Summary
Older individuals and people with HIV (PWH) were prioritized for COVID-19 vaccination, yet comprehensive studies of the immunogenicity of these vaccines and their effects on HIV reservoirs are not available. Our study on 68 PWH and 23 HIV-negative participants aged 55 and older post-three vaccine doses showed equally strong anti-spike IgG responses in serum and saliva through week 48 from baseline, while PWH salivary IgA responses were low. PWH had diminished live-virus neutralization responses after two vaccine doses, which were ‘rescued’ post-booster. Spike-specific T cell immunity was enhanced in PWH with normal CD4+ T cell count, suggesting Th1 imprinting. The frequency of detectable HIV viremia increased post-vaccination, but vaccines did not affect the size of the HIV reservoir in most PWH, except those with low-level viremia. Thus, older PWH require three doses of COVID-19 vaccine for maximum protection, while individuals with unsuppressed viremia should be monitored for adverse reactions from HIV reservoirs.
Subject areas: Immunology, Virology
Graphical abstract

Highlights
-
•
Third SARS-CoV-2 vaccine dose rescued diminished nAb responses in older PWH
-
•
PWH had diminished spike IgA responses in saliva after vaccination
-
•
Th1 imprinting from HIV enhances spike T cell immunity in PWH with normal CD4 count
-
•
HIV reservoir did not grow post-vaccine, except in PWH with unsuppressed viremia
Immunology; Virology
Introduction
People with HIV infection (PWH) may be at a higher risk of severe symptoms and mortality related to COVID-19.1,2,3 In-depth studies demonstrate a nuanced role of HIV infection in COVID-19 outcomes, arguing that co-morbidities and low CD4+ T cell count are better predictors of outcomes than the HIV diagnosis per se.4,5,6 Considering that not all PWH restore their immunity to pre-infection levels despite receiving combination antiretroviral therapy (cART), immunodeficiency may pose a serious risk in the context of COVID-19 disease. Thus, vaccine-mediated prevention of SARS-CoV-2 infection and/or severe disease is essential in this vulnerable population. There is a concern, however, that immunodeficiency may result in impaired responses to COVID-19 immunization, as has been reported for other vaccines in PWH7,8,9 and for COVID-19 vaccines in other immunocompromised groups.10,11,12,13,14 The risk of inadequate responses may be even higher for older individuals due to their aging immune system. Since the introduction of cART in 1995, life expectancy of PWH has increased to ages beyond 50.15 The incidence of HIV infection has also increased in older individuals.16 Given the concern of relative immunodeficiency in treated HIV and the effects of age on immunity, it is unclear whether living with HIV into older age affects responses to COVID-19 vaccines.
Despite cART effectiveness, HIV suppression is often insufficient to completely dampen the heightened immune activation, which itself has a negative impact on morbidity and mortality – but has also been linked to destabilized HIV reservoirs.17 A significant gap in our knowledge is whether COVID-19 vaccines may exacerbate ongoing immune dysregulations that can affect the dynamics of HIV reservoirs, as has been the case with some other vaccines.18,19,20 Previous work also demonstrated that influenza vaccines could disproportionately increase the HIV proviral burden in influenza-specific CD4+ T cells of cART-treated PWH with suppressed viral loads (VL), suggesting the ability of vaccination to affect HIV reservoirs.19 For this reason, understanding how COVID-19 vaccines affect immune activation and HIV reservoirs is paramount.
Given these concerns, we performed a longitudinal observational study to understand how PWH aged 55 and older respond to three COVID-19 vaccine doses across different arms of the immune system, compared to HIV-negative individuals. We also determined whether these vaccines destabilize HIV reservoirs, which has been a major concern for PWH.
Results
Study protocol, timeline, participants
Baseline characteristics of study participants and clinical information are provided in Tables S1 and S2, Table 1, and Figures 1A and 1B. The study visits are defined in Figure 1C and STAR Methods. In total, we recruited 91 participants: 23 HIV-negative individuals and 68 PWH on cART, including 42 immunological responders, IR (for definitions see STAR Methods), 20 immunological non-responders, INR, five PWH with low-level viremia, LLV, and one long-term non-progressor, LTNP. At the time of screening, all PWH, except the LLVs, had had viremia suppressed to undetectable levels (below 40 copies/mL) for 5–30 years.
Table 1.
Baseline characteristics of study participants
| Variable | HIV– (n = 23) | HIV+ (n = 68) | p | IRb (n = 42) | INR (n = 20) | LLV (n = 5) | LTNP (n = 1) |
|---|---|---|---|---|---|---|---|
| Age | 0.982 | ||||||
| Median (IQR) | 62.0 (58.0, 70.0) | 63.0 (58.0, 69.0) | 62.0 (57.0, 68.0) | 63.0 (60.5, 67.5) | 66.0 (63.0, 70.0) | 76.0 (76.0, 76.0) | |
| Range | (55.0, 77.0) | (54.0, 85.0) | (54.0, 85.0) | (55.0, 78.0) | (62.0, 73.0) | (76.0, 76.0) | |
| CD4 counta | – | ||||||
| Median (IQR) | 527 (364, 665) | 635 (519, 833) | 316 (252, 410) | 511 (325, 565) | 597 (597, 597) | ||
| Range | (74.0, 1784) | (347, 1784) | (74.0, 502) | (322, 764) | (597, 597) | ||
| CD4 nadir | – | ||||||
| Median (IQR) | 148 (65.0, 240) | 210 (110, 290) | 31.0 (10.0, 130) | 91.0 (70.0, 120) | 470 (470, 470) | ||
| Range | (0.0, 1090) | (0.0, 1090) | (0.0, 200) | (10.0, 220) | (470, 470) | ||
| CD4 percentagea | – | ||||||
| Median (IQR) | 30.0 (22.0, 38.5) | 36.7 (29.8, 42.9) | 19.9 (17.6, 24.8) | 24.7 (17.4, 26.1) | 35.2 (35.2, 35.2) | ||
| Range | (12.9, 62.3) | (21.2, 62.3) | (12.9, 30.1) | (15.3, 48.2) | (35.2, 35.2) | ||
| CD4/CD8 ratioa | – | ||||||
| Median (IQR) | 0.9 (0.5, 1.2) | 1.1 (0.9, 1.5) | 0.5 (0.4, 0.6) | 0.5 (0.3, 0.5) | 1.0 (1.0, 1.0) | ||
| Range | (0.2, 6.1) | (0.4, 6.1) | (0.2, 1.1) | (0.3, 1.9) | (1.0, 1.0) | ||
| HIV subgroup, n (%) | – | ||||||
| Immune responders (IR) | 42 (61.8) | ||||||
| Immune non-responders (INR) | 20 (29.4) | ||||||
| Low-level viremics (LLV) | 5 (7.4) | ||||||
| Long-term non-progressors (LTNP) | 1 (1.5) | ||||||
| Vaccine D1/D2, n (%) | 0.766 | ||||||
| mRNA/mRNA | 13 (56.5) | 35 (51.5) | 21 (50.0) | 11 (55.0) | 2 (40.0) | 1 (100) | |
| ChAdOx1/ChAdOx1 | 9 (39.1) | 27 (39.7) | 17 (40.5) | 7 (35.0) | 3 (60.0) | 0 (0.0) | |
| ChAdOx1/mRNA | 1 (4.3) | 6 (8.8) | 4 (9.5) | 2 (10.0) | 0 (0.0) | 0 (0.0) | |
| Vaccine D3, n (%) | 1.000 | ||||||
| None | 0 (0.0) | 1 (1.5) | 1 (2.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| BNT162b2 | 19 (82.6) | 54 (79.4) | 29 (69.0) | 20 (100) | 5 (100) | 0 (0.0) | |
| mRNA-1273 | 4 (17.4) | 13 (19.1) | 12 (28.6) | 0 (0.0) | 0 (0.0) | 1 (100) | |
| Time lapse between D1 and D2 (days) | 0.819 | ||||||
| Median (IQR) | 63.0 (53.0, 65.0) | 60.0 (54.0, 71.0) | 61.0 (53.0, 74.0) | 58.0 (52.5, 67.5) | 65.0 (56.0, 75.0) | 59.0 (59.0, 59.0) | |
| Range | (30.0, 78.0) | (28.0, 98.0) | (28.0, 92.0) | (28.0, 98.0) | (56.0, 84.0) | (59.0, 59.0) | |
| Time lapse between D3 and V9 (days) | 0.076 | ||||||
| Median (IQR) | 118 (107, 130) | 130 (112, 150) | 121 (110, 135) | 152 (138, 165) | 125 (112, 133) | 117 (117, 117) | |
| Range | (97.0, 189) | (82.0, 180) | (82.0, 179) | (96.0, 179) | (110, 180) | (117, 117) | |
| N+ and/or post-COVID-19, n (%) | 6 (26.1) | 10 (14.7) | 7 (16.7) | 1 (5.0) | 2 (40.0) | 0 (0.0) | |
| First visit number with N+/post-COVID-19, n (%) | |||||||
| None | 17 (73.9) | 58 (85.3) | 35 (83.3) | 19 (95.0) | 3 (60.0) | 1 (100) | |
| V1 | 1 (4.3) | 1 (1.5) | 1 (2.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| V4 | 0 (0.0) | 1 (1.5) | 0 (0.0) | 1 (5.0) | 0 (0.0) | 0 (0.0) | |
| V8 | 0 (0.0) | 1 (1.5) | 1 (2.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| V8a | 1 (4.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| V8b | 0 (0.0) | 2 (2.9) | 1 (2.4) | 0 (0.0) | 1 (20.0) | 0 (0.0) | |
| V9 | 4 (17.4) | 5 (7.4) | 4 (9.5) | 0 (0.0) | 1 (20.0) | 0 (0.0) |
p values are based on Fisher’s exact test for categorical variables and on Wilcoxon sum rank test for continuous variables. Related to Figures 1A and 1B, Table S1, and STAR Methods.
If baseline data were not available, the data at screening were used instead (n = 24).
IR = PWH who are immunological responders, INR = immunological non-responders, LLV = PWH with low-level viremia, LTNP = long-term non-progressors.
Figure 1.
Study protocol, timeline, participants
(A) HIV status of study participants. PWH: IR – immunological responders, INR – immunological non-responders, LLV – PWH with low-level viremia, LTNP – long-term non-progressors. See also Tables 1, S1.
(B) Age distribution. See also Tables 1, S1.
(C) The timeline showing study visits (in blue, gray, or green) and clinically administered COVID-19 vaccine doses D1-3 (red). See also Tables 1, S1, and S2.
The age of participants ranged between 55 and 85 years, with median age in PWH being 63 (IQR 58–69), and 62 in the HIV− cohort (IQR 58–70). One participant was a female (IR), the rest were males; 79 were Caucasian (86.8%), four were Latino (4.4%), and the rest included Black, Middle Eastern, South Asian (two individuals each, or 2.2%), one East Asian and one Native Brazilian (1.1% each).
As this was an observational study, participants had a variable vaccine regimen of one to three COVID-19 vaccines that had been approved in Ontario, Canada, by the time this study commenced (Tables 1, S1): an adenoviral vector vaccine ChAdOx1 and two mRNA vaccines, BNT162b2 and mRNA-1273.21,22,23,24,25,26 We did not observe significant differences in D1 and D2 vaccine regimens between PWH and HIV− (Table 1); all participants received an mRNA vaccine booster (D3). Importantly, the time interval between D1 and D2 was similar for PWH and HIV−, as was the one between D3 and V9 (Table 1).
Eighty-nine participants completed the protocol (Table S2). Volunteer OM5128 dropped out after the primary endpoint visit (V8), and CIRC0319 died shortly before the last study visit. This death was not related to the study or vaccination.
Rates of breakthrough SARS-CoV-2 infection increased with the emergence of Omicron VOC, but were not higher in PWH
Assessment of vaccine efficiency was beyond the scope of this study. In order to discriminate between vaccine-induced changes and those from potential SARS-CoV-2 infection, we determined potentially convalescent samples based on clinically confirmed SARS-CoV-2 infection (by PCR or rapid antigen test) and tested all sera at six study visits with ELISA for anti-N (nucleocapsid) IgG. We did not observe higher rates of breakthrough infection in PWH than in HIV-negative participants – rather, the trend was the opposite (Table 1). One PWH and one HIV− (2.2%) had anti-N IgG in sera at baseline, i.e., had possibly been infected before getting vaccinated. Fourteen more (15.7% of the remaining 89) had a breakthrough infection between D1 and V9, including nine PWH (13.4%) and five HIV− individuals (22.7%). Interestingly, most breakthrough infections occurred after the third vaccine dose (11/16, or 68.8%), i.e., at the time of the spread of the more contagious Omicron variants from late 2021 through early 2022. The samples we presumed convalescent were further considered in sensitivity tests and excluded from some analyses.
Three doses of COVID-19 vaccines elicit equally high levels of serum anti-spike/RBD IgG that increase with each dose
Neutralizing Abs (nAbs) against SARS-CoV-2 spike protein, particularly its receptor-binding domain (RBD), are a major effector in protection against this infection. We used ELISA assay to assess longitudinally anti-RBD (Figure 2; Table S3) and anti-spike (Figure S1; Table S4) IgG responses in sera at eight timepoints before and after each of the three vaccine doses. At each timepoint, we analyzed differences between total PWH and HIV−, IR and HIV−, INR and HIV−, and IR vs. INR (Tables S5 and S6). IgG levels were not normally distributed and were log-transformed to satisfy the required assumption for the regression analysis. Formal comparisons of loge IgG levels between the groups were based on mixed effects linear regression. We also analyzed longitudinal changes in IgG levels within groups and subgroups between adjacent timepoints (Tables S7 and S8).
Figure 2.
Three doses of COVID-19 vaccines elicit equally high levels of serum anti-RBD IgG that increase with each dose
The top panel shows a violin plot with medians for IgG concentrations (BAU/mL) in HIV− (blue), total PWH (green), IRs (cyan), INRs (purple); N+ and/or post-COVID-19 samples are excluded. The bottom panel shows adjusted loge mean differences between PWH and HIV− individuals, based on mixed effects linear regression. p values for loge mean differences between HIV+ and HIV− samples at each timepoint are shown at the top of the bottom panel: p < 0.001 (∗∗∗), p < 0.01 (∗∗), p < 0.05 (∗), p ≥ 0.05 (ns = ‘not significant’). p values for within-group/subgroup changes from the preceding timepoint for each pair of adjacent timepoints are shown at the top of the top panel and are color-coded accordingly. p values for IR vs. INR differences are shown below each pair at the bottom of the top panel. The lower limit of detection, LLOD (3.02 BAU/mL), the upper limit of quantification, ULOQ (4,454 BAU/mL), and the seropositivity threshold (31 BAU/mL) are shown as dashed horizontal lines. LLV participants are shown as red dots. Vaccination timepoints (D1, D2, D3) are indicated with red arrowheads. See also Figure S1, Tables S3, S5, and S7.
We observed clear RBD and spike IgG responses in PWH and HIV− participants that were boosted by each subsequent dose (Figures 2, S1), with little difference in Ab levels between the two groups (Tables S5 and S6). For RBD IgG, the median baseline levels were below the seropositivity threshold of 30.97 BAU/mL in all groups, while at 4 weeks after D1 (V4), the median value in PWH went up to 45.6 BAU/mL (V4 mean vs. V1 p < 0.001) and 64.5 in HIV− (V4 mean vs. V1 p < 0.001). At 2 weeks post-D2, the median levels jumped to 893 in PWH (V6 mean vs. V4a p < 0.001) vs. 1,169 in HIV− (V6 mean vs. V4a p < 0.001). Finally, the RBD IgG peaked at 4 weeks after D3 (V8b) at 2,655 BAU/mL in PWH (V8b mean vs. V8a p < 0.001) vs. 1,983 in HIV− (V8b mean vs. V8a p < 0.001). The differences between PWH and HIV− means at these three timepoints were not statistically significant (Figure 2; Tables S3; S5; and S7). The same pattern was observed for spike IgG (Figure S1; Tables S4; S6, and S8). Of note, at V8b, RBD IgG responses in IRs were significantly higher than in the HIV− group, with adjusted log difference in mean being 0.89 (p = 0.022, with N+/post-COVID-19 samples excluded), which was the only significant difference in serum IgG responses that we observed between PWH and HIV− participants.
An increase in serum IgG levels post-vaccine was followed by decay, as seen at V4a and V8-8a (Figure 2, S1). Interestingly, the change from the peak values at V8b to the last study visit (V9) suggest a slower decay of Abs following D3 – see the model below for more details.
No significant statistical differences were observed between IRs and INRs, although INRs showed a trend for lower IgG levels at most timepoints.
At 48 weeks (V9), after three vaccine doses, we registered the following median IgG concentrations in sera: for spike, 916 BAU/mL in PWH vs. 919 in HIV− (p = 0.624), and for RBD, 706 BAU/mL in PWH vs. 752 in HIV− (p = 0.198).
PWH had lower SARS-CoV-2 nAb titers after two vaccine doses than HIV− individuals, with responses ‘rescued’ by the booster
We performed live SARS-CoV-2 microneutralization (MN) to assess the neutralization capacity of sera following COVID-19 vaccination of PWH and HIV− individuals at baseline, V8 (after two vaccine doses) and V9 (after three vaccine doses). Baseline 50% neutralization titers (NT50) were summarized as dichotomized variables (0 vs. > 0) given that most data were zero (Table S9). As expected, no differences were observed at V1. Post-baseline data were non-normal even after log-transformation, and the intergroup/subgroup comparisons were based on adjusted quantile regression (Tables S10 and S11).
In contrast to spike and RBD IgG responses, at 24 weeks, we observed markedly lower SARS-CoV-2 neutralization titers in PWH vs. HIV− participants after two COVID-19 vaccine doses (p < 0.001). The median NT50 in PWH was 82.9 vs. 535 in HIV−, with N+/post-COVID-19 samples excluded (Figure 3A; Tables S10 and S11).
Figure 3.
PWH had lower SARS-CoV-2 nAb titers after two vaccine doses than HIV– individuals, with responses ‘rescued’ by the booster
(A) Live SARS-CoV-2 50% neutralization titers (NT50) in HIV− individuals (blue), total PWH (green), IRs (cyan), INRs (purple); the horizontal bars show median titers. The bottom panel shows adjusted median differences between PWH and HIV− participants at each timepoint (based on quantile regression), with p values shown at the top: p < 0.001 (∗∗∗), p < 0.01 (∗∗), p < 0.05 (∗), p ≥ 0.05 (ns). p values for within-group/subgroup changes from V8 to V9 are shown at the top of the top panel and are color-coded accordingly. Vaccination timepoints (D1-2, D3) are indicated with red arrowheads. See also Figure S2, Tables S9–S11.
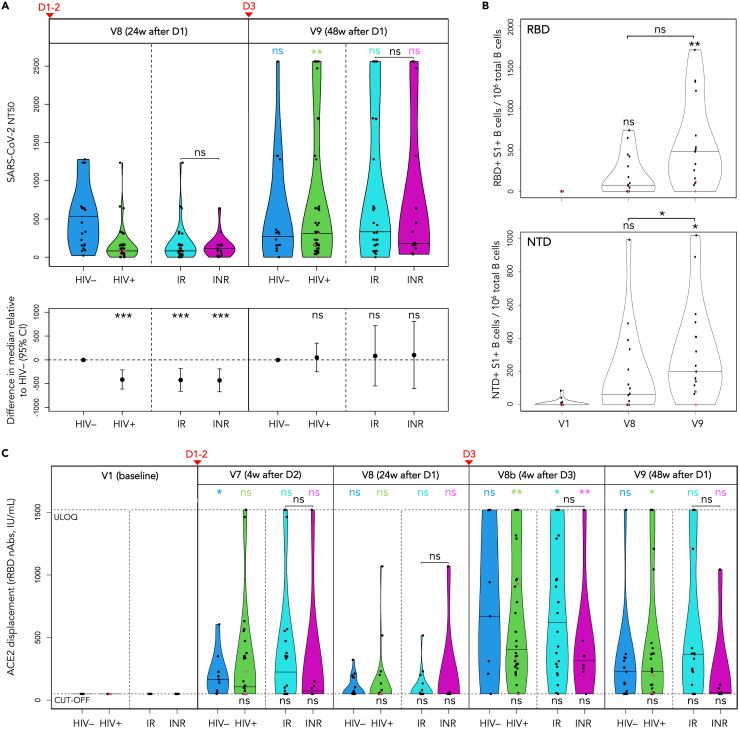
(A–C) LLV participants are shown as red dots. (B) Changes in frequency of RBD/NTD-specific B cells in selected PWH (n = 17) following COVID-19 vaccination expressed as the number of RBD+S1+ and NTD+S1+ B cells per 106 total B cells, based on spectral flow cytometry data. The horizontal bars show median frequencies. p values are based on quantile regression. See also Figures S2A and S2B, Table S12.
(C) Changes in concentrations of anti-RBD nAbs in sera (IU/mL) following COVID-19 vaccination: in HIV− participants (blue), total PWH (green), IRs (cyan), INRs (purple). The responses are measured with snELISA based on the ability of sera to displace rACE2 and analyzed by adjusted quantile regression. The horizontal bars show median concentrations; the neutralization cut-off (54.8 IU/mL) and ULOQ (1,520 IU/mL) are shown as dashed horizontal lines. p values for differences between PWH and HIV− participants at each time point are shown at the bottom. p values for within-group/subgroup changes from the preceding timepoint for each pair of adjacent timepoints are shown at the top and are color-coded accordingly. Vaccination timepoints (D1-2, D3) are indicated with red arrowheads. See also Figure S2C, Tables S13 and S14.
At 48 weeks, the pattern changed considerably, with PWH responses having been ‘rescued’ by the D3 booster: the median NT50 value increased in PWH from 82.9 to 309 (p = 0.009); the change in HIV− was not statistically significant (p = 0.858). The adjusted median difference between PWH and HIV− individuals, as a result, decreased from −413 at V8 (p < 0.001) to 49.1 at V9 (p = 0.745).
The difference between IR and INR was not significant at V8. The V9 increase, however, was more pronounced in the former, albeit not significant. Therefore, IRs contributed the most to the ‘rescue’ of PWH responses post-D3 (Table S10): median 332 in IR vs. 177 in INR. The adjusted median difference between IRs and INRs at V9 had a wide 95% CI and was not significant (Table S11). LLVs and our only LTNP showed approximately a two-fold increase in their NT50 titers from V8 to V9 (Table S10). However, formal comparisons involving these subgroups were not performed due to the small sample size.
Third vaccine dose boosts spike-specific B cells
The discrepancy that we observed in PWH between total spike/RBD IgG responses and neutralizing titers in sera prompted us to evaluate vaccine-induced B cell responses. We focused on circulating B cells targeting RBD and NTD (N-terminal domain) – two domains within the S1 subunit of the SARS-CoV-2 spike that represent mutational hotspots and antigenic supersites for nAbs.27,28,29,30,31,32 The responses were measured by spectral flow cytometry at baseline, V8 and V9 in selected PWH with similar D3-V9 intervals (n = 17) and reported as frequencies: the number of RBD+S1+ and NTD+S1+ B cells per 106 total B cells (Table S12; Figure 3B). HIV-negative participants were not included due to a limited number of specimens collected from this cohort.
Similarly to the NT50 titers, we only observed a moderate increase in frequencies of RBD+S1+ and NTD+S1+ B cells from V1 to V8 (Figure 3B; Table S12). The third dose significantly boosted the RBD+S1+ and NTD+S1+ B cells, with median frequencies increasing from 73.6 at V8 to 480 at V9 for RBD, and from 62.6 to 200 for NTD. The change from baseline to V9 was significant for both domains (p = 0.008 and p = 0.034, respectively), whereas the change from V8 to V9 was only significant for the NTD+S1+ B cells (p = 0.036).
Frequencies of RBD+S1+ and NTD+S1+ B cells showed moderate positive correlation with NT50 titers (Figures S1A and S1B): ρ = 0.46 and ρ = 0.54, respectively (p < 0.001). Interestingly, the only participant without either response (RBD or NTD) at V9 was the LLV individual OM5094 who also had negligible or undetectable NT50 titers at V8 and V9.
RBD surrogate neutralization ELISA reaffirms the benefits of the D3 booster, points to weaker responses in INRs
We performed snELISA to estimate longitudinal changes in the concentration of anti-RBD nAbs based on the capacity of sera to prevent the binding of the labeled ACE2 receptor (the mediator of the SARS-CoV-2 cell entry) to immobilized RBD.33,34
All V1 data were below the neutralization cut-off of 54.8 IU/mL (Tables S13 and S14; Figure 3C). In line with assays described above, the boosting effect from D3 was confirmed by the ACE2 displacement. With regards to responses in PWH at V8 and V9, the observed pattern was similar to that of NT50 titers, with much higher nAb concentrations at V9, which reaffirms the critical role of the booster. Likewise, we note little difference in median V9 values between PWH and HIV− participants: 232 IU/mL vs. 228, respectively. Spearman correlation test for snELISA and NT50 datasets for the two timepoints (Figure S2C) showed a moderate positive correlation for the V8 dataset (ρ = 0.56, p < 0.001) and strong for V9 (ρ = 0.79, p < 0.001).
Higher responses at V7 vs. V8, and V8b vs. V9 demonstrate a relatively fast decay of RBD nAbs, particularly after D1-2. Neutralizing Abs targeting RBD peaked at 4 weeks post-D3 (V8b), and the difference between PWH and HIV− was not statistically significant (Tables S13 and S14): the median value in PWH increased from undetectable at V8 to 403 IU/mL at V8b (p = 0.007).
We did not observe statistically significant differences between IRs and INRs, but a clear trend for weaker median responses in the latter was evident: 225 IU/mL vs. 73.0 at V7, 621 vs. 315 at V8b, and 367 vs. 59.9 at V9, respectively. Interestingly, the responses in the LLV participants were among the weakest, often undetectable (Figure 3C; Table S13).
Strong anti-spike IgG responses to three mRNA vaccine doses in saliva contrast with weak anti-spike IgA responses, with little effect from the booster on IgA
Mucosal immunity of the upper respiratory tract and oral cavity, particularly its IgA component, is a first-line defense against SARS-CoV-2, but one with many unknowns.35,36 We performed ELISA to look at longitudinal changes in anti-spike IgG and IgA levels in saliva at several timepoints following three doses of mRNA COVID-19 vaccines in 31 participants (Tables S15–S18). The results were expressed as %AUC (area under the curve) of the AUC of a pooled positive control of acute and convalescent saliva. Comparisons were based on Wilcoxon rank-sum test (intergroup) or quantile regression (within-group/subgroup). Individuals who had at least one dose of adenoviral vaccine were not included, as such vaccines apparently elicit weaker mucosal anti-SARS-CoV-2 immunity when administered intramuscularly.37
The median baseline levels of anti-spike IgG in the HIV− group, 0.65%AUC, were just above the positive cut-off of 0.35%AUC (Table S15), being significantly higher than in PWH (p = 0.025). Of note, we only analyzed saliva from seven HIV− participants due to a limited number of mRNA-vaccinated HIV− age-matched individuals. Post-baseline IgG responses to each vaccine dose were quite strong in PWH and HIV− (Figure 4, top), without significant differences between the two groups (Tables S15–S17). The IgG levels peaked at 4 weeks post-D3 (V8b) at 119%AUC in PWH and 136%AUC in HIV−, decreasing by V9 to median 48.1%AUC in PWH (p = 0.023), whereas the HIV− decrease to 95.9%AUC was not significant.
Figure 4.
Strong anti-spike IgG responses to three mRNA vaccine doses in saliva contrast with weak anti-spike IgA responses, with little effect from the booster on IgA
Longitudinal changes in levels of spike-specific IgG (top) and IgA (bottom) Abs in saliva following COVID-19 vaccination in HIV− participants (blue), total PWH (green), IRs (cyan), INRs (purple). The values are expressed as %AUC (percentage of the area under the curve of the positive control of acute and convalescent COVID-19 samples whose average response is plotted as 100% and marked with black horizontal dashed lines); the horizontal bars show median responses. p values for differences between PWH and HIV− participants, and those between IRs and INRs, are based on Wilcoxon rank-sum test and are shown below their respective violin plots or above each IR/INR pair: p < 0.001 (∗∗∗), p < 0.01 (∗∗), p < 0.05 (∗), p ≥ 0.05 (ns). p values for within-group/subgroup changes from the preceding timepoint for each pair of adjacent timepoints are shown at the top of each panel and are color-coded accordingly (based on quantile regression). LLV participants are shown as red dots. Seropositivity thresholds, 0.35%AUC for IgG and 4.06%AUC for IgA, are shown as red dashed lines. Vaccination timepoints (D1, D2, D3) are indicated with red arrowheads. See also Tables S15–S18.
Unexpectedly, we observed relatively high baseline anti-spike IgA responses in HIV− participants, with a median of 17.1%AUC – well above the positive cut-off of 4.06%AUC (Figure 4, bottom; Table S16). In fact, IgA levels in this group remained at similarly high levels at all timepoints, with medians ranging between 10.3%AUC at V8 and 20.5%AUC at V9 (Table S18). High baseline levels of anti-spike IgA (and to a lesser extent IgG) differed from previously studied younger cohorts.36,38 In PWH, the V1 IgA responses did not exceed the cut-off. Consistent with previous findings,36 the IgA responses in PWH were modest relative to the positive control, with median values slightly exceeding the seropositivity threshold at V4, V5, and V8, peaking at 7.62%AUC at V8b. At the last study visit (V9), the IgA levels in PWH dropped below the cut-off to 3.83%AUC (p = 0.007), compared to 20.5%AUC in HIV− (p = 0.039 for PWH vs. HIV−).
We observed a trend for weaker spike IgA responses in INRs, compared to IRs, but these differences were not statistically significant (Figure 4, bottom).
IRs mount strong T cell responses to two and three vaccine doses, outperforming HIV− individuals, while INRs show modest responses
We used cytokine enzyme-linked immune absorbent spot assay (ELISpot) to compare spike-specific T cell immunity in PWH and HIV− participants following three doses of COVID-19 vaccines at baseline, 24 and 48 weeks post-D1 (V8, V9). Because T cell spike-specific responses demonstrate a strong Th1 shift,39 we measured them as the frequency of cells that secrete IFN-γ, IL-2 or both – ‘dual’ (Figure 5), following stimulation with spike peptides. The values were expressed as the number of spot-forming cells (SFC) per 106 PBMCs.
Figure 5.
IRs mount strong T cell responses to two and three vaccine doses, outperforming HIV– individuals, while INRs show modest responses
Changes in anti-spike T cell responses in HIV− participants (blue), total PWH (green), IRs (cyan), and INRs (purple) following COVID-19 vaccination are determined with ELISpot as frequencies of cells secreting IFN-γ (top), IL-2 (middle) or both (‘Dual’, bottom) in response to stimulation with SARS-CoV-2 spike peptide pool and expressed as spot-forming cells (SFC) per 106 PBMC. The horizontal bars show median frequencies. p values for differences between PWH and HIV− participants, and those between IRs and INRs, are based on Wilcoxon rank-sum test and are shown below their respective violin plots or above each IR/INR pair: p < 0.001 (∗∗∗), p < 0.01 (∗∗), p < 0.05 (∗), p ≥ 0.05 (ns). p values for within-group/subgroup changes from the preceding timepoint for each pair of adjacent timepoints are shown at the top of each panel and are color-coded accordingly (based on quantile regression). LLV participants are shown as red dots. Vaccination timepoints (D1-2, D3) are indicated with red arrowheads. See also Tables S19 and S20.
The cytokine responses differed in magnitude: IL-2>IFN-γ>dual (Tables S19 and S20). For IL-2, we observed a dramatic shift in median SFC frequency from baseline to V8: zero to 92.5 in PWH (p = 0.005), and 1.9 to 41.3 in HIV− (p < 0.001). For IFN-γ, the change was more subtle: zero to 12.5 in PWH (p = 0.02), and zero to 11.3 in HIV− (not significant). The dual responses at V8 only showed a minor change from baseline.
Dose 3 boosted T cell responses further, but the change from V8 was moderate, compared to the change from V1 to V8 (Figure 5). Perhaps, most notable were the stronger responses in PWH compared to HIV− participants – at V8 and V9 for IL-2, and at V9 for IFN-γ and dual, with similar observations also made recently with flow cytometry.40 The difference between IRs and HIV− was even more profound (Table S19; Figure 5), as were the changes within IRs (Table S20; Figure 5). The median IL-2 responses at V8 were 123 in IR vs. 41.3 in HIV− (p = 0.009), vs. 92.5 in total PWH (p = 0.196). The V9 difference was even greater: 135 in IR vs. 43.8 in HIV− (p < 0.001), vs. 103 in total PWH (p = 0.027). The same pattern was observed for IFN-γ and dual.
INRs had significantly lower responses than IRs at V8 and V9 for both cytokines. For IL-2, the means compared as 10.6 in INRs vs. 92.5 in IRs at V8 (p = 0.016); at V9, 12.5 in INRs vs. 135 in IRs (p = 0.001). INRs also showed a trend for weaker responses than in HIV− individuals.
The size of intact HIV reservoir in peripheral CD4+ T cells does not increase after three COVID-19 vaccine doses, with a possible exception of PWH with unsuppressed viremia
We used IPDA, or Intact Proviral DNA Assay,41 to assess the influence of two (V8) and three (V9) doses of COVID-19 vaccines on the size of HIV reservoir measured as the number of intact HIV proviruses per 106 CD4+ T cells and analyzed by quantile regression. The pattern and magnitude of changes varied among PWH (Figure 6, left). For total PWH, the median frequency of intact proviruses changed from 90.9 at baseline to 122 at V8, and to 95.0 at V9 (Table S21). Median changes between the three timepoints were subtle and statistically not significant.
Figure 6.
The size of intact HIV reservoir in peripheral CD4+ T cells does not increase after three COVID-19 vaccine doses, with a possible exception of PWH with unsuppressed viremia
The violin plot (left) shows IPDA-derived frequencies of intact HIV proviruses (with medians) per 106 CD4+ T cells analyzed by quantile regression. The dots representing the same person are connected, and LLV participants are shown in red; ns = ‘not significant’ (p ≥ 0.05). Vaccination timepoints (D1-2, D3) are indicated with red arrowheads. IPDA values for LLV participants on the original scale. See also Table S21.
No significant changes or differences were observed within or between IRs and INRs. By contrast, we saw a dramatic increase in the frequency of intact HIV proviruses from baseline to V9 in all three LLV participants tested: by 35.5% in OM5019, 70.9% in OM5094, and 175% in OM5004, which is a mean increase of 93.7% (Figure 6, right).
Destabilization of the intact HIV reservoir may trigger viral replication, which could then result in detectable VL. For most timepoints in this study, the number of VL ‘blips’ above 40 copies/mL was limited to one or two participants (Table S22; Figure S3). After D2, however, we observed an increase from one such case at V5 (1/39, or 2.6%) to three at V6 (3/40, or 7.5%), and five at V7 (5/43, or 11.6%), with most cases being IRs. Only one of the latter five participants had a noticeable IPDA increase at V8. In total, we observed 24 such blips in 14 non-LLV PWH post-D1 over the course of the study. Only three of them (OM5085, OM5005, CIRC0041) had episodes of detectable HIV viremia over the period of two years preceding the study – one blip per person. It should be noted that the frequency of tests was lower before the study and varied among participants.
HIV gag/nef-specific T cells do not change in frequency following COVID-19 vaccination
As another measure of potential changes in the HIV reservoir, we assessed frequencies of gag- and nef-specific T cells by ELISpot, estimating a number of cells secreting IFN-γ, IL-2 or both (dual) following stimulation with gag or nef peptide pools and expressed as SFC per 106 PBMC (Figure S4). Contrary to the recent finding of increased frequencies of nef-specific CD8+ T cells in a cohort of 13 cART-treated PWH following two doses of the BNT162b2 mRNA vaccine,42 we did not observe such changes for nef- or gag-specific T cells after two or three vaccine doses, even after excluding non-mRNA vaccine data (not shown). None of the changes were significant, including those at the level of subgroups (Table S23).
CD4+ T cell count in PWH increased transiently after the primary vaccine dose
The only significant change in CD4-related clinical parameters that we observed in PWH throughout the study and which is noteworthy was a transient increase in CD4+ T cell count at V2, or 10 days post-D1 (Figures S5–S7): the median count increased from 527 cells/μL at baseline to 597 at V2 (Table S24). This is consistent with an expectation of immune activation following the primary vaccine dose. The mean change from V1 to V2 was even more pronounced in IRs (Tables S25 and S26): 47.9 for total PWH (p = 0.008) vs. 73.0 for IR (p = 0.001).
For CD4+ T cell percentage (Figure S7), we observed a small but statistically significant transient increase in INRs at 4 weeks post-D3 (V8b), with estimated mean change of 1.7% from V8a (p = 0.013). No changes in CD4+/CD8+ ratio were statistically significant (Tables S25 and S26).
Longitudinal modeling of humoral spike/RBD-specific IgG responses
Figure 7 details longitudinal model-fit responses for serum anti-spike and anti-RBD IgG, as well as RBD+S1+ and NTD+S1+ B cells, sorted by HIV status. Overall, little difference in median responses was found (Table S27). For individual fit examples, see Figure S8.
Figure 7.
Longitudinal modelling of humoral spike/RBD-specific IgG responses
(A) Serum anti-spike IgG longitudinal model (Equation 1d) mean fit prediction as a function of HIV status. Solid lines show IgG dynamics from the primary COVID-19 vaccine series (D1-2) and booster (D3). Dashed lines extend the trend in IgG decay from D1-2.
(B and C) Model-predicted spike IgG production rates, μAspike (B), and decay rates, γAspike (C). For (A–C), see also Figures 9, S8, Tables S4, S6, S8, and S27. The lower error bars for the IQR boxplots with medians are computed by Q1 (25th percentile) multiplied by 1.5 times the IQR, and the top bar is Q3 (75th percentile) multiplied by 1.5 times the IQR.
(D) Serum anti-RBD IgG longitudinal model (Equation 1e) mean fit prediction as a function of HIV status. Also see Figure S8. Model predictions for PWH and HIV-negative participants in (A) and (D) are similar; both are shown as equivalent and near-overlapping time-dependent trends. The error bars are computed as in (B and C).
(E and F) Model-predicted RBD IgG production rates, μARBD (E), and decay rates, γARBD (F). For (D–F), see also Figures 9, S8, Tables S3, S5, S7, S27.
(G) Longitudinal model (Equations 1f and 1g) mean fit predictions for RBD+S1+ and NTD+S1+ B cells, respectively, in PWH. Shaded regions illustrate the range in dosage time intervals where the B cell data were measured. See also Figure 9.
(H) A boxplot of individual model-predicted production rates for spike-specific B cells. The error bars are computed as in (B, C, E, and F).
For PWH and HIV− participants, spike IgG production for D1-D2 was almost identical, 3.18 and 3.17 BAU/mL·d, respectively, and decreased to 0.643 and 0.625 BAU/mL·d post-D3. RBD IgG production for D1-D2 is 2.4 BAU/mL·d and decreased post-D3 to 0.81 BAU/mL·d. IgG spike and RBD half-life values for D2 are 28.9 ± 7.2 and 32.1 ± 4.3 days, respectively, where errors are the relative standard error (Table S27). Estimated half-lives of spike and RBD IgG increased substantially post-D3 to 80 ± 22.4 and 63 ± 15 days, respectively. For distributions of serum IgG production and decay parameter estimates, see Figures 7B and 7C (spike) and Figures 7E and 7F (RBD).
Figure 7G shows mean longitudinal fit results for RBD+S1+ and NTD+ S1+ B cells in PWH: both are found to strictly increase over the study period of ∼340 days and display no decay in response. Thus, no estimate of half-life of B cells is possible in this study. The median production rate for RBD+S1+ B cells was higher, 0.28 days−1, vs. 0.21 days−1 for NTD+S1+ (Figure 7H). From equations Equations 1f and 1g we can also estimate the doubling time, TD, defined as the estimated time it takes for a population to double in size at a particular time. For NTD+S1+, we estimate TD to be 40 days post-D1, 97 days post-D2, and 377 days post-D3.
Longitudinal modeling of cytokine responses in T cells
Figure 8 details longitudinal model-fit responses for IFN-γ and IL-2. Total PWH and IR IFN-γ mean responses are elevated relative to HIV− (Figure 8A). In contrast, INR IFN-γ mean responses are found to fall slightly below the HIV− response. PWH and IR IL-2 mean responses are elevated, whereas INR IL-2 responses fall slightly below the HIV− response (Figure 8B).
Figure 8.
Longitudinal modelling of cytokine responses in T cells
(A-B) Longitudinal model (Equations 1g and 1h) mean fit predictions for anti-spike cytokine T cell responses as a function of HIV status. Shaded regions are the dosage time intervals across all individuals. Panels (A) and (B) are IFN-γ (Equation 1g), and IL-2 (Equation 1h) three-dose predictions as a function of days since COVID-19 vaccine D1, respectively.
(C-D) Boxplots of production rate model estimates sorted by HIV status for IFN-γ (C) and IL-2 (D). For (A-D), see also Figures 9, S8, Table S28. The lower error bars for the IQR boxplots with medians are computed by Q1 (25th percentile) multiplied by 1.5 times the IQR, and the top bar is Q3 (75th percentile) multiplied by 1.5 times the IQR.
Figures 8C and 8D displays boxplots of individual IFN-γ and IL-2 production rate estimates (summarized in Table S28). For IFN-γ production, the median response in PWH was 78% higher than in HIV− individuals, and it was also significantly more heterogeneous. IR and INR individuals had median IFN-γ production rates of 0.12 days−1 and 0.01 days−1, respectively, which suggests a 91.7% slower response in INR relative to IR. For IL-2 production, HIV− individuals had a median production rate of 0.12 days−1, which was narrowly distributed with an SD of 0.05 days−1. The median IL-2 production rate amongst PWH was 0.23 days−1, which is a 92% higher rate relative to HIV−, but also with a higher SD of 0.18 days−1. IRs display a faster IL-2 response than HIV−, with a median production rate of 0.27 days−1, vs. 0.06 days−1 in INRs, suggesting a 77.8% slower response in the latter.
Among all vaccine doses, the lowest TD for all participants were observed post-D1, suggesting rapid population growth. For IL-2, TD were approximately equal post-D1, 45–48 days for all PWH. IFN-γ TD was 162 days for HIV− individuals, while 150 and 47 days for INR and IR, respectively. The doubling time suggests that IRs mounted the IL-2 response faster than HIV− and INR individuals post-D1. For D2 and D3, the estimated TD increased substantially in PWH, suggesting saturation of IFN-γ and IL-2 responses. No decline was observed in IFN-γ and IL-2 longitudinal median responses across the ∼340-day study period. Figure 9 details the vaccination model for within-host immunization.
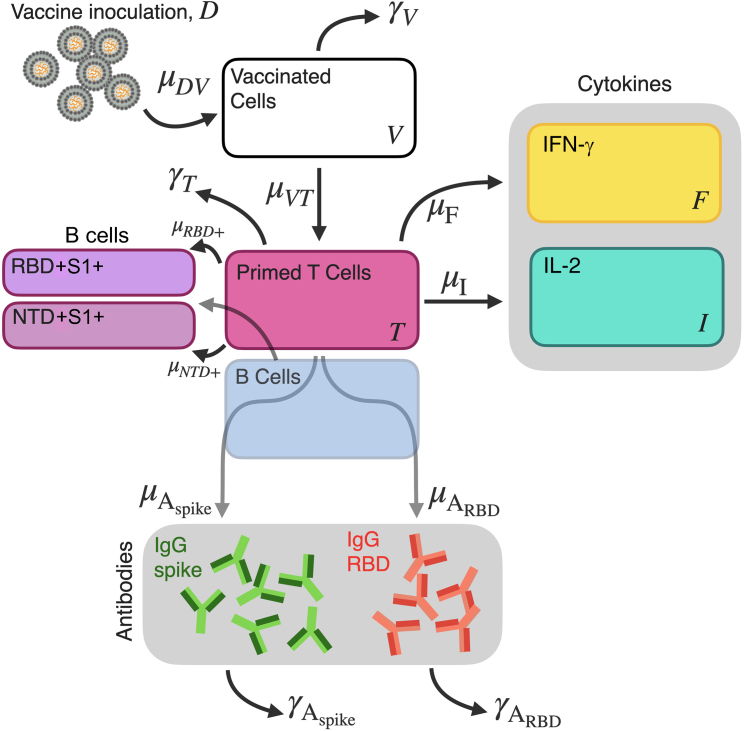
Figure 9.
Vaccination model for within-host immunization
A schematic diagram of COVID-19 vaccine inoculation, T cell stimulation, cytokine, IgG, and spike-specific B cell production model ((Equation 1a), (Equation 1b), (Equation 1c), (Equation 1d), (Equation 1e), (Equation 1f), (Equation 1g), (Equation 1h), (Equation 1i)).
Discussion
Recent COVID-19 vaccine trials showed that age had an impact on immunogenicity after one dose of BNT162b2,24 but this effect diminished with two doses.25 Phase 3 mRNA-1273 studies showed lower vaccine efficacy in older age groups,23 and differences between binding and neutralizing Ab responses were also reported.43 In the ChAdOx1 phase 2/3 trial, nAb titers were relatively similar after the second dose across all age groups, but still noticeably lower in participants aged 56–69.21 Older age was also associated with lower T cell responses for ChAdOx1, but the efficacy was not affected.21,22
Studies on PWH in COVID-19 vaccine trials were limited,23,25 but they showed strong initial immune responses after two vaccine doses.44,45,46,47,48,49 However, these studies often excluded the most vulnerable subgroups with low CD4+ T cell count, unsuppressed viremia, older individuals, or had small sample sizes. Meanwhile, weaker responses to the vaccine have been reported for some PWH with lower11,50,51,52 or even preserved CD4+ T cell count,44 although age discrepancies between the study groups might have played a role in the latter. In some cases, untreated PWH with uncontrolled viremia and low CD4+ T cell count did not seroconvert after two vaccine doses.53
Only a few studies have analyzed immune responses in PWH following three doses of COVID-19 vaccines, but initial reports on individuals with preserved CD4+ T cell count are optimistic.54 In our study of cART-treated PWH in the older age group, we showed that each vaccine dose, regardless of its type, induced binding IgG Abs for spike and RBD in plasma and spike IgG in saliva at levels similar in PWH and age-matched HIV-negative individuals. Each dose boosted IgG, which peaked at four weeks after the third dose. Importantly, production and decay rates of plasma IgG did not vary as a function of HIV status, while their half-lives increased post-D3 from 28.9 to 80 days for anti-spike IgG and from 32.1 to 63 days for anti-RBD IgG, indicating a beneficial role of the booster. The two-dose half-life results compare well to previous two-dose IgG half-life measurements for BNT162b2 and mRNA-1273 which estimate a half-life of 16 days,55 and are similar to IgG half-life of 21 days for mild SARS-CoV-2 cases.56
Unlike RBD-binding Abs, serum neutralizing responses were significantly lower in PWH than in HIV− participants after two vaccine doses, but a rapid decay was observed in both groups. This difference could have several explanations. First, not every Ab detected in the binding assays is a nAb. Second, neutralization capacity of sera may result from nAbs to other SARS-CoV-2 spike domains, such as NTD. Most studies focus on RBD, which is the primary target for nAbs. However, NTD, which facilitates the virus’s entry into the host cell, proved to be another mutational hotspot and antigenic supersite that represents an important target for nAbs.27,28,29,30,31 Indeed, the positive correlation that we observed between the frequency of spike-specific B cells and neutralizing titers in PWH was stronger for NTD+S1+ B cells than for RBD+S1+.
The suboptimal neutralizing responses in PWH with two vaccine doses were ‘rescued’ by the booster: PWH and HIV− participants had similar NT50 and anti-RBD nAb concentrations at the last study visit. These changes were mirrored by frequencies of spike-specific B cells in PWH. It is important to note, however, that the quality and breadth of neutralization may be potentially different between these populations, especially in the context of escape mutations of the SARS-CoV-2 virus.
Mucosal immunity of the upper respiratory tract is the first line of defense against SARS-CoV-2, particularly its IgA component,35,36 and the least understood in the context of COVID-19 vaccination. We demonstrated that the pattern of longitudinal changes in anti-spike IgG responses in the saliva of mRNA-vaccinated participants mirrors spike/RBD IgG responses in sera. Conversely, the amplitude of IgA responses was very low and did not improve with D2 or D3. This echoes an earlier finding by co-authors of this study on HIV-negative individuals for two doses of mRNA COVID-19 vaccine.36 HIV− individuals showed relatively high baseline spike-specific IgA levels which remained virtually unchanged throughout the study. One possible explanation could be a cross-reactivity with Abs to seasonal coronaviruses, after repeated exposure with older age. Alternatively, these Abs may have arisen from exposure to SARS-CoV-2 that did not result in productive infection. Both scenarios would suggest weaker and/or less durable salivary IgA responses to coronaviruses in PWH, particularly in INRs. It has been established previously that blood plasmablasts in healthy HIV-negative individuals predominantly produce IgA.57 In PWH, however, the proportion of IgA-producing plasmablasts is reduced,58 resulting in reduced HIV-specific IgA both in plasma and in mucosae.59,60 Of note, the bias toward IgG-secreting plasma cells is not unique for HIV infection and was also reported by co-authors of this study for other conditions that cause inflammation in the mucosae.61 The underlying mechanism affecting IgA production in PWH is yet unknown, however, one hypothesis suggests the ability of the HIV nef protein to interfere with B-cell signaling and Ab class switching,62,63,64 which could be the reason behind the pattern observed in the present study. Such poor mucosal IgA response to current intramuscular COVID-19 vaccine regimens may be affecting protection against SARS-CoV-2, especially in PWH, indicating the need for more advanced vaccine regimens. An intranasal adenoviral booster following an intramuscular mRNA priming could be the alternative.65
CD4+ T cell responses to COVID-19 vaccines demonstrate a strong Th1 shift to TNF, IFN-γ and IL-2,39 with expression patterns varying between different cells.66 A recent report suggests that most PWH mount a response after two vaccine doses regardless of their CD4+ T cell count.66 Our study demonstrates enhanced spike-specific T cell immunity in PWH who are IRs, even when compared to HIV− participants, suggesting Th1 imprinting from pre-existing HIV infection. In INRs, the trend is opposite, with significantly weaker IL-2, IFN-γ and dual responses than in IRs or HIV−, indicating a relative defect in spike-specific Th1 immunity.
PWH showed heterogeneity for IFN-γ and IL-2 production rates, with IRs surpassing even HIV− individuals, whereas in INRs they were the lowest. Mean doubling times for both cytokines increased post-D3 by a factor of 3–4, suggesting a near-saturated response post-booster. No decay in responses was observed for PWH or HIV− over the study period. However, the frequency values for cytokine-producing T cells were increasing significantly slower in INRs and predicted to saturate at about an order of magnitude lower median value than in IRs and HIV− individuals. Our observations suggest the importance of stratification of PWH in similar studies according to their immune status. The effect of Th1 imprinting on coronavirus infections will require further study.
Limited studies reported a transient increase in HIV VL and decrease in CD4+ T cell count in some PWH immediately after COVID-19 vaccination, which could potentially be deleterious for individuals with immunodeficiency.44 We only observed a brief increase in CD4+ T cell count in PWH post-D1. Concerning HIV VL, however, we did register a gradual transient increase in the proportion of PWH with detectable VL post-vaccination. These changes in a subset of participants may not be incidental in light of the recent finding of ex vivo activation of HIV transcription by mRNA COVID-19 vaccines through the RIG-I/TLR–TNF–NFκB pathway.42
Influenza vaccines have been shown to cause an increase in the HIV proviral burden in influenza-specific CD4+ T cells of cART-treated PWH with suppressed VL.19 In our study, we did not find significant changes in the size of HIV reservoir in peripheral CD4+ T relative to the baseline after two or three COVID-19 vaccine doses, except in three LLV participants who had a mean increase of 93.7% in the frequency of intact HIV proviruses after the booster. The small LLV sample size may not allow a generalization, but this finding could be pointing at potential risks for PWH with unsuppressed viremia, and therefore should not be discounted. That said, a recent finding suggests that persistent low-level viremia can be driven by non-infectious virions lacking envelope that are induced from CD4+ T cell clones harboring proviruses with 5′-leader defects.67 Thus, mRNA vaccines may simply be increasing clonal populations of cells that transcribe defective virions. The only other study (to the best of our knowledge) that addressed the issue of stability of the HIV reservoir in the context of COVID-19 vaccination reported a lack of significant changes in 11 PWH after two vaccine doses.42 The same study reported an increase in HIV-nef-specific T cell responses in 13 cART-treated PWH with suppressed viremia following two doses of BNT162b2, and a significant increase in cell-associated (but not extracellular) HIV RNA post-D3 in another cohort of eight PWH, suggesting a degree of adverse reaction to the vaccine from HIV reservoirs.42 Contrary to these findings, we did not observe changes in HIV nef or gag responses after two or three vaccine doses (n = 28–29). This discrepancy might be due to the nature and timing of responses measured. In our analysis, we looked at total T cell responses, whereas the only significant difference observed by Stevenson and co-authors42 was in the frequency of granzyme-B-producing cells. Like us, this group did not report significant changes in nef-specific IFN-γ responses. Their sampling timeline had the median of 17 and 16 days post-D1 and D2, respectively,42 as opposed to our study’s 109 and 130 days post-D2 and D3, respectively. Therefore, we could have missed a potential transient increase in the frequency of nef-specific T cells. Overall, we can conclude that a limited-scale destabilization of HIV reservoirs may, indeed, occur in response to COVID-19 vaccines, which may not necessarily affect the VL or proviral burden. These negative outcomes may evidently be more likely in unsuppressed HIV viremia.
In summary, cART-treated PWH aged 55 and older had diminished SARS-CoV-2 neutralization with two COVID-19 vaccine doses, but the third dose ‘rescued’ the responses. Further studies are needed to determine whether these rescued responses show similar ability to neutralize subsequent variants compared to HIV-negative individuals. PWH had lower levels of spike-specific IgA in saliva post-vaccination, which could affect protection. Enhanced anti-spike T cell immunity in this population, except for INRs, indicates Th1 imprinting from pre-existing HIV infection. Finally, COVID-19 vaccines did not affect the size of intact HIV reservoir in most PWH, except those with persistent low-level viremia, but were associated with increased viral blips. The latter findings indicate potential risks for some PWH, especially in unsuppressed HIV viremia.
Limitations of the study
One limitation of our study is that the majority of our participants were male due to recruitment from a clinic with a large MSM population. Further, a relatively small number of specimens collected from HIV-negative individuals did not allow us to estimate changes in frequencies of RBD/NTD-specific B cells in this cohort and make comparisons with PWH. Our finding of increased HIV reservoir post-D3 was made on a small number of PWH with unsuppressed viremia (n = 3) and, thus, requires a confirmatory study on a larger sample of cART-treated PWH with persistent HIV viremia.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human anti-IgG secondary mAb, HRP | NRC Canada; Colwill et al., 202233 | Cat#IgG#5-HRP |
| Llama anti-RBD neutralizing mAb (VHH-72) | NRC Canada; Colwill et al., 202233 | Cat#VHH72-hFc1X7 |
| SARS-CoV-2 Nucleocapsid IgG (HC2003) | GenScript | Cat#A02039 |
| Mouse anti-human CD45 BUV805 | BD Biosciences | Cat#612891; RRID: AB_2870179 |
| Mouse anti-human CD19 BV650 | BD Biosciences | Cat# 563226; RRID: AB_2744313 |
| Mouse anti-human CD20 APC-H7 | BD Biosciences | Cat#560734; RRID: AB_1727449 |
| Mouse anti-human CD10 BV510 | BD Biosciences | Cat#563032; RRID: AB_2737964 |
| Mouse anti-human IgG PE-Cy7 | BD Biosciences | Cat#561298; RRID: AB_10611712 |
| Mouse anti-human CD11c BUV395 | BD Biosciences | Cat#563787; RRID: AB_2744274 |
| Mouse anti-human CD3 BV570 | Biolegend | Cat#300436; RRID: AB_2562124 |
| Mouse anti-human IgD BV605 | Biolegend | Cat#348232; RRID: AB_2563337 |
| Mouse anti-human IgM BV711 | Biolegend | Cat#314540; RRID: AB_2687215 |
| Mouse anti-human CD27 BV785 | Biolegend | Cat#302832; RRID: AB_2562674 |
| Mouse anti-human CD21 PE/Dazzle594 | Biolegend | Cat#354922; RRID: AB_2739536 |
| Mouse anti-human CD38 APC/Fire810 | Biolegend | Cat#356644; RRID: AB_2860936 |
| Mouse anti-human CD71 Alexa Fluor 700 | Biolegend | Cat#334130; RRID: AB_2888770 |
| Mouse anti-human IgA VioBlue | Miltenyi Biotec | Cat#130-113-479; RRID: AB_2726166 |
| Goat anti-human IgG, HRP | Southern Biotech | Cat#2044-05; RRID:AB_2795673 |
| Goat anti-human IgA, HRP | Southern Biotech | Cat#2053-05; RRID:AB_2795715 |
| Mouse anti-human IFN-γ mAb (1-D1K) | Mabtech | Cat#3420-3-1000; RRID:AB_907282 |
| Mouse anti-human IL-2 mAbs (MT2A91/2C95) | Mabtech | Cat#3445-3-1000 |
| Anti-human IFN-γ mAb (7-B6-1), ALP | Mabtech | Cat#3420-9A-1000 |
| Anti-human IL-2 mAb (MT8G10), biotin | Mabtech | Cat#3445-6-1000 |
| Bacterial and virus strains | ||
| SARS-CoV-2: Wuhan-Hu-1: SB2: P3 | Dr. Samira Mubareka, Sunnybrook Health Science Centre, Toronto, Canada | N/A |
| Biological samples | ||
| Human peripheral blood samples from PWH who are SARS-CoV-2 vaccine recipients | Collected at Maple Leaf Medical Clinic, Toronto, Canada | N/A |
| Human peripheral blood samples from HIV-negative recipients of SARS-CoV-2 vaccine | Collected at Maple Leaf Medical Clinic, Toronto, Canada | N/A |
| Human saliva samples from SARS-CoV-2 vaccine recipients | Collected at Maple Leaf Medical Clinic, Toronto, Canada | N/A |
| Human saliva samples from SARS-CoV-2 acute and convalescent individuals (positive controls) | Dr. Allison McGeer, Mount Sinai Hospital, Toronto, Canada | N/A |
| Human saliva samples collected prior to the SARS-CoV-2 pandemic (negative controls) | Self-collected in salivettes by participants of the CORIPREV cohort and mailed to University of Toronto | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ficoll-Paque PLUS | Cytiva | Cat#17144003 |
| FBS | Wisent Bioproducts | Cat#080450 |
| RPMI 1640 cell culture medium | Wisent Bioproducts | Cat#350-030-CL |
| DMEM cell culture medium | Gibco | Cat#11995-065 |
| HEPES buffer | Wisent Bioproducts | Cat#350-050-EL |
| L-glutamine | Wisent Bioproducts | Cat#609-065-EL |
| Penicillin-Streptomycin | Wisent Bioproducts | Cat#450-201-EL |
| D-PBS | Wisent Bioproducts | Cat#311-425-CL |
| Sterile water, molecular-grade | Wisent Bioproducts | Cat#809-115-CL |
| DMSO | Sigma-Aldrich | Cat#D2650-100ML |
| SARS-CoV-2 Spike glycoprotein SmT1 | NRC Canada; Colwill et al., 202233 | Cat#SMT1-1 |
| SARS-CoV-2 RBD (331-521) | NRC Canada; Colwill et al., 202233 | N/A |
| SARS-CoV-2 Nucleocapsid | NRC Canada; Colwill et al., 202233 | Cat#NCAP-1 |
| Blocker BLOTTO | ThermoFisher Scientific | Cat#37530 |
| BLOTTO solution | BioShop | Cat#SKI400.500 |
| PBS-TWEEN tablets | MilliporeSigma | Cat#524653 |
| TWEEN-20 | BioShop | Cat#TWN510 |
| SuperSignal ELISA Pico Chemiluminescent Substrate | ThermoFisher Scientific | Cat#37069 |
| Biotinylated SARS-CoV-2 rACE2 | Dr. James Rini, University of Toronto, Toronto, Canada | N/A |
| Biotinylated SARS-CoV-2 rRBD | Dr. James Rini, University of Toronto, Toronto, Canada | N/A |
| Biotinylated SARS-CoV-2 rSpike | Dr. James Rini, University of Toronto, Toronto, Canada | N/A |
| Streptavidin-Peroxidase polymer, ultrasensitive | MilliporeSigma | Cat#S2438 |
| SARS-CoV-2 S1 WT | Biolegend | Cat#793806 |
| SARS-CoV-2 S2P WT | AcroBiosystems | Cat#SPN-C82E9 |
| SARS-CoV-2 RBD WT | AcroBiosystems | Cat#SPD-C82E9 |
| SARS-CoV-2 NTD WT AVI-biotin | Vaccine Research Center, NIH; Teng et al., 200268 | N/A |
| PepMix SARS-CoV-2 spike peptide master pool | JPT | Cat#PM-WCPV-S-1 |
| HIV-1 Nef region peptide pool | NIH HIV Reagent Program | Cat#ARP-12822 |
| HIV-1 Gag region peptide pool | NIH HIV Reagent Program | Cat#ARP-12437 |
| Human cytomegalovirus, Epstein-Barr virus, and influenza virus peptide pool (CEF) | NIH HIV Reagent Program | Cat#ARP-9808 |
| CEFTA peptide pool | Mabtech | Cat#3617-1 |
| Staphylococcal enterotoxin B (SEB) | Millipore-Sigma | Cat#S4881 |
| Zombie NIR fixable Live/Dead dye | Biolegend | Cat#423105 |
| Streptavidin PE | ThermoFisher Scientific | Cat#S21388 |
| Streptavidin PE-Cy5.5 | ThermoFisher Scientific | Cat#SA1018 |
| Streptavidin BV421 | Biolegend | Cat#405225 |
| Streptavidin Alexa Fluor 488 | ThermoFisher Scientific | Cat#S32354 |
| Streptavidin-HRP | Mabtech | Cat#3310-9-1000 |
| Brilliant Stain Buffer Plus | BD Biosciences | Cat#566385 |
| BD FACS Lysing Solution 10X concentrate | BD Biosciences | Cat#349202 |
| Permeabilizing Solution 2 | BD Biosciences | Cat#347692 |
| 3,3′,5,5′tetramethylbenzidine (TMB) substrate solution | ThermoFisher Scientific | Cat#00-4201-56 |
| ddPCR Supermix for Probes (no dUTPs) | Bio-Rad | Cat#1863024 |
| XhoI restriction enzyme | ThermoFisher Scientific | Cat#ER0692 |
| Critical commercial assays | ||
| Vector Blue Substrate Kit, AP | Vector Laboratories | Cat#SK-5300 |
| Vector NovaRED Substrate Kit, HRP | Vector Laboratories | Cat#SK-4800 |
| EasySep Human CD4+ T cell Enrichment Kit | STEMCELL Technologies | Cat#19052 |
| DNeasy Blood & Tissue Kit | QIAGEN | Cat#69506 |
| Experimental models: Cell lines | ||
| Vero-E6 | ATCC | Cat#CRL-1586; RRID:CVCL_YZ66 |
| Huh-7 | Dr. Sonya McParland, University of Toronto, Toronto, Canada | RRID:CVCL_0336 |
| Oligonucleotides | ||
| See Table S25 for primers | Integrated DNA Technologies | N/A |
| See Table S25 for probes | ThermoFisher Scientific | N/A |
| Software and algorithms | ||
| GraphPad Prism v8 | GraphPad | RRID: SCR_002798 |
| SpectroFlo v3.0.1 | Cytek Biosciences | N/A |
| FlowJo v10.8.1 | FlowJo | RRID: SCR_008520 |
| BioSpot Pro v7.0.23.3 | CTL | N/A |
| QuantaSoft v1.7.4.0917 | Bio-Rad | Cat#1864011 |
| QuantaSoft Analysis Pro v1.0.596 | Bio-Rad | N/A |
| MonolixSuite v2020R1 | LIXOFT | N/A |
| SAS 9.4 | SAS Institute Inc. | RRID:SCR_008567 |
| Other | ||
| Vacutainer SST blood collection tubes | BD | Cat#367988 |
| Vacutainer ACD Solution A blood collection tubes | BD | Cat#364606 |
| Salivette | Sarstedt | Cat#51.1534 |
| CellDrop BF cell counter | DeNovix | N/A |
| SepMate-50 | STEMCELL Technologies | Cat#85450 |
| LUMITRAC 600 microplates | Greiner | Cat#781074 |
| MaxiSorp plates | ThermoFisher Scientific | Cat#460372 |
| Streptavidin-coated microplates | ThermoFisher Scientific | Cat#436014 |
| MultiScreen filter plates | MilliporeSigma | Cat#MSIPS4W10 |
| EnVision XCite 2105 multimode plate reader | Perkin Elmer | Cat# 2105-0010 |
| Cytek Aurora spectral cytometer | Cytek Biosciences | N/A |
| Multiskan FC microplate photometer | ThermoFisher Scientific | Cat#51119000 |
| ImmunoSpot S3 analyzer | CTL | N/A |
| Microplates for ddPCR | Bio-Rad | Cat#12001925 |
| Automated droplet generator | Bio-Rad | Cat#1864101 |
| DG32 automated droplet generator cartridges | Bio-Rad | Cat#1864108 |
| C1000 Touch thermal cycler | Bio-Rad | Cat#1851197 |
| QX200 droplet reader | Bio-Rad | Cat#1864003 |
| PX1 PCR plate sealer | Bio-Rad | Cat#1814000 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Vitaliy Matveev (vitaliy.matveev@utoronto.ca).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Study design
This was a single-site prospective non-randomized observational study (CTNPT 045) where PWH and HIV-negative individuals were followed over 48 weeks with blood draws and saliva sampling, with evaluations taken at screening, baseline, and 12 additional timepoints (Figure 1) preceding or following three doses of one (or a combined regimen) of the following COVID-19 vaccines: an adenoviral vector vaccine ChAdOx1 (Oxford University, AstraZeneca) and two mRNA vaccines, BNT162b2 (Pfizer-BioNTech) and mRNA1273 (Moderna).
The screening visit took place within 90 days prior to the baseline visit. The baseline visit (V1) occurred up to 24 hours prior to the first COVID-19 vaccine dose (D1). The second, third and fourth study visits (V2, V3, V4) are 10, 20 and 28 days following D1, respectively. As extended time intervals between the first and the second (D2) vaccine doses had been adopted in Ontario, we introduced additional study visit up to three days prior to D2 – Visit 4a (V4a). Visits 5, 6 and 7 (V5, V6, V7) took place one, two and four weeks after D2, respectively. Our primary endpoint, Visit 8 (V8), occurred at a 24-week mark (six months) following D1. Visit 8a (V8a) took place within three days preceding the third vaccine dose (D3), which was then followed by Visits 8b and 8c (V8b, V8c) at four and 16 weeks post-D3. The final study visit, Visit 9 (V9), corresponds to 48 weeks (one year) after D1.
Study participants
A total of 91 participants aged 55 and over were recruited for the study through Maple Leaf Medical Clinic in Toronto, Canada, including 68 PWH and 23 HIV-negative individuals (Figures 1A and 1B; Table S1). We initially approached 73 eligible PWH, five of whom declined to participate. Next, we approached 28 HIV-negative individuals in the matching age range. Five of them decline to participate. In total, 91 out of 101 approached candidates (90.1%) volunteered and signed an informed consent form. Due to the rapid vaccine rollout in Ontario for the older demographic, baseline samples could only be obtained from 57 participants, including 45 PWH and 12 HIV-negative individuals. An additional 34 participants joined the study at Visit 8, including 23 PWH and 11 HIV–. All PWH had been on cART for at least one year.
Our PWH subgroup definitions are as follows. Immunological responders (IR): undetectable viral load (VL) for at least one year, CD4+ T-cell count > 500 cells/μL and any CD4+/CD8+ ratio, or CD4+ T-cell count = 350-500 cells/μL and CD4+/CD8+ ratio ≥ 0.7. Immunological non-responders (INR): undetectable VL for at least one year, CD4+ T-cell count = 350-500 cells/μL and CD4+/CD8+ ratio < 0.7, or CD4+ T-cell count < 350 cells/μL and any CD4+/CD8+ ratio. PWH with low-level viremia (LLV): cART-adherent, VL > 40 copies/mL at least three times a year for at least three consecutive years, any CD4+ T-cell count, any CD4+/CD8+ ratio. Long-term non-progressors (LTNP): no decline for at least five years without cART, VL < 5,000 copies/mL (our participant had low or undetectable VL from 1987 – when he was diagnosed with HIV, to 2011 – when he elected to start cART), CD4+ T cell count > 500 cells/μL.
Sixty out of 91 participants had either completed V8a, or it was not scheduled for them because their V8 fell within seven days preceding D3. For most participants in the additional cohort recruited at V8, the V8a and V8b visits were not scheduled. For those participants for whom V8c and V9 fell within two weeks of each other, these two timepoints were combined into one study visit (Table S2). We allowed a three-day window for each study visit on each side of the target date. For the datasets appearing as ‘Out-of-window data excluded’, we excluded samples outside of the three-day window from the target date (±7 days for V8 and V9). Samples deviating by more than 14 days from the target date were excluded from all analyses. Three participants received a fourth dose before the final study visit, so their V9 samples were excluded from the datasets.
Study approval
All study participants provided informed written consent. The study protocol and consent form were approved by the University of Toronto Research Ethics Board (RIS #40713) and Sinai Health REB (21-0223-E). The study was conducted in accordance with the protocol, applicable regulations, and guidelines for Good Clinical Practice (GCP), Health Canada’s regulations, and the Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans (TCPS 2.0).
Method details
Blood and saliva sampling
Vaccination was not provided as a part of this study. The timeline of the study protocol is shown on Figure 1C. At the screening visit, we performed routine blood work (hematology, biochemistry) for all participants and HIV test for HIV-negative individuals. PWH were assayed for CD4+ T-cell count, VL, and CD4+/CD8+ ratio, which were also recorded retrospectively, along with CD4+ T-cell nadir. CD4+ and CD8+ T-cell count and percentage, CD4+/CD8+ ratio and VL were assessed at each study visit. For each participant, blood was collected in a single SST tube (8.5 mL/tube), and in up to 10 ACD tubes (at V1, V4a, and V8-9; 8.5 mL/tube). Saliva was collected in a Salivette (Sarstedt, Germany) at V1, V4-5, and V8-9. Specimens were processed immediately upon sampling.
Serum isolation
SST tubes with whole clotted blood were centrifuged at 1,200×g for 10 minutes. Sera were then aspirated, aliquoted in 0.5 mL increments and frozen at –80°C.
PBMC isolation
PBMC were isolated by centrifugation using Ficoll-Paque PLUS (Cytiva) as previously described.69 The cell suspension was diluted 1:1 with a freezing medium and aliquoted for storage at –150°C.
Saliva processing
Salivette tubes were centrifuged at 1,200×g for 10 minutes at room temperature (RT). Saliva was collected from the bottom section, aliquoted in cryovials up to 1 mL per vial, and frozen at –80°C.
Antibody detection in serum (ELISA)
An automated ELISA assay was used to detect total IgG Ab levels to full-length spike trimer, RBD and nucleocapsid as previously described.33,38 Briefly, 384-well microplates were pre-coated with spike (SmT1), RBD or N, supplied by the National Research Council of Canada (NRC). The main steps were blocking in Blocker BLOTTO (ThermoFisher Scientific), incubation with 1:160 or 1:2,560 sera dilutions, incubation with human anti-IgG fused to HRP (IgG#5 by NRC), and incubation with ELISA Pico Chemiluminescent Substrate (ThermoFisher Scientific). Chemiluminescence was read on an EnVision 2105 Multimode Plate Reader (Perkin Elmer). Raw values were normalized to a synthetic standard on each plate (VHH72-Fc by NRC for spike/RBD or an anti-N IgG from Genscript, #A02039). The relative ratios were further converted to BAU/mL using the WHO International Standard 20/136 as a calibrant.33,70 Seropositivity thresholds were determined for the 1:160 dilution using 3×SD from the mean of controls.33
Live SARS-CoV-2 microneutralization (MN)
A MN assay to measure 50% neutralizing titers (NT50) in the sera was performed as described previously.71 Briefly, heat-inactivated sera were serially diluted and incubated with 100 TCID50 of the wild type Wuhan-Hu-1 SARS-CoV-2 virus (a gift from Samira Mubareka, Sunnybrook Health Science Centre) in serum-free DMEM, then added onto the VeroE6 cells (a clone of a cell line derived from a kidney tissue of the African green monkey, ATCC Cat#CRL-1586) in 96-well plates. After 1h, inoculums were removed and DMEM with 2% FBS was added to the cells. The cells were incubated for 5 days and checked daily for cytopathic effects (CPE). NT50 titer was defined as the highest dilution factor of the serum that protected 50% of cells from CPEs and calculated by the four-parameter logistic regression in GraphPad Prism 8.
B-cell frequency (spectral flow cytometry)
We measured frequencies of RBD- and NTD-specific B cells (RBD+S1+ and NTD+S1+, respectively) among total B cells by spectral flow cytometry using a 19-color subset (Table S29) of a previously described 21-color panel.72 Briefly, the biotinylated spike proteins were tetramerized with fluorescently labeled streptavidin at a molar ratio of 4:1 as described previously,73 with minor modifications.72 Cryopreserved PBMCs were thawed and stained with Zombie NIR Fixable Viability Dye, followed by staining with a cocktail of mAbs and four fluorochrome-conjugated spike proteins in staining buffer (2% FBS/PBS) supplemented with Brilliant Stain Buffer Plus (BD Biosciences). The stained cells were fixed (Lysing Solution, BD Biosciences) and acquired on an Aurora spectral cytometer using SpectroFlo Software v3.0.1 (Cytek Biosciences). The analysis was performed with FlowJo v10 (BD Biosciences). The CIRC0041 baseline plot is shown as an example of our gating strategy (Figure S9).
ACE2 displacement assay (snELISA)
The rACE2 displacement assay was performed as previously described,33,34 except assay plates were MaxiSorp (ThermoFisher Scientific). Briefly, the 384-well plates were coated with RBD (supplied by NRC), blocked with BSA in PBS-T, incubated with 1:10 or 1:40 sera dilutions, then with biotinylated ACE2 (supplied by James Rini, University of Toronto), and treated with Streptavidin-Peroxidase Polymer Ultrasensitive (Sigma-Aldrich). Incubation with ELISA Pico Chemiluminescent Substrate and reading on the EnVision were performed as in direct detection. All values were normalized to sample-free blanks on the same plate. The resulting relative ratios (RR) were converted to IU/mL using the WHO International Standard 20/136 as a calibrant33 and the following formula: log2(IU/mL at sample dilution d)=RR/(-0.2308)+5.7898+log2(d). Neutralization threshold was determined for the 1:10 dilution as 2×MAD (median absolute deviation) from the mean of blanks.
Antibody detection in saliva (ELISA)
ELISA assays for anti-spike IgA and IgG in saliva were performed in 96-well plates pre-coated with streptavidin as described earlier.36,38 Briefly, the plates were coated with biotinylated SARS-CoV-2 spike or PBS (controls) and incubated with saliva samples diluted with a BLOTTO solution (BioShop) at 1:5, 1:10, 1:20. After washing, the plates were incubated with HRP-conjugated goat anti-human IgG and IgA secondary Abs (Southern Biotech, IgG: 2044-05, IgA: 2053-05) and developed by the TMB substrate (ThermoFisher). The reaction was stopped with 1N H2SO4, and the plates were read on the Thermo Multiskan FC spectrophotometer. Raw OD450 values for the PBS controls were subtracted from the sample values for each dilution. Each adjusted value was used to calculate the area under the curve (AUC) which was then normalized to the AUC of the positive control of pooled saliva from COVID-19 acute and convalescent participants. The normalized AUC was also converted to %AUC of the positive control. Pooled early pandemic saliva from 10 individuals with a median bin age range of 30-39 years was used as a negative control whose integrated scores were calculated as described previously.36 Seropositivity values for each isotype (0.35% for IgG, 4.06% for IgA) were calculated as average integrated scores of pre-pandemic samples (0.086% for IgG, 0.85% for IgA) plus 2×SD. Only samples from mRNA-vaccinated individuals were used (n=31).
T-cell cytokine responses (dual-color ELISpot)
Ex vivo ELISpot assay was performed as previously described.69 Briefly, 96-well MultiScreen plates (Millipore) were coated with capture mAbs to IFN-γ (Mabtech #1-D1K) and IL-2 (Mabtech #MT2A91/2C95) and blocked with R-10 medium. PBMCs were plated at 200,000 cells/well and stimulated with one of the following peptide pools: SARS-CoV-2 spike (JPT #PM-WCPV-S-1), HIV-1 nef or HIV-1 gag (NIH HIV Reagent Program #ARP-12822, #ARP-12437). Positive controls included SEB (Sigma-Aldrich), CEF (NIH HIV Reagent Program), and CEFTA (Mabtech). Washed plates were incubated with alkaline-phosphatase-conjugated mAb for IFN-γ (Mabtech #7-B6-1) and with biotinylated mAb for IL-2 (Mabtech #MT8G10), then incubated with streptavidin-HRP and developed with Vector Blue for ALP and Vector NovaRed for HRP (Vector Laboratories). The plates were read with ImmunoSpot S3 Analyzer (CTL), and the data were processed with BioSpot v7.0.23.3 Pro (CTL). The mean number of spots in the negative-control (DMSO) wells were subtracted from peptide-stimulated wells, and the results were expressed as spot-forming cells (SFC) per 106 PBMC.
Intact Proviral DNA assay (IPDA)
The assay was run in 96-well plates (Bio-Rad) as described previously,41,74 with minor adjustments to a 22-μL reaction volume and four technical replicates (wells) per HIV and RPP30 reaction per sample. CD4+ T cells were isolated from PBMCs with the EasySep Human CD4+ T cell Enrichment Kit (STEMCELL Technologies). Genomic DNA was isolated from a median of 3.65 million CD4+ T cells (IQR 2.48-4.98) with the DNeasy Blood & Tissue Kit (QIAGEN). Each HIV reaction contained 825 ng of gDNA, and each RPP30 reaction contained 8.25 ng of gDNA. Genomic DNA was combined with ddPCR Supermix for Probes (no dUTPs, Bio-Rad), primers (900 nM final, IDT), probes (250 nM final, ThermoFisher Scientific), and XhoI restriction enzyme (0.5 U/μL, ThermoFisher Scientific). For sequences, see Table S30. For participants OM5365, CIRC0324, OM5005, and OM5016 we used an alternative primer/probe mix74 as the IPDA Ψ failed (Alt Ψ in Table S30). Droplets were generated with Automated Droplet Generator (Bio-Rad), and the PCR was run as described earlier.41,74 The plates were read on a QX200 Droplet Reader (Bio-Rad) and analysed with QuantaSoft v.1.7.4.0917 and QuantaSoft Analysis Pro v.1.0.596 (Bio-Rad). Replicate wells were merged prior to analysis. DNA from JLat-8.4 cells, which harbour a single HIV provirus per cell, was used as a positive control. HIV– DNA from Huh-7 cells and no-template controls were also used.
Quantification and statistical analysis
Vaccination model for within-host immunization
To model the vaccine inoculation and subsequent within-host immunization, we used a simplified compartmental model previously shown to accurately determine long-term IgG decay and capture cytokine dynamics of the BNT162b2 and mRNA-1273 vaccines,55 determine significant immunological timescales from these mRNA-based vaccines,75 and explore differing prime-booster strategies of adenovirus-based ChAdOx1 vaccine in humans.76 The model is briefly described as follows. The initial concentration of the vaccine (BNT162b2, mRNA-1273, or ChAdOx1) in the host following the inoculation is D0. Upon inoculation, the host begins to produce vaccinated cells, V, through the interaction term μVD. The amount of vaccine concentration, D, is strictly decreasing within the host for all t > t0. Vaccinated cells produce antigen, and are not modelled as cell-type specific. The priming dynamics of naïve T cells into antigen-primed T cells is captured by the term μTV, loss of primed T-cell secretion activity is γT. Increase in IFN-γ and IL-2 cytokine-producing T cells is modelled by the proliferation terms μF and μI, respectively. Plasma B cells are primed by antigen-specific T cells.77 However, in this work longitudinal B cell trajectories are absent. In the absence of plasma B cell longitudinal data, we model the production of IgG values as linearly proportional to primed antigen-specific T cells and thus implicitly consider the plasma B cell response. Generation of memory B cells is linked to a T-dependent response. We further consider the development of anti-RBD (RBD+S1+) and anti-NTD (NTD+S1+) memory B cells through proliferation terms μRBD+ and μNTD+, respectively. The full model used in this work ((Equation 1a), (Equation 1b), (Equation 1c), (Equation 1d), (Equation 1e), (Equation 1f), (Equation 1g), (Equation 1h), (Equation 1i)) is described by nine equations, whereby Equations 1d–1i are fit to longitudinal anti-spike IgG, anti-RBD IgG, RBD+ and NTD+ B cells, IFN-γ, and IL-2 data sets, respectively. A schematic of the model is shown in Figure 9.
| (Equation 1a) |
| (Equation 1b) |
| (Equation 1c) |
| (Equation 1d) |
| (Equation 1e) |
| (Equation 1f) |
| (Equation 1g) |
| (Equation 1h) |
| (Equation 1i) |
Parameter assumptions, estimations, initial conditions, and sensitivity analysis for the model
Previous findings in mice have shown that mRNA-LNP vaccines are translated locally at an intramuscular injection site for approximately 10 days.55 In this work we, therefore, fix the translation half-life to approximately 10 days upon receiving a vaccine dose, leading to a fixed γv of 0.07 d–1. Initial inoculation (Dose 1, D1), and subsequent dosages (Dose 2 and 3, D2-3) are assumed to be of value D0=2 and are in arbitrary units of concentration. Thus, we do not consider differences in dosage concentrations between vaccine types.
All fits to clinical data using (Equation 1a), (Equation 1b), (Equation 1c), (Equation 1d), (Equation 1e), (Equation 1f), (Equation 1g), (Equation 1h), (Equation 1i) were performed in Monolix v2020R1 using non-linear mixed effects. All individuals and all individual data points for three doses are used in the longitudinal modelling assay; data points acquiring post-Dose-4 are removed from the analysis. IgG best-fit parameters are obtained in a dose-dependent two-stage fitting process whereby the data is separated into two parts: baseline to pre-Dose-3, and pre-Dose-3 through to end of study timepoint V9. A set of IgG fit parameters are therefore obtained for each of the two fits. Cytokine data is included in the IgG fits. However, these are not reported on as the errors and uncertainties in the cytokine-respective two-stage fitted parameters were unable to be estimated due to the lack of data resolution within the time window for the two-stage fits. We note that the production and decay rates estimated from the longitudinal modelling approach will be affected by the IgG saturation values which limit the dynamical range in observed BAU/mL. Therefore, our production and decay rates likely underestimate and overestimate the true value, respectively.
A single-stage fitting process is used to estimate robust longitudinal cytokine and memory B cell dynamics: these fitted parameters are estimated through a fit to all Dose-3 clinical data present for all time points, V1 through to V9, resulting in a single set of cytokine model parameters. The cytokine and memory B cell data resolution do not support dose-dependent parameter estimate as done with the IgG. All random effects, and relative standard error on random effects from the model results are shown in Table S31.
We performed structural and practical sensitivity analysis to characterize the response of our model outputs to variation in the fitted parameters.78 Latin hypercube sampling (LHC) and partial rank correlation coefficient (PRCC)79 are employed to study the structural identifiability of the longitudinal model outcomes. For a particular model parameter, PRCC values close to the maximum value of 1.0 indicate model output is highly sensitive to variation in that parameter, with values greater than 0.5 considered significant.80 LHC PRCC analysis results are shown in Figure S10. We also report random effects and relative standard error on random effects to estimate practical data-driven identifiability in model outcomes (Table S31). Practical identifiability analysis quantifies how well-constrained the estimated model parameters are by the data sets to which they are being fit.78 Both structural and practical demonstrate no identifiability issues with any parameters reported in this work for IgG, memory B cells, or cytokines.
Statistical analysis
Comparison of serum IgG levels was done by mixed effects linear regression adjusted for the same set of variables. Serum IgG levels were not normally distributed and were loge-transformed for analysis. Baseline live SARS-CoV-2 neutralization titers were summarized as dichotomized variable given that most values were zero. Comparison between groups was based on Chi-square test or Fisher’s exact test as appropriate. Post-baseline comparison between groups was performed with quantile regression adjusted for age, vaccine regimen for D1 and D2 (mRNA/mRNA, ChAdOx1/ChAdOx1 and ChAdOx1/mRNA), and time between D1 and D2. For comparison at V9, we further adjusted for time between D3 and V9. For saliva ELISA and T cell ELISpot, the sample sizes were too small for a proper adjusted comparison between groups. Unadjusted comparison was based on Wilcoxon rank sum test. Analyses were conducted using SAS 9.4 (SAS Institute Inc., Cary, NC).
Acknowledgments
This study was supported by the Juan and Stefania Speck research fund and CITF grant 253488 (MO), CTN grant CTNPT045 (VAM, MO), and OHTN grant EFP-1032-IP (CK). M.O. is thankful to Samira Mubareka (University of Toronto, Sunnybrook Health Science Center) for providing the SARS-CoV-2 virus. A-C.G., K.C., and J.G. thank James Rini (University of Toronto) for supplying recombinant proteins. Convalescent and acute saliva was kindly provided by Dr. Allison McGeer (Sinai Health/University of Toronto). A-C.G. and K.C. are grateful to Yves Durocher (NRCC) for ELISA reagents. S.M. and L.K. are grateful to Drs. I-Ting Teng and Peter Kwong (Vaccine Research Center, NIAID, NIH) for providing the NTD protein. Huh-7 cells are provided by Dr. Sonya McParland (University of Toronto). The robotics equipment at the Network Biology Collaborative Center, Lunenfeld-Tanenbaum Research Institute, is supported by Canada Foundation for Innovation, the Ontarian Government, Genome Canada and Ontario Genomics (OGI-139). C.S.K. and J.M.H. acknowledge funding from NSERC, CIHR, NSERC EIDM, and the York Research Chair Program. R.K. acknowledges a CIHR grant MM1-174919. S.M. would like to acknowledge funding from Intramural Research Program of the Division of Intramural Research, NIAID, NIH. RBJ acknowledges grant UM1AI164565 from NIAID, NIH. The authors are grateful to all study participants for their time, dedication and samples provided.
Author contributions
Conceptualization: V.A.M., E.B., Y.C., R.R., R.K., T.W.C., C.K., and M.O. Funding Acquisition: V.A.M., C.K., and M.O. Project Administration and/or Data Curation: V.A.M., E.Z.M., E.B., G.C., S.C., A.I., and C.K. Methodology: V.A.M., G.C., Y.C., R.R., R.K., J.M.H., S.M., J.S., J.G., A-C.G., T.W.C., C.K., and M.O. Investigation: E.Z.M., E.B., S.C., A.I., S.G., P.S., H.S., R.L., P.B., V.A.M., L.W., S.S.M., G.M., M.D-B., T.T., L.K., F.Q., and A.P. Validation: V.A.M., K.C., J.H.W., F.Q., A.P., D.C.C.Jr., R.B.J., J.M.H., S.M., J.S., J.G., and A-C.G. Formal Analysis: T.L. and C.S.K. Mathematical Modeling: C.S.K. Supervision: V.A.M., K.C., J.H.W., J.M.H., S.M., J.S., J.G., and A-C.G. Writing – Original draft: V.A.M., C.S.K., and T.L. Writing – Review and Editing: All authors.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: September 14, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107915.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Tesoriero J.M., Swain C.A.E., Pierce J.L., Zamboni L., Wu M., Holtgrave D.R., Gonzalez C.J., Udo T., Morne J.E., Hart-Malloy R., et al. COVID-19 Outcomes Among Persons Living With or Without Diagnosed HIV Infection in New York State. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2020.37069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Western Cape Department of Health in collaboration with the National Institute for Communicable Diseases South Africa. Davies M.-A., Hussey H., Ismail M., Morden E., Vundle Z., Zweigenthal V. Risk Factors for Coronavirus Disease 2019 (COVID-19) Death in a Population Cohort Study from the Western Cape Province, South Africa. Clin. Infect. Dis. 2021;73:e2005–e2015. doi: 10.1093/cid/ciaa1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaskaran K., Rentsch C.T., MacKenna B., Schultze A., Mehrkar A., Bates C.J., Eggo R.M., Morton C.E., Bacon S.C.J., Inglesby P., et al. HIV infection and COVID-19 death: a population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. Lancet. HIV. 2021;8:e24–e32. doi: 10.1016/S2352-3018(20)30305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigel K., Swartz T., Golden E., Paranjpe I., Somani S., Richter F., De Freitas J.K., Miotto R., Zhao S., Polak P., et al. Coronavirus 2019 and People Living With Human Immunodeficiency Virus: Outcomes for Hospitalized Patients in New York City. Clin. Infect. Dis. 2020;71:2933–2938. doi: 10.1093/cid/ciaa880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann C., Casado J.L., Härter G., Vizcarra P., Moreno A., Cattaneo D., Meraviglia P., Spinner C.D., Schabaz F., Grunwald S., Gervasoni C. Immune deficiency is a risk factor for severe COVID-19 in people living with HIV. HIV Med. 2021;22:372–378. doi: 10.1111/hiv.13037. [DOI] [PubMed] [Google Scholar]
- 6.Dandachi D., Geiger G., Montgomery M.W., Karmen-Tuohy S., Golzy M., Antar A.A.R., Llibre J.M., Camazine M., Díaz-De Santiago A., Carlucci P.M., et al. Characteristics, Comorbidities, and Outcomes in a Multicenter Registry of Patients With Human Immunodeficiency Virus and Coronavirus Disease 2019. Clin. Infect. Dis. 2021;73:e1964–e1972. doi: 10.1093/cid/ciaa1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Chaer F., El Sahly H.M. Vaccination in the Adult Patient Infected with HIV: A Review of Vaccine Efficacy and Immunogenicity. Am. J. Med. 2019;132:437–446. doi: 10.1016/j.amjmed.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Kernéis S., Launay O., Turbelin C., Batteux F., Hanslik T., Boëlle P.Y. Long-term immune responses to vaccination in HIV-infected patients: a systematic review and meta-analysis. Clin. Infect. Dis. 2014;58:1130–1139. doi: 10.1093/cid/cit937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geretti A.M., Doyle T. Immunization for HIV-positive individuals. Curr. Opin. Infect. Dis. 2010;23:32–38. doi: 10.1097/QCO.0b013e328334fec4. [DOI] [PubMed] [Google Scholar]
- 10.Deepak P., Kim W., Paley M.A., Yang M., Carvidi A.B., Demissie E.G., El-Qunni A.A., Haile A., Huang K., Kinnett B., et al. Effect of Immunosuppression on the Immunogenicity of mRNA Vaccines to SARS-CoV-2 : A Prospective Cohort Study. Ann. Intern. Med. 2021;174:1572–1585. doi: 10.7326/M21-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haidar G., Agha M., Bilderback A., Lukanski A., Linstrum K., Troyan R., Rothenberger S., McMahon D.K., Crandall M.D., Sobolewksi M.D., et al. Prospective Evaluation of Coronavirus Disease 2019 (COVID-19) Vaccine Responses Across a Broad Spectrum of Immunocompromising Conditions: the COVID-19 Vaccination in the Immunocompromised Study (COVICS) Clin. Infect. Dis. 2022;75:e630–e644. doi: 10.1093/cid/ciac103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moor M.B., Suter-Riniker F., Horn M.P., Aeberli D., Amsler J., Möller B., Njue L.M., Medri C., Angelillo-Scherrer A., Borradori L., et al. Humoral and cellular responses to mRNA vaccines against SARS-CoV-2 in patients with a history of CD20 B-cell-depleting therapy (RituxiVac): an investigator-initiated, single-centre, open-label study. Lancet. Rheumatol. 2021;3:e789–e797. doi: 10.1016/S2665-9913(21)00251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massarweh A., Eliakim-Raz N., Stemmer A., Levy-Barda A., Yust-Katz S., Zer A., Benouaich-Amiel A., Ben-Zvi H., Moskovits N., Brenner B., et al. Evaluation of Seropositivity Following BNT162b2 Messenger RNA Vaccination for SARS-CoV-2 in Patients Undergoing Treatment for Cancer. JAMA Oncol. 2021;7:1133–1140. doi: 10.1001/jamaoncol.2021.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grupper A., Rabinowich L., Schwartz D., Schwartz I.F., Ben-Yehoyada M., Shashar M., Katchman E., Halperin T., Turner D., Goykhman Y., et al. Reduced humoral response to mRNA SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients without prior exposure to the virus. Am. J. Transplant. 2021;21:2719–2726. doi: 10.1111/ajt.16615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhavan K.P., Kampalath V.N., Overton E.T. The aging of the HIV epidemic. Curr. HIV AIDS Rep. 2008;5:150–158. doi: 10.1007/s11904-008-0023-3. [DOI] [PubMed] [Google Scholar]
- 16.Paul S.M., Martin R.M., Lu S.E., Lin Y. Changing trends in human immunodeficiency virus and acquired immunodeficiency syndrome in the population aged 50 and older. J. Am. Geriatr. Soc. 2007;55:1393–1397. doi: 10.1111/j.1532-5415.2007.01295.x. [DOI] [PubMed] [Google Scholar]
- 17.Ruggiero A., De Spiegelaere W., Cozzi-Lepri A., Kiselinova M., Pollakis G., Beloukas A., Vandekerckhove L., Strain M., Richman D., Phillips A., et al. During Stably Suppressive Antiretroviral Therapy Integrated HIV-1 DNA Load in Peripheral Blood is Associated with the Frequency of CD8 Cells Expressing HLA-DR/DP/DQ. EBioMedicine. 2015;2:1153–1159. doi: 10.1016/j.ebiom.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen-Quick A., Chaillon A., Yek C., Zanini F., Jordan P., Ignacio C., Caballero G., Gianella S., Smith D. Influenza Vaccination Can Broadly Activate the HIV Reservoir During Antiretroviral Therapy. J. Acquir. Immune Defic. Syndr. 2018;79:e104–e107. doi: 10.1097/QAI.0000000000001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones R.B., Kovacs C., Chun T.W., Ostrowski M.A. Short communication: HIV type 1 accumulates in influenza-specific T cells in subjects receiving seasonal vaccination in the context of effective antiretroviral therapy. AIDS Res. Hum. Retrovir. 2012;28:1687–1692. doi: 10.1089/AID.2012.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negredo E., Domingo P., Sambeat M.A., Rabella N., Vázquez G. Effect of pneumococcal vaccine on plasma HIV-1 RNA of stable patients undergoing effective highly active antiretroviral therapy. Eur. J. Clin. Microbiol. Infect. Dis. 2001;20:287–288. doi: 10.1007/s100960100470. [DOI] [PubMed] [Google Scholar]
- 21.Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., Voysey M., Aley P.K., Angus B., Babbage G., et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2021;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falsey A.R., Sobieszczyk M.E., Hirsch I., Sproule S., Robb M.L., Corey L., Neuzil K.M., Hahn W., Hunt J., Mulligan M.J., et al. Phase 3 Safety and Efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 Vaccine. N. Engl. J. Med. 2021;385:2348–2360. doi: 10.1056/NEJMoa2105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zafack J., Warshawsky B., Forbes N., Ismail S.J., Montroy J., Nunn A., Krishnan R., Wong E., Stirling R., Salvadori M., et al. An Advisory Committee Statement (ACS); 2022. Initial Guidance on a Second Booster Dose of COVID-19 Vaccines in Canada. National Advisory Committee on Immunization (NACI) [Google Scholar]
- 27.Amraei R., Yin W., Napoleon M.A., Suder E.L., Berrigan J., Zhao Q., Olejnik J., Chandler K.B., Xia C., Feldman J., et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2020.06.22.165803. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seyran M., Takayama K., Uversky V.N., Lundstrom K., Palù G., Sherchan S.P., Attrish D., Rezaei N., Aljabali A.A.A., Ghosh S., et al. The structural basis of accelerated host cell entry by SARS-CoV-2dagger. FEBS J. 2021;288:5010–5020. doi: 10.1111/febs.15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCallum M., Marco A.D., Lempp F., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.01.14.426475. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dicken S.J., Murray M.J., Thorne L.G., Reuschl A.K., Forrest C., Ganeshalingham M., Muir L., Kalemera M.D., Palor M., McCoy L.E., et al. Characterisation of B.1.1.7 and Pangolin Coronavirus Spike Provides Insights on the Evolutionary Trajectory of SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.03.22.436468. Preprint at. [DOI] [Google Scholar]
- 31.Lam S.D., Waman V.P., Orengo C., Lees J. Insertions in the SARS-CoV-2 Spike N-Terminal Domain May Aid COVID-19 Transmission. bioRxiv. 2021 doi: 10.1101/2021.12.06.471394. Preprint at. [DOI] [Google Scholar]
- 32.Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., Wang Y., Hong Q., Wang S., Zhao Q., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colwill K., Galipeau Y., Stuible M., Gervais C., Arnold C., Rathod B., Abe K.T., Wang J.H., Pasculescu A., Maltseva M., et al. A scalable serology solution for profiling humoral immune responses to SARS-CoV-2 infection and vaccination. Clin. Transl. Immunology. 2022;11 doi: 10.1002/cti2.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., Budylowski P., Dupuis A.P., 2nd, Girardin R.C., Rathod B., et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5 doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.H., Oliveira T.Y., Oren D.A., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheikh-Mohamed S., Isho B., Chao G.Y.C., Zuo M., Cohen C., Lustig Y., Nahass G.R., Salomon-Shulman R.E., Blacker G., Fazel-Zarandi M., et al. Systemic and mucosal IgA responses are variably induced in response to SARS-CoV-2 mRNA vaccination and are associated with protection against subsequent infection. Mucosal Immunol. 2022;15:799–808. doi: 10.1038/s41385-022-00511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nahass G.R., Salomon-Shulman R.E., Blacker G., Haider K., Brotherton R., Teague K., Yiu Y.Y., Brewer R.E., Galloway S.D., Hansen P., et al. Intramuscular SARS-CoV-2 vaccines elicit varying degrees of plasma and salivary antibody responses as compared to natural infection. medRxiv. 2021 doi: 10.1101/2021.08.22.21262168. Preprint at. [DOI] [Google Scholar]
- 38.Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexandrova Y., Yero A., Mboumba Bouassa R.S., Comeau E., Samarani S., Brumme Z.L., Hull M., Crawley A.M., Langlois M.A., Angel J.B., et al. SARS-CoV-2 Vaccine-Induced T-Cell Response after Three Doses in People Living with HIV on Antiretroviral Therapy Compared to Seronegative Controls (CTN 328 COVAXHIV Study) Viruses. 2023;15 doi: 10.3390/v15020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruner K.M., Wang Z., Simonetti F.R., Bender A.M., Kwon K.J., Sengupta S., Fray E.J., Beg S.A., Antar A.A.R., Jenike K.M., et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019;566:120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson E.M., Terry S., Copertino D., Leyre L., Danesh A., Weiler J., Ward A.R., Khadka P., McNeil E., Bernard K., et al. SARS CoV-2 mRNA vaccination exposes latent HIV to Nef-specific CD8(+) T-cells. Nat. Commun. 2022;13:4888. doi: 10.1038/s41467-022-32376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy I., Wieder-Finesod A., Litchevsky V., Biber A., Indenbaum V., Olmer L., Huppert A., Mor O., Goldstein M., Levin E.G., et al. Immunogenicity and safety of the BNT162b2 mRNA COVID-19 vaccine in people living with HIV-1. Clin. Microbiol. Infect. 2021;27:1851–1855. doi: 10.1016/j.cmi.2021.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woldemeskel B.A., Karaba A.H., Garliss C.C., Beck E.J., Wang K.H., Laeyendecker O., Cox A.L., Blankson J.N. The BNT162b2 mRNA Vaccine Elicits Robust Humoral and Cellular Immune Responses in People Living With Human Immunodeficiency Virus (HIV) Clin. Infect. Dis. 2022;74:1268–1270. doi: 10.1093/cid/ciab648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brumme Z.L., Mwimanzi F., Lapointe H.R., Cheung P.K., Sang Y., Duncan M.C., Yaseen F., Agafitei O., Ennis S., Ng K., et al. Humoral immune responses to COVID-19 vaccination in people living with HIV receiving suppressive antiretroviral therapy. NPJ Vaccines. 2022;7:28. doi: 10.1038/s41541-022-00452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruddy J.A., Boyarsky B.J., Werbel W.A., Bailey J.R., Karaba A.H., Garonzik-Wang J.M., Segev D.L., Durand C.M. Safety and antibody response to the first dose of severe acute respiratory syndrome coronavirus 2 messenger RNA vaccine in persons with HIV. AIDS. 2021;35:1872–1874. doi: 10.1097/QAD.0000000000002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madhi S.A., Koen A.L., Izu A., Fairlie L., Cutland C.L., Baillie V., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 in people living with and without HIV in South Africa: an interim analysis of a randomised, double-blind, placebo-controlled, phase 1B/2A trial. Lancet. HIV. 2021;8:e568–e580. doi: 10.1016/S2352-3018(21)00157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frater J., Ewer K.J., Ogbe A., Pace M., Adele S., Adland E., Alagaratnam J., Aley P.K., Ali M., Ansari M.A., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 in HIV infection: a single-arm substudy of a phase 2/3 clinical trial. Lancet. HIV. 2021;8:e474–e485. doi: 10.1016/S2352-3018(21)00103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noe S., Ochana N., Wiese C., Schabaz F., Von Krosigk A., Heldwein S., Rasshofer R., Wolf E., Jonsson-Oldenbuettel C. Humoral response to SARS-CoV-2 vaccines in people living with HIV. Infection. 2022;50:617–623. doi: 10.1007/s15010-021-01721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nault L., Marchitto L., Goyette G., Tremblay-Sher D., Fortin C., Martel-Laferrière V., Trottier B., Richard J., Durand M., Kaufmann D., et al. Covid-19 vaccine immunogenicity in people living with HIV-1. Vaccine. 2022;40:3633–3637. doi: 10.1016/j.vaccine.2022.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balcells M.E., Le Corre N., Durán J., Ceballos M.E., Vizcaya C., Mondaca S., Dib M., Rabagliati R., Sarmiento M., Burgos P.I., et al. Reduced Immune Response to Inactivated Severe Acute Respiratory Syndrome Coronavirus 2 Vaccine in a Cohort of Immunocompromised Patients in Chile. Clin. Infect. Dis. 2022;75:e594–e602. doi: 10.1093/cid/ciac167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Touizer E., Alrubayyi A., Rees-Spear C., Fisher-Pearson N., Griffith S.A., Muir L., Pellegrino P., Waters L., Burns F., Kinloch S., et al. Failure to seroconvert after two doses of BNT162b2 SARS-CoV-2 vaccine in a patient with uncontrolled HIV. Lancet. HIV. 2021;8:e317–e318. doi: 10.1016/S2352-3018(21)00099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lapointe H.R., Mwimanzi F., Cheung P.K., Sang Y., Yaseen F., Umviligihozo G., Kalikawe R., Speckmaier S., Moran-Garcia N., Datwani S., et al. People with HIV receiving suppressive antiretroviral therapy show typical antibody durability after dual COVID-19 vaccination, and strong third dose responses. J. Infect. Dis. 2023;227:838–849. doi: 10.1093/infdis/jiac229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korosec C.S., Farhang-Sardroodi S., Dick D.W., Gholami S., Ghaemi M.S., Moyles I.R., Craig M., Ooi H.K., Heffernan J.M. Long-term durability of immune responses to the BNT162b2 and mRNA-1273 vaccines based on dosage, age and sex. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-25134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox R.J., Brokstad K.A. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat. Rev. Immunol. 2020;20:581–582. doi: 10.1038/s41577-020-00436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mei H.E., Yoshida T., Sime W., Hiepe F., Thiele K., Manz R.A., Radbruch A., Dörner T. Blood-borne human plasma cells in steady state are derived from mucosal immune responses. Blood. 2009;113:2461–2469. doi: 10.1182/blood-2008-04-153544. [DOI] [PubMed] [Google Scholar]
- 58.Buckner C.M., Moir S., Ho J., Wang W., Posada J.G., Kardava L., Funk E.K., Nelson A.K., Li Y., Chun T.W., Fauci A.S. Characterization of plasmablasts in the blood of HIV-infected viremic individuals: evidence for nonspecific immune activation. J. Virol. 2013;87:5800–5811. doi: 10.1128/JVI.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raux M., Finkielsztejn L., Salmon-Céron D., Bouchez H., Excler J.L., Dulioust E., Grouin J.M., Sicard D., Blondeau C. Comparison of the distribution of IgG and IgA antibodies in serum and various mucosal fluids of HIV type 1-infected subjects. AIDS Res. Hum. Retrovir. 1999;15:1365–1376. doi: 10.1089/088922299310070. [DOI] [PubMed] [Google Scholar]
- 60.Schneider T., Zippel T., Schmidt W., Zeitz M., Ullrich R. Secretory immunity in HIV infection. Pathobiology. 1998;66:131–138. doi: 10.1159/000028009. [DOI] [PubMed] [Google Scholar]
- 61.Buckner C.M., Moir S., Kardava L., Ho J., Santich B.H., Kim L.J.Y., Funk E.K., Nelson A.K., Winckler B., Chairez C.L., et al. CXCR4/IgG-expressing plasma cells are associated with human gastrointestinal tissue inflammation. J. Allergy Clin. Immunol. 2014;133:1676–1685.e5. doi: 10.1016/j.jaci.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu W., Santini P.A., Sullivan J.S., He B., Shan M., Ball S.C., Dyer W.B., Ketas T.J., Chadburn A., Cohen-Gould L., et al. HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat. Immunol. 2009;10:1008–1017. doi: 10.1038/ni.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiao X., He B., Chiu A., Knowles D.M., Chadburn A., Cerutti A. Human immunodeficiency virus 1 Nef suppresses CD40-dependent immunoglobulin class switching in bystander B cells. Nat. Immunol. 2006;7:302–310. doi: 10.1038/ni1302. [DOI] [PubMed] [Google Scholar]
- 64.Shacklett B.L. Mucosal Immunity in HIV/SIV Infection: T Cells, B Cells and Beyond. Curr. Immunol. Rev. 2019;15:63–75. doi: 10.2174/1573395514666180528081204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lapuente D., Fuchs J., Willar J., Vieira Antão A., Eberlein V., Uhlig N., Issmail L., Schmidt A., Oltmanns F., Peter A.S., et al. Protective mucosal immunity against SARS-CoV-2 after heterologous systemic prime-mucosal boost immunization. Nat. Commun. 2021;12:6871. doi: 10.1038/s41467-021-27063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao Y., Cai C., Wullimann D., Niessl J., Rivera-Ballesteros O., Chen P., Lange J., Cuapio A., Blennow O., Hansson L., et al. Immunodeficiency syndromes differentially impact the functional profile of SARS-CoV-2-specific T cells elicited by mRNA vaccination. Immunity. 2022;55:1732–1746.e5. doi: 10.1016/j.immuni.2022.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emery A., Joseph S.B., Swanstrom R. Nonsuppressible viremia during HIV-1 therapy meets molecular virology. J. Clin. Invest. 2023;133 doi: 10.1172/JCI167925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teng I.T., Nazzari A.F., Choe M., Liu T., Oliveira de Souza M., Petrova Y., Tsybovsky Y., Wang S., Zhang B., Artamonov M., et al. Molecular probes of spike ectodomain and its subdomains for SARS-CoV-2 variants, Alpha through Omicron. PLoS One. 2022;17 doi: 10.1371/journal.pone.0268767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin J., Law R., Korosec C.S., Zhou C., Koh W.H., Ghaemi M.S., Samaan P., Ooi H.K., Matveev V., Yue F., et al. Longitudinal Assessment of SARS-CoV-2-Specific T Cell Cytokine-Producing Responses for 1 Year Reveals Persistence of Multicytokine Proliferative Responses, with Greater Immunity Associated with Disease Severity. J. Virol. 2022;96 doi: 10.1128/jvi.00509-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mattiuzzo, G., Bentley, E.M., Hassall, M., Routley, S., Richardson, S., Bernasconi, V., Kristiansen, P., Harvala, H., Roberts, D., Semple, M.G., et al. (2020). Establishment of the WHO International Standard and Reference Panel for anti-SARS-CoV-2 antibody. Expert Committee on Biological Standardization WHO/BS/2020.2403.
- 71.Law J.C., Koh W.H., Budylowski P., Lin J., Yue F., Abe K.T., Rathod B., Girard M., Li Z., Rini J.M., et al. Systematic Examination of Antigen-Specific Recall T Cell Responses to SARS-CoV-2 versus Influenza Virus Reveals a Distinct Inflammatory Profile. J. Immunol. 2021;206:37–50. doi: 10.4049/jimmunol.2001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buckner C.M., Kardava L., El Merhebi O., Narpala S.R., Serebryannyy L., Lin B.C., Wang W., Zhang X., Lopes de Assis F., Kelly S.E.M., et al. Interval between prior SARS-CoV-2 infection and booster vaccination impacts magnitude and quality of antibody and B cell responses. Cell. 2022;185:4333–4346.e14. doi: 10.1016/j.cell.2022.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kardava L., Sohn H., Youn C., Austin J.W., Wang W., Buckner C.M., Justement J.S., Melson V.A., Roth G.E., Hand M.A., et al. IgG3 regulates tissue-like memory B cells in HIV-infected individuals. Nat. Immunol. 2018;19:1001–1012. doi: 10.1038/s41590-018-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kinloch N.N., Ren Y., Conce Alberto W.D., Dong W., Khadka P., Huang S.H., Mota T.M., Wilson A., Shahid A., Kirkby D., et al. HIV-1 diversity considerations in the application of the Intact Proviral DNA Assay (IPDA) Nat. Commun. 2021;12:165. doi: 10.1038/s41467-020-20442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moyles I.R., Korosec C.S., Heffernan J.M. Determination of significant immunological timescales from mRNA-LNP-based vaccines in humans. medRxiv. 2022 doi: 10.1101/2022.07.25.22278031. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farhang-Sardroodi S., Korosec C.S., Gholami S., Craig M., Moyles I.R., Ghaemi M.S., Ooi H.K., Heffernan J.M. Analysis of Host Immunological Response of Adenovirus-Based COVID-19 Vaccines. Vaccines. 2021;9:861. doi: 10.3390/vaccines9080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McHeyzer-Williams L.J., McHeyzer-Williams M.G. Antigen-specific memory B cell development. Annu. Rev. Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 78.Sher A., Niederer S.A., Mirams G.R., Kirpichnikova A., Allen R., Pathmanathan P., Gavaghan D.J., van der Graaf P.H., Noble D. A Quantitative Systems Pharmacology Perspective on the Importance of Parameter Identifiability. Bull. Math. Biol. 2022;84:39. doi: 10.1007/s11538-021-00982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKay M.D., Beckman R.J., Conover W.J. A Comparison of Three Methods for Selecting Values of Input Variables in the Analysis of Output from a Computer Code. Technometrics. 1979;21:239–245. doi: 10.2307/1268522. [DOI] [Google Scholar]
- 80.Wu J., Dhingra R., Gambhir M., Remais J.V. Sensitivity analysis of infectious disease models: methods, advances and their application. J. R. Soc. Interface. 2013;10 doi: 10.1098/rsif.2012.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.