SUMMARY
Spontaneous transcription and translation of HIV can persist during suppressive anti-retroviral therapy (ART). The quantity, phenotype and biological relevance of this spontaneously “active” reservoir remains unclear. Using multiplexed single-cell RNAflow-FISH, we detect active HIV transcription in 14/18 people with HIV on suppressive ART, with a median of 28/million CD4+ T cells. While these cells predominantly exhibit abortive transcription, p24-expressing cells are evident in 39% of participants. Phenotypically diverse, active reservoirs are enriched in central memory T cells and CCR6- and activation marker-expressing cells. The magnitude of the active reservoir positively correlates with total HIV-specific CD4+ and CD8+ T cell responses and with multiple HIV-specific T cell clusters identified by unsupervised analysis. These associations are particularly strong with p24-expressing active reservoir cells. Single-cell vDNA sequencing shows that active reservoirs are largely dominated by defective proviruses. Our data suggest that these reservoirs maintain HIV-specific CD4+ and CD8+ T responses during suppressive ART.
eTOC blurb
Dubé et al. identify phenotypically diverse HIV-infected cells that spontaneously express viral RNA, and occasionnally protein, during antiretroviral treatment. Despite carrying defective proviruses, active reservoirs correlate with HIV-specific CD4+ and CD8+ T cell responses. These results suggest that ongoing expression of viral genes maintain HIV-specific immune responses during suppressive ART.
Graphical Abstract
INTRODUCTION
The persistence of HIV represents a fundamental challenge to achieving a cure. When ART is interrupted in people with HIV (PWH), rare persisting infected cells can fuel viral rebound1. Recent advances provided sensitive evaluations of the size of the HIV reservoir2. CD4+ T cells bearing HIV genomes3,4 are one order of magnitude more abundant than those with the ability to transcribe multiply-spliced RNA5,6, themselves another order of magnitude more frequent than those producing p247–9. These differences may reflect stepwise stages of blocks in viral transcription and translation10,11. Detecting viral reservoirs based on either viral RNA (vRNA) or protein typically involves ex vivo stimulation with latency reversal agents (LRAs) that induce viral gene expression. This approach provided valuable information on inducible reservoirs poised for reactivation. Current antiretroviral therapies do not target HIV transcription nor translation, therefore spontaneous viral gene transcription10,12–17 and translation18–20 can persist during ART. The definite quantification, single-cell phenotyping, and biological relevance of these “active” reservoir cells are not established.
CD4+ and CD8+ T cells are increasingly recognized as essential actors in the control of SIV and HIV infections21–26. Consequently, anti-HIV immunity is expected to play an important role in cure strategies and to contribute to purging reservoirs, exerting immunosurveillance of residual virus and/or supporting the development of broadly neutralizing HIV-specific antibodies27,28. HIV-specific CD4+ and CD8+ T cells can be detected during ART, although their magnitude and functions present notable interindividual heterogeneity29–32. The mechanisms involved in the persistence of such HIV-specific immunity during suppressive ART are not entirely understood but are unlikely to result from ongoing residual viral replication, an actively debated concept33–36. Conversely, active reservoirs that lead to protein expression from a fraction of the largely dominant pool of defective proviruses and low-level virion release from an even smaller proportion of active reservoirs with intact genomes could maintain and shape anti-HIV CD4+ and CD8+ T cell responses during ART14,37–40.
Herein, we quantified and phenotyped viral reservoirs spontaneously expressing viral RNA and the p24 protein in primary clinical samples directly ex vivo. We found associations between active reservoirs and HIV-specific CD4+ and CD8+ T cells, supporting that low level viral gene expression by spontaneous reservoirs is sufficient to maintain anti-HIV adaptive immunity.
RESULTS
Spontaneous vRNA-expressing reservoirs are detectable in a majority of ART-suppressed PWH.
We previously used a multiplexed HIV RNAflow-FISH assay to characterize viral reservoirs induced ex vivo7–9. A previous digital-droplet study on bulk CD4+ T cells showed expression during latency of “long-LTR” abortive transcripts10,41. Therefore, to maximize detection of viral reservoir cells that spontaneously express vRNA without ex vivo stimulation, termed active reservoir, we adapted our multiplexed vRNA detection strategy and focused on 5’ HIV genes (Figure 1A). As described before9, a first-step analysis provided an inclusive detection of viral transcripts, consisting of an exonRNA probe set targeting exon sequences, including a portion of the 5’LTR region, present on all viral transcripts42. The second step of detection added stringency and ensure specificity. To this end, two additional probesets were generated: 1) a gagRNA probeset, detecting full-length genomic transcripts but also shorter abortive or defective transcripts containing at least a portion of the gag gene9; 2) a polRNA detecting transcripts that elongated beyond the gene gag. We also stained intracellular p24 to assess viral translation.
Figure 1: Spontaneous vRNA expression by HIV reservoirs in ART-suppressed PWH.
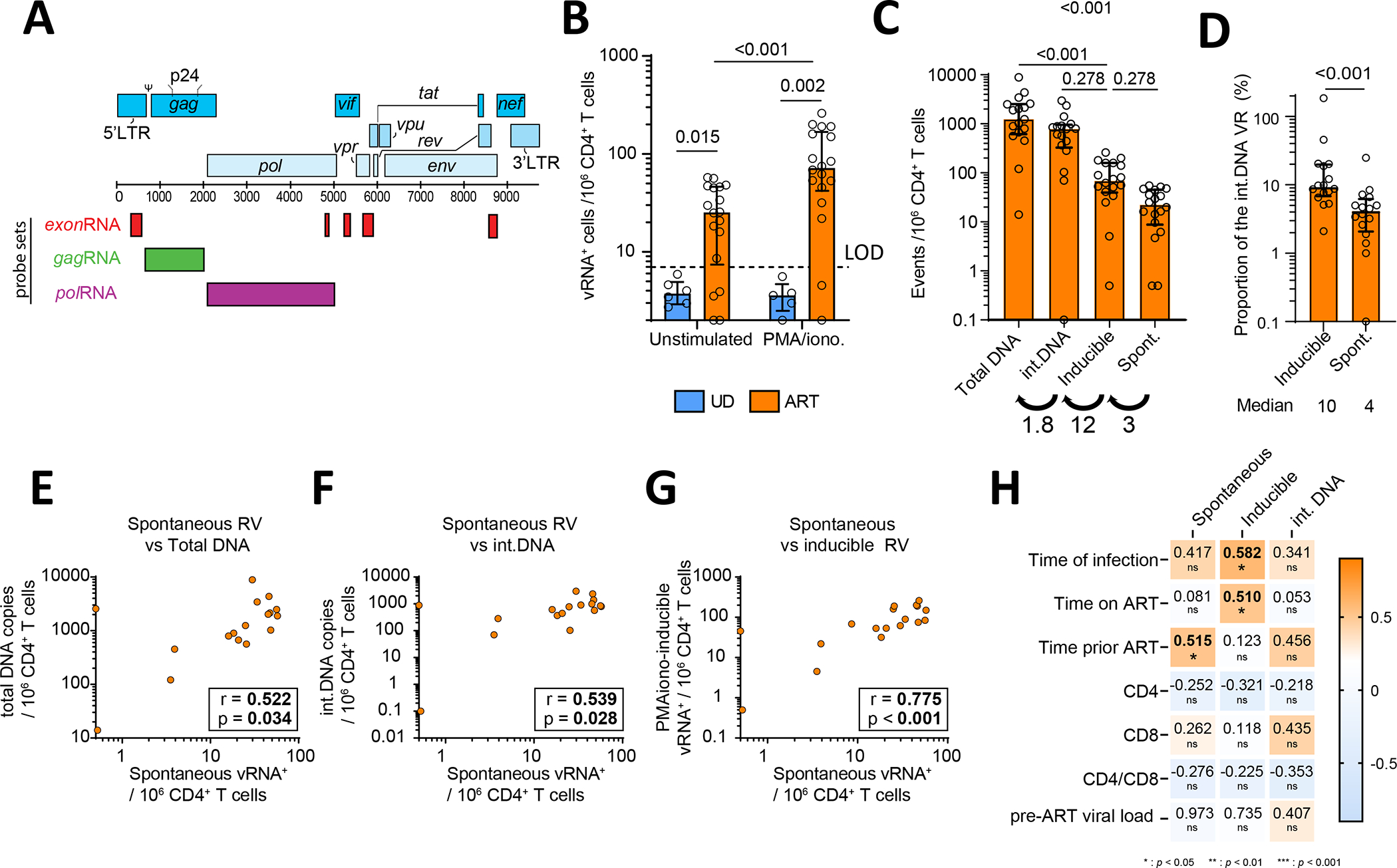
(A) vRNA probeset designs. (B) Quantification of vRNA+ cells in purified CD4+ T cells from ART-treated PWH and uninfected (UD) as controls. Cells were either unstimulated or treated ex vivo with PMA/ionomycin for 15h. Two statistical tests are shown: Mann-Whitney for cohort comparisons and Wilcoxon between stimulations. (C) Comparison between four different types of viral reservoir measurements. Numbers below indicate the median fold increase between groups. (D) The proportion of inducible and spontaneous vRNA+ reservoirs using integrated DNA as denominator. A Wilcoxon test was performed, shown above. Median values are shown below. (E-G) Correlations between (E) spontaneous reservoirs and total DNA, (F) spontaneous reservoirs and integrated DNA, (G) spontaneous and inducible reservoirs, with r and p values from Spearman tests. (H) Correlations between reservoirs metrics and indicated clinical parameters. CD4 and CD8 stand for CD4+ and CD8+ T cell clinical counts. Values in the heat map indicate r values, with p values underneath: * <0.05, ** <0.01, *** <0.001. The shade of colors indicates the r value. N=18. In B and D, the histograms indicate the median, and the error bars illustrate the interquartile range
We used this adapted HIV RNAflow assay to quantify the viral reservoirs in 18 PWH on ART for >3 years (median =10 years, see clinical characteristics in Table S1). To reveal inducible viral reservoirs, CD4+ T cells isolated from PBMCs were treated 15h with PMA/ionomycin. To identify viral reservoirs able to spontaneously express HIV RNA and/or protein, we left CD4+ T cells unstimulated after isolation. We identified HIV-infected cells by Boolean ORgating9, therefore included any cells either exonRNA+gagRNA+ or exonRNA+polRNA+ and exonRNA+p24+ into a single non-overlapping population expressing any combination of the viral genes assessed (henceforth termed vRNA+; Figure S1A). CD4+ T cells from 6 uninfected donors (UD) served as specificity controls. We set a positive detection threshold at 7 events/106 CD4+ T cells corresponding to the mean detection in UD plus twice the standard deviation, rounded up (mean + 2*SD). Based on these criteria, we detected spontaneous reservoirs in 78% (14/18) of PWH on suppressive ART, with a median of 28 active vRNA+ cells/106 CD4+ (Figure 1B). This level of detection represents a compromise compared to a limiting dilution RT-qPCR assay, sacrificing some sensitivity for higher throughput and the ability to maintain phenotyping capability (Figure S1F,G). PMA/ionomycin-inducible reservoirs could be detected in 89% (16/18) of participants, with a significantly higher median (79 vRNA+ /106 CD4+). Reservoirs induced by Panobinostat and Ingenol-3-angelate (termed LRA) reached an even higher level (325 induced vRNA+ /106 CD4+) (Figure S1B). By comparison, the frequency of CD4+ T cells harboring integrated HIV DNA was 12-fold higher than PMA/ionomycin-inducible reservoirs. In contrast, inducible reservoirs were 3-fold more frequent than spontaneous reservoirs (Figure 1C). We calculated that a median of 10% of the CD4+ T cells harboring integrated HIV DNA produced viral transcripts upon stimulation, whereas 4% for spontaneously expressed vRNA+ (Figure 1D). Active vRNA+ reservoirs correlated with total and integrated HIV DNA (Figure 1EF), whereas PMA/ionomycin-induced reservoirs showed strong trends (Figure S1CD). We found strong associations between reservoir cells with spontaneous viral expression and both PMA/ionomycin (Figure 1G) and LRA-inducible (Figure S1E) reservoirs.
We next tested for associations between integrated HIV DNA, inducible and spontaneously active vRNA+ reservoirs, and clinical features (Figure 1H). The time of infection correlated with the size of inducible reservoirs, with a positive trend with the magnitude of the spontaneous reservoir. There was no association between the spontaneous reservoir and time on ART, while there was an association for inducible reservoirs. The level of spontaneous vRNA expression, but not inducibility, was associated with a longer duration of untreated infection. CD4+ T cell counts, CD4/CD8 ratios and pre-ART viral loads did not correlate with either inducible or spontaneous reservoirs.
Globally, these data highlighted the existence of spontaneous vRNA+ reservoirs in most PWH on suppressive ART. Spontaneous and inducible reservoirs were strongly associated, suggesting a robust relationship.
Spontaneously active reservoirs are enriched in short abortive and defective gagRNA+ transcripts.
We next sought to characterize these active reservoirs based on p24 (Figure 2A), gagRNA, and polRNA co-expression (Figure 2B). The expression of p24 was used to assess viral protein expression8,9, whereas gagRNA and polRNA defined the processivity of the transcription. This bilayered analysis created 8 theoretical subpopulations (Figure 2C, S2AB). Among these, single exonRNA+ cells could not be interpreted because of the insufficient specificity of single-parameter detection9. The prevalence of the remaining seven theoretical populations is summarized in Figures 2D and S2A. Unstimulated samples from ART-suppressed individuals were homogeneously enriched in p24− gagRNA+ polRNA− cells (Figure S2C). This population corresponded to short abortive or deleted transcripts (gagRNA+) described before9,10. PMA/ionomycin-induced reservoirs presented a more heterogeneous profile: although p24− gagRNA+ polRNA− cells were frequent, other more processive populations could also be detected, such as p24− gagRNA+ polRNA+ (elongated transcripts not sustaining translation, termed vRNADP for “double gagRNA+ polRNA+ positive”) > p24− gagRNA− polRNA+ (likely deleted transcripts, termed polRNA+) > p24+ gagRNA+ polRNA+ (translation-competent reservoirs, termed p24+). The pattern of active reservoirs was similar to combinatory LRA (panabinostat+ingenol), with strong expression of gagRNA and little expression of polRNA and marginal p24 translation (Figure S2A,C)9,10. In a subset of participants, the polRNA probeset was substituted by a more distal nefRNA probeset9. We detected rare nefRNA+ cells among active reservoirs and none that co-expressed gag and nef genes (Figure S2B), further suggesting unproductive transcription aborting in the 5’ portion of the viral genome. Spontaneous p24-expressing cells represented a rare fraction (Figure 2E). These cells could be detected at low frequencies in 7/18 HIV+ participants, with a median frequency of 0.44 p24+vRNA+ cell/106 CD4+ (Figure 2F). These active p24+ cells did not correlate with translation-competent reservoirs identified after PMA/ionomycin-stimulation (Figure 2G).
Figure 2: Spontaneously active reservoirs are enriched in short vRNA transcripts.

(A-B) Gating strategy to assess: (A) p24 expression in vRNA+ cells, and (B) gagRNA and polRNA co-expression in p24− or p24+ vRNA+ cells. (C) List of the theoretical vRNA+ populations. (D) Donut charts presenting the median proportions of each vRNA+ subpopulation for spontaneous or PMA/ionomycin-induced reservoirs, as colored in C. Numbers in the donut holes represent the median vRNA+ / 106 CD4+. (E) The proportion of p24+ cells in spontaneous compared to PMA/ionomycin-induced vRNA+ reservoirs. The frequency of participants with p24+ cell detection is indicated underneath. (F) Compared frequencies of spontaneous vRNA+ and p24+ cells. The median of the 7 participants in which p24 was spontaneously detected is shown. (G) Correlation between p24+ cells in unstimulated and PMA/ionomycin-induced conditions. (H) Violin plots showing total single-cell fluorescence intensities in 18 participants. (E,F) The bars indicate the median. The error bars illustrate the interquartile range. Results from Wilcoxon tests are shown above. N=18 (excepted for panel E, where n=16 because <5 vRNA+ events). (I) Representative maximal intensity of confocal microscopy projections from Z-stacks of vRNA− and vRNA+ sorted cells. (J) Number of spots per cell for exonRNA (left) and gagRNA (right) probes. The results from a Mann-Whitney test are shown above. False-positive rates are indicated by the dashed lines.
We used cytometric fluorescent intensities (FI) to obtain a semi-quantitative measurement of RNA copies / cell43 and assessed the viral transcription and translation levels / infected cell. We used this metric to determine if the level of viral transcription could define the success of viral translation. To avoid biases due to a low number of viral reservoir cells in some PWH, we concatenated all events per condition. Since exonRNA is expressed in all vRNA+ cells, we used it as a surrogate of global transcriptional activity. In both PMA/ionomycin- or LRA-induced reservoirs, the single-cell exonRNA, gagRNA, and polRNA expression presented a skewed distribution composed of a low-yield bulky population and a tail spreading about one order of magnitude higher (Figure 2H, S2E). P24-expressing PMA/ionomycin-induced vRNA+ cells were almost exclusively found in these tails of high transcriptional activity (Figure S2F). Consistent with the rare p24 expression in active reservoirs, such a high-yield tail was essentially absent in spontaneous reservoirs. Instead, exonRNA and gagRNA could be detected at low levels, whereas polRNA expression remained comparably marginal (Figure 2H). In contrast to spontaneous p24+ cells, the levels of CD4 on the surface of spontaneous vRNA+ cells were comparable to vRNA-cells, suggesting suboptimal or no expression of Nef and Vpu proteins (Figure S2G). These data demonstrate the low-yield, unprocessed nature of the active reservoirs.
We next used confocal fluorescence microscopy to count the number of HIV RNA foci per vRNA+ cell, as done previously8. We sorted vRNA+ and vRNA− cells, imaged and manually enumerated foci per cell (Figure S2H and Figure 2I). The conservative false-positive rates (assuming Gaussian distribution, mean foci/cell in vRNA− cells + 3SD) were established at 1.1 foci/cell for exonRNA and 5.1 foci/cell for gagRNA (Figure 2J). Conversely, vRNA+ cells containing down to 5 foci were clearly detected for both probes, but few vRNA+ cells were below this cutoff. The median detection reached 16 foci for exonRNA, and 14 foci / cell for gagRNA, both significantly higher than their respective false-positive rate. ExonRNA+ and gagRNA+ foci / cell correlated significantly (Figure S2I). Taken together, these results suggest that our approach can reliably identify spontaneously active reservoirs with a cutoff of 5 vRNA copies/cell.
To determine the proviral features of spontaneously vRNA+ cells, we next performed near full-length sequencing of using a modified FLIPS assay9,44(Figure 3). We obtained a total of 40 amplicons from three ART-treated participants. From these 40 amplicons there was 36 distinct sequences, and 3 small clones (Figure 3A). Most sequences harbored large deletions spanning the entire env region. Deletions in pol were also common (Figure 3B). We found two sequences with an inversion. Seven amplicons were near null length: one was hypermutated, two had early stop codons, and two bore packaging signal defects. No intact proviral sequence were found in our limited sampling. This indicates that spontaneous viral transcription is mainly fueled by defective proviruses.
Figure 3: Near full-length single-cell vDNA sequencing of spontaneously active viral reservoirs identifies underlying proviral defects.
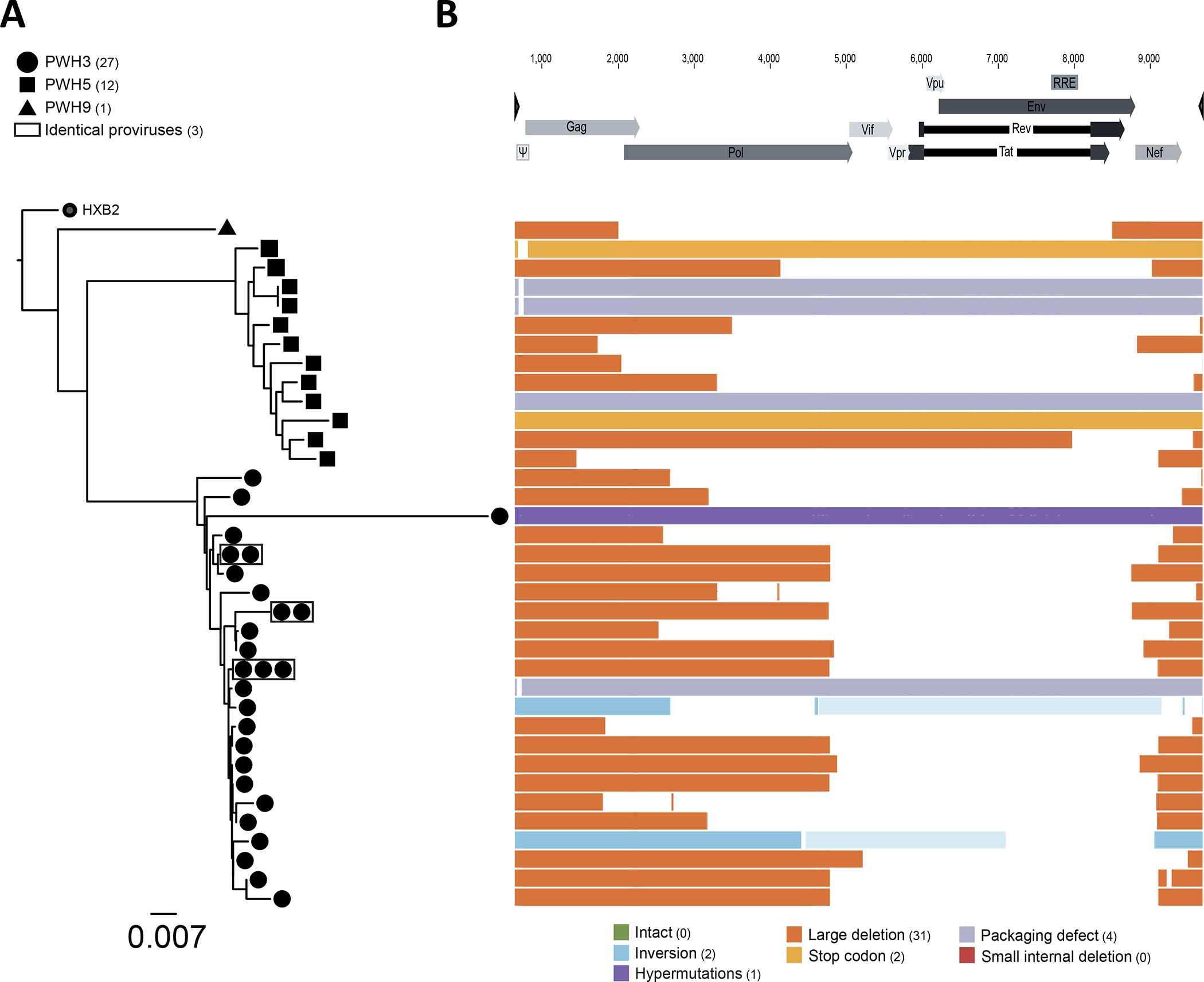
Purified CD4+ T cells from 3 ART-treated participants were co-stained by multiplexed HIV-1 RNAflow-FISH. vRNA+ cells were individually sorted for nested PCR amplification and near full-length sequencing. (A) Phylogenetic trees for the three participants PWH3, PWH5 and PWH9 built from the entire amplified area sequenced based on maximum likelihood. Sequences with 100% identity are boxed in gray. (B) List of the different defects found in the 36 proviral sequences, aligned on the HxB2 genome. The type of defect is color coded, based on the legend presented below.
Spontaneously active viral reservoirs are phenotypically diverse.
LRA stimulation can alter cell surface marker expression, thus biasing phenotyping9. As measuring spontaneous cells does not require an in vitro stimulation, the experimental approach used allows reliable phenotyping of viral reservoir cells. We performed unsupervised analyses of high-dimensional flow cytometric phenotyping data to avoid priori-defined marker combinations (Figure 4). We examined features that were previously found enriched in HIV-infected cells either during ART or during untreated chronic infection (Table S2): chemokine receptors involved in tissue homing [(CXCR5 for Tfh; CXCR3 for Th1; CCR6 for Th17/Th22, CCR4 for Th2 (also expressed on Th17/Th22 and Tregs; reviewed in 45) gut homing markers (integrin β7 and CD103, reviewed in 46); activation markers (CD38, HLA-DR, ICOS); cell cycle/proliferation (Ki67); inhibitory checkpoint (PD-1); and memory and differentiation markers (CD45RA, CCR7, and CD27). To avoid excessive background noise in phenotype profiling, we focused the analyses on participants with >5 vRNA+ cells and with spontaneous reservoirs above the level of positivity (14/18 participants).
Figure 4: Spontaneously active viral reservoirs are phenotypically diverse.
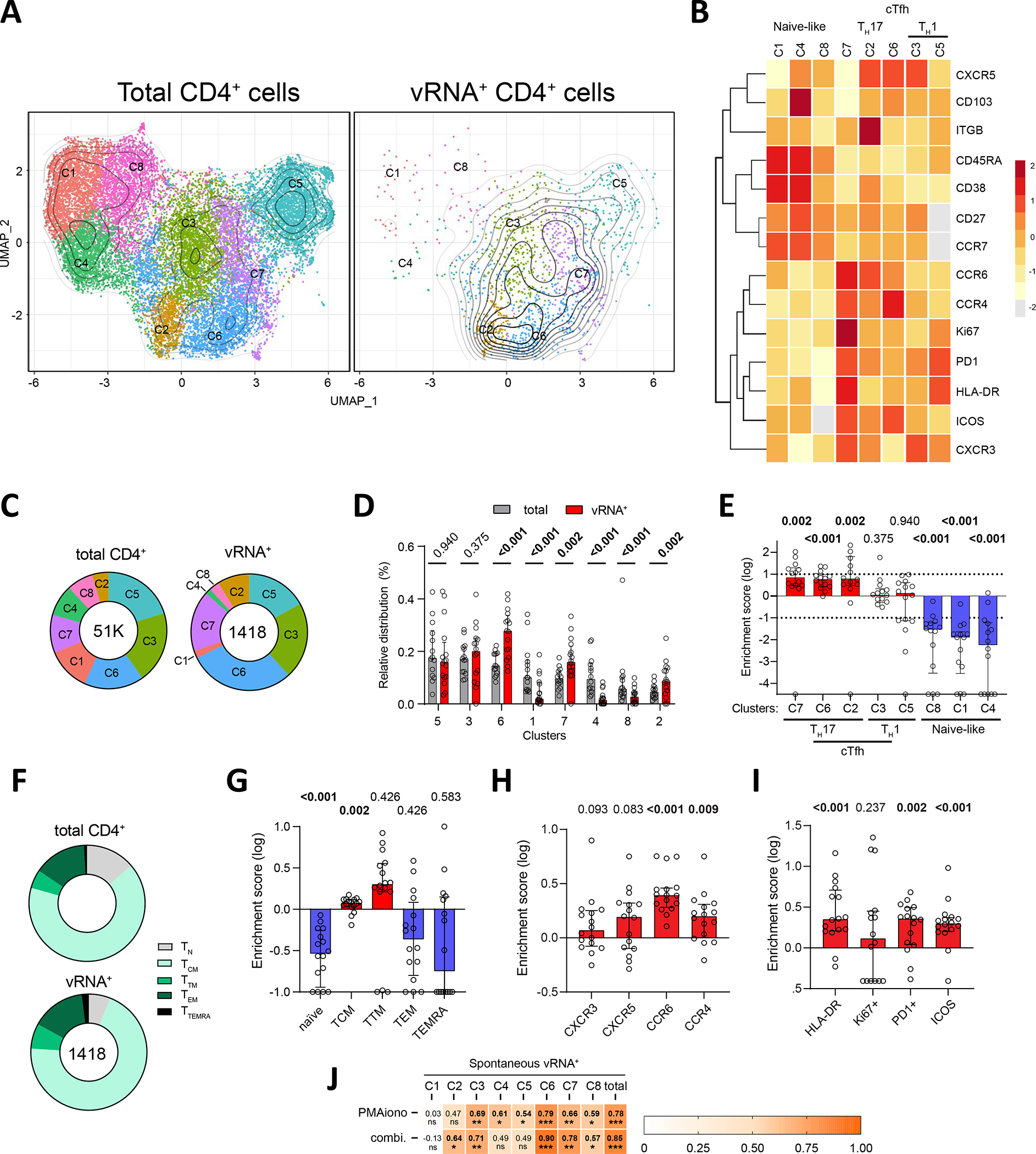
(AB) Global Uniform manifold approximation and projection (UMAP) for dimension reduction map of all 1418 vRNA+ cells. These cells were embedded among downsampled 3000 CD4+ per donor to help identify stable population clusters. (A) All 8 clusters are identified. (B) Individual CD4 (left) and vRNA+ (right) UMAP are shown. Contours show the density of vRNA+ cells. (C) Heat map showing an unsupervised hierarchical clustering of the 8 clusters, defined by MFI. (D) The donut charts present the proportion of each cluster for the indicated population, with color matching with panel A. The total number of events used to generate the plot are indicated in the donut holes. (E) The histogram presents the proportion of each cluster, with a side-by-side comparison between CD4 and vRNA+ cells. Wilcoxon tests are shown. (F) The enrichment score for each cluster is calculated as the log of the ratios between vRNA+ / CD4 cluster proportions. (G) Donut charts presenting the median proportions of each memory vRNA+ subpopulation for spontaneous or PMA/ionomycin-induced reservoirs. (H-J) Enrichment scores for univariate analysis of (H) memory, (I) polarization and (J) activation subsets. The enrichment scores were calculated as in (F). In EF and H-J. (K) Correlations between PMA/ionomycin or Panobinostat+ingenol inducible reservoirs and each active vRNA+ cluster. Values and the shade of color indicate r values. P values are shown underneath: * p <0.05, ** p <0.01, *** p <0.001. In E,F,H-J, the histograms indicate the median, and the error bars illustrate the interquartile range.
We illustrated the distribution of clustered populations by the uniform manifold approximation and projection (UMAP) algorithm47. To help define clear phenotypic clusters, all identified vRNA+ cells were concatenated and downsampled to 3,000 autologous CD4+ T cells per participant. Cluster identification was performed using Phenograph48. 8 clusters were defined based on distinct profiles of relative marker expression (Figure 4A–C, S3A). Active vRNA+ reservoir cells were found in all 8 clusters (Figure 4D,E). However, compared to total CD4+ T cells, vRNA+ cells were sparsely found in clusters C1, C4, and C8 (Figure 4B,D,E) enriched in CD45RA, CCR7, and CD27, consistent with a naïve phenotype (Figure 4C). CCR7 and CD27 expression was moderate in CD45RA-negative clusters C2, C3, C5, C6 and C7, suggesting that these clusters are composed of mixed populations of central memory (TCM), transitory (TTM), and effector (TEM) cells defined by other phenotypic markers (Figure 4C). We calculated enrichment scores to evaluate the relative over- or under-representation of active vRNA+ reservoir cells among the identified clusters (Figure 4F). This analysis confirmed the under-representation of active vRNA+ reservoir cells in the naïve-like clusters C1, C4, and C8. Univariate analysis relying on CD45RA, CD27, and CCR7 confirmed this finding, and further showed a significant enrichment of active vRNA+ reservoir cells in TCM and a trend for TTM (Figure 4G,H, S3B,C). The paucity of naïve active vRNA+ reservoir cells is consistent with the significant negative association between the prevalence of naïve CD4+ T cells and the frequency of active reservoirs (Figure S3D).
Active reservoirs were enriched in clusters C2, C6 and C7 defined by the expression of CCR6 and/or CCR4, suggesting preferential Th17 or Th22 differentiation. C2 and C6 were also enriched in CXCR5 expression, a marker enriched in follicular helper (Tfh) cells, but this chemokine receptor was not expressed by cells clustering in C7. C7 also included relatively rare cells with strong features of activation, including higher FI for ICOS, HLA-DR, Ki67, and PD-1 (Figure S3A). Although, we detected frequent active vRNA+ reservoir cells in the Th1-like CXCR3-enriched clusters C5 and C3, these subsets were not enriched in infected cells compared to the global CD4+ T cell population.
We next analyzed the polarization, integrin and activation marker expression of the active reservoirs through univariate analyses (Figure 4IJ, S4E–L). We focused these analyses on CD45RA− vRNA+ cells to avoid biasing our phenotyping given the near complete absence of vRNA+ cells in naïve populations. Active vRNA+ reservoirs could express any of the four chemokine receptors tested (Figure S3E), with no clear dominance for one over another (Figure S3F). Confirming the previous unsupervised findings, significant enrichments were observed for CCR6+ and CCR4+ cells, with trends for CXCR5 and CXCR3 (Figure 4I, S3G). CCR6 stood out as the most consistently enriched chemokine receptor among all tested participants (increase in all 16/18 participants). A trend for enrichment of vRNA+ events in β7-integrin+ cells was observed, but not in CD103+ cells, nor CD32a, another marker previously associated with viral transcription49 (Figure S3H–J). HLA-DR and Ki67 expression was infrequent in vRNA+ cells (2–15%) (Figure S3K,L). ICOS and PD-1 expression was more heterogeneous, with frequencies reaching a much higher level in some participants (2–43%). Irrespective of their range, all these activation markers except for Ki67, appeared significantly enriched in active vRNA+ reservoir cells compared to the global CD4+ T cells (Figure 4J).
We next tested if the magnitude of the inducible reservoir correlated with specific spontaneous vRNA+ reservoir clusters (Figure 4K). Several correlations were observed at varying degrees. The connections appeared particularly strong and significant with C3, C6, and C7, which corresponded to the enriched CCR6/CCR4+ vRNA+ active reservoirs. Because inducible reservoirs could not be reliably phenotyped, we could not relate active reservoirs to inducible reservoir subsets.
These multivariate and univariate analyses provide complementary portraits of peripheral blood active vRNA+ cell phenotypes. These cells tend to be memory CD4+ T cells, particularly TCM. Compared to other memory CD4+ T cells, they are polarized, with a consistent enrichment for CCR6, a marker associated with Th17 and Th22 differentiation, and are more frequently activated than the global CD4+ T cell population.
Spontaneously active reservoirs are associated with HIV-specific CD4+ and CD8+ T cell responses.
We next tested whether infected cells spontaneously expressing vRNA+ could fuel adaptive cellular immunity against HIV during ART. We used a TCR-dependent activation induced marker (AIM) assay that broadly identifies antigen-specific T cells29,50,51 (Table S3). We complemented these data with functional profiling by intracellular cytokine staining (ICS) (flow cytometry panels: Table S4). The AIM assay was previous shown to allow a broader capture of antigenic responses than standard ICS, even in the context of CD4+ T cell dysfunction29,50.
The AIM assay involved a 15-h incubation of autologous PBMCs with an overlapping peptide pool spanning the coding sequences of either Gag, Pol, Env, or Nef. In CD4+ T cells, specificity for these peptides was inferred by the upregulation of CD69, CD40L, 4–1BB, and OX-40 upon stimulation, compared to unstimulated controls, whereas CD69 and 4–1BB co-expression was used for CD8+ T cells. We used an AND/OR Boolean combination gating to assess the total frequencies of antigen-specific CD4+ and CD8+ T cells50,51,52 (Figure S4A). Cells co-expressing at least one pair of AIM were deemed HIV-specific. The significant increases compared to unstimulated conditions confirmed the assay’s specificity (Figure S4B).
Effector CD4+ and CD8+ T cell functions were measured by a 6-h ICS using the same stimulation conditions as for the AIM assays. We focused on IFN-γ, IL-2 and TNF expression. We also defined total cytokine+ CD4+ and CD8+ T cell responses by an OR Boolean gating strategy (Figure S4C). Most participants showed cytokine+ CD4+ and CD8+ T cell responses, although cytokine+ CD8+ T responses were smaller and more frequently undetectable (Figure S4D).
Gag, Pol, Env, and Nef responses were all readily detectable (Figure 5A). We used the sum of the net responses to each peptide pool to quantify “total” HIV responses. We next correlated total HIV, Gag, Pol, Env, and Nef responses to cells harboring integrated HIV DNA, PMA/ionomycin-inducible, and spontaneous reservoirs (Figure 5B). There was no significant correlation between AIM+ CD4+ and CD8+ T cell responses and integrated DNA (Figure 5B,C). In contrast, inducible reservoirs were strongly associated with total HIV-specific CD4+ T cell responses defined by AIM (Figure 5B,D). Similar association existed when considering Gag, Pol, Env and Nef-specific CD4+ T cells. A strong correlation was also found between the magnitude of the spontaneous reservoir and total HIV-specific CD4+ T cell responses (Figure 5B,E). The correlation with Nef was particularly strong (r =0.591), that with Gag (r =0.375) was the weakest. The association between active reservoir cells and total HIV-specific CD8+ T cell responses was weaker than with CD4, yet there was a trend. Individual peptide analyses revealed a significant correlation of the active reservoir cells with Env-specific CD8+ T cell responses (Figure 5B,F, r =0.522). Correlations with Gag, Pol or Nef-specific CD8+ T cell responses were weaker and did not reach significance.
Figure 5: Associations between spontaneously active reservoirs and HIV-specific CD4+ and CD8+ T cell responses.

(A) Net magnitude of HIV-specific CD4+ and CD8+ T cells by AIM assay. (B) Heat map reporting the associations between AIM+ CD4+ and CD8+ T responses and integrated DNA, inducible and spontaneous reservoirs. (C-E) Correlations between net AIM+ CD4+ responses and (C) integrated DNA, (D) inducible reservoirs, (E) spontaneous reservoirs. (F) Correlation between net AIM+ CD8+ T cell responses and spontaneous reservoirs. (G) Net magnitude of HIV-specific CD4+ and CD8+ T cells defined by the ICS. (H) Heat map reporting the associations between cytokine+ CD4+ and CD8+ T responses and integrated DNA, inducible and spontaneous reservoirs. (A,B,G,H) Peptide pools used to stimulate PMBCs are indicated. “HIV” responses were inferred by the sum of Gag, Pol, Env, and Nef net responses. In A and G, net magnitudes after background subtraction are shown. The bars indicate the median, and the error bars illustrate the interquartile range. The results from a Friedman test are shown above. In B and H, R and p (Spearman) values are shown. p <0.05, ** p <0.01, *** p <0.001. N=16 (1 participant had <5 vRNA+ cells, therefore could not be phenotyped).
Cytokine+ Gag-specific responses were also slightly higher for CD4+ and CD8+ T cell responses (Figure 5G). The correlation between reservoir metrics and ICS was weaker than with AIM (Figure 5H), possibly due to the lower sensitivity of functional assays. Total HIV-specific CD4+ T cells correlated with inducible reservoirs, and a strong trend was observed with active reservoirs. For both inducible and active reservoirs, strong correlations were observed with Env-specific cytokine+ CD4+ T cells (Figure 5H). The other correlations were weaker and only Gag-specific cytokine+ CD4+ T cells, for inducible reservoirs, reached significance. No correlation was observed with CD8+ T cell effector functions (Figure 5H). These data suggest that active reservoirs and HIV-specific immune responses, particularly Thelper responses, are connected.
A subset of active reservoirs displays stronger links to HIV-specific CD4+ and CD8+ T cell responses.
Next, we performed an unsupervised analysis to characterize HIV-specific CD4+ T cells. To account for the inherent phenotypic diversity of circulating CD4+ T cells and avoid a priori defined marker combinations, we performed an unsupervised analysis of the high-dimensional flow cytometric phenotyping data including total (Gag+Pol+Env+Nef) HIV-specific CD4+ T cells. We used chemokine receptors, activation markers and a key immune checkpoint (CXCR5; CXCR3; CCR6; CD38 and HLA-DR; and PD-1). The various clusters were represented using UMAP, and clusters were identified by Phenograph. 15 clusters were identified (Figure 6A) based on distinct profiles of relative marker expression (Figure S5AB). Each of these clusters represented 3–12% of total responses, with the largest C2 and C4 clusters representing no more than 13% of the total population examined (Figure 6B). To simplify the analysis, we grouped these clusters into 6 “superclusters” defined by the expression of chemokine receptors (Figure 6AB, S5B). The superclusters, ranked by decreasing frequencies, were characterized by the expression of i) CCR6; ii) none of the tested chemokine receptor; iii) CXCR3; iv) CXCR3 and CCR6, v) CXCR5 and vi) CXCR5 and CXCR3. Based on previous studies53,54, these superclusters would correspond respectively to i) TH17, ii) unpolarized cells, iii) TH1, iv) non-conventional TH1 (TH1*), v) cTfh and vi) a TH1 subset of cTfh. The frequencies of HIV-specific superclusters in total CD4+ T cells showed great variability among participants (Figure S5C), indicating that HIV-specific CD4+ T responses can adopt very different profiles during ART.
Figure 6: A subset of active reservoirs display strong links to HIV-specific CD4+ and CD8+ T cell responses.

(A) Global Uniform manifold approximation and projection (UMAP) for dimension reduction map of HIV-specific AIM+ CD4 T cells. 15 clusters were identified and regrouped in 6 superclusters. (B) Median proportions of each cluster, regrouped by superclusters. (C-D) Heat map showing correlations between (C) vRNA+ cluster frequencies and net magnitudes of AIM+ CD4+, regrouped by superclusters, or (D) vRNA+ cluster frequencies and net magnitudes of AIM+ CD8+ T cells. P values from Spearman test are shown, with significance underneath. * : p <0.05, ** p <0.01, *** p <0.001. (E-F) Correlations between the inducible or spontaneous vRNA+ reservoir and the magnitude of cells in (E) AIM+ HIV-specific CCR6+ CD4+ T supercluster, (F) AIM+ HIV-specific CXCR3+ CD4+ T supercluster. (G-H) Correlations between the inducible or spontaneous p24+ reservoir and the magnitude of cells in (G) total AIM+ HIV-specific CD4+, and (H) total AIM+ HIV-specific CD8+ T cells. Spearman tests were performed. R and p values are shown. (IJ) Histogram comparing median HIV-specific AIM+ (I) CD4 and (J) CD8 T responses in people where p24+ cells were detectable in peripheral blood (n=5) or were not (n=11). Mann-Whitney tests are shown above. Error bars indicate the interquartile range. N=16 (1 participant had <5 vRNA+ cells, therefore could not be phenotyped).
As our results showed relationships between active reservoirs and immune responses, we next examined how associations varied between types of viral reservoirs and subpopulations of HIV-specific AIM+ CD4+ and CD8+ T cells. Our first layer of analysis focused on total HIV-specific CD4+ T superclusters (Figure 6C). We found a strong correlation between the magnitude of both inducible or spontaneous vRNA+ cells and total AIM+ CD4+ T cell responses (Figure 6C). For the spontaneous reservoirs, correlations with CCR6+ AIM+ CD4+ T cell responses was particularly strong (Figure 6C,E), and reached a trend with CXCR3+ AIM+ CD4+ T cell responses (Figure 6C,F). Inducible vRNA+ cells showed significant correlations with total and PD1+ AIM+ CD8+ T responses (Figure 6D). The correlations between spontaneous vRNA+ cells and total and PD1+ AIM+ CD8+ T responses showed trends. Associations with HLA-DR+ and CD38+ responses were weaker.
We observed a significant correlation between active p24+ reservoirs and total AIM+ specific CD4+ T cells (Figure 6C,G). The r value (r =0.720) was particularly strong for the CXCR3+ supercluster (Figure 6C). We also noted strong correlations of total and activated AIM+ specific CD8 T cells with active p24+ reservoirs (Figure 6D,H). Correlations were lost with PMA/ionomycin-induced p24+ reservoirs, indicating that, critically, only spontaneously p24-expressing cells can shape HIV-specific CD4+ and CD8+ T cell responses. We divided our cohort based on whether p24+ could or could not be detected among vRNA+ cells (Figure 2F). HIV-specific CD4+ and CD8+ T cell responses were significantly higher in the p24+ group (Figure 6I,J). There were marked differences between the two groups for CXCR3+ and CXCR3+ CXCR5+ superclusters, suggesting that protein expression is a prerequisite for their maintenance.
Together, these results demonstrate important relationships between spontaneous vRNA+ and – especially - p24-expressing viral reservoirs and magnitude and function of HIV-specific CD4+ and CD8+ T cell responses.
DISCUSSION
In this study, we characterized HIV reservoirs spontaneously expressing viral transcripts, as detected at the single-cell level by RNAflow-FISH. We identified these active reservoir cells in most PWH investigated, only 3-fold lower than the PMA/ionomycin-inducible reservoirs. We phenotyped these active reservoirs without the confounding impact of LRAs. Integrated analyses demonstrated the heterogeneous nature of active vRNA+ reservoirs. Certain features were enriched, such as high expression of CCR6, a marker of Th17 and Th22 cells. We found associations between active viral reservoirs and HIV-specific CD4+ and CD8+ T cell responses during suppressive ART. CCR6+ HIV-specific CD4+ T cells seemed particularly connected to various active reservoirs. HIV-specific CD4+ and CD8+ T cell responses were higher in the few participants in which active p24+ reservoir cells could be detected, suggesting that maintaining those immune responses requires spontaneous viral translation.
We and others previously showed how the RNAflow-FISH assay could identify reactivated viral reservoirs at the single-cell level8,9,55. By adapting a multigene HIV-specific probeset design, we detected viral reservoirs with great sensitivity and resolution without ex vivo latency reversal. Spontaneous and inducible vRNA+ reservoirs correlated, but were found at lower frequencies. One possible explanation for this relationship is that they represent two facets of the same cell: latency reversal could exacerbate pre-existing, very low-yield transcriptional activity possibly missed by our experimental technique. These cells may be poised for higher transcriptional activity upon induction, as suggested by the higher MFI for the vRNA signal in induced vs spontaneously active cells. The low transcriptional activity may be due to i) suboptimal Tat-mediated transactivation during latency56; ii) the shortness of the abortive transcripts, consistent with data previously obtained by digital droplet PCR10,41, that may make them less sensitive to detection by a branched DNA amplification technique; and iii) the requirement for staining with two HIV RNA probesets can reduce sensitivity of detection. Alternatively, a portion of the PMA/ionomycin inducible reservoirs may have been in a deeper state of latency and fully silent prior to stimulation57–59, therefore only revealed after pharmacological reactivation. Spontaneous viral gene expression was also characterized by a shorter transcription and rare translation of Gag. These observations are consistent with previous reports and suggest transcriptional10 and post-transcription blocks60 to viral gene expression.
Another notable finding of our study is that active reservoirs are not confined to any specific CD4+ T subset. This finding is consistent with recent reports correlating viral transcriptomic and genomic properties60,61. All CD4+ clusters we analyzed were populated to some degree with vRNA+ reservoirs. Yet, compared to total peripheral CD4+ T cells, active reservoir cells profiles were: i) rarely naïve and mostly TCM; ii) enriched in CCR6, suggesting a preferential Th17 and Th22 polarization, iii) enriched in activation and exhaustion markers such as HLA-DR, ICOS, and PD-1, and iv) enriched with cell proliferation marker Ki67. Although modest, enrichment in activation/exhaustion and proliferation markers is consistent with homeostatic62–64 and antigen-driven proliferation reported during ART34,65. Although we observed some enrichment of activation markers in active reservoirs, the vast majority of spontaneous reservoir cells were still found in cells that did not express activation markers, indicating that T cell activation is not a prerequisite for spontaneous viral gene expression. The enrichment of HIV-infected cells we have observed in TCM, CCR6, and to a lesser extent, CXCR5 and CXCR3 are consistent with previous reports using univariate analyses62,66–71. These enrichments may reflect preferential replication of HIV-1 in specific anatomic compartments before ART initiation, such as the intestinal mucosa (hence the enrichment in the gut homing and Th17 marker CCR6)8,72,73 and germinal centers of lymph nodes (hence the enrichment in the Tfh marker CXCR5)8,72,73. Some tissues and microenvironments (e.g. gastrointestinal tract) may also be more permissive to viral transcription74,75, and this would be mirrored in the circulation when these cells egress from tissues into the blood. We cannot exclude that residual replication occurs in tissues where penetration of antivirals may be suboptimal. However, to date, no direct evidence has shown that ongoing viral replication contributes to viral reservoir persistence in PWH receiving suppressive ART33,35,36.
An association was previously reported between cytotoxic Nef-specific CD8+ T cell responses and HIV DNA and RNA76. Using single-cell approaches, we now find multiple strong positive correlations between active reservoirs and the magnitude of Pol, Env, Nef and to a lesser extent, Gag-specific CD4+ and CD8+ T cell responses, indicating a broader relationship than initially anticipated. The correlations were always stronger with HIV RNA than with HIV DNA, suggesting that some gene expression is necessary to reveal these relationships. Detailed analyses showed that the strength of these correlations varied among the pairs of viral and immune clusters assessed, suggesting that viral reservoirs do not all influence anti-HIV immune responses equally. HIV-specific CCR6+ CD4+ clusters were particularly well correlated with active reservoir cells. While this correlation suggests tissue-specific interface between active reservoirs and HIV-specific T cells, putatively the gut, further investigations will be necessary to deeply decipher connections between reservoirs and HIV-specific immune responses.
The positivity of the relationship suggests that spontaneous reservoirs are sustaining HIV-specific CD4 and CD8 T cells rather than HIV-specific cells controlling the reservoirs. In this later case, a negative association would have been expected. However, spontaneously active reservoirs that still persist after >3 post-ART initiation and reservoir selection by the immune system are likely inherently more resistant to cell death. Consistent with this, recent studies showed that persistent reservoirs adopt a pro-survival gene expression profile60,77,78. How viral transcription may drive immune responses remains a key question. The magnitude of HIV-specific CD4+ and CD8+ T cell responses were lower in participants with no spontaneous p24-expressing active reservoir. In contrast, these responses were robust in participants in which p24-expressing vRNA+ reservoir cells could be detected. These findings suggest that viral protein expression, although rare, is the driving force keeping HIV reservoirs and HIV-specific immune responses closely related. It will be important to determine whether spontaneous expression of Gag and other HIV proteins is more frequent in specific tissues, possibly the gut due to proinflammatory microenvironment more favorable for provirus activation or in anatomic compartments harboring a weaker immune surveillance79. Alternatively, sporadic Gag expression in peripheral blood, perhaps during transient challenges such as acute illnesses could possibly contribute to the antigen-stimulated maintenance of HIV-specific immunity. If this is the case, active vRNA+ reservoir cells in the circulation may mirror tissue reservoir cells that are prone to produce proteins and subsequently cognate peptides presented by MHC molecules to T cells.
Inducible proviruses can bear not only minor9,17, but extensive defects predicted to prevent viral replication. We showed here that active reservoirs are mostly defective as well. Yet, reservoirs expressing transcripts from these defective proviruses may still allow some translation37,38,80, depending on where the defect is located. Our work supports that such defective proviruses can also maintain HIV-specific CD4+ and CD8+ T responses in a chronic state of expansion, activation, and exhaustion during ART.
Our study has some limitations. Only Caucasian males were included mostly due to the epidemiology of the PWH population in Montreal and, to some extent, the more frequent difficulties for peripheral vein access to perform leukaphereses in females. It will be important to conduct such studies in women and in other ethnicities, as immune responses may vary with sex and genetic background. ART was initiated in most participant’s years after HIV acquisition; investigations of PWH who initiated therapy early would provide valuable complementary information. While the assay used allows high-parameter profiling of active reservoir cells, our conservative detection limit (about 5 vRNA copies/cell)8,43 missed the lowest levels of gene transcription. Other approaches indeed indicated even higher magnitudes of spontaneous reservoirs81. Studies of gut and other lymphoid tissues will be important to gain a deeper understanding of the immunovirological dynamics involved. The relatively small size of the cohort studied did not allow us to reliably rank the strength of the associations between active reservoir clusters and HIV-specific T cell superclusters.
Finally, our study may have notable implications for HIV cure strategies. Approaches considered include Env-specific broadly neutralizing antibodies. The fraction of active reservoirs that are competent for Env expression will impact the potential effectiveness of such therapies to eliminate these cells. While the replication-competent HIV reservoir is the primary target of HIV cure, our data highlight the pathophysiologic relevance of other fractions of the HIV reservoirs, likely contributing to the deleterious effect of immune activation. Research efforts should also consider the putatively negative impact of spontaneous reservoirs in the design of interventions aiming at clinical benefit for PWH on suppressive ART.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and fulfilled by the lead contact, Daniel E. Kaufmann (daniel.kaufmann@chuv.ch).
Materials availability
All unique reagents generated during this study are available from the Lead contact upon a material transfer agreement (MTA).
Data and code availability
The published article includes all datasets generated and analyzed for this study. We developed R codes scripted to perform unsupervised analysis of B and T cells from SARS-CoV-2 naïve and previously infected individuals. All original codes have been deposited at Github and are publicly available as of publication. URL link is listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact Author upon request (daniel.kaufmann@chuv.ch).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| UCHT1 (BUV395) [Human anti-CD3] | BD Biosciences | Cat#563546; Lot:9058566; RRID:AB_2744387 |
| UCHT1 (BUV496) [Human anti-CD3] | BD Biosciences | Cat#612941; Lot:1022424; RRID:AB_2870222 |
| L200 (BV711) [Human anti-CD4] | BD Biosciences | Cat#563913; Lot:03000025; RRID:AB_2738484 |
| SK3 (BB630) [Human anti-CD4] | BD Biosciences | Cat#624294 CUSTOM; Lot:0289566 |
| RPA-T8 (BV570) [Human anti-CD8] | Biolegend | Cat#301037; Lot:B281322; RRID:AB_10933259 |
| M5E2 (BUV805) [Human anti-CD14] | BD Biosciences | Cat#612902; Lot:0262150; RRID:AB_2870189 |
| M5E2 (BV480) [Human anti-CD14] | BD Biosciences | Cat#746304; Lot : 9133961; RRID:AB_2743629 |
| 3G8 (BV650) [Human anti-CD16] | Biolegend | Cat#302042; Lot:B323847; RRID:AB_2563801 |
| HIB19 (APC-eFluor780) [Human anti-CD19] | Thermo Fisher Scientific | Cat#47-0199; Lot:2145095; RRID:AB_1582231 |
| HIB19 (BV480) [Human anti-CD19] | BD Biosciences | Cat#746457; Lot:1021649; RRID:AB_2743759 |
| HI100 (PerCP Cy5.5) [Human anti-CD45RA] | BD Biosciences | Cat#563429; Lot:8332746; RRID:AB_2738199 |
| NCAM16.2 (BUV737) [Human anti-CD56] | BD Biosciences | Cat#564448; Lot:8288818; RRID:AB_2744432 |
| FN50 (PerCP-eFluor710) [Human anti-CD69] | Thermo Fisher Scientific | Cat#46-0699-42; Lot:1920361; RRID:AB_2573694 |
| FN50 (BV650) [Human anti-CD69] | Biolegend | Cat# 310934; Lot:B303462; RRID:AB_2563158 |
| H4A3 (BV786) [Human anti-CD107A] | BD Biosciences | Cat#563869; Lot:8144866; RRID:AB_2738458 |
| ACT35 (APC) [Human anti-CD134 (OX40)] | BD Biosciences | Cat#563473; Lot:1015537; RRID:AB_2738230 |
| 4B4-1 (PE-Dazzle 594) [Human anti-CD137 (4-1BB)] | Biolegend | Cat# 309826; Lot:B253152; RRID:AB_2566260 |
| TRAP1 (BV421) [Human anti-CD154 (CD40L)] | BD Biosciences | Cat#563886; Lot:9037850; RRID:AB_2738466 |
| TRAP1 (PE) [Human anti-CD154 (CD40L)] | BD Biosciences | Cat#555700; Lot:7086896; RRID:AB_396050 |
| J25D4 (BV421) [Human anti-CD185 (CXCR5)] | Biolegend | Cat# 356920; Lot:B325837; RRID:AB_2562303 |
| B27 (PECy7) [Human anti-IFN-γ] | BD Biosciences | Cat#557643; Lot:8256597; RRID:AB_396760 |
| MQ1-17H12 (PE-Dazzle594) [Human anti-IL-2] | Biolegend | Cat#500344; Lot:B2261476; RRID:AB_2564091 |
| JES3-9D7 (PE) [Human anti-IL-10] | BD Biosciences | Cat#554498; Lot:8198773; RRID:AB_395434 |
| eBio64CAP17 (eFluor660) [Human anti-IL-17A] | Thermo Fisher Scientific | Cat#50-7179-42; Lot:2151998; RRID:AB_11149126 |
| Mab11 (Alexa Fluor 488) [Human anti-TNF-α] | Biolegend | Cat#502915; Lot:B285221; RRID:AB_493121 |
| Mab11 (APC) [Human anti-TNF-α] | BD Biosciences | Cat#562084 Lot: 0350627 RRID:AB_10893226 |
| LIVE/DEAD Fixable dead cell | Thermo Fisher Scientific | L34960 |
| Biological samples | ||
| SARS-CoV-2 naïve donor blood samples | N/A | |
| SARS-CoV-2 prior infection donor blood samples | N/A | |
| Chemicals, peptides, and recombinant proteins | ||
| PepMix™ SARS-CoV-2 (Spike Glycoprotein) | JPT | Cat#PM-WCPV-S-1 |
| Staphylococcal Enterotoxin B (SEB) | Toxin technology | Cat#BT202 |
| Software and algorithms | ||
| Flow Jo v10.8.0 | Flow Jo | https://www.flowjo.com |
| GraphPad Prism v8.4.1 | GraphPad | https://www.graphpad.com |
| R studio v4.1.0 | R studio | https://rstudio.com |
| R codes scripted | Github | https://github.com/otastet/Nayrac_et_al |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics Statement.
All work was conducted following the Declaration of Helsinki regarding informed consent and approval by an appropriate institutional board. Blood samples were obtained from donors who consented to participate in this research project at CHUM (CE13.019).
Experimental Model and Subject Details.
Participants and Samples.
Leukaphereses were obtained from study participants at the McGill University Health Centre, Montreal, Quebec, Canada, and at Centre Hospitalier de l’Université de Montréal (CHUM), Quebec, Canada. The study was approved by the respective IRBs, and written informed consent was obtained from all participants before enrolment. Uninfected donors (UD) are free of HIV-1 infection. Treated subjects (ART) were on antiretrovirals with controlled viremia (<40 vRNA copies/mL). Participants’ characteristics are summarized in Table S1. PBMCs were isolated by the Ficoll density gradient method and stored in liquid nitrogen until use.
Method Details
Total and integrated DNA measures.
The quantifications of total and integrated HIV-1 DNA were determined as previously described82.
CD4+ T cell preparation for HIV reservoir profiling
Frozen PBMCs were thawed in cold heat-inactivated Fetal Calf Serum (FCS; Seradigm) before CD4+ T-cells isolation. CD4+ T-cells were isolated by negative magnetic bead selection (StemCell). Purified CD4+ T cells were resuspended at 2×106/mL in RPMI (Gibco by Life Technologies) supplemented with penicillin/streptomycin (Gibco by Life Technologies), 10% heat-inactivated FCS, and ARV (Maraviroc [10μM] + Raltegravir [0.2μM] + Tenofovir [5μM]) + Emtricitabine [10μM] and seeded into 24-well plates (all obtained through the NIH AIDS Reagent Program). After a rest of 2h at 37°C, 5% CO2, the cells were either left unstimulated or stimulated with PMA/ionomycin (162 nM PMA, 705 nM Ionomycin, Sigma) for 15-h. Alternatively, cells were either left unstimulated or stimulated with 30nM panobinostat (Selleck Chem) complemented with 25nM ingenol-3-angelate (Sigma). 10–15×106 purified CD4+ T-cells were used per condition.
HIV-1 RNAflow-FISH assay.
The HIV-1 RNAflow-FISH assay was performed as previously described and per the manufacturer’s instructions8,9,83,84. All buffers and fixation reagents were provided with the kit, except flow cytometry staining (1% FCS/PBS). Briefly, negatively purified CD4+ T cells were harvested after stimulation and stained first with Fixable Viability Dye (20 min, 4°C, Fixable LiveDead, eBioscience), FcR blocked, followed by a mix containing a brilliant stain buffer (BD Biosciences) and the surface markers for memory (CD45RA) and gut homing (integrin β7, CD103) phenotype, activation markers (HLA-DR, ICOS) and inhibitory checkpoint (PD-1) as well as for CD4+ T-cells detection (CD3 and CD4) and CD8/NK/B cells and macrophages exclusions (CD8, CD56, CD14, CD19, CD16) (30 min, 4°C). Anti-CXCR5, CCR6, CCR4, CXCR3, CCR7 and CD27 were added at 37°C 15 min before stimulation to stain chemokine receptors. Samples were fixed, permeabilized, and labeled intracellularly for the activation marker Ki67 and the structural HIV-1 p24 protein with the anti-p24 clone KC57 antibody (30 min RT followed by 30 min 4°C, Beckman Coulter). HIV-1 RNA probing was performed using the PrimeFlow RNA Assay (ThermoFisher). HIV-1 RNA was labeled using HIV-1 gagRNA (20 pairs of “ZZ” probes), HIV-1 exonRNA (21 pairs of “ZZ” probes), and HIV-1 polRNA (6 pairs of “ZZ” probes) probe sets, all designed based on a consensus B HIV sequence. The probes were diluted 1:5 in diluent and hybridized to the target mRNAs for 2 hrs at 40°C. Samples were washed to remove excess probes and stored overnight in the presence of RNAsin. Signal amplification was achieved by sequential 1.5 hr at 40°C incubations with the pre-amplification and amplification mix. Amplified mRNAs were labeled with fluorescently tagged probes for 1h at 40°C. The complete list of antibodies used is presented in Table S2 for the panel. Samples were acquired on a FACSymphony™ (BD Biosciences) and analyzed using FlowJo (BD, V10.8.0). Unspecific binding of the fluorescent-labeled branched probe in the multiplex kit can lead to a low level of false-positive background noise, which, if present, is detected across all the four channels corresponding to the types of labeled probes (AF488, AF594, AF647, AF750). To decrease background noise, we thus left the AF594 channel vacant and excluded false-positive events based on fluorescence in this channel before further gating. Gates were set on the HIV-uninfected donor control or unstimulated control where appropriate (See gating strategy, Figure S1). Because HIV infection can downregulate CD4, and because RNAflow-FISH was performed on negatively purified CD4+ T cells, no CD4 gating was applied during the analysis. To calculate the frequency of vRNA+ cells per 106, we directly used as the denominator the counts of cells after the dump exclusion gating.
Limiting Dilution Assay.
CD4+ T cells were isolated from PBMC by negative selection using magnetic beads (StemCell). After 18h hours of resting, the cells were distributed in limiting dilutions in a 96-well plate, with 11 replicates for each of the following dilutions: 100000, 50000, 16667, 5556 cells per well. The plate was spun at 300xg for 5 min, and the cell pellets were resuspended in Lysis/Binding solution (Magmax 96 Total RNA Isolation Kit, Life Technologies). Ca-RNA was extracted in plate using Magmax 96 Total RNA Isolation Kit (Life Technologies) following manufacturer’s instructions. A nested RT-qPCR was performed to amplify LTRgag caRNA for all the replicates of each dilution. A 1-step RT and pre-amplification step was carried out by adding 9uL of extracted RNA (corresponding to 18000, 9000, 3000 and 1000 cell-equivalent depending on the dilution) to a mix of 6.25uL of Taq-1-Path master mix (Applied Biosystems) and 800 nM of LTRgag specific primers82. The PCR cycles were as follow: 15 min at 53°C, 2 min at 95°C, and 18 cycles of 15 s at 95°C and 2 min at 60°C. The pre-amplified product was used to perform a real-time PCR as previously described82. Positive wells at each dilution were counted and the maximum likelihood method was used to calculate the frequency of cells with LTRgag RNA (http://bioinf.wehi.edu.au/software/elda).
Microscopy.
CD4+ T cells from one uninfected and one ART-treated PWH were isolated, rested 2h and reactivated with LRA (30nM panobinostat (Selleck Chem) and 25nM ingenol-3-angelate (Sigma) for 15h. Cells were collected and stained with a viability dye (Fixable Live/Dead (eF780, eBioscience) and with antibodies against surface CD8, CD14 and CD19 (BV510). mRNA Flow FISH was performed as described above and sorted with a BD FACS Aria. Single, CD8/14/19- T cells were sorted into two populations based on exonRNA AF647 and gagRNA AF488 staining (Figure S2H). Prior to microscopy analysis, nuclei were stained (DAPI, 1ng/ml, 2 min RT), directly loaded in ibidi μ-Slide VI 0.4 microscopy chambers and imaged using a Zeiss Axio Observer.Z1 inverted spinning disk confocal microscope coupled to an Evolve camera (EMCCD, 512×512, 16bit, 1.2x adapter) and ZEN blue software (version 2012). Images were acquired with an alpha Plan-Apochromat 100x/1.46 Oil DIC (UV) M27 objective. Excitation was performed with a 639nm, a 488nm and a 405nm solid state lasers for AF647, AF488 and DAPI respectively. Emission was collected through Chroma filters: DBP 527/54 + 645/60 for AF488 and AF647 and a DBP 460/30 + 590/30 for DAPI. Z-stacks were performed to image whole cells with a step size of 0.220μm. Final resolution is 0.133μm × 0.133μm × 0.220μm in xyz. Brightfield images were acquired with the same modalities but with a LED white light illumination, no filter and without Zstack. Fiji was used for all image analysis and facilitate counting. DAPI staining was used to define the nuclear compartment and the “Find Maxima” command was used to identify and count HIV RNA foci.
Single-cell near full-length PCR.
Unstimulated CD4+ T-cells from 3 ART-treated donors were stained in HIV-1 RNAflow-FISH assays using HIV-1 gagRNA, exonRNA, and polRNA probes. Single vRNA+ cells were sorted in 12-wells PCR strips containing 8μL of DirectPCR Lysis Reagent (Viagen Biotech) with 0.4mg/mL proteinase K. The PCR strips were subsequently incubated at 55°C for 1h for cell lysis followed by 15 min at 85°C to inactivate proteinase K. Single sorted cells were subjected to near full-length amplification using a modified FLIPS assay9,44. HIV-1 genomes were pre-amplified using Invitrogen Platinum SuperFi II MasterMix with 0.2μM of each primer44. 30μl of PCR mix was added directly to the lysed cells for a 25 cycles 3-steps PCR protocol as recommended by the manufacturer. The pre-amplified products were diluted 1:3 with Tris-HCl 0.5μM pH 8.0 and subjected to a nested PCR with 5μL of pre-amplified product, 2X of Platinum SuperFi II PCR Mix and 0.2μM of each primer in a 30μL final volume44. This second amplification consists in 30 cycles and follows the manufacturer’s instructions. The length of the sequences obtained were verified on a 0.8% agarose gel and the amplicons were individually barcoded for PacBio Sequel II sequencing (DNA Link, South Korea), The demultiplex barcodes analysis was powered by the Lima PacBio software v2.0.0. High-quality phased consensus sequences representing near full HIV-1 genome sequences with high fidelity and without reconstruction have been generated with the LAA PacBio algorithm v2.4.2. For each individual, sequences obtained were aligned using Multiple Alignment using Fast Fourier Transform (MAFFT) with strategy E-INS-i and Scoring matrix for nucleotide sequences of 1PAM/ k=2 (online https://mafft.cbrc.jp/alignment/server/ or with Geneious Prime (v2021.1.1) plugging). Trees were built with iqtree2 using Maximum-Likelihood tree GTR+I+G model, with 1000 bootstraps, and then visualized with Figtree (v1.4.4). Clonality was evaluated with diversity of sequences in Geneious Prime, and sequences with 0 nucleotide difference were considered clonal. Integrity was assessed using both HIVDatabase QCtool (https://www.hiv.lanl.gov/content/sequence/QC/index.html) and ProseqIT (https://psd.cancer.gov/tools/pvs_annot.php). Finally, Psi defects were confirmed manually by visualization in Geneious Prime of this portion of the sequence. Genebank accession numbers for single-cell sequences are provided in Table S5.
Activation-induced marker (AIM) assay.
The AIM assay was reported before29,30,50,51. PBMCs were thawed and rested for 3h in 96-well flat-bottom plates in RPMI 1640 supplemented with HEPES, penicillin and streptomycin and 10% FBS. 2×106 PBMCs and stimulated with Gag or Pol or Env or Nef peptide 15-mers pools (0.5 μg/ml per peptide) spanning the complete amino acid sequence of each HIV protein (JPT) for 15h at 37 °C and 5% CO2. CD40-blocking antibody was added to prevent CD40L downregulation following activation. In addition, CXCR3, CCR6, and CXCR5 antibodies were added to the culture 15 min before stimulation. A DMSO-treated condition served as a negative control and Staphylococcus enterotoxin B SEB-treated condition (0.5 μg/ml) as a positive control. Cells were stained for viability dye for 20 min at 4 °C, FcR receptors were blocked using an FcR block antibody, and then surface markers (CD3, CD4, CD8, CD45RA, CD69, OX40, 41BB, CD40L, PD1, HLA-DR) (30 min, 4 °C). Abs used are listed in the Table S3. Cells were fixed using 2% paraformaldehyde for 15min at 4 °C before acquisition on Symphony cytometer (BD Biosciences). Analyses were performed using FlowJo v10.8.0 software.
Intracellular Cytokine Staining (ICS).
The previously described ICS assay was adapted to study HIV-specific T cells29,51,85. PBMCs were thawed and rested for 2-h in RPMI 1640 medium supplemented with 10% FBS, Penicillin-Streptomycin (Thermo Fisher Scientific, Waltham, MA), and HEPES (Thermo Fisher Scientific, Waltham, MA). 1.7×106 PBMCs were stimulated with Gag or Pol or Env or Nef peptide pools (0.5 μg/ml per peptide; JPT) for 15h at 37 °C and 5% CO2. Cell stimulation was carried out for 6h at 5% CO2 at 37 °C. Brefeldin A and monensin (BD Biosciences, San Jose, CA) was added 1h after stimulation. DMSO-treated cells served as a negative control and SEB as a positive control. Cells were stained for Aquavivid viability marker (Thermo Fisher Scientific, Waltham, MA) for 20 min at 4 °C, then surface markers (CD4, CD3, CD8, CD14, CD19; 30 min, 4 °C), followed by intracellular Detection of cytokines (IFNγ, Il-2, and TNF-α) using the IC Fixation/Permeabilization kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol before acquisition on a LSRII flow cytometer (BD Biosciences). Analysis was performed using FlowJo v10.8.0 software. Abs used are listed in Table S4.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis.
Symbols represent biologically independent samples from uninfected and HIV-infected under suppressive ART. Wilcoxon, Mann-Whitney, and Friedman with Dunn’s post-test were generated using GraphPad Prism version 8.4.3 (GraphPad, San Diego, CA). P values <0.05 were considered significant. P values are indicated for each comparison assessed. Fold differences were calculated per participant, then median of these fold differences was calculated. Median values were used to generate donut charts. Each median value was normalized to obtain a total of 100%. For descriptive correlations, Spearman’s R correlation coefficient was applied. For graphical representation on a log scale (but not for statistical tests), null values were arbitrarily set at the minimum values for each assay.
Software scripts and visualization.
Graphics and pie charts were generated using GraphPad PRISM version 8.4.1 and ggplot2 (v3.3.3) in R (v4.1.0). Heat maps were generated in GraphPad PRISM version 8.4.1. Uniform manifold approximation and projection (UMAP) was performed using package M3C (v1.14.0) on gated FCS files loaded through the flowCore package (v2.4.0). For reservoir phenotyping, all vRNA+ events were loaded along with 3000 downsampled autologous CD4+ T cells per participant, for a total of 1,418 vRNA+ cells + 51,000 autologous CD4+ T cells. For the AIM analysis, samples were downsampled to a comparable 300 cells per peptide (Gag, Pol, Env and Pol) pool tested, therefore 1200 peptide-specific cells per participant, and a grand total of 20,400 HIV-specific cells. This number was chosen to avoid biases due to larger responses in certain participants. Scaling and logicle transformation of the flow cytometry data were applied using the FlowSOM86 R package (v2.0.0). Clustering was achieved using Phenograph (v0.99.1) with the hyperparameter k (number of nearest neighbors) set to 150). R code scripted for this paper was adapted from https://github.com/otastet/Nayrac_et_al with the parameters described above. We obtained an initial 18 AIM+ clusters. After careful examination, we regrouped these clusters into 6 larger superclusters based on similar chemokine receptor expression. For vRNA+ and CD4+ T cell phenotyping, only participants with >5 events were analyzed.
Supplementary Material
Highlights.
Spontaneously active HIV RNA+ and protein+/− reservoirs exist in people with HIV on ART
These are enriched in central memory, CCR6 and activation marker expressing cells
Proviruses integrated in these active reservoir cells are mostly defective
Spontaneously active reservoirs correlate with HIV-specific CD4+ and CD8+ T cells
ACKNOWLEDGMENTS
We thank Josée Girouard, Angie Massicotte (CUSM), and all study participants for their invaluable role in this project; Dr. Dominique Gauchat, Dr. Gaël Dulude, Philippe St-Onge, and the CRCHUM Flow Cytometry Platform; Dr. Olfa Debbeche and the CRCHUM BSL3 platform; Aurélie Cleret-Buhot and the Microscopy platform. The following reagents were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID: Maraviroc (ARP-11580), Raltegravir (ARP-11680), Tenofovir (ARP-10199), and Emtricitabine (ARP-10071).
FUNDING
This study was supported by the Canadian Institutes for Health Research (CIHR grant #152977), the US National Institutes of Health (R01AI143411 to J.D.E, N.C and D.E.K) and the Réseau Fonds de la recherche Québec-Santé (FRQ-S) SIDA & Maladies infectieuses and thérapies cellulaires. D.E.K is a FRQS Merit Research Scholar. N.C is a FRQ-S Research Scholar awardee. G.S is supported by a FRQS doctorate fellowship and by a scholarship from the Department of Microbiology, Infectious Disease and Immunology of the University of Montreal. J.D.E is supported by the Oregon National Primate Research Center grant award P51OD011092.
Footnotes
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
REFERENCES
- 1.Davey RT Jr., Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, et al. (1999). HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proceedings of the National Academy of Sciences of the United States of America 96, 15109–15114. 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter AE, O’Doherty U, and Kaufmann DE (2018). Beyond the replication-competent HIV reservoir: transcription and translation-competent reservoirs. Retrovirology 15, 18. 10.1186/s12977-018-0392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaebler C, Lorenzi JCC, Oliveira TY, Nogueira L, Ramos V, Lu CL, Pai JA, Mendoza P, Jankovic M, Caskey M, and Nussenzweig MC (2019). Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. The Journal of experimental medicine 216, 2253–2264. 10.1084/jem.20190896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, et al. (2019). A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125. 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, Richman DD, O’Doherty U, Palmer S, Hecht FM, et al. (2015). A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine 2, 874–883. 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasternak AO, and Berkhout B (2018). What do we measure when we measure cell-associated HIV RNA. Retrovirology 15, 13. 10.1186/s12977-018-0397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, Leyre L, Routy JP, Kaufmann DE, and Chomont N (2019). Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS pathogens 15, e1007619. 10.1371/journal.ppat.1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, Massanella M, Brassard N, Alsahafi N, Delgado GG, et al. (2016). Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals. Cell host & microbe 20, 368–380. 10.1016/j.chom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sannier G, Dube M, Dufour C, Richard C, Brassard N, Delgado GG, Pagliuzza A, Baxter AE, Niessl J, Brunet-Ratnasingham E, et al. (2021). Combined single-cell transcriptional, translational, and genomic profiling reveals HIV-1 reservoir diversity. Cell reports 36, 109643. 10.1016/j.celrep.2021.109643. [DOI] [PubMed] [Google Scholar]
- 10.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, and Wong JK (2018). HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Science translational medicine 10. 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohammadi P, di Iulio J, Munoz M, Martinez R, Bartha I, Cavassini M, Thorball C, Fellay J, Beerenwinkel N, Ciuffi A, and Telenti A (2014). Dynamics of HIV latency and reactivation in a primary CD4+ T cell model. PLoS pathogens 10, e1004156. 10.1371/journal.ppat.1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L Jr., Ingerman MJ, Witek J, Kedanis RJ, Natkin J, DeSimone J, and Pomerantz RJ (1999). Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. Jama 282, 1627–1632. 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 13.Gunthard HF, Havlir DV, Fiscus S, Zhang ZQ, Eron J, Mellors J, Gulick R, Frost SD, Brown AJ, Schleif W, et al. (2001). Residual human immunodeficiency virus (HIV) Type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years. The Journal of infectious diseases 183, 1318–1327. 10.1086/319864. [DOI] [PubMed] [Google Scholar]
- 14.Halvas EK, Joseph KW, Brandt LD, Guo S, Sobolewski MD, Jacobs JL, Tumiotto C, Bui JK, Cyktor JC, Keele BF, et al. (2020). HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. The Journal of clinical investigation 130, 5847–5857. 10.1172/JCI138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishizaka A, Sato H, Nakamura H, Koga M, Kikuchi T, Hosoya N, Koibuchi T, Nomoto A, Kawana-Tachikawa A, and Mizutani T (2016). Short Intracellular HIV-1 Transcripts as Biomarkers of Residual Immune Activation in Patients on Antiretroviral Therapy. Journal of virology 90, 5665–5676. 10.1128/JVI.03158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer M, Gunthard HF, Opravil M, Joos B, Huber W, Bisset LR, Ott P, Boni J, Weber R, and Cone RW (2000). Residual HIV-RNA levels persist for up to 2.5 years in peripheral blood mononuclear cells of patients on potent antiretroviral therapy. AIDS research and human retroviruses 16, 1135–1140. 10.1089/088922200414974. [DOI] [PubMed] [Google Scholar]
- 17.White JA, Wu F, Yasin S, Moskovljevic M, Varriale J, Dragoni F, Camilo Contreras A, Duan J, Zheng MY, Tadzong NF, et al. (2023). Clonally expanded HIV-1 proviruses with 5’-Leader defects can give rise to nonsuppressible residual viremia. The Journal of clinical investigation. 10.1172/JCI165245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferdin J, Goricar K, Dolzan V, Plemenitas A, Martin JN, Peterlin BM, Deeks SG, and Lenassi M (2018). Viral protein Nef is detected in plasma of half of HIV-infected adults with undetectable plasma HIV RNA. PloS one 13, e0191613. 10.1371/journal.pone.0191613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passaes C, Delagreverie HM, Avettand-Fenoel V, David A, Monceaux V, Essat A, Muller-Trutwin M, Duffy D, De Castro N, Wittkop L, et al. (2021). Ultrasensitive Detection of p24 in Plasma Samples from People with Primary and Chronic HIV-1 Infection. Journal of virology 95, e0001621. 10.1128/JVI.00016-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, Zuck P, Goh SL, Milush JM, Vohra P, Wong JK, Somsouk M, Yukl SA, Shacklett BL, Chomont N, et al. (2021). Gag p24 Is a Marker of Human Immunodeficiency Virus Expression in Tissues and Correlates With Immune Response. The Journal of infectious diseases 224, 1593–1598. 10.1093/infdis/jiab121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, et al. (2016). CD8(+) Lymphocytes Are Required for Maintaining Viral Suppression in SIV-Infected Macaques Treated with Short-Term Antiretroviral Therapy. Immunity 45, 656–668. 10.1016/j.immuni.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chevalier MF, Julg B, Pyo A, Flanders M, Ranasinghe S, Soghoian DZ, Kwon DS, Rychert J, Lian J, Muller MI, et al. (2011). HIV-1-specific interleukin-21+ CD4+ T cell responses contribute to durable viral control through the modulation of HIV-specific CD8+ T cell function. Journal of virology 85, 733–741. 10.1128/JVI.02030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray GE, Huang Y, Grunenberg N, Laher F, Roux S, Andersen-Nissen E, De Rosa SC, Flach B, Randhawa AK, Jensen R, et al. (2019). Immune correlates of the Thai RV144 HIV vaccine regimen in South Africa. Science translational medicine 11. 10.1126/scitranslmed.aax1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tebas P, Jadlowsky JK, Shaw PA, Tian L, Esparza E, Brennan AL, Kim S, Naing SY, Richardson MW, Vogel AN, et al. (2021). CCR5-edited CD4+ T cells augment HIV-specific immunity to enable post-rebound control of HIV replication. The Journal of clinical investigation 131. 10.1172/JCI144486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuyishime S, Haut LH, Kurupati RK, Billingsley JM, Carnathan D, Gangahara S, Styles TM, Xiang Z, Li Y, Zopfs M, et al. (2018). Correlates of Protection Against SIVmac251 Infection in Rhesus Macaques Immunized With Chimpanzee-Derived Adenovirus Vectors. EBioMedicine 31, 25–35. 10.1016/j.ebiom.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, et al. (2010). The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557. 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moysi E, Petrovas C, and Koup RA (2018). The role of follicular helper CD4 T cells in the development of HIV-1 specific broadly neutralizing antibody responses. Retrovirology 15, 54. 10.1186/s12977-018-0437-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trautmann L (2016). Kill: boosting HIV-specific immune responses. Current opinion in HIV and AIDS 11, 409–416. 10.1097/COH.0000000000000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niessl J, Baxter AE, Morou A, Brunet-Ratnasingham E, Sannier G, Gendron-Lepage G, Richard J, Delgado GG, Brassard N, Turcotte I, et al. (2020). Persistent expansion and Th1-like skewing of HIV-specific circulating T follicular helper cells during antiretroviral therapy. EBioMedicine 54, 102727. 10.1016/j.ebiom.2020.102727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiss S, Baxter AE, Cirelli KM, Dan JM, Morou A, Daigneault A, Brassard N, Silvestri G, Routy JP, Havenar-Daughton C, et al. (2017). Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PloS one 12, e0186998. 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren JA, Zhou S, Xu Y, Moeser MJ, MacMillan DR, Council O, Kirchherr J, Sung JM, Roan NR, Adimora AA, et al. (2020). The HIV-1 latent reservoir is largely sensitive to circulating T cells. eLife 9. 10.7554/eLife.57246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas AS, Jones KL, Gandhi RT, McMahon DK, Cyktor JC, Chan D, Huang SH, Truong R, Bosque A, Macedo AB, et al. (2017). T-cell responses targeting HIV Nef uniquely correlate with infected cell frequencies after long-term antiretroviral therapy. PLoS pathogens 13, e1006629. 10.1371/journal.ppat.1006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mok HP, Norton NJ, Hirst JC, Fun A, Bandara M, Wills MR, and Lever AML (2018). No evidence of ongoing evolution in replication competent latent HIV-1 in a patient followed up for two years. Scientific reports 8, 2639. 10.1038/s41598-018-20682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, et al. (2016). Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proceedings of the National Academy of Sciences of the United States of America 113, 1883–1888. 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Zyl GU, Katusiime MG, Wiegand A, McManus WR, Bale MJ, Halvas EK, Luke B, Boltz VF, Spindler J, Laughton B, et al. (2017). No evidence of HIV replication in children on antiretroviral therapy. The Journal of clinical investigation 127, 3827–3834. 10.1172/JCI94582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vancoillie L, Hebberecht L, Dauwe K, Demecheleer E, Dinakis S, Vaneechoutte D, Mortier V, and Verhofstede C (2017). Longitudinal sequencing of HIV-1 infected patients with low-level viremia for years while on ART shows no indications for genetic evolution of the virus. Virology 510, 185–193. 10.1016/j.virol.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Imamichi H, Smith M, Adelsberger JW, Izumi T, Scrimieri F, Sherman BT, Rehm CA, Imamichi T, Pau A, Catalfamo M, et al. (2020). Defective HIV-1 proviruses produce viral proteins. Proceedings of the National Academy of Sciences of the United States of America 117, 3704–3710. 10.1073/pnas.1917876117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, Capoferri AA, Beg SA, Huang SH, Karandish S, et al. (2017). Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell host & microbe 21, 494–506 e494. 10.1016/j.chom.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonetti FR, Zhang H, Soroosh GP, Duan J, Rhodehouse K, Hill AL, Beg SA, McCormick K, Raymond HE, Nobles CL, et al. (2021). Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. The Journal of clinical investigation 131. 10.1172/JCI145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiegand A, Spindler J, Hong FF, Shao W, Cyktor JC, Cillo AR, Halvas EK, Coffin JM, Mellors JW, and Kearney MF (2017). Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proceedings of the National Academy of Sciences of the United States of America 114, E3659–E3668. 10.1073/pnas.1617961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moron-Lopez S, Xie G, Kim P, Siegel DA, Lee S, Wong JK, Price JC, Elnachef N, Greenblatt RM, Tien PC, et al. (2021). Tissue-specific differences in HIV DNA levels and mechanisms that govern HIV transcription in blood, gut, genital tract and liver in ART-treated women. Journal of the International AIDS Society 24, e25738. 10.1002/jia2.25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karn J, and Stoltzfus CM (2012). Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harbor perspectives in medicine 2, a006916. 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porichis F, Hart MG, Griesbeck M, Everett HL, Hassan M, Baxter AE, Lindqvist M, Miller SM, Soghoian DZ, Kavanagh DG, et al. (2014). High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry. Nature communications 5, 5641. 10.1038/ncomms6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dufour C, Richard C, Pardons M, Massanella M, Ackaoui A, Murrell B, Routy B, Thomas R, Routy JP, Fromentin R, and Chomont N (2023). Phenotypic characterization of single CD4+ T cells harboring genetically intact and inducible HIV genomes. Nature communications 14, 1115. 10.1038/s41467-023-36772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strazza M, and Mor A (2017). Consider the chemokines: a review of the interplay between chemokines and T cell subset function. Discovery medicine 24, 31–39. [PMC free article] [PubMed] [Google Scholar]
- 46.Barczyk M, Carracedo S, and Gullberg D (2010). Integrins. Cell and tissue research 339, 269–280. 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nature biotechnology. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 48.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdel-Mohsen M, Kuri-Cervantes L, Grau-Exposito J, Spivak AM, Nell RA, Tomescu C, Vadrevu SK, Giron LB, Serra-Peinado C, Genesca M, et al. (2018). CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells. Science translational medicine 10. 10.1126/scitranslmed.aar6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morou A, Brunet-Ratnasingham E, Dube M, Charlebois R, Mercier E, Darko S, Brassard N, Nganou-Makamdop K, Arumugam S, Gendron-Lepage G, et al. (2019). Altered differentiation is central to HIV-specific CD4(+) T cell dysfunction in progressive disease. Nature immunology 20, 1059–1070. 10.1038/s41590-019-0418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niessl J, Baxter AE, Mendoza P, Jankovic M, Cohen YZ, Butler AL, Lu CL, Dube M, Shimeliovich I, Gruell H, et al. (2020). Combination anti-HIV-1 antibody therapy is associated with increased virus-specific T cell immunity. Nature medicine 26, 222–227. 10.1038/s41591-019-0747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nayrac M, Dube M, Sannier G, Nicolas A, Marchitto L, Tastet O, Tauzin A, Brassard N, Lima-Barbosa R, Beaudoin-Bussieres G, et al. (2022). Temporal associations of B and T cell immunity with robust vaccine responsiveness in a 16-week interval BNT162b2 regimen. Cell reports 39, 111013. 10.1016/j.celrep.2022.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, Fernandez B, Kelderman S, Schumacher TN, Corti D, et al. (2015). T cell immunity. Functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science 347, 400–406. 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 54.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, et al. (2011). Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 34, 108–121. 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grau-Exposito J, Luque-Ballesteros L, Navarro J, Curran A, Burgos J, Ribera E, Torrella A, Planas B, Badia R, Martin-Castillo M, et al. (2019). Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations. PLoS pathogens 15, e1007991. 10.1371/journal.ppat.1007991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sodroski J, Rosen C, Wong-Staal F, Salahuddin SZ, Popovic M, Arya S, Gallo RC, and Haseltine WA (1985). Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science 227, 171–173. 10.1126/science.2981427. [DOI] [PubMed] [Google Scholar]
- 57.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, and Siliciano RF (2013). Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551. 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SM, Jiang C, Lian X, Chowdhury FZ, et al. (2019). Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. The Journal of clinical investigation 129, 988–998. 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon KJ, Timmons AE, Sengupta S, Simonetti FR, Zhang H, Hoh R, Deeks SG, Siliciano JD, and Siliciano RF (2020). Different human resting memory CD4(+) T cell subsets show similar low inducibility of latent HIV-1 proviruses. Science translational medicine 12. 10.1126/scitranslmed.aax6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark IC, Mudvari P, Thaploo S, Smith S, Abu-Laban M, Hamouda M, Theberge M, Shah S, Ko SH, Perez L, et al. (2023). HIV silencing and cell survival signatures in infected T cell reservoirs. Nature. 10.1038/s41586-022-05556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun W, Gao C, Hartana CA, Osborn MR, Einkauf KB, Lian X, Bone B, Bonheur N, Chun TW, Rosenberg ES, et al. (2023). Phenotypic signatures of immune selection in HIV-1 reservoir cells. Nature. 10.1038/s41586-022-05538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, et al. (2009). HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nature medicine 15, 893–900. 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, Kumar MR, Beg SA, Capoferri AA, Ray SC, et al. (2018). Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proceedings of the National Academy of Sciences of the United States of America 115, E2575–E2584. 10.1073/pnas.1720665115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DI, Keele BF, Ho YC, Siliciano JD, and Siliciano RF (2017). Proliferation of latently infected CD4(+) T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. The Journal of experimental medicine 214, 959–972. 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mendoza P, Jackson JR, Oliveira TY, Gaebler C, Ramos V, Caskey M, Jankovic M, Nussenzweig MC, and Cohn LB (2020). Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. The Journal of experimental medicine 217. 10.1084/jem.20200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, et al. (2014). HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nature medicine 20, 139–142. 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo HH, Hua S, Chen HR, Ouyang Z, et al. (2017). Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. The Journal of clinical investigation 127, 2689–2696. 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, et al. (2014). Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4(+) memory T Cells. Nature communications 5, 5407. 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gosselin A, Wiche Salinas TR, Planas D, Wacleche VS, Zhang Y, Fromentin R, Chomont N, Cohen EA, Shacklett B, Mehraj V, et al. (2017). HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. Aids 31, 35–48. 10.1097/QAD.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pardons M, Fromentin R, Pagliuzza A, Routy JP, and Chomont N (2019). Latency-Reversing Agents Induce Differential Responses in Distinct Memory CD4 T Cell Subsets in Individuals on Antiretroviral Therapy. Cell reports 29, 2783–2795 e2785. 10.1016/j.celrep.2019.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banga R, Procopio FA, Ruggiero A, Noto A, Ohmiti K, Cavassini M, Corpataux JM, Paxton WA, Pollakis G, and Perreau M (2018). Blood CXCR3(+) CD4 T Cells Are Enriched in Inducible Replication Competent HIV in Aviremic Antiretroviral Therapy-Treated Individuals. Frontiers in immunology 9, 144. 10.3389/fimmu.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, and Pantaleo G (2013). Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. The Journal of experimental medicine 210, 143–156. 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, Grandvaux N, Boulassel MR, Routy JP, and Ancuta P (2011). Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. Journal of immunology 186, 4618–4630. 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- 74.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, et al. (2017). Defining total-body AIDS-virus burden with implications for curative strategies. Nature medicine 23, 1271–1276. 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, Kim P, Chen TH, Milush J, Hunt PW, et al. (2018). Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS pathogens 14, e1007357. 10.1371/journal.ppat.1007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stevenson EM, Ward AR, Truong R, Thomas AS, Huang SH, Dilling TR, Terry S, Bui JK, Mota TM, Danesh A, et al. (2021). HIV-specific T cell responses reflect substantive in vivo interactions with antigen despite long-term therapy. JCI insight 6. 10.1172/jci.insight.142640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang SH, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, Patel S, Bollard CM, Cruz CR, Karandish S, et al. (2018). Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. The Journal of clinical investigation 128, 876–889. 10.1172/JCI97555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ren Y, Huang SH, Patel S, Alberto WDC, Magat D, Ahimovic D, Macedo AB, Durga R, Chan D, Zale E, et al. (2020). BCL-2 antagonism sensitizes cytotoxic T cell-resistant HIV reservoirs to elimination ex vivo. The Journal of clinical investigation 130, 2542–2559. 10.1172/JCI132374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang S, Chan CN, Rovira-Clave X, Chen H, Bai Y, Zhu B, McCaffrey E, Greenwald NF, Liu C, Barlow GL, et al. (2022). Combined protein and nucleic acid imaging reveals virus-dependent B cell and macrophage immunosuppression of tissue microenvironments. Immunity 55, 1118–1134 e1118. 10.1016/j.immuni.2022.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, Fauci AS, and Lane HC (2016). Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America 113, 8783–8788. 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Einkauf KB, Osborn MR, Gao C, Sun W, Sun X, Lian X, Parsons EM, Gladkov GT, Seiger KW, Blackmer JE, et al. (2022). Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 185, 266–282 e215. 10.1016/j.cell.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vandergeeten C, Fromentin R, Merlini E, Lawani MB, DaFonseca S, Bakeman W, McNulty A, Ramgopal M, Michael N, Kim JH, et al. (2014). Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. Journal of virology 88, 12385–12396. 10.1128/JVI.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baxter AE, Niessl J, Morou A, and Kaufmann DE (2017). RNA flow cytometric FISH for investigations into HIV immunology, vaccination and cure strategies. AIDS research and therapy 14, 40. 10.1186/s12981-017-0171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dube M, and Kaufmann DE (2022). Single-Cell Multiparametric Analysis of Rare HIV-Infected Cells Identified by Duplexed RNAflow-FISH. Methods in molecular biology 2407, 291–313. 10.1007/978-1-0716-1871-4_20. [DOI] [PubMed] [Google Scholar]
- 85.Brunet-Ratnasingham E, Morou A, Dube M, Niessl J, Baxter AE, Tastet O, Brassard N, Ortega-Delgado G, Charlebois R, Freeman GJ, et al. (2022). Immune checkpoint expression on HIV-specific CD4+ T cells and response to their blockade are dependent on lineage and function. EBioMedicine 84, 104254. 10.1016/j.ebiom.2022.104254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quintelier K, Couckuyt A, Emmaneel A, Aerts J, Saeys Y, and Van Gassen S (2021). Analyzing high-dimensional cytometry data using FlowSOM. Nature protocols 16, 3775–3801. 10.1038/s41596-021-00550-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated and analyzed for this study. We developed R codes scripted to perform unsupervised analysis of B and T cells from SARS-CoV-2 naïve and previously infected individuals. All original codes have been deposited at Github and are publicly available as of publication. URL link is listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact Author upon request (daniel.kaufmann@chuv.ch).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| UCHT1 (BUV395) [Human anti-CD3] | BD Biosciences | Cat#563546; Lot:9058566; RRID:AB_2744387 |
| UCHT1 (BUV496) [Human anti-CD3] | BD Biosciences | Cat#612941; Lot:1022424; RRID:AB_2870222 |
| L200 (BV711) [Human anti-CD4] | BD Biosciences | Cat#563913; Lot:03000025; RRID:AB_2738484 |
| SK3 (BB630) [Human anti-CD4] | BD Biosciences | Cat#624294 CUSTOM; Lot:0289566 |
| RPA-T8 (BV570) [Human anti-CD8] | Biolegend | Cat#301037; Lot:B281322; RRID:AB_10933259 |
| M5E2 (BUV805) [Human anti-CD14] | BD Biosciences | Cat#612902; Lot:0262150; RRID:AB_2870189 |
| M5E2 (BV480) [Human anti-CD14] | BD Biosciences | Cat#746304; Lot : 9133961; RRID:AB_2743629 |
| 3G8 (BV650) [Human anti-CD16] | Biolegend | Cat#302042; Lot:B323847; RRID:AB_2563801 |
| HIB19 (APC-eFluor780) [Human anti-CD19] | Thermo Fisher Scientific | Cat#47-0199; Lot:2145095; RRID:AB_1582231 |
| HIB19 (BV480) [Human anti-CD19] | BD Biosciences | Cat#746457; Lot:1021649; RRID:AB_2743759 |
| HI100 (PerCP Cy5.5) [Human anti-CD45RA] | BD Biosciences | Cat#563429; Lot:8332746; RRID:AB_2738199 |
| NCAM16.2 (BUV737) [Human anti-CD56] | BD Biosciences | Cat#564448; Lot:8288818; RRID:AB_2744432 |
| FN50 (PerCP-eFluor710) [Human anti-CD69] | Thermo Fisher Scientific | Cat#46-0699-42; Lot:1920361; RRID:AB_2573694 |
| FN50 (BV650) [Human anti-CD69] | Biolegend | Cat# 310934; Lot:B303462; RRID:AB_2563158 |
| H4A3 (BV786) [Human anti-CD107A] | BD Biosciences | Cat#563869; Lot:8144866; RRID:AB_2738458 |
| ACT35 (APC) [Human anti-CD134 (OX40)] | BD Biosciences | Cat#563473; Lot:1015537; RRID:AB_2738230 |
| 4B4-1 (PE-Dazzle 594) [Human anti-CD137 (4-1BB)] | Biolegend | Cat# 309826; Lot:B253152; RRID:AB_2566260 |
| TRAP1 (BV421) [Human anti-CD154 (CD40L)] | BD Biosciences | Cat#563886; Lot:9037850; RRID:AB_2738466 |
| TRAP1 (PE) [Human anti-CD154 (CD40L)] | BD Biosciences | Cat#555700; Lot:7086896; RRID:AB_396050 |
| J25D4 (BV421) [Human anti-CD185 (CXCR5)] | Biolegend | Cat# 356920; Lot:B325837; RRID:AB_2562303 |
| B27 (PECy7) [Human anti-IFN-γ] | BD Biosciences | Cat#557643; Lot:8256597; RRID:AB_396760 |
| MQ1-17H12 (PE-Dazzle594) [Human anti-IL-2] | Biolegend | Cat#500344; Lot:B2261476; RRID:AB_2564091 |
| JES3-9D7 (PE) [Human anti-IL-10] | BD Biosciences | Cat#554498; Lot:8198773; RRID:AB_395434 |
| eBio64CAP17 (eFluor660) [Human anti-IL-17A] | Thermo Fisher Scientific | Cat#50-7179-42; Lot:2151998; RRID:AB_11149126 |
| Mab11 (Alexa Fluor 488) [Human anti-TNF-α] | Biolegend | Cat#502915; Lot:B285221; RRID:AB_493121 |
| Mab11 (APC) [Human anti-TNF-α] | BD Biosciences | Cat#562084 Lot: 0350627 RRID:AB_10893226 |
| LIVE/DEAD Fixable dead cell | Thermo Fisher Scientific | L34960 |
| Biological samples | ||
| SARS-CoV-2 naïve donor blood samples | N/A | |
| SARS-CoV-2 prior infection donor blood samples | N/A | |
| Chemicals, peptides, and recombinant proteins | ||
| PepMix™ SARS-CoV-2 (Spike Glycoprotein) | JPT | Cat#PM-WCPV-S-1 |
| Staphylococcal Enterotoxin B (SEB) | Toxin technology | Cat#BT202 |
| Software and algorithms | ||
| Flow Jo v10.8.0 | Flow Jo | https://www.flowjo.com |
| GraphPad Prism v8.4.1 | GraphPad | https://www.graphpad.com |
| R studio v4.1.0 | R studio | https://rstudio.com |
| R codes scripted | Github | https://github.com/otastet/Nayrac_et_al |


