Abstract
Mitochondria adapt to changing cellular environments, stress stimuli, and metabolic demands through dramatic morphological remodeling of their shape, and thus function. Such mitochondrial dynamics is often dependent on cytoskeletal filament interactions. However, the precise organization of these filamentous assemblies remains speculative. Here, we apply cryogenic electron tomography to directly image the nanoscale architecture of the cytoskeletal-membrane interactions involved in mitochondrial dynamics in response to damage. We induced mitochondrial damage via membrane depolarization, a cellular stress associated with mitochondrial fragmentation and mitophagy. We find that, in response to acute membrane depolarization, mammalian mitochondria predominantly organize into tubular morphology that abundantly displays constrictions. We observe long bundles of both unbranched actin and septin filaments enriched at these constrictions. We also observed septin-microtubule interactions at these sites and elsewhere, suggesting that these two filaments guide each other in the cytosolic space. Together, our results provide empirical parameters for the architecture of mitochondrial constriction factors to validate/refine existing models and inform the development of new ones.
Significance
This study used cryo-electron tomography on thin samples prepared using cryofocused ion beam milling to generate three-dimensional (3D) reconstructions (tomograms) of mitochondria in cells in response to membrane depolarization. The authors found that the majority of mitochondria exhibited tubular morphologies with constricted membranes. Using semi-automated segmentation workflows and quantitative analyses, the authors demonstrated that long, linear filaments of actin and septin were both preferentially enriched as parallel bundles around synchronously constricting inner and outer mitochondrial membranes. Overall, this study reveals for the first time native 3D architecture of constricting mitochondria along with cytoskeletal filaments at the nanoscale resolution, thus providing new insight into the ultrastructural arrangement of molecular components that mediate mitochondrial membrane constriction.
Introduction
Mitochondria are double-membraned, semiautonomous organelles that perform critical metabolomic, signaling, and biogenesis pathways in cells, including ATP generation via oxidative phosphorylation, biogenesis of amino acids, regulation of ion homeostasis, and initiation of apoptosis (1). Their ability to carry out these diverse and essential roles is directly linked to their dynamic behavior (2,3,4). As a network, mitochondria dynamically change shape through the processes of fission and fusion; and as individual organelles, the architecture of their outer and inner membranes remodel in response to varying metabolic demands, stress signaling, and apoptotic stimuli. Mitochondrial dynamics/remodeling enables eukaryotic cells to maintain a healthy population of mitochondria by sequestering damaged components to specific mitochondrial regions and selectively degrading them in a process called mitophagy (5). Collectively, these processes help to maintain cellular health during conditions of stress. Understandably, disruptions to mitochondrial dynamics are associated with etiologically diverse diseases including neurodegenerative disorders (6,7), underscoring the importance of understanding how these processes are orchestrated during cellular homeostasis and how they break down to cause disease pathologies.
Recent work has implicated the cytoskeleton in mitochondrial membrane remodeling in response to damage. Mitochondrial damage involving dissipation of the proton gradient using the pharmacological uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) causes a rapid accumulation of actin filaments into “clouds'' surrounding depolarized mitochondria (8). This in turn causes the highly interconnected network of mitochondria to fragment via extensive fission in addition to other kinds of remodeling that create circularized mitochondrial morphologies (9,10), finally leading to widespread mitophagy (11). This damage-induced actin polymerization depends on the actin nucleating factor, Arp2/3 (9). On the other hand, calcium-triggered actin polymerization has also been shown to remodel mitochondria in an INF2-dependent manner (12,13); INF2 is an ER-associated actin-nucleating factor that belongs to the Formin family of proteins (14). Previous work showed that septins (particularly Sept2 and Sept7) also localize to mitochondria and facilitate fragmentation upon FCCP-induced mitochondrial membrane depolarization (15) and upon S. flexneri infection (16). Both types of cytoskeletal filaments seem to function in conjunction with DRP1, a mitochondrial fission factor (15,16,17,18,19). Importantly, the precise organization of these filaments, their interaction with mitochondria, and their role in mediating mitochondrial membrane remodeling remain unclear, due to a lack of high-resolution structural information.
In this study, we established a workflow to directly visualize the native nanoscale architecture of the cytoskeletal-membrane interactions involved in mediating mitochondrial dynamics. We activated mitochondrial dynamics using FCCP in mouse embryonic fibroblasts (MEFs) and imaged them using cryogenic electron tomography (cryo-ET). We demonstrate that mitochondria exposed to acute FCCP treatment displayed several morphologies, including the previously noted circularized shapes and tubular shapes with varying degree of constriction. We frequently observed the ER and two distinct types of filaments associated with constricting mitochondria, including thin (6–8 nm) actin filaments and thicker (10 nm) filaments that we determined to be septin filaments through genetic knockdown. We developed and applied several filament analysis tools and discovered that both actin and septin filaments were preferably enriched at mitochondrial constrictions and formed closely associated, parallel bundles. Our results demonstrate that mitochondrial membrane dynamics triggered by membrane depolarization promotes association with cytoskeletal filaments, suggesting that these interactions are important for damage-induced mitophagy.
Materials and methods
Cell growth
Adherent MEFs (gifts of D. Chan, California Institute of Technology and G. Shadel of Salk Institute of Biological Studies), bone osteosarcoma cells (U2OS) (gift of R.L. Wiseman, The Scripps Research Institute), and rat insulinoma cells (INS-1E) (gift of P. Maechler, Université de Genève) were grown in a humidified 37°C incubator with a constant supply of 5% CO2. Cells were cultured in high-glucose (L-glutamine+) Dulbecco’s modified Eagle’s medium (Caisson Labs, Smithfield, UT, USA, DML09) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA, cat. no. 10437028), 1 mM sodium pyruvate (Thermo Fisher Scientific, cat. no. 11360070), 100 units/mL penicillin, and 100 μg/mL streptomycin. Plasmids for shRNA-mediated knockdown of Sept2 were maintained in MEF cells using 3 μg/mL puromycin (gift of D. Chan). For cryogenic focused ion beam (cryo-FIB) milling and cryo-ET, cells were grown on 200-mesh gold R2/2 London Finder Quantifoil grids (Quantifoil Micro Tools, Jena, Germany). Before seeding cells, these grids were coated with 0.1 mg/mL human fibronectin (PromoCell, Heidelberg, Germany, cat. no. C-43060) by floating them on fibronectin droplets on parafilm for approximately 15–30 min. Cells were grown to a density of approximately two to three per grid square over a period of 1–2 days.
Gene knockdown
A knockdown cell line for Sept2 was created in MEF cells using retroviral transduction with guidance from C.-S. Shin and reagents kindly gifted by D. Chan, both at California Institute of Technology. In brief, an shRNA fragment for Sept2 (a hairpin-forming sequence derived from the Sept2 targeting sequence of 5′ GCAGTTTGAACGCTACCTACA 3′) was inserted into pRetroX-H1 and confirmed by sequencing. For delivery of this shRNA plasmid into MEF cells, retroviruses (packaged with this plasmid) were produced by cotransfecting 293T cells with the retrovirus packaging vector, pCL-Eco2.1, and the shRNA plasmid. Viruses were harvested from supernatant by centrifugation (to remove cell debris) and filtration through a 0.45-μm syringe filter. MEF cells were subsequently infected with these retroviruses at a low confluence (∼10%) in the presence of 4 μg/mL polybrene to enhance virus-host interaction; after the addition of viruses to cells, low-speed centrifugation was performed to increase infection efficiency. MEF cells successfully transduced with the shRNA plasmid were selected using 3 μg/mL puromycin and maintained in the presence of this drug. Sept2 knockdown was confirmed by western blotting using a rabbit monoclonal primary antibody against Sept2 (EPR12123) (1:500–1:2000 dilution, Abcam, Cambridge, MA, USA, cat. no. ab179436) and a goat anti-rabbit IgG (H+L) secondary antibody conjugated with horseradish peroxidase (Peroxidase AffiniPure) (1:10,000 dilution, Jackson ImmunoResearch, PA, USA, cat. no 111-035-003). A mouse anti-α-tubulin primary antibody (monoclonal) (Sigma-Aldrich, St. Louis, MO, USA, cat. no. T6199) and a goat anti-mouse secondary antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch, cat. no 115-056-003) were used to quantify α-tubulin in the same samples as loading control. The band intensities representing Sept2 protein expression level were quantified and normalized to the band intensities representing α-tubulin. The intensities of protein bands from three different experiments were quantified using the ImageJ Gel Analysis program as illustrated here (https://imagej.nih.gov/ij/docs/menus/analyze.html#gels and https://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/). As shown in Fig. S6 A, there is significantly less Sept2 expression relative to α-tubulin in shRNA-mediated knockdown. The p value was calculated using the Mann-Whitney rank test, a nonparametric alternative to the independent Student’s t-test.
Confocal light microscopy
Experimentation
To capture mitochondrial remodeling by cytoskeletal filaments by cryo-ET, we first conducted live-cell light microscopy in MEF cells. Our goal was to use light microscopy to determine the optimal time frame in which the majority of mitochondrial networks in FCCP-treated cells exhibited an intermediate network morphology. We reasoned that this time frame contained cells actively undergoing mitochondrial fission, thus maximizing our chances to capture mitochondrial constriction events by cryo-ET. For these experiments, MEF cells were grown on glass bottom MatTek dishes (MatTek Life Sciences, Ashland, MA, USA) and exposed to FCCP in the dish via media exchange. This experimental setup allowed us to start imaging mitochondrial networks (using MitoTracker) within these cells immediately after FCCP treatment by fluorescence microscopy. Imaging was performed at the Caltech Biological Imaging Facility on a Zeiss (Blackwood, NJ, USA) LSM800 microscope equipped with a large environmental chamber to maintain the temperature at 37°C and a smaller insert module that helped maintain both the temperature and a CO2 level of 5%. MEF cells were first treated with 200 nM of MitoTracker Red CMXRos (Thermo Fisher Scientific, cat. no. M7512) to fluorescently label mitochondria. Cells were treated in full medium (no HEPES) for 15 min at 37°C and washed twice using the same medium (no dye) before treatment with 10 μM FCCP. For experiments involving visualization of F-actin, cells were treated with FITC-phalloidin (Enzo Life Sciences, Ann Arbor, MI, USA, cat. no. ALX-350-268-MC01). In brief, post-FCCP treatment, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 15 min, washed twice with Hanks’ balanced salt solution (HBSS), incubated in acetone for 4 min at −20°C, washed twice again with HBSS, stained with 220 nM of FITC-phalloidin for 20 min, and finally washed twice with HBSS. Both bright-field and fluorescence imaging were performed using an LD C-Apochromat 40× water-immersion objective with a numerical aperture of 1.1, and images were recorded using photomultiplier tubes (PMTs) (for bright-field image) and GaAsP-PMT (for fluorescence). FITC-phalloidin and MitoTracker Red were imaged using diode lasers at 488 and 561 nm, respectively, operating at ∼1.5–2% of its maximum power. The maximum power for the laser lines was 500 mW at the source but measured to be ∼750 μW at the level of the objective lens for the 488-nm laser and ∼500 μW for the 561-nm laser. Mitochondrial morphology was monitored approximately every 2 min for up to 30 min after FCCP treatment. A scan speed of 7 was used for imaging cells. The pixel size for imaging was set at 0.312 μm (0.156 μm at a zoom factor of 2), while the image sizes were fixed at 512 × 512 pixels.
Quantification of mitochondrial morphology
For analysis of mitochondrial network morphologies, we collected images of cells with fluorescently labeled mitochondria at various time points after FCCP treatment. We blinded these images and scored the cells for containing primarily hyperfused, intermediate, or fragmented mitochondrial morphology (see Fig. S2 A for examples of mitochondrial morphology classifications). A total of 174 cells distributed across different time points were analyzed for this purpose.
Plunge freezing
Cells were cultured on electron microscopy grids that were placed at the bottom of MatTek dishes similar to those used for our light microscopy experiment. Cells on grids were similarly exposed to 10 μM FCCP via media exchange, and plunge frozen at different time points after FCCP exposure for subsequent cryo-FIB milling and cryo-ET. For plunge freezing, we used a mixture of liquid ethane/propane (20) and a Vitrobot Mark IV (FEI, Hillsboro, OR). The Vitrobot was set to 95–100% relative humidity at 37°C and blotting was done manually from the back side of the grids using Whatman filter paper strips. In the few cases where cryo-ET was performed on peripheral cell regions without cryo-FIB milling (INS-1E cells)), Au fiducials (Ted Pella, Redding, CA, USA, cat.no. 15703) resuspended in 1% bovine serum albumin were added to the grids just before blotting and plunge freezing. Plunge-frozen grids were subsequently loaded into Krios autogrid cartridges (Thermo Fisher Scientific). EM cartridges containing frozen grids were stored in liquid nitrogen and maintained at ≤−170°C throughout storage, transfer, and cryo-EM imaging. Our best efforts to achieve a similar time frame of FCCP exposure in our cryo-ET samples (as was established using fluorescence microscopy earlier) yielded cells that were predominantly plunge frozen at 3–6 min after drug treatment. Since this time comfortably fell within our goal, we did not feel the need to further optimize it.
Cryo-FIB milling
These experiments were performed at two different facilities: (1) at the Transmission Electron Microscopy Facility for material science at Caltech using a Versa 3D DualBeam scanning electron microscope (Thermo Fisher Scientific) equipped with a cryo-stage and a cryo-transfer system (PP3010 from Quorum Technologies, East Sussex, UK) and (2) The Scripps Research Institute Hazen Cryo-EM Microscopy Suite using an Aquilos 1 cryo-DualBeam FIB/SEM. Plunge-frozen cells were first coated with a 5–15-nm platinum layer using a sputter coater. Ideally positioned cells were first identified on the grids (preferably in the middle of the grid squares) using the electron beam (5 kV for observing topology and 20 kV for observing grid bars). Lamellae were milled at an angle of ∼15° with respect to the horizontal (corresponding to 22° on the cryo-stage) or less. After tilting the stage to the desired angle, material above and below the target volume was milled away by progressively stepping down the FIB current (at a constant voltage of 30 kV) as we approached the target volume: starting from 300 to 100 pA for removing material up to ∼1.5 μm from the target volume and finishing the process with a polishing step using 10–30 pA (21,22). The electron beam was used intermittently to monitor the milling process. Different voltages ranging from 5 to 20 kV helped discern the topology of cells and their positioning on the grids. We prepared more than 100 lamellae in this manner for this study.
Cryo-ET
Cryo-ET was performed using a Krios G3i 300 kV FEG cryo-TEM (Thermo Fisher Scientific) at the Caltech CryoEM Facility and using a similar instrument at The Scripps Research Institute Hazen Cryo-EM Microscopy Suite. The microscopes were equipped with a 4k x 4k K2 Summit direct detector (Gatan, Pleasanton, CA, USA) operated in electron counting mode. For data collected at the Caltech CryoEM Facility, an energy filter was used to increase the contrast at both medium and higher magnifications with a slit width of 50 and 20 eV, respectively. In addition, defocus values of close to negative 100 and negative 1–8 μm were used to boost the contrast (in the lower spatial resolution range) at the medium and higher magnifications, respectively. Magnifications typically used on the Krios were 3600× (or 4800×; in the medium range) and 26,000× (in the higher range), corresponding to pixel sizes of 4.2/3.1 nm and 5.38 Å, respectively. For data collected at the Caltech CryoEM Facility, a Volta phase plate was optionally used to further improve contrast at higher magnifications in certain cases. SerialEM software (23) was used for all imaging. The order of imaging steps was as follows. First, a full grid montage at a low magnification (close to 100×) was acquired to locate the milled lamella on a grid. Mitochondria were then identified in the lamella by their characteristic appearance (double membrane and matrix morphology) in projection images. Once the areas of interest were identified and marked, anchor maps were used to revisit these locations and collect tilt series in an automated fashion. Each tilt series was collected from negative 60° to positive 60° with an increment of 2° using the low-dose functions of tracking and focusing. The cumulative dose of each tilt series ranged between 80 and 150 e−/Å2. Once acquired, tilt series were binned into 1k x 1k arrays before alignment and reconstruction into 3D tomograms with the IMOD software package (24) and tomo3D (25). Tilt series were aligned using 10 nm Au fiducials (for unmilled samples) or patch tracking (for milled samples) in IMOD, while reconstructions were performed using SIRT in tomo3D.
Segmentations and quantifications
Quantification of mitochondrial morphology
We collected an initial cryo-ET data set consisting of 31 tilt series of cryo-FIB milled cells treated for 3–6 min with FCCP before vitrification. Two researchers classified the morphology of individual mitochondria as either tubular, spheroid, spherical, or ambiguous (see Fig. S1 for all examples and their assigned mitochondrial morphology classifications). An ambiguous designation was given in cases in which only a portion of the mitochondria was visible within the field of view and therefore an accurate description of mitochondrial morphology could not be assigned. A total of 9 cells distributed across different time points between 3 and 6 min of FCCP treatment were analyzed.
Segmentation of mitochondrial and ER membranes
Preliminary segmentations were performed using IMOD before the final segmentations were performed using Amira (Thermo Fisher Scientific), in both cases, manually. The final segmentations of a total of 22 mitochondrial constriction sites along 18 different mitochondrial profiles are included in this study.
Segmentation of actin, septin, and microtubule filaments
For all figures (except Fig. S4 C), actin and septin filaments were segmented using automated detection and tracing using the XTracing for the Amira extension package, which implements methods described by Rigort et al. (26). We optimized the parameters for these detection approaches to be specific for each filament type (e.g., actin and septin). Separate cylinder correlation procedures were performed to detect actin filaments (cylinder length, 500; angular sampling, 5; mask cylinder radii, 70; outer radius, 40; inner radius, 0) and septin filaments (cylinder length, 400; angular sampling, 5; mask cylinder radii, 100; outer radius, 60; inner radius, 0). Next, interactive thresholding was performed on the output of the cylinder correlation to identify ideal thresholds where all parts of filaments are visible. These threshold values were used to determine the parameters for minimum and continuation correlation values for the trace correlation lines procedure (direction coefficient, 0.2; minimum distance, 60; minimum length, 2500; search cone, 300; search angle, 30). This resulted in comprehensive coverage of the target filaments, plus some false positives, which we removed by manual verification using Filament editor in Amira. The assignment of each corresponding filament segmentation was visually inspected by one researcher initially (D.A.G.) and then verified by two other independent researchers (S.K.M. and B.A.B.). In very few instances where the assignment was ambiguous, we opted to leave these filaments out; however, this likely constitutes less than 5% of the entire data set. It is noteworthy that there were a few false actin filament ends generated by incomplete automated segmentation but they did not affect our analyses. For Fig. S4 C, actin and septin filaments were segmented manually (the signal/noise in the tomogram did not allow for exhaustive segmentation of these filaments). The segmentations therefore were not analyzed further but used for qualitative assessments. Microtubules were traced manually using the manual filament tracer option in the Amira Filament editor window.
Quantification of filaments at mitochondrial constriction sites
A region around mitochondria up to 50 nm from the outer mitochondrial membrane (OMM) was analyzed to quantify association of filaments with mitochondria. This region was first classified into constriction and nonconstriction zones in a semiautomated fashion as follows. The principal axis of each mitochondrion was computed by principal-component analysis on the coordinates of all of its segmented voxels. This principal axis was subsequently divided into 100 sections along its length and mitochondrial volumes constituting each section were plotted against the position of those sections along the principal axis. The resulting plot helped to visualize the constriction profile for each mitochondrion, which in turn helped to manually classify the surrounding region into constriction and nonconstriction zones as shown in Figs. 2 and 3. The abundance of a certain type of filament in each of the zones was computed as the percentage occupancy for that filament type in the total number of voxels contained in that zone. After repeating the procedure for 18 mitochondrial profiles, the p value was calculated using the Wilcoxon signed rank test, a nonparametric alternative to the paired Student’s t-test.
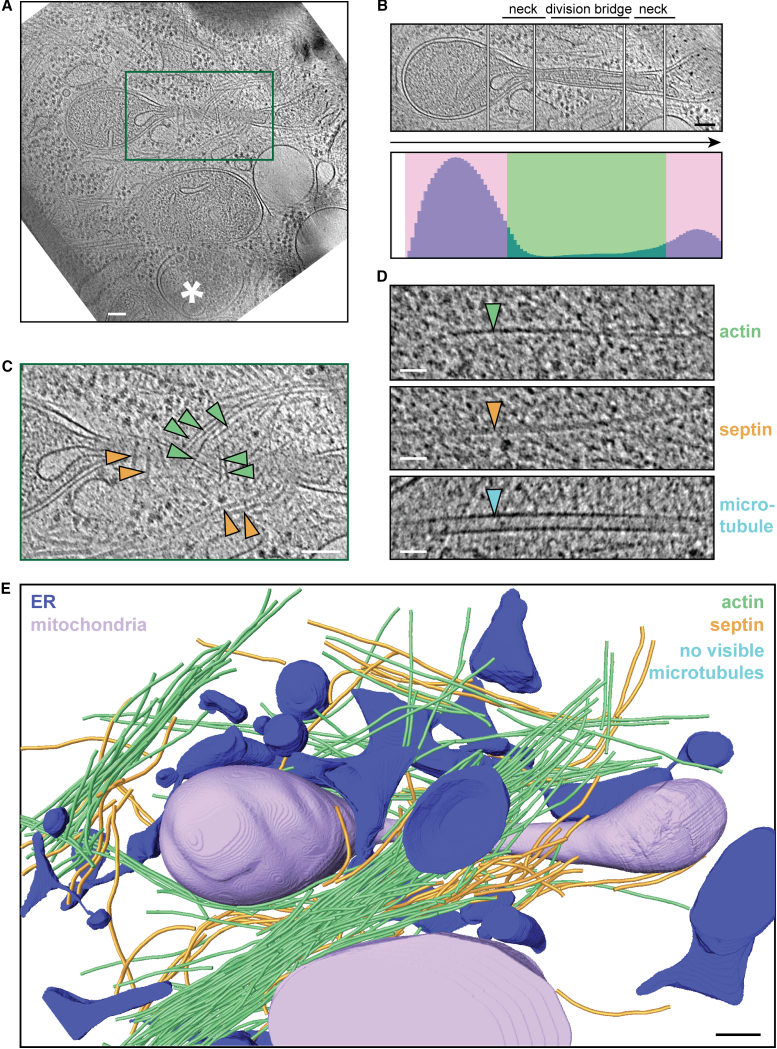
Figure 2.
Ultrastructure of mitochondrial constriction sites. (A) Slice through a 3D tomogram of a dividing mitochondrion in an FCCP-treated MEF cell. The asterisk indicates an autolysosome-like organelle. (B) Montage of slices at different heights from the tomogram in (A) capturing the central axis of the dividing mitochondrion. The corresponding constriction profile is shown below, depicting the number of voxels contained inside the outer mitochondrial membrane (OMM) in each of 100 sections along its principal axis. This profile allows us to visually classify constriction (green) and nonconstriction (pink) zones. (C) Enlargement of the boxed region in (A) highlighting filaments visible at the constriction site. (D) Cytoskeletal filaments distinguishable in our tomograms: actin (green arrowhead; 7–10 nm diameter), septin (orange arrowhead; 10 nm diameter), and microtubules (teal arrowhead; 24 nm diameter). (E) 3D segmented model of the tomogram in (A) showing OMM (purple), endoplasmic reticulum (ER) (blue), actin filaments (green), septin filaments (orange), and microtubule (teal). Scale bars, 100 nm (A–C and E) and 20 nm (D).
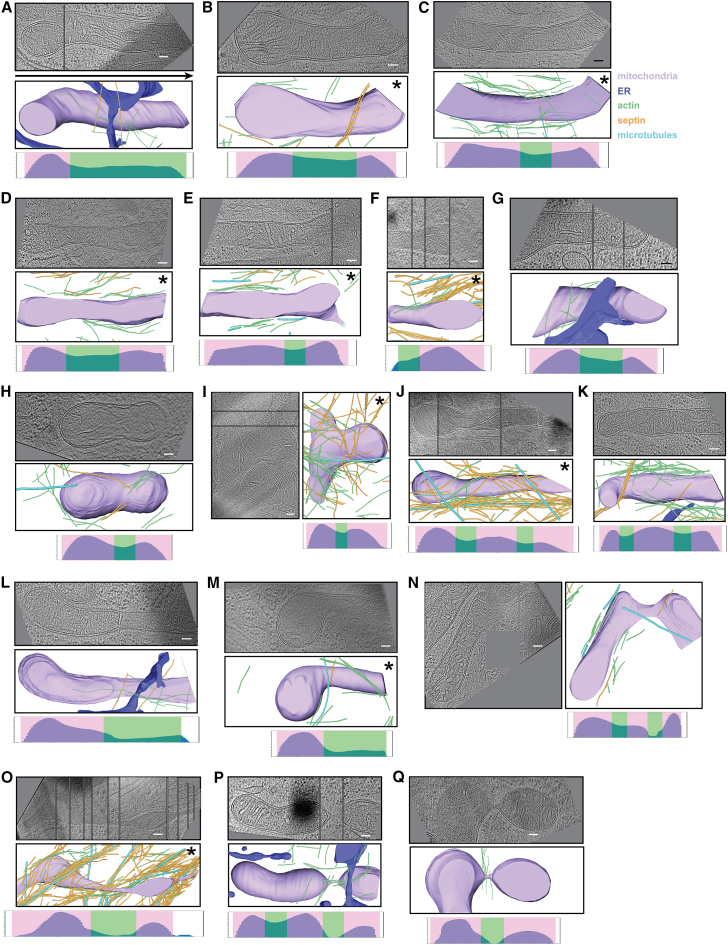
Figure 3.
Gallery of mitochondrial constrictions. (A–Q) Examples of constricting mitochondria observed in FCCP-treated MEF cells (n = 5 cells), arranged approximately in the order of degree of constriction. In each case, a central tomographic slice (or a montage of multiple slices to capture the central axis) is shown, along with a 3D segmentation and constriction profile. Color scheme is the same as in Fig. 2. Asterisks indicate that ER was not segmented. Scale bars, 100 nm.
Quantification of interfilament distances and orientations
For each segmented tomogram, filaments in each class (microtubule, septin, and actin) were subdivided into 4-nm segments using linear interpolation, and both the position and orientation of these segments were recorded as numpy arrays. For each segment on each filament, the distance was measured to every segment on every other filament of the same class and every filament of a different class. The distance as well as the calculated relative orientation of the nearest overall segment for each class was recorded. These measurements were aggregated for all filaments of each class from all tomograms (n = 13, 11, 10, and 11 tomograms for actin-actin, septin-septin, septin-microtubule, and actin-microtubule interactions, respectively) to generate the two-dimensional (2D) histograms in Fig. 4.
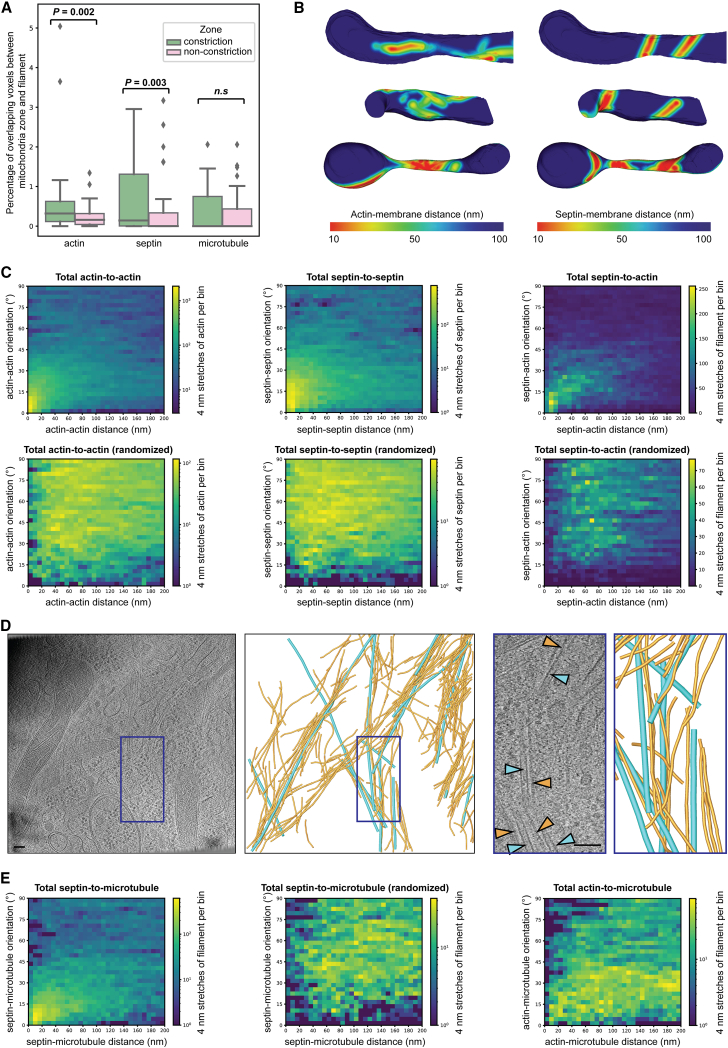
Figure 4.
Cytoskeletal filament architecture at mitochondrial constrictions. (A) Box plots showing the quantification of filament distribution in constriction and nonconstriction zones from 18 mitochondrial profiles. The box limits denote the upper and lower quartiles while the whiskers denote the 1.5× interquartile range. Diamonds show the outliers. The p values were calculated using the Wilcoxon signed rank test. These plots show a significant enrichment of actin and septin filaments, but not microtubules. (B) 3D segmented models of the OMM of the mitochondria shown in Fig. 3L (top), Fig. 3K (middle), and Fig. 2 (bottom). Surfaces are colored according to the proximity of actin (left) and septin (right) filaments. (C) 2D histograms of the relative orientations and distances between neighboring segments of actin (left panels; from 13 tomograms) and septin (middle panels; from 11 tomograms), showing a strong correlation compared with simulated negative controls with randomized positions and orientations. Right panels: 2D histograms of the relative orientations and distances between septin filaments and actin filaments (from 11 tomograms), showing a correlation compared with simulated negative controls with randomized positions and orientations. However, this actin-septin correlation was reduced compared with actin-actin or septin-septin correlations. (D) Slice from a tomogram and 3D segmented model showing close parallel association between septin filaments (orange) and microtubules (teal). The boxed region is enlarged on the right. (E) 2D histograms of the relative orientations and distances between microtubules and septin filaments (left panels; from 10 tomograms) or actin filaments (right panel; from 11 tomograms). Septin, but not actin, filaments show a strong positional and orientational correlation compared with the randomized negative control. Scale bars, 100 nm.
As a negative control for filament-filament interactions, the same analyses were run with randomized filament positions and orientations. To do this, after interpolation, all of the filaments were aligned by their first segment and then randomized such that the first segment was oriented ±30° relative to the growth plane (but otherwise unconstrained). The starting orientations of the filaments were then randomized and, to maintain constant total filament density, they were wrapped using a repeating boundary condition, with distance measurements only considered within the tomogram rather than across boundaries. Each filament maintained the same shape as before randomization, thus enabling randomization while maintaining physiological curvatures.
Interpolation and distance measurement were calculated with optimized functions in scipy (27) and using numpy arrays (28) to maximize performance, allowing comparison of hundreds of microns of interacting filaments in minutes on a single core of a consumer laptop CPU. Plotting was performed with matplotlib (https://doi.org/10.1109/MCSE.2007.55). The python scripts to run these analyses are available at https://github.com/Grotjahnlab/measure_models and are free to use or modify with a BSD license.
Generation of heatmaps for OMM proximities to filaments
Before distance calculations, all segmented models were converted to surfaces in Amira using either the “Generate surface” function for voxel-based OMM segmentations or the “Extract surface from spatial graph” function for filament segmentation in Amira. The “surface distance” function in Amira was used to calculate the distance between segmented surface models of the OMM (surface 1) and cytoskeletal filaments (surface 2). For visual representation of the surface proximity heatmaps, the OMM was colored by setting the “color field” to the output from the “surface distance” calculation, and the “color map” to “Physics.” The range of the color map was adjusted to reflect appropriate minimum and maximum distances.
Quantification of abundance of 10-nm filaments (septin) in wild-type and Sept2 knockdown cells
Automated detection, tracing, and segmentation of these cytoskeletal filaments was performed using the XTracing for Amira extension package using the same parameters for septin filaments (as described above) for both wild-type (n = 14 tomograms) and Sept2 knockdown (n = 14 tomograms) cells. The total length of these filaments was calculated and divided by the total volume of the tomogram to calculate their total abundance in wild-type and Sept2 knockdown cells. The p value was calculated using the Mann-Whitney rank test.
Quantification of intermembrane distance between IMMs and OMMs
These quantifications were performed using IMOD. The long axis was identified for each of the mitochondria and mitochondrial width (diameter of the OMM; perpendicular to the long axis) was measured at different positions along the long axis. The central tomogram section for each of these positions along the mitochondria was used for this purpose and distances were measured center-to-center. Using the same central section, two measurements were made for the intermembrane distance (one on either side of the mitochondrion) at each of the positions mentioned above. These mitochondrial width measurements were plotted against their corresponding intermembrane distances (average of the two values) from a total of 15 mitochondrial constriction profiles in the boxplot-cum-beeswarm plot shown in Fig. 5 C. The p value was calculated using the Mann-Whitney rank test.
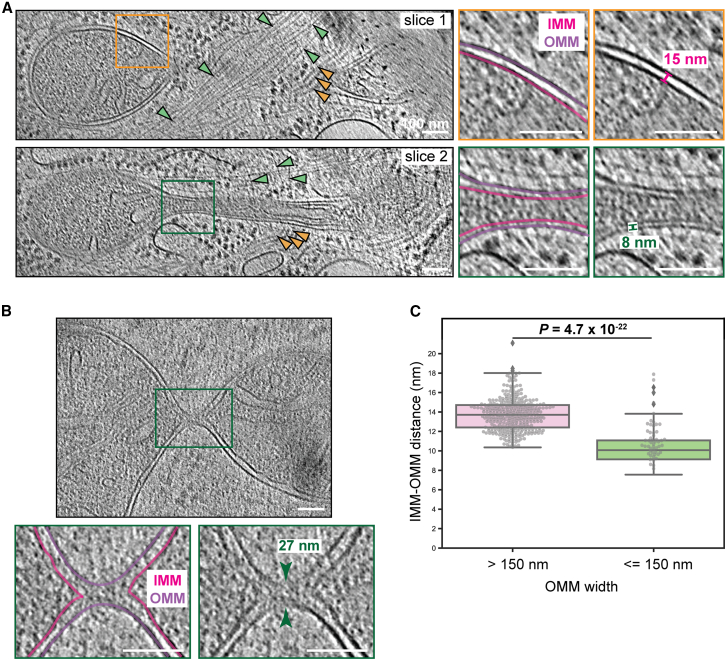
Figure 5.
Synchronous constriction of mitochondrial membranes. (A) Tomogram slices at different heights through the constricting mitochondrion shown in Fig. 2. Actin and septin filaments are indicated by green and orange arrowheads, respectively. Enlargements of the boxed regions are shown at right, highlighting the spacing between the IMM (pink) and OMM (purple). (B) Tomogram slice of the constricting mitochondrion shown in Fig. 3Q. An enlargement of the boxed region is shown below, highlighting complete fission of the IMM (pink) but not OMM (purple). (C) Box and beeswarm plots of IMM-OMM distances along 15 mitochondrial constriction profiles measured at multiple positions where the total width (measured from the center of the OMM on either side) was either >150 nm (pink) or ≤150 nm (green). In the box plots, the box limits denote the upper and lower quartiles while the whiskers denote the 1.5× interquartile range. Diamonds show the outliers. The p value was calculated using the Mann-Whitney rank test. Scale bars, 100 nm.
Results
Capturing cytoskeleton interactions with mitochondria using cryo-ET after membrane depolarization
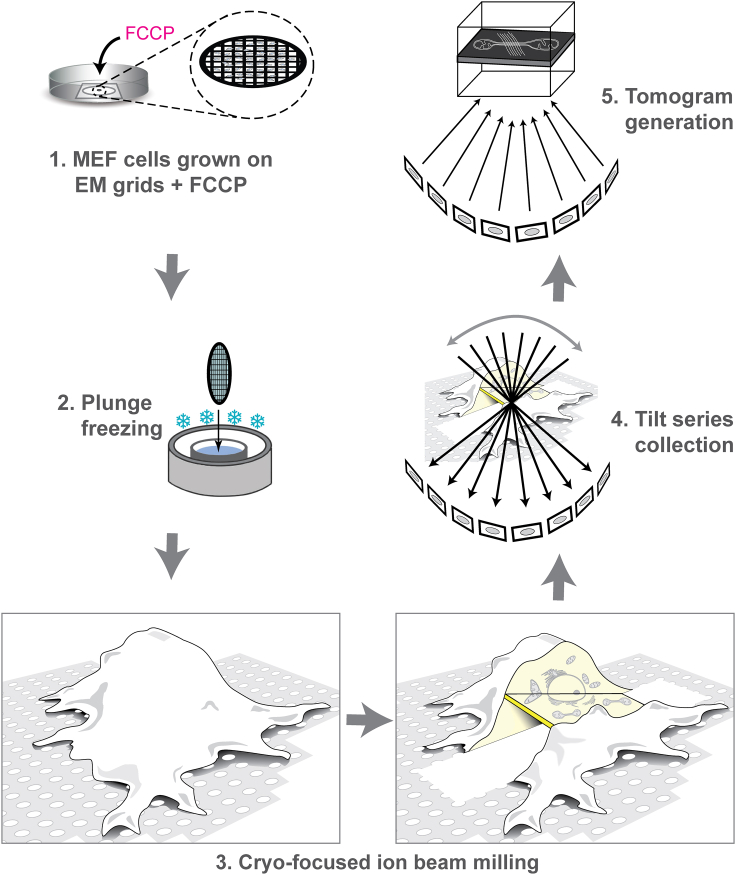
Previous work using fluorescence light microscopy and room temperature transmission electron microscopy showed that depolarization of mitochondrial membrane potential causes mitochondrial shape changes including fragmentation and circularization, as well as accumulation of actin and septin filaments near mitochondria (8,9,10,15,16). However, these approaches are unable to preserve and capture the nanoscale architecture of cytoskeletal filaments and their interactions with mitochondrial membranes. Cryo-ET is complementary to these other techniques in that it can preserve mitochondrial membranes in the process of remodeling along with the interacting cytoskeleton in their near-native state and reveal their artifact-free 3D architecture at nanometer-scale resolution in situ. A previous study showed abundant filamentous actin associating with depolarized mitochondria in ∼5 min after FCCP treatment (8). Therefore, in our experiments, we treated MEF cells with FCCP for 3–6 min on EM grids before vitrifying them (by plunge freezing) for cryo-ET. One major obstacle for directly imaging this sample is its thickness. Volumes thicker than ∼1 μm are electron-opaque, and ideal samples are <200 nm thick (29). In the case of our adherent cells, this corresponds to only the thinnest peripheral regions, a small fraction of the total volume while most of the mitochondrial population is off limits. Moreover, FCCP treatment induces a shift in mitochondria localization from the periphery to the perinuclear region. We therefore used cryo-FIB milling to create ∼100- to 300-nm-thick electron-transparent lamellae in the perinuclear region of vitrified cells before subjecting them to cryo-ET. Our overall workflow is summarized in Fig. 1.
Figure 1.
Experimental workflow imaging mitochondrial fission by cryo-ET. (1) Mammalian cells were grown on fibronectin-coated EM grids for 24–48 h and treated with FCCP for 3–6 min to induce mitochondrial fission. (2) Samples were then plunge frozen and (3) cryo-FIB milled to create thin lamellae randomly positioned near the middle of the cell. (4) Lamellae were imaged by cryo-ET to create tilt series, sets of projection images obtained by iteratively tilting the sample that are used to (5) computationally reconstruct a 3D volume (a tomogram). Images in (3) are adapted from (22).
Using our workflow, we captured 31 tomograms containing 46 mitochondria from 9 distinct cells (Fig. S1) and observed varying mitochondrial morphologies. The vast majority of mitochondria exhibited a tubular (n = 28 mitochondria) morphology while a subset of the population exhibited additional morphologies, including spheroid (n = 8 mitochondria), spherical (n = 4 mitochondria), and ambiguous (unassigned; n = 6 mitochondria) shapes (Fig. S1). Interestingly, the tubular population showed regions where the outer and inner membranes (OMM and IMM, respectively) were simultaneously constricted and showed associations with cytoskeletal filaments. Consistent with these observations, our live-cell fluorescence microscopy experiments (performed on MEF cells grown on glass and treated with FCCP) showed a progressive reduction in hyperfused mitochondria and a concomitant increase in fragmented mitochondria (Fig. S2). Furthermore, staining with FITC-phalloidin revealed the accumulation of actin filaments in the vicinity of mitochondria, especially in the earlier time points of 2–5 min (Fig. S2 B).
Tubular mitochondria exhibit constrictions that were associated with cytoskeletal filaments upon membrane depolarization
To further characterize cytoskeletal interactions with mitochondria, we collected an additional cryo-ET data set from FCCP-treated MEF cells using a Volta phase plate for enhanced contrast and focused on tubular mitochondria (Figs. 2 and 3). We then carefully analyzed 18 tubular profiles of mitochondria that displayed varying degrees of constrictions from 5 distinct cells. We frequently observed both rough and smooth ER compartments organized nearby these constriction sites. In a few cases, the ER was closely positioned within 10 nm of these sites (e.g., Fig. 2 E), but we did not observe ER completely encircling any of them. These proximal ER membranes often appeared as though they were derived from a more expansive pool of the organelle tens or hundreds of nanometers away from these sites suggesting that the ER-mitochondria interaction is non-random and somehow spatially regulated. We also frequently observed what appeared to be autolysosomes in the vicinity of mitochondrial constriction sites (marked with an asterisk in Fig. 2 A). These may be involved in the mitophagy that follows FCCP-induced fission events (30).
As mentioned previously, we also observed extensive association of cytoskeletal filaments with the constriction sites (Figs. 2 C and 3). These filaments were of three distinct types, visually distinguishable by the human eye as well as by diameter-based automated segmentation (Figs. 2 D and S3, A–D). Actin filaments were thin and, due to their helical nature, exhibited a seemingly variable diameter in tomogram slices, between 7 and 10 nm. In unbinned tomograms, these filaments even displayed helical arrangement of monomer-like densities (Fig. S3 D), reminiscent of actin filaments previously published from neurons (31) (Fig. S3 E). Microtubules had a much thicker and uniform diameter of 24 nm. The third class, which exhibited an intermediate thickness of 10 nm, were clearly distinct from the other two types (Figs. 2 D and S3, A–D). Importantly, they showed subtle but definitive differences from actin filaments including their high-resolution features, apparent lack of helicity, and thicker profile.
Our tomograms of another FCCP-treated cell type, namely U2OS cells, showed similar mitochondrial constrictions and associations with the ER and cytoskeletal filaments (Fig. S4 A). Moreover, untreated MEF and INS-1E cells showed a similar ultrastructural architecture at mitochondrial constrictions (Fig. S4 B,C). These observations together suggest that our observations in FCCP-treated MEFs represent a more universal response to mitochondrial depolarization and are likely relevant for other physiological contexts of mitochondrial constriction.
Bundles of long unbranched actin filaments are associated with mitochondrial constrictions
Actin filaments are known to play a key role in mitochondrial dynamics, although its relevant architecture at mitochondrial membranes during depolarization-induced membrane remodeling remains unclear. In all tomograms of constricting mitochondria, we observed abundant actin filaments near constriction sites (Figs. 2, 3, S3, and S5). To quantify this association, we demarcated “constriction zones'' radiating 50 nm out from the OMM at mitochondrial constrictions. We observed significant enrichment of actin filaments in these zones compared with neighboring regions around the mitochondria (Figs. 4 A and S5). By contrast, there was no similar enrichment of microtubules (Figs. 4 A and S5). Interestingly, we rarely found free actin filament ends within the constriction zones, or even beyond in the 1–2-μm field of view of the tomograms (apart from some discontinuities due to minor errors in segmentation), suggesting that these were predominantly long filaments, nucleated elsewhere in the cell. Previously, Arp2/3, a major nucleator of actin branching, was shown to be involved in mitochondrial remodeling triggered by carbonyl cyanide m-chlorophenylhydrazone, another uncoupler of oxidative phosphorylation like FCCP (9). The actin filaments we observed, however, were linear with no observable branching, suggesting that Arp2/3 is not organizing F-actin networks locally at constriction sites.
Actin filaments were oriented mostly obliquely or perpendicular to the long axis of the constricting mitochondria, and were closely associated with the OMM, often within 10 nm (Fig. 4 B). They also displayed a tendency to bundle, a phenomenon we quantified by measuring the distances and relative orientations in 3D between short (4 nm) segments of neighboring actin filaments. The resulting heatmaps revealed a tight correlation in orientation of adjacent filaments, consistent with bundling (Fig. 4 C). As a negative control, we simulated random positions and orientations for the same actin filaments, which caused the relationship to disappear (Fig. 4 C).
Septin filaments associate with mitochondrial constriction necks and interact with microtubules
As mentioned above, we observed another type of filament that was 10 nm wide associating with mitochondrial constriction sites in our tomograms. We hypothesized that Sept2, a septin known to localize to mitochondrial membranes upon FCCP-induced membrane depolarization (15), is important for the formation of these 10-nm-wide filaments. Knocking down Sept2 protein expression in MEF cells (Fig. S6 A) caused a significant decrease in the abundance of these filaments in tomograms (Fig. S6 B), suggesting that these filaments likely contain Sept2, although it is possible that they may also contain additional septin filament subtypes. Interestingly, Sept2 knockdown also altered mitochondrial morphology. The mitochondrial matrix of FCCP-treated Sept2 knockdown cells, unlike that of FCCP-treated wild-type cells, lost its granular appearance (instead appearing smoothly textured; Fig. S6 C) and retained solid-state calcium phosphate granules (indicated by yellow arrows in Fig. S6 C) that were previously characterized by Wolf et al. (32).
While septins have been implicated in mitochondrial dynamics, their precise role is unknown. Mammalian septins—which fall into four families, Sept2, Sept3, Sept6, and Sept7—assemble into linear apolar filaments composed of a repeating octameric unit (33), which further associate with one another to form higher-order assemblies (34,35). We observed numerous long, unbranched septin filaments in our tomograms of constriction sites (Figs. 2, 3, and S3). Like actin filaments, septin filaments were present from the earliest stages of constriction and persisted throughout the process, missing only from the final stage, in which the IMM had separated and only a bridge of OMM remained connected (Fig. 3, P and Q). Similar to actin filaments, no septin filament ends were detected near the constriction sites, suggesting that they were long filaments that originated far from the constricting mitochondria, were arranged perpendicular (Fig. 3, A, B, K, L, M, O, and Q) or oblique/parallel (Fig. 3, C–F, J, and N) to the long axis of the mitochondria, were significantly enriched in constriction zones (Figs. 4 A and S5), and closely associated (≤10 nm) with the OMM (Fig. 4 B). Also similar to F-actin, septin filaments formed tight bundles of parallelly arranged filaments (Fig. 4 C). These septin bundles often partially tracked the local curvature of the OMM independent of the mitochondrial long axis or stage of constriction (e.g., Figs. 2 E, 3, A, B, H, K, and 4 B), consistent with their capacity to bind/sense curved membranes (36). However, we did not observe them to completely encircle constrictions. Although these filaments did not show any preference for a particular stage of constriction, in mitochondria with long division bridges we observed preferential localization of filament bundles to the “neck” regions on either side (or, if both were not visible in the tomogram, at least one side) of the bridge (Fig. 4 B). This localization pattern was notably distinct from that of F-actin (Fig. 4 B).
Intriguingly, we noticed septin filaments interacting with microtubules (e.g., Fig. 3, F, J, and O) both inside and outside constriction zones. When we mapped all septin filaments and microtubules in our tomograms, they exhibited a strong tendency to interact laterally (Fig. 4 D). We observed a strong correlation between the orientation and proximity of adjacent septin filaments and microtubules, confirming this interaction (Fig. 4 E). No similar relationship was observed between actin filaments and microtubules in our tomograms (Fig. 4 E). It is possible that this septin-microtubule interaction is important for microtubule-based transport in conjunction with mitochondrial remodeling. We also observed septin filaments interacting with actin in our tomograms (e.g., Fig. 3, E, F, I, J, and O). Upon quantification, we observed a slight correlation between the orientation and proximity of adjacent septin and actin filaments (Fig. 4 D). However, this relationship is not as strong as that observed between septin filaments and microtubules, indicating a stronger preference for the latter.
IMMs and OMMs constrict synchronously
We next analyzed the relationship between the IMM and the OMM during mitochondrial constriction in the context of FCCP-induced depolarization. While, of course, IMM constriction must precede that of the OMM, the extent to which they are coordinated is unknown (12,37). Two competing models can be considered—the IMM and OMM could be simultaneously constricted and severed by forces originating outside the OMM or, alternatively, the IMM could be constricted independent of the OMM via a dedicated mechanism (12,38). Our tomograms resolved both membranes well at various stages of constriction and revealed a high degree of synchrony between them (Figs. 3 and 5 A). We observed coordinated constriction until the IMM was brought into contact, pinching off and sealing the new compartments on each side (Fig. 5 B). At this point, the remaining neck of the OMM was ∼25 nm wide (Fig. 5 B). Interestingly, we observed a change in the distance between the IMM and OMM at later stages of constriction (Fig. 5 A). We quantified this effect by measuring the intermembrane distance at multiple points along 15 constriction sites. We found that in early (light) constrictions, the intermembrane distance remained unchanged (∼14 nm). However, in deeper constriction profiles with a width of 150 nm or less, this distance decreased to ∼10 nm (Fig. 5 C).
Discussion
In this study, we sought to understand how cytoskeletal filaments may facilitate mitochondrial membrane remodeling in the context of membrane depolarization. Using FCCP treatment followed by cryo-FIB milling and cryo-ET, we captured tomograms of mitochondria exhibiting several morphologies including circularization that was previously reported to be induced upon membrane depolarization via treatment with FCCP (9,10). The majority of mitochondria exhibited tubular shapes with constricted regions along their length that were preferentially associated with ER, actin filaments, and septin filaments. Since these cytosolic factors were previously implicated in large-scale mitochondrial fission and fragmentation upon membrane depolarization (8,13,15,39), we likely captured intermediates of this process. Our snapshots could represent either successful or “frustrated” fission events as previously demonstrated for the constrictions of the mitochondrial inner compartment using time-lapse fluorescence microscopy (38). Even if the constrictions do not proceed to complete membrane scission, it is possible that they could sequester damaged portions of mitochondria during mitophagy. Alternatively, they could contribute to mitochondrial circularization since this morphology often exhibited regions of thin mitochondrial matrix. Regardless of the purpose or the outcome for such dynamic mitochondrial constrictions, our work revealed several common themes underlying their association with cytosolic factors, shedding light on possible mechanisms.
Contrary to a handoff model in which various factors act sequentially, all three factors—ER, actin filaments, and septin filaments—exhibited no clear preference for any particular stage of constriction. This observation is also consistent with previous studies that showed no complete abrogation of fission with the loss of any one of these factors, suggestive of significant overlaps in their function. Previous studies using fluorescence microscopy and biochemical assays found that ER-mitochondria contact sites coincide with sites of mitochondrial fission, mtDNA replication, and mitophagy (39,40,41), implicating ER in mitochondrial constriction. In some reports, ER tubules are proposed to wrap around the mitochondria, contributing to force generation and initiating constriction on their own (42). In our tomograms, however, we observed only partial ER coverage around mitochondrial constrictions. This suggests that the wrapping of ER is likely highly dynamic and short lived to have been captured by our snapshots or is highly context specific.
Several reports have implicated actin filaments in membrane remodeling in distinct physiological contexts although their precise role in mitochondrial dynamics is still unclear. In the context of fission mediated by calcium release (e.g., stimulation with either histamine or ionomycin), actin is nucleated by two formins, an ER-specific isoform of INF2 (12,13) and a formin-domain-containing, OMM-localized Spire1C (43). In contrast, depolarization-induced (e.g., stimulation with FCCP) dynamics is facilitated by the nucleator, Arp2/3. Given the role of actin in force generation for other cellular processes such as cell migration via lamellipodia formation, at least two models, which are not mutually exclusive, have been proposed for actin-mediated force generation in the context of mitochondrial constriction. In one model, linear actin filaments nucleated at the ER polymerize toward the mitochondrion, applying an inward force onto the OMM surface (43). In the other model, ER nucleates and organizes actin filaments circumferentially around a mitochondrion. Bipolar myosin molecules then constrict these actin assemblies by walking on filaments with opposing polarities, thereby applying force onto the trapped mitochondrion (44). Interestingly, a recent platinum replica electron microscopy study revealed neither kind of assemblies for actin filaments and instead reported an interstitial network of filaments around constricting mitochondria, along with a stochastic distribution of myosin II (45). The authors proposed that bipolar myosin activity might produce heterogeneous deformations in the actin network that exert force onto the trapped mitochondria. One consistent theme for all these models is the inward force generation onto the OMM from the cytosol.
Our use of cryo-FIB milling and cryo-ET, which better preserve and reveal cellular membranes and protein macromolecules, revealed the native arrangement of actin at mitochondrial constriction sites to test these models. Consistent with the platinum replica electron microscopy study, we observed actin filaments neither encircling constriction sites nor originating from ER at contact sites upon FCCP treatment. Moreover, the enhanced resolution of our tomograms enabled additional observations on actin filaments at the constriction sites. They displayed abundant bundling, a behavior that is likely important for assembling the filament network at potential constriction sites for subsequent remodeling and force generation. They were also significantly enriched close (≤10 nm) to the OMM, suggesting direct interactions with the constricting mitochondria. Finally, they were unbranched in the vicinity of constrictions and were long, predominantly not originating at nearby ER membranes. These observations suggest that the Arp2/3-dependent nucleation activity, although possibly important for increasing the overall availability of actin filaments, is not locally controlled at mitochondrial constrictions. Interestingly, actin filaments are known to bind Drp1 and enhance its oligomerization at constriction sites (18) while Drp1 increases actin filament bundling (17,18), which we abundantly observe in our tomograms. Thus, another plausible model for actin-mediated constriction is via the recruitment and activity of Drp1. Further studies are required to test these models and to determine other unifying or unique architectural features of actin in distinct physiological contexts of mitochondrial constrictions and triggers for fission.
Septins were recently discovered to participate in depolarization-induced (15) and Shigella infection-induced (16) mitochondrial remodeling, but the details of their involvement, including any interplay with actin, remain unknown. In our tomograms, we observed bundles of long, unbranched, Sept2-dependent filaments that were visibly distinct from actin filaments and concentrated at constriction sites in conjunction with actin filaments upon FCCP treatment. Their arrangement—prominent localization to the neck region of division bridges—differed from that of actin filaments, suggesting distinct roles. We also noted several instances where septin filaments appear to follow the local curvature of mitochondrial membranes (e.g., Figs. 2 E, 3, A, B, H, K, and 4 B), although they do not fully encircle the constricting mitochondria. Septins are known to sense micron-scale membrane curvature (36,46), including at the poles of rod-shaped bacteria (47). A recent study showed that short septin filaments closely associated with the outer membrane of mycobacteria and S. flexneri (∼16–20 nm) as irregularly distributed bundles preferentially arranged perpendicular to the long axis (48). In this study, we observed similar close associations of unbranched bundles of septin filaments with mitochondrial membranes. However, in contrast to the irregular distribution of short filaments around the bacteria observed in the previous study, we observed long septin bundles preferentially positioned close to the neck of constrictions in some instances (Fig. 4 B). It is therefore possible that the septin filaments we observed in our tomograms are sensing membrane curvature; if not for driving remodeling directly, they are perhaps used to reinforce curvature by physically preventing the OMM from relaxing back to a larger diameter. Consistent with such a role, septins are known to participate in another membrane deformation process: cytokinesis in yeast (49,50). In that system, septins organize into diverse assemblies including long linear filaments, circumferential rings, and a gauze-like meshwork. However, the precise role in membrane remodeling for these arrangements and the linear septin filaments we observed in the context of mitochondrial constrictions are currently unclear. In addition to potentially driving membrane constriction, septins may also recruit Drp1 to assist in the process (15,16). Furthermore, the altered mitochondrial morphology in Sept2 knockdown cells suggests additional functions in mitochondrial homeostasis.
Interestingly, our tomograms revealed septin-microtubule and septin-actin interactions (although the latter was comparatively weaker). These interactions may bring microtubules to the sites of constriction and provide a link between actin and microtubules to mediate their polymerization dynamics at mitochondrial constrictions (similar to a phenomenon recently observed in neuronal growth cones) (51). It is known that septins can spatially guide microtubules and regulate their plus-end dynamics, and this was attributed to the presence of septin 9 (52,53,54). Our observations therefore suggest that septin filaments in our tomograms may contain Sept9. Septin-microtubule interaction has been shown to modulate microtubule motor activity, which can in turn be used for trafficking functions (55); for example, it can help deliver fission components, move divided mitochondria apart after fission or deliver damaged mitochondria undergoing mitophagy to autophagosomes/autolysosomes. A previous study found pieces of mitochondria transported away from the sites of mitochondrial damage for mitophagy following fission (56), consistent with our observations of autolysosomes near constriction sites.
Finally, our imaging was able to resolve the inner and outer membranes of mitochondria with sufficient resolution during constriction. Our observations here of synchronous membrane constriction and compression of the intermembrane space at the later stages of constriction are consistent with a model in which FCCP-induced IMM constriction is driven predominantly by OMM constriction via flexible protein spacers between the two membranes such as the mitochondrial intermembrane space bridging complex (57). This also suggests a change in forces or kinetics at the later stages, perhaps associated with recruitment of known late-acting factors such as the dynamin proteins. Alternatively, if IMM constricts via a dedicated mechanism independent of OMM (12,38), it is unclear how synchronization is achieved. Ultimately, when the channel inside the IMM becomes sufficiently narrow (on the order of a few nanometers), the IMM fuses, presumably followed by a similar process in the OMM to complete fission.
Conclusion
Together, this study has demonstrated cryo-ET’s ability to reveal mitochondrial membrane-cytoskeletal interactions at high resolution. Our work lays the groundwork for cellular cryo-ET to be an integral part of structural and functional studies on mitophagy and mitochondrial membrane dynamics in distinct physiological contexts or pharmacological treatments. We can determine coordination between various factors and derive plausible models for force generation by integrating cryo-ET with complementary experimental and computation tools including correlative light and electron microscopy and coarse-grained molecular dynamics simulations.
Author contributions
Conceptualization, S.K.M., D.A.G., W.Y.Y., and G.J.J.; methodology, S.K.M. and D.A.G., sample preparation, S.K.M., D.A.G., M.M., and W.Y.Y.; data collection, S.K.M., D.A.G., M.M., and W.Y.Y.; segmentation, S.K.M., D.A.G., and M.J.D.; data analysis, S.K.M., D.A.G., X.Z., B.A.B., M.M., and M.H.H.; software/code development, S.K.M., X.Z., and B.A.B.; figure generation, S.K.M., D.A.G., X.Z., and B.A.B.; writing, S.K.M., D.A.G., X.Z., and M.H.H.; editing. S.K.M., D.A.G., X.Z., B.A.B., Y.-W.C., X.M., W.Y.Y., and G.J.J.; supervision, D.A.G., Y.-W.C., M.X., and G.J.J.
Acknowledgments
We thank S. Chen and A. Malyutin at the California Institute of Technology cryo-EM facility and Bill Anderson at The Scripps Research Institute electron microscopy facility for microscope support, and Jean-Christophe Ducom at The Scripps Research Institute for computational support. We thank A. Collazo and S. Wilbert for technical assistance with confocal microscopy. We thank Catherine Oikonomou for her critical input on the manuscript. The confocal imaging was performed at the Biological Imaging Facility at Caltech, and the cryo-EM imaging was performed at the Beckman Institute Resource Center for Transmission Electron Microscopy at Caltech and the Scripps Research Institute Hazen Cryo-EM Microscopy Suite. this work was supported by NIH grant P50-AI150464 (to G.J.J.), the Nadia’s Gift Foundation Innovator Award from the Damon Runyon Cancer Foundation (DRR-65-21 to D.A.G.), NIH grant R01GM134020 and NSF grants DBI-1949629 and NSF IIS-2007595 (to M.X.), a David and Lucile Packard Fellowship for Science and Engineering (2019-69645) and NIH grants RM1GM136511 and R01GM134020 (to Y.-W.C.), and a Philadelphia Center off-campus study program award to M.H.H.
Declaration of interests
The authors declare no competing interests.
Editor: Carolyn Moores.
Footnotes
Shrawan Kumar Mageswaran and Danielle Ann Grotjahn contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.07.030.
Contributor Information
Danielle Ann Grotjahn, Email: grotjahn@scripps.edu.
Grant J. Jensen, Email: grant_jensen@byu.edu.
Supporting material
References
- 1.Osellame L.D., Blacker T.S., Duchen M.R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metabol. 2012;26:711–723. doi: 10.1016/j.beem.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 3.Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 4.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 6.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serasinghe M.N., Chipuk J.E. Mitochondrial Fission in Human Diseases. Handb. Exp. Pharmacol. 2017;240:159–188. doi: 10.1007/164_2016_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., Xu S., et al. Karbowski M. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol. 2015;208:109–123. doi: 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung T.S., Ji W.K., et al. Chakrabarti R. Two distinct actin filament populations have effects on mitochondria, with differences in stimuli and assembly factors. J. Cell Sci. 2019;132 doi: 10.1242/jcs.234435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazono Y., Hirashima S., et al. Ohta K. Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fission-independent manner. Sci. Rep. 2018;8:350. doi: 10.1038/s41598-017-18582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti R., Ji W.K., et al. Higgs H.N. INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol. 2018;217:251–268. doi: 10.1083/jcb.201709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korobova F., Ramabhadran V., Higgs H.N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chhabra E.S., Higgs H.N. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- 15.Pagliuso A., Tham T.N., et al. Stavru F. A role for septin 2 in Drp1-mediated mitochondrial fission. EMBO Rep. 2016;17:858–873. doi: 10.15252/embr.201541612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sirianni A., Krokowski S., et al. Mostowy S. Mitochondria mediate septin cage assembly to promote autophagy of Shigella. EMBO Rep. 2016;17:1029–1043. doi: 10.15252/embr.201541832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatch A.L., Ji W.K., et al. Higgs H.N. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol. Biol. Cell. 2016;27:3109–3121. doi: 10.1091/mbc.E16-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji W.K., Hatch A.L., et al. Higgs H.N. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015;4 doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smirnova E., Griparic L., et al. van der Bliek A.M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tivol W.F., Briegel A., Jensen G.J. An improved cryogen for plunge freezing. Microsc. Microanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigort A., Villa E., et al. Plitzko J.M. Integrative approaches for cellular cryo-electron tomography: correlative imaging and focused ion beam micromachining. Methods Cell Biol. 2012;111:259–281. doi: 10.1016/B978-0-12-416026-2.00014-5. [DOI] [PubMed] [Google Scholar]
- 22.Villa E., Schaffer M., et al. Baumeister W. Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography. Curr. Opin. Struct. Biol. 2013;23:771–777. doi: 10.1016/j.sbi.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 25.Agulleiro J.I., Fernandez J.J. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- 26.Rigort A., Günther D., et al. Hege H.C. Automated segmentation of electron tomograms for a quantitative description of actin filament networks. J. Struct. Biol. 2012;177:135–144. doi: 10.1016/j.jsb.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Virtanen P., Gommers R., et al. van Mulbregt P., SciPy 10 Contributors SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris C.R., Millman K.J., et al. Oliphant T.E. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro P.P. Quantitative Cryo-Electron Tomography. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.934465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vives-Bauza C., Zhou C., et al. Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hylton R.K., Heebner J.E., et al. Swulius M.T. Cofilactin filaments regulate filopodial structure and dynamics in neuronal growth cones. Nat. Commun. 2022;13:2439. doi: 10.1038/s41467-022-30116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf S.G., Mutsafi Y., et al. Fass D. 3D visualization of mitochondrial solid-phase calcium stores in whole cells. Elife. 2017;6 doi: 10.7554/eLife.29929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertin A., McMurray M.A., et al. Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Booth E.A., Sterling S.M., et al. Thorner J. Effects of Bni5 Binding on Septin Filament Organization. J. Mol. Biol. 2016;428:4962–4980. doi: 10.1016/j.jmb.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia G., 3rd, Bertin A., et al. Nogales E. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J. Cell Biol. 2011;195:993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridges A.A., Jentzsch M.S., et al. Gladfelter A.S. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J. Cell Biol. 2016;213:23–32. doi: 10.1083/jcb.201512029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagliuso A., Cossart P., Stavru F. The ever-growing complexity of the mitochondrial fission machinery. Cell. Mol. Life Sci. 2018;75:355–374. doi: 10.1007/s00018-017-2603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho B., Cho H.M., et al. Sun W. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun. 2017;8 doi: 10.1038/ncomms15754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman J.R., Lackner L.L., et al. Voeltz G.K. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis S.C., Uchiyama L.F., Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 2016;353:aaf5549. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowland A.A., Voeltz G.K. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manor U., Bartholomew S., et al. Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 2015;4 doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatch A.L., Gurel P.S., Higgs H.N. Novel roles for actin in mitochondrial fission. J. Cell Sci. 2014;127:4549–4560. doi: 10.1242/jcs.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C., Svitkina T.M. Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat. Cell Biol. 2019;21:603–613. doi: 10.1038/s41556-019-0313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMurray M.A. The long and short of membrane curvature sensing by septins. J. Cell Biol. 2019;218:1083–1085. doi: 10.1083/jcb.201903045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krokowski S., Lobato-Márquez D., et al. Mostowy S. Septins Recognize and Entrap Dividing Bacterial Cells for Delivery to Lysosomes. Cell Host Microbe. 2018;24:866–874.e4. doi: 10.1016/j.chom.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobato-Márquez D., Xu J., et al. Mostowy S. Mechanistic insight into bacterial entrapment by septin cage reconstitution. Nat. Commun. 2021;12:4511. doi: 10.1038/s41467-021-24721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertin A., McMurray M.A., et al. Nogales E. Three-dimensional ultrastructure of the septin filament network in Saccharomyces cerevisiae. Mol. Biol. Cell. 2012;23:423–432. doi: 10.1091/mbc.E11-10-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ong K., Wloka C., et al. Bi E. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat. Commun. 2014;5:5698. doi: 10.1038/ncomms6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakos K., Alam M.N.A., et al. Spiliotis E.T. Septins mediate a microtubule-actin crosstalk that enables actin growth on microtubules. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2202803119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowen J.R., Hwang D., et al. Spiliotis E.T. Septin GTPases spatially guide microtubule organization and plus end dynamics in polarizing epithelia. J. Cell Biol. 2011;194:187–197. doi: 10.1083/jcb.201102076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakos K., Rosenberg M., Spiliotis E.T. Regulation of microtubule plus end dynamics by septin 9. Cytoskeleton (Hoboken) 2019;76:83–91. doi: 10.1002/cm.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sellin M.E., Stenmark S., Gullberg M. Mammalian SEPT9 isoforms direct microtubule-dependent arrangements of septin core heteromers. Mol. Biol. Cell. 2012;23:4242–4255. doi: 10.1091/mbc.E12-06-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spiliotis E.T., Kesisova I.A. Spatial regulation of microtubule-dependent transport by septin GTPases. Trends Cell Biol. 2021;31:979–993. doi: 10.1016/j.tcb.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J.Y., Yang W.Y. Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nat. Commun. 2013;4:2428. doi: 10.1038/ncomms3428. [DOI] [PubMed] [Google Scholar]
- 57.Ott C., Ross K., et al. Kozjak-Pavlovic V. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.