Abstract
The C-terminal CaaX sequence (cysteine-aliphatic-aliphatic-any of several amino acids) is subject to isoprenylation on the conserved cysteine and is estimated to occur in 1–2% of proteins within yeast and human proteomes. Recently, non-canonical CaaX sequences in addition to shorter and longer length CaX and CaaaX sequences have been identified that can be prenylated. Much of the characterization of prenyltransferases has relied on the yeast system because of its genetic tractability and availability of reporter proteins, such as the a-factor mating pheromone, Ras GTPase, and Ydj1 Hsp40 chaperone. To compare the properties of yeast and human prenyltransferases, including the recently expanded target specificity of yeast farnesyltransferase, we have developed yeast strains that express human farnesyltransferase or geranylgeranyltransferase-I in lieu of their yeast counterparts. The humanized yeast strains display robust prenyltransferase activity that functionally replaces yeast prenyltransferase activity in a wide array of tests, including the prenylation of a wide variety of canonical and non-canonical human CaaX sequences, virus encoded CaaX sequences, non-canonical length sequences, and heterologously expressed human proteins HRas and DNAJA2. These results reveal highly overlapping substrate specificity for yeast and human farnesyltransferase, and mostly overlapping substrate specificity for GGTase-I. This yeast system is a valuable tool for further defining the prenylome of humans and other organisms, identifying proteins for which prenylation status has not yet been determined.
Keywords: CaaX, farnesylation, geranylgeranylation
Summary Statement:
We report yeast engineered to express human prenylation enzymes with which prenylation can be investigated for established and novel CaaX sequences associated with proteins involved in human disease.
Introduction
Post-translational modification of the C-terminal CaaX1 sequence is important for regulating the localization and function of many proteins, including the Ras GTPases often cited as archetypical CaaX proteins (Campbell and Philips, 2021; Cox et al., 2015; Ravishankar et al., 2023). CaaX proteins are functionally involved in disease states, such as cancer, Alzheimer’s disease and viral infections, including Hepatitis D and SARS-CoV2 (Jeong et al., 2022; Marakasova et al., 2017; Ring et al., 2022; Soveg et al., 2021; Wickenhagen et al., 2021).
The C-terminal CaaX sequence has been traditionally defined as a Cysteine (C), two aliphatic amino acids (a1a2), and one of several amino acids (X). The first step in CaaX modification is covalent attachment of a farnesyl (C15) or geranylgeranyl (C20) isoprene lipid to the Cysteine. farnesyltransferase (FTase) generally targets CaaX sequences, while geranylgeranyltransferase-I (GGTase-I) targets the subset of CaaL/I/M sequences (Hartman et al., 2005). CaaX sequences with an aliphatic amino acid at the a2 position commonly follow the canonical modification pathway that involves three steps: initial isoprenylation, proteolytic removal of aaX, and carboxymethylation of the exposed prenylated cysteine. In yeast, the Ras2 GTPase and a-factor mating pheromone are highly studied examples of canonically modified CaaX proteins. In addition, CaaX sequences lacking a1 and a2 aliphatic residues can be farnesylated (Kim et al., 2023). These so-called shunted CaaX sequences retain their last three amino acids, resulting in a biochemically distinct C-terminus compared to canonically modified CaaX proteins (Hildebrandt et al., 2016b). The yeast Ydj1 HSP40 protein is a recently characterized example of a shunted CaaX protein.
Farnesylation and geranylgeranylation of CaaX sequences is accomplished by heterodimeric farnesyltransferase (FTase) and geranylgeranyltransferase-I (GGTase-I), respectively. The enzymes share an α subunit while the β subunits provide isoprenoid specificity and substrate recognition (Lane and Beese, 2006; Maurer-Stroh et al., 2003). Human FTase (HsFTase) is composed of HsFNTA and HsFNTB subunits; the orthologous subunits of yeast Saccharomyces cerevisiae FTase (ScFTAse) are ScRam2 and ScRam1 (Figure 1A). Human GGTase-I (HsGGTase-I) is composed of HsFNTA and HsPGGT1B subunits, while the orthologous subunits of yeast GGTase-I (ScGGTase-I) are ScRam2 and ScCdc43. In yeast, RAM1 is not an essential gene (i.e., ram1Δ strains are viable), whereas RAM2 and CDC43 are essential, raising the possibility that GGTase-I is more critical to yeast life processes relative to FTase.
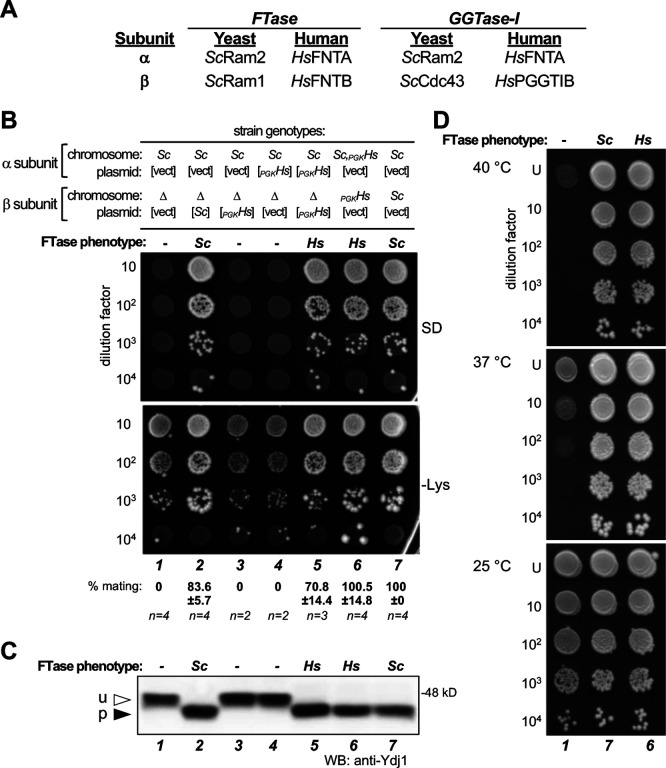
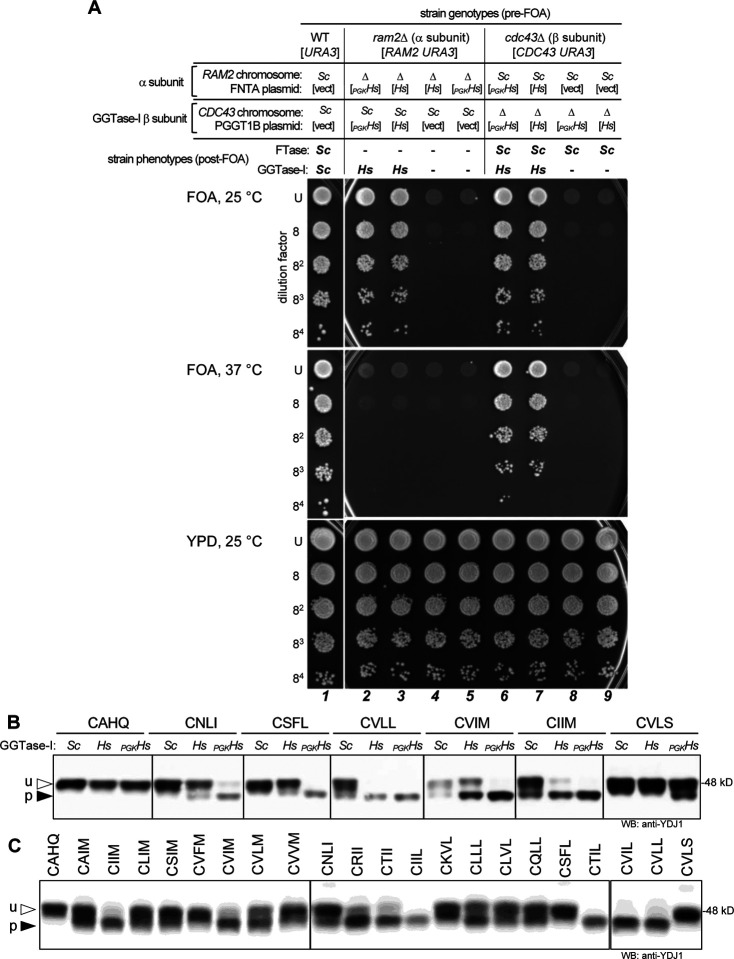
Figure 1. Interspecies complementation studies of FTase subunits.
HsFTase and ScFTase subunits are not interchangeable, but co-expression of both α and β subunits of HsFTase can restore FTase activity in the absence of ScFTase. A) Relationships between yeast and human prenyltransferase subunits. B) The mating assay was used to assess FTase dependent production of a-factor. MATa haploid strains were engineered to express yeast (Sc) and/or human FTase α and β subunits (PGKHs), where subunits were encoded either on plasmids (indicated by brackets) or from chromosomal loci (no brackets). MATa strains were mixed with MATα cells (SM1068), mixes subject to 10-fold dilutions, and dilution mixtures spotted onto minimal media (SD) and SC-lysine (-Lys) media. Growth on SD indicates diploid formation, which is a direct indicator of a-factor mating pheromone production. Growth on -Lys reflects the input of MATa cells and reflects both unmated haploid and mated diploid cells; unmated haploid cells (ram1Δ) grow less well on -Lys due to the absence of FTase activity. Quantitative mating test results are reported below each lane relative to the WT strain (7). Strains used were yWS3276 (1), yWS3277 (2), yWS3278 (3), yWS3408 (4), yWS3280 (5), yWS3282 (6), and yWS3283 (7). C) Gel-shift analysis of Ydj1 using the same strains described in panel B. Total cell lysates were prepared and equivalent protein amounts analyzed by Western blot using anti-Ydj1 antibody. u – unprenylated Ydj1; p – prenylated Ydj1. D) Thermotolerance test of the FTase-deficient strain compared to wildtype and humanized FTase strains described in panel B. Strains used were yWS3276 (1), yWS3282 (6), and yWS3283 (7).
There are 1207 annotated human proteins possessing a CaaX sequence (i.e., Cysteine followed by any 3 amino acids at the C-terminus) within UniProtKB/Swiss-Prot representing 680 unique sequences among the 8000 possible CaaX amino acid combinations. There are also 375 viral proteins possessing a CaaX sequence that could potentially utilize host enzymes for isoprenylation (Table 1). Determining the prenylation status of candidate CaaX sequences has depended on a variety of methods and prediction algorithms, each with its own limitations. Methods for case-by-case verification of protein prenylation include radiolabeling with 3H-mevalonate, gel mobility analysis, localization studies and mass spectrometry (Anderegg et al., 1988; Hancock et al., 1989; Hildebrandt et al., 2016b; Michaelson et al., 2005; Ravishankar et al., 2023). Methods for systematic verification have relied on metabolic labeling with reagents compatible with click chemistry, peptide arrays, and genetic approaches (Kho et al., 2004; Kim et al., 2023; Rashidian et al., 2013; Storck et al., 2019; Suazo et al., 2018; Suazo et al., 2021; Wang et al., 2014). Differentiating FTase and GGTase-I target specificity has mostly relied on in vitro and genetic approaches (Hougland et al., 2010; Kim et al., 2023; Stein et al., 2015). Independent of the methods employed, it remains a distinct challenge to confirm the predictive prenylation of candidate CaaX sequences, especially across species where conservation of target specificity has not necessarily been confirmed.
Table 1.
Summary of number of proteins identified in UniProtKB/Swiss-Prot database with C-terminal CaaX sequences and with shorter and longer CaaX sequences.
| Search String | Human | Non-human Eukaryotes | Virusesa | Total | |
|---|---|---|---|---|---|
| Cxxx> | 1207 | 5047 | 375 | 6629 | |
| Cxxxx> (omitted CCxxx>) | 941 | 3582 | 462 | 4985 | |
| Cxx> (omitted CCxx>) | 989 | 4206 | 269 | 5464 | |
| TOTAL | 3137 | 12835 | 1106 | 17078 |
Bacteriophage sequences were manually subtracted from the search.
In this study, we have developed yeast strains expressing HsFTase and HsGGTase-I that functionally replace the yeast prenyltransferases in a wide array of tests. Results with a universal reporter for both farnesylation and geranylgeranylation indicate highly overlapping target specificity across species for each prenyltransferase. We also demonstrate the utility of these strains for testing the in vivo prenylation of heterologously expressed human proteins. These humanized strains provide a valuable new resource for protein prenylation research, especially for target specificity studies of the human prenyltransferase in a genetically amenable cell-based system. This resource is expected to enhance the identification and characterization of novel prenyltransferase targets, including those harboring atypical CaaX sequences.
Results
Interspecies complementation analysis of FTase subunits
As a key first step toward developing a yeast system to express HsFTase, complementation studies were performed to assess the functional equivalence of yeast and human FTase subunits. Codon optimized genes encoding HsFNTB and HsFNTA were introduced into yeast and evaluated for the ability to complement for loss of yeast FTase β subunit Ram1 (i.e., ram1Δ) using an assay that measures production of the farnesylated a-factor mating pheromone (Figure 1B). No a-factor was produced by ram1Δ yeast, while complementation with plasmid-encoded Ram1 restored a-factor production to near normal levels as measured using an a-factor dependent yeast mating assay (i.e., 0% and 83.6% mating relative to WT, respectively). As expected, plasmid encoded HsFNTA did not complement for activity since this strain only carries copies of FTase α subunits (yeast and human). Many human orthologs of yeast proteins can substitute for their yeast counterpart, yet HsFNTB was unable to do so, implying that either the yeast Ram2 and HsFNTB subunits do not interact or a hybrid FTase complex (Ram2-HsFNTB) is non-functional. In fact, a-factor was produced only when the human FTase α and β subunits were co-expressed. The observed mating activity was slightly below wildtype levels when HsFTase subunits were plasmid-encoded (70.8% of WT) and at wildtype levels when subunits were integrated into the genome (100.5% of WT). These phenotypes were derived using the strong constitutive phosphoglycerate kinase promoter (i.e., PPGK1) to drive expression of HsFTase subunits. Expression of HsFNTA and HsFNTB from their orthologous yeast promoters restored mating to less than 1% of wildtype (Supplemental Figure S1).
To confirm the ability of yeast expressed HsFTase to fully prenylate other CaaX proteins, the extent of endogenous Ydj1 farnesylation was determined by gel-shift analysis (Figure 1C). Farnesylated Ydj1 has faster gel mobility than unfarnesylated Ydj1 when analyzed by SDS-PAGE and immunoblot. Using the same strains evaluated for a-factor production, farnesylation of Ydj1 was evident only when ScFTase or HsFTase dimeric complex was present. Farnesylation of Ydj1 by HsFTase was qualitatively complete. When combined with the analysis of a-factor production, these results indicate that HsFTase activity is unlikely to be limiting at the cellular level. In addition, we confirmed the ability of chromosomally integrated HsFTase to reverse the temperature sensitive growth phenotype of ram1Δ yeast (Figure 1D). Together, the results of these complementation studies indicate that HsFTase activity can be engineered to mimic the ScFTase activity levels in a cell-based system.
HsFTase expressed in yeast modifies human Ras CaaX sequences
Our complementation studies indicated that HsFTase can modify yeast proteins harboring either a canonical CaaX sequence (a-factor; CVIA) or a shunted CaaX sequence (Ydj1; CASQ). To extend this observation to other protein contexts and canonical sequences, the farnesylation of the yeast Ras2 GTPase (ScRas2) was evaluated using the chromosomally integrated HsFTase yeast strain (Supplemental Figure S2A). ScRas2 has 65.6% and 64.4% identity to human NRas and HRas, respectively. In yeast and humans, Ras localization to the plasma membrane depends on canonical modification of the CaaX sequence (Boyartchuk et al., 1997; Michaelson et al., 2005; Ravishankar et al., 2023). Indeed, GFP-ScRas2 with its natural CaaX sequence (CIIS) was localized to the plasma membrane when ScFTase or HsFTase was expressed (Figure 2A; Sc and Hs, respectively). GFP-ScRas2 was mislocalized in the absence of FTase activity (ram1Δ) or when harboring a CaaX cysteine to serine substitution mutation (SIIS), as has been previously observed (Dong et al., 2003; Ravishankar et al., 2023).
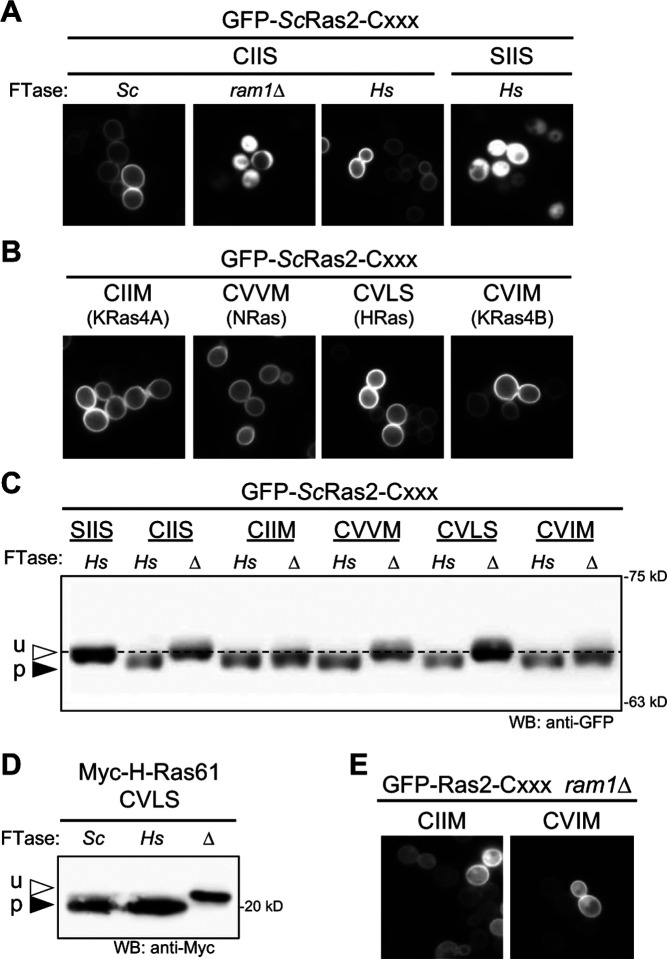
Figure 2. The humanized FTase strain fully modifies yeast and human Ras CaaX sequences.
GFP-Ras2 was used as a reporter to evaluate CaaX sequences derived from yeast Ras2 (CIIS) and human proteins KRas4A (CIIM), NRas (CVVM), HRas (CVLS), and KRas4B (CVIM). A) The localization of GFP-ScRas2 produced in wildtype (Sc), humanized (Hs), and FTase-deficient (ram1Δ) yeast strains was determined by fluorescence microscopy. GFP-ScRas2-SIIS is a mutant that cannot be prenylated and is cytosolically localized. B) The localization of GFP-ScRas2-CaaX variants encoding human Ras CaaX sequences produced in the humanized FTase strain was determined by fluorescence microscopy. The source of the CaaX sequence is indicated below each specific sequence. C) Western blot analysis of GFP-ScRas2-CaaX variants produced in humanized (Hs) or FTase-deficient (Δ) yeast strains. Total cell lysates were prepared, and equivalent protein amounts analyzed by SDS-PAGE and Western blot using anti-GFP antibody. u –unprenylated GFP-ScRas2; p – prenylated GFP-ScRas2. The dashed line was aligned with unprenylated GFP-Ras2 to serve as a visual reference. The plasmids used in panels A-C were pWS1735 (GFP-ScRas2), pWS1889 (GFP-ScRas2-SIIS), pWS1997 (GFP-ScRas2-CIIM), pWS1998 (GFP-ScRas2-CVVM), pWS1999 (GFP-ScRas2-CVLS), and pWS2000 (GFP-ScRas2-CVIM). pWS1889 encodes a double cysteine to serine mutation; the upstream cysteine is typically palmitoylated but was mutated to avoid creation of a non-canonical length CaaaX sequence. Yeast strains used were BY4741 (wildtype ScFTase), yWS3220 (HsFTase), and yWS3202 (ram1Δ). D) Myc-HRas61 (p-05547) was produced in wildtype (Sc; yWS2544), humanized FTase (Hs; yWS3186), or FTase-deficient (Δ; yWS3209) yeast strains. HRas61 is the Q61L oncogenic derivative of human HRas (Adari et al., 1988; Farnsworth et al., 1991). E) The localization of GFP-ScRas2-CaaX variants (CIIM and CVIM) that undergo alternate prenylation by ScGGTase-I in the absence of FTase (i.e., ram1Δ strain background).
We utilized the GFP-ScRas2 reporter to further evaluate the ability of HsFTase to recognize CaaX sequences associated with human Ras orthologs: HsKRas4A (CIIM), HsNRas (CVVM), HsHRas (CVLS), and HsKRas4B (CVIM) (Figure 2B). All the sequences directed GFP-ScRas2 to the plasma membrane, indicating that they are farnesylated by HsFTase. As additional confirmation of prenylation by HsFTase, the mobilities of GFP-ScRas2-CaaX variants were analyzed by gel-shift assay (Figure 2C). Like farnesylated Ydj1, prenylated ScRas2 migrates faster relative to unprenylated protein when analyzed by SDS-PAGE. GFP-ScRas2 exhibited faster mobility relative to protein produced in the absence of FTase (i.e., ram1Δ) in the context of its wildtype sequence (CIIS) and two human Ras CaaX sequences (CVVM and CVLS; CIIM and CVIM are discussed below), indicative of complete prenylation. Gel-shift studies were also used to examine the farnesylation of heterologously expressed human oncogenic protein Myc-HRas61 (CVLS) that was heterologously expressed in the humanized yeast strain (Adari et al., 1988; Farnsworth et al., 1991). It too was completely prenylated (Figure 2D).
Some CaaX sequences, including those associated with KRas4A (CIIM), KRas4B (CVIM) and NRas (CVVM), are alternately prenylated by HsGGTase-I in the absence of HsFTase activity, which is often established in systems through the action of FTase inhibitors (Mohammed et al., 2016; Whyte et al., 1997). Consistently, GFP-ScRas2-CaaX variants harboring CIIM and CVIM sequences exhibited mobility shifts of less magnitude in the gel-shift assay (Figure 2C), which we attribute to alternate prenylation by yeast GGTase-I in the absence of FTase (ram1Δ). To bolster this conclusion, we determined that GFP-ScRas2-CaaX variants CIIM and CVIM exhibited plasma membrane localization in the FTase-deficient ram1Δ strain, which would be predicted if they were geranylgeranylated (Figure 2E). Combined, these observations suggest that ScGGTase-I also has the opportunistic ability to modify certain CaaX sequences in the absence of FTase activity (i.e., ram1Δ), resulting in geranylgeranylated products having similar although not identical mobility as the farnesylated products.
HsFTase expressed in yeast modifies the non-canonical human DNAJA2 CAHQ sequence
The ability of ScFTase to modify the non-canonical CaaX sequence associated with Ydj1 (CASQ) and other non-canonical sequences has been reported (Berger et al., 2018; Hildebrandt et al., 2016b; Kim et al., 2023; Ravishankar et al., 2023). The complementation studies described above indicate that HsFTase can also modify the Ydj1 CASQ sequence (see Figure 1). To further investigate the breadth of sequences recognized by HsFTase, we used thermotolerance and gel-shift assays to evaluate a set of Ydj1-CaaX variants representing sequences that are well characterized in terms of modification status. In the thermotolerance assay, yeast lacking Ydj1 (vector) or an unmodifiable Ydj1(SASQ) are unable to grow at 40 °C because Ydj1 farnesylation is required for efficient growth at elevated temperatures (Figure 3A). All the remaining Ydj1-CaaX variants exhibited growth profiles that were nearly identical whether farnesylation was supported by ScFTase or HsFTase. Yeast expressing Ydj1 with prenylation-only sequences (CASQ and CAHQ) supported the most robust thermotolerance; these sequences were derived from yeast Ydj1 and human DNAJA2, respectively. The prenylation-only status of CAHQ was previously based on homology and prediction algorithms (Hildebrandt et al., 2016b; Kim et al., 2023) and is confirmed here by the observation that CAHQ supports thermotolerance similar to CASQ and is resistant to proteolysis by the CaaX proteases (Supplemental Figure S3). Yeast expressing Ydj1 with canonical CaaX sequences (CVIA and CTLM) exhibited partial growth under this condition as has been previously observed (Hildebrandt et al., 2016b); these sequences were derived from yeast a-factor and Gγ Ste18, respectively. Side-by-side SDS-PAGE analysis of the Ydj1-CaaX variants revealed that both canonical and prenylation-only CaaX sequences are fully prenylated by both ScFTase and HsFTase (Figure 3B). Additional support for the ability of HsFTase to modify these sequences is evident when comparing the mobilities of Ydj1-CaaX variants in the absence of FTase (i.e., ram1Δ) (Figure 3C).
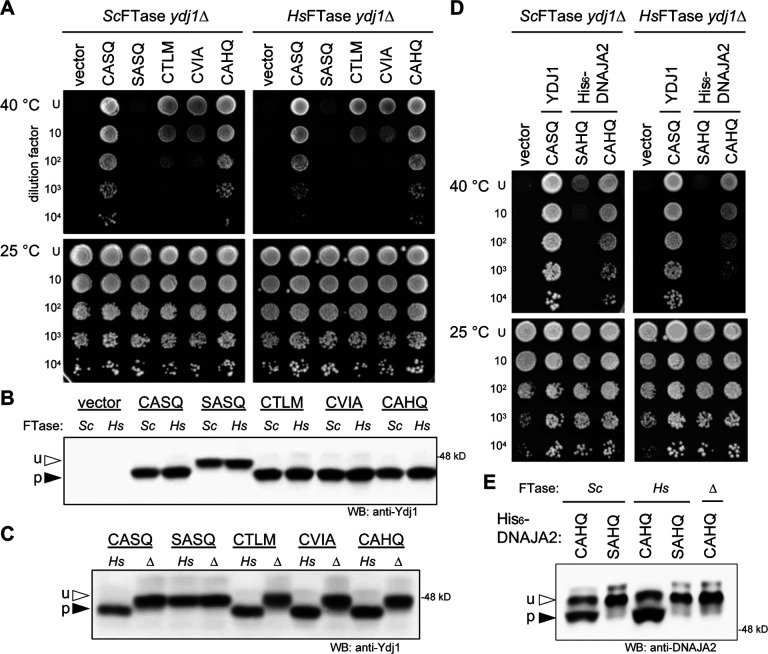
Figure 3. The humanized FTase strain modifies HSP40-CaaX variants.
A Ydj1-CaaX reporter was used to evaluate CaaX sequences from yeast HSP40 Ydj1 (CASQ), an unmodifiable sequence (SASQ), yeast Ste18 (CTLM), yeast a-factor (CVIA), and human HSP40 DNAJA2 (CAHQ). A) The thermotolerance assay was used to determine prenyl modification of Ydj1. Strains were engineered to express ScFTase (left cultured to equivalent density, cultures subject to 10-fold dilutions, and dilution mixtures spotted e cultured to equivalent density, cultures subject to 10-fold dilutions, and dilution mixtures spotted onto rich media (YPD) at either room temperature (RT) or elevated temperature (40 °C). Growth at 40 °C reflects farnesylation of Ydj1. B) Gel-shift analysis of Ydj1-CaaX variants. Total cell lysates prepared using the strains described in panel A were analyzed by SDS-PAGE and Western blot using anti-Ydj1 antibody. u – unprenylated Ydj1; p – prenylated Ydj1. C) Comparison of Ydj1-CaaX variant gel mobilities in the absence (Δ) and presence of HsFTase (Hs). D) Thermotolerance analysis of strains producing native Ydj1 (CASQ), plasmid-encoded human His6-DNAJA2 (CAHQ), and an unmodifiable sequence (SAHQ). Strains were evaluated as described in panel A. E) Gel-shift analysis of human His6-DNAJA2 and His6-DNAJA2-SAHQ produced in wildtype (Sc), humanized FTase (Hs), and FTasedeficient (Δ) yeast strains. Total cell lysates were prepared and analyzed as described in panel B. For all panels the strains used were yWS2544 (ScFTase ydj1Δ), yWS3186 (HsFTase ydj1Δ; see Supplemental Figure S2B), and yWS3209 (ram1Δ ydj1Δ). Plasmids used were pRS316 (vector), pWS942 (YDJ1), pWS1132 (YDJ1-SASQ), pWS1246 (YDJ1-CTLM), pWS1286 (YDJ1-CVIA), pWS1495 (YDJ1-CAHQ), pWS1424 (His6-DNAJA2), and pWS2256 (His6-DNAJA2-SAHQ).
The observations made using Ydj1 as a reporter were also evident for HsDNAJA2 (CAHQ) that was heterologously expressed in the humanized yeast strain. HsDNAJA2 has 46% identity to Ydj1 and can complement some Ydj1-associated defects (Whitmore et al., 2020). HsDNAJA2 supported thermotolerance in the context of both ScFTase and HsFTase strains, and this phenotype was prenylation dependent because DNAJA2-SAHQ did not support thermotolerance (Figure 3D). By gel-shift analysis, both ScFTase and HsFTase modified HsDNAJA2 (Figure 3E). These results further extend the utility of the humanized yeast strain for studies of heterologously expressed human CaaX proteins. Moreover, these observations support the hypothesis that ScFTase and HsFTase have highly similar sequence recognition profiles that are likely broader than the CaaX consensus sequence.
HsFTase expressed in yeast modifies a range of human and viral CaaX protein sequences
Studies on the prenylation of CaaX proteins in most systems is hindered by several factors ranging from difficulty in odetection of low abundance proteins, to lack of phenotypic readouts, to labor and time costs for case-by-case studies. Many of these hurdles can be overcome using the Ydj1 reporter and genetically tractable yeast system described in this study. To support the utility of our approach for investigations of HsFTase specificity, Ydj1-CaaX variants harboring the CaaX sequences of known prenylated human proteins were evaluated by gel-shift assay. Each variant was evaluated in the presence and absence of HsFTase to ensure that gel-shifts were due to HsFTase and to assess whether alternate prenylation was possible. The CaaX sequences evaluated were from chaperone proteins (Nap1L1, DNAJA1 and Pex19), chromosome stability proteins (CENPE, CENPF and Spindly), nuclear lamins (Prelamin A and Lamin B), Ras-like proteins (Rab38, KRas4A, KRas4B and HRas) and Prickle1 (Figure 4A and Supplemental Table S5) (Ashar et al., 2000; Kho et al., 2004; Kim et al., 1990; Leung et al., 2007; Moudgil et al., 2015; Onono et al., 2010; Palsuledesai et al., 2014; Storck et al., 2019; Strutt et al., 2013; Varela et al., 2008). Apart from the Rab38-associated sequence (CAKS), every sequence was completely modified by HsFTase. CAKS is reported to be underprenylated in the context of Rab38, which is consistent with our findings (Kohnke et al., 2013). Sequences ending in methionine (CSIM, CAIM, CLIM, CIIM and CVIM) were fully modified in the presence of HsFTase yet partially modified in the absence of FTase (ram1Δ), suggesting that ScGGTase-I contributes to their prenylation. These observations are consistent with studies reporting that ScFTase, HsFTase, and HsGGTase-I recognize CaaM sequences, which we propose is a property that extends to ScGGTase-I as well (Caplin et al., 1994; Zhang et al., 1997).
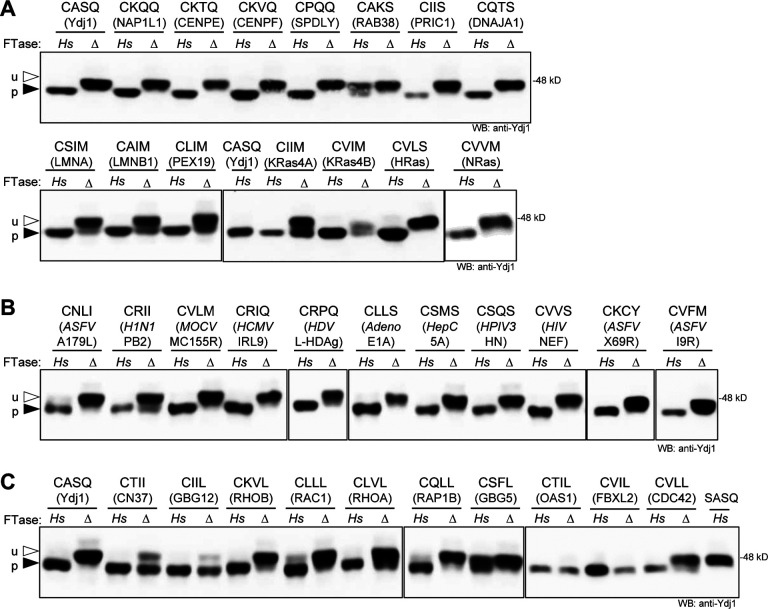
Figure 4. The humanized FTase yeast strain modifies Ydj1-CaaX variants with CaaX sequences occurring in human and viral proteins.
Ydj1-CaaX variants were produced in the HsFTase (Hs) and FTase deficient (Δ) strains and evaluated by gel-shift assay as described in Figure 3B. The source of the CaaX sequence is indicated below each specific sequence. A) Analysis of Ydj1-CaaX variants based on the indicated human proteins. B) Analysis of Ydj1-CaaX variants based on mammalian viral proteins. Virus abbreviations are human influenza virus (H1N1), Molluscum contagiosum virus (MOCV), human cytomegalovirus (HCMV), Hepatitis Delta Virus (HDV), human adenovirus 1 (Adeno), human Hepatitis C virus (HepC), human parainfluenza virus 3 (HPIV3), Human immunodeficiency virus 1 (HIV), and African swine fever virus (ASFV). C) Analysis of Ydj1 with CaaL/I sequences based on the indicated human proteins. Strains used were yWS3186 (Hs) and yWS3209 (Δ). Plasmids used are listed in Supplemental Table S3. See Supplemental Table S1 for gel quantification.
The Ydj1 reporter system was also used to evaluate the prenylation of CaaX sequences associated with human and swine viral proteins (Figure 4B and Supplemental Table S5). The importance of prenylation in viral infection is an emerging line of inquiry for infectious disease therapy but documentation of viral proteins prenylation is limited (Einav and Glenn, 2003; Marakasova et al., 2017). All of the viral CaaX sequences evaluated were modified by HsFTase. These sequences were associated with influenza virus H1N1 PB2 (CRII); Molluscum contagiosum virus subtype 1 MC155R (CVLM); human cytomegalovirus protein IRL9 (CRIQ); Hepatitis Delta Virus L-HDAg (CRPQ); human adenovirus1 E1A (CLLS); hepatitis C virus non-structural 5A protein (CSMS); human parainfluenza virus 3 hemagglutinin-neuraminidase (CSQS); HIV NEF protein (CVVS); and African swine fever virus proteins Bcl-2 homolog A179L (CNLI), X69R (CKCY) and I9R (CVFM). Of these, only hepatitis L-HDAg has been biochemically characterized as being prenylated (Glenn et al., 1992). Several others are inferred to be prenylated based on suppression of viral replication by statins (Pronin et al., 2021).
We also evaluated the prenylation status of CaaL/I/M sequences associated with prenylated human proteins. Sequences adhering to consensus motif are considered targets of GGTase-I or interchangeably by FTase or GGTase-I (Krzysiak et al., 2010; Trueblood et al., 1993; Zhang et al., 1994). The CaaX sequences evaluated were from small GTPases (CDC42, RAC1, RAP1B, RHOA and RHOB), G-protein gamma subunits (GBG5 and GBG12), myelin protein CN37, F-box protein FBXL2 and anti-viral protein OAS1 p46 (De Angelis and Braun, 1994; Katayama et al., 1991; Kawata et al., 1990; Kilpatrick and Hildebrandt, 2007; Onono et al., 2010; Soveg et al., 2021; Storck et al., 2019; Wang et al., 2005; Wickenhagen et al., 2021). All Ydj1-CaaL/I/M variants appeared to be fully prenylated in the presence of HsFTase, with exception of CSFL that does not adhere to the consensus sequence due to non-aliphatic amino acids at the a1 and a2 positions. Of note, a few sequences with non-aliphatic a1 amino acids were fully prenylated (i.e., CTII, CKVL, and CQLL). Comparing band mobilities in the presence and absence of HsFTase indicated that several sequences were prenylated in a predominantly HsFTase-dependent manner (CKVL, CLLL, CLVL, CQLL, CSFL, and CVLL). For the remaining sequences, a smaller portion of the population exhibited a mobility shift in the absence of HsFTase (CTII, CIIL, CTIL and CVIL), suggesting that these sequences are either naturally geranylgeranylated naturally farnesylated but strongly subject to alternate prenylation when FTase is compromised (Figure 4C).
HsFTase modifies non-canonical length sequences
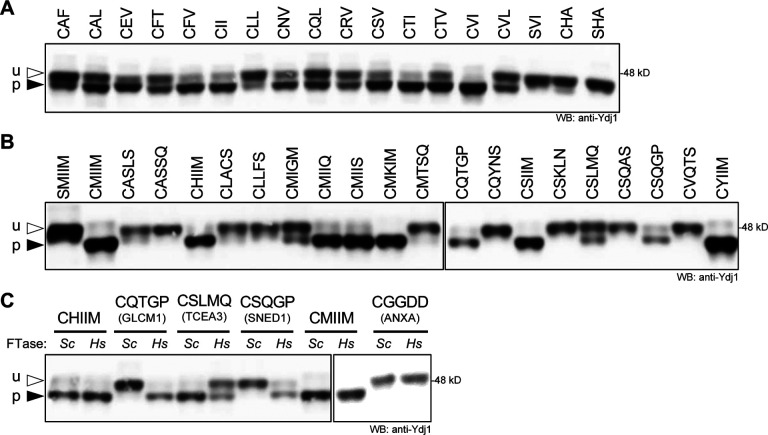
The substrate recognition profile of FTase has recently been expanded to certain sequences one amino acid longer or shorter than the tetrapeptide CaaX sequence (i.e., CaX and CaaaX). This expanded specificity was first identified using a combination of yeast genetics and in vitro biochemical analyses involving mammalian and yeast FTase (Ashok et al., 2020; Blanden et al., 2018; Schey et al., 2021). Additional CaaaX sequences subject to farnesylation were subsequently identified by in vitro screening of synthetic peptide libraries and by searches of the human proteome ((Blanden et al., 2018; Schey et al., 2021) and this study). To extend these observations to HsFTase, previously studied Ydj1-CaX and -CaaaX variants were expressed in the humanized FTase yeast strain and analyzed by gel-shift assay (Figure 5). One additional CaX (CHA) sequence was also evaluated due to its association with SARS-CoV2 ORF7b.
Figure 5. The humanized FTase yeast strain modifies certain non-canonical length sequences.
Ydj1-CaaX variants with A) shorter CaX and B) longer CaaaX sequences were produced in the humanized FTase strain and evaluated by gel-shift assay as described in Figure 3. u – unprenylated Ydj1; p – prenylated Ydj1. C) Comparison of Ydj1-CaaaX variant gel mobilities in the presence of ScFTase (Sc) and HsFTase (Hs). The source of the CaaaX sequence is indicated below each specific sequence. Total cell lysates were prepared and evaluated as described in Figure 3B. Strains used were yWS3186 (HsFTase ydj1Δ) and yWS2544 (ScFTase ydj1Δ). Plasmids used are listed in Supplemental Table S3. See Supplemental Table S1 for gel quantification.
The 16 CaX sequences evaluated exhibited varying degrees of farnesylation ranging from >90% (CVI, CTI) to <25% (CLL and CHA), with half exhibiting >50% farnesylation (8 of 16) (Figure 5A and Supplemental Table S1). The farnesylation patterns were similar to that observed for ScFTase. The very weak modification of Ydj1-CHA, which was quantified to be ~8% of the population, was not evident with SHA, consistent with the cysteine-dependent nature of this modification, but it is unclear whether such a low level of modification it is likely to be biologically significant for the biology of SARS-CoV2.
The 20 CaaaX sequences evaluated also exhibited varying degrees of farnesylation ranging from >90% (e.g., CMIIM) to unfarnesylated (e.g., CASSQ), with nearly half exhibiting >50% farnesylation (9 of 20) (Figure 5B). In most cases, ScFTase and HsFTase reactivities toward Ydj1-CaaX sequences have been highly similar, with the extent of modification being occasionally different for partially modified sequences (Supplemental Table S1). Here, however, dramatic differences were observed for a few CaaaX sequences. Two sequences were modified by HsFTase but not ScFTase: CQTGP (associated with GLCM1) and CSQGP (SNED1). One sequence was fully modified by ScFTase but <50% modified by HsFTase: CSLMQ (TCEA3) (Figure 5C). The species specificity differences observed with these CaaaX sequences may indicate subtle differences in the FTase substrate binding pockets of these enzymes that warrants future investigation.
This survey of CaX and CaaaX sequences reveals that HsFTase can indeed modify non-canonical length sequences in a cell-based system, albeit only a few exhibit full modification. For the sequences associated with eukaryotic or viral proteins (see Supplemental Table S5), it remains to be determined whether any are modified in their native context. Considering that there are 988 CaX and 941 CaaaX human proteins in the UniProtKB/Swiss-Prot database (Table 1), it is possible that some of these will be prenylated.
The humanized FTase yeast strain can be used to identity novel HsFTase target sequences
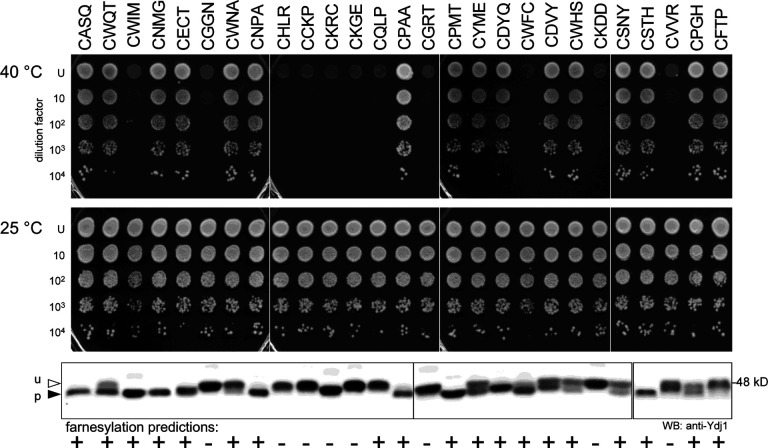
Two genetic screens, utilizing plasmid libraries based on distinct CaaX protein reporters (i.e., Ras and Ydj1), have comprehensively evaluated CaaX sequence space for reactivity with ScFTase (Kim et al., 2023; Stein et al., 2015). These genetic strategies are fully compatible with the humanized FTase strain developed in this study and could be used to query CaaX sequence space recognized by HsFTase. As proof of principle of this possibility, we performed a miniscreen by transforming the Ydj1-CaaX plasmid library into the humanized FTase strain and recovered a random sampling of thermotolerant and temperature sensitive colonies from which plasmids were recovered, sequenced, and evaluated by thermotolerance and gel-shift assays (Figure 6 and Supplemental Table S1). The results reveal a correlation between the ability to support thermotolerance and being farnesylated by HsFTase. The reciprocate is true for temperature sensitivity and lack of farnesylation, except for the sequences CWIM and CWFC, which were fully farnesylated yet temperature sensitive. The observations for CWIM and CWFC are consistent with the profile of canonical CaaX sequences that temper the ability of Ydj1 to support thermotolerance (i.e., fully-modified; farnesylated, cleaved, and carboxylmethylated) (Berger et al., 2018; Hildebrandt et al., 2016b).
Figure 6. Miniscreen using the humanized FTase strain for identifying prenylatable CaaX sequences.
The humanized FTase yeast strain (yWS3186) was transformed with a plasmid library encoding all 8000 Ydj1-CaaX variants. Plasmids were recovered and sequenced from a randomly selected population of thermotolerant and temperature-sensitive colonies. The plasmids were individually retransformed into yWS3186 and resultant strains evaluated by thermotolerance (upper panels), and gel-shift assays (lower panel) as described in Figure 3. Predictions for the farnesylation of each sequence by ScFTase are indicated at the bottom of the figure. Predictions were derived using Heat Map prediction scores reported in Kim et al (Kim et al., 2023); HM score >3 predicts farnesylation, HM score <3 predicts not farnesylated. See Supplemental Table S1 for gel quantification.
Studies of ScFTase specificity have led to the development of predictive models for prenylation (Berger et al., 2022; Kim et al., 2023). The more recent of these models was used to predict the farnesylation status (positive and negative) of the randomly sampled CaaX sequences. The predictions correlated well with empirical observations for positive and negative modification by HsFTase with the majority of sequences (20 of 26) being accurately predicted using rigorous thresholds (>50% modification as determined by band quantitation was required for positive classification; <10% modification was required for negative classification). For classification purposes, CKRC was judged to be unmodified, consistent with its negative prediction, because the gel-shift associated with this sequence also occurred in the absence of FTase (i.e., ram1Δ background), indicating that it is not farnesylated (Supplemental Figure S5). Of the outlier sequences, some were predicted to be modified, and indeed exhibited partial modification, but the extent of their modification was below the threshold for positive classification (i.e., CWNA, CWHS, CSNY and CPGH). By contrast, two outlier sequences that were predicted to be modified were unmodified (i.e., CQLP and CFTP), perhaps indicating that a terminal proline is unfavorable for farnesylation by HsFTase, which has been previously reported (Kim et al., 2023; Moores et al., 1991).
Among the randomly sampled sequences, only CPAA was found to exist in the human proteome when querying the UniprotKB/SwissProt database. It is associated with two proteins: the ER-localized DNase-1-like protein (P49184) and the extracellular laminin alpha subunit (Q16363). While CPAA was reactive with HsFTase, the subcellular locations of these human proteins likely disqualify them from being substrates of cytosolic FTase. Nevertheless, our results suggest ~77% accuracy when using a farnesylation prediction model developed using ScFTase specificity data to predict modification by HsFTase (Kim et al., 2023). The substrate specificities of ScFTase and HsFTase, although highly similar, are not identical, which prompts the need for future studies aimed at fully evaluating CaaX sequence space in the context of HsFTase. Such studies are now possible with our humanized FTase yeast strain, and such investigations may better refine the target profile of HsFTase, potentially leading to the discovery of novel CaaX proteins.
Interspecies complementation analysis of GGTase-I subunits
To develop a yeast system for expressing HsGGTase-I, studies were first performed to assess the functional equivalence of ScGGTase-I and HsGGTase-I subunits. Because GGTase-I activity is essential for yeast viability, strains lacking a ScGGTase-I subunit can only be propagated when complemented by plasmid-encoded copy of the missing gene (e.g., ram2Δ [URA3 RAM2]). These strain backgrounds were further modified to introduce plasmid-encoded copies of one or both orthologous HsGGTase-I subunit genes FNTA and PGGT1B under different promoter conditions. The strains were cultured, serially diluted, and spotted onto media containing 5-fluroorotic acid (5FOA) that counter selects for the URA3-marked plasmids encoding RAM2 or CDC43 in the parent strains. This strategy forced strains to contain either species matched or mismatched subunits of GGTase-I in the absence or presence of FTase (ram2Δ and cdc43Δ backgrounds, respectively).
The 5FOA analysis revealed, as observed for FTase, that neither FNTA nor PGGT1B alone could complement for the absence of its yeast ortholog ram2Δ or cdc43Δ, respectively, indicating that interspecies subunits do not form functional GGTase-I complexes (Figure 7A, 5FOA, 25 °C). This was observed whether FNTA and CDC43 (conditions 4 and 5) or Ram2 and PGGT1B (8 and 9) were co-expressed, and the result did not change when human subunits were over-expressed using the PGK1 strong constitutive promoter (5 and 8). Only when strains producing both HsGGTase-I subunits, whether expressed from the yeast orthologous promoters (3 and 7) or the PGK1 promoter (2 and 6), was growth equivalent to wildtype yeast (1). This observation contrasts with the FTase situation where both HsFTase subunits had to be expressed from the PGK1 promoter for functional complementation (Supplemental Figure S1). As additional confirmation that yeast growth was specifically due to HsGGTase-I activity, we observed that HsGGTase-I supported growth on 5FOA at 37 °C in the cdc43Δ but not ram2Δ background (Figure 7A, 37 °C). FTase activity is required for growth at higher temperature (He et al., 1991). The cdc43Δ strains (6 and 7) grew because they retain endogenous FTase activity (i.e., Ram2/Ram1 complex), while the ram2Δ strains (2 and 3) failed to grow because they lack FTase activity (i.e., Ram1 subunit only). Thus, it can be inferred that growth on 5FOA at 25 °C in the ram2Δ background can be attributed entirely to HsGGTase-I.
Figure 7. Interspecies complementation studies of GGTase-I subunits.
HsGGTase-I and ScGGTase-I subunits are not interchangeable, but co-expression of both α and β subunits of HsGGTase-I in yeast can restore GGTase-I activity in the absence of ScGGTase-I. A) Viability assay to assess GGTase-I-dependent growth. The yeast genetic backgrounds have only one of the naturally encoded ScGGTase-I subunits encoded on the chromosome, with the presence (Sc) and absence (Δ) of the specific subunit indicated above the panel. Because GGTase-I activity is essential for yeast viability, the other subunit is plasmid-encoded. The strains also contain plasmid(s) encoding HsGGTase-I FNTA and/or PGGT1B subunits (indicated by brackets) driven by either the orthologous yeast gene promoter (Hs) or the PGK1 promoter (PGKHs). Select strains were also transformed with empty vectors ([vect]) so that all strains had the same selectable markers. The pre-FOA strain genotypes are indicated at the top of the panel. Strains were cultured to the same density, serially diluted, and the dilution series mixtures spotted onto YPD and 5FOA media. Growth on YPD media provides an assessment for the quality of the serial dilutions. 5FOA media counter selects for the URA3 gene and effectively eliminates the URA3-marked plasmid, resulting in the post-FOA phenotypes indicated at the top of the panel. Growth on 5FOA at 25 °C indicates the presence of functional GGTase-I (2, 3, 6 and 7). Growth on 5FOA at 37 °C is also dependent on functional FTase, which requires α and FTase β subunits of the same species (6 and 7). Strains used were yWS3481 (1), yWS3388 (2), yWS3414 (3), yWS3639 (4), yWS3287 (5), yWS3387 (6), yWS3413 (7), yWS3285 (8), and yWS3638 (9). B) Gel-shift analysis of Ydj1-CaaX variants produced in yeast expressing ScGGTase-I (Sc) or plasmid-encoded HsGGTase-I (Hs or PGKHs). Total cell lysates were prepared and analyzed as described in Figure 3B. Strains used were yWS3209 (Sc), yWS3451 (Hs), and yWS3169 (PGK1Hs). C) The humanized GGTase-I strain allows prenylation of Ydj1-CaaM/I/L variants taken from human proteins. The expression of GGTase-I subunits in this strain was driven from the orthologous yeast gene promoters to better match the natural in vivo activity of yeast GGTase-I. Total cell lysates were prepared and analyzed as described in Figure 3B. The strain used was yWS3451. The plasmids in used in this figure are listed in Supplemental Table S3. See Supplemental Table S1 for gel quantification.
HsGGTase-I expressed in yeast modifies both canonical and non-canonical CaaX sequences
Evidence suggests that GGTase-I may have broader substrate specificity at the a1 and a2 positions akin to what has been observed for yeast and mammalian FTase (Kawata et al., 1990; Kilpatrick and Hildebrandt, 2007; Lebowitz et al., 1997; Onono et al., 2010). We investigated this issue more thoroughly using a three-plasmid system to examine geranylgeranylation of plasmid-encoded Ydj1-CaaX variants in the context of HsGGTase-I (Figure 7B). The strains evaluated all lack endogenous FTase and Ydj1, but express either endogenous GGTase-I (Sc) or plasmid-encoded HsGGTase-I subunits from the orthologous RAM2 and CDC43 promoters (Hs) or constitutive PGK1 promoters (PGKHs). Gel-shift analysis revealed that neither ScGGTase-I nor HsGGTase-I were able to modify Ydj1 harboring the CAHQ sequence that is associated with the Ydj1 human ortholog DNAJA2 that is strictly a farnesylated sequence (Andres et al., 1997). Other CaaX sequences (CNLI, CSFL, CVLL, CVIM, CIIM and CVLS) exhibited no or limited modification by ScGGTase-I, yet were better modified by HsGGTase-I. This was more evident when HsGGTase-I subunit pairs were expressed using the strong PGK1 promoter. These observations suggest that HsGGTase-I expressed in this yeast system is either more active or has a different specificity than ScGGTase-I, and that HsGGTase-I indeed has the potential to modify certain non-canonical CaaL/I/M sequences (e.g., CNLI and CSFL).
To further survey the geranylgeranylation potential of CaaX sequences, we evaluated a test set of sequences that included 22 sequences that mostly matched the CaaL/I/M consensus. For this analysis, we used the yeast system producing HsGGTase-I from orthologous yeast promoters to reduce the potential of over-expression artifacts (Supplemental Figure S2C). Most sequences exhibited a gel-shift indicative of modification, but only a few exhibited complete modification (i.e., CIIL, CTIL, CVIL, CVLL) or near-complete modification (CIIM, CVIM, CTII) (Figure 7C). Within this data set, we observed that the a1 position influenced prenylation by HsGGTaseI-1. This is evident when comparing the CxIM set of sequences: CSIM, CAIM, and CLIM (~50% modified) vs. CIIM and CVIM (100% modified). A similar observation was made when comparing CxLL sequences: CLLL and CQLL (~50% modified) vs. CVLL (100% modified). Sequences where the X position did not match the CaaL/I/M consensus (CAHQ, CVLS) were unmodified by HsGGTase-I.
Human lamins and Pex19 CaaX sequences have distinct CaaX protease profiles
The ability to humanize yeast for investigations of CaaX protein modifications can be extended to studies of the human CaaX protease Rce1. This protease cleaves CaaX proteins that follow the canonical modification pathway, which are subject to the coupled modifications of proteolysis and carboxymethylation. Rce1 prefers to cleave prenylated CaaX sequences having an aliphatic amino acid at a2, and this substrate specificity is generally conserved among orthologs (Mokry et al., 2009; Plummer et al., 2006). The Ste24 protease has also been referred to as a CaaX protease, but its primary role is in protein quality control, and its substrates do not need to be prenylated, unlike Rce1 (Ast et al., 2016; Boyartchuk et al., 1997; Hildebrandt et al., 2016a; Runnebohm et al., 2020).
In yeast, the a-factor mating pheromone is a useful reporter of CaaX proteolysis that can be coupled with expression of human Rce1 and ZMPSte24 for studies of target specificity in a cell-based model (Mokry et al., 2009; Plummer et al., 2006). Such studies can be used to help resolve the CaaX protease preferences of medically relevant human proteins. For example, defects in the proteolytic processing of Prelamin A (CSIM) are associated with progeria-like diseases (Barrowman et al., 2012b). Prelamin A is subject to two proteolytic events that yield Lamin A: one within the CaaX sequence and another 15 amino acids from the C-terminus. Both cleavage events have been attributed to ZMPSte24 (Barrowman et al., 2012a; Quigley et al., 2013). With a humanized CaaX protease yeast model, however, we observe that a-factor-CSIM is cleaved by HsRce1, not ZMPSte24, in accordance with more recent observations (Figure 8) (Berger et al., 2022; Nie et al., 2020). We extended these studies to evaluate the cleavage of the Lamin B CaaX sequence (CAIM), which was cleaved by both HsRce1 and ZMPSte24. This result suggests that FTase inhibitors (FTIs) may impact the CaaX processing of both lamins, while Rce1 inhibitors are likely to differentially impact Prelamin A and Lamin B processing. We also examined cleavage of the human peroxisomal chaperone Pex19 CaaX sequence.
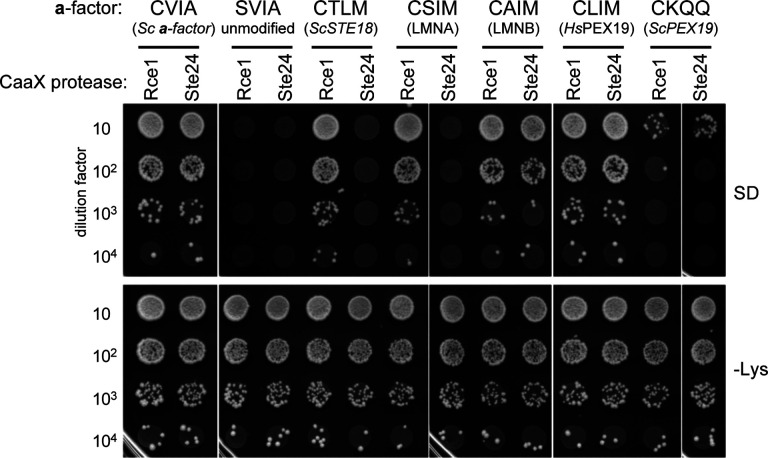
Figure 8. Prenylation studies can be coupled with CaaX proteolysis studies to better understand CaaX PTMs.
The mating assay was performed as described in Figure 1B using MATa strains that carry plasmid-encoded a-factor-CaaX variants and plasmid-encoded HsRCE1 (Rce1) or ZMPSTE24 (Ste24) CaaX proteases. The source of the CaaX sequence is indicated below each specific sequence. The strain background for this experiment lacks endogenous a-factor (MFA1 and MFA2) and yeast CaaX protease genes (yWS164; MATa mfa1Δ mfa2Δ rce1Δ ste24Δ). MATa strains were mixed with MAT03b1 cells (SM1068), mixes subject to 10-fold dilutions, and dilution mixtures spotted onto minimal media (SD) and SC-lysine (-Lys) media. Growth on SD indicates diploid formation, which is a direct indicator of a-factor mating pheromone production. Growth on –Lys reflects the input of MATa cells. Plasmids are listed in Supplemental Table S3.
Farnesylation of Pex19 is critical for its interaction with client proteins, and mutations of Pex19 unrelated to its CaaX sequence are associated with Zellweger syndrome (Emmanouilidis et al., 2017; Matsuzono et al., 1999). Human Pex19 (CLIM) and yeast Pex19 (CKQQ) have strikingly different CaaX sequences despite moderate homology (i.e., 21% identity; 48% similarity) (Madeira et al., 2022). Both sequences are fully prenylated in the context of either HsFTase or ScFTase (Figure 4A and Supplemental Figure S4). Using a-factor-CaaX variants harboring these sequences, CLIM was observed to be cleaved by both HsRCE1 and ZMPSte24, whereas the non-canonical CKQQ sequence was not. This result suggests that Pex19 proteins have evolved to have C-termini with distinct biophysical properties: canonical for human Pex19 and only isoprenylated for yeast Pex19. The underlying reasons for these differences remain unknown. Nonetheless, our studies with lamins and Pex19 fully support the utility of the humanized CaaX protease model system for expanding studies of CaaX protein post-translational modifications (PTMs) beyond the initial prenylation step. Such studies could prove useful for predicting the potential effects of inhibiting the Rce1 CaaX protease, which would impact a narrower range of CaaX protein targets than prenyltransferase inhibitors.
Discussion
We report the development of yeast strains that can be used for systematic characterization of prenylation by human FTase and GGTase-I. The humanized FTase strain, with subunits FNTA and FNTB integrated into the yeast genome, was phenotypically equivalent to yeast expressing native FTase in four functional tests: 1) production of a-factor mating pheromone, 2) Ras2 localization 3) Ydj1-based thermotolerance, and 4) Ydj1 farnesylation. Thus, the humanized FTase yeast strain provides in vivo FTase activity levels compatible for relatively rapid characterization of candidate CaaX sequences and heterologously expressed human CaaX proteins that may be targets for farnesylation. Moreover, the humanized FTase yeast strain provides a cell-based system that overcomes concerns associated with in vitro farnesylation assays that depend on chemically modified substrates and unnatural enzyme and substrate amounts. For example, the annexin A2 CGGDD extended CaaaX sequence has been reported as farnesylated with methods involving metabolic labeling with an azido-farnesyl analog (Kho et al., 2004), but this sequence is unmodified by human FTase in our cell-based system (Figure 5C). The convenience of the yeast system for heterologous protein expression makes it ideal for coupling with other methods, such as mass-spectrometry, for confidently validating the prenylation status of candidate CaaX proteins.
Limited comparative data exists for the specificities of mammalian FTase relative to ScFTase. In vitro, rat FTase and ScFTase and exhibit similar specificities when evaluated against a CVa2X peptide library (Wang et al., 2014). Our in vivo results confirm and further extend such observations to reveal that HsFTase expressed in yeast and native ScFTase have a striking degree of substrate conservation (see Supplemental Figure S4, Supplemental Table S1). This is most evident in the ability of both enzymes to similarly prenylate canonical and prenylation-only CaaX sequences. Thus, HsFTase also appears to possess the broadened ability to modify non-canonical sequences (i.e., sequences without aliphatic amino acids at the a1 and a2 positions), which was initially reported for ScFTase (Berger et al., 2018). These findings indicate that farnesylation prediction algorithms developed using ScFTase should work reasonably well for predicting HsFTase specificity (Kim et al., 2023). The conservation between human and yeast FTases extends to prenylation of non-standard length sequences that are one amino acid shorter or longer than the conventional tetrapeptide CaaX sequence, although species specific differences in substrate recognition of longer CaaaX sequences was observed.
The humanized GGTase-I strain, with subunits FNTA and PGGT1B expressed from low copy plasmids and driven by the orthologous yeast promoters, was phenotypically equivalent to yeast expressing native GGTase-I in supporting viability. The humanized GGTase-I strain, however, displayed more robust activity than endogenous yeast GGTase-I in modifying Ydj1-CaaL/I/M variants. This observation could be due to higher GGTase-I enzyme levels and/or subtle specificity differences between yeast and human enzymes. Human GGTase-I prenylated a wide range of CaaL/I/M sequences and did not prenylate CaaS/Q sequences, in alignment with expectations. One feature that was conserved across species was the ability of GGTase-I to alternatively prenylate normally farnesylated sequences. This typically occurs when FTase activity is reduced through action of FTIs, and in our case, when the FTase activity was genetically ablated. The observation that yeast and human FTase and GGTase-I can alternatively prenylate Ras sequences CIIM and CVIM when using Ydj1 or GFP-Ras2 reporters (Supplemental Figure S6) demonstrates that recognition of these sequences by both prenyltransferases is transferable and evolutionarily conserved between yeast and human enzymes, and likely across other species as well.
The scope of potentially prenylated proteins in protein databases is expansive with over 6000 annotated proteins in eukaryotes and viruses ending in Cxxx. When expanded to 3-mer and 5-mer length sequences, this number is over 15,000 (see Table 1). Not accounted for in this table, is the potential number of prokaryotic CaaX proteins. While prokaryotes lack protein prenylation machinery, there is evidence that some pathogenic bacteria, such as Legionella pneumophila and Salmonella typhimurium, encode CaaX proteins that are prenylated by host enzymes as part of their infectious life cycle (Ivanov et al., 2010; Reinicke et al., 2005). Thus, a reliable prediction algorithm and reporter systems to identify and verify the subpopulation of CaaX sequences that are likely to be modified by either FTase or GGTase-I will be a key step toward defining the prenylome across many different organisms.
Viral genomes encode many CaaX proteins that might be prenylated by human FTase and/or GGTase-I. We have evaluated several such sequences using our system, with results confirming instances of previously reported prenylation or indicating compatibility for prenylation. For example, we observed that the CaaX sequence of Hepatitis Delta Virus Large Delta antigen (L-HDAg; CRPQ) is modified by human and yeast FTases, but not by GGTase-I. Moreover, we observed that the CRPQ sequence is neither cleaved nor carboxylmethylated, consistent with its non-canonical sequence (see Figure 4 and Supplemental Figures 2 and 3). Prenylation of L-HDAg is required for virion assembly, and the FTI Lonafarnib is currently in Phase 3 clinical trials for use in combination treatments for Hepatitis D infections (Khalfi et al., 2023; Koh et al., 2015; Yardeni et al., 2022). We also determined that the CaaX sequence of human cytomegalovirus protein of unknown function IRL9 (CRIQ) can be farnesylated, cleaved and carboxymethylated. The modifications of IRL9 have not been previously reported, so it is a good candidate for additional investigations related to its prenylation status and impact of PTMs to virion formation. Other viral CaaX sequences CRII (Influenza virus H1N1 protein PB2), CVLM (Molluscum contagiosum virus protein MC155R) and CNLI (ASFV protein A179L) were substrates of both HsFTase and HsGGTase-I in our system. If these sequences are prenylated in their native context, dual prenyltransferase inhibitors might have better utility for blocking modification. Given that all the viral CaaX sequences evaluated in our study were targets for prenylation, it is prudent to consider the role that the prenylation of viral proteins may have on viral propagation and infection. The ability to readily assess the prenylation status of viral CaaX sequences afforded by our cell-based system could provide the necessary evidence for new investigations of viral biology or viral therapies in human and veterinary medicine involving FTIs, geranylgeranyltransferase inhibitors (GGTIs), dual prenyltransferase inhibitors (DPIs), and possibly CaaX protease and ICMT inhibitors as well.
Our system is well-suited for assessing the prenylation status of human proteins as supported by the modification of heterologously expressed human HRas61 and DNAJA2. An important caveat to consider is that SDS-PAGE gel-shifts do not universally occur for all prenylated proteins, which is why Ydj1 is such a great reporter protein for testing CaaX sequences. Unfortunately, our system does not exclude the possibility of a CaaX sequence being reactive in the context of Ydj1, but unreactive its native context, for a myriad of reasons (e.g., structurally inaccessible C-terminus, localization of protein inaccessible to cytosolic prenyltransferases, alternate cysteine modifications such as disulfide bonding, etc.). Therefore, positive results with our system should be coupled with additional validation of prenylation in native contexts. Nonetheless, the ease and convenience of the humanized FTase and GGTase-I yeast strains that we have developed are an important and valuable new resource for initial investigations into evaluating the prenylation potential of a wide array of CaaX sequences.
Materials and Methods
Yeast strains:
The humanized prenyltransferase strains are summarized in Supplemental Figure S2. All yeast strains used in this study are listed in Supplemental Table S2. Detailed descriptions of strain constructions can be found in the Supplemental Methods file. Yeast were typically cultured in standard liquid and solid yeast media unless otherwise noted. For expression of Myc-HRas61 from the inducible MET25 promoter (p-05547) in methionine auxotroph strains (i.e., yWS2544 and yWS3186), yeast were first cultured in SC-leucine containing 20 μg/ml methionine to late log phase, washed, and resuspended in SC-leucine containing 2 μg/ml methionine to allow for both growth and induction of the MET25 promoter.
New strains were typically created for this study by standard genetic manipulations starting with commercially available haploid and heterozygous diploid genomic deletions. KANR and NATR marked gene replacements were confirmed by growth on YPD containing G418 (200 μg/ml; Research Products International) or nourseothricin (100 μg/ml; GoldBio), respectively. All gene replacements were further checked by PCR to confirm the presence of the knockout at the correct locus and absence of the wild-type open reading frame. Plasmids were introduced into strains via a lithium acetate-based transformation procedure (Elble, 1992). Yeast sporulation was carried out in a solution of 2% potassium acetate, 0.25% yeast extract, and 0.1% dextrose. Spore enrichment and random spore analysis followed published methods (Rockmill et al., 1991). For the humanized FTase strains (yWS3186 and yWS3220), PPGK1-FNTB was integrated at the RAM1 locus, replacing the open reading frame, using a loop-in loop out strategy, and PPGK1-FNTA was integrated at the his3Δ1 locus by homologous recombination using a HIS3-based integrative plasmid (Schey et al., 2023).
Plasmids:
The plasmids used in this study are listed in Supplemental Table S3. Detailed descriptions of plasmid construction can be found in the Supplemental Methods file. Plasmids were analyzed by diagnostic restriction digest and DNA sequencing (Eurofins Genomics, Louisville, KY) to verify the entire open reading frame and surrounding sequence. Plasmids recovered from the Ydj1-CaaX Trimer20 library are described elsewhere (Kim et al., 2023). New plasmids encoding Ydj1-CaaX and a-factor-CaaX (encoded by MFA1 gene) variants were made by PCR-directed plasmid based recombinational cloning as previously described (Berger et al., 2018; Oldenburg et al., 1997). Plasmids with GFP-Ras2-CaaX variants were generated by QuikChange site-directed mutagenesis. Plasmids encoding HsFTase and HsGGTase-I subunits were created in multiple steps. First, the open reading frames (ORFs) and flanking 5´ and 3śequences of yeast prenyltransferase subunits were PCR-amplified from strain BY4741 and subcloned into the multicloning sites of appropriate pRS series vectors (Sikorski and Hieter, 1989). In parallel, synthetic cDNAs for human FNTA, FNTB and PGGT1B that were codon optimized for S. cerevisiae were subcloned into the PstI and XhoI sites of pBluescriptII KS(−); all cDNAs were commercially obtained (GenScript). The human ORFs were engineered to also have 39 bp of flanking sequence on both the 5´ and 3énds that equivalently matched the flanking sequences of the orthologous yeast ORFs. Next, DNA fragments encoding the human ORFs and yeast flanking sequences were used with recombination-based methods for direct gene replacement of the plasmid-encoded yeast ORFs. The PGK1 promoter was amplified and used to replace the orthologous yeast promoters by similar recombination-based methods as described in Supplemental Materials. To change selectable markers, the various FTase and GGTase-I promoter-containing segments were subcloned as XhoI-SacI fragments into different pRS vector backbones by conventional ligation-based cloning.
Temperature sensitivity assay:
Thermotolerance assays were performed as previously described (Blanden et al., 2018; Hildebrandt et al., 2016b). In brief, plasmid-transformed strains were cultured in appropriate selective synthetic drop-out (SC-) liquid media to saturation (25 °C, 24 hours); strains without plasmids were cultured in SC complete media. Cultures were serially diluted into H2O (10-fold dilutions), and dilutions replica pinned onto YPD plates. Plates were incubated (25 °C for 72 hours; 37 °C for 48 hours; 40 °C and 41 °C for 72 hours) then digitally imaged using a Canon flat-bed scanner (300 dpi; grayscale; TIFF format).
Yeast lysate preparations and immunoblots:
Cell-mass equivalents of log-phase yeast (A600 0.95–1.1) cultured in appropriate selective SC- liquid media were harvested by centrifugation, washed with water, and processed by alkaline hydrolysis and TCA precipitation (Kim et al., 2005). Total protein precipitates were resuspended in urea-containing Sample Buffer (250 mM Tris, 6 M urea, 5% β-mercaptoethanol, 4% SDS, 0.01% bromophenol blue, pH 8), and analyzed by SDS-PAGE (9.5%) followed by immunoblotting. Blots were processed according to standard protocols. Antibodies used for Western blots were rabbit anti-Ydj1 polyclonal (1:10,000 dilution; gift from A. Caplan); mouse anti-c-Myc 9E10 monoclonal (1:1000 dilution, Santa Cruz Biotech. #sc-40); mouse anti-GFP B-2 monoclonal (1:500, Santa Cruz Biotech. # sc-9996); mouse anti-DnaJA2(7) monoclonal (1:500 dilution, Santa Cruz Biotech. sc-136515); mouse anti-HIS his.h8 monoclonal (1:1000 dilution, VWR #101981–852); goat HRP-anti-rabbit and goat HRP-anti-mouse (1:1000 dilutions, Kindle Biosciences, LLC #R1006 and R1005, respectively). Immunoblots were developed with ECL reagent (ProSignal Pico ECL Spray or Kindle Biosciences KwikQuant Western Blot Detection Kit) and images were digitally captured using the KwikQuant Imager system (Kindle Biosciences). Adobe Photoshop was used for cropping and rotating of images; no other image adjustments were applied.
Prenylation of Ydj1-CaaX was quantified using ImageJ and multiple exposures having good dynamic range of band intensities. Percent prenylation was calculated as the intensity of the lower (prenylated band) divided by total intensity of the prenylated and unprenylated (upper) bands. Data presented in Supplemental Table S1 represent the averages of biological replicates. For duplicate samples, values are reported as an average and associated range. For multiplicate samples, the values are reported as an average and standard error of the mean (SEM).
Yeast mating assay:
Qualitative and quantitative yeast mating assays were performed as detailed previously (Berger et al., 2018). In brief, MATa test strains and the MATα strain (IH1793) were cultured to saturation at 25 °C in appropriate selective SC- media and YPD liquid media, respectively. Cultures were normalized by dilution with fresh media (A600 1.0), mixed 1:9 (MATa:MATα) in individual wells of a 96-well plate, and cell mixtures subject to 10-fold serial dilution using the MATα cell suspension as the diluent. For qualitative analyses, each dilution series was replica pinned onto SC-lysine and minimal SD solid media. For quantitative analysis, a portion of a dilution mixture was spread onto SC-lysine and SD plates in duplicate, and colonies counted after plate incubation (72 hours, 30 °C). The SC-lysine cell count reports on the total number of MATa haploid cells initially in the mixture while the SD cell count reports on the number of mating events (i.e., diploids). Mating efficiencies were normalized to an a-factor (MFA1) positive control within each experiment.
Microscopy:
Imaging was performed as detailed previously (Ravishankar et al., 2023). In brief, yeast transformed with CEN URA3 PYdj1-GFP-RAS2-CaaX variants were cultured to late log (A600 0.8–1) in SC-uracil liquid media and viewed using a Zeiss Axio Observer microscope equipped with fluorescence optics (Plan Apochromat 63X/1.4 N.A objective). Images were captured using AxioVision software and minor image adjustments performed using Adobe Photoshop.
Humanized yeast miniscreen for identification of Ydj1-CaaX variants:
yWS3186 was transformed with the Ydj1-CaaX plasmid library (pWS1775), plated onto SC-uracil solid media, and incubated 4 days at 25 °C. Single colonies of varying size were individually cultured and subject to the temperature sensitivity assay described above except that 3 spots of 20-fold serial dilutions were prepared. Growth at 41 °C was scored on a scale of 0 to 5 relative to control strains yWS3311 (Ydj1-CASQ, score of 5) and yWS3312 (Ydj1-SASQ, score of 0). Plasmids were recovered from candidates scoring 0 (n=11) and candidates scoring 5 (n=15), sequenced to identify the CaaX sequence, and retransformed into yWS3186 for retesting by the temperature sensitivity assay (10-fold dilutions) and SDS-PAGE gel-shift assay.
5-fluroorotic acid (5FOA) assay:
BY4741, yWS3106 and yWS3109 were transformed with combinations of HIS3 and LEU2 based plasmids carrying HsFNTA, HsPGGT1B, and/or empty vector plasmids such that all strains had the same selectable markers. Strains cultured on SC-histidine-leucine-uracil solid media were used to inoculate SC-histidine-leucine liquid media. After incubation (25 °C, 24 hours), cultures were normalized with fresh media (A600 2.0), added to wells of a 96 well plate, serial diluted 8-fold with H2O as the diluent, and the dilution series pinned onto SC complete solid media containing 1 mg/ml 5-fluroorotic acid (5FOA).
Search for CaaX sequences:
Scan Prosite (https://prosite.expasy.org/scanprosite/) was used to search for CaaX sequences in Homo sapiens and viruses in the UniProtKB/Swiss-Prot database using the search strings “CXXX>”, “C{C}XXX>”, or “{C}CXX>” (search performed 07/12/2023). For certain CaaaX and CaX and sequences that were studied in previous publications but have no human or virus associated protein in the annotated UniProtKB/SwissProt database, the search was expanded UniProtKB/TrEMBL database, inclusive of all eukaryotes.
Acknowledgements:
We thank Dr. Avrom Caplan (City College of New York) for anti-Ydj1 antibody, Dr. Jun Wang (Institute Pasteur of Shanghai, Chinese Academy of Sciences) for discussions on ASFV CaaX proteins, and Schmidt lab members for constructive feedback and technical assistance.
Funding:
This research was supported by Public Health Service grant GM132606 from the National Institute of General Medical Sciences (WKS).
Funding Statement
This research was supported by Public Health Service grant GM132606 from the National Institute of General Medical Sciences (WKS).
Footnotes
Competing interests: none
the term CaaX will refer to all C-terminal tetrapeptide sequences whether or not they fit the traditional consensus sequence.
Data Availability:
Strains and plasmids generated for this study are available upon request.
References
- Adari H., Lowy D. R., Willumsen B. M., Der C. J. and McCormick F. (1988). Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science 240, 518–21. [DOI] [PubMed] [Google Scholar]
- Anderegg R., Betz R., Carr S., Crabb J. and Duntze W. (1988). Structure of Saccharomyces cerevisiae mating hormone a-factor: identification of S-farnesyl cysteine as a structural component. J Biol Chem 263, 18236–18240. [PubMed] [Google Scholar]
- Andres D. A., Shao H., Crick D. C. and Finlin B. S. (1997). Expression cloning of a novel farnesylated protein, RDJ2, encoding a DnaJ protein homologue. Arch Biochem Biophys 346, 113–24. [DOI] [PubMed] [Google Scholar]
- Ashar H. R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W. R. and Kirschmeier P. (2000). Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J Biol Chem 275, 30451–7. [DOI] [PubMed] [Google Scholar]
- Ashok S., Hildebrandt E. R., Ruiz C. S., Hardgrove D. S., Coreno D. W., Schmidt W. K. and Hougland J. L. (2020). Protein Farnesyltransferase Catalyzes Unanticipated Farnesylation and Geranylgeranylation of Shortened Target Sequences. Biochemistry 59, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast T., Michaelis S. and Schuldiner M. (2016). The protease Ste24 clears clogged translocons. Cell 164, 103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J., Hamblet C., Kane M. S. and Michaelis S. (2012a). Requirements for efficient proteolytic cleavage of prelamin A by ZMPSTE24. PLoS One 7, e32120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J., Wiley P. A., Hudon-Miller S. E., Hrycyna C. A. and Michaelis S. (2012b). Human ZMPSTE24 disease mutations: residual proteolytic activity correlates with disease severity. Hum Mol Genet 21, 4084–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B. M., Kim J. H., Hildebrandt E. R., Davis I. C., Morgan M. C., Hougland J. L. and Schmidt W. K. (2018). Protein isoprenylation in yeast targets COOH-terminal sequences not adhering to the CaaX consensus. Genetics 210, 1301–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B. M., Yeung W., Goyal A., Zhou Z., Hildebrandt E. R., Kannan N. and Schmidt W. K. (2022). Functional classification and validation of yeast prenylation motifs using machine learning and genetic reporters. PLoS One 17, e0270128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanden M. J., Suazo K. F., Hildebrandt E. R., Hardgrove D. S., Patel M., Saunders W. P., Distefano M. D., Schmidt W. K. and Hougland J. L. (2018). Efficient farnesylation of an extended C-terminal C(x)3X sequence motif expands the scope of the prenylated proteome. J Biol Chem 293, 2770–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyartchuk V. L., Ashby M. N. and Rine J. (1997). Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275, 1796–800. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P. and Boeke J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–32. [DOI] [PubMed] [Google Scholar]
- Cadinanos J., Varela I., Mandel D., Schmidt W. K., Díaz-Perales A., López-Otín C. and JMP F. (2003). AtFACE-2, a prenylated-protein protease from Arabidopsis thaliana related to Ras converting enzymes. J Biol Chem 278, 42091–7. [DOI] [PubMed] [Google Scholar]
- Campbell S. L. and Philips M. R. (2021). Post-translational modification of RAS proteins. Curr Opin Struct Biol 71, 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplin B. E., Hettich L. A. and Marshall M. S. (1994). Substrate characterization of the Saccharomyces cerevisiae protein farnesyltransferase and type-I protein geranylgeranyltransferase. Biochim Biophys Acta 1205, 39–48. [DOI] [PubMed] [Google Scholar]
- Cox A. D., Der C. J. and Philips M. R. (2015). Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin Cancer Res 21, 1819–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis D. A. and Braun P. E. (1994). Isoprenylation of brain 2’,3’-cyclic nucleotide 3’phosphodiesterase modulates cell morphology. J Neurosci Res 39, 386–97. [DOI] [PubMed] [Google Scholar]
- Dong X., Mitchell D. A., Lobo S., Zhao L., Bartels D. J. and Deschenes R. J. (2003). Palmitoylation and plasma membrane localization of Ras2p by a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol Cell Biol 23, 6574–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einav S. and Glenn J. S. (2003). Prenylation inhibitors: a novel class of antiviral agents. J Antimicrob Chemother 52, 883–6. [DOI] [PubMed] [Google Scholar]
- Elble R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques 13, 18–20. [PubMed] [Google Scholar]
- Emmanouilidis L., Schutz U., Tripsianes K., Madl T., Radke J., Rucktaschel R., Wilmanns M., Schliebs W., Erdmann R. and Sattler M. (2017). Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. Nat Commun 8, 14635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth C. L., Marshall M. S., Gibbs J. B., Stacey D. W. and Feig L. A. (1991). Preferential inhibition of the oncogenic form of RasH by mutations in the GAP binding/”effector” domain. Cell 64, 625–33. [DOI] [PubMed] [Google Scholar]
- Glenn J. S., Watson J. A., Havel C. M. and White J. M. (1992). Identification of a prenylation site in delta virus large antigen. Science 256, 1331–3. [DOI] [PubMed] [Google Scholar]
- Hancock J. F., Magee A. I., Childs J. E. and Marshall C. J. (1989). All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57, 1167–77. [DOI] [PubMed] [Google Scholar]
- Hartman H. L., Hicks K. A. and Fierke C. A. (2005). Peptide specificity of protein prenyltransferases is determined mainly by reactivity rather than binding affinity. Biochemistry 44, 15314–24. [DOI] [PubMed] [Google Scholar]
- He B., Chen P., Chen S. Y., Vancura K. L., Michaelis S. and Powers S. (1991). RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc Natl Acad Sci USA 88, 11373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E. R., Arachea B. T., Wiener M. C. and Schmidt W. K. (2016a). Ste24p Mediates Proteolysis of Both Isoprenylated and Non-prenylated Oligopeptides. J Biol Chem 291, 14185–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E. R., Cheng M., Zhao P., Kim J. H., Wells L. and Schmidt W. K. (2016b). A shunt pathway limits the CaaX processing of Hsp40 Ydj1p and regulates Ydj1p-dependent phenotypes. eLife 5, e15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland J. L., Hicks K. A., Hartman H. L., Kelly R. A., Watt T. J. and Fierke C. A. (2010). Identification of novel peptide substrates for protein farnesyltransferase reveals two substrate classes with distinct sequence selectivities. J Mol Biol 395, 176–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S. S., Charron G., Hang H. C. and Roy C. R. (2010). Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J Biol Chem 285, 34686–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong A., Auger S. A., Maity S., Fredriksen K., Zhong R., Li L. and Distefano M. D. (2022). In Vivo Prenylomic Profiling in the Brain of a Transgenic Mouse Model of Alzheimer’s Disease Reveals Increased Prenylation of a Key Set of Proteins. ACS Chem Biol 17, 2863–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama M., Kawata M., Yoshida Y., Horiuchi H., Yamamoto T., Matsuura Y. and Takai Y. (1991). The posttranslationally modified C-terminal structure of bovine aortic smooth muscle rhoA p21. J Biol Chem 266, 12639–45. [PubMed] [Google Scholar]
- Kawata M., Farnsworth C. C., Yoshida Y., Gelb M. H., Glomset J. A. and Takai Y. (1990). Posttranslationally processed structure of the human platelet protein smg p21B: evidence for geranylgeranylation and carboxyl methylation of the C-terminal cysteine. Proc Natl Acad Sci USA 87, 8960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfi P., Kennedy P. T., Majzoub K. and Asselah T. (2023). Hepatitis D virus: Improving virological knowledge to develop new treatments. Antiviral Res 209, 105461. [DOI] [PubMed] [Google Scholar]
- Kho Y., Kim S. C., Jiang C., Barma D., Kwon S. W., Cheng J., Jaunbergs J., Weinbaum C., Tamanoi F., Falck J. et al. (2004). A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci USA 101, 12479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick E. L. and Hildebrandt J. D. (2007). Sequence dependence and differential expression of Ggamma5 subunit isoforms of the heterotrimeric G proteins variably processed after prenylation in mammalian cells. J Biol Chem 282, 14038–47. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Hildebrandt E. R., Sarkar A., Yeung W., Waldon R. A., Kannan N. and Schmidt W. K. (2023). A comprehensive in vivo screen of yeast farnesyltransferase activity reveals broad reactivity across a majority of CXXX sequences. G3 (Bethesda) 7, jkad094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R., Rine J. and Kim S. H. (1990). Prenylation of mammalian Ras protein in Xenopus oocytes. Mol Cell Biol 10, 5945–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lapham A., Freedman C., Reed T. and Schmidt W. (2005). Yeast as a tractable genetic system for functional studies of the insulin-degrading enzyme. J Biol Chem 280, 27481–27490. [DOI] [PubMed] [Google Scholar]
- Koh C., Canini L., Dahari H., Zhao X., Uprichard S. L., Haynes-Williams V., Winters M. A., Subramanya G., Cooper S. L., Pinto P. et al. (2015). Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: a proof-of-concept randomised, double-blind, placebo-controlled phase 2A trial. Lancet Infect Dis 15, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnke M., Delon C., Hastie M. L., Nguyen U. T., Wu Y. W., Waldmann H., Goody R. S., Gorman J. J. and Alexandrov K. (2013). Rab GTPase prenylation hierarchy and its potential role in choroideremia disease. PLoS One 8, e81758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnankutty R. K., Kukday S. S., Castleberry A. J., Breevoort S. R. and Schmidt W. K. (2009). Proteolytic processing of certain CaaX motifs can occur in the absence of the Rce1p and Ste24p CaaX proteases. Yeast 26, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak A. J., Aditya A. V., Hougland J. L., Fierke C. A. and Gibbs R. A. (2010). Synthesis and screening of a CaaL peptide library versus FTase reveals a surprising number of substrates. Bioorg Med Chem Lett 20, 767–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane K. T. and Beese L. S. (2006). Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res 47, 681–99. [DOI] [PubMed] [Google Scholar]
- Lebowitz P. F., Casey P. J., Prendergast G. C. and Thissen J. A. (1997). Farnesyltransferase inhibitors alter the prenylation and growth-stimulating function of RhoB. J Biol Chem 272, 15591–4. [DOI] [PubMed] [Google Scholar]
- Leung K. F., Baron R., Ali B. R., Magee A. I. and Seabra M. C. (2007). Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J Biol Chem 282, 1487–1497. [DOI] [PubMed] [Google Scholar]
- Madeira F., Pearce M., Tivey A. R. N., Basutkar P., Lee J., Edbali O., Madhusoodanan N., Kolesnikov A. and Lopez R. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50, W276–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakasova E. S., Eisenhaber B., Maurer-Stroh S., Eisenhaber F. and Baranova A. (2017). Prenylation of viral proteins by enzymes of the host: Virus-driven rationale for therapy with statins and FT/GGT1 inhibitors. Bioessays 39, 1700014. [DOI] [PubMed] [Google Scholar]
- Matsuzono Y., Kinoshita N., Tamura S., Shimozawa N., Hamasaki M., Ghaedi K., Wanders R. J., Suzuki Y., Kondo N. and Fujiki Y. (1999). Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci USA 96, 2116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S., Washietl S. and Eisenhaber F. (2003). Protein prenyltransferases. Genome Biol 4, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Ali W., Chiu V. K., Bergo M., Silletti J., Wright L., Young S. G. and Philips M. (2005). Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell 16, 1606–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed I., Hampton S. E., Ashall L., Hildebrandt E. R., Kutlik R. A., Manandhar S. P., Floyd B. J., Smith H. E., Dozier J. K., Distefano M. D. et al. (2016). 8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells. Bioorg Med Chem 24, 160–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry D. Z., Manandhar S. P., Chicola K. A., Santangelo G. M. and Schmidt W. K. (2009). Heterologous expression studies of Saccharomyces cerevisiae reveal two distinct trypanosomatid CaaX protease activities and identify their potential targets. Eukaryot Cell 8, 1891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores S. L., Schaber M. D., Mosser S. D., Rands E., O’Hara M. B., Garsky V. M., Marshall M. S., Pompliano D. L. and Gibbs J. B. (1991). Sequence dependence of protein isoprenylation. J Biol Chem 266, 14603–14610. [PubMed] [Google Scholar]
- Moudgil D. K., Westcott N., Famulski J. K., Patel K., Macdonald D., Hang H. and Chan G. K. (2015). A novel role of farnesylation in targeting a mitotic checkpoint protein, human Spindly, to kinetochores. J Cell Biol 208, 881–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L., Spear E., Babatz T. D., Quigley A., Dong Y. Y., Chu A., Rotty B., Chalk R., Mukhopadhyay S. M. M., Burgess-Brown N. A. et al. (2020). A new paradigm for Prelamin A proteolytic processing by ZMPSTE24: the upstream SŶLL cleavage occurs first and there is no CaaX processing by ZMPSTE24. bioRxiv doi: 10.1101/2020.05.13.093849 . [DOI] [Google Scholar]
- Oldenburg K. R., Vo K. T., Michaelis S. and Paddon C. (1997). Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 25, 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onono F. O., Morgan M. A., Spielmann H. P., Andres D. A., Subramanian T., Ganser A. and Reuter C. W. (2010). A tagging-via-substrate approach to detect the farnesylated proteome using two-dimensional electrophoresis coupled with Western blotting. Mol Cell Proteomics 9, 742–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsuledesai C. C., Ochocki J. D., Markowski T. W. and Distefano M. D. (2014). A combination of metabolic labeling and 2D-DIGE analysis in response to a farnesyltransferase inhibitor facilitates the discovery of new prenylated proteins. Mol Biosyst 10, 1094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer L. J., Hildebrandt E. R., Porter S. B., Rogers V. A., McCracken J. and Schmidt W. K. (2006). Mutational analysis of the Ras converting enzyme reveals a requirement for glutamate and histidine residues. J Biol Chem 281, 4596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronin A. V., Narovlyansky A. N. and Sanin A. V. (2021). New Approaches to the Prevention and Treatment of Viral Diseases. Arch Immunol Ther Exp (Warsz) 69, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley A., Dong Y., Pike A., Dong L., Shrestha L., Berridge G., Stansfeld P., Sansom M., Edwards A., Bountra C. et al. (2013). The structural basis of ZMPSTE24-dependent laminopathies. Science 339, 1604–7. [DOI] [PubMed] [Google Scholar]
- Rashidian M., Dozier J. K. and Distefano M. D. (2013). Enzymatic labeling of proteins: techniques and approaches. Bioconjug Chem 24, 1277–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravishankar R., Hildebrandt E. R., Greenway G., Asad N., Gore S., Dore T. M. and Schmidt W. K. (2023). Specific Disruption of Ras2 CAAX Proteolysis Alters Its Localization and Function. Microbiol Spectr 11, e0269222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinicke A. T., Hutchinson J. L., Magee A. I., Mastroeni P., Trowsdale J. and Kelly A. P. (2005). A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J Biol Chem 280, 14620–7. [DOI] [PubMed] [Google Scholar]
- Ring J., Tadic J., Ristic S., Poglitsch M., Bergmann M., Radic N., Mossmann D., Liang Y., Maglione M., Jerkovic A. et al. (2022). The HSP40 chaperone Ydj1 drives amyloid beta 42 toxicity. EMBO Mol Med 14, e13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Lambie E. J. and Roeder G. S. (1991). Spore enrichment. Methods Enzymol 194, 146–9. [DOI] [PubMed] [Google Scholar]
- Runnebohm A. M., Richards K. A., Irelan C. B., Turk S. M., Vitali H. E., Indovina C. J. and Rubenstein E. M. (2020). Overlapping function of Hrd1 and Ste24 in translocon quality control provides robust channel surveillance. J Biol Chem 295, 16113–16120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schey G., Hildebrandt E., Wang Y., Diwan S., Passetti H., Potts G., Sprague-Getsy A., Leoni E., Hougland J., Beese L. et al. (2023). Library Screening, In Vivo Confirmation, and Structural and Bioinformatic Analysis of Pentapeptide Sequences as Sub-strates for Protein Farnesyltransferase. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schey G. L., Buttery P. H., Hildebrandt E. R., Novak S. X., Schmidt W. K., Hougland J. L. and Distefano M. D. (2021). MALDI-MS Analysis of Peptide Libraries Expands the Scope of Substrates for Farnesyltransferase. Int J Mol Sci 22, 12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker D. D., Lashkari D. A., Morris D., Mittmann M. and Davis R. W. (1996). Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nature Genetics 14, 450–6. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S. and Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soveg F. W., Schwerk J., Gokhale N. S., Cerosaletti K., Smith J. R., Pairo-Castineira E., Kell A. M., Forero A., Zaver S. A., Esser-Nobis K. et al. (2021). Endomembrane targeting of human OAS1 p46 augments antiviral activity. eLife 10, e71047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein V., Kubala M. H., Steen J., Grimmond S. M. and Alexandrov K. (2015). Towards the systematic mapping and engineering of the protein prenylation machinery in Saccharomyces cerevisiae. PLoS One 10, e0120716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storck E. M., Morales-Sanfrutos J., Serwa R. A., Panyain N., Lanyon-Hogg T., Tolmachova T., Ventimiglia L. N., Martin-Serrano J., Seabra M. C., Wojciak-Stothard B. et al. (2019). Dual chemical probes enable quantitative system-wide analysis of protein prenylation and prenylation dynamics. Nat Chem 11, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H., Thomas-MacArthur V. and Strutt D. (2013). Strabismus promotes recruitment and degradation of farnesylated prickle in Drosophila melanogaster planar polarity specification. PLoS Genetics 9, e1003654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suazo K. F., Hurben A. K., Liu K., Xu F., Thao P., Sudheer C., Li L. and Distefano M. D. (2018). Metabolic Labeling of Prenylated Proteins Using Alkyne-Modified Isoprenoid Analogues. Curr Protoc Chem Biol 10, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suazo K. F., Park K. Y. and Distefano M. D. (2021). A Not-So-Ancient Grease History: Click Chemistry and Protein Lipid Modifications. Chem Rev 121, 7178–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C., Ohya Y. and Rine J. (1993). Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol Cell Biol 13, 4260–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I., Pereira S., Ugalde A. P., Navarro C. L., Suarez M. F., Cau P., Cadinanos J., Osorio F. G., Foray N., Cobo J. et al. (2008). Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat Med 14, 767–72. [DOI] [PubMed] [Google Scholar]
- Wang C., Gale M. Jr., Keller B. C., Huang H., Brown M. S., Goldstein J. L. and Ye J. (2005). Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell 18, 425–34. [DOI] [PubMed] [Google Scholar]
- Wang Y. C., Dozier J. K., Beese L. S. and Distefano M. D. (2014). Rapid analysis of protein farnesyltransferase substrate specificity using peptide libraries and isoprenoid diphosphate analogues. ACS Chem Biol 9, 1726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore A., Freeny D., Sojourner S. J., Miles J. S., Graham W. M. and Flores-Rozas H. (2020). Evaluation of the Role of Human DNAJAs in the Response to Cytotoxic Chemotherapeutic Agents in a Yeast Model System. Biomed Res Int 2020, 9097638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte D. B., Kirschmeier P., Hockenberry T. N., Nunez-Oliva I., James L., Catino J. J., Bishop W. R. and Pai J. K. (1997). K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 272, 14459–64. [DOI] [PubMed] [Google Scholar]
- Wickenhagen A., Sugrue E., Lytras S., Kuchi S., Noerenberg M., Turnbull M. L., Loney C., Herder V., Allan J., Jarmson I. et al. (2021). A prenylated dsRNA sensor protects against severe COVID-19. Science 374, eabj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardeni D., Heller T. and Koh C. (2022). Chronic hepatitis D-What is changing? J Viral Hepat 29, 240–251. [DOI] [PubMed] [Google Scholar]
- Zhang F. L., Diehl R. E., Kohl N. E., Gibbs J. B., Giros B., Casey P. J. and Omer C. A. (1994). cDNA cloning and expression of rat and human protein geranylgeranyltransferase type-I. J Biol Chem 269, 3175–3180. [PubMed] [Google Scholar]
- Zhang F. L., Kirschmeier P., Carr D., James L., Bond R. W., Wang L., Patton R., Windsor W. T., Syto R., Zhang R. et al. (1997). Characterization of Ha-ras, N-ras, KiRas4A, and Ki-Ras4B as in vitro substrates for farnesyl protein transferase and geranylgeranyl protein transferase type I. J Biol Chem 272, 10232–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids generated for this study are available upon request.