Abstract
Macroautophagy is a cellular quality control process that degrades proteins, protein aggregates and damaged organelles. Autophagy plays a fundamental role in cancer, where in the presence of stressors (e.g. nutrient starvation, hypoxia, mechanical pressure), tumour cells activate it to degrade intracellular substrates and provide energy. Cell-autonomous autophagy in tumour cells and cell-nonautonomous autophagy in the tumour microenvironment and in the host converge on mechanisms that modulate metabolic fitness, DNA integrity and immune escape and, consequently, support tumour growth. In this review we will discuss insights into the tumour-modulating roles of autophagy in different contexts and reflect on how future studies using physiological culture systems may help to understand the complexity and open new therapeutic avenues.
Introduction
Macroautophagy is a cellular process that captures and degrades intracellular proteins and organelles to maintain cellular metabolism and homeostasis. Under physiological conditions, basal autophagy is essential for the removal of protein aggregates and damaged organelles that could be toxic for the cells1–4. Organism-wide autophagy-deficient adult mice displayed disrupted glucose homeostasis, lipid accumulation, neurodegeneration, muscle atrophy and liver dysfunction, ultimately reducing lifespan5. In contrast, overexpression of the essential autophagy gene Atg5 in mice induced moderate autophagy and prolonged survival6. Therefore, autophagy could be considered a crucial quality control system in mammalian cells.
Given its role in regulating key cellular processes, autophagy is involved in the pathophysiology of many diseases, including neurodegeneration and cancer. Its role in cancer, the focus of the present review, has been debated with studies showing either tumour suppressing or promoting functions7. For instance, autophagy inhibition enhanced the formation of benign neoplasms in the liver8, suggesting on the one hand, that autophagy is important to suppress tumour initiation and on the other hand also implying it could promote the progression to malignant stages. Accordingly, other findings confirmed this dual role in genetically engineered mouse models (GEMM) of pancreatic ductal adenocarcinoma (PDAC)9. In established tumours, autophagy levels are higher in poorly vascularized areas where nutrients and oxygen are scarce10. The dependency on autophagy can vary between different tumour types; for example, pancreatic cancer cells are highly dependent on autophagy to grow and support metabolic demands11. Interestingly, the activation of autophagy is not limited to the tumour cell itself, but also occurs in stromal cells, immune cells and even in distant organs of the tumour-bearing host12. Emerging evidence indicates that autophagy in these other cell types and organs is similarly important to promote and maintain tumour growth, therefore, connecting autophagy in the tumour to autophagy in the microenvironment and, more globally, in the host. This article reviews the role of autophagy in the tumour and its macro- and micro-environment and aims to provide an overview of a complex and open-ended field of study.
The molecular underpinnings of autophagy
Autophagy can be divided into three main types. The first type is microautophagy in which the lysosomal membrane engulfs restricted parts of cellular organelles13–15. The second type is chaperone-mediated autophagy in which substrates destined for degradation are recognized by the chaperone protein HSC70 and trafficked to the lysosomal machinery16–18. The last and most commonly studied type is macroautophagy (hereafter referred as autophagy) in which parts of the cytoplasm and organelles are sequestrated into double-membrane vesicles called autophagosomes. After maturation, the autophagosome fuses with lysosomes to deliver cytoplasmic materials destined for degradation by lysosomal enzymes19. This bulk autophagy process, also called the canonical autophagy pathway, is believed to occur in response to nutrient deprivation where cells non-selectively degrade cytoplasmic entities to provide metabolites and maintain survival (the role of autophagy in supporting metabolic pathways has been reviewed in20,21). However, there are more selective forms of autophagy that target specific proteins or organelles to support precise homeostatic needs. Such selective autophagy pathways have been described for mitochondria (mitophagy), peroxisome (pexophagy), nucleus (nucleophagy), ribosome (ribophagy), endoplasmic reticulum (reticulophagy), protein aggregates (aggrephagy), lipid droplets (lipophagy), cytosolic iron storing complex ferritin (ferritinophagy) and pathogens (xenophagy). Molecular mechanisms underlying canonical and non-canonical autophagy as well as organelle-specific autophagy have been reviewed in more detail elsewhere2,22–24.
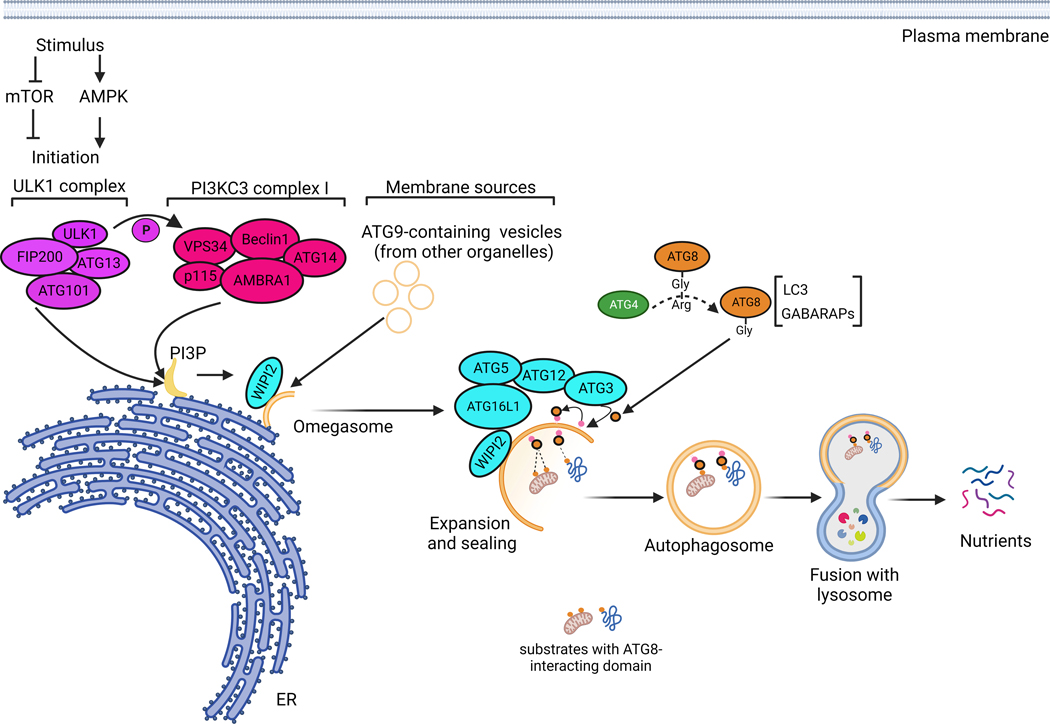
Autophagy is initiated by the ULK1 complex (ULK1/ATG13/FIP200/ATG101) and class III PI3K (PI3KC3) complex I (VPS34/BECLIN 1/ATG14/AMBRA1/p115)25,26. Both complexes translocate to the endoplasmic reticulum (ER) to form a pre-autophagosomal structure (PAS). The ULK1 complex phosphorylates components of the PI3KC3 complex, which in turn initiates the production of local phosphatidylinositol-3-phosphate (PI3P). These PI3P-enriched ER subdomains are coined with the term omegasome and can be identified by the presence of double-FYVE-containing protein 1 (DFCP1); the function of this factor is still unclear27,28. Additional sources of membranes from different organelles can be shuttled by ATG9 to the site of phagophore nucleation and seem to be important for effective autophagy initiation29–32. During phagophore expansion, most nascent ubiquitin-like ATG8s, such as LC3 and GABARAPs, are processed at their C-termini by the cysteine protease ATG433. The resulting glycine-exposed C-terminus is activated by ATG7 and ATG3 and finally conjugated to a major membrane phospholipid, phosphatidylethanolamine (PE). ATG3-mediated LC3 lipidation is stimulated by the ATG5-ATG12-ATG16L1-WIPI2 complex34–36. The resulting lipidated membrane-bound form of LC3 (LC3-II) is not only important for the interaction with specific substrates destined for degradation, but also for sealing and maturation of autophagosomes (Figure 1).
Figure 1. Molecular mechanisms of autophagy.
In response to environmental stresses, autophagy initiation is thought to take place at the endoplasmic reticulum membranes, where the ULK1 complex activates the PI3KC3 complex by phosphorylation. The PI3KC3 complex initiates the production of phosphatidylinositol-3-phosphate (PI3P) on ER subdomains forming a structure called the omegasome. In addition to PI3P provided by PI3KC3 complex, ATG9-containing small vesicles, originating from the membranes of other organelles, are involved in autophagy initiation. From the omegasome structure, a series of reactions involving a large panel of ATGs will take place to allow autophagosome elongation, sealing, maturation and fusion with lysosomes. ATG4, including ATG4B, cleaves Pro-ATG8 forms, such as LC3 and GABARAPL, to allow their conjugation into major phospholipids on the forming autophagosome. This lipidation reaction is catalysed by a complex involving WIPI2, ATG16L1, ATG5, ATG12, ATG7 and ATG3. The lipidation of ATG8s, such as LC3, is important for autophagosome elongation and maturation, but also for the interaction with cargos destined for autophagic degradation; it is therefore a major and critical step in the autophagic process.
The initiation of autophagy occurs in response to various stresses, the most important and well-characterized is starvation from amino acids, hormones and growth factors37. In this scenario, it has been proposed that the activation of autophagy is controlled by two major pathways, AMPK and mTOR. Upon a drop in nutrient levels, mTOR is inactivated and AMPK promotes autophagy by phosphorylating ULK1 on different serine residues, including ser777. In contrast, in the presence of sufficient nutrients, mTOR activity is high and prevents ULK1 activation by phosphorylation at ser75738. However, this “simple” model of duality between AMPK and mTOR does not apply to all circumstances. For example, pancreatic cancer cells, which are known to be metabolically plastic, induce autophagy in the presence of active mTOR signalling39; the activation of autophagy in this model is attributed to upregulated expression of core autophagy-lysosomal genes by the MiT/TFE family of transcription factors39. Interestingly, although mTOR is inactivated during autophagy initiation in various animal species, it is reactivated during periods of prolonged starvation to promote lysosome recycling and homeostasis40. Therefore, mTOR and AMPK may play various roles throughout the initiation, progression and termination steps of autophagy. Other pathways (e.g. Ca2+-cAMP-PKA) may be involved depending on the type of stimulus and its duration 41. Since most studies on signalling pathways governing autophagy were performed using starvation as a stimulus, it will be important for future studies to focus on other stimuli, such as hypoxia, matrix stiffness as well as mechanical and osmotic pressure, all of which are relevant to cancer pathophysiology.
The complex cellular interactions governing autophagy
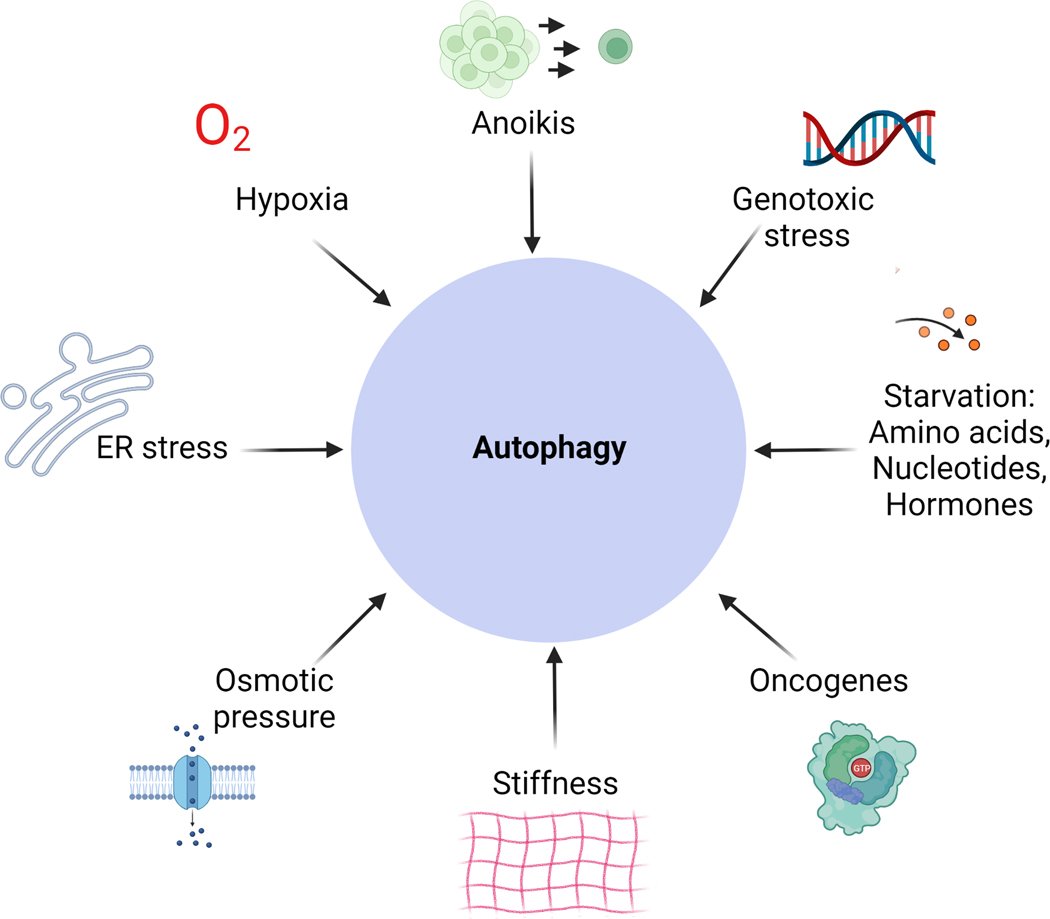
As autophagy in the tumour, the tumour microenvironment and in the host are important in promoting tumour progression, defining the potential physiological inputs that contribute to this process is a challenge that needs to be addressed in the cancer field. Some of these stressors have been already proposed and include nutrients and oxygen deprivation, anoikis, mechanical and osmotic pressure, ER stress and DNA damage42,43 (Figure 2). Nutrient deprivation has been extensively studied in the field and reviewed elsewhere2,44, hence we will focus briefly on the “less-studied” stressors that activate autophagy. ECM degradation (resulting in reduced ECM density) was shown to modulate glucose uptake in a large panel of cancer cell lines, however whether ECM detachment, nutrient uptake and autophagy constitute a continuous survival axis in cancer cells deserves further investigation45. Paradoxically, the mechanical pressure induced by cell interaction with stiff matrix and cell-to-cell contact also activates autophagy via the YAP/TAZ pathway46. This contrasting example of ECM attachment versus detachment illustrates the complexity of cellular signalling governing autophagy activation.
Figure 2. Cellular responses modulating autophagy.
A distinct set of stressors can activate autophagy in cancer cells. They do not act isolated from each other and may co-exist in the cell to activate autophagy, resulting in a highly complex signalling network.
The accumulation of unfolded proteins in the cytoplasm is a major inducer of ER stress and autophagy. Indeed, PERK-mediated phosphorylation of Eukaryotic Initiation Factor 2α (eIF2α), allows the specific translation of activating transcription factor 4 (ATF4), which in turn promotes the expression of a set of core autophagy genes47–49. ER stress promotes the release of calcium ions (Ca2+) into the cytoplasm, leading to the activation of Ca2+-dependent proteases, including Calpain. Calpain induces autophagy through mechanisms that are not yet fully elucidated and autophagy inducers, such as rapamycin, fail to induce autophagosome formation in cells lacking Calpain50–52. Ca2+ ions can also induce autophagy by inhibiting mTORC1 activity through the CAMKKβ/AMPK pathway53.
Other interesting inducers of autophagy are osmotic pressure and fluctuations in pH and temperature. Applying osmotic pressure on cancer cells using gradients of sorbitol or mannitol induced autophagy through the mTOR pathway, which was dependent on the polycystin-2 membrane channel, a sensor for osmotic balance54. However, more physiologic models are needed to better understand the link between osmotic pressure and autophagy in cancer. Changes in pH are a well-known characteristic of the tumour microenvironment, where cancer cells survive in a hypoxic and acidic milieu55,56. In cancer cells, acute change in extracellular pH has been shown to modulate autophagy flux in two opposite directions, with an alkaline milieu promoting autophagy, contrary to an acidic milieu where autophagy is reduced57. However, once cancer cells have adapted to their acidic environment they activate autophagy flux and become even more resistant to autophagy inhibition58. It was found that only the non-acidic normoxic areas of tumours were sensitive to autophagy inhibition by chloroquine58, providing a potential explanation for the limited benefit of chloroquine in the clinic as a single agent.
Autophagy in tumour cells
Cell-autonomous autophagy is an important process that tumour cells employ to preserve their metabolic fitness and reduce DNA damage, as demonstrated in various tumour types including melanoma, glioblastoma, breast, lung, pancreas, prostate and intestinal cancers9,59–66 . A major focus was to determine the exact autophagy-derived substrates that cancer cells use to survive intense stress episodes and it was shown that autophagy maintains glutamate, α-ketoglutarate and nucleotide pools67, which serve to fuel mitochondrial TCA cycle and preserve functional mitochondria68. Other studies have pointed to the importance of autophagy, and more specifically, mitophagy, in optimizing lipid catabolism and fatty acid oxidation59,69,70. Inhibition of autophagy in a model of lung adenocarcinoma, driven by KrasG12D mutation and lack of Trp53 expression, resulted in the accumulation of defective mitochondria and the conversion of adenomas and adenocarcinomas to begin oncocytomas. In the absence of Atg7, mitochondria were unable to oxidize fatty acids (defective respiration) and ultimately accumulated in oncocytomas, suggesting a possible role of mitophagy in maintaining mitochondrial quality and lipid energy homeostasis in the tumour69. Recent evidence indicates that defective mitochondrial respiration following autophagy inhibition may also be due to the alteration of iron metabolism, which is essential for the function of iron sulphur clusters involved in electron transfer within the mitochondrial respiratory chain71,72. Mechanistically, autophagy controls intracellular iron availability through NCOA4 that acts as a cargo receptor mediating ferritin degradation by autophagy to increase free iron in the cell73. Interestingly, mitophagy not only regulates mitochondrial quality, but also mitochondrial mass to optimize tumour progression74. In a model of KRAS-driven pancreatic cancer, authors showed that mutant KRAS induces the expression of BNIP3L, which in turn induces mitophagy in cancer cells, leading to reduced glucose consumption and increased redox buffering. BNIP3L deletion reduces mitophagy and induces the accumulation of mitochondria, which in turn enhance glucose consumption and reactive oxygen species production74. The increase in mitochondrial mass resulted in a decrease in cell proliferation under nutrient scarce conditions, leading to a delay in tumour progression in vivo. These studies and many others placed autophagy as a central node in modulating mitochondrial oxidative phosphorylation and, subsequently, buffering intracellular oxidative damages67,75. In a recent study, we provided direct evidence for the role of autophagy in regulating redox homeostasis in pancreatic cancer cells; namely, by demonstrating that autophagy maintains intracellular cysteine levels through recycling and targeting the cystine transporter SLC7A11 at the plasma membrane76,77.
Through preserving the global metabolic and redox balance in the cell, autophagy protects DNA integrity. Indeed, autophagy inhibition increases DNA damage, micronuclei formation, chromosome instability and aneuploidy78–81. Interestingly, autophagy-competent cells were shown to use the error-free homologous DNA repair process, whereas autophagy-deficient cells rely on the error-prone non-homologous end joining repair82; however, the exact mechanisms underlying the switch in DNA repair processes are not completely understood (Figure 3). In response to genotoxic stress, chaperone-mediated autophagy is increased to degrade checkpoint kinase 1 (CHK1)83, which in the nucleus compromises cell cycle progression and delays the DNA repair response83. Following DNA damage, histone2A is ubiquitinated by the E3 ubiquitin ligase RNF168, an essential step for the recruitment of DNA repair factors. In autophagy-deficient cells, the autophagic cargo p62 accumulates in the cell and binds to RNF168, thereby preventing its chromatin-ubiquitinating activity and the subsequent activation of the DNA repair response84. These two studies reveal distinct mechanisms linking autophagy and DNA integrity. Oncogene-induced replication stress activates the DNA damage response followed by autophagy activation85. The latter is needed for effective recovery from replication stress and to enhance the speed of the replication fork, possibly through maintaining intracellular nucleotide pools85. Indeed, knockout of Atg5 and Atg7 induces spontaneous replication stress and DNA damage85. Therefore, autophagy can promote tumour progression through multiple mechanisms, including by controlling mitochondrial function and mass, as well as ensuring DNA stability and replication.
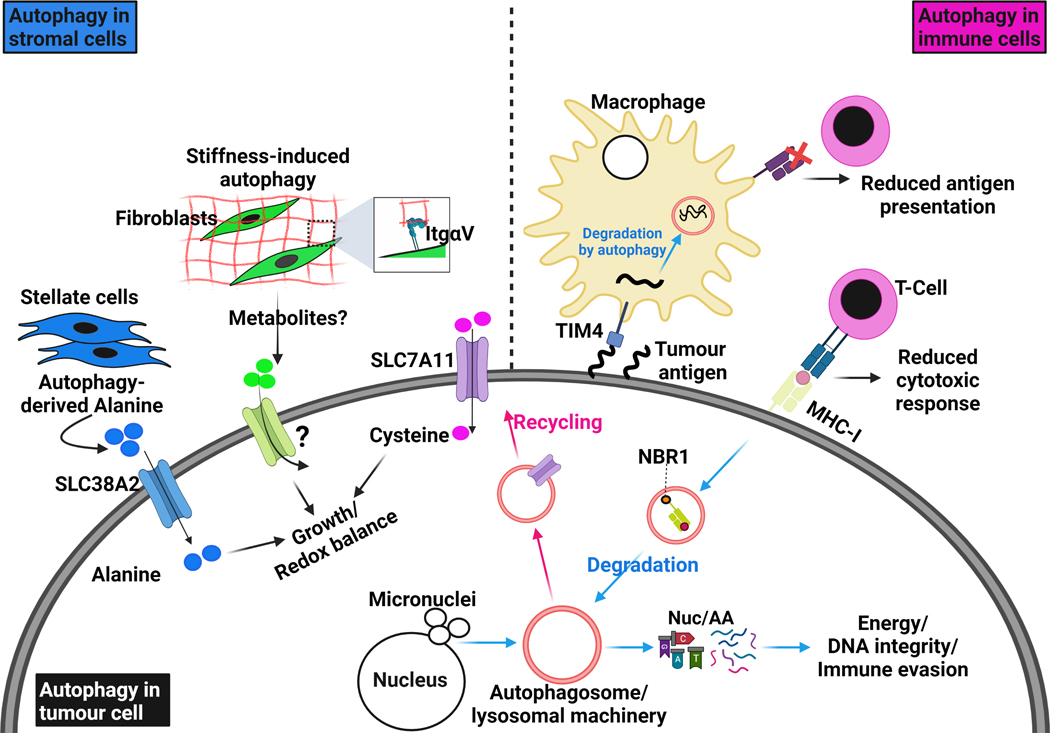
Figure 3. Autophagy in the tumour and surrounding microenvironment.
Autophagy is involved in complex crosstalk between tumour cells and their surrounding microenvironment. Cell-autonomous autophagy in tumour cells is important to relieve replication stress and maintain DNA stability by removing damaged chromosomes and micronuclei, which are the result of aberrant proliferation. It also degrades and recycles essential components for redox homeostasis like the cystine transporter SLC7A11. As energy demands in tumour cells are progressively increasing, they produce factors that activate the stromal component in their microenvironment. Tumour-stimulated stromal cells activate autophagy to release amino acids, such as alanine, which in turn are up-taken by tumour cells through specific transporters, like SLC38A2, to fuel their metabolic networks. Integrin-mediated interaction with stiff extracellular matrix (ECM) directly activates autophagy in stromal cells, such as fibroblasts and stellate cells, and provides tumour cells with growth advantages. More studies are still needed to understand whether ECM stiffness modulates nutrients release from stromal cells. Therefore, autophagy in tumour cells and stromal cells provides the former with sufficient amino acids and nucleotide pools to survive intense episodes of stress. The degradation of surface proteins such as MHC-I by autophagy allows tumour cells to escape anti-tumor immunity; MHC-I degradation and delivery to autophagosomes is dependent on the cargo NBR1. Interestingly, autophagy in immune cells can also sense and degrade tumour antigens through TIM4 and reduce their presentation to cytotoxic T-cells. All these examples illustrate the autophagy-dependent crosstalk that exists between tumour cells and their surrounding microenvironment.
The use of autophagy by tumour cells goes beyond the above-mentioned pathways and includes aspects related to immune surveillance. In a recent study, pancreatic cancer cells were found to target the MHC-I molecule through the cargo receptor NBR-1, for degradation by the autophagosome/lysosomal system86,87. This degradation process was dependent on canonical autophagy, but not on LC3-associated phagocytosis (LAP). Indeed, genetic and pharmacologic autophagy inhibition enhanced anti-tumour immune response and sensitized tumours to immunotherapy87. In addition, breast tumour cells escape immune killing by activating autophagy to degrade granzyme B produced by natural killer (NK) cells88. Another study found that hypoxia-induced autophagy in tumour cells protects them against the cytolytic activity of T-cells89. Lung tumours with a high mutational burden were shown to have increased autophagy to compromise immunoproteasome activity and antigen presentation and the inhibition of autophagy using ULK1 inhibitors was found to synergize with PD-1 blockade to reduce tumour growth90. In a mammary tumour model, the ULK1 complex member FIP200 has been shown to interact with 5-Azacytidine induced 2 (AZI2) to inhibit Tank binding kinase 1 (TBK1)-derived interferon response91. Accordingly, FIP200 deletion resulted in enhanced interferon signalling and increased CD8+ T-cell infiltration91. Functional genetic screens, in vitro and in vivo, revealed the involvement of core autophagy genes in a conserved pathway allowing cancer cell evasion from cytotoxic T lymphocytes and mediating resistance to TNF and IFN-γ in multiple syngeneic tumour models92. Another set of CRISPR screens revealed that autophagy in pancreatic tumour cells serves as a determinant factor to escape CD8+ T cell-mediated killing; specifically, autophagy rendered tumour cells more resistant to TNF-α-induced cell death93. Taken together, these studies emphasize the importance of cell-autonomous autophagy for tumour survival; however, emerging evidence indicate that cell-nonautonomous autophagy in the tumour microenvironment is also determinant for tumour cell fate (Figure 3).
Autophagy in the tumour microenvironment
The tumour microenvironment is composed of stromal, immune, nerve and endothelial cells. Stromal cells, such as fibroblasts, are the most prevalent type and are responsible for excessive intra-tumoral collagen and extracellular matrix (ECM) deposition, a characteristic shared by many aggressive cancers. This fibrotic stromal reaction, also called desmoplasia, is dependent on autophagy in stromal cells94; it stiffens the tumour microenvironment, thereby limiting the accessibility of cancer cells to nutrients and oxygen. In this austere milieu, the crosstalk between the stromal compartment and tumour cells becomes crucial for survival. Interestingly, using co-cultures of human pancreatic cancer and stellate cells tumour-derived factors were found to stimulate autophagy flux in stellate cells, which in turn released the non-essential amino acid (NEAA) alanine at high concentrations. This study showed that when essential nutrients like glucose and glutamine are scarce in the microenvironment, tumour cells shift their dependency on NEAA to fuel TCA cycle and lipid biosynthesis95. In 2020, a follow-up study identified SLC38A2 as the main transporter expressed by pancreatic cancer cells to mediate the import of stellate cells-derived alanine96; however, whether the expression of SLC38A2 in pancreatic cancer cells is by itself regulated by autophagy warrants further investigation. Autophagy in pancreatic stellate cells has been shown to be associated with reduced overall survival in patients and inhibition of autophagy in stellate cells reduced primary tumour growth and liver metastasis in mice97. In addition to direct cell-cell communication, the stiffness of the ECM has been shown to be an important driver of autophagy within fibroblasts and ECM stiffness was shown to activate AMPK by stabilizing its protein half-life via surface Integrin-αV98. Accordingly, stiffness-induced autophagy in stellate cells conferred a growth advantage for pancreatic tumour cells when co-injected into mouse pancreata98.
Endothelial cells constitute an important component of the tumour microenvironment, and execute autophagy to modulate intra-tumour vasculature. Indeed, ablation of Atg5 specifically in the murine endothelial compartment provoked the formation of immature and tortuous blood vessels within melanoma tumours, leading to a reduction in tumour growth99. However, autophagy inhibition in endothelial cells did not affect the metastatic potential of cancer cells99, suggesting that autophagy in different cell types may preferentially promote either tumour growth and/or metastasis. Different studies highlighted the responsiveness of tumour endothelial cells to activators of the stimulator of interferon genes (STING) pathway. Intra-tumour injection of STING agonists cGAMP or RR-CDA increased the expression of genes related to type-I interferon and angiogenesis in endothelial cells, which led to enhanced CD8+ immune cell infiltration100,101. Accordingly, combination of STING activation with immune checkpoint blockade induced a complete regression of melanoma, colon and breast tumours100,101. Since pro-autophagy proteins and cargoes including ATG12, ATG5 and p62 can bind elements in the STING pathway and target them for degradation102–104, it is tempting to speculate that autophagy in endothelial cells can modulate immune cell activation at least partly through inhibiting the STING pathway. This possibility deserves further investigations.
Autophagy is an important physiological process for immune cell differentiation, survival and fitness105–107 but in the context of cancer has been shown to mediate immunosuppressive actions to facilitate tumour progression. For example, autophagy is essential for the survival and function of the immunosuppressive regulatory T-cells (Tregs)108. Other groups found that ablation of essential core autophagy genes in T-cells provoked a significant increase in their tumour killing capacities109. In addition, autophagy inhibition elicits antitumour immune response by favouring the polarization of the anti-inflammatory M2 macrophage to the proinflammatory M1 phenotype110,111; cytokines released by M1 macrophages promote a more effective chemotaxis, leading to enhanced T-cell intra-tumour infiltration. Interestingly, tumour cells undergoing chemotherapy produce factors that induce the expression of the surface glycoprotein TIM-4 on tumour-associated macrophages112. TIM-4 interacts with AMPK to activate autophagy in macrophages upon uptake of dying tumour cells, therefore leading to reduced antigen presentation and increased tumour resistance to conventional chemotherapies112. These studies clearly show that autophagy in stromal and immune cells is highly connected to cell-autonomous autophagy in tumour cells and autophagy at both levels seems to be essential for successful tumour progression (Figure 3).
Autophagy, metastasis and cancer stem cells
Autophagy in tumour cells and their microenvironment impacts the ability of tumour cells to disseminate into secondary organs, for example by promoting the release of pro-migratory factors, including interleukine-6, matrix metalloproteinase 2 (MMP2) and WNT5A113. It is also involved in the biogenesis and release of exosomes, which may facilitate cancer cell metastasis114. In addition, autophagy has been shown to either promote or supress epithelial-to-mesenchymal transition (EMT)115, a process preceding cancer cell extravasation from the primary tumour to distant organs. Two studies in breast cancer and glioblastoma have shown that the master EMT regulators TWIST and SNAIL can be degraded by autophagy through interaction with BECLIN 1116,117. Another study in squamous cell carcinoma demonstrated that autophagy inhibition resulted in accumulation of p62, which in turn bound to TWIST to stabilize its expression and promote metastasis118. On the other hand, knockout studies of different core autophagy genes found the reversed EMT phenotype, also known as, mesenchymal-to-epithelial transition (MET)113,119.
Following extravasation from the primary tumour site, cancer cells need to survive matrix detachment in the bloodstream, a process termed anoikis, which was shown to be dependent on autophagy120. Upon detachment, cells upregulate protein kinase R (PKR)–like endoplasmic reticulum kinase (PERK), which in turn activates AMPK and inhibits mTOR signalling, leading to autophagy induction. Interestingly, the dynamics of PERK signalling to AMPK and mTOR were only observed during anoikis and could not be recapitulated using ER stress inducers120. Other mechanisms of resistance to anoikis via autophagy have been reported such as the activation of the NF-κB (nuclear factor-κB) pathway 121 and the autophagic degradation of RHOA, which regulates anoikis by modulating cytoskeleton dynamics122. Indeed, the role of autophagy in anoikis resistance and metastasis has been validated in different cancer types, including hepatocellular carcinoma and ovarian cancer123–125. In addition, a link between autophagy and cancer stemness was established. Given their ability to self-renew, cancer stem cells are thought to be important for tumour aggressiveness and metastasis initiation126 (reviewed in 127). Accordingly, autophagy promotes cancer stem cell-like phenotypes by increasing the number of CD44-positive over CD24-negative cells and supporting their ability to form mammospheres in breast cancer models128,129. In addition to the role of autophagy in solid tumours, selective autophagy forms such as AMPK/FIS1-mediated mitophagy have been shown to promote leukaemia stem cell self-renewal ability, which is essential to drive the genesis of acute myeloid leukaemia130. Mechanistically, it was found in a model of hepatic cancer that mitophagy degrades p53 through its association with PINK1, thereby reducing p53 phosphorylation by PINK1 and its translocation into the nucleus, where p53 binds to NANOG promoter to prevent the expression of OCT4 and SOX2131. Therefore, bulk or selective autophagy positively regulate stemness and self-renewal ability of cancer stem cells. While there is convincing evidence to support a role for autophagy in driving cancer cell migration and invasion, the role of autophagy as a metastasis-promoter or suppressor seems to be dependent on tumour stage. Using a mammary tumour model, where autophagy was inhibited at different stages of tumour progression, autophagy was found to reduce primary tumour growth, but enhance spontaneous metastasis132. This phenotype was dependent on the accumulation of NBR1 cargo, as genetic ablation of the latter in autophagy-deficient cells reversed the spontaneous increase in metastasis132. Similarly, ablation of RUBICON induced autophagy, which in turn decreased the metastatic index in mice. Interestingly, inhibition of autophagy, at the lysosomal level, with chloroquine in this study had no impact on metastasis as compared to the genetic knockout of Atg5 and Atg12132. In contrast, another study showed that both chloroquine and tumour-specific Atg5 deletion reduce lung metastasis in a melanoma model, although the effects of both interventions on the tumour microenvironment were different99. The discrepancy in the results of these two studies could be due to the difference in tumour types (breast cancer and melanoma) and/or the genetic background of animals. They also suggest that different mechanisms may operate following genetic autophagy targeting versus pharmacological inhibition of lysosome function in the context of metastasis; an important detail that should be considered in the design of translational studies.
Autophagy in organs distant from the primary tumour
In addition to autophagy in tumour cells and their microenvironment, it is remarkable to note that host autophagy (in other tissues/organs) has also a critical impact on tumour progression and growth. Early studies using organism-wide autophagy deficient animals, showed that neonates lacking essential autophagy genes Atg5 and Atg7 only survive for few hours after birth133. In adults, autophagy-deficient mice survive for two to three months, but experience excessive muscle wasting, liver damage and inflammation, neurodegeneration and adipose tissue lipolysis, leading to premature death5. Fasting was lethal in host autophagy-deficient adult mice due to hypoglycaemia, emphasizing a global role for autophagy in regulating systemic metabolism and tissue metabolic homeostasis5. Using a complementary approach, some groups engineered mice overexpressing moderate levels of Atg56. In these animals, the increase in autophagy levels was associated with resistance to oxidative stress, improved lean mass and extended overall survival6. Although the induction of autophagy through Atg5 overexpression is clear, it is possible that Atg5 may exert some autophagy-independent functions that could have contributed to the observed phenotype134. All these prominent effects of autophagy on organism physiology raised the question on whether or not host autophagy has an impact on tumour development. Interestingly, whole-organism deletion of autophagy (Atg7 knockout model), reduced autochthonous KrasG12D- and Trp53−/−- driven lung cancers, even more potently than tumour-specific autophagy inhibition5. These findings in the same transgenic background were recently corroborated by showing that transient systemic Atg5 deletion restricts tumour growth by reducing glucose and lactate uptake135. Consequently, the reduction in carbon sources slowed down the activity of essential metabolic pathways required for tumour growth, such as TCA cycle and serine biosynthesis135. To specifically show the contribution of host autophagy to tumour growth, autophagy-competent cancer cells were subcutaneously implanted into mice with competent or deficient host autophagy136. The study showed a clear host-autophagy dependent decrease of tumour growth in different cancer types, including melanoma, urothelial carcinoma and lung cancer136. This was accompanied with systemic metabolic alterations, where a drop in arginine levels was most remarkable136. Dysfunction or alteration in de novo arginine synthesis makes tumour cells dependent on exogenous arginine uptake and occurs in many human cancers that are known as arginine auxotrophs137 and indeed, the subset of tumour allografts that regressed in autophagy-deficient hosts were arginine auxotrophs too136. The low levels of circulating arginine were due to increased circulating levels of the hepatic enzyme arginase-1 (ARG1), which degrades arginine. Indeed, the inhibition of autophagy in the host or specifically in the liver, both induced hepatic stress leading to the release of ARG1 from hepatocytes into the bloodstream and the resulting degradation of arginine136. As expected, an arginine-rich diet partially rescued circulating arginine levels and tumour growth in autophagy-deficient mice. These findings indicate that host autophagy is critical to supply arginine-auxotrophic tumours with arginine136. In another study, the same group showed that host autophagy has an immunomodulatory function on the anti-tumour immune response138. The authors describe the effect to be specific for tumours with high mutational burden. Organism-wide deletion of Atg7 reduced tumour growth by enhancing T-cell infiltration. Single-cell transcriptomics and histology analysis showed increased tumour infiltration of macrophages, dendritic cells and T-cell populations138. This response was dependent on STING and the resulting activation of interferon-γ signalling. It has been shown that mitophagy mitigates STING signalling through clearing mitochondrial DNA139. Therefore, it is possible that host autophagy inhibition impairs the clearance of mitochondrial or genomic DNA, which leads to STING activation. There may also be a possible role of tumour endothelial cells in the observed phenotype. As mentioned above, the endothelial compartment in the tumour is known to be responsive to STING agonists and is capable to effectively activate the interferon response to promote T-cell infiltration. Interestingly, liver-specific autophagy inhibition recapitulated the different phenotypes observed in organism-wide autophagy inhibition138, highlighting the prominent role of hepatic autophagy in remodelling metabolic and immune aspects in the primary tumour. In another example, using a doxycycline-inducible Atg4B dominant negative mouse model (refer to Figure 1 regarding the role of ATG4), our laboratory has previously shown that inhibition of host autophagy delays tumour take140. This effect is likely, in part, due to a reshaping of the tumour microenvironment, which hampered metabolic crosstalk between tumour and stellate cells140. Given the widespread expression of the Atg4b dominant negative, it is highly probable that autophagy inhibition in distant tissues is also contributing to this phenotype. In line with the Atg4b dominant negative model, removal of Atg13 in eye tumour cells or in their surrounding environment (epithelial cells) in Drosophila Melanogaster impaired autophagy and reduced tumour cell growth and similar data were obtained with Atg14 deletion141. Interestingly, supplementation with Atg13 DNA in the host or specifically in the tumour-bearing organ rescued autophagy and tumour growth, suggesting that both cell-autonomous and host autophagy are required for tumour growth and invasiveness. Mechanistically, tumour cells increased interleukin 6 (IL6) production, which possibly acted in an autocrine manner to increase reactive oxygen species generation within tumour cells, which in turn induced autophagy in the microenvironment. In a series of allograft experiments, the authors transplanted autophagy-competent or -deficient tumour cells into hosts with functional or inactive autophagy and found that systemic autophagy had a great impact on the growth capability of tumours141. In the same model it was shown that autophagy in host organs increases as tumour progresses and muscle wasting occurs142. The full-body removal of either Atg13 or Atg14, decreased tumour growth and reduced muscle atrophy, weight and motility loss, and body fat alterations. Specific expression of Atg13 in the tumour and the tumour microenvironment rescued tumour growth, but failed to induce muscle wasting, indicating that systemic body wasting was only dependent on systemic autophagy, while tumour growth depended on both cell-autonomous and host autophagy. This global wasting state was associated with an increase in circulating amino acids, including arginine and glutamine, which was reversed in autophagy-deficient flies. Early in the wasting process, autophagy-competent hosts showed depletion of glycogen stores and an increase in haemolymph sugar (trehalose), which was normalized in autophagy-deficient counterparts142. Therefore, similarly to what has been described in mouse models, host autophagy can control the systemic metabolism and mobilization of nutrients in tumour-bearing hosts. It is worth mentioning that the mobilization of nutrients into the circulation preceded the exponential growth of tumour, suggesting that it may contribute to accelerate tumour progression142. Using two food regimens with different carbon isotopes, the authors show that amino acids, lipids and sugars can be mobilized from host tissues to the tumour in an autophagy-dependent fashion. The findings paint a picture where the progressive loss of body biomass, orchestrated by autophagy, serves to fuel tumour growth and progression. Together, the data support a clear role for systemic or host autophagy in promoting tumour progression and cancer cachexia (Figure 4). In an attempt to provide a translational context, another team inhibited systemic autophagy intermittently to mimic what would happen in a clinical setting of cancer therapy135 and was found to extend the survival of mice and allow recovery of normal organs, while maintaining its inhibitory effects on lung tumour growth135. An analogous intermittent autophagy inhibition approach using the aforementioned Atg4B dominant negative system in pancreatic cancer, showed similar effects in mouse models9. Whether or not these findings could be recapitulated by hydroxychloroquine treatment requires further investigation, given the known potency issues of hydroxychloroquine (as recently reviewed recently in 43) and the fact that it targets the lysosome and is not selective for autophagy.
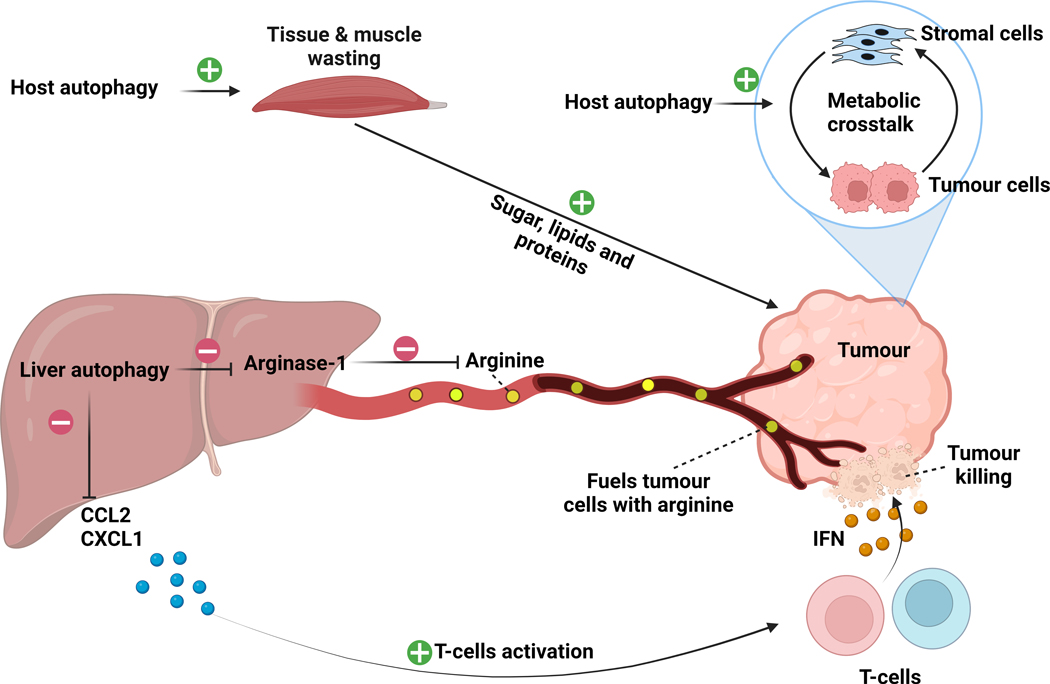
Figure 4. Autophagy in the tumour-bearing host.
Inhibition of host autophagy by expressing a dominant negative version of Atg4B impairs tumour growth by disrupting the metabolic crosstalk between tumour cells and stromal cells and may have other systemic effects in various tissues. In drosophila melanogaster, host autophagy has been shown to induce tissue and muscle wasting to fuel tumour cells with sugars, lipids and proteins. In another model, liver-specific inhibition of autophagy, through genetic Atg7 ablation, induced a stress response in the liver, leading to the release of Arginase-1 into the circulation. High circulating Arginase-1 promoted the degradation of the amino acid arginine that is necessary for the growth of a subset of arginine auxotroph tumours. In parallel, liver-specific autophagy inhibition induced the production and release of pro-inflammatory cytokines from the liver into the circulation. This pro-inflammatory state activated the CD4+ and CD8+ T-cell immune response, resulting in increased tumour killing through the IFN-γ pathway. Green positive sign indicates activation. Red negative sign indicates inhibition.
Modulation of autophagy in cancer treatment
In addition to driving the progression of many human cancers, autophagy has been shown to be elevated in response to cancer therapies, suggesting that it may also serve as a resistance mechanism. The inhibition of autophagy using hydroxychloroquine in combination with chemotherapeutic agents (e.g. gemcitabine, nab-paclitaxel, carboplatin, paclitaxel) showed an improvement in progression-free survival in lung cancer patients and improved the response to chemotherapy in lung and pancreatic cancers143,144; the addition of hydroxychloroquine in the neoadjuvant setting may also facilitate tumour resection due to local tumour shrinkage143,145. Conversely, in some circumstances, induction of autophagy in combination with chemotherapy may also be beneficial to enhance the recruitment of immune cells into the tumour bed and promote tumour killing146. The inhibition of the ERK pathway has been proposed as a treatment for RAS-mutant cancers where the RAS-RAF-MEK-ERK pathway is hyperactivated and drives cancer initiation and progression147,148. Three independent studies found that inhibition of the ERK pathway leads to autophagy activation149–151. Concurrent inhibition of the ERK pathway with trametinib and autophagy with hydroxychloroquine led to a drastic regression of tumour growth in patient-derived xenograft models of pancreatic cancer, melanoma and colorectal cancer149,150. The combination was also successful to reduce pancreatic and metastatic liver lesions in a patient, along with normalization of the circulating levels of cancer antigen 19–9149. Multiple ongoing clinical trials are assessing this approach in patients. In addition, the combination of dabrafenib (BRAF inhibitor), trametinib and hydroxychloroquine was safe and produced a high response rate in BRAF V600-mutant melanoma patients152. A recent study revealed that autophagy may also be a resistance mechanism in response to the CDK4/6 inhibitor-based therapy, suggesting that adding hydroxychloroquine may provide a greater therapeutic benefit in this context153. Using a genetic CRISPR loss-of-function screen, insulin-like growth factor 1 receptor (IGF1R) was identified as a sensitizer to hydroxychloroquine treatment154and dual inhibition of IGF1R and ERK pathways increased dependency on autophagy154. Accordingly, the inhibition of autophagy, IGFR1 and ERK pathways led to a reduction in survival in cancer cell lines and organoids154. Pancreatic tumours are known to be particularly refractory to immunotherapy. We have recently found that hydroxychloroquine treatment combined with dual immune checkpoint blockade (anti-PD1 and anti-CTLA4) induced a drastic regression of pancreatic tumours87. Combining chemotherapy, hydroxychloroquine, and anti-CTLA4 is currently being assessed in pancreatic cancer patients. Similar data were obtained in melanoma models, where inhibition of palmitoyl-protein thioesterase 1 by hydroxychloroquine synergized with anti-PD1 therapy to impair tumour growth and improve survival in mice155. Therefore, modulation of autophagy levels in combination with existing clinically-approved drugs has therapeutic benefit in preclinical and clinical settings. Optimizing these combination approaches, developing robust biomarkers of responsiveness, and bringing forward more potent and selective autophagy inhibitors will be critical for future success.
Conclusive remarks and future perspectives
Here, we have reviewed early and recent literature on the role of autophagy in cancer, highlighting a role of autophagy on three main levels: 1) in the tumour, 2) in the tumour microenvironment and 3) in the host. Autophagy and/or mitophagy in tumour cells is important to maintain functional mitochondrial metabolism, redox balance and DNA integrity. In the tumour microenvironment, autophagy in stromal cells is crucial for tumour supply with nutrients. Autophagy in both immune and tumour cells converge into mechanisms allowing tumour immune tolerance and, therefore, to the establishment of immune evasion. Even autophagy in organs distant from the primary tumour has significant impact on systemic metabolism and inflammation, which can shape the microenvironment of the tumour itself.
The physiological stressors that modulate tumour autophagy in vivo are various and complex. Depending on the location in the tumour, tumour cells are exposed to distinct stressors, resulting insubstantial heterogeneity in autophagy levels, directly impacting cell metabolism and growth. This heterogeneity may also explain why many conventional therapies are not very successful in treating patients with aggressive cancers. Previous studies have helped to understand the role of autophagy particularly in the context of nutrient starvation, while the impact of other environmental factors on autophagy has not been well characterized. Although culture models of nutrient starvation helped to elucidate the mechanistic underpinning of the autophagic machinery156,157, addressing its role in the cancer setting requires a more physiological environment. For example, 3D culture platforms may better mimic the behaviour of tumour cells and help to define which environmental factors modulate and induce the autophagic activity in tumour cells. Importantly, 3D organoids may be cultured in media with more physiological metabolite concentrations compared to the supraphysiological levels of glucose and metabolites in cell culture 158. The successful use of such models to identify key physiological autophagy regulators may hold the promise of optimally modulating autophagy levels in the tumour. Such discoveries can be aided by sophisticated tools, such as functional genomics in 3D conditions, and may pave the way for more targeted, autophagy-focused anticancer therapies.
Acknowledgements
We apologize for the omission of any primary references. This work was supported by NCI Grants P01CA117969, R35CA232124, P30CA016087-38, 1R01CA251726-01A1; the Lustgarten Foundation, and SU2C to A.C.K.
Footnotes
Competing Interest
A.C.K. has financial interests in Vescor Therapeutics and is an inventor on patents pertaining to KRAS- regulated metabolic pathways and redox control pathways in pancreatic cancer, targeting GOT1 as a therapeutic approach, targeting alanine transport, and the autophagic control of iron metabolism. A.C.K. is on the scientific advisory board of Rafael/Cornerstone Pharmaceuticals, and is advisor for OncoRev, and has been a consultant for Deciphera and Abbvie. The other authors declare no competing interests. M.A. is postdoctoral fellow at New York University Langone Health.
References
- 1.Mizushima N. & Komatsu M. Autophagy: renovation of cells and tissues. Cell 147, 728–741, doi: 10.1016/j.cell.2011.10.026 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Yoshimori T. & Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27, 107–132, doi: 10.1146/annurev-cellbio-092910-154005 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Levine B. & Kroemer G. Autophagy in the pathogenesis of disease. Cell 132, 27–42, doi: 10.1016/j.cell.2007.12.018 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B. & Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 176, 11–42, doi: 10.1016/j.cell.2018.09.048 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karsli-Uzunbas G. et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4, 914–927, doi: 10.1158/2159-8290.CD-14-0363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyo JO et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun 4, 2300, doi: 10.1038/ncomms3300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White E. The role for autophagy in cancer. J Clin Invest 125, 42–46, doi: 10.1172/JCI73941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takamura A. et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25, 795–800, doi: 10.1101/gad.2016211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang A. et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov 4, 905–913, doi: 10.1158/2159-8290.CD-14-0362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degenhardt K. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64, doi: 10.1016/j.ccr.2006.06.001 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S. et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25, 717–729, doi: 10.1101/gad.2016111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poillet-Perez L. & White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev 33, 610–619, doi: 10.1101/gad.325514.119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Duve C. & Wattiaux R. Functions of lysosomes. Annu Rev Physiol 28, 435–492, doi: 10.1146/annurev.ph.28.030166.002251 (1966). [DOI] [PubMed] [Google Scholar]
- 14.Ahlberg J. & Glaumann H. Uptake--microautophagy--and degradation of exogenous proteins by isolated rat liver lysosomes. Effects of pH, ATP, and inhibitors of proteolysis. Exp Mol Pathol 42, 78–88, doi: 10.1016/0014-4800(85)90020-6 (1985). [DOI] [PubMed] [Google Scholar]
- 15.Schuck S. Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 133, doi: 10.1242/jcs.246322 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Sahu R. et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 20, 131–139, doi: 10.1016/j.devcel.2010.12.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dice JF Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 15, 305–309, doi: 10.1016/0968-0004(90)90019-8 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Arias E. & Cuervo AM Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol 23, 184–189, doi: 10.1016/j.ceb.2010.10.009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abada A. & Elazar Z. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep 15, 839–852, doi: 10.15252/embr.201439076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimmelman AC & White E. Autophagy and Tumor Metabolism. Cell Metab 25, 1037–1043, doi: 10.1016/j.cmet.2017.04.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Encarnacion-Rosado J. & Kimmelman AC Harnessing metabolic dependencies in pancreatic cancers. Nat Rev Gastroenterol Hepatol 18, 482–492, doi: 10.1038/s41575-021-00431-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Codogno P, Mehrpour M. & Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol 13, 7–12, doi: 10.1038/nrm3249 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Sica V. et al. Organelle-Specific Initiation of Autophagy. Mol Cell 59, 522–539, doi: 10.1016/j.molcel.2015.07.021 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Kaushik S. & Cuervo AM The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19, 365–381, doi: 10.1038/s41580-018-0001-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosokawa N. et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20, 1981–1991, doi: 10.1091/mbc.E08-12-1248 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosokawa N. et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5, 973–979, doi: 10.4161/auto.5.7.9296 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Hamasaki M. et al. Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393, doi: 10.1038/nature11910 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Nascimbeni AC et al. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J 36, 2018–2033, doi: 10.15252/embj.201797006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki SW et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci U S A 112, 3350–3355, doi: 10.1073/pnas.1421092112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karanasios E. et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci 126, 5224–5238, doi: 10.1242/jcs.132415 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Manifava M. et al. Dynamics of mTORC1 activation in response to amino acids. Elife 5, doi: 10.7554/eLife.19960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura T. et al. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J 36, 1719–1735, doi: 10.15252/embj.201695189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slobodkin MR & Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem 55, 51–64, doi: 10.1042/bse0550051 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Nishimura T. et al. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep 14, 284–291, doi: 10.1038/embor.2013.6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuma A, Mizushima N, Ishihara N. & Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277, 18619–18625, doi: 10.1074/jbc.M111889200 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y. & Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem 285, 1508–1515, doi: 10.1074/jbc.M109.053520 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxton RA & Sabatini DM mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371, doi: 10.1016/j.cell.2017.03.035 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Kim J, Kundu M, Viollet B. & Guan KL AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13, 132–141, doi: 10.1038/ncb2152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perera RM et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365, doi: 10.1038/nature14587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946, doi: 10.1038/nature09076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grisan F. et al. PKA compartmentalization links cAMP signaling and autophagy. Cell Death Differ 28, 2436–2449, doi: 10.1038/s41418-021-00761-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy 4, 351–353, doi: 10.4161/auto.5523 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Amaravadi RK, Kimmelman AC & Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 9, 1167–1181, doi: 10.1158/2159-8290.CD-19-0292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell RC, Yuan HX & Guan KL Autophagy regulation by nutrient signaling. Cell Res 24, 42–57, doi: 10.1038/cr.2013.166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan WJ et al. Extracellular Matrix Remodeling Regulates Glucose Metabolism through TXNIP Destabilization. Cell 175, 117–132 e121, doi: 10.1016/j.cell.2018.08.017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pavel M. et al. Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat Commun 9, 2961, doi: 10.1038/s41467-018-05388-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yorimitsu T, Nair U, Yang Z. & Klionsky DJ Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281, 30299–30304, doi: 10.1074/jbc.M607007200 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rzymski T. et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29, 4424–4435, doi: 10.1038/onc.2010.191 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Rouschop KM et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 120, 127–141, doi: 10.1172/JCI40027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gozuacik D. et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ 15, 1875–1886, doi: 10.1038/cdd.2008.121 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Hyrskyluoto A, Reijonen S, Kivinen J, Lindholm D. & Korhonen L. GADD34 mediates cytoprotective autophagy in mutant huntingtin expressing cells via the mTOR pathway. Exp Cell Res 318, 33–42, doi: 10.1016/j.yexcr.2011.08.020 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Madden DT, Egger L. & Bredesen DE A calpain-like protease inhibits autophagic cell death. Autophagy 3, 519–522, doi: 10.4161/auto.4052 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verfaillie T, Salazar M, Velasco G. & Agostinis P. Linking ER Stress to Autophagy: Potential Implications for Cancer Therapy. Int J Cell Biol 2010, 930509, doi: 10.1155/2010/930509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pena-Oyarzun D. et al. Hyperosmotic stress stimulates autophagy via polycystin-2. Oncotarget 8, 55984–55997, doi: 10.18632/oncotarget.18995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corbet C. & Feron O. Tumour acidosis: from the passenger to the driver’s seat. Nat Rev Cancer 17, 577–593, doi: 10.1038/nrc.2017.77 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Corbet C. et al. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab 24, 311–323, doi: 10.1016/j.cmet.2016.07.003 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Xu T, Su H, Ganapathy S. & Yuan ZM Modulation of autophagic activity by extracellular pH. Autophagy 7, 1316–1322, doi: 10.4161/auto.7.11.17785 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pellegrini P. et al. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies. Autophagy 10, 562–571, doi: 10.4161/auto.27901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo JY & White E. Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy 9, 1636–1638, doi: 10.4161/auto.26123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strohecker AM et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov 3, 1272–1285, doi: 10.1158/2159-8290.CD-13-0397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie X, Koh JY, Price S, White E. & Mehnert JM Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov 5, 410–423, doi: 10.1158/2159-8290.CD-14-1473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santanam U. et al. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev 30, 399–407, doi: 10.1101/gad.274134.115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei H. et al. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25, 1510–1527, doi: 10.1101/gad.2051011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gammoh N. et al. Suppression of autophagy impedes glioblastoma development and induces senescence. Autophagy 12, 1431–1439, doi: 10.1080/15548627.2016.1190053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levy J. & Romagnolo B. Autophagy, microbiota and intestinal oncogenesis. Oncotarget 6, 34067–34068, doi: 10.18632/oncotarget.5966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levy J. et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol 17, 1062–1073, doi: 10.1038/ncb3206 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Guo JY et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev 30, 1704–1717, doi: 10.1101/gad.283416.116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo JY et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25, 460–470, doi: 10.1101/gad.2016311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo JY et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27, 1447–1461, doi: 10.1101/gad.219642.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135, doi: 10.1038/nature07976 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santana-Codina N. et al. NCOA4-mediated ferritinophagy is a pancreatic cancer dependency via maintenance of iron bioavailability for iron-sulfur cluster proteins. Cancer Discov, doi: 10.1158/2159-8290.CD-22-0043 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ravichandran M. et al. Coordinated transcriptional and catabolic programs support iron dependent adaptation to RAS-MAPK pathway inhibition in pancreatic cancer. Cancer Discov, doi: 10.1158/2159-8290.CD-22-0044 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mancias JD, Wang X, Gygi SP, Harper JW & Kimmelman AC Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109, doi: 10.1038/nature13148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Humpton TJ et al. Oncogenic KRAS Induces NIX-Mediated Mitophagy to Promote Pancreatic Cancer. Cancer Discov 9, 1268–1287, doi: 10.1158/2159-8290.CD-18-1409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Y. & White E. Autophagy suppresses TRP53/p53 and oxidative stress to enable mammalian survival. Autophagy 16, 1355–1357, doi: 10.1080/15548627.2020.1765522 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukhopadhyay S. et al. Autophagy is required for proper cysteine homeostasis in pancreatic cancer through regulation of SLC7A11. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2021475118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mukhopadhyay S. & Kimmelman AC Autophagy is critical for cysteine metabolism in pancreatic cancer through regulation of SLC7A11. Autophagy 17, 1561–1562, doi: 10.1080/15548627.2021.1922984 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mathew R. et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 21, 1367–1381, doi: 10.1101/gad.1545107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathew R. & White E. Why sick cells produce tumors: the protective role of autophagy. Autophagy 3, 502–505, doi: 10.4161/auto.4605 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karantza-Wadsworth V. et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21, 1621–1635, doi: 10.1101/gad.1565707 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karantza-Wadsworth V. & White E. Role of autophagy in breast cancer. Autophagy 3, 610–613, doi: 10.4161/auto.4867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu EY et al. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proc Natl Acad Sci U S A 112, 773–778, doi: 10.1073/pnas.1409563112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park C, Suh Y. & Cuervo AM Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun 6, 6823, doi: 10.1038/ncomms7823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y. et al. Autophagy Regulates Chromatin Ubiquitination in DNA Damage Response through Elimination of SQSTM1/p62. Mol Cell 63, 34–48, doi: 10.1016/j.molcel.2016.05.027 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Vanzo R. et al. Autophagy role(s) in response to oncogenes and DNA replication stress. Cell Death Differ 27, 1134–1153, doi: 10.1038/s41418-019-0403-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamamoto K, Venida A, Perera RM & Kimmelman AC Selective autophagy of MHC-I promotes immune evasion of pancreatic cancer. Autophagy 16, 1524–1525, doi: 10.1080/15548627.2020.1769973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto K. et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105, doi: 10.1038/s41586-020-2229-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baginska J. et al. Granzyme B degradation by autophagy decreases tumor cell susceptibility to natural killer-mediated lysis under hypoxia. Proc Natl Acad Sci U S A 110, 17450–17455, doi: 10.1073/pnas.1304790110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Noman MZ et al. Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res 71, 5976–5986, doi: 10.1158/0008-5472.CAN-11-1094 (2011). [DOI] [PubMed] [Google Scholar]
- 90.Deng J. et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1 mutant lung cancer. Nat Cancer 2, 503–514, doi: 10.1038/s43018-021-00208-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okamoto T. et al. FIP200 Suppresses Immune Checkpoint Therapy Responses in Breast Cancers by Limiting AZI2/TBK1/IRF Signaling Independent of Its Canonical Autophagy Function. Cancer Res 80, 3580–3592, doi: 10.1158/0008-5472.CAN-20-0519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lawson KA et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature 586, 120–126, doi: 10.1038/s41586-020-2746-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu XG et al. Functional Genomics In Vivo Reveal Metabolic Dependencies of Pancreatic Cancer Cells. Cell Metab 33, 211–221 e216, doi: 10.1016/j.cmet.2020.10.017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rudnick JA et al. Autophagy in stromal fibroblasts promotes tumor desmoplasia and mammary tumorigenesis. Genes Dev 35, 963–975, doi: 10.1101/gad.345629.120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sousa CM et al. Erratum: Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 540, 150, doi: 10.1038/nature19851 (2016). [DOI] [PubMed] [Google Scholar]
- 96.Parker SJ et al. Selective Alanine Transporter Utilization Creates a Targetable Metabolic Niche in Pancreatic Cancer. Cancer Discov 10, 1018–1037, doi: 10.1158/2159-8290.CD-19-0959 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Endo S. et al. Autophagy Is Required for Activation of Pancreatic Stellate Cells, Associated With Pancreatic Cancer Progression and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology 152, 1492–1506 e1424, doi: 10.1053/j.gastro.2017.01.010 (2017). [DOI] [PubMed] [Google Scholar]
- 98.Hupfer A. et al. Matrix stiffness drives stromal autophagy and promotes formation of a protumorigenic niche. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2105367118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maes H. et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell 26, 190–206, doi: 10.1016/j.ccr.2014.06.025 (2014). [DOI] [PubMed] [Google Scholar]
- 100.Demaria O. et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A 112, 15408–15413, doi: 10.1073/pnas.1512832112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang H. et al. STING activation reprograms tumor vasculatures and synergizes with VEGFR2 blockade. J Clin Invest 129, 4350–4364, doi: 10.1172/JCI125413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Takeshita F, Kobiyama K, Miyawaki A, Jounai N. & Okuda K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy 4, 67–69, doi: 10.4161/auto.5055 (2008). [DOI] [PubMed] [Google Scholar]
- 103.Wu Y. et al. Selective autophagy controls the stability of transcription factor IRF3 to balance type I interferon production and immune suppression. Autophagy 17, 1379–1392, doi: 10.1080/15548627.2020.1761653 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prabakaran T. et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J 37, doi: 10.15252/embj.201797858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Puleston DJ et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife 3, doi: 10.7554/eLife.03706 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Riffelmacher T. et al. Autophagy-Dependent Generation of Free Fatty Acids Is Critical for Normal Neutrophil Differentiation. Immunity 47, 466–480 e465, doi: 10.1016/j.immuni.2017.08.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Phadwal K. et al. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy 8, 677–689, doi: 10.4161/auto.18935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wei J. et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol 17, 277–285, doi: 10.1038/ni.3365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.DeVorkin L. et al. Autophagy Regulation of Metabolism Is Required for CD8(+) T Cell Anti-tumor Immunity. Cell Rep 27, 502–513 e505, doi: 10.1016/j.celrep.2019.03.037 (2019). [DOI] [PubMed] [Google Scholar]
- 110.Frazier JP et al. Multidrug Analyses in Patients Distinguish Efficacious Cancer Agents Based on Both Tumor Cell Killing and Immunomodulation. Cancer Res 77, 2869–2880, doi: 10.1158/0008-5472.CAN-17-0084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen D. et al. Publisher Correction: Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat Commun 9, 1808, doi: 10.1038/s41467-018-04169-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baghdadi M. et al. TIM-4 glycoprotein-mediated degradation of dying tumor cells by autophagy leads to reduced antigen presentation and increased immune tolerance. Immunity 39, 1070–1081, doi: 10.1016/j.immuni.2013.09.014 (2013). [DOI] [PubMed] [Google Scholar]
- 113.Lock R, Kenific CM, Leidal AM, Salas E. & Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov 4, 466–479, doi: 10.1158/2159-8290.CD-13-0841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Villarroya-Beltri C. et al. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun 7, 13588, doi: 10.1038/ncomms13588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marsh T, Tolani B. & Debnath J. The pleiotropic functions of autophagy in metastasis. J Cell Sci 134, doi: 10.1242/jcs.247056 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lv Q. et al. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res 72, 3238–3250, doi: 10.1158/0008-5472.CAN-11-3832 (2012). [DOI] [PubMed] [Google Scholar]
- 117.Catalano M. et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol Oncol 9, 1612–1625, doi: 10.1016/j.molonc.2015.04.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiang L. et al. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc Natl Acad Sci U S A 111, 9241–9246, doi: 10.1073/pnas.1322913111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Li J. et al. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis 34, 1343–1351, doi: 10.1093/carcin/bgt063 (2013). [DOI] [PubMed] [Google Scholar]
- 120.Avivar-Valderas A. et al. Regulation of autophagy during ECM detachment is linked to a selective inhibition of mTORC1 by PERK. Oncogene 32, 4932–4940, doi: 10.1038/onc.2012.512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen N. & Debnath J. IkappaB kinase complex (IKK) triggers detachment-induced autophagy in mammary epithelial cells independently of the PI3K-AKT-MTORC1 pathway. Autophagy 9, 1214–1227, doi: 10.4161/auto.24870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ma Z, Myers DP, Wu RF, Nwariaku FE & Terada LS p66Shc mediates anoikis through RhoA. J Cell Biol 179, 23–31, doi: 10.1083/jcb.200706097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kiyono K. et al. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res 69, 8844–8852, doi: 10.1158/0008-5472.CAN-08-4401 (2009). [DOI] [PubMed] [Google Scholar]
- 124.Peng YF et al. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy 9, 2056–2068, doi: 10.4161/auto.26398 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Cai Q, Yan L. & Xu Y. Anoikis resistance is a critical feature of highly aggressive ovarian cancer cells. Oncogene 34, 3315–3324, doi: 10.1038/onc.2014.264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Malanchi I. et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 481, 85–89, doi: 10.1038/nature10694 (2011). [DOI] [PubMed] [Google Scholar]
- 127.Nazio F, Bordi M, Cianfanelli V, Locatelli F. & Cecconi F. Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death Differ 26, 690–702, doi: 10.1038/s41418-019-0292-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cufi S. et al. Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle 10, 3871–3885, doi: 10.4161/cc.10.22.17976 (2011). [DOI] [PubMed] [Google Scholar]
- 129.Wolf J. et al. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res 15, R109, doi: 10.1186/bcr3576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pei S. et al. AMPK/FIS1-Mediated Mitophagy Is Required for Self-Renewal of Human AML Stem Cells. Cell Stem Cell 23, 86–100 e106, doi: 10.1016/j.stem.2018.05.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu K. et al. Mitophagy Controls the Activities of Tumor Suppressor p53 to Regulate Hepatic Cancer Stem Cells. Mol Cell 68, 281–292 e285, doi: 10.1016/j.molcel.2017.09.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marsh T. et al. Autophagic Degradation of NBR1 Restricts Metastatic Outgrowth during Mammary Tumor Progression. Dev Cell 52, 591–604 e596, doi: 10.1016/j.devcel.2020.01.025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kuma A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036, doi: 10.1038/nature03029 (2004). [DOI] [PubMed] [Google Scholar]
- 134.Li S. et al. A nonautophagic role of ATG5 in regulating cell growth by targeting c-Myc for proteasome-mediated degradation. iScience 24, 103296, doi: 10.1016/j.isci.2021.103296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Khayati K. et al. Transient Systemic Autophagy Inhibition is Selectively and Irreversibly Deleterious to Lung Cancer. Cancer Res, doi: 10.1158/0008-5472.CAN-22-1039 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Poillet-Perez L. et al. Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573, doi: 10.1038/s41586-018-0697-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patil MD, Bhaumik J, Babykutty S, Banerjee UC & Fukumura D. Arginine dependence of tumor cells: targeting a chink in cancer’s armor. Oncogene 35, 4957–4972, doi: 10.1038/onc.2016.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Poillet-Perez L. et al. Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nat Cancer 1, 923–934, doi: 10.1038/s43018-020-00110-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sliter DA et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262, doi: 10.1038/s41586-018-0448-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Yang A. et al. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer Discov 8, 276–287, doi: 10.1158/2159-8290.CD-17-0952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Katheder NS et al. Microenvironmental autophagy promotes tumour growth. Nature 541, 417–420, doi: 10.1038/nature20815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khezri R. et al. Host autophagy mediates organ wasting and nutrient mobilization for tumor growth. EMBO J 40, e107336, doi: 10.15252/embj.2020107336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zeh HJ et al. A Randomized Phase II Preoperative Study of Autophagy Inhibition with High-Dose Hydroxychloroquine and Gemcitabine/Nab-Paclitaxel in Pancreatic Cancer Patients. Clin Cancer Res 26, 3126–3134, doi: 10.1158/1078-0432.CCR-19-4042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Malhotra J. et al. Phase Ib/II study of hydroxychloroquine in combination with chemotherapy in patients with metastatic non-small cell lung cancer (NSCLC). Cancer Treat Res Commun 21, 100158, doi: 10.1016/j.ctarc.2019.100158 (2019). [DOI] [PubMed] [Google Scholar]
- 145.Karasic TB et al. Effect of Gemcitabine and nab-Paclitaxel With or Without Hydroxychloroquine on Patients With Advanced Pancreatic Cancer: A Phase 2 Randomized Clinical Trial. JAMA Oncol 5, 993–998, doi: 10.1001/jamaoncol.2019.0684 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Michaud M. et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334, 1573–1577, doi: 10.1126/science.1208347 (2011). [DOI] [PubMed] [Google Scholar]
- 147.Sanclemente M. et al. c-RAF Ablation Induces Regression of Advanced Kras/Trp53 Mutant Lung Adenocarcinomas by a Mechanism Independent of MAPK Signaling. Cancer Cell 33, 217–228 e214, doi: 10.1016/j.ccell.2017.12.014 (2018). [DOI] [PubMed] [Google Scholar]
- 148.Blasco RB et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663, doi: 10.1016/j.ccr.2011.04.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kinsey CG et al. Publisher Correction: Protective autophagy elicited by RAF-->MEK-->ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 25, 861, doi: 10.1038/s41591-019-0433-3 (2019). [DOI] [PubMed] [Google Scholar]
- 150.Bryant KL et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 25, 628–640, doi: 10.1038/s41591-019-0368-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee CS et al. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc Natl Acad Sci U S A 116, 4508–4517, doi: 10.1073/pnas.1817494116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mehnert JM et al. BAMM (BRAF Autophagy and MEK Inhibition in Melanoma): A Phase I/II Trial of Dabrafenib, Trametinib, and Hydroxychloroquine in Advanced BRAFV600-mutant Melanoma. Clin Cancer Res 28, 1098–1106, doi: 10.1158/1078-0432.CCR-21-3382 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goodwin CM et al. Combination Therapies with CDK4/6 Inhibitors to Treat KRAS-mutant Pancreatic Cancer. Cancer Res, doi: 10.1158/0008-5472.CAN-22-0391 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Stalnecker CA et al. Concurrent Inhibition of IGF1R and ERK Increases Pancreatic Cancer Sensitivity to Autophagy Inhibitors. Cancer Res 82, 586–598, doi: 10.1158/0008-5472.CAN-21-1443 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sharma G. et al. PPT1 inhibition enhances the antitumor activity of anti-PD-1 antibody in melanoma. JCI Insight 5, doi: 10.1172/jci.insight.133225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kirkin V. History of the Selective Autophagy Research: How Did It Begin and Where Does It Stand Today? J Mol Biol 432, 3–27, doi: 10.1016/j.jmb.2019.05.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakatogawa H, Suzuki K, Kamada Y. & Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10, 458–467, doi: 10.1038/nrm2708 (2009). [DOI] [PubMed] [Google Scholar]
- 158.Cantor JR et al. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 169, 258–272 e217, doi: 10.1016/j.cell.2017.03.023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]