Abstract
Specifically targeted antimicrobial peptides (STAMPs) are novel alternatives to antibiotics, whereas the development of STAMPs for colonic infections is hindered by limited de novo design efficiency and colonic bioavailability. In this study, we report an efficient de novo STAMP design strategy that combines a traversal design, machine learning model, and phage display technology to identify STAMPs against Clostridium perfringens. STAMPs could physically damage C. perfringens, eliminate biofilms, and self-assemble into nanoparticles to entrap pathogens. Further, a gut-targeted engineering particle vaccine (EPV) was used for STAMPs delivery. In vivo studies showed that both STAMP and EPV@STAMP effectively limited C. perfringens infections and then reduced inflammatory response. Notably, EPV@STAMP exhibited stronger protection against colonic infections than STAMPs alone. Moreover, 16S ribosomal RNA sequencing showed that both STAMPs and EPV@STAMP facilitated the recovery of disturbed gut microflora. Collectively, our work may accelerate the development of the discovery and delivery of precise antimicrobials.
Precision peptide antimicrobials, designed and delivered for bacterial colonic infections, offer targeted treatment options.
INTRODUCTION
The rise in antimicrobial resistance (AMR) poses a major threat to human health worldwide. In 2019 alone, 1.27 million people died directly from AMR, with an additional 4.95 million deaths attributed to bacterial AMR (1). As the spread of severe acute respiratory syndrome coronavirus 2 continues, the global crisis of AMR has further exacerbated, resulting in more preventable deaths owing to complications in patient treatment (2). Antibiotic-associated diarrhea (AAD) is a serious complication of AMR, often stemming from dysbacteriosis, which subsequently predisposes individuals to opportunistic infections (3). While Clostridium difficile has been extensively studied as the main causative pathogen of AAD, it accounts for merely up to 25% of all AAD cases (4). Other Clostridium spp., such as Clostridium perfringens, which causes up to 15% of AAD and is involved in the development of inflammatory bowel disease, deserve attention as well (3, 5). C. perfringens is one of the most common causes of food poisoning, with the Centers for Disease Control estimating that the bacterium causes nearly 1 million illnesses in the United States annually (6). Furthermore, AAD can exacerbate the colonization and overgrowth of C. perfringens, leading to a more protracted and severe disease manifestation (3, 7).
Antimicrobial peptides (AMPs) are promising antibiotic alternatives, demonstrating diverse bioactive functions such as antibacterial, antibiofilm, and anti-inflammatory activities, while posing a low risk of drug resistance (mainly attributed to their mechanism of physically killing bacteria) (8). As important components of immune defense, intestinal AMPs can regulate microbial communities during dysbiosis and infection, which is essential for maintaining gut homeostasis and preventing inflammation (9). Moreover, AMPs resistance genes infrequently participate in horizontal transfer in the gut microbiota (10). Using AMPs as therapeutics for intestinal infections presents two primary challenges: (i) Broad-spectrum AMPs may eliminate pathogens along with host commensal bacteria, causing unpredictable disruptions in the microbiota and potential adverse effects (11–14), and (ii) exogenous AMPs encounters limited colonic bioavailability due to restricted routes of administration and the complex intestinal environment, potentially impeding direct antibacterial effects of AMPs (15, 16).
The development of design strategies for specifically targeted AMPs (STAMPs) is urgently required to expand the discovery of precision antimicrobials for personalized treatment of infectious diseases (13, 14). However, the design of STAMPs is difficult to design, and a stable or universal design system remains elusive. Now, the hybrid peptide strategy is the main approach used for STAMPs design, but it encounters several challenges: (i) de novo design of antimicrobial domains—low transferability design strategies and uncertainty in unknown-sequence activity constraint AMPs design and discovery; (ii) selection of targeting domains—the usage and quantity of existing peptide pheromones cannot meet the increasing demand for targeting peptides (17–20). Furthermore, the new targeting peptides obtained via previous phage display technologies might enhance the antibacterial activity of hybrid peptides rather than their specificity (12).
Biomaterials has shown great potential in enhancing the bioavailability of AMPs by sustaining their release at the infection site and increasing their stability and activity to reduce potentially adverse systemic effects (21). Delivery vehicles such as nanoparticles and hydrogels have been used for the treatment of infections in various organs, including the bone (22–26), lung (27), wound (28–33), urinary tract (34, 35), and eyes (36–38). However, to our knowledge, there have been no reports on gut-targeted biomaterials for the delivery of AMPs in the treatment of intestinal infections.
To address the aforementioned challenges, we have established an efficient and straightforward system for screening STAMPs and delivering them using gut-targeted nanoparticles for the treatment of C. perfringens intestinal infections. Using a free-experience in silico design strategy, we generated five highly active AMPs, termed MLamPs. The specifically targeting peptide from phage display libraries conferred specific anti–C. perfringens activity to MLamPs. Among them, the broad-spectrum AMPs F6P1 and specific AMPs F6P6 were selected, both demonstrating potent in vitro antibacterial, antibiofilm, targeting, self-assemble properties. These two hybrid AMPs (HAMPs) have good in vivo biocompatibility and mainly targeted the gut. Then, gut-targeted engineering particle vaccine (EPV) was used to deliver HAMPs for the treatment of C. perfringens infection (CPI) and antibiotic-induced persistent CPI. Subsequently, we evaluated their in vivo efficacy and investigated the impact on the gut flora community. Our results provide an insight into the design and delivery of STAMPs to treat intestinal infections.
RESULTS AND DISCUSSION
Use of traversal design and ML to develop AMPs
To address the challenges of AMP design, we initially acquired fundamental parameter information, such as amino acid composition, using database filtering technology (DFT) (39). We then used a traversal design strategy to generate and filter the potential AMP sequences. The activity levels of these sequences were predicted using a high-performance machine learning (ML) model and then further synthesized and verified (Fig. 1A).
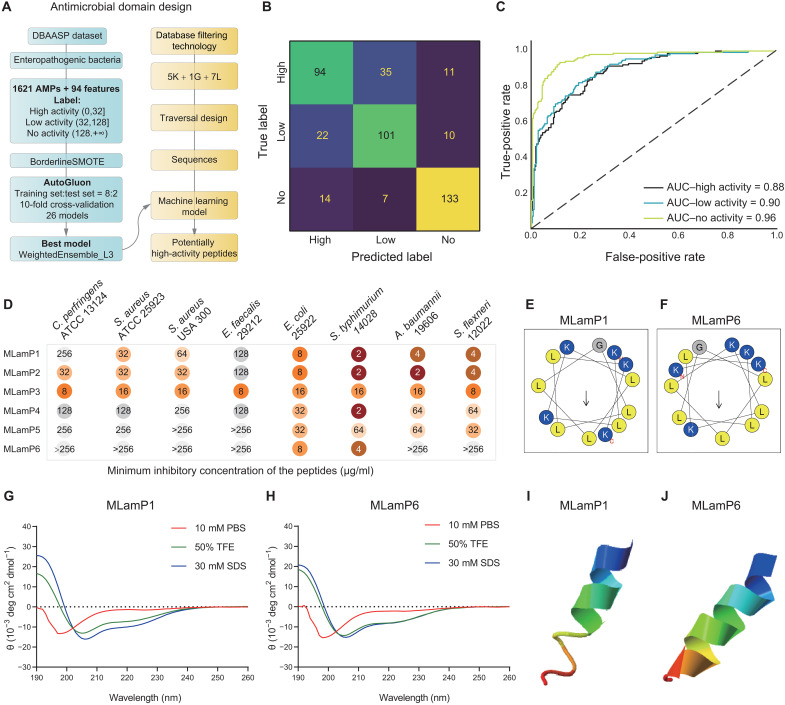
Fig. 1. Design, screening, antimicrobial activity, and structure of AMPs derived from traversal design and ML.
(A) Flow chart of AMPs design. (B and C) Confusion matrix and ROC curve of ML model (WeightedEnsemble_L3; ROC-AUC = 0.912; accuracy = 76.8%). (D) Minimum inhibitory concentration (MIC) of the MLamPs against Gram-positive pathogens (Clostridium perfringens, Staphylococcus aureus, and Enterococcus faecalis) and Gram-negative pathogens (Escherichia coli, Salmonella typhimurium, Acinetobacter baumannii, and Shigella flexneri). Each test was performed in three independent experiments. (E and F) Helical wheel projections of MLamP1 and MLamP6. (G and H) Circular dichroism (CD) spectra of MLamP1 and MLamP6. The peptides were dissolved in phosphate-buffered saline (PBS) (pH = 7.4), 30 mM SDS, and 50% trifluoroethyl alcohol (TFE). All CD measurements were performed with an average of three replicates. α-Helical content of AMPs is listed in table S5. (I and J) three-dimensional (3D) structure of MLamP1 and MLamP6, both of which have an α-helix structure. ATCC, American Type Culture Collection.
The DFT yielded a 13-residue peptide comprising five K, one G, and seven L residues. We generated 10,296 enumerable sequences, after layer-by-layer screening for hydrophobic moment (n = 2214), tilt angle (n = 280), propensity to in vitro aggregation (n = 42), and helix content (n = 9), six sequences meeting the requirements were lastly identified (fig. S1). The Database of Antimicrobial Activity and Structure of Peptides (DBAASP) database was used to construct a dataset for ML model (40). Monomeric AMPs without unusual amino acids and bonds and with known minimum inhibitory concentration (MIC) for enteric pathogens were screened out. In total, 1621 AMPs were considered, and the AMPs were divided into three groups: high activity (0 μg/ml < MIC ≤32 μg/ml), low activity (32 μg/ml < MIC ≤128 μg/ml), and no activity (MIC >128 μg/ml). After the oversampling algorithm was used to balance the samples in each class, we randomly split the dataset into 80% training data and 20% test data. AutoGluon was used to establish the ML prediction models. A total of 26 models with 10-fold cross-validation were tested, of which WeightedEnsemble_L3 provided the best performance (table S1). This model achieved a receiver operating characteristic curve–area under the curve (ROC-AUC) of 0.912 on the test data (Fig. 1, B to C). Using inactive peptides as the external test set, the prediction accuracy reached 90%. In addition, the importance of 94 features of AMPs was ranked, and tilt angle, angle subtended by the hydrophobic residues, propensity to in vitro aggregation, amphiphilicity, hydrophobicity, net charge, amino acid composition, and secondary structure were found to have important roles in the antimicrobial activity (table S2). Existing models (41–46) were used to optimize algorithms for the identification of AMPs, whereas our model focused on the activity level of AMPs. We believe that the prediction of activity levels can accelerate the mining of potent AMPs targeted against specific pathogens. To this end, the method that applies both traversal design and ML was referred to as free-experience design (FED). The original intention of our proposed FED was to fill the gap between amino acids and AMPs and reduce the influence of circumscribed experience; hence, the uncertainty observed in other design models for sequence acquisition and increase the AMPs design success rate.
The six peptides screened using the traversal design were predicted to have a high antimicrobial activity (probability >55%) and were tested using MIC assays (table S3 and Fig. 1D). As shown in Fig. 1D, MLamP3 exhibited broad-spectrum antimicrobial activity, but other MLamPs primarily acted against Gram-negative (G−) pathogens such as Escherichia coli, Salmonella typhimurium, Acinetobacter baumannii, and Shigella flexneri. These results showed that our proposed design system was feasible and that MLamPs had antibacterial activity (success rate > 80%). MLamP1 and MLamP6 were screened as the antibacterial domains of the HAMPs since they were active against G− bacteria and inactive against C. perfringens. Helical wheel projections showed that MLamP1 and MLamP6 were imperfectly amphipathic AMPs (Fig. 1, E and F). AMPs with imperfect amphipathicity exhibit lower cytotoxicity and higher selectivity than AMPs with perfect amphipathicity (47–49). Similarly, both MLamP1 and MLamP6 exhibited low hemolytic activity and cytotoxicity (table S4).
Circular dichroism spectroscopy was used to determine the secondary structures of AMPs in different environments (Fig. 1, G and H). The content of the secondary structure was quantified using the K2D3 algorithm (50). The helix ratio increased as MLamP1 and MLamP6 transitioned from the aqueous environment to the anionic membrane environment and hydrophobic environment (table S5). The conformations of MLamP1 and MLamP6 were environmentally responsive, suggesting that AMPs can switch from random coils to α helices when approaching or penetrating the bacterial membranes. To further understand the structure of the AMPs, Iterative Threading ASSEmbly Refinement (I-TASSER) was used to predict the three-dimensional structure, which indicated that the third structure of MLamP1 and MLamP6 was helical (Fig. 1, I and J) (51).
Phage display selection of specially targeted peptides
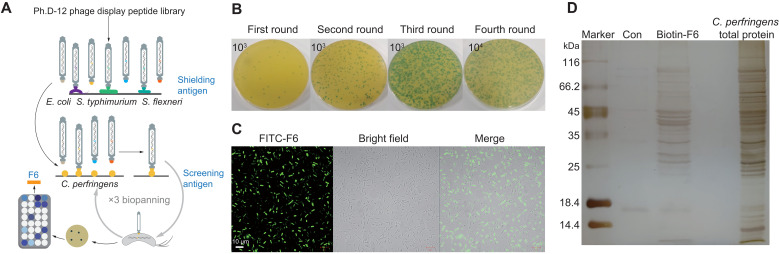
We hypothesized that the use of anti-G− AMPs as antibacterial domain-fused targeting peptides, identified with phage display with subtract biopanning, could increase the generation of STAMPs against C. perfringens (Fig. 2A). After four rounds of biopanning using a 12-peptide random peptide library, ~800 phage clones were selected and tested for affinity with C. perfringens and shielding antigen [E. coli O157:H7, S. typhimurium American Type Culture Collection (ATCC) 14028, and S. flexneri ATCC 12022] affinity (Fig. 2B). The clones that bound to the target bacterium with high affinity and did not or weakly recognized the shielded antigen were selected. Of them, eight peptides were acquired via sequencing (table S6). F6 was selected because of its high affinity and specificity for C. perfringens. Using confocal laser scanning microscopy (CLSM) showed that fluorescein isothiocyanate (FITC)–F6 could bind to C. perfringens (Fig. 2C). We further identified the potential binding targets for the F6 peptide in C. perfringens via the pull-down assays. Compared with that in the control group, biotin-F6 pulled down 13 differential proteins, comprising 12 cytoplasmic proteins and 1 membrane protein (Fig. 2D and table S7). Conclusively, F6 was selected as the targeting peptide for the construction of HAMPs.
Fig. 2. Biopanning and screening of targeting peptides.
(A) Schematic illustration of the phage display technology. (B) Process of four-round biopanning. (C) CLSM images of Clostridium perfringens treated with FITC-F6. Experiments were performed in triplicate with similar results; one representative image is shown. Scale bars, 10 μm. (D) Pull-down of C. perfringens total proteins that interacted with the biotin-F6. Total proteins and biotin-F6 were mixed and pulled down through conjugation with streptavidin-coupled magnetic beads. Proteins recovered from the pull-down experiment were resolved via SDS–polyacrylamide gel electrophoresis and stained with silver staining. Lane 1 is the molecular weight marker; lane 2 is the control group used streptavidin-coupled magnetic beads; lane 3 is F6-captured proteins from the total proteins; lane 4 is the total proteins of C. perfringens. The bands excised from the gel to identify by mass spectrometry (table S7).
In vitro antimicrobial activity, structure, and mechanism of HAMPs
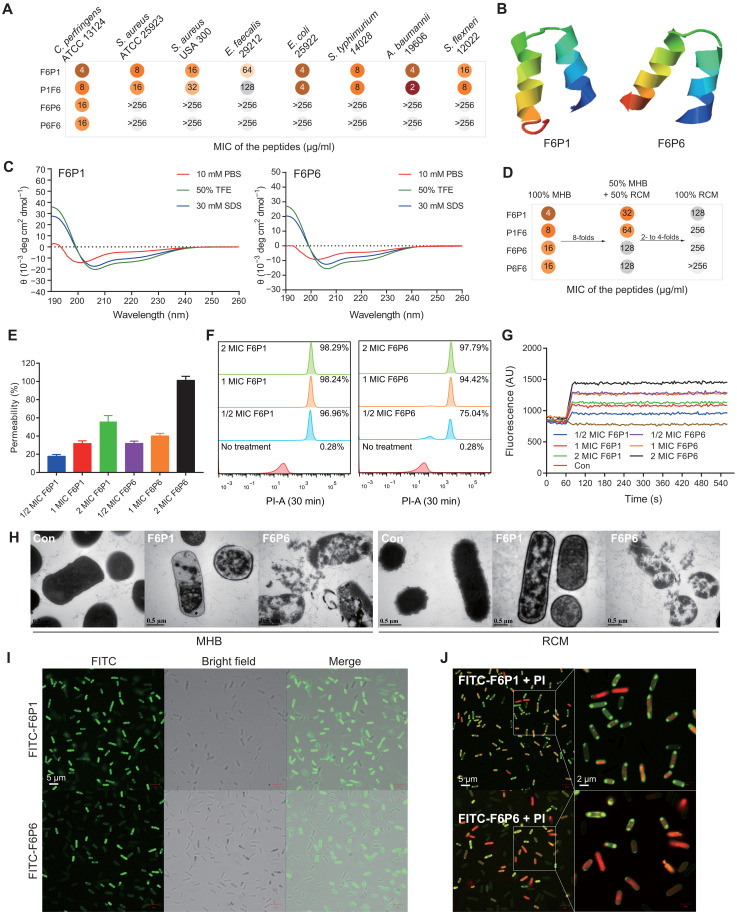
HAMPs were constructed by joining the F6 and MLamP1 or MLamp6 via a short linker (GGG) (table S8). We evaluated the MIC of HAMPs against different species of G− and G+ bacterial species (Fig. 3A). F6 successfully reversed the antibacterial activity of MLamP1 (from 256 to 4 μg/ml) and MLamP6 (from >256 to 16 μg/ml) against C. perfringens. F6P1 and P1F6 were broad-spectrum AMPs, whereas F6P6 and P6F6 were specific AMPs. We evaluated the cytotoxicity and hemolytic activity of the HAMPs that showed good biocompatibility (table S9). On the basis of the cell selection index, we selected F6P1 and F6P6 for follow-up studies. F6P1 and F6P6 were also maintained environment-responsive helical conformation (Fig. 3, B and C, and table S10). Moreover, we found that the MIC of HAMPs increased several fold when the ratio of reinforced Clostridium medium (RCM), which is rich in glucose, was increased in the culture medium (Fig. 3D). We considered that abundant nutrients, especially carbohydrates, exacerbated C. perfringens proliferation and spore germination (52). Both 1× MIC F6P1 and F6P6 could rapidly reduced the load of C. perfringens cultivated in Mueller-Hinton Broth (MHB) within 15 min, and the pathogen was entirely killed within 2 hours (fig. S2A). In a high-glucose environment, C. perfringens metabolized more gelatinous metabolites, experienced exacerbated proliferation and spore germination, and produced thicker capsules (fig. S2, B to D). We further assessed the variations in the antibacterial activity of HAMPs against C. perfringens cultured under four different conditions (details in fig. S2, E and F). Specifically, we used flow cytometry to analyze the growth patterns of C. perfringens and the propensity of HAMPs to disrupt bacteria of varying morphologies (figs. S3 to S4). Above results showed that HAMPs seems to have a stronger ability to destroy spores than rhabditiform C. perfringens, but HAMPs preferentially destroy rhabditiform C. perfringens. Thus, early escaped free spores will transform into rhabditiform bacteria, which will consume more HAMPs.
Fig. 3. Design, antimicrobial activity, and bactericidal action of HAMPs.
(A) MIC of the HAMPs.Clostridium perfringens was cultured in RCM, and other bacteria were cultured in MHB. The bacterial solution was diluted in MHB for MIC assays. Each test was performed in three independent experiments. (B) 3D structure of F6P1 and F6P6, both of which have an α-helix structure. (C) CD spectra of F6P1 and F6P6 in PBS (pH = 7.4), 30 mM SDS, and 50% TFE. (D) MIC of the HAMPs against C. perfringens cultivated in different medium (100% MHB, 50% MHB + 50% RCM, and 100% RCM). (E to G) Mechanistic tests, including N-phenyl-1-naphthyl amine (NPN), propidium iodide (PI), and DiSC3-5 assays, showing the effects of HAMPs on cell wall permeabilization and membrane permeability and depolarization. C. perfringens was cultured in MHB. Dosage-dependent increase in signals was observed. Data are presented as the means ± SEM. n = 3 or 4 biologically independent samples for each assay. (H) Transmission electron microscopy (TEM) images of untreated or HAMPs-treated C. perfringens cultured in MHB (left) or RCM (right). Scale bars, 0.5 μm. (I and J) Fluorescence image of C. perfringens treated with 2× MIC FITC-HAMPs (I) and additional 10 μM PI (J). Scale bars, 5 μm. AU, arbitrary unit.
We further explored the predominant mechanism of the bactericidal action of HAMPs by examining their effects on cell wall permeabilization, membrane permeability, and membrane depolarization, transmission electron microscopy (TEM), and CLSM assay. We used N-phenyl-1-naphthyl amine (NPN), a hydrophobic and sensitive fluorescent probe, to measure cell wall permeabilization. As shown in Fig. 3E, HAMPs penetrated cell wall of C. perfringens in a dose-dependent manner; F6P6 showed a stronger ability to damage the cell wall than F6P1. Membrane disruption was measured using propidium iodide (PI) uptake. Flow cytometric analysis showed that HAMPs can rapidly destroy the bacterial membrane within 30 min. When the concentrations of peptides F6P1 and F6P6 approached 1× MIC, they caused the death of ~98 and ~94% of C. perfringens cells, respectively (Fig. 3F and fig. S5A). Membrane depolarization was measured using the DiSC3-5 cationic dye. The extent of inner membrane depolarization in HAMPs-treated C. perfringens increased in a concentration-dependent manner. F6P1 and F6P6 at 1× MIC increased membrane depolarization levels by approximately 30 and 40%, respectively (Fig. 3G and fig. S5B). Using TEM, we further explored the morphological changes in bacteria treated with 2× MIC HAMPs. As shown in Fig. 3H, the control group bacteria exhibited a clear capsule and cell wall, complete membrane structure, and dense cytoplasm; the F6P1-treated bacteria showed no capsule, a mildly attenuated cell wall, membrane rupture, and cytoplasmic vacuole formation. By contrast, F6P6-treated bacteria showed a completely damaged bacterial morphology with capsule and cell wall dissolution, membrane destruction, and cytoplasm leakage. CLSM showed that FITC-HAMPs could target and localize to the C. perfringens membrane to facilitate the entry of PI into the cytoplasm, suggesting that the HAMPs had both targeting ability and antibacterial activity (Fig. 3, I and J). Collectively, these results indicated that HAMPs targeted C. perfringens, disintegrated the capsule and cell wall, disrupted membrane integrity, increased membrane depolarization, and promoted cytoplasmic efflux to kill the bacterium. F6P1 tended to disrupt bacterial membranes, whereas F6P6 was inclined to disrupt capsules and bacterial walls, which could also explain the difference in their MICs. Overall, F6P6 caused more serious and thorough morphological damage to C. perfringens than F6P1, but F6P1 was more likely to kill C. perfringens than F6P6 at the same concentration.
Moreover, C. perfringens can form biofilms that adhere to host wounds and the intestinal epithelium and help the pathogens to evade host immunity and antimicrobial agents (53). Antibiofilm assays were conducted to test the biofilm-disrupting properties of HAMPs. HAMPs demonstrated the ability to inhibit biofilm formation and remove pre-established biofilms (fig. S6).
Bacterial aggregation and self-assembly of HAMPs
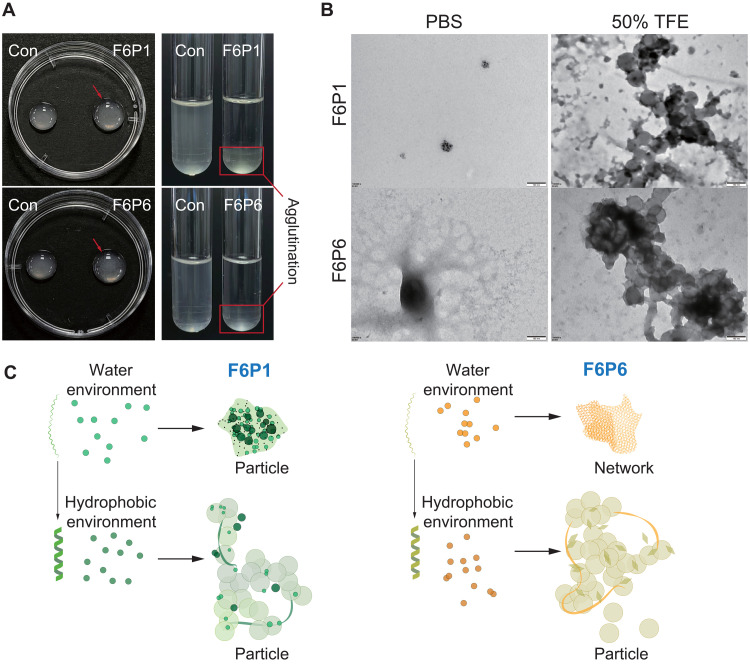
The phenomenon of bacterial agglutination was observed in MIC and antibiofilm assays. We tested the agglutination effect of the HAMPs on planktonic bacteria with a high inoculation concentration and found that the HAMPs could agglutinate bacteria (Fig. 4A). We used 1-anilino-8-naphthalene sulfonate (ANS) fluorescence assay to measure the self-assembling ability and critical aggregation concentration (CAC) of the HAMPs. The ANS fluorescence intensity increased with increasing peptide concentration and aggregation strength, indicating that the HAMPs aggregated to form a supramolecular particle (fig. S7). The CAC of F6P1 and F6P6 was 4.40 and 18.14 μg/ml, respectively. As shown in fig. S8, F6P1 and F6P6 form nanoparticles with a diameter of 428 ± 48 nm and 254 ± 28 nm, respectively. However, the size and morphology of HAMPs in an aqueous environment are not uniform. The zeta potential of F6P1 and F6P6 was 8.63 and 23.20 mV, respectively. TEM showed that F6P1 and F6P6 formed nanoparticles and nanonetworks in an aqueous environment, respectively (Fig. 4B). Previous studies have shown that the structure of HAMPs is environmentally responsive; thus, we investigated their ability to self-assemble in other environments. The CAC of F6P1 and F6P6 dropped to 0.5 and 0.8 μg/ml in an anionic membrane environment and 3.6 and 5.6 μg/ml in a hydrophobic environment, respectively (fig. S9). In a hydrophobic environment, HAMPs formed α helices and self-assembled into vacuolar nanoparticles (Fig. 4C). In addition, the activity of self-assembled AMPs against planktonic bacteria may be associated with their CAC (fig. S10). The MIC value for planktonic bacteria became close to that of CAC. Previous studies have also found that the MICs of AMPs were close to their CACs (54, 55). Together, these results indicated that a balance relationship exists between the antibacterial and entrapment ability, and the antibacterial activity gradually transitioned to bacterial agglutination as the concentration of HAMPs increased.
Fig. 4. Bacterial aggregation and self-assembly of HAMPs.
(A) Entrapment phenomenon caused by HAMPs. HAMPs (128 μg/ml) agglutinated 108 CFU bacteria in 100 μl of PBS in 1 hour (left). HAMPs (128 μg/ml) agglutinated 107 CFU bacteria in 2 ml of PBS in 8 hours (right). (B) TEM images of HAMPs at a concentration of 128 μg/ml in water or hydrophobic environment. Scale bars, 100 nm. (C) Schematic illustration of the self-assembly of HAMPs.
In vivo biocompatibility and biodistribution of HAMPs
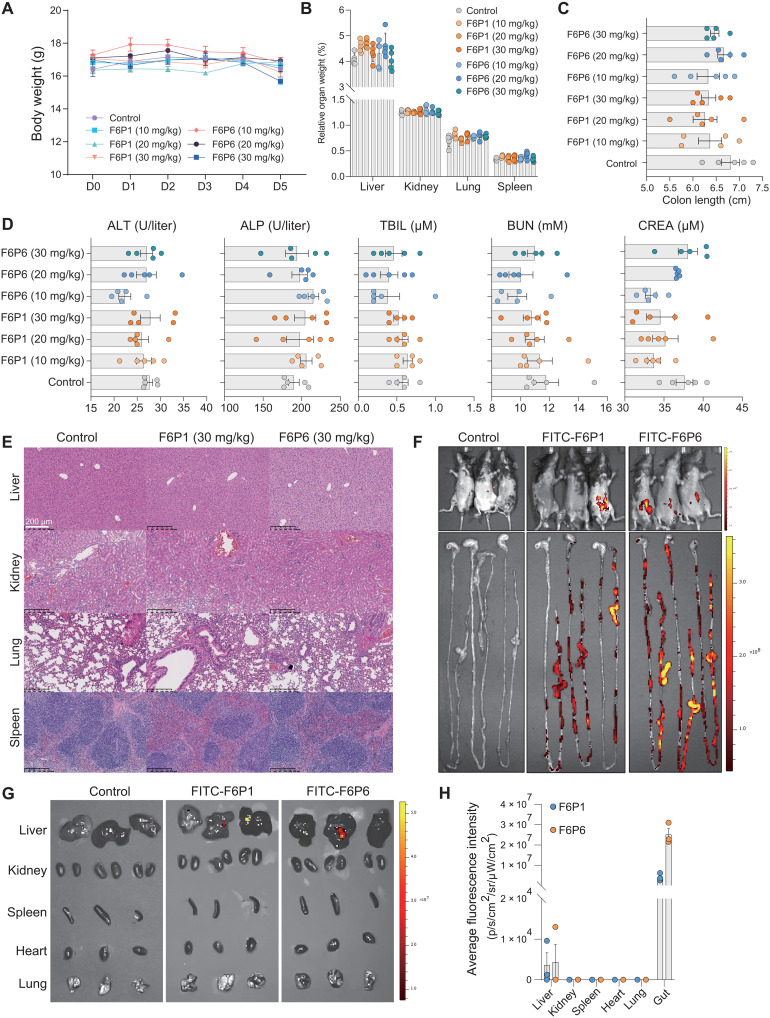
To investigate the in vivo toxicity of HAMPs, we intraperitoneally injected F6P1 or F6P6 (10, 20, or 30 mg/kg) into C57BL/6 mice for 5 days and evaluated the potential adverse reactions of HAMPs through body weight, relative organ weight, colon length, organ histological examination, and liver and renal function-related indexes, including alanine transaminase, alkaline phosphatase, total bilirubin, blood urea nitrogen, and creatinine. As shown in Fig. 5 (A to D), no significant differences were observed in body weight, relative organ weight, and colon length. Serum indicators were maintained at normal physiological levels, indicating that the HAMPs did not induce hepatotoxicity or nephrotoxicity. Hematoxylin and eosin (H&E) staining also showed no evident tissue abnormalities in any treatment groups (Fig. 5E and fig. S11). Together, the HAMPs F6P1 and F6P6 did not induce primary toxic irritation, showed good biocompatibility in vivo, and provided a dosage basis for preclinical testing. Using in vivo imaging systems, we investigated the biodistribution of the HAMPs and found that 12 hours after intraperitoneal delivery, FITC-HAMPs were distributed in the abdomen, enriched in the gut, and had remnants in the liver (Fig. 5, F to H). The fluorescence intensity of intestinal tissues was 7.4-logs higher than that of other tissues, indicating that HAMPs were highly selective and targeted to the gut. Thus, F6P1 and F6P6 may be gut-targeted AMPs.
Fig. 5. Biocompatibility and biodistribution of HAMPs in vivo.
(A to C) Changes in body weight, relative organ weight, and colon length of C57BL/6 mice after administration of HAMPs (0, 10, 20, or 30 mg/kg). PBS treatment group was regarded as a control. Data are presented as the means ± SEM. n = 5 mice for per group. (D) Renal and liver function-related indexes of mice following administration of HAMPs for 5 days. Data are presented as the means ± SEM. n = 5 biologically independent samples. In (B) to (D), the differences between the groups were determined using one-way analysis of variance (ANOVA) followed by Tukey's post hoc analysis. No significant difference between the groups (P > 0.05) was observed. (E) Histopathological morphology analysis of the liver, kidney, lung, and spleen in mice following administration of HAMPs (30 mg/kg) for 5 days. Scale bars, 200 μm. Histopathological morphology analysis of other doses of HAMPs is shown in fig. S11. (F to H) Mice and organs were imaged via in vivo imaging system at 12 hours after intraperitoneal injection of FITC-HAMPs (5 mg/kg); the intestine showed strong signals compared with other organs. PBS treatment as control. Data are presented as the means ± SEM. n = 3 mice for per group. ALT, alanine transaminase; ALP, alkaline phosphatase; TBIL, total bilirubin; BUN, blood urea nitrogen; CREA, creatinine.
In vivo efficacy of HAMPs and EPV@HAMPs
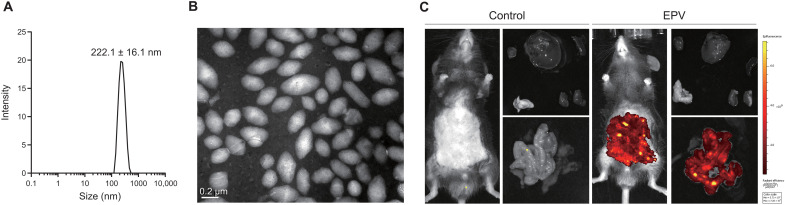
To improve the colonic bioavailability of HAMPs, we designed gut-targeted nanoparticles, termed EPV, that are coated with biotin to bind intestinal lymphoid cells and dendritic cells. The diameter and zeta potential of EPV were 222.1 ± 16.1 nm and −30.3 mV, respectively (Fig. 6A). TEM results showed EPV was the mesoporous fusiform nanoparticles (Fig. 6B). As shown in Fig. 6C, EPV had a targeting biodistribution in intestine after 2-day injection. Here, we used EPV to deliver HAMPs to treat CPI. In addition, the results of MIC and antibiofilm assays showed that EPV enhanced the antibacterial activity and bacterial agglutination ability of HAMPs, which we hypothesized that EPV might increase the local concentration of HAMPs around bacteria (fig. S12).
Fig. 6. Characterization of gut-targeted EPV.
(A) Diameter of EPV. (B) TEM images of EPV. Scale bar, 0.2 μm. (C) Modified EPV targeting biodistribution in intestine after 2-day injection. PBS treatment as control. Experiments (B to C) were performed in triplicate with similar results; one representative image is shown.
Food-borne CPI is a self-limiting disease usually resolved within 24 hours; however, CPI after systemic antibiotic administration aggravates the colonization and overgrowth of C. perfringens, which causes more serious and intractable diseases (3). Therefore, CPI mice pretreated with antibiotics was used to evaluate in vivo activity of HAMPs and EPV@HAMPs. In addition, there were no significant differences observed in the bacterial load in both mice feces and colonic tissues between mice infected with C. perfringens cultured in RCM and MHB (fig. S13).
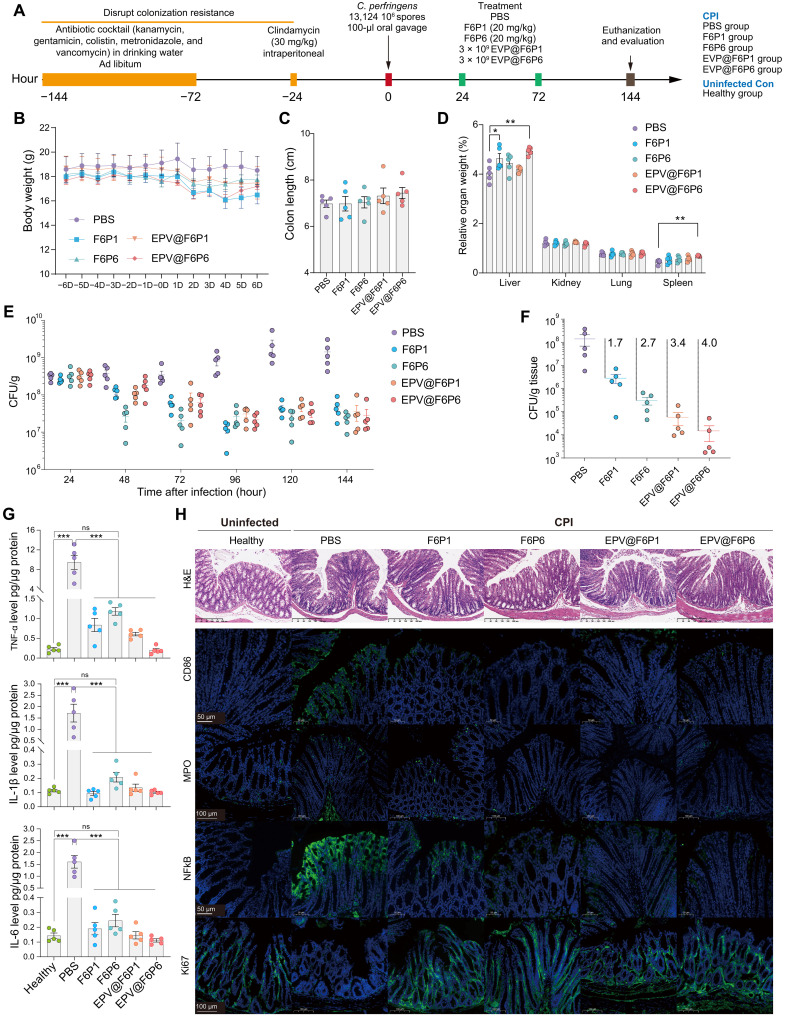
Subsequent to antibiotic cocktail and clindamycin treatment in mice, mice were orally administered high-inoculum C. perfringens cultured in RCM. HAMPs (20 mg/kg) and EPV@HAMPs (~3 × 109 particles) were injected intraperitoneally at 24 and 72 hours after infection, and the phosphate-buffered saline (PBS) treatment group was used as a control (Fig. 7A). During the 6-day assay period, no significant differences in the body weight and colon length of the mice were observed among all groups (Fig. 7, B and C). As shown in Fig. 7D, compared with the control group, EPV@F6P6 group had significantly increased relative liver and spleen weight, whereas the F6P1 group had significantly increased relative liver weight.
Fig. 7. In vivo efficacy of HAMPs and EPV@HAMPs against CPI.
(A) Experimental design for CPI and treatment. CPI mice were injected HAMPs intraperitoneally, EPV@HAMPs, or PBS, and PBS treatment group was regarded as a control. Healthy mice were used as uninfected control for enzyme-linked immunosorbent assay and immunofluorescence assays. (B to D) Body weight, colon length, and relative organ weight of mice. Data are presented as the means ± SEM. n = 5 mice per group. (E and F) Bacterial load of Clostridium perfringens in feces (E) and colon tissue (F) of infected mice. Data are presented as the means ± SEM. Each point represents a mouse. n = 5 for each group. (G) Levels of tumor necrosis factor–α (TNF-α), interleukin-1β (IL-1β), and IL-6 in colon tissues of healthy and CPI mice treated with PBS and HAMPs and EPV@HAMPs. (H) Histopathological H&E staining of colon tissues and immunofluorescence analysis of colonic cryosections. Colon tissues were stained with CD86, myeloperoxidase (MPO), nuclear factor κB (NFκB), or Ki67 antibody, whereas nuclei were stained with DAPI. Scale bars, 50, 100, or 200 μm. In (C), (D), and (G), the differences between the groups were determined using one-way ANOVA followed by Tukey's post hoc analysis. *P < 0.05, **P < 0.01, and ***P < 0.001. ns, not significant.
After gavage of 106 colony-forming units (CFU) C. perfringens, fecal samples were collected separately every day to quantify the C. perfringens load. We found that 24 hours after infection, C. perfringens in the feces was ~3 × 108 CFU/g and peaked at ~2 × 109 CFU/g at 120 hours, indicating that C. perfringens had successfully colonized and overgrown in the colon of inoculated mice. HAMPs and EPV@HAMPs suppressed the marked overgrowth of C. perfringens, and the bacterial load decreased by 1.5-log (up to 97% killing efficacy) in the feces of treated mice compared with that in the control (Fig. 7E). Both HAMPs and EPV@HAMPs reduced the colonic C. perfringens load; EPV@HAMPs reduced the load by over 1.3-log than the corresponding HAMPs (Fig. 7F). F6P6 reduced the C. perfringens load in the feces and colon more rapidly and efficiently than the broad-spectrum F6P1. For the organ translocation of C. perfringens, we only detected C. perfringens (~104 CFU/g) in the liver of the control group, but not in the HAMPs and EPV@HAMPs groups (fig. S14). These results show that HAMPs and EPV@HAMPs reduced the load of C. perfringens in the gut to mitigate C. perfringens intestinal infection and lower the risks of bacterial colon invasion and translocation. This undoubtedly provides more time for the treatment of Clostridial infections. Moreover, the levels of the proinflammatory factors tumor necrosis factor–α (TNF-α), interleukin-6 (IL-6), and IL-1β and C. perfringens alpha toxin significantly decreased in the serum and colon of treated mice compared with PBS-treated mice (Fig. 7G and fig. S15).
We next conducted an in-depth analysis to determine how HAMPs and EPV@HAMPs protected mice from CPI. The pathological changes in the colon were evaluated using H&E staining, which indicated that the treatment with HAMPs and EPV@HAMPs reduced inflammatory cell infiltration in the colon (Fig. 7H). Immunofluorescence analysis showed that, compared with the infected control group, the HAMP and EPV@HAMP groups had reduced CD86, nuclear factor κB (NFκB), and myeloperoxidase expression and increased Ki67 expression with alleviated colitis signs (Fig. 7H). These findings are consistent with the results obtained from the colon infection and enzyme-linked immunosorbent assay assays, indicating that the administration of HAMPs and EPV@HAMPs led to a reduction in bacterial burden, resulting in a lower inflammatory response and toxin levels in treated mice.
We further investigated the effects of HAMPs and EPV@HAMPs on inflammatory cytokine levels and colonic tight junction proteins in healthy mice. Compared to the PBS-treated group, both HAMPs and EPV@HAMPs showed no significant impact on the levels of inflammatory cytokines in the colon and serum (fig. S16). Immunofluorescence analysis revealed that neither HAMPs nor EPV@HAMPs influenced the expression of CD86 and NFκB in colon tissues, while EPV@HAMPs increased the expression of tight junction proteins such as Occludin and zonula occludens-1 (ZO-1) (fig. S17). These findings suggest that EPV@HAMPs treatment enhanced protection to the colon compared to HAMPs alone in CPI, potentially by enhancing the integrity of the intestinal barrier to resist bacterial invasion.
16S rRNA sequencing analysis of gut microbiota
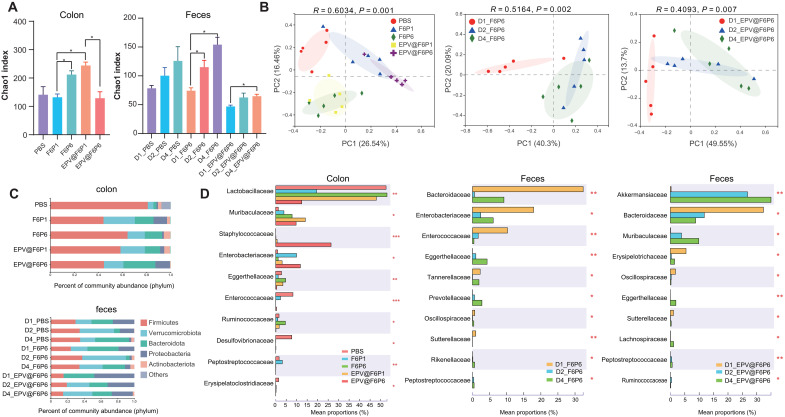
To investigate the effects of and differences between broad- spectrum and specific AMPs on the composition and abundance of the gut microbiota, we used 16S ribosomal RNA (rRNA) sequencing technology to further analyze colonic and fecal bacteria in mice. Both community richness and α-diversity of gut microbiota in mice-administered F6P6 were significantly higher than those of mice-administered F6P1, suggesting that specific AMPs were more conducive to the remodeling of disturbed microflora. The Chao1 index of EPV@F6P1 was significantly higher than that of F6P1, possibly since EPV alleviated the direct effect of F6P1 on the microbiota (Fig. 8A and fig. S18). F6P6 and EPV@F6P6 significantly increased the richness of fecal microbiota with the increase in duration or dosage. Furthermore, the treatment of HAMPs and EPV@HAMPs significantly altered the composition of colonic microflora in CPI mice (Fig. 8B). Community composition and species differences between samples at the phylum and family level (Fig. 8, C and D, and fig. S19) suggested that HAMPs and EPV@HAMPs increased the abundance of microbial consortium of mucosal sugar utilizers such as Akkermansiaceae and Muribaculaceae as well as the probiotics Lactobacillaceae and decreased harmful bacteria that exacerbate gut inflammation, such as Clostridiaceae, Enterobacteriaceae, Desulfovibrionaceae, Sutterellaceae, Erysipelatoclostridiaceae, and Bacteroidaceae (56–60). Random forest and network analyses revealed important bacterial and related networks of F6P6 and EPV@F6P6 (fig. S20). Both F6P6 and EPV@F6P6 reduced the microbiota functions related to carbohydrate transport and metabolism, which could reduce the nutrient supply of C. perfringens (fig. S21). Moreover, C. perfringens produces histotoxic gas through glucose fermentation, which is a hallmark of CPI. Together, HAMPs and EPV@HAMPs inhibited the colonization and overgrowth of C. perfringens via four main ways: (i) direct bactericidal effect, (ii) increased the abundance of probiotics to competitively inhibit pathogens, (iii) increased the abundance of symbiotic bacteria with the same niche as C. perfringens that compete for the growth space of C. perfringens, and (iv) decreased the nutrient sources of C. perfringens.
Fig. 8. 16S rRNA sequencing analysis of gut microbiota regulated by HAMPs and EPV@HAMPs.
(A) Chao1 index of observed operational taxonomic units showing the α-diversity of the microbial community in the colon (left) and feces (right) of CPI mice. D1, D2, and D4 represent the fecal samples collected after 0, 1, and 2 administrations of F6P6 or EPV@F6P6, respectively. The Kruskal-Wallis test was applied for statistical analysis. *P < 0.05. (B) Principal coordinate analysis showing the β-diversity of the colonic contents (left) and F6P6-treated group (middle) and EPV@F6P6-treated group (right). Each point represents a mouse. n = 5 for each group. The significance of clustering was determined using analysis of similarities (ANOSIM). (C) Community histogram showing the microbial compositional profiling at the phylum level. n = 5 for each group. (D) Relative abundance of microbiota that is significantly altered at the family level. The Kruskal-Wallis test was applied for statistical analysis. *P < 0.05, **P < 0.01, and ***P < 0.001.
Treatment of persistent CPI with F6P6 and EPV@F6P6
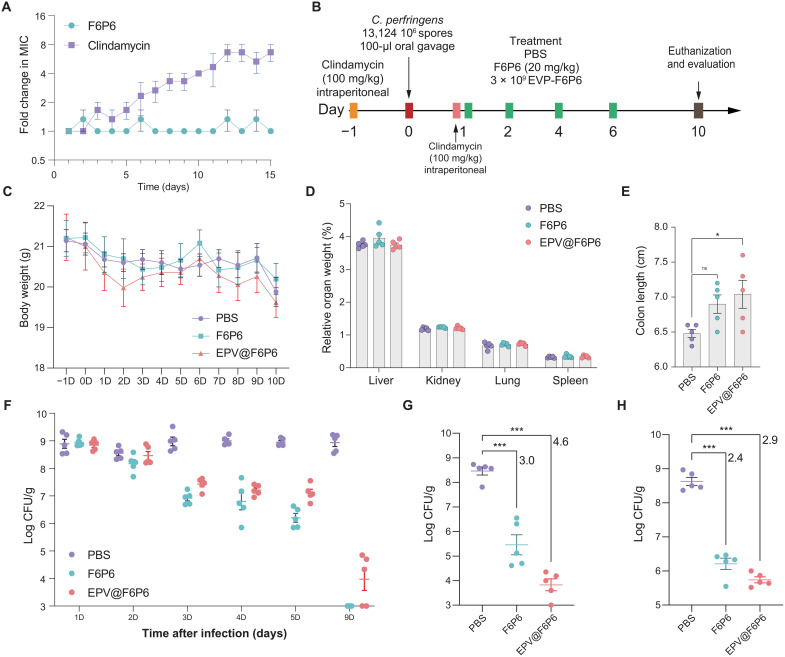
We used a clindamycin-treated CPI model to investigate the ability of F6P6 and EPV@F6P6 to protect against antibiotic-induced CPI. Clindamycin is an alternative to penicillin for the treatment of CPI; however, it increases the risk of Clostridium spp. overgrowth. Before that, we found no spontaneous resistance development to F6P6, which was attributed to its mechanism of action of destroying bacterial wall and membranes (Fig. 9A). Conversely, clindamycin showed an eightfold increase in the MIC during serial passages at sub-MIC concentrations.
Fig. 9. F6P6 and EPV@F6P6 treat persistent CPI.
(A) Resistance development of C. perfringens to the drugs at sub-MIC concentrations. Data are presented as the means ± SEM. n = 3. (B) Experimental design for persistent CPI and treatment. (C to E) Body weight, relative organ weight, and colon length of mice. Data are presented as the means ± SEM. n = 5 mice per group. (F to H) Bacterial load of C. perfringens in feces (F), colonic contents (G), and colon tissues (H) of infected mice. Data are presented as the mean ± SEM. Each point represents a mouse. n = 5 for each group. In (E), (G), and (H), the differences between the groups were determined using one-way ANOVA followed by Tukey's post hoc analysis. *P < 0.05 and ***P < 0.001. ns, not significant.
We constructed a persistent CPI model in which clindamycin was administered after establishing the model but before antimicrobial therapy (Fig. 9B). In this assay, 103 to 104 CFU C. perfringens/g feces were considered to be the end point as C. perfringens constitutes the normal human and animal gut flora at up to 103 CFU/g feces (61). We observed no significant differences in the body weight and relative organ weight among any groups (Fig. 9, C and D). Colonic shortening occurs as a result of colonic inflammation, which leads to swelling and affects the length of the colon (62). Compared with that in the control group, EPV@F6P6 significantly increased colon length (Fig. 9E). C. perfringens in feces was approximately 109 CFU/g at 24 hours after infection. The control group maintained high fecal counts (~109 CFU/g) of C. perfringens, whereas the treatment groups displayed sterilization beginning at 1 day after treatment, with 100% of the F6P6-treated or 40% of the EPV@F6P6-treated mice free of infection 8 days after treatment (Fig. 9F). As shown in Fig. 9 (G and H) and fig. S22, F6P6 and EPV@F6P6 significantly reduced the bacterial load in the colon contents and colonic tissues and lessen the infiltration of inflammatory cells. Collectively, these results suggested that F6P6 and EPV@F6P6 exhibit promising efficacy in treating antibiotic-induced persistent CPIs and that EPV@F6P6 might be more protective to the colon than F6P6.
Limitations of this study
Our study has several limitations. First, ML models may struggle with peptides that have identical amino acid compositions and similar parameters. Therefore, in this work, we substitute sequences obtained from an exhaustive design approach for the full library sequences to improve design efficiency. Second, the pull-down assay did not find a specific target protein, but according to the CLSM results of F6 and the hybrid peptides, the target protein was mainly distributed at both ends and the C. perfringens membrane. Third, limited by the colonic infection model, antimicrobials reduce bacterial burden to reduce inflammatory response. Future studies can explore the anti-inflammatory or immunomodulatory activities of antimicrobials in noninfectious colitis models.
In this study, we propose an approach for the design and delivery of STAMPs against enteric pathogens and difficult-to-treat intestinal infections. F6P6 and EPV@F6P6 are promising drug candidates for treating CPI given their activity against planktonic bacteria and biofilms, difficult to develop resistance, efficacy against persistent CPI, and ability to optimize gut flora composition and target the colon. Our study provides an insight into the development of STAMP capable of combating intestinal infections caused by other pathogens. Engineered bacteria-expressing STAMPs may have clinical implication in preventing and treating intestinal infections, which we intend to develop in future work.
MATERIALS AND METHODS
Study design
The goal of this study is to enhance the design efficiency and delivery of STAMPs via nanoparticles to treat intestinal infections. Initially, we propose a ML-based FED to expedite AMPs design, and the efficiency of the design is validated using MIC assays. A broad-spectrum AMP P1 and a narrow-spectrum AMP P6 are considered as antimicrobial structural domains due to their high cell selectivity. Broad- and narrow-spectrum AMPs were used to hybridize the targeting peptide from a phage display library. Then, according to the antimicrobial spectra and activity of the HAMPs, broad-spectrum and specific HAMPs were screened. During the process, we explored the apparent patterns of C. perfringens' defenses against HAMPs. We further explored the predominant mechanism of the bactericidal action of HAMPs using various in vitro assays, including NPN, PI, and DiSC3-5, along with TEM and CLSM. We observed the entrapment properties of the HAMPs and tested the self-assembling ability of HAMPs in water and a hydrophobic environment using ANS and TEM assays.
The in vivo biocompatibility and biodistribution of HAMPs were evaluated using renal and liver function-related indexes, histopathological morphology, and in vivo imaging systems. To enhance the bioavailability and targeting of HAMPs, we designed a gut-targeted EPV. The in vivo efficacy of HAMPs and EPV@HAMPs was accessed in a CPI mouse model. In addition, we evaluated the effects of HAMPs and EPV@HAMPs on the levels of serum and colitis inflammatory factors in healthy mice, as well as their impact on the tight junction proteins in the colon. Furthermore, 16S rRNA sequencing was used to determine the composition and distribution of the gut microbiota regulated by HAMPs and EPV@HAMPs. Last, the in vivo activity of F6P6 and EPV@F6P6 was validated in mice with persistent CPI.
Animals
All mice (female C57BL/6) were obtained and maintained in pathogen-free conditions at the Laboratory Animal Centre of Zhejiang University and provided with water and a standard laboratory diet ad libitum, unless otherwise indicated. All animal studies were performed in compliance with the Guide for the Care and Use of Laboratory Animals and adopted the protocol that has been approved by the Medical Experimental Animal Care Commission of Zhejiang University (ZJU20220164).
Statistical analysis
Statistics were processed and analyzed by using GraphPad Prism 9.3.1 (GraphPad, USA). Thereinto, quantitative data are expressed as means ± SEM, whereas categorical data are expressed as numbers (percentages). The nonparametric Mann-Whitney U test and analysis of variance (ANOVA) followed by Tukey's post hoc analysis were used to compare the differences in two or more groups. The GraphPad Prism was used for graph plotting. Statistical significance was set at P < 0.05.
Acknowledgments
We thank the Medical Experimental Animal Care Commission of Zhejiang University for the help of animal experiments. We are grateful to Analysis Center of Agrobiology and Environmental Sciences of Zhejiang University for the support of TEM assays. We thank Y. Li (Analysis Center of Agrobiology and Environmental Science of Zhejiang University) for technical assistance on laser confocal microscopy. We thank Wuhan Gene Create Biological Engineering Co. Ltd. for the support of the phage display assays and pull-down assays.
Funding: This work was supported by the National Natural Science Foundation of China (grant nos. U21A20249 and 32022079), Zhejiang Provincial Key R&D Program of China (grant no. 2021C02008), and China Agriculture Research System of MOF and MARA (CARS-35).
Author contributions: Conceptualization: B.X. Methodology: B.X., W.S., L.W., T.C., X.J., Z.J., R.Y., P.Z., and C.Y. Investigation: B.X. Visualization: B.X. Funding acquisition: Y.W. and M.L.J. Project administration: Y.W. and M.L.J. Supervision: Y.W. and M.L.J. Writing—original draft: B.X. Writing—review and editing: B.X., W.S., and C.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The data shown in the images are stored in data S1. The relevant dataset, code, and model of ML are available on Dryad (DOI: 10.5061/dryad.31zcrjds7) and GitHub (https://github.com/xubocheng/Anti_Cp.git). The sequence data were then deposited in the Sequence Read Archive under the accession number PRJNA857195.
Supplementary Materials
This PDF file includes:
Experimental Section
Figs. S1 to S23
Tables S1 to S10
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Murray C. J., Ikuta K. S., Sharara F., Swetschinski L., Aguilar G. R., Gray A., Han C., Bisignano C., Rao P., Wool E., Johnson S. C., Browne A. J., Chipeta M. G., Fell F., Hackett S., Haines-Woodhouse G., Hamadani B. H. K., Kumaran E. A. P., McManigal B., Agarwal R., Akech S., Albertson S., Amuasi J., Andrews J., Aravkin A., Ashley E., Fr S. B., Baker B., Basnyat A., Bekker R., Bender A., Bethou J., Bielicki S., Boonkasidecha J., Bukosia C., Carvalheiro C., Castañeda-Orjuela V., Chansamouth S., Chaurasia S., Chiurchiù F., Chowdhury A. J., Cook B. C., Cressey T. R., Criollo-Mora E., Cunningham M., Darboe S., Day N. P. J., De Luca M., Dokova K., Dramowski A., Dunachie S. J., Eckmanns T., Eibach D., Emami A., Feasey N., Fisher-Pearson N., Forrest K., Garrett D., Gastmeier P., Giref A. Z., Greer R. C., Gupta V., Haller S., Haselbeck A., Hay S. I., Holm M., Hopkins S., Iregbu K. C., Jacobs J., Jarovsky D., Javanmardi F., Khorana M., Kissoon N., Kobeissi E., Kostyanev T., Krapp F., Krumkamp R., Kumar A., Kyu H. H., Lim C., Limmathurotsakul D., Loftus M. J., Lunn M., Ma J., Mturi N., Munera-Huertas T., Musicha P., Mussi-Pinhata M. M., Nakamura T., Nanavati R., Nangia S., Newton P., Ngoun C., Novotney A., Nwakanma D., Obiero C. W., Olivas-Martinez A., Olliaro P., Ooko E., Ortiz-Brizuela E., Peleg A. Y., Perrone C., Plakkal N., Ponce-de-Leon A., Raad M., Ramdin T., Riddell A., Roberts T., Robotham J. V., Roca A., Rudd K. E., Russell N., Schnall J., Scott J. A. G., Shivamallappa M., Sifuentes-Osornio J., Steenkeste N., Stewardson A. J., Stoeva T., Tasak N., Thaiprakong A., Thwaites G., Turner C., Turner P., van Doorn H. R., Velaphi S., Vongpradith A., Vu H., Walsh T., Waner S., Wangrangsimakul T., Wozniak T., Zheng P., Sartorius B., Lopez A. D., Stergachis A., Moore C., Dolecek C., Naghavi M., Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laxminarayan R., The overlooked pandemic of antimicrobial resistance. Lancet 399, 606–607 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larcombe S., Hutton M. L., Lyras D., Involvement of bacteria other than Clostridium difficile in antibiotic-associated diarrhoea. Trends Microbiol. 24, 463–476 (2016). [DOI] [PubMed] [Google Scholar]
- 4.McFarland L. V., Antibiotic-associated diarrhea: Epidemiology, trends and treatment. Future Microbiol. 3, 563–578 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Banaszkiewicz A., Kdzielska J., Gawrońska A., Pituch H., Obuch-Woszczatyński P., Albrecht P., Młynarczyk G., Radzikowski A., Enterotoxigenic Clostridium perfringens infection and pediatric patients with inflammatory bowel disease. J. Crohns Colitis 8, 276–281 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Foodborne, Waterborne, and Environmental Diseases, Prevention, Prevent Illness from C. perfringens (2023);www.cdc.gov/foodsafety/diseases/clostridium-perfringens.html.
- 7.Carman R. J., Clostridium perfringens in spontaneous and antibiotic-associated diarrhoea of man and other animals. Rev. Med. Microbiol. 8, S46 (1997). [Google Scholar]
- 8.Wang J., Dou X., Song J., Lyu Y., Zhu X., Xu L., Li W., Shan A., Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 39, 831–859 (2019). [DOI] [PubMed] [Google Scholar]
- 9.L. K. Chung, M. Raffatellu, in Seminars in Cell and Developmental Biology (Elsevier, 2019), vol. 88, pp. 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kintses B., Méhi O., Ari E., Számel M., Györkei Á., Jangir P. K., Nagy I., Pál F., Fekete G., Tengölics R., Nyerges Á., Likó I., Bálint A., Molnár T., Bálint B., Vásárhelyi B. M., Bustamante M., Papp B., Pál C., Phylogenetic barriers to horizontal transfer of antimicrobial peptide resistance genes in the human gut microbiota. Nat. Microbiol. 4, 447–458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert R., Qi F., Yarbrough D. K., He J., Anderson M. H., Shi W., Adding selectivity to antimicrobial peptides: Rational design of a multidomain peptide against Pseudomonas spp. Antimicrob. Agents Chemother. 50, 1480–1488 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H., Jang J. H., Kim S. C., Cho J. H., Development of a novel hybrid antimicrobial peptide for targeted killing of Pseudomonas aeruginosa. Eur. J. Med. Chem. 185, 111814 (2020). [DOI] [PubMed] [Google Scholar]
- 13.de la Fuente-Nunez C., Torres M. D., Mojica F. J., Lu T. K., Next-generation precision antimicrobials: Towards personalized treatment of infectious diseases. Curr. Opin. Microbiol. 37, 95–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei M., Jayaraman A., Van Deventer J. A., Lee K., Engineering selectively targeting antimicrobial peptides. Annu. Rev. Biomed. Eng. 23, 339–357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu J. N., Traverso G., Foundations of gastrointestinal-based drug delivery and future developments. Nat. Rev. Gastroenterol. Hepatol. 19, 219–238 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Zeeshan M., Ali H., Khan S., Khan S. A., Weigmann B., Advances in orally-delivered pH-sensitive nanocarrier systems; an optimistic approach for the treatment of inflammatory bowel disease. Int. J. Pharm. 558, 201–214 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Qiu X.-Q., Wang H., Lu X.-F., Zhang J., Li S.-F., Cheng G., Wan L., Yang L., Zuo J.-Y., Zhou Y.-Q., Wang H.-Y., Cheng X., Zhang S.-H., Ou Z.-R., Zhong Z.-C., Cheng J.-Q., Li Y.-P., Wu G. Y., An engineered multidomain bactericidal peptide as a model for targeted antibiotics against specific bacteria. Nat. Biotechnol. 21, 1480–1485 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Qiu X.-Q., Zhang J., Wang H., Wu G. Y., A novel engineered peptide, a narrow-spectrum antibiotic, is effective against vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 49, 1184–1189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao R., Teng D., Wang X., Xi D., Zhang Y., Hu X., Yang Y., Wang J., Design, expression, and characterization of a novel targeted plectasin against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 97, 3991–4002 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Xu L., Shao C., Li G., Shan A., Chou S., Wang J., Ma Q., Dong N., Conversion of broad-spectrum antimicrobial peptides into species-specific antimicrobials capable of precisely targeting pathogenic bacteria. Sci. Rep. 10, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalelkar P. P., Riddick M., Garcia A. J., Biomaterial-based antimicrobial therapies for the treatment of bacterial infections. Nat. Rev. Mater. 7, 39–54 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X., Al-Baadani M. A., He H., Cai L., Wu Z., Yao L., Wu X., Wu S., Chen M., Zhang H., Liu J., Antibacterial and osteogenesis performances of LL37-loaded titania nanopores in vitro and in vivo. Int. J. Nanomedicine 14, 3043–3054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G., Huang T., Wang Y., Wang H., Li Y., Yu K., Dong L., Sustained release of antimicrobial peptide from self-assembling hydrogel enhanced osteogenesis. J. Biomater. Sci. Polym. Ed. 29, 1812–1824 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Chen R., Willcox M. D., Ho K. K. K., Smyth D., Kumar N., Antimicrobial peptide melimine coating for titanium and its in vivo antibacterial activity in rodent subcutaneous infection models. Biomaterials 85, 142–151 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Kazemzadeh-Narbat M., Noordin S., Masri B. A., Garbuz D. S., Duncan C. P., Hancock R. E. W., Wang R., Drug release and bone growth studies of antimicrobial peptide-loaded calcium phosphate coating on titanium. J. Biomed. Mater. Res. B Appl. Biomater. 100, 1344–1352 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Yuan X., Ouyang L., Luo Y., Sun Z., Yang C., Wang J., Liu X., Zhang X., Multifunctional sulfonated polyetheretherketone coating with beta-defensin-14 for yielding durable and broad-spectrum antibacterial activity and osseointegration. Acta Biomater. 86, 323–337 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Kwon E. J., Skalak M., Bertucci A., Braun G., Ricci F., Ruoslahti E., Sailor M. J., Bhatia S. N., Porous silicon nanoparticle delivery of tandem peptide anti-infectives for the treatment of Pseudomonas aeruginosa lung infections. Adv. Mater. 29, 1701527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kłodzińska S. N., Pletzer D., Rahanjam N., Rades T., Hancock R. E. W., Nielsen H. M., Hyaluronic acid-based nanogels improve in vivo compatibility of the anti-biofilm peptide DJK-5. Nanomed. Nanotechnol. Biol. Med. 20, 102022 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Maiden M. M., Zachos M. P., Waters C. M., Hydrogels embedded with melittin and tobramycin are effective against Pseudomonas aeruginosa biofilms in an animal wound model. Front. Microbiol. 10, 1348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puthia M., Butrym M., Petrlova J., Strömdahl A. C., Andersson M. Å., Kjellström S., Schmidtchen A., A dual-action peptide-containing hydrogel targets wound infection and inflammation. Sci. Transl. Med. 12, eaax6601 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Liu M., Liu T., Zhang X., Jian Z., Xia H., Yang J., Hu X., Xing M., Luo G., Wu J., Fabrication of KR-12 peptide-containing hyaluronic acid immobilized fibrous eggshell membrane effectively kills multi-drug-resistant bacteria, promotes angiogenesis and accelerates re-epithelialization. Int. J. Nanomedicine 14, 3345–3360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi F., Qian Y., Shao N., Zhou R., Zhang S., Lu Z., Zhou M., Xie J., Wei T., Yu Q., Liu R., Practical preparation of infection-resistant biomedical surfaces from antimicrobial β-peptide polymers. ACS Appl. Mater. Interfaces 11, 18907–18913 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Moosazadeh Moghaddam M., Eftekhary M., Erfanimanesh S., Hashemi A., Fallah Omrani V., Farhadihosseinabadi B., Lasjerdi Z., Mossahebi-Mohammadi M., Chauhan N. P. S., Seifalian A. M., Gholipourmalekabadi M., Comparison of the antibacterial effects of a short cationic peptide and 1% silver bioactive glass against extensively drug-resistant bacteria, Pseudomonas aeruginosa and Acinetobacter baumannii, isolated from burn patients. Amino Acids 50, 1617–1628 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Yu K., Lo J. C. Y., Yan M., Yang X., Brooks D. E., Hancock R. E. W., Lange D., Kizhakkedathu J. N., Anti-adhesive antimicrobial peptide coating prevents catheter associated infection in a mouse urinary infection model. Biomaterials 116, 69–81 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Lim K., Saravanan R., Chong K. K. L., Goh S. H. M., Chua R. R. Y., Tambyah P. A., Chang M. W., Kline K. A., Leong S. S. J., Anhydrous polymer-based coating with sustainable controlled release functionality for facile, efficacious impregnation, and delivery of antimicrobial peptides. Biotechnol. Bioeng. 115, 2000–2012 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Cole N., Hume E. B. H., Vijay A. K., Sankaridurg P., Kumar N., Willcox M. D. P., In vivo performance of melimine as an antimicrobial coating for contact lenses in models of CLARE and CLPU. Invest. Ophthalmol. Vis. Sci. 51, 390–395 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Dutta D., Kamphuis B., Ozcelik B., Thissen H., Pinarbasi R., Kumar N., Willcox M. D. P., Development of silicone hydrogel antimicrobial contact lenses with Mel4 peptide coating. Optom. Vis. Sci. 95, 937–946 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Dutta D., Vijay A. K., Kumar N., Willcox M. D., Melimine-coated antimicrobial contact lenses reduce microbial keratitis in an animal model. Invest. Ophthalmol. Vis. Sci. 57, 5616–5624 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Mishra B., Wang G., Ab initio design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 134, 12426–12429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirtskhalava M., Amstrong A. A., Grigolava M., Chubinidze M., Alimbarashvili E., Vishnepolsky B., Gabrielian A., Rosenthal A., Hurt D. E., Tartakovsky M., DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 49, D288–D297 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma R., Shrivastava S., Kumar Singh S., Kumar A., Saxena S., Kumar Singh R., AniAMPpred: Artificial intelligence guided discovery of novel antimicrobial peptides in animal kingdom. Brief. Bioinform. 22, bbab242 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Wang C., Garlick S., Zloh M., Deep learning for novel antimicrobial peptide design. Biomolecules 11, 471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capecchi A., Cai X., Personne H., Köhler T., van Delden C., Reymond J.-L., Machine learning designs non-hemolytic antimicrobial peptides. Chem. Sci. 12, 9221–9232 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres M. D., Cao J., Franco O. L., Lu T. K., de la Fuente-Nunez C., Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano 15, 2143–2164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vishnepolsky B., Gabrielian A., Rosenthal A., Hurt D. E., Tartakovsky M., Managadze G., Grigolava M., Makhatadze G. I., Pirtskhalava M., Predictive model of linear antimicrobial peptides active against gram-negative bacteria. J. Chem. Inf. Model. 58, 1141–1151 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Das P., Sercu T., Wadhawan K., Padhi I., Gehrmann S., Cipcigan F., Chenthamarakshan V., Strobelt H., dos Santos C., Chen P.-Y., Yang Y. Y., Tan J. P. K., Hedrick J., Crain J., Mojsilovic A., Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat. Biomed. Eng. 5, 613–623 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Wang J., Chou S., Yang Z., Yang Y., Wang Z., Song J., Dou X., Shan A., Combating drug-resistant fungi with novel imperfectly amphipathic palindromic peptides. J. Med. Chem. 61, 3889–3907 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Hawrani A., Howe R. A., Walsh T. R., Dempsey C. E., Origin of low mammalian cell toxicity in a class of highly active antimicrobial amphipathic helical peptides. J. Biol. Chem. 283, 18636–18645 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Zhu X., Dong N., Wang Z., Ma Z., Zhang L., Ma Q., Shan A., Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater. 10, 244–257 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Louis-Jeune C., Andrade-Navarro M. A., Perez-Iratxeta C., Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 80, 374–381 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Yang J., Zhang Y., I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J., Paredes-Sabja D., Sarker M. R., McClane B. A., Clostridium perfringens sporulation and sporulation-associated toxin production. Microbiol. Spectr. 4, 4–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vidal J. E., Shak J. R., Canizalez-Roman A., The CpAL quorum sensing system regulates production of hemolysins CPA and PFO to build Clostridium perfringens biofilms. Infect. Immun. 83, 2430–2442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu L., Xu K., Wang H., Tan P. K. J., Fan W., Venkatraman S. S., Li L., Yang Y.-Y., Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat. Nanotechnol. 4, 457–463 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Tan P., Tang Q., Xu S., Zhang Y., Fu H., Ma X., Designing self-assembling chimeric peptide nanoparticles with high stability for combating piglet bacterial infections. Adv. Sci. 9, e2105955 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhai Q., Feng S., Arjan N., Chen W., A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 59, 3227–3236 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Pereira F. C., Wasmund K., Cobankovic I., Jehmlich N., Herbold C. W., Lee K. S., Sziranyi B., Vesely C., Decker T., Stocker R., Warth B., von Bergen M., Wagner M., Berry D., Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat. Commun. 11, 1–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu H., Cai Z., Wang F., Hong L., Deng L., Zhong J., Wang Z., Cui W., Colon-targeted adhesive hydrogel microsphere for regulation of gut immunity and flora. Adv. Sci. 8, e2101619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rowan F., Docherty N. G., Murphy M., Murphy B., Coffey J. C., O’Connell P. R., Desulfovibrio bacterial species are increased in ulcerative colitis. Dis. Colon Rectum 53, 1530–1536 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Khan S., Waliullah S., Godfrey V., Khan M. A. W., Ramachandran R. A., Cantarel B. L., Behrendt C., Peng L., Hooper L. V., Zaki H., Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci. Transl. Med. 12, eaay 6218 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Modi N., Wilcox M. H., Evidence for antibiotic induced Clostridium perfringens diarrhoea. J. Clin. Pathol. 54, 748–751 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu S., Zhao W., Lan P., Mou X., The microbiome in inflammatory bowel diseases: From pathogenesis to therapy. Protein Cell 12, 331–345 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental Section
Figs. S1 to S23
Tables S1 to S10
Legend for data S1
Data S1