Abstract
Obesity-induced inflammation plays a substantial role in the development of insulin resistance and type 2 diabetes. The altered gut flora in obesity can also contribute to metabolic dysregulation and systemic inflammation. However, it remains unclear how dysregulation of systemic inflammation in obesity affects the gut microbiome. We hypothesized that colchicine’s systemic anti-inflammatory effects in obesity would be associated with improvements in gut microbial diversity. We conducted a secondary analysis of a double-blind randomized placebo-controlled trial, in which 40 adults with obesity, high C-reactive protein (CRP) (≥2.0 mg/L), insulin resistance (homeostatic model of insulin resistance: HOMA-IR ≥2.6 mg/L), and metabolic syndrome (MetS) were randomized to three months of colchicine 0.6 mg or placebo tablets twice daily. Serum and stool samples were collected at baseline and final visit. Gut microbiota composition was characterized from stool DNA by dual-index amplification and sequencing of 16S ribosomal RNA. Pre- and post-intervention stool samples were available for 15 colchicine- and 12 placebo-treated subjects. Circulating high sensitivity CRP (hsCRP), interleukin-6, resistin, white blood count, and neutrophils were significantly decreased in the colchicine arm as compared to placebo. However, changes in stool microbiome alpha diversity, as assessed by the Chao1, Shannon, and Pielou indices, were not significant between groups. Amplicon sequence variant counts were unchanged among all examined phyla or families. Oscillibacter was the only genus to demonstrate even a nominally significant change. Among adults with obesity and MetS, colchicine significantly improved systemic inflammation. However, this anti-inflammatory effect was not associated with significant changes in the gut microbiome. Further studies are warranted to investigate this relationship.
Keywords: microbiome, colchicine, metabolic syndrome, obesity, inflammation
INTRODUCTION
Obesity, as defined by a body mass index (BMI) ≥30 kg/m2, is found in over 40% of US adults and is associated with significant medical comorbidities, such as metabolic syndrome (MetS), type 2 diabetes mellitus, and cardiovascular disease [1]. Although the progression from obesity to its related sequelae are multifactorial, murine models and human studies indicate that chronic inflammation likely plays a significant role [2,3,4].
Recent literature has also identified that the altered gut flora and decreased microbial diversity in obesity is associated with increased systemic inflammation [5,6,7]. Changes to the microbiome through fecal transplantation or diet may reduce this inflammatory state while simultaneously improving insulin resistance and body weight [8,9,10]. However, it is not well understood whether the reverse is true; namely whether reducing systemic inflammation in obesity can effectuate positive changes in the gut microbiome.
Colchicine, an anti-inflammatory medication commonly used in the management of gout, has recently garnered significant interest in the management of cardiometabolic disease. Prospective clinical trials have demonstrated that colchicine can ameliorate insulin resistance, decrease systemic inflammation, and reduce cardiovascular events in at-risk individuals [11,12,13,14]. However, colchicine’s effects on the microbiome in individuals with obesity and MetS are unknown.
Herein, we describe a secondary analysis of a randomized controlled trial examining colchicine’s metabolic effects in adults with obesity and MetS to study colchicine’s effect on the gut microbiome. Given that adiposity, dyslipidemia, and inflammation are associated with low gut microbiome diversity [15, 16], we hypothesized that colchicine treatment would increase markers of gut microbiome alpha-diversity. We also explored whether colchicine use significantly changed populations in any specific bacterial taxa as compared to placebo and whether changes in these populations were correlated with changes in particular markers of inflammation.
MATERIALS AND METHODS
Study design
The details of this single-center, double-blind, randomized, placebo-controlled trial have been described previously [11]. Subjects were randomized in a 1:1 ratio to receive colchicine 0.6 mg (Spectrum Chemical Corp, New Brunswick, NJ, USA) or identically-appearing placebo capsules twice-daily for three months. The primary outcome was change in insulin sensitivity. For the pilot study, it was estimated that a total sample size of 40 subjects (20 colchicine and 20 placebo) would have 80% power to detect a moderate effect size in the difference in change in insulin sensitivity between groups [11]. No power calculation was conducted for the exploratory microbiome analyses presented herein. The study protocol was approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board, and participants provided written consent for participation. No participant, investigator, or other staff interacting with participants was aware of study group assignment during the trial.
Participants
A convenience sample of 40 adults with obesity (BMI ≥30 kg/m2) and MetS were studied at the NIH Clinical Research Center between 2014 and 2018. At baseline, participants were required to have evidence for chronic inflammation (high-sensitivity C-reactive protein [hsCRP] ≥2.0 mg/L) and insulin resistance (Homeostatic Model of Insulin Resistance [HOMA-IR] ≥2.6). Subjects with significant chronic medical conditions, such as diabetes mellitus or cardiovascular disease, or taking medications affecting glucose homeostasis (e.g., metformin, insulin), lipids/cholesterol (e.g., statins, fibrates), body weight, or inflammation (e.g., glucocorticoids, non-steroidal anti-inflammatory drugs: NSAIDs) were excluded. Exclusion criteria for premenopausal female participants included irregular menses, pregnancy, or breastfeeding.
Stool microbiome measurements
Stool samples were collected from participants at baseline and at week 12. From each available sample, 50 mg aliquots were made after homogenization, and DNA was extracted from each sample using the Qiagen MagAttract PowerMicrobiome DNA/RNA EP Kit on an Eppendorf automated liquid handling system. A dual-index amplification and sequencing approach was taken to assess the composition of microbial communities from the given samples using the V4 region of the 16S ribosomal RNA gene (16S rRNA) on the Illumina MiSeq Platform.
16S amplicon libraries were prepared using 10.5 µL DNA (10 ng/µl) as starting material in a 96-well polymerase chain reaction (PCR) plate proceeding with PCR I setup and cleanup, followed by PCR II setup and cleanup using AMPure XP Beads. Library QC included quantification of the final amplicon library using KAPA quantitative PCR (qPCR). The individual sample libraries were diluted and pooled at a concentration of 12 nM, to make the final library pool. The final amplicons were normalized to 11 pm, spiked-in with 15% phiX control library and sequenced on the MiSeq instrument.
Read pairs were trimmed for quality and adapter and primer sequences using BBDuk v38.34. The pairs were subsequently processed with the dada2 R package v1.10 with taxonomy assigned using the SILVA database v132. Samples with inadequate amplicon sequence variant (ASV) counts as evaluated by rarefaction plots were excluded. Paired samples were then analyzed for alpha diversity and differential abundance. Specific taxa were excluded from analyses if >25% of samples had absent ASV counts.
To assess the alpha diversity, three indices were calculated using the vegan R package v2.5: Chao1 to estimate the total number of different species in a given sample (richness), Pielou to estimate the overall evenness of the distribution of relative abundances across the different species in the sample, and Shannon, a measure of both richness and evenness [17, 18].
Laboratory measurements
Blood samples were drawn at baseline and after 3 months of study drug. Peripheral blood was collected after overnight fasting in serum tubes and centrifuged for 10 min at 3,500 rpm. Obtained serum was immediately stored at –80°C until further analysis without being exposed to freeze-thaw cycles, according to NIH Center for Human Immunology protocols (https://chi.niaid.nih.gov/web/new/our-research/sop.html). HsCRP was assessed by the NIH CRC clinical laboratory on a Roche Cobas 6000 analyzer (Roche Diagnostics, Indianapolis, IN, USA). Glycoprotein acetyls (GlycA) concentrations were measured with a Vantera Clinical Analyzer using the LipoProfile-3 algorithm (LabCorp, Burlington, NC, USA) as described previously [19]. Serum proteomic analysis for interleukin-6 (IL-6) and resistin used the SOMAscan 1.3k Assay (SomaLogic, Boulder, CO, USA) as previously described [20].
Statistical analysis
Assessment of differences between treatment arms (calculated as end-treatment – baseline value) was conducted using Student’s unpaired t-test for normally distributed data or Mann Whitney U test for nonnormally distributed data. Pearson and Spearman correlation analyses were used for normally and nonnormally distributed data, respectively. Two-sided significance tests were performed for all analyses. As this was an exploratory secondary analysis, statistical significance was defined at an FDR-adjusted p-value of 0.05. SPSS v27.0 (IBM Corp, Armonk, NY, USA) was used for statistical analyses.
RESULTS
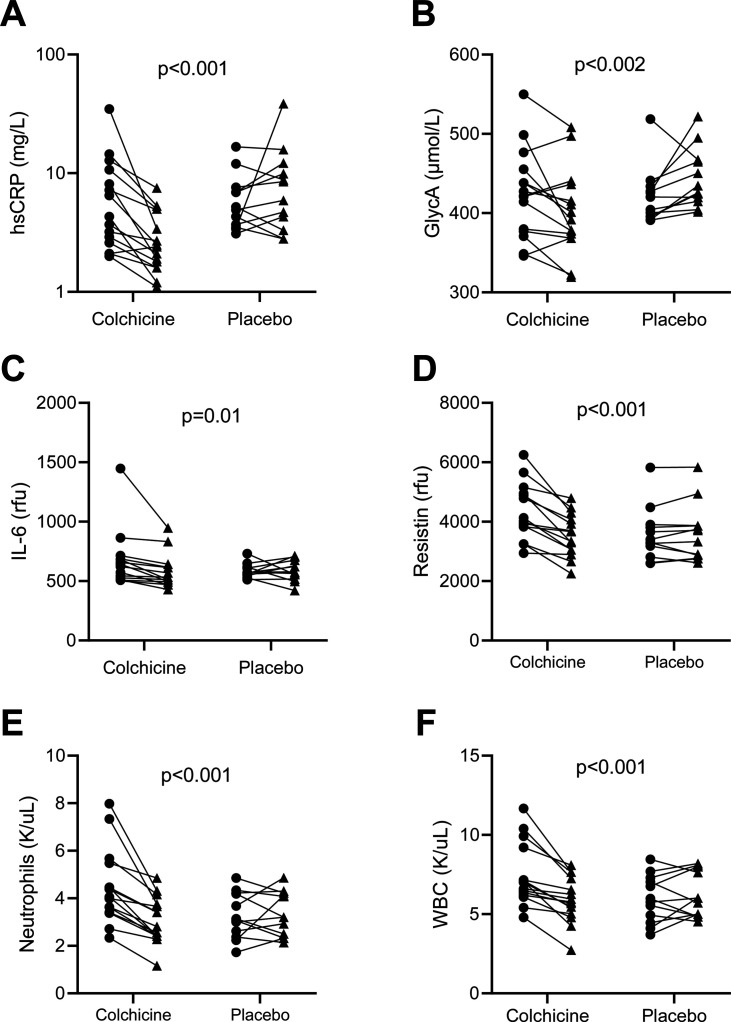
Of the 40 participants included in the primary efficacy analyses, adequate stool sample collection for both pre- and post-intervention time points were available for 27 participants (colchicine n=15, placebo n=12). Baseline characteristics were similar between groups (Table 1). The 13 participants that did not provide an adequate stool sample showed statistically significant differences in several sociodemographic variables at baseline from the 27 participants studied (Supplementary Table 1). Similar to the primary efficacy analyses [11], systemic levels of inflammation in our subset of 27 subjects were significantly decreased in the colchicine arm as compared to placebo, including hsCRP (mean ± SD: −3.35 ± 3.12 vs. 0.79 ± 2.69 mg/L; p<0.001), GlycA (−23.50 ± 38.31 vs. 23.31 ± 35.72 μmol/L; p<0.002), IL-6 (−72.89 ± 57.01 vs. 1.36 ± 94.28 rfu; p=0.01), resistin (−812.92 ± 545.11 vs. 28.50 ± 249.72 rfu; p<0.001), white blood cell count (WBC) (−1.52 ± 1.13 vs. 0.19 ± 1.05 K/uL; p<0.001), and neutrophils (−1.18 ± 0.57 vs. 0.06 ± 0.84 K/uL; p<0.001) (Fig. 1).
Table 1. Baseline characteristics.
| Variable | Colchicine (n=15) | Placebo (n=12) |
|---|---|---|
| Age (years) | 50.2 ± 6.4 | 47.3 ± 6.2 |
| Sex (n, %) | ||
| Male | 3 (20%) | 2 (16.7%) |
| Female | 12 (80%) | 10 (83.3%) |
| Race (n, %) | ||
| White | 8 (53.3%) | 4 (33.3%) |
| Black | 3 (20%) | 3 (25%) |
| Multiracial/Unknown/Other | 4 (26.7%) | 5 (41.7%) |
| BMI (kg/m2) | 38.2 ± 3.1 | 39.5 ± 3.9 |
| Hemoglobin A1c (%) | 5.7 ± 0.2 | 5.4 ± 0.2 |
| hsCRP (mg/L) | 7.8 ± 4.1 | 6.5 ± 2.2 |
| GlycA (μmol/L) | 424.3 ± 27.5 | 420.4 ± 19.3 |
| IL-6 (rfu) | 677.3 ± 123.2 | 586.3 ± 32.2 |
| Resistin (rfu) | 4,350.7 ± 485.4 | 3,573.3 ± 486.0 |
| Neutrophils (K/uL) | 4.5 ± 0.8 | 3.3 ± 0.5 |
| WBC (K/uL) | 7.4 ± 1.0 | 6.0 ± 0.8 |
Baseline characteristics of randomized participants for whom stool samples for microbiome analysis were available. Values presented in mean ± SD format except where otherwise noted. BMI: body mass index; hsCRP: high-sensitivity C-reactive protein; GlycA: glycoprotein acetyls; IL-6: interleukin-6; rfu: relative fluorescence units; WBC: white blood cells.
Fig. 1.
Changes in inflammatory markers. Solid lines connect samples from the same subject between Week 0 (circle) and Week 12 (triangle) (A) hsCRP, (B) GlycA, (C) IL-6, (D) Resistin, (E) Neutrophils, and (F) WBC. The p-values were for comparison of changes between groups per inflammatory marker. Data were analyzed using the unpaired Student’s t-test. hsCRP: high-sensitivity C-reactive protein; GlycA: glycoprotein acetyls; IL-6: interleukin-6; WBC: white blood cells.
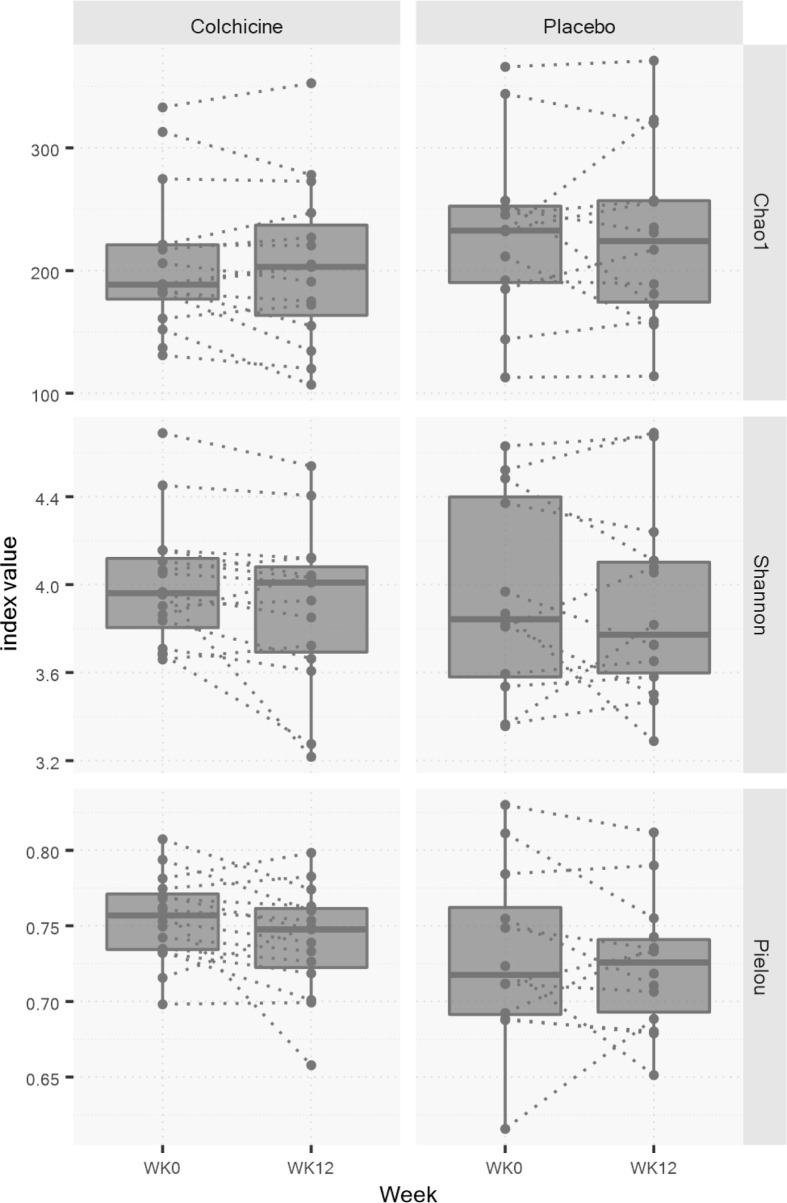
Alpha diversity was assessed using the Chao1, Shannon and Pielou indices to compare Week 12 (final visit) and Week 0 (baseline visit) for each group. No statistically significant differences in the Chao1 (−6.37 ± 23.84 vs. −0.74 ± 44.23; p=0.68), Shannon (−0.10 ± 0.19 vs. −0.04 ± 0.30; p=0.53), or Pielou (−0.01 ± 0.03 vs. −0.01 ± 0.04; p=0.57) indices were seen between the colchicine and placebo groups over time (Fig. 2).
Fig. 2.
Changes in alpha-diversity indices. Dotted lines connect samples from the same subject between weeks 0 and 12. Boxes represent the range between the first and third quartiles. The horizontal line in the box displays the median value. No significant differences were found in any alpha-diversity index between the colchicine and placebo groups over the study duration. Data were analyzed using the unpaired Student’s t-test.
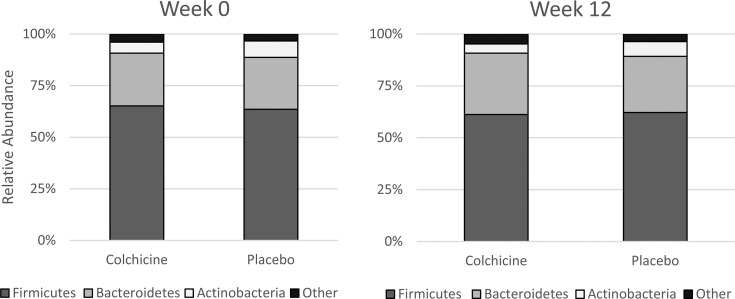
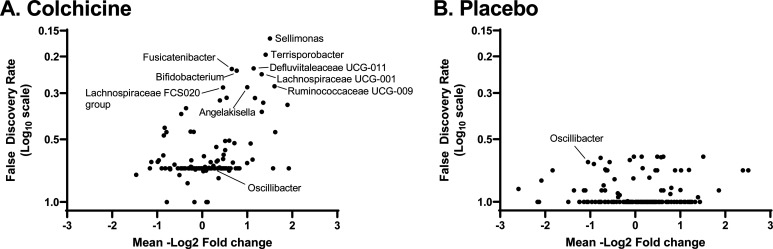
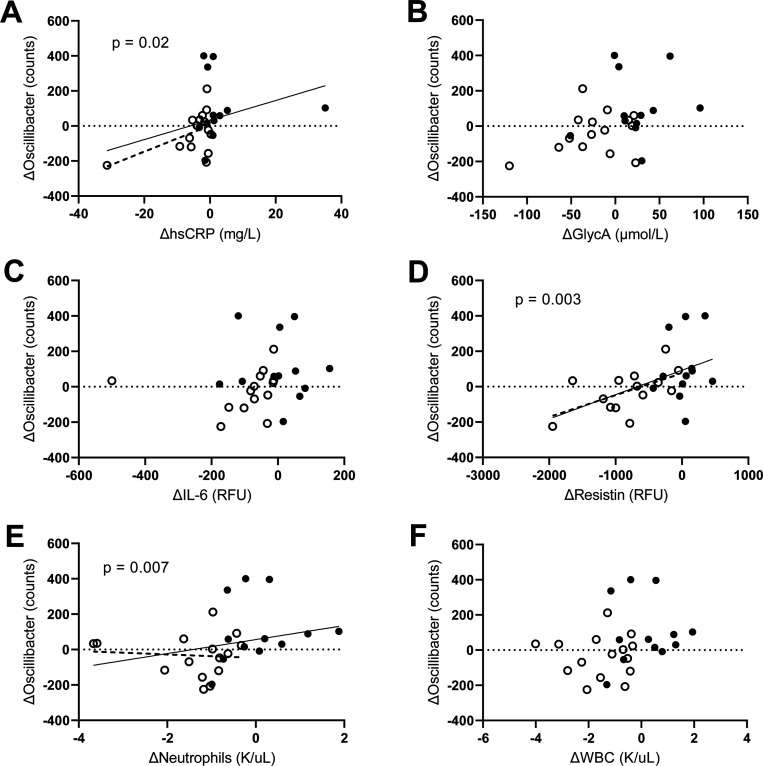
Although ASV counts, converted to relative abundance, for phyla, classes, or families were not significantly changed between groups (Fig. 3), there was a trend towards nominal significance in several genera and species (Fig. 4). ASV counts demonstrated a potential change in relative abundance of one genus, Oscillibacter, with colchicine having a nominally-significant (but not FDR-corrected significant) decrease in counts as compared to placebo (−33.5 ± 118.4 vs. 102.5 ± 183.9 counts; p=0.04). Changes in Oscillibacter counts were nominally significantly associated with changes in concentrations of the inflammatory markers hsCRP (ρ=0.433, p=0.02), neutrophils (ρ=0.508, p=0.007), and resistin (ρ=0.554, p=0.003), while correlations with GlycA (ρ=0.370, p=0.057) and WBC (ρ=0.355, p=0.07) trended towards but did not reach nominal statistical significance (Fig. 5). No significant correlations were seen with changes in Oscillibacter amplicon counts and IL-6 (ρ=0.294, p=0.15).
Fig. 3.
Phyla compositions at baseline and final visits between groups. Counts were not significantly changed for any phyla between groups over time.
Fig. 4.
Changes in the abundance of gut microbial populations at the genus level from baseline (week 0) to final visit (week 12). Each point represents a different microbiome bacterial genus. Descriptions of statistical tests used per microbial variable is included in Supplementary Table 2.
Fig. 5.
Correlations of changes in Oscillibacter amplicon counts versus changes in inflammatory markers (A) hsCRP, (B) GlycA, (C) IL-6, (D) Resistin, (E) Neutrophils, and (F) WBC. Oscillibacter counts were significantly associated with ΔhsCRP, Δresistin, and Δneutrophils.
Solid line: entire cohort, dotted line: colchicine-treated group. Open circles: colchicine, closed circles: placebo. Data were analyzed using Spearman’s rank correlation. hsCRP: high-sensitivity C-reactive protein; GlycA: glycoprotein acetyls; IL-6: interleukin-6; WBC: white blood cells.
DISCUSSION
To our knowledge, this is the first study examining the effects of colchicine on the gut microbiome in humans. In this study, we observed that despite significantly reducing levels of systemic inflammation in adults with obesity and MetS, colchicine had no significant effects on gut microbiome alpha-diversity measures. Furthermore, colchicine had limited effect on specific taxonomic populations, with only one genus demonstrating even a nominally-significant change over the three-month study.
Previous research in mice has also suggested that colchicine may not have appreciable effects on the microbiome. In a recent study by Shi et al. [21], no significant changes to alpha diversity of the gut microbiome, as measured by the Observed species index, Shannon index, or Simpson index, were seen at lower daily doses of colchicine (0.1 mg/kg or 0.5 mg/kg). Only once toxic doses of colchicine (2.5 mg/kg/day) were administered, were significant decreases in alpha-diversity observed [21]. However, this study was conducted in chow fed mice, so it is unclear whether these effects would be similar in mouse models of obesity.
Similarly, few studies have previously investigated colchicine’s effects on the human microbiome. In a study of subjects with Behcet’s disease (BD), individuals on powerful systemic immunosuppressant medications (e.g., cyclosporin A, azathioprine, prednisone) did not have significantly different salivary microbiome measures from those on colchicine [22]. However, an unpublished study investigating genital and oral microbiome populations in BD suggested that colchicine use may have significant effects on genital microbial abundance [23]. A study evaluating individuals with gout found that anti-inflammatory drug use as a whole, which included colchicine in some subjects, did not impact alpha-diversity as measured by the Shannon index, but affected the populations of several specific gut species [24]. Although anti-inflammatory medications do not seem to significantly affect microbial diversity [25], it is well documented that the reverse is true; namely, improving the diversity and composition of the gut microbiome can reduce systemic inflammation [8,9,10].
In our study, only one taxonomic rank, the genus Oscillibacter, was found to be nominally significantly decreased in the colchicine group versus the placebo group, but the effect appeared mostly driven by an increase of Oscillibacter in the placebo group. Interestingly, changes in Oscillibacter counts were positively correlated with changes in multiple measures of inflammation, including hsCRP, resistin, and neutrophil count. However, as correlation does not imply causation, it is unclear whether colchicine’s anti-inflammatory effects directly influenced changes in Oscillibacter counts, other mechanisms (e.g. colchicine’s microtubule effects) instead contributed to changes in Oscillibacter counts, or whether the changes seen in Oscillibacter counts were simply spurious findings.
Our findings contrast with previous cross-sectional studies which have suggested that Oscillibacter counts may be associated with metabolic health. For example, a decreasing predominance of Oscillibacter has been found in individuals with obesity [26]. Conversely, increasing Oscillibacter valericigenes populations have been seen with increased adherence to Mediterranean diet, as well as in individuals on low carbohydrate/high protein diets [27, 28].
The findings from our study are limited by their conduct as a secondary analysis as well as the small sample size, which may have hampered the ability to identify significant differences between groups. The strengths of the study design include the conduct of a prospective randomized placebo-controlled trial.
In conclusion, our results indicate that administration of colchicine in adults with obesity and MetS did not result in significant changes in microbial diversity or abundance of particular bacterial taxa in the stool microbiome, despite significant improvements in systemic inflammation. Thus, colchicine’s previously described cardiometabolic benefits are unlikely to be due to any significant salutary effects on the gut microbiome. Additional studies are warranted to further explore this relationship.
FUNDING AND CONFLICT OF INTEREST
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) grant number ZIA-HD00641. The authors declare no conflict of interest for this research. JAY reports unrelated grant funds to NICHD supporting his research from Soleno Therapeutics, Rhythm Pharmaceuticals, Hikma Pharmaceuticals, and Versanis Bio. The opinions and assertions expressed herein are those of the authors and are not to be construed as reflecting the views of the National Institutes of Health.
Supplementary Material
REFERENCES
- 1.Wilson PW, D’Agostino RB, Parise H, Sullivan L, Meigs JB. 2005. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation 112: 3066–3072. [DOI] [PubMed] [Google Scholar]
- 2.Bonora E, Kiechl S, Willeit J, Oberhollenzer F, Egger G, Targher G, Alberiche M, Bonadonna RC, Muggeo M. 1998. Prevalence of insulin resistance in metabolic disorders: the Bruneck Study. Diabetes 47: 1643–1649. [DOI] [PubMed] [Google Scholar]
- 3.van der Heijden RA, Sheedfar F, Morrison MC, Hommelberg PP, Kor D, Kloosterhuis NJ, Gruben N, Youssef SA, de Bruin A, Hofker MH, Kleemann R, Koonen DP, Heeringa P. 2015. High-fat diet induced obesity primes inflammation in adipose tissue prior to liver in C57BL/6j mice. Aging (Albany NY) 7: 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goossens GH. 2017. The metabolic phenotype in obesity: fat mass, body fat distribution, and adipose tissue function. Obes Facts 10: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE. 2010. Obesity and the human microbiome. Curr Opin Gastroenterol 26: 5–11. [DOI] [PubMed] [Google Scholar]
- 6.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres-Fuentes C, Schellekens H, Dinan TG, Cryan JF. 2017. The microbiota-gut-brain axis in obesity. Lancet Gastroenterol Hepatol 2: 747–756. [DOI] [PubMed] [Google Scholar]
- 8.Liou AP, Paziuk M, Luevano JM, Jr, Machineni S, Turnbaugh PJ, Kaplan LM. 2013. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 5: 178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aron-Wisnewsky J, Clément K, Nieuwdorp M. 2019. Fecal microbiota transplantation: a future therapeutic option for obesity/diabetes? Curr Diab Rep 19: 51. [DOI] [PubMed] [Google Scholar]
- 10.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O, MetaHIT consortium. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- 11.Demidowich AP, Levine JA, Onyekaba GI, Khan SM, Chen KY, Brady SM, Broadney MM, Yanovski JA. 2019. Effects of colchicine in adults with metabolic syndrome: a pilot randomized controlled trial. Diabetes Obes Metab 21: 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nidorf SM, Fiolet ATL, Eikelboom JW, Schut A, Opstal TSJ, Bax WA, Budgeon CA, Tijssen JGP, Mosterd A, Cornel JH, Thompson PL, LoDoCo2 Investigators.2019. The effect of low-dose colchicine in patients with stable coronary artery disease: the LoDoCo2 trial rationale, design, and baseline characteristics. Am Heart J 218: 46–56. [DOI] [PubMed] [Google Scholar]
- 13.Tardif JC, Kouz S, Waters DD, Bertrand OF, Diaz R, Maggioni AP, Pinto FJ, Ibrahim R, Gamra H, Kiwan GS, Berry C, López-Sendón J, Ostadal P, Koenig W, Angoulvant D, Grégoire JC, Lavoie MA, Dubé MP, Rhainds D, Provencher M, Blondeau L, Orfanos A, L’Allier PL, Guertin MC, Roubille F. 2019. Efficacy and safety of low-dose colchicine after myocardial infarction. N Engl J Med 381: 2497–2505. [DOI] [PubMed] [Google Scholar]
- 14.Nidorf SM, Fiolet ATL, Mosterd A, Eikelboom JW, Schut A, Opstal TSJ, The SHK, Xu XF, Ireland MA, Lenderink T, Latchem D, Hoogslag P, Jerzewski A, Nierop P, Whelan A, Hendriks R, Swart H, Schaap J, Kuijper AFM, van Hessen MWJ, Saklani P, Tan I, Thompson AG, Morton A, Judkins C, Bax WA, Dirksen M, Alings M, Hankey GJ, Budgeon CA, Tijssen JGP, Cornel JH, Thompson PL, LoDoCo2 Trial Investigators. 2020. Colchicine in patients with chronic coronary disease. N Engl J Med 383: 1838–1847. [DOI] [PubMed] [Google Scholar]
- 15.Ejtahed HS, Angoorani P, Soroush AR, Hasani-Ranjbar S, Siadat SD, Larijani B. 2020. Gut microbiota-derived metabolites in obesity: a systematic review. Biosci Microbiota Food Health 39: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida N, Watanabe S, Yamasaki H, Sakuma H, Takeda AK, Yamashita T, Hirata KI. 2022. Average gut flora in healthy Japanese subjects stratified by age and body mass index. Biosci Microbiota Food Health 41: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demidowich AP, Wolska A, Wilson SR, Levine JA, Sorokin AV, Brady SM, Remaley AT, Yanovski JA. 2019. Colchicine’s effects on lipoprotein particle concentrations in adults with metabolic syndrome: a secondary analysis of a randomized controlled trial. J Clin Lipidol 13: 1016–1022.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demidowich AP, Levine JA, Apps R, Cheung FK, Chen J, Fantoni G, Patel TP, Yanovski JA, CHI Consortium 2020. Colchicine’s effects on metabolic and inflammatory molecules in adults with obesity and metabolic syndrome: results from a pilot randomized controlled trial. Int J Obes 44: 1793–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Li J, Yang P, Niu Z, Wei L, Chen L, Gao L. 2020. Colchicine increases intestinal permeability, suppresses inflammatory responses, and alters gut microbiota in mice. Toxicol Lett 334: 66–77. [DOI] [PubMed] [Google Scholar]
- 22.Coit P, Mumcu G, Ture-Ozdemir F, Unal AU, Alpar U, Bostanci N, Ergun T, Direskeneli H, Sawalha AH. 2016. Sequencing of 16S rRNA reveals a distinct salivary microbiome signature in Behçet’s disease. Clin Immunol 169: 28–35. [DOI] [PubMed] [Google Scholar]
- 23.Senusi AA, Ogunkolade WB, Sandionigi A, Fortune F. 2022. Genital and oral microbiome and Behçet’s disease activity. Research Square. [Google Scholar]
- 24.Chu Y, Sun S, Huang Y, Gao Q, Xie X, Wang P, Li J, Liang L, He X, Jiang Y, Wang M, Yang J, Chen X, Zhou C, Zhao Y, Ding F, Zhang Y, Wu X, Bai X, Wu J, Wei X, Chen X, Yue Z, Fang X, Huang Q, Wang Z, Huang R. 2021. Metagenomic analysis revealed the potential role of gut microbiome in gout. NPJ Biofilms Microbiomes 7: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers MAM, Aronoff DM. 2016. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect 22: 178.e1–178.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thingholm LB, Rühlemann MC, Koch M, Fuqua B, Laucke G, Boehm R, Bang C, Franzosa EA, Hübenthal M, Rahnavard A, Frost F, Lloyd-Price J, Schirmer M, Lusis AJ, Vulpe CD, Lerch MM, Homuth G, Kacprowski T, Schmidt CO, Nöthlings U, Karlsen TH, Lieb W, Laudes M, Franke A, Huttenhower C. 2019. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe 26: 252–264.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosés C, Cuevas-Sierra A, Quintana S, Riezu-Boj JI, Martínez JA, Milagro FI, Barceló A. 2021. Gut microbiota bacterial species associated with Mediterranean diet-related food groups in a Northern Spanish population. Nutrients 13: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E, Kim DB, Park JY. 2016. Changes of mouse gut microbiota diversity and composition by modulating dietary protein and carbohydrate contents: a pilot study. Prev Nutr Food Sci 21: 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.