Abstract
We have analyzed amino acid substitutions at position G190 in the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1). The mutation G190E, which is responsible for resistance to certain nonnucleoside inhibitors, results in RT that has significantly less polymerase activity and that is less processive than wild-type RT. Its kinetic profile with respect to dGTP and poly(rC) · oligo(dG) is significantly altered compared to that of wild-type RT. The combination of either of the mutations L74V or V75I with the G190E mutation appears to be compensatory and mitigates many of the deleterious effects of the G190E mutation.
There has been considerable progress in developing effective methods to treat human immunodeficiency virus type 1 (HIV-1) infections. This progress has relied on combination therapies; in general, the most effective combinations involve a protease inhibitor and two reverse transcriptase (RT) inhibitors, usually nucleoside analogs (11, 12). One of the problems with these nucleoside analogs is that they are relatively toxic. A second class of RT inhibitors, the nonnucleosides, are not only potent but also relatively nontoxic. However, the usefulness of both classes of inhibitors is limited by the selection of drug-resistant variants (see references 8, 24, and 26 for reviews), a problem that is particularly acute for the nonnucleoside RT inhibitors. Both the available data that describe the dynamics of viral replication (15, 30) and the analyses of these data, principally by Coffin (7), imply that, in a patient, the virus is exceptionally well adapted. This implies that viral variants, including drug-resistant variants, are to either a large or small degree less fit than the wild type. If this were not true, the variant would eventually replace the wild-type virus. This suggests that to be a useful therapeutic, a drug must be broadly effective against all of the viral variants that replicate reasonably efficiently. Put another way, a good drug selects relatively weak viral variants.
This is the test that the nonnucleoside inhibitors fail: in general, these drugs select for viral variants that replicate quite well. However, two classes of nonnucleoside inhibitors, (S)-4-isopropoxycarbonyl-6-methoxy-3-(methylthiomethyl)-3,4-dihydroquinoxaline-2(1H)-thione (HBY 097) and (alkylamino)piperidine bis(heteroaryl)piperizine (AAP-BHAP), select a viral variant that carries the RT mutation G190E and whose replication appears to be substantially impaired (1, 9, 19–22, 25). Other mutants with changes at this residue, i.e., with mutation G190A or G190S, are resistant to the nonnucleoside inhibitor neviripine and appear to be relatively unimpaired (25, 27, 30). However, RTs carrying the mutation G190E are deficient in polymerase activity, and viruses carrying this mutation replicate poorly compared to wild-type viruses (1, 6, 9, 19–22, 25). It is interesting to note that, at least with the inhibitor HBY 097, the G190E mutation is generated only under strong selective pressure, i.e., high levels of inhibitor (22). Low levels of the inhibitor select other mutations, including G190A (22). The G190S mutation has not been isolated in response to treatment with HBY 097 or AAP-BHAP derivatives, suggesting that this mutation does not confer significant resistance to these inhibitors.
Continued passage of viruses carrying the G190E mutation in the presence of nonnucleoside inhibitor HBY 097 selects for viruses that have additional changes in RT. What is surprising is the nature of these secondary mutations, which are located at either amino acid L74 or amino acid V75 (21). Mutations at these positions are associated with resistance to nucleoside analogs: L74V and V75T are associated with resistance to ddI (2′,3′-dideoxyinosine) and d4T (2′,3′-didehydro-2′,3′-dideoxythymidine), respectively (23, 29). We have suggested that resistance to nucleoside analogs involves the repositioning of the nucleic acid substrate (4). Based on this idea, Kleim et al. (21) proposed that the G190E mutation may adventitiously reposition the nucleic acid substrate, which might explain the poor polymerase activity of this mutant enzyme, and that the secondary mutations at either position 74 or 75 compensate by returning the nucleic acid to a position more similar to that of the wild-type RT. We wished to test this hypothesis experimentally and to examine the interactions between the G190E mutation and the mutations at L74 and V75. The mutations G190E, L74V-G190E, V75I-G190E, G190A, and G190S were analyzed for their effects on polymerase activity and RNase H activity.
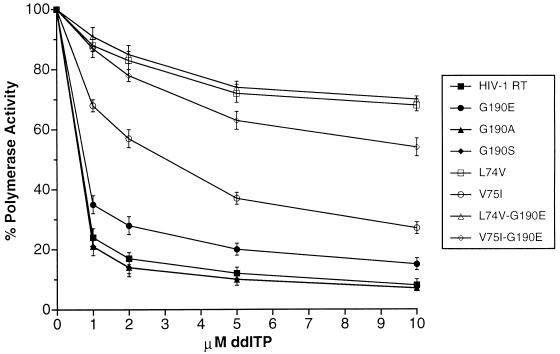
Since the mutations at L74 and V75 confer resistance to nucleoside inhibitors (8, 23, 24, 26, 29), it is possible that the L74V-G190E and V75I-G190E double mutants will be resistant to nucleoside analogs as well as to HBY 097. Viruses carrying the mutations V75I or V75L (V75I/L) and G190E do not have increased resistance to analogs d4T and ddA (dideoxyadenosine). However, viruses containing the double mutation L74V/I-G190E were more resistant to d4T and ddA (21). Heterodimeric HIV-1 RTs containing various mutations at G190 were expressed in E. coli, purified, and assayed for their polymerase activities (Table 1) and their sensitivities to the nucleoside analog ddI triphosphate (ddITP) (Fig. 1).
TABLE 1.
RNA-dependent DNA polymerase activities of the mutant heterodimersa
| Heterodimerb | Activity (%) |
|---|---|
| Wild type | 100 |
| G190A | 170 |
| G190S | 120 |
| G190E | 20 |
| L74V-G190E | 100 |
| V75I-G190E | 105 |
| L74V | 100 |
| V75I | 100 |
Individual mutations were prepared in RT(66) clones (13, 17) by the method of BspMI cassette mutagenesis as previously described (2, 3). p66-p51 heterodimers were generated and purified as previously described (4). The polymerase assays were done as previously described (4). The reactions used equal amounts of purified p66-p51 heterodimers (2.0 ng) and poly(rC) · oligo(dG) as the substrate. The amount of labeled polymer collected for the wild-type heterodimer was considered 100% activity, and the amounts of labeled polymer collected for the various mutant heterodimers were normalized to this value.
The heterodimers other than the wild type are designated by the mutations they carry.
FIG. 1.
Inhibition of RNA-dependent DNA polymerase activity by ddITP. The inhibition assays were done as previously described (4). The reactions used equal amounts of purified p66-p51 heterodimers (2.0 ng) and used poly(rC) · oligo(dG) as the substrate. ddITP was added to the indicated final concentrations. The amount of labeled polymer collected in the absence of the ddITP inhibitor was considered 100% activity, and the amount of labeled polymer collected in the presence of ddITP was normalized to this value. The error bars indicate the variation between duplicate assays.
The G190A and G190S HIV-1 RT variants have an increased level of polymerase activity (Table 1) and appear to be as sensitive to ddITP as wild-type HIV-1 RT (Fig. 1). The G190A mutant has been reported to have 75% of the activity of wild-type RT in bacterial extracts; however, this does not provide an accurate measure of the specific activity of the enzyme (6). The G190E HIV-1 RT variant has a significantly lower polymerase activity than wild-type RT (Table 1). This is in agreement with results obtained by other groups (1, 6, 9, 19–22, 25). The G190E variant appears to be slightly more resistant to ddITP than wild-type RT. The L74V mutation can be selected by ddI, and it is not surprising that a variant RT carrying this mutation is resistant to ddITP; the L74V-G190E double mutant is as resistant to ddITP as the L74V single mutant. The V75I mutant shows a moderate level of resistance to ddITP (Fig. 1). Interestingly, the V75I-G190E double mutant has a much higher level of resistance to ddITP than either the V75I or the G190E single mutant (Fig. 1). We believe that this result can be most easily explained in terms of an effect on the position of the template primer.
Polymerases have two substrates, nucleotides and the template primer; kinetic analysis was done with each of the substrates being the variable reactant. We first examined the kinetic parameters for the various RTs with poly(rC) · oligo(dG) template primer as the variable substrate. The Km of wild-type RT for substrate poly(rC) · oligo(dG) was measured as 2.29 μg/ml, which is in good agreement with the value of 2.0 μg/ml obtained by Hizi et al. (14). The G190A mutant has a much higher Km for poly(rC) · oligo(dG) than does wild-type RT, indicating that, relative to wild-type RT, this enzyme requires higher levels of template primer to achieve the maximal catalytic rate (Table 2). However, the catalytic constant (kcat) of the G190A mutant is significantly higher than that of wild-type RT, indicating that the G190A mutant is able to catalyze more reactions per unit time than wild-type RT (Table 2). For the G190A mutant, these two differences tend to balance each other, and the G190A mutant has a catalytic efficiency that is only slightly lower than that of wild-type RT (Table 2). The G190S mutant has a somewhat lower Km for poly(rC) · oligo(dG) than wild-type RT but also has a lower kcat (Table 2). The overall catalytic efficiency is approximately the same as that of wild-type RT.
TABLE 2.
Kinetic analysis with poly(rC) · oligo(dG) as the variable substratea
| Enzymeb | Vmax | Km | kcat (s−1) | kcat/Km |
|---|---|---|---|---|
| Wild type | 270 ± 4 | 2.29 ± 0.02 | 0.265 ± 0.004 | 0.116 ± 0.003 |
| G190A | 572 ± 8 | 5.14 ± 0.03 | 0.561 ± 0.008 | 0.109 ± 0.002 |
| G190S | 235 ± 5 | 1.94 ± 0.02 | 0.230 ± 0.005 | 0.119 ± 0.003 |
| G190E | 22 ± 4 | 0.23 ± 0.02 | 0.022 ± 0.004 | 0.096 ± 0.003 |
| L74V-G190E | 285 ± 5 | 2.72 ± 0.03 | 0.279 ± 0.005 | 0.103 ± 0.003 |
| V75I-G190E | 286 ± 4 | 2.74 ± 0.02 | 0.280 ± 0.004 | 0.102 ± 0.002 |
| L74V | 269 ± 5 | 2.01 ± 0.01 | 0.264 ± 0.005 | 0.131 ± 0.004 |
| V75I | 254 ± 4 | 2.33 ± 0.04 | 0.249 ± 0.004 | 0.107 ± 0.004 |
The poly(rC) · oligo(dG) kinetic assay was similar to that described by Hizi et al. (14). All assays were carried out in triplicate, and the results are the averages of the three assays ± the error range of the three assays. The Km and Vmax values were determined from the double reciprocal (Lineweaver-Burk) plots of the starting concentrations of the variable component [poly(rC) · oligo(dG)] versus the initial velocities of dGTP incorporation at each concentration. Vmax is in units of picomoles of dGTP incorporated in 10 min; Km is in units of micrograms of poly(rC) · oligo(dG) per milliliter.
Enzymes other than the wild type are designated by the mutations they carry.
Interestingly, the G190E mutant has a very low Km for poly(rC) · oligo(dG), indicating that the enzyme binds this RNA-DNA substrate tightly. However, this template primer binding does not appear to be in a configuration that is favorable for catalysis since the kcat of the G190E mutant is quite low (10% of the level of wild-type RT), indicating that catalysis proceeds relatively slowly (Table 2). As discussed above, Kleim et al. have suggested that the G190E mutation may adventitiously reposition the nucleic acid substrate (21). Our results with poly(rC) · oligo(dG) support this conjecture.
The L74V-G190E and V75I-G190E double mutants have nearly identical kinetic parameters with poly(rC) · oligo(dG), and those of both differ significantly from those of the G190E mutant (Table 2). The combination of either the L74V or V75I mutation with the G190E mutation increases the rate of catalysis compared to that seen for an RT carrying only the G190E mutation (Table 2). Both L74V-G190E and V75I-G190E double mutants have Km and kcat values for poly(rC) · oligo(dG) that are somewhat higher than those of the wild-type enzyme, and both enzymes have catalytic efficiencies that are slightly lower than that of the wild-type RT.
We then examined the kinetic parameters with dGTP as the variable substrate. The Km for wild-type RT with dGTP was 3.55 μM dGTP (Table 3), in good agreement with the value of 3.1 μM dGTP obtained by Hizi et al. (14). The G190A mutant has a lower Km for dGTP than does wild-type RT and has a higher kcat (Table 3). These two factors combine to give the G190A mutant a catalytic efficiency that is approximately twice that of wild-type RT when dGTP is the limiting substrate (Table 3). The G190S mutant also has a lower Km for dGTP than does the wild-type enzyme. However, its kcat is somewhat lower than that of the wild-type RT, yielding a catalytic efficiency that is slightly higher than that of the wild-type RT (Table 3).
TABLE 3.
Kinetic analysis with dGTP as the variable substratea
| Enzymeb | Vmax | Km | kcat (s−1) | kcat/Km |
|---|---|---|---|---|
| Wild type | 198 ± 2 | 3.55 ± 0.02 | 0.194 ± 0.002 | 0.055 ± 0.001 |
| G190A | 293 ± 3 | 2.42 ± 0.03 | 0.287 ± 0.003 | 0.119 ± 0.002 |
| G190S | 172 ± 3 | 2.29 ± 0.03 | 0.169 ± 0.003 | 0.074 ± 0.002 |
| G190E | 68 ± 2 | 10.46 ± 0.09 | 0.067 ± 0.002 | 0.006 ± 0.001 |
| L74V-G190E | 184 ± 4 | 5.18 ± 0.03 | 0.180 ± 0.004 | 0.035 ± 0.001 |
| V75I-G190E | 179 ± 3 | 5.27 ± 0.02 | 0.175 ± 0.003 | 0.033 ± 0.001 |
| L74V | 170 ± 3 | 3.31 ± 0.02 | 0.167 ± 0.003 | 0.051 ± 0.001 |
| V75I | 158 ± 4 | 5.23 ± 0.04 | 0.155 ± 0.004 | 0.030 ± 0.001 |
The dGTP kinetic assays were similar to that described by Hizi et al. (14). All assays were carried out in triplicate, and the results are averages of the three assays ± the error range of the three assays. The Km and Vmax values were determined from the double reciprocal (Lineweaver-Burk) plots of the starting concentrations of the variable component (dGTP) versus the initial velocities of dGTP incorporation at each dGTP concentration. Vmax is in units of picomoles of dGTP incorporated in 10 min; Km is in units of micromolar dGTP.
Enzymes other than the wild type are designated by the mutations they carry.
The G190E mutant has a Km for dGTP that is approximately three times higher than that seen for wild-type RT (Table 3), suggesting that the G190E mutant binds dGTP less efficiently than does wild-type RT. This may be due in part to altered template primer positioning. The polymerase active site is composed of both the end of the template primer and protein components (4). If the position of the nucleic acid is shifted, the incoming nucleotide may not bind as efficiently to the active site. Once the nucleotide is bound, the G190E variant does not appear to add the nucleotide to the growing primer strand as efficiently as wild-type RT. The number of reaction processes each G190E active site catalyzes per unit time (kcat) is only approximately one-third that measured for wild-type HIV-1 RT; this lower turnover number may also reflect the suboptimal position of the template primer described above. If dGTP is the variable substrate, the catalytic efficiency of the G190E mutant is only 10% that of wild-type RT (Table 3).
The L74V-G190E and V75I-G190E double mutants have similar kinetics with dGTP. The presence of the L74V or V75I mutation reverses most of the deleterious effects of the G190E mutation (Table 3). The double mutants have a Km for dGTP which, while not as low as that of wild-type RT, is significantly lower than the Km measured for the G190E mutation alone (Table 3). The kcat and the catalytic efficiency for the double mutants are also greatly improved compared to those for the RT carrying only the G190E mutation (Table 3).
Wild-type HIV-1 RT and the seven mutant HIV-1 RTs were assayed for their processivities, i.e., their abilities to extend a DNA primer hybridized to a DNA template. The presence of an excess of unlabeled poly(rC) · oligo(dG) insures that there is only one round of polymerization in these assays. Any RT that falls off the original template primer will rebind to the poly(rC) · oligo(dG). The longest major extension product produced by wild-type HIV-1 RT is approximately 350 bp; in this assay, the L74V RT variant is virtually indistinguishable from wild-type RT (Fig. 2). The V75I RT variant was significantly less processive than either wild-type RT or the L74V RT variant (Fig. 2). The largest major extension product for the V75I variant was approximately 200 bp. However, the overall pattern of the smaller extension products is similar to that seen for wild-type RT.
FIG. 2.
Processivities of the RT variants compared to that of wild-type RT. The processivity assay was done as previously described (5). The substrate was [32P]ATP-labeled M13 sequencing primer hybridized to single-stranded M13 DNA. All of the assays used equal amounts of p66-p51 heterodimer (1.0 μg). Two separate processivity reactions were done for each RT sample, and these two samples were fractionated on adjacent lanes of the gel. The molecular weight marker is 32P-labeled MspI-digested pBR322 DNA. The sizes of the marker bands are indicated on the left side of the figure.
HIV-1 RTs carrying the nonnucleoside inhibitor resistance mutations G190A and G190S have different processivities (Fig. 2). The G190A mutant generates longer extension products than does wild-type RT; the longest major extension product is approximately 620 bp long. The G190S mutant is less processive than wild-type RT; the longest major extension product is about 150 bp long; the G190S mutant makes more of the shorter extension products than does wild-type RT (Fig. 2).
The G190E mutant is the least processive of the enzymes we tested. Compared to wild-type RT, the G190E mutant makes a large amount of short extension products (Fig. 2). This is in agreement with data obtained by Fan et al. (9) for RNA-dependent DNA polymerization. While the processivity of the G190E mutant is significantly impaired relative to that of the wild-type RT, the G190E variant was able to extend the primer for longer distances than might be expected for an enzyme with such a low level of polymerase activity. As an example, the G190S mutant, which has a higher polymerase activity than the G190E mutant, is only marginally more processive (Fig. 2). A possible explanation is the tight binding measured for the template primer poly(rC) · oligo(dG) (Table 2). Tighter binding of the G190E variant to nucleic acid may allow the enzyme to catalyze a substantial number of polymerization steps before disassociating from the template primer (Fig. 2).
Interestingly, the combination of either the L74V mutation or the V75I mutation with the G190E mutation appears to reverse some of the deleterious effects of the G190E mutation (Fig. 2). The extension products are much longer, and the pattern of products produced by the L74V-G190E and V75I- G190E RTs is similar to that seen for RTs carrying only the V75I mutation. However, the processivities of the L74V- G190E and V75I-G190E enzymes are still significantly lower than that of wild-type RT (Fig. 2). This improved processivity is probably due to more efficient polymerization since the L74V-G190E and V75I-G190E variants do not have the low Kms for poly(rC) · oligo(dG) that the G190E variant has (Table 2).
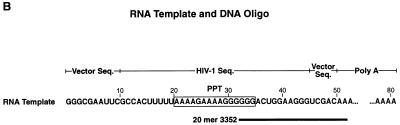
If the G190E mutation repositions the nucleic acid, it is possible that the mutation would have some effect on the RNase H activity. We have found that the ability of the RNase H of HIV-1 RT to cleave RNA-DNA substrates containing the polypurine tract from the HIV-1 genome can be used to monitor interaction(s) between the polymerase domain of HIV-1 RT and nucleic acid (9a). We have used one such substrate (Fig. 3B) to compare the RNase H activities of wild-type HIV-1 RT and the various mutants. There are two major sites of RNase H cleavage. One family of cleavage sites is approximately 17 nucleotides from the 5′ end of the primer oligonucleotide (template position 49) and are designated the −17 cleavages (10). From crystallographic analysis, it has been noted that there are approximately 17 or 18 nucleotides between the polymerase active site and the RNase H active site (18). The −17 cleavages appear to represent the first cleavages made after HIV-1 RT binds to the template primer. The second family of cleavage sites are approximately eight nucleotides from the 5′ end of the primer oligonucleotide. The −8 cleavages appear to occur only after the −17 cleavages have been made. It is not clear what mechanism is responsible for the −8 cleavages, although it has been suggested that this is the result of processive cleavage (10). The 81-base-long RNA substrate was labeled by the incorporation of α-32P-labeled uridine and hybridized to a DNA oligonucleotide (Fig. 3B). The −17 cleavages of the 81-nucleotide starting template will produce two sets of fragments: one family of fragments will be approximately 49 nucleotides in length and will be radioactively labeled, while the second family of fragments will not be radioactively labeled since they contain no uridine residues. The −8 cleavages will produce labeled fragments approximately 39 nucleotides long. Both the amount of full-length RNA template remaining after cleavage and the amount of −17 and −8 cleavage products generated can be used as measures of RNase H activity. By both measures the RNase H activity of the G190E mutant is significantly reduced (Fig. 3A) relative to that of wild-type RT. These data are in agreement with those of Fan et al. (9), who also showed that the G190E mutant had a reduced RNase H activity. Since the G190E mutant has a lower Km for nucleic acid than does wild-type RT (Table 2), the reduction in RNase H activity cannot be simply due to a reduction in the binding of the nucleic acid substrate. However, repositioning of the nucleic acid could result in reduced RNase H activity if the RNA strand of the RNA-DNA heteroduplex was moved away from the RNase H active site. If the combination of either of the secondary mutations (L74V or V75I) with the G190E mutation causes the nucleic substrate to be bound in a configuration more like that of wild-type RT, we would expect that the RNase H activity would also be restored to near-wild-type levels. The data match this prediction (Fig. 3A), providing additional support for the idea that the effects we have measured are the result of the mispositioning of the nucleic acid substrate (for the G190E mutant) and the repositioning to a more nearly wild-type configuration (for the L74V-G190E and V75I-G190E mutants) of the nucleic acid substrate.
FIG. 3.
(A) RNase H activities of HIV-1 RT mutants compared to that of wild-type RT. An α-32P-uridine-labeled RNA template (81 nucleotides in length) was hybridized to a 20-base oligonucleotide (the sequences of the RNA and the DNA are given in panel B). Approximately 50,000 cpm of RNA (100 ng) and 40 ng of the DNA oligonucleotide were used in each assay. The upper band represents full-length template RNA; the middle band represents the template remaining after the −17 cleavage. The lower band represents the template remaining after the −8 cleavage. The sizes of the markers (which are DNA) do not precisely correspond to the sizes of the RNA digestion products. (B) Schematic diagram of the template primer used in the RNase H activity assay. The RNA template was labeled with [α-32P]UTP. Oligonucleotide 3352 is complementary to the sequence between base 33 and base 52 of the RNA; the −17 cuts center on base 49 of the RNA; the −8 cuts center on base 39.
In summary, the amino acid substitution G190E has significant deleterious effects on both the polymerase and RNase H activities of HIV-1 RT. The hypothesis that the G190E mutation repositions the nucleic acid is well supported by the data presented here. Normally, in the absence of a nonnucleoside inhibitor, the nonnucleoside drug binding pocket does not exist; the space that would be occupied by the drug is filled by the side chains of the surrounding amino acids including Y181, Y188, and W229 (16, 28). It may well be that inserting a hydrophilic (and potentially charged) residue into this environment causes a partial expansion of the pocket region, not only because the side chain of the glutamic acid residue is much larger than the hydrogen of glycine but also because its hydrophilic nature might make the large aromatic groups move away. This could lead to a distortion of the position of β12-β13, which we believe plays a critical role in properly positioning the primer strand. The mutations G190A and G190S replace the hydrogen of glycine with a small hydrophobic methyl group and an uncharged hydrophilic group, respectively. These substitutions do not have the large deleterious effects seen with the G190E mutation, which may be due to the fact that these side groups do not lead to as large a distortion in the pocket area. Based on this line of argument, we do not believe that either the G190A or the G190S mutation causes significant repositioning of the template primer.
As expected from their behavior in the context of the virus, mutations at amino acid residues L74 and V75 appear to be compensatory and to mitigate the deleterious effects of the G190E mutation. Unlike the G190E variant, which had a low level of polymerase activity, the L74V-G190E and V75I-G190E variants have polymerase activities similar to that of wild-type RT (Tables 1, 2, and 3; Fig. 2). Interestingly, these double mutants still retain considerable resistance to nucleoside analogs, suggesting that combination therapy with HBY 097 and ddI will not be effective (Fig. 1).
This proposal that mutations in the nucleoside binding pocket (like G190E) can influence template primer positions explains the data presented here and could provide an explanation for some otherwise unexplained interactions between mutations that confer resistance to nucleoside analogs and nonnucleoside inhibitors. We suggest that the central theme in such interactions is the proximity of the drug binding pocket and the primer grip motif (especially β12-β13). This would suggest that certain mutations that cause resistance to nonnucleoside inhibitors adventitiously distort the interaction of RT with the template primer. Since these interactions are adventitious, their effects on the positioning of the nucleic acid and, by extension, on the resistance to nucleotide analogs are, at our current state of knowledge, unpredictable. The effects can be additive (G190E and L74V or V75I mutations) or negative (Y181C and T215Y/F mutations) (24). However, this model does give us some hope, not only that the interactions of mutations that confer resistance to nucleoside analogs and nonnucleoside inhibitors can be rationally explained but also that, when structural data that describe these positioning effects more accurately (precisely) become available, it should also be possible to use this information to predict how some of the resistance mutations interact and, by extension, to predict which drug combinations will be effective.
Acknowledgments
We thank Pat Clark and Peter Frank for protein purification, J.-P. Kleim, E. Arnold, and their colleagues for many helpful discussions, and Hilda Marusiodis for preparation of the manuscript.
The research was sponsored by the National Cancer Institute, DHHS, under contract with ABL, and the National Institute of General Medical Sciences.
REFERENCES
- 1.Balzarini J, Karlsson A, Meichsner C, Paessens A, Reiss G, de Clercq E, Kleim J-P. Resistance pattern of human immunodeficiency virus type 1 reverse transcriptase to quinoxaline S-2720. J Virol. 1994;68:7986–7992. doi: 10.1128/jvi.68.12.7986-7992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer P L, Ferris A L, Hughes S H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 1992;66:1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer P L, Ferris A L, Hughes S H. Mutational analysis of the fingers domain of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:7533–7537. doi: 10.1128/jvi.66.12.7533-7537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer P L, Tantillo C, Jacobo-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E A, Hughes S H. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type-1 (HIV-1) reverse transcriptase. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 6.Chao S-F, Chan V L, Juanka P, Kaplan A H, Swanstrom R, Hutchison C A., III Mutational sensitivity patterns define crucial residues in the palm subdomain of the reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 1995;23:803–810. doi: 10.1093/nar/23.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 8.De Clercq E. HIV resistance to reverse transcriptase inhibitors. Biochem Pharmacol. 1994;47:155–169. doi: 10.1016/0006-2952(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 9.Fan N, Rank K B, Slade D E, Poppe S M, Evans D B, Kopta L A, Olmstead R A, Thomas R C, Tarpley W G, Sharma S K. A drug resistance mutation in the inhibitor binding pocket of human immunodeficiency virus type 1 reverse transcriptase impairs DNA synthesis and RNA degradation. Biochemistry. 1996;35:9737–9745. doi: 10.1021/bi9600308. [DOI] [PubMed] [Google Scholar]
- 9a.Gao, H.-Q. et al. Unpublished data.
- 10.Ghosh M, Howard K J, Cameron C E, Benkovic S J, Hughes S H, Le Grice S F J. Truncating α-helix E′ of p66 human immunodeficiency virus reverse transcriptase modulates RNase H function and impairs DNA strand transfer. J Biol Chem. 1995;270:7068–7076. doi: 10.1074/jbc.270.13.7068. [DOI] [PubMed] [Google Scholar]
- 11.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohn A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 12.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 13.Hizi A, McGill C, Hughes S H. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci USA. 1988;85:1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hizi A, Tal R, Shaharabany M, Loya S. Catalytic properties of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J Biol Chem. 1991;266:6230–6239. [PubMed] [Google Scholar]
- 15.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 16.Hsiou Y, Ding J, Das K, Clark A D, Jr, Hughes S H, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 17.Hughes S H, Ferris A, Hizi A. Analysis of the reverse transcriptase of human immunodeficiency virus expressed in Escherichia coli. In: Laver W G, Air G M, editors. Use of X-ray crystallography in the design of antiviral agents. San Diego, Calif: Academic Press, Inc.; 1990. pp. 297–307. [Google Scholar]
- 18.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleim J P, Bender R, Billhardt U M, Meichsner C, Riess G, Rosner M, Winkler I, Paessens A. Activity of a novel quinoxaline derivative against human immunodeficiency virus type 1 reverse transcriptase and viral replication. Antimicrob Agents Chemother. 1993;37:1659–1664. doi: 10.1128/aac.37.8.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleim J P, Bender R, Kirsch R, Meichsner C, Paessens A, Riess G. Mutational analysis of residue 190 of human immunodeficiency virus type 1 reverse transcriptase. Virology. 1994;200:696–701. doi: 10.1006/viro.1994.1233. [DOI] [PubMed] [Google Scholar]
- 21.Kleim J P, Rosner M, Winkler I, Paessens A, Kirsch R, Hsiou Y, Arnold E, Riess G. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleim J P, Winkler I, Rosner M, Kirsch R, Rubsamen-Waigmann H, Paessens A, Riess G. In vitro selection for different mutational patterns in HIV-1 reverse transcriptase using high and low selective pressure of the nonnucleoside reverse transcriptase inhibitor HBY 097. Virology. 1997;231:112–118. doi: 10.1006/viro.1997.8513. [DOI] [PubMed] [Google Scholar]
- 23.Lacey S F, Larder B A. A novel mutation (V75T) in the human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob Agents Chemother. 1994;38:1428–1432. doi: 10.1128/aac.38.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellors J W, Larder B A, Schinazi R F. Mutations in HIV-1 reverse transcriptase and protease associated with drug resistance. Int Antivir News. 1995;3:8–13. [Google Scholar]
- 25.Olmsted R A, Slade D E, Kopta L A, Poppe S M, Poel T J, Newport S W, Rank K B, Biles C, Morge R A, Dueweke T J, Yagi Y, Romero D L, Thomas R C, Sharma S K, Tarpley W G. (Alkylamino)piperidine bis(heteroaryl)piperizine analogs are potent, broad-spectrum nonnucleoside reverse transcriptase inhibitors of drug-resistant isolates of human immunodeficiency virus type 1 (HIV-1) and select for drug-resistant variants of HIV-1IIIB with reduced replication phenotypes. J Virol. 1996;70:3698–3705. doi: 10.1128/jvi.70.6.3698-3705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richman D D. Resistance of clinical isolates of human immunodeficiency virus to antiretroviral agents. Antimicrob Agents Chemother. 1993;37:1207–1213. doi: 10.1128/aac.37.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richman D D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S A, Sullivan J, Cheeseman S, Barringer K, Pauletti D, Shih C-K, Myers M, Griffin J. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodgers D W, Bamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouch C, Harrison S C. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 30.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]