Abstract
Directed transport of cellular components is often dependent on the processive movements of cytoskeletal motors. Myosin 2 motors predominantly engage actin filaments of opposing orientation to drive contractile events and are therefore not traditionally viewed as processive. However, recent in vitro experiments with purified nonmuscle myosin 2 (NM2) demonstrated myosin 2 filaments could move processively. Here, we establish processivity as a cellular property of NM2. Processive runs in central nervous system-derived CAD cells are most apparent on bundled actin in protrusions that terminate at the leading edge. We find that processive velocities in vivo are consistent with in vitro measurements. NM2 makes these processive runs in its filamentous form against lamellipodia retrograde flow, though anterograde movement can still occur in the absence of actin dynamics. Comparing the processivity of NM2 isoforms, we find that NM2A moves slightly faster than NM2B. Finally, we demonstrate that this is not a cell-specific property, as we observe processive-like movements of NM2 in the lamella and subnuclear stress fibers of fibroblasts. Collectively, these observations further broaden NM2 functionality and the biological processes in which the already ubiquitous motor can contribute.
Significance
Nonmuscle myosin 2 (NM2) is well known for its ability to interact with actin filaments of opposing orientations to drive contractile events and is not considered a processive motor. However, NM2 should be able to move processively along actin under the right conditions. Here, we have identified cells where NM2 readily undergoes processive movements. We thoroughly characterize these events, describe the actin conditions that are necessary for them to occur, compare processive movements between the NM2A and NM2B isoforms, and identify NM2 processive behavior in additional cell types. In addition to revealing a novel mechanism of how NM2 filaments are distributed throughout a cell, this study implies a more complex role for NM2 in regulating actin structures than previously thought.
Introduction
The ability for actin- and microtubule-based molecular motors to processively “walk” along tracks, typically carrying cargo attached to their tails, is critical throughout physiology (1,2,3,4,5). A simplified motor mechanochemical cycle includes ATP hydrolysis, track binding, release, a power stroke, track unbinding, and nucleotide exchange (6). Processive motors are often dimers whose ability to walk along a cytoskeletal filament is dependent on the coordinated mechanochemical cycle and track binding of the motor pair to prevent the motor from diffusing away (7,8). The fraction of each mechanochemical cycle a motor spends in its track-bound state, also known as its duty ratio, is thus critical for processivity. Most processive motors have a high duty ratio, spending the majority of their mechanochemical cycle in the track-bound state (9), though increasing the number of motors in an ensemble can enhance the synergistic processivity of low-duty-ratio motors (10).
Myosin 2s are low-duty-ratio motors (11,12) that are the dominant contractile motor proteins in all cells (13,14). The myosin 2 superfamily includes striated myosin 2 (cardiac and skeletal), smooth muscle myosin 2, and nonmuscle myosin 2 (NM2). Mammalian cells express up to three isoforms of nonmuscle myosin 2 (NM2A, NM2B, and NM2C) that are derived from three distinct genes (MYH9, MYH10, and MYH14, respectively) (15,16,17). All myosin 2s form hexameric “monomers” that consist of two myosin heavy chains (MHCs), two essential light chains, and two regulatory light chains (RLCs) (18). This holoenzyme is referred to as a monomer because they dynamically polymerize, or assemble, into bipolar filaments with motor domains at opposing ends. Myosin 2 filaments contain between 15 and 300 monomers depending on the isoform (19,20,21). Although NM2 monomers are generally considered nonprocessive, NM2 filaments that contain many monomers could remain track-bound for extended periods because of the high probability that at least one of its motor heads is in contact with the actin filament at any given time. Indeed, NM2A and NM2B filament processivity has been observed in vitro (22), though this required the presence of crowding agents for NM2A. Although experiments have hinted at the possibility (23), no discrete cellular observations or characterization for NM2 processivity on actin filaments, to our knowledge, has been reported.
The lack of apparent NM2 processivity in cells could be attributed to the architecture of most actin networks. If the bipolar filament is presented with actin filaments of opposing orientation, then both sets of motors can engage the respective actin filament, hydrolyze ATP, and produce force. This is the basic mechanism of contractile force generation that drives muscle contraction (24,25), cell division (26), and many more myosin 2-dependent processes. However, if a bipolar filament is presented with a single actin filament, or a parallel actin bundle, then one set of motors can efficiently engage the actin, whereas the opposing motors on the opposite end of the filament are less likely to engage. In this case, the engaging motors can dominate, and processive movements are possible. Interestingly, even the poorly engaging motors could contribute to the processive movement, as they have sufficient flexibility in the proximal tail region to engage and move actin in the “wrong” orientation (27,28). Muscle myosin 2s, especially striated, are unlikely to encounter parallel actin bundles, as the sarcomere is vitally assembled with individual actin “thin filaments” in opposing orientation to drive contraction (29). However, because NM2s are expressed in such a diverse array of cell types, they are more likely to encounter parallel actin bundles (30,31) that could support processive movements.
Here, we document the processive movement of NM2 along actin filaments in living cells. Using multiple tagging approaches, we demonstrate processive filamentous NM2 that is independent of actin dynamics but is dependent on actin architecture. We also observe subtle kinetic differences between NM2 isoforms consistent with previous in vitro experiments. Finally, we show that processive-like movements occur in multiple cell types, demonstrating that this is not a cell-specific NM2 feature.
Materials and methods
Cell culture and transfection
Cath.-a-differentiated (CAD) cells were purchased from Sigma-Aldrich and cultured in DMEM/F12 medium (Gibco) supplemented with 8% fetal calf serum, 1% L-glutamine, and 1% penicillin-streptomycin. 2–4 h before imaging, CAD cells were plated on coverslips coated overnight at 4°C with 10 μg/mL laminin (Sigma-Aldrich). DMEM/F12 medium without phenol red (Gibco) supplemented with 15 mM HEPES was used for live-cell imaging. Cell lines were also routinely tested for mycoplasma using the Universal Detection Kit (ATCC). PFN1 KO cells were generated with CRISPR-Cas9 as previously described (32). CAD cells were transfected with plasmid DNA via electroporation as previously described (32) or with LipoD293 (SignaGen, “Hard-To-Transfect Mammalian Cell” protocol). Cells expressing HaloTag constructs were incubated overnight with 10–100 nM Janelia Fluor 646 HaloTag ligand (Promega) (33). EGFP-NM2A knockin mouse embryonic fibroblasts (MEFs) were generated from mice (34) and isolated and cultured as previously described (35).
Plasmids
NM2A-Halo (pHalo-N1-NM2A) was generated by swapping HaloTag for mApple in pmApple-N1-NM2A, which was previously described (36). NM2B-Halo (pHalo-N1-NM2B) was previously described (35). EGFP-NM2A was a gift from Dr. Thomas Egelhoff and is available at Addgene (https://www.addgene.org/11347/). RLC-iRFP was generated by removing FTractin from pLV-3x-iRFP670-FTractin (gift from Dr. John A. Hammer, NHLBI/NIH, Bethesda, MD) and introducing a gBlock (IDT) containing RLC coding sequence using Gibson cloning. FTractin-mNeon-Green was a gift from M. Schell (Uniformed Services University, Bethesda, MD) (37). FTractin-mApple was previously described (36).
Antibodies
Antibodies were as follows: NMIIA (a.a.1936-1950, ECM Biosciences) rabbit polyclonal antibody used at 1:2000, NMIIB (clone A-3, SC-3769-42, Santa Cruz Biotechnology) mouse monoclonal antibody used at 1:2000, and GFP (clone B-2, SC-9996, Santa Cruz Biotechnology) mouse monoclonal antibody used at 1:2000.
Imaging
NM2 isoform analyses (immunostaining and NM2A-Halo and NM2B-Halo imaging), jasplakinolide and latrunculin (JL) experiments, and ventral lamellar EGFP-NM2A primary MEF imaging were done on a Zeiss Airyscan 880 Microscope using a Plan-Apochromat 63x/1.4 Oil DIC M27 objective and 633-nm laser. Raw data was processed using Zen software with automated processing strength. Subnuclear stress fiber imaging in EGFP-NM2A primary MEF was done using total internal reflection fluorescence structured-illumination microscopy (TIRF-SIM), as previously described (36). To decrease the background artifact inherent in SIM processing, TIRF-SIM data were blurred once with “Smooth” in Fiji/ImageJ (38).
Imaging of CAD cells expressing RLC-iRFP, EGFP-NM2A, and NM2A-Halo in PFN1 KO/OE experiments was performed using a Nikon CSU-W1 SoRa spinning disk confocal microscope using a 100× 1.49NA SR objective and a Hamamatsu Fusion BT Camera. Notably, the reduced spatial resolution and additional fluorophores in the RLC-3x-iRFP670 imaging make it more difficult to observe discrete doublets indicative of bipolar filaments. All images from these data sets were acquired in SoRa mode with the 2.8× magnifier. Movies used for NM2-Halo tracking were acquired with 2 × 2 binning to enhance signal/noise and increase acquisition speed to 5 Hz. All data from the SoRa had background noise removed using Denoise.ai (NIS-Elements, Nikon), a trained neural network that uses deep learning to estimate and remove the noise component of an image. Images acquired using 1 × 1 binning were also deconvolved in NIS-Elements using the Blind Deconvolution algorithm. Before analysis, images were exported into Fiji/ImageJ and corrected for photobleaching using the histogram matching algorithm.
Inhibitors were used at the following concentrations: Y27632 (EMD Millipore #68801), 10 μM; LatrunculinB (EMD Millipore #428020), 1.25 μM; Jasplakinolide (EMD Millipore #420127), 2 μM. Drug treatments were prepared at a 2× solution in L-15 imaging media and then added to the wells at a 1:1 dilution while imaging.
Western blotting
To quantify isoform ratios for NM2A and NM2B in CAD cells, whole-cell lysates were collected from untransfected, EGFP-NM2A expressing, and EGFP-NM2B expressing cells. These lysates were subject to polyacrylamide gel electrophoresis with 7.5% Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad). Gels were run long enough to achieve separation of EGFP-tagged NM2 from endogenous NM2 (4 h at 180 v). Three Western blots were performed for each experiment with anti-NM2A, anti-GFP, and ant-NM2B antibodies. Identical volumes of each whole-cell lysate were loaded for each Western blot. From the first anti-NM2A Western blot, we observe the ratio between endogenous NM2A to EGFP-NM2A. From the anti-GFP Western blot, we observe the ratio between the same amount of EGFP-NM2A to EGFP-NM2B. Finally, from the anti-NM2B blot, we observe the ratio between the same amount of EGFP-NM2B to endogenous NM2B. Therefore, we can compare endogenous NM2A to endogenous NM2B using EGFP-NM2A and EGFP-NM2B as intermediates, with the final endogenous NM2A/NM2B ratio obtained using the three ratios: NM2A/EGFP-NM2A:EGFP-NM2A/EGFP-NM2B:EGFP-NM2B/NM2B. Notably, this approach can be performed with any two proteins with an EGFP tag.
Quantification of myosin motion
All tracking analysis was performed in python. The code and example data can be found at https://github.com/OakesLab. Briefly, images were first filtered with a 15-pixel-square Laplacian of Gaussian filter with a standard deviation of 2, to emphasize the myosin filaments. Filaments were then tracked using the trackpy software package (https://github.com/soft-matter/trackpy) with the following relevant parameters: feature_size = 11 pixels, memory = 2, and separation = 3. As trackpy doesn’t use subpixel localization, the resulting tracks were filtered using a running window average over 2.5 s (translating to five to eight frames depending on the imaging frequency). The resulting tracks were further filtered to only consider tracks with a pathlength of at least 900 nm (e.g., the length of 3 myosin filaments) to ensure that we were only considering persistent motion. Although tracks shorter than this certainly occurred, we restricted measurements to this range to ensure we could have confidence in the measurements of the particle velocities. Due to our inability to track motion in dense regions, and the potential for filaments moving in planes outside our resolution, our tracking analysis did not capture 100% of the filaments. Therefore, whereas the number of retrograde tracks outnumbered anterograde tracks in every sample, we do not make an estimate of the percentage of processive filaments. To determine the direction of the flow, we first defined the image intensity center of mass. A vector drawn from the center of the image to the center of mass determined the mean direction of retrograde flow. The direction of the myosin movement was defined as the vector drawn from the first position in the smoothed track to the last. By calculating the dot product of these two vectors, we were able to define an angle, θ, relative to the mean direction of flow. We considered any angle to be retrograde flow, whereas any angle was considered to be anterograde flow (essentially creating a 120° cone in each direction). Tracks with angles between these two directions were not considered in our analysis, as they were predominantly moving parallel to the edge of the cell.
Mean values for both retrograde and anterograde flow rates were determined by fitting a gaussian to the histogram of the pooled data. The data are reported as the mean of the gaussian fit the standard deviation of the gaussian curve.
Quantification of NM2A-Halo anterograde events
The number of NM2A-Halo anterograde events in lamellipodia protrusions was quantified by drawing an 8 × 8 μm region of interest against the leading edge and manually counting all puncta that moved >1 μm in the anterograde direction during the duration of the movie. Three 8 × 8 μm regions were analyzed per movie and then averaged so that each movie received a single score. To quantify the density of NM2A-Halo puncta at the leading edge, all puncta within a 1-μm region of interest placed against the cell edge in lamellipodia protrusions were manually counted in frames 1, 100, and 200. The number of puncta was divided by the area that was analyzed for each region, and then all puncta density values were averaged so that each movie received a single score. All manual counting was performed by researchers who were blinded to the experimental conditions and molecules being imaged.
Statistical analysis
Biological and experimental replicates were as follows.
Fig. 1E = 10 cells from three independent repeats.
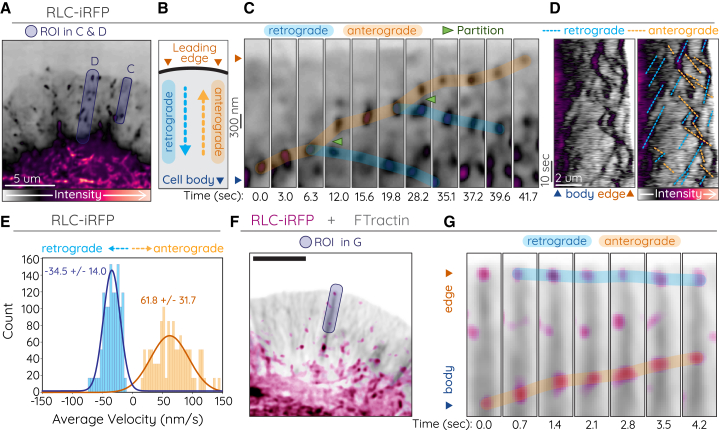
Figure 1.
Processive-like anterograde movements by RLC
(A–E) CAD cell expressing RLC-iRFP. Purple translucent regions in (A) indicate ROI for (C) and (D). (A) corresponds to t = 0 s, and (C) corresponds to 0.0–41.7 s in Video S1. (B) Cartoon depicting location of cell body and leading edge and direction of retrograde (dotted blue arrow) and anterograde (dotted orange arrow) movements. (C) Time-lapse of anterograde (orange highlight) and retrograde (blue highlight) motion of RLC puncta. Partitioning filaments are indicated by green arrows. (D) Duplicate kymographs without (left) or with (right) retrograde and anterograde movements indicated. (E) Histograms of velocity measurements from automated tracking of RLC-iRFP retrograde and anterograde movements in protrusions (see Table 1). (F and G) CAD cell expressing RLC-iRFP (magenta) and FTractin-Neon (gray). (F) corresponds to t = 13.0 s, and (G) corresponds to 13.0–17.9 s of Video S3. Blue translucent ROI in (F) indicates region used for time-lapse in (G). (G) Selected time points demonstrating retrograde (blue highlight) and anterograde (orange highlight) motion along an actin bundle. Imaging was performed on Nikon CSU-W1 SoRa spinning disk.
Fig. 3G = 17 cells from three independent repeats.
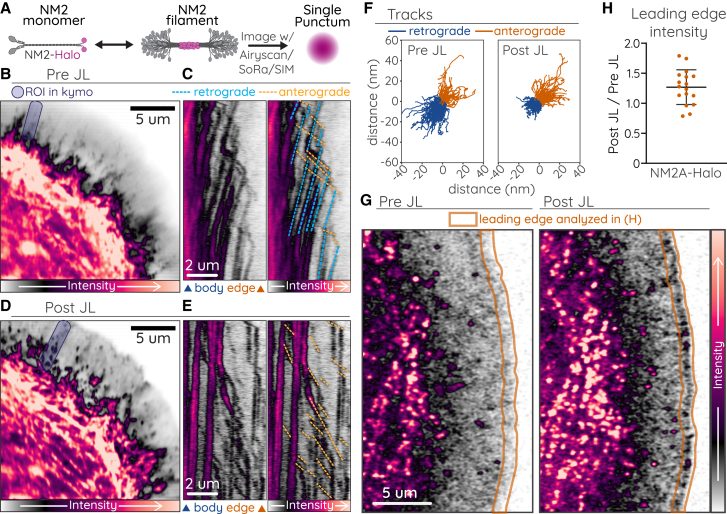
Figure 3.
NM2 anterograde movements independent of actin dynamics
(A) Cartoon of C-terminal HaloTag on an NM2 monomer, filament, and imaged with high-resolution microscopy. (B and D) CAD cell expressing NM2A-Halo imaged before (B) or after (D) treatment with jasplakinolide/latrunculin (JL) cocktail. Purple shaded boxes indicate ROIs for (C) and (E). (C and E) Kymographs shown without (left) and with (right) retrograde (blue dotted lines) and anterograde (orange dotted lines) movements indicated. (F) Rose plot of individual retrograde (blue) and anterograde (orange) tracks detected in a single cell before and after JL treatment. (G) Example lamellar regions of NM2A-Halo used for quantification of leading edge intensity (orange outline) in (H) before and 45 s after JL treatment. (H) Ratio of NM2A-Halo intensity within 1 μm of the leading edge before and 45 s after JL treatment. Imaging was performed on Zeiss 880 Aiscan.
Fig. 4G = 11 cells from three independent repeats.
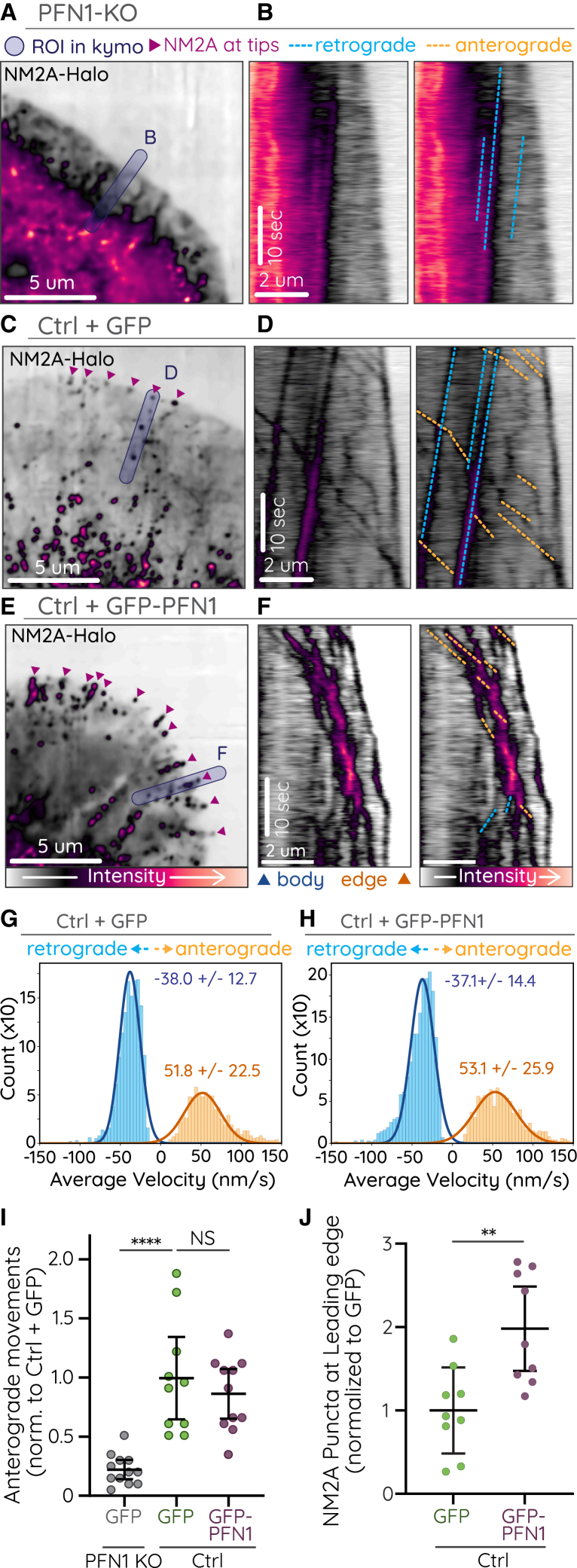
Figure 4.
Processive NM2 is dependent on lamellipodia architecture
NM2A-Halo was imaged in PFN1-KO CAD cells, CAD cells expressing GFP, or CAD cells overexpressing GFP-PFN1. (A–F) Representative images (A, C, E) and duplicate kymographs (B, D, F) with (right) and without (left) annotation of anterograde (orange dotted lines) and retrograde (blue dotted lines) NM2A movements in PFN1 KO (A and B), ctrl + GFP (C and D) and ctrl + GFP-PFN1 (E and F) cells. (G and H) Histograms of velocity measurements from automated tracking of NM2A-Halo puncta in control and PFN1 overexpressing cells (see Table 1). (I) Plot of the number of NM2A-Halo anterograde movements in the lamellipodia protrusions of PFN1 KO, control, and PFN1 overexpressing cells. Only anterograde events that initiated within 8 m of the leading edge were counted. Each dot indicates one cell. Error bars depict mean ± SD. p-values were calculated from a one-way ANOVA followed by Tukey's multiple comparisons post-hoc test. ∗∗∗∗, p <0.0001; NS, not significant. (J) Number of NM2A-Halo puncta within 1 m of the leading edge of PFN1 KO, control, and PFN1 overexpressing cells. Each dot indicates one cell. Error bars depict mean ± SD. p-values were calculated from a student's t-test. ∗∗, p < 0.01. Imaging was performed on Nikon CSU-W1 SoRa spinning disk.
Fig. 4H = 12 cells from three independent repeats.
Fig. 4I = 10+ cells for each condition from three independent repeats.
Fig. 4J = 9 cells for each condition from three independent repeats.
Fig. 5F = 21 cells from three independent repeats.
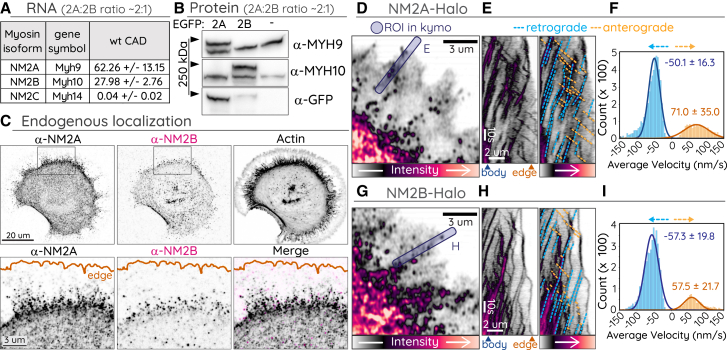
Figure 5.
Differential expression, localization, and velocity of NM2 isoforms
(A) Relative RNA expression of NM2A MHC genes. (B) Whole-cell lysates of untransfected CAD cells or cells overexpressing EGFP-NM2A or -NM2B were subject to Western blotting with the indicated antibody. The NM2A/NM2B ratio was calculated using anti-GFP blot as intermediate (see materials and methods). (C) CAD cells fixed and immunostained with indicated NM2 isoform-specific antibody and phalloidin (F-actin). Box in top row indicates ROI for insets in bottom row, which includes merge (rightmost panel). (D–I) CAD cells expressing NM2A-Halo (D–F) or NM2B-Halo (G–I). Blue boxes indicate ROI for kymographs in (E) and (H). (E and H) Kymographs shown with (right) and without (left) retrograde (blue dotted lines) and anterograde (orange dotted lines) movements indicated. (F and I) Histograms of velocity measurements from automated tracking of NM2A-Halo and NM2B-Halo retrograde and anterograde movements in protrusions (see Table S1). Imaging was performed on Zeiss 880 Airyscan.
Fig. 5I = 12 cells from two independent repeats.
To compare the number of anterograde NM2A movements in lamellipodia protrusions of PFN1 KO, control, and PFN1 overexpressing cells (Fig. 4 I), we used one-way ANOVA followed by Tukey’s multiple comparison test. To compare the number of NM2A puncta at the leading edge of control and PFN1 overexpressing cells (Fig. 4 J), we used a Student's t-test. Statistical analysis was performed using Prism (GraphPad). To compare the distributions of myosin 2A and 2B in both retrograde and anterograde flows (Fig. 5 F and I), we used a Kolmogorov-Smirnov test. Briefly, the cumulative distributions functions of the distributions were calculated and compared using the stats module from scipy. p-values less than 0.05 were considered significant. The anterograde distributions were found to be significantly different.
Results
Tagging RLC reveals NM2 processive-like anterograde movements in CAD cells
CAD cells are a central nervous system-derived cell line (39) that form broad actin-based lamellar protrusions, reminiscent of the leading edge of a neuronal growth cone. These lamellae contain both Arp-dependent actin mesh networks and Mena/VASP-dependent linear parallel actin bundles (32). To observe all NM2 localization and dynamics in these protrusions, we expressed RLC with a C-terminal 3x-iRFP670 tag (RLC-iRFP; Fig. 1 A). We observed filaments throughout the cell body, along transverse arcs, and nascent filament clusters in the protrusion (Fig. 1 A). As expected, the majority of these NM2 clusters moved retrograde toward the cell center, coupled with actin retrograde flow (Fig. 1 B–E, blue indicators; Video S1). However, unexpectedly, we also observed a subset of NM2 anterograde movements toward the leading edge (Fig. 1 B–E, orange indicators). These anterograde-moving puncta mostly originated from dense regions in the posterior protrusion, though they also occasionally partitioned off of existing NM2 filaments (Fig. 1 C, green arrows; Video S2). Anterograde puncta often reached the leading edge as in Fig. 1 C. Using automated tracking analysis, we measured the kinetics of these retrograde and anterograde movements, finding retrograde velocities of 35 nm/s in magnitude (Fig. 1 E, blue) and anterograde velocities of 60 nm/s (Fig. 1 E, orange). These values are consistent with actin retrograde movements in these cells (32,40) and are on a similar scale to in vitro processive measurements for NM2A and NM2B (120 nm/s and 40 nm/s, respectively) (22), especially when accounting for retrograde flow. To determine the underlying actin architecture on which these anterograde movements were occurring, we co-expressed RLC-iRFP with the filamentous actin reporter FTractin tagged with mNeon-Green (FTractin-Neon; Fig. 1 F–G, Videos S3 and S4). We readily observed anterograde RLC movements (magenta) on thick actin bundles (gray) that terminate at the leading edge. This architecture is consistent with parallel actin bundles assembled by the Mena/VASP family (32).
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 3 Hz, and playback is at 30 fps.
Cell was imaged at 3 Hz, and playback is at 30 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 1.5 Hz, and playback is at 30 fps.
Cell was imaged at 1.4 Hz, and playback is at 15 fps.
Tagging MHC reveals NM2 processive-like anterograde movements in CAD cells
Although these observations are consistent with processive NM2, RLCs are promiscuous and can bind other members of the myosin superfamily (41,42,43,44). We sought, therefore, to confirm that these anterograde movements were indeed filamentous NM2 movements by expressing NM2A with an N-terminal EGFP (EGFP-NM2A). Similar to the RLC, we observed filamentous structures in the cell body with nascent filaments appearing in the protrusion (Fig. 2 A). We again observed both retrograde and anterograde movements (Fig. 2 B and C; Video S5), and we observed filaments partitioning while moving anterograde (Fig. 2 B, green arrows), before eventually reaching the leading edge or stalling and moving retrograde.
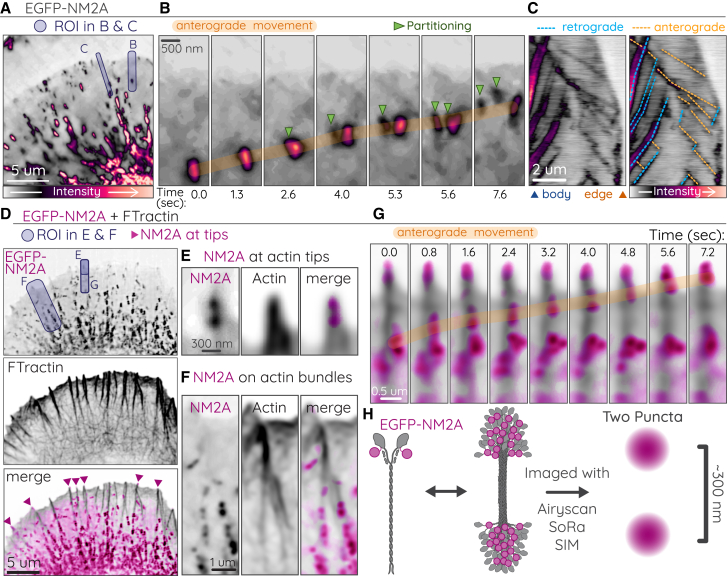
Figure 2.
Processive-like anterograde movements by filamentous NM2A
(A–C) A representative CAD cell expressing EGFP-NM2A. Purple shaded boxes indicate ROIs for (B) and (C). (A) corresponds to t = 0 s, and (B) corresponds to 1.6–9.2 s of Video S5. (B) Time-lapse of anterograde-moving NM2 filament (orange highlight) that partitions twice (green arrows). (C) Kymograph of EGFP-NM2A (left) with retrograde (blue dotted lines) and anterograde (orange dotted lines) movements indicated (right). Blue arrow and orange arrow below left kymograph indicate cell body and leading edge, respectively. (D) CAD cell expressing EGFP-NM2A (magenta) and FTractin-mScarlet (gray). Purple shaded boxes indicate ROIs for (E), (F), and (G). (D)–(F) correspond to t = 0 s, and (G) corresponds to 27.6–34.8 s in Video S6. (E) NM2A filament clearly localizing at a protrusive actin tip. (F) NM2A filaments accumulating along actin bundles. (G) Time-lapse anterograde movements of NM2A (orange highlight). (H) Cartoon of N-terminal EGFP tag on NM2A monomer, filament, and filament imaged with high-resolution microscopy. Imaging was performed on Nikon CSU-W1 SoRa spinning disk.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 3 Hz, and playback is at 40 fps.
Previous EM studies have observed activated monomeric NM2 in cells (45), and NM2 is known to associate with various vesicular organelles that undergo directed transport (46,47,48,49). To confirm that our anterograde movements were filamentous NM2 structures moving on actin, we imaged both EGFP-NM2A and FTractin-mScarlet in CAD protrusions (Fig. 2 D–F; Video S6). N-terminal tagging of NM2A coupled with high-resolution imaging reveals two puncta 300 nm apart, indicative of bipolar filamentous NM2 (Fig 2 H) (35,50). We readily observed these bipolar filaments at the tips of short actin protrusions (Fig. 2 E), on actin bundles (Fig. 2 F), and moving anterograde along actin bundles (Fig. 2 G). Collectively these different tagging approaches of the NM2 holoenzyme demonstrate the anterograde movements of filamentous NM2 along actin bundles near the leading edge, often referred to as prefilopdial bundles or microspikes (32,51).
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 1.25 Hz, and playback is at 15 fps.
NM2 moves anterograde independent of actin dynamics
We next sought to isolate movement of the NM2 filaments from the underlying actin dynamics. We used a small-molecule inhibitor cocktail of JL to stall actin dynamics, similar to previous work (52), and we monitored NM2 by expressing NM2A with a C-terminal HaloTag (NM2A-Halo). This places all fluorophores into a single punctum at the middle of the bipolar filament (Fig. 3 A), in contrast to N-terminal tags that create two puncta at opposing ends of the bipolar filament (Fig. 2 H). This C-terminal HaloTag provides higher signal-to-noise to closely monitor filament dynamics via kymographs or automated tracking. Before JL treatment, we observed both retrograde and anterograde movements of NM2A-Halo (Fig. 3 B, C, and F; Video S7). After JL treatment, NM2A-Halo retrograde flow was largely abrogated, as evidenced by the vertical lines in the kymograph and reduced movements detected in automated tracking (Fig. 3 D–F). This is consistent with NM2 retrograde movements being dependent on actin dynamics. After JL treatment, however, we still observed robust anterograde movements (Fig. 3 D–F; Video S7). In addition, inhibition of retrograde flow increased the leading edge localization of NM2 (Fig. 3 G and H). Collectively, these data illustrate that the anterograde movements we observe are processive NM2 filaments and that NM2 can move processively in the absence of actin dynamics.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 2 Hz, and playback is at 30 fps.
Processive NM2 is dependent on actin architecture
Previous studies in CAD cells demonstrated that knocking out the monomeric actin binding protein profilin 1 (PFN1 KO) disrupts lamellipodia and prefilopodial actin bundling, whereas overexpression of profilin enhances actin bundles at the expense of Arp2/3-dendritic networks (32). We therefore used different levels of profilin 1 expression to determine if altering the lamellipodia actin network affected NM2 anterograde movements. When NM2A-Halo was expressed in PFN1 KO CAD cells, it appeared largely diffuse throughout the cell, with little apparent retrograde flow and few discernible processive anterograde movements (Fig. 4 A and B; Video S8). This lack of NM2 architecture is not surprising, considering the extent to which PFN1-KO disrupts actin structures in these cells (32). However, it does support a model that appropriate actin architecture in the form of linear arrays is required for processive NM2 anterograde runs. Indeed, anterograde-moving NM2A-Halo labeled filaments were readily observed in membrane protrusions in CAD cells overexpressing PFN1 (Fig. 4 E; Video S8). Though the number of initiated anterograde events and run velocity were no different than controls (GFP; Fig. 4 G–I), we did observe a significantly higher number of anterograde-moving NM2A puncta that reached the leading edge (Fig. 4 J). We also observed persistent localization of these puncta at the tips of short actin spikes, likely prefilopodial bundles (Fig. 4 C; magenta arrows). The PFN1 overexpression data suggest that dendritic actin networks inhibit the ability of NM2A to move processively along prefilopodia actin bundles, consistent with previous reports (53,54,55). This model could explain why NM2A anterograde movements have not been reported in lamellipodia that are more dependent on Arp2/3 networks, like those found in migratory fibroblasts.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 5 Hz, and playback is at 30 fps.
Characterization of NM2 isoform processivity
To better understand processivity of NM2 isoforms, we evaluated the endogenous RNA, protein, and localization in CAD cells. For simplicity, we use NM2A, NM2B, and NM2C terminology here whereas experimentally observing the respective MHCs (MHC 2A, MHC 2B, and MHC 2C from Myh9, Myh10, and Myh14). Previous RNAseq data (32) demonstrated that NM2A and NM2B were transcribed at moderate levels, with 2:1 ratio of NM2A/NM2B (mean = 2.20 ± 0.28), whereas NM2C was barely detected (Fig. 5 A). Western blotting of CAD whole-cell lysates demonstrated similar expression levels of NM2A and NM2B (Fig. 5 B). Correlative blots with EGFP-NM2A and EGFP-NM2B intermediates revealed endogenous NM2A dominant over NM2B expression (see materials and methods), again with an 2:1 ratio of NM2A/NM2B (mean = 1.95 ± 0.66). Immunostaining of CAD cells also revealed robust NM2A signal with clear puncta extending into the protrusion, whereas NM2B signal was less continuous throughout the cell, with less signal above background in the protrusion (Fig. 5 C). This is consistent with previous localization in polarized cells, where NM2A extends more peripheral and NM2B remains more centripetal and rearward (56).
In addition to localization differences, NM2 isoforms were found to possess kinetic differences in processivity in vitro (22). To determine if we could detect similar differences in processivity kinetics in cells, we expressed NM2A-Halo or NM2B-Halo with a C-terminal HaloTag. When NM2A-Halo was expressed in CAD cells, we again observed robust processivity in the protrusions, with many runs reaching the leading edge (Fig. 5 D–E; Video S9). When NM2B-Halo was expressed in CAD cells, we observed processive runs but with clear differences relative to NM2A. NM2B runs again initiated from the dense cell body or from partitioning events off of retrograde moving NM2 clusters. However, the runs often stalled in the mid-protrusion region before reaching the leading edge (Fig. 5 G and H). Average velocity measurements revealed a mild but significant difference between the isoforms, with NM2A running over 70 nm/s and NM2B running under 60 nm/s (Fig. 5 F and I; Table S1).
Scale bar represents 2 μm, and time is in mm:ss. Cell was imaged at 2 Hz, and playback is at 30 fps.
Processive NM2 in fibroblasts
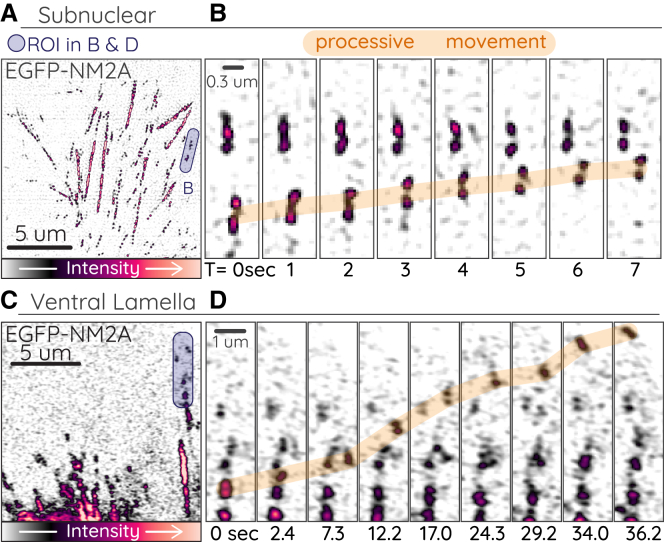
Although the actin architecture and morphology of CAD cells provide opportunity for readily observable processive NM2 movements, we hypothesized that similar movements could and should still be occurring in diverse cell types, if perhaps less frequently. To explore this possibility, we imaged primary MEF cells with NM2A endogenously labeled with EGFP (EGFP-NM2A). When sampled with high spatial and temporal frequency, we observed processive NM2 movements in both subnuclear stress fibers (Fig. 6 A and B; Video S10) and ventral stress fibers in the lamellar protrusions (Fig. 6 C and D; Video S11). The subnuclear movements were directed toward both the anterior and posterior of the cell, whereas the discernible lamellar movements were directed exclusively anterior. This is consistent with expected polarity of actin bundles within these regions of a polarized fibroblast (30). Importantly, these observations with endogenously labeled NM2 argue against the possibility that NM2 overexpression in earlier CAD cell experiments was solely responsible for processive movements.
Figure 6.
Processive EGFP-NM2A in fibroblasts
(A–D) Primary EGFP-NM2A MEF cells imaged with DeltaVision OMX SR TIRF-SIM (total internal reflection fluorescence structured-illumination microscopy) (subnuclear stress fibers; A and B) or Zeiss 880 Airyscan (ventral stress fibers; C and D). Purple boxes in (A) and (C) indicate ROI for time-lapses in (B) and (D), where processive movements are indicated with orange highlight. (A) corresponds to t = 19 s, and (B) corresponds to 19–26 s in Video S10. (C) corresponds to t = 298 s, and (D) corresponds to 298–334 s in Video S11.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 1 Hz, and playback is at 20 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 0.4 Hz, and playback is at 20 fps.
Discussion
We conclude that the anterograde movements we observe for NM2 are indeed processive for the following reasons: first, they travel against the direction of actin retrograde flow; second, the velocity of movement is consistent with in vitro measurements of NM2 processivity (22); third, they often travel along actin bundles that terminate at the leading edge, the exact actin architecture where NM2 processivity would theoretically thrive; and finally, stalling actin dynamics did not inhibit movement. These results, in combination with the previous in vitro work by Sellers and colleagues, establish processivity as a property of NM2 filaments. Our collective observations provoke a range of questions, from the biophysical nature to the biological relevance of processive filaments.
Previous studies have used a simple formula to estimate the likelihood of synergistic processivity for an ensemble of motors:
| (1) |
where is the duty ratio of a filamentous ensemble, r is the duty ratio of an individual motor, and n is the number of motors in a filamentous ensemble (22,57). As increases, the likelihood of processivity increases. Melli et al. calculated NM2A could be processive with a duty ratio of 0.05 and 50 motors per side of the bipolar filament (25 “monomers” per side with two motors per monomer) (22). Considering NM2A filaments mature with 30 monomers per filament (30 motors per side) (19), only two mature filaments would be required to create a processive ensemble. Previous EM studies (53,58) demonstrate the existence of filament stacks in the lamella, with multiple filaments in register. If many lamellar filamentous structures are indeed small subresolution stacks with at least two filaments, this would provide sufficient motors to maintain binding to the actin track and enable NM2A to move processively toward the leading edge.
A similar theoretical approach was taken by Nagy et al. who calculated that NM2B would be processive with a duty ratio of 0.22 and 10–12 motors per side of the bipolar filament (57), although previous contrasting work suggested NM2B monomer processivity (59). This theoretical 10–12 NM2B motors is easily attained in a single filament. However, the increased duty ratio of NM2B relative to NM2A should lead to more motors engaged with filamentous actin, especially if there are multiple mature filaments in an ensemble in a dense actin network. We hypothesize that this increased actin binding explains our observation that NM2B filaments often struggle to reach the leading edge, as they are more easily entangled in the lamellar actin network. In addition, the moderate decrease in NM2B velocity relative to NM2A could be explained by this increased NM2B duty ratio and decreased ATPase kinetics (60), adding drag to the ensemble with decreased motor cycling.
In reality, our kinetic measurements are complicated by a number of factors. The cellular actin architecture is more complex than the in vitro assays. Cellular actin is decorated with an array of actin binding proteins, unlike the pure actin filaments in vitro. Any of these actin binding proteins could potentially enhance or inhibit processive NM2. Cellular NM2 filaments are in dynamic equilibrium with monomeric NM2 and could be bound to numerous binding partners (see below). Perhaps most importantly, NM2 isoforms have the ability to co-assemble into mixed filaments (36,45). The cell types analyzed in this study are skewed toward NM2A expression over NM2B. Our immunostaining of endogenous isoforms, consistent with previous observations (56), demonstrates spatial sorting of NM2 filaments, with NM2B restricted to central regions in the cell body and NM2A extending more peripherally. Therefore, our kinetic measurements of peripheral EGFP-NM2A processivity are likely to be measuring filaments largely composed of NM2A, whereas EGFP-NM2B processive filaments in more centripetal regions are likely to be mixed NM2A/NM2B filaments. When NM2A and NM2B were mixed 2:1 in vitro and allowed to co-polymerize, the velocity was indistinguishable from NM2B alone (22), suggesting that relatively small numbers of NM2B motors can significantly alter filamentous biophysical and kinetic properties. Therefore, we hypothesize that the EGFP-NM2B kinetics we measure for mixed NM2A/NM2B filaments are revealing the underlying properties of NM2B processivity. More sophisticated experimental approaches are required to further dissect these concepts.
A curious question is “why has NM2 processivity not been previously reported?” Certainly, there has been an abundance of live-cell imaging of NM2. We suspect a confluence of factors can explain this discrepancy. Both subcellular conditions and experimental approach must be optimal. First, there must be a relatively high ratio between parallel actin bundles and other actin structures (antiparallel bundles, mesh networks, etc.). This excludes many stress fibers, transverse arcs, contractile rings, cortices, and lamellipodia. Second, actin retrograde flow must be significantly slower than the velocity of NM2 processive movements in order for the NM2 processive movements to be observable. This is especially true for NM2B, which has slower velocities in vitro and in cells. Considering retrograde flow in some immune cell lamellipodia can be 100 nm/s (61), many anterograde NM2 movements would be difficult to observe. Third, NM2 density must be sufficiently low to observe filament dynamics. Many regions of nonmuscle cells contain dense actomyosin networks, and discerning the movements of individual filaments or small filamentous ensembles is beyond the resolution of most light microscopy. Considering these caveats, we suggest that protrusions with highly bundled parallel actin, as can be found in neuronal growth cones, filopodia, stereocilia, invadopodia, etc., are the most likely subcellular environments where NM2 processivity might be apparent. Consistently, previous immunostaining of neuronal growth cones demonstrated ample NM2 signal in peripheral structures and filopodia (62,63). It is certainly possible that single actin filaments are also sufficient for processivity, as was observed in vitro (22). However, there are not many cellular environments where single actin filaments of significant length do not encounter other actin filaments, making parallel bundles the most likely tracks. It will also be interesting to determine which of these specialized actin structures actually enable NM2 processivity and if actin binding and bundling proteins can modulate NM2 processivity. This is especially true for tropomyosins, which are required for processivity of certain class V myosins (64). Finally, concerning experimental approach, temporal sampling frequency needs to be sufficiently high (1–2 Hz) so that processive movements can be revealed. Studies that observe NM2 dynamics in slower cellular processes (e.g., migration, cytokinesis, epithelial dynamics, etc.) by sampling with longer intervals are likely to miss any processive movements.
The most outstanding question remains as to the biological function of processive NM2. One obvious answer is that as a processive motor NM2 could act as a cargo transporter. Numerous NM2 binding partners have been identified (65,66,67,68), and NM2 has been localized to various vesicle and organelle populations (exocytic vesicles, lytic granules, lipid droplets, Golgi vesicles, etc.) (46,47,48,49). It is possible that these or other cargos are being transported to the distal regions of these parallel actin structures. However, as other specialized processive myosins exist with this capability (e.g., myosins V, X, and XV (31,69)), it is unclear why the cell would rely on NM2 for this function. One also wonders if processive NM2 in these structures would compete with or complement the function of these other myosins. Another potential model for the functional relevance of NM2 processivity is that it serves to localize NM2 itself into peripheral structures with parallel actin bundles. This model, however, seems inefficient compared with the numerous phosphorylation assembly mechanisms that are used throughout cell biology (65,70). In addition, signaling pathways exist that specifically inhibit NM2 assembly at the leading edge (54). It would be interesting to test if these pathways are turned off in CAD cells or during PFN1 overexpression. Manipulation of NM2 to dissect these models is likely to prove challenging. Studies that broadly inhibit NM2 function throughout the cell via knockdown/knockout or small-molecule inhibitors (e.g., blebbistatin) would be difficult to interpret, as they would disrupt the entirety of the actomyosin network. Photomanipulation approaches that can target NM2 specifically in protrusions (e.g., iLID-based inhibitor (71)) might provide the spatio-temporal precision required. Finally, correlative imaging with NM2 and potential cargos could also prove insightful but requires identifying potential cargos to make it practical.
In conclusion, the identification of NM2 processivity forces us to broaden the potential functional roles for NM2 in cell physiology. Although its dominant function is to drive contractility, its ability to function as a monomer, to drive actin expansion, to cross-link actin, and to move processively should not be overlooked.
Author contributions
E.A.V., P.W.O., and J.R.B. designed the research. E.A.V., M.A.Q., J.J.T., L.D.T., A.C., and J.R.B. performed experiments. M.A.Q., L.D.T., B.A.C., M.L.J., P.W.O., and J.R.B. analyzed data. E.A.V., M.A.Q., J.J.T., P.W.O., and J.R.B. wrote the manuscript.
Acknowledgments
This work is dedicated to the memory of Dr. Ken Jacobson. Research reported in this publication was supported by the Maximizing Investigators’ Research Award (MIRA) (R35) from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under grant number R35GM137959 to E.A.V. and R35GM138183 to J.R.B., and Graduate Research Fellowship (GRFP) from the National Science Foundation (NSF) under grant number DGE-1842190 to M.A.Q. P.W.O. is supported in part by a National Science Foundation CAREER Award #2000554 and National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) (Award # P01-AI02851).
Declaration of interests
The authors declare no competing interests.
Editor Name: Kathleen Trybus.
Footnotes
Eric A. Vitriol, Melissa A. Quintanilla, and Joseph J. Tidei contributed equally to this work.
Patrick W. Oakes and Jordan R. Beach contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.05.014.
Contributor Information
Eric A. Vitriol, Email: evitriol@augusta.edu.
Patrick W. Oakes, Email: poakes@luc.edu.
Jordan R. Beach, Email: jbeach1@luc.edu.
Supporting material
Data availability
The data that support the findings of this study are available upon reasonable request from the corresponding authors (Vitriol, Oakes, Beach). All code, along with example data, used to automatically analyze myosin motion is available at https://github.com/OakesLab.
References
- 1.Svoboda K., Schmidt C.F., et al. Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 2.Svoboda K., Block S.M. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 3.Vallee R.B., Wall J.S., et al. Shpetner H.S. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 1988;332:561–563. doi: 10.1038/332561a0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Khan S., Sheetz M.P. Single cytoplasmic dynein molecule movements: characterization and comparison with kinesin. Biophys. J. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta A.D., Rock R.S., et al. Cheney R.E. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 6.De La Cruz E.M., Ostap E.M. Relating biochemistry and function in the myosin superfamily. Curr. Opin. Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Veigel C., Wang F., et al. Molloy J.E. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 8.Robblee J.P., Olivares A.O., de la Cruz E.M. Mechanism of nucleotide binding to actomyosin VI: evidence for allosteric head-head communication. J. Biol. Chem. 2004;279:38608–38617. doi: 10.1074/jbc.M403504200. [DOI] [PubMed] [Google Scholar]
- 9.De La Cruz E.M., Wells A.L., et al. Sweeney H.L. The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schindler T.D., Chen L., et al. Bryant Z. Engineering myosins for long-range transport on actin filaments. Nat. Nanotechnol. 2014;9:33–38. doi: 10.1038/nnano.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris D.E., Warshaw D.M. Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 1993;268:14764–14768. [PubMed] [Google Scholar]
- 12.Uyeda T.Q., Kron S.J., Spudich J.A. Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 13.Sweeney H.L., Holzbaur E.L.F. Muscle contraction. Cold Spring Harbor Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quintanilla M.A., Hammer J.A., Beach J.R. Non-muscle myosin 2 at a glance. J. Cell Sci. 2023;136 doi: 10.1242/jcs.260890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toothaker L.E., Gonzalez D.A., et al. Tenen D.G. Cellular myosin heavy chain in human leukocytes: isolation of 5’ cDNA clones, characterization of the protein, chromosomal localization, and upregulation during myeloid differentiation. Blood. 1991;78:1826–1833. [PubMed] [Google Scholar]
- 16.Simons M., Wang M., et al. Weir L. Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ. Res. 1991;69:530–539. doi: 10.1161/01.res.69.2.530. [DOI] [PubMed] [Google Scholar]
- 17.Leal A., Endele S., et al. Rautenstrauss B. A novel myosin heavy chain gene in human chromosome 19q13.3. Gene. 2003;312:165–171. doi: 10.1016/s0378-1119(03)00613-9. [DOI] [PubMed] [Google Scholar]
- 18.Brito C., Sousa S. Non-Muscle Myosin 2A (NM2A): structure, regulation and function. Cells. 2020;9:1590. doi: 10.3390/cells9071590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billington N., Wang A., et al. Sellers J.R. Characterization of three full-length human nonmuscle myosin II paralogs. J. Biol. Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederman R., Pollard T.D. Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J. Cell Biol. 1975;67:72–92. doi: 10.1083/jcb.67.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasquez C.G., Heissler S.M., et al. Martin A.C. Drosophila non-muscle myosin II motor activity determines the rate of tissue folding. Elife. 2016;5 doi: 10.7554/eLife.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melli L., Billington N., et al. Sellers J.R. Bipolar filaments of human nonmuscle myosin 2-A and 2-B have distinct motile and mechanical properties. Elife. 2018;7 doi: 10.7554/eLife.32871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaub S., Bohnet S., et al. Verkhovsky A.B. Comparative maps of motion and assembly of filamentous actin and myosin II in migrating cells. Mol. Biol. Cell. 2007;18:3723–3732. doi: 10.1091/mbc.E06-09-0859. http://www.molbiolcell.org/cgi/content/abstract/18/10/3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huxley H., Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 25.Huxley A.F., Niedergerke R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- 26.Straight A.F., Cheung A., et al. Mitchison T.J. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 27.Toyoshima Y.Y., Toyoshima C., Spudich J.A. Bidirectional movement of actin filaments along tracks of myosin heads. Nature. 1989;341:154–156. doi: 10.1038/341154a0. [DOI] [PubMed] [Google Scholar]
- 28.Sellers J.R., Kachar B. Polarity and velocity of sliding filaments: control of direction by actin and of speed by myosin. Science. 1990;249:406–408. doi: 10.1126/science.2377894. [DOI] [PubMed] [Google Scholar]
- 29.Huxley H.E. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J. Mol. Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 30.Cramer L.P., Siebert M., Mitchison T.J. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 1997;136:1287–1305. doi: 10.1083/jcb.136.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weck M.L., Grega-Larson N.E., Tyska M.J. MyTH4-FERM myosins in the assembly and maintenance of actin-based protrusions. Curr. Opin. Cell Biol. 2017;44:68–78. doi: 10.1016/j.ceb.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skruber K., Warp P.V., et al. Vitriol E.A. Arp2/3 and Mena/VASP require profilin 1 for actin network assembly at the leading edge. Curr. Biol. 2020;30:2651–2664.e5. doi: 10.1016/j.cub.2020.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimm J.B., English B.P., et al. Lavis L.D. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Conti M.A., et al. Adelstein R.S. Mouse models of MYH9-related disease: mutations in nonmuscle myosin II-A. Blood. 2012;119:238–250. doi: 10.1182/blood-2011-06-358853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beach J.R., Bruun K.S., et al. Hammer J.A. Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat. Cell Biol. 2017;19:85–93. doi: 10.1038/ncb3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beach J.R., Shao L., et al. Hammer J.A., 3rd Nonmuscle myosin II isoforms coassembly in living cells. Curr. Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schell M.J., Erneux C., Irvine R.F. Inositol 1,4,5-trisphosphate 3-kinase A associates with F-actin and dendritic spines via its N terminus. J. Biol. Chem. 2001;276:37537–37546. doi: 10.1074/jbc.M104101200. [DOI] [PubMed] [Google Scholar]
- 38.Schindelin J., Arganda-Carreras I., et al. Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Y., Wang J.K., et al. Chikaraishi D.M. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vitriol E.A., McMillen L.M., et al. Zheng J.Q. Two functionally distinct sources of actin monomers supply the leading edge of lamellipodia. Cell Rep. 2015;11:433–445. doi: 10.1016/j.celrep.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guzik-Lendrum S., Heissler S.M., et al. Sellers J.R. Mammalian myosin-18A, a highly divergent myosin. J. Biol. Chem. 2013;288:9532–9548. doi: 10.1074/jbc.M112.441238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bird J.E., Takagi Y., et al. Friedman T.B. Chaperone-enhanced purification of unconventional myosin 15, a molecular motor specialized for stereocilia protein trafficking. Proc. Natl. Acad. Sci. USA. 2014;111:12390–12395. doi: 10.1073/pnas.1409459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Z., Ma X.-N., et al. Li X.-D. Mouse myosin-19 is a plus-end-directed, high-duty ratio molecular motor. J. Biol. Chem. 2014;289:18535–18548. doi: 10.1074/jbc.M114.569087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maliga Z., Junqueira M., et al. Hyman A.A. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat. Cell Biol. 2013;15:325–334. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- 45.Shutova M.S., Spessott W.A., et al. Svitkina T. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr. Biol. 2014;24:1958–1968. doi: 10.1016/j.cub.2014.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milberg O., Shitara A., et al. Weigert R. Concerted actions of distinct nonmuscle myosin II isoforms drive intracellular membrane remodeling in live animals. J. Cell Biol. 2017;216:1925–1936. doi: 10.1083/jcb.201612126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carisey A.F., Mace E.M., et al. Orange J.S. Nanoscale dynamism of actin enables secretory function in cytolytic cells. Curr. Biol. 2018;28:489–502.e9. doi: 10.1016/j.cub.2017.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfisterer S.G., Gateva G., et al. Ikonen E. Role for formin-like 1-dependent acto-myosin assembly in lipid droplet dynamics and lipid storage. Nat. Commun. 2017;8 doi: 10.1038/ncomms14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miserey-Lenkei S., Chalancon G., et al. Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat. Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- 50.Fenix A.M., Taneja N., et al. Burnette D.T. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol. Biol. Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dimchev G., Amiri B., et al. Schur F.K. Computational toolbox for ultrastructural quantitative analysis of filament networks in cryo-ET data. J. Struct. Biol. 2021;213 doi: 10.1016/j.jsb.2021.107808. [DOI] [PubMed] [Google Scholar]
- 52.Peng G.E., Wilson S.R., Weiner O.D. A pharmacological cocktail for arresting actin dynamics in living cells. Mol. Biol. Cell. 2011;22:3986–3994. doi: 10.1091/mbc.E11-04-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verkhovsky A.B., Svitkina T.M., Borisy G.G. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asokan S.B., Johnson H.E., et al. Bear J.E. Mesenchymal chemotaxis requires selective inactivation of myosin II at the leading edge via a noncanonical PLCgamma/PKCalpha pathway. Dev. Cell. 2014;31:747–760. doi: 10.1016/j.devcel.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muresan C.G., Sun Z.G., et al. Murrell M.P. F-actin architecture determines constraints on myosin thick filament motion. Nat. Commun. 2022;13:1–16. doi: 10.1038/s41467-022-34715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolega J. Cytoplasmic dynamics of myosin IIA and IIB: spatial ’sorting’ of isoforms in locomoting cells. J. Cell Sci. 1998;111:2085–2095. doi: 10.1242/jcs.111.15.2085. [DOI] [PubMed] [Google Scholar]
- 57.Nagy A., Takagi Y., et al. Sellers J.R. Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J. Biol. Chem. 2013;288:709–722. doi: 10.1074/jbc.M112.424671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verkhovsky A.B., Borisy G.G. Non-sarcomeric mode of myosin II organization in the fibroblast lamellum. J. Cell Biol. 1993;123:637–652. doi: 10.1083/jcb.123.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norstrom M.F., Smithback P.A., Rock R.S. Unconventional processive mechanics of non-muscle myosin IIB. J. Biol. Chem. 2010;285:26326–26334. doi: 10.1074/jbc.M110.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang F., Kovacs M., et al. Sellers J.R. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J. Biol. Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 61.Yi J., Wu X.S., et al. Hammer J.A., III Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol. Biol. Cell. 2012;23:834–852. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rochlin M.W., Itoh K., et al. Bridgman P.C. Localization of myosin II A and B isoforms in cultured neurons. J. Cell Sci. 1995;108:3661–3670. doi: 10.1242/jcs.108.12.3661. [DOI] [PubMed] [Google Scholar]
- 63.Hur E.-M., Yang I.H., et al. Zhou F.-Q. Engineering neuronal growth cones to promote axon regeneration over inhibitory molecules. Proc. Natl. Acad. Sci. USA. 2011;108:5057–5062. doi: 10.1073/pnas.1011258108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hodges A.R., Krementsova E.B., et al. Trybus K.M. Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr. Biol. 2012;22:1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dulyaninova N.G., Malashkevich V.N., et al. Bresnick A.R. Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–6876. doi: 10.1021/bi0500776. [DOI] [PubMed] [Google Scholar]
- 66.Du M., Wang G., et al. Rudland P.S. S100P dissociates myosin IIA filaments and focal adhesion sites to reduce cell adhesion and enhance cell migration. J. Biol. Chem. 2012;287:15330–15344. doi: 10.1074/jbc.M112.349787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Even-Faitelson L., Ravid S. PAK1 and aPKCzeta regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol. Biol. Cell. 2006;17:2869–2881. doi: 10.1091/mbc.E05-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kothari P., Srivastava V., et al. Robinson D.N. Contractility kits promote assembly of the mechanoresponsive cytoskeletal network. J. Cell Sci. 2019;132 doi: 10.1242/jcs.226704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Titus M.A. Myosin-driven intracellular transport. Cold Spring Harbor Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amano M., Ito M., et al. Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J. Biol. Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto K., Miura H., et al. Aoki K. Optogenetic relaxation of actomyosin contractility uncovers mechanistic roles of cortical tension during cytokinesis. Nat. Commun. 2021;12:7145. doi: 10.1038/s41467-021-27458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 3 Hz, and playback is at 30 fps.
Cell was imaged at 3 Hz, and playback is at 30 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 1.5 Hz, and playback is at 30 fps.
Cell was imaged at 1.4 Hz, and playback is at 15 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 3 Hz, and playback is at 40 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 1.25 Hz, and playback is at 15 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 2 Hz, and playback is at 30 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 5 Hz, and playback is at 30 fps.
Scale bar represents 2 μm, and time is in mm:ss. Cell was imaged at 2 Hz, and playback is at 30 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 1 Hz, and playback is at 20 fps.
Scale bar represents 5 μm, and time is in mm:ss. Cell was imaged at 0.4 Hz, and playback is at 20 fps.
Data Availability Statement
The data that support the findings of this study are available upon reasonable request from the corresponding authors (Vitriol, Oakes, Beach). All code, along with example data, used to automatically analyze myosin motion is available at https://github.com/OakesLab.