Abstract
Sponges are among the earliest branching extant animals. As such, genetic data from this group are valuable for understanding the evolution of various traits and processes in other animals. However, like many marine organisms, they are notoriously difficult to sequence, and hence, genomic data are scarce. Here, we present the draft genome assembly for the North Atlantic deep-sea high microbial abundance species Geodia barretti Bowerbank 1858, from a single individual collected on the West Coast of Sweden. The nuclear genome assembly has 4,535 scaffolds, an N50 of 48,447 bp and a total length of 144 Mb; the mitochondrial genome is 17,996 bp long. BUSCO completeness was 71.5%. The genome was annotated using a combination of ab initio and evidence-based methods finding 31,884 protein-coding genes.
Keywords: Geodia barretti, Porifera, Tetractinellida, Sweden, symbionts, metagenome-assembled genome
Introduction
Sponges (phylum Porifera) hold an evolutionarily important position as one of the earliest animal lineages (Redmond and McLysaght 2021; Schultz et al. 2023). Sponges do not produce organs but feature instances of true epithelial tissue, a hallmark of metazoans (Leys and Riesgo 2012). Their body features cells of varying complexity and fate (Musser et al. 2021), and only recently, major breakthroughs in sponge cell cultures were made (Conkling et al. 2019). Among the focal aspects of research on sponges are their remarkable microbiota and chemical diversities (Thomas et al. 2016; Calado et al. 2022).
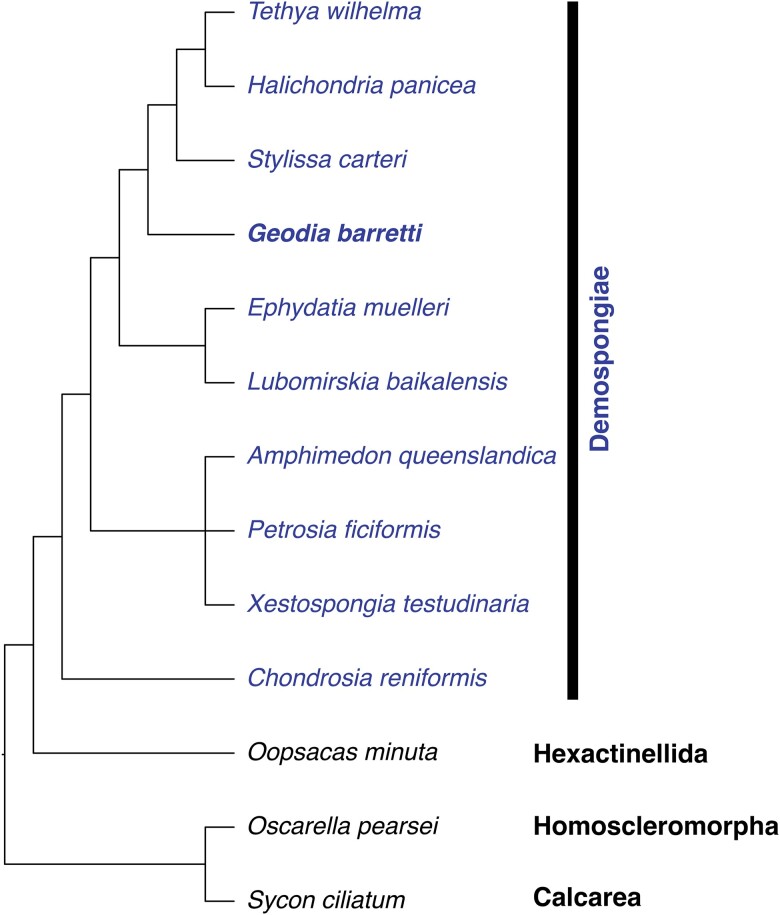
In the past, these 2 aspects complicated access to their genomes, as DNA extracted from sponges is contaminated by microbial DNA and by compounds binding DNA (Marshall and Barrows 2004) and potentially interfering with sequencing. Workarounds like producing a genome from DNA of thousands of larvae which naturally have a lower abundance of microbial symbionts (Srivastava et al. 2010) or whole-genome amplification (WGA) from single cells (Ryu et al. 2016) both yielded highly fragmented assemblies. Recent sequencing strategies (long reads, synthetic long reads, and/or Hi-C) yield chromosome-level assemblies, as in the case of Ephydatia muelleri (Kenny et al. 2020) as well as Petrosia ficiformis and Chondrosia reniformis (McKenna et al. 2021). However, 13 years after the first sponge genome, there were only 12 sponge genomes available, of over 9,500 described species of sponges (Fig. 1; Table 1) (de Voogd et al. 2023).
Fig. 1.
Cladogram of all sponge species with genomes published to date, drawn after (Morrow and Cárdenas 2015; Plese et al. 2021).
Table 1.
Sponge genomes and their respective repositories.
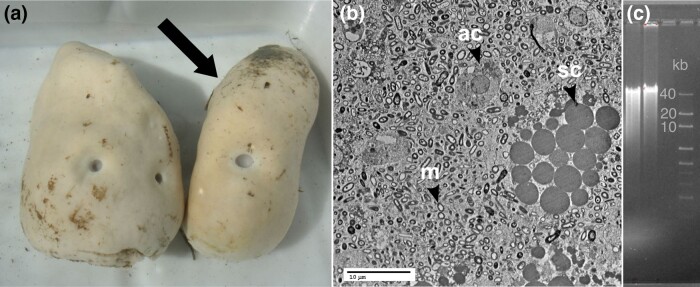
Geodia barretti Bowerbank 1858 (Fig. 2a) is a widespread North Atlantic deep-sea demosponge found in depths of 30–2,000 m (Cárdenas et al. 2013) and, thus, would represent 1 of the few genomes of a deep-sea animal. As a high microbial abundance (HMA) sponge, G. barretti hosts an outstanding density and diversity of microbes (Fig. 2b) with an average of 3 × 1010 microbes/cm3 (Hoffmann et al. 2006; Leys et al. 2018) from over 400 prokaryotic amplicon sequence variants across 17 phyla (Radax et al. 2012; Steffen et al. 2022). The characterization as “HMA” (highly abundant and diverse microbiota) and “LMA” (lowly abundant and species-poor microbiota) sponges has been recognized since the 1970s and is a species specific attribute, but its significance to the organisms is not yet fully elucidated (Vacelet 1975; Hentschel et al. 2003). This microbiota partly accounts for its richness in natural products (Erngren et al. 2021; Steffen et al. 2021) with still many unknown bioactive metabolites. Finally, G. barretti is a key species of sponge grounds, deep-sea habitats characterized by mass accumulation of sponges (Klitgaard and Tendal 2004; Cárdenas et al. 2013). Sponge grounds are considered vulnerable marine ecosystems (VMEs) and as such G. barretti is part of the VME indicator species list (ICES 2019). Its physiology has been extensively studied to understand the many ecosystem services it provides (Cárdenas and Rapp 2013; Koutsouveli, Cárdenas, Santodomingo, et al. 2020; Maier et al. 2020; Rooks et al. 2020; Bart et al. 2021) as well as to investigate its resilience to human activities (Kutti et al. 2015; Colaço et al. 2022) including climate change (Strand et al. 2017). Therefore, producing a genomic resource for this species is not only valuable for conservation efforts but also for a more general understanding of deep-sea benthic ecosystems.
Fig. 2.
a) G. barretti specimens freshly collected on 4 May 2016, Kosterfjord, Sweden: left (UPSZMC 184976) and right (UPSZMC 184975) with arrow, the specimen chosen for whole-genome sequencing. b) TEM image of G. barretti (UPSZMC 184975) showing 2 types of sponge archeocyte cells archaeocytes (ac) and spherulous cells (sc). All other smaller bodies are microbial symbionts (m). Image courtesy of V. Koutsouveli and A. Riesgo, NHM London. c) DGGE image of DNA prior to PacBio library production. Image courtesy of SciLifeLab Uppsala.
Methods and materials
Sampling
The specimen of G. barretti (Tetractinellida, Astrophorina, Geodiidae) (Fig. 2a) was collected on 4 May 2016 with a basket fixed to an ROV on board the R/V Nereus in the Kosterfjord National Park, Sweden, West of Yttre Vattenholmen (58.876233, 11.101483) at 96-m depth. The sample was identified on board by P. Cárdenas. Small-tissue sections were immediately flash-frozen in liquid nitrogen, while larger pieces were frozen at −20°C, and the rest was kept as a voucher and fixed in ethanol 96% (ethanol changed twice). The voucher is stored in 96% ethanol at the Museum of Evolution, Uppsala, Sweden, under museum number UPSZMC 184975. G. barretti is a gonochoric species, but the sex of the specimen could not be determined since it was not reproducing at the time of its collection and did not contain any observable larvae as it is oviparous (= “specimen NR_1” in Koutsouveli, Cárdenas, Conjeco, et al. 2020). During this same reproduction study, transmission electron microscope (TEM) pictures were made from UPSZMC 184975, confirming the high abundance of microbes (Fig. 2b).
DNA extraction and sequencing
DNA extraction for whole-genome sequencing was impeded by rapidly degrading DNA and chemical contamination coisolated with the DNA. We obtained the best results by macerating flash-frozen tissue in 0.2 M EDTA pH 8.0 and straining the dissociated cells through a filter (40 µm, Nalgene) before extracting DNA using a traditional chloroform/isoamyl alcohol partitioning protocol (Dharamshi et al. 2022). The resulting DNA was assayed by NanoDrop (passing range: 260/280, 1.8–2.2; 260/230, 2.0–2.2) and denaturing gradient gel electrophoresis (Fig. 2c), and suitable fractions were sequenced on PacBio (RSII and Sequel) and Illumina (HiSeqX) platforms, all performed by the SNP&SEQ Technology Platform, SciLifeLab Uppsala, Sweden.
Data processing
RNAseq data preparation
Poly-A selected RNA-sequencing (RNAseq) data of UPSZMC 184975 and 6 other G. barretti individuals were used for identification of sponge contigs and gene annotation (Koutsouveli, Cárdenas, Santodomingo, et al. 2020). For identification of sponge contigs in the subsequent whole-genome assembly, the RNAseq data were de novo assembled using Trinity (Haas et al. 2013) with default parameters. For annotation, the same RNAseq data were reassembled with fastp (Chen et al. 2018), hisat2 (Kim et al. 2015), and StringTie (Pertea et al. 2015) with the genome assembly in order to avoid a high number of small unsupported genes and erroneous transcripts.
Assembly
PacBio data were assembled with Flye using the “-meta” flag (Kolmogorov et al. 2020) and polished with the Illumina short reads using one round of Pilon (Walker et al. 2014).
To remove contamination (i.e. nonsponge contigs/scaffolds) and only keep sponge contigs, 2 different strategies were combined: taxonomic identification of contigs and contig coverages with RNAseq data. First, taxonomic classification of the contigs was performed by both contigtax (https://github.com/NBISweden/contigtax, v0.5.9) and BlobTools (Laetsch and Blaxter 2017). In order to run the latter, the short reads were aligned to the assembly with BWA (Li and Durbin 2009); and using the assembled contigs, we performed both a blastn search (Altschul et al. 1990) against the nt database and a DIAMOND blastx search (Buchfink et al. 2015) against the nr database. The obtained BAM file and BLAST output files were used as input files for BlobTools. We kept all contigs annotated as “Eukaryota” by both contigax and BlobTools as a first noncontaminated set for the sponge assembly. Second, 3 of the de novo built transcriptomes were mapped on the full assembly using gmap (Wu et al. 2016). The contigs that on average for the 3 transcriptomes had at least 20% of their length mapped by transcripts were also added to the noncontaminated set of contigs. Coverage was calculated by mapping short and long reads to the genome with BWA and minimap2 (Li 2018), respectively. The coverage per position was extracted from the resulting BAM file using samtools depth (Li et al. 2009) and the average and median across all positions was calculated.
Annotation
All parts of the annotation workflow (annotation preprocessing, transcript assembly, ab initio training, and functional annotation) were performed with 4 Nextflow pipelines from https://github.com/NBISweden/pipelines-nextflow. For annotation preprocessing, all nucleotides were changed to uppercase to prevent interpretation as repeats. The Ns at start or end of contigs are trimmed out to avoid problems when submitting data to public archives. Repeats were masked using RepeatModeler package. Candidate repeats modeled by RepeatModeler were vetted against our protein set (minus transposons) to exclude any nucleotide motif stemming from low-complexity coding sequences. From the repeat library, identification of repeat sequences present in the genome was performed using RepeatMasker (Smit et al. 2013) and RepeatRunner (Smith et al. 2007).
The genome was annotated using the MAKER package (Holt and Yandell 2011), and both evidence-based (using the transcriptomes and gene sets) and ab initio approaches (optimize_augustus.pl) were performed in several (3) iterations until the number of false positive predictions clearly decreased. The annotation quality is given by the annotation edit distance (AED) provided in the gff file. The functional annotation was performed with an in-house pipeline (https://github.com/NBISweden/pipelines-nextflow/tree/master/subworkflows/functional_annotation) based on BLAST and InterProScan. tRNAs were annotated by tRNAscan, and only those with an AED < 1 are reported. The mtDNA was recovered in a single contig and annotated by lifting over annotations from the mtDNA of Geodia neptuni (AY320032) with Geneious v 8.1.9 at a similarity threshold of 75%; ORFs were annotated using genetic code 4. The annotations were adapted to EMBL format using a gff conversion tool (Norling et al. 2018).
Comparative genomics
For placing this sponge genome in its context, all other available sponge genome assemblies known to us were downloaded from NCBI GenBank or their respective repositories (Table 1). After initial analysis, 6 assemblies were excluded. Six genomes of Aplysina aerophoba under BioProject PRJEB24804 appear uncharacteristically small (largest assembly is 3 Mb) and mainly consist of microbial symbiont sequences (D. Sipkema, pers. comm.). In addition, the alternate pseudohaplotype of P. ficiformis (GCA_947044245.1) was excluded as it is only 12% complete according to BUSCO, as was C. reniformis (GCA_947172445.1). For Amphimedon queenslandica, there are currently 3 genome assemblies available: the original first sponge genome “v1.0” (GCA_000090795.1) (Srivastava et al. 2010), superseded by a second version “v1.1” (GCA_000090795.2), followed by a third assembly from the same research group “UQ_AmQuee_3” (GCA_016292275.1) but from a different/unrelated specimen and sequencing project. Although there is no publication for this third genome assembly, it was included here since it seemed highly complete.
Genome completeness was assessed with BUSCO and metazoa_odb10 (Simão et al. 2015). For identification of biosynthetic gene clusters (BGCs), genomes were vetted by antiSMASH (bacterial version) using prodigal-m as gene finder with default parameters (Blin et al. 2021). All figures were created in R (R Core Team 2016) using the packages within tidyverse.
Reproducibility
GitHub repositories and versions
AGAT repository: agat 0.6.2, commit 338be8; GAAS repository: gaas 1.2.0, commit 9af467.
Nextflow pipeline repository: commit 612364.
Tool versions
Flye (2.4.2), Pilon (1.22), BUSCO (5.2.2/5.3.1), gmap (2018-02-12), Trinity (2.11.0), antiSMASH (5.2.1), BWA (0.7.8, 0.7.17), contigtax (v 0.5.9) with UniRef90 database (v2019_11), BlobTools (v1.1.1), minimap2 (2.4), samtools (1.9), fastp (0.20.0), hisat2 (2.1.0), StringTie (2.0), RepeatModeler package (1.0.11), RepeatMasker (4.0.9_p2), RepeatRunner, MAKER package (3.01.02), exonerate (2.4.0), BLAST (2.9.0), Bioperl (1.7.2), Augustus (3.3.3), TRNAscan-se (1.3.1), Snap (version 2013_11_29), GeneMark-ET (4.3), GeneMark (ES Suite version 4.48_3.60_lic), InterProScan (5.30–69.0), Infernal (1.1.2), Prokka (1.11), and R (v4.2.1) using tidyverse (1.3.1) packages for visualization.
Databases versions
UniProt Swiss-Prot database (downloaded on 2020-12; 563 972 proteins), Rfam version 14.4.
Results and discussion
The G. barretti genome assembly
We generated 2,364,732 (2.4 M) reads with long-read technologies (PacBio RSII, Sequel) for an average coverage of 18.82632 (19×, median 14×) and 427,393,248 (427.4 M) reads with short-read technology (Illumina HiSeqX) for an average coverage of 339.9417 (340×, median 256). Sponge DNA degraded in PacBio library production and hence long-read data output was low. The reason for this breakdown of sponge DNA is currently unknown.
Assessing the initial metagenomic assembly from BlobTools results, at the super kingdom level, the reads mapping the metagenome assembly were classified as Eukaryota (37.2%) and bacteria (37.2%). 20.5% of the mapped reads were annotated as “no-hit.” At the phylum level, the mapped reads were mostly classified as Porifera (28.3%), “no-hit” (20.5%), “other” (16.3%), Proteobacteria (15.3%), and Chordata (4.3%) (Supplementary Figs. 1 and 2); 6.9% of the mapped reads were annotated as Candidatus Poribacteria, one of the most abundant microorganisms in demosponges (Lafi et al. 2009) including in G. barretti (Radax et al. 2012; Steffen et al. 2022), although its abundance is often greatly underestimated due to 16S rRNA primer biases (Steinert et al. 2017; Steffen et al. 2022). In total, the mapped reads covered 66 phyla, showing the complexity of microbial communities residing within marine sponges. Similar results are observed with contigtax. At the super kingdom level, more than half of the contigs were annotated as “unclassified,” a third as bacteria, and only 12% as Eukaryota. Among the contigs annotated as bacteria, the majority (30%) belong to the phylum Candidatus Poribacteria. Other major categories are Proteobacteria (19%) and Chloroflexi (14%), 2 phyla that have been identified as frequent and abundant symbionts in sponge microbiota (Thomas et al. 2016) including in G. barretti (Radax et al. 2012; Steinert et al. 2017; Steffen et al. 2022). Among the eukaryotic contigs, 72% are annotated as Porifera. For comparison, Supplementary Figs. 3 and 4 contain the final assembly evaluated with BlobTools showing a decrease in contribution of foreign sequences to the genome assembly.
The genome assembly has a length of 144,789,364 bp (144.7 Mb) across 4,535 contigs. There are 110 Ns in the genome, and it has a GC content of 49.3%. According to BUSCO (v. 5.3.1 metazoa_odb10), the genome is 71.5% complete with 3.6% duplicates of single-copy orthologs. N50 length is 48,446 (contig size ranging from minimum 1,002 bp and median 22,605 bp to maximum 495,233 bp). For comparison, the haploid genome size was estimated to be 127 Mb based on a C-value of 0.13 pg measured by Feulgen image analysis densitometry (FIAD) (Ryan Gregory and Darren Kelly, pers. comm.). The excess sequences could be due to noncollapsed heterozygous regions in the sponge genome and/or incorporation of microbial symbiont sequences in the genome. As part of the genome assembly, we recovered the mitochondrial genome in 1 chromosome. The mtDNA was circular and had a length of 17,996 bp and the characteristic synteny of tetractinellid mtDNA (Plese et al. 2021): rnl-cox2-atp8-atp6-cox3-cob-atp9-nad4-nad6-nad3-nad4L-cox1-nad1-nad2-nad5-rrns. Beyond the sponge genome, the sequencing data are in fact metagenomic data, and we invite the use of it for exploration of the microbial and viral communities of G. barretti UPSZMC 184975, which was beyond the scope of our work.
In the assembly, RepeatMasker annotated 117,982 repeats with a total size of 26,945.75 kb or 18.61% of the genome (mean 228.47 bp). RepeatRunner annotated 1,043 repeats with a total size of 631.47 kb or 0.44% of the genome (mean 605.43 bp). The difference in results is to be expected as the 2 programs are complementary. RepeatMasker identifies repeats based on similarity to known repeats, whereas RepeatRunner identifies highly divergent repeats.
The genome annotation contains 31,884 protein-coding genes. The BUSCO scores together with the number of annotated protein-coding genes suggest that protein-coding genes are well represented in the assembled sequence. There were 66,936 mRNAs (as there were several isoforms per gene) with an average of 7.2 exons per mRNA, an average exon length of 244 bp, and an average coding sequence length of 1,122 bp. Of those genes, 27,544 were functionally annotated, as were 59,664 of the mRNAs. The genome contained 156 tRNAs with an AED < 1.
Comparison with currently available sponge genomes
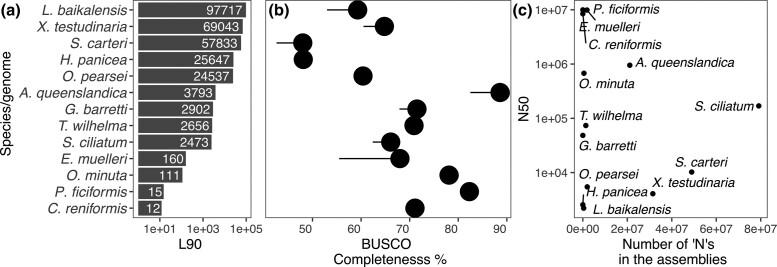
It is worthwhile noting that about half of the 12 previously published sponge genomes are not deposited in widely used databases such as NCBI GenBank or ENA but in other data repositories. Therefore, all currently valid download links are summarized in Table 1. To place our genome in its context, we summarized technical assembly metrics in Table 2 and biological metrics in Table 3 and visualize a subset in Fig. 3.
Table 2.
Various genome assembly metrics.
| Contiguity | BUSCO scores | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Shortest contig | Longest contig | Assembly size | contig number | count of N's | N(G)50 | L90 | C% | S% | D% | F% | M% |
| A. queenslandica (1) | 633 | 1,888,931 | 164,262,607 | 13,133 | 21,126,302 | 123,180 | 3,793 | 88.6 | 82.5 | 6.1 | 3.9 | 7.5 |
| A. queenslandica (2) | 314 | 4,599,197 | 167,703,827 | 3,871 | 22,857,408 | 950,481 | 268 | 89.1 | 83.5 | 5.6 | 3.7 | 7.2 |
| C. reniformis | 6,754,398 | 10,413,042 | 117,372,766 | 14 | 5,600 | 8,459,200 | 12 | 71.1 | 70.4 | 0.6 | 12.2 | 16.8 |
| E. muelleri | 282 | 34,737,626 | 322,619,961 | 1,434 | 1,864,700 | 9,883,643 | 160 | 68 | 55.5 | 12.6 | 9.3 | 22.6 |
| G. barretti | 1,002 | 495,233 | 144,789,364 | 4,535 | 110 | 48,447 | 2,902 | 71.5 | 67.9 | 3.6 | 12.6 | 15.9 |
| H. panicea | 949 | 50,219 | 73,970,439 | 32,385 | 0 | 2,555 | 25,647 | 48.1 | 47.7 | 0.4 | 28.1 | 23.8 |
| L. baikalensis | 500 | 124,926 | 209,989,122 | 135,191 | 373,641 | 2,213 | 97,717 | 59.3 | 53 | 6.3 | 19.6 | 21.1 |
| O. minuta | 1002 | 1,926,057 | 61,460,524 | 365 | 501,094 | 676,369 | 111 | 78.1 | 77.3 | 0.8 | 4.6 | 17.3 |
| O. pearsei | 100 | 107,672 | 57,775,306 | 67,767 | 1,985,416 | 5,457 | 24,537 | 60.4 | 59.7 | 0.6 | 16 | 23.6 |
| P. ficiformis | 4,740,285 | 63,279,122 | 191,074,932 | 18 | 15,000 | 9,942,894 | 15 | 82.3 | 81.4 | 0.8 | 6.5 | 11.2 |
| S. carteri | 800 | 965,826 | 418,920,728 | 97,497 | 48,953,456 | 10,236 | 57,833 | 48 | 42.6 | 5.5 | 28.5 | 23.5 |
| S. ciliatum | 1,001 | 1,380,240 | 357,509,570 | 7,780 | 79,115,821 | 169,232 | 2,473 | 66.1 | 62.4 | 3.8 | 14.9 | 19 |
| T. wilhelma | 1,004 | 659,656 | 125,670,620 | 5,936 | 1,516,047 | 73,701 | 2,656 | 70.9 | 69.2 | 1.7 | 12.7 | 16.5 |
| X. testudinaria | 800 | 1,572,474 | 257,935,546 | 97,640 | 31,482,608 | 4,078 | 69,043 | 64.8 | 60.5 | 4.3 | 14.8 | 20.4 |
N(G)50, size of the smallest contig that, with all larger contigs, sums up to over half of the assembly length; L90, number of contigs to span 90% of the genome assembly; BUSCO, total BUSCOs searched are 954; values given in the table are the percentages of the total BUSCOs that were identified respectively in each genome. C, complete BUSCOs; S, complete and single-copy BUSCOs; D, complete and duplicated BUSCOs; F, fragmented BUSCOs; M, missing BUSCOs. A. queenslandica 1 is GCF_000090795.2; A. queenslandica 2 is GCA_016292275.1. Species listed are in alphabetical order. In bold the genome reported herein.
Table 3.
Number of BGCs according to antiSMASH and biological classification (type) as “HMA” or “LMA” species.
| Species | Number of BGCs | Type | GC% | Gene number/CDS | % repetitive |
|---|---|---|---|---|---|
| A. queenslandica (1) | 1 | LMA | 35.5 | 30,327 | 43 |
| A. queenslandica (2) | 2 | LMA | 35.8 | — | — |
| C. reniformis | 1 | HMA | 37.3 | — | — |
| E. muelleri | 10 | LMA | 43.2 | 39,245 | 47 |
| G. barretti | 21 | HMA | 49.3 | 31,844 | 19.05 |
| H. panicea | 4 | LMA | 42.2 | — | — |
| L. baikalensis | 69 | LMA | 43.8 | — | — |
| O. minuta | 4 | LMA | 35.7 | 16,413 | — |
| O. pearsei | 3 | LMA | 43.5 | 9,823 | — |
| P. ficiformis | 3 | HMA | 33.9 | — | — |
| S. carteri | 15 | LMA | 43.9 | 26,967 | — |
| S. ciliatum | 9 | LMA | 47.0 | — | — |
| T. wilhelma | 3 | LMA | 39.9 | 37,416 | — |
| X. testudinaria | 133 | HMA | 49.9 | 22,337 | — |
GC % was calculated from individual nucleotide counts. The numbers of genes/CDS and the percentage of repetitive sequences were taken from the respective publications. Species are listed in alphabetical order. In bold the genome reported herein.
Fig. 3.
a) L90, number of contigs that cumulatively cover 90% of the assembly and b) the corresponding BUSCO completeness (C). The tail indicates the number of duplicate BUSCOs. c) The N50 compared to the number of Ns in the assemblies.
Assembly sizes in sponges range from 58 to 419 Mb (Table 2), which is in the range of genome sizes reported in the literature. Using FIAD and flow cytometry across a set of 75 sponge species, Tethya actinia and an unknown Dictyonellidae had the smallest genome with 39.1 Mb, while Mycale laevis 616.1–694.4 Mb and Placospongia intermedia 528.1–782.4 Mb had the largest genomes (Jeffery et al. 2013). In terms of difference between the size of genome assembly and genome size measured from cells, the assembly for Xestospongia testudinaria is almost 60% larger than the genome size estimated by flow cytometry (161.37 vs 258 Mb assembly). Indeed, both X. testudinaria and Stylissa carteri assemblies are hologenomes, and this excess sequence could indicate significant microbial contamination and/or high heterozygosity.
We selected a set of frequently used metrics to place the genome assembly of G. barretti in context with the other sponge genomes available (Table 2). The first set includes various metrics to express contiguity, i.e. the degree of fragmentation in the assembly. Ideally, the number of contigs should be the number of chromosomes (haploid), which is the case for the genomes of C. reniformis and P. ficiformis. The genome assembly of E. muelleri represents 23 chromosomes in 24 scaffolds but opted to also include a number of unplaced contigs, thus increasing the total number of contigs.
The second set of metrics in Table 2 was produced by BUSCO, a tool approximating biological completeness of a genome assembly by assessing the presence or absence of near universal single-copy genes (orthologs) (Simão et al. 2015). The numbers in this table may deviate from the values given in the original publications as there are pronounced differences between different versions of BUSCO and different reference gene sets. For comparability, we computed the metrics again, all with the same version of the program (v. 5.3.1, metazoa_odb_10, 2021-02-24). Overall completeness (C) frequently used to describe assemblies ranged from 48 to 89%. Generally, while higher values are better, some genes may truly be absent thus not allowing for a 100% completeness. Genes (BUSCOs) may also escape detection due to technical limitations, mainly due to gene prediction difficulties for highly derived lineages. The 2 chromosome-level assemblies for instance score “only” 71.1 and 82.3% complete. This “completeness” is the sum of single (S) and duplicate (D) BUSCOs identified. The number of duplicate BUSCOs is an important parameter as it can be indicative of whether diploid genomes were correctly and consistently collapsed to a haploid assembly, which can be an issue in organisms with high heterozygosity and/or when assembling long reads (Guiglielmoni et al. 2021). The highest number of duplicate single-copy orthologs (12.6%) was detected in E. muelleri assembly.
For the sponge genome assemblies published to date, different strategies were employed to isolate the starting material, DNA. Most of the genomes are “single origin” meaning that all DNA was extracted from a single individual: C. reniformis, Halichondria panicea, Lubomirskia baikalensis, Oscarella pearsei, P. ficiformis, S. carteri, Tethya wilhelma (D. Erpenbeck and W. Francis, pers. comm.), X. testudinaria, and G. barretti, the genome presented herein. However, several assemblies are based on DNA extracted from several (Sycon ciliatum (M. Adamska, pers. comm.), Oopsacas minuta (E. Renard, pers. comm.) to thousands of individuals (A. queenslandica (Srivastava et al. 2010; B. Degnan, pers. comm.). Most frequently, adult biomass (“tissue”) was extracted (C. reniformis, H. panicea, L. baikalensis, O. minuta, P. ficiformis, S. carteri, S. ciliatum, T. wilhelma, X. testudinaria, and G. barretti), but in some cases, whole larvae were extracted instead (A. queenslandica (Srivastava et al. 2010; B. Degnan, pers. comm.; O. pearsei). Notably, sponge biology allows for further alternative strategies to obtain DNA or biological material. The specimen of E. muelleri was grown from a gemmule (a clonal structure for dispersion) under sterile conditions (Kenny et al. 2020). T. wilhelma also has a form of clonal reproduction called budding. Sampling a bud is thus an easy way of sampling the sponge without harming it.
For most of the assemblies, DNA isolated from the sponges was sufficient. However, in case of single larvae (O. pearsei) and cells (S. carteri and X. testudinaria), WGAs were performed prior to sequencing. Whether to have sequencing reads derived from (1) a single individual as compared to (2) several individuals or (3) WGA DNA matters as differences due to individual variation and amplification errors can lead to more fragmented assemblies. However, these strategies are a trade-off to avoid overwhelming microbial contamination in the sequencing reads (Fig. 2b). One of the crucial aspects of sponge biology is their pervasive association with microbial symbionts (prokaryotes: bacteria, and archaea), especially in their sessile adult stage (Vacelet 1975; Leys et al. 2018). Recognizing this association has led to the classification of sponge species as HMA or LMA sponges. Typically, HMA also implies high microbial diversity and vice versa. Larvae also contain microbial symbionts, albeit to a greatly reduced extent (Björk et al. 2019). These microbial symbionts affect sponge genome sequencing in several ways. Extracting DNA from sponges inevitably leads to contamination with microbial DNA. This can be a challenge in the assembly process and lead to contamination and fragmentation in the resulting genome. Identification of microbial sequences can be difficult as there is a lack of both bona fide sponge sequences in databases as well as genomes of deep-sea microbes. At the same time, a locus with similarity to microbial sequences can also originate from horizontal gene transfer (HGT), which was previously shown in the A. queenslandica genome (Conaco et al. 2016). Conversely, bacterial genes coding for eukaryote-like proteins, present in sponge microsymbionts (Reynolds and Thomas 2016), could potentially be mistaken for true sponge genes. This means that in silico decontamination is challenging. Overall, the most successful contiguous assemblies have so far leveraged Hi-C (C. reniformis, P. ficiformis, and E. muelleri) and/or some form of (synthetic) long reads (G. barretti, O. minuta, and T. wilhelma).
Terpene BGCs have been lately discovered in several genomes of octocoral species (Burkhardt et al. 2022; Scesa et al. 2022) and sponges (Wilson et al. 2023). We therefore analyzed all 13 genomes with antiSMASH to identify possible BGCs: results ranged from 1–2 BGCs (A. queenslandica and C. reniformis) to 133 BGCs (X. testudinaria) (Table 3). While we did not determine whether these gene clusters originate from contamination or genuine cases of HGT, they highlight once more the close association of the sponges with their microbes and raise the possibility that BGCs are relatively widespread in sponge genomes. Interestingly, 1–10 BGCs were still found in chromosome-level assemblies (C. reniformis, P. ficiformis, and E. muelleri), which hints at genuine cases of BGC transferred to the sponge, deserved to be further studied. In the G. barretti genome, 21 BGCs were detected (Supplementary Table 1): 1 arylpolyene, 1 betalactone, 1 nonribosomal peptide synthetase (NRPS), 5 NRPS-like, and 13 terpene clusters. This is the third highest number only exceeded by L. baikalensis with 69 and X. testudinaria with 133 BGCs (Table 2). A full list of all BGCs is included in Supplementary Table 1. Overall, there seems to be no correlation between HMA/LMA status and the number of BGCs detected. It is worth noting that the majority of BGCs in L. baikalensis and X. testudinaria start at the first position of a short contig which indicates that the BGC is likely truncated and/or incomplete. The high numbers of BGCs for these 2 species might thus be an overestimation due to counting the same BGC multiple times.
To conclude, the G. barretti genome is the first genome for the Tetractinellida order, the second most speciose order of demosponges with over 1,100 species (de Voogd et al. 2023). All 4 deep-sea sponge genomes published so far are glass sponges (class Hexactinellida) (Francis et al. 2023; Santini et al. 2023; Schultz et al. 2023), with 3 of them published after submission of this study; the G. barretti genome is the first deep-sea genome of a demosponge. This genome will firmly establish the North Atlantic G. barretti as a prominent deep-sea sponge species for future studies, allowing the generation of new hypotheses about multicellularity, immunity, chemistry, and symbiont/cell recognition and interaction, which could be tested in vitro, thanks to the successful G. barretti cell line and CRISPR/Cas12a gene-editing system (Hesp et al. 2020, 2023).
Supplementary Material
Acknowledgments
We acknowledge the crew of the R/V Nereus (Tjärnö Marine Laboratory, Sweden) for their help with collection of the specimen. Thank you to Olga Vinnere Pettersson (NGI Uppsala) for discussions and trouble-shooting for DNA extraction and genome sequencing. We thank Björn Nystedt (NBIS/SciLifeLab) for help with bioinformatic analyses, Stephan Nylinder (NBIS) for help publishing our data on ENA. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project SNIC 2017-7-396, SNIC 2021-22-311, and SNIC 2022-22-411. The authors thank T. Ryan Gregory and Darren Kelly for genome size estimation with flow cytometry. Thank you to the Riesgo Lab (Natural History Museum, London) for making G. barretti transcriptomes available to us early in this project. We thank the anonymous reviewer for improving an earlier version of this manuscript.
Contributor Information
Karin Steffen, Pharmacognosy, Department of Pharmaceutical Biosciences, Uppsala University, Uppsala 751 24, Sweden.
Estelle Proux-Wéra, Department of Biochemistry and Biophysics, National Bioinformatics Infrastructure Sweden, Science for Life Laboratory, Stockholm University, Solna SE-17121, Sweden.
Lucile Soler, Department of Medical Biochemistry and Microbiology, National Bioinformatics Infrastructure Sweden (NBIS), Science for Life Laboratory, Uppsala University, Uppsala 752 37, Sweden.
Allison Churcher, Department of Molecular Biology, National Bioinformatics Infrastructure Sweden, Science for Life Laboratory, Umeå University, Umeå 901 87, Sweden.
John Sundh, Department of Biochemistry and Biophysics, National Bioinformatics Infrastructure Sweden, Science for Life Laboratory, Stockholm University, Solna SE-17121, Sweden.
Paco Cárdenas, Pharmacognosy, Department of Pharmaceutical Biosciences, Uppsala University, Uppsala 751 24, Sweden.
Data availability
The data associated with this study are deposited at the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB58046. Raw reads from PacBio RS II are ERR10902930, and from PacBio Sequel are ERR10857208, ERR10857206, ERR10857204, ERR10857202, and Illumina HiSeq X Ten ERR10857169. Previously published RNAseq data (BioProject: PRJNA603347, SRA: SRS6083072) include 4 individuals from the Norwegian and Barents seas, sequenced with ScriptSeq V2 (ROV6_3, trawl_5, trawl_6, and trawl_8) and 3 individuals from Sweden sequenced with TruSeq v2 [Geodia_01 (UPSZMC 184975), Geodia_02 (UPSZMC 184976), and Geodia_03 (UPSZMC 184977)] (Koutsouveli, Cárdenas, Santodomingo, et al. 2020).
Supplemental material available at G3 online.
Funding
K.S. and P.C. have received support from the H2020 EU Framework Program for Research and Innovation Project SponGES (Grant Agreement no. 679849). This document reflects only the authors’ view, and the Executive Agency for Small and Medium-sized Enterprises (EASME) is not responsible for any use that may be made of the information it contains. P.C. acknowledges WABI long-term bioinformatic support from NBIS (National Bioinformatics Infrastructure Sweden). K.S. acknowledges KVA (BS2017-0037) and Inez Johansson Stipendium (Uppsala University) for funding parts of this work. E.P-.W., J.S., and A.C. are financially supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab.
Literature cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bart MC, Hudspith M, Rapp HT, Verdonschot PFM, de Goeij JM. A deep-sea sponge loop? Sponges transfer dissolved and particulate organic carbon and nitrogen to associated fauna. Front Mar Sci. 2021;8:604879. doi: 10.3389/fmars.2021.604879. [DOI] [Google Scholar]
- Björk JR, Díez-Vives C, Astudillo-García C, Archie EA, Montoya JM. Vertical transmission of sponge microbiota is inconsistent and unfaithful. Nat Ecol Evol. 2019;3(8):1172–1183. doi: 10.1038/s41559-019-0935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49(W1):W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerbank JS. On the anatomy and physiology of the Spongiadae. Part I. On the spicula. Philos Trans R Soc Lond. 1858;148(2):279–332. https://www.jstor.org/stable/8b2ca25f-f017-3d4f-bb87-13f97f1c3a5b?seq=58. [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Burkhardt I, de Rond T, Chen PY-T, Moore BS. Ancient plant-like terpene biosynthesis in corals. Nat Chem Biol. 2022;18(6):664–669. doi: 10.1038/s41589-022-01026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado R, Mamede R, Cruz S, Leal MC. Updated trends on the biodiscovery of new marine natural products from invertebrates. Mar Drugs. 2022;20(6):389. doi: 10.3390/md20060389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas P, Rapp HT. Disrupted spiculogenesis in deep-water Geodiidae (Porifera, Demospongiae) growing in shallow waters. Invertebr Biol. 2013;132(3):173–194. doi: 10.1111/ivb.12027. [DOI] [Google Scholar]
- Cárdenas P, Rapp HT, Klitgaard AB, Best M, Thollesson M, Tendal OS. Taxonomy, biogeography and DNA barcodes of Geodia species (Porifera, Demospongiae, Tetractinellida) in the Atlantic boreo-arctic region. Zool J Linn Soc. 2013;169(2):251–311. doi: 10.1111/zoj.12056. [DOI] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaço A, Rapp HT, Campanyà-Llovet N, Pham CK. Bottom trawling in sponge grounds of the Barents Sea (Arctic Ocean): a functional diversity approach. Deep Sea Res Part I: Oceanogr Res Pap. 2022;183:103742. doi: 10.1016/j.dsr.2022.103742. [DOI] [Google Scholar]
- Conaco C, Tsoulfas P, Sakarya O, Dolan A, Werren J, Kosik KS. Detection of prokaryotic genes in the Amphimedon queenslandica genome. Thomas T, editor. PLoS One. 2016;11(3):e0151092. doi: 10.1371/journal.pone.0151092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkling M, Hesp K, Munroe S, Sandoval K, Martens DE, Sipkema D, Wijffels RH, Pomponi SA. Breakthrough in marine invertebrate cell culture: sponge cells divide rapidly in improved nutrient medium. Sci Rep. 2019;9(1):17321. doi: 10.1038/s41598-019-53643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Voogd NJ, Alvarez B, Boury-Esnault N, Carballo JL, Cárdenas P, Díaz MC, Dohrmann M, Downey R, Hajdu E, Hooper JNA, et al. 2023. World Porifera database. World Porifera database. doi: 10.14284/359. [accessed 2018 Oct 1].http://www.marinespecies.org/porifera. [DOI]
- Dharamshi JE, Gaarslev N, Steffen K, Martin T, Sipkema D, Ettema TJG. Genomic diversity and biosynthetic capabilities of sponge-associated chlamydiae. ISME J. 2022;16(12):2725–2740. doi: 10.1038/s41396-022-01305-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erngren I, Smit E, Pettersson C, Cárdenas P, Hedeland M. The effects of sampling and storage conditions on the metabolite profile of the marine sponge Geodia barretti. Front Chem. 2021;9:662659. doi: 10.3389/fchem.2021.662659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato S, Adamski M, Bergum B, Guder C, Jordal S, Leininger S, Zwafink C, Rapp H, Adamska M. Genome-wide analysis of the sox family in the calcareous sponge Sycon ciliatum: multiple genes with unique expression patterns. EvoDevo. 2012;3(1):14. doi: 10.1186/2041-9139-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis WR, Eitel M, Vargas S, Adamski M, Haddock SHD, Krebs S, Blum H, Erpenbeck D, Wörheide G. The genome of the contractile demosponge Tethya wilhelma and the evolution of metazoan neural signalling pathways. Genomics. [accessed 2022 Nov 7]2017. http://biorxiv.org/lookup/doi/10.1101/120998.
- Francis WR, Eitel M, Vargas S, Garcia-Escudero CA, Conci N, Deister F, Mah JL, Guiglielmoni N, Krebs S, Blum H, et al. The genome of the reef-building glass sponge Aphrocallistes vastus provides insights into silica biomineralization. R Soc open sci. 2023;10(6):230423. doi: 10.1098/rsos.230423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiglielmoni N, Houtain A, Derzelle A, Van Doninck K, Flot J-F. Overcoming uncollapsed haplotypes in long-read assemblies of non-model organisms. BMC Bioinform. 2021;22(1):303. doi: 10.1186/s12859-021-04118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNAseq: reference generation and analysis with trinity. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Fieseler L, Wehrl M, Gernert C, Steinert M, Hacker J, Horn M. Microbial diversity of marine sponges. In: Müller WEG, editor. Sponges (Porifera). Progress in Molecular and Subcellular Biology. Vol. 37. Berlin, Heidelberg: Springer; 2003. p. 59–88. doi: 10.1007/978-3-642-55519-0_3. [DOI] [PubMed] [Google Scholar]
- Hesp K, Flores Alvarez JL, Alexandru A-M, van der Linden J, Martens DE, Wijffels RH, Pomponi SA. CRISPR/Cas12a-mediated gene editing in Geodia barretti sponge cell culture. Front Mar Sci. 2020;7:599825. doi: 10.3389/fmars.2020.599825. [DOI] [Google Scholar]
- Hesp K, Van Der Heijden JME, Munroe S, Sipkema D, Martens DE, Wijffels RH, Pomponi SA. First continuous marine sponge cell line established. Sci Rep. 2023;13(1):5766. doi: 10.1038/s41598-023-32394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann F, Rapp HT, Reitner J. Monitoring microbial community composition by fluorescence in situ hybridization during cultivation of the marine cold-water sponge Geodia barretti. Mar Biotechnol. 2006;8(4):373–379. doi: 10.1007/s10126-006-5152-3. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICES . Report of the ICES/NAFO Joint Working Group on Deep-water Ecology (WGDEC). doi:10.17895/ICES.PUB.5567. [accessed 2023 Feb 15]. 2019. https://ices-library.figshare.com/articles/_/18621755.
- Jeffery NW, Jardine CB, Gregory TR. A first exploration of genome size diversity in sponges. Genome. 2013;56(8):451–456. doi: 10.1139/gen-2012-0122. [DOI] [PubMed] [Google Scholar]
- Kenny NJ, Francis WR, Rivera-Vicéns RE, Juravel K, de Mendoza A, Díez-Vives C, Lister R, Bezares-Calderón LA, Grombacher L, Roller M, et al. Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat Commun. 2020;11(1):3676. doi: 10.1038/s41467-020-17397-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, Plese B, Riesgo A, Itskovich VB. Symbiosis, selection, and novelty: freshwater adaptation in the unique sponges of Lake Baikal. Mol Biol Evol. 2019;36(11):2462–2480. doi: 10.1093/molbev/msz151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitgaard AB, Tendal OS. Distribution and species composition of mass occurrences of large-sized sponges in the northeast Atlantic. Prog Oceanogr. 2004;61(1):57–98. doi: 10.1016/j.pocean.2004.06.002. [DOI] [Google Scholar]
- Kolmogorov M, Bickhart DM, Behsaz B, Gurevich A, Rayko M, Shin SB, Kuhn K, Yuan J, Polevikov E, Smith TPL, et al. Metaflye: scalable long-read metagenome assembly using repeat graphs. Nat Methods. 2020;17(11):1103–1110. doi: 10.1038/s41592-020-00971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsouveli V, Cárdenas P, Conejero M, Rapp HT, Riesgo A. Reproductive biology of Geodia Species (Porifera, Tetractinellida) from boreo-Arctic north-Atlantic deep-sea sponge grounds. Front Mar Sci. 2020;7:595267. doi: 10.3389/fmars.2020.595267. [DOI] [Google Scholar]
- Koutsouveli V, Cárdenas P, Santodomingo N, Marina A, Morato E, Rapp HT, Riesgo A. The molecular machinery of gametogenesis in Geodia demosponges (Porifera): evolutionary origins of a conserved toolkit across animals. Mol Biol Evol. 2020;37(12):3485–3506. doi: 10.1093/molbev/msaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutti T, Bannister RJ, Fosså JH, Krogness CM, Tjensvoll I, Søvik G. Metabolic responses of the deep-water sponge Geodia barretti to suspended bottom sediment, simulated mine tailings and drill cuttings. J Exp Mar Biol Ecol. 2015;473:64–72. doi: 10.1016/j.jembe.2015.07.017. [DOI] [Google Scholar]
- Laetsch DR, Blaxter ML. BlobTools: interrogation of genome assemblies. F1000Res. 2017;6:1287. doi: 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- Lafi FF, Fuerst JA, Fieseler L, Engels C, Goh WWL, Hentschel U. Widespread distribution of Poribacteria in Demospongiae. Appl Environ Microbiol. 2009;75(17):5695–5699. doi: 10.1128/AEM.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys SP, Kahn AS, Fang JKH, Kutti T, Bannister RJ. Phagocytosis of microbial symbionts balances the carbon and nitrogen budget for the deep-water boreal sponge Geodia barretti. Limnol Oceanogr. 2018;63(1):187–202. doi: 10.1002/lno.10623. [DOI] [Google Scholar]
- Leys SP, Riesgo A. Epithelia, an evolutionary novelty of metazoans. J Exp Zool Part B: Mol Dev Evol. 2012;318(6):438–447. doi: 10.1002/jez.b.21442. [DOI] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier SR, Kutti T, Bannister RJ, Fang JK-H, van Breugel P, van Rijswijk P, van Oevelen D. Recycling pathways in cold-water coral reefs: use of dissolved organic matter and bacteria by key suspension feeding taxa. Sci Rep. 2020;10(1):9942. doi: 10.1038/s41598-020-66463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KM, Barrows LR. Biological activities of pyridoacridines. Nat Prod Rep. 2004;21(6):731. doi: 10.1039/b401662a. [DOI] [PubMed] [Google Scholar]
- McKenna V, Archibald JM, Beinart R, Dawson MN, Hentschel U, Keeling PJ, Lopez JV, Martín-Durán JM, Petersen JM, Sigwart JD, et al. The aquatic symbiosis genomics project: probing the evolution of symbiosis across the tree of life. Wellcome Open Res. 2021;6:254. doi: 10.12688/wellcomeopenres.17222.1. [DOI] [Google Scholar]
- Morrow C, Cárdenas P. Proposal for a revised classification of the Demospongiae (Porifera). Front Zool. 2015;12(7):1–27. doi: 10.1186/s12983-015-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Schippers KJ, Nickel M, Mizzon G, Kohn AB, Pape C, Ronchi P, Papadopoulos N, Tarashansky AJ, Hammel JU, et al. Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science. 2021;374(6568):717–723. doi: 10.1126/science.abj2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc Natl Acad Sci USA. 2012;109(32):13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norling M, Jareborg N, Dainat J. EMBLmyGFF3: a converter facilitating genome annotation submission to European Nucleotide Archive. BMC Res Notes. 2018;11(1):584. doi: 10.1186/s13104-018-3686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNAseq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plese B, Kenny NJ, Rossi ME, Cárdenas P, Schuster A, Taboada S, Koutsouveli V, Riesgo A. Mitochondrial evolution in the Demospongiae (Porifera): phylogeny, divergence time, and genome biology. Mol Phylogenet Evol. 2021;155:107011. doi: 10.1016/j.ympev.2020.107011. [DOI] [PubMed] [Google Scholar]
- Radax R, Rattei T, Lanzen A, Bayer C, Rapp HT, Urich T, Schleper C. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ Microbiol. 2012;14(5):1308–1324. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. https://www.R-project.org/. [Google Scholar]
- Redmond AK, McLysaght A. Evidence for sponges as sister to all other animals from partitioned phylogenomics with mixture models and recoding. Nat Commun. 2021;12(1):1783. doi: 10.1038/s41467-021-22074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds D, Thomas T. Evolution and function of eukaryotic-like proteins from sponge symbionts. Mol Ecol. 2016;25(20):5242–5253. doi: 10.1111/mec.13812. [DOI] [PubMed] [Google Scholar]
- Rooks C, Fang JK-H, Mørkved PT, Zhao R, Rapp HT, Xavier JR, Hoffmann F. Deep-sea sponge grounds as nutrient sinks: denitrification is common in boreo-Arctic sponges. Biogeosciences. 2020;17(5):1231–1245. doi: 10.5194/bg-17-1231-2020. [DOI] [Google Scholar]
- Ryu T, Seridi L, Moitinho-Silva L, Oates M, Liew YJ, Mavromatis C, Wang X, Haywood A, Lafi FF, Kupresanin M, et al. Hologenome analysis of two marine sponges with different microbiomes. BMC Genom. 2016;17(1):1–11. doi: 10.1186/s12864-016-2501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini S, Schenkelaars Q, Jourda C, Duchesne M, Belahbib H, Rocher C, Selva M, Riesgo A, Vervoort M, Leys SP, et al. The compact genome of the sponge Oopsacas minuta (Hexactinellida) is lacking key metazoan core genes. BMC Biol. 2023;21(1):139. doi: 10.1186/s12915-023-01619-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scesa PD, Lin Z, Schmidt EW. Ancient defensive terpene biosynthetic gene clusters in the soft corals. Nat Chem Biol. 2022;18(6):659–663. doi: 10.1038/s41589-022-01027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DT, Haddock SHD, Bredeson JV, Green RE, Simakov O, Rokhsar DS. Ancient gene linkages support ctenophores as sister to other animals. Nature. 2023;618(7963):110–117. doi: 10.1038/s41586-023-05936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-4.0. http://www.repeatmasker.org. 2013.
- Smith CD, Edgar RC, Yandell MD, Smith DR, Celniker SE, Myers EW, Karpen GH. Improved repeat identification and masking in Dipterans. Gene. 2007;389(1):1–9. doi: 10.1016/j.gene.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466(7307):720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K, Indraningrat AAG, Erngren I, Haglöf J, Becking LE, Smidt H, Yashayaev I, Kenchington E, Pettersson C, Cárdenas P, et al. Oceanographic setting influences the prokaryotic community and metabolome in deep-sea sponges. Sci Rep. 2022;12(1):3356. doi: 10.1038/s41598-022-07292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K, Laborde Q, Gunasekera S, Payne CD, Rosengren KJ, Riesgo A, Göransson U, Cárdenas P. Barrettides: a peptide family specifically produced by the deep-sea sponge Geodia barretti. J Nat Prod. 2021;84(12):3138–3146. doi: 10.1021/acs.jnatprod.1c00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert G, Rohde S, Janussen D, Blaurock C, Schupp PJ. Host-specific assembly of sponge-associated prokaryotes at high taxonomic ranks. Sci Rep. 2017;7(1):2542. doi: 10.1038/s41598-017-02656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand R, Whalan S, Webster NS, Kutti T, Fang JKH, Luter HM, Bannister RJ. The response of a boreal deep-sea sponge holobiont to acute thermal stress. Sci Rep. 2017;7(1):1660. doi: 10.1038/s41598-017-01091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehlow BW, Schuster A, Francis WR, Canfield DE. Metagenomic data for Halichondria panicea from Illumina and nanopore sequencing and preliminary genome assemblies for the sponge and two microbial symbionts. BMC Res Notes. 2022;15(1):135. doi: 10.1186/s13104-022-06013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, Olson JB, Erwin PM, López-Legentil S, Luter H, et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun. 2016;7:11870. doi: 10.1038/ncomms11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacelet J. Etude en microscopie electronique de l’association entre bacteries et spongiaires du genre Verongia (Dictyoceratida). J Microsc Biol Cell. 1975;23:271–288. [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. Wang J, editor. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, De Rond T, Burkhardt I, Steele TS, Schäfer RJB, Podell S, Allen EE, Moore BS. Terpene biosynthesis in marine sponge animals. Proc Natl Acad Sci USA. 2023;120(9):e2220934120. doi: 10.1073/pnas.2220934120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Reeder J, Lawrence M, Becker G, Brauer MJ . GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. In: Mathé E, Davis S, editors. Statistical Genomics. Vol. 1418. New York (NY): Springer; 2016 (Methods in Molecular Biology). p. 283–334. doi: 10.1007/978-1-4939-3578-9_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with this study are deposited at the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB58046. Raw reads from PacBio RS II are ERR10902930, and from PacBio Sequel are ERR10857208, ERR10857206, ERR10857204, ERR10857202, and Illumina HiSeq X Ten ERR10857169. Previously published RNAseq data (BioProject: PRJNA603347, SRA: SRS6083072) include 4 individuals from the Norwegian and Barents seas, sequenced with ScriptSeq V2 (ROV6_3, trawl_5, trawl_6, and trawl_8) and 3 individuals from Sweden sequenced with TruSeq v2 [Geodia_01 (UPSZMC 184975), Geodia_02 (UPSZMC 184976), and Geodia_03 (UPSZMC 184977)] (Koutsouveli, Cárdenas, Santodomingo, et al. 2020).
Supplemental material available at G3 online.