Abstract
Membrane cholesterol-rich domains have been shown to be important for regulating a range of membrane protein activities. Low-density lipoprotein receptor (LDLR)-mediated internalization of cholesterol-rich LDL particles is tightly regulated by feedback mechanisms involving intracellular sterol sensors. Since LDLR plays a role in maintaining cellular cholesterol homeostasis, we explore the role that membrane domains may have in regulating LDLR activity. We expressed a fluorescent LDLR-mEGFP construct in HEK293T cells and imaged the unligated receptor or bound to an LDL/DiI fluorescent ligand using total internal reflection fluorescence microscopy. We studied the receptor’s spatiotemporal dynamics using fluorescence fluctuation analysis methods. Image cross correlation spectroscopy reveals a lower LDL-to-LDLR binding fraction when membrane cholesterol concentrations are augmented using cholesterol esterase, and a higher binding fraction when the cells are treated with methyl-β-cyclodextrin) to lower membrane cholesterol. This suggests that LDLR’s ability to metabolize LDL particles is negatively correlated to membrane cholesterol concentrations. We then tested if a change in activity is accompanied by a change in membrane localization. Image mean-square displacement analysis reveals that unligated LDLR-mEGFP and ligated LDLR-mEGFP/LDL-DiI constructs are transiently confined on the cell membrane, and the size of their confinement domains increases with augmented cholesterol concentrations. Receptor diffusion within the domains and their domain-escape probabilities decrease upon treatment with methyl-β-cyclodextrin, consistent with a change in receptor populations to more confined domains, likely clathrin-coated pits. We propose a feedback model to account for regulation of LDLR within the cell membrane: when membrane cholesterol concentrations are high, LDLR is sequestered in cholesterol-rich domains. These LDLR populations are attenuated in their efficacy to bind and internalize LDL. However, when membrane cholesterol levels drop, LDL has a higher binding affinity to its receptor and the LDLR transits to nascent clathrin-coated domains, where it diffuses at a slower rate while awaiting internalization.
Significance
We propose a new, rapid-response, regulatory model for the low-density lipoprotein receptor (LDLR) that is based on its membrane localization. Using image correlation analysis on fluorescence images taken from fixed HEK293T cells, we detect that LDL-LDLR binding fractions are dependent on membrane cholesterol levels. Furthermore, fluorescence fluctuation analyses of live-cell experiments reveal a membrane cholesterol-dependent change in LDLR’s confinement and transport dynamics. Our proposed model features a negative-feedback loop where LDLR is sequestered within cholesterol-rich reservoirs in a low-functional form and, as membrane cholesterol concentrations decrease, is released from the reservoirs as active receptors. We discuss the possible reasons for changes in the receptor’s membrane dynamics, localization, and the implications of this proposed model for the regulation of cellular cholesterol homeostasis.
Introduction
A high concentration of low-density lipoprotein (LDL) particles in the blood sera is one of the causative agents of coronary artery disease (1). Hepatic LDL receptor (LDLR), a membrane protein that is the primary regulator of LDL, metabolizes up to 75% of LDL particles (2,3). In fact, homozygous mutations of the LDLR in hypercholesterolemia patients have been linked to a 5- to 13-fold increase in plasma LDL concentration compared with their wild-type counterparts (4). Therefore, there are important efforts to classify and understand the regulatory mechanisms that underlie LDLR expression and function.
Intracellular cholesterol concentrations tightly control LDLR expression on the cell membrane to maintain cellular cholesterol homeostasis (3,5). When cholesterol levels in the endoplasmic reticulum fall below 5% of the total lipid content, the sterol response element-binding protein 2 (SREBP2) pathway is activated, promoting the transcription of LDLR mRNA (6). The SREBP2 pathway will also promote the transcription of proprotein convertase subtilisin/kexin type 9 (PCSK9), a secreted protein that binds to LDLR on the cell membrane and reroutes the receptor to lysosomal degradation. PCSK9 is coregulated with LDLR to quickly downregulate cell surface LDLR, acting as a break to prevent accumulation of LDL (3,7). Conversely, an excess of intracellular cholesterol activates the liver X receptor (LXR) pathway and promotes the transcription of the inducible degrader of LDLR (IDOL). IDOL is a E3 ubiquitin ligase that catalyzes the ubiquitination of LDLR’s cytoplasmic domain and targets it to lysosomal degradation (8). Since total cellular cholesterol plays a key role in LDLR expression and function, we explored the roles that membrane cholesterol may have in regulating LDLR transport and localization on the cell membrane.
Cholesterol-rich domains on the cellular membrane, sometimes called lipid rafts, are rigid and ordered regions that mediate specific interactions between saturated and glycosylated lipids with cholesterol and other sphingolipids. Lipid rafts can form domains that range in size from approximately 10 nm to greater than 300 nm in diameter and have been shown to play an important role in regulating receptor and signal transduction proteins, such as those related to immune cell regulation, host-pathogen interfaces, and cancer signaling (9).
Recent studies suggest that membrane cholesterol concentrations can regulate the spatiotemporal regulation of LDLR expression and function. Using a Triton X solubilization assay, Ivaturi et al. (10) demonstrated that, if the cellular membrane is supplemented with extra cholesterol, the population of LDLR shifts to Triton-insoluble regions of the cell membrane, usually associated with cholesterol-rich domains. They further show that incubation with LDL particles lowers uptake after 30 min of incubation. Moreover, Sorrentino et al. (11) show that IDOL-dependent degradation of LDLR selectively occurs in cholesterol-rich regions of the cell membrane. Similar experiments showed the LDLR-related protein (LRP1), a structurally similar protein to LDLR (12), to be associated with lipid rafts (13).
In this study, we use total internal reflection fluorescence (TIRF) microscopy to explore the role that cholesterol-rich domains may play on the function and lateral movement of unligated LDLR and LDL-bound LDLR fluorescent constructs on fixed and living human embryonic kidney 293T (HEK293T) cells. Since the advent of fluorescence microscopy, there have been many analysis techniques developed that have facilitated an understanding of the spatiotemporal regulation of membrane proteins. Ghosh and Webb initiated studies that pioneered the automated tracking of single fluorescent particles by using a fluorescently labeled DiI-LDL construct to track membrane transport and measure the temperature-dependent diffusion coefficients of an internalization-deficient LDLR (14). Single-particle tracking has since been widely used to study membrane confinement, and has revealed membrane domains that are dependent on lateral membrane protein interactions (15), cytoskeleton “fences” that trap membrane proteins (16,17), or lipid rafts (18,19). Single fluorescent particle tracking requires the low-density labeling of target receptor proteins and bright, photostable fluorophores to allow tracking with minimal photobleaching or blinking to resolve sufficiently long trajectories for accurate analysis (20,21).
Fluorescence fluctuation analysis methods provide an alternative way to study the transport dynamics and interactions of labeled membrane receptor proteins at expressed or labeled at higher surface densities. The first of these techniques, fluorescence correlation spectroscopy (FCS), was first described by Magde et al. (22) in 1972 and relies on an autocorrelation analysis of fluorescence intensity fluctuations arising from changes in the number of fluorescent molecules excited within a small focal spot of an excitation laser beam within the sample. The fluorescence fluctuations can be due to fluorophore transport dynamics, chemical reaction kinetics, or photophysical processes. Transport parameters and the concentrations of fluorescent molecules are measured by calculating a time correlation function from the fluctuation time series and fitting the time correlation function with an appropriate fit model. There have been many extensions of FCS that have been used to study membrane domains and compartmentalization. Among them, stimulated emission depletion-FCS acquires FCS correlation curves at super-resolved spatial scales, allowing the analysis to be sensitive to nanoscale compartmentalization on the plasma membrane (23,24). Furthermore, imaging FCS and binned-imaging FCS (ITIR-FCS and bimFCS) use a camera-based approach and treat pixels or binned pixels as the FCS observation value. In these techniques, the characteristic diffusion time changes with the observation area as pixels are binned together, and the trend reveals information about the trapping of proteins on the cell membrane, such as the proteins’ diffusive mode at different spatial scales (25,26,27,28,29).
Image correlation spectroscopy (ICS) is an imaging analog of FCS (30). It entails the spatial and temporal correlation of fluorescence fluctuations sampled in an image time series via fluorescence microscopy and technique variants measure molecular transport, concentrations, and molecular interactions within a cell. The fluorescence fluctuations in the image series can be analyzed in space (ICS), time (TICS), or a combination (spatiotemporal ICS [STICS]). Spatial ICS is commonly used to determine fluorophore densities on the cell membrane, while its two-color counterpart, image cross correlation spectroscopy (ICCS) is used to measure the fraction of colocalized molecules labeled with two different fluorophores imaged at two different emission wavelengths (31,32,33). Both TICS and STICS have been used to measure protein diffusion, flow, and immobilization fractions on the cell membrane (34,35). Work by Di Rienzo et al. extended STICS analysis to classify membrane protein diffusion as free, confined, or transiently confined by calculating an image-averaged mean-square displacement (iMSD) of the labeled proteins as measured from the time-dependent spreading of the space-time correlation function peak fit variance (36).
In this study, we imaged HEK293T cells expressing LDLR-mEGFP either in its unligated form or bound with LDL-DiI ligand. We use STICS/iMSD to measure LDLR’s lateral mobility and trapping in putative lipid domains on the cell membrane in normal growth conditions at 37°C, and how it changes when membrane cholesterol concentrations are altered. Spatial ICS was applied to measure the LDLR protein’s density at the cell membrane after treatment with drugs that change membrane cholesterol levels. Two-channel spatial ICCS analysis was also applied to quantify the fraction of LDLR-mEGFP bound to LDL-DiI under different membrane cholesterol concentrations. From these results, we postulate that LDLR’s membrane environment changes as a function of the cholesterol concentrations, and that these membrane environments function to regulate LDLR’s ability to bind its LDL ligand. We propose that cholesterol-rich domains act as LDLR reservoirs that sequester and attenuate LDLR activity when cholesterol concentrations are high. However, after membrane cholesterol depletion, LDLR has a higher binding affinity for LDL particles and the receptor-ligand complexes now become confined within smaller domains with slower diffusive transport; most likely clathrin-coated pits, where it can efficiently metabolize the LDL particles via the canonical clathrin-mediated endocytosis. Once LDL particles internalize and release cholesterol molecules, cholesterol concentrations can be replenished on the cell membrane. This model accounts for a negative feedback mechanism that regulates LDLR activity based on membrane cholesterol concentrations and may help to maintain cellular cholesterol homeostasis in mammalian cells.
Materials and methods
Cell culture
The regular tissue culture media to passage HEK293T cells was made from Dulbecco’s modified Eagle’s medium supplemented with 0.02 M HEPES buffer solution, 2 mM L-glutamine, 10% fetal bovine serum (FBS), 100 units/mL penicillin/streptomycin, 0.1 mM nonessential amino acids (Gibco, Waltham, MA). Cells were cultured on 100 mm tissue culture dishes (VWR, Randor, PA) and maintained in a humidified incubator with 5% CO2 at 37°C.
Cellular treatments
Before imaging, cells were plated onto poly-L-lysine-coated 35 mm glass-bottom plates (Matsunami Glass, Bellingham, WA). At 24 h after plating, cells were transfected with pRK5 plasmids (Addgene, Watertown, MA) encoding LDLR-mEGFP protein constructs (Genescript, Piscataway, NJ). Cells were starved for 24 h before imaging by incubating in regular culturing medium without FBS added.
For ligand-bound and -unbound receptor imaging experiments, the cell medium was supplemented with either 2 mM methyl-β-cyclodextrin (MβCD) (Sigma-Aldrich, St. Louis, MO) to reduce membrane cholesterol (37) or 0.2 units/mL cholesterol esterase (CE) (Sigma-Aldrich) for 2 h to augment membrane cholesterol (38). Negative control cells were incubated for another 2 h in regular FBS-negative media. Unbound receptor was imaged at the 2 h incubation point. For ligand-receptor complex imaging, treatment medium was replaced with FBS-negative culturing medium supplemented with 5 mg/mL fluorescent LDL-DiI complex (Invitrogen Technologies, Eugene, OR) for 1 h before imaging. For fixed cell experiments, samples were incubated at room temperature with 4% paraformaldehyde for 10 min and then washed with Dulbecco’s phosphate buffer solution three times before imaging (Gibco).
TIRF imaging and analysis
All imaging was performed using a home-built TIRF microscope that was constructed around an inverted Olympus IX71 microscope base (Olympus, Tokyo, Japan) equipped with a TIRF 100×, 1.49 NA oil immersion objective lens (Nikon, Tokyo, Japan). The LDLR-mEGFP fluorescence was excited using an OBIS 488 nm LS laser and the LDL-DiI was excited using an OBIS 561 nm LS laser (Coherent, Santa Clara, CA). The excitation and emission wavelengths were separated using a TRF69901-EM - ET - 405/588/561 nm Laser Triple Band Set for TIRF application filter cube (Chroma, Bellows Falls, VT). Fluorescence signals were imaged on an Andor iXON Ultra 888 EMCCD camera (SnowHouse Solutions, Lac Beauport, Quebec, Canada). Live-cell experiments were performed at 37°C using a temperature-controlled enclosure around the imaging space. Fixed-cell experiments were performed at room temperature, and only one frame was used for analysis from each cell imaged in the fixed samples.
ICS analysis
All image correlation analysis was carried out on standard PC computers using MATLAB R2019 or MATLAB R2020 (The MathWorks Natick, MA). Code for ICS analysis can be found at https://github.com/stevekochscience/Image-Correlation-Spectroscopy. Regions of interest for ICS analysis were picked around homogenous fluorescence areas of the image.
General ICS theory
ICS-related techniques, including their spatial (39), temporal (40), two-color (41), and spatiotemporal (35) variants, have been thoroughly reported previously. Here, we present a general outline of the techniques used in this study.
ICS is based on correlation analysis of fluorescence intensity fluctuations that are recorded within images collected with a fluorescence microscope. The two-dimensional image acquired at each time point is a result of the convolution between a point emitter (in this case, the mEGFP protein fluorophore or the DiI dye) and the point-spread function (PSF) of the fluorescence microscope. The image pixel size is smaller than the PSF size so the imaged diffraction spot spreads over several pixels, which encodes spatial correlation between adjacent pixels. The intensity value of each pixel in the image matrix will represent the integration of photons (photoelectrons) collected by the optics and detected by the image detector over the image acquisition time. The intensity of each pixel will vary based on the number of emitting fluorescent molecules detected within each PSF-sized focal volume (30), which is the source of the signal fluctuations, in addition to fluctuations due to noise sources.
Most of the techniques in this study can be represented as simplified variants of a generalized spatiotemporal correlation function:
| (1) |
where the angular brackets in the numerator symbolize spatial correlation followed by temporal correlation, ξ and η are the spatial lag (pixel shift) variables and τ is the temporal lag (image frame shift). The angular brackets in the denominator indicate spatial intensity averages (r space) over the region of interest at times t and t + τ. The subscripts a and b refer to images collected in different wavelength detection channels on the microscope. In the case of the analysis being performed on the same channel, we can simplify a = b, which is an autocorrelation function (30). The fluorescence intensity fluctuation of each pixel, δi(x,y,t), is defined as its difference from the mean intensity,
| (2) |
where i is the intensity of pixel x, y at time t, and the angular brackets represent the spatial average intensity of the image region of interest at image frame time t. The different forms of ICS are limits of Eq. 1 with correlations over different lag variables and one or two detection channels and this will dictate which fit model is applied, and the parameters (dynamic or static) extracted from the fit.
Receptor density assays using spatial ICS
Spatial image correlation was used to measure receptor densities in the cell membrane. This technique depends on a two-dimensional spatial correlation analysis of the image for a fixed time (single frame). Eq. 3 is a discrete approximation to the spatial autocorrelation function.
| (3) |
Where Δx and Δy are the discrete pixel lag shifts in the correlation calculation and X and Y are the number of pixels spanning the region of interest in a single image frame. We fit this correlation function using a nonlinear least-squares fit of a two-dimensional Gaussian with three free fitting parameters,
| (4) |
The fitting variables are the zero-lags amplitude, g(0,0,0), the e−2 decay radius of the correlation function, ω0, and offset g∞, which accounts for incomplete decay of the correlation function due to long-range spatial correlations. The mean number of independent fluorescent particles <N> per focal area is inversely proportional to the zero lags amplitude g(0,0,0) if image background is corrected for (38). The correlation area can be calculated from the ω0 radius and, therefore, we can calculate the cluster density (CD) of fluorescent particles per unit area:
| (5) |
Ligand binding assays using ICCS
Spatial ICCS was used to measure the interaction fraction of LDL-DiI particle binding to LDLR-mEGFP receptors (41). In these assays, cells were incubated with 4% PFA and imaged sequentially on the TIRF microscope using different detection channels (a = LDLR-mEGFP detection channel and b = LDL-DiI detection channel). The normalized spatial cross correlation function is given by Eq. 6:
| (6) |
We fit Eq. 6 with the same two-dimensional Gaussian from Eq. 4, with the caveat that the zero-lag variable (gab(0,0,0)) represents the amplitude of the cross correlation function. The average density of colocalized independently fluorescing particles within a beam area is given by Eq. 7.
| (7) |
where gaa(0,0,0) and gbb(0,0,0) are the respective best fit autocorrelation amplitudes in the two independent detection channels. The fraction of bound receptors (M1) is then calculated by dividing Na,b with the number density of the receptor, as calculated from the spatial autocorrelation function of channel a (Eq. 4):
| (8) |
Confinement assays using STICS and iMSD
To find the diffusive mode and diffusion parameters of the LDL receptor in its bound and unbound forms, we performed an STICS/iMSD analysis on image series acquired at 20 ms/frame for 50 frames. We start by defining with the general STICS formula,
| (9) |
STICS was first described by Hebert et al. (35) and was used to measure either isotropic diffusion or directed velocity dynamics of fluorescent particles. More recently, Di Rienzo et al. (36) adapted STICS in the context of Fick’s second law and the FCS diffusion laws method (42) to determine the diffusive and confined transport nature of membrane proteins. We fit our STICS autocorrelation functions with a nontranslating two-dimensional Gaussian with three fit parameters,
| (10) |
where gd is the zero-lag variable and decays with time (gd(τ) = (4πDτ)−1) and the Gaussian standard deviation term σ(τ) represents the average two-dimensional iMSD of the fluorescently labeled receptors as a function of time. The first lag of σ2(0) is proportional to the e−2 radius of the microscope’s PSF.
We recorded the iMSD value for each time lag and fit the iMSD versus time plots according to three different diffusion models (36). For a protein that undergoes free diffusion on the cell membrane, the iMSD plot is linear in time:
| (11) |
where D is the diffusion coefficient of the labeled membrane protein. If the membrane protein is totally confined within membrane domains, the mean-square displacement will approach a horizontal asymptote at longer lag times:
| (12) |
where L is the length of the confinement area and τc is the characteristic diffusion time for the protein within the small, confined area, related to the microscopic diffusion coefficient of the protein as .
Finally, the intermediate case involves a local diffusion within the confinement domains (Dmicro) coupled with a slower and longer spatial range diffusion, Dmacro, upon escape from domain confinement. This confined diffusion is modeled by Eq. 13:
| (13) |
In this model, the diffusion within the confinement domains is defined by . The Dmacro parameter is correlated to the probability of escape from the confinement domains, such that a higher macro diffusion coefficient is related to higher escape probabilities (36). Most image time series presented in this study fit best with the latter model (Eq. 13) based on comparison of fit r-squared values.
Principal-component analysis
The purpose of principal-component analysis (PCA) is to minimize the dimensionality of a complex multivariate data set while preserving its variance to uncover any underlying trends. If the data set consists of p samples, denoted by X = (, …, ), and n observations performed for each sample. Performing PCA will create a new set of uncorrelated variables, Y = (, …, ), generated by taking linear combinations of the data in X, known as the principal components. The elements in Y explain successively decreasing variation in the original data set. The maximization of variance in PCA stems from the assumption that samples whose observations contain the greatest variance also contain the dynamics of interest in the system. Mathematical formalism on the derivation and application of PCA is given in (43) and (44). PCA on this data set was performed using RStudio, version 1.4.1717, using prcomp with the correlation matrix (45). PCA results are visualized as a 2D biplot. This is a combination of a score plot (scatterplot of the contributions of each sample on the plane defined by the selected principal components) and a loading plot (plot of the observations as vectors whose magnitude and direction characterize their influence on the principal components that define the plane).
Results
To examine the plasma membrane regulation of LDLR and LDL bound to the membrane receptors, we genetically fused LDLR to monomeric enhanced green fluorescent protein (mEGFP) on a pRK5 plasmid and transfected it into human embryonic kidney cells (HEK293T). Importantly, for LDL-DiI imaging, we are only capturing LDL-DiI that are bound to the receptors in the cell membrane. LDL particles are 22–29 nm in diameter (46) and free extracellular LDL particles would diffuse too quickly to be resolved on our EMCCD camera based on the finite frame rate and theoretical Stokes-Einstein estimates of the diffusion coefficients. However, when they bind to receptors in the membrane, they are slowed down significantly and the ligand-receptor complex can be imaged using our TIRF microscope and camera.
LDLR expression levels on the cell membrane are regulated by cholesterol-dependent domains
LDLR expression on the cell membrane is turned on by a transcriptional activation pathway mediated by SREBP2 (3,6). Once on the cell membrane, LDLR can be targeted to degradation by a PCSK9-dependent internalization route via clathrin-coated pits (47,48), or poly-ubiquitinated by the IDOL E3 ubiquitin ligase within lipid rafts and internalized in a multivesicular body mechanism (3,11,49). These pathways are intrinsically linked to sterol concentrations within the cell, with sterol-sensing SREBP2 and LXR transcription factors controlling LDLR, PCSK9, and IDOL expression. However, these different LDLR internalization pathways depend on LDLR localization within different plasma membrane domains that have differing cholesterol concentrations (50), namely, clathrin-coated pits or cholesterol-rich lipid rafts.
Therefore, we performed experiments to test if the membrane localization of LDLR influenced its expression on the cell membrane in a cholesterol-dependent way, independent of transcriptional regulatory factors. To do so, we genetically introduced the LDLR-mEGFP fluorescent construct to a pRK5 plasmid. This construct does not contain any transcriptional binding sites associated with the SREBP2 or LXR pathways and, therefore, regulation of LDLR-mEGFP expression on the cell membrane should be solely dependent on post-transcriptional regulatory mechanisms.
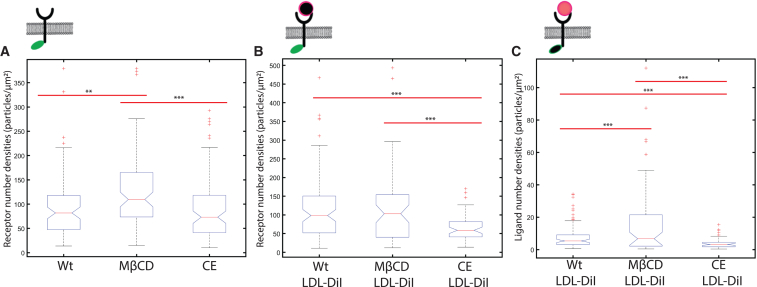
First, we imaged LDLR-mEGFP in the absence of its LDL-ligand and analyzed the images with spatial ICS. Representative spatial correlation functions are shown in Fig. S1 and the cell population mean receptor number densities calculated from the ICS best fit parameters are shown in Fig. 1. We found that cells with a lower levels of membrane cholesterol express significantly more LDLR receptors on the plasma membrane (Fig. 1 A). Treatment with MβCD causes the targeted disappearance of cholesterol-rich lipid raft domains (51,52), and directly disrupts IDOL-mediated degradation of LDLR (11).
Figure 1.
Membrane cholesterol regulates LDLR and LDLR-LDL membrane densities. Spatial ICS analysis was performed on (A) unligated LDLR-mEGFP or (B and C) LDLR-mEGFP/LDL-DiI TIRF microscopy images to measure fluorescent particle densities at the membrane. The cartoon on the top of each graph illustrates the fluorescent construct that was imaged. (A) Shows unligated LDLR-mEGFP densities after treatment with MβCD or CE. (B) Shows densities for the receptor in the presence of LDL-DiI (i.e., imaged LDLR-mEGFP/LDL-DiI in the green channel) and (C) shows ligand density when bound to the cell membrane (i.e., LDLR-mEGFP/LDL-DiI imaged in the red channel). Outliers ± three standard deviations from the mean were removed. ∗∗p < 0.01, ∗∗∗p < 0.001 using a single-tail Student's t-test. n > 30 cells used for each treatment, with 1–3 regions analyzed per cell. To see this figure in color, go online.
However, when cells are incubated with LDL-DiI, we find a significant decrease in the cell surface LDLR population when also incubated with CE (Fig. 1 B). Adding cholesterol-rich LDL-DiI particles to the samples is known to increase the cholesterol in the cell and on the cell membrane (53). Therefore, additional membrane cholesterol provided by CE treatment along with an abundance of cholesterol being internalized from the bound LDL particles likely leads to an increased number or size of cholesterol-rich regions on the membrane, and an activation of IDOL-mediated degradation of LDLR. The MβCD treatment showed no significant change to the LDLR density for this sample. It is likely that the cholesterol removed by cyclodextrin was replenished by the increased metabolism of cholesterol-rich LDL-DiI particles.
From these results, we sought to explore if the number density LDL-DiI ligand bound to the cell membrane varies with different cholesterol concentrations (Fig. 1 C). We find that there are 50% more LDL-DiI particles measured per unit area on the cell membrane when cells are treated with 2 mM MβCD for 2 h, while 25% fewer particles bind when the membrane cholesterol is upregulated by CE treatment. This can be attributed to changes in the receptor density on the cell membrane. However, it is notable that when cholesterol is depleted on the cell membrane, there is a higher incidence of LDL binding, and vice versa. Since LDL internalization accounts for a significant amount of cholesterol being introduced into the cell, this reveals an inherent negative feedback loop of cholesterol metabolism on the cell membrane: 1) high membrane cholesterol, as induced by CE treatment, lowers LDLR concentrations and LDL binding on the cell membrane, lowering the downstream uptake of LDL particles; on the other hand, 2) low membrane cholesterol leads to high LDLR concentrations on the cell membrane and high LDL-DiI particle binding to the cell membrane receptors, thereby increasing the amount of LDL being metabolized by the cell, and replenishing the membrane cholesterol.
LDLR-mEGFP/LDL-DiI binding affinity is inhibited in cholesterol-rich domains
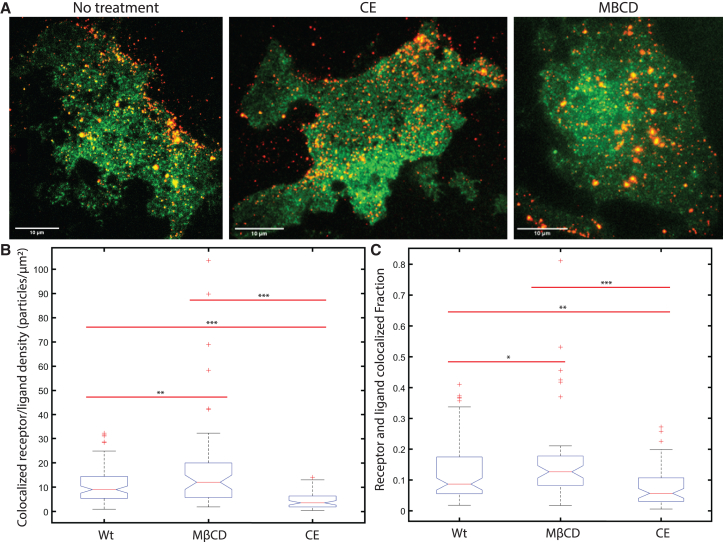
Following on from the ligand binding experiments shown in Fig. 1, B and C, we were curious to examine if the changes in ligand density were due to changes in the receptor concentration within the cell membrane or if there was a change in the receptor-ligand binding affinity. For each fixed cell, imaging was performed sequentially using a 488 nm excitation laser followed by a 561 nm excitation laser. The images were overlaid for cells under each experimental condition (no treatment, MβCD, and CE) and representative images of the LDLR-mEGFP/LDL-DiI ligated cells are displayed in Fig. 2 A. Two-color spatial ICCS analysis was performed on the image pairs (Fig. S2) and we calculated a colocalized density of 9 particles/μm2 of LDLR-mEGFP/LDL-DiI with an increase of 33% binding density when treated with MβCD and a decrease of 60% colocalization when treated with CE (Fig. 2 B). These values were normalized relative to the number of receptors detected as calculated via spatial ICS autocorrelation of the green receptor channel and shows an average of 8.7% of receptor bound to ligand for the negative control sample. This value increased to 12.6% when cells were incubated with MβCD and decreased to 5.7% colocalization when samples were incubated with CE (Fig. 2 C).
Figure 2.
Spatial ICCS reveals a membrane cholesterol-dependent regulation of LDLR’s ligand binding affinity. A spatial cross correlation analysis was performed on cells incubated with LDL-DiI ligand for 1 h and fixed with 4% PFA. LDLR-mEGFP and LDL-DiI were imaged sequentially. (A) Shows a representative overlay of receptor and ligand images under the different treatment conditions. The data in (B) represent the density of colocalized particles and (C) the fraction of receptor that is bound to ligand on the cell membrane. Outliers ± three standard deviations from the mean were removed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 using a single-tail Student's t-test. n > 30 cells used for all samples, with 1–3 regions analyzed per cell. Scale bars are 10 μm. To see this figure in color, go online.
These data indicate that LDLR is attenuated in its LDL-binding activity in cholesterol-rich conditions on the cell membrane, while it is more actively binding LDL when the membrane is in a cholesterol-depleted condition. We speculate that this may be due to LDLR conformational changes as it transits different membrane environments, such as cholesterol-rich and cholesterol-poor regions of the cell membrane. This hypothesis could be tested via single-molecule Förster resonance energy transfer, but is not considered in this study.
LDLR membrane transport dynamics are regulated by cholesterol concentrations in the cell membrane
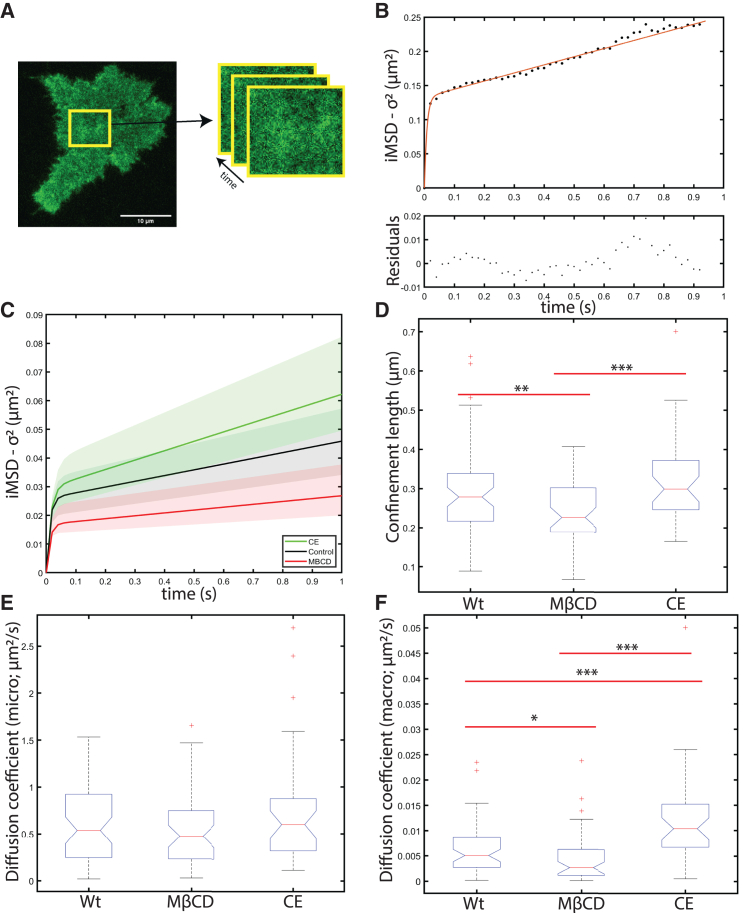
Protein residency in different membrane domains often dictates their lateral mobility. Using STICS and iMSD, we explored the range of diffusion transport that LDLR-mEGFP exhibits within the cell membrane under different conditions of membrane cholesterol expression. As described in the materials and methods section, we calculated a space-time correlation function for a region of interest of the TIRF-based images and measured the time-dependent broadening of the space-time autocorrelation standard deviation following the iMSD method (Fig. 3 A). We fit this curve with three models of transport: free diffusion, confined diffusion, and confined diffusion with transient escape from confinement domains (Eqs. 11, 12, and 13). We consistently found that the transient confinement was the best model for LDLR membrane transport under the treatment conditions we imaged (Fig. 3 B). In part, the confined dynamics of LDLR likely arise from some of the receptor population that is trapped in clathrin-coated pits. However, when we modulate cholesterol levels using either MβCD or CE, we find that the size of LDLR domains and the receptor’s lateral mobility changes and is dependent on membrane cholesterol levels (Fig. 3 C).
Figure 3.
LDLR-mEGFP resides transiently in cholesterol-dependent domains on the cell membrane. The STICS-iMSD analysis was used to examine unligated LDLR-mEGFP dynamics. (A) Displays a representative sample cell and an example region over which the spatiotemporal analysis was carried out. (B) Shows a representative iMSD versus time plot for the first 50 frames of analysis. It shows an initial steep slope followed by an inflection point and a smaller magnitude slope at lag times after the inflection point. This is a hallmark signature of transiently confined diffusion. (C) Bar whisker plots of the median iMSD fit from the measured iMSD confinement parameters. The solid line represents the median fit for cells treated with CE (green), MβCD (red), or the negative control (black), and the shaded region represents the 95% confidence intervals. (D and E) Plot the confinement parameters as fit with Eq. 13. (D) Confinement diameter, (E) Dmacro, and (F) Dmicro. Outliers ± three standard deviations from the mean were removed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 using a single-tail Student's t-test. n > 37 cells, with 1–3 regions analyzed per cell. Scale bars, 10 μm. To see this figure in color, go online.
From the iMSD fits we extract three parameters: 1) the average size of the domain, 2) the Dmicro, which is related to the short spatial range diffusion within the confinement domains, and 3) the Dmacro, which is related to the longer spatial range diffusion, and its value is proportional to the probability that the receptor will escape its confinement regions (36). Our data show that LDLR in HEK293T cells is transiently confined within domains that have a median of 279 ± 120 nm in diameter. The measured size of the confinement domains is similar upon augmentation of cholesterol with CE treatment (295 ± 93 nm), and decreases to a median of 225 ± 81 nm when cholesterol is sequestered from the membrane using MβCD (Fig. 3 D). A decrease in the domain size suggests that either the average size of the cholesterol-rich domains is becoming smaller, or that the receptors are shifting to other smaller membrane domains. Notably, while cholesterol-rich domains have been documented to span upward of 300 nm in diameter depending on cell type (9,54), clathrin-coated pits have an upper size limit of about 200 nm (55,56). Our results are consistent with the LDLR proteins switching from cholesterol-rich regions to clathrin-coated domains under cholesterol depletion conditions.
The MβCD treatment also leads to a decreased Dmacro coefficient (Fig. 3 F), indicating that depleted membrane cholesterol levels are related to a lower probability that a given receptor will escape from its domain, so the LDLR is found within stronger confinement domains. The efficient internalization of receptors in clathrin-coated domains require them to have long residency times relative to other membrane regions. This slow escape of receptors from clathrin-coated pits has been attributed to clathrin’s “rugged energy landscape” (57). Therefore, the decrease in domain escape probability for receptors when cells are depleted of cholesterol may be attributed to a shift in receptor populations to clathrin-coated domains in the absence of cholesterol-rich regions.
LDLR-mEGFP transport and confinement are more sensitive to membrane cholesterol concentrations when ligated to LDL-DiI
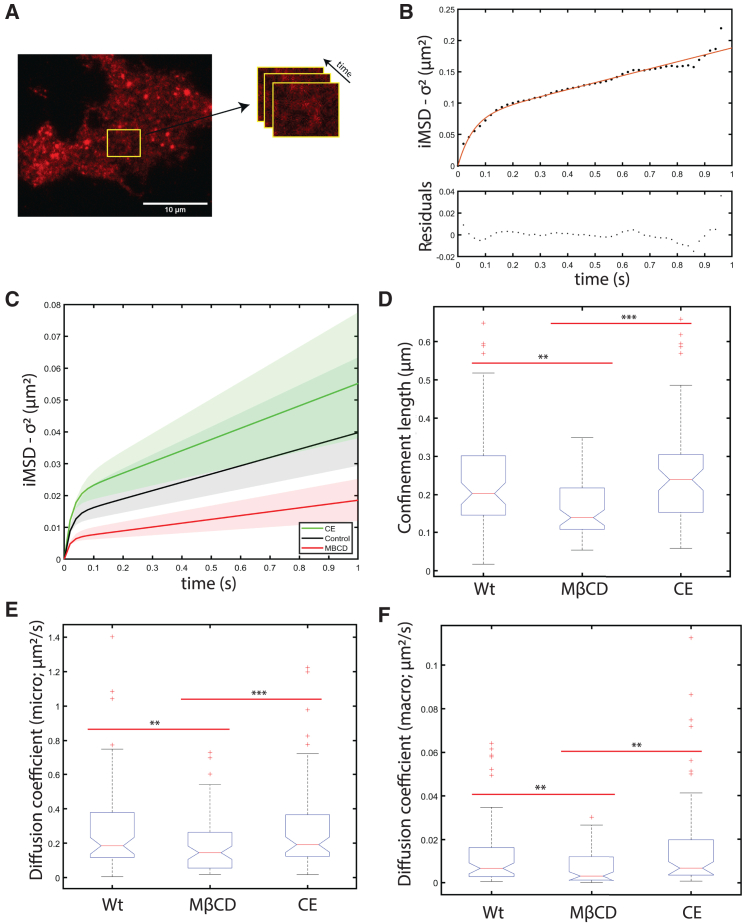
We were curious to see if LDLR-mEGFP’s membrane transport and confinement dynamics under different membrane cholesterol conditions were also changing when it was bound to its LDL-DiI ligand. To do this, the same experimental and imaging protocol was followed as shown in Fig. 4 but, additionally, after incubation with CE or MβCD we incubated with 5 mg/mL LDL-DiI for 1 h at 37°C before the live-cell imaging. For this set of experiments, we only excited the sample with 561 nm monochromatic laser light, meaning that only LDL-DiI particles that were bound to the membrane LDLR were resolved by the camera. We assume that only the LDL-DiI particles that are ligated to LDLR in the membrane are imaged and detected for these samples. The single DiI emission channel was recorded to preserve the same imaging frame time resolution on this single camera TIRF system since it requires sequential imaging and laser line cycling to capture (nonsimultaneously) in the two channels.
Figure 4.
LDLR-mEGFP/LDL-DiI complexes are also detected within confined domains. The LDLR-mEGFP/LDL-DiI complex membrane transport was studied using STICS-iMSD analysis. (A and B) Show a representative image and the calculated iMSD versus lag time data for a region of interest. Tau refers to the temporal lag of the respective ACF. (C) Illustrates the curves with the median parameters as extracted from the model representing confined diffusion with transient escapes for untreated cells (in black), CE treated cells (in green), and MβCD-treated cells (in red). Solid lines represent the fit with the median parameters and the shaded background represents the 95% confidence intervals. The data distribution for (D) the length of the confined region, (E) the Dmicro, and (F) the Dmacro are shown. Outliers ± three standard deviations from the mean were removed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 using a single-tail Student's t-test. n > 37 cells, with 1–3 regions analyzed per cell. Scale bar, 10 μm. To see this figure in color, go online.
We found that the iMSD-measured transport modes of LDLR/LDL-DiI were consistently best fit with the transiently confined diffusive model as the unligated receptor (Figs. 4, A, B and S3). The median parameters calculated from the iMSD analysis are shown in Fig. 4 C. Following the same trend as the receptor in its unligated state, the diameter of the median confined region (203 ± 141 nm, untreated) decreases by approximately 31% when treated with MβCD (140 ± 78 nm) and increases 20% when treated with CE (245 ± 130 nm) (Fig. 4 D). Similarly, consistent with the unligated receptor, receptors in cells depleted of membrane cholesterol have slower long-range diffusion than control cells (Dmacro; Fig. 4 F). Our model proposes that MβCD treatment shifts these receptor-ligand populations to rough energy landscape clathrin-coated domains while CE segregates LDLR/LDL-DiI populations to cholesterol-rich reservoirs.
Finally, the Dmicro variable describes the diffusion of the receptors and the receptor-ligand systems within the domains. If LDLR is transiting different domains, we would expect the receptor to have lower diffusion coefficients in regions with higher energy barriers, such as clathrin-coated pits (57). Fig. 4 E shows that cholesterol depletion decreases the Dmicro for the receptor-ligand complex, indicating a possible change in the lipid domain environment of the receptor.
LDL binding to LDLR shifts receptor populations to smaller domains with slower transport
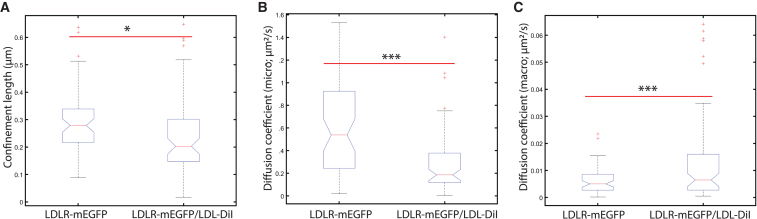
We noted that Figs. 3 and 4D–F, show a ligand-dependent change in the membrane transport and confinement of LDLR-mEGFP in cells untreated with cholesterol-modifying agents. In a direct comparison between the ligated and unligated receptor in the negative control cells, our data show that receptor-ligand complexes are present in domains that are 27% smaller than their unligated counterpart (Fig. 5 A). Furthermore, the Dmicro of the ligand-receptor complex is 66% lower (Fig. 5 B). The smaller domain size and lower diffusion coefficients appear to be indicative of a change in state from cholesterol-rich domains to clathrin-coated pits after LDL ligand binding. Our iMSD measurements are consistent with ligand binding, shifting the LDLR-LDL population to clathrin-coated pits, priming the LDL-DiI ligand for receptor-mediated endocytosis, as is well established for the LDLR system.
Figure 5.
LDL-binding shifts receptor populations to smaller domains with lower receptor mobility. The membrane regulation of the receptor was tested either unligated or bound to LDL-DiI ligand. The data shown represent the negative control samples from the STICS-iMSD results from Figs. 3 and 4. Unligated LDLR-mEGFP is imaged in the green channel and the bound LDLR-mEGFP/LDL-DiI is imaged in the red channel. The data plotted compare (A) the confinement lengths of the receptors, (B) the Dmicro, and (C) the Dmacro. Outliers ± three standard deviations from the mean were removed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 using a single-tail Student's t-test. n > 37 cells, with 1–3 regions analyzed per cell. To see this figure in color, go online.
Interestingly, we also find that the probability that the receptor escapes from the membrane domains is higher when bound to ligand, as compared with when the receptor is in its unligated form (Fig. 5 C). This would occur if LDL binding triggers a change in localization, away from cholesterol-rich regions, effectively creating a higher escape probability and larger Dmacro coefficient. This could also explain why ligand-bound receptors are more sensitive to treatment than unbound LDLR. Unbound LDLR favors cholesterol-rich domain localization and adding cholesterol will result in a minimal increase in these domains. However, if the ligand-receptor complex’s equilibrium shifts to favor clathrin-coated pits, there is more available receptor-ligand complex available to be sequestered within lipid domains as the membrane cholesterol content increases after treatment with CE.
PCA reveals trends related to a cholesterol-dependent regulation of LDLR spatiotemporal dynamics and expression
We have measured four separate variables (domain diameters, Dmicro, Dmacro, and particle densities) tested under six different conditions (ligated and unligated receptors treated with MβCD, CE, or a negative control). From these data, we have drawn correlations between these variables under different conditions to understand LDLR expression and membrane regulation. To illustrate these correlations and better understand their trends and connections, we analyzed them using PCA. PCA is a multivariate analysis and dimension reduction technique that takes data from a matrix of variables that are tested under different conditions to extract the relevant information (58).
From the matrix given, PCA generates variables known as “principal components” that are defined as combinations of the original variables. The first principal component explains the largest variance, and therefore it contains the factors that cause the greatest dynamic change in the data. The second, third, etc., principal components are calculated with decreasing amounts of variance.
Presented in Fig. 6, we show two identical PCA biplots, with the experimental conditions grouped in separate ways. The arrows represent the correlations of the variables while the point markers are the experimental conditions. Our first two principal components can explain 99% of the variance from the data. In Fig. 6 A we separate the data into two groups, one group representing the conditions for the ligated receptor (in blue) and for the unligated receptor (in green). Interestingly, whether or not the receptor is ligated appears to be explained by the first principal component; where the ligated conditions are represented by a positive first principal component and the unligated conditions are shown in the negative first principal component.
Figure 6.
Principal-component analysis (PCA) illustrates trends in LDLR-mEGFP membrane regulation. A matrix with the samples along the rows (ligated LDLR-mEGFP/LDL-DiI or unligated LDLR-mEGFP negative control or treated with CE or MβCD), and measured variables for each sample along the columns (number density [from the spatial ICS data] or domain size, Dmicro, Dmacro [from iMSD data]) was constructed. The data were centered and scaled before performing PCA. The percent variance explained by a given principal component is shown on the axes labels. The observables are shown as loading vectors pinned to the origin in blue. Their projection on a given principal component (PC) axis is representative of the weight they have on the PC. The angle between a pair of observables describes the correlation between them. For angles less than 90°, there is a positive correlation increasing in strength as 0° is approached. For angles larger than 90°, there is a negative correlation increasing in strength as 180° is approached. At 90°, there is no correlation. The samples are shown as a score plot via scatter points in black. In centered and scaled data sets, the Euclidean distance between sample scores describes their similarity to one another. The samples were then grouped according to their experimental conditions. (A) Groups the samples based on LDLR-mEGFP’s ligation state. Unligated LDLR is explained by a negative first PC, highlighted in green, while the LDLR/LDL complex is explained by a positive first PC, in blue. (B) The same PCA biplot as (A) but grouped by treatment conditions. The group in red shows the samples treated with CE; shown in orange are the negative control samples, and in yellow are the samples treated with MβCD. A decrease in membrane cholesterol content is associated with a more negative second PC. To see this figure in color, go online.
The directions of the arrows correlate with the position of the markers. In other words, the unligated receptor is characterized by a larger domain diameter, a more rapid diffusion within its domain, and a greater number density compared with the receptor-ligand complex. This illustration shows how ligand binding affects the spatial and temporal dynamics of LDLR, and LDL binding is correlated with variables that we believe to be associated to a transition from cholesterol-rich domains to clathrin-coated pits with a smaller domain diameter and a lower intradomain diffusion coefficient.
We then separated the data into three groups related to the relative cholesterol concentration on the membrane—whether the cell membrane was augmented in cholesterol (CE, in red), reduced in cholesterol (MβCD, in yellow), or untreated (in orange) (Fig. 6 B). Changes in cholesterol concentration appear to be well explained by the second principal component, where a higher membrane cholesterol concentration is correlated with a greater second principal component.
Following the trends, higher membrane cholesterol concentrations are indicative of a higher probability of escape from membrane domains, and larger domain lengths, while lower membrane cholesterol concentrations are correlated with a higher number density in both unligated receptors and receptor-ligand complexes. Corroborating the trends in the fluorescence fluctuation analysis, low cholesterol concentrations are correlated with variables that we believe are indicative of residence in clathrin-coated pits, such as a lower domain escape probability (Dmacro) and a smaller domain size.
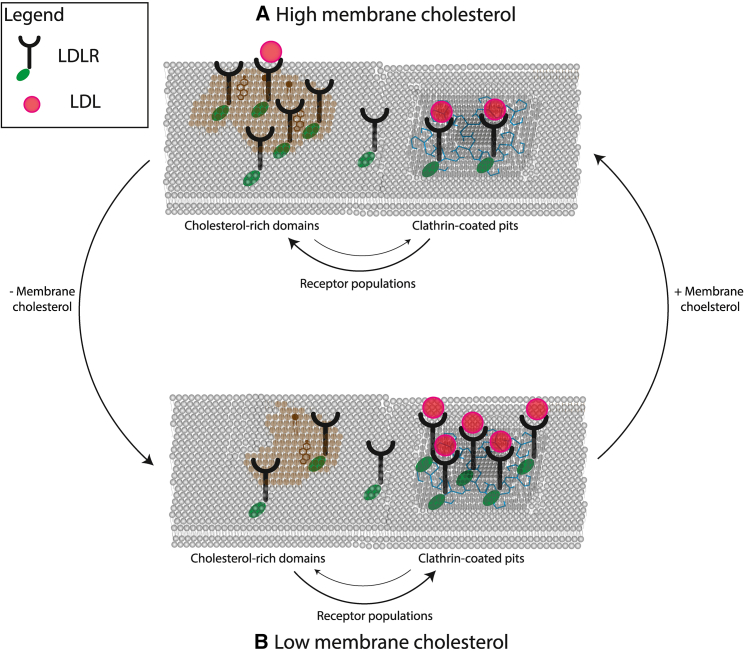
A proposed model for membrane cholesterol-mediated regulation of LDLR
In this study, we have shown that changes in cholesterol concentration within the cell membrane modulate the transport dynamics and membrane expression of LDLR in its unligated form and when ligated to LDL. Interpreting our results in context of the previous literature showing LDLR transport and confinement in clathrin-coated pits and in lipid rafts, we postulate that cholesterol-rich membrane domains form reservoirs of LDLR that are attenuated in their ability to bind LDL ligand, and that are susceptible to a higher rate of degradation (Fig. 7 A). When cholesterol concentrations are high, this mechanism functions to prevent too much exogenous cholesterol from internalizing via clathrin-mediated endocytosis. However, when cholesterol levels on the membrane begin to decrease, these cholesterol domains dissociate and release LDLs, which can then bind their LDL particle ligands with higher affinity before or after transit to clathrin-coated domains. Once in coated pit domains they are internalized by the cell via the canonical clathrin-coated pit-mediated endocytosis pathway (Fig. 7 B).
Figure 7.
A schematic of the negative feedback loop regulating membrane cholesterol homeostasis via LDLR domain localization. An illustration of the proposed model in this thesis. (A) When membrane cholesterol levels are high, LDLR is sequestered in cholesterol-rich domains, forming a reservoir of LDLR that is attenuated in their ability to bind and metabolize LDL, and increasing ubiquitin-dependent degradation of LDLR. Less LDL being internalized may lead to membrane cholesterol levels decreasing, thereby dissociating cholesterol-rich domains and (B) permitting LDLR to transit to clathrin-coated pit domains, where they can efficiently bind and internalize LDL particles. LDLR-mediated endocytosis of LDL particles may cause membrane cholesterol concentrations to increase, reforming the cholesterol-rich domains found in (A). To see this figure in color, go online.
To summarize these findings, we present a four-point model of LDLR regulation via cholesterol rich domains in HEK293T cells.
-
(1)
LDLR expression on the cell membrane is regulated by membrane cholesterol concentrations; whereas high cholesterol on the membrane targets LDLR to cholesterol-rich reservoirs such as lipid rafts where it can undergo IDOL-mediated degradation, low cholesterol concentration targets LDLR to clathrin-coated pits, where it may be internalized and be degraded or recycled.
-
(2)
Unligated LDLR on HEK293T cells is sequestered preferentially into cholesterol-dependent domains (as presented in the discussions of Figs. 3, D and F, and 6).
-
(3)
LDL-binding shifts LDLR away from cholesterol-rich domains. A comparison of the ligand-bound and unligated receptors (Fig. 3 versus Fig. 4) shows that bound receptors are more sensitive to changes in cholesterol, suggesting a ligand-dependent shift in the equilibrium state between domains. Furthermore, Fig. 6 shows a ligand-dependent transition of the receptor to smaller domains with slower receptor transport, suggesting a transition to clathrin-coated pits.
-
(4)
LDLR membrane expression and its binding affinity is attenuated in these cholesterol-rich domains.
Discussion
Cholesterol homeostasis is one of the most extensively studied human metabolic processes. The feedback mechanisms associated with the LDLR-dependent regulation of LDL have been deeply explored at the transcriptional level. In this study, we present a series of tissue culture experiments and imaging-based fluorescence fluctuation analyses that reveal details about the cholesterol-domain-dependent regulation of LDLR on the cell membrane.
Based on this work we hypothesized a model that accounts for an alternative regulatory mechanism for cellular cholesterol homeostasis featuring a negative feedback loop that is dependent on membrane cholesterol concentrations. When LDL particles are internalized, they are trafficked toward lysosomal organelles where the particles are degraded into their constituents (59). The canonical regulatory pathways for LDLR depend on sterol-sensing elements within the cell. For instance, the SREBP2 pathway is inactivated by SCAP, a protein that senses cholesterol concentrations in the ER membrane (3,60). LXR proteins are also sterol sensors in the cytoplasm that dampen cellular cholesterol intake and promote its excretion when levels are too high. Notably, before cholesterol is trafficked intracellularly, it must first be transported to the plasma membrane, where it is then redistributed (59). Therefore, cholesterol domains on the plasma membrane may act as a fast, front-line sensor, keeping LDLR in less-active states, while a high degree of LDL particles are being endocytosed, and then release LDLR into more active states as membrane cholesterol is internalized or otherwise depleted.
For a complete discussion of our hypothesized model, we must discuss its limitations and caveats. As with all fluorescence-based experiments, fluorescent proteins may have an impact on protein function; however, the LDLR fluorescent protein constructs have been used in HEK293 cells and similar cultured cell lines and shown not to impair normal trafficking of LDLR and PCSK9 within the cells (61). We present a model where LDLR is localized in either cholesterol-dependent domains or clathrin-coated pits. The plasma membrane is vast and heterogeneous, and other domains may be playing a role in LDLR’s mobility and transient localization. We excluded consideration of LDLR localization in membrane-disordered regions (i.e., referring to the regions of the plasma membrane that are neither ordered cholesterol domains nor clathrin-coated pits). We rationalized this based on the diffusion measurements as calculated by iMSD (Figs. 3 and 4). We noticed that cholesterol depletion lowers LDLR’s diffusion coefficient within domains as well as its probability from escaping the domains (Figs. 3, F, E and 4 F). This was unexpected, as cholesterol is known to induce rigidity and order to its membrane domains, which should be accompanied by a lower diffusion coefficient of its resident macromolecules relative to the disordered plasma membrane (62,63). From these observations, we postulated that LDLR may be transitioning from cholesterol-dependent domains, likely large lipid rafts, to clathrin-coated domains, which are expected to have a larger diffusive barrier (57). Furthermore, our model assumes that there are short transit times in the disordered region of the membrane as LDLR shifts between these two domains, which were likely outside of the time resolution of our imaging and fluorescence fluctuation measurements. Another potential limitation with the study is that we assume that LDLR-mEGFP dynamics are in two dimensions. Previous analysis of FCS measurements on freely diffusing proteins have revealed that membrane topography may result in the FCS curves following a trapped-diffusion model (64,65). We explored the impact that membrane topography might have in our spatial ICS density measurements and found that deep invaginations (∼200 nm) if present, would decrease the ICS-measured particle density on the cell membrane, and that this systematic error becomes more significant for higher particle densities (Fig. S4).
This model is consistent with our experimental data, but future work would be needed to confirm some of its postulates. Notably, a series of colocalization experiments of LDLR-mEGFP with a lipid-raft marker and a clathrin-coated pit marker could help identify the proportion of receptors in each domain and how the equilibrium of the receptors shifts in response to the different cholesterol concentrations. Further single-particle tracking experiments may be able to detect LDLR transiting between domains; however, high statistical sampling would be needed for such experiments.
Another caveat in the model is that all the experiments were performed on embryonic kidney cells. This was because this cell line is known to contain very low levels of endogenous LDLR (66) and, therefore, we knew that our imaging and analysis included almost the full population LDLR that was expressed on the cell membrane. However, immunoblotting experiments in different studies have shown that the fraction of LDLR localized to lipid rafts may be cell type dependent (10,11). Furthermore, most of the receptor-mediated internalization of LDLR takes place in the liver (67). It would therefore be of value to perform these studies in various cell lines, including hepatocytes, to test if this model holds for LDLR membrane regulation in other cell types.
A continued exploration of the regulatory mechanisms that underlie cholesterol homeostasis is invaluable to our understanding of human health and disease, as well as the many biological implications that come with the regulation. Biophysical image analysis techniques have provided a unique view into how we can begin to understand cholesterol metabolism from the perspective of its membrane dynamics and confinement and possible regulatory mechanisms in live cells.
Author contributions
S.V.M., J.M., D.F., and P.W.W. designed the study. J.M. performed plasmid cloning. S.V.M. and J.P. performed the experiments and data analysis. A.M. performed and interpreted the PCA analysis. S.V.M., J.M., D.F., and P.W.W. interpreted the results. S.V.M. and P.W.W. wrote the first draft, which was reviewed and edited by all authors.
Acknowledgments
This work was supported by grants to P.W.W. from the National Sciences and Engineering Research Council of Canada (NSERC) and to P.W.W. and D.F. from the Canadian Institute for Advanced Research (CIFAR). S.V.M. was a recipient a studentship from the Centre de Recherche en Biologie Structurale (CRBS) at McGill University. We would also like to thank Thomas Mosher for his help in developing the iMSD code.
Declaration of interests
The authors declare no competing interests in this work.
Editor: H. Raghuraman.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.08.005.
Supporting material
References
- 1.CARDIoGRAMplusC4D Consortium. Deloukas P., et al. Samani N.J. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown M.S., Goldstein J.L. How LDL Receptors Influence Cholesterol and Atherosclerosis. Sci. Am. 1984;251:58–66. doi: 10.1038/scientificamerican1184-58. [DOI] [PubMed] [Google Scholar]
- 3.Yang H.-x., Zhang M., et al. Liao D.F. Cholesterol in LDL receptor recycling and degradation. Clin. Chim. Acta. 2020;500:81–86. doi: 10.1016/j.cca.2019.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Bilheimer D.W., Stone N.J., Grundy S.M. Metabolic studies in familial hypercholesterolemia: evidence for a gene-dosage effect in vivo. J. Clin. Invest. 1979;64:524–533. doi: 10.1172/JCI109490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein J.L., Brown M.S. Regulation of low-density lipoprotein receptors: implications for pathogenesis and therapy of hypercholesterolemia and atherosclerosis. Circulation. 1987;76:504–507. doi: 10.1161/01.cir.76.3.504. [DOI] [PubMed] [Google Scholar]
- 6.Radhakrishnan A., Goldstein J.L., et al. Brown M.S. Switch-like Control of SREBP-2 Transport Triggered by Small Changes in ER Cholesterol: A Delicate Balance. Cell Metabol. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubuc G., Chamberland A., et al. Prat A. Statins Upregulate PCSK9, the Gene Encoding the Proprotein Convertase Neural Apoptosis-Regulated Convertase-1 Implicated in Familial Hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 8.Zelcer N., Hong C., et al. Tontonoz P. LXR Regulates Cholesterol Uptake Through Idol-Dependent Ubiquitination of the LDL Receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sezgin E., Levental I., et al. Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivaturi S., Wooten C.J., et al. Lopez D. Distribution of the LDL receptor within clathrin-coated pits and caveolae in rat and human liver. Biochem. Biophys. Res. Commun. 2014;445:422–427. doi: 10.1016/j.bbrc.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 11.Sorrentino V., Nelson J.K., et al. Zelcer N. The LXR-IDOL axis defines a clathrin-caveolae-and dynamin-independent endocytic route for LDLR internalization and lysosomal degradation. J. Lipid Res. 2013;54:2174–2184. doi: 10.1194/jlr.M037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lillis A.P., Mikhailenko I., Strickland D.K. Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability. J. Thromb. Haemostasis. 2005;3:1884–1893. doi: 10.1111/j.1538-7836.2005.01371.x. [DOI] [PubMed] [Google Scholar]
- 13.Wu L., Gonias S.L. The low-density lipoprotein receptor-related protein-1 associates transiently with lipid rafts. J. Cell. Biochem. 2005;96:1021–1033. doi: 10.1002/jcb.20596. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh R.N., Webb W.W. Automated detection and tracking of individual and clustered cell surface low density lipoprotein receptor molecules. Biophys. J. 1994;66:1301–1318. doi: 10.1016/S0006-3495(94)80939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daumas F., Destainville N., et al. Salomé L. Confined Diffusion Without Fences of a G-Protein-Coupled Receptor as Revealed by Single Particle Tracking. Biophys. J. 2003;84:356–366. doi: 10.1016/S0006-3495(03)74856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusumi A., Sako Y. Cell surface organization by the membrane skeleton. Curr. Opin. Cell Biol. 1996;8:566–574. doi: 10.1016/s0955-0674(96)80036-6. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara T., Ritchie K., et al. Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietrich C., Yang B., et al. Jacobson K. Relationship of Lipid Rafts to Transient Confinement Zones Detected by Single Particle Tracking. Biophys. J. 2002;82:274–284. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C.Y., Huang J.Y., Lo L.-W. Unraveling the impact of lipid domains on the dimerization processes of single-molecule EGFRs of live cells. Biochim. Biophys. Acta. 2015;1848:886–893. doi: 10.1016/j.bbamem.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt T., Schütz G.J., et al. Schindler H. Imaging of single molecule diffusion. Proc. Natl. Acad. Sci. USA. 1996;93:2926–2929. doi: 10.1073/pnas.93.7.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manzo C., Garcia-Parajo M.F. A review of progress in single particle tracking: from methods to biophysical insights. Rep. Prog. Phys. 2015;78 doi: 10.1088/0034-4885/78/12/124601. [DOI] [PubMed] [Google Scholar]
- 22.Magde D., Elson E., Webb W.W. Thermodynamic fluctuations in a reacting system—measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett. 1972;29:705–708. [Google Scholar]
- 23.Mueller V., Ringemann C., et al. Eggeling C. STED Nanoscopy Reveals Molecular Details of Cholesterol- and Cytoskeleton-Modulated Lipid Interactions in Living Cells. Biophys. J. 2011;101:1651–1660. doi: 10.1016/j.bpj.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller V., Honigmann A., et al. Eggeling C. In: Tetin S.Y., editor. Vol. 519. Academic Press; 2013. Chapter One - FCS in STED Microscopy: Studying the Nanoscale of Lipid Membrane Dynamics; pp. 1–38. (Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 25.Veerapathiran S., Wohland T. The imaging FCS diffusion law in the presence of multiple diffusive modes. Methods. 2018;140–141:140–150. doi: 10.1016/j.ymeth.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Sankaran J., Shi X., et al. Wohland T. ImFCS: A software for Imaging FCS data analysis and visualization. Opt Express. 2010;18:25468–25481. doi: 10.1364/OE.18.025468. [DOI] [PubMed] [Google Scholar]
- 27.Huang H., Simsek M.F., et al. Pralle A. Effect of Receptor Dimerization on Membrane Lipid Raft Structure Continuously Quantified on Single Cells by Camera Based Fluorescence Correlation Spectroscopy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin W., Simsek M.F., Pralle A. Quantifying spatial and temporal variations of the cell membrane ultra-structure by bimFCS. Methods. 2018;140–141:151–160. doi: 10.1016/j.ymeth.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng X.W., Bag N., Wohland T. Characterization of Lipid and Cell Membrane Organization by the Fluorescence Correlation Spectroscopy Diffusion Law. Chimia. 2015;69:112–119. doi: 10.2533/chimia.2015.112. [DOI] [PubMed] [Google Scholar]
- 30.Kolin D.L., Wiseman P.W. Advances in Image Correlation Spectroscopy: Measuring Number Densities, Aggregation States, and Dynamics of Fluorescently labeled Macromolecules in Cells. Cell Biochem. Biophys. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- 31.Petersen N.O., Höddelius P.L., et al. Magnusson K.E. Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophys. J. 1993;65:1135–1146. doi: 10.1016/S0006-3495(93)81173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown C.M., Petersen N.O. An image correlation analysis of the distribution of clathrin associated adaptor protein (AP-2) at the plasma membrane. J. Cell Sci. 1998;111:271–281. doi: 10.1242/jcs.111.2.271. [DOI] [PubMed] [Google Scholar]
- 33.Costantino S., Comeau J.W.D., et al. Wiseman P.W. Accuracy and dynamic range of spatial image correlation and cross-correlation spectroscopy. Biophys. J. 2005;89:1251–1260. doi: 10.1529/biophysj.104.057364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiseman P.W., Squier J.A., et al. Wilson K.R. Two-photon image correlation spectroscopy and image cross-correlation spectroscopy. J. Microsc. 2000;200:14–25. doi: 10.1046/j.1365-2818.2000.00736.x. [DOI] [PubMed] [Google Scholar]
- 35.Hebert B., Costantino S., Wiseman P.W. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Rienzo C., Gratton E., et al. Cardarelli F. Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc. Natl. Acad. Sci. USA. 2013;110:12307–12312. doi: 10.1073/pnas.1222097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilangumaran S., HOESSLI D.C. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 1998;335:433–440. doi: 10.1042/bj3350433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu-Arish A., Pandzic E., et al. Wiseman P.W. Cholesterol Modulates CFTR Confinement in the Plasma Membrane of Primary Epithelial Cells. Biophys. J. 2015;109:85–94. doi: 10.1016/j.bpj.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiseman P.W., Petersen N.O. Image correlation spectroscopy. II. Optimization for ultrasensitive detection of preexisting platelet-derived growth factor-β receptor oligomers on intact cells. Biophys. J. 1999;76:963–977. doi: 10.1016/S0006-3495(99)77260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolin D.L., Costantino S., Wiseman P.W. Sampling Effects, Noise, and Photobleaching in Temporal Image Correlation Spectroscopy. Biophys. J. 2006;90:628–639. doi: 10.1529/biophysj.105.072322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comeau J.W.D., Kolin D.L., Wiseman P.W. Accurate measurements of protein interactions in cells via improved spatial image cross-correlation spectroscopy. Mol. Biosyst. 2008;4:672–685. doi: 10.1039/b719826d. [DOI] [PubMed] [Google Scholar]
- 42.Wawrezinieck L., Rigneault H., et al. Lenne P.F. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jolliffe I.T. Springer; 2002. Principal Component Analysis for Special Types of Data. [Google Scholar]
- 44.Everitt B., Hothorn T. Springer Science & Business Media; 2011. An Introduction to Applied Multivariate Analysis with R. [Google Scholar]
- 45.Team R. In: RStudio: Integrated Development Environment for R. RStudio, editor. PBC; 2021. [Google Scholar]
- 46.Sacks F.M., Campos H. Low-Density Lipoprotein Size and Cardiovascular Disease: A Reappraisal. J. Clin. Endocrinol. Metab. 2003;88:4525–4532. doi: 10.1210/jc.2003-030636. [DOI] [PubMed] [Google Scholar]
- 47.Nassoury N., Blasiole D.A., et al. Seidah N.G. The Cellular Trafficking of the Secretory Proprotein Convertase PCSK9 and Its Dependence on the LDLR. Traffic. 2007;8:718–732. doi: 10.1111/j.1600-0854.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Huang Y., et al. Cohen J.C. Molecular characterization of proprotein convertase subtilisin/kexin type 9-mediated degradation of the LDLR. J. Lipid Res. 2012;53:1932–1943. doi: 10.1194/jlr.M028563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scotti E., Calamai M., et al. Tontonoz P. IDOL stimulates clathrin-independent endocytosis and multivesicular body-mediated lysosomal degradation of the low-density lipoprotein receptor. Mol. Cell Biol. 2013;33:1503–1514. doi: 10.1128/MCB.01716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirkham M., Parton R.G. Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim. Biophys. Acta. 2005;1745:273–286. doi: 10.1016/j.bbamcr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Giocondi M.-C., Milhiet P.E., et al. Le Grimellec C. Use of Cyclodextrin for AFM Monitoring of Model Raft Formation. Biophys. J. 2004;86:861–869. doi: 10.1016/s0006-3495(04)74161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai M., Zhao W., et al. Wang H. Direct Evidence of Lipid Rafts by in situ Atomic Force Microscopy. Small. 2012;8:1243–1250. doi: 10.1002/smll.201102183. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein J.L., Brown M.S. Progress in understanding the LDL receptor and HMG-CoA reductase, two membrane proteins that regulate the plasma cholesterol. J. Lipid Res. 1984;25:1450–1461. [PubMed] [Google Scholar]
- 54.Honigmann A., Mueller V., et al. Eggeling C. STED microscopy detects and quantifies liquid phase separation in lipid membranes using a new far-red emitting fluorescent phosphoglycerolipid analogue. Faraday Discuss. 2013;161:77–89. doi: 10.1039/c2fd20107k. [DOI] [PubMed] [Google Scholar]
- 55.Rejman J., Oberle V., et al. Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heuser J.E., Anderson R.G. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J. Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Licht S.S., Sonnleitner A., et al. Schultz P.G. A rugged energy landscape mechanism for trapping of transmembrane receptors during endocytosis. Biochemistry. 2003;42:2916–2925. doi: 10.1021/bi026059v. [DOI] [PubMed] [Google Scholar]
- 58.Abdi H., Williams L.J. Principal component analysis. WIREs. Comp. Stat. 2010;2:433–459. [Google Scholar]
- 59.Trinh M.N., Brown M.S., et al. Lu F. Last step in the path of LDL cholesterol from lysosome to plasma membrane to ER is governed by phosphatidylserine. Proc. Natl. Acad. Sci. USA. 2020;117:18521–18529. doi: 10.1073/pnas.2010682117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goedeke L., Fernández-Hernando C. Regulation of cholesterol homeostasis. Cell. Mol. Life Sci. 2012;69:915–930. doi: 10.1007/s00018-011-0857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poirier S., Hamouda H.A., et al. Mayer G. Trafficking Dynamics of PCSK9-Induced LDLR Degradation: Focus on Human PCSK9 Mutations and C-Terminal Domain. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niemelä P.S., Ollila S., et al. Vattulainen I. Assessing the nature of lipid raft membranes. PLoS Comput. Biol. 2005;preprint:e34. doi: 10.1371/journal.pcbi.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simons K., Vaz W.L.C. Model Systems, Lipid Rafts, and Cell Membranes. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 64.Gesper A., Wennmalm S., et al. Parmryd I. Variations in plasma membrane topography can explain heterogenous diffusion coefficients obtained by fluorescence correlation spectroscopy. Front. Cell Dev. Biol. 2020;8:767. doi: 10.3389/fcell.2020.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adler J., Shevchuk A.I., et al. Parmryd I. Plasma membrane topography and interpretation of single-particle tracks. Nat. Methods. 2010;7:170–171. doi: 10.1038/nmeth0310-170. [DOI] [PubMed] [Google Scholar]
- 66.Lara S., Alnasser F., et al. Dawson K.A. Identification of Receptor Binding to the Biomolecular Corona of Nanoparticles. ACS Nano. 2017;11:1884–1893. doi: 10.1021/acsnano.6b07933. [DOI] [PubMed] [Google Scholar]
- 67.Brown M.S., Goldstein J.L. Receptor-mediated endocytosis: insights from the lipoprotein receptor system. Proc. Natl. Acad. Sci. USA. 1979;76:3330–3337. doi: 10.1073/pnas.76.7.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.