Abstract
Juvenile polyposis syndrome (JPS) is a hereditary hamartomatous polyposis syndrome characterized by gastrointestinal juvenile polyps and increased risk of gastrointestinal cancer. Germline pathogenic variants are detected in SMAD4 or BMPR1A, however in a significant number of patients with JPS, the etiology is unknown. From Danish registers, and genetic department and laboratories, we identified all patients in Denmark with a clinical diagnosis of JPS and/or a pathogenic variant in BMPR1A or SMAD4. In patients where no variant had been detected, we performed genetic analysis, including whole genome sequencing. We collected clinical information on all patients to investigate the phenotypic spectrum. Sixty-six patients (mean age 40 years) were included of whom the pathogenic variant was unknown in seven patients. We detected a pathogenic variant in SMAD4 or PTEN in additional three patients and thus ≈ 95% of patients had a pathogenic germline variant. Endoscopic information was available in fifty-two patients (79%) and of these 31 (60%) fulfilled the clinical criteria of JPS. In 41 patients (79%), other types of polyps than juvenile had been removed. Our results suggest that almost all patients with a clinical diagnosis of JPS has a pathogenic variant in mainly BMPR1A, SMAD4, and more rarely PTEN. However, not all patients with a pathogenic variant fulfil the clinical criteria of JPS. We also demonstrated a wide clinical spectrum, and that the histopathology of removed polyps varied.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10689-023-00338-z.
Keywords: Juvenile, Polyp, Cancer, Syndrome, Hereditary
Introduction
Juvenile polyposis syndrome (JPS) is a hereditary hamartomatous polyposis syndrome. The incidence has been estimated to 1:100,000–160,000 [1]. The syndrome is characterized by gastrointestinal juvenile polyps (JPs), mainly the colon, rectum, and stomach as well as an increased risk of colorectal (CRC) and gastric cancer (GC) [2, 3]. The inheritance pattern is autosomal dominant, and symptoms typically develops in adolescence.
The diagnosis is clinical and based on the Jass criteria, where one or more of the following must be fulfilled: (1) more than five juvenile polyps of the colorectum, (2) multiple juvenile polyps throughout the gastrointestinal tract and/or (3) any number of juvenile polyps with a family history of JPS [4].
In four studies from 2007 to 2021, a pathogenic germline variant was identified in SMAD4 or BMPR1A in 45–60% of patients fulfilling the criteria of JPS, suggesting that variants in other genes may be causative [1, 5–8]. However, in the study by Latchford published in 2012, the detection rate was 82%, leaving open the possibility that the lower detection rate in other studies have other causes [1]. The identification of the predisposing gene variant is important as surveillance should be tailored based on genotype and because patients can have the possibility of prenatal diagnostics including pre-implantation genetic diagnostics.
Patients with a pathogenic variant in SMAD4 (SMAD4-related JPS) often present with an additional phenotype not seen in patients with a pathogenic variant in BMPR1A (BMPR1A-related JPS) or in patients with JPS with unknown etiology [9]. This additional phenotype includes symptoms of hereditary hemorrhagic telangiectasia (HHT) (recurrent epistaxis, AV-malformations primarily in the lungs, liver and brain, besides skin/mucosal telangiectasias) [10]. The patients also have an increased risk of aortic root dilatation [11, 12]. Additionally, patients with SMAD4-related JPS have a more severe gastric phenotype including increased risk of massive polyposis, GC, and characteristic endoscopic features of the gastric mucosa [13].
In this study, we included patients that fulfilled the clinical criteria of JPS and/or had a pathogenic variant in SMAD4 or BMPR1A. We performed genetic analysis, including WGS, to detect the genetic etiology in as many patients as possible. Furthermore, we also collected clinical information to describe the phenotypic spectrum of the syndrome.
Materials and methods
Identification of patients
Patients were identified from The Danish Pathology Register; a search was performed using the Danish version of the Systematized Nomenclature of Medicine (SNOMED) diagnostic codes for “hamartomatous polyp” and “juvenile polyp.”
In addition, all Danish genetic departments and laboratories were asked to identify patients with a clinical diagnosis of JPS and/or a pathogenic variant (PV) in BMPR1A or SMAD4. A variant was classified as a PV if it was classified as “pathogenic” or “likely pathogenic” according to the guidelines of American College of Medical Genetics (ACMG) [14]. Information on family members were also collected, and relatives with signs or symptoms of JPS (early CRC/GC, juvenile polyps and/or HHT symptoms) were included.
The study was approved by The Danish Patient Safety Authority (journal no. 31-1521-329), the Regional Danish Data Protection Agency (journal-no.: P-2020-557/P-2020-696, The Capital Region of Denmark and the National Scientific and regional Scientific Ethics Committee (no-2105809/H-16030776).
Inclusion
A patient was included if the Jass criteria were fulfilled and/or if the patient were heterozygous for a PV in BMPR1A or SMAD4. Patients at all ages as well as deceased patients were included.
Genetic analysis If the etiology was not known, patients were contacted and offered genetic counselling and (re-)testing. Genetic analyses were performed on DNA extracted from peripheral blood. Primarily, a custom-made gene panel including BMPR1A, PTEN and SMAD4, as well as other genes associated with an increased risk of polyposis and colorectal cancer (APC, AXIN2, GREM1, MLH1, MSH2, MLH3, MSH6, MUTYH, NTHL1, PMS2, POLD1, POLE, STK11), was analyzed by NGS (Illumina Technology). The sequencing analysis enabled detection of single nucleotide variants in the coding regions as well as in the first and last 50 bp of the intronic regions, along with detection of copy number variants (CNVs). If a PV was not revealed, WGS was performed with Illumina Technology and sequencing to a median depth of at least 30×.
Clinical data Clinical information on all patients, both deceased and alive, was retrieved from Danish registers and medical records throughout the country. Data were collected from relevant departments, including surgical, pediatric, and oncological departments.
Study period Information on each person was retrieved from birth to death or until December 31st, 2021.
Statistics The point prevalence for 2021 was calculated based on the total Danish population retrieved from Statistics Denmark (5,843,347 residents) pr. 31st of December 2021. Descriptive data are presented in absolute numbers and proportions (%). Probabilities of cancer were estimated with Kaplan–Meier analysis. For patients who had been diagnosed with cancer, the time to event was not truncated at any age. For cancer probability, the follow-up time in the model were the time between date of birth and 31.12.2021, date of death, date of loss of follow up or date of first cancer diagnosis, whichever came first. The statistical software R version 4.2.1 (R Foundation for Statistical Computing, Vienna, Austria) was used in the analysis.
Results
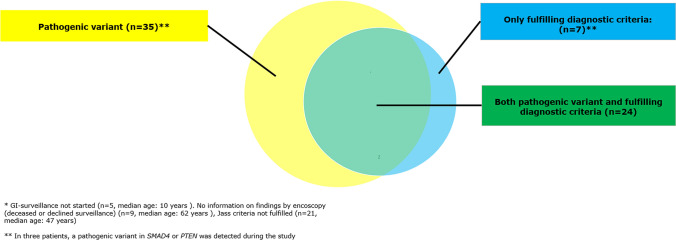
Sixty-six patients (34 males) fulfilled the inclusion criteria. Seven patients were included based on a clinical diagnosis of JPS, only, while a PV had been detected in BMPR1A or SMAD4 in 59 patients (Fig. 1) of which 24 patients also fulfilled the clinical criteria. Thereby, 35 patients were included based on a detected PV alone. These patients had been genetically tested because a PV had been detected in a family member. Average age at the time of study was 40 years. Thirteen patients were deceased. Five patients were under the age of 12 years, where GI-surveillance begins, and 9 patients had not had a GI-endoscopy—either because they were diagnosed posthumously or because they declined surveillance. The estimated point prevalence of patients being heterozygous for a PV in SMAD4 or BMPR1A and/or a clinical diagnosis of JPS was 1:110.000.
Fig. 1.
Patients carrying a pathogenic variant and/or fulfilling clinical criteria at study inclusion (n = 66)
Genetic analysis
In seven non-related patients, no PV had previously been identified. These patients were all identified in The Danish Pathology Register, and they all fulfilled the first Jass criterion (more than 5 juvenile colorectal polyps).
All seven patients were contacted. One patient did not respond to our inquiry, while the remaining six patients consented participation. By analyzing the NGS panel of polyposis associated genes, two different PVs in SMAD4 were detected in the two patients who did not follow JPS-surveillance, while it was not possible to detect a PV in the remaining four patients. WGS was then performed, and in one of these an intronic pathogenic variant in PTEN was detected. The patient was diagnosed with macrocephaly and multinodular non-toxic goiter; however, he did not fulfill the clinical criteria of Cowden syndrome. He had multiple colorectal polyps removed since the age 33. The polyps were mainly described as hamartomatous juvenile polyps, but in some cases, they were difficult to distinguish from an inflammatory polyp. Therefor he fulfilled the clinical criteria of JPS.
In three other patients, no PV or variant(s) of unknown significance were detected despite meticulous analysis of polyposis-associated genes, including BMPR1A, SMAD4, and PTEN. One of these patients had six juvenile polyps removed at 11–13 years of age and had not had any other polyps detected since (last follow-up at 25 years of age). The two other patients both had app. 10–20 colonic juvenile polyps removed and one was also diagnosed with gastric and duodenal hamartomatous polyps and had a history of epistaxis. None of these three patients had a relative with JPS. We observed no indication of mosaicism, as DNA from polyp tissue were analyzed with the NGS panel in two of these patients. In the third patient the quality of DNA in the polyp was not sufficient for genetic analysis.
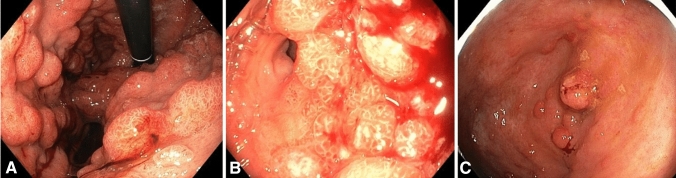
Both patients in whom we detected a PV in SMAD4 had 10–20 colonic polyps removed over a period of 15–20 years, that had shown inflammatory or hyperplastic histopathology with only few polyps suspected to be juvenile. Upon detection of the PVs in SMAD4 they were recommended additional upper GI surveillance. Both were then diagnosed with massive polyposis (Fig. 2) and subsequently had a gastrectomy. A summary of the patients included, and the genetic findings is presented in Table 1.
Fig. 2.
Endoscopic pictures of polyps in patients with Juvenile Polyposis Syndrome and pathogenic variants in SMAD4. A, B upper GI-tract, C colon
Table 1.
Information on patients and families, at the end of the study, stratified by genetic findings
| No PV | BMPR1A | SMAD4 | PTEN | Total | |
|---|---|---|---|---|---|
| Number of patients (male:female) | 4* (2:2) | 20 (10:10) | 41 (21:20) | 1 (1:0) | 66 (34:32) |
| Number of families | 4* | 7 | 15 | 1 | 27 |
| Mean age | 34 years | 48 years (11–73 years) | 40 years (2–78 years) | 59 years | 40 years (2–78 years) |
| Deceased patients | 0 | 4 | 9 | 0 | 13 |
| Mean age at death | – | 59 years (37–86 years) | 54 years (25–77 years) | – | 57 years |
| Family history: yes/no/unknown | 0/4/0 | 15/2/4 | 30/7/4 | 0/1/0 | 45/13/8 |
*In one patient/family genetic analyses were not performed
Genotype
Among the 65 patients where genetic analysis was performed, a PV was detected in BMPR1A, SMAD4 and/or PTEN in 62 patients (95%). One patient had a deletion of both BMPR1A and PTEN, and one patient had a PV in PTEN, only. In the 3 patients who fulfilled the clinical criteria and in which a blood sample had been subjected to genetic analyses including WGS, no relevant variant was detected. The gene variants detected are given in supplementary Table 1.
GI-manifestations
Information on GI-manifestations in patients who had endoscopic examinations (n = 52) is described in Table 2. Thirty-one patients (60%) fulfilled the clinical criteria of JPS with over five juvenile polyps in the GI-tract, and of these a PV could be found in 27 (87%). All 52 patients had one or more polyps removed. About 20% received a hemicolectomy or colectomy either because of massive polyposis or because of CRC. Gastrectomy was performed in 14% of the patients because of massive polyposis or GC—all of these had a PV in SMAD4.
Table 2.
Manifestations of the GI tract
| No PV | BMPR1A | SMAD4 | PTEN | Total | |
|---|---|---|---|---|---|
| Fulfilling clinical criteria (more than five colorectal juvenile polyps) | 4 (100%) | 7 (50%) | 19 (58%) | 1 | 31 (60%) |
| Patients without any juvenile polyps | 0 | 3 (21%) | 4 (9%) | 0 | 7 (13%) |
| Patient with adenomas, hyperplastic or inflammatory polyps in addition to juvenile polyps | 3 (75%) | 10 (71%) | 28 (88%) | 1 | 41 (79%) |
| Polyposis | |||||
| Gastric | 1 (25%) | 4 (29%) | 29 (70%) | 0 | 34 (65%) |
| Colorectal | |||||
| < 20 polyps | 3 (75%) | 6 (38%) | 17 (52%) | 1 | 25 (48%) |
| 20–100 polyps | 1 (25%) | 6 (43%) | 10 (30%) | 17 (33%) | |
| > 100 polyps | 0 | 2 (14%) | 6 (18%) | 8 (15%) | |
| Surgery | |||||
| Gastrectomy | 0 | 0 | 9 | 0 | 9 (14%)* |
| Colectomy or hemicolectomy | 0 | 4 | 10 | 0 | 14 (21%)* |
| Other | Resection of sigmoid colon | – | – | – |
*Percent of the total cohort
Table 2 shows that 41 patients (79%) on whom endoscopic information was available, had other types of polyps than juvenile removed. These included adenomas, hyperplastic and/or inflammatory polyps. Seven patients (13%) had not had any juvenile polyps removed but other types of polyps (mean age at end of follow-up: 47 years). The number of polyps varied between families, but also within families. Dysplasia in the juvenile polyp were detected in a minority of patients (under 10) and included dysplasia in both colonic and gastric polyps, However, no juvenile polyps were found with malignant alterations.
Genotype–phenotype correlation
Most patients with SMAD4-related JPS had HHT-manifestations including epistaxis, telangiectasias and AV-malformations (mainly pulmonary), this was not noted in patients with PV in BMPR1A and/or PTEN (see Supplementary Table 2). Some patients with PV in BMPR1A had gastric polyps. However, gastric involvement was more frequent and more massive in patients with SMAD4-related JPS.
Cancer
Seventeen patients (26%) had been diagnosed with cancer as presented in Table 3. The mean age at diagnosis was 48 years of age, however the age at diagnosis varied from 20 to 72 years. GC was diagnosed in two patients—both with SMAD4-related JPS—and both under 50 years at diagnosis. Pancreatic and oesophageal cancer were seen in patients with pathogenic BMPR1A variants. No cases of small bowel cancer were noted.
Table 3.
Cancer occurrence
| No PV | BMPR1A | SMAD4 | Total | |
|---|---|---|---|---|
| Total number of cancers | 0 | 5 (24%) | 12 (29%) | 17 (26%) |
| Average age all cancers | – | 49 years (31–64 years) | 47 years (20–72 years) | 48 years (20–72 years) |
| Type of cancer | ||||
| Colorectal | – | 3 (14%) | 7 (11%) | 10 (15%) |
| Gastric | – | 0 | 2 (5%) | 2 (3%) |
| Small bowel | – | 0 | 0 | 0 |
| Pancreatic | – | 1 (5%) | 0 | 1 (2%) |
| Other | – | 1 (5%, Oesophageal) | 3 (7%, lung, lymphoma) | 4 (6%) |
The probability of cancer was calculated over a total of 2782 person years. The cumulative probability of cancer at the age of 40 years was 12.6% [95% CI (3.1–22.2)], and at age 70 years 49.2% (95% CI 28.4–77.1). The probability of cancer tended to be higher in men [63%, 95% CI (25.0–100.0)], than female [43%, 95% CI (28.4–77.1)] at age 70 years.
Discussion
This study is a nationwide study including all 66 Danish patients who fulfilled the clinical criteria of JPS and/or had a PV in SMAD4 or BMPR1A. We found that almost 90% of patients with a clinical diagnosis of JPS had a PV in either BMPR1A or SMAD4. Endoscopy had been performed in 52 patients, and 21 (40%) of these did not fulfill the Jass criteria. Furthermore, the removed polyps frequently were other types than juvenile.
SMAD4 and BMPR1A: the only genes associated with JPS?
Previous studies have reported a relatively low variant detection rate when analyzing SMAD4 or BMPR1A indicating genetic heterogeneity [1, 5–8] (see Table 4). Our study as well as others suggest that PVs in PTEN are found in an additional (small) number of patients [7, 15]. The implementation of NGS, including WGS, did not add significantly to our detection rate.
Table 4.
Detection of gene variants in JPS
| Study | Number of patients/families fulfilling clinical Jass criteria | Technique | Genes investigated | Variant detection rate in SMAD4 or BMPR1A (per family) | BMPR1A | SMAD4 | PTEN | BMPR1A/PTEN deletion |
|---|---|---|---|---|---|---|---|---|
| Aretz et al. [7] | 65/65 | SS + MLPA | PTEN*, BMPR1A, SMAD4, | 39 (60%) | 16 (25%) | 23 (35%) | 2 (5%) | – |
| Van Hattem [8] | 29/27 | SS + MLPA | PTEN, BMPR1A, SMAD4, ENG | 13/27 (48%) | 4 (15%) | 7 (26%) | – | 2 (7%) |
| Calva-Cerqueira [5] | 102/102 | SS + MLPA | BMPR1A, SMAD4 | 45 (45%) | 22 (23%) | 22 (22%) | – | 1 (1%) |
| Latchford et al. [1] | 31/17 | NR | NR | 14 (82%) | 9 (29%) | 19 (61%) | – | – |
| MacFarland [6] | 118/? | Sequencing and CNV analysis | BMPR1A, SMAD4 | 54 (46%)** | 24 (20%) | 27 (23%) | – | 3 (3%) |
| Present study | 31/27 | NGS panel + WGS | See text | 23 (85%) | 6 (19%) | 19 (61%) | 1 (3%) | 1 (3%) |
SS Sanger sequencing, NR not reported
*In 60% of the patients, PTEN was not analyzed
**Per patient
The reason for the discrepancy in the variant detection rate between our study and most others is uncertain. Possibly, it could be caused by differences in methods used for identification and inclusion of patients. The mode of inclusion in our study is quite robust, as inclusion was based on data from The Danish Pathology Register: The register comprises data from all histopathological examinations carried out in Denmark since 1997, and for some departments even earlier. Although the significantly higher detection rate is unexplained, our findings, along with the findings by Latchford et al. suggest that in most cases, PJS is caused by a PV in either SMAD4 or BMPR1A.
We observed no indication of mosaicism, and to our knowledge this has neither been reported in other studies. Yet, mosaicism for variants in APC and STK11 has been reported [16–18], and it seems unlikely that mosaicism do not occur in patients with JPS. Possibly the phenotype in patients being mosaic for a variant in SMAD4 or BMPR1A is so “mild” that it typically is overlooked.
Varying histopathology leads to underdiagnosis
The high variant detection rate indicates that the Jass criteria [4] are a strong indicator of a pathogenic variant. However, our findings also suggest that there is a risk of missing patients with PVs in SMAD4 or BMPR1A if using these criteria alone: only 60% of the patients with a PV in one of these genes, who had endoscopic investigations, fulfilled the Jass criteria. Thus, one can only speculate that JPS in general is underdiagnosed, and that the calculated incidence is an underestimation. This also suggests that the threshold for performing genetic analysis should be rather low.
The histopathology of the colorectal polyps varied greatly—an observation that has also been recognized in other studies although at a smaller scale [15, 19, 20]. All patients with a PV in SMAD4 or BMPR1A had polyps removed and, furthermore, in approximately 15% of the patients no juvenile polyps (but other types) had been removed. This emphasizes that there should be an awareness, in the clinical setting, that JPS can present with a broad spectrum of polyps. This also indicates that SMAD4 and BMPR1A should be included in the panel of genes analyzed in all patients suspected to suffer from a genetic predisposition to intestinal polyposis. However, the varying histopathology can also be due to misclassification of polyps in the first place as have been reported before [7, 21]. In general, it can be difficult to distinguish different types of polyps from each other and especially juvenile polyps can resemble inflammatory polyps.
The consequences of the varying histopathology were evident in the two patients who had 10–20 colonic polyps removed over a period of 15–20 years. The polyps had shown various histopathology with only few polyps suspected to be juvenile. When participating in our study, both patients were found to have a PV in SMAD4 and were recommended additional upper GI surveillance. Both were then diagnosed with massive gastric polyposis and subsequently had a gastrectomy.
Wide clinical spectrum leading to suboptimal surveillance
In general, we observed that the clinical spectrum of patients with a PV in SMAD4 or BMPR1A was very wide: from massive polyposis (both gastric and colorectal) to very few colorectal polyps. Our findings also support the general conception of a difference in the phenotype of SMAD4-related JPS and BMPR1A-related JPS: We diagnosed more massive gastric involvement and HHT-manifestations in SMAD4-related JPS (Supplementary table 2) compared to BMPR1A-related JPS.
The highly variable phenotype suggests that the current surveillance guidelines impose a risk of overtreatment. According to most guidelines, surveillance of the GI tract should begin at age 12–15 years and examinations should be repeated every 2–3 years [22, 23]. Most agree that surveillance in JPS patients is indicated to reduce the risk of getting and dying from cancer as well as to reduce the morbidity caused by polyposis. However, there is a lack of long-term studies that can support the effect of surveillance. We calculated a probability of cancer at age 70 years to be approximately 50%, but various estimates have been reported and basically, a lack of knowledge concerning the pathophysiological mechanisms makes it difficult to tailor optimal surveillance [24–27]. As it is, a “one-size-fits-all” program does not seem to fit all. Possibly one could achieve a better balance between the “costs” (i.e. inconvenience of attending surveillance and the risk of overtreatment) and benefits for the individual patient by adjusting the surveillance program by the gene variant causing the predisposition and the personal history of the patient.
Strength and limitations of the study
A limitation of this study is the relatively low number of patients included and a risk of overlooking patients with a mild phenotype. Also, the facts that we ascertained from various sources, that we ascertained both on phenotype and on genotype, and that the patients had various ages at the end of follow-up, may have limited the generalizability of our observations.
However, it is a major strength that our study is nationwide and conducted in Denmark, where the health care system offers services, without out-of-the pocket expenses, to all citizens, where all citizens are identifiable by the unique social security number, and where registration in nation-wide registers, such as the Danish Pathology Register, is mandatory. This made it possible to identify the patient’s medical data and histopathological samples across registers and departments throughout the country, several years back.
Conclusion
In most patients, JPS is caused by a PV in BMPR1A or SMAD4, and the threshold for genetic analysis should be lowered. Not all patients with a PV in BMPR1A or SMAD4 fulfil the Jass-criteria, mainly because polyps show various histopathology. Consequently, the incidence of JPS is probably underestimated. Long-term studies and further investigations into the underlying molecular mechanism are needed to improve surveillance strategies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Marius Kløvgaard for statistical support.
Author contributions
AMJ: Designed, analyzed data and conducted the study. Drafted the paper. TOH: Performed genetic analysis and analyzed genetic data. LBG: Performed genetic analysis and analyzed genetic data. NQ: Collected surgical data and data on polyps in the Southern part of Denmark. Critically reviewed the paper. L-LC: Identified patients, critically reviewed the paper. CKL: Identified patients, collected genetic and surgical data, critically reviewed the paper. KL: Collected surgical data and data on polyps in the Western part of Denmark. Critical reviewed the paper. LTC: Collected surgical data and data on polyps in the Eastern part of Denmark. KR: Identified patients, collected genetic and surgical data, critically reviewed the paper. PMT: Identified patients, collected genetic data, critically reviewed the paper. BB: Performed genetic analysis and analyzed genetic data. LS: Collected surgical and genetic data in the Northern part of Denmark: critically reviewed the paper. JGK: Supervised the project, collected and analyzed data. Reviewed the paper.
Funding
Open access funding provided by National Hospital. This project was funded by Research Foundation from the University Hospital of Copenhagen, Rigshospitalet, Denmark.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Latchford AR, Neale K, Phillips RK, Clark SK. Juvenile polyposis syndrome: a study of genotype, phenotype, and long-term outcome. Dis Colon Rectum. 2012;55:1038–1043. doi: 10.1097/DCR.0b013e31826278b3. [DOI] [PubMed] [Google Scholar]
- 2.Brosens LA, van Hattem A, Hylind LM, Iacobuzio-Donahue C, Romans KE, Axilbund J, et al. Risk of colorectal cancer in juvenile polyposis. Gut. 2007;56:965–967. doi: 10.1136/gut.2006.116913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howe JR, Mitros FA, Summers RW. The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann Surg Oncol. 1998;5:751–756. doi: 10.1007/BF02303487. [DOI] [PubMed] [Google Scholar]
- 4.Jass JR, Williams CB, Bussey HJ, Morson BC. Juvenile polyposis—a precancerous condition. Histopathology. 1988;13:619–630. doi: 10.1111/j.1365-2559.1988.tb02093.x. [DOI] [PubMed] [Google Scholar]
- 5.Calva-Cerqueira D, Dahdaleh FS, Woodfield G, Chinnathambi S, Nagy PL, Larsen-Haidle J, et al. Discovery of the BMPR1A promoter and germline mutations that cause juvenile polyposis. Hum Mol Genet. 2010;19:4654–4662. doi: 10.1093/hmg/ddq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacFarland SP, Ebrahimzadeh JE, Zelley K, Begum L, Bass LM, Brand RE, et al. Phenotypic differences in juvenile polyposis syndrome with or without a disease-causing SMAD4/BMPR1A variant. Cancer Prev Res (Phila) 2021;14(2):215–222. doi: 10.1158/1940-6207.CAPR-20-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aretz S, Stienen D, Uhlhaas S, Stolte M, Entius MM, Loff S, et al. High proportion of large genomic deletions and a genotype phenotype update in 80 unrelated families with juvenile polyposis syndrome. J Med Genet. 2007;44:702–709. doi: 10.1136/jmg.2007.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hattem WA, Brosens LA, de Leng WW, Morsink FH, Lens S, Carvalho R, et al. Large genomic deletions of SMAD4, BMPR1A and PTEN in juvenile polyposis. Gut. 2008;57:623–627. doi: 10.1136/gut.2007.142927. [DOI] [PubMed] [Google Scholar]
- 9.Blatter R, Tschupp B, Aretz S, Bernstein I, Colas C, Evans DG, et al. Disease expression in juvenile polyposis syndrome: a retrospective survey on a cohort of 221 European patients and comparison with a literature-derived cohort of 473 SMAD4/BMPR1A pathogenic variant carriers. Genet Med. 2020;22(9):1524–1532. doi: 10.1038/s41436-020-0826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jelsig AM, Kjeldsen A, Christensen LL, Bertelsen B, Karstensen JG, Brusgaard K, et al. Hereditary haemorrhagic telangiectasia in Danish patients with pathogenic variants in SMAD4: a nationwide study. J Med Genet. 2022 doi: 10.1136/jmg-2022-108766. [DOI] [PubMed] [Google Scholar]
- 11.Teekakirikul P, Milewicz DM, Miller DT, Lacro RV, Regalado ES, Rosales AM, et al. Thoracic aortic disease in two patients with juvenile polyposis syndrome and SMAD4 mutations. Am J Med Genet A. 2013;161A(1):185–191. doi: 10.1002/ajmg.a.35659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heald B, Rigelsky C, Moran R, LaGuardia L, O’Malley M, Burke CA, et al. Prevalence of thoracic aortopathy in patients with juvenile polyposis syndrome-hereditary hemorrhagic telangiectasia due to SMAD4. Am J Med Genet A. 2015 doi: 10.1002/ajmg.a.37093. [DOI] [PubMed] [Google Scholar]
- 13.Jelsig AM, Qvist N, Bertelsen B, Christensen LL, Grossjohan H, Lautrup CK, et al. Distinct gastric phenotype in patients with pathogenic variants in SMAD4: a nationwide cross-sectional study. Endosc Int Open. 2022;10(12):E1537–E1543. doi: 10.1055/a-1954-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanich PP, Pearlman R, Hinton A, Gutierrez S, LaDuca H, Hampel H, et al. Prevalence of germline mutations in polyposis and colorectal cancer-associated genes in patients with multiple colorectal polyps. Clin Gastroenterol Hepatol. 2019;17(10):2008–15.e3. doi: 10.1016/j.cgh.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Spier I, Drichel D, Kerick M, Kirfel J, Horpaopan S, Laner A, et al. Low-level APC mutational mosaicism is the underlying cause in a substantial fraction of unexplained colorectal adenomatous polyposis cases. J Med Genet. 2016;53(3):172–179. doi: 10.1136/jmedgenet-2015-103468. [DOI] [PubMed] [Google Scholar]
- 17.Jelsig AM, Bertelsen B, Forss I, Karstensen JG. Two cases of somatic STK11 mosaicism in Danish patients with Peutz-Jeghers syndrome. Fam Cancer. 2020 doi: 10.1007/s10689-020-00191-4. [DOI] [PubMed] [Google Scholar]
- 18.Butel-Simoes GI, Spigelman AD, Scott RJ, Vilain RE. Low-level parental mosaicism in an apparent de novo case of Peutz-Jeghers syndrome. Fam Cancer. 2019;18:109–112. doi: 10.1007/s10689-018-0093-3. [DOI] [PubMed] [Google Scholar]
- 19.O'Riordan JM, O'Donoghue D, Green A, Keegan D, Hawkes LA, Payne SJ, et al. Hereditary mixed polyposis syndrome due to a BMPR1A mutation. Colorectal Dis. 2010;12:570–573. doi: 10.1111/j.1463-1318.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- 20.Rosner G, Petel-Galil Y, Laish I, Levi Z, Kariv R, Strul H, et al. Adenomatous polyposis phenotype in BMPR1A and SMAD4 variant carriers. Clin Transl Gastroenterol. 2022;13(10):e00527. doi: 10.14309/ctg.0000000000000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jelsig AM, Brusgaard K, Hansen TP, Qvist N, Larsen M, Bojesen A, et al. Germline variants in Hamartomatous Polyposis Syndrome-associated genes from patients with one or few hamartomatous polyps. Scand J Gastroenterol. 2016;51:1118–1125. doi: 10.1080/00365521.2016.1174880. [DOI] [PubMed] [Google Scholar]
- 22.Monahan KJ, Bradshaw N, Dolwani S, Desouza B, Dunlop MG, East JE, et al. Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom Cancer Genetics Group (UKCGG) Gut. 2020;69(3):411–444. doi: 10.1136/gutjnl-2019-319915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boland CR, Idos GE, Durno C, Giardiello FM, Anderson JC, Burke CA, et al. Diagnosis and management of cancer risk in the gastrointestinal hamartomatous polyposis syndromes: recommendations from the U.S. Multi-Society Task Force on Colorectal Cancer. Gastrointest Endosc. 2022;95(6):1025–47. doi: 10.1016/j.gie.2022.02.044. [DOI] [PubMed] [Google Scholar]
- 24.Järvinen HFK. Familial juvenile polyposis coli; increased risk of colorectal cancer. Gut. 1984;25:792–800. doi: 10.1136/gut.25.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howe JRMF, Summers RW. The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann Surg Oncol. 1998;5(8):751–756. doi: 10.1007/BF02303487. [DOI] [PubMed] [Google Scholar]
- 26.Coburn MCPV, DeLuca FG, Bland KI. Malignant potential in intestinal juvenile polyposis syndromes. Ann Surg Oncol. 1995;2(5):386–391. doi: 10.1007/BF02306370. [DOI] [PubMed] [Google Scholar]
- 27.Brosens LAA, van Hattem A, Hylind LM, Iacobuzio-Donahue C, Romans KE, Axilbund J, Cruz-Correa M, Tersmette AC, Offerhaus GJ, Giardiello FM. Risk of colorectal cancer in juvenile polyposis. Gut. 2007;56:965–7. doi: 10.1136/gut.2006.116913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.