Abstract
Background & Aims
Circadian rhythms play significant roles in immune responses, and many inflammatory processes in liver diseases are associated with malfunctioning molecular clocks. However, the significance of the circadian clock in autoimmune hepatitis (AIH), which is characterised by immune-mediated hepatocyte destruction and extensive inflammatory cytokine production, remains unclear.
Methods
We tested the difference in susceptibility to the immune-mediated liver injury induced by concanavalin A (ConA) at various time points throughout a day in mice and analysed the effects of global, hepatocyte, or myeloid cell deletion of the core clock gene, Bmal1 (basic helix–loop–helix ARNT-like 1), on liver injury and inflammatory responses. Multiple molecular biology techniques and mice with macrophage-specific knockdown of Junb, a Bmal1 target gene, were used to investigate the involvement of Junb in the circadian control of ConA-induced hepatitis.
Results
The susceptibility to ConA-induced liver injury is highly dependent on the timing of ConA injection. The treatment at Zeitgeber time 0 (lights on) triggers the highest mortality as well as the severest liver injury and inflammatory responses. Further study revealed that this timing effect was driven by macrophage, but not hepatocyte, Bmal1. Mechanistically, Bmal1 controls the diurnal variation of ConA-induced hepatitis by directly regulating the circadian transcription of Junb and promoting M1 macrophage activation. Inhibition of Junb in macrophages blunts the administration time-dependent effect of ConA and attenuates liver injury. Moreover, we demonstrated that Junb promotes macrophage inflammation by regulating AKT and extracellular signal-regulated kinase (ERK) signalling pathways.
Conclusions
Our findings uncover a critical role of the Bmal1–Junb–AKT/ERK axis in the circadian control of ConA-induced hepatitis and provide new insights into the prevention and treatment of AIH.
Impact and Implications
This study unveils a critical role of the Bmal1–Junb–AKT/ERK axis in the circadian control of ConA-induced liver injury, providing new insights into the prevention and treatment of immune-mediated hepatitis, including autoimmune hepatitis (AIH). The findings have scientific implications as they enhance our understanding of the circadian regulation of immune responses in liver diseases. Furthermore, clinically, this research offers opportunities for optimising treatment strategies in immune-mediated hepatitis by considering the timing of therapeutic interventions.
Keywords: Circadian rhythm, Bmal1, Junb, Macrophage, Autoimmune hepatitis
Graphical abstract
Highlights
-
•
The susceptibility to ConA-induced liver injury is highly dependent on the timing of ConA administration.
-
•
Depletion of Bmal1 in macrophages eliminates the administration time-dependent effect and protects against ConA-induced acute liver injury.
-
•
Junb is a direct target gene of Bmal1 in macrophages.
-
•
Bmal1 regulates M1 macrophage activation via Junb–AKT/ERK signalling pathways and controls immune-mediated hepatitis in a circadian manner.
Introduction
The circadian clock is a daily timing system that coordinates the internal physiological processes with periodic environmental stimuli.1 Although circadian rhythms were initially generated by the Earth’s rotation about 2.5 billion years ago,2,3 organisms evolved self-sustained mechanisms, letting them persist even in the absence of daily environmental changes.4 At the molecular level, these rhythms are typically controlled by a series of clock components, forming a transcriptional autoregulatory feedback loop.5 The core mammalian ‘clock genes’ mainly include the activation factors Bmal1 (basic helix–loop–helix ARNT-like 1) and Clock and inhibitors Periods and Cryptochromes, although Bmal1 is the only one whose sole deletion results in a complete loss of many physiological and behavioural rhythms in mice.6
The transcriptional autoregulatory feedback loop exists in almost every cell in mammals. In terms of the number of oscillating genes, the liver is topping the list of most major tissues in mice.7,8 To date, intensive studies have reported the essential roles of the circadian clock in maintaining liver homoeostasis, including the regulation of energy metabolism, inflammatory responses, and virus infection.9 Disruption of the clock system has been associated with many liver diseases, such as fatty liver, cirrhosis, and cancer.10,11 Interestingly, the replication of the HBV and the consequent viral hepatitis are also controlled by the circadian clock. Among the core clock components, REV-ERB regulates HBV and HDV particle entry, and Bmal1 directly binds to HBV DNA, increasing the activity of the HBV promoter activity and thus promoting the production of viral particles in the liver.12,13 On the contrary, Bmal1 depletion inhibits the replication of the HCV via disturbing the lipid signalling pathways.14
Autoimmune hepatitis (AIH), characterised by immune-mediated destruction of the liver and the production of cytokines, is another progressive inflammatory hepatopathy that could lead to hepatic cirrhosis or tumour.15,16 Different from other common liver diseases described above that were well known to be implicated with circadian rhythms, AIH was not investigated specifically in the aspect of biological clocks, although inflammatory cytokines are under circadian control and various immune responses are dependent on the time of day.17,18
Concanavalin A (ConA)-induced liver injury is a well-established experimental model for acute immune-mediated hepatitis, including AIH. In the present study, we aimed to clarify whether the hepatopathy of this model and cytokine production are regulated by the circadian clock. Moreover, we examined the role of Bmal1, particularly the hepatocyte and macrophage Bmal1, in the diurnal variation of ConA-induced liver injury, and provided a potential mechanism by which Bmal1 regulates acute immune-mediated hepatitis.
Materials and methods
Animals
Wild-type (WT) C57BL/6 mice were purchased from Liaoning Changsheng Biotechnology (Benxi, Liaoning, China). Adult-life Bmal1 global knockout mice (Bmal1fl/fl-EsrCre+ mice treated with tamoxifen, iKO), myeloid cell Bmal1 knockout mice (Bmal1fl/fl-LysMCre+/+, Bmal1ΔMye), and their relative control mice were generated as we previously described.19,20 For specific knockdown of Bmal1 in hepatocytes, the Bmal1fl/fl mice were injected with adeno-associated virus (AAV)-TBG-Cre or AAV-TBG-Vector (Hanheng Biotechnology, Shanghai, China) via the tail vein for 21 days. Male mice at 10–12 weeks old were used in all experiments, during which all mice were maintained under 12-h light/12-h dark conditions with ad libitum access to food and water unless specified. All animal studies were performed in accordance with the guidelines approved by the Institutional Animal Care and Use Committee of Dalian University of Technology, Dalian Medical University, or Shanghai University of Medicine and Health Sciences.
ConA injection
A single dose (20 mg/kg or 12 mg/kg) of ConA (Sigma, C2010) was i.v. injected through the tail vein at Zeitgeber time 0 (ZT0) (lights on), ZT6, ZT12 (lights off), or ZT18. Mice were sacrificed before or after ConA treatment at indicated time points to obtain the serum and liver samples.
Macrophage depletion in mice
Clodronate liposome-mediated macrophage depletion was performed as previously described. Briefly, the WT mice were i.p. injected with 200 μl of clodronate liposomes (LIPOSOMA, C-010) or an equal volume of PBS liposomes. Forty-eight hours later, mice were further treated with ConA injection, and the depletion of macrophages was evaluated by immunofluorescence staining of F4/80 or CD68 in the liver sections.
Deletion of Junb in macrophages
To obtain macrophage-specific Junb knockdown mice, the WT mice were randomly assigned to two groups, and recombinant AAV vectors containing either scramble-short-hairpin RNA (shRNA) or Junb-shRNA sequences under the control of the macrophage-specific promoter F4/80 (Hanheng Biotechnology, Shanghai, China) were injected through the tail vein. After 28 days, the mice received ConA injection, and the extent of Junb knockdown was assessed in macrophages using quantitative PCR (qPCR) and Western blot analysis. This approach allowed for specific targeting of Junb expression in macrophages and evaluation of its effects on subsequent experiments. The shRNA sequences for Junb were as follows: 5′-GCATCAAAGTGGAGCGAAAGC-3′.
Additional information related to materials and methods is available in the Supplementary information.
Results
Effect of ConA administration time on mortality and liver injury
The lethality of ConA (20 mg/kg, i.v.) varies with the timing of treatment (Fig. S1A). Among the four groups we used, the mice treated with ConA at ZT0 (lights on) and ZT12 (lights off) have the highest and the lowest mortality 24 h after the treatment, respectively, and the groups ZT6 and ZT18 were in the middle. Therefore, to achieve obvious administration time-dependent effects, we treated mice with ConA at ZT0 or ZT12 for all subsequent experiments.
First, we weighed ConA-treated mice every 6 h for 4 days and recorded death events over time. As shown in Fig. 1A and B, compared with that in the ZT12 group, the decrease in body weight of mice in the ZT0 group is more evident (Fig. 1A), which is concomitant with a lower survival rate (Fig. 1B). Next, we used the same dose to treat another batch of mice and sacrificed them before (0 h) and after (12, 24, and 96 h) ConA injection for liver and serum collection. Gross hepatic morphology showed obvious light rough nodules on the surface of the livers in the ZT0 group, which was peaking at 24 h (Fig. 1C). By contrast, the ZT12 group has much milder pathological changes at all time points we examined. Consistently, the liver injury-caused release of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels was lower in the ZT12 group than in the ZT0 group (Fig. 1D and Fig. S1B). These results indicated that the deterioration effects of ConA are highly dependent on the time point when it is administered, but not on the sampling time.
Fig. 1.
Effect of ConA administration time on mortality and liver injury.
(A–D) Wild-type mice were maintained in the regular 12-h light/12-h dark cycles and treated with ConA (20 mg/kg body weight, i.v.) at ZT0 (lights on) or ZT12 (lights off). (A) Percentage of body weight loss following ConA treatment for 96 h (n = 14–15). (B) Survival curve of mice following ConA treatment for 96 h (n = 14–15). (C) Representative photographs of the livers following ConA treatment for 12, 24, or 96 h. (D) Serum AST and ALT levels of mice following ConA treatment for 24 h. (E–I) Wild-type mice were treated with PBS or ConA (12 mg/kg body weight, i.v.) at ZT0 or ZT12 for 24 h. (E) Representative images of H&E (the dotted lines mark liver damage areas) and TUNEL staining of the livers. Scale bar = 20 μm. (F) Quantification of liver damage areas and TUNEL-positive cells. (G) Western blot analysis of the protein expression and quantification of cleaved caspase-3 and caspase-3 in the liver. ELISA analysis of the (H) serum and (I) hepatic contents of inflammatory cytokines. Data are presented as mean ± SEM. ∗p <0.05, ∗∗p <0.01, and ∗∗∗p <0.001. Statistical significance was determined using the unpaired Student t test in (A), the log-rank test in (B), and two-way ANOVA in (D) and (F–I). ALT, alanine aminotransferase; AST, aspartate aminotransferase; ConA, concanavalin A; iNOS, inducible nitric oxide synthase; TNF, tumour necrosis factor; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labelling; ZT, Zeitgeber time.
To avoid high mortality, we used a lower dose of ConA (12 mg/kg) in all subsequent experiments for the mechanism study (Fig. S1C). Such dose-induced milder liver injury without death events occurred during the whole experiment. Similar to the results using high doses, the gross observation of the livers revealed severer hepatic injury in mice of the ZT0 group (Fig. S1D). Consistently, serum AST and ALT were less induced in the ZT12 group than in the ZT0 group (Fig. S1E). Then, we used H&E staining and TUNEL (terminal deoxynucleotidyl transferase dUTP nick-end labelling) staining to assess whether there was necrosis or apoptosis in the injured livers (Fig. 1E). H&E staining showed larger necrotic areas in the ZT0 group. Similarly, TUNEL-positive cells (apoptotic cells) were significantly more numerous in the ZT0 group (Fig. 1F).
Moreover, we examined the expression of an apoptosis-related protein, cleaved Caspase-3 in liver lysate, and found higher induction in the ZT0 group than in the ZT12 group (Fig. 1G). Because ConA-induced hepatitis is immune-related, we examined the mRNA expression levels of several pro-inflammatory factors, tumour necrosis factor (TNF)-α, IL-6, inducible nitric oxide synthase (iNOS), and IL-1β in the liver (Fig. S1F) and their protein levels in the serum (Fig. 1H) and liver lysate (Fig. 1I) and found that all these factors were more obviously induced in the ZT0 group than in the ZT12 group.
Administration time-dependent effect of ConA is controlled by the endogenous circadian clock
To investigate whether the administration time-dependent effect of ConA is mediated by the endogenous circadian clock, or by the cycling light exposures, we first repeated the above experiments with mice maintained under constant darkness for more than 1 week (Fig. 2A) to eliminate the effects of environmental light, and the ConA injection time is referred to as CT0 (or CT12), which corresponds to ZT0 (or ZT12) on the regular light/dark cycle. Consistent with the previous conclusion, we found highly similar effects in serum levels of AST and ALT (Fig. 2B) and mRNA levels of TNF-α, IL-6, and iNOS in the liver (Fig. S2), that is, milder induction of these factors in mice treated with ConA at rest-to-active transit (CT12).
Fig. 2.
Administration time-dependent effect of ConA is controlled by the endogenous circadian clock.
(A) Schematic diagram of the study design on ConA-induced liver injury in wild-type mice under constant darkness conditions. (B) Serum AST and ALT levels. (C–H) Bmal1fl/fl (Ctrl) and Bmal1fl/fl-EsrCre+ (iKO) mice were treated with ConA at ZT0 or ZT12 for 24 h. (C) Schematic diagram of the study design. (D) Representative images of liver photographs, H&E (the dotted line marks liver damage areas), and TUNEL staining. Scale bar = 20 μm. (E) Quantification of liver damage areas and TUNEL-positive cells. (F) Serum AST and ALT levels. (G) qPCR analysis of the mRNA expression of inflammatory cytokines in the liver. (H) ELISA analysis of the hepatic contents of inflammatory cytokines. Data are presented as mean ± SEM. ∗p <0.05, ∗∗p <0.01, and ∗∗∗p <0.001 for the indicated comparisons by two-way ANOVA. ALT, alanine aminotransferase; AST, aspartate aminotransferase; Bmal1, basic helix–loop–helix ARNT-like 1; ConA, concanavalin A; CT, ConA injection time; Ctrl, control; DD, constant darkness; iKO, Bmal1 global knockout mice; iNOS, inducible nitric oxide synthase; LD, light–dark; ns, no statistical difference; qPCR, quantitative PCR; TNF, tumour necrosis factor; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labelling; ZT, Zeitgeber time.
It is well established that nutrient availability plays a crucial role in regulating peripheral circadian rhythms, particularly in the liver.9,21 Therefore, we next investigated whether dietary intake could impact the diurnal variation of ConA-induced acute liver injury. We conducted experiments on mice following a regular light/dark cycle and subjected them to different feeding conditions: ad libitum, daytime-restricted feeding, or nighttime-restricted feeding before and during ConA treatment (Fig. S3A). Serum and liver samples were collected 24 h after ConA injection at either ZT0 or ZT12. The data presented in Fig. S3B and C demonstrate that serum levels of AST and ALT, as well as mRNA levels of inflammatory cytokines in the liver, consistently exhibited lower levels in the ZT12 group than in the ZT0 group. Notably, these differences were not influenced by any specific feeding behaviour.
Finally, we used tamoxifen-inducible Bmal1 global knockout mice (iKO)19 to investigate whether the loss of endogenous circadian rhythms will eliminate the timing effect of ConA. qPCR results confirmed the knockout efficacy in the liver, and the hepatocytes and hepatic non-parenchymal cells (Fig. S4A and B). Wheel running activity results confirmed the loss of circadian behaviour in iKO mice under constant darkness conditions (Fig. S4C), which is comparable with our previous results.6,19 As expected, the results showed that Bmal1 deletion did abolish the diurnal variations between the ZT0 and ZT12 groups in hepatic pathology (Fig. 2C–E), serum aminotransferases, and the levels of inflammatory cytokines in the liver (Fig. 2F–H). Moreover, the iKO mice were grossly healthier than Ctrl mice irrespective of ConA administration time, and the above pathological and molecular changes were much milder in the iKO mice, particularly for the ZT0 groups, suggesting that lack of the core clock gene is protective to ConA-induced liver injury. Taken together, our data strongly implied that the administration time-dependent effect of ConA is controlled by the endogenous circadian clock, but not food intake or external light exposure.
Depletion of Bmal1 in hepatocytes failed to affect ConA-induced liver injury
Interestingly, ConA induction significantly reduced Bmal1 expression in hepatocytes (Fig. 3A and B). However, the expression level of Bmal1 in hepatic non-parenchymal cells was contrarily elevated after ConA treatment (Fig. 3C and D). We then meant to dissect the contribution of Bmal1 in hepatocytes and hepatic non-parenchymal cells to ConA-induced liver injury. The Bmal1fl/fl mice were injected with AAV-TBG-Cre to selectively knock out Bmal1 in hepatocytes but retain Bmal1 expression in hepatic non-parenchymal cells (Fig. S5A–D). After that, the mice were subjected to ConA-induced hepatitis as before (Fig. 3E). Surprisingly, unlike the global Bmal1 deletion, hepatocyte-specific Bmal1 depletion failed to protect the liver injury and inflammatory responses induced by ConA, and the administration time-dependent effect of ConA-induced hepatic injury was not affected as well (Fig. 3F–J), implying that the expression of Bmal1 in hepatocytes is likely to be dispensable during the pathogenesis of ConA-induced autoimmune hepatitis.
Fig. 3.
Effect of ConA in mice with hepatocyte-specific Bmal1 depletion.
(A, B) qPCR and Western blot analysis of the expression of Bmal1 in the hepatocytes of the WT mice following ConA treatment for 24 h. (C, D) qPCR and Western blot analysis of the expression of Bmal1 in the hepatic non-parenchymal cells of the WT mice following ConA treatment for 24 h. (E) Schematic diagram of the study design on ConA-induced liver injury in AAV-TBG-Cre-mediated hepatocyte-specific depletion of Bmal1 in mice (Bmal1ΔHep) under regular 12-h light/12-h dark cycles. (F) Representative images of liver photographs, H&E staining (the dotted line marks liver damage areas), and TUNEL assay. Scale bar = 20 μm. (G) Quantification of liver damage areas and TUNEL-positive cells. (H) Serum AST and ALT levels. (I) qPCR analysis of the mRNA expression of inflammatory cytokines in the liver. (J) ELISA analysis of the hepatic contents of inflammatory cytokines in the liver. Data are presented as mean ± SEM. ∗p <0.05, ∗∗p <0.01, and ∗∗∗p <0.001. Statistical significance was determined by unpaired Student’s t test in (A)–(D) and two-way ANOVA in (G)–(J). ALT, alanine aminotransferase; AST, aspartate aminotransferase; Bmal1, basic helix–loop–helix ARNT-like 1; ConA, concanavalin A; iNOS, inducible nitric oxide synthase; LD, light–dark; ns, no statistical difference; qPCR, quantitative PCR; TNF, tumour necrosis factor; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labelling; WT, wild-type; ZT, Zeitgeber time.
Macrophage depletion protects the liver from ConA-induced injury irrespective of the administration time of ConA
Macrophages and T cells play an essential role in the pathogenesis of ConA-induced hepatitis,22,23 and their numbers exhibit significant circadian rhythms in many immune-mediated inflammatory diseases.18 To investigate whether they mediate the circadian difference in ConA-induced hepatitis, we examined the number of CD86 (a marker for M1-type macrophage; Fig. S6A), CD206 (a marker for M2-type macrophage; Fig. S6B), and CD3 and CD4 (markers for T cells; Fig. S6C and D) positive cells in the liver by immunofluorescence. Although the results showed no significant difference in CD206, CD3, or CD4 staining between the ZT0 and ZT12 groups, the abundance of CD86-positive cells in the ZT0 group was significantly higher than that in the ZT12 group. Consistently, the expression of M1 macrophage signature genes (TNF-α, IL-6, iNOS, and IL-1β) was also higher in the ZT0 group (Fig. S1F), suggesting that macrophages, particularly M1 macrophages, may be the major inflammatory cells mediating the administration time-dependent effect of ConA in hepatitis. Furthermore, mice were treated with clodronate liposomes to deplete macrophages and then subjected to ConA treatment (Fig. S7A). The depletion was confirmed by immunofluorescence of pan-macrophage markers, F4/80, and CD68 (Fig. S7B and C). H&E and TUNEL staining showed that the deletion of macrophages remarkedly protected the liver from ConA-induced hepatopathy (Fig. S7D and E). Serum aminotransferases (Fig. S7F) and the mRNA expression of pro-inflammatory molecules (Fig. S7G) were less induced as well. Notably, macrophage depletion also completely abolished the diurnal variation of ConA induction on liver injury and inflammatory response.
Bmal1 deletion in myeloid cells protects the liver from ConA-induced injury irrespective of the administration time of ConA
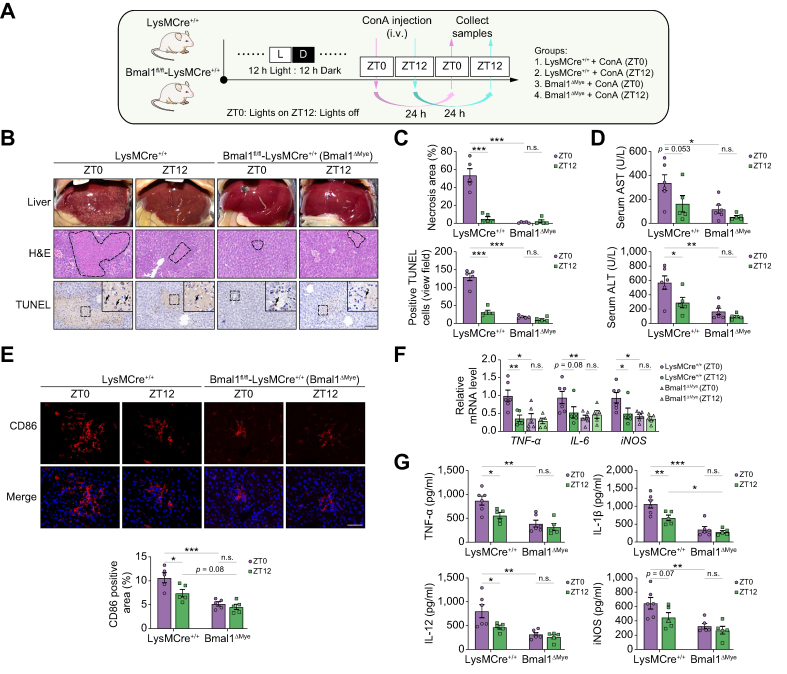
The results of global Bmal1 deletion, hepatocyte-specific Bmal1 deletion, and macrophage depletion described above indicate a possibility that macrophage Bmal1 may mediate the pathological effect of ConA in a time-dependent manner. To prove our hypothesis, we used myeloid cells, particularly macrophage Bmal1 deletion mice (Bmal1fl/fl-LysMCre+/+, Bmal1ΔMye),24 to repeat the study of ConA-induced hepatitis. First, the expression of Bmal1 in the isolated hepatocytes, hepatic non-parenchymal cells, and peritoneal macrophages was examined to confirm the macrophage-specific Bmal1 depletion (Fig. S8A and B). Next, we treated these mice and their littermate controls with ConA at ZT0 and ZT12 to induce autoimmune hepatitis (Fig. 4A). Similar to the iKO mice and macrophage depletion mice, the diurnal variation of ConA-induced liver injury disappeared in the Bmal1ΔMye mice (Fig. 4B). Moreover, the Bmal1ΔMye mice also developed milder hepatopathy revealed by H&E and TUNEL staining (Fig. 4B and C) and the decreased levels of serum AST and ALT (Fig. 4D). Meanwhile, the staining of an M1 macrophage marker, CD86 (Fig. 4E), and the levels of pro-inflammatory molecules in the liver, TNF-α, IL-6, iNOS, IL-12, and IL-1β (Fig. 4F and G), were also lowered by Bmal1 deletion in myeloid cells. Thus, Bmal1 deletion in myeloid cells may protect ConA-induced liver injury irrespective of the ConA administration time.
Fig. 4.
Administration time-dependent effect of ConA in myeloid cell Bmal1-deficient mice.
(A) Schematic diagram of the study design on ConA-induced liver injury in LysMCre+/+ and Bmal1fl/fl-LysMCre+/+ (Bmal1ΔMye) mice under regular 12-h light/12-h dark cycles. (B) Representative images of liver photographs, H&E staining (the dotted line marks liver damage areas), and TUNEL assay. Scale bar = 20 μm. (C) Quantification of liver damage areas and TUNEL-positive cells. (D) Serum AST and ALT levels. (E) Representative immunofluorescence images and the quantification of M1 macrophage marker CD86 in the livers. (F) qPCR analysis of the mRNA expression of inflammatory cytokines in the liver. (G) ELISA analysis of the hepatic contents of inflammatory cytokines in the liver. Data are presented as mean ± SEM. ∗p <0.05, ∗∗p <0.01, and ∗∗∗p <0.001 for the indicated comparisons by two-way ANOVA. ALT, alanine aminotransferase; AST, aspartate aminotransferase; Bmal1, basic helix–loop–helix ARNT-like 1; ConA, concanavalin A; iNOS, inducible nitric oxide synthase; LD, light–dark; ns, no statistical difference; qPCR, quantitative PCR; TNF, tumour necrosis factor; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labelling; ZT, Zeitgeber time.
Transcriptomics analysis on the effects of Bmal1 deletion in ConA-induced hepatitis
To understand the molecular mechanism of Bmal1 and the circadian clock in the pathogenesis of ConA-induced hepatitis, we performed RNA sequencing (RNA-seq) using liver samples of the iKO mice and their littermate controls (Ctrl) treated with ConA at ZT0 or ZT12. Principal component analysis (Fig. 5A) and Pearson correlation analysis (Fig. S9A) showed that the reproducibility of samples in the same group was very good. We compared the differential gene expression of the four groups and found that the Ctrl mice injected with ConA at ZT0 had a distinct transcriptomic profile compared with other groups (Fig. 5B). This gross expression pattern is consistent with the severity of ConA-induced hepatitis.
Fig. 5.
Transcriptomics analysis on the effects of Bmal1 deletion in ConA-induced hepatitis.
(A–F) Bmal1fl/fl (Ctrl) and Bmal1fl/fl-EsrCre+ (iKO) mice were treated with ConA at ZT0 or ZT12 for 24 h, and the liver samples were collected for RNA-seq analysis. (A) Principal component analysis. (B) Heatmap of DEGs. (C) Volcano plot of the DEGs between the ZT0-Ctrl and iKO groups. The green dots represent genes upregulated in the iKO group, the red dots represent genes downregulated in the iKO group, and the black dots represent non-DEGs. (D) KEGG enrichment analysis showed the top pathways enriched by the downregulated genes in the iKO group. (E) KEGG enrichment analysis showed the top pathways enriched by the upregulated genes in the iKO group. (F) Volcano plot of the DEGs related to TNF signalling pathway between the ZT0-Ctrl and iKO groups. (G) Analysis of the circadian expression patterns of Junb in the livers of wild-type mice (n = 3 for each time point). (H) Immunofluorescence staining of the expression and co-localisation of Bmal1 (red), Junb (green), CD86 (pink), and DAPI (blue) in the liver sections from Ctrl and iKO mice with or without 24-h ConA treatment. Scale bar = 20 μm. Bmal1, basic helix–loop–helix ARNT-like 1; ConA, concanavalin A; Ctrl, control; DEG, differentially expressed gene; iKO, Bmal1 global knockout mice; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen activated protein kinase; PC, principal component; PI3K, phosphoinositide 3-kinase; RNA-seq, RNA sequencing; TGF, transforming growth factor; TNF, tumour necrosis factor; Zt, Zeitgeber time.
According to volcano plot analysis, there were 2,906 differentially expressed genes between the ZT0-Ctrl and ZT12-Ctrl groups, with the number of genes upregulated and downregulated in the ZT12-Ctrl group being 1,299 and 1,607 (Fig. S9B), respectively. However, only 90 differentially expressed genes were detected between the ZT0-iKO and ZT12-iKO groups (Fig. S9C). Therefore, we combined the two iKO groups for the following analysis. Thus, compared with those in the ZT0-Ctrl group, in total 2,339 differentially expressed genes were detected in the iKO mice, of which 1,090 genes were markedly downregulated and 1,249 genes were upregulated (Fig. 5C). Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis identified that most of the downregulated pathways were associated with immune and inflammatory responses (Fig. 5D), such as transforming growth factor-beta signalling pathway, cytokine-cytokine receptor interaction, and TNF signalling pathway, which is consistent with the in vivo less inflammatory phenotypes. By contrast, the upregulated genes in iKO livers were mainly enriched in metabolism-related pathways, including drug metabolism, steroid hormone biosynthesis, amino acid metabolism, and glutathione metabolism (Fig. 5E), implying that the liver function was improved in the iKO mice. Notably, KEGG pathway analysis showed similar results for the comparison between the ZT0-Ctrl and ZT12-Ctrl groups (Fig. S9D and E).
We next focused on the TNF signalling pathway, which has been reported to be highly related to immune-mediated hepatisis25,26 and was significantly enriched and downregulated in both the iKO and ZT12-Ctrl groups as compared with the ZT0-Ctrl group. Therefore, the differentially expressed genes of the TNF signalling pathway are presented in Fig. 5F, and there were in total 20 genes downregulated in the ZT12-Ctrl and iKO groups (Fig. S10A). Consistent with the findings of RNA-seq, the significant downregulation of genes in the TNF signalling pathway was mostly independently validated by qPCR analysis (Fig. S10B). Interestingly, among the validated downregulated genes, Junb is an important transcription factor that may regulate the expression of other genes in the TNF signalling pathway, such as Ptgs2, Cxcl2, Cxcl3, Cxcl10, Ccl2, Il6, Socs3, and Mmp9,[27], [28], [29], [30] and it has been reported that Junb is involved in ConA-induced acute liver injury and other immune-mediated hepatisis.31 Moreover, we found that the expression of Junb mRNA exhibited robust circadian rhythmicity in the liver (Fig. 5G). Thus, here we hypothesised that the prevented liver injury and blunted diurnal variation in the iKO mice might result from inhibition of Junb expression and rhythms. To test this hypothesis, we first analysed the expression and cellular localisation of Bmal1 and Junb in liver tissues of ConA-induced hepatitis by immunofluorescence staining. The results showed that upon ConA induction, the expression levels of Bmal1 and Junb were particularly increased and localised in CD86-positive macrophages within the liver. However, after Bmal1 deletion, the signalling of Junb was weakened (Fig. 5H). To further investigate this, hepatic macrophages were extracted from both ConA-treated Ctrl mice and iKO mice, and the mRNA expression levels of Bmal1 and Junb were examined. qPCR results demonstrated that deletion of Bmal1 markedly attenuated ConA-induced upregulation of Junb in hepatic macrophages (Fig. S11).
These data suggest that Junb is a potential regulatory target of Bmal1 in macrophages and Bmal1 may control the diurnal variation of ConA-induced hepatitis by regulating macrophage activation via Junb.
Junb was identified as a new target of Bmal1 in macrophages
Next, we aimed to detect whether Junb would be a direct target of Bmal1 in macrophages. We isolated bone marrow-derived macrophages from the iKO and Ctrl mice. After synchronisation using dexamethasone, we observed clear circadian rhythms in mRNA levels of both Bmal1 (Fig. 6A) and Junb (Fig. 6B) in Ctrl cells, whereas iKO cells displayed dampened rhythms and overall lower mRNA levels. Next, we used the JASPAR database (jaspar.genereg.net) to analyse the DNA sequence of the murine Junb promoter and found a potential binding site for Bmal1 in the region near the transcription start sites (Fig. 6C). Chromatin immunoprecipitation (ChIP) assay and subsequent qPCR results indicated that direct binding of Bmal1 to the region embracing the potential binding site was enhanced (Fig. 6D and E and Fig. S12). Furthermore, a luciferase reporter assay using a WT Junb promoter sequence (Fig. 6F) and the sequence with a mutation at the binding site (Fig. 6G) revealed activation and blocking of the activation of Junb transcription by Bmal1/Clock, respectively. These results indicated that Junb is a direct target of Bmal1 in macrophages via binding to the potential E-box-like element.
Fig. 6.
Junb was identified as a new target of Bmal1 in macrophages.
(A, B) BMDMs were cultured from Bmal1fl/fl (Ctrl) and Bmal1fl/fl-EsrCre+ (iKO) mice and synchronised. RNA was extracted at 4-h intervals for 36 h, and Bmal1 and Junb mRNA expression levels were measured by qPCR. (C) Schematic of the predictive binding sequence of Bmal1 at the Junb promoter region 2,000 bp upstream of the transcription start site from the JASPAR database (jaspar.genereg.net). (D) The representative ChIP-PCR and (E) quantitative data show that Bmal1 binds to the Junb promoter in RAW264.7 cells. (F, G) The dual-luciferase assay shows the effects of Bmal1/Clock co-transfection on wt-Junb and mut-Junb promoter activity (n = 8 in each group). Data are presented as mean ± SEM. ∗p <0.05, ∗∗p <0.01, and ∗∗∗p <0.001. Statistical significance was determined using two-way ANOVA in (A) and (B), one-way ANOVA in (E), and the unpaired Student t test in (F) and (G). Bmal1, basic helix–loop–helix ARNT-like 1; BMDM, bone marrow-derived macrophage; ChIP, chromatin immunoprecipitation; Ctrl, control; iKO, Bmal1 global knockout mice; ns, no statistical difference; qPCR, quantitative PCR.
Junb knockdown in macrophages protects the liver from ConA-induced injury
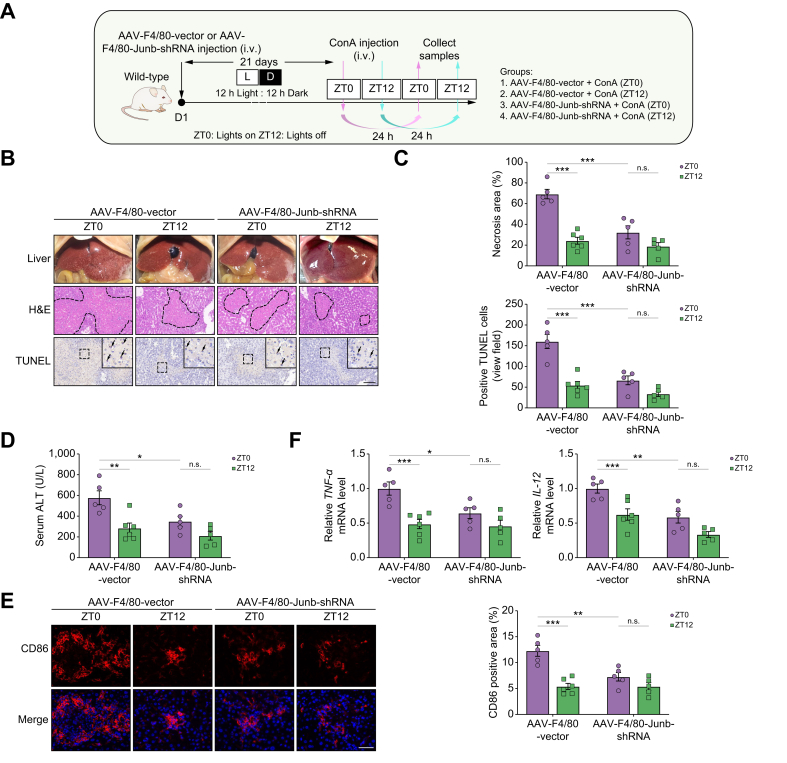
Subsequently, we used an AAV viral vector containing the macrophage-specific promoter F4/80 and Junb shRNA sequence to selectively knock down Junb in macrophages. The mice were then subjected to ConA-induced hepatitis (Fig. 7A). After 28 days of AAV injection, the decrease in Junb mRNA and protein expression in peritoneal macrophages was confirmed (Fig. S13A and B). Similar to the Bmal1ΔMye mice, Junb knockdown in macrophages dramatically alleviated the severe liver injury caused by ConA. In addition, the diurnal variation of the injury was abolished in the Junb knockdown mice (Fig. 7B–D). Furthermore, the infiltration and time-dependent variation of CD86+ M1 macrophages (Fig. 7E) and the secretion of pro-inflammatory cytokines (Fig. 7F) were significantly attenuated in knockdown mice. Similar to the in vivo phenotypes, the knockdown of Junb remarkedly reduced ConA-induced CD86 and TNF-α expression in RAW264.7 cells (Fig. S14 and Fig. 8A and B). Taken together, the knockdown of Junb in macrophages may protect against ConA-induced liver injury by reducing M1 macrophage activation and alleviating inflammatory responses.
Fig. 7.
Junb knockdown in macrophages protects the liver from ConA-induced injury.
(A) Schematic diagram of the study design on ConA-induced liver injury in AAV-F4/80-mediated knockdown Junb in macrophage from mice under regular 12-h light/12-h dark cycles. (B) Representative images of liver photographs, H&E staining (the dotted line marks liver damage areas), and TUNEL assay. Scale bar = 20 μm. (C) Quantification of liver damage areas and TUNEL-positive cells. (D) Serum ALT levels. (E) Representative immunofluorescence images and the quantification of CD86 staining. (F) qPCR analysis of the mRNA expression of inflammatory cytokines in the liver. Data are presented as mean ± SEM. ∗p <0.05, ∗∗p <0.01, and ∗∗∗p <0.001 for the indicated comparisons by two-way ANOVA. AAV, adeno-associated virus; ALT, alanine aminotransferase; ConA, concanavalin A; LD, light–dark; ns, no statistical difference; qPCR, quantitative PCR; shRNA, short hairpin RNA; TNF, tumour necrosis factor; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labelling; ZT, Zeitgeber time.
Junb activated M1 macrophages via AKT and ERK signalling pathways
Lastly, we further explored the signalling pathway that contributes to Bmal1/Junb axis-mediated liver inflammatory responses. It has been reported that the AKT and extracellular signal-regulated kinase (ERK) signalling pathways are critical to the pro-inflammatory response of M1 macrophages.32,33 Moreover, Junb could regulate the activity of AKT and ERK signalling pathways.29,34 Indeed, phosphoinositide 3-kinase (PI3K)–AKT and mitogen activated protein kinase (MAPK) signalling pathways were significantly affected by Bmal1 deletion in ConA-induced livers (Fig. 5D), and AKT and ERK are the most classical signalling molecules in the PI3K–AKT and MAPK signalling pathways, respectively. Therefore, we examined whether Junb regulates M1 macrophage activation through these pathways. As shown in Fig. 8C and D and Fig. S15, ConA treatment time-dependently induced AKT and ERK phosphorylation in RAW264.7 cells, although this induction was significantly inhibited by Junb silencing. In addition, Western blot confirmed higher hepatic expression levels of p-ERK and p-AKT in the ZT0 group than in the ZT12 group, whereas knockdown of Junb in macrophages and deletion of Bmal1 eliminated this time-dependent difference and lowered the levels of p-ERK and p-AKT (Fig. 8E–H). Collectively, these data suggested that the Bmal1/Junb axis-mediated diurnal control of ConA-induced liver inflammatory response is at least partly dependent on the activation of AKT and ERK signalling pathways.
Fig. 8.
Junb activated M1 macrophages via AKT and ERK signalling pathways.
(A) The mRNA expression levels of Bmal1, Junb, CD86, and TNF-α in RAW264.7 cells treated with ConA (50 μg/ml) for the indicated time. (B) qPCR analysis of the mRNA expression of CD86 and TNF-α in RAW264.7 cells transfected with or without Junb siRNA for 24 h. (C, D) Western blot analysis and the quantification of the protein expression of p-AKT, AKT, p-ERK, and ERK in RAW264.7 cells transfected with or without Junb siRNA for 24 h. (E, F) Western blot analysis and the quantification of the protein expression of p-AKT, AKT, p-ERK, and ERK in the livers of the AAV-F4/80-Vector and AAV-F4/80-Junb-shRNA mice following ConA treatment. (G, H) Western blot analysis and the quantification of the protein expression of p-AKT, AKT, p-ERK, and ERK in the livers of the Ctrl and iKO mice following ConA treatment. Data are presented as mean ± SEM. ∗p <0.05, ∗∗p <0.01, and ∗∗∗p <0.001. Statistical significance was determined by one-way ANOVA in (A), (B), and (D) and by two-way ANOVA in (F) and (H). AAV, adeno-associated virus; Bmal1, basic helix–loop–helix ARNT-like 1; ConA, concanavalin A; Ctrl, control; ERK, extracellular signal-regulated kinase; iKO, Bmal1 global knockout mice; ns, no statistical difference; qPCR, quantitative PCR; shRNA, short hairpin RNA; TNF, tumour necrosis factor; ZT, Zeitgeber time.
Discussion
Extensive studies demonstrated that disruption of the circadian rhythms is implicated in various major liver diseases, including fatty liver, cirrhosis, and cancer.10,11 Here, by studying a well-known immune-mediated liver injury model that is induced by i.v. injection of ConA, we further found that the severity of hepatitis was dependent on the administration time of ConA and this timing effect was eliminated by the removal of the core clock component Bmal1 or knocking down its direct targeting gene Junb in macrophages in mice. More importantly, these mice developed milder hepatitis than controls that have an intact clock system, suggesting that the macrophage Bmal1/Junb axis may serve as a promising target for immune-mediated liver injury. Furthermore, we identified that AKT and ERK signalling pathways mediated the role of the Bmal1/Junb axis in the pathogenesis of ConA-induced liver injury.
First, we confirmed the temporal variation in ConA-induced hepatotoxicity on WT mice, which can also be observed in some medicines that have side effects on the liver in a dosing time-dependent manner, such as acetaminophen,35 Fuzi,36 and brucine.37 Contrary to a lipopolysaccharide-induced sepsis model,38 administration of ConA at the start of the rest phase, but not the active phase, was more harmful to mice under both regular light/dark cycles and constant darkness conditions, implicating that there is a drug specificity and this timing effect is not mediated by the light/dark cycle. We also excluded the effect of timing of food intake by subjecting mice to either daytime- or nighttime-restricted feeding. Furthermore, by maintaining the adult-life Bmal1 knockout mice under regular light/dark conditions, which allowed the mice to exhibit day/night variations in behavioural activities but lacked endogenous circadian rhythms,6,19 we found that the difference in hepatic injury between different time points of ConA administration completely disappeared in these mice. These findings strongly support the notion that the endogenous circadian clock, rather than light exposure or feeding status, predominantly contributes to the diurnal variation of ConA-induced hepatitis. However, considering that feeding is a key regulator of the liver,9,21 further validation with additional feeding/fasting schedules may be warranted to fully elucidate the role of feeding in the observed hepatic injury. Moreover, it remains unclear whether the diurnal variation of ConA-induced hepatitis is influenced by glucocorticoids, which are known to be crucial to the circadian control of immunity.39,40 Further investigations are needed to determine the potential involvement of glucocorticoids in this context.
T cells and macrophages are the main immune cells that mediate ConA-induced hepatitis, and their function was known to be regulated by circadian rhythms.41 It has been shown that Bmal1 is involved in the development of immune-related diseases by regulating T cells42 and macrophages.18 Here, we confirmed the significant increase of these two cell types in the injured liver, whereas the temporal variation was obvious in macrophages only. This fluctuation of macrophages that contributes to the administration time-dependent effect of ConA was further confirmed by the removal of macrophages. It is well known that macrophages acquire distinct functional phenotypes in response to various stimuli or under different pathophysiological conditions.43 Classical M1 (pro-inflammatory) and alternative M2 (anti-inflammatory) activation of macrophages represent two extremes of a dynamic changing state of macrophage activation.44 Our study revealed that the number of M1 macrophages, but not that of M2-type macrophages, significantly varied between time points with higher levels in the liver of mice treated with ConA at ZT0. The secretion of inflammatory cytokines showed similar pattern as well, implicating the involvement of M1 macrophages in the timing effect of ConA. As these cytokines damage contiguous tissue and inhibit the proliferation of surrounding cells,45 the findings that depletion of total macrophages attenuated hepatopathy further suggested the primary contribution of M1 macrophages.
Given that the core clock component Bmal1 plays a fundamental role in the circadian immune responses in macrophages38 and both global Bmal1 deletion and macrophage depletion in the present study eliminated the timing effect of ConA in mice, it is reasonable to propose an essential role of macrophage Bmal1 in these processes. As expected, results with the myeloid Bmal1 knockout mice strongly supported the finding that the myeloid Bmal1 mediated the timing effect. Although, unlike macrophages, neutrophils were also myeloid lineage originated and increased in the liver by ConA administration, and they did not vary in the number between the ZT0 and ZT12 groups (data not shown). Therefore, the timing effect of ConA was mainly attributed to the clocks in macrophages. It is notable that mice with selective knockdown of Bmal1 in hepatocytes failed to affect either the circadian variation or the damaging effect of ConA-induced liver injury, indicating that the role of Bmal1 in macrophages is dominant over its function in hepatocytes in this setting. We acknowledge that the AAV-TBG-Cre-mediated depletion we used resulted in only approximately 50% depletion in Bmal1 protein expression. This partial depletion may not fully reflect the actual impact of hepatocyte Bmal1 deficiency on ConA-induced liver injury. Therefore, an alternative strategy using Cre/loxP-mediated hepatocyte-specific Bmal1 depletion may provide additional valuable insights to verify the role of Bmal1 in ConA-induced liver injury. Moreover, the opposite regulation of Bmal1 in hepatocytes compared with non-parenchymal liver cells upon ConA induction suggests a cell-specific role of Bmal1 in immune-mediated liver injury, although the underlying mechanisms remain unclear. Moreover, accumulating evidence indicates that the function of Bmal1 varies across different cell types. For instance, Bmal1 in hepatocytes prevents atherogenesis,46 whereas myeloid Bmal1 is deleterious in this particular condition.20 Therefore, it is crucial to comprehend the distinct regulatory mechanisms underlining Bmal1 function in response to ConA exposure within various cell types. Furthermore, investigating the role of Bmal1 in other cell types, such as T cells, is necessary for future studies to gain a comprehensive understanding of its function.
In addition to eliminating the timing effect, myeloid Bmal1 deletion also alleviated ConA-induced liver injury. Clock proteins, especially Bmal1, as a transcription factor, regulate the expression of various immune-related genes.47,48 To reveal the molecular mechanism of the particular AIH model in the current study, we performed RNA-seq using liver samples from the iKO and control mice treated with ConA at ZT0 or ZT12 to screen differentially expressed genes. The most downregulated pathways in the iKO group were associated with immune and inflammatory responses. Among them, a transcription factor, Junb, from the TNF signalling pathway, was significantly higher expressed in the ZT0-Ctrl mice than in the other three groups. This pattern is highly consistent with their phenotypes. Importantly, the expression level of Junb mRNA exhibited rhythmicity throughout 24 h in the WT liver with peak and trough at ZT0 and ZT12, respectively, which is similar to Bmal1. Further investigation by ChIP suggested Junb as a direct target of Bmal1 in macrophages. Moreover, specific knockdown of Junb in macrophages dampened the phenotypic variations between ZT0 and ZT12 and alleviated ConA-induced liver injury, which highly mimics the effect of Bmal1 deletion in macrophages.
Junb performs an essential role in liver regeneration, liver ischaemia–reperfusion injury, and hepatocellular carcinoma.49,50 It regulates the expression of various cytokines in a tissue- or cell-specific manner. Mice lacking Junb in hepatocytes displayed a mild increase in ConA-induced liver injury. However, globally knocking down Junb in the liver inhibits interferon-γ secretion through natural killer T/natural killer cells, thus protecting hepatocytes from ConA injury.31 These results may indicate a dominant role of Junb in immune cells over hepatocytes. Furthermore, studies have indicated that Junb is strongly expressed in macrophages and plays a central role in macrophage activation.27,[51], [52], [53] With the aim of a more detailed view of the molecular changes in the diurnal variation of ConA-induced liver injury, we sought to investigate the molecular mechanisms of Junb-regulated M1 macrophage activity. It is well known that AKT and ERK signalling pathways are the key factors in the pro-inflammatory response in M1 macrophages.32,33 In addition, Junb could affect cartilage degeneration in osteoarthritis by regulating ERK phosphorylation.34 Junb plays a crucial role in pancreatic beta cell apoptosis by regulating AKT phosphorylation.54 Therefore, we proposed the hypothesis that Junb may regulate M1 macrophage activation through AKT and ERK signalling pathways in ConA-induced hepatitis. Indeed, KEGG pathway analysis revealed that activation of the PI3K–AKT and MAPK pathway might be related to the diurnal variation of ConA-induced liver injury. In vitro, we found that ConA could promote the activation of M1 macrophage and the secretion of various cytokines. However, knocking down Junb inhibits the phosphorylation of AKT and ERK, which ultimately reduces the activation of M1 macrophages and the secretion of cytokines. In vivo, ERK and AKT phosphorylation in the liver of the ZT0 group was higher than the ZT12 group, whereas Junb knockdown in macrophages and Bmal1 knockout eliminated this difference. Thus, the evidence demonstrated that Bmal1 in macrophages could regulate the diurnal variation of ConA-induced hepatitis and the molecular mechanism is that Bmal1 could directly bind to the promoter of Junb to regulate M1 macrophage activation through the AKT and ERK signal pathways. M1 macrophages could produce large amounts of pro-inflammatory cytokines that induce inflammatory responses and apoptosis in hepatocytes through direct contact with hepatocytes, which exacerbates liver injury and even liver failure.
Altogether, our results showed that Bmal1 could regulate macrophage activation by targeting Junb–AKT/ERK pathway, leading to the circadian variation in ConA-induced hepatitis. This temporal difference in outcomes based on the time of infection should have broader relevance for other circadian-gated immune-mediated inflammatory diseases. The molecular clock acted as a timed regulator of Junb activity in macrophages, which greatly affected the inflammasome output through macrophage activation. Given the importance of macrophages in innate immunity, it is distinctly possible that many aspects of the innate immune system mediated by macrophages could be under circadian regulation. Therefore, the activation of the Bmal1–Junb–AKT/ERK axis might also explain the variations of circadian rhythms in inflammatory diseases caused by macrophage activation, such as COVID-19,[55], [56], [57], [58] respiratory tract infections,59 atherosclerosis,20,60 and sepsis.61 Further understanding of the interaction between Junb and Bmal1 not only could facilitate an effective target for the prevention and improvement of AIH but could also provide insight into the temporal control of inflammation. Meanwhile, it might reveal opportunities for chronotherapies in the treatment of circadian-gated immune-mediated inflammatory diseases.
Financial support
This study was supported by the grants from the National Key Research and Development Program of China (#2019YFA0802400) and the National Natural Science Foundation of China (32171165).
Authors’ contributions
Conceived and designed the studies: ZL, GY, LC. Assisted with animal experiments: JZ, SL, HW, JL. Participated in cell experiments: BR, ZB, JL, MG. Wrote the manuscript draft: ZL. Revised the manuscript: GY, LC. Reviewed the manuscript: all authors. Guarantors of the article: GY, LC.
Data availability statement
The authors confirm that the data supporting the findings of this study within the article and/or supplementary materials are available upon request.
Conflicts of interest
The authors have declared that no conflict of interest exists.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We thank Garret A. FitzGerald (University of Pennsylvania, Philadelphia, PA, USA) and C.A. Bradfield (University of Wisconsin, Madison, WI, USA) for generously providing us Bmal1fl/fl mice.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100856.
Contributor Information
Guangrui Yang, Email: guangrui.yang@hotmail.com.
Lihong Chen, Email: lihong@dmu.edu.cn.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Partch C.L., Green C.B., Takahashi J.S. Molecular architecture of the mammalian circadian clock. Trends Cel Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dvornyk V., Vinogradova O., Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci U S A. 2003;100:2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loudon A.S. Circadian biology: a 2.5 billion year old clock. Curr Biol. 2012;22:R570–R571. doi: 10.1016/j.cub.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Harmer S.L., Panda S., Kay S.A. Molecular bases of circadian rhythms. Annu Rev Cel Dev Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 5.Yang G., Paschos G., Curtis A.M., Musiek E.S., McLoughlin S.C., Fitzgerald G.A. Knitting up the raveled sleave of care. Sci Transl Med. 2013;5:212rv213. doi: 10.1126/scitranslmed.3007225. [DOI] [PubMed] [Google Scholar]
- 6.Yang G., Chen L., Zhang J., Ren B., FitzGerald G.A. Bmal1 deletion in mice facilitates adaptation to disrupted light/dark conditions. JCI Insight. 2019;4 doi: 10.1172/jci.insight.125133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mure L.S., Le H.D., Benegiamo G., Chang M.W., Rios L., Jillani N., et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018;359 doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahara Y., Shibata S. Circadian rhythms of liver physiology and disease: experimental and clinical evidence. Nat Rev Gastroenterol Hepatol. 2016;13:217–226. doi: 10.1038/nrgastro.2016.8. [DOI] [PubMed] [Google Scholar]
- 10.Crespo M., Leiva M., Sabio G. Circadian clock and liver cancer. Cancers. 2021;13:3631. doi: 10.3390/cancers13143631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherji A., Bailey S.M., Staels B., Baumert T.F. The circadian clock and liver function in health and disease. J Hepatol. 2019;71:200–211. doi: 10.1016/j.jhep.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang X., Forde D., Tsukuda S., D'Arienzo V., Mailly L., Harris J.M., et al. Circadian control of hepatitis B virus replication. Nat Commun. 2021;12:1658. doi: 10.1038/s41467-021-21821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skrlec I., Talapko J. Hepatitis B and circadian rhythm of the liver. World J Gastroenterol. 2022;28:3282–3296. doi: 10.3748/wjg.v28.i27.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang X., Magri A., Hill M., Lai A.G., Kumar A., Rambhatla S.B., et al. The circadian clock components BMAL1 and REV-ERBα regulate flavivirus replication. Nat Commun. 2019;10:377. doi: 10.1038/s41467-019-08299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heneghan M.A., Yeoman A.D., Verma S., Smith A.D., Longhi M.S. Autoimmune Hepatitis Lancet. 2013;382:1433–1444. doi: 10.1016/S0140-6736(12)62163-1. [DOI] [PubMed] [Google Scholar]
- 16.Gao B., Jeong W.I., Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 17.Wang C., Lutes L.K., Barnoud C., Scheiermann C. The circadian immune system. Sci Immunol. 2022;7 doi: 10.1126/sciimmunol.abm2465. [DOI] [PubMed] [Google Scholar]
- 18.Scheiermann C., Kunisaki Y., Frenette P.S. Circadian control of the immune system. Nat Rev Immunol. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G., Chen L., Grant G.R., Paschos G., Song W.L., Musiek E.S., et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med. 2016;8:324ra316. doi: 10.1126/scitranslmed.aad3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang G., Zhang J., Jiang T., Monslow J., Tang S.Y., Todd L., et al. Bmal1 deletion in myeloid cells attenuates atherosclerotic lesion development and restrains abdominal aortic aneurysm formation in hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2020;40:1523–1532. doi: 10.1161/ATVBAHA.120.314318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinke H., Asher G. Circadian clock control of liver metabolic functions. Gastroenterology. 2016;150:574–580. doi: 10.1053/j.gastro.2015.11.043. [DOI] [PubMed] [Google Scholar]
- 22.Tiegs G., Hentschel J., Wendel A. A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J Clin Invest. 1992;90:196–203. doi: 10.1172/JCI115836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heymann F., Hamesch K., Weiskirchen R., Tacke F. The concanavalin A model of acute hepatitis in mice. Lab Anim. 2015;49:12–20. doi: 10.1177/0023677215572841. [DOI] [PubMed] [Google Scholar]
- 24.Chen L., Yang G., Monslow J., Todd L., Cormode D.P., Tang J., et al. Myeloid cell microsomal prostaglandin E synthase-1 fosters atherogenesis in mice. Proc Natl Acad Sci U S A. 2014;111:6828–6833. doi: 10.1073/pnas.1401797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valaydon Z., Pellegrini M., Thompson A., Desmond P., Revill P., Ebert G. The role of tumour necrosis factor in hepatitis B infection: jekyll and Hyde. Clin Transl Immunol. 2016;5:e115. doi: 10.1038/cti.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laidlaw S.M., Marukian S., Gilmore R.H., Cashman S.B., Nechyporuk-Zloy V., Rice C.M., et al. Tumor necrosis factor inhibits spread of hepatitis C virus among liver cells, independent from interferons. Gastroenterology. 2017;153:566–578.e565. doi: 10.1053/j.gastro.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontana M.F., Baccarella A., Pancholi N., Pufall M.A., Herbert D.R., Kim C.C. JUNB is a key transcriptional modulator of macrophage activation. J Immunol. 2015;194:177–186. doi: 10.4049/jimmunol.1401595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomard T., Michaud H.A., Tempé D., Thiolon K., Pelegrin M., Piechaczyk M. An NF-κB-dependent role for JunB in the induction of proinflammatory cytokines in LPS-activated bone marrow-derived dendritic cells. PLoS One. 2010;5:e9585. doi: 10.1371/journal.pone.0009585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunha D.A., Gurzov E.N., Naamane N., Ortis F., Cardozo A.K., Bugliani M., et al. JunB protects β-cells from lipotoxicity via the XBP1-AKT pathway. Cell Death Differ. 2014;21:1313–1324. doi: 10.1038/cdd.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rylski M., Amborska R., Zybura K., Michaluk P., Bielinska B., Konopacki F.A., et al. JunB is a repressor of MMP-9 transcription in depolarized rat brain neurons. Mol Cel Neurosci. 2009;40:98–110. doi: 10.1016/j.mcn.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Thomsen M.K., Bakiri L., Hasenfuss S.C., Hamacher R., Martinez L., Wagner E.F. JUNB/AP-1 controls IFN-γ during inflammatory liver disease. J Clin Invest. 2013;123:5258–5268. doi: 10.1172/JCI70405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neamatallah T. Mitogen-Activated Protein Kinase Pathway: a critical regulator in tumor-associated macrophage polarization. J Microsc Ultrastruct. 2019;7:53–56. doi: 10.4103/JMAU.JMAU_68_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou D., Huang C., Lin Z., Zhan S., Kong L., Fang C., et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192–197. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Lin Z., Miao J., Zhang T., He M., Wang Z., Feng X., et al. JUNB–FBXO21–ERK axis promotes cartilage degeneration in osteoarthritis by inhibiting autophagy. Aging Cell. 2021;20 doi: 10.1111/acel.13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y.C., Lee S.J. Temporal variation in hepatotoxicity and metabolism of acetaminophen in mice. Toxicology. 1998;128:53–61. doi: 10.1016/s0300-483x(98)00046-8. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z., Lin Y., Gao L., Zhou Z., Wang S., Dong D., et al. Circadian clock regulates metabolism and toxicity of Fuzi (lateral root of Aconitum carmichaeli Debx) in mice. Phytomedicine. 2020;67 doi: 10.1016/j.phymed.2019.153161. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Z., Lin Y., Gao L., Yang Z., Wang S., Wu B. Cyp3a11 metabolism-based chronotoxicity of brucine in mice. Toxicol Lett. 2019;313:188–195. doi: 10.1016/j.toxlet.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Curtis A.M., Fagundes C.T., Yang G., Palsson-McDermott E.M., Wochal P., McGettrick A.F., et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A. 2015;112:7231–7236. doi: 10.1073/pnas.1501327112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimba A., Ikuta K. Glucocorticoids regulate circadian rhythm of innate and adaptive immunity. Front Immunol. 2020;11:2143. doi: 10.3389/fimmu.2020.02143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimba A., Ejima A., Ikuta K. Pleiotropic effects of glucocorticoids on the immune system in circadian rhythm and stress. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.706951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Man K., Loudon A., Chawla A. Immunity around the clock. Science. 2016;354:999–1003. doi: 10.1126/science.aah4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutton C.E., Finlay C.M., Raverdeau M., Jo Early, DeCourcey J., Zaslona Z., et al. Loss of the molecular clock in myeloid cells exacerbates T cell-mediated CNS autoimmune disease. Nat Commun. 2017;8:1923. doi: 10.1038/s41467-017-02111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang N., Liang H., Zen K. Molecular mechanisms that influence the macrophage M1–M2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan X., Bradfield C.A., Hussain M.M. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat Commun. 2016;7 doi: 10.1038/ncomms13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hergenhan S., Holtkamp S., Scheiermann C. Molecular interactions between components of the circadian clock and the immune system. J Mol Biol. 2020;432:3700–3713. doi: 10.1016/j.jmb.2019.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver A.C., Arjona A., Walker W.E., Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36:251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan P., Zhou B., Ma Y., Wang A., Hu X., Luo Y., et al. Tracking the important role of JUNB in hepatocellular carcinoma by single-cell sequencing analysis. Oncol Lett. 2020;19:1478–1486. doi: 10.3892/ol.2019.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu J.C., Bravo R., Taub R. Interactions among LRF-1, JunB, c-Jun, and c-Fos define a regulatory program in the G1 phase of liver regeneration. Mol Cel Biol. 1992;12:4654–4665. doi: 10.1128/mcb.12.10.4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I., et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fontana M.F., Baccarella A., Kellar D., Oniskey T.K., Terinate P., Rosenberg S.D., et al. Myeloid expression of the AP-1 transcription factor JUNB modulates outcomes of type 1 and type 2 parasitic infections. Parasite Immunol. 2015;37:470–478. doi: 10.1111/pim.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Locati M., Mantovani A., Sica A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv Immunol. 2013;120:163–184. doi: 10.1016/B978-0-12-417028-5.00006-5. [DOI] [PubMed] [Google Scholar]
- 54.Cunha D.A., Gurzov E.N., Naamane N., Ortis F., Cardozo A.K., Bugliani M., et al. JunB protects beta-cells from lipotoxicity via the XBP1-AKT pathway. Cel Death Differ. 2014;21:1313–1324. doi: 10.1038/cdd.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meidaninikjeh S., Sabouni N., Marzouni H.Z., Bengar S., Khalili A., Jafari R. Monocytes and macrophages in COVID-19: friends and foes. Life Sci. 2021;269 doi: 10.1016/j.lfs.2020.119010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knoll R., Schultze J.L., Schulte-Schrepping J. Monocytes and macrophages in COVID-19. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosyreva A., Dzhalilova D., Lokhonina A., Vishnyakova P., Fatkhudinov T. The role of macrophages in the pathogenesis of SARS-CoV-2-associated acute respiratory distress syndrome. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.682871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sefik E., Qu R., Junqueira C., Kaffe E., Mirza H., Zhao J., et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature. 2022;606:585–593. doi: 10.1038/s41586-022-04802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirato K., Sato S. Macrophage meets the circadian clock: implication of the circadian clock in the role of macrophages in acute lower respiratory tract infection. Front Cel Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.826738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huo M., Huang Y., Qu D., Zhang H., Wong W.T., Chawla A., et al. Myeloid Bmal1 deletion increases monocyte recruitment and worsens atherosclerosis. FASEB J. 2017;31:1097–1106. doi: 10.1096/fj.201601030R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen K.D., Fentress S.J., Qiu Y., Yun K., Cox J.S., Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study within the article and/or supplementary materials are available upon request.