Abstract
MicroRNAs (miRNAs) are small non-coding RNA species involved in diverse physiological processes, including immunity. Accumulating evidence suggests that miRNA-induced gene silencing plays a significant role in the regulation of the intestinal immune system by the gut commensal microbiota. This review aims to provide an overview of the intestinal miRNA-mediated crosstalk between the gut microbiota and the host intestinal immune system. First, we describe the role of miRNAs in regulating the intestinal immune system. Then we describe the effect of the gut microbiota on intestinal miRNA expression. Subsequently, we describe the role of miRNAs in the modulation of the intestinal immune system by the gut microbiota. Finally, we describe the effect of host miRNAs on the gut microbiota. Although the entire picture of this complex crosstalk remains unclear, efforts to unravel it will contribute significantly to developing new strategies for preventing and treating intestinal immune disorders such as inflammatory bowel disease.
Keywords: microRNAs (miRNAs), gut microbiota, intestinal immune system
INTRODUCTION
MicroRNAs (miRNAs) are small (~22 nucleotides) single-stranded non-coding RNA species involved in diverse physiological processes by regulating gene expression [1, 2]. They are transcribed by RNA polymerase II as much longer sequences named primary miRNAs (pri-miRNAs) that consist of a hairpin with long single-stranded terminal regions [3]. Pri-miRNAs are trimmed by the RNase III enzyme Drosha and essential cofactor DiGeorge syndrome critical region 8 (DGCR8) into 70- to 90-nucleotide sequences, hairpin-shaped pre-miRNAs, in the cell nucleus [4]. Pre-miRNAs are then transported into the cytoplasm by the RanGTP-dependent nuclear transport receptor Exportin-5 [5, 6] and further processed to mature miRNAs by another RNase III enzyme, Dicer [7,8,9,10,11]. The mature miRNA is then incorporated into the RNA-induced silencing complex (RISC). It binds to the specific 3′ untranslated region (UTR) of the target mRNA, which results in the silencing of gene expression by two distinct mechanisms: mRNA degradation or translational repression [12]. Due to the partial complementarity of miRNAs with the 3′ UTR of their target mRNAs, a single miRNA can target multiple mRNAs, and multiple miRNAs can target a single mRNA. In addition, mammalian miRNAs are known to regulate approximately 30% of all protein-coding genes [13]. This complex regulatory network makes miRNAs important in regulating diverse physiological processes, including immunity.
Studies in the last two decades have revealed that the gut commensal microbiota modulates the development and function of the host intestinal immune system. Early studies demonstrated that germ-free (GF) mice showed extensive defects in the development of intestinal immunity and the integrity of the intestinal immune response, suggesting a close relationship between gut microbiota and the intestinal immune system. Subsequent studies revealed the cellular and molecular mechanisms by which metabolites and cellular constituents of gut microbes influence the intestinal immune system. We encourage readers to refer to the many other excellent reviews on this subject [14,15,16,17,18,19,20,21,22,23]. More recently, gene silencing by miRNAs has attracted attention to the mechanism for gut microbiota regulation of the intestinal immune system [24,25,26]. This review aimed to overview miRNA roles in the crosstalk between the gut commensal microbiota and intestinal immune system. The miRNAs involved in the immune function of intestinal epithelial cells are covered in other reviews [27, 28].
ROLE OF miRNAs IN THE REGULATION OF THE INTESTINAL IMMUNE SYSTEM
Innate immunity
The innate immune system involves macrophages, dendritic cells, granulocytes, and natural killer cells. It plays a role as the first line of defense by providing fast non-specific responses upon immunological stimulation. In addition, the innate immune system interacts with and controls the adaptive immune system. Previous studies have shown the roles of miRNAs in regulating the intestinal innate immune system.
miR-29 has been reported to play a role in regulating dendritic cell function in the intestine. Brain et al. [29] showed that miR-29, in response to intracellular microbial sensor nucleotide-binding oligomerization domain containing 2 (NOD2), downregulated interleukin (IL)-23 by targeting IL-12p40 mRNA directly and IL-23p19 mRNA indirectly in intestinal dendritic cells. Thus, miR-29 was suggested to suppress the proinflammatory mode of intestinal dendritic cells. Indeed, the authors also showed that experimental colitis was exacerbated in miR-29-deficient mice with elevated IL-23 in the intestine.
miR-146b is reportedly involved in regulating macrophage polarization in the intestine. Peng et al. [30] showed that IL-10 and lipopolysaccharide (LPS) induced miR-146b expression in macrophages and that the expression of miR-146b was impaired in IL-10-deficient macrophages. In addition, they showed that miR-146b and interferon regulatory factor 5 (IRF5) mRNA could occupy the same RISC and that the transfection of miR-146b mimic decreased LPS-induced IRF5 protein expression and M1 macrophage activation, suggesting targeting of IRF5 mRNA by miR-146b. Furthermore, miR-146b-deficient mice exhibited enhanced M1 macrophage polarization. From these findings, the authors proposed that the IL-10-miR-146b-IRF5 axis plays an essential role in the modulation of M1 macrophage activation in the intestine.
miR-223 has been shown to function as a regulator of intestinal macrophages and dendritic cells. Zhou et al. [31] demonstrated that miR-223-deficient mice had intestinal macrophages and dendritic cells with a solid proinflammatory phenotype. In addition, CCAAT/enhancer binding protein β (C/EBPβ) mRNA was identified as the target of miR-223. Thus, it was suggested that miR-223 suppresses the proinflammatory phenotype in intestinal macrophages and dendritic cells by directly targeting C/EBPβ mRNA. Neudecker et al. [32] reported that miR-223 deficient mice showed exacerbation of experimental colitis and activation of the nucleotide-binding domain leucine-rich-containing family pyrin domain-containing-3 (NLRP3) inflammasome. In addition, mice with a deletion of the miR-223 binding site in the NLRP3 3′ UTR also showed colitis exacerbation and NLRP3 activation. Furthermore, miR-223 mimic administration attenuated the colitis.
Other miRNAs, such as miR-20a, miR-24, miR-34a, miR-150, miR-155, and miR-183, reportedly play a role in regulating the function of innate immune cells, including neutrophils, innate lymphoid cells, and natural killer cells [28]. Further studies are needed to test whether these miRNAs function in the intestinal innate immune system.
Adaptive immunity
Recent studies have revealed that miRNA-induced gene silencing is vital in regulating the intestinal adaptive immune system.
Takahashi et al. [33] showed that miR-10a, which is highly expressed in regulatory T (Treg) cells, is induced by retinoic acid and transforming growth factor-β (TGF-β) and attenuates the conversion of inducible Treg cells into follicular helper T cells by targeting B cell leukemia/lymphoma (Bcl) 6 mRNA and nuclear receptor co-repressor 2 (Ncor2) mRNA in the small intestinal Peyer’s patches. They also showed that miR-10a limited the differentiation of helper T (Th)17 cells; thus, miR-10a is likely to have an anti-inflammatory function. A more recent study by Yang et al. [34] showed contrasting findings: miR-10a suppressed IL-10 production in the intestinal CD4+ T cells by targeting the Prdm1 gene, which encodes transcription factor Blimp1, and mice with deficient miR-10a in CD4+ T cells were more resistant to intestinal inflammation induced by dextran sulfate sodium (DSS). In the large intestine, Wang et al. [35] showed that miR-34a targets IL-6 receptor mRNA and IL-23 receptor mRNA to suppress Th17 cell differentiation and proliferation and target chemokine (C-C motif) ligand 22 (CCL22) mRNA to inhibit Th17 recruitment to the epithelium. By employing inflammatory bowel disease (IBD) model mice exhibiting systemic elevation of tumor necrosis factor-α (TNF-α), Sanctuary et al. [36] observed that miR-106a increased in response to TNF-α to reduce Treg cell function by targeting IL-10 mRNA. Conversely, miR-106a deficiency promoted Treg cell induction and IL-10 production and attenuated intestinal inflammation. Ge et al. [37] demonstrated that miR-125a is downregulated in the colon of IBD patients, which is associated with suppressing proinflammatory cytokine production by targeting transcription factor E26 avian leukemia oncogene 1, 5′ domain (ETS-1) mRNA in CD4+ T cells. The authors also showed that miR-125a deficiency exacerbated trinitrobenzene sulphonic acid-induced colitis in mice. The role of miR-155 in T-cell response with regard to intestinal inflammation is controversial. Das et al. [38] demonstrated that miR-155 is involved in TGF-β-induced suppression of intestinal T-cell activation, such as IL-2 and interferon-γ (IFN-γ) production, by targeting IL-2-inducible T-cell kinase mRNA. In contrast, Chao et al. [39] observed that mice with overexpression of miR-155 in Treg cells exhibit spontaneous autoimmunity and exacerbation of DSS-induced colitis. In addition, miR-155 targeted the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) mRNA in Treg cells to suppress the regulatory function of Treg cells. Concerning miR-221 and miR-222, Mikami et al. [40] demonstrated an essential role of these miRNAs in regulating intestinal Th17 cells. They found that miR-221 and miR-222 targeted MAF bZIP transcription factor (MAF) mRNA and IL-23 receptor mRNA to suppress intestinal Th17 cell expansion in response to IL-23. T cell-specific loss of miR-221 and miR-222 exacerbated DSS-induced colitis in mice. Thus, it was suggested that miR-221 and miR-222 act as negative feedback regulators downstream of IL-23 to modulate proinflammatory Th17 cell response in the intestine.
In addition to T cells, the function of intestinal B cells also involves miRNA silencing. Casali et al. [41] showed that miR-146a targets small mothers against decapentaplegic (Smad)2, Smad3, and Smad4 mRNA, which results in reduced class-switch recombination to immunoglobulin A (IgA). In miR-146a-deficient mice, increased IgA+ B cells were observed in the intestine.
Although previous studies have proposed that some miRNAs are involved in the regulation of intestinal immunity, the molecular mechanisms, i.e., their target mRNAs, remain to be determined under intestinal inflammation conditions in particular. In addition, several controversial observations concerning the function of each miRNA have been reported, as described above, which are probably due to the complex regulatory network of miRNA-induced gene silencing. Nevertheless, identification of miRNA-induced gene silencing involved in the regulation of intestinal immunity would lead to the development of novel therapeutic strategies for IBD.
EFFECT OF THE GUT MICROBIOTA ON INTESTINAL miRNA EXPRESSION
The gut commensal microbiota modulates the development and function of the host intestinal immune system. Given that miRNAs play a significant role in regulating the intestinal immune system, as described above, the influence of the gut microbiota on the intestinal immune system may be mediated by intestinal miRNAs. If this is the case, the expression profile of miRNAs in the intestine should be influenced by the gut microbiota. Indeed, some previous studies have investigated whether the gut microbiota affects the expression of miRNAs in the intestine.
Dalmasso et al. [42] compared miRNA expression profiles in ileal and colonic mucosal tissues between GF mice and GF mice colonized with microbiota isolated from specific pathogen-free (SPF) mice by using a microarray that contained 656 miRNAs. The authors found that one (miR-298) and three (miR-128, miR-200c-5p, miR-342-5p) miRNAs were upregulated by colonization in the ileum and colon, respectively. In addition, five (miR-465c-5p, miR-466d-3p, miR-466d-5p, miR-665, miR-683) miRNAs were downregulated by colonization in the colon. By employing two approaches, i.e., in silico prediction of target genes and a DNA microarray, in parallel, they found that several genes were regulated by miRNAs with altered expression due to colonization. For instance, the mRNA of an ATP-binding cassette transporter, ATP-binding cassette sub-family C member 3 (ABCC3), was identified as a target of miR-665 in the colon. Thus, the gut commensal microbiota could promote the expression of ABCC3 by downregulating miR-665 in the colon.
A similar study was reported by Singh et al. [43]. The authors compared miRNA expression profiles in cecal tissue between GF mice and conventionally raised (CV) mice by multiplex reverse transcription polymerase chain reaction (RT-PCR) assay, which can analyze 585 miRNAs. They found that, among the 334 miRNAs detected, 16 miRNAs were differentially expressed between GF and CV mice. Through in silico target prediction and network analysis, they identified 2,755 putative target genes of differently expressed miRNAs and found 34 genes related to intestinal barrier function. From these findings, they proposed that miRNA-induced gene silencing contributes to the regulation of intestinal barrier function by the gut commensal microbiota.
Aoki et al. [44] also compared miRNA expression profiles in the whole colon between GF and SPF mice. Their microarray analysis identified 48 miRNAs as differently expressed miRNAs. Upon analyzing putative target genes and their functions, the authors suggested that many of the predicted target genes were related to GTPases and nerves.
The miRNAs that were differently expressed between GF mice and CV or SPF mice were not necessarily similar among these three studies. This may be due to differences in the mouse strains and tissues examined. In addition, considering that the gut microbiota composition differs depending on the breeder, it is likely that the effects of the gut microbiota on miRNA expression differ between different studies.
Unfortunately, these studies analyzed miRNAs isolated from whole or mucosal tissues of the intestine. The intestinal tissues are composed of a heterogeneous population of cells. To elucidate the roles of miRNAs in the gut microbiota regulation of intestinal function, including immune response, it is necessary to analyze miRNA expression profiles in specific intestinal cell types in the intestine. For instance, Nakata et al. [45] isolated epithelial cells from the small and large intestines of GF and CV mice and compared the miRNA expression profiles. The authors identified miR-21-5p as highly expressed miRNA in the epithelial cells of both the small and large intestines of CV mice as compared with GF mice. They also found that miR-21-5p upregulated ADP ribosylation factor 4 (ARF4) by silencing known targets phosphatase and tensin homolog (PTEN) mRNA and programmed cell death 4 (PDCD4) mRNA, which resulted in increased epithelial permeability. Thus, investigating miRNAs in specific cell types could reveal more precisely the roles of miRNAs in regulating intestinal function. Recent progress in cell sorting and single-cell RNA sequencing should enable us to uncover the effect of the gut microbiota on miRNA expression in specific cell types in the intestine.
ROLE OF miRNAs IN THE MODULATION OF THE INTESTINAL IMMUNE SYSTEM BY THE GUT MICROBIOTA
The gut commensal microbiota has been shown to modulate the development and function of the host intestinal immune system. In recent years, gene silencing by miRNAs has attracted attention to the mechanism for gut microbiota regulation of the intestinal immune system. However, studies regarding this topic have just begun.
Xue et al. [46] examined the expression level of miR-10a in epithelial cells, lamina propria dendritic cells, T cells, and B cells isolated from GF and SPF mice. They found that epithelial and dendritic cells in SPF mice expressed lower levels of miR-10a than GF mice, suggesting negative regulation of miR-10a by the gut microbiota. An in silico analysis predicted IL-12/IL-23p40 mRNA as a target of miR-10a, and this was experimentally demonstrated by employing transfection with miR-10a in murine macrophage cell line RAW264.7. They also found that ligands for Toll-like receptor (TLR)1/2, TLR4, TLR5, TLR9, and NOD2 downregulated the expression of miR-10a in dendritic cells, suggesting that gut microbiota regulation of miR-10a in intestinal dendritic cells is mediated by the TLR/myeloid differentiation primary response gene 88 (MyD88) signaling pathway. Considering that IL-12/IL-23p40 is a crucial molecule for innate immune responses to commensal bacteria [47], this study demonstrated the significant role of miR-10a in modulation of the intestinal innate immune system by the gut microbiota.
NOD2, an intracellular microbial sensor, has also been identified as a target of miR-10a in intestinal dendritic cells. Wu et al. [48] observed that the expression of miR-10a in the inflamed mucosal tissues of IBD patients was decreased, while that of NOD2, as well as IL-12/IL-23p40, was increased. They also found that the expression of miR-10a was reduced by supplementation with Escherichia coli and various TLR ligands in human monocyte-derived dendritic cells in vitro. Furthermore, by employing transfection with miR-10a in human monocyte-derived dendritic cells, NOD2 mRNA was identified as a target of miR-10a. As described above, miR-10a reportedly attenuates the conversion of inducible Treg cells into follicular helper T cells and limits the differentiation of Th17 cells [33]. Wu et al. [48] thus examined whether miR-10a is involved in the phenotypic changes in helper T cells in IBD patients. They found that overexpression of miR-10a inhibited the response of Th1 and Th17 cells in CD4+ T cells isolated from IBD patients in vitro. Therefore, it was revealed that miR-10a plays a significant role in regulating both innate and adaptive immunities in response to the gut microbiota.
Xue et al. [49] investigated the expression of miR-107 in experimental colitis mice since miR-107 reportedly regulates the innate immune response to microbes [50]. They observed reduced expression levels of miR-10a in CD11c+ myeloid cells, i.e., dendritic cells and macrophages, and epithelial cells in the inflamed tissues of colitis mice. They also found that the intestinal expression of miR-107 was higher in GF mice than in SPF mice. Furthermore, IL-23p19 mRNA was identified as a target of miR-107. Because IL-23p19 is a critical molecule in the regulation of innate immunity in response to the gut microbiota [51], it was suggested that miR-107, by targeting IL-23p19 mRNA, plays a significant role in regulating innate intestinal immunity in response to the gut microbiota. In other words, under intestinal inflammatory conditions, the gut microbiota may reduce miR-10a expression in intestinal dendritic cells and macrophages. This upregulates IL-23p19, which in turn results in further enhancement of the innate immune response.
Recently, we compared the expression profiles of miRNAs and mRNAs in lamina propria leukocytes (LPLs) isolated from the large intestines of GF and SPF mice [52]. We employed microarray analyses with mouse miRNA and mRNA oligo chips containing 1,900 and 23,474 probes, respectively. We found that the expression levels of miR-148a-3p, miR-192-5p, miR-194-5p, and miR-200 family members, i.e., miR-141-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, and miR-429-3p, were higher in SPF mice than in GF mice. A combination of in silico and gene expression analyses suggested that BCL11B, ETS-1, guanylate binding protein 7 (GBP7), signal transducer and activator of transcription 5B (STAT5B), and zinc finger E-box binding homeobox 1 (ZEB1) are targets of miR-200 family members. By western blot analysis, we found that the protein expression of BCL11B and ETS-1, but not ZEB1, in large intestinal LPLs was significantly lower in SPF mice than in GF mice. Because BCL11B, ETS-1, and ZEB1 are transcription factors involved in regulating IL-2 production in T cells [53,54,55], we measured IL-2 production in cultured LPLs isolated from the large intestine. Upon stimulation with phorbol 12-myristate 13-acetate and ionomycin, IL-2 production was lower in the LPLs isolated from SPF mice than in those isolated from GF mice. From these findings, we suggested that miR-200 family members are involved in gut microbiota regulation of IL-2 production in large intestinal LPLs by targeting BCL11b and ETS-1.
As described, comparisons between GF mice and CV or SPF mice suggested that the existence of the gut commensal microbiota influences intestinal miRNA expression [42,43,44,45, 52]. We next examined whether a change in the composition of the gut commensal microbiota alters the expression of miRNAs [56]. It is well known that consuming indigestible oligosaccharides, including fructooligosaccharides (FOS), influences the gut microbiota composition. Indeed, dietary supplementation with 1-kestose (KES), the smallest constituent of FOS, altered the gut microbiota composition in mice, rats, dogs, and humans [57,58,59,60]. In particular, KES supplementation increased the population of bifidobacteria. We therefore tested whether the consumption of KES influences the miRNA expression profiles in large intestinal LPLs of mice. By microarray analysis followed by RT-PCR validation, we found that KES consumption increased the levels of miR-205-5p, miR-200 family members, and miR-192/215 family members, i.e., miR-192-5p, miR-194-5p, and miR-215-5p. We also observed that intragastric administration of Bifidobacterium pseudolongum isolated from mouse feces increased the levels of miR-182-5p, miR-194-5p, and miR-200a-3p and tended to increase the levels of miR-200b-3p, miR-215-5p, and miR-429-3p. These results suggest that dietary KES influences miRNA expression in large intestinal LPLs, which may be associated with an increased population of B. pseudolongum. Considering that diet is a major determinant of gut microbiota composition and function [61, 62], it is possible that miRNA-induced gene silencing may contribute to dietary modulation of intestinal immune function.
The mechanisms by which gut microbes influence the expression of intestinal miRNAs remain unclear. Regarding our findings suggesting that gut commensal microbes promote the expression of miR-200 family members in large intestinal LPLs [52], our initial prediction was that gut microbial metabolites and/or cellular constituents might directly activate these miRNAs. However, our preliminary experiments showed that supplementation with short-chain fatty acids (SCFAs), gut microbial fermentation products of indigestible carbohydrates, failed to alter the levels of miR-200 family members in cultured LPLs isolated from the large intestines of SPF mice [52]. In addition, LPLs isolated from the large intestines of GF mice were cultured with fecal extracts prepared from the cecal contents of GF and SPF mice. The levels of miR-200 family members were unaffected not only by SCFAs but also by fecal extracts. We therefore suspect that an indirect action of gut commensals, such as through epithelial cells, may alter the expression of miR-200 family members in large intestinal LPLs. Further studies are needed to elucidate the cellular and molecular mechanisms by which gut commensal microbes influence intestinal miRNA expression.
EFFECT OF HOST miRNAs ON THE GUT MICROBIOTA
The host animal contributes to the maintenance of gut microbiota homeostasis through antimicrobial peptides and IgA antibodies secreted into the intestinal lumen. Intriguingly, Liu et al. [63] showed the possibility that host-derived miRNAs might alter gut microbiota composition and function. By employing intestinal epithelial cell-specific Dicer-deficient mice, the authors demonstrated that miRNAs in the intestinal lumen are derived from intestinal epithelial cells and that miRNA deficiency in the lumen alters the gut microbiota composition. They also showed that host-derived miRNAs are incorporated into gut microbes, i.e., E. coli and Fusobacterium nucleatum, where they regulate gene transcription and affect the growth of the microbes.
Another study by Liu et al. showed more specific miRNA regulation of gut microbes, which is clinically relevant [64]. Because the authors previously reported that multiple sclerosis (MS) patients have an altered structure of the gut microbiota [65], they performed fecal transplantation from experimental autoimmune encephalomyelitis (EAE) mice, a model of human MS, into naïve mice before induction of EAE and found that fecal transplantation ameliorated the symptoms of EAE in the recipient mice [64]. Taking into consideration that host-derived miRNAs in the intestinal lumen could influence the gut microbiota composition [63], the authors administered miRNAs isolated from EAE mice to naïve mice before induction of EAE. They found that miRNA administration ameliorated the disease in the recipient mice and identified miR-30d-5p as a responsible miRNA. In addition, they discovered that miR-30d-5p regulates the expression of the β-galactosidase gene in Akkermansia muciniphila, which results in the expansion of this microbe in the gut. Finally, the authors showed that A. muciniphila suppresses EAE symptoms by increasing splenic Treg cells.
Likewise, Santos et al. also reported the modulation of specific gut microbes by host-derived miRNA [66]. They previously observed that miR-21-deficient mice exhibited protection against bile duct ligation (BDL)-induced liver injury [67]. The authors then showed that miR-21-deficient mice had an altered gut microbiota structure characterized by an increased abundance of Lactobacillus spp. and that supplementation of miR-21 promoted the growth of cultured Limosilactobacillus reuteri [66]. They also found that administration of L. reuteri ameliorated BDL-induced liver injury in mice. Thus, it was suggested that host-derived miR-21 suppresses BDL-induced liver injury by increasing the abundance of intestinal Lactobacillus spp.
CONCLUDING REMARKS
As described, the gut commensal microbiota is a regulator of the intestinal immune system. In addition, miRNA-induced gene silencing is also involved in regulating the intestinal immune system. Furthermore, the gut commensal microbiota influences intestinal miRNA expression. Some evidence suggests that miRNA-induced gene silencing plays a significant role in the regulation of the intestinal immune system by the gut microbiota. On the other hand, host-derived miRNAs secreted into the intestinal lumen could influence gut microbiota composition and function. Therefore, host miRNAs mediate the bidirectional gut commensal microbiota-host axis. In addition, it is worthwhile to note that diet may affect intestinal miRNA expression and release in a direct and/or gut microbiota-mediated indirect manner. In fact, Tarallo et al. reported that fecal miRNA profiles are associated with specific diets [68]. The authors investigated fecal miRNA profiles by small RNA-seq in fecal samples obtained from vegans, vegetarians, and omnivores and found 49 miRNAs differentially expressed among different dietary habit subjects. Hence, diet would be an environmental factor that affects intestinal miRNA-mediated crosstalk between the gut microbiota and host (Fig. 1). However, further studies are required to elucidate the entire picture of this complex crosstalk. For instance, the target genes of miRNAs that are assumed to be involved in this crosstalk and the mechanisms by which gut microbes influence the expression of intestinal miRNAs should be clarified. In addition, how host-derived miRNAs regulate bacterial gene expression remains to be elucidated. Nevertheless, efforts to unravel this crosstalk will contribute significantly to developing new strategies for preventing and treating intestinal immune disorders such as IBD.
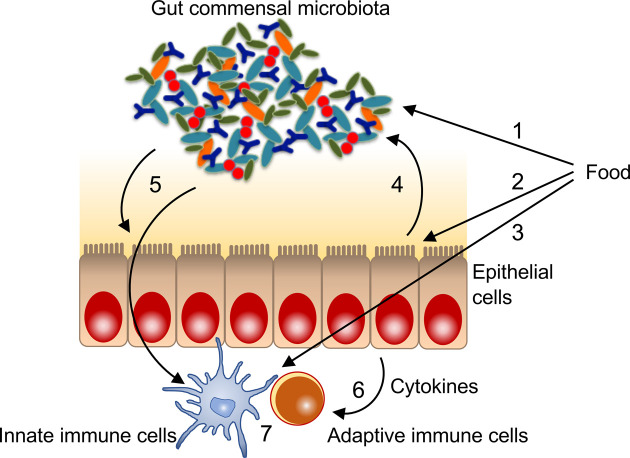
Fig. 1.
A putative bidirectional gut microbiota-intestinal immunity axis mediated by microRNAs (miRNAs). 1. Dietary modulation of gut microbiota composition and function, 2. dietary modulation of miRNA expression in intestinal epithelial cells, 3. dietary modulation of miRNA expression in intestinal immune cells, 4. host-derived miRNA modulation of gut microbiota composition and function, 5. gut microbiota modulation of miRNA expression in intestinal epithelial cells and immune cells, 6. cytokine-mediated modulation of miRNA expression in immune cells, 7. miRNA-induced gene silencing in immune cells.
CONFLICT OF INTEREST
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- 1.Ambros V. 2004. The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- 5.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. 2004. Nuclear export of microRNA precursors. Science 303: 95–98. [DOI] [PubMed] [Google Scholar]
- 6.Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- 8.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- 9.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- 10.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight SW, Bass BL. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian MR, Sonenberg N. 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19: 586–593. [DOI] [PubMed] [Google Scholar]
- 13.Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114. [DOI] [PubMed] [Google Scholar]
- 14.Cerf-Bensussan N, Gaboriau-Routhiau V. 2010. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol 10: 735–744. [DOI] [PubMed] [Google Scholar]
- 15.Honda K, Littman DR. 2016. The microbiota in adaptive immune homeostasis and disease. Nature 535: 75–84. [DOI] [PubMed] [Google Scholar]
- 16.Hooper LV, Macpherson AJ. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10: 159–169. [DOI] [PubMed] [Google Scholar]
- 17.Ivanov II, Honda K. 2012. Intestinal commensal microbes as immune modulators. Cell Host Microbe 12: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee WJ, Hase K. 2014. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol 10: 416–424. [DOI] [PubMed] [Google Scholar]
- 19.Ohno H. 2020. The impact of metabolites derived from the gut microbiota on immune regulation and diseases. Int Immunol 32: 629–636. [DOI] [PubMed] [Google Scholar]
- 20.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi D, Kimura S, Hase K. 2021. Intestinal immunity: to be, or not to be, induced? That is the question. Int Immunol 33: 755–759. [DOI] [PubMed] [Google Scholar]
- 22.Tanoue T, Atarashi K, Honda K. 2016. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol 16: 295–309. [DOI] [PubMed] [Google Scholar]
- 23.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 24.Bi K, Zhang X, Chen W, Diao H. 2020. MicroRNAs regulate intestinal immunity and gut microbiota for gastrointestinal health: a comprehensive review. Genes (Basel) 11: 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masotti A. 2012. Interplays between gut microbiota and gene expression regulation by miRNAs. Front Cell Infect Microbiol 2: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runtsch MC, Round JL, O’Connell RM. 2014. MicroRNAs and the regulation of intestinal homeostasis. Front Genet 5: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belcheva A. 2017. MicroRNAs at the epicenter of intestinal homeostasis. BioEssays 39: 1600200. [DOI] [PubMed] [Google Scholar]
- 28.Dhuppar S, Murugaiyan G. 2022. miRNA effects on gut homeostasis: therapeutic implications for inflammatory bowel disease. Trends Immunol 43: 917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brain O, Owens BM, Pichulik T, Allan P, Khatamzas E, Leslie A, Steevels T, Sharma S, Mayer A, Catuneanu AM, Morton V, Sun MY, Jewell D, Coccia M, Harrison O, Maloy K, Schönefeldt S, Bornschein S, Liston A, Simmons A. 2013. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity 39: 521–536. [DOI] [PubMed] [Google Scholar]
- 30.Peng L, Zhang H, Hao Y, Xu F, Yang J, Zhang R, Lu G, Zheng Z, Cui M, Qi CF, Chen C, Wang J, Hu Y, Wang D, Pierce S, Li L, Xiong H. 2016. Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine 14: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou H, Xiao J, Wu N, Liu C, Xu J, Liu F, Wu L. 2015. MicroRNA-223 regulates the differentiation and function of intestinal dendritic cells and macrophages by targeting C/EBPβ. Cell Rep 13: 1149–1160. [DOI] [PubMed] [Google Scholar]
- 32.Neudecker V, Haneklaus M, Jensen O, Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS, Gerich ME, Mack M, Robertson AAB, Cooper MA, Furuta GT, Dinarello CA, O’Neill LA, Eltzschig HK, Masters SL, McNamee EN. 2017. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med 214: 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciumè G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O’Shea JJ. 2012. TGF-β and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol 13: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang W, Chen L, Xu L, Bilotta AJ, Yao S, Liu Z, Cong Y. 2021. MicroRNA-10a negatively regulates CD4+ T cell IL-10 production through suppression of Blimp1. J Immunol 207: 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Wang E, Wang Y, Mines R, Xiang K, Sun Z, Zhou G, Chen KY, Rakhilin N, Chao S, Ye G, Wu Z, Yan H, Shen H, Everitt J, Bu P, Shen X. 2018. miR-34a is a microRNA safeguard for Citrobacter-induced inflammatory colon oncogenesis. eLife 7: e39479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanctuary MR, Huang RH, Jones AA, Luck ME, Aherne CM, Jedlicka P, de Zoeten EF, Collins CB. 2019. miR-106a deficiency attenuates inflammation in murine IBD models. Mucosal Immunol 12: 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge Y, Sun M, Wu W, Ma C, Zhang C, He C, Li J, Cong Y, Zhang D, Liu Z. 2019. MicroRNA-125a suppresses intestinal mucosal inflammation through targeting ETS-1 in patients with inflammatory bowel diseases. J Autoimmun 101: 109–120. [DOI] [PubMed] [Google Scholar]
- 38.Das LM, Torres-Castillo MD, Gill T, Levine AD. 2013. TGF-β conditions intestinal T cells to express increased levels of miR-155, associated with down-regulation of IL-2 and itk mRNA. Mucosal Immunol 6: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao G, Li X, Ji Y, Zhu Y, Li N, Zhang N, Feng Z, Niu M. 2019. MiR-155 controls follicular Treg cell-mediated humoral autoimmune intestinal injury by inhibiting CTLA-4 expression. Int Immunopharmacol 71: 267–276. [DOI] [PubMed] [Google Scholar]
- 40.Mikami Y, Philips RL, Sciumè G, Petermann F, Meylan F, Nagashima H, Yao C, Davis FP, Brooks SR, Sun HW, Takahashi H, Poholek AC, Shih HY, Afzali B, Muljo SA, Hafner M, Kanno Y, O’Shea JJ. 2021. MicroRNA-221 and -222 modulate intestinal inflammatory Th17 cell response as negative feedback regulators downstream of interleukin-23. Immunity 54: 514–525.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casali P, Li S, Morales G, Daw CC, Chupp DP, Fisher AD, Zan H. 2021. Epigenetic modulation of class-switch DNA recombination to IgA by miR-146a through downregulation of Smad2, Smad3 and Smad4. Front Immunol 12: 761450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, Sitaraman SV, Merlin D. 2011. Microbiota modulate host gene expression via microRNAs. PLoS One 6: e19293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh N, Shirdel EA, Waldron L, Zhang RH, Jurisica I, Comelli EM. 2012. The murine caecal microRNA signature depends on the presence of the endogenous microbiota. Int J Biol Sci 8: 171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoki A, Aoki R, Yatagai M, Kawasumi T. 2021. Comparative analysis of microRNA expression profiles in the colons of specific pathogen-free mice and germ-free mice. Biosci Biotechnol Biochem 85: 1869–1872. [DOI] [PubMed] [Google Scholar]
- 45.Nakata K, Sugi Y, Narabayashi H, Kobayakawa T, Nakanishi Y, Tsuda M, Hosono A, Kaminogawa S, Hanazawa S, Takahashi K. 2017. Commensal microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4. J Biol Chem 292: 15426–15433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue X, Feng T, Yao S, Wolf KJ, Liu CG, Liu X, Elson CO, Cong Y. 2011. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J Immunol 187: 5879–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langrish CL, McKenzie BS, Wilson NJ, de Waal Malefyt R, Kastelein RA, Cua DJ. 2004. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev 202: 96–105. [DOI] [PubMed] [Google Scholar]
- 48.Wu W, He C, Liu C, Cao AT, Xue X, Evans-Marin HL, Sun M, Fang L, Yao S, Pinchuk IV, Powell DW, Liu Z, Cong Y. 2015. miR-10a inhibits dendritic cell activation and Th1/Th17 cell immune responses in IBD. Gut 64: 1755–1764. [DOI] [PubMed] [Google Scholar]
- 49.Xue X, Cao AT, Cao X, Yao S, Carlsen ED, Soong L, Liu CG, Liu X, Liu Z, Duck LW, Elson CO, Cong Y. 2014. Downregulation of microRNA-107 in intestinal CD11c(+) myeloid cells in response to microbiota and proinflammatory cytokines increases IL-23p19 expression. Eur J Immunol 44: 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hennessy EJ, Sheedy FJ, Santamaria D, Barbacid M, O’Neill LA. 2011. Toll-like receptor-4 (TLR4) down-regulates microRNA-107, increasing macrophage adhesion via cyclin-dependent kinase 6. J Biol Chem 286: 25531–25539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trinchieri G, Pflanz S, Kastelein RA. 2003. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses. Immunity 19: 641–644. [DOI] [PubMed] [Google Scholar]
- 52.Ohsaka F, Karatsu Y, Kadota Y, Tochio T, Takemura N, Sonoyama K. 2021. Gut commensals suppress interleukin-2 production through microRNA-200/BCL11B and microRNA-200/ETS-1 axes in lamina propria leukocytes of murine large intestine. Biochem Biophys Res Commun 534: 808–814. [DOI] [PubMed] [Google Scholar]
- 53.Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, Avram D. 2006. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood 108: 2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsao HW, Tai TS, Tseng W, Chang HH, Grenningloh R, Miaw SC, Ho IC. 2013. Ets-1 facilitates nuclear entry of NFAT proteins and their recruitment to the IL-2 promoter. Proc Natl Acad Sci USA 110: 15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Lee S, Teh CE, Bunting K, Ma L, Shannon MF. 2009. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int Immunol 21: 227–235. [DOI] [PubMed] [Google Scholar]
- 56.Ohsaka F, Honma D, Kadota Y, Tochio T, Sonoyama K. 2023. Consumption of 1-kestose upregulates microRNA-200 and -192/215 families in lamina propria leukocytes of the murine large intestine. J Nutr Sci Vitaminol (Tokyo) 69: 150–154. [DOI] [PubMed] [Google Scholar]
- 57.Tatsuoka M, Osaki Y, Ohsaka F, Tsuruta T, Kadota Y, Tochio T, Hino S, Morita T, Sonoyama K. 2022. Consumption of indigestible saccharides and administration of Bifidobacterium pseudolongum reduce mucosal serotonin in murine colonic mucosa. Br J Nutr 127: 513–525. [DOI] [PubMed] [Google Scholar]
- 58.Tochio T, Kitaura Y, Nakamura S, Sugawa C, Takahashi M, Endo A, Shimomura Y. 2016. An alteration in the cecal microbiota composition by feeding of 1-kestose results in a marked increase in the cecal butyrate content in rats. PLoS One 11: e0166850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ide K, Shinohara M, Yamagishi S, Endo A, Nishifuji K, Tochio T. 2020. Kestose supplementation exerts bifidogenic effect within fecal microbiota and increases fecal butyrate concentration in dogs. J Vet Med Sci 82: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tochio T, Kadota Y, Tanaka T, Koga Y. 2018. 1-Kestose, the smallest fructooligosaccharide component, which efficiently stimulates Faecalibacterium prausnitzii as well as bifidobacteria in humans. Foods 7: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, Wang J, Imhann F, Brandsma E, Jankipersadsing SA, Joossens M, Cenit MC, Deelen P, Swertz MA, Weersma RK, Feskens EJ, Netea MG, Gevers D, Jonkers D, Franke L, Aulchenko YS, Huttenhower C, Raes J, Hofker MH, Xavier RJ, Wijmenga C, Fu J, LifeLines cohort study. 2016. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL. 2016. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19: 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu S, Rezende RM, Moreira TG, Tankou SK, Cox LM, Wu M, Song A, Dhang FH, Wei Z, Costamagna G, Weiner HL. 2019. Oral administration of miR-30d from feces of MS patients suppresses MS-like symptoms in mice by expanding Akkermansia muciniphila. Cell Host Microbe 26: 779–794.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, Cook S, Tankou S, Stuart F, Melo K, Nejad P, Smith K, Topçuolu BD, Holden J, Kivisäkk P, Chitnis T, De Jager PL, Quintana FJ, Gerber GK, Bry L, Weiner HL. 2016. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun 7: 12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santos AA, Afonso MB, Ramiro RS, Pires D, Pimentel M, Castro RE, Rodrigues CMP. 2020. Host miRNA-21 promotes liver dysfunction by targeting small intestinal Lactobacillus in mice. Gut Microbes 12: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afonso MB, Rodrigues PM, Simão AL, Gaspar MM, Carvalho T, Borralho P, Bañales JM, Castro RE, Rodrigues CMP. 2018. miRNA-21 ablation protects against liver injury and necroptosis in cholestasis. Cell Death Differ 25: 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tarallo S, Ferrero G, De Filippis F, Francavilla A, Pasolli E, Panero V, Cordero F, Segata N, Grioni S, Pensa RG, Pardini B, Ercolini D, Naccarati A. 2022. Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals. Gut 71: 1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]