/− mice. Based on an unbiased integrative analysis across all WT and Sel1l−/− T cells, we annotated 13 clusters of T-cell populations according to the most salient identified cell markers (Fig. S5H) [26–28]. In particular, we revealed 2 naïve T-cell subsets, namely, CD4+ naïve T cells (Cd4+Sell+Lef1+) and CD8+ naïve T cells (Cd8+Sell+Ccr7+); 3 memory T-cell subsets, namely, CD4+ TEMs (Cd4+Cd28+Tcf7+), CD8+ TCMs (Cd8+Tcf7+Sell+), and CD8+ TEMs (Cd8+Prf1+Ccl5+); Tregs (Cd4+Foxp3+Il2ra+); Th1_like cells (Cd4+Stat1+); Th17 cells (Cd4+Il17a+); proliferating CD4+ T cells (Cd4+Mki67+); CD8+ CTLs (Cd8+Prf1+Ccl5+); γδT cells (Cd3e+Aif1+); and NKT cells (Cd3e+Tbx21+) (Fig. 5A). Notably, the proportions of CD4+ naïve T cells were comparable between WT and Sel1l−/− CD4+ T cells, which was in line with our flow cytometry analysis (Fig. 5B, C). In addition, we noticed striking differences in terms of the proportions and diverse clusters between WT and Sel1l−/− CD8+ T-cell subsets, with the particular loss of naïve CD8+ T-cell subsets and accumulation of the memory CD8+ population (Fig. 5A). Indeed, the proportions of naïve CD62L+CD44−CD8+ T cells in the peripheral blood, spleen and pLNs were evidently reduced, whereas the frequencies of both central memory CD62L+CD44+ and activated/effector-like CD62L−CD44+ populations were markedly increased (Figs. 5B and S5A, C), suggesting that SEL1L deficiency disturbed naïve CD8 T-cell homeostasis and differentiation. However, the frequencies of distinct subsets of CD4+ T cells were comparable in peripheral blood, spleen and pLNs between WT and Sel1l−/− mice (Figs. 5B and S5A, C). These data suggest that the SEL1L-mediated ERAD pathway is selectively required for the maintenance of naïve CD8+ T-cell homeostasis, which was further validated with a competitive BM transplantation experiment (Figs. 5C and S5B). Next, based on the surface expression of the canonical T-cell activation marker CD44, we aimed to determine at which stage of T-cell development the proportions of T cells with an activated phenotype increased. The CD44+ population was significantly increased in peripheral CD8+ T cells rather than DP or SP8+ thymocytes in Sel1l−/− mice, whereas the CD44+ population was comparable between WT and Sel1l−/− mice in peripheral CD4+ T cells and DP and SP4+ thymocytes (Fig. 5D). Similar observations were obtained in the competitive BM transplantation experiment (Fig. 5E). As Sel1l−/− mice displayed decreased naïve but increased memory CD8+ T-cell numbers in the periphery, we next investigated whether loss of SEL1L may regulate CD8+ T-cell differentiation status. We sorted naïve CD8+ and naïve CD4+ T cells (as controls) and transferred these naïve T cells into recipients (Fig. 5F). As expected, on day 30 after transfer, almost all adoptively transferred naïve Sel1l−/− CD8+ T cells (CD62L+CD44−), but ~50% of naïve WT CD8+ T cells, had acquired CD44 expression. On the other hand, comparable percentages of naïve Sel1l−/− CD4+ T cells (75%) and WT CD4+ T cells (65%) differentiated into the CD44+ phenotype (Fig. 5G). Taken together, these observations strongly suggest that SEL1L is indispensable for the maintenance of homeostasis in peripheral naïve CD8+ but not CD4+ T cells.
Fig. 5.
SEL1L is indispensable for the maintenance of peripheral naïve CD8+ T cells. A Schematic diagram of the experimental design, scRNA-seq, and data analysis. Pooled T cells from three mice were sorted from 8-week-old WT and Sel1l−/− mice and subjected to single-cell RNA sequencing by 10x Genomics. UMAP projections of splenic T-cell profiles in WT and Sel1l−/− mice are shown. B Representative flow cytometry plots showing the percentages of naïve (CD44− CD62L+), central memory (CD44+ CD62L+) and effector memory (CD44+ CD62L−) populations in splenic CD8+ and CD4+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 5; Sel1l−/−, n = 5). C Representative flow cytometry plots showing the frequencies of naïve, central memory and effector memory populations in CD8+ and CD4+ T cells in spleens from mice transplanted with WT to Sel1l−/− whole bone marrow cells at the indicated time points (n = 3). D, E Representative flow cytometry plots and MFI of CD44 in thymic SP8+ and SP4+ T cells and splenic CD8+ and CD4+ T cells (pCD8+ and pCD4+) in 8-week-old WT and Sel1l−/− mice (WT, n = 5; Sel1l−/−, n = 5) (E) and mice transplanted with WT to Sel1l−/− whole bone marrow cells at the indicated time points (n = 3) (F). F Schematic diagram of the cotransfer model of WT and Sel1l−/− naïve CD8+ or CD4+ T cells. The recipient mice (CD45.2, irradiated with 6 Gy) were administered equal amounts of sorted naïve T cells from WT (CD45.1) and Sel1l−/− (CD45.1/CD45.2) mice. After 1 month, the spleens were harvested and analyzed by flow cytometry. G Representative flow cytometry plots and MFI of CD44 in splenocyte-transferred naïve CD8+ and CD4+ T cells (recipient for CD4+ T cells, n = 5; recipient for CD8+ T cells, n = 6). H Representative flow cytometry plots showing the MFI of TCRβ in three subpopulations of splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 5; Sel1l−/−, n = 4). I Representative flow cytometry plots showing the MFI of CD5 in three subpopulations of splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 5; Sel1l−/−, n = 4). The data are shown as the mean ± SD, and statistical significance was determined by unpaired two-tailed Student’s t test (B–E, H, I) and paired two-tailed Student’s t test (G). The data are representative of at least three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001)
Naïve T cells are dependent on tonic TCR signaling for homeostasis maintenance [29]. Tonic TCR signaling occurs constitutively upon the binding of T-cell receptors to self-peptide–MHC molecules (pMHC) to enforce naïve T-cell quiescence [30]. CD5 surface expression on SP thymocytes and peripheral T cells is recognized to reflect tonic TCR signaling intensity [31]. Consistent with the increased proportion of CD44+CD8+ T cells in the absence of SEL1L, we demonstrated that the expression levels of the TCRβ chain and CD5 were particularly decreased in CD8+ rather than CD4+ T cells (Fig. 5H, I). In addition, we found that the expression of CD3, an important surface molecule that stabilizes TCRα and TCRβ on the cell surface and carries out the signaling function of the TCR complex, was significantly decreased in naïve Sel1l−/− CD8+ T cells but slightly diminished in naïve Sel1l−/− CD4+ T cells (Fig. S5D). Since Sel1l knockout occurs at the double-positive stage of thymocytes, we further measured the surface expression of TCRβ, CD5 and CD3 in immature DP thymocytes, mature SP thymocytes and peripheral T cells to determine at which stage tonic TCR signaling was attenuated. TCRβ, CD5, and CD3 surface expression was comparable in DP cells between WT and Sel1l−/− mice (Fig. S5E–G). TCRβ and CD3 levels were slightly reduced in mature SP8+ cells and significantly diminished in peripheral CD8+ T cells in Sel1l−/− mice compared with their WT counterparts (Fig. S5E, G). CD5 surface expression was decreased in SP8+ cells and peripheral CD8+ T cells in Sel1l−/− mice compared with their WT counterparts (Fig. S5F). However, the expression of TCRβ, CD5, and CD3 in mature SP4 and peripheral CD4+ T cells was comparable between WT and Sel1l−/− mice (Fig. S5E–G). These data suggested that SEL1L deficiency particularly triggered dysregulated tonic TCR signaling in mature CD8+ T cells.
SEL1L deficiency fuels mTORC1 activation by increasing IL-15 receptor signaling
In addition to upregulated CD44 expression, impaired homeostasis is often coupled with a dysregulated cell cycle and intracellular signaling cascade. Naïve T cells are quiescent, and it is well known that the TSC-mTORC1 signaling axis represents an important checkpoint to maintain naïve T-cell homeostasis [32, 33]. Our previous study and others have demonstrated that TSC1 deficiency leads to exit of quiescence and spontaneous activation of naïve CD8+ T cells, which are also susceptible to apoptosis [34, 35]. However, deletion of Raptor, a key component of the mTORC1 complex, impairs the activation, proliferation and antigen-specific immune responses of CD8+ T cells [36]. In addition, both tonic TCR signaling and cytokine signaling are tightly linked to the mTORC1 pathway to maintain T-cell quiescence [37, 38]. Therefore, we sought to determine whether the mTORC1 signaling pathway was impacted by SEL1L deficiency. In contrast to WT CD8+ T cells, the levels of phosphorylated p70 S6K and 4EBP1, two major downstream functional readouts of mTORC1 activation, were upregulated in Sel1l−/− CD8+ T cells (Fig. 6A). Consistently, Sel1l−/− CD8+ T cells exhibited enlarged cell sizes and more neosynthetic proteins, as indicated by an increased median fluorescence intensity of the forward scatter area (FSC-A) (Fig. 6B) and anti-puromycin staining (Fig. 6C). c-MYC can also act as downstream of mTOR signaling and as a key regulator of the expression of multiple amino acid transporters [39]. In contrast to the low expression of c-MYC in unprimed WT CD8+ T cells, significant upregulation of c-MYC was observed at both the transcriptional and translational levels in the absence of SEL1L (Figs. 6D, E and S6A). Amino acid uptake is essential for proper T-cell activation and acquisition of effector functions. Consistent with the increase in c-MYC expression, we noticed significantly increased transcription of several amino acid transporters, including Slc3a2, Slc7a1, Slc7a5 and Slc7a11, in Sel1l−/− naïve CD8+ T cells (Fig. 6F). CD98 is a key transporter of multiple types of amino acids that supports T-cell activation and proliferation, and we found that surface CD98 expression was drastically increased in splenic CD8+ T cells of Sel1l−/− mice under steady-state conditions (Fig. 6G). In addition, sterol-responsive element binding protein 1 (SREBP1) and SREBP2 are key regulators of lipid and cholesterol biosynthesis, and SREBP1 and SREBP2 expression is controlled by mTORC1 signaling [40]. Hence, we measured the expression of SREBPs and noticed a drastic increase in Srebp1 and Srebp2 mRNA expression in naïve Sel1l−/− CD8+ T cells (Fig. 6H). This corresponded to a marked increase in lipid uptake in Sel1l−/− CD8+ Tcm and Tem cells (Fig. 6I, J). In addition, we demonstrated that Sel1l−/− CD8+ T cells harbored increased intracellular neutral lipid and cholesterol levels (Fig. 6K, L). However, glucose uptake as measured by 2-NBDG staining was significantly decreased in Sel1l−/− naïve CD8+ T cells compared with their WT counterparts (Fig. 6M), which suggests that SEL1L deficiency may induce the metabolic switch from glucose metabolism to fatty acid and amino acid oxidation (FAO/AAO). Altogether, these observations suggest that SEL1L deficiency leads to elevated mTORC1 activation in CD8+ T cells, which reshapes intracellular metabolism and profoundly impairs peripheral CD8+ T-cell homeostasis.
Fig. 6.
SEL1L deficiency fuels mTORC1 activation and triggers metabolic reprogramming. A Western blot of the expression of p-p70 S6K, p70S6K, p4EBP1, 4-EBP1 and GAPDH in unprimed CD8+ T cells from 8-week-old WT and Sel1l−/− mice. The results are representative of two independent biological replicates. Each lane shows the results of pooled CD8+ T cells from three mice. B Representative flow cytometry plots showing the MFI of FSC-A in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 3). C Representative flow cytometry plots showing the MFI of anti-puromycin in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 3). D Western blot of the expression of c-MYC and β-actin with the relative intensity (c-MYC normalized to β-actin) in unprimed CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 5). E Representative flow cytometry plots showing the MFI of c-MYC in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 6; Sel1l−/−, n = 6). F Total RNA was extracted from naïve WT and Sel1l−/− CD8+ T cells. RT‒PCR analysis was performed to determine the amino acid transporter messenger RNA level normalized to that of B2M. (n = 3 mice; combined from two independent repeats.). G Representative flow cytometry plots showing the MFI of CD98 in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 4; Sel1l−/−, n = 4). H Total RNA was extracted from naïve WT and Sel1l−/− CD8+ T cells. RT‒PCR analysis of Srebp1 and Srebp2 messenger RNA levels normalized to that of β-actin (WT, n = 3; Sel1l−/−, n = 3). I Representative flow cytometry plots showing the MFI of BODIPY-C12 in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 5). J Representative flow cytometry plots showing the MFI of BODIPY-C16 in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 4). Representative flow cytometry plots showing the MFI of BODIPY493/503 (K) and filipin III (L) in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 3). M Representative flow cytometry plots showing the MFI of 2-NBDG in splenic CD8+ T cells from 8-week-old WT and Sel1l−/− mice (WT, n = 4; Sel1l−/−, n = 4). The data are shown as the mean ± SD, and statistical significance was determined by unpaired two-tailed Student’s t test. The data are representative of at least three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001)
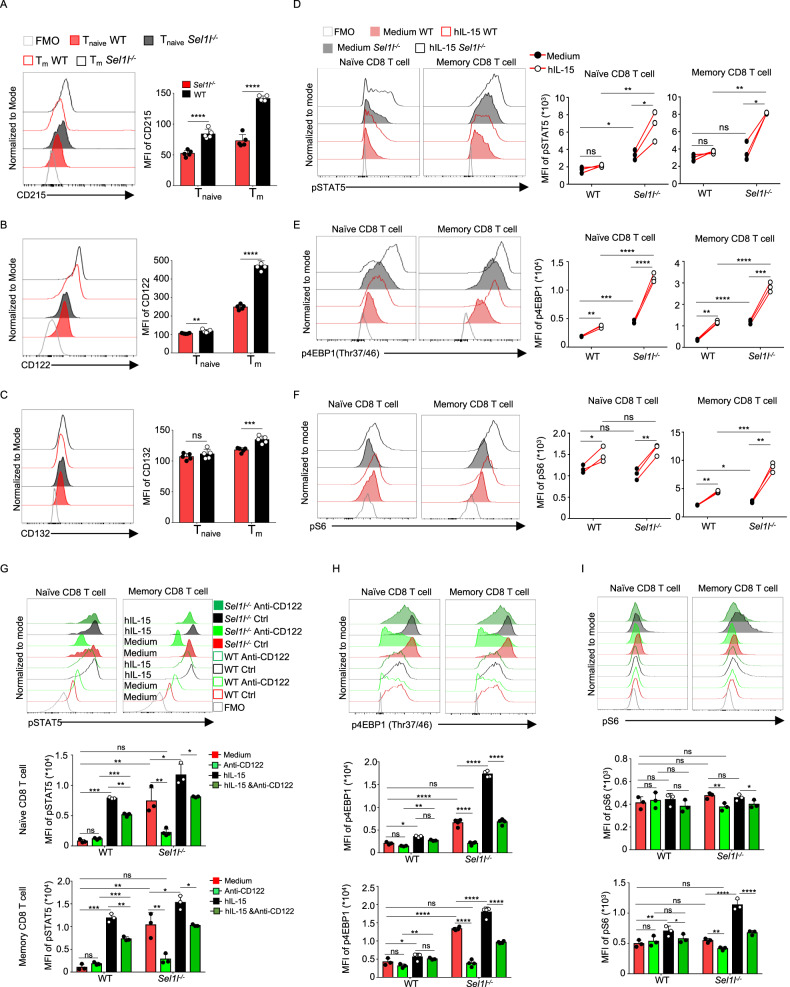
Previous studies have demonstrated that IL-15/IL-15 receptor (IL-15R) signaling triggers mTORC1 activation in NK and CD8+ T cells [41–43]. To further evaluate whether elevated mTORC1 activity in Sel1l−/− CD8+ T cells is due to dysregulated IL-15/IL-15R signaling, we measured the surface expression of distinct IL-15R subunits and their responsiveness to IL-15. Surprisingly, we found that the expression of CD215, the α subunit of the IL-15 receptor and the β subunit of IL-15R (CD122) was evidently upregulated, whereas the γ subunit of IL-15R (CD132) remained unchanged, in Sel1l−/−CD8+ T cells (Fig. 7A–C). Consistent with the increased expression of IL-15Rα and IL-15Rβ, IL-15 treatment significantly increased the phosphorylation of STAT5 at Tyr694 in Sel1l−/− CD8+ compared to WT CD8+ T cells (Fig. 7D). Moreover, we further showed that IL-15 treatment activated mTORC1 downstream signaling, as evidenced by increased phosphorylation of mTOR at Ser2448 (Fig. S6J), phosphorylation of 4EBP1 at Thr37/46 (Fig. 7E) and Ser65 (Fig. S6H) and phosphorylation of S6 ribosomal protein at Ser235/236 (Fig. 7F), and c-MYC expression was also upregulated (Fig. S6B) in Sel1l−/− CD8+ T cells. Further demonstrating that increased mTORC1 activity in Sel1l−/− CD8+ T cells is dependent on high IL-15/IL-15R signaling, inhibition of IL-15/IL-15R signaling with a CD122-blocking antibody decreased the phosphorylation of STAT5, 4EBP1 and S6 in Sel1l−/− CD8+ T cells (Fig. 7G–I). In addition, blocking IL-15/IL-15R signaling completely suppressed the phosphorylation of STAT5, mTOR, 4EBP1 and the S6 ribosomal protein in Sel1l−/− CD8+ T cells in response to IL-15 treatment (Fig. 7G–I). Next, we explored at which stage of T-cell development IL-15Rα and IL-15Rβ surface expression is dysregulated in Sel1l−/− T cells. The expression levels of both IL-15Rα and IL-15Rβ on DP cells and SP8+ T cells were comparable between WT and Sel1l−/− mice; however, IL-15Rα and IL-15Rβ expression was significantly augmented in peripheral CD8+ T cells from Sel1l−/− mice compared with their WT counterparts (Fig. S7A, C), indicating that SEL1L deficiency specifically upregulated IL-15Rα and IL-15Rβ expression in peripheral CD8+ T cells. Therefore, our results demonstrate that SEL1L deficiency promotes the mTORC1 signaling cascade by increasing IL-15 receptor expression.
Fig. 7.
SEL1L deficiency fuels mTORC1 activation by increasing IL-15 receptor signaling. Flow cytometric analysis showing CD215 (A), CD122 (B) and CD132 (C) expression in WT and Sel1l−/− CD8+ T cells (WT, n = 5; Sel1l−/−, n = 5). Representative flow cytometry plots showing the MFI of pSTAT5 (Tyr694) (D), p4EBP1 (Thr37/46) (E) and pS6 (Ser235/236) (F) in CD8+ T-cell subsets from 8-week-old WT and Sel1l−/− mice treated for 10 h with hIL-15 (WT, n = 3; Sel1l−/−, n = 3). Representative flow cytometry plots showing the MFI of pSTAT5 (Tyr694) (G), p4EBP1 (Thr37/46) (H) and pS6 (Ser235/236) (I) in CD8+ T cells from 8-week-old WT and Sel1l−/− mice treated for 10 h with hIL-15 and anti-CD122 antibody (WT, n = 3; Sel1l−/−, n = 3). The data are shown as the mean ± SD, and statistical significance was determined by unpaired two-tailed Student’s t test (A–C, G–I) and paired two-tailed Student’s t test (D–F). The data are representative of at least three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001)
IRE1α accumulation in Sel1l−/− CD8+ T cells governs metabolic reprogramming
Given that increased IL-15/IL-15R signaling is linked to mTORC1 activation in Sel1l−/− CD8+ T cells, we investigated how the IL-15 receptor was upregulated. As IRE1α is a substrate of SEL1L-Hrd1 ERAD and controls mTOR activation and metabolic homeostasis, we explored whether IRE1α affects IL-15/IL-15R-mTORC1 signaling in Sel1l−/− CD8+ T cells [44–46]. We found that Ern1 deletion specifically decreased the expression of the IL-15Rα chain (CD215) rather than the β (CD122) and γ chains (CD132) in Sel1l−/− CD8+ T cells (Fig. 8A–C). This implied that IRE1α accumulation led to the increased expression of IL-15Rα and thus promoted IL-15 receptor signaling in Sel1l−/− CD8+ T cells. Next, we further explored the activation status of mTORC1 signaling and responsiveness to IL-15 in DKO (Sel1l−/−Ern1−/−) CD8+ T cells. Although Ern1 deficiency did not reduce phosphorylation of STAT5, mTOR, 4EBP1 or the S6 ribosomal protein or the expression of c-MYC in Sel1l−/− CD8+ T cells under homeostatic conditions, IL-15 treatment failed to trigger phosphorylation of STAT5, mTOR, 4EBP1 and the S6 ribosomal protein and upregulation of c-MYC in DKO CD8+ T cells, especially naïve CD8+ T cells (Figs. 8D–G and S8A, N). These data strongly supported the idea that Ern1 deficiency inhibited the response of Sel1l−/− CD8+ T cells, especially naïve CD8+ T cells, to IL-15 by decreasing IL-15Rα surface expression. Moreover, we noticed that the cell size (FSC-A), neosynthetic protein numbers and CD98 expression were significantly decreased in splenic CD8+ T cells from DKO mice compared with Sel1l−/− CD8+ T cells (Fig. 9A–C). Similarly, lipid uptake (BODIPY C12; BODIPY C16) was markedly decreased in DKO CD8+ T cells compared with Sel1l−/− CD8+ T cells (Fig. 9D, E). Consistently, DKO CD8+ T cells harbored markedly lower intracellular neutral lipid and cholesterol levels than Sel1l−/− CD8+ T cells (Fig. 9F, G). In addition, glucose uptake was significantly increased in DKO naïve CD8+ T cells compared with Sel1l−/− CD8+ T cells (Fig. 9H), which further supports the notion that the Sel1l deficiency-induced metabolic switch may be largely dependent on IRE1α accumulation. Taken together, these findings demonstrate that IRE1α accumulation causes hyperactive responses of Sel1l−/− CD8+ T cells to IL-15 by increasing IL-15Rα expression and thus augmenting mTORC1 signaling.
Fig. 8.
Ern1 deficiency limits mTORC1 activation in Sel1l−/− CD8+ T cells by decreasing IL-15 receptor signaling. Representative flow cytometry analysis of CD215 (A), CD122 (B) and CD132 (C) expression in splenic CD8+ T cells from 6-week-old WT, Sel1l−/− and DKO mice (WT, n = 6; Sel1l−/−, n = 5; DKO, n = 3). Representative flow cytometry plots showing the MFI of pSTAT5 (D), p4EBP1 (E), pS6 (F) and c-MYC (G) in CD8+ T cells from 7-week-old WT and Sel1l−/− mice treated with hIL-15 for 10 h (WT, n = 3; Sel1l−/−, n = 3). The data are shown as the mean ± SD, and statistical significance was determined by paired two-tailed Student’s t test (D–G) and one-way ANOVA followed by the SNK-q post hoc test (A–C). The data are representative of three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; *p < 0.001; ****p < 0.0001)
Fig. 9.
Ern1 deficiency remodels cellular metabolism in Sel1l−/− CD8+ T cells. A Representative flow cytometry plots showing the MFI of FSC-A in splenic CD8+ T cells from ~7-week-old WT, Sel1l−/− and DKO mice (WT, n = 3; Sel1l−/−, n = 3; DKO, n = 3). B Representative flow cytometry plots showing the MFI of anti-puromycin in splenic CD8+ T cells from ~7-week-old WT, Sel1l−/− and DKO mice (WT, n = 6; Sel1l−/−, n = 5; DKO, n = 3). C Representative flow cytometry plots showing the MFI of CD98 in splenic CD8+ T cells from ~7-week-old WT, Sel1l−/− and DKO mice (WT, n = 3; Sel1l−/−, n = 3; DKO, n = 3). Representative flow cytometry plots showing the MFI of BODIPY-C12 (D) and BODIPY-C16 (E) in splenic CD8+ T cells from 6-week-old WT, Sel1l−/− and DKO mice (WT, n = 6; Sel1l−/−, n = 5; DKO, n = 3). Representative flow cytometry plots showing the MFI of BODIPY 493/503 (F) and filipin III (G) in splenic CD8+ T cells from 6-week-old WT, Sel1l−/− and DKO mice (WT, n = 6; Sel1l−/−, n = 5; DKO, n = 3). H Representative flow cytometry plots showing the MFI of 2-NBDG in splenic CD8+ T cells from 6-week-old WT, Sel1l−/− and DKO mice (WT, n = 6; Sel1l−/−, n = 5; DKO, n = 3). The data are shown as the mean ± SD, and statistical significance was determined by one-way ANOVA followed by the SNK-q post hoc test. Data are representative of three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; *p < 0.001; ****p < 0.0001)
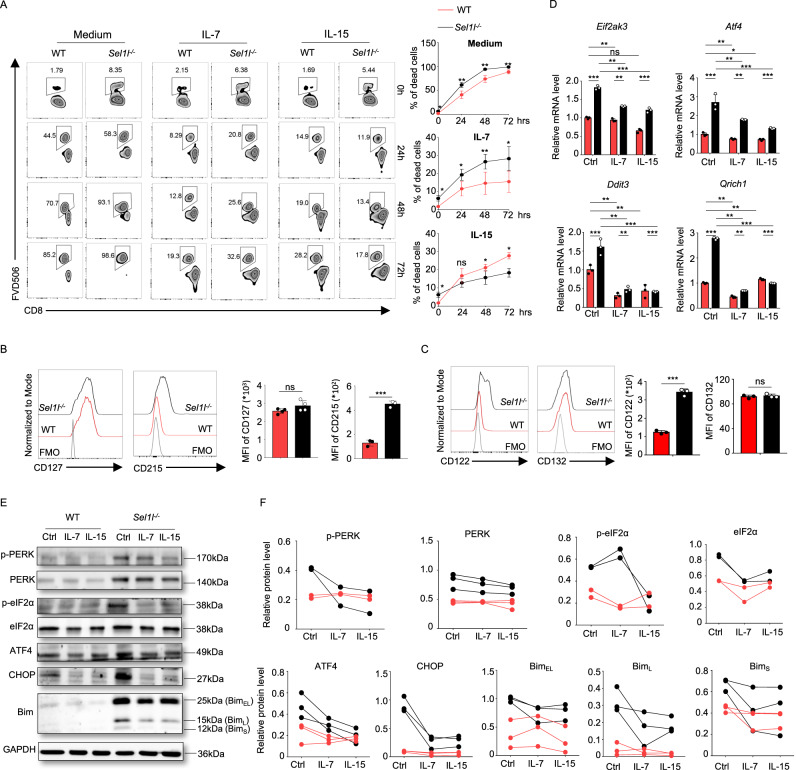
IL-7 and IL-15 rescue the survival of Sel1l−/− CD8+ T cells by blocking PERK/ATF4/CHOP/Bim signaling
IL-7 and IL-15 are two key cytokines for maintaining T-cell survival and homeostasis in vivo [47, 48]. As expected, WT CD8+ T cells underwent massive cell death in vitro, which was largely reversed by addition of either IL-7 or IL-15 (Fig. 10A). As described previously, Sel1l−/− CD8+ T cells displayed more striking survival defects than their WT counterparts. IL-7 administration significantly improved the cell survival of Sel1l−/− CD8+ T cells, although it remained lower than that of their WT counterparts (Fig. 10A). Notably, IL-15 was able to fully reverse the survival defects of Sel1l−/−CD8+ T cells (Fig. 10A). This was likely attributable to the increased expression of the IL-15Rα subunit (CD215) and common β chain (CD122) of IL-2 and IL-15 in Sel1l−/−CD8+ T cells (Fig. 10B, C). Furthermore, we found that the expression levels of the IL-7 receptor α subunit CD127 (Fig. 10B), the IL-2 receptor α subunit CD25 (Fig. S12A) and the common γ chain of IL-2, IL-7 and IL-15 (Fig. 10C) were comparable between WT and Sel1l−/−CD8+ T cells. Given that the expression of the common β chain of the IL-2 and IL-15 receptors, CD122, is increased in Sel1l−/− CD8+ T cells, IL-2 treatment partially rescue the survival of Sel1l−/− naïve and CD8+ Tcm cells, although the efficacy was much lower than that of IL-15 (Fig. S12B, C). In addition, we noticed that IL-15 treatment better rescued the survival of Sel1l−/− CD8+ Tcm cells than naïve CD8+ T cells (Fig. S12D). Intriguingly, IL-7 and IL-15 treatment greatly reduced the expression of several ER stress response genes (Figs. 10D and S11A–C), particularly PERK signaling-associated genes, such as Eif2ak3, ATF4, Ddit3, and Qrich1, in both WT and Sel1l−/− CD8+ T cells (Fig. 10D). Consistently, the protein expression of multiple ER stress regulators, such as p-PERK, PERK, p-eFI2α, eFI2α, ATF4, CHOP and Bim, in Sel1l−/− CD8+ T cells was also greatly reduced upon IL-7 and IL-15 treatment (Fig. 10E, F). Additionally, IL-7 and IL-15 treatment reduced the expression of ER stress response genes in Sel1l−/− CD4+ T cells (Fig. S11D, E). In contrast, IL-15 treatment failed to rescue the survival of DKO CD8+ T cells (Fig. S12E–G). Altogether, these findings strongly suggested that Sel1l−/− CD8+ T cells underwent severe ER stress and gradually lost their survival ability, an effect that was able to be largely reversed by IL-7 and IL-15.
Fig. 10.
IL-7 and IL-15 rescue the survival of Sel1l−/− CD8+ T cells through inhibition of PERK/ATF4/CHOP/Bim signaling. A Representative flow cytometry plots and quantification of dead cells in sorted WT and Sel1l−/− CD8+ T cells cultured with or without IL-7 (10 ng/ml) and IL-15 (10 ng/ml) for 24, 48 and 72 h separately. Representative flow cytometry plots showing the MFI of CD127 and CD215 (B) and CD122 and CD132 (C) in splenic CD8+ T cells from ~7-week-old WT and Sel1l−/− mice (WT, n = 3; Sel1l−/−, n = 3). D RT‒qPCR analysis of PERK signaling-associated messenger RNA levels normalized to that of β-actin. The data are representative of three independent biological replicates using primers for Eif2ak3, ATF4, Ddit3, and Qrich1 in splenic WT and Sel1l−/– CD8+ T cells treated with IL-7 and IL-15 for 24 h. E, F Lysates from WT and Sel1l−/− CD8+ T cells treated with IL-7 and IL-15 for 24 h were subjected to immunoblotting with p-PERK, PERK, p-eFI2α, eFI2α, ATF4, CHOP, Bim and GAPDH antibodies (E), and the relative intensity of two or three independent biological replicates was calculated (ER stress effector molecules normalized to GAPDH) (F). The WT and Sel1l−/− lanes show the results of pooled CD8+ T cells from four mice. The data are shown as the mean ± SD, and statistical significance was determined by unpaired two-tailed Student’s t test (A–D) and paired two-tailed Student’s t test (F). The data are representative of at least three independent experiments. (n.s. not significant; *p < 0.05; **p < 0.01; ****p < 0.0001)
Discussion
ERAD is an important pathway that helps ensure protein homeostasis by degrading misfolded or aggregated proteins. In the present study, we revealed an indispensable role of SEL1L in preserving CD8+ T-cell survival, homeostasis and metabolism. Genetic loss of SEL1L in T cells led to markedly decreased numbers of mature T cells and disrupted the homeostasis of peripheral CD8+ T cells. We further demonstrated that SEL1L deficiency induced aberrant activation of the PERK-ATF4-CHOP-Bim signaling cascade and excessive ER stress in unprimed CD8+ T cells, which severely impaired CD8+ T-cell survival. In addition, SEL1L deletion in CD8+ T cells prompted mTORC1 activation by increasing IL-15 receptor signaling and triggered evident metabolic remodeling. Furthermore, we revealed that IRE1α deletion in Sel1l−/− CD8+ T cells decreased the expression of the IL-15 receptor α chain and mTORC1 signaling and reversed the shift toward dysregulated metabolism.
Consistent with previous studies showing that SEL1L deletion impairs cell survival [13, 14, 49], we noticed that Sel1l−/− CD8+ T cells displayed evident survival defects under basal conditions. In addition, we demonstrated that apoptosis, pyroptosis, and ferroptosis, rather than necroptosis and autophagic cell death, were critically involved in the survival defects induced by SEL1L deficiency. Importantly, PERK inhibition rescued Sel1l−/− CD8+ T cells from death, which is consistent with a recent study [50]. In addition, we first demonstrated that PERK inhibition decreases cleaved Caspase3 or GSDMD levels and may alleviate apoptosis or pyroptosis in Sel1l−/− CD8+ T cells. Moreover, we observed that SEL1L deletion triggered DNA damage and G1/S arrest, which may, at least in part, have contributed to the disrupted homeostatic proliferation of CD8+ T cells. Consistent with the evident survival defects of Sel1l−/− T cells, we also observed increased levels of intracellular ROS. DNA damage in Sel1l−/− CD8+ T cells may be related to increased intracellular ROS generation, which needs further investigation.
Notably, we observed that SEL1L deficiency triggered severe loss of the naïve CD8+ T-cell population and increased expression of CD44, CD98 and several other amino acid transporters, indicating that naïve CD8+ T-cell homeostasis was disrupted in the absence of SEL1L. Naïve T-cell homeostasis is tightly controlled at multiple levels. Accumulating evidence suggests that mTORC1 activation leads to exit of naïve T cells from quiescence and disrupted homeostasis [34, 35, 51]. Consistent with a previous study [12], we found that mTORC1 signaling activity was significantly elevated in unprimed Sel1l−/− CD8+ T cells. These findings support the notion that loss of the naïve CD8+ T-cell population and an increase in the memory CD8+ T-cell population in the absence of SEL1L are likely associated with mTORC1 activation. Stress-inducing ATF4 upregulation often results in amino acid uptake as well as nonessential amino acid biosynthesis. Indeed, ATF4 expression was elevated in the absence of SEL1L. However, further investigation is required to determine how the SEL1L-ATF4 axis regulates amino acid metabolism in CD8+ T cells by altering the amino acid sensing or transportation system.
SEL1L deficiency led to PERK and IRE1α accumulation in CD8+ T cells. In particular, we found that PERK inhibition significantly rescued Sel1l−/− CD8+ T cells from cell death, but deletion of IRE1α further aggravated the survival defects of Sel1l−/− CD8+ T cells. Of note, IRE1α accumulation is certainly responsible for elevated mTORC1 activation and metabolic shifts in Sel1l−/− CD8+ T cells via control of the IL-15 receptor α chain. The distinct branch of the UPR induced by SEL1L deletion likely accounts for either dysregulated survival or homeostasis. However, the precise signaling pathways and key mediators of the aforementioned biological processes should be further investigated, which will extend our knowledge about ER stress or ERAD in CD8+ T-cell biology.
IL-7 and IL-15 are critical cytokines that maintain T-cell survival and homeostasis. Sel1l−/− naïve CD8+ T cells display striking survival defects both in vivo and in vitro. Notably, we demonstrated that IL-7 and IL15 inhibited the intense ER stress response and PERK activation-induced survival defects by markedly reducing the expression of multiple key regulators of the ER stress response at both the protein and mRNA levels. Importantly, IL-15 supplementation in cell culture almost fully reversed the survival defects of Sel1l−/− CD8+ T cells, although Sel1l−/− naïve CD8+ T cells showed a slightly reduced survival rate even in the presence of IL-7. This was associated with increased expression of the α and β subunits of the IL15 receptor. However, the specific mechanism by which IL-7 and IL-15 inhibit ER stress in CD8+ T cells awaits further investigation.
In addition, it remains intriguing to further elucidate the differential regulatory effects of SEL1L on CD8+ and CD4+ T cells. On the one hand, the levels of key molecules involved in tonic TCR signaling, such as TCRβ and CD3, are significantly reduced in Sel1l−/− CD8+ T cells but not in CD4+ T cells, which may lead to disrupted naïve T-cell homeostasis. On the other hand, it is possible that the elevated expression of multiple subunits of the IL-15 receptor on CD8+ T cells contributes to the basal activation of mTORC1 and the c-MYC signaling cascade under steady-state conditions, whereas CD4+ T cells barely express the IL-15 receptor. In addition, CD8+ T cells seem to have a low threshold and are often more prone to respond to homeostatic cytokines or metabolic cues. Further investigations are needed to determine whether ATF4 or other key factors of the ER stress response can directly regulate the upstream open reading frames (uORFs) at the 5′ UTRs of key regulatory genes, such as the IL-15 receptor, in Sel1l−/− CD8+ T cells. Overall, our present study reveals an important role of the SEL1L and ERAD pathways in preserving CD8+ T-cell quiescence and homeostasis.
Materials and methods
Mice
Sel1lflox/flox mice were generated as previously described [5]. T-cell-specific Sel1l-conditional-knockout mice were obtained by crossing Sel1lflox/flox mice with CD4-Cre transgenic mice, and the mouse line was maintained on a C57BL/6 background. C57BL/6N mice were purchased from Vital River Laboratory (Beijing, China). Sel1lflox/flox/Ern1flox/flox:CD4-Cre mice were generated by crossing Sel1lflox/flox:CD4-Cre with Ern1flox/flox mice. All mice were maintained in the Center for Experimental Animals at the Suzhou Institute of Systems Medicine (Suzhou, China). All mice were housed in a specific pathogen-free facility, and all experiments were approved by the Animal Ethics Committee (ISM-IACUC-0055).
In vivo assay
For the competitive bone marrow transplantation experiments, CD45.2+ recipient mice were lethally irradiated (10 Gy, treated with two equal doses 4 h apart) and injected intravenously with 2.5 × 106 CD45.1+ BM cells (Sel1l−/−) with an equal number of CD45.1/2 competitor BM cells (WT). For naïve T-cell differentiation, CD45.2 recipient mice were irradiated with 6 Gy and transferred with equal proportions of naïve CD8+ or CD4+ T cells (0.8–1 × 106 cells) from WT (CD45.1) and Sel1l−/− (CD45.1/2) mice the next day. To assess competitive survival, homeostatic proliferation and the cell cycle of mature CD8+ T cells, CD45.2 recipient mice were irradiated with 5 Gy, and equal proportions of CD8+ T cells (0.8–1 × 106 cells) from WT (CD45.1) and Sel1l−/− (CD45.1/2) mice were transferred the next day. To assess the effect of PERK inhibition on Sel1l−/− CD8+ T-cell numbers and differentiation, the PERK inhibitor GSK2606414 was formulated in 40% PGE300 and 5% Tween-80 in saline and administered once every 3 days (80 mg/kg) for 44 days starting from 3 days after birth. For the PERK inhibitor rescue experiment, sublethally irradiated recipients (CD45.2+) were cotreated with 5 × 105 WT (CD45.1/2) and Sel1l–/− (CD45.1) naïve CD8+ T cells labeled with CFSE and then administered GSK2606414 intraperitoneally once every 2 days (80 mg/kg) for 10 days.
Flow cytometry
Thymocytes, splenocytes and lymph node cells were obtained by gently grinding these organs. Erythrocytes were removed with 1×RBC (red blood cell) Lysing Buffer (BioLegend, 420301). Peripheral blood was obtained by tail vein bleeding. Single-cell suspensions of thymocytes, splenocytes and lymph node cells were obtained by filtering 40 μm strainers. For cell analysis, 1 ~ 6 × 106 cells were resuspended in 100 µl of FACS buffer (PBS, pH 7.2, 2% FBS in PBS), blocked with anti-mouse FcR mAb (BD Biosciences, clone 2.4G2) and stained with Fixable Viability Dye eFluorTM 506 (1:1000, Invitrogen, 65-0866-18) in PBS at 4 °C for 20 min. Then, these cells were washed two times with cold FACS buffer and incubated with fluorochrome-conjugated mAbs at 4 °C for 30 min. The samples were acquired by an LSR Fortessa (Becton Dickinson, CA), and the acquired data were analyzed with FlowJo 8 software.
For intracellular Foxp3 and Ki67 staining, cells were fixed with fixation buffer (Invitrogen, 00-5223-56, 00-5123-43) for 20 min at 4 °C after surface marker staining. Then, the cells were washed twice with intracellular staining permeabilization wash buffer (1×, Invitrogen, 00-8333-56) and stained with anti-FoxP3-Alexa Fluor 488 (BioLegend, clone MF-14) in permeabilization wash buffer for 25 min at 4 °C. The cells were then washed and resuspended in FACS buffer for FCM detection.
T-cell isolation and activation
CD8+ T cells were isolated from the spleens of WT or Sel1l−/− mice with a MojoSort™ Mouse CD8 T-Cell Isolation Kit (Biolegend, 480035) and cultured in RPMI-1640 medium (Gibco, 11875-519) containing 10% FBS (HyClone, SH30396.03), 2 mM glutamine (Gibco, 25030081), 1 mM pyruvate (Gibco, 11360-070), 10 mM HEPES (Bioind, 03-025-1B), 1NEAAs (Gibco, 11140-050), 100 units penicillin‒streptomycin (HyClone, SV30010) and 50 M -mercaptoethanol (Sigma‒Aldrich, 444203). For naïve T-cell activation, cells were cultured at 1 × 106 cells/ml in 24-well or 48-well plates with hIL-2 (10 ng/ml, PeproTech) and coated with anti-CD3 (Invitrogen, 16-0031-86) and anti-CD28 (Invitrogen, 16-0281-86) antibodies for the indicated durations in RPMI-1640 complete medium.
Western blot analysis
Cells were collected and washed twice with cold PBS and then lysed for 30 min on ice in M-PER™ Mammalian Protein Extraction Reagent (Thermo, #78501) together with phosphatase inhibitors (Sigma, #04906837001) and a protease inhibitor cocktail (Sigma, #11697498001) as described previously. Equivalent protein concentrations were subjected to 8–12% SDS‒PAGE (Bio-Rad, Hercules, CA). Rabbit anti-BIP (CST, #3177, 1:1000), rabbit anti-IRE1α (CST #3294, 1:1000), rabbit anti-P-PERK (CST #3179, 1:250), rabbit anti-PERK (CST #3192, 1:1000), rabbit anti-P-eIF2α (CST #3398, 1:1000), rabbit anti-eIF2α (CST #5324, 1:1000), rabbit anti-ATF4 (CST #11815, 1:1000), rabbit anti-Caspase-3 (CST #9662, 1:1000), rabbit anti-Bim (CST #, 1:1000), rabbit anti-BAX (CST #, 1:1000), rabbit anti-BAX (CST #, 1:1000), rabbit anti-BCL-2 (CST #, 1:1000), rabbit anti-BCL-xL (CST #, 1:1000), rabbit anti-p-CHK1 (CST #, 1:1000), rabbit anti-CHK1 (CST #, 1:1000), rabbit anti-γH2AX (CST #, 1:1000), rabbit anti-GAPDH (CST #5174, 1:1000), mouse anti-CHOP (CST #2895, 1:1000), and mouse anti-p53 (CST #, 1:1000) were purchased from Cell Signaling Technology. Rabbit anti-SEL1L (#ab78298, 1:1000), rabbit anti-GSDMD (ab209845, 1:1000), rabbit anti-MLKL (phospho S345), rabbit anti-glutathione peroxidase 4 (ab125066, 1:1000) and rabbit anti-GSDME (ab215191, 1:1000) were purchased from Abcam. Rabbit anti-LC-3 (PM036, 1:1000) and rabbit anti-p62 (PM045, 1:1000) were purchased from MBL International Corporation. An actin antibody was obtained from ABclonal (#AC026, 1:100,000), and goat anti-rabbit IgG-HRP (Absin, abs20040, 1:5000), and goat anti-mouse IgG-HRP (Absin, abs20039, 1:5000) were obtained from Absin.
Real-time PCR analysis
RNA was extracted from T cells as indicated by using TRIzol (TIANGEN, #dp424) and reversed-transcribed into cDNA (TAKARA, #RR036A) following the manufacturer’s protocol. RT‒PCR was performed with SYBR Green Master Mix (Vazyme, #Q311-00) on an RT‒PCR machine (Roche LC480). The sequences of the primers used are listed in Table S1.
Detection of pSTAT5 and mTORC1 signaling activity
CD3+ T cells were sorted from the spleens of WT and Sel1l−/− mice with a Mouse CD3 T-Cell Isolation Kit (BioLegend, #480031). The sorted T cells were treated with 10 μg/ml human IL-15 (hIL-15) (PeproTech, #200-15-10UG) for 10–12 h and then stained with Fixable Viability Dye eFluorTM 506 (1:1000) in PBS at 4 °C for 20 min. The stained cells were resuspended in ~4% formaldehyde, fixed for 15 min at room temperature (20–25 °C) and then washed by centrifugation with excess 1× PBS. The supernatant was discarded. The fixed cells were permeabilized with ice-cold 90% methanol for a minimum of 30 min on ice or stored at −20 °C in 90% methanol. After permeabilization, the cells were washed twice with excess 1× PBS and incubated with diluted primary antibody (0.5% BSA PBS buffer) for 1 h at room temperature or overnight at 4 °C. The cells were stained with a diluted fluorochrome-conjugated secondary antibody (Jackson ImmunoResearch, # 711-545-152) for 30 min at room temperature, then washed and resuspended in 1× PBS and then analyzed on a flow cytometer.
Detection of CD8+ T-cell survival in vitro
CD3+ T cells were sorted from the spleens of WT and Sel1l−/− mice with a Mouse CD3 T-Cell Isolation Kit. The cells were treated with 1 mM NAC (Sigma-Aldrich, #A9165-5G), 20 μM Z-VAD-FMK (MedChemExpress, #HY-16658B), 10 μM MCC950 (MedChemExpress, #HY-12815A), 10 μM NEC-1 s (MedChemExpress, #HY-14622A), disulfiram (MedChemExpress, #HY-B0240), 1 μM GSK2606414 (MedChemExpress, #HY-18072), 100 nM ISRIB (MedChemExpress, #HY-12495), 50 μM STF083010 (MedChemExpress, #HY-15845), 1.2 μM Ceapin-A7 (MedChemExpress, #HY-108434), 0.5 mM TAUDC (MedChemExpress, #HY-19696), 10 ng/ml human IL-7 (Novoprotein, #CX47), and 10 ng/ml human IL-15 (PeproTech, #200-15) for 24 h and then harvested for staining with FVD506, anti-mouse CD4 and anti-mouse CD8 antibodies for 25 min at 4 °C. The cells were then washed and resuspended in FACS buffer for flow analysis. The following inhibitors were dissolved in DMSO (Sigma, #D2650) and used: Z-VAD-FMK (10 mM), GSK2606414 (2 mM), ISRIB (1 mM), STF083010 (100 mM), ceapin-A7 (10 mM), MCC950 (50 mM), and NEC-1s (50 mM). NAC (500 mM) and TAUDC (200 mM) were dissolved in distilled water and used.
Glucose, fatty acid uptake and lipid content measurement
To determine glucose uptake, cells were cultured in glucose-free RPMI medium containing 100 μM 2-NBDG (Invitrogen, #N13195) at 37 °C for 30 min and then washed with FACS buffer for surface staining. To determine lipid uptake, cells were incubated with 0.5 μM BODIPY FL C12 (Invitrogen, #C3927MP) or BODIPY FL C16 (Invitrogen, # D3821) at 37 °C for 20 min and then washed with FACS buffer for surface staining. For detection of lipid content and cholesterol content, after permeabilization and fixation, cells were stained using BODIPY 493/503 (Invitrogen, # D2191) at a final concentration of 2 μM for measuring lipids or Filipin (Sigma, # SAE0087) at a final concentration of 20 μg/ml for measuring cholesterol content, together with other intracellular proteins.
Apoptosis assay
Cells were first stained for surface markers at 4 °C for 25 min. Then, the cells were washed two times with cold PBS and stained with 7AAD (BD Biosciences, #51-68981E) and Annexin V-Alexa Fluor 488 (Life Technologies, #V13245) in Annexin V binding buffer (1×, BD Biosciences, # 51-66121E) for 20 min at room temperature. The cells were used immediately for analysis.
CPD labeling and proliferation assay
For CPD labeling, isolated splenic T cells were washed twice with PBS to remove any serum and stained with Cell Proliferation Dye eFluor 670 (5 µM, Invitrogen, #65-0840). The cells were incubated for 10 min at 37 °C in the dark. Then, the cells were resuspended in 4–5 volumes of cold complete medium (containing ≥10% serum) and incubated on ice for 5 min to stop labeling. This step was followed by washing the cells 3 times with complete medium. The CPD-labeled T cells were cultured with complete T-cell culture medium and separately stained with surface markers at 48, 72 and 96 h for flow cytometry analysis.
Cell cycle analysis
Cells were first subjected to viability staining with Fixable Viability Dye eFluor 506 (eBioscience, #65-0866-18) for 20 min on ice and then stained with CD4-FITC and CD8-PE for 25 min on ice. Then, the cells were fixed with fixation buffer (Invitrogen, fixation/permeabilization: fixation/permeabilization diluent = 1:3) for 20 min at 4 °C and stained with anti-Ki67-Percp-cy5.5 (BioLegend, clone 16A8) in permeabilization wash buffer (1×, Invitrogen) for 25 min at 4 °C. Finally, the cells were washed twice with intracellular staining permeabilization wash buffer and stained with DAPI (Beyotime Biotechnology, #C1002) in distilled water for 15 min at room temperature. An LSR Fortessa was immediately used for analysis.
Transmission electron microscopy
CD8+ T cells from spleens and LNs were sorted. The sorted 5 × 107 CD8+ T cells were fixed in precooled fixation buffer (2.5% glutaraldehyde, 0.1 M phosphate buffer (PB), pH 7.4) overnight at 4 °C. After being washed with PBS three times, the cells were postfixed in 1% osmium tetroxide in PBS for 2 h, dehydrated and embedded in Spurr’s resin according to the standard procedure. Ultrathin sections were stained with uranyl acetate and lead citrate. Endoplasmic reticulum (ER) morphology was imaged by using a Hitachi HT-7800 transmission electron microscope (TEM) (v01.20) and an AMT-XR81DIR camera. For quantitation, regions of the ER in the TEM images were selected using the multiple AOI menu and analyzed under the count and data collector menu in Image-Pro Plus 6.0 software.
ROS measurement
ROS were assessed by DCFH-DA (Beyotime Biotechnology, #S0033S) staining. In brief, cells were washed with PBS (1×) and incubated with 10 µM DCFH-DA at 37 °C in the dark for 30 min. After incubation, cells were stained with surface markers. Finally, the cells were resuspended in FACS buffer for flow cytometry analysis.
Lipid peroxidation measurement
Lipid peroxidation was detected by BODIPY™ 581/591 C11 (Invitrogen, #D3861) staining. First, cells were washed with PBS (1×) and incubated with 1.5 µM BODIPY™ 581/591 C11 at 37 °C in the dark for 30 min. This step was followed by cell surface staining. Finally, the cells were resuspended in FACS buffer for further analysis. In addition, as the end-product of peroxidation of polyunsaturated lipids, malondialdehyde (MDA) was also measured with an MDA assay kit (Abcam, #ab118970). A total of 1 × 106 sorted WT and Sel1l−/− CD8+ T cells were homogenized on ice in 300 μl of MDA lysis buffer containing 3 μl of BHT (100×). The samples were centrifuged at 13,000 × g for 10 min to remove insoluble material. Next, 200 μl of the supernatant from each homogenized sample was placed into a microcentrifuge tube, and 600 μl of TBA solution was added into each vial containing standard and sample. The MDA-TBA adduct was incubated at 95 °C for 60 min, and then 300 μl of 1-butanol was added to extract the MDA-TBA adduct from the 800 μl reaction mixture. Finally, 1-butanol was removed by heating on a hot block at 55 °C. The residue containing the MDA-TBA adduct was dissolved in 200 μl of water and then transferred to a 96-well plate for analysis.
Single-cell collection, library construction and sequencing
Spleen tissues were dissociated into single-cell suspensions. Briefly, spleens were first processed with the flat end of a syringe in a 100 mm culture dish containing 5 ml of cold FACS buffer (2% FBS in PBS) and then passed through a 40 μm cell strainer into a 15 ml tube. Second, the cells were centrifuged to remove the supernatant. The cell pellets were treated with RBC lysis buffer to remove red blood cells. After washing with cold FACS sorting buffer, the sorted CD3+ cells from 3 WT or Sel1l−/− mice were pooled, and cells with a viability higher than 90% were used for 10× genomics scRNA-seq.
Furthermore, the cell suspensions were loaded into Chromium microfluidic chips with 3’ chemistry and barcoded with a 10× Chromium Controller (10× Genomics). RNA from the barcoded cells was subsequently reverse-transcribed, and sequencing libraries were constructed with reagents from a Chromium Single-Cell 3’ v3 reagent kit (10× Genomics) according to the manufacturer’s instructions. Then, sequencing was performed with an Illumina HiSeq 2000 according to the manufacturer’s instructions (Illumina). Cell Ranger (V4.0.0, https://support.10xgenomics.com/) was used to process scRNA-seq data and generate matrix data containing gene counts for each cell per sample. Briefly, the 10× sequencing data were mapped to the mouse genome, which was downloaded from 10× Genomics, and the unique molecular identifier (UMI) matrix of each cell was generated by using the Cell Ranger count pipeline.
Single-cell RNA-seq data analysis
The gene expression matrices of all samples were imported into Seurat (version 4.0.1) v4 [52] and merged for subsequent analyses. The following filtering steps were carried out to exclude low-quality cells: cells with fewer than 500 and more than 8000 detected genes were discarded, and cells with a high fraction of mitochondrial genes (>10%) were removed. As a result, a total of 17,040 cells (WT: 9,015; Sel1l−/−: 8025) with a median of 2286 genes were included in the further analyses. We applied the SCTransform workflow (https://satijalab.org/seurat/) to analyze the scRNA-seq data with the default parameters, which replaced the “NormalizeData”, “ScaleData” and “FindVariableFeatures” functions. Then, we performed a principal component analysis (PCA) dimensionality reduction (RunPCA) and selected the first 30 PCs to construct a shared nearest neighbor (SNN) graph (FindNeighbors). To visualize the clustering results, nonlinear dimensional reduction was performed with the UMAP method, and cluster biomarkers were found with the FindAllMarkers function from the Seurat package.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed for each cell subpopulation using the scaled gene expression matrix and GSEA package (V.4.1) available in the Molecular Signatures Database (MSigDB, https://www.gseamsigdb.org/gsea/downloads.jsp) with the default parameters.
Supplementary information
Acknowledgements
LZ was in part supported by the National Key R&D Program of China (2022YFA0807300), the National Natural Science Foundation of China (82271775 and 81971466), the Natural Science Foundation Outstanding Youth Fund of Jiangsu Province (BK20220049) and the CAMS Innovation Fund for Medical Sciences (CIFMS 2021-I2M-1-061, 2021-I2M-1-047 and 2022-I2M-2-004). BZ was in part supported by the Innovation Capability Support Program of Shaanxi 2021TD-38. JZ was in part supported by a Translational Research Grant of NCRCH (2020ZKZC04) and the National Natural Science Foundation of China (82071765). WL was supported by the Natural Science Foundation of China (NSFC 31900645). We thank Prof. Yonghong Wan from McMaster University, Canada, for his critical reading of the manuscript and helpful discussions.
Author contributions
LZ and YG conceived the experiments; LZ, BZ and JZ designed the experiments; and YG, WL and ZW performed most of the experiments. CZ performed bioinformatics analysis of the single-cell RNA-seq data. YH, KT and XL helped maintain the mouse line and perform PCR analysis. WZ, QL and YL helped with data interpretation and valuable discussions. LZ, YG and WL wrote the manuscript. All authors provided intellectual input to the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yafeng Gao, Wenhui Li, Zhenghao Wang, Cangang Zhang.
Contributor Information
Jinping Zhang, Email: j_pzhang@suda.edu.cn.
Baojun Zhang, Email: bj.zhang@mail.xjtu.edu.cn.
Lianjun Zhang, Email: zlj@ism.cams.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-023-01078-x.
References
- 1.Klaips CL, Jayaraj GG, Hartl FU. Pathways of cellular proteostasis in aging and disease. J Cell Biol. 2018;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christianson JC, Carvalho P. Order through destruction: how ER-associated protein degradation contributes to organelle homeostasis. EMBO J. 2022;41:e109845. doi: 10.15252/embj.2021109845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–72. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 4.Krshnan L, van de Weijer ML, Carvalho P. Endoplasmic reticulum–associated protein degradation. Cold Spring Harb Perspect Biol. 2022;14:a041247. doi: 10.1101/cshperspect.a041247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sha H, Sun S, Francisco AB, Ehrhardt N, Xue Z, Liu L, et al. The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab. 2014;20:458–70. doi: 10.1016/j.cmet.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–82. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S, Shi G, Han X, Francisco AB, Ji Y, Mendonça N, et al. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc Natl Acad Sci USA. 2014;111:E582–91. doi: 10.1073/pnas.1318114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi G, Somlo D, Kim GH, Prescianotto-Baschong C, Sun S, Beuret N, et al. ER-associated degradation is required for vasopressin prohormone processing and systemic water homeostasis. J Clin Investig. 2017;127:3897–912. doi: 10.1172/JCI94771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim GH, Shi G, Somlo DR, Haataja L, Song S, Long Q, et al. Hypothalamic ER-associated degradation regulates POMC maturation, feeding, and age-associated obesity. J Clin Investig. 2018;128:1125–40. doi: 10.1172/JCI96420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya A, Sun S, Wang H, Liu M, Long Q, Yin L, et al. Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. EMBO J. 2020;37:e99277. doi: 10.15252/embj.201899277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Z, Torres M, Sha H, Halbrook CJ, Van den Bergh F, Reinert RB, et al. Endoplasmic reticulum-associated degradation regulates mitochondrial dynamics in brown adipocytes. Science. 2020;368:54–60. doi: 10.1126/science.aay2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Inoki A, Fan K, Mao F, Shi G, Jin X, et al. ER-associated degradation preserves hematopoietic stem cell quiescence and self-renewal by restricting mTOR activity. Blood. 2020;136:2975–86. doi: 10.1182/blood.2020007975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L, Liu X, Peng F, Zhang W, Zheng L, Ding Y, et al. Protein quality control through endoplasmic reticulum-associated degradation maintains haematopoietic stem cell identity and niche interactions. Nat Cell Biol. 2020;22:1162–9. doi: 10.1038/s41556-020-00581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Yu J, Xu L, Umphred-Wilson K, Peng F, Ding Y, et al. Notch-induced endoplasmic reticulum-associated degradation governs mouse thymocyte beta-selection. Elife. 2021;10:e69975. doi: 10.7554/eLife.69975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprent J, Surh CD. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat Immunol. 2011;12:478–84. doi: 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–62. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Yan G, Elbadawi M, Efferth T. Multiple cell death modalities and their key features (Review) World Acad Sci J. 2020;2:39–48. [Google Scholar]
- 18.Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Y, Luo Y, Wu Y, Sun Y, Zhao L, Xue Z, et al. SEL1L-HRD1 endoplasmic reticulum-associated degradation controls STING-mediated innate immunity by limiting the size of the activable STING pool. Nat Cell Biol. 2023;25:726–39. doi: 10.1038/s41556-023-01138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang J, Qi L. Quality control in the endoplasmic reticulum: crosstalk between ERAD and UPR pathways. Trends Biochem Sci. 2018;43:593–605. doi: 10.1016/j.tibs.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao SS, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal. 2014;21:396–413. doi: 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94:909–50. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Zhang L, Zhou L, Lei Y, Zhang Y, Huang C. Redox signaling and unfolded protein response coordinate cell fate decisions under ER stress. Redox Biol. 2019;25:101047. doi: 10.1016/j.redox.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–49. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Hu H, Tian M, Ding C, Yu S. The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front Immunol. 2019;9:3083. doi: 10.3389/fimmu.2018.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Tabula Muris. Nature. 2018;562:367–72. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C, Lei L, Yang X, Ma K, Zheng H, Su Y, et al. Single-cell sequencing reveals antitumor characteristics of intratumoral immune cells in old mice. J Immunother Cancer. 2021;9:e002809. doi: 10.1136/jitc-2021-002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JK, Klinger M, Benjamin J, Xiao Y, Erle DJ, Littman DR, et al. Impact of the TCR signal on regulatory T cell homeostasis, function, and trafficking. PLoS ONE. 2009;4:e6580. doi: 10.1371/journal.pone.0006580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaud G, Lesourne R, Love PE. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol. 2018;18:485–97. doi: 10.1038/s41577-018-0020-8. [DOI] [PubMed] [Google Scholar]
- 31.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188:2301–11. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–38. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werlen G, Jain R, Jacinto E. MTOR signaling and metabolism in early T cell development. Genes. 2021;12:728. doi: 10.3390/genes12050728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Zhang H, Li L, Xiao Y, Rao E, Miao Z, et al. TSC1/2 signaling complex is essential for peripheral naïve CD8+ T cell survival and homeostasis in mice. PLoS ONE. 2012;7:e30592. doi: 10.1371/journal.pone.0030592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–97. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–56. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi YJ, Lee H, Kim JH, Kim SY, Koh JY, Sa M, et al. CD5 suppresses IL-15-induced proliferation of human memory CD8(+) T cells by inhibiting mTOR pathways. J Immunol. 2022;209:1108–17. doi: 10.4049/jimmunol.2100854. [DOI] [PubMed] [Google Scholar]
- 38.Chapman NM, Boothby MR, Chi H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol. 2020;20:55–70. doi: 10.1038/s41577-019-0203-y. [DOI] [PubMed] [Google Scholar]
- 39.Dong Y, Tu R, Liu H, Qing G. Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct Target Ther. 2020;5:124. doi: 10.1038/s41392-020-00235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marçais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol. 2014;15:749–57. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Y, van Hoef V, Zhang X, Wennerberg E, Lorent J, Witt K, et al. IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells. Blood. 2016;128:1475–89. doi: 10.1182/blood-2016-02-698027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sowell RT, Goldufsky JW, Rogozinska M, Quiles Z, Cao Y, Castillo EF, et al. IL-15 complexes induce migration of resting memory CD8 T cells into mucosal tissues. J Immunol. 2017;199:2536–46. doi: 10.4049/jimmunol.1501638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun S, Shi G, Sha H, Ji Y, Han X, Shu X, et al. IRE1α is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat Cell Biol. 2015;17:1546–55. doi: 10.1038/ncb3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S, Xing Y, Liu Y. Emerging roles for the ER stress sensor IRE1α in metabolic regulation and disease. J Biol Chem. 2019;294:18726–41. doi: 10.1074/jbc.REV119.007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Huang Z, Zhang H, Lu J, Wei Y, Yang Y, et al. IRE1-mTOR-PERK axis coordinates autophagy and ER stress-apoptosis induced by P2X7-mediated Ca(2+) influx in osteoarthritis. Front Cell Dev Biol. 2021;9:695041. doi: 10.3389/fcell.2021.695041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace DL, Bérard M, Soares MV, Oldham J, Cook JE, Akbar AN, et al. Prolonged exposure of naïve CD8+ T cells to interleukin-7 or interleukin-15 stimulates proliferation without differentiation or loss of telomere length. Immunology. 2006;119:243–53. doi: 10.1111/j.1365-2567.2006.02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan JT, Dudl E, LeRoy E, Murray R, Sprent J, Weinberg KI, et al. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc Natl Acad Sci USA. 2001;98:8732–7. doi: 10.1073/pnas.161126098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q, Yang X, Long G, Hu Y, Gu Z, Boisclair YR, et al. ERAD deficiency promotes mitochondrial dysfunction and transcriptional rewiring in human hepatic cells. J Biol Chem. 2020;295:16743–53. doi: 10.1074/jbc.RA120.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dils AT, Correa LO, Gronevelt JP, Liu L, Kadiyala P, Li Q, et al. The endoplasmic reticulum associated degradation adaptor Sel1L regulates T cell homeostasis and function. J Immunol. 2021. 10.1101/2021.05.22.445275.
- 51.O'brien TF, Gorentla BK, Xie D, Srivatsan S, McLeod IX, He YW, et al. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. Eur J Immunol. 2011;41:3361–70. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.