Abstract
Imaging two or more fluorescent biosensors in the same living cell can reveal the spatiotemporal coordination of protein activities. However, using multiple Förster resonance energy transfer (FRET) biosensors together is challenging due to toxicity and the need for orthogonal fluorophores. Here we generate a biosensor component that binds selectively to the activated conformation of three different proteins. This enabled multiplexed FRET with fewer fluorophores, and reduced toxicity. We generated this MultiBinder (MB) reagent for the GTPases RhoA, Rac1, and Cdc42 by combining portions of the downstream effector proteins Pak1 and Rhotekin. Using FRET between mCherry on the MB and YPet or mAmetrine on two target proteins, the activities of any pair of GTPases could be distinguished. The MB was used to image Rac1 and RhoA together with a third, dye-based biosensor for Cdc42. Quantifying effects of biosensor combinations on the frequency, duration, and velocity of cell protrusions and retractions demonstrated reduced toxicity. Multiplexed imaging revealed signaling hierarchies between the three proteins at the cell edge where they regulate motility.
Significance
Cell behavior is governed by the coordination of multiple protein activities in space and time. This coordination can be characterized with high resolution by studying two protein activities in the same live cell. However, FRET biosensors typically require the use of two fluorophores for each protein activity, complicating imaging, and the use of multiple biosensors can be toxic to cells. Here we generate a single protein, bearing one fluorophore, which binds specifically to the activated conformation of three different target proteins. Multiplexed imaging with this reagent facilitates multiplexing and reduces cell perturbation. Correlation analysis of simultaneous GTPase imaging revealed localized signaling interactions governing cell motility.
Introduction
We describe here an approach to simultaneously image two protein activities in the same living cell. Multiple studies have shown that biosensors provide valuable information about the spatiotemporal dynamics of protein activity (1,2,3,4). More recently, the correlation of different molecular activities, revealed by using two biosensors together, has provided information on the functional coupling between signaling events. Imaging multiple biosensors based on Förster resonance energy transfer (FRET) has been challenging because this typically requires spectrally orthogonal FRET fluorophore pairs (1,5,6). Furthermore, two or more biosensors increase toxic effects. With low abundance proteins, maintaining normal cell behavior requires that even one biosensor is imaged at levels barely providing sufficient signal/noise; further lowering biosensor concentrations to reduce toxicity is often not feasible.
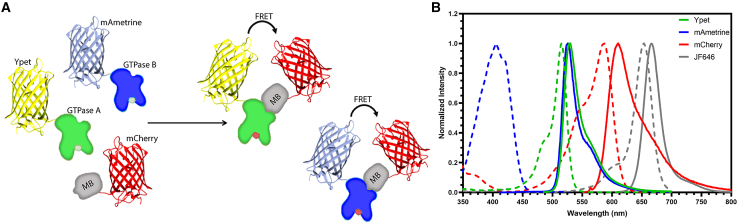
We describe here a method that ameliorates these problems by using a single “affinity reagent” to bind the activated conformation of multiple target proteins. Affinity reagents (ARs) are small proteins that bind selectively to a specific target protein conformation. They have been derived from downstream molecules (4,7,8), screening of protein scaffolds (9,10), or antibody fragments (9,11). When one fluorophore is placed on the AR and the other on the target protein, binding of the AR to the activated target generates FRET. We produced an AR named MultiBinder (MB) that interacts selectively with the active conformation of three different GTPases (Fig. 1 A). By appropriate use of fluorescent proteins, we found that the MB could generate a different FRET signal during co-imaging of any 2 GTPases. Using a single AR to monitor two different activities required less of the fluorescence wavelength spectrum and reduced toxicity. The set of fluorophores utilized in this study are shown in Fig. 1 B. We simultaneously monitored two GTPase activities using MB in FRET biosensors while also monitoring a third activity using a biosensor based on a long wavelength dye.
Figure 1.
Simultaneously reporting the activation of three different GTPases. (A) MultiBinder (MB) binds to the activated conformation of two different GTPases. Binding produces FRET for each GTPase, but with different excitation spectra. (B) Fluorescence excitation (dashed) and emission spectra (solid) of the FRET donors mAmetrine (blue) and Ypet (green) and the FRET acceptor mCherry (red). The orthogonal, near-IR fluorophore JF646 is used in an intensiometric biosensor (9) that is combined with the FRET biosensors.
We targeted the Rho family of GTPases because we could build on prior biosensor designs (2,3,4,7) and because simultaneous imaging of these GTPases could shed light on feedback interactions and other aspects of GTPase signaling that require live cell visualization (12,13). Rho family GTPases are key regulators of the cytoskeleton and other cell functions (13,14,15,16). They are monomeric enzymes that are active when bound to GTP and turn themselves off through hydrolysis of GTP to GDP. Their activity is spatiotemporally controlled by over 100 different upstream regulators impacting this “GTPase cycle” (12). Computational methods have been developed to dissect signaling circuitry using GTPase biosensor data (2,3,17).
To make an AR that can bind all three of the canonical Rho family GTPases—RhoA, Rac1, and Cdc42—we combined different portions of these proteins’ downstream effectors. There are naturally occurring proteins that can bind Cdc42 and Rac1, but we know of none that can interact with RhoA and Rac1 or other specific GTPase pairs. We modified published biosensor designs for the three individual GTPases by incorporating the MB and by using fluorescence wavelengths suitable for multiplexing. This led to the successful proof-of-principle application shown in Fig. 1. Quantitative analysis of cell motility showed that the MB substantially reduced cell perturbation when imaging two GTPases in the same cell and revealed signaling relationships between GTPase activation events.
Materials and methods
Molecular cloning
Primers were purchased from Integrated DNA Technologies (IDT, Morrisville, NC), and the sequence of interest was confirmed by sequencing (Azenta/Genewiz, Morrisville, NC). Sequences of interest were amplified by PCR from other vectors and then assembled by restriction cloning (restriction enzymes from New England Biolabs, Ipswitch, MA); commercially available kits (Qiagen, Germantown, MD, and Thermo Fisher Scientific, Waltham, MA) were used for gel band purification and DNA extraction. Chimeras were purchased from IDT as ultramers, amplified, restricted, and ligated to the vectors. Single point mutations for sequences of CA GTPases were performed by assembly PCR.
Cell culture
HEK-293t, Cos7, and mouse embryonic fibroblasts (MEF), the latter with a TetR gene (G418 resistant), were cultured in DMEM media (Corning, Corning, NY) supplemented with 10% FBS (GeminiBio, Sacramento, CA) and 1 mM GlutaMAX (Corning, Corning, NY) and split every 2–3 days (70%–80% confluency) by trypsinization (Corning, Corning, NY). Cells were kept below passage 30. Cos7 cells were transiently transfected for bleed-through (BT) corrections by mixing 3 μL of Fugene6 (Promega, Madison, MA) with 100 ng of DNA for 15–30 min in OptiMEM (Gibco, Grand Island, NY). This was added dropwise.
MEF stable cell lines
Stable, tet-off MEF cell lines were generated using the piggybac system (18). MEF cells were seeded in six-well plates and allowed to reach 70%–80% confluency before transfection. 5 μL of TransIT-X2 (Mirus Bio, Madison, WI), 500 ng of transposase (in a pUC19 vector) (18), and 500 ng of the piggybac vector (containing a tet recognition sequence upstream of the gene of interest and different resistance markers downstream) were incubated for 15–30 min in 100 μL of OptiMEM and then added dropwise to the wells containing 2 mL of DMEM. The medium was removed and replaced ∼4 h after addition. Cells were passed to a T-25 flask when they reached near confluency; doxycycline was added 1 day after transfections and the first dose of the resistance antibiotic (either puromycin, hygromycin, or blasticidin, depending on the selection in the desired vector) 2 days after transfection. Antibiotic concentration was doubled every 2 days, reaching a maximum at 1 week, and maintained for ∼1.5 weeks. The concentration gradients for the different selection antibiotics were as follows: 1) hygromycin (Corning, Corning, NY): 200–600 μg/mL; 2) blasticidin (Gold Biotechnology, Olivette, MO): 0.5–5 μg/mL; and 3) puromycin (Gold Biotechnology, Olivette, MO) 0.5–5 μg/mL. Cells were then either sorted at the UNC FACS facility and/or aliquoted and frozen in 90% DMEM and 10% DMSO for 1 week at −80○C and then moved to liquid nitrogen for storage. Antibiotic resistance and plasmids used for stable cell line creation are summarized in Table S1 in the supporting material.
Biosensor characterization
HEK-293t cells were seeded in 96-well plates coated with poly-lysine (3 × 105 cells/mL) 48 h before imaging. 24 h before imaging, cells were transfected using Lipofectamine and Plus reagents (Thermo Fischer Scientific, Waltham, MA) in a 1:1:1 ratio of DNA/Lipo/plus. Cells were transfected with the different biosensor variants either alone, for constitutively active (CA), or with increasing amounts of different regulators in serum-deprived DMEM during 4 h, after which FBS was added (10% final). CA variants of the biosensors were Rac1-Q61L, RhoA-Q63L, and Cdc42-Q61L. The following guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanosine nucleotide dissociation inhibitor (GDI) were used: Dbl, DH-PH domain of Vav2 (191–573), Tiam (C1199), Tim, Asef (full-length), p50RhoGAP (full-length), RacGAP1 (full-length), and RhoGDI-1 (full-length). All samples were brought to 200 ng total DNA by adding an empty vector (pTriEx) as needed, using 50 ng of GTPase biosensor and 160 ng of the regulators. When two regulators were used together (GDI and GAP) the amount was halved for each regulator. Each assay was performed in triplicate, and each individual plate contained a mock control (empty DNA carrier), donor, and acceptor alone for BT corrections. Rac1 experiments were based on two ARs from earlier published work (2,4,8), AR PBD75 and AR PBD92, which differ in length (see Table S1). They were indistinguishable in assays of upstream regulation and of dynamics in cells. The stable cell line piggybac construct for Rac1 was made with PBD75. The plate data shown in this study was based on PBD92. On the imaging day, cells were starved for ∼2 h at 37°C in Hanks' Balanced Salt solution (HBSS, Thermo Fischer Scientific, Waltham, MA) media containing 1% FBS and 1 mM HEPES (Gibco, Grand Island, NY), and 30 min before imaging, plates were allowed to reach thermal equilibrium at room temperature for ∼45 min. Each well was imaged at four different positions using an ORCA-Flash4.0 V2+ sCMOS camera (Hamamatsu Photonics, Bridgewater, NJ), an inverted IX81 Olympus epifluorescence microscope, Metamorph acquisition software (Molecular Devices, Silicon Valley, CA), mercury arc lamp illumination, and an Olympus UPLFLN U Plan Fluorite 10x objective. For all biosensors, a 445/505/580 ET series (Chroma Technology Corporation, Bellows Falls, VT) dichroic filter was used, with the following excitation and emission bandpass filters: 1) ET436/20X (Chroma Technology, Bellows Falls, VT) and ET535/30M (Chroma Technology, Bellows Falls, VT) for mAmetrine; 2) ET500/20X (Chroma Technology, Bellows Falls, VT) and ET535/30M for Ypet, 3) FF575/15 (Semrock, Rochester, NY) and 605/15 (Semrock, Rochester, NY) for mScarlet and BP-585/35 (Carl Zeiss, White Plains, NJ); and 4) FF01-647/57 (Semrock, Rochester, NY) for mCherry and TagRFP-T. Images were analyzed using MATLAB (The MathWorks, Natick, MA). The intensity from each field of view was summed for each channel and background subtracted using values from mock-transfected wells; corrected FRET values were determined by subtracting the individual, weighted contributions of donor and acceptor channels calculated from wells transfected with only donor or acceptor respectively; FRET/donor and FRET efficiency was then calculated for each position and averaged from at least three independent measurements.
Single-cell biosensor imaging
Biosensor expression was induced 48 h before imaging by washing the cells, trypsinizing, seeding in a T-25 flask, and re-washing ∼1 h after. On the imaging day, cells were maintained in DMEM and ∼2 h before imaging seeded on coverslips that had been coated 24 h before with fibronectin (Sigma-Aldrich, St. Louis, MO). Cells were imaged in F-12 Ham’s media (Caisson Labs, Smithfield, UT) supplemented with 5% FBS and 1 mM HEPES. For dual and triple biosensor imaging, media was degassed with Argon before adding FBS, HEPES, 100 μm Trolox (Sigma-Aldrich, St. Louis, MO), 0.5 mM Ascorbate (Sigma-Aldrich, St. Louis, MO), and Oxyfluor (Sigma-Aldrich, St. Louis, MO) at 1:100 dilution. Open chambers were used for single biosensor imaging and closed chambers for dual and triple biosensor imaging. Biosensor imaging of Ypet-mScarlet variants was as previously described (3,19). All remaining imaging was performed using a different microscope (3), described below. Briefly, cells were imaged using two ORCA-Flash4.0 sCMOS cameras (Hamamatsu Photonics, Bridgewater, NJ), an inverted IX81 Olympus epifluorescence microscope, mercury arc lamp illumination, and a 40x 1.3NA Silicon oil objective (UPLSAPO40XS, Olympus, Center Valley, PA). For single and dual biosensor imaging, we used a two-camera adapter (TuCam, Andor Technology, South Windsor, CT) separating donor, FRET, and acceptor emissions; for triple biosensor imaging, we used a W-VIEW GEMINI (Hamamatsu Photonics, Bridgewater, NJ) image splitter between the two-camera adapter and each camera. The following excitation and emission filter combinations were used for single and dual biosensor imaging: 1) FF01-514/3 (Semrock, Rochester, NY) and FF01-535/22 (Semrock, Rochester, NY) for Ypet; 2) FF01-405/10 (Semrock, Rochester, NY) or FF01-409/32 (Semrock, Rochester, NY) and FF01-535/22 for mAmetrine; and 3) FF01-586/15 (Semrock, Rochester, NY) or FF01-561/14 (Semrock, Rochester, NY) and FF01-617/73 for mScarlet and mCherry. FF410/504/582/669 (Semrock, Rochester, NY) or ZT442/514/561m (Chroma Technology, Bellows Falls, VT) and FF580-FDi01 (Semrock, Rochester, NY) beam splitters were used at the microscope and two-camera adapter respectively. For triple biosensor imaging, the following excitation and emission filter combinations were used: 1) FF01-405/10 and FF01-514/3, for mAmetrine and Ypet respectively, and FF01-535/22; 2) FF01-561/14 and FF01-617/73 for mCherry; and 3) FF01-640/15 (Semrock, Rochester, NY) and FF01-685/40 for JF646. mAmetrine, Ypet, and mCherry were excited using a ZT442/514/561m dichroic at the microscope and imaged in a single camera using an FF580-FDi01 in the W-VIEW GEMINI to separate donor from FRET/acceptor channels; JF646 was excited using ZT445/514/561/640 (Chroma Technology, Bellows Falls, VT) and imaged in half the chip of a single camera. An FF01-640-FDi01 (Semrock, Rochester, NY) was placed in the two-camera adapter to separate the emission of the dual-chain biosensors with UBD from the ratiometric Cdc42 sensor. In some cases when imaging Ypet-mScarlet variants, we used a filter wheel coupled to the long wavelength emission path and a 1.2x lens on the short wavelength path to account for the different light paths between both cameras.
Biosensor imaging analysis and BT corrections
Single-cell biosensor imaging data were processed as previously described (2,3) with small modifications. Briefly, dark current for each image was subtracted from all channels; donors (mAmetrine and Ypet) were aligned to FRET/acceptor channels using a registration file created from images of a 2-μm fluorescent bead slide obtained before and after the experiments. All channels were then background subtracted. Donor channels were used to generate a mask applied to all remaining channels. BT corrections were then applied to the FRET channel. Donor, corrected FRET, and acceptor were photobleach corrected. BT contributions from donor and acceptor fluorescence to the FRET channel were determined from images of Cos7 cells transiently transfected with either donor (mAmetrine or Ypet) or acceptor (mCherry or mScarlet), imaged using the same microscope optics and illumination conditions used for the real samples. Background subtraction was applied to all channels to correct for different background values at different excitation wavelengths. For imaging of a single, dual-chain biosensor, corrected FRET was calculated as follows: , where α and β are respectively donor and acceptor individual emissions in the FRET channel divided by their emission in the appropriate channel. For imaging of two dual-chain biosensors utilizing a single AR, FRET between Ypet (subscript 1) and mCherry was BT corrected following the same formula as above; for FRET between mAmetrine (subscript 2) and mCherry, mAmetrine, emission was corrected before photobleaching correction as follows: ; γ values ranged from 0.02 to 0.001 depending on relative exposure times, neutral density, and bandpass filters utilized. Contributions to the FRET channel from Ypet emission upon excitation with mAmetrine wavelengths were negligible under our imaging conditions and therefore not considered for BT correction. Addition of the JF646 channel for triple biosensor imaging did not require additional BT corrections. Although the SNAPsense biosensor allowed orthogonal dye labeling with the HALO-Tag (e.g., with JF722 (20)), we chose to bypass this secondary label to reduce perturbation, utilizing instead the donor (Ypet) signal from RhoA for volume normalization.
Cross correlation of edge velocity with biosensor activity
The pipelines for this analysis were published previously (17). In short, cross correlation analysis shows the strength of coupling between motion and biosensor activity represented by a peak magnitude at a time lag, which indicates by how much signal fluctuations precede (negative lags) or succeed (positive lag) edge motion fluctuations. Cross correlation curves were measured for individual probing windows and then averaged within each cell. The curves for multiple cells allowed us to compute a standard confidence interval (±2× SEM) about the mean correlation curve reflecting the level of cell-to-cell variation across multiple rounds of experiments.
Hidden Markov modeling of edge velocity
To identify clean states of cell edge protrusion and retraction in the edge velocity maps, hidden Markov modeling (HMM) was applied using the R package “depmixS4” (21). The modeling breaks the time series of each local cell edge sector into distinct motion states determined by the average and variance of the data within each state. The seed was initialized before model fitting by the function set.seed() for consistent output. For consistency of state selection across multiple cells, the velocity time series data were normalized to zero mean and standard deviation of 1 and then concatenated into a single long vector with missing values removed. The single vector was saved as a depmix object and computed using the fit() function without setting initial estimates state transition probabilities. The model fitting was iteratively updated based on the state selections of the previously fitted model until there was no improvement in model likelihood. Our predetermined number of states was eight, consistent with previous work published in Welf et al. (22). The states were then ordered by the average speed, from positive to negative. This information was used to determine protrusion/retraction events in the analysis detailed below.
Definition of protrusion and retraction events using the states
For consistent definition of protrusion/retraction events, we utilized the HMM states derived from the pooled velocity data described above. Among the eight states, four showed positive average velocity values, and four showed negative average velocity values. The positive velocity states were pooled in phases of protrusion and stored in an event database describing each event by a cell index, window index, average protrusion speed, protrusion duration, standard deviation of speed, and maximum speed. These variables served further filtering of valid events: we considered a protrusion event as valid if it had a minimal duration of 30 s and average speed above 2 nm/s. The duration requirement of 30 s was equivalent to the requirement that the HHMI classified three or more consecutive frames in one of the four positive velocity states. Moreover, 30 s approximately matches the duration of half of the average main lobe of the correlation between biosensor activity and edge movement (see, for example, Fig. 2 J–M), which is equivalent to half the duration of an average protrusion-retraction cycle. A 2 nm/s average speed over 30 s is equivalent to a displacement of ½ pixel or more over a protrusion event. Any displacement less than ½ pixel cannot be reliably detected by the cell contour tracker. Retraction events were selected analogously as those with less than –2 nm/s and a duration over 30 s. We extracted three features for every cell: average duration, average speed, and average frequency of valid protrusion/retraction events. The frequency was calculated as the number of protrusions for the whole observation period divided by window number and observation time for proper comparison. These features were then grouped by experimental condition and compared using t-tests for statistical significance.
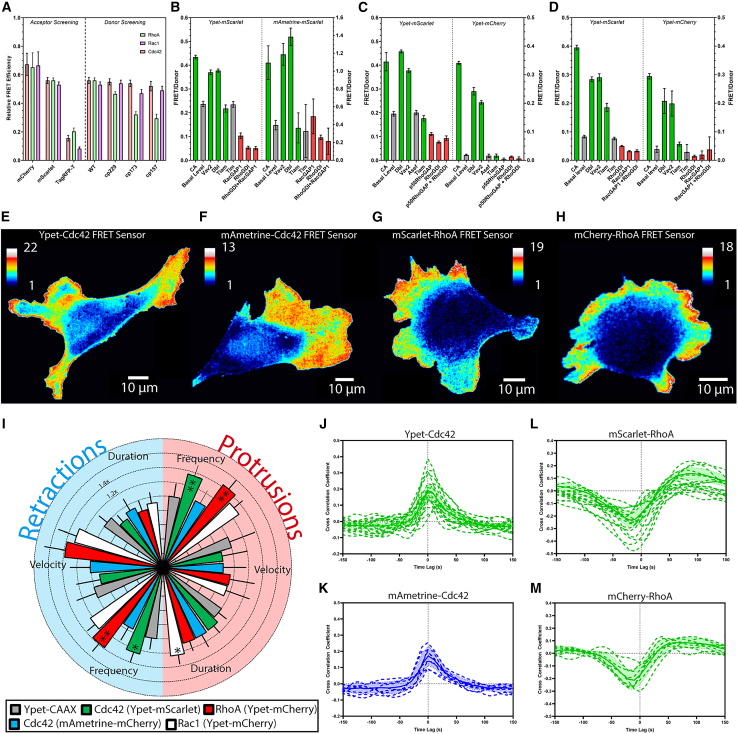
Figure 2.
Development and testing of red-shifted GTPase biosensors. (A) Donor and acceptor optimization for dual-chain FRET GTPase biosensors (n ≥ 3). FRET efficiency of the Cdc42 (B), RhoA (C), and Rac1 (D) biosensors alone (Basal Level, gray) or when exposed to upstream activators (green), inhibitors (red), upstream regulators not specific for the tested GTPase (gray), and activating point mutations (green, CA) (error bars SD, n ≥ 3). Activity maps of Cdc42 (E and F) and RhoA (G and H) biosensors in randomly moving MEFs. Pseudocolor scales indicate ratio values relative to the lowest values (materials and methods). (I) Radial plot summarizing parameters of protrusion (light red, right side) and retraction (light blue, left side) from MEFs expressing biosensors, normalized to Ypet-CAAX (negative control) (error bars 95% CI). Data statistically different from the negative control are marked as ∗ (p-value < 0.05) and ∗∗ (p-value < 0.01); see additional information and alternate representation in Fig. S1 in the supporting material. (J–M) Cross correlation of biosensor activity and edge velocity, showing the lag between changes in protrusion velocity and GTPase activation, for Cdc42 (J and K) and RhoA (L and M) (n ≥ 7, error bands 95% CI).
Analysis of activation hierarchy between GTPases
The capacity of the MB for imaging two signaling activities at a time allowed us to assess the directional influence of one GTPase (referred to as the effector) on another GTPase (referred to as the target). We quantified the strength of this influence by the likelihood for the target signal to increase after a transient interaction with its putative effector (Text S1 in the supporting material). This likelihood had to be controlled for the likelihood of spontaneous target increase with only weak or no effector interaction. Therefore, we generated two groups of interaction events: Group 1 containing strong interaction events and Group 2 containing weak interaction events. As detailed below, this event selection was performed based on the local GTPase time series recorded for each probing window in a particular layer. The analysis was applied separately to probing windows in Layer 1 and Layer 2. Collecting the events in each group, we computed the success/fail odds of target signal increase and determined the odds ratio that Group 1 contains more successes than Group 2. This would indicate that strong effector/target signal interactions systematically elevated the target signal. Statistical significance was derived by application of a natural log transformation of the odds ratio setting under the null hypothesis of no difference between the Group 1 and Group 2 success/fail odds, i.e., a natural log of zero. This was then followed by a Student's t-test for comparison of the log-transformed odds ratios of individual cells.
Selection of effector/target signal interaction events by hidden Markov modeling
We considered time periods with highly correlated fluctuations between two GTPases in the same probing window as periods where the two signals may be functionally interacting. To identify periods of local correlation between two GTPases, we used an HMM to determine interaction states defined by a pairing of intercept and slope in linear regression model between effector GTPase activity A and target GTPase activity B. The predetermined number of interaction states was six and was ordered by the average target signal activity level from high to low. For the definition of an interaction event, we utilized the HMM states 3 and 4 with a target signal close to zero in Z-scores. With this choice, interaction events could yield either increased target signal activations or the target signal would stay similar or drop compared with the level before the event. The correlation values vary depending on the duration of the interaction event. Short interaction events with high correlation can be a measurement error, and extremely long interaction events with low correlation can still show statistical significance. We tested the statistical significance of the correlation value by the Student's t-test and applied false discovery rate control for the p-values measured within each cell (using the function “qvalue” within R) (23). After elimination of insignificant and falsely discovered events, we formed Group 1 of strong effector/target signal interactions as events with correlation above 0.4. Group 2 encompassed events with a correlation level in the interval [–0.4, 0.4]. The threshold of 0.4 was chosen such that Group 1 and 2 had approximately the same number of events, allowing for balanced computation of the odds ratio. See Text S1 for additional information.
Results and discussion
We first generated red-shifted versions of our previously published FRET biosensors for Cdc42, RhoA, and Rac1 (3). The domains CBD (from Wiscott-Aldrich syndrome protein), RBD (from Rhotekin), and PBD (from Pak1) were used for Cdc42, RhoA, and Rac1, respectively (4,7,8). The new biosensors were optimized by testing different FRET donors and acceptors and circular permutants of the donor (Fig. 2 A).
We tested for specificity and response to upstream activators using previously published assays (Fig. 2 B–D) (3,24). The new biosensors were based on FRET from Ypet to mCherry (RhoA and Rac1), mAmetrine to mScarlet (Cdc42), and Ypet to mScarlet (Cdc42, RhoA, and Rac1). To examine the biosensors’ response in living cells, we generated MEFs stably expressing biosensors under the control of a dox-inducible piggybac vector (18) (Fig. 2 E–H). Cell-to-cell variation in donor/acceptor ratio was reduced by expressing the two chains of each biosensor as one gene, with two ribosomal skip sequences between them (p2a and t2a (25)), enabling ∼100% chain separation during translation.
We examined cell toxicity by comparing the frequency, duration, and velocity of protrusions and retractions in cells expressing biosensor versus the membrane marker YFP-CAAX (used for edge segmentation and as a control). Only five of 30 comparisons showed statistically significant differences (Figs. 2I and S1 in the supporting material). Cells were most sensitive to the expression of the RhoA biosensor, consistent with prior studies (26). Further analysis of RhoA sensors showed that perturbation and peak correlation were independent of biosensor expression in the range we used (Fig. S2 in the supporting material). This indicated that the biosensors could be used at concentrations sufficient to image activity without producing significant changes in cell behavior (Fig. S2).
To establish a baseline for studies of perturbation during multiplexing, we examined the correlation between edge velocity and local biosensor activity (Figs. 2 J–M and S3 in the supporting material). These correlations have been used extensively to study protrusion signaling (2,3) and have served as a measure of toxicity (3). The new versions of the RhoA and Cdc42 biosensors produced correlations like those previously seen in MDA-MB-231 cells (3) (Figs. 2 J–M, and S4 in the supporting material; Videos S1, S2, S3, and S4 in the supporting material). In the current study, Cdc42 activity was strongly correlated with edge velocity, with no time lag (Fig. S3 A–D; Videos S1 and S2). In earlier MEF studies (2), Cdc42 was maximally active ∼50 s after protrusion; however, qualitatively, both data sets are in agreement that Cdc42 activity increases in concert with cell edge advancement. For RhoA, biosensors showed a tighter lobe of negative correlation with edge motion at a time lag of −25 s, followed by a much wider positive lobe peaking at lag of ∼100 s (Fig. S3 E–H; Videos S3 and S4). As discussed before in the context of MDA-MB-231 cells (3), these correlation behaviors suggest that RhoA activity increases primarily during cell edge retraction. This deviates from earlier MEF studies (2), where RhoA was positively correlated with edge motion at ∼0 s time lag (2), a behavior that led to the suggestion that RhoA signals increased in concert with cell edge advancement. We noted that compared with these earlier studies, the peak edge velocity values had increased by a factor of about three (current study, ≥50 nm/s, earlier ≤15 nm/s) (Fig. S5 in the supporting material). There are a number of experimental factors, including cell culture media, substrate coating, and passage number, that may lead to such shifts. However, in an analysis of local velocity dynamics and correlation peak value, we found that cell edge sectors from the new data that had velocity variations comparable to the slower moving cells analyzed in Machacek et al. (2) systematically displayed a positive correlation between RhoA activity and edge motion (Fig. S5 E). On average, the time lag was shifted to −20 s. That is, in these sectors, increased RhoA activity tended to lead edge advancement (Fig. S5 F), whereas in the majority of faster moving sectors, increased RhoA signaling led retraction. Thus, MEFs in this new data set display differential RhoA signaling regimes that relate to distinct levels of cell edge dynamics.
With the red-shifted biosensors and their correlation behavior in hand, we set out to produce an AR that could bind the activated conformation of Rac1, RhoA, and Cdc42. This was achieved by engineering an optimized chimera of the PBD AR (which binds Cdc42 and Rac1) and the RBD AR (which binds RhoA) (Fig. 3 A). PBD, based on p21-activated kinase (PAK), has some β-pleated sheet character but is mostly unstructured, whereas RBD, based on Rhotekin, is a coiled-coil domain of interacting alpha helices (Fig. 3). PBD/RBD chimeras were generated in three sequential optimization rounds, with binding affinity measured indirectly via FRET efficiency (Fig. 3 D; Table S2 in the supporting material; Text S2 in the supporting material). The binding of different PBD/RBD chimeras (Table S2) to each GTPase was assessed using a high-content screening assay (24) (Fig. 3 D). This confirmed that the highly conserved CRIB domain in PBD (“ISLPSDFEHTIHVG") (32) was essential for Cdc42 and Rac1 binding, and it revealed a previously unidentified “RQMALSL” domain for RhoA binding by RBD. The final, optimized MB AR (C3.5) was a 136-a.a. peptide composed of a 53-a.a. minimal binding motif for Rac1 and Cdc42 and an 83-a.a. binding domain for RhoA. The binding affinities of the MB were 80%–90% those of the proven, previously published ARs for Rac1, RhoA, and Cdc42.
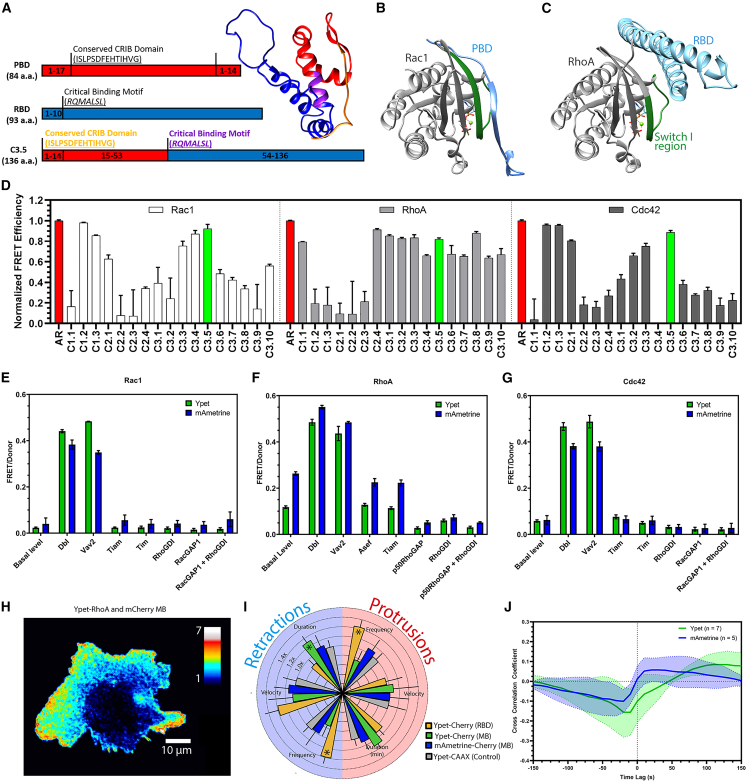
Figure 3.
Development and testing of the MultiBinder. (A) Schematic representations of PBD (red), RBD (blue), and their combination to make MB (C3.5), highlighting essential binding domains (purple and orange); MB model obtained using I-Tasser (25). (B and C) Ribbon representations of crystal structures relevant to our affinity reagents (27,28,29). (B) Rac3 with PBD from Pak1 (PDB: 2QME) (30) and (C) RhoA with RBD from protein kinase N (PDB:1CXZ) (31). ARs (PBD and RBD) are blue, GTPases gray, and the Switch I region (GTPase motif that binds the ARs) is green. (D) PBD-RBD chimeras were screened by comparing FRET efficiencies, normalized to the AR of the published biosensors (red) (see Fig. S1 and Table S2 in the supporting material). The final chimera C3.5, MB, is colored green (error bars SD, n ≥ 3). (E–G) FRET response of Ypet- (green) or mAmetrine-mCherry (blue) biosensors to upstream regulators (error bars SD, n ≥ 3). (H) RhoA activity map in a randomly moving MEF, monitored using Ypet-mCherry RhoA and the MB. (I) Radial plot comparing parameters of protrusion (light red, right side) and retraction (light blue, left side) normalized to the negative control Ypet-CAAX for cells expressing RhoA biosensor variants (error bars 95% CI, Fig. S5C–H). (J) The cross correlation between RhoA activity and local edge velocity, determined using MB-mCherry and measured in small probing windows 1.3 μm wide and 1.3 μm deep. This shows the time lag between GTPase activation and changes in protrusion velocity (Ypet: green, n = 5; mAmetrine: blue, n = 5; error bands 95% CI).
MB-based biosensors showed the same response to upstream regulators as those based on the original AR, using Ypet or mAmetrine FRET donors (Fig. 3 E–G). Using RhoA with MB (Figs. 3 H and S6 A in the supporting material), we checked cell perturbation by MB (Figs. 3 I and S6 B–G; Video S5 in the supporting material). As for the biosensors above, cells showed similar or lower perturbation than with the original RBD-based RhoA biosensor (Fig. 3 I). Correlations qualitatively agreed with prior measurements (Figs. 3 J and S6 H). Together, these data showed that MB could replace previously published, successful ARs.
Finally, we examined the ability of MB to carry out the multiplexing experiment shown in Fig. 1 A (Text S1). To test if MB could be used to simultaneously image the conformational state of two GTPases, we stably expressed mAmetrine-labeled Cdc42 or Rac1 in MEF cells already stably expressing Ypet-RhoA and mCherry-MB. We chose to use a single gene copy of the MB in anticipation of the final study using three biosensors. This produced weaker correlation values, but GTPase activation dynamics and their correlation with edge dynamics were like those obtained earlier with published ARs (Fig. 4 A–C, Fig. S7 in the supporting material and Video S6 in the supporting material). We were able to image all three canonical GTPases in the same cell using MB for RhoA and Rac1 and combining these with a Cdc42 biosensor based on an environment-sensing dye (33). For the dye-based biosensor, a SNAP tag was attached to Cdc42, and this was covalently labeled within the cell with the dye JF646, whose fluorescence responded to Cdc42 conformational changes.
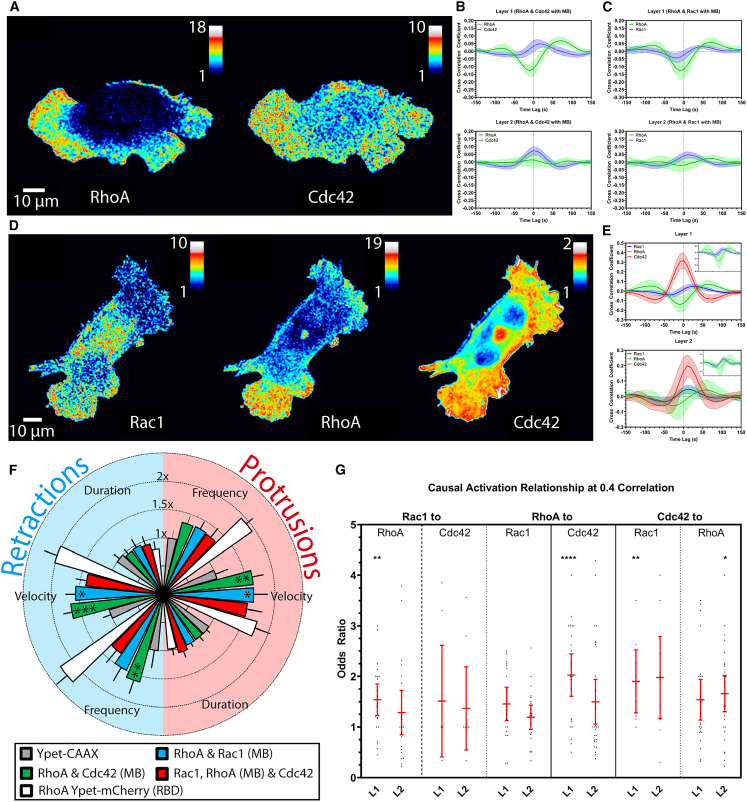
Figure 4.
Application of MultiBinder to imaging and analyzing multiplex signals in the same cell. (A and B) RhoA and Cdc42 activity maps and cross correlations plots for randomly migrating MEFs, obtained using Ypet-RhoA, mAmetrine-Cdc42, and MB-Cherry (n = 17). (C) Cross correlation plots for simultaneously imaged RhoA and Rac1 activities (n = 13). (D and E) Rac1, RhoA, and Cdc42 activity mapped in the same MEF using Ypet-RhoA, mAmetrine-Rac1, MB-Cherry, and the SNAPsense biosensor (n = 8). Cross correlation analyses in (B), (C), and (E) were performed in two layers of probing windows of 1.3 μm width. Layer 1 covered 0–1.3 μm from the cell edge, and Layer 2 covered 1.3–2.6 μm from the edge. (F) Radial plot of protrusion (light red, right side) and retraction (light blue, left side) parameters normalized to the negative control Ypet-CAAX (n = 11) for dual and triple imaging with Ypet-RhoA mCherry-RBD as a positive control (statistical comparisons shown to negative control; see Fig. S8 for full analysis). Error bands 95% CI for (B), (C), and (E). (G) Odds ratio values of GTPase activation events at 0.4 correlation threshold from combined dual and triple imaging datasets (error bars SD).
Simultaneous imaging of Rac1, RhoA and Cdc42 produced activation patterns and cross correlation like those observed earlier using individual biosensors (Figs. 4 D and E and S8; Video S7 in the supporting material). To assess toxic effects, we compared various combinations of two biosensors, and the three biosensors together, with the Rho biosensor shown earlier to cause the greatest perturbation increase. Assessment of protrusion frequency, velocity, and duration indicated that the combination of two or three biosensors was less perturbing than the single RhoA biosensor (Figs. 4 F and S9).
We exploited the unique multiplexing capability of MB to test the signaling hierarchy of the GTPases, examining their mutual regulation. For this, we implemented a statistical paradigm that examines whether two signals are in an effector/target relation; i.e., the activation of one signal systematically shifts the activation of another signal. In practical terms, we classify two signals as functionally connected if the odds for the target to increase in activation upon an interaction with a putative effector is significantly higher than the odds for no change or a decrease. We considered interaction events between the effector and target as periods during which the readout of two GTPase activities locally correlated above a threshold of 0.4. As expected, the majority of GTPase pairings showed no significant odds for a positive change; i.e., there was no indication that GTPases activated one another (Fig. 4 G). The exception to this was an activation hierarchy that occurred next to the cell edge (Layer 1), in which RhoA affected Cdc42 and Cdc42 affected Rac1. This is consistent with the previously published activation kinetics of these proteins (2). The hierarchy was diminished at distances >1.4 μm from the cell edge (Layer 2), indicating that these signaling interactions are narrowly confined.
There are no known direct interactions between GTPases. The uncovered transduction hierarchy is likely caused by a cascade of signaling intermediaries. MB can provide a valuable tool to identify such intermediaries using knockdown screens. The ability to visualize and correlate the activation of multiple GTPase activities will be relevant to a host of cell behaviors.
Author contributions
F.M.P. initiated this study. Experimental work was carried out by F.M.P., B.G., and D.A. Computational analysis was carried out by J.H. D.M. and N.P. provided input on imaging experiments and use of SNAPsense, respectively. The study was directed by K.H. and G.D. The manuscript was written by F.P. and K.H., with extensive input from J.H. and G.D.
Acknowledgments
The authors thank Luke Lavis (HHMI Janelia Research Campus), for providing Janelia Fluor 646 dye, and Mauro Calabrese (University of North Carolina at Chapel Hill) for the piggybac vectors. This work was funded by the National Institute of General Medical Sciences of the National Institutes of Health (N.K.P., F32GM120958; G. D. R01CA252826; K.M.H. R35-GM122596). J.H. and G.D. thank the Human Frontiers in Science Program (LT000911/2018-C) for financial support.
Declaration of interests
The authors declare no competing interests.
Editor: Les Loew.
Footnotes
Frederico M. Pimenta and Jaewon Huh contributed equally to this work
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.04.020.
Supporting material
References
- 1.Greenwald E.C., Mehta S., Zhang J. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 2018;118:11707–11794. doi: 10.1021/acs.chemrev.8b00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machacek M., Hodgson L., et al. Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marston D.J., Vilela M., et al. Hahn K.M. Multiplexed GTPase and GEF biosensor imaging enables network connectivity analysis. Nat. Chem. Biol. 2020;16:826–833. doi: 10.1038/s41589-020-0542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pertz O., Hodgson L., et al. Hahn K.M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 5.Lin W., Mehta S., Zhang J. Genetically encoded fluorescent biosensors illuminate kinase signaling in cancer. J. Biol. Chem. 2019;294:14814–14822. doi: 10.1074/jbc.REV119.006177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris M.C. Fluorescent biosensors - probing protein kinase function in cancer and drug discovery. Biochim. Biophys. Acta. 2013;1834:1387–1395. doi: 10.1016/j.bbapap.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Hodgson L., Pertz O., Hahn K.M. Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol. 2008;85:63–81. doi: 10.1016/S0091-679X(08)85004-2. [DOI] [PubMed] [Google Scholar]
- 8.Kraynov V.S., Chamberlain C., et al. Hahn K.M. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 9.Gulyani A., Vitriol E., et al. Hahn K.M. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat. Chem. Biol. 2011;7:437–444. doi: 10.1038/nchembio.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kummer L., Hsu C.-W., et al. Plückthun A. Knowledge-based design of a biosensor to quantify localized ERK activation in living cells. Chem. Biol. 2013;20:847–856. doi: 10.1016/j.chembiol.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szent-Gyorgyi C., Schmidt B.F., et al. Waggoner A. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat. Biotechnol. 2008;26:235–240. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- 12.Cherfils J., Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 13.Hanna S., El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cell. Signal. 2013;25:1955–1961. doi: 10.1016/j.cellsig.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Abreu-Blanco M.T., Verboon J.M., Parkhurst S.M. Coordination of Rho family GTPase activities to orchestrate cytoskeleton responses during cell wound repair. Curr. Biol. 2014;24:144–155. doi: 10.1016/j.cub.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chircop M. Rho GTPases as regulators of mitosis and cytokinesis in mammalian cells. Small GTPases. 2014;5 doi: 10.4161/sgtp.29770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffe A.B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 17.Azoitei M.L., Noh J., et al. Danuser G. Spatiotemporal dynamics of GEF-H1 activation controlled by microtubule- and Src-mediated pathways. J. Cell Biol. 2019;218:3077–3097. doi: 10.1083/jcb.201812073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schertzer M.D., Thulson E., et al. Calabrese J.M. A piggyBac-based toolkit for inducible genome editing in mammalian cells. RNA. 2019;25:1047–1058. doi: 10.1261/rna.068932.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B., Hobson C.M., et al. Superfine R. VIEW-MOD: a versatile illumination engine with a modular optical design for fluorescence microscopy. Opt Express. 2019;27(14):19950–19972. doi: 10.1364/OE.27.019950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm J.B., Tkachuk A.N., et al. Lavis L.D. A general method to optimize and functionalize red-shifted rhodamine dyes. Nat. Methods. 2020;17:815–821. doi: 10.1038/s41592-020-0909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visser I., Speekenbrink M. depmixS4: an R package for hidden markov models. J. Stat. Softw. 2010;36:1–21. doi: 10.18637/jss.v036.i07. [DOI] [Google Scholar]
- 22.Welf E.S., Miles C.E., et al. Danuser G. Actin-membrane release initiates cell protrusions. Dev. Cell. 2020;55:723–736.e8. doi: 10.1016/j.devcel.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storey J.D. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 24.Slattery S.D., Hahn K.M. A high-content assay for biosensor validation and for examining stimuli that affect biosensor activity. Curr. Protoc. Cell Biol. 2014;65:14.15.1–31. doi: 10.1002/0471143030.cb1415s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J.H., Lee S.R., et al. Choi S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tkach V., Bock E., Berezin V. The role of RhoA in the regulation of cell morphology and motility. Cell Motil Cytoskeleton. 2005;61:21–33. doi: 10.1002/cm.20062. [DOI] [PubMed] [Google Scholar]
- 27.Bishop A.L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348(Pt 2):241–255. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1221060/pdf/10816416.pdf [PMC free article] [PubMed] [Google Scholar]
- 28.Dvorsky R., Blumenstein L., et al. Ahmadian M.R. Structural insights into the interaction of ROCKI with the Switch regions of RhoA. J. Biol. Chem. 2004;279:7098–7104. doi: 10.1074/jbc.M311911200. [DOI] [PubMed] [Google Scholar]
- 29.Reid T., Furuyashiki T., et al. Narumiya S. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J. Biol. Chem. 1996;271:13556–13560. doi: 10.1074/jbc.271.23.13556. [DOI] [PubMed] [Google Scholar]
- 30.Ugochukwu E., Yang X., et al. Doyle D. RCSB Protein Data Bank; 2007. Crystal Structure of Human RAC3 in Complex with CRIB Domain of Human P21-Activated Kinase 1 (PAK1). 12/18/2019. [DOI] [Google Scholar]
- 31.Maesaki R., Ihara K., Hakoshima T., et al. The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1. Mol. Cell. 1999;4:793–803. doi: 10.1016/S1097-2765(00)80389-5. [DOI] [PubMed] [Google Scholar]
- 32.Burbelo P.D., Drechsel D., Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J. Biol. Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 33.Pinkin N.K., Liu B., Hahn K.M., et al. Using SNAP-tag for facile construction of dye-based biosensors in living cells. bioRxiv. 2020 doi: 10.1101/2020.07.16.206748. Preprint at. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.