Abstract
The gyrA and parC genes of 31 clinical isolates of Enterococcus faecalis, including fluoroquinolone-resistant isolates, were partially sequenced and analyzed for target alterations. Topoisomerase IV may be a primary target in E. faecalis, but high-level fluoroquinolone resistance was associated with simultaneous alterations in both GyrA and ParC.
Although it depends on the bacterial species, resistance to quinolones appears to be due mainly to alterations in the GyrA subunit of DNA gyrase or in the ParC subunit of topoisomerase IV. DNA gyrase is a primary target of fluoroquinolones in gram-negative bacteria such as Escherichia coli (9, 11, 22), Neisseria gonorrhoeae (2), and Klebsiella pneumoniae (3), whereas topoisomerase IV is a primary target in gram-positive bacteria such as Staphylococcus aureus (4, 5, 14) and Streptococcus pneumoniae (12, 15, 20). Enterococcus faecalis, which is a gram-positive organism, is one of the nosocomial pathogens that causes serious infections, including urinary tract infections. The number of quinolone-resistant clinical isolates of E. faecalis has been gradually increasing with increased use of fluoroquinolones (13). It has already been demonstrated that quinolone resistance in E. faecalis is associated with alterations of GyrA (10, 21). To date, there have been no reports of a relationship between quinolone resistance and alterations in ParC. Thus, we became interested in assessing whether topoisomerase IV is a primary target in E. faecalis as well as S. aureus and S. pneumoniae. In the present study, we sequenced the regions of the gyrA and parC genes of E. faecalis that are analogous to the quinolone resistance-determining regions (QRDRs) of the E. coli gyrA gene (23) and we assessed the possible association of alterations of GyrA and ParC with fluoroquinolone resistance.
To characterize the parC gene of E. faecalis, we sequenced the region corresponding to the QRDR of the E. coli gyrA gene in the type strain of E. faecalis, ATCC 19433. A 248-bp DNA fragment was amplified from chromosomal DNA of the type strain by PCR with two primers (5′-AAACCTGTTCAGCGCCGCAT-3′ and 5′-TCGGTGTAACGCATTGCCGC-3′), which were derived from nucleotide positions 140 to 159 and 371 to 390, respectively, of the E. coli parC gene (8).
We randomly selected 53 clinical isolates of E. faecalis obtained from 1994 through 1996 from patients with urinary tract infections who had not received antibiotic treatment before attending our clinic. We examined the isolates for susceptibilities to fluoroquinolones (norfloxacin, ofloxacin, and ciprofloxacin) by the twofold agar dilution method (2). MICs were defined as the lowest concentrations of drug that completely inhibited visible growth of the inoculum after incubation for 18 h at 37°C.
The amino acid sequence of the 208-bp fragment between the primers, which we determined, showed 84, 77, and 41% identities with those of the equivalent regions in ParC of S. pneumoniae (16), S. aureus (4), and E. coli (8), respectively (Fig. 1). The identity between GyrA and ParC of E. faecalis was found to be 60% in the regions analogous to the QRDR of the E. coli gyrA gene (10, 19, 23).
FIG. 1.
Comparison of ParC sequences of E. faecalis (EFEPARC), S. pneumoniae (SPNPARC) (16), S. aureus (SAUPARC) (4), and E. coli (ECOPARC) (8). Asterisks, amino acids identical to those in EFEPARC; arrowheads, amino acid positions involved in quinolone resistance.
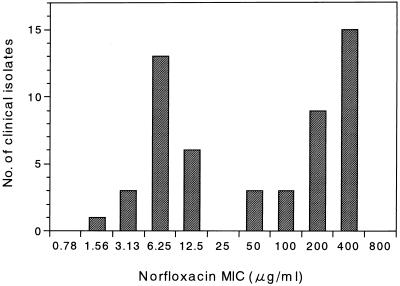
Figure 2 shows the distribution of the MICs of norfloxacin for the 53 clinical isolates. They could be clearly assigned to the following two categories: 30 isolates for which the MICs of norfloxacin were ≥50 μg/ml were classified as quinolone resistant, and the remaining 23 isolates for which the MICs of norfloxacin were ≤12.5 μg/ml were classified as quinolone susceptible. The type strain ATCC 19433 could be assigned to the quinolone-susceptible category. From these 53 isolates, we selected 31 with various levels of fluoroquinolone resistance. For the 31 isolates, sequences of the regions including the QRDRs of the gyrA and parC genes were determined. To amplify a DNA fragment including the QRDR of the gyrA gene, two oligonucleotide primers from nucleotide positions 163 to 182 and 358 to 377 (E. coli coordinates) were synthesized on the basis of the sequence of the E. faecalis gyrA gene reported by Korten et al. (10). Amplification of the region including the QRDR of the parC gene was performed as described above.
FIG. 2.
Distributions of MICs of ciprofloxacin for 53 clinical isolates of E. faecalis.
Mann-Whitney’s U test was used for statistical analysis. A P value of <0.05 was regarded as statistically significant.
Table 1 shows a summary of the analysis of the 31 clinical isolates of E. faecalis for an association of alterations in GyrA and ParC with susceptibilities to fluoroquinolones. Twelve isolates in the fluoroquinolone-susceptible category had no alterations in the QRDR of either GyrA or ParC. Three isolates had AGC rather than AGT for serine at position 80. Conversely, of 19 isolates in the quinolone-resistant category, 17 isolates had single amino acid changes in both GyrA and ParC. One of the 19 isolates had a single amino acid change in GyrA and a double change in ParC. The remaining isolate had a single amino acid change in ParC with no alterations in GyrA. The ATC (Ile) and ATT (Ile) at codon 80 in ParC observed in this study are likely to have arisen by one-base changes from AGC (Ser) and AGT (Ser), respectively. This would suggest that the drug-resistant mutants must be derived from at least two different parental strains. The 18 isolates with alterations in both GyrA and ParC in the fluoroquinolone-resistant category were significantly more norfloxacin, ofloxacin, and ciprofloxacin resistant than the 12 isolates in the fluoroquinolone-susceptible category (P < 0.001).
TABLE 1.
Alterations in GyrA and ParC in clinical isolates of E. faecalis
| Strain | MIC (μg/ml)a
|
Amino acid (codon) at the positions indicatedb
|
|||||
|---|---|---|---|---|---|---|---|
| NFLX | OFLX | CPFX | GyrA
|
ParC
|
|||
| 83 | 87 | 80 | 84 | ||||
| Type strain | 6.25 | 3.13 | 1.56 | Ser (AGT) | Glu (GAA) | Ser (AGT) | Glu (GAA) |
| F26 | 6.25 | 1.56 | 1.56 | − | − | − | − |
| B21, B86 | 6.25 | 3.13 | 1.56 | − | − | Ser (AGC) | − |
| 138 | 6.25 | 3.13 | 1.56 | − | − | − | − |
| F163 | 6.25 | 6.25 | 1.56 | − | − | Ser (AGC) | − |
| F120 | 6.25 | 6.25 | 1.56 | − | − | − | − |
| B146, F112 | 6.25 | 6.25 | 3.13 | − | − | Ser (AGC) | − |
| 103, 118, B3, and B16 | 12.5 | 6.25 | 3.13 | − | − | − | − |
| B214 | 50 | 12.5 | 3.13 | − | − | Arg (CGC) | − |
| F51 | 50 | 25 | 25 | − | Lys (AAA) | Arg (AGA) | − |
| 139 | 50 | 25 | 25 | Arg (AGA) | − | Ile (ATC) | − |
| 102 | 100 | 25 | 12.5 | Asn (AAT) | − | Ile (ATC) | − |
| F219 | 100 | 50 | 50 | Arg (AGA) | − | Ile (ATC) | − |
| D61 | 100 | 100 | 50 | Ile (ATT) | − | Ile (ATT) | − |
| F121 | 200 | 50 | 50 | − | Gly (GGA) | Ile (ATT) | − |
| B159 and F159 | 200 | 100 | 50 | Ile (ATT) | − | Ile (ATC) | − |
| F77 | 200 | 200 | 100 | Ile (ATT) | − | Ile (ATC) | − |
| B452 | 200 | 400 | 50 | Ile (ATT) | − | Ile (ATT) | Ala (GCA) |
| F185 and B281 | 400 | 100 | 100 | Arg (AGA) | − | Ile (ATC) | − |
| B242 | 400 | 100 | 100 | Arg (AGA) | − | Ile (ATT) | − |
| B17 | 400 | 100 | 100 | Ile (ATT) | − | Ile (ATC) | − |
| 114 | 400 | 200 | 100 | Ile (ATT) | − | Ile (ATT) | − |
| F32 | 400 | 200 | 100 | Ile (ATT) | − | Ile (ATC) | − |
| B150 | 400 | 400 | 100 | Ile (ATT) | − | Ile (ATC) | − |
| F192 | 400 | 400 | 100 | Ile (ATT) | − | Ile (ATT) | − |
NFLX, norfloxacin; OFLX, ofloxacin; CPFX, ciprofloxacin.
−; identical to type strain.
We did not undertake this study to prove that one-step mutants with low-level resistance to fluoroquinolones always carry a mutation in the parC gene but not in the gyrA gene or that mutation in the parC gene is a prerequisite for mutations in the gyrA gene to occur, nor have we conducted transformation experiments. We are fully aware that this study includes some limitations, such as the contributions of alterations in GyrB and ParE (6, 16, 18) or active efflux of drugs (1, 7). However, we found the existence of an isolate with an alteration in ParC but not in GyrA, and the level of fluoroquinolone resistance in this isolate was between that of the isolates with alterations in neither GyrA nor ParC and that of the isolates with alterations in both GyrA and ParC. These findings suggest that topoisomerase IV is a primary target of fluoroquinolones in E. faecalis as well as in other gram-positive bacteria such as S. aureus (4, 5, 14) and S. pneumoniae (12, 15, 20), although a recent study showed that either gyrase or topoisomerase IV can be the primary target, depending on the structure of the fluoroquinolone (17). The present study also suggests that the simultaneous presence of alterations in GyrA and ParC is commonly associated with the development of higher-level fluoroquinolone resistance in clinical isolates of E. faecalis.
Nucleotide sequence accession number.
The partial sequence of the E. faecalis parC gene reported here appears in the DDBJ, EMBL, and GenBank nucleotide sequence database under the accession no. AB005036.
Acknowledgments
We thank Kyoko Hirata for technical assistance.
REFERENCES
- 1.Baranova N N, Neyfakh A A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deguchi T, Yasuda M, Nakano M, Ozeki S, Ezaki T, Saito I, Kawada Y. Quinolone-resistant Neisseria gonorrhoeae: correlation of alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV with antimicrobial susceptibility profiles. Antimicrob Agents Chemother. 1996;40:1020–1023. doi: 10.1128/aac.40.4.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, Nishino Y, Ishihara S, Ban Y, Kawada Y. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1997;41:699–701. doi: 10.1128/aac.41.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito H, Yoshida H, Bogaki-Shonai M, Niga T, Hattori H, Nakamura S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2014–2023. doi: 10.1128/aac.38.9.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaatz G W, Seo S M, Ruble C A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1086–1094. doi: 10.1128/aac.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 9.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korten V, Huang W M, Murray B E. Analysis by PCR and direct DNA sequencing of gyrA mutations associated with fluoroquinolone resistance in Enterococcus faecalis. Antimicrob Agents Chemother. 1994;38:2091–2094. doi: 10.1128/aac.38.9.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumagai Y, Kato J-I, Hoshino K, Akasaka T, Sato K, Ikeda H. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob Agents Chemother. 1996;40:710–714. doi: 10.1128/aac.40.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz R, de la Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakanishi N, Yoshida S, Wakebe H, Inoue M, Mitsuhashi S. Mechanisms of clinical resistance to fluoroquinolones in Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1053–1059. doi: 10.1128/aac.35.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng E Y, Trucksis M, Hooper D. Quinolone resistance mutations in topoisomerase IV: relationship to flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1994;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan X-S, Ambler J, Mehtar S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan X-S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perichon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanberg S L, Wang J C. Cloning and sequencing of the Escherichia coli gyrA gene coding for the A subunit of the DNA gyrase. J Mol Biol. 1986;197:729–736. doi: 10.1016/0022-2836(87)90479-7. [DOI] [PubMed] [Google Scholar]
- 20.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tankovic J, Mahjoubi F, Courvalin P, Duval J, Leclercq R. Development of fluoroquinolone resistance in Enterococcus faecalis and role of mutation in the DNA gyrase gyrA gene. Antimicrob Agents Chemother. 1996;40:2558–2561. doi: 10.1128/aac.40.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vila J, Ruiz J, Goñi P, Jimenez de Anta M T. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:491–493. doi: 10.1128/aac.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]