Abstract
Many strains of Pseudomonas aeruginosa are resistant to the antibiotics cerulenin and thiolactomycin, potent inhibitors of bacterial fatty acid biosynthesis. A novel yeast Flp recombinase-based technique was used to isolate an unmarked mexAB-oprM deletion encoding an efflux system mediating resistance to multiple antibiotics in P. aeruginosa. The experiments showed that the MexAB-OprM system is responsible for the intrinsic resistance of this bacterium to cerulenin and thiolactomycin. Whereas thiolactomycin was not a substrate of the MexCD-OprJ pump expressed in a Δ(mexAB-oprM) nfxB mutant, cerulenin was efficiently effluxed by the MexCD-OprJ system. It was also found that the MexAB-OprM system is capable of efflux of irgasan, a broad-spectrum antimicrobial compound used in media selective for Pseudomonas.
Clinical isolates of Pseudomonas aeruginosa are characterized by their frequent resistance to antibiotics, including cross-resistance to chemically unrelated compounds. Recent studies showed that the major cause of cross-resistance is the existence of multidrug efflux pumps (19, 21, 24, 26, 30). These pumps can pump out a wide range of compounds, and it is often difficult to discern any common structural features among the substrates. To date, three such efflux systems have been described for P. aeruginosa, MexA-MexB-OprM (hereafter, MexAB-OprM), MexC-MexD-OprJ (hereafter, MexCD-OprJ), and MexE-MexF-OprN, respectively (for reviews, see references 24 and 26). These three systems are members of the RND-type family of multidrug efflux systems in gram-negative bacteria. This family is characterized by a transporter (e.g., MexB), a linker protein (e.g., MexA), and a putative outer membrane channel (e.g., OprM). The MexAB-OprM system appears to mediate efflux of a very wide range of compounds (tetracycline, chloramphenicol, fluoroquinolones, β-lactams [except carbapenems], novobiocin, erythromycin, fusidic acid, rifampin, etc.). The system is overexpressed in many carbenicillin-resistant (Cbr) clinical isolates of P. aeruginosa, and the multidrug-resistant phenotype of many such isolates can now be explained as a consequence of the expression of this system. The additional MexAB-OprM homologs are very similar in structure and function, but there are significant differences in the specificities of the systems. For example, whereas the MexCD-OprJ system efficiently extrudes “fourth-generation” cephems, it does not pump carbenicillin or carbapenems.
Emerging resistance to existing antibiotics has prompted investigations into the hitherto unexplored P. aeruginosa fatty acid biosynthetic (Fab) pathway for potential antimicrobial targets. The hypothesis is that this pathway is an excellent candidate for targeting antimicrobial agents since it plays a pivotal role in providing metabolic precursors for several important cellular functions, including cell wall biogenesis (phospholipids, lipopolysaccharide, and lipoproteins) (4), rhamnolipid synthesis (25), and synthesis of the acylated homoserine lactones required for virulence factor gene expression (23, 27, 28, 32).
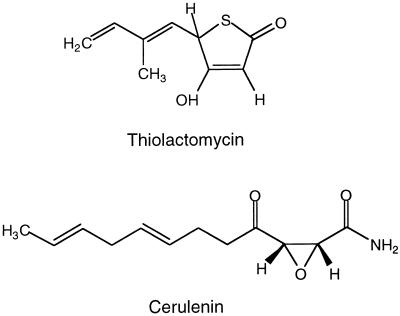
Several of the Fab proteins are the targets of inhibitors of fatty acid synthesis (FAS), including cerulenin (Cer) and thiolactomycin (TLM) (Fig. 1), which specifically target the fatty acid synthases (for a review, see reference 4). Cer is a fungal product that irreversibly inhibits at least two of the three Escherichia coli FASs, as well as yeast and mammalian FASs (5, 40). Unlike Cer, TLM specifically inhibits dissociated or type II bacterial and plant FASs but not multifunctional or type I yeast and mammalian FASs (9, 10, 16, 38). Thus, only TLM is of therapeutic interest.
FIG. 1.
Structures of the fatty acid synthase inhibitors TLM and Cer.
Kawahara et al. (17) reported that P. aeruginosa is intrinsically resistant to Cer. Cer-susceptible mutants that simultaneously became hypersusceptible to several other antibiotics, especially carbenicillin and tetracycline, could be isolated. Growth of the susceptible mutant was severely inhibited by 50 μg of Cer per ml.
TLM exhibits broad antibacterial action (9, 22, 38), with some exceptions (1, 8). Hayashi and coworkers (10) reported that some strains of P. aeruginosa were hypersusceptible to TLM (they were completely inhibited by TLM at ∼0.5 μg/ml) and that this hypersusceptibility coincided with that to several other antibiotics, including carbenicillin and tetracycline. Although these findings indicated the feasibility of using TLM as an anti-Pseudomonas drug, subsequent studies on the antibacterial action of TLM focused on E. coli (10, 16, 39), mainly since until very recently nothing was known about the FAS system in pseudomonads and since the mechanism(s) of TLM resistance remained not understood.
Mutational analyses of the multidrug resistance (MDR) efflux systems are hampered by the fact that mutations in these systems generally lead to drug hypersensitivity (20, 30). This precludes application of conventional mutational strategies that usually include tagging of plasmid-borne genes with antibiotic resistance markers, followed by their return into the chromosome (35, 36). In the case of efflux systems, subsequent analyses of their roles in MDR make it desirable to have mutants devoid of any drug markers. Although the previously described sacB-based technique has proven to be very useful for the introduction of marked and unmarked mutations into the chromosome (13, 36), despite repeated attempts, I was unable to use this technique to return the unmarked K337 Δ(mexAB-oprM) mutation (29) into the PAO1 chromosome. I therefore decided to isolate the desired Δ(mexAB-oprM) mutation by tagging it with a selectable antibiotic marker followed by its in vivo excision (3, 14, 18).
In this paper, I describe the application of a novel gene replacement method for investigations of the role of efflux in resistance to the FAS inhibitors Cer and TLM.
MATERIALS AND METHODS
Bacterial strains and growth media.
The P. aeruginosa strains used in this study are listed in Table 1 and were derived as follows. Among the PAO1 derivatives, PAO196 is PAO1 with Δ(mexAB-oprM)::Gmr-GFP (gentamicin resistant [Gmr] and expressing green fluorescent protein [GFP] from the gentamicin gene promoter) (this study) and PAO200 contains an unmarked Δ(mexAB-oprM) and is derived from PAO196 by Flp-mediated excision of the Gmr-GFP markers (this study). Among the K337 derivatives, K337 Δ(mexAB-oprM) (29) and K337 Δ(mexAB-oprM) nfxB (29) were previously derived from K337 (30). The Δ(mexAB-oprM) deletion in K337 is slightly smaller than the one described in this study and was derived by deletion of an internal 4,103-bp SacII fragment, followed by integration of the resulting unmarked deletion into the K337 chromosome (29). For growth of P. aeruginosa strains, the rich media used were Luria-Bertani (LB) agar (31), RB (rich broth) agar (11), or Pseudomonas isolation agar (PIA; Difco, Detroit, Mich.), and VBMM (Vogel-Bonner minimal medium) (33) was used as the minimal medium. E. coli strains were grown on LB medium. Unless indicated otherwise, antibiotics were used in the selection media at the following concentrations (per milliliter): for E. coli, ampicillin (Sigma, St. Louis, Mo.) was used at 100 μg and gentamicin (ICN, Costa Mesa, Calif.) was used at 10 μg; for P. aeruginosa, carbenicillin (Foothills Hospital Pharmacy, Calgary, Alberta, Canada) was used at 500 μg and gentamicin was used at 200 μg. Cer (Sigma) and TLM (synthesized as described previously [38] in the Mycobacteriology Research Laboratories at Colorado State University) were used at the concentrations specified in Table 2 and the legend to Fig. 3.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Reference or origin |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Prototroph | 15 |
| PAO196 | PAO1 with Δ(mexAB-oprM)::Gmr-GFP | This study |
| PAO200 | PAO1 with unmarked Δ(mexAB-oprM) | This study |
| K337a | Same as ML5087 (ilv-220 thr-9001 leu-9001 met-9011 pur-67 aphA) | 30 |
| E. coli SM10 | Kmr; mobilizer strain (thi-1 thr leu tonA lacY supE recA::RP4-2Tc::Mu) | 6 |
| Plasmidsb | ||
| pAK1900 | Apr; broad-host-range cloning vector | 30 |
| pEX100T | Apr; sacB+oriT+; gene replacement vector | 35 |
| pFLP | Apr; source of yeast Flp recombinase | 14 |
| pUC18 | Apr; general purpose cloning and expression vector | 41 |
| pUCP21T | Apr; mobilizable broad-host-range cloning vector | 36 |
| pRSP01 | Apr; mexA+mexB+oprM+ (8.5-kb chromosomal HindIII fragment cloned into pAK1900) | 29 |
| pRSP14 | Apr; Δ(mexAB-oprM) (pRSP01 with 4.1-kb SacII deletion) | 29 |
| pPS807 | Apr; Δ(mexAB-oprM) (1.8-kb HindIII-KpnI fragment from pRSP14 cloned between the same sites of pUC18) | This study |
| pPS809 | Apr; PCR-amplified 4.5-kb fragment from pPS807 ligated to 1.8-kb blunt-ended Gmr-GFP fragment from pPS858) | This study |
| pPS858 | Apr Gmr; source of Gmr-GFP Gmr-conferring fragment flanked by FRT sites | 14 |
| pPS951 | Apr Gmr; subcloning of a 5.9-kb blunt-ended HindIII-KpnI fragment from pPS809 into the SmaI site of pEX100T | This study |
| pPS952 | Apr; mexA+mexB+oprM+ (8.5-kb chromosomal HindIII fragment from pRSP01 cloned into the same site of pUCPT21T) | This study |
For a detailed description of other K337 derivatives used in this study, see Materials and Methods.
Details on the construction of recombinant plasmids are presented in Materials and Methods.
TABLE 2.
Susceptibilities of P. aeruginosa strains to selected antimicrobial agents
| Strain | MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|
| Cb | Tc | Gm | Cer | TLM | |
| PAO1 | 75 | 7.5 | 1–5 | >100 | >100 |
| PAO200 | 0.75 | <0.5 | 0.5 | 7.5 | 25–50b |
| PAO200/pUCP21T | NDc | <0.5 | ND | 10 | 25 |
| PAO200/pPS952 | ND | 10 | ND | >100 | >100 |
| K337 Δ(mexAB-oprM) | 0.5 | <0.5 | ND | 50 | 25 |
| K337 Δ(mexAB-oprM) nfxBd | 0.5 | 25 | ND | >100 | 25 |
Susceptibilities to antimicrobial agents were tested as described in Materials and Methods. Cb, carbenicillin; Tc, tetracycline; Gm, gentamicin.
Inhibition levels were somewhat dependent with different batches of TLM.
ND, not determined.
This nfxB strain expresses the MexCD-OprJ efflux system.
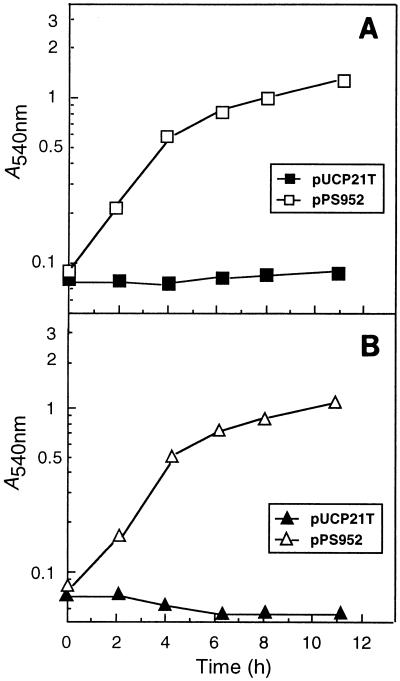
FIG. 3.
Complementation of PAO200 with a cloned mexA+-mexB+-oprM+ operon. Cells of strain PAO200 containing the vector pUCP21T (closed symbols) or pPS952 (open symbols) were grown overnight in RB medium (11) supplemented with 200 μg of carbenicillin per ml and were inoculated into carbenicillin-free RB medium to an initial absorbance at 540 nm of 0.05 to 0.08 (time zero). Cultures were shaken at 37°C. At the indicated times, samples were withdrawn and the absorbance at 540 nm was recorded. Cultures contained either 50 μg of Cer per ml (A) or 50 μg of TLM per ml (B).
Construction of recombinant plasmids.
Restriction enzymes and T4 DNA ligase were used as recommended by the supplier (Gibco-BRL, Gaithersburg, Md.). DNA fragments were blunt-ended with T4 DNA polymerase in the presence of 100 μM deoxynucleoside triphosphates (31). Small-scale isolations of plasmid DNA from E. coli and DNA transformations were done as described previously (37). Restriction fragments were eluted from agarose gels by using the Geneclean procedure (Bio 101, Inc., San Diego, Calif.). Plasmid pPS951 was derived in several steps. First, a 1.8-kb HindIII-KpnI fragment from pRSP14 (29) containing the N-terminal 172 mexA codons and codons 104 to 395 of oprM (GenBank accession no. L11616) was subcloned between the same sites of pUC18 (41) to form pPS807. Next, two oligonucleotides were designed to introduce unique EcoRV sites at positions corresponding to codon 165 of mexA and codon 128 of oprM. These primers were used in a reverse PCR to prime synthesis from pPS807 DNA in a 50-μl reaction mixture containing 1× Taq+ buffer (Stratagene, La Jolla, Calif.), 200 μM (each) deoxynucleoside triphosphate, 10 pmol of each primer, ∼10 pmol of pPS807 DNA, and 5 U of Taq+ (Stratagene). The reaction mixtures were subjected to the following cycles: 1 cycle at 96°C for 5 min; 35 cycles of 95°C for 1 min, 57°C for 20 s, and 72°C for 4.5 min; and a final extension at 72°C for 5 min. The 4.5-kb PCR product was eluted from an agarose gel. After digestion with EcoRV and gel purification, the resulting fragment was ligated to an 1,812-bp GFP-containing and Gmr-conferring SacI fragment from pPS858 (14) to yield plasmid pPS809 [Δ(mexAB-oprM)::Gmr-GFP]. The mutated region was then subcloned as a blunt-ended HindIII-KpnI fragment into the SmaI site of the gene replacement vector pEX100T (35) to form pPS951. Plasmid pPS952 was constructed by ligating a blunt-ended mexA+-mexB+-oprM+ HindIII fragment from pRSP14 (29, 30) into the blunt-ended PstI site of pUCP21T (36), resulting in transcription of the mexA+-mexB+-oprM+ operon from its own promoter.
Gene replacement.
For gene replacement, the previously described sacB-based strategy (35) was used, selecting Gmr colonies on VBMM-gentamicin medium after conjugal transfer of pPS951 from E. coli SM10 (6). Sucrose-resistant colonies were obtained on LB medium containing 5% sucrose and 15 μg of gentamicin per ml. Deletion of the chromosomally integrated Gmr-GFP markers by Flp recombinase-catalyzed excision was achieved by conjugally transferring Flp-expressing, nonreplicative pFLP (14) from E. coli SM10 into the Gmr-GFP strain and plating dilutions of the recipient cells at 42°C on VBMM plates. The cells growing on these plates were then tested for the loss of the Gmr marker.
Genomic Southern analyses.
Chromosomal DNA was isolated by a miniprep procedure (2). Nylon membranes containing electrophoretically separated genomic DNA fragments were probed with biotinylated DNA by previously described procedures (12). The gentamicin probe was derived by labelling a 850-bp gel-purified fragment from pUCGM (34), and the mex probe was obtained by labelling the 1.8-kb insert of pPS807 (this study) containing the N-terminal mexA-coding sequence and sequences internal to oprM.
Antibiotic susceptibility studies.
Susceptibilities to antimicrobial agents were tested as described previously (30), with minor modifications. One-milliliter cultures of LB medium containing 100, 75, 50, 25, 10, 5, 2.5, 1, and 0 μg of each antimicrobial agent were inoculated with 5 × 106 logarithmically (absorbance at 540 nm, ∼0.8 to 1.0) growing organisms. Growth was assessed visually after 18 h of incubation at 37°C. The MIC was defined as the lowest concentration of antimicrobial agent that inhibited visible growth. Plasmid-containing strains were pregrown in LB medium containing 200 μg of carbenicillin per ml, and susceptibilities to antimicrobial agents were then tested in the absence of carbenicillin.
RESULTS AND DISCUSSION
Construction of an unmarked Δ(mexAB-oprM) mutant.
A defined pPS951-borne Δ(mexAB-oprM) mutation was constructed as described in Materials and Methods, and the deletion was returned to the P. aeruginosa chromosome as illustrated in Fig. 2A. After conjugal transfer of the nonreplicative pPS951 from E. coli SM10 into PAO1, merodiploids were obtained by selecting for Gmr. From these, colonies having undergone the deletion marked with Δ1 in Fig. 2A were selected as sucrose resistant, Gmr, and Cbs. The unmarked Δ(mexAB-oprM) mutant PAO200 was then derived from the Gmr-GFP integrant PAO196 by Flp-catalyzed excision of the Gmr-GFP markers. During its transient expression in the recipient, Flp recombinase acted at the Flp recombination target (FRT) sites to catalyze excision of the Gmr-GFP element (marked with Δ2 in Fig. 2A) at low but detectable frequencies (0.1 to 0.5%), leaving behind a short FRT-containing sequence (3, 14). It should be noted that although the GFP marker has proven to be useful for monitoring the loss of the entire Gmr-GFP cassette in E. coli, it did not prove to be useful during these particular experiments due to the intense fluorescence of P. aeruginosa cells grown on VBMM.
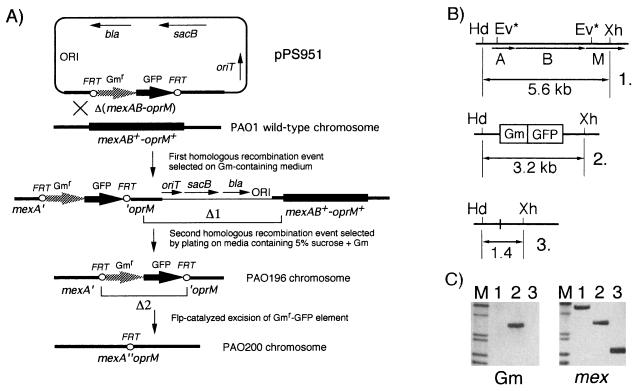
FIG. 2.
Strategy for isolation of an unmarked Δ(mexAB-oprM) mutation. (A) For gene replacement, the previously described sacB-based strategy (35) was used, as detailed in Materials and Methods. Colonies having undergone the deletion marked Δ1 were screened as sucrose resistant, Gmr, and Cbs. The unmarked Δ(mexAB-oprM) mutant was then derived from the Gmr-GFP integrant by Flp-catalyzed excision (Δ2) of the Gmr-GFP markers. Abbreviations: bla, β-lactamase structural gene; GFP, green fluorescent protein structural gene; ORI, ColE1-derived origin of replication; oriT, origin of transfer; sacB, levansucrase structural gene. (B) Genomic organization of the PAO1 mexA+-mexB+-oprM+ region (B-1) and of Δ(mexAB-oprM) mutants PAO196 (B-2) and PAO200 (B-3). Ev* mark the positions of the artificially generated EcoRV sites used for deletion of the intervening 4.2 kb of DNA and replacement with a 1.8-kb Gmr-GFP fragment. The relative positions of HindIII (Hd) and XhoI (Xh) sites, as well as the lengths of HindIII-XhoI fragments expected after digestion of the respective chromosomal DNAs, are shown. (C) Genomic Southern analysis. Nylon membranes containing electrophoretically separated genomic DNA fragments from the isolates depicted in panel B were probed either with a biotinylated DNA fragment from pPS807 (panel labeled mex) or with a Gmr fragment (panel labeled Gm), as described in Materials and Methods. The DNAs in lanes 1, 2, and 3 correspond to the HindIII-XhoI fragments from the strains 1, 2, and 3, respectively, described for panel B. Lane M contained (top to bottom) biotinylated λ HindIII fragments (6.3, 4.3, 2.4, and 2.04 kb) and biotinylated φX HaeIII fragments (1.35 and 1.08 kb).
Successful execution of the steps labelled Δ1 and Δ2 in Fig. 2A was monitored by colony PCR analysis with primers specific for the gentamicin resistance gene (data not shown) and by genomic Southern analysis (Fig. 2B and C). From the results presented in Fig. 2C it is evident that both deletion events produced the desired restriction patterns. Probing with a mexA- and oprM-specific probe (panel labeled mex in Fig. 2C) revealed deletion of a 4.2-kb region from the PAO1 wild-type chromosome in both the Δ(mexAB-oprM)::Gmr-GFP insertion mutant PAO196 (lane 2) and the Δ(mexAB-oprM) mutant PAO200 (lane 3). The size of the 5.6-kb HindIII-XhoI fragment observed in wild-type PAO1 (lane 1) was reduced to 3.2 kb (5.6 kb minus 4.2 kb of genomic DNA plus 1.8 kb of the Gmr-GFP fragment) (lane 2) in the insertion mutant PAO196 and to 1.4 kb (lane 3) in the unmarked deletion mutant PAO200. Probing with a probe specific for the gentamicin resistance gene (panel labeled Gm in Fig. 2C) revealed the presence of the 1.8-kb Gmr-FRT cassette only in the Δ(mexAB-oprM)::Gmr-GFP insertion mutant PAO196 on a 3.2-kb HindIII-XhoI fragment (lane 2). As expected, the sequences encoding gentamicin resistance were absent from wild-type PAO1 genomic DNA (lane 1), and they were deleted from the excision mutant PAO200 (lane 3).
The results suggest that the experimental strategy described herein will facilitate studies aimed at elucidation of the modes of action of the efflux systems of P. aeruginosa and other pathogenic bacteria. More generally, unmarked efflux pump mutants will enable the search and design of new antimicrobial agents that are no longer substrates of the efflux systems, while maintaining their inhibitory effects.
Susceptibility of efflux pump mutants to FAS inhibitors.
Susceptibility studies revealed that PAO200 was hypersusceptible to all of the antibiotics tested except gentamicin; the pattern of susceptibility to gentamicin was marginally altered (Table 2). Transformation with the mexA+-mexB+-oprM+ plasmid pPS952 restored resistance to tetracycline, Cer, and TLM to the levels found in PAO1 (Table 2 and Fig. 3). These experiments demonstrated that the MexAB-OprM efflux system was indeed responsible for the previously observed intrinsic resistance of some P. aeruginosa strains to Cer and TLM, as well as the concomitant cross-resistance to other antibiotics.
Preliminary experiments with K337 Δ(mexAB-oprM) nfxB, i.e., a strain expressing the MexCD-OprJ pump, indicated that whereas Cer is a substrate of the MexCD-OprJ pump, TLM is not effluxed by this system (Table 2). As observed with PAO200 (Fig. 3B), growth of K337 Δ(mexAB-oprM) nfxB in RB medium was completely inhibited by 50 μg of TLM per ml. In contrast to PAO200, which showed no visible growth in medium containing 50 μg of Cer per ml (Fig. 3A), K337 Δ(mexAB-oprM) was intrinsically more resistant to this antimicrobial agent (levels of growth inhibition were 53 and 80% with 50 and 100 μg of Cer per ml, respectively). Since strain K337 Δ(mexAB-oprM) nfxB was resistant to >100 μg Cer per ml, it can be concluded that this antimicrobial agent is efficiently extruded by the MexCD-OprJ pump. Clearly, since the PAO1 and K337 strain backgrounds display different levels of intrinsic resistance, the experiments described above will have to be repeated with a PAO200 nfxB strain.
Although the results indicate a major role of efflux systems in resistance to FAS inhibitors, additional resistance mechanisms, i.e., target alterations, probably exist, and their contributions to resistance, if any, will need to be further evaluated. In E. coli, the two known mechanisms contributing to TLMr are efflux via the major facilitator-type EmrAB system (7) and FAS I (FabB) target overproduction (39).
MexAB-OprM-mediated efflux is required for growth of wild-type P. aeruginosa on Pseudomonas isolation agar.
In the course of the studies on FAS inhibitors it was discovered that strains PAO196 and PAO200 were no longer able to grow on PIA. Growth on medium with the same formulation except that it contained no irgasan indicated that this was due to the susceptibility of the mutants to the broad-spectrum antimicrobial irgasan (also known as triclosan) present in PIA, which in wild-type P. aeruginosa is apparently pumped out of the cell via the MexAB-OprM efflux system. Transformation with the mexA+-mexB+-oprM+ plasmid pPS952 restored the ability of PAO200 to grow on PIA. By streaking the same strain on PIA, irgasan-resistant mutants could be isolated at high frequencies. Since the MIC of tetracycline (∼0.5 μg/ml) for these mutants was still low, none of the other known efflux systems seems to be responsible for the irgasan resistance in these mutant strains. In contrast to PAO200, strain K337 Δ(mexAB-oprM) grew normally on PIA, again indicating a hitherto unknown mechanism besides efflux contributing to irgasan resistance. In light of these observations, the usefulness of this medium for the differentiation of Pseudomonas strains may be limited to MexAB-OprM efflux system-expressing organisms, while MexAB-OprM efflux pump-negative or MexAB-OprM-nonexpressing mutants may be missed when this medium is used for screening. As with the FAS inhibitors, the role of other P. aeruginosa efflux pumps in irgasan resistance, if any, has yet to be elucidated. In light of the present findings, the general use of PIA medium for differentiation purposes should therefore be reevaluated. In contrast, wild-type and efflux pump mutants grew equally well on agar base selective for Pseudomonas (also called cetrimide agar) (Difco), whose active ingredient, cetyltrimethylammonium bromide, did not seem to be a substrate for the MexAB-OprM pump. These results would indicate that cetrimide agar is the more reliable choice for use as a selective medium for Pseudomonas.
ACKNOWLEDGMENTS
I am indebted to K. Poole for the gift of bacterial strains and plasmids and to G. S. Besra from the Mycobacteriology Research Laboratories at Colorado State University for the synthesis and generous gift of TLM.
Financial support was provided by start-up funds from the Department of Microbiology at Colorado State University and by a grant from the CSU College of Veterinary Medicine and Biomedical Sciences.
REFERENCES
- 1.Arimura N, Kaneda T. Type selective inhibition of microbial fatty acid synthases by thiolactomycin. Arch Microbiol. 1993;160:158–161. doi: 10.1007/BF00288719. [DOI] [PubMed] [Google Scholar]
- 2.Barcack G J, Chandler M S, Redfield R J, Tomb J-F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 3.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 4.Cronan J E, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 5.D’Agnolo G, Rosenfeld I S, Awaya J, Omura S, Vagelos P R. Inhibition of fatty acid biosynthesis by the antibiotic cerulenin. Biochim Biophys Acta. 1973;326:155–166. doi: 10.1016/0005-2760(73)90241-5. [DOI] [PubMed] [Google Scholar]
- 6.De Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5 and Tn10-derived transposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 7.Furukawa H, Tsay J-T, Jackowski S, Takamura Y, Rock C O. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol. 1993;175:3723–3729. doi: 10.1128/jb.175.12.3723-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamada S, Fujiwara T, Shimauchi H, Ogawa T, Nishihara T, Koga T, Nehashi T, Matsuno T. Antimicrobial activities of thiolactomycin against gram-negative anaerobes associated with periodontal disease. Oral Microbiol Immunol. 1990;5:340–345. doi: 10.1111/j.1399-302x.1990.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi T, Yamamoto O, Sasaki H, Kawaguchi A, Okazaki A. Mechanism of action of the antibiotic thiolactomycin: inhibition of fatty acid synthesis of Escherichia coli. Biochem Biophys Res Commun. 1983;115:1108–1113. doi: 10.1016/s0006-291x(83)80050-3. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi T, Yamamoto O, Sasaki H, Okazaki H. Inhibition of fatty acid synthesis by the antibiotic thiolactomycin. J Antibiot. 1984;37:1456–1461. doi: 10.7164/antibiotics.37.1456. [DOI] [PubMed] [Google Scholar]
- 11.Henry M F, Cronan J E. A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell. 1992;70:671–679. doi: 10.1016/0092-8674(92)90435-f. [DOI] [PubMed] [Google Scholar]
- 12.Hoang T, Williams S, Schweizer H P. Molecular genetic analysis of the region containing the essential Pseudomonas aeruginosa asd gene encoding aspartate-β-semialdehyde dehydrogenase. Microbiology. 1997;143:899–907. doi: 10.1099/00221287-143-3-899. [DOI] [PubMed] [Google Scholar]
- 13.Hoang T T, Schweizer H P. Fatty acid biosynthesis in Pseudomonas aeruginosa: cloning and characterization of the fabAB operon encoding β-hydroxyacyl-acyl carrier protein dehydratase (fabA) and β-ketoacyl-acyl carrier protein synthase I (fabB) J Bacteriol. 1997;179:5326–5332. doi: 10.1128/jb.179.17.5326-5332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang, T. T., and H. P. Schweizer. Unpublished data.
- 15.Holloway B W, Zhang C. Genetic maps. In: O’Brien S J, editor. Locus maps of complex genomes. 5th ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 2.71–2.78. [Google Scholar]
- 16.Jackowski S, Murphy C M, Cronan J E, Rock C O. Acetoacetyl-acyl carrier protein synthase. A target for the antibiotic thiolactomycin. J Biol Chem. 1989;264:7624–7629. [PubMed] [Google Scholar]
- 17.Kawahara K, Uchida K, Aida K. Isolation and partial characterization of a cerulenin-sensitive mutant of Pseudomonas aeruginosa. J Antibiot. 1983;36:1329–1335. doi: 10.7164/antibiotics.36.1329. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, De Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X Z, Ma D, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta-lactam resistance. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 22.Miyakawa S, Suzuki K, Noto T, Harada Y, Okazaki H. Thiolactomycin, a new antibiotic. IV. Biological properties and chemotherapeutic activity in mice. J Antibiot. 1982;35:411–419. doi: 10.7164/antibiotics.35.411. [DOI] [PubMed] [Google Scholar]
- 23.Moré M, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsner U A. Genetics and biochemistry of Pseudomonas aeruginosa rhamnolipid biosurfactant synthesis. Ph.D. dissertation. Zurich, Switzerland: Swiss Federal Institute of Technology; 1993. [Google Scholar]
- 26.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson J, Gray K, Passador L, Tucker K, Eberhard A, Iglewski B, Greenberg E. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson J, Passador L, Iglewski B, Greenberg E. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole, K. 1996. Personal communication.
- 30.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Greenberg E P. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer H P. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweizer H P. Small broad-host-range gentamycin resistance cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 35.Schweizer H P, Hoang T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 36.Schweizer H P, Klassen T R, Hoang T. Improved methods for gene analysis and expression in Pseudomonas. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: American Society for Microbiology; 1996. pp. 229–237. [Google Scholar]
- 37.Schweizer H P, Po C. Cloning and nucleotide sequence of the glpD gene encoding sn-glycerol-3-phosphate dehydrogenase from Pseudomonas aeruginosa. J Bacteriol. 1994;176:2184–2193. doi: 10.1128/jb.176.8.2184-2193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slayden R A, Lee R E, Armour J W, Cooper A M, Brennan P J, Besra G S. Antimycobacterial action of thiolactomycin: an inhibitor of fatty acid and mycolic acid synthesis. Antimicrob Agents Chemother. 1996;40:2813–2819. doi: 10.1128/aac.40.12.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsay J T, Rock C O, Jackowski S. Overproduction of beta-ketoacyl-acyl carrier protein synthase I imparts thiolactomycin resistance to Escherichia coli K-12. J Bacteriol. 1992;174:508–513. doi: 10.1128/jb.174.2.508-513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vance D E, Goldberg I, Mitsuhashi O, Bloch K, Omura S. Inhibition of fatty acid synthetases by the antibiotic cerulenin. Biochem Biophys Res Commun. 1972;48:649–656. doi: 10.1016/0006-291x(72)90397-x. [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]