Abstract
Undifferentiated intestinal stem cells (ISCs), particularly those marked by Lgr5, are crucial for maintaining homeostasis and resolving injury. Lgr5+ cells in the crypt base constantly divide, pushing daughter cells upward along the crypt axis, where they differentiate into a variety of specialized cell types. This process requires coordinated execution of complex transcriptional programs, which allow for the maintenance of undifferentiated stem cells while permitting differentiation of the wide array of intestinal cells necessary for homeostasis. Thus, disrupting these programs may negatively impact homeostasis and response to injury. Previously, members of the myeloid translocation gene (MTG) family have been identified as transcriptional co-repressors that regulate stem cell maintenance and differentiation programs in multiple organ systems, including the intestine. One MTG family member, myeloid translocation gene related 1 (MTGR1), has been recognized as a crucial regulator of secretory cell differentiation and response to injury. However, whether MTGR1 contributes to the function of ISCs has not yet been examined. Here, using Mtgr1−/− mice, we have assessed the effects of MTGR1 loss on ISC biology and differentiation programs. Interestingly, loss of MTGR1 increased the total number of cells expressing Lgr5, the canonical marker of cycling ISCs, suggesting higher overall stem cell numbers. However, expanded transcriptomic analyses revealed MTGR1 loss may instead promote stem cell differentiation into transit-amplifying cells at the expense of cycling ISC populations. Furthermore, ex vivo intestinal organoids established from Mtgr1 null were found nearly completely unable to survive and expand, likely due to aberrant ISC differentiation, suggesting that Mtgr1 null ISCs were functionally deficient as compared to WT ISCs. Together, these results identify a novel role for MTGR1 in ISC function and suggest that MTGR1 is required to maintain the undifferentiated state.
Introduction
The intestinal epithelium is under constant metabolic, mechanical, and microbial stress and, as such, is in a continual state of regeneration and renewal. While models for cell function in the gut are evolving, it has been repeatedly demonstrated that intestinal stem cells (ISCs) are key to maintaining intestinal homeostasis and response to injury1. Maintenance of intestinal health and regeneration requires the orchestrated execution of stem cell, early progenitor, and differentiation programs, usually via coordinated activation and suppression of transcriptional circuits, to ensure proper stem cell function and balanced downstream lineage allocation2.
To accommodate the constant cellular turnover of the intestinal epithelium, ISCs rapidly proliferate in the crypt base, pushing their daughter cells out of the crypt and up the intestinal villus while undergoing terminal differentiation into an assortment of secretory and absorptive cell types. These highly proliferative crypt base ISCs, termed crypt base columnar cells (CBCs), are identified by their expression of the gene, leucine-rich repeat containing G-protein coupled receptor 5 (Lgr5)1. Lgr5 expression is highly regulated, most notably by Wnt and Notch signaling pathways, and is suppressed upon exit of the stem cell niche and activation of cellular differentiation programs3. Interestingly, while WNT pathway activation in CBCs upregulates Lgr5 expression, LGR5 protein also functionally promotes Wnt signaling and ISC biology by serving as the receptor for the potent Wnt agonist, R-spondin4. As proliferation continues, daughter cells from CBCs move further up the crypt base into the transit amplifying (TA) compartment. Here, cells proliferate even more rapidly than parental CBCs, yet simultaneously begin lineage commitment and terminal differentiation.
In addition to Lgr5 + CBCs, other populations of ISCs have been identified, many of which are less proliferative than CBCs at homeostasis but are “activated” as part of intestinal regenerative programs. These cell populations are denoted by expression of specific genes, such as Bmi1, mTert, Hopx, Lrig1, and Clu; often reside higher in the crypt base than CBCs in the + 4/+5 position; and may have begun the process of lineage commitment5,6. However, functional compensation by these slowly cycling ISC populations may still require dedifferentiation and interconversion to CBCs, in which ISCs gain expression of Lgr5 and take up residence in the crypt base. Interestingly, the ability to reconstitute the CBC population has even been noted in committed progenitors of the secretory and absorptive lineages7–10. Thus, coordinating ISC function and differentiation among differentiated cell types and different ISC populations is a far from linear process, yet remains a crucial component of both intestinal homeostasis and injury responses.
MTGR1 (Cbfa2t2) is a member of the three-protein myeloid translocation gene (MTG) family of transcriptional co-repressors, which were originally identified in translocation fusion proteins driving acute myeloid leukemia11. MTGs, also including MTG16 and MTG8, serve as scaffolding proteins that orchestrate the formation of repression complexes containing histone deacetylases, other co-repressor proteins, and DNA binding factors, thereby modifying chromatin state of key loci12. In the intestine, MTGs are known to serve important functions, as MTG-deficient mice display a range of unprovoked intestinal phenotypes. For example, loss of MTG8 (Mtg8−/−) results in deletion of the midgut, while loss of MTG16 or MTGR1 (Mtg16−/− and Mtgr1−/−) alters intestinal proliferation, apoptosis, migration, and lineage specification13–16. Mtgr1−/− mice are also exquisitely sensitive to dextran sodium sulfate (DSS)-induced injury, with marked depletion of viable, regenerative crypts post-injury17. Finally, we have previously identified MTGR1 as a key regulator of intestinal differentiation into the secretory cell fate, as Mtgr1−/− mice have greatly reduced numbers of Paneth, goblet, and enteroendocrine cells15,18.
Interestingly, work aiming to determine signaling pathways by which MTGs mediate these functions have uncovered roles for MTGs in modulating Wnt and Notch signaling pathways18,19. Despite contributions to these key ISC-associated signaling pathways, the exact role of MTGR1 in ISC biology remains incompletely understood. Here, we have assessed the impact of MTGR1 deficiency on stem cell identity and function in the small intestine both in vivo and ex vivo. Together, these studies uncover a crucial role for MTGR1 in maintaining proper ISC identity and function and expand our knowledge of the mechanisms regulating intestinal differentiation and regeneration.
Methods and Materials
Mice
Mtgr1−/− mice were previously established and characterized15. Lgr5-EGFP-IRES-creERT2 mice were a generous gift from Dr. Robert Coffey1. Male and female age-matched, littermate WT and Mtgr1−/− mice were used for all experiments. Mice were cohoused and maintained on standard chow with 12-hour light/dark cycles. All in vivo procedures were carried out in accordance with protocols approved by the Vanderbilt Institutional Animal Care and Use Committee.
Enteroid culture
Enteroids were established from 8–12-week old WT and Mtgr1−/− mice as previously described18,20. Crypts were counted and equal numbers plated in growth factor reduced Matrigel® (Corning) and overlaid with “ENR” mini-gut culture media. (Advanced Dulbecco’s modified Eagle’s medium–F-12 [Gibco], 100 U/ml penicillin, 100 μg/ml streptomycin, 1X N2 [Gibco], 1X B27 [Gibco], 1X Glutamax [Gibco], 1mM HEPES [Gibco], 20% R-spondin conditioned media [generated from R-spondin-expressing cells generously provided by Dr. Jeff Whitsett, Cincinnati Children’s Hospital], 10% Noggin conditioned media [generated from Noggin-expressing cells generously provided by Dr. G.R. van den Brink, described in 21], and 50ng/ml EGF [R&D Systems]). Media at plating was supplemented as noted in the text with various inhibitors, described in Supplemental Table S1. To determine plating efficiency, the number of viable enteroids was assessed at day 1 post-plating and normalized to the number of crypts plated. Viability was determined by daily enteroid counts and normalized to the number of enteroids established on day 1. ImageJ software (version 1.51) was used to measure organoid size and count crypt buds.
Enteroid hMTGR1 addback
GFP and human MTGR1 were cloned into the pLEX-307 vector (a gift from David Root, Addgene plasmid 41392). pLEX-307-GFP and pLEX-307-MTGR1 were transfected into HEK 293T cells (ATCC CRL-3216) along with psPAX2 and pMD2.g (gifts from Didier Trono, Addgene plasmids 12260 and 12259). After 48 hours, supernatants were collected and viral particles were concentrated by overnight centrifugation at 9,500g at 4°C. Pelleted lentiviral particles were resuspended in mouse Intesticult media (StemCell Technologies) supplemented with 10μM Y-27632 (Tocris) and mixed with duodenal crypt isolations from WT or Mtgr1−/− mice. Crypt/virus mixtures were incubated for 2 hours at 37°C prior to washing and plating in Matrigel plugs overlaid with ENR media supplemented with CHIR 99021 (3μM, Tocris) and Y-27632. After 4 days, CHIR 99021 and Y-27632 were removed. Viability and gene expression were assessed at day 7 post-plating.
Immunohistochemistry
For in vivo analysis, mice were sacrificed and intestinal tissue was “Swiss-rolled” prior to fixation in 10% neutral-buffered formalin. Intestinal samples were then paraffin-embedded and 5 μm sections were cut by the Vanderbilt Translational Pathology Shared Resource (TPSR). For enteroid staining, cultures were collected and fixed as described previously22. Briefly, enteroids were collected and fixed in 10% neutral buffered formalin on ice for 20 min. After washing, the enteroid pellet was resuspended in 2% agarose dissolved in PBS. The agarose was allowed to solidify for 10 min, placed in 70% ethanol, and paraffin-embedded.
For all fluorescent immunohistochemistry (IHC), antigen retrieval consisted of 10 minutes boiling in 10 mM sodium citrate (pH 6) prior to blocking. After blocking, samples were incubated with antibodies against E-cadherin (BD Biosciences, 1:500), Ki67 (Abcam, 1:1000), phospho-histone H3 (Millipore, 1:400), cleaved caspase-3 (Cell Signaling Technology, 1:400), or β-catenin (BD Biosciences, 1:500) overnight at 4°C. Secondary antibodies conjugated to 488 or 594 Alexafluor dyes (Invitrogen) were incubated for 2 hours at room temperature. Nuclear staining was done with ProLong Gold antifade reagent with DAPI (Invitrogen). Staining was visualized with a Nikon Eclipse E800 microscope and Zyla SCMOS camera. Images were processed and quantified using Nikon NIS-Elements Basic Research software.
qRT-PCR analysis
Freshly isolated murine small intestinal crypts were collected and homogenized in TRIzol reagent (Thermo Fisher) using a 21g needle. RNA was isolated using the Rneasy Mini Kit (Qiagen) with on-column DNAse digestion. cDNA was synthesized using the qScript cDNA synthesis kit (Quantabio). qPCR reactions were run using PerfeCTa SYBR Green SuperMix ROX (Quantabio) and primers designated in Supplemental Table S2. For analysis of human MTGR1, probes for MTGR1 (Hs00602520_m1, Thermo Fisher) and Gapdh (Mm99999915_g1, Thermo Fisher) were used in conjunction with TaqMan Universal Master Mix II (Thermo Fisher). All samples were run in triplicate and target gene expression was analyzed using the delta–delta Ct method normalized to GAPDH.
RNA Scope
High-resolution RNA in situ hybridization was performed using the RNAscope® Multiplex Fluorescent V2 assay or RNAscope® 2.5HD Assay – Brown, according to the manufacturer’s instructions (ACDBio). Antigen retrieval was performed under standard pretreatment conditions as specified by the manufacturer. Probes were directed against mouse Cbfa2t2 (#434601), Lgr5 (#312171), or Clu (#427891). Fluorescent assay samples were mounted with ProLong Gold antifade reagent with DAPI and imaged as described above.
RNA-sequencing
For RNA-sequencing, small intestine crypts were isolated from 3 WT and 3 Mtgr1−/− mice. Following crypt isolation, a portion of the samples were collected and homogenized in TRIzol reagent while the remaining crypts were plated for enteroid culture. After 24 hours, half of the plated enteroids were collected for RNA extraction, while the remaining enteroids were cultured for an additional 48 hours and harvested at 72 hours post-plating. Additional samples were collected from passaged WT and Mtgr1−/− enteroids, and RNA for all was isolated as described above. For RNA-sequencing studies, mRNA enrichment and cDNA library preparation was performed by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) facility utilizing the Illumina Tru-seq stranded mRNA sample prep kit. Sequencing was performed at Single-Read 50 HT bp on the Illumina HiSeq 2500. After adapter trimming by Cutadapt23, RNA-seq reads were aligned to the mm10 genome using STAR24 and quantified by featureCounts25. Differential analyses were performed by DESeq226, which determined the log2 fold changes, Wald test p-values, and adjusted p-value (FDR) by the Benjamini-Hochberg procedure. The significantly changed genes were assessed with an FDR < 0.05. Gene set enrichment analysis (GSEA) was performed using GSEA software version 4.03 maintained by the Broad Institute27,28.
Single Cell RNA-sequencing
Mouse ileal tissues were used to generate single-cell RNA-seq data, following a methodology similar to previous studies29,30. In brief, mouse tissues were incubated in a chelating buffer composed of 20mM HEPES and 3mM EDTA in DPBS for 1.25hrs. Tissue was then transferred to 10ml of PBS and shook vigorously for 2–3 minutes to liberate crypts, which were then passed through a 70μm filter and washed by centrifugation at 300g for 5 minutes and resuspension in PBS. Isolated crypts were pelleted and resuspended in a solution of cold active protease (5 mg/ml Protease from Bacillus licheniformis and 2.5 mg/mL Dnase in PBS) and incubated for 25 minutes at 4°C. Following this, the tissues were gently pipetted 10–20 times to obtain single cells and passed through a 70μm filter into a clean tube. Isolated cells were pelleted by centrifugation at 700g for 5 min and washed in PBS containing 0.02% BSA three times before final resuspension in PBS containing 15% Optiprep and filtered using a 40μm flowmi filter. The resulting cell suspensions underwent filtration, washing, and quality inspection before being loaded onto inDrops for microfluidic capture. The inDrops scRNA-seq procedure was carried out according to a modified protocol31,32. Single-cell libraries were prepared for sequencing as detailed in previous documentation33. These libraries, each containing an estimated 2000–3000 cell transcriptomes, were then sequenced on the Novaseq6000 platform, generating approximately 125 million reads per library. Data were processed according to an established pipeline34 and filtered using dropkick35.
Electron Microscopy
Specimens were processed for transmission electron microscopy (TEM) and imaged in the Vanderbilt Cell Imaging Shared Resource: Research Electron Microscopy facility. Briefly, enteroid-containing Matrigel plugs were fixed in 2.5% glutaraldehyde in 0.1M cacodylate buffer, pH7.4 at room temperature (RT) for 1 hour, then transferred to 4°C overnight. The samples were washed in 0.1M cacodylate buffer, incubated for 1 hour in 1% osmium tetraoxide at RT, and washed with 0.1M cacodylate buffer. The samples were then dehydrated through a graded ethanol series followed by 3 exchanges of 100% ethanol. Next, the samples were incubated for 5 minutes in 100% ethanol and propylene oxide (PO) followed by 2 exchanges of pure PO. Samples were then infiltrated with 25% Epon 812 resin and 75% PO for 30 minutes at RT. Next, they were infiltrated with Epon 812 resin and PO [1:1] for 1 hour at RT, then overnight at RT. The next day, the samples went through a [3:1] (resin: PO) exchange for 3–4 hours and were incubated with pure epoxy resin overnight. Samples were then incubated in 2 more changes of pure epoxy resin and allowed to polymerize at 60°C for 48 hours.
500–1000 nm-thick sections were cut for ultra-structure identification. Then, 70–80 nm ultra-thin sections were cut from the region of interest, collected on 300-mesh copper grids, and post-stained with 2% uranyl acetate followed by Reynold’s lead citrate. Samples were subsequently imaged on the Philips/FEI Tecnai T12 electron microscope.
Live cell imaging
Crypts were plated according to standard protocols and immediately placed in the incubation chamber of the EVOS® FL Auto Cell Imaging System. Organoids were imaged every 15 minutes over 5 days with periodic location updates to maintain image focus.
Statistics
Unless noted, statistical analysis was performed in Graphpad Prism 8 Software using Student’s t-test (unpaired, two-tailed) for single timepoints, or two-way ANOVA and Sidak’s multiple comparison post-test for time course analyses. Samples were excluded if determined to be statistical outliers based on “robust regression and outlier removal” (ROUT) analysis. For all studies, center values represent experimental mean, error is represented by standard error of the mean, and P < 0.05 is considered significant.
Results
MTGR1 is widely expressed in the intestine.
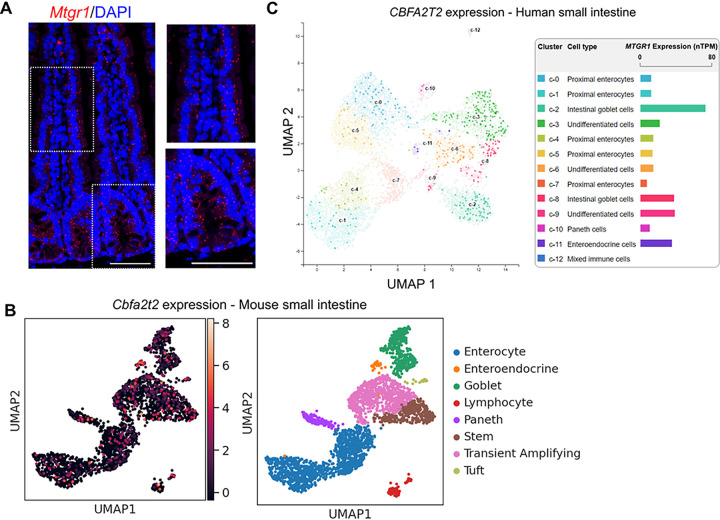
As multiple epithelial cell types exist in the small intestine, cellular expression patterns can be used to infer genetic contributions to cell type-specific functions. To define MTGR1 expression patterns within the intestinal crypt, we first utilized in situ staining methods to spatially visualize Mtgr1 transcripts in the murine small intestine (Fig. 1A). As previously reported, Mtgr1 expression was dispersed throughout the intestinal crypt and did not appear to be specifically localized to distinct cell populations16. As in situ staining does not easily allow for overlay with multiple cell lineage markers, we next investigated Mtgr1 expression in specific cell types via single-cell RNA-sequencing (scRNA-seq) of the murine ileum29,36. These results confirm widespread Mtgr1 expression in various intestinal cells that was not restricted to specific cellular lineages (Fig. 1B and Supplemental Fig. 1). Similar results were observed in the human small intestine (Fig. 1C). Here, we queried publicly available scRNA-seq data from the Human Protein Atlas (GSE125970), and again, MTGR1 expression was observed in multiple differentiated and undifferentiated cell types37,38. Thus, Mtgr1 is widely expressed throughout the intestinal epithelium.
Figure 1. MTGR1 is widely expressed in the small intestine.
(A) Mtgr1 mRNA assessed by RNAScope in the WT small intestine. Results representative of 3 independent experiments. Dotted lines indicate inset areas. Scale bar = 100μm. (B) Uniform manifold approximation and projections (UMAPs) showing Mtgr1 (Cbfa2t2) expression in the murine jejunum by scRNA-sequencing (left) and associated cell clusters (right). n=2 mice. (C) Human MTGR1 (CBFA2T2) expression was queried from the Human Protein Atlas scRNA-sequencing data. MTGR1expression is visualized by UMAP (left) and bar graphs (right) in various intestinal cell types.
MTGR1 loss dysregulates ISC populations in vivo.
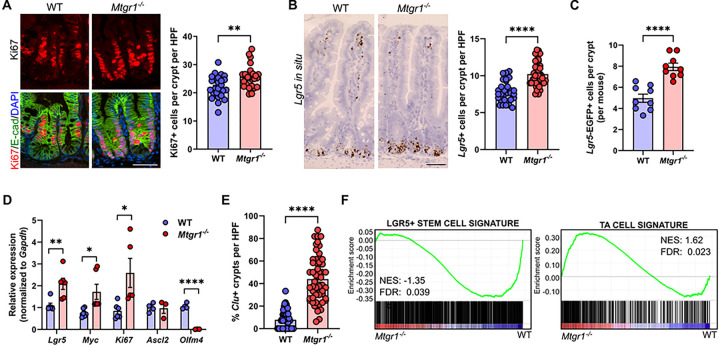
Next, we directly investigated the function of MTGR1 in ISC biology. Interestingly, higher levels of cellular proliferation have been reported in mice globally lacking MTGR1 (Mtgr1−/−), which may be due to loss of MTGR1-mediated downregulation of Wnt pathway genes via TCF4 repression15,19,39. Here, we first confirmed the expansion of proliferative cells in the crypts of Mtgr1−/− mice via quantifying Ki67 expression (Fig. 2A). We next hypothesized that this increase in proliferation may be associated with higher numbers of LGR5 + ISCs, due to their role in maintaining proliferation in the intestine at baseline15,19,39. Lgr5-expressing cells in the small intestine were identified by in situ hybridization. Here, we determined that Mtgr1−/− mice indeed had higher numbers of Lgr5 + cells per crypt as compared to WT mice (Fig. 2B). This was further confirmed by intercross of Mtgr1−/− mice with the Lgr5-Cre-EGFP reporter line (Fig. 2C), which expresses EGFP from the Lgr5 locus. Analysis of these mice again showed increased numbers of Lgr5-EGFP + cells in Mtgr1−/− versus WT Lgr5-Cre-EGFP mice. Higher levels of Lgr5, Ki67, and the Wnt target transcript Myc were also observed in Mtgr1−/− crypts by qPCR (Fig. 2D). Based on these results, it seems likely that the LGR5 + CBC population and overall ISC function may increase when MTGR1 is lost.
Figure 2. MTGR1 loss deregulates intestinal stem cell programs and promotes transit amplifying identity.
(A) Immunofluorescent staining for Ki67 (red), E-cadherin (green), and nuclei (DAPI, blue) in the small intestine of 8–12-week-old WT and Mtgr1−/− mice. n=4 mice per genotype, >20 high-powered fields (HPFs) er mouse. (B) Lgr5 mRNA expression was visualized in the WT and Mtgr1−/− small intestine by RNAscope. N = 3–4 mice per genotype, 12 HPFs per mouse. (C) Mtgr1−/− mice were intercrossed with the Lgr5-cre-EGFP reporter strain and Lgr5-EGFP was assessed by immunofluorescence. Quantification shows the number of GFP positive cells in each reporter positive crypt, per mouse. N = 9 mice per genotype. (D) qPCR of stem cell markers Lgr5, Myc, Ki67, Ascl2, and Olfm4 in intestinal crypt isolates from Mtgr1−/− and WT mice (n = 3–6 mice per genotype). Results were normalized to Gapdh and represented as fold change over WT expression. (E) Clusterin (Clu) mRNA expression was visualized in the WT and Mtgr1−/− small intestine by RNAscope. Quantification shows the percent of crypts with Clu expression per high powered field. n = 2 mice per genotype, > 20 HPFs per mouse. (F) Crypts were collected from WT and Mtgr1−/− mice and assessed by RNA-sequencing. Gene set enrichment analysis was performed for gene sets associated with Lgr5+ stem cells (left) and transit amplifying cell (TA, left) populations. NES = normalized enrichment score. *P<0.05, **P<0.01, ****P<0.0001, Student’s t test (A-E), significance indicated by FDR q value (F). Scale bars = 100μm.
While Lgr5 is often regarded as the canonical identifier of CBCs and a robust ISC marker, we next assayed for expression of Ascl2 and Olfm4, which are highly expressed in CBCs along with Lgr540,41 (Fig. 2D). Interestingly, although Lgr5 transcript and Lgr5-EGFP + cells were consistently increased with MTGR1 loss, neither Ascl2 nor Olfm4 mirrored these changes. Here, we observed that Ascl2 transcript remained unchanged in Mtgr1−/− crypts, while Olfm4 was nearly undetectable in Mtgr1−/− crypts. Similarly, survey of markers associated with non-CBC ISC populations revealed further differences, such as greater numbers of Clusterin (Clu)-expressing cells (Fig. 2E)6.
As cell identity and function are complex, we next isolated WT and Mtgr1−/− crypts and expanded our transcriptomic analysis of MTGR1-dependent changes in intestinal cell types through bulk RNA sequencing. Differential expression data was then analyzed by gene set enrichment analysis (GSEA), and results from WT and Mtgr1−/− crypts were compared to gene sets established from prototypic Lgr5-expressing ISCs (Fig. 2F and Supplemental Fig. 2)9. Despite increased expression of Lgr5 itself in the setting of MTGR1 loss, total canonical Lgr5 + ISC gene signatures were significantly de-enriched in crypts collected from Mtgr1−/− mice. Instead, Mtgr1−/− samples were enriched for genes which indicated an expansion of TA populations, which is also consistent with increased proliferation noted in Mtgr1−/− crypts (Fig. 2A). Taken together, these results indicate that while more cells in Mtgr1−/− crypts express Lgr5, these cells may instead resemble more differentiated TA cells, rather than true Lgr5 + ISCs.
MTGR1 is required for enteroid viability.
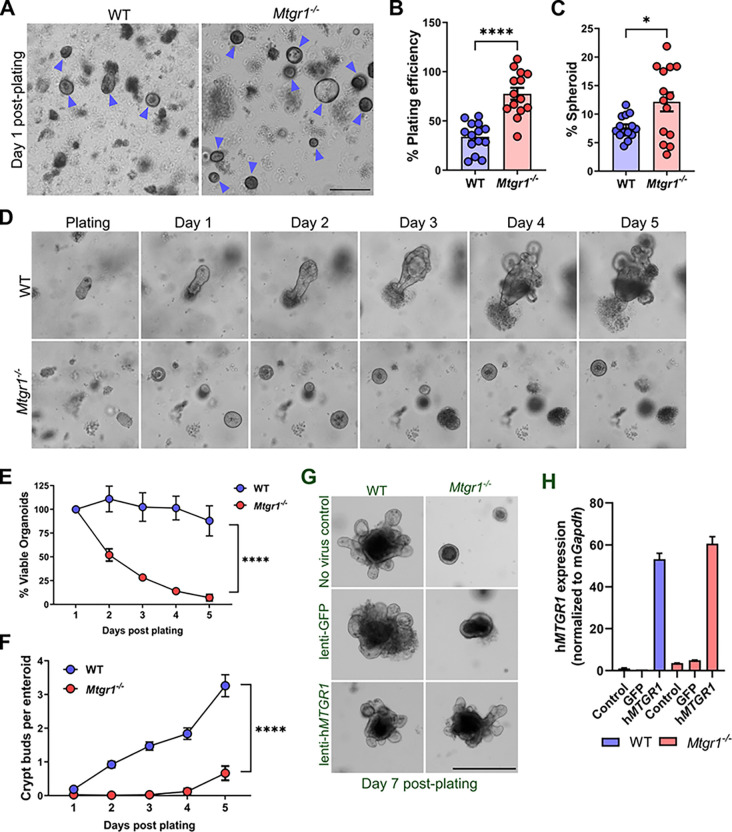
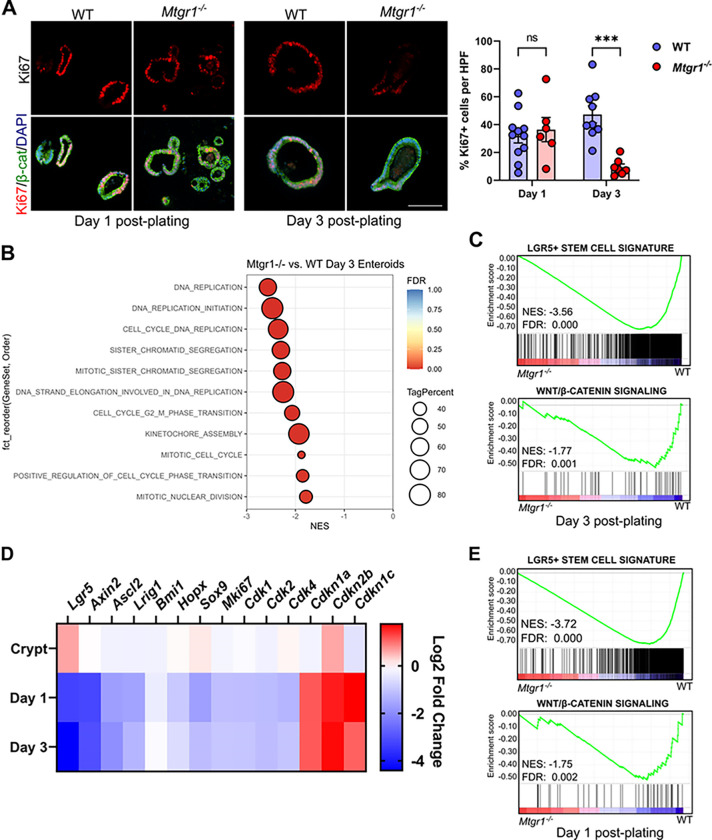
In our in vivo analysis, loss of MTGR1 appeared to deregulate ISC-associated genes and promote TA-associated differentiation. Thus, we questioned whether Mtgr1−/− ISCs were fully functional as multipotent intestinal stem cells. We next specifically tested whether MTGR1 loss affected overall ISC function using the small intestinal organoid or “enteroid” system. Since enteroids rely on ISCs for their establishment and growth, enteroid formation efficiency can be used to assess general stem cell function and fitness3. Here, enteroids were established from duodenal crypts harvested from WT and Mtgr1−/− mice, and enteroid formation efficiency was assessed after 24 hours in culture (Figs. 3A and 3B). By dividing the number of enteroids formed by the number of crypts plated, we noted an approximately 2-fold enhancement of enteroid formation in the setting of MTGR1 loss. We also observed higher percentages of Mtgr1−/− enteroids with a cystic, spheroid morphology (Fig. 3C), a phenotype associated with increased Wnt tone20 ,compared to WT enteroids. Taken together, these results suggest that MTGR1-deficient ISCs are indeed initially functionally competent, despite de-enrichment of a canonical Lgr5 transcriptional profile.
Figure 3. MTGR1 loss increases initial plating efficiency but MTGR1 is required for enteroid survival.
(A) Crypts were isolated from WT and Mtgr1−/− mice and plated as intestinal enteroids. Representative images of organoids at day 1 post-plating with enteroids marked by blue arrows. Scale bar = 200μm (left), representative of 4 independent experiments. (B) Quantification of overall plating efficiency (enteroids established divided by crypts plated) and (C) percentage of enteroids with cystic morphology calculated per well at day 1 post-plating. n = 14 wells per genotype. (B) 5-day timelapse imaging of WT and Mtgr1−/− enteroids. (E) Average enteroid viability post-plating, shown as the percent viable enteroids remaining from day 1. N ≥ 12 wells per genotype. (F) Quantification of crypt budding post-plating. n > 65 organoids per genotype for early timepoints (day 1–3), n > 5 for later timepoints (day 4–5). (G) WT and Mtgr1−/− crypts were transduced with lentiviral GFP or human MTGR1 and plated to allow organoid formation. Representative images at day 7 post-infection/plating. Scale bars = 500μm. Transduced enteroids were collected at day 7 post-infection for mRNA analysis of human MTGR1. Results were normalized to Gapdh and shown as fold change over WT non-transduced controls. *P<0.05, ****P<0.0001, Student’s t test (B-C), two-way ANOVA (E-F).
While initial enteroid formation was augmented in Mtgr1−/− cultures, we observed striking viability defects in Mtgr1−/− enteroids within 48 hours of initial plating. Daily imaging (Fig. 3D) and viable enteroid counts (Fig. 3E) revealed that Mtgr1−/− cultures failed almost completely by day 5 post-plating. While WT enteroids formed crypt buds by day 3, Mtgr1−/− enteroids rarely developed crypt buds, even in the structures that survived until day 5 (Fig. 3F). These findings were also confirmed by live cell imaging, which showed no morphological changes in Mtgr1−/− enteroids throughout the 5-day period, until organoid death (Supplemental videos SV1 and SV2). Importantly, restoration of MTGR1 expression via lentiviral transduction rescued Mtgr1−/− enteroids and restored branching morphology, confirming the MTGR1 dependency of this phenotype (Fig. 3G and 3H). Thus, MTGR1 appears to be required for ex vivo enteroid survival and expansion.
Inhibition of programmed cell death does not rescue Mtgr1 −/− viability.
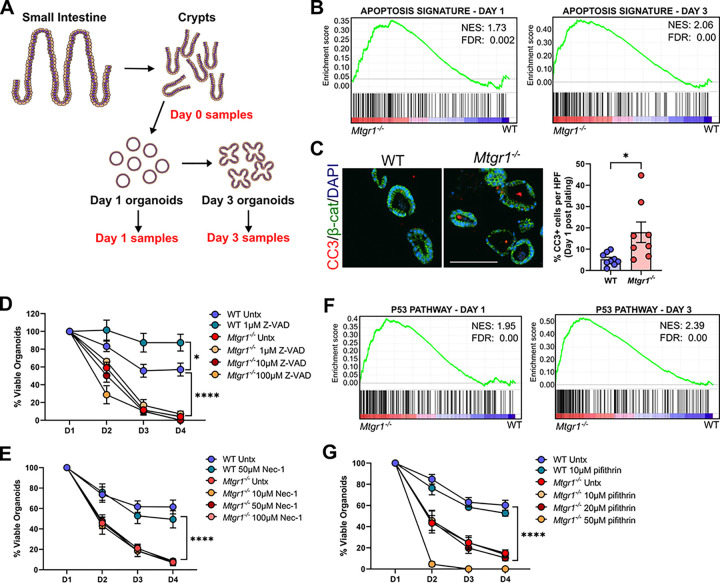
We next aimed to determine the mechanism driving the viability loss in Mtgr1−/− enteroids. As MTGR1 is a transcriptional co-repressor, we again utilized a bulk RNA-sequencing approach to broadly investigate MTGR1-dependent changes in gene expression. Briefly, crypts were isolated from age-matched WT and Mtgr1−/− mice, and mRNA was collected at the time of crypt isolation (day 0), or at day 1 and day 3 post-plating to yield matched RNA sets of crypts, day 1 enteroids, and day 3 enteroids (Fig. 4A). After RNA-sequencing, differential expression profiles were generated and analyzed using GSEA27,28.
Figure 4. MTGR1 loss increases enteroid apoptosis, but effects cannot be rescued by apoptosis inhibition.
(A) Schematic of RNA-sequencing experiment of crypts and enteroids. n = 3 mice per genotype. (B) GSEA of “Hallmark” collection apoptosis-related genes in Mtgr1−/− enteroids at day 1 (left) and day 3 (right) post-plating. NES = normalized enrichment score. (C) Enteroids were fixed and embedded at day 1 post-plating, and apoptotic cells were marked by immunofluorescent staining against cleaved caspase-3 (CC3, red). β-catenin (green) and DAPI (blue) were used for co-staining. Quantification shown as percent CC3-positive cells per high powered field (HPF). Scale bar = 200μm. n = 8–9 HPFs per genotype. (D) Enteroids were plated and overlaid with media containing indicated concentrations of the caspase inhibitor, Z-VAD-FMK, or (E) the necrosis inhibitor, necrostatin. Enteroids were counted daily and normalized to day 1 numbers. n ≥ 6 wells per condition. (F) GSEA of “Hallmark” collection p53 pathway-related genes in Mtgr1−/− enteroids at day 1 (left) and day 3 (right) post-plating. (G) Enteroids were plated and overlaid with media containing the p53 inhibitor, pifithrin, as indicated. n ≥ 3 wells per condition. *P<0.05, ****P<0.0001, Student’s t test (C) or two-way ANOVA (D, E, G), significance indicated by FDR q value (B, F).
Due to the rapid loss of established cultures, we hypothesized that MTGR1 loss may aberrantly activate programed cell death pathways to drive the observed decline in enteroid viability. Indeed, GSEA analysis from the Hallmark gene set collection identified a significant enrichment in apoptosis-associated genes in Mtgr1−/− enteroids at both day 1 and day 3 post-plating (Fig. 4B). Mtgr1−/− enteroids collected at day 1 post-plating also displayed higher numbers of apoptotic cells as compared to WT enteroids (Fig. 4C) as measured by fluorescent immunohistochemistry (IHC) against cleaved caspase-3. However, inhibiting apoptosis using the cell-permeable pan-caspase inhibitor, Z-VAD-FMK, failed to improve survival of Mtgr1−/− enteroids (Fig. 4D), even at concentrations which improved viability in WT cultures42. Likewise, inhibition of necroptosis, whose dysregulation has been noted in intestinal inflammatory diseases43–45, had no effect on Mtgr1−/− enteroid viability (Fig. 4E). Finally, we assessed the impact of p53 inhibition, as p53-related gene sets were also positively enriched in Mtgr1−/− samples by GSEA (Fig. 4F). As with Z-VAD-FMK, inhibition of p53-dependent apoptosis with pifithrin-α had no effect on Mtgr1−/− enteroid survival (Fig. 4G)46. Thus, despite increases in apoptosis, inhibition of known cell death mechanisms is insufficient to rescue Mtgr1−/− enteroid viability.
Proliferation and ISCs are lost in MTGR1-deficient enteroids.
Due to constant cell clearance, actively cycling stem cells and high levels of proliferation are necessary to maintain intestinal cell populations47. Thus, rather than aberrant apoptosis, we next hypothesized that the viability defect in Mtgr1−/− enteroids may instead be due to reduced proliferation and/or depletion of ISCs. To determine cell proliferation, sections from enteroids embedded at day 1 and day 3 post-plating were assessed by Ki67 IHC (Fig. 5A). Although we observed similar numbers of proliferating cells in day 1 enteroids, by day 3, the enteroid cultures established from Mtgr1−/− mice displayed a drastic, nearly 80% reduction in Ki67 + cells. Cell cycle- and proliferation-associated genes were also highly downregulated in Mtgr1−/− enteroids by day 3 (Fig. 5B), as well as ISC-associated genes and signaling pathways (Fig. 5C), as determined by GSEA. Interestingly, while numbers of Ki67 + cells were similar between WT and Mtgr1−/− enteroids at day 1 post-plating, proliferation-, ISC-, and Wnt-associated genes were still significantly downregulated at this early timepoint (Fig. 5D, 5E, and Supplemental Table S3). Mtgr1−/− enteroids, at either day 1 or day 3 post-plating, also demonstrated significant upregulation of the cell cycle inhibitors Cdkn1a, Cdkn1c, Cdkn2b. These results indicate that viability defects in Mtgr1−/− enteroids may arise from proliferation defects and the inability to maintain cycling ISC populations ex vivo.
Figure 5. MTGR1 loss decreases proliferation and stem cell-associated gene expression during organoid maturation.
(A) WT and Mtgr1−/− enteroids were fixed at day 1 and day 3 post-plating and proliferative cells were marked via immunohistochemistry for Ki67 (red). β-catenin (green) and DAPI (blue) were used for co-staining. Quantification shown as percent Ki67-positive cells per high powered field (HPF). Scale bar = 200μm. n≥7 fields per genotype per timepoint. (B) Gene set enrichment analysis (GSEA) of day 3 RNA-sequencing results using cell cycle-related gene sets queried from the Gene Ontology collection. NES = normalized enrichment score. Tag % = the percentage of gene hits before (for positive ES) or after (for negative ES) the peak in the running ES, indicating the percentage of genes contributing to the ES. (C) GSEA of day 3 RNA-sequencing results with intestinal stem cell- (top) and Wnt-associated (bottom) gene sets. (D) Heatmap of RNA-sequencing results of stem cell, cyclin dependent kinases, and cyclin-dependent kinase inhibitors from crypt, day 1, and day 3 results. Represented as the Log2 fold change of Mtgr1−/− results as compared to WT at that timepoint. (E) GSEA of day 1 RNA-sequencing results with intestinal stem cell- (top) and Wnt-associated (bottom) gene sets. ns = nonsignificant, ***P<0.001, Student’s t test (A), significance indicated by FDR q value (B, C, E).
MTGR1 loss promotes absorptive enterocyte differentiation.
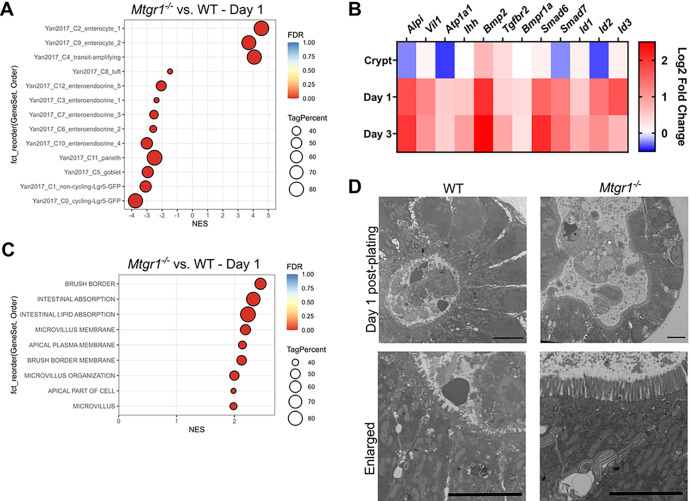
After expansion in the TA zone and exit from the intestinal crypt, most ISC-derived cells rapidly undergo differentiation into non-proliferative cell lineages48. As stem and proliferative cell populations are not maintained in Mtgr1−/− enteroids, and their loss appears unlikely due to programed cell death, we hypothesized that the loss of enteroid viability may be due to augmented ISC differentiation into non-proliferative cells. Indeed, this would result in failed ISC amplification, inability to maintain enteroid cultures, and eventual death of terminally differentiated cells. Therefore, we next broadly surveyed differentiated intestinal cell types using GSEA. In agreement with the data presented in Fig. 5 and the results from intestinal crypts (Fig. 2F), cycling Lgr5 + cells again appeared to be greatly depleted, and were the most reduced cell population in Mtgr1−/− enteroids at both day 1 and day 3 post-plating (Fig. 6A and Supplemental Fig. 3). Conversely, more differentiated TA populations were greatly increased in Mtgr1−/− enteroids, as were fully differentiated populations of enterocytes.
Figure 6. MTGR1 loss promotes absorptive enterocyte differentiation.
(A) Gene set enrichment analysis (GSEA) of Mtgr1−/− day 1 organoid RNA-sequencing results using gene sets representing intestinal epithelial cell types. NES = normalized enrichment score. Tag % = the percentage of gene hits before (for positive ES) or after (for negative ES) the peak in the running ES, indicating the percentage of genes contributing to the ES. Significance indicated by FDR q value. (B) Heatmap of RNA-sequencing results of enterocyte- and differentiation-associated genes in crypts, day 1 enteroids, and day 3 enteroids. Represented as the log2 fold change of Mtgr1−/− results as compared to WT at that timepoint. (C) GSEA of day 1 Mtgr1−/− RNA-sequencing results using gene sets representing microvilli and brush border biology queried from the Gene Ontology collection. (D) Representative electron microscopy images from WT and Mtgr1−/− day 1 enteroid samples. Scale bar = 5μm.
To more clearly define whether MTGR1 loss indeed accelerates intestinal differentiation, we next determined the expression of genes associated with villi and enterocyte differentiation pathways. We first surveyed classical markers of the enterocyte lineage, such as intestinal alkaline phosphatase (Alpi), as well as genes associated with BMP and IHH pathways, as these work in opposition to Wnt-mediated signaling in order to promote differentiation3. In nearly all cases, these differentiation-associated genes were significantly enriched in Mtgr1−/− enteroids as compared to matched WT samples (Fig. 6B and Supplemental Table S4). GSEA analysis also determined significant enrichment of genes associated with features of absorptive enterocytes, such as the brush border, microvilli, and intestinal absorption (Fig. 6C). Finally, the cellular structure of day 1 enteroids was investigated using transmission electron microscopy (TEM). Comparison of WT and Mtgr1−/− enteroids illustrates an expansion of the apical cell surface as well as more pronounced and mature microvilli (Fig. 6D). Altogether, these results indicate that Mtgr1−/− ISCs are likely further differentiated at baseline than WT ISCs, and upon ex vivo culture, differentiation is so accelerated that proliferative cells are lost entirely to the absorptive enterocyte lineage, to the point that cultures cannot be maintained.
Secretory cells promote the survival of Mtgr1 −/− enteroids.
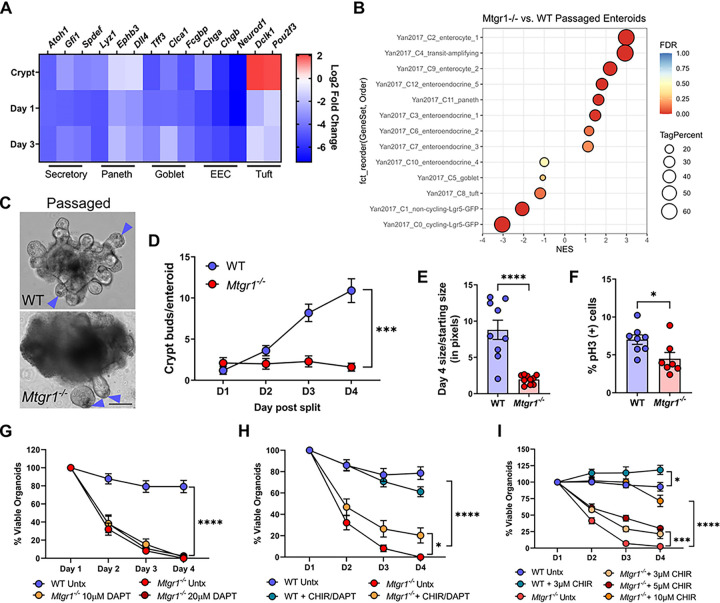
Previous research has indicated that MTGR1 is necessary for the differentiation of multiple secretory lineages in the small intestine, including Paneth, goblet, and enteroendocrine cells (EECs)15,18. Thus, we next confirmed the loss of these cell types in Mtgr1−/− crypts and enteroid cultures. A query of RNA-seq results indeed showed downregulation of genes associated with general secretory cell differentiation and those enriched in the Paneth, goblet, and EEC lineages (Fig. 7A, Supplemental Fig. 2, and Supplemental Table S5). Interestingly, loss of MTGR1 had the opposite effect on the tuft cell lineage in vivo, which was increased in the intestinal crypts; however, this same cell population appeared to be downregulated in Mtgr1−/− enteroids. Together, these results further define the variable effect of MTGR1 on intestinal cell differentiation.
Figure 7. High-WNT, but not secretory differentiation, rescues Mtgr1 null enteroids.
(A) Heatmap of RNA-sequencing results of secretory-associated genes from crypts, day 1 enteroids, and day 3 enteroids. Results represented as the log2 fold change of Mtgr1−/− samples as compared to WT at that timepoint. (B) Gene set enrichment analysis (GSEA) of passaged Mtgr1−/− enteroids using gene sets representing intestinal epithelial cell types. NES = normalized enrichment score. Tag % = the percentage of gene hits before (for positive ES) or after (for negative ES) the peak in the running ES, indicating the percentage of genes contributing to the ES. Significance indicated by FDR q value. (C)Representative images showing passaged WT enteroids and Mtgr1−/− enteroids. Passaged WT and Mtgr1−/− enteroids both had discernible Paneth cells in the crypt base (arrows). Scale bar = 200μm. (D) Quantification of crypt buds per Passaged enteroids post-split, n=10 enteroids per genotype. (E) Passaged enteroids were imaged at day 1 and day 4 post-passage and enteroid area measured via ImageJ. Change in size was calculated by dividing day 4 measurements by those taken at day 1. n = 10 enteroids per genotype (F) Passaged Mtgr1−/− enteroids were fixed and stained with phospho-histone H3 (pH3) to mark proliferative cells. Quantification shown as percent pH3-positive cells per high powered field (HPF). n ≥ 7 fields per genotype. (G). Enteroids were plated and overlaid with media containing indicated concentrations of the gamma secretase inhibitor, DAPT. Enteroids were counted daily and normalized to day 1 numbers. n = 7–8 wells per condition. (H) WT and Mtgr1−/− enteroids were plated and supplemented with 3μM of CHIR-99021 (CHIR) and 10μM DAPT. Enteroids were counted daily and normalized to day 1 numbers. n = 8 wells per condition. (I) WT and Mtgr1−/− enteroids were plated with CHIR as indicated. Enteroid numbers were assessed daily and normalized to day 1 results. n ≥ 4 wells per condition. *P<0.05, ***P<0.001, ****P<0.0001, significance indicated by FDR q value (A, B), two-way ANOVA (D, G, H, I), and Student’s t test (E, F).
Secretory lineages, specifically Paneth cells, are crucial regulators of CBCs and provide Wnt ligands that maintain ISC stemness, and crypts from mice lacking Paneth cells cannot form enteroid cultures without Wnt supplementation49–52. Thus, the loss of Mtgr1−/− CBCs, and by extension enteroid cultures, may be due to the lack of Paneth cells. While the majority of enteroids from Mtgr1−/− mice died by day 5, a small number of surviving enteroids occasionally could be maintained and passaged. We hypothesized that these few “passaged” Mtgr1−/− enteroids may provide clarity regarding mechanisms that could rescue MTGR1-dependent growth defects. To this end, we established and analyzed two independent Mtgr1−/− enteroid lines by RNA-sequencing to determine how these surviving cells differed from Mtgr1−/− cultures that subsequently died. Cell type analysis via GSEA found no deficiencies in Paneth or EEC populations in passaged Mtgr1−/− enteroids in comparison to WT enteroids (Fig. 7B), and Paneth cells could often be distinguished in the crypt base (Fig. 7C). Therefore, Paneth cells may drive the survival of passaged Mtgr1−/− cultures. However, despite the presence of Paneth cells, passaged Mtgr1−/− enteroids still displayed striking alterations in morphology, an inability to form enteroid buds (Fig. 7D), and little expansion in size over time (Fig. 7E). As observed in early enteroids at day 1 and day 3 post-plating, passaged Mtgr1−/− enteroids also showed significant reductions in ISC populations by GSEA (Fig. 7B) and proliferation by immunofluorescent IHC against the proliferative marker phospho-histone H3 (pH3, Fig. 7F). Thus, while Paneth cells likely aid the survival of passaged Mtgr1−/− enteroids, these results suggest that the presence of Paneth cells alone may not be sufficient to rescue growth, morphology, or ISC populations in the setting of MTGR1 loss.
Mtgr1 −/− enteroids can be rescued by high Wnt activity.
As our investigation of passaged Mtgr1−/− enteroid lines showed that Paneth cells may assist ISC survival in the context of MTGR1 loss, we next employed various combinations of small molecules to promote secretory cell differentiation and survival of Mtgr1−/− ISCs. First, we utilized DAPT, a γ-secretase inhibitor that has been shown to increase secretory cell numbers ex vivo at the expense of absorptive lineages. However, γ-secretase inhibitor treatment failed to rescue Mtgr1−/− enteroid growth ex vivo (Fig. 7G), even when treatment was begun in vivo prior to and continuing through enteroid establishment (data not shown). Next, we combined DAPT with the Wnt pathway agonist, CHIR 99021, as this combination should greatly promote Paneth cell differentiation53. These studies revealed only a modest induction of enteroid survival (Fig. 7H). Finally, we investigated whether Wnt pathway activation alone was sufficient to maintain Mtgr1−/− ISCs. Surprisingly, low-dose CHIR 99021 treatment (3μM), a concentration sufficient to promote WT ISCs and mimic the effects of recombinant Wnt3a, only had a modest effect on Mtgr1−/− enteroid survival (Fig. 7I)54. This could be overcome by increasing the concentration of CHIR 99021 (10 μM) to sustain survival and growth of Mtgr1−/− ISCs ex vivo. All together, these results indicate that while MTGR1 loss increases cell proliferation and Lgr5 expression, the overall functionality and differentiation state of Mtgr1−/− ISCs is severely compromised. While previously regarded as a transcription factor regulating secretory/absorptive lineage specification, these data indicate that MTGR1 is necessary for maintaining overall dedifferentiation and stem cell status in the small intestine.
Discussion
In this study, we establish that MTGR1 is widely expressed in the intestine and is required for the proper function of ISCs. While MTGR1 loss led to increased proliferation and Lgr5 + stem cell numbers, a prototypic Lgr5 + cell transcriptional profile was absent. Instead, our data indicate that Mtgr1−/− ISCs were more fully differentiated and may not wholly function as bona-fide stem cells. In support of this, expression of Clusterin, an ISC-associated gene normally indicative of injury response and loss of Lgr5 + CBCs, was increased, as were genes associated with the small intestinal TA compartment. Cells from MTGR1-deficient crypts also demonstrated defective stem cell faculty, as they failed to proliferate in ex vivo enteroid cultures in association with increased apoptosis, cell cycle arrest, aberrant absorptive lineage differentiation, and loss of Lgr5 + ISCs as noted by transcriptomic profiling. Indeed, survival of Mtgr1−/− enteroids was possible through high Wnt pathway activity, indicating that strong stem cell-promoting signals correct the intrinsic defects of MTGR1 deficiency. Thus, MTGR1 loss significantly impacted survival and differentiation of multiple intestinal cell types, identifying a novel and crucial role of this gene in ISC function.
MTGR1 belongs to the Myeloid Translocation Gene family, member of which have been widely implicated in stem cell maintenance and lineage commitment in the hematopoietic system and beyond. In the homeostatic intestine, Mtg16−/− and Mtgr1−/− mice both display increased proliferation and altered secretory lineage allocation15,16,18,39,55,56. However, while the MTG family members share significant sequence homology and generally similar phenotypes, they can also exhibit diverse effects in the gut, particularly in specific cell types. For example, while loss of MTGR1 depleted nearly all intestinal secretory cells, loss of MTG16 appeared to regulate more subtle cell fate decisions between goblet cells and EECs. Meanwhile, ISC-specific roles have been investigated for some, but not all MTG family members. In the small intestine, it has recently been reported that MTG16 and MTG8 are both enriched in + 4/+5 cells, where they repress ISC-specific genes to control exit from the stem cell niche16. On the other hand, enrichment of MTGR1 was not observed in ISCs, which instead was broadly expressed throughout the intestinal crypt. Both Mtg8 and Mtg16 were also reported to be negatively regulated by the ISC-associated Notch signaling pathway; however, Notch activity did not appear to similarly regulate Mtgr116. Finally, unlike the results described herein from Mtgr1−/− crypts, neither loss of Mtg8 nor Mtg16 was incompatible with enteroid culture and ISC maintenance ex vivo16,55,57. Thus, each member of the MTG family appears to play crucial, distinct roles in intestinal biology and differentiation. Indeed, our data suggests Mtgr1 is uniquely required among its family members to maintain small intestinal ISC function.
While single genes may contribute to ISC function, our results also underscore the complexity of various stem cell populations and caution against the use of single genetic markers as an indicator of cell abundance or function. Nor, as others have pointed out, does the expression of ISC markers truly indicate facultative stem cell function, as not all cells positive for ISC markers will demonstrate lineage tracing even in the setting of injury58. Here, while Lgr5 + cells were increased as measured by qRT-PCR, RNAscope, and intercross with Lgr5-EGFP-IRES-creERT2 mice, 58we observed functional Lgr5 + stem cells to be greatly depleted in the setting of Mtgr1 loss, by expanding ISC analysis to include a larger and more complex genetic signature. Furthermore, despite heightened expression of Lgr5, these cells were not able to maintain self-renewal ex vivo and rapidly differentiated into absorptive cells. Thus, true ISC function may be vastly more complicated than implied by their collection of ISC markers, and how the interplay of multiple proteins and signaling pathways contributes to stem cell function remains to be fully understood.
Interestingly, MTGR1 is well-poised to act as a master regulator of intestinal differentiation and lineage allocation. Although MTGs lack enzymatic activity and DNA-binding capability, they act as scaffolds facilitating transcription factor and epigenetic modifier complex assembly. As such, they associate with basic helix-loop-helix (bHLH) transcription factors, including tissue- and cell type-specific bHLH transcription factors and E proteins (E2A, HEB, and E2–2), which coordinate hematopoietic differentiation with cell cycle exit to repress their transcriptional activity. Importantly, genetic specificity is imparted via interactions with transcription factors bound to specific enhancers. Similar to hematopoietic stem cell and differentiation, transcriptional networks also regulate intestinal programs. Of note, studies to date have implicated MTGR1 in two such transcriptional networks known as key regulators of ISC identity and function: Wnt and Notch3. Indeed, our previous research has determined that MTGR1 can compete with β-catenin for TCF4 occupancy and, in doing so, suppresses Wnt transcriptional targets19. MTGR1 can also suppress Notch targets via interactions with CSL, a key Notch effector18. Despite the ability of MTGR1 to repress ISC-related signaling pathways, which one may expect to augment ISC fitness upon MTGR1 loss, we instead noted that MTGR1 deficiency clearly abrogates ISC function. Thus, further investigation of the function of MTGR1 in ISC-related signaling cascades and identification of bona-fide MTGR1 genomic targets, using methodology such as CUT&RUN, ATAC-seq, and scRNA-seq, will be critical in furthering our understanding of how MTGR1 contributes to crypt and intestinal biology.
In conclusion, our studies more fully elucidate the functional contributions of MTGR1 to small intestinal homeostasis and have identified a novel role for MTGR1 in maintaining Lgr5 + CBC populations and the dedifferentiated state. While loss of MTGR1 increased Lgr5 expression and LGR5 + cell numbers, Mtgr1−/− ISCs displayed widespread dysregulation of ISC programs and were functionally deficient as compared to WT ISCs. Instead, Mtgr1−/− ISCs were primed for cell cycle exit and underwent rapid aberrant differentiation into absorptive enterocytes. Together, these findings indicate that MTGR1 is required for stem cell maintenance in the intestinal epithelium.
Acknowledgements:
We thank past and present members of the Williams and Hiebert Labs for their critical insights into this manuscript. We would also like to thank the VUMC Translational Pathology Shared Resource (supported by NCI/NIH Cancer Center Support Grant P30CA068485), the Vanderbilt Technologies for Advanced Genomics (VANTAGE), and Vanderbilt Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, DK59637 and EY08126) for assistance with experiments and analysis.
Grant Support:
Financial support includes National Institutes of Health (R01DK099204 to CSW, R01CA255446 to SWH, K01DK123495 to SPS, F30DK120149 to REB, F31CA232272 to JMP, F30DK096718 to BP, R01DK103831 to KSL); Office of Medical Research, Department of Veterans Affairs (1I01BX001426 to CSW); Crohn’s and Colitis Foundation (623541 to CSW and 662877 to SPS); the Royal Netherlands Academy of Arts and Sciences (Academy Ter Meulen Grant to J.J); and the Prince Bernhard Cultural Foundation (Cultural Foundation Grant to J.J.)
Footnotes
Disclosures: The authors declare no competing interest.
Contributor Information
Christopher Williams, Vanderbilt University Medical Center.
Rachel Brown, Vanderbilt University.
Yue Zhao, Vanderbilt University.
Jing Wang, Vanderbilt University Medical Center.
Zhengyi Chen, Vanderbilt University.
Koral Blunt, Vanderbilt University.
Jennifer Pilat, Vanderbilt University.
Bobak Parang, Vanderbilt University.
Yash Choksi, Vanderbilt University Medical Center.
Ken Lau, Vanderbilt University.
Scott Hiebert, Vanderbilt University.
Sarah Short, University of Iowa.
Justin Jacobse, Vanderbilt.
Yanwen Xu, Vanderbilt.
Yilin Yang, Vanderbilt.
Jeremy Goettel, Vanderbilt.
References
- 1.Barker N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007, doi: 10.1038/nature06196 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bankaitis E. D., Ha A., Kuo C. J. & Magness S. T. Reserve Stem Cells in Intestinal Homeostasis and Injury. Gastroenterology 155, 1348–1361, doi: 10.1053/j.gastro.2018.08.016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beumer J. & Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 143, 3639–3649, doi: 10.1242/dev.133132 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ruffner H. et al. R-Spondin potentiates Wnt/beta-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One 7, e40976, doi: 10.1371/journal.pone.0040976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vega P. N., Lau K. S. & Goldenring J. R. Not All Insults Are Created Equal for Awakening Dormant Stem Cell Abilities. Cell Mol Gastroenterol Hepatol 7, 619–621, doi: 10.1016/j.jcmgh.2018.12.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayyaz A. et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125, doi: 10.1038/s41586-019-1154-y (2019). [DOI] [PubMed] [Google Scholar]
- 7.Davidson L. A. et al. Alteration of colonic stem cell gene signatures during the regenerative response to injury. Biochim Biophys Acta 1822, 1600–1607, doi: 10.1016/j.bbadis.2012.06.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan K. S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A 109, 466–471, doi: 10.1073/pnas.1118857109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan K. S. et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell 21, 78–90 e76, doi: 10.1016/j.stem.2017.06.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tetteh P. W. et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell 18, 203–213, doi: 10.1016/j.stem.2016.01.001 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Calabi F. & Cilli V. CBFA2T1, a gene rearranged in human leukemia, is a member of a multigene family. Genomics 52, 332–341, doi: 10.1006/geno.1998.5429 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Rossetti S., Hoogeveen A. T. & Sacchi N. The MTG proteins: chromatin repression players with a passion for networking. Genomics 84, 1–9, doi: 10.1016/j.ygeno.2004.02.011 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Calabi F., Pannell R. & Pavloska G. Gene targeting reveals a crucial role for MTG8 in the gut. Mol Cell Biol 21, 5658–5666, doi: 10.1128/MCB.21.16.5658-5666.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonough E. M. et al. MTG16 is a tumor suppressor in colitis-associated carcinoma. JCI Insight 2, doi: 10.1172/jci.insight.78210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amann J. M. et al. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Mol Cell Biol 25, 9576–9585, doi: 10.1128/MCB.25.21.9576-9585.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baulies A. et al. The Transcription Co-Repressors MTG8 and MTG16 Regulate Exit of Intestinal Stem Cells From Their Niche and Differentiation Into Enterocyte vs Secretory Lineages. Gastroenterology 159, 1328–1341 e1323, doi: 10.1053/j.gastro.2020.06.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez J. A. et al. Deletion of Mtgr1 sensitizes the colonic epithelium to dextran sodium sulfate-induced colitis. Gastroenterology 131, 579–588, doi: 10.1053/j.gastro.2006.06.009 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Parang B. et al. The transcriptional corepressor MTGR1 regulates intestinal secretory lineage allocation. FASEB J 29, 786–795, doi: 10.1096/fj.14-254284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore A. C. et al. Myeloid translocation gene family members associate with T-cell factors (TCFs) and influence TCF-dependent transcription. Mol Cell Biol 28, 977–987, doi: 10.1128/MCB.01242-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy V. K. et al. BVES Regulates Intestinal Stem Cell Programs and Intestinal Crypt Viability after Radiation. Stem Cells 34, 1626–1636, doi: 10.1002/stem.2307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heijmans J. et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep 3, 1128–1139, doi: 10.1016/j.celrep.2013.02.031 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Thompson J. J. et al. Blood vessel epicardial substance (BVES) reduces LRP6 receptor and cytoplasmic -catenin levels to modulate Wnt signaling and intestinal homeostasis. Carcinogenesis, doi: 10.1093/carcin/bgz007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3, doi: 10.14806/ej.17.1.200 (2011). [DOI] [Google Scholar]
- 24.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y., Smyth G. K. & Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Banerjee A. et al. Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 159, 2101–2115 e2105, doi: 10.1053/j.gastro.2020.08.029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simmons A. J. & Lau K. S. Dissociation and inDrops microfluidic encapsulation of human gut tissues for single-cell atlasing studies. STAR Protoc 3, 101570, doi: 10.1016/j.xpro.2022.101570 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201, doi: 10.1016/j.cell.2015.04.044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arceneaux D. et al. A contamination focused approach for optimizing the single-cell RNA-seq experiment. iScience 26, 107242, doi: 10.1016/j.isci.2023.107242 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southard-Smith A. N. et al. Dual indexed library design enables compatibility of in-Drop single-cell RNA-sequencing with exAMP chemistry sequencing platforms. BMC Genomics 21, 456, doi: 10.1186/s12864-020-06843-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B., Ramirez-Solano M. A., Heiser C. N., Liu Q. & Lau K. S. Processing single-cell RNA-seq data for dimension reduction-based analyses using open-source tools. STAR Protoc 2, 100450, doi: 10.1016/j.xpro.2021.100450 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heiser C. N., Wang V. M., Chen B., Hughey J. J. & Lau K. S. Automated quality control and cell identification of droplet-based single-cell data using dropkick. Genome Res 31, 1742–1752, doi: 10.1101/gr.271908.120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herring C. A. et al. Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut. Cell Syst 6, 37–51 e39, doi: 10.1016/j.cels.2017.10.012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsson M. et al. A single-cell type transcriptomics map of human tissues. Sci Adv 7, doi: 10.1126/sciadv.abh2169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y. et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med 217, doi: 10.1084/jem.20191130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farmer T. E., Williams C. S., Washington M. K. & Hiebert S. W. Inactivation of the p19(ARF) tumor suppressor affects intestinal epithelial cell proliferation and integrity. J Cell Biochem 104, 2228–2240, doi: 10.1002/jcb.21779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan K. S. & Kuo C. J. Ascl2 reinforces intestinal stem cell identity. Cell Stem Cell 16, 105–106, doi: 10.1016/j.stem.2015.01.014 (2015). [DOI] [PubMed] [Google Scholar]
- 41.van der Flier L. G., Haegebarth A., Stange D. E., van de Wetering M. & Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17, doi: 10.1053/j.gastro.2009.05.035 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Slee E. A. et al. Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem J 315 (Pt 1), 21–24, doi: 10.1042/bj3150021 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunther C. et al. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature 477, 335–339, doi: 10.1038/nature10400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welz P. S. et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 477, 330–334, doi: 10.1038/nature10273 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Werts A. D. et al. A Novel Role for Necroptosis in the Pathogenesis of Necrotizing Enterocolitis. Cell Mol Gastroenterol Hepatol 9, 403–423, doi: 10.1016/j.jcmgh.2019.11.002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Komarov P. G. et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285, 1733–1737, doi: 10.1126/science.285.5434.1733 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15, 19–33, doi: 10.1038/nrm3721 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Creamer B., Shorter R. G. & Bamforth J. The turnover and shedding of epithelial cells. I. The turnover in the gastro-intestinal tract. Gut 2, 110–118, doi: 10.1136/gut.2.2.110 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418, doi: 10.1038/nature09637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farin H. F., Van Es J. H. & Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529 e1517, doi: 10.1053/j.gastro.2012.08.031 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Durand A. et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A 109, 8965–8970, doi: 10.1073/pnas.1201652109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cray P., Sheahan B. J. & Dekaney C. M. Secretory Sorcery: Paneth Cell Control of Intestinal Repair and Homeostasis. Cell Mol Gastroenterol Hepatol 12, 1239–1250, doi: 10.1016/j.jcmgh.2021.06.006 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boonekamp K. E., Dayton T. L. & Clevers H. Intestinal organoids as tools for enriching and studying specific and rare cell types: advances and future directions. J Mol Cell Biol 12, 562–568, doi: 10.1093/jmcb/mjaa034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin X. et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods 11, 106–112, doi: 10.1038/nmeth.2737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poindexter S. V. et al. Transcriptional corepressor MTG16 regulates small intestinal crypt proliferation and crypt regeneration after radiation-induced injury. Am J Physiol Gastrointest Liver Physiol 308, G562–571, doi: 10.1152/ajpgi.00253.2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown R. E. et al. MTG16 regulates colonic epithelial differentiation, colitis, and tumorigenesis by repressing E protein transcription factors. JCI Insight 7, doi: 10.1172/jci.insight.153045 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Short S. P. et al. Kaiso is required for MTG16-dependent effects on colitis-associated carcinoma. Oncogene 38, 5091–5106, doi: 10.1038/s41388-019-0777-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson N. M. et al. Autophagic state prospectively identifies facultative stem cells in the intestinal epithelium. EMBO Rep 23, e55209, doi: 10.15252/embr.202255209 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]