Abstract
The microtubule (MT) cytoskeleton and its dynamics play an important role in cell migration. Depletion of the microtubule-severing enzyme Fidgetin-like 2 (FL2), a regulator of MT dynamics at the leading edge of migrating cells, leads to faster and more efficient cell migration. Here we examine how siRNA knockdown of FL2 increases cell motility. Förster resonance energy transfer biosensor studies shows that FL2 knockdown decreases activation of the p21 Rho GTPase, RhoA, and its activator GEF-H1. Immunofluorescence studies reveal that GEF-H1 is sequestered by the increased MT density resulting from FL2 depletion. Activation of the Rho GTPase, Rac1, however, does not change after FL2 knockdown. Furthermore, FL2 depletion leads to an increase in focal adhesion kinase activation at the leading edge, as shown by immunofluorescence studies, but no change in actin dynamics, as shown by fluorescence recovery after photobleaching. We believe these results expand our understanding of the role of MT dynamics in cell migration and offer new insights into RhoA and Rac1 regulation.
Significance
Cell migration is an important element of both human development and disease, and the process relies heavily on the microtubule cytoskeleton. The regulation of microtubule dynamics is an essential component of cell migration, as dynamic microtubules help to coordinate the actin cytoskeleton and cellular adhesion at the leading edge. Depletion of the microtubule-severing enzyme Fidgetin-like 2 (FL2) has been shown to enhance cell migration, which is of particular interest as a potential wound-healing therapeutic. Understanding how FL2 fits into the greater process of cell migration further expands our understanding of the role of microtubule-severing enzymes and microtubule dynamics therein.
Introduction
Cell motility plays an important role in human development and disease (1,2,3). Modifiers of cell motility have therapeutic potential in wound healing (4), angiogenesis (5), and neuronal repair (6). Therefore, a more comprehensive understanding of the mechanisms and modifications of cell motility in adherent cells would be valuable for the development of new pharmacological technologies.
Microtubules, actin, and focal adhesions are all crucial components of cell migration, providing polarity, structure, and adhesion. The roles of actin and focal adhesions are the most widely studied aspects of adherent cell motility (1,2,7,8). The actin cytoskeleton, which is composed of F-actin filaments consisting of G-actin monomers, drives the cyclical protrusion of the actin-based lamellipodium at the front of moving cells (7,8). Focal adhesions (FAs), multiprotein complexes that form at the junction of the cell cortex and the extracellular matrix (7), create traction for cells as they move. However, it is becoming clear that the microtubule cytoskeleton serves to connect these various facets of adherent cell motility. Microtubules (MTs) are polarized polymers consisting of α- and β-tubulin subunits that extend from the centromeres in the cell interior out to the cell cortex (1,7). Directional protrusion of the leading edge and overall cell polarity during motility are determined by MT dynamics and orientation (7,9). MTs locally regulate Rho GTPases at the leading edge, which coordinate the cytoskeletal and adhesive components of cell motility, including lamellipodial actin and focal adhesion dynamics (10,11). There is also substantial interplay between MTs and FAs. In fact, FAs, which consist of transmembrane integrins bound to a host of intracellular structural proteins such as vinculin and paxillin, include a variety of cytoskeletal regulators (12,13). In particular, in addition to regulators such as focal adhesion kinase (FAK) (14,15), MT contact with FAs mediates their turnover (1,3,13).
A subset of the MTs that regulate motility at the leading edge undergo a behavior known as dynamic instability, wherein the plus-end, or the distal end facing the cortex, stochastically shifts between phases of growth and shrinkage. This dynamicity is essential for these MTs to regulate key cyclical pathways in motility, as well as to probe the intracellular space in order to direct cellular orientation (3). Dynamic instability has also been shown to influence leading edge processes to regulate membrane protrusion and motility (1,3). As a result, the precise and timely regulation of MT dynamics at the leading edge is crucial for efficient motility.
One class of MT regulators that locally modulate MT dynamics are the microtubule-severing enzymes (MSEs) (16,17), important in cell motility, cell division, and neurogenesis. Although MSEs function in a variety of contexts, the most recently identified, Fidgetin-like 2 (FL2), has been shown to localize to the leading edge, where it regulates MT dynamics and cell migration (4). Like the other MSEs, FL2 contains an N-terminal MT-interacting and trafficking domain, which binds to the MT lattice, and a C-terminal AAA ATPase domain, which uses ATP hydrolysis to pull individual tubulin dimers free. Repeated iterations of this behavior result in MT severing (4,16,17). FL2 putatively severs specifically the dynamic MTs. FL2 depletion therefore leads to a significant increase in both local MT density and dynamicity, and as a result, cells migrated both more quickly and more directionally in in vitro scratch assays after siRNA-mediated FL2 knockdown (KD) (4). All of these effects were successfully rescued by transfection with a GFP-tagged FL2 construct (4).
FL2’s impact on the MT cytoskeleton and cell migration has been translated into multiple animal models of tissue repair: our previous work has shown that siRNA-mediated FL2 KD can expedite wound healing in murine excision and burn wound models (18), enhance cardiac output after myocardial infarction (5), and induce neuronal regeneration of severed or crushed peripheral nerves (6). Although previous work has shown that FL2 KD promotes cell migration in adherent cells (4,5), the intervening steps in this pathway remained elusive. The therapeutic potential of this MSE as a target for tissue repair underscores the need to elucidate the cellular processes that link FL2 to cell motility regulation. Here, we show that FL2’s MT-severing activity releases GEF-H1, an activator of the p21 family of small GTPase RhoA, sequestered by dynamic MTs at the leading edge. This highly localized GEF-H1 activation of RhoA shifts the balance and polarization of RhoA and Rac1 activation kinetics and directly impacts FA turnover, as opposed to F-actin polymerization, to regulate cell motility.
Materials and Methods
Experimental models and subject details
U2OS cell lines
All U2OS cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 1% GlutaMax, and 1% penicillin/streptomycin in a humidified incubator at 37°C and 5% CO2. Maintenance medium for U2-OS cells stably expressing Förster resonance energy transfer (FRET) biosensors was additionally supplemented with 0.1% puromycin, 0.1% doxycycline; 1% G418 was also added to medium after cells were thawed from liquid nitrogen.
U2OS cells are female and acquired from ATCC.
Method details
Transfection and siRNA treatment
For all transfections, cells were counted using a hemocytometer, then nucleofected using Lonza 4-D Nucleofector X Unit, following the manufacturer’s protocols. Unless otherwise specified, for FL2 knockdown studies, cells were transfected with 100 pmol negative control siRNA (Sigma-Aldrich, St. Louis, MO, USA; SIC002 or SIC001) or FL2 siRNA (Dharmacon, Lafayette, CO, USA; ON-TARGETplus Set of four, LO-030094-01-0002).
Förster resonance energy transfer
Stable cell line production
U2OS cell lines stably expressing either the RhoA, Rac1, or GEF-H1 FRET biosensors under the tetracycline-inducible promoter (tet-OFF, Clontech, Kyoto, Japan) were made and used as previously described (19). RhoA (20) and Rac1 (21) FRET biosensors have been previously described (22). GEF-H1 FRET biosensor (23) was generously gifted by Dr. Klaus Hahn.
Transfection and FL2 knockdown
8 × 105 cells were nucleofected with negative control or FL2 siRNA (as described above) and grown in a 15-cm cell culture dish (USA Scientific, Ocala, FL, USA; CC7682-4875) without doxycycline, puromycin, or G418 to induce biosensor expression 48 h before imaging. After 24 h, transfected cells were trypsinized a second time then replated at a 1:2 mixture ratio with untreated U2OS cells in a 35-mm MatTek dish (MatTek, Ashland, MA, USA; P35G-1.5-14-C). Monolayer was scratched with a 200-ul Pipette tip 12–14 h before imaging for RhoA and Rac1 experiments and 4 h before imaging for GEF-H1 experiments. For GEF-H1 biosensor FRET experiments, cells were additionally transfected with pTriEX-mScarlet-NES plasmid. pTriEX-mScarlet-NES was produced by synthesizing and inserting a full length codon optimized mScarlet (24) at NcoI/BamHI sites in pTriEX-4 (Novagen, Sigma-Aldrich) backbone, by using the primer pair: 5′-GGATTATATAATTATATAAACCATGGTGAGCAAGGGCGAGGCCGTGAT-3′ and 5′-CGTTAATATATTAATATGGATCCCTTGTACAGCTCGTCCATGCCGCCGG-3′, followed by a nuclear export sequence LQLPPLERLTL taken from human HIV-1 Rev protein (25), by ligating a small linker constructed from primer pair: 5′-GATCCCTACAGCTACCACCACTAGAGCGACTAACACTATAAC-3′ and 5′-TCGAGTTATAGTGTTAGTCGCTCTAGTGGTGGTAGCTGTAGG-3′ at BamHI/XhoI sites in the pTriEX-4 (Novagen, Sigma-Aldrich) backbone. Cytoplasmic mScarlet fluorescence was used to define the cell edge to account for the GEF-H1 expression pattern not necessarily coinciding with the actual cell edge location.
Live cell imaging
Cells expressing RhoA, Rac1, or GEF-H1 biosensors were imaged as previously described (19). Cells were washed once with 1× PBS then imaged in Ham’s F12 media +3% FBS, argon gas sparged, and supplemented with 1:100 diluted Oxyfluor reagent (Oxyrase, Mansfield, OH, USA; OF-0005) and 10 mM sodium lactate. In all experiments, cells were illuminated using 100 W Hg arc lamp, through excitation bandpass filters: ET436/20X (Chroma Technology, Bellows Falls, VT, USA) for CFP, ET500/20× (Chroma Technology) for YFP, FF575/25 (Semrock, Rochester, NY, USA) for mScarlet excitations, and ET480/40M (Chroma Technology) for CFP, ET535/30M (Chroma Technology) for YFP-FRET, and FF628/32 (Semrock) for mScarlet emissions, and imaged using a 40×/1.3 NA-DIC objective and at 2 × 2 camera binning on a pair of PrimeBSI-Express sCMOS cameras (Photometrics, Tucson, AZ, USA), resulting in an effective pixel size in object plane of 309 nm. Cells expressing RhoA or Rac1 biosensors were imaged at 5-s intervals for 20 min, and a 0.3-ND filter was used in the excitation path of the CFP channel to attenuate the excitation light intensity, and 0.1-ND filter was used in the FRET emission path to balance the relative intensities of FRET versus the CFP at the two sCMOS cameras for simultaneous image acquisition. Cells expressing GEF-H1 biosensor were imaged at 10-s intervals for 20 min; a 0.3-ND filter was used in the CFP excitation path for intensity attenuation, and a 0.1-ND filter was used for the mScarlet excitation attenuation. In the GEF-H1 biosensor imaging, a 0.1-ND filter was used in the FRET emission path to balance the relative intensities of FRET versus the CFP at the two sCMOS cameras during simultaneous image acquisition. Images were acquired using MetaMorph ver. 7.10.4 software (RRID:SCR_002368).
Western blotting
Whole-cell lysates were obtained from cells transfected with 140 pmol of either negative control siRNA (Sigma-Aldrich; SIC002) or FL2 siRNA (Dharmacon; J-030094-09) as described above. Cells were lysed in ice-cold RIPA buffer then frozen 48 h after siRNA treatment was used. A standard gel electrophoresis and Western blotting protocol was followed (26). Blots were imaged on iBright CL1500 imaging system. FL2 band density was normalized to the housekeeping gene GAPDH using the iBright Image Analysis 1.4.0 software. The following antibodies were used: mouse anti-FL2 antibody purified in-house (as described in (27)), mouse anti-GAPDH (Fitzgerald Industries International, Acton, MA, USA; 10R-G109a, RRID:AB_1285808), and peroxidase-labeled anti-mouse IgG (SeraCare, Milford, MA, USA; 5220-0341, RRID:AB_2891080).
Immunofluorescence
1 × 105 cells were nucleofected as described above then grown on glass coverslips in a 24-well plate cell culture plate (Thermo Fisher Scientific, Waltham, MA, USA; 3527) for 48 h. Monolayer was scratched with a 200-μL Pipette tip 6 h before fixation. For actin experiments, cells were fixed for 10 min on ice with a fixative solution (4% paraformaldehyde, 0.1% Triton X, and 0.15% glutaraldehyde in BRB80). For all other experiments, cells were fixed with methanol at −20°C for 20 min. A standard immunolabeling protocol was then followed (26). The following antibodies and dilutions were used: 1:400 mouse anti-tubulin (NeoMarkers, Thermo Fisher Scientific; MS-581-P1), 1:400 rabbit anti-GEF-H1 (Abcam, Cambridge, UK; ab155785, RRID:AB_2818944), 1:400 rabbit anti-FAK (Invitrogen, Waltham, MA, USA; 44625G), 1:400 rabbit anti-phospho-FAK (Y397) (Invitrogen; 700255), 1:800 Alexa Fluor 488 anti-mouse IgG (LifeTechnologies, Thermo Fisher Scientific; A21202), 1:800 Alexa Fluor 568 anti-mouse IgG (Invitrogen; A10037), 1:800 Alexa Fluor 568 anti-rabbit IgG (Invitrogen; A10042), and 1:800 Alexa Fluor 488 anti-rabbit IgG (Invitrogen; A21206). Actin was labeled by either 1:200 Alexa Fluor 488 phalloidin (Invitrogen; A12379) or 1:200 Alexa Fluor 568 phalloidin (Invitrogen; A12380).
Cells were imaged using a 4D spinning-disk confocal microscope (PerkinElmer, Waltham, MA, USA) with either a 60× (1.4 NA) objective (actin) or a 100× (1.4 NA) objective (GEF-H1, FAK, p-FAK) and a digital camera (Orca ER; Hamamatsu, Shizuoka, Japan). Z planes = 0.2 μm, and Z stack = 2 μm. Laser intensity and exposure time were kept consistent across conditions within a single experiment. Images were acquired using the Volocity 6.2.2 (2013) software (RRID:SCR_002668).
Fluorescence recovery after photobleaching
Cells were transfected with siRNA (as described above) and F-Tractin-eGFP plasmid (Addgene, Watertown, MA, USA; #58743, a gift from Dr. Dyche Mullins) then plated onto 35-mm MatTek dishes 48 h before imaging. Monolayer was scratched with a 200-μL Pipette tip 16 h before imaging.
Cells were imaged in phenol-free Leibovitz-15 medium supplemented with 10% FBS, 1% GlutaMax, and 1% penicillin/streptomycin. Imaging was performed using a Zeiss LSM 5 Live duo-scan laser scanning microscope with 63× (1.4 NA) objective at a controlled temperature of 37°C. Images were acquired and analyzed using ZEN 2009 software. The experiment sequence was as follows: 20 acquisition steps at 60% laser intensity then 1 bleaching step at 100% laser intensity at wavelength 488 nm. Bleaching region of interest (ROI) was defined as a circle of diameter 4.7 μm. Fluorescence recovery images were acquired every 0.0667 s for 180 s.
Quantification and statistical analysis
Förster resonance energy transfer
Briefly, images were processed and analyzed as follows. Raw images were background subtracted, shade corrected, and camera noise filtered. Flat-field correction and pixel-by-pixel alignment (28) were performed before being processed for ratiometric analysis (19). Corrected FRET images were then registered to corrected CFP (cyan fluorescent protein) images (29) before being analyzed as described in Machacek et al. (30). Leading edge motion was tracked using a previously published algorithm (31). Since GEF-H1 localization at the leading edge is not coincident with the actual cell edge location, mScarlet-NES was expressed to mark the cell cytoplasm and visualize the cell cortex in order to track the leading edge using the edge tracking tool (31). The ROI window was 6 × 3 pixels, which was determined to be diffusion-limited by analysis of diffusion radius within the time-lapse time frame (30). ROI windows were moved backward so that Pearson’s correlations were performed at specific distances proximal to the edge (30) (Fig. S2 A). Cross correlation functions were determined between local edge motion at the leading edge to the corresponding local average FRET biosensor readouts (FRET/donor ratio) to edge protrusion at various distances away from the edge (Fig. S2 A) using MATLAB function xcov (RRID:SCR_001622). As the GEF-H1 FRET biosensor is an inverse FRET biosensor (23), FRET biosensor data were processed as a donor/FRET ratio instead. Otherwise, analyses were conducted as was done for the RhoA and Rac1 biosensor experiments. Individual cross correlation functions were pooled between all measurement windows at the same distance from the leading edge and smooth-spline fitted between all conditions (30). Average cross correlation functions were calculated, and 95% confidence intervals were estimated by a nonparametric bootstrap method (32).
Immunofluorescence and colocalization analyses
All images were analyzed using FIJI software (RRID:SCR_002285) and statistical analysis performed using GraphPad Prism (RRID:SCR_002798). ROIs were drawn around the first 5.6 μm of the leading edge of cells along the scratch. Integrated density fluorescence values were background subtracted to get a CTCF (corrected total cell fluorescence) value. CTCF values were then normalized to the average control condition CTCF for that experiment.
For the FAK and p-FAK IF experiments, ROIs were drawn around each individual FA within the ROI defined above. CTCF values were calculated for each individual FA and then normalized to the average CTCF of all the FAs of the control condition for that experiment.
For the colocalization experiments, all images for a single experiment were set to the minimum and maximum fluorescence values of the entire data set (both control and FL2 KD conditions) then converted to 8-bit images. ROIs around the leading edge as described above were drawn. Colocalization within these ROIs was analyzed by the FIJI plugin Coloc2 (33), and the Spearman correlation values were compared between control and FL2 siRNA-treated cells using Welch’s t-test.
Fluorescence recovery after photobleaching
Mean fluorescence values over time were extracted using FIJI for three ROIs: bleaching area (ROIbleach, defined above), background (ROIbkgd, defined as an area within the scratch), and the cell interior (ROIcell, defined as an area in the cytoplasm away from the bleaching ROI). ROI sizes and shapes were kept constant throughout the data set.
FRAP images were analyzed using an R script (RRID:SCR_001905) to produce t1/2 and mobile fraction. The half-time to recovery (t1/2) indicates how quickly the fluorescence is recovering (in this case, how quickly the actin is polymerizing). The mobile fraction indicates how much the fluorescence recovers (in this case, how much actin is in the dynamic pool and actively polymerizing). The process was adapted from Giakoumakis et al. (34).
First, ROIbleach and ROIcell fluorescence values were background subtracted using ROIbkgd fluorescence values. Background-subtracted ROIbleach fluorescence values were 1) normalized to the pre-bleach fluorescence level of the bleached area and 2) corrected for photobleaching due to imaging using the background-subtracted ROIcell fluorescence values. Corrected ROIbleach fluorescence values were additionally normalized for bleaching efficiency such that the mean pre-bleach fluorescence value was 1 and the initial post-bleach fluorescence value was 0. Finally, a least-squares regression fit was applied to the corrected values using R’s NL2SOL algorithm. The starting values of the algorithm parameters were α = 0.5 and β = 0.563, and the maximum number of fitting iterations was set to 1000 steps (35). The parameters of the best-fit curve that was determined from this algorithm were used to calculate the t1/2 and the mobile fraction. The t1/2 and mobile fraction values were compared between control and FL2 siRNA-treated cells using Welch’s t-test on GraphPad Prism.
Results
Fidgetin-like 2 knockdown polarizes RhoA, but not Rac1, activation at the leading edge
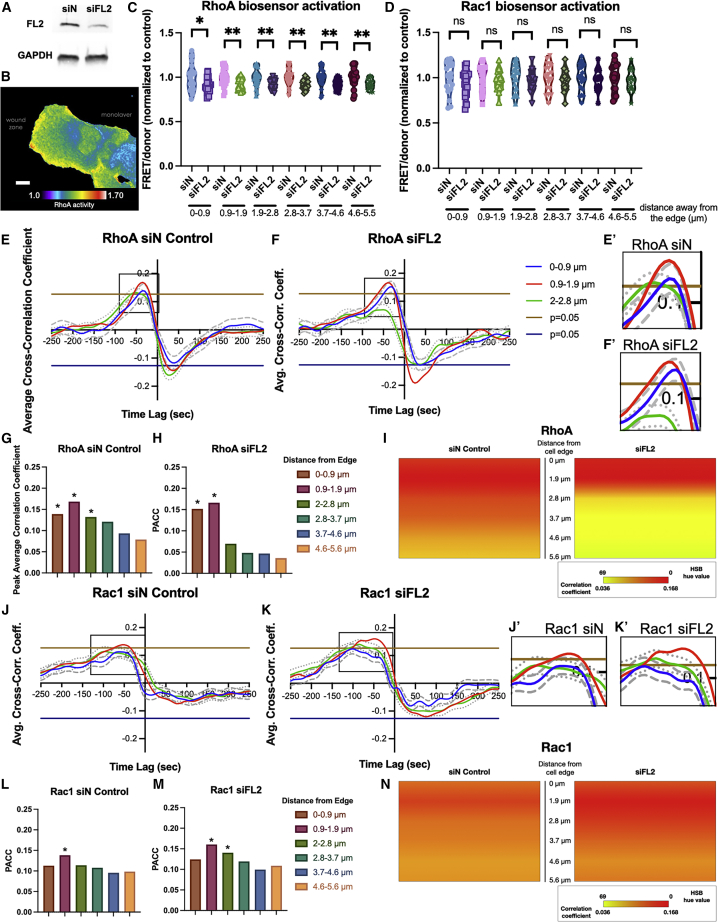
The RhoA and Rac1 GTPases function as master signaling regulators in adherent cell motility (36) and control the dynamics of actin and focal adhesion turnover (11,37,38). Classically, RhoA and Rac1 act antagonistically through reciprocal inhibition; however, more recent evidence suggests this reciprocity is more nuanced than previously believed (3,30). Both RhoA and Rac1 are activated by guanine nucleotide exchange factors (GEFs), some of which are regulated by MT and FA dynamics (1,7,9,13,23,39,40). FL2 depletion has been shown to enhance cell migration, but the role of RhoA and Rac1 activity in this pathway remained unknown. U2OS cells stably expressing either RhoA or Rac1 FRET biosensors (22) were transfected with either control or FL2-targeting siRNA, which resulted in a 60%–70% knockdown of endogenous FL2 after 48 h, confirmed by Western blot (Fig. 1 A). Knockdown of FL2 by siRNA increased MT density at the leading edge, which corroborated previous work in U2OS cells (4) and served as a secondary confirmation of FL2 depletion (Fig. S1 A). At 48 h after transfection, the monolayer was scratched to induce directional cell motility. After 12–14 h, biosensor activities of the cells migrating into the wound space were imaged (Fig. 1 B; Videos S1 and S2). Average FRET biosensor readouts (FRET/donor ratios), which represent Rho GTPase activation, were measured within 0.9-μm increments up to 5.6 μm distal from the edge (Fig. S1 B). The leading edge and the interior of the lamellipodium do not necessarily behave uniformly due to the presence of microtubules and FAs, so GTPase activation in these different regions is of interest. This 5.6-μm region of the leading edge was analyzed in all subsequent experiments. The overall RhoA activity levels were significantly reduced after FL2 depletion, particularly beyond the first 0.9 μm from the leading edge (Fig. 1 C). On the other hand, Rac1 activation levels at the leading edge did not significantly change after FL2 KD (Fig. 1 D).
Figure 1.
RhoA but not Rac1 activation becomes increasingly polarized at the leading edge after FL2 KD. (A) Representative Western blot confirming FL2 siRNA KD in U2OS cells. (B) Pseudocolor map of RhoA activity as indicated by the RhoA FRET biosensor in a representative siN control-treated U2OS cell. (C and D) FRET/donor ratios of either (C) RhoA or (D) Rac1 biosensors activities were extracted for the first time point at each distance away from the edge (Fig. S1B, up to 5.6 μm) after control or FL2 siRNA knockdown. Mean ratios were plotted then compared between control and FL2 KD conditions using Welch’s t-test. (∗p < 0.05.) Data points represent averages of the individual windows in individual cells. (E and F, J and K) Curves showing correlation between (E–F) RhoA or (J–K) Rac1 FRET biosensor activation and edge velocity relative to edge protrusion initiation (x = 0) after either (E, J) control siRNA or (F, K) FL2 siRNA treatment. Curves plotted are spline fits from pooled correlation coefficients of individual windows from cells (See also Fig. S2A–D). Individual curves represent various distances (up to 2.8 μm) away from the cell edge (Fig. S1B). Dashed and dotted lines show 95% confidence intervals, estimated by a nonparametric bootstrap method. Horizontal lines indicate significance of Pearson’s correlation coefficient at p < 0.05. (E: n = 440 windows, 21 cells; F: 462 windows, 21 cells; G: n = 401 windows, 17 cells; H: 334 windows, 17 cells). (E′ and F′ and J′ and K′) Insets show higher magnification of (E′–F′) RhoA or (J′–K′) Rac1 peak correlation coefficients. (G–I and L–N) (G–H, L–M) Bar graphs showing peak average correlation coefficient (PACC) irrespective of time of (G, H) RhoA or (L, M) Rac1 FRET biosensor activation to edge protrusion at various distances (up to 5.6 μm) (Fig. S1B) away from the cortex after (G, L) control siRNA or (H, M) FL2 siRNA treatment. (∗p < 0.05.) (I and N) Quantitative schematic representing the spatial polarization of (I) RhoA or (N) Rac1 involvement in edge protrusion in the first 5.6 μm away from the leading edge of (G) and (L) (left) and (H) and (M) (right). To see this figure in color, go online.
Linear pseudocolor map indicates RhoA activity. Images taken in 5-s intervals for 20-min time-lapse on a confocal microscope using a 40× oil objective. Scale bar indicates 10 μm. Video plays at 20 frames per sec (fps)
Linear pseudocolor map indicates RhoA activity. Images taken in 5-s intervals for 20-min time-lapse on a confocal microscope using a 40× oil objective. Scale bar indicates 10 μm. Video plays at 20 frames per sec (fps)
RhoA activity increases contractility, whereas Rac1 activation is associated with lamellipodial protrusiveness (10,11). Although Rac1 activity did not change after FL2 KD, the reduction in RhoA activity may allow for a stronger effect of Rac1 promotion of leading edge protrusion, which would encourage greater cell motility. Therefore, the effect of these changes in RhoA but not Rac1 activation on leading edge protrusion was determined. For the leading edge biosensor analysis, a previously described morphodynamics analysis package (30) was used, which determined the kinetic and kinematic coupling of the leading edge motion to the FRET biosensor readouts (30,31). Cross correlation coefficient functions between the leading edge velocities and the changes in the FRET biosensor signals in each cell were calculated at the edge and in 0.9-μm increments up to 5.6 μm distal from the edge (Fig. S1 B). The average cross correlation coefficients were then plotted against their time lags from edge protrusion (Figs. 1 E, F, J, K, and S2 A–D). This allowed for the correlation of Rho GTPase activity and cell edge velocity with respect to both time and space. Moreover, RhoA and Rac1 coordination in the lamellipodium has previously been shown to have distinct lamellipodial sublocalizations both spatially and temporally: Rac1 activation peaks at the edge just after the initiation of a protrusion, and RhoA activation peaks just behind Rac1 at the start of a protrusion (30). The appearance of distinct peaks in the average cross correlation functions pointed to a cyclical nature of the edge fluctuations, as evidenced by the edge velocity autocorrelation functions (Fig. S1 C). The peak correlation coefficient at each distance increment (Fig. S1 B) away from the edge was also plotted (Fig. 1 G, H, L, and M). Finally, the peak correlation values were depicted visually by applying a pseudocolor scale adjusted linearly (Fig. 1 I and N).
FL2 depletion significantly altered the spatial activation patterns of RhoA at the leading edge. In cells treated with control siRNA, RhoA activation was significantly correlated to edge motion up to 2.8 μm away and particularly tightly correlated just behind the leading edge (0.9–1.9 μm). The peak cross correlation coefficient gradually decreased beyond the 2.8-μm distance from the edge (Fig. 1 E, G, and I). In contrast, after FL2 siRNA-mediated KD, average cross correlation coefficient peaks were dramatically abrogated beyond the first 1.9 μm away from the edge (Fig. 1 F and H), resulting in a strongly polarized, forward bias to the coupling of RhoA activity to the cell edge motion (Fig. 1 I). This suggests that FL2 KD enhances cell motility by reducing RhoA activation and producing an overall forward asymmetry of RhoA activity coupling to cell edge motion (reflected in the schematics in Fig. 1 I). As a result, the leading edge maintains a balance of Rac1-mediated protrusiveness and RhoA-mediated contractility, and the lamellipodial body is governed mostly by Rac1 activity due to the polarized decrease in correlation between RhoA activity and edge dynamics, which all together leads to an increase in cell migration.
In contrast to RhoA, Rac1’s correlation with leading edge protrusion was not as asymmetric. In control-treated cells, Rac1 FRET biosensor activation was strongly tied to leading edge protrusion throughout the analyzed region, though it was only significantly correlated within 0.9–1.9 μm behind the leading edge (Figs. 1 J, L, N, and S2 C). In FL2-depleted cells, cross correlation values increased uniformly across the analyzed region relative to control cells (Fig. 1 K and M), though the difference was not significant (Fig. S1 D); Rac1 activity remained significantly correlated to edge protrusion in roughly the same area (Figs. 1 J, K, and S2 D). Importantly, the same spatial patterns of Rac1-protrusion coupling remained in both control and FL2 depletion conditions (Fig. 1 N). These results indicate that the increase in cell migration after FL2 depletions occurs as a result of decreased RhoA, but not Rac1, activation at the leading edge.
FL2 KD leads to increased sequestration of the RhoA activator GEF-H1 by MTs
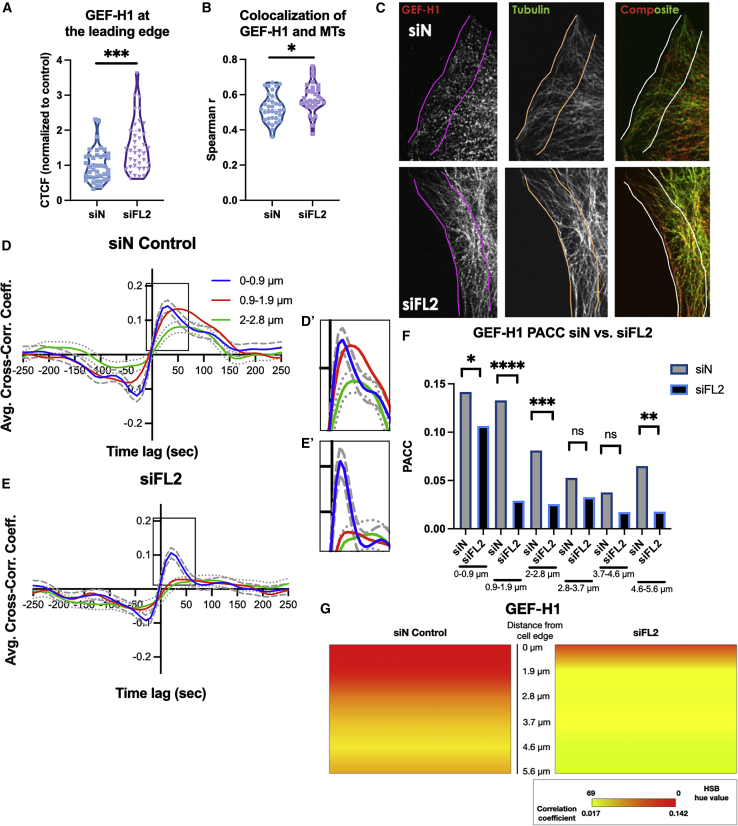
The decrease in RhoA activation after depletion of the MT regulator FL2 pointed to the involvement of a regulator of RhoA activity, controlled by an MT-FL2 mechanism. We turned to GEF-H1, an activator of RhoA (39,40), which is known to be involved in RhoA’s regulation of both focal adhesion turnover and leading edge dynamics (39,40). Moreover, GEF-H1 is itself sequestered and inactivated by MTs (30,39), allowing for tight and localized control of RhoA activation at the leading edge (40). To determine whether the decrease in RhoA activation was due to MT sequestration of GEF-H1, immunofluorescence was used to determine GEF-H1 localization during a scratch assay after FL2 KD. Immunofluorescence analysis revealed both an increase in GEF-H1 at the leading edge (Fig. 2 A and C) as well as stronger colocalization with MTs (Fig. 2 B and C) after FL2 depletion. This suggested greater sequestration of GEF-H1 by the increased density of MTs found locally at the leading edge after FL2 depletion (Fig. S1 A).
Figure 2.
FL2 depletion increases GEF-H1 sequestration by MTs at the leading edge. (A) Violin plot of GEF-H1 fluorescence intensity after control or FL2 siRNA treatment (n = 41,39 cells; Welch’s t-test, ∗∗∗p < 0.001). Data points represent individual cells. (B) Violin plot of Spearman r correlation of GEF-H1 and tubulin immunofluorescence (indicating colocalization) after control or FL2 siRNA treatment. (n = 33, 32 cells; Welch’s t-test, ∗p < 0.05) Datapoints represent individual cells. (C) Representative immunofluorescence images of U2OS cells along a scratch stained for GEF-H1 (red) and tubulin (green). Colored lines indicate 5.6-μm-wide region analyzed in (A) and (B). (D and E) Curves showing correlation between GEF-H1 FRET biosensor activation and edge velocity relative to edge protrusion initiation (x = 0) after either (D) control or (E) FL2 siRNA treatment. Curves plotted are spline fits from pooled correlation coefficients of individual cells. Individual curves represent various distances (up to 2.8 μm) away from the cell edge (Fig. S1B). Dashed and dotted lines show 95% confidence intervals, estimated by a nonparametric bootstrap method. Horizontal lines indicate significance of Pearson’s correlation coefficient at p < 0.05. (D: n = 341 windows, 22 cells; E: 457 windows, 25 cells). (D′–E′) Insets show higher magnification of peak correlation coefficients. All experiments performed in triplicate. (F) Bar graphs showing PACC of GEF-H1 FRET biosensor activation irrespective of time to edge protrusion at various distances (up to 5.6 μm) (Fig. S1B) away from the cortex after control or FL2 siRNA treatment. (n = 341 windows/22 cells (siN control), 457 windows/25 cells (siFL2); Student's t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). (G) Quantitative schematic representing correlation of local GEF-H1 FRET biosensor activation to leading edge protrusion at various distances away from the leading edge (up to 5.6 μm) (Fig. S1B) (341 windows, 22 cells (siN control); 457 windows, 25 cells (siFL2)). To see this figure in color, go online.
Next, changes in GEF-H1 activation due to the MT sequestration after FL2 depletion were assessed. U2OS cells stably expressing a GEF-H1 biosensor (23) were treated with either control or FL2-targeting siRNA. Cells were imaged and analyzed similarly to the RhoA and Rac1 biosensors. In cells treated with control siRNA, GEF-H1 activation was most tightly correlated with edge protrusion up to 1.9 μm away from the cortex. The peak cross correlation coefficient decreased beyond the 1.9 μm distance from the edge (Fig. 2 D, D′, and S2 E). Importantly, it followed the general pattern of spatial correlation found in the FRET experiments with its substrate RhoA. Similarly, after FL2 siRNA-mediated KD, which increased GEF-H1 sequestration by MTs, peak cross correlation coefficients significantly decreased (Figs. 2 E, E′, F, and S2 F). Together, these data strongly suggest a link between FL2’s microtubule-severing activity and its effects on RhoA GTPase involvement in leading edge protrusion during cell migration.
FA turnover, but not actin polymerization, increases after FL2 KD
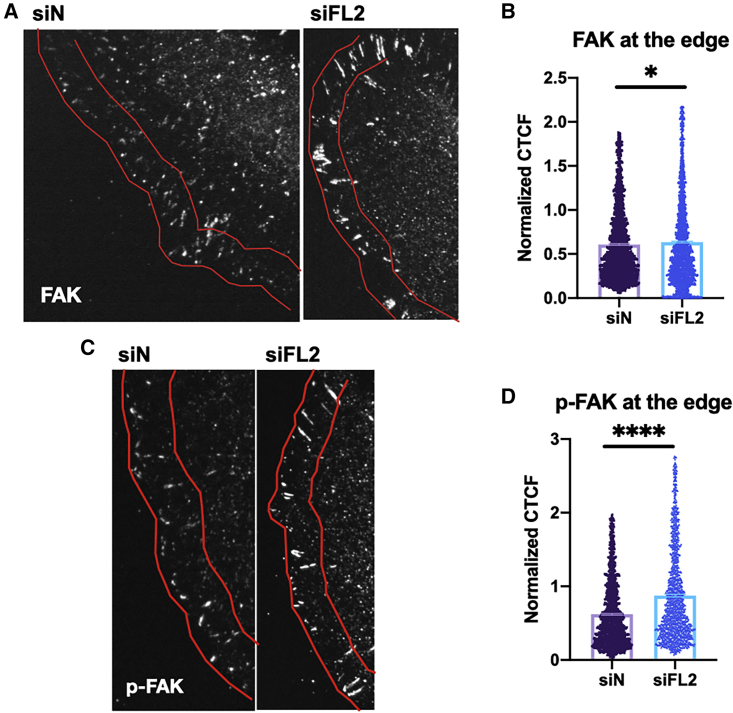
FAs, which in previously published work have shown an increase in size after FL2 depletion (4,27), are known to be regulated by RhoA (37): RhoA promotes FA maturation (10), and its constitutive activity leads to decreased FA turnover (41). Although increased FA size can often impede cell movement, FA size at the leading edge after FL2 KD increases to the optimal size that allows for greater cortical traction without impeding movement (4,27). To determine if there is an accompanying increase in FA turnover to maintain sufficient FA dynamics, the effect of FL2 siRNA-mediated knockdown on FAK localization at the leading edge was determined using immunofluorescence. FAK, which is activated by phosphorylation (p-FAK) (14,15), is one of the determinants of FA breakdown (3,13,15,42). Both FAK (Fig. 3 A and B) and p-FAK (phosphorylated at Y397, indicating FAK activation (43)) (Fig. 3 C and D) localization to the leading edge increased significantly after FL2 depletion. Corroborating previous work (4), there was a significant increase in the size (Fig. S3 C and D) but not the number (Fig. S3 E and F) of FAs, as defined by FAK and p-FAK staining.
Figure 3.
FL2 knockdown results in increased focal adhesion turnover at the leading edge. (A–D) (A and C) Representative immunofluorescence images of U2OS cells along a scratch stained for (A) FAK or (C) p-FAK. Red lines indicate 5.6-μm-wide region of leading edge analyzed in (B and D). (B and D) Scatterplots of fluorescence intensity of (B) FAK and (D) p-FAK. Data points represent individual FAs. (B: n = 2569,3057; D: n = 1458,1426, mean ± SEM; Welch’s t-test, ∗p < 0.05, ∗∗∗∗p < 0.0001). To see this figure in color, go online.
Since actin at the leading edge can also be modulated by RhoA (37), immunofluorescence was used to assess changes in actin density after FL2 depletion. Cells were treated with either control or FL2 siRNA, scratched to induce migration, and then fixed and stained using phalloidin. Phalloidin staining in the 5.6-μm-wide region analyzed did not significantly change after FL2 KD (Fig. S3 A). Lamellipodial actin is often more dynamic in cells that are actively migrating, as the lamellipodium constantly shifts and probes the environment (8). FRAP was utilized to determine whether FL2 depletion affected actin dynamics rather than density. Cells were transfected with F-Tractin-eGFP (an F-action probe (44)) and either control or FL2 siRNA before the monolayer was scratched to induce migration. Actin dynamics were not significantly affected by FL2 depletion: the half-time to recovery was not altered by FL2 KD (Fig. S3 B). Altogether, these data suggest that FL2 depletion enhances cell migration through an increase in FA turnover at the leading edge rather than changes in lamellipodial actin dynamics.
Discussion
Fidgetin-like 2 regulates cell migration via RhoA and FAK activation
The data described here suggest a model in which FL2’s MT-severing activity suppresses cell migration by broadly enhancing RhoA activation at the leading edge through release of GEF-H1 from dynamic MTs. Activated RhoA in spatially expansive regions at and away from the leading edge reduces the relative signaling asymmetry and initiates signaling pathways that result in edge retraction and more stable FAs (9). Our results indicate how FL2 determines the relative forward bias of the RhoA signaling pathway to control leading edge protrusion dynamics and adhesion strength. RhoA and Rac1 activation are regulated reciprocally, as are their downstream effectors (10). Both RhoA and Rac1 activation patterns under control treatment reflected previous studies with other adherent cells (23,30,40). Although GEF-H1 activation at the leading edge reflected the results reported in Azoitei et al. (23) spatially, the temporal coordination with edge protrusion did not. This may be due to a difference in cell types, as the temporal correlation of RhoA activation with edge protrusion was also different (23). Discrepancies in substrate likely also contributed to the differences reported (23). Together, Rac1 and RhoA reciprocal activation helps to generate the lamellipodial protrusion-retraction cycle (30) as well as the FA assembly-disassembly cycle (10,23,30,45) that are essential for efficient cell migration.
FL2 depletion disrupts the Rac1-RhoA balance via GEF-H1 to fine-tune cell migration. GEF-H1 and MT dynamic instability already go hand in hand: stochastic switches between growth and shrinkage bind and release GEF-H1 to cyclically activate RhoA during the retraction phase of edge dynamics (1,40). FL2 depletion increases the density of specifically dynamic MTs, which are crucial for regulation of cell movement (4). These MTs bind and sequester a portion of the GEF-H1 population so that it cannot be activated, particularly just behind the leading edge. This leads to a clearly polarized decrease in local RhoA activation (Fig. 4) in the same region. Not only is RhoA overall less activated here, but its activity is less correlated with leading edge dynamics. It must be noted that we cannot rule out the effect of GEF-H1 on other Rho isoforms, such as RhoC. Future work may elucidate the role of these Rho isoforms in FL2’s regulation of cell migration.
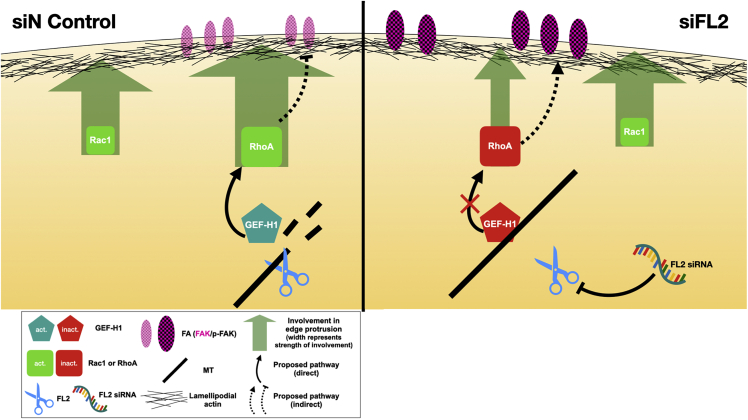
Figure 4.
Model of proposed pathway. Left: in the control condition, FL2 severs microtubules at the leading edge, releasing GEF-H1 to activate RhoA, which regulates FA turnover via FAK (pink) and p-FAK (black checked pattern) to suppress cell motility. Wider solid green arrows indicate involvement in edge protrusion dynamics, and solid and dashed black arrows indicate the pathway FL2 initiates directly and indirectly, respectively. Red and green indicate inactivation and activation, respectively, for GEF-H1 and the Rho GTPases. Right: after FL2 siRNA knockdown, the intact MTs continue to sequester GEF-H1 and suppress this pathway, leading to an associated increase in FA turnover via FAK and p-FAK to promote cell migration. To see this figure in color, go online.
For a Figure360 author presentation of Figure 4, see https://doi.org/10.1016/j.bpj.2022.12.018.
The decrease in RhoA activation seen at the leading edge after FL2 depletion leads to both a shift in FA size (4) as well as recruitment of FAK, a marker of FA disassembly. FAK is normally recruited to FAs in its auto-inhibited state where it binds to both integrins and structural proteins, such as paxillin (3). FAK activation, indicated by Y397 phosphorylation, opens the kinase domain to phosphorylate and disassemble structural proteins of the FA (15). Rapid FA turnover and FAK phosphorylation are inversely correlated to RhoA activity (9,46), as FAK is responsible for breaking down FAs in preparation for the forward lamellipodial surge (9). The exact mechanism by which decreased RhoA activity in these cells leads to increased FAK recruitment and phosphorylation at the leading edge remains to be elucidated (3). Regardless, the increased FAK activation seen after FL2 depletion, which is required for increased FA turnover, likely accommodates the concurrent increase in FA size (3,45).
Although decreased RhoA activity is associated with FA turnover, increased Rac1 activity is associated with dendritic actin polymerization (11). Consistent with the lack of significant change in Rac1 biosensor activation after FL2 KD, actin density and dynamics at the leading edge also did not appear to change significantly.
Altogether, these results show that FL2 depletion enhances cell migration primarily through a mechanism involving decreased RhoA activity and optimized FA dynamics at the leading edge (Fig. 4).
Nuance is the name of the game: Fidgetin-like 2 knockdown reflects the subtle, finely tuned nature of efficient cell motility
FL2’s regulation of cell migration demonstrates the nuanced and delicate balance required for such a complex cellular process. The transient depletion of this hyper-localized enzyme significantly enhances in vitro cell migration as well as in vivo tissue repair in multiple different therapeutic contexts without any apparent risk of hypermotility. The strictly reciprocal relationship between Rac1 and RhoA is turning out to be an oversimplification (30), and this is corroborated by our findings. Depletion of FL2 has distinct, noninverse effects on Rac1 and RhoA activation kinetics at the leading edge, which optimizes migration. The increase in FA size after FL2 KD is also specifically tuned for more efficient movement: FA area increased to the optimal size for strong traction without being too large to disassemble quickly (4).
Further exploration into the relationship between FL2 and the actin cytoskeleton at the leading edge is still necessary. Although immunofluorescence and FRAP did not reveal any significant effect on actin after FL2 KD, these techniques only offer broad characterizations of the actin population. Delving into more specific and nuanced features of actin dynamics is likely to reveal more about its involvement in FL2 regulation and activity during cell migration. Deeper investigations into this unusual enzyme will expand upon current understanding of the complex cell migration machinery as well as provide a biological foundation for a promising therapeutic.
Author contributions
Conceptualization, D.S. and K.S. Methodology, L.H. Software, L.H. and S.S. Formal analysis & writing – Original Draft, K.S. Investigation, K.S. and A.K. Resources & supervision, D.S. and L.H. Writing – review & editing, K.S., D.S., and L.H. Visualization, K.S. and L.H.; Funding acquisition, L.H. and D.S.
Acknowledgments
We thank the Albert Einstein College of Medicine Analytical Imaging and Flow Cytometry Core Facilities, funded in part by the NCI Cancer Center grant P30CA013330. This work was supported by the NIH (5R01GM109909 and 1R01DK109314 to D.S., R35GM136226 to L.H., and 5T32GM007491). L.H. is an Irma T. Hirschl Career Scientist.
Declaration of interests
The authors declare no competing interests.
Editor: Kinneret Keren.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.12.018.
Contributor Information
Louis Hodgson, Email: louis.hodgson@einsteinmed.edu.
David J. Sharp, Email: david.sharp@einsteinmed.edu.
Supporting material
References
- 1.Garcin C., Straube A. Microtubules in cell migration. Essays Biochem. 2019;63:509–520. doi: 10.1042/ebc20190016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shellard A., Mayor R. All roads lead to directional cell migration. Trends Cell Biol. 2020;30:852–868. doi: 10.1016/j.tcb.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Stehbens S., Wittmann T. Targeting and transport: how microtubules control focal adhesion dynamics. J. Cell Biol. 2012;198:481–489. doi: 10.1083/jcb.201206050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charafeddine R.A., Makdisi J., et al. Sharp D.J. Fidgetin-like 2: a microtubule-based regulator of wound healing. J. Invest. Dermatol. 2015;135:2309–2318. doi: 10.1038/jid.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kramer A.H. Ph.D. Albert Einstein College of Medicine; 2019. Fidgetin-like 2: A Developmental Gene as a Promising Target to Induce Adult Tissue Repair. [Google Scholar]
- 6.Baker L., Tar M., et al. Sharp D.J. Fidgetin-like 2 negatively regulates axonal growth and can be targeted to promote functional nerve regeneration. JCI Insight. 2021;6:e138484. doi: 10.1172/jci.insight.138484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hohmann T., Dehghani F. The cytoskeleton-A complex interacting meshwork. Cells. 2019;8:362. doi: 10.3390/cells8040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ananthakrishnan R., Ehrlicher A. The forces behind cell movement. Int. J. Biol. Sci. 2007;3:303–317. doi: 10.7150/ijbs.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhshi T.K., Wernike D., Piekny A. Microtubules and actin crosstalk in cell migration and division. Cytoskeleton (Hoboken) 2014;71:1–23. doi: 10.1002/cm.21150. [DOI] [PubMed] [Google Scholar]
- 10.Guan X., Guan X., et al. Jiao Z. Rho GTPases and related signaling complexes in cell migration and invasion. Exp. Cell Res. 2020;388:111824. doi: 10.1016/j.yexcr.2020.111824. [DOI] [PubMed] [Google Scholar]
- 11.Lawson C.D., Ridley A.J. Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 2018;217:447–457. doi: 10.1083/jcb.201612069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadry Y.A., Calderwood D.A. Chapter 22: structural and signaling functions of integrins. Biochim. Biophys. Acta Biomembr. 2020;1862:183206. doi: 10.1016/j.bbamem.2020.183206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seetharaman S., Etienne-Manneville S. Microtubules at focal adhesions - a double-edged sword. J. Cell Sci. 2019;132:jcs232843. doi: 10.1242/jcs.232843. [DOI] [PubMed] [Google Scholar]
- 14.Tapial Martínez P., López Navajas P., Lietha D. FAK structure and regulation by membrane interactions and force in focal adhesions. Biomolecules. 2020;10:179. doi: 10.3390/biom10020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer M.S., Baumann F., et al. Lietha D. Structural and mechanistic insights into mechanoactivation of focal adhesion kinase. Proc. Natl. Acad. Sci. USA. 2019;116:6766–6774. doi: 10.1073/pnas.1820567116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp D.J., Ross J.L. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 2012;125:2561–2569. doi: 10.1242/jcs.101139. https://www.ncbi.nlm.nih.gov/pubmed/22595526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNally F.J., Roll-Mecak A. Microtubule-severing enzymes: from cellular functions to molecular mechanism. J. Cell Biol. 2018;217:4057–4069. doi: 10.1083/jcb.201612104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Rourke B.P., Kramer A.H., et al. Sharp D.J. Fidgetin-like 2 siRNA enhances the wound healing capability of a surfactant polymer dressing. Adv. Wound Care. 2019;8:91–100. doi: 10.1089/wound.2018.0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiering D., Hodgson L. Multiplex imaging of Rho family GTPase activities in living cells. Methods Mol. Biol. 2012;827:215–234. doi: 10.1007/978-1-61779-442-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertz O., Hodgson L., et al. Hahn K.M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 21.Moshfegh Y., Bravo-Cordero J.J., et al. Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat. Cell Biol. 2014;16:574–586. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donnelly S.K., Bravo-Cordero J.J., Hodgson L. Rho GTPase isoforms in cell motility: don't fret, we have FRET. Cell Adhes. Migrat. 2014;8:526–534. doi: 10.4161/cam.29712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azoitei M.L., Noh J., et al. Danuser G. Spatiotemporal dynamics of GEF-H1 activation controlled by microtubule- and Src-mediated pathways. J. Cell Biol. 2019;218:3077–3097. doi: 10.1083/jcb.201812073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bindels D.S., Haarbosch L., et al. Gadella T.W.J., Jr. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods. 2017;14:53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- 25.Wen W., Meinkoth J.L., et al. Taylor S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 26.O'Rourke B.P., Gomez-Ferreria M.A., et al. Sharp D.J. Cep192 controls the balance of centrosome and non-centrosomal microtubules during interphase. PLoS One. 2014;9:e101001. doi: 10.1371/journal.pone.0101001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charafeddine R.A. Ph.D. Albert Einstein College of Medicine; 2015. Fidgetin-like 2: A Novel Microtubule Based Regulator of Wound Healing and Vasculogenesis. [Google Scholar]
- 28.Bhalla R.M., Hülsemann M., et al. Hodgson L. Multiplex imaging of rho GTPase activities in living cells. Methods Mol. Biol. 2021;2350:43–68. doi: 10.1007/978-1-0716-1593-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodgson L., Shen F., Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr. Protoc. Cell Biol. 2010;Chapter 14 doi: 10.1002/0471143030.cb1411s46. Chapter 14:Unit 14.11.Unit 14 11 1-26-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machacek M., Hodgson L., et al. Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machacek M., Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efron B., Tibshirani R.J. Chapman & Hall; New York: 1993. An Introduction to the Bootstrap. [Google Scholar]
- 33.Schindelin J., Arganda-Carreras I., et al. Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giakoumakis N.N., Rapsomaniki M.A., Lygerou Z. Analysis of protein kinetics using fluorescence recovery after photobleaching (FRAP) Methods Mol. Biol. 2017;1563:243–267. doi: 10.1007/978-1-4939-6810-7_16. [DOI] [PubMed] [Google Scholar]
- 35.Koulouras G., Panagopoulos A., et al. Lygerou Z. EasyFRAP-web: a web-based tool for the analysis of fluorescence recovery after photobleaching data. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky508. W467-w472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wojnacki J., Quassollo G., et al. Cáceres A. Rho GTPases at the crossroad of signaling networks in mammals: impact of Rho-GTPases on microtubule organization and dynamics. Small GTPases. 2014;5:e28430. doi: 10.4161/sgtp.28430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salloum G., Jaafar L., El-Sibai M. Rho A and Rac1: antagonists moving forward. Tissue Cell. 2020;65:101364. doi: 10.1016/j.tice.2020.101364. [DOI] [PubMed] [Google Scholar]
- 38.El-Sibai M., Pertz O., et al. Backer J.M. RhoA/ROCK-mediated switching between Cdc42- and Rac1-dependent protrusion in MTLn3 carcinoma cells. Exp. Cell Res. 2008;314:1540–1552. doi: 10.1016/j.yexcr.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krendel M., Zenke F.T., Bokoch G.M. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat. Cell Biol. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- 40.Nalbant P., Chang Y.C., et al. Bokoch G.M. Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol. Biol. Cell. 2009;20:4070–4082. doi: 10.1091/mbc.e09-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z.G., Lambert C.A., et al. Aumailley M. Effects of constitutively active GTPases on fibroblast behavior. Cell. Mol. Life Sci. 2006;63:82–91. doi: 10.1007/s00018-005-5416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stehbens S., Pemble H., et al. Wittmann T. Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol. 2012;504:293–313. doi: 10.1016/B978-0-12-391857-4.00015-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cary L.A., Chang J.F., Guan J.L. Stimulation of cell migration by overexpression of focal adhesion kinase and its association with Src and Fyn. J. Cell Sci. 1996;109:1787–1794. doi: 10.1242/jcs.109.7.1787. [DOI] [PubMed] [Google Scholar]
- 44.Yamashiro S., Taniguchi D., et al. Watanabe N. Convection-induced biased distribution of actin probes in live cells. Biophys. J. 2019;116:142–150. doi: 10.1016/j.bpj.2018.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridley A. Rho GTPases. Integrating integrin signaling. J. Cell Biol. 2000;150:F107–F109. doi: 10.1083/jcb.150.4.f107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren X.D., Kiosses W.B., et al. Schwartz M.A. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J. Cell Sci. 2000;113:3673–3678. doi: 10.1242/jcs.113.20.3673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linear pseudocolor map indicates RhoA activity. Images taken in 5-s intervals for 20-min time-lapse on a confocal microscope using a 40× oil objective. Scale bar indicates 10 μm. Video plays at 20 frames per sec (fps)
Linear pseudocolor map indicates RhoA activity. Images taken in 5-s intervals for 20-min time-lapse on a confocal microscope using a 40× oil objective. Scale bar indicates 10 μm. Video plays at 20 frames per sec (fps)