Abstract
Exosomes are membrane-bound extracellular vesicles released following the fusion of multivesicular bodies (MVBs) with the cell membrane. Exosomes transport diverse molecules, including proteins, lipids, DNA and RNA, and regulate distant intercellular communication. Noncoding RNA (ncRNAs) carried by exosomes regulate cell-cell communication in tissues, including adipose tissue. This review summarizes the action mechanisms of ncRNAs carried by exosomes on adipocyte differentiation and modulation of adipogenesis by exosomal ncRNAs. This study aims to provide valuable insights for developing novel therapeutics.
Keywords: exosomes, ncRNA, adipogenesis, lipid metabolism, obesity-related metabolic diseases
1 Introduction
Non-coding RNAs (ncRNAs) are RNA molecules that do not encode proteins. ncRNAs act as regulators of gene expression at various levels, including the transcriptional, post-transcriptional and translational levels. ncRNAs include micro-RNAs (miRNAs), long-chain non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) (Kapranov et al., 2007). miRNAs are highly conserved small ncRNAs (Holoch and Moazed, 2015). On the other hand, lncRNAs are poorly conserved ncRNAs with a length above 200 nt. LncRNAs act as competitive endogenous RNA (ceRNA) by binding to complementary binding sites on miRNA, thus regulating target gene expression by binding to the 3′UTR region (He et al., 2020). circRNAs are highly conserved ncRNAs with a circular structure formed by covalent binding of the 3′ end and the 5′ end after back-splicing. circRNAs also regulate gene and protein expression by serving as miRNA and RNA-binding protein sponges (Aufiero et al., 2019). ncRNAs are associated with various diseases and have diagnostic and therapeutic value (Wang et al., 2021). Genomic DNA can be transcripted into coding RNA or non-coding RNA. Approximately two percent of the human genome is made up of protein-coding genes (Kaikkonen and Adelman, 2018). Non-coding RNAs are not translated into proteins (Adams et al., 2017). Non-coding RNAs can be divided into long-chain noncoding rNA (lncRNAs, >200 nucleotides), medium-chain ncRNAs (20-200 nucleotides), and short-chain ncRNAs (<20 nucleotides). Short-chain ncRNAs can be further divided into piwi-interacting RNA, small interfering RNAs (siRNAs), and miRNAs. Medium-chain ncRNAs include small-nuclear RNA involved in transcript splicing in protein synthesis, nucleolar RNA involved in ribosomal RNA modification, transcription start site (TSS)-related RNA, and promoter-related miRNA. miRNAs play an important regulatory role at different stages of lipid metabolism (Aryal et al., 2017).
Studies have demonstrated that lncRNAs regulate various physiological and pathological processes, including growth and development, hematopoietic processes, cell proliferation and apoptosis, tumorigenesis, cell metabolism, genomic imprinting, chromatin modification, infection, and immune response (Karreth et al., 2015; Wang et al., 2016; Kasagi et al., 2017). lncRNAs interact with the target protein-coding genes through a highly complicated RNA regulatory network. Mutations or abnormal expressions of lncRNAs are closely linked to various diseases.
Valadi et al. demonstrated that exosomes could transport miRNAs and mRNAs into other cells (Valadi et al., 2007). Furthermore, miRNAs, lncRNAs, and circRNAs were shown to be encapsulated and transported intercellularly. When exosome ncRNAs undergo tissue-specific changes due to stimulation from various internal and external factors, they can cause organ dysfunction, aging, and disease. Previous studies have shown that exosomal ncRNAs participate in the pathogenesis and development of infectious, autoimmune, metabolic, neurodegenerative, cardiovascular, and neurodegenerative diseases. Exosomal ncRNAs also show differential expression in different cells or under different physiological or pathological conditions. Therefore, exosome ncRNAs serve as potential diagnostic and therapeutic biomarkers.
On the other hand, different biological processes, such as differentiation, proliferation, apoptosis, metabolism, immune responses, and tumorigenesis, are influenced by miRNAs (Alvarez-Garcia and Miska, 2005; Schickel et al., 2008). Yan et al. (2016) showed differential expression of miRNAs between the plasma of patients with diabetes and controls. miRNAs are differentially expressed in different organs related to metabolism, including the liver, pancreatic islets, and adipose tissue. In recent years, significant advances have been made in understanding the regulation of adipocyte differentiation (Esau et al., 2004; Kajimoto et al., 2006; Cheung et al., 2008; Lin et al., 2009). Several protein-coding genes, mRNAs, and microRNAs are associated with lipid metabolism and adipocyte differentiation.
However, gene expression profiles and functions of lncRNAs during adipocyte differentiation remain to be fully analyzed. Sun et al. (2013) identified 175 lncRNAs which were significantly dysregulated during adipocyte differentiation by profiling transcriptomes of primary brown and white adipocytes, precursor adipocytes and mature adipocytes. They found that C/EBPα and PPARγ could bind to the promoter regions of many lncRNAs. It was also revealed that inhibition of ten lncRNAs by RNAi significantly inhibited lipid droplet formation. Few studies have investigated the expression profile of non-coding RNAs in human adipose-derived stem cells (hADSCs). The regulatory mechanisms and pathways of adipocyte differentiation by ncRNAs remain unstudied. A recent study revealed that adipose tissue-derived exosomes could regulate physiological signal transduction and metabolism in adipose tissue and other peripheral tissues (Wang et al., 2022). Adipose tissue-derived exosomes could contribute to the development of obesity and related metabolic syndromes. Adipose tissue can regulate the metabolic homeostasis of various tissues and organs by secreting exosomes (Mathieu et al., 2019). Furthermore, exosomes derived from various tissues can disproportionately affect lipid accumulation and metabolism in adipose tissue. Previous studies demonstrated that ncRNAs regulate cell differentiation, epigenetics, and cell cycle regulation. In addition, ncRNAs can modulate adipocyte differentiation and adipogenesis by targeting transcription factors and signaling molecules. In this review, we outline the exosome production pathways and the molecular regulatory mechanisms of ncRNAs carried by adipogenic exosomes. The review aims to provide valuable insights for treating metabolic diseases by targeting exosome production.
2 Exosomes in regulating adipocyte differentiation and lipid metabolism
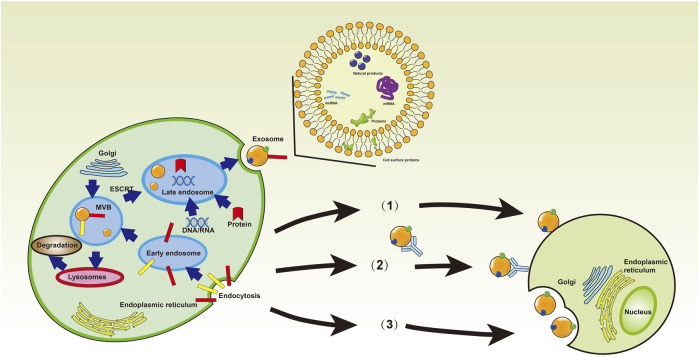
After fusion of intracellular multivesicular bodies (MVBs) with the cell membrane, exosomes are discharged into the extracellular environment (Simpson et al., 2009; Mathivanan et al., 2010). Exosomes are crucial to maintaining human health. Therefore, understanding their role in diseases can provide valuable insights for developing effective therapies. Recent studies have shown that exosomes regulate human diseases by regulating immune responses, oxidative stress, autophagy, gut microbiome, and cell cycle (Baixauli et al., 2014; Muller et al., 2016; Real et al., 2018; Anel et al., 2019; Elashiry et al., 2020; Xu et al., 2020; Liu et al., 2021; Shen et al., 2021; Wang et al., 2021; Xing et al., 2021; Jahangiri et al., 2022; Yan et al., 2023). Exosomes form through the inward budding of early endosomal membranes (Figure 1), which mature into multivesicular bodies (MVBs). Multivesicular bodies are involved in endocytosis and transport of cellular material (Borges et al., 2013; Bebelman et al., 2018). Multivesicular bodies have two possible fates, including fusion with lysosomes, resulting in acidification and degradation of their contents, and fusion with the cytoplasmic membrane, releasing the endoluminal vesicles outside the cell and eventually forming exosomes (Raposo and Stoorvogel, 2013). Understanding the yield and protein composition of exosomes will improve our understanding of the molecular mechanisms underlying their formation. The exosomal formation mechanism involves two pathways, including the ESCRT (endosomal sorting complex required for transport) dependent pathway and the ESCRT independent pathway (Figure 2) (Dreyer and Baur, 2016). The ESCRT-dependent pathway comprises four protein complexes (ESCRT-0, -I, -II, -III), which regulate exosome formation and transport, with ESCRT-0 mediating substrate recognition and sorting, ESCRT-I and ESCRT-II mediating inward budding of endosomal membranes, and ESCRT-III shearing the bud neck to form MVB (Figure 3) (Hurley and Hanson, 2010; Hanson and Cashikar, 2012; Zeng et al., 2018).
FIGURE 1.
The forming process and structure of the exosomes. Donor cells secrete exosomes into the extracellular space, and exosomes can carry various cargos and interact with recipient cells through endocytosis, or direct membrane fusion, or receptor-ligand interfaces [Reprinted by permission from John Wiley and Sons Ltd. Journal of cellular and molecular medicine, Xing et al. (2021), copyright 2021].
FIGURE 2.
Mechanisms for biogenesis and secretion of exosomes. Exosome-producing MVBs travel along the secretory pathway, move to the cell periphery, fuse with the plasma membrane, and release their ILVs into the extracellular environment. MVBs can interact dynamically with other organelles or compartments. The exosome formation from MVBs proceeds via ESCRT-dependent and ESCRT-independent pathways [Reprinted by permission from Macmillan Publishers Ltd. Nature Reviews Immunology, Robbins and Morelli (2014), copyright 2014].
FIGURE 3.
Global protein/ncRNA regulatory networks during brown/beige adipogenesis. Brown and beige adipocytes have different progenitors and are affected differently either by intrinsic factors or extrinsic factors that indirectly regulate key transcription factors [Reprinted by permission from Elsevier Inc. Advances in clinical chemistry, Zeng et al. (2018), copyright 2018].
Exosomes have been found in various body fluids, including blood, urine, cerebrospinal fluid, joint effusion, breast milk, and saliva (Nafar et al., 2022). Exosomes carry various molecules, including DNA, mRNA, non-coding RNA, lipids, cytokines, and proteins, and are crucial for cellular communication and information transfer (Mathieu et al., 2019). One previous study revealed that exosomes play an important role in modifying the extracellular microenvironment, immune regulation, homeostasis, and are incorporated into a variety of physiological and pathological processes (Raposo et al., 1996). Exosomes are mediators of intercellular communications as they deliver miRNA and other contents to recipient cells, and miRNA-containing exosomes can be absorbed by neighboring cells via endocytosis, thus influencing the phenotype of neighboring cells (Bruns et al., 2017). Exosomes can promote tumor progression and drug resistance in cancer by inhibiting the host immune response. Of note, exosomes released by tumor cells are implicated in resistance-associated secretory phenotype (RASP) by which immune escape is established. Heat shock proteins (HSPs) carried by exosomes can be co-transported with oncogenic factors to promote tumor development and escape to the immune system resulting in immunostimulatory effects (Taha et al., 2019). On the other hand, exosomes derived from platelets activated by thrombin may promote the survival, multiplication, and chemotaxis of hematopoietic cells as well as the release of pro-inflammatory cytokines (such as myeloperoxidase and superoxide dismutase) from monocytes, thus triggering local and systemic inflammation (Yasaka et al., 1981). Exosomes can be used as drug delivery vehicles as they are non-immunogenic, have good biocompatibility, and have efficient transport (Kim et al., 2018; Hou et al., 2019).
Exosomes have different biological characteristics and may exert different effects depending on their contents, tissue micro-environment, and recipient cells. Mature adipocyte-derived exosomes show a greater capacity for adipocyte differentiation than adipose stem cell-derived exosomes. Furthermore, adipocyte-derived exosomes can promote the differentiation of adipose-derived stem cells (ADSCs) (Dai et al., 2017). The repair and regenerative functions of adipose stem cell-derived exosomes mainly rely on the delivery of self-packaged proteins, lipids, mRNAs and miRNAs, and the required nutrients and inflammatory mediators (Cabral et al., 2018). Adipocyte-derived exosomes can promote insulin resistance and type II diabetes by affecting immune cells through paracrine pathways (Deng et al., 2009; Connolly et al., 2015; Ying et al., 2017). Studies have demonstrated that adipocyte-derived exosomes can influence foam cell formation and polarization, resulting in atherosclerosis (Kranendonk et al., 2014; Li X et al., 2019).
2.1 Negative regulation of adipocyte differentiation by exosomes
Wang et al. (2017) revealed that tumor-derived exosomes decrease the formation of lipid droplets and lower the mRNA expression of the adipogenic transcription factor, peroxisome proliferator-activated receptor γ (PPARγ, an adipocyte-specific marker), and lipoprotein lipase. They also demonstrated that pulmonary tumor-derived exosomes suppress adipogenic differentiation of ADSCs via the TGFβ signaling pathway.
2.2 Positive regulation of adipocyte differentiation by exosomes
Exosomes can induce their differentiation into mature adipocytes and cause lipid deposition (Wan et al., 2019). Adipocyte-derived exosomes (Ad-Exo) contain miRNA, RNA, and proteins that determine the lineage of human bone marrow mesenchymal stem cells (hMSCs). Ad-Exo can enhance ECM-induced differentiation of hMSCs to adipocytes. Narayanan revealed that the combination of osteoblast/adipocyte ECM and exosomes induced the expression of lineage-specific genes at the early differentiation stage. Their findings also showed that hMSCs differentiated on ECM of osteoblasts with adipogenic exosomes could express adipogenic genes (Narayanan et al., 2018).
Interfering ADSCs with adipose tissue-derived exosomes caused upregulated expression of PPARγ, adipocyte-specific fatty acid binding protein 2 (aP2), and lipocalin and an increase in the number of mature cells (Dai et al., 2017). However, exosomes released by lipopolysaccharide-activated macrophages did not affect the final maturation and differentiation of preadipocytes and fat storage (De Silva et al., 2018).
2.3 Regulation of lipid metabolism by adipose-derived exosomes
ADSCs-derived exosomes activate M2 macrophage polarization, express high levels of tyrosine hydroxylase, and enhance the expression of related thermogenic genes through a signal transduction and transcriptional activator 3-related pathway. Thus, they promote white adipose tissue browning, restore uncoupling protein 1 (UCP1)-dependent energy expenditure and ultimately reduce the effects of insulin resistance, dyslipidemia and hepatic steatosis caused by obesity (Zhao et al., 2018). A high expression of miRNA-155 in adipose tissue macrophages collected from obese mice effectively inhibit white fat browning and thermogenesis, reduce brown adipose tissue, as well as inhibits its function (Chen et al., 2013; Zhang et al., 2019).
A previous study showed a higher expression of Ad-Exo, in obese mice compared with non-obese mice (Lazar et al., 2016). Ad-Exo promotes fatty acid oxidation. Ad-Exo can increase the lipid content in adipose tissue macrophages (ATMs), and deliver triglycerides to macrophages, resulting in lipid accumulation in macrophages. Ad-Exo can induce the expression of lipid metabolism-related genes (Flaherty et al., 2019). Furthermore, mature adipocyte-derived exosomes are necessary for adipogenesis and rely on transient receptor potential mucolipin 1 (TRPML1) mediated lysosomal cytokinesis. Mature adipocyte-derived exosomes stimulate adipogenesis in a paracrine and autocrine manner. The expression of endogenous TRPML1 increased during the maturation and differentiation of preadipocytes. Notably, lipid synthesis reduced in the absence of TRPML1 (Kim et al., 2019).
In patients with obesity-related cardiovascular disease, Zhang found increased levels of cystatin C and CD14 (Zhang et al., 2016). Exosomes with high concentrations of cystatin C and CD14 have been linked to a higher risk of myocardial infarction and mortality. The expression of cystatin C in plasma exosomes was also positively associated with low-grade inflammatory response, low HDL-cholesterol levels and metabolic syndrome. Contrarily, a negative correlation was noted between the expression of CD14 and adipose tissue abundance, dyslipidemia, and a lower risk of type 2 diabetes (Zhang et al., 2016). Exosomes contribute to the onset of diabetes and its associated complications. Therefore, exosomes can act as novel therapeutic targets as well as biomarkers for early detection and staging of diabetes (Cianciaruso et al., 2017).
3 Regulation of adipogenic differentiation by exosome-derived ncRNAs
3.1 Regulation of adipogenic differentiation by miRNAs in exosomes
Mature miRNA can mediate mRNA degradation or suppress protein translation by binding with specific ribonucleoprotein AGO (Argonaute) to generate RNA-induced silencing complex (RISC). RISC recognizes target genes via complementary binding of miRNA seed sequences to mRNA 3′UTR or ORF regions (Bartel, 2004; Shenoy and Blelloch, 2014). miRNA can regulate mRNA expression by competitively binding to RNA-binding proteins (Wilczynska and Bushell, 2015). In the study by Son et al., it was found that miRNA can regulate adipogenesis by interacting with adipocyte differentiation-related transcription factor and key signaling molecules (Son et al., 2014). Table 1 shows various miRNAs and target genes that regulate adipocyte differentiation.
TABLE 1.
The miRNAs associated with adipogenesis.
| microRNA | Target | Function | Experiment model | References |
|---|---|---|---|---|
| miR-8 | TCF | Proadipogenic | MSCs, ST2 | Kennell et al. (2008) |
| miR-14 | P38MAPK | Antiadipogenic | Adipose tissue | Xu et al. (2003) |
| miR-17-5p | BMPR2, BMP2 | Proadipogenic | ADSCs | Li et al. (2013) |
| miR-17-92 | Rb2/p130 | Proadipogenic | 3T3L1 | Wang et al. (2008) |
| miR-21 | TGFBR2 | Proadipogenic | MSC, 3T3-L1, hASCs | Kim et al. (2009) |
| miR-22 | HDAC6 | Antiadipogenic | ADSCs | Huang et al. (2012) |
| miR-27a | PPARγ | Antiadipogenic | 3T3L1 | Kim et al. (2010) |
| miR-27b | PPARγ | Antiadipogenic | hMADS | Karbiener et al. (2009) |
| miR-29 | AKT | Antiadipogenic | 3T3L1 | He et al. (2007) |
| miR-31 | C/EBPα | Antiadipogenic | MSCs | Tang et al. (2009) |
| miR-33b | EBF1 | Antiadipogenic | PSPA | Taniguchi et al. (2014) |
| miR-103 | MEF2D | Proadipogenic | 3T3L1 | Li et al. (2015) |
| miR-106a | BMP2 | Proadipogenic | ADSCs | Li et al. (2013) |
| miR-128 | ABCA1, ABCG1 | Proadipogenic | HEK293T, HepG2, MGF7 | Adlakha et al. (2013) |
| miR-135a-5p | APC | Antiadipogenic | 3T3L1 | Chen et al. (2014a) |
| miR-137 | CDC42 | Antiadipogenic | ADSCs | Shin et al. (2014) |
| miR-138 | EID-1 | Antiadipogenic | MSCs | Yang et al. (2011) |
| miR-139-5p | NOTCH1, IRS1 | Antiadipogenic | 3T3L1 | Mi et al. (2015) |
| miR-143 | ERK5 | Proadipogenic | 3T3L1, MSCs | Esau et al. (2004); Oskowitz et al. (2008); Takanabe et al. (2008) |
| PTN, ORP8 | Glucose homeotatsis ↓ | 3T3L1, MSCs | Jordan et al. (2011); Yi et al. (2011) | |
| miR-146b | SIRT1 | Proadipogenic | 3T3L1 | Ahn et al. (2013) |
| miR-155 | CREB | Antiadipogenic | 3T3L1 | Liu et al. (2011) |
| C/EBPβ | Antiadipogenic | Adipose tissue | Liu et al. (2011) | |
| miR-183 | LRP6 | Proadipogenic | 3T3L1 | Chen et al. (2014b) |
| miR-204-5p | DVL3 | Proadipogenic | ADSCs | He et al. (2015) |
| miR-210 | TCF7L2 | Proadipogenic | 3T3L1 | Qin et al. (2010) |
| miR-224 | EGR2, ACSL4 | Antiadipogenic | 3T3L1 | Peng et al. (2013) |
| miR-302 | CDKN1A | Proadipogenic | ASDCs | Kim et al. (2014) |
| miR-320 | PI3K | Antiadipogenic | 3T3L1 | Ling et al. (2009) |
| miR-326 | C/EBPα | Antiadipogenic | MSCs | Tang et al. (2009) |
| miR-335 | MEST | Proadipogenic | 3T3L1, MSCs | Zhu et al. (2014) |
| miR-363 | E2F3 | Antiadipogenic | ADSCs | Chen et al. (2014) |
| miR-375 | ERK1/2 | Proadipogenic | 3T3L1 | Ling et al. (2011) |
| miR-378a-3p | MAPK1 | Proadipogenic | 3T3L1 | Huang et al. (2015) |
| miR-448 | KLF5 | Proadipogenic | 3T3L1, MSCs | Kinoshita et al. (2010) |
| miR-486-5p | SIRT1 | Antiadipogenic | ADSCs | Kim et al. (2012) |
| miR-540 | PPARγ | Antiadipogenic | ADSCs | Chen et al. (2015a) |
| miR-548d-5p | PPARγ | Proadipogenic | hBMSCs | Sun et al. (2014) |
| miR-561 | HSD11B1 | Antiadipogenic | A549, HepG2 | Han et al. (2013) |
| miR-579 | HSD11B1 | Antiadipogenic | A549, HepG2 | Han et al. (2013) |
Note: MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; MSC, mesenchymal stem cell; PI3K, phosphoinositide 3-kinase; BMPR, bone morphogenetic protein receptor; HDAC, histone deacetylase; EBF1, early B-cell factor 1; MEF2, Myocyte Enhancer Factor 2; ABCA, ATP-binding cassette transporter A1; CDC, cell division cycle; NOTCH, neurogenic locus notch homolog protein; SIRT, silent information regulator; LRP, low density lipoprotein receptor related protein; DVL, dishevelled segment polarity protein; EGR, early growth response protein; CDKN, cyclin-dependent kinase inhibitor; PTN, pleiotrophin; ORP, oxysterol-binding protein-related protein; MEST, mesoderm specific transcript; HSD11b1, 11-Beta-Hydroxysteroid Dehydrogenase Type 1; cAMP, cyclic adenosine monophosphate; PKA, protein kinase A; CREB, cAMP, response element-binding; TCF, T-cell-specific transcription factor; TGF-β, transforming growth factor β; TGFBR2, TGF-β, receptor 2; hASC, human adipose tissue-derived stem cell; HMGA2, high mobility group AT-hook2; PPAR, peroxisome proliferator-activated receptor; hMADS, human multipotent adipose-derived stem; C/EBP, CCAAT/enhancerbinding protein; EID, EP300 interacting inhibitor of differentiation 1; KLF, Kruppel-like factor; BMSC, bone marrow stromal cell.
The three different types of adipocytes are referred to as white adipose tissue (WAT), brown adipose tissue (BAT), and beige adipose tissue. The WAT has increased white adipocytes. Mature white adipocytes contain a large lipid droplet that secrets some adipocytic factors, including leptin and adiponectin) (Cristancho and Lazar, 2011). Brown adipose tissue contains brown adipocytes. Classic brown adipocytes contain several small lipid droplets and mitochondria in the cytoplasm (Cristancho and Lazar, 2011). On the other hand, beige adipose tissue was discovered in recent years. Beige adipocytes develop in WAT under certain conditions, including long-term cold stimulation or following treatment with β3-adrenergic agonists (Harms and Seale, 2013).
PPARγ and C/EBPs are significant transcriptional regulators involved in adipocyte differentiation. miRNAs can interact with these transcription factors to regulate cell differentiation (Son et al., 2014). Adipose tissue macrophage-derived exosomes, miR-155 and miR-27, can prevent adipocyte differentiation by suppressing the expression of PPAR-γ. BAT mass decrease in mice upon overexpression of miR-155 (Chen et al., 2013). MiR-155 contained in milk exosomes may therefore reduce BAT differentiation. The miR-155 and miR-27 can regulate in vivo and in vitro insulin sensitivity (Ying et al., 2017). Furthermore, miR-27a and miR-130a can bind to the 3′UTR of PPARγ, resulting in the downregulation of PPARγ (Kang et al., 2013; Liu et al., 2021). The expression of miR-27a and miR-130a was downregulated during the differentiation of 3T3-L1. Upregulated expression of miR-27a and miR-130a is associated with downregulated expression of PPARγ and decreased adipocyte differentiation. In summary, miR-27a and miR-130a could inhibit adipocyte differentiation by targeting PPARγ. MiRNA-29b, another signature miR of cow’s milk exosomes, is also involved in adipogenesis (Bian et al., 2015). The C/EBPs transcription factor family is essential for the differentiation of adipocytes. C/EBPβ and C/EBPδ are induced early following exposure of preadipocytes to differentiation culture media, followed by induction of C/EBPα expression. C/EBPα acts as a transcriptional activator of multiple adipocyte genes and promotes adipocyte differentiation (Gregoire et al., 1998). The 3′-UTR of C/EBPα has two miR-326 binding sites (Feng et al., 2020). It has been reported that miR-326 binds to C/EBPα, thereby downregulating its expression and reducing adipogenic differentiation of hASCs. miR-31 can inhibit adipocyte differentiation by targeting C/EBPα, during differentiation of human mesenchymal stem cells (MSCs) into adipocytes (Sun et al., 2009; Tang et al., 2009).
miRNAs are involved in the regulation of adipocyte differentiation by the PI3K/Akt signaling pathway. In the PI3K/Akt signaling pathway, Akt is known as a downstream effector of c-Met. The miRNA-206 can prevent adipocyte differentiation by suppressing c-Met expression and reducing Akt phosphorylation (Tang et al., 2017). Additionally, by specifically targeting IRS1, miR-139-5p may have an adverse regulatory effect on adipocyte differentiation, which is a key member of the IRS1/PI3K/Akt insulin signaling pathway (Mi et al., 2015).
Adipocytes are derived from Mesenchymal stem cells (MSCs). MSCs first differentiate into adipoblasts, which then form precursor adipocytes that undergo clonal proliferation, growth arrest and terminal differentiation to form mature adipocytes. During the differentiation of MSCs into mature adipocytes, the pRB-E2F, MAPK, SMAD/TGFβ, WNT signaling pathways, and CCAAT/enhancer-binding proteins (C/EBPs) and peroxisome proliferator-activated receptors (PPRs) are activated. Esau discovered that the gene mitogen-activated protein kinase 5 (MAP2K5) is a target of miR-143, which can facilitate adipocyte differentiation (Esau et al., 2004). A previous study showed that miR-143 promoted adipocyte differentiation by inhibiting the expression of Pleiotrophin (PTN) (Yi et al., 2011). A study by Chen showed that transfection of miR-143 into hADSCs at different stages of differentiation induced varied effects (Chen et al., 2015b). Transfection during the mitotic proliferation phase prevented adipocyte differentiation, whereas transfection that occurred during the growth arrest or terminal differentiation stage facilitated differentiation. Additionally, miR-143 can regulate adipocyte differentiation by targeting the MAPK signaling pathway. Adipose tissues from obese people and mice given a high-fat diet had higher levels of exosome-derived miR-148a (Shi et al., 2015). miR-17-92 promotes adipocyte differentiation, whereas miR-363 inhibits adipocyte differentiation via the pRB-E2F signaling pathway (Wang et al., 2008; Chen et al., 2014). A previous study showed upregulated expression of miR-17-92 and downregulated expression of Rb2/P130 in hormone-stimulated 3T3L1 precursor adipocytes. The downregulated expression of Rb2/P130 caused reduced dimerization with the transcription factor E2F (E2F), thereby increasing free E2F4 and E2F5. Further, the free E2F4 and E2F5 activated the pRB-E2F signaling pathway, thus promoting the cells entry into the next cycle (Wang et al., 2008). Another study (Chen et al., 2014), exploring the miRNA expression profile of rat ADSCs during differentiation into mature adipocytes showed that miR-363 activated the retinoma-forming signaling pathway pRB-E2F by post-transcriptionally suppressing the translation of its target gene E2F3. This suppressed the expression of cyclin E (CYCE) and prevented the transition of cells from G1 to S phase, thereby inhibiting the clonal proliferation. The study showed downregulated expression of C/EBPα, which suppressed terminal cell differentiation.
Adipocyte differentiation is affected by miR-21, a miRNA that is a component of milk exosomes (Kim et al., 2012; Kang et al., 2013; Mei et al., 2013; Guglielmi et al., 2017). Kim showed that upregulated expression of miR-21 promoted adipogenic differentiation in hADSCs. In contrast, the inhibition of miR-21 suppressed adipogenic differentiation (Kim et al., 2009). The upregulation of miR-21 inhibited transforming growth factor beta receptor 2 (TGFβR2), which decreased the phosphorylation of Mothers against decapentaplegic homolog 3 (SMAD3), a downstream signaling molecule of the TGFβ/SMAD signaling pathway. miR-21 modulates the TGFβ/SMAD signaling pathway by inhibiting TGFβR2, a positive regulator of white adipocyte differentiation.
miRNAs can also bind to mRNAs of key molecules in signal transduction pathways and indirectly regulate adipocyte differentiation by modulating cell signaling. The Wnt/β-catenin signaling pathway is a key regulator of adipocyte differentiation. Activating Wnt pathway can inhibit adipocyte differentiation (Prestwich and Macdougald, 2007). miR-450a-5p promoted adipogenic differentiation of ADSCs in mice by targeting Wnt1-inducible signaling pathway protein 2 (WISP2) (Zhang et al., 2017). Chen et al. (2014) revealed that miR-344 could activate the Wnt pathway and inhibit adipocyte differentiation by binding to the 3′UTR region of GSK3-β (Glycogen synthase kinase 3-β), leading to the downregulation of GSK3-β expression and upregulation of β-catenin expression, the downstream effectors of GSK3-β. Chen showed that miR-183 promotes the differentiation of 3T3-L1 precursor adipocytes by targeting LRP6 (Low-density lipoprotein receptor-related protein 6), a Wnt signaling pathway molecule (Chen et al., 2014b).
A recent study showed that treating 3T3-L1 preadipocytes with TNF-α upregulated miR-155, which inhibited adipocyte differentiation by binding to the 3′UTR region of CREB (cAMP-response element binding protein) (Liu et al., 2011; Mi et al., 2015 also showed that miR-139-5p might prevent adipocyte differentiation by reducing Notch1 and IRS1 expression as well as preventing 3T3-L1 differentiation.
The transcriptional activation of adipocyte-specific function genes is closely related to adipocyte differentiation. Multiple transcription factors are decisive in the differentiation of precursor adipocytes to mature adipocytes and gene transcription (Prestwich and MacDougald, 2007). PGC-1α, PRDM16, PPARγ and C/EBPs are essential transcription factors for brown adipogenesis (Figure 4) (Prestwich and MacDougald, 2007; Chen et al., 2018). miR-133 downregulation by cold stimulation was shown to promote brown adipocytes differentiation as miR-133 can downregulate PRDM16 expression by binding to its 3′UTR region (Trajkovski et al., 2012; Yin et al., 2013). Sun et al. demonstrated significantly upregulated expression of miR-193b-365 during brown adipocyte differentiation. miR-193b-365 can bind to myogenesis-related cytokines, Igfbp5 (Insulin-like growth factor binding protein 5) and Cdon to promote brown adipocyte differentiation and inhibit muscle formation (Sun et al., 2011).
FIGURE 4.
White and brown adipogenesis are affected by pro- and anti-adipogenic lncRNAs. LncRNAs regulate the fate of adipocytes derived from pluripotent stem cells. Brown adipocytes have many small lipid droplets and are abundant in mitochondrion compared to mature white adipocytes that contain TG in a solitary lipid droplet. Besides, LncRNAs modulate the browning of white adipocytes [Reprinted by permission from Elsevier Inc. Cellular signaling, Chen et al. (2018), copyright 2018].
miR-17-92 promotes preadipocyte differentiation by targeting Rb2/p130 mRNA. As preadipocytes undergo cell fusion and growth inhibition, the cell reenters the cell cycle under the stimulation of hormones. The cell begins to differentiate upon clonal expansion (Wang et al., 2008). In this process, cell transition from G1 to S phase depends on transcription factor E2F. The binding of Rb2/p130 to E2F protein inhibits the function of E2F, thereby promoting cell differentiation. miR-17-92 binds to Rb2/p130 mRNA and reduces the expression of Rb2/p130 protein by blocking its translation, thus promoting adipocyte differentiation. Additionally, miR-27a and miR-27b also inhibit adipocyte differentiation by targeting PPARγ expression (Kim et al., 2010).
Evidence show that miRNAs can regulate the browning of white adipocytes, thereby affecting beige fat formation (Fu et al., 2014; Hu et al., 2015). Lentivirus-mediated diet-induced downregulation of miR-34a in obese mice was shown to reduce blood lipid levels, increase mitochondrial copy number, and oxidative function in adipose tissue, as well as significantly increase the expression of beige fat marker proteins, CD137 and UCP-1 (Fu et al., 2014). miR-34a could increase the expression of fibroblast growth factor-1 receptor (FGFR1) by binding to its 3′UTR region. Downregulated expression of miR-34a was associated with upregulated expression of FGFR1 and enhanced signaling through the MAPK signaling pathway, causing increased phosphorylation of the extracellular regulated kinase (ERK) and increased beige fat formation. Downregulation of miR-34a can inhibit beige adipocyte differentiation by increasing PGC-1α deacetylation and improving the transcriptional activity of PGC-1α (Fu et al., 2014).
miR-122 is an abundantly expressed miRNA in the liver, and is engaged in the maintenance of liver homeostasis and lipid metabolism. Esau discovered that miR-122 deletion is associated with downregulation of several genes related to lipid metabolism, including acetyl-CoA carboxylase alpha (ACACA) and fatty acid synthase (FASN) (Esau et al., 2006). Additionally, Krützfeldt et al. (2005) found decreased plasma cholesterol levels in mice treated with antagomir-122. Overexpression of miR-122 was associated with increased transcription of several genes related to cholesterol biosynthesis and enhanced cholesterol synthesis. Exosomes from adipose tissue have considerably high levels of miR-122 (Huang et al., 2022), miR-103 (Li et al., 2015), miR-146b (Ahn et al., 2013), and miR-148a (He et al., 2018), which modulate adipogenesis.
MiRNA-33 is a crucial post-transcriptional regulator of intracellular cholesterol homeostasis and affects multiple pathways of lipid metabolism. miR-33 inhibits fatty acid β-oxidation by post-transcriptionally repressing the target genes, Carnitine O-octanoyltransferase (CROT), Trifunctional protein, beta subunit (HADHB) and Carnitine palmitoyltransferase 1A (CPT1A). The miR-33 regulates cholesterol efflux by targeting the ATP-binding cassette transporter A1 (ABCA1) (Esau et al., 2006; Iliopoulos et al., 2010). Also, miR-33 can inhibit negative regulation of SREBP proteins such as insulin receptor substrate 2 (IRS2), AMP-activated protein kinase alpha 1 subunit (AMPKA1) or AMP-activated protein kinase alpha 1 catalytic subunit (PRKAA1) and deacetylase 6 (SIRT6) (Ramirez et al., 2011). Cancer-associated cachexia patients exhibit elevated expression levels of miR-410-3p in their subcutaneous adipose tissues and serum exosomes, leading to significant suppression of adipogenesis and lipid synthesis (Sun et al., 2021). Exosomal miR-425-3p in tumor cells inhibit preadipocyte proliferation, prevent adipogenic differentiation, and promote adipocyte lipolysis and browning of white adipocytes (Liu et al., 2022). Furthermore, miR-27 (Ling et al., 2009; Yi et al., 2011; Zhu et al., 2014), miR-30c (Irani and Hussain, 2015), miR-122 (He et al., 2015), miR-144 (Ling et al., 2011), miR-168a (Zhang et al., 2012), miR-223 (Vickers et al., 2014a), miR-302a (Meiler et al., 2015), miR-370 (Iliopoulos et al., 2010), miR-378 (Gerin et al., 2010) and miR-758 (Ramirez, et al., 2011) can also exert regulatory effects on lipid metabolism.
3.2 LncRNA from exosome regulate adipogenic differentiation
To date, lncRNAs have been implicated in the regulation of various biological processes including growth and development, and other metabolic processes in the form of inducible, signaling, and scaffold molecules from epigenetic, transcriptional, and post-transcriptional regulation (Schmitz et al., 2016). Table 2 and Figure 2 present the lncRNAs which exert different functions during adipogenesis. The exosome acts as a transport mediator, encapsulating lncRNAs, and promoting their transit and function in target cells. Exosome-derived lncRNAs may influence adipocyte growth via pre- and post-transcriptional regulation of epigenetic regulation, competitive binding of miRNAs, and lipid metabolism by targeting inflammatory responses.
TABLE 2.
The long noncoding RNA associated with adipogenesis.
| LncRNA | Target | Function | Experiment model | References |
|---|---|---|---|---|
| HOTAIR | PPARγ | Proadipogenic | Primary preadipocytes from Human | Rinn et al. (2007); Gupta et al. (2010); Divoux et al. (2014) |
| ADINR | C/EBPα | Proadipogenic | Human MSCs | Xiao et al. (2015) |
| SRA | PPARγ | Proadipogenic | 3T3L1 | Xu et al. (2010); Liu S et al. (2014) |
| ADNCR | miRNA-204 | Antiadipogenic | 3T3L1, ADSCs | Li et al. (2016) |
| NEAT1 | PPARγ2 | Proadipogenic | 3T3L1, ADSCs | Cooper et al. (2014); Gernapudi et al. (2016) |
| HoxA-AS3 | Ezh2, Runx2 | Proadipogenic | BMSCs | Zhu et al. (2016) |
| PU.1 AS | PU.1 mRNA | Proadipogenic | 3T3L1 | Pang et al. (2013) |
| SlincRAD | PPARγ | Proadipogenic | 3T3L1 | Yi et al. (2013) |
| Lnc RNA H19 | miR-188 | Antiadipogenic | BMSCs | Huang et al. (2016) |
| Lnc RAP-1 | HnRNP-U | Proadipogenic | Primary predipocytes from mice | Quinn and Chang (2016) |
| U90926 | PPARγ2 | Antiadipogenic | 3T3L1 | Chen et al. (2017) |
| Blinc1 | EBF2 | Proadipogenic | Primary predipocytes from mice | Zhao et al. (2014), Mi et al. (2017) |
| Gm15290 | miR-27b, PPARγ | Proadipogenic | Murine primary Preadipocytes, HEK293 | Liu et al. (2017) |
| MIR31HG | Fabp4 | Proadipogenic | ADSCs | Huang et al. (2017) |
| Paral1 | RBM14 | Proadipogenic | 3T3L1 | Firmin et al. (2017) |
| MEG3 | miR-140-5p | Antiadipogenic | ADSCs | Li et al. (2017) |
| PVT1 | PPARγ | Proadipogenic | 3T3L1 | Zhang et al. (2020) |
Note: SRA, steroid receptor activator; NEAT1, nuclear-enriched abundant transcript 1; ADINR, adipogenic differentiation induced noncoding RNA; ADNCR, adipocyte differentiation-associated long noncoding RNA; Blnc1, brown fat enriched lncRNA, 1; ATGL, adipose triglyceride lipase; MSC, mesenchymal stem cell; PI3K, phosphoinositide 3-kinase; HOTAIR, HOX, transcript antisense intergenic RNA; BMPR, bone morphogenetic protein receptor; HDAC, histone deacetylase; EBF1, early B-cell factor 1; MEF2, Myocyte Enhancer Factor 2; ABCA, ATP-binding cassette transporter A1; EZH2, Enhancer of Zeste homolog 2; NOTCH, neurogenic locus notch homolog protein; SIRT, silent information regulator; LRP, low density lipoprotein receptor related protein; DVL, dishevelled segment polarity protein; EGR, early growth response protein; CDKN, cyclin-dependent kinase inhibitor; hnRNP, heterogeneous nuclear ribonucleoprotein; FABP4, Fatty Acid-Binding Protein 4; RBM14, RNA, Binding Motif Protein 14; PPAR, peroxisome proliferator-activated receptor; C/EBP, CCAAT/enhancer binding protein; BMSC, bone marrow stromal cell.
Gene imprinting is an epigenetic regulatory mechanism (Patten et al., 2014; Tucci et al., 2019), and lncRNA-H19 is a newly discovered lncRNA that inhibits adipogenic differentiation of BMSCs, and functions via epigenetic modification of histone deacetylases (Huang et al., 2016). lncRNA H19 overexpression inhibits obesity, improves insulin sensitivity, and promotes mitochondrial biosynthesis (Schmidt et al., 2018).
Insulin-like growth factor 2 (IGF 2) is an important imprinting gene, highly conserved in vertebrates (Smits et al., 2008), and is closely related to individual lean meat percentage, backfat thickness as well as other productive properties. Methylation occurs in the ICR region, one of the key pathways responsible for controlling gene imprinting. Additionally, exosome lncRNA may affect the adipogenic capacity of an individual by modulating IGF2 expression, thereby disrupting lipid accumulation (Hark et al., 2000; Smits et al., 2008).
Recent studies have shown that exosome lncRNA competitively binds with miRNA and circRNA to co-regulate gene expression (Jalali et al., 2013), ensuring a stable expression of lipid metabolism-related target genes. The sponge effect of lncRNA has now been used to bind miRNA in order to analyze its molecular structure and regulatory mechanism (Tong et al., 2019; Yao et al., 2019). Additionally, MALAT1 is linked to the occurrence of metabolic diseases and is involved in lipid metabolism in the liver (Liu JY et al., 2014; Yan et al., 2014).
Silencing MALAT1 in ob/ob mice abolishes the palmitate-induced increase in nuclear SREBP-1c protein and hepatic lipid accumulation by inhibiting stearoyl-Coenzyme A desaturase 1 (SCD1), FAS, Acetyl-CoA carboxylase 1 (ACC1) and ATP citrate lyase (ACLY), which are the major targets of SREBP-1c (Lee et al., 2013). MALAT1 may enhance lipid accumulation via a mechanism involving contact with SREBP-1c to increase the level of nuclear SREBP-1c protein. Notably, lncHR1 is implicated in the transcriptional inhibition of SREBP-1c expression (Li D et al., 2017). Similar to lncHR1, lncRNA Gm16551 suppresses SREBP1c in the mouse liver (Yang et al., 2016). Gm16551 in the liver has been found to inhibit lipid biosynthesis. The increased triglyceride circulation and adipogenic gene expression caused by SREBP1c are inhibited by Gm16551 overexpression, which increases the possibility that Gm16551 might interfere with SREBP1c functions.
Peroxisome proliferators-activated receptors (PPARs) can regulate lipid metabolism, including subtypes PPAR α, β, and γ. Among them, PPARγ is primarily expressed in the immune system and modulates glucose and lipid metabolism. Ligand of PPARγ not only improves insulin resistance and blood lipid, immune regulation and anti-inflammation, but also induce anti-tumor cell proliferation and promotes cell differentiation. It has been reported to alter various biological behaviors of tumor cells such as metastasis and invasion. By influencing the signaling pathway regulated by NFκB and activator protein-1, PPARγ can effectively inhibit the activation and transcription of target gene promoters, achieving its intended purpose. Genes containing peroxisome proliferator responsive element (PPRE) structure include caproyl coenzyme A synthetase, lipoprotein lipase (LPL), insulin receptor substrate-2 (IRS-2), leptin and tumor necrosis factor-α (TNF-α) (Lapsys et al., 2000). PPARγ is related to the incidence and progression of multiple diseases, including diabetes, obesity, and hypertension. Specifically, PPARγ is a key factor in adipocyte differentiation and has attracted much attention in recent years.
Steroid receptor RNA activator (SRA) was the first lncRNA implicated in adipogenesis to be discovered (Xu et al., 2010). By binding to PPARγ, SRA in adipose tissue increases its transcriptional activity and promotes the differentiation of 3T3-L1 preadipocytes. It can regulate the adipocyte cell cycle, insulin-related signaling pathway, and TNF-α signaling pathway (Xu et al., 2010). In the liver, SRA can also promote lipid metabolism via promoter activity suppression of adipose triglyceride lipase (ATGL) by targeting transcription factor FoxO1, thereby downregulating ATGL expression to promote hepatic steatosis (Chen et al., 2016). During the maturation and differentiation of preadipocytes, NEAT1 expression is upregulated, and its activation is necessary for adipogenesis; this suggests that it is a positive regulator of lipogenic differentiation (Pang et al., 2013). Cooper et al. (2014) reported that lncRNA NEAT1 expression temporally fluctuates during the differentiation of 3T3-L1 adipocytes into adipocytes and modulates multiple splicing of PPARγ mRNA, a major transcription factor in adipogenesis. Similarly, other lncRNAs, U90926, Paral1, Gm15290, and Plnc1, also regulate adipogenesis by targeting key adipogenesis transcription factor PPARγ (Chen et al., 2017; Firmin et al., 2017; Liu et al., 2017; Zhu et al., 2019).
LncRNA U90926 is a class of adipose tissue-specific lncRNAs whose expression is downregulated with 3T3-L1 differentiation, its overexpression reduces 3T3-L1 adipogenic differentiation capacity, whereas its knockdown has the opposite effect (Chen et al., 2017). lncRNA-U90926 knockdown in 3T3-L1 cells inhibits the transcriptional activity of the PPARγ promoter, thereby suppressing 3T3-L1preadipocyte differentiation (Chen et al., 2017). Gm15290 overexpression significantly promotes adipocyte differentiation, whereas its interference significantly suppresses adipocyte differentiation; lnc RNA Gm15290 upregulates PPARγ expression level to promote adipogenic, thereby affecting lipid accumulation in the mouse model (Liu et al., 2017). Firmin et al. (2017) found upregulated Paral1 expression during adipogenic differentiation, which increased the transcription activity of PPARγ by synergistically activating RNA binding motif protein 14 (RBM14). In turn, PPARγ upregulated Paral1 expression and its binding level to RBM14. This feedback loop mechanism promotes adipogenesis.
LncRNA IMFNCR promotes adipogenic differentiation of adipocytes in muscle by regulating PPARγ expression through competitively repressing miRNA-128-3p and miRNA-27b-3p (Khalil et al., 2009). Of note, lncRNA-GAS5 is an important gene that regulates adipocyte proliferation and growth. GAS5 can regulate the expression level of cAMP via PDE4B and function on DNL-related enzymes (e.g., ACC1, FAS) and those related to mitochondrial function (e.g., PGC-1α) through phosphorylation of CREB downstream of Camp; thus, this enhances lipid de novo synthesis, inhibits mitochondrial oxidative function, and eventually promotes lipid accumulation in hepatocytes (Xu et al., 2022). The lncRNA GAS5 suppresses the inhibitory effect of miR-18a on connective tissue growth factor (CTGF), thereby reducing adipogenic differentiation of MSCs (Li M et al., 2018).
Adipocyte differentiation-associated long noncoding RNA (ADINR) is a newly discovered ncRNA that regulates adipogenesis (Xiao et al., 2021). ADINR could specifically bind to PA1 during adipocyte differentiation, recruiting methylates lysine and MLL3/4 histone methyltransferase complex, increasing H3K4 (histone H3 lysine4) methylation levels at the C/EBPα site, promoting C/EBPα expression and accelerating adipogenesis. Labeled genes related to adipocyte differentiation were downregulated after ADINR silencing, resulting in reduced lipid accumulation, and suppressed adipogenic differentiation (Xiao et al., 2021).
ADNCR is a lncRNA that is reported to be significantly downregulated during adipocyte adipogenic differentiation (Li et al., 2016). ADNCR, as a competing endogenous RNA (ceRNA) of miR-204, can dependently and competitively upregulate the expression of a target gene, a silent information regulator of transcription 1 (SIRT1), suppressing adipocyte differentiation.
Adiponectin AS is another negative regulator is, whose expression is upregulated during adipose differentiation and plays a negative regulatory role. It forms RNA double-stranded aggregates during the translocation from the nucleus to the cytoplasm, which inhibit the translation of AS mRNA and cause a reduction of body weight, adipose tissue content, and hepatic triglycerides in DIO (diet-induced-obese) mice, thus acting as a barrier in adipogenic differentiation (Cai et al., 2018).
HOTAIR, a lncRNA is involved in preadipocyte differentiation and can promote adipocyte differentiation in abdominal preadipocytes (Divoux et al., 2014). When HoxA-AS3 expression is suppressed in bone marrow mesenchymal stem cells (BMSCs), the formation of fat and fat marker gene expression decreases, implying that HoxA-AS3 is important in adipogenesis (Zhu et al., 2016). LncRNA MIR31HG decreases the enrichment of active histone markers H3K4me3 and AcH3 in the promoter of adipogenesis-related genes FABP4, thereby inhibiting its expression and adipogenesis (Huang et al., 2017).
3.3 Regulation of adipogenic differentiation by circRNAs in exosomes
circRNA is a type of RNA with a covalent closed-loop structure; circRNA from adipose tissue exosomes can function in an autocrine pathway (Shah et al., 2018). circRNA, which is enriched in exosomes, is important for inter-cell communication (Shang et al., 2020). Due to the absence of a 5′ or 3′ end, circRNA is resistant to nuclease-mediated degradation and is thought to be more stable than linear RNA (Salzman et al., 2012). Current studies on the mechanism of exosome circRNA focus on its capacity to effectively act as a miRNA sponge (Zhang et al., 2020; Zhang et al., 2020). Since circRNA is abundant in miRNA binding sites, it can reduce the repressive effect of miRNA on target genes and increase the expression level of those genes via competitive binding to miRNA (Ebert et al., 2007). circRNA can also regulate pre-mRNA by targeting alternative splicing or transcription (Ashwal-Fluss et al., 2014). Table 3 presents the circular RNAs which exert different functions during adipogenesis.
TABLE 3.
The circular RNA associated with adipogenesis.
| circRNA | Origin | Target | Function | References |
|---|---|---|---|---|
| circARF3 | Adipose tissue (mice) | miR-103 | Alleviate adipose inflammation | Zhang et al. (2019) |
| circNrxn2 | Adipose tissue (mice) | miR-103 | Promotes WAT browning | Zhang et al. (2019) |
| circRNA_26852 | Adipose tissue (pig) | ssc-miR-486, ssc-miR-874 | Regulates adipogenic differentiation | Li A et al. (2018) |
| circSAMD4A | Visceral adipose tissue | miR-138-5p | Regulates preadipocytes differentiation | Liu et al. (2020) |
| circFUT10 | Adipose tissue (bovine) | miR-let-7c | Inhibits adipocyte differentiation | Jiang et al. (2020) |
| hsa-circRNA9227-1 | HPA-v/adipocytes | hsa-miR-665 | Regulates adipogenesis | Sun et al. (2020) |
| circRNA_0046367 | HepG cells | miR-34a, PPARα | Regulates lipid metabolism | Guo et al. (2017a) |
| circScd1 | NAFLD mouse | JAK2/STAT5 pathway | Regulates lipid metabolism | Li P et al. (2019) |
Note: circARF3, ADP-ribosylation factor 3; circSAMD4A, sterile alpha motif domain containing 4A; BMI, body mass index; HPA-v, human preadipocytes from visceral fat tissue; ITIH5, inter-alpha-trypsin inhibitor heavy chain; LPL, lipoprotein lipase; JAK, janus protein tyrosine kinases; STAT, signal transducer and activator of transcription; NAFLD, nonalcoholic fatty liver disease; WAT, white adipose tissue.
circH19 secreted by hADSCs can bind to polypyrimidine tract-binding protein 1 (PTBP1), suppressing the translocation of sterol regulatory element-binding protein 1 (SREBP1) from the cytoplasm to the nucleus, consequently inhibiting cell differentiation and lipid accumulation (Park et al., 2012; Oliva-Olivera et al., 2016; Zhu et al., 2020). Adipose tissue exosome circRNA realizes its function through paracrine. Patients with obesity often have a slower rate of wound healing, than healthy individuals, and circRNA Circ_0075932, which is a single exon exosome secreted by his/her adipose tissues, can bind to pumilio homolog 2 (PUM2), promoting the expression of serine/threonine protein kinase, activating nuclear factor kappa beta (NF-kB) pathway to induce inflammation and apoptosis, eventually accelerating healing (Zhang X et al., 2019).
Circ FUT10 competitively represses the expression of target gene let-7c and can inhibit adipogenic differentiation in adipocytes by disrupting the expression of its target genes PPARγ coactivator-1α (PGC1α) (Jiang et al., 2020). On the other hand, circ SAMD4A promotes adipocyte differentiation by acting as a “sponge” for miR-138-5p, thereby upregulating the expression of enhancer of zeste homologue 2 (EZH2), whereas adipocyte differentiation becomes significantly inhibited after circ SAMD4A knockdown (Liu et al., 2020).
In high-fat diet (HFD)-induced obese mice, mmu_circ_0000529 (the homologous mouse circRNA for circSAMD4A) knockdown alleviated weigh gain, lowered food intake, reduced body fat, and increased energy consumption. Additionally, it improved insulin sensitivity and glucose tolerance (Liu et al., 2020). In vitro tests demonstrated that circSAMD4A can bind to miR-138-5p and function as a miRNA sponge to subsequently regulate the expression of EZH2 (Liu et al., 2020). circSAMD4A was significantly upregulated in obese people compared to lean people, and its expression level was associated with a poor prognosis for obese patients (Liu et al., 2020). Functional analysis revealed that circSAMD4A overexpression can regulate preadipocyte differentiation and accurately forecast the development of obesity in humans.
Exosomal plasma from osteoporotic patients demonstrated a higher expression level of has_circ_0006859 than that from healthy individuals, and has_circ_0006859 can act by inhibiting miR-431-5p expression, subsequent upregulation of rock1 (rho-associated coiled-coil containing protein kinase 1), as well as increasing the probability of hBMSCs to differentiate into adipocytes rather than osteoblast (Zhi et al., 2021). Guo et al. (2017b) constructed a regulation network of circRNA-miRNA-mRNA to analyze the mechanism of metabolism by applying high-fat stimulation to build HepG2 fatty liver. Consequently, circRNA_021412 and miR-1972 showed weakened inhibition on LPIN1. LPIN1 induced down-expression of Long-chain-fatty-acid-CoA ligase (ACSL), thus resulting in fatty liver. This evidence confirms that circRNA is a key regulatory factor in fatty liver, and transcription-dependent regulation in metabolic pathways is partially caused by circRNA_021412/miR-1972/LPIN1 signal.
During in vivo and in vitro steatosis of hepatocytes, the normalized expression of circRNA-0046367 can facilitate PPARs to return to normal expression, furtherly improving lipid metabolism disorder, thereby effectively preventing hepatocytes steatosis from lipid peroxidation (Guo et al., 2017a). Therefore, circRNA-0046367 is a potential therapeutic intervention for speroxidative damage.
Accumulating evidence suggests that circRNAs are strongly connected to non-alcoholic fatty liver disease (NAFLD), a condition characterized by a variety of factors including hepatic lipid accumulation, insulin resistance (IR), adipose tissue, mitochondrial dysfunction, a high-fat diet, obesity, a chronic inflammatory state (Huang et al., 2017; Liu et al., 2017).
circRNA genome-wide abnormal regulation is responsible for hepatocytes steatosis (Guo et al., 2017b). For instance, CircScd1 downregulation could damage Janus kinase2/signal transducer and activator of the transcription 5 pathway, causing fatty steatosis in NAFLD (Li P et al., 2019). circRNA_021412/miR-1972/LPIN1 signal might be a key factor in circRNA-related fatty acid metabolic disorder, resulting in increased adipogenesis. Meanwhile, circRNA_0046366 could help normalize the PPAR signal to suppress hepatocyte fat steatosis. Bioinformatic and functional analyses revealed that circRNA_0046366 is a miR-34a-specific antagonist. Because of its multi-targeting characteristics, miR-34 promotes an antagonistic effect on adipose-related miRNA/mRNA interactions based on competitive binding (Li et al., 2011; Dekkers et al., 2018; Gulsin et al., 2018; Lee et al., 2018). As the only target of circRNA_0046366, miR-34a modulates the lipid metabolism of hepatocytes. circRNA_0046366 expression suppresses the level of hepatocytes TG, and circRNA_0046366 is important in hepatocyte fat steatosis. Disruption of the circRNA_0046366/miR-34a/PPARα signal may be a new epigenetic basis of hepatocyte fat steatosis, suggesting that circRNA_0046366 is a potential treatment target for hepatocytes steatosis (Guo et al., 2018). Abnormal genomic regulation of circRNA is often thought to cause hepatocytes steatosis (Dekkers et al., 2018). Transcriptional regulation is thought to be involved in the metabolism pathway. circRNA_021412/miR-1972/LPIN1 signal may be the key factor affecting circRNA-related fatty acid metabolic disorder, resulting in adipogenesis. As indicated, circTshz2-1 and circArhgap5-2 are key adipogenic regulators in vitro. Also, in vivo silencing of circArhgap5-2 inhibits lipid accumulation and downregulation of adipogenesis markers, suggesting that circArhgap5-2 is important to preserving lipid biosynthesis and metabolism in the whole transcription process of adipocytes (Arcinas et al., 2019).
4 Role of exosomal ncRNAs in diseases
As newly discovered means in inter-cellular communications, exosomes modulate the progression of different human diseases (e.g., cancer, metabolic diseases). They comprise growth factors, protein, lipids, DNA, miRNA, lncRNA, and circRNAs (Li C et al., 2021). Once released to the extracellular environment, exosomes bind to receptors and deliver carried ncRNAs, inducing functional reactions and phenotype changes (Figure 5) (Zhang et al., 2016). Exosomal analyses focus on how they regulate inter-cellular signal transduction, immune system function, functional development, and differentiation in the tumor microenvironment. Additionally, the application of exosomal ncRNAs is continually expanding on clinical diagnosis and treatment.
FIGURE 5.
Roles of exosomes in obesity-related disorders. MSC-derived exosomes reduce insulin resistance by promoting M2 macrophage polarization and repressing M1 macrophage polarization. Lipid droplets enlarge after ingestion by tiny adipocytes from exosomes. Insulin resistance in adipocytes and liver develops as a result of exosomes from the adipose tissue of obese people activating monocyte differentiation into macrophages and triggering the release of inflammatory factors. Exosomes released from dysfunctional and hypertrophic adipocytes can impair the functional ability of vascular endothelial cells. Exosomes produced from adipose tissue contain miRNA, necessary for modulating gene expression in adipose tissue and other cells. Adipose tissue-derived exosomes that contain various miRNAs are partially upregulated, while others are downregulated [Reprinted by permission from John Wiley and Sons Ltd. Cell Proliferation, Zhang et al. (2016), copyright 2016].
4.1 Exosomal ncRNAs and insulin resistance
IR is among the crucial sources of Type 2 Diabetes Mellitus (T2DM) (Qiu et al., 2018). It contributes to the development of Alzheimer’s disease and gestational diabetes mellitus (GDM). Insulin resistance refers to insulin-induced tissue damage, such as facilitating glucose absorption and inhibiting glycogen metabolism in adipocytes and skeletal muscle cells. Besides its role in glucose metabolism, insulin also affects the metabolism of proteins and lipids (Qiu et al., 2018). A knockout of insulin β-cell exosome-derived β linc1 (β-cellintergenic non-coding RNA1) gene causes insulin secretion disorder in adult mice and can impair islet cell maturation as well as differentiation in embryonic mice (Arnes et al., 2016).
Adipose tissues play a significant role in insulin resistance, and exosomes derived from adipose tissue may mediate this process. Exosomes secreted by the adipose tissues of mice can induce macrophage cell activation, where retinol-binding protein 4 (RBP4) is the key activation factor (Deng et al., 2009). Additionally, the exosomes linked to obesity deliver macrophages to the liver and adipose tissues, where macrophages release TNF-α and IL-6, causing insulin resistance. Song et al. (2018) uncovered that insulin-resistant adipocyte-derived exosomes (IRADE) treated macrophages could promote insulin resistance in adipose tissues by lowering insulin receptor substrate-1 (IRS-1) and hormone-sensitive lipase (HSL) expression.
Active substances in exosomes modulate inter-cellular insulin signal transduction. Pancreatic β-cells secrete miR-29 family members (miR-29s) via exosomes to regulate glucose homeostasis by manipulating glucose output in the liver, thereby inhibiting insulin signaling in the liver, whereas blocking miR-29s expression in pancreatic β-cells can reverse (HFD)-induced insulin resistance (Li J et al., 2021). Adipose tissue macrophages (ATM) of mice with obesity can secrete exosomes transferring exosomes with miRNA into insulin target cells, which can evaluate FFA levels in the blood, damaging insulin sensitivity, and enhancing insulin resistance (Ying et al., 2017). Additional studies have confirmed that exosomes from ATM of normal mice can reduce insulin resistance in obese mice, whereas exosomes with miR-15 from ATM of obese mice can cause insulin resistance (Ying et al., 2017).
Adipocyte-derived exosomes can promote macrophage activation by accelerating M1 polarization and inhibiting M2 polarization, before activating insulin resistance. Serum exosomes from patients with T2DM and insulin resistance exhibit markedly upregulated miR-222 expression levels. High miR-222 expression in mouse adipose tissue-derived exosomes can aggravate insulin resistance in the liver and skeletal muscle of obese mice on a high-fat diet by suppressing insulin receptor substrate 1 (IRS1) expression (Li et al., 2020).
As endocrine factors, ADSC-derived exosomes (ADSC-Exos) can disrupt the metabolism of corresponding target organs and functions by biologically active vesicular molecules. In the liver, miRNAs carried by ADEVs could regulate PPARγ and FGF-21 expression and are involved in glucose tolerance and insulin sensitivity regulation (Thomou et al., 2017). In cases of obesity, additional organs may also generate IR-related exosomes. miR-130a-3p-containing exosomes secreted by the liver can improve glucose tolerance by inhibiting PH domain leucine-rich phosphatase 2 (PHLPP2) expression in adipocytes; On the other hand, in the case of specific knockdown of miR-130a-3p, mice with obesity showed a notable increase in blood glucose level as well as a decrease in glucose tolerance and insulin sensitivity in vivo (Wu et al., 2020). This indicates that miRNAs secreted by hepatic exosomes can significantly reduce insulin resistance. Pancreatic islets are involved in the regulation of insulin sensitivity and can act also produce insulin. For instance, miR-26a from pancreatic β-cell could improve insulin sensitivity with preserved β-cell function.
miR-27a and miR-320 expressions in plasma exosomes are noticeably higher in T2DM patients than in healthy individuals, making it possible for exosomes with specific bioactive substances to act as serum biomarkers of T2DM (Zhang et al., 2018). Notably, the Dicer enzyme is an RNA endonuclease; Thomou et al. (2017) used Cre-Lox recombination to construct a Dicer-KO mouse. In contrast with wild mice, the quantity of white adipose tissues in the experimental mice was reduced, causing insulin resistance, adipose tissue inflammation, and dyslipidemia; consequently, miRNA expression in its circulating exosomes becomes significantly downregulated. Transplanting adipocyte-derived exosomes with multiple miRNAs, particularly miR-99b, can restore circulating miRNA, reducing FGF21mRNA expression and its 3′-UTR receptor activity, eventually regulating lipid metabolism in multiple tissues (Thomou et al., 2017). Considering the molecular regulatory mechanism, in which lncRNA can function for the competitive binding of ceRNA to miRNA, it is hypothesized that exosome-derived lncRNA may activate pro-inflammatory factors via its abnormal expression, potentially causing abnormal glucolipid metabolism including insulin resistance or alteration of glucose tolerance in the body.
5 Exosomal ncRNAs as a diagnostic biomarker of human diseases
Biomarkers are molecules used for disease detection and/or prognostic prediction. An effective biomarker exhibits high sensitivity and specificity, good stability, and anti-interference. Exosomes are broadly spread in different bodily fluids with various sources and can easily be utilized and stably detected. Various human diseases possess various expression levels of exosomal ncRNAs, including cancers and metabolic diseases, laying a foundation for ncRNAs as a biomarker in early diagnosis.
Abnormal expression of exosome-derived lncRNA is responsible for disordered lipid metabolism (Khalil et al., 2009; Yin et al., 2013; Zhu et al., 2019). Exosomes are characterized by targeted delivery and can protect lncRNA from interferences from exogenous substances. As a result, lncRNA can be effectively maintained in target cells and function normally. Thus, analysis of the regulation of lipid metabolism through the perspective of exosomes may reveal important biomakers. Future studies should attempt to predict fat deposition sites in livestock and even regulate lipid accumulation.
With continuing research on the function of exosomal lncRNA molecular markers, there is increasing evidence on the application of exosome-derived lncRNAs as biomarkers to indirectly regulate lipid metabolism. Koeck et al. (2014) isolated exosomes from the adipose tissue of patients with obesity, and co-cultured it with hepatocytes in vitro. As previously stated, when exosomes are transported into hepatocytes, the expression of enzymes, including TIMP-1 and other bio-active factors, is altered, causing a disorder in the NAFLD related transforming growth factor β (TGFβ) pathway, and ultimately lipid metabolism disorder.
lncRNA has been shown to affect NAFLD symptom formation (Chi et al., 2021). Researchers analyzed the expression pattern of lncRNA-MEG3, as well as its regulatory function on triglyceride and adipogenesis-related genes, using free fatty acids induced HepG2 cells and HFD-mice to establish biological models in vitro and in vivo (Chi et al., 2021). Downregulation of lncRNA-MEG3 suppresses adipogenesis-related genes, whereas MEG3 over-expression reduces lipid accumulation in cells. The hidden molecular mechanism could be that lncRNA-MEG3 competitively binds to miR-21 with LRP6 to suppress the mTOR pathway and stimulate lipid accumulation in cells, resulting in NAFLD. The results demonstrated that exosome-derived lncRNA-MEG3 might be used as a NAFLD biomarker.
Exosomes can be used as detection markers and treatment targets to alleviate insulin resistance. A useful test that reveals the general physical state is liquid biopsy. Because of their sensitivity to changes in human psychological and pathological states, exosomes have the potential to act as a biomarker in insulin resistance. Exosomes have a membrane structure that prevents inner molecular degeneration, allowing their use in detecting nuclear acid changes in patients. With a deeper understanding of exosomes, we will learn more about their benefit as a liquid biopsy tool, particularly in early diagnosis of insulin resistance, where exosome is the most promising liquid biopsy method. Notably, potential exosomal biomarkers, including Let-7b, miR-144-5p, and miR-34a carried by exosomes in plasma (Jones et al., 2017) and miR-20b-5p carried by exosomes in serum (Katayama et al., 2019), can all be candidates in diagnosing insulin resistance. Exosomes regulate the incidence and development of diabetes and its associated complications, and thus it can be used as an early detection and staging biomarker of diabetes mellitus, and as promising treatment targets in diabetes.
6 Disease treatment based on exosomal ncRNAs (metabolic diseases and obesity/diabetes)
Exosome, a potential biomarker and treatment target, is a clinically promising carrier for therapeutic agents. Exosomes, ncRNAs mimics, and inhibitors can be prepared through separation and purification, followed by exosome surface modification to improve target specificity, then used as a carrier to transport ncRNAs to target tissue or organ. However, there is no standardized protocol for the processing and characterization of exosomes, and further studies are needed to address this issue (Lai et al., 2022). Advances in biomedical materials and molecular targeted therapy may also help uncover the role of additional exosomal ncRNAs.
Exosomes have also shown an advantage in disease diagnosis, and the ncRNAs stored inside are closely associated with disease progression, acting as a novel biomarker for diseases such as diabetes. Nevertheless, the interaction network between bioactive molecules in exosomes and their receptors remains unclear. Research on exosomes as a novel diagnostic and treatment tool remains in its infancy and warrants substantial efforts before clinical application.
7 Conclusion and outlook
Exosomal ncRNAs have a high potential as a biomarker in multiple human diseases and therapeutic medicine. However, with a focus on early disease diagnosis markers, exosome suppression, and drug delivery, ncRNAs carried by exosomes have recently provided novel ideas and methods in function and related mechanisms of adipogenesis, development and energy metabolism to prevent obesity. Nevertheless, research on exosomes and the lipid metabolism pathway is currently limited. Thus, to ensure that exosomes are a direct source of relevant biomarkers to broaden the source of the exosome remain unresolved. The pharmacokinetics, pharmacodynamics, and toxicity of exosomal ncRNAs remain to be clinically validated. Also, the regulation of exosomal ncRNAs in psychological and pathological process warrant additional analysis. Because adipose tissues interact with other organs or tissues of the body (central nervous system, important metabolic organs, and immune system), the dynamics of adipocyte lipid droplets and regulation of lipid metabolism by external nutrients, signals, and stress states merit additional investigation and exploration. Research on exosome origin, synthesis and secretion, intake, and other mechanisms is necessary to accelerate the development of diagnostic and prognostic biomarkers of disease.
Acknowledgments
We are grateful to Dr. Lixin Guo for her excellent work, assistance and stimulating discussion.
Funding Statement
This work was supported by Science and Technology Department of Jilin Province (YDZJ202201ZYTS532); Health Research Talents Special Project of Jilin Province (2021SCZ31).
Author contributions
CL, XL, and ZK contributed to conception and design of the study. CL organized the database. XL performed the statistical analysis. CL wrote the first draft of the manuscript. HL, XL, and ZK wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adams B. D., Parsons C., Walker L., Zhang W. C., Slack F. J. (2017). Targeting noncoding RNAs in disease. J. Clini. Invest. 127, 761–771. 10.1172/JCI84424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlakha Y. K., Khanna S., Singh R., Singh V. P., Agrawal A., Saini N. (2013). Pro-apoptotic miRNA-128-2 modulates ABCA1, ABCG1 and RXRα expression and cholesterol homeostasis. Cell Death Dis. 4, e780. 10.1038/cddis.2013.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J., Lee H., Jung C. H., Jeon T. I., Ha T. Y. (2013). MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. EMBO Mol. Med. 5, 1602–1612. 10.1002/emmm.201302647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia I., Miska E. A. (2005). MicroRNA functions in animal development and human disease. Development 132, 4653–4662. 10.1242/dev.02073 [DOI] [PubMed] [Google Scholar]
- Anel A., Gallego-Lleyda A., de Miguel D., Naval J., Martínez-Lostao L. (2019). Role of exosomes in the regulation of T-cell mediated immune responses and in autoimmune disease. Cells 8, 154. 10.3390/cells8020154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcinas C., Tan W., Fang W., Desai T. P., The D. C. S., Degirmenci U., et al. (2019). Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat. Metab. 1, 688–703. 10.1038/s42255-019-0078-z [DOI] [PubMed] [Google Scholar]
- Arnes L., Akerman I., Balderes D. A., Ferrer J., Sussel L. (2016). βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes. Dev. 30, 502–507. 10.1101/gad.273821.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal B., Singh A. K., Rotllan N., Price N., Fernández-Hernando C. (2017). MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 28, 273–280. 10.1097/MOL.0000000000000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Aufiero S., Reckman Y. J., Pinto Y. M., Creemers E. E. (2019). Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 16, 503–514. 10.1038/s41569-019-0185-2 [DOI] [PubMed] [Google Scholar]
- Baixauli F., López-Otín C., Mittelbrunn M. (2014). Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 20 (5), 403. 10.3389/fimmu.2014.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bebelman M. P., Smit M. J., Pegtel D. M., Baglio S. R. (2018). Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 188, 1–11. 10.1016/j.pharmthera.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Bian Y., Lei Y., Wang C., Wang J., Wang L., Liu L., et al. (2015). Epigenetic regulation of miR-29s affects the lactation activity of dairy cow mammary epithelial cells. J. Cell. Physiol. 230, 2152–2163. 10.1002/jcp.24944 [DOI] [PubMed] [Google Scholar]
- Borges F. T., Reis L. A., Schor N. (2013). Extracellular vesicles: structure, function, and potential clinical uses in renal diseases. Braz. J. Med. Biol. Res. 46, 824–830. 10.1590/1414-431X20132964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns H., Böttcher M., Qorraj M., Fabri M., Jitschin S., Dindorf J., et al. (2017). CLL-cell-mediated MDSC induction by exosomal miR-155 transfer is disrupted by vitamin D. Leukemia 31, 985–988. 10.1038/leu.2016.378 [DOI] [PubMed] [Google Scholar]
- Cabral J., Ryan A. E., Griffin M. D., Ritter T. (2018). Extracellular vesicles as modulators of wound healing. Adv. Drug Deliv. Rev. 129, 394–406. 10.1016/j.addr.2018.01.018 [DOI] [PubMed] [Google Scholar]
- Cai R., Sun Y., Qimuge N., Wang G., Wang Y., Chu G., et al. (2018). Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 420–432. 10.1016/j.bbalip.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Chen C., Cui Q., Zhang X., Luo X., Liu Y., Zuo J., et al. (2018). Long non-coding RNAs regulation in adipogenesis and lipid metabolism: emerging insights in obesity. Cell. Signal. 51, 47–58. 10.1016/j.cellsig.2018.07.012 [DOI] [PubMed] [Google Scholar]
- Chen C., Peng Y., Peng Y., Peng J., Jiang S. (2014a). miR-135a-5p inhibits 3T3-L1 adipogenesis through activation of canonical Wnt/β-catenin signaling. J. Mol. Endocrinol. 52, 311–320. 10.1530/JME-14-0013 [DOI] [PubMed] [Google Scholar]
- Chen C., Xiang H., Peng Y., Peng J., Jiang S. (2014b). Mature miR-183, negatively regulated by transcription factor GATA3, promotes 3T3-L1 adipogenesis through inhibition of the canonical Wnt/β-catenin signaling pathway by targeting LRP6. Cell. Signal. 26, 1155–1165. 10.1016/j.cellsig.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Chen G., Yu D., Nian X., Liu J., Koenig R. J., Xu B., et al. (2016). LncRNA SRA promotes hepatic steatosis through repressing the expression of adipose triglyceride lipase (ATGL). Sci. Rep. 6, 35531. 10.1038/srep35531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang S., Chen L., Chen Y., Wu M., Zhang Y., et al. (2014c). MicroRNA-344 inhibits 3T3-L1 cell differentiation via targeting GSK3β of Wnt/β-catenin signaling pathway. FEBS Lett. 588, 429–435. 10.1016/j.febslet.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Chen J., Ke S., Zhong L., Wu J., Tseng A., Morpurgo B., et al. (2018). Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem. Pharmacol. 152, 94–103. 10.1016/j.bcp.2018.03.019 [DOI] [PubMed] [Google Scholar]
- Chen J., Liu Y., Lu S., Yin L., Zong C., Cui S., et al. (2017). The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int. J. Obes. 41, 299–308. 10.1038/ijo.2016.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chen Y., Zhang S., Ye L., Cui J., Sun Q., et al. (2015a). MiR-540 as a novel adipogenic inhibitor impairs adipogenesis via suppression of PPARγ. J. Cell. Biochem. 116, 969–976. 10.1002/jcb.25050 [DOI] [PubMed] [Google Scholar]
- Chen L., Cui J., Hou J., Long J., Li C., Liu L. (2014d). A novel negative regulator of adipogenesis: microRNA-363. Stem Cells 32, 510–520. 10.1002/stem.1549 [DOI] [PubMed] [Google Scholar]
- Chen L., Hou J., Ye L., Chen Y., Cui J., Tian W., et al. (2015b). MicroRNA-143 regulates adipogenesis by modulating the MAP2K5–ERK5 signaling. Sci. Rep. 4, 3819. 10.1038/srep03819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Siegel F., Kipschull S., Haas B., Fröhlich H., Meister G., et al. (2013). miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 4, 1769. 10.1038/ncomms2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung O., Puri P., Eicken C., Contos M. J., Mirshahi F., Maher J. W., et al. (2008). Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 48, 1810–1820. 10.1002/hep.22569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Gong Z., Xin H., Wang Z., Liu Z. (2021). Correction to: long noncoding RNA lncARSR promotes nonalcoholic fatty liver disease and hepatocellular carcinoma by promoting YAP1 and activating the IRS2/AKT pathway. J. Transl. Med. 19, 438. 10.1186/s12967-021-02759-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciaruso C., Phelps E. A., Pasquier M., Hamelin R., Demurtas D., Alibashe Ahmed M., et al. (2017). Primary human and rat β-cells release the intracellular autoantigens GAD65, IA-2, and proinsulin in exosomes together with cytokine-induced enhancers of immunity. Diabetes 66, 460–473. 10.2337/db16-0671 [DOI] [PubMed] [Google Scholar]
- Connolly K. D., Guschina I. A., Yeung V., Clayton A., Draman M. S., Von Ruhland C., et al. (2015). Characterisation of adipocyte-derived extracellular vesicles released pre- and post-adipogenesis. J. Extracell. Vesicles 4, 29159. 10.3402/jev.v4.29159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D., Carter G., Li P., Patel R., Watson J., Patel N. (2014). Long Non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARγ2 splicing during adipogenesis in 3T3-L1 cells. Genes 5, 1050–1063. 10.3390/genes5041050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho A. G., Lazar M. A. (2011). Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 12, 722–734. 10.1038/nrm3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Yu M., Zhang Y., Tian W. (2017). Exosome-like vesicles derived from adipose tissue provide biochemical cues for adipose tissue regeneration. Tissue Eng. Part A 23, 1221–1230. 10.1089/ten.TEA.2017.0045 [DOI] [PubMed] [Google Scholar]
- De Silva N., Samblas M., Martínez J. A., Milagro F. I. (2018). Effects of exosomes from LPS-activated macrophages on adipocyte gene expression, differentiation, and insulin-dependent glucose uptake. J. Physiol. Biochem. 74, 559–568. 10.1007/s13105-018-0622-4 [DOI] [PubMed] [Google Scholar]
- Dekkers C. C. J., Gansevoort R. T., Heerspink H. J. L. (2018). New diabetes therapies and diabetic kidney disease progression: the role of SGLT-2 inhibitors. Curr. Diab. Rep. 18, 27. 10.1007/s11892-018-0992-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Poliakov A., Hardy R. W., Clements R., Liu C., Liu Y., et al. (2009). Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 58, 2498–2505. 10.2337/db09-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divoux A., Karastergiou K., Xie H., Guo W., Perera R. J., Fried S. K., et al. (2014). Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation: lncRNA HOTAIR in gluteal adipose tissue. Obesity 22, 1781–1785. 10.1002/oby.20793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer F., Baur A. (2016). “Biogenesis and functions of exosomes and extracellular vesicles,” in Lentiviral vectors and exosomes as gene and protein delivery tools. Editor Federico M. (New York: Springer New York; ), 201–216. 10.1007/978-1-4939-3753-0_15 [DOI] [PubMed] [Google Scholar]
- Ebert M. S., Neilson J. R., Sharp P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 4, 721–726. 10.1038/nmeth1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElashiryElashiry M. M. M., Elsayed R., Rajendran M., Auersvald C., Zeitoun R., et al. (2020). Dendritic cell derived exosomes loaded with immunoregulatory cargo reprogram local immune responses and inhibit degenerative bone disease in vivo . Extracell. vesicles. 9, 1795362. 10.1080/20013078.2020.1795362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., et al. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98. 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Esau C., Kang X., Peralta E., Hanson E., Marcusson E. G., Ravichandran L. V., et al. (2004). MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279, 52361–52365. 10.1074/jbc.C400438200 [DOI] [PubMed] [Google Scholar]
- Feng Y., Zhou L., Peng Y., Yang Y., Fan T., Jiang X., et al. (2020). The role of miR‐326 in adipogenic differentiation of human adipose‐derived stem cells by targeting C/EBPα in vitro . Anat. Rec. 303, 2054–2060. 10.1002/ar.24281 [DOI] [PubMed] [Google Scholar]
- Firmin F. F., Oger F., Gheeraert C., Dubois-Chevalier J., Vercoutter-Edouart A. S., Alzaid F., et al. (2017). The RBM14/CoAA-interacting, long intergenic non-coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARγ. Sci. Rep. 7, 14087. 10.1038/s41598-017-14570-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty S. E., Grijalva A., Xu X., Ables E., Nomani A., Ferrante A. W. (2019). A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science 363, 989–993. 10.1126/science.aaw2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T., Seok S., Choi S., Huang Z., Suino-Powell K., Xu H. E., et al. (2014). MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell. Biol. 34, 4130–4142. 10.1128/MCB.00596-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I., Bommer G. T., McCoin C. S., Sousa K. M., Krishnan V., MacDougald O. A. (2010). Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 299, E198–E206. 10.1152/ajpendo.00179.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernapudi R., Wolfson B., Zhang Y., Yao Y., Yang P., Asahara H., et al. (2016). MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol. Cell. Biol. 36, 30–38. 10.1128/MCB.00702-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire F. M., Smas C. M., Sul H. S. (1998). Understanding adipocyte differentiation. Physiol. Rev. 78, 783–809. 10.1152/physrev.1998.78.3.783 [DOI] [PubMed] [Google Scholar]
- Guglielmi V., D’Adamo M., Menghini R., Cardellini M., Gentileschi P., Federici M., et al. (2017). MicroRNA 21 is up-regulated in adipose tissue of obese diabetic subjects. Nutr. Healthy. Aging. 4, 141–145. 10.3233/NHA-160020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsin G. S., Swarbrick D. J., Hunt W. H., Levelt E., Graham-Brown M. P. M., Parke K. S., et al. (2018). Relation of aortic stiffness to left ventricular remodeling in younger adults with type 2 diabetes. Diabetes 67, 1395–1400. 10.2337/db18-0112 [DOI] [PubMed] [Google Scholar]
- Guo X. Y., Chen J. N., Sun F., Wang Y. Q., Pan Q., Fan J. G. (2017a). circRNA_0046367 prevents hepatoxicity of lipid peroxidation: an inhibitory role against hepatic steatosis. Oxid. Med. Cell. Longev. 2017, 3960197–3960216. 10.1155/2017/3960197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. Y., He C. X., Wang Y. Q., Sun C., Li G. M., Su Q., et al. (2017b). Circular RNA profiling and bioinformatic modeling identify its regulatory role in hepatic steatosis. Biomed. Res. Int. 2017, 5936171. 10.1155/2017/5936171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. Y., Sun F., Chen J. N., Wang Y. Q., Pan Q., Fan J. G. (2018). circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. WJG 24, 323–337. 10.3748/wjg.v24.i3.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. A., Shah N., Wang K. C., Kim J., Horlings H. M., Wong D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Staab-Weijnitz C. A., Xiong G., Maser E. (2013). Identification of microRNAs as a potential novel regulatory mechanism in HSD11B1 expression. J. Steroid Biochem. Mol. Biol. 133, 129–139. 10.1016/j.jsbmb.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Cashikar A. (2012). Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28, 337–362. 10.1146/annurev-cellbio-092910-154152 [DOI] [PubMed] [Google Scholar]
- Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., Tilghman S. M. (2000). CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489. 10.1038/35013106 [DOI] [PubMed] [Google Scholar]
- Harms M., Seale P. (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263. 10.1038/nm.3361 [DOI] [PubMed] [Google Scholar]
- He A., Zhu L., Gupta N., Chang Y., Fang F. (2007). Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 21, 2785–2794. 10.1210/me.2007-0167 [DOI] [PubMed] [Google Scholar]
- He D., Zheng J., Hu J., Chen J., Wei X. (2020). Long non-coding RNAs and pyroptosis. Clin. Chim. Acta 504, 201–208. 10.1016/j.cca.2019.11.035 [DOI] [PubMed] [Google Scholar]
- He H., Cai M., Zhu J., Xiao W., Liu B., Shi Y., et al. (2018). miR-148a-3p promotes rabbit preadipocyte differentiation by targeting PTEN. Vitro. Cell. Dev. Biol. Anim. 54, 241–249. 10.1007/s11626-018-0232-z [DOI] [PubMed] [Google Scholar]
- He H., Chen K., Wang F., Zhao L., Wan X., Wang L., et al. (2015). miR-204-5p promotes the adipogenic differentiation of human adipose-derived mesenchymal stem cells by modulating DVL3 expression and suppressing Wnt/β-catenin signaling. Int. J. Mol. Med. 35, 1587–1595. 10.3892/ijmm.2015.2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Moazed D. (2015). RNA-mediated epigenetic regulation of gene expression. Rev. Genet. 16, 71–84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou R., Li Y., Sui Z., Yuan H., Yang K., Liang Z., et al. (2019). Advances in exosome isolation methods and their applications in proteomic analysis of biological samples. Anal. Bioanal. Chem. 411, 5351–5361. 10.1007/s00216-019-01982-0 [DOI] [PubMed] [Google Scholar]
- Hu F., Wang M., Xiao T., Yin B., He L., Meng W., et al. (2015). miR-30 Promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 64, 2056–2068. 10.2337/db14-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Wang J., Xie W., Lyu Q., Wu J., He J., et al. (2015). MiR-378a-3p enhances adipogenesis by targeting mitogen-activated protein kinase 1. Biochem. Biophys. Res. Commun. 457, 37–42. 10.1016/j.bbrc.2014.12.055 [DOI] [PubMed] [Google Scholar]
- Huang S., Wang S., Bian C., Yang Z., Zhou H., Zeng Y., et al. (2012). Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic Differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 21, 2531–2540. 10.1089/scd.2012.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chen J., Ren Y., Fan L., Xiang W., He X. (2022). Exosomal miR‐122 promotes adipogenesis and aggravates obesity through the VDR/SREBF1 axis. Obesity 30, 666–679. 10.1002/oby.23365 [DOI] [PubMed] [Google Scholar]
- Huang Y., Jin C., Zheng Y., Li X., Zhang S., Zhang Y., et al. (2017). Knockdown of lncRNA MIR31HG inhibits adipocyte differentiation of human adipose-derived stem cells via histone modification of FABP4. Sci. Rep. 7, 8080. 10.1038/s41598-017-08131-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zheng Y., Jin C., Li X., Jia L., Li W. (2016). Long non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci. Rep. 6, 28897. 10.1038/srep28897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H., Hanson P. I. (2010). Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat. Rev. Mol. Cell Biol. 11, 556–566. 10.1038/nrm2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Drosatos K., Hiyama Y., Goldberg I. J., Zannis V. I. (2010). MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J. Lipid Res. 51, 1513–1523. 10.1194/jlr.M004812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani S., Hussain M. M. (2015). Role of microRNA-30c in lipid metabolism, adipogenesis, cardiac remodeling and cancer. Curr. Opin. Lipidol. 26, 139–146. 10.1097/MOL.0000000000000162 [DOI] [PubMed] [Google Scholar]
- Jahangiri B., Saei A. K., Obi P. O., Asghari N., Lorzadeh S., Hekmatirad S., et al. (2022). Exosomes, autophagy and ER stress pathways in human diseases: cross-regulation and therapeutic approaches. Biochim. Biophys. Acta. Mol. Basis. Dis. 10, 166484. 10.1016/j.bbadis.2022.166484 [DOI] [PubMed] [Google Scholar]
- Jalali S., Bhartiya D., Lalwani M. K., Sivasubbu S., Scaria V. (2013). Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS ONE 8, e53823. 10.1371/journal.pone.0053823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Li H., Yang J., Shen X., Song C., Yang Z., et al. (2020). circRNA profiling reveals an abundant circFUT10 that promotes adipocyte proliferation and inhibits adipocyte differentiation via sponging let-7. Mol. Ther. Nucleic Acids 20, 491–501. 10.1016/j.omtn.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A., Danielson K. M., Benton M. C., Ziegler O., Shah R., Stubbs R. S., et al. (2017). Macartney-coxson, miRNA signatures of insulin resistance in obesity: miRNA signatures of insulin resistance in obesity. Obesity 25, 1734–1744. 10.1002/oby.21950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. D., Krüger M., Willmes D. M., Redemann N., Wunderlich F. T., Brönneke H. S., et al. (2011). Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 13, 434–446. 10.1038/ncb2211 [DOI] [PubMed] [Google Scholar]
- Kaikkonen M. U., Adelman K. (2018). Emerging roles of non-coding RNA transcription. Trends biochem. Sci. 43, 654–667. 10.1016/j.tibs.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Kajimoto K., Naraba H., Iwai N. (2006). MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA 12, 1626–1632. 10.1261/rna.7228806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Yan L.-M., Zhang W.-Y., Li Y.-M., Tang A.-Z., Ou H.-S. (2013). Role of microRNA-21 in regulating 3T3-L1 adipocyte differentiation and adiponectin expression. Mol. Biol. Rep. 40, 5027–5034. 10.1007/s11033-013-2603-6 [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D. A., Duttagupta R., Willingham A. T., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- Karbiener M., Fischer C., Nowitsch S., Opriessnig P., Papak C., Ailhaud G., et al. (2009). microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 390, 247–251. 10.1016/j.bbrc.2009.09.098 [DOI] [PubMed] [Google Scholar]
- Karreth F. A., Reschke M., Ruocco A., Ng C., Chapuy B., Léopold V., et al. (2015). The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo . Cell 161, 319–332. 10.1016/j.cell.2015.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasagi Y., Oki E., Ando K., Ito S., Iguchi T., Sugiyama M., et al. (2017). The expression of CCAT2, a novel Long noncoding RNA transcript, and rs6983267 single-nucleotide polymorphism genotypes in colorectal cancers. Oncology 92, 48–54. 10.1159/000452143 [DOI] [PubMed] [Google Scholar]
- Katayama M., Wiklander O. P. B., Fritz T., Caidahl K., El-Andaloussi S., Zierath J. R., et al. (2019). Circulating exosomal miR-20b-5p is elevated in type 2 diabetes and could impair insulin action in human skeletal muscle. Diabetes 68, 515–526. 10.2337/db18-0470 [DOI] [PubMed] [Google Scholar]
- Kennell J. A., Gerin I., MacDougald O. A., Cadigan K. M. (2008). The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 105, 15417–15422. 10.1073/pnas.0807763105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 106, 11667–11672. 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Shin K. K., Lee A. L., Kim Y. S., Park H. J., Park Y. K., et al. (2014). MicroRNA-302 induces proliferation and inhibits oxidant-induced cell death in human adipose tissue-derived mesenchymal stem cells. Cell Death Dis. 5, e1385. 10.1038/cddis.2014.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Haney M. J., Zhao Y., Yuan D., Deygen I., Klyachko N. L., et al. (2018). Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine 14, 195–204. 10.1016/j.nano.2017.09.011 [DOI] [PubMed] [Google Scholar]
- Kim M. S., Muallem S., Kim S. H., Kwon K. B., Kim M. S. (2019). Exosomal release through TRPML1-mediated lysosomal exocytosis is required for adipogenesis. Biochem. Biophys. Res. Commun. 510, 409–415. 10.1016/j.bbrc.2019.01.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Kim A. Y., Lee H. W., Son Y. H., Lee G. Y., Lee J. W., et al. (2010). miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 392, 323–328. 10.1016/j.bbrc.2010.01.012 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Hwang S. H., Lee S. Y., Shin K. K., Cho H. H., Bae Y. C., et al. (2012). miR-486-5p induces replicative senescence of human adipose tissue-derived mesenchymal stem cells and its expression is controlled by high glucose. Stem Cells Dev. 21, 1749–1760. 10.1089/scd.2011.0429 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Hwang S. J., Bae Y. C., Jung J. S. (2009). MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 27, 3093–3102. 10.1002/stem.235 [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Ono K., Horie T., Nagao K., Nishi H., Kuwabara Y., et al. (2010). Regulation of adipocyte differentiation by activation of serotonin (5-HT) receptors 5-HT2AR and 5-HT2CR and involvement of microRNA-448-mediated repression of KLF5. Mol. Endocrinol. 24, 1978–1987. 10.1210/me.2010-0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck E. S., Iordanskaia T., Sevilla S., Ferrante S. C., Hubal M. J., Freishtat R. J., et al. (2014). Adipocyte exosomes induce transforming growth factor beta pathway dysregulation in hepatocytes: A novel paradigm for obesity-related liver disease. J. Surg. Res. 192, 268–275. 10.1016/j.jss.2014.06.050 [DOI] [PubMed] [Google Scholar]
- Kranendonk M. E. G., Visseren F. L. J., van Balkom B. W. M., Nolte-’t Hoen E. N. M., van Herwaarden J. A., de Jager W., et al. (2014). Human adipocyte extracellular vesicles in reciprocal signaling between adipocytes and macrophages. Obesity 22, 1296–1308. 10.1002/oby.20679 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., et al. (2005). Silencing of microRNAs in vivo with “antagomirs”. Nature 438, 685–689. 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- Lin Q., Gao Z., Alarcon R. M., Ye J., Yun Z. (2009). A role of miR-27 in the regulation of adipogenesis. FEBS J. 276, 2348–2358. 10.1111/j.1742-4658.2009.06967.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J. J., Chau Z. L., Chen S., Hill J. J., Korpany K. V., Liang N., et al. (2022). Exosome processing and characterization approaches for research and technology development. Adv. Sci. 9, 2103222. 10.1002/advs.202103222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapsys N. M., Kriketos A. D., Lim-Fraser M., Poynten A. M., Lowy A., Furler S. M., et al. (2000). Expression of genes involved in lipid metabolism correlate with peroxisome proliferator-activated receptor γ expression in human skeletal muscle. J. Clin. Endocrinol. Metab. 85, 4293–4297. 10.1210/jcem.85.11.6973 [DOI] [PubMed] [Google Scholar]
- Lazar I., Clement E., Dauvillier S., Milhas D., Ducoux-Petit M., LeGonidec S., et al. (2016). Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: A novel mechanism linking obesity and cancer. Cancer Res. 76, 4051–4057. 10.1158/0008-5472.CAN-16-0651 [DOI] [PubMed] [Google Scholar]
- Lee M., Lee S., Kang J., Yang H., Jeong E., Kim H., et al. (2013). Salicortin-derivatives from Salix pseudo-lasiogyne twigs inhibit adipogenesis in 3T3-L1 cells via modulation of C/EBPα and SREBP1c dependent pathway. Molecules 18, 10484–10496. 10.3390/molecules180910484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. Y., Hsiao P. J., Huang J. C., Hsu W. H., Chen S. C., Shin S. J. (2018). Association between metabolic syndrome and microvascular and macrovascular disease in type 2 diabetic mellitus. Am. J. Med. Sci. 355, 342–349. 10.1016/j.amjms.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Li A., Huang W., Zhang X., Xie L., Miao X. (2018). Identification and characterization of CircRNAs of two pig breeds as a new biomarker in metabolism-related diseases. Cell Physiol. biochem. 47, 2458–2470. 10.1159/000491619 [DOI] [PubMed] [Google Scholar]
- Li C., Ni Y. Q., Xu H., Xiang Q. Y., Zhao Y., Zhan J. K., et al. (2021). Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Sig. Transduct. Target. Ther. 6, 383. 10.1038/s41392-021-00779-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Cheng M., Niu Y., Chi X., Liu X., Fan J., et al. (2017). Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int. J. Biol. Sci. 13, 349–357. 10.7150/ijbs.16635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Song H., Shuo L., Wang L., Xie P., Li W., et al. (2020). Gonadal white adipose tissue-derived exosomal MiR-222 promotes obesity-associated insulin resistance. Aging 12, 22719–22743. 10.18632/aging.103891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li T., Wang S., Wei J., Fan J., Li J., et al. (2013). miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res. 10, 313–324. 10.1016/j.scr.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Li J., Zhang Y., Ye Y., Li D., Liu Y., Lee E., et al. (2021). Pancreatic β cells control glucose homeostasis via the secretion of exosomal miR‐29 family. J. Extracell. Vesicles 10, e12055. 10.1002/jev2.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Liu Z., Zhang Z., Liu G., Sun S., Sun C. (2015). miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol. Chem. 396, 235–244. 10.1515/hsz-2014-0241 [DOI] [PubMed] [Google Scholar]
- Li M., Sun X., Cai H., Sun Y., Plath M., Li C., et al. (2016). Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim. Biophys. Acta Gene Regul. Mech. 1859, 871–882. 10.1016/j.bbagrm.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Li M., Xie Z., Wang P., Li J., Liu W., Tang S., et al. (2018). The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis. 9, 554. 10.1038/s41419-018-0627-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Shan K., Liu Y., Zhang Y., Xu L., Xu L. (2019). CircScd1 promotes fatty liver disease via the janus kinase 2/signal transducer and activator of transcription 5 pathway. Dig. Dis. Sci. 64, 113–122. 10.1007/s10620-018-5290-2 [DOI] [PubMed] [Google Scholar]
- Li W. Q., Chen C., Xu M. D., Guo J., Li Y. M., Xia Q. M., et al. (2011). The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats: miR-34 family is upregulated in hepatic fibrosis. FEBS J. 278, 1522–1532. 10.1111/j.1742-4658.2011.08075.x [DOI] [PubMed] [Google Scholar]
- Li X., Ballantyne L. L., Yu Y., Funk C. D. (2019). Perivascular adipose tissue-derived extracellular vesicle miR-221-3p mediates vascular remodeling. FASEB J. 33, 12704–12722. 10.1096/fj.201901548R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Jin C., Chen S., Zheng Y., Huang Y., Jia L., et al. (2017). Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol. Cell. Biochem. 433, 51–60. 10.1007/s11010-017-3015-z [DOI] [PubMed] [Google Scholar]
- Ling H. Y., Ou H. S., Feng S. D., Zhang X. Y., Tuo Q. H., Chen L. X., et al. (2009). CHANGES in microRNA (miR) profile and effects of miR-320 in insulin-resistant 3T3-L1 adipocytes. Clin. Exp. Pharmacol. Physiol. 36, e32–e39. 10.1111/j.1440-1681.2009.05207.x [DOI] [PubMed] [Google Scholar]
- Ling H. Y., Wen G. B., Feng S. D., Tuo Q. H., Ou H. S., Yao C. H., et al. (2011). MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signalling. Clin. Exp. Pharmacol. Physiol. 38, 239–246. 10.1111/j.1440-1681.2011.05493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Pan W., Zhuang S., Tang Y., Zhang H. (2022). Cancer cell-derived exosomal miR-425-3p induces white adipocyte atrophy. Adipocyte 11, 487–500. 10.1080/21623945.2022.2108558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Y., Yao J., Li X. M., Song Y. C., Wang X. Q., Li Y. J., et al. (2014). Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 5, e1506. 10.1038/cddis.2014.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Sheng L., Miao H., Saunders T. L., MacDougald O. A., Koenig R. J., et al. (2014). SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J. Biol. Chem. 289, 13000–13009. 10.1074/jbc.M114.564658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yang Y., Wu J. (2011). TNFα-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochem. Biophys. Res. Commun. 414, 618–624. 10.1016/j.bbrc.2011.09.131 [DOI] [PubMed] [Google Scholar]
- Liu W., Ma C., Yang B., Yin C., Zhang B., Xiao Y. (2017). LncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat deposition and contribute to body weight gain in mice. Biochem. Biophys. Res. Commun. 493, 1168–1175. 10.1016/j.bbrc.2017.09.114 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu H., Li Y., Mao R., Yang H., Zhang Y., et al. (2020). Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 10, 4705–4719. 10.7150/thno.42417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang M., Liang Y., Wang C., Naruse K., Takahashi K. (2021). Treatment of oxidative stress with exosomes in myocardial ischemia. Int. J. Mol. Sci. 9 (22), 1729. 10.3390/ijms22041729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M., Martin-Jaular L., Lavieu G., Théry C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17. 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- Mathivanan S., Ji H., Simpson R. J. (2010). Exosomes: extracellular organelles important in intercellular communication. J. Proteomics. 73, 1907–1920. 10.1016/j.jprot.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Mei Y., Bian C., Li J., Du Z., Zhou H., Yang Z., et al. (2013). miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J. Cell. Biochem. 114, 1374–1384. 10.1002/jcb.24479 [DOI] [PubMed] [Google Scholar]
- Meiler S., Baumer Y., Toulmin E., Seng K., Boisvert W. A. (2015). MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. ATVB 35, 323–331. 10.1161/ATVBAHA.114.304878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L., Chen Y., Zheng X., Li Y., Zhang Q., Mo D., et al. (2015). MicroRNA-139-5p suppresses 3T3-L1 preadipocyte differentiation through notch and IRS1/PI3K/Akt insulin signaling pathways: MICRORNA-139-5p suppresses 3T3-L1 preadipocyte. J. Cell. Biochem. 116, 1195–1204. 10.1002/jcb.25065 [DOI] [PubMed] [Google Scholar]
- Mi L., Zhao X. Y., Li S., Yang G., Lin J. D. (2017). Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol. Metab. 6, 101–110. 10.1016/j.molmet.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L., Mitsuhashi M., Simms P., Gooding W. E., Whiteside L. (2016). Tumor-derived exosomes regulate expression of immune function-related genes in human t cell subsets. Sci. Rep. 6 (1), 20254. 10.1038/srep20254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafar S., Nouri N., Alipour M., Fallahi J., Zare F., Tabei S. M. B. (2022). Exosome as a target for cancer treatment. J. Investig. Med. 70, 1212–1218. 10.1136/jim-2021-002194 [DOI] [PubMed] [Google Scholar]
- Narayanan K., Kumar S., Padmanabhan P., Gulyas B., Wan A. C. A., Rajendran V. M. (2018). Lineage-specific exosomes could override extracellular matrix mediated human mesenchymal stem cell differentiation. Biomaterials 182, 312–322. 10.1016/j.biomaterials.2018.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva-Olivera W., Coín-Aragüez L., Lhamyani S., Clemente-Postigo M., Alcaide Torres J., Bernal-López M. R., et al. (2016). Adipogenic impairment of adipose tissue-derived mesenchymal stem cells in subjects with metabolic syndrome: possible protective role of FGF2. J. Clin. Endocrinol. Metab. 102, 478–487. 10.1210/jc.2016-2256 [DOI] [PubMed] [Google Scholar]
- Oskowitz A. Z., Lu J., Penfornis P., Ylostalo J., McBride J., Flemington E. K., et al. (2008). Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc. Natl. Acad. Sci. U. S. A. 105, 18372–18377. 10.1073/pnas.0809807105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang W. J., Lin L. G., Xiong Y., Wei N., Wang Y., Shen Q. W., et al. (2013). Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation: knockdown of PU.1 AS lncRNA inhibits adipogenesis. J. Cell. Biochem. 114, 2500–2512. 10.1002/jcb.24595 [DOI] [PubMed] [Google Scholar]
- Park H. T., Lee E. S., Cheon Y. P., Lee D. R., Yang K. S., Kim Y. T., et al. (2012). The relationship between fat depot-specific preadipocyte differentiation and metabolic syndrome in obese women: preadipocyte and metabolic syndrome. Clin. Endocrinol. 76, 59–66. 10.1111/j.1365-2265.2011.04141.x [DOI] [PubMed] [Google Scholar]
- Patten M. M., Ross L., Curley J. P., Queller D. C., Bonduriansky R., Wolf J. B. (2014). The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity 113, 119–128. 10.1038/hdy.2014.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Xiang H., Chen C., Zheng R., Chai J., Peng J., et al. (2013). MiR-224 impairs adipocyte early differentiation and regulates fatty acid metabolism. Int. J. Biochem. Cell Biol. 45, 1585–1593. 10.1016/j.biocel.2013.04.029 [DOI] [PubMed] [Google Scholar]
- Prestwich T. C., Macdougald O. A. (2007). Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr. Opin. Cell Biol. 19, 612–617. 10.1016/j.ceb.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L., Chen Y., Niu Y., Chen W., Wang Q., Xiao S., et al. (2010). A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical wnt/beta-catenin signaling pathway. BMC Genomics 11, 320. 10.1186/1471-2164-11-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q. Y., Zhang B. L., Zhang M. Z., Wu J. H., Zhou J. W., Liang Z., et al. (2018). Combined influence of insulin resistance and inflammatory biomarkers on type 2 diabetes: A population-based prospective cohort study of inner mongolians in China. Biomed. Environ. Sci. 31, 300–305. 10.3967/bes2018.038 [DOI] [PubMed] [Google Scholar]
- Quinn J. J., Chang H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Ramirez C. M., Dávalos A., Goedeke L., Salerno A. G., Warrier N., Cirera-Salinas D., et al. (2011). MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. ATVB 31, 2707–2714. 10.1161/ATVBAHA.111.232066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., et al. (1996). B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172. 10.1084/jem.183.3.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Stoorvogel W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real J. M., Ferreira L. R. P., Esteves G. H., Koyama F. C., Dias M. V. S., Bezerra-Neto J. E., et al. (2018). Exosomes from patients with septic shock convey miRNAs related to inflammation and cell cycle regulation: new signaling pathways in sepsis? Crit. Care. 15 (22), 68. 10.1186/s13054-018-2003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P. D., Morelli A. E. (2014). Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195–208. 10.1038/nri3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J., Gawad C., Wang P. L., Lacayo N., Brown P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7, e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickel R., Boyerinas B., Park S. M., Peter M. E. (2008). MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27, 5959–5974. 10.1038/onc.2008.274 [DOI] [PubMed] [Google Scholar]
- Schmidt E., Dhaouadi I., Gaziano I., Oliverio M., Klemm P., Awazawa M., et al. (2018). LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 9, 3622. 10.1038/s41467-018-05933-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz S. U., Grote P., Herrmann B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 73, 2491–2509. 10.1007/s00018-016-2174-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R., Patel T., Freedman J. E. (2018). Circulating extracellular vesicles in human disease. N. Engl. J. Med. 379, 2180–2181. 10.1056/NEJMc1813170 [DOI] [PubMed] [Google Scholar]
- Shang A., Gu C., Wang W., Wang X., Sun J., Zeng B., et al. (2020). Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol. Cancer 19, 117. 10.1186/s12943-020-01235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Jia Y. A., Wang X., Zhang J., Liu K., Jing W., et al. (2021). Exosomes from adipose-derived stem cells alleviate the inflammation and oxidative stress via regulating Nrf2/HO-1 axis in macrophages. Free. Radic. Bio. Med. 165, 54–66. 10.1016/j.freeradbiomed.2021.01.023 [DOI] [PubMed] [Google Scholar]
- Shenoy A., Blelloch R. H. (2014). Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat. Rev. Mol. Cell Biol. 15, 565–576. 10.1038/nrm3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Zhang M., Tong M., Yang L., Pang L., Chen L., et al. (2015). miR-148a is associated with obesity and modulates adipocyte differentiation of mesenchymal stem cells through Wnt signaling. Sci. Rep. 5, 9930. 10.1038/srep09930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K. K., Kim Y. S., Kim J. Y., Bae Y. C., Jung J. S. (2014). miR-137 controls proliferation and differentiation of human adipose tissue stromal cells. Cell Physiol. biochem. 33, 758–768. 10.1159/000358650 [DOI] [PubMed] [Google Scholar]
- Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009). Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283. 10.1586/epr.09.17 [DOI] [PubMed] [Google Scholar]
- Smits G., Mungall A. J., Griffiths-Jones S., Smith P., Beury D., Matthews L., et al. (2008). Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat. Genet. 40, 971–976. 10.1038/ng.168 [DOI] [PubMed] [Google Scholar]
- Son Y. H., Ka S., Kim Y. A., Kim J. B. (2014). Regulation of adipocyte differentiation via microRNAs. Endocrinol. Metab. 29, 122–135. 10.3803/EnM.2014.29.2.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M., Han L., Chen F., Wang D., Wang F., Zhang L., et al. (2018). Adipocyte-derived exosomes carrying sonic hedgehog mediate M1 macrophage polarization-induced insulin resistance via ptch and PI3K pathways. Cell Physiol. biochem. 48, 1416–1432. 10.1159/000492252 [DOI] [PubMed] [Google Scholar]
- Sun D., Ding Z., Shen L., Yang F., Han J., Wu G. (2021). miR-410-3P inhibits adipocyte differentiation by targeting IRS-1 in cancer-associated cachexia patients. Lipids. Health. Dis. 20, 115. 10.1186/s12944-021-01530-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Wang J., Pan Q., Yu Y., Zhang Y., Wan Y., et al. (2009). Characterization of function and regulation of miR-24-1 and miR-31. Biochem. Biophys. Res. Commun. 380, 660–665. 10.1016/j.bbrc.2009.01.161 [DOI] [PubMed] [Google Scholar]
- Sun J., Wang Y., Li Y., Zhao G. (2014). Downregulation of PPARγ by miR-548d-5p suppresses the adipogenic differentiation of human bone marrow mesenchymal stem cells and enhances their osteogenic potential. J. Transl. Med. 12, 168. 10.1186/1479-5876-12-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Goff L. A., Trapnell C., Alexander R., Lo K. A., Hacisuleyman E., et al. (2013). Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 110, 3387–3392. 10.1073/pnas.1222643110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xie H., Mori M. A., Alexander R., Yuan B., Hattangadi S. M., et al. (2011). Mir193b–365 is essential for brown fat differentiation. Nat. Cell. Biol. 13, 958–965. 10.1038/ncb2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Sun X., Chu W., Yu S., Dong F., Xu G. (2020). CircRNA expression profiles in human visceral preadipocytes and adipocytes. Mol. Med. Rep. 21, 815–821. 10.3892/mmr.2019.10886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha E. A., Ono K., Eguchi T. (2019). Roles of extracellular HSPs as biomarkers in immune surveillance and immune evasion. Int. J. Mol. Sci. 20, 4588. 10.3390/ijms20184588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanabe R., Ono K., Abe Y., Takaya T., Horie T., Wada H., et al. (2008). Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem. Biophys. Res. Commun. 376, 728–732. 10.1016/j.bbrc.2008.09.050 [DOI] [PubMed] [Google Scholar]
- Tang R., Ma F., Li W., Ouyang S., Liu Z., Wu J. (2017). miR-206-3p inhibits 3T3-L1 cell adipogenesis via the c-Met/PI3K/Akt pathway. Int. J. Mol. Sci. 18, 1510. 10.3390/ijms18071510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. F., Zhang Y., Li X. Y., Li C., Tian W., Liu L. (2009). Expression of miR-31, miR-125b-5p, and miR-326 in the adipogenic differentiation process of adipose-derived stem cells. OMICS 13, 331–336. 10.1089/omi.2009.0017 [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Nakajima I., Chikuni K., Kojima M., Awata T., Mikawa S. (2014). MicroRNA-33b downregulates the differentiation and development of porcine preadipocytes. Mol. Biol. Rep. 41, 1081–1090. 10.1007/s11033-013-2954-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomou T., Mori M. A., Dreyfuss J. M., Konishi M., Sakaguchi M., Wolfrum C., et al. (2017). Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455. 10.1038/nature21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Wang M., Dai Y., Bao D., Zhang J., Pan H. (2019). LncRNA HOXA-AS3 sponges miR-29c to facilitate cell proliferation, metastasis, and EMT process and activate the MEK/ERK signaling pathway in hepatocellular carcinoma. Hum. gene Ther. Clin. Dev. 30, 129–141. 10.1089/humc.2018.266 [DOI] [PubMed] [Google Scholar]
- Trajkovski M., Ahmed K., Esau C. C., Stoffel M. (2012). MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell. Biol. 14, 1330–1335. 10.1038/ncb2612 [DOI] [PubMed] [Google Scholar]
- Tucci V., Isles A. R., Kelsey G., Ferguson-Smith A. C., Tucci V., Bartolomei M. S., et al. (2019). Genomic imprinting and physiological processes in mammals. Cell 176, 952–965. 10.1016/j.cell.2019.01.043 [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 9, 654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Vickers K. C., Landstreet S. R., Levin M. G., Shoucri B. M., Toth C. L., Taylor R. C., et al. (2014a). MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. U. S. A. 111, 14518–14523. 10.1073/pnas.1215767111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z., Chen X., Gao X., Dong Y., Zhao Y., Wei M., et al. (2019). Chronic myeloid leukemia‐derived exosomes attenuate adipogenesis of adipose derived mesenchymal stem cells via transporting miR‐92a‐3p. J. Cell. Physiol. 234, 21274–21283. 10.1002/jcp.28732 [DOI] [PubMed] [Google Scholar]
- Wang D., Ming X., Xu J., Xiao Y. (2021). Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol. Oncol. 39 (3), 390–400. 10.1002/hon.2874 [DOI] [PubMed] [Google Scholar]
- Wang H., Tang J., Ding Y., Guo F. (2021). Exploring associations of non-coding RNAs in human diseases via three-matrix factorization with hypergraph-regular terms on center kernel alignment. Brief. Bioinform. 22, bbaa409. 10.1093/bib/bbaa409 [DOI] [PubMed] [Google Scholar]
- Wang J., Li L., Zhang Z., Zhang X., Zhu Y., Zhang C., et al. (2022). Extracellular vesicles mediate the communication of adipose tissue with brain and promote cognitive impairment associated with insulin resistance. Cell Metab. 34, 1264–1279.e8. 10.1016/j.cmet.2022.08.004 [DOI] [PubMed] [Google Scholar]
- Wang Q., Li Y. C., Wang J., Kong J., Qi Y., Quigg R. J., et al. (2008). miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl. Acad. Sci. U. S. A. 105, 2889–2894. 10.1073/pnas.0800178105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li X., Xu M., Wang J., Zhao R. C. (2017). Reduced adipogenesis after lung tumor exosomes priming in human mesenchymal stem cells via TGFβ signaling pathway. Mol. Cell Biochem. 435, 59–66. 10.1007/s11010-017-3056-3 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang X. J., Ji Y. X., Zhang P., Deng K. Q., Gong J., et al. (2016). The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 22, 1131–1139. 10.1038/nm.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A., Bushell M. (2015). The complexity of miRNA-mediated repression. Cell Death Differ. 22, 22–33. 10.1038/cdd.2014.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Dong T., Chen T., Sun J., Luo J., He J., et al. (2020). Hepatic exosome-derived miR-130a-3p attenuates glucose intolerance via suppressing PHLPP2 gene in adipocyte. Metabolism 103, 154006. 10.1016/j.metabol.2019.154006 [DOI] [PubMed] [Google Scholar]
- Xiao T., Liu L., Li H., Sun Y., Luo H., Li T., et al. (2015). Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Rep. 5, 856–865. 10.1016/j.stemcr.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Liu L., Li H., Sun Y., Luo H., Li T., et al. (2021). Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Rep. 16, 1006–1008. 10.1016/j.stemcr.2021.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Tan J., Miao Y., Lv Y., Zhang Q. (2021). Crosstalk between exosomes and autophagy: A review of molecular mechanisms and therapies. J. Cell. Mol. Med. 25 (5), 2297–2308. 10.1111/jcmm.16276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Gerin I., Miao H., Vu-Phan D., Johnson C. N., Xu R., et al. (2010). Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE 5, e14199. 10.1371/journal.pone.0014199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy S. Y., Guo M., Hay B. A. (2003). The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13, 790–795. 10.1016/s0960-9822(03)00250-1 [DOI] [PubMed] [Google Scholar]
- Xu S., Wang Y., Li Z., Hua Q., Jiang M., Fan X. (2022). LncRNA GAS5 knockdown mitigates hepatic lipid accumulation via regulating MiR-26a-5p/PDE4B to activate cAMP/CREB pathway. Front. Endocrinol. 13, 889858. 10.3389/fendo.2022.889858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. C., Lin Y. S., Zhang L., Lu Y., Sun Y. L., Fang Z. G., et al. (2020). MicroRNAs of bone marrow mesenchymal stem cell-derived exosomes regulate acute myeloid leukemia cell proliferation and apoptosis. Chin. Med. J. Engl. 5, 2829–2839. 10.1097/CM9.0000000000001138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Tao Z. F., Li X. M., Zhang H., Yao J., Jiang Q. (2014). Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 55, 941–951. 10.1167/iovs.13-13221 [DOI] [PubMed] [Google Scholar]
- Yan S., Wang T., Huang S., Di Y., Huang Y., Liu X., et al. (2016). Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol. 53, 693–702. 10.1007/s00592-016-0837-1 [DOI] [PubMed] [Google Scholar]
- Yan Y., Li K., Jiang J., Jiang L., Ma X., Ai F., et al. (2023). Perinatal tissue-derived exosomes ameliorate colitis in mice by regulating the Foxp3 + Treg cells and gut microbiota. Stem. Cell. Res. Ther. 20, 43. 10.1186/s13287-023-03263-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Li P., Yang W., Ruan X., Kiesewetter K., Zhu J., et al. (2016). Integrative transcriptome analyses of metabolic responses in mice define pivotal LncRNA metabolic regulators. Cell Metab. 24, 627–639. 10.1016/j.cmet.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Bian C., Zhou H., Huang S., Wang S., Liao L., et al. (2011). MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 20, 259–267. 10.1089/scd.2010.0072 [DOI] [PubMed] [Google Scholar]
- Yao X., Liu C., Liu C., Xi W., Sun S., Gao Z. (2019). lncRNA SNHG7 sponges miR‐425 to promote proliferation, migration, and invasion of hepatic carcinoma cells via Wnt/β‐catenin/EMT signalling pathway. Cell biochem. Funct. 37, 525–533. 10.1002/cbf.3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasaka T., Mantich N. M., Boxer L. A., Baehner R. L. (1981). Functions of human monocyte and lymphocyte subsets obtained by countercurrent centrifugal elutriation: differing functional capacities of human monocyte subsets. J. Immunol. 127, 1515–1518. 10.4049/jimmunol.127.4.1515 [DOI] [PubMed] [Google Scholar]
- Yi C., Xie W., Li F., Lv Q., He J., Wu J., et al. (2011). MiR-143 enhances adipogenic differentiation of 3T3-L1 cells through targeting the coding region of mouse pleiotrophin. FEBS Lett. 585, 3303–3309. 10.1016/j.febslet.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Yi F., Yang F., Liu X., Chen H., Ji T., Jiang L., et al. (2013). RNA-seq identified a super-long intergenic transcript functioning in adipogenesis. RNA Biol. 10, 991–1001. 10.4161/rna.24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pasut A., Soleimani V. D., Bentzinger C. F., Antoun G., Thorn S., et al. (2013). MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 17, 210–224. 10.1016/j.cmet.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J. B., et al. (2017). Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell 171, 372–384. 10.1016/j.cell.2017.08.035 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Ren K., Zhu X., Zheng Z., Yi G. (2018). Long noncoding RNAs: advances in lipid metabolism. Adv. Clin. Chem. 87, 1–36. 10.1016/bs.acc.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Zhang B., Yang Y., Xiang L., Zhao Z., Ye R. (2019). Adipose-derived exosomes: A novel adipokine in obesity-associated diabetes. J. Cell. Physiol. 234, 16692–16702. 10.1002/jcp.28354 [DOI] [PubMed] [Google Scholar]
- Zhang H., Liu J., Qu D., Wang L., Wong C. M., Lau C. W., et al. (2018). Serum exosomes mediate delivery of arginase 1 as a novel mechanism for endothelial dysfunction in diabetes. Proc. Natl. Acad. Sci. U. S. A. 115, E6927–E6936. 10.1073/pnas.1721521115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., et al. (2012). Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22, 107–126. 10.1038/cr.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang D., Qin Z. Y., Li J., Shen Z. Y. (2020). The role and possible mechanism of long noncoding RNA PVT1 in modulating 3T3-L1 preadipocyte proliferation and differentiation. IUBMB Life 72, 1460–1467. 10.1002/iub.2269 [DOI] [PubMed] [Google Scholar]
- Zhang N., Nan A., Chen L., Li X., Jia Y., Qiu M., et al. (2020). Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer 19, 101. 10.1186/s12943-020-01221-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. F., Gao C., Huang X. Y., Lu J. C., Guo X. J., Shi G. M., et al. (2020). Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 19, 110. 10.1186/s12943-020-01222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhang Z., Xia T., Liu C., Sun C. (2019). circNrxn2 promoted WAT browning via sponging miR-103 to relieve its inhibition of FGF10 in HFD mice. Mol. Ther. Nucleic Acids 17, 551–562. 10.1016/j.omtn.2019.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen L., Xiao B., Liu H., Su Y. (2019). Circ_0075932 in adipocyte-derived exosomes induces inflammation and apoptosis in human dermal keratinocytes by directly binding with PUM2 and promoting PUM2-mediated activation of AuroraA/NF-κB pathway. Biochem. Biophys. Res. Commun. 511, 551–558. 10.1016/j.bbrc.2019.02.082 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu M., Dai M., Chen C., Tang Q., Jing W., et al. (2017). miR-450a-5p within rat adipose tissue exosome-like vesicles promotes adipogenic differentiation by targeting WISP2. J. Cell Sci. 130, 1158–1168. 10.1242/jcs.197764 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu M., Tian W. (2016). Physiological and pathological impact of exosomes of adipose tissue. Cell Prolif. 49, 3–13. 10.1111/cpr.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang T., Feng R., Huang H., Xia T., Sun C. (2019). circARF3 alleviates mitophagy-mediated inflammation by targeting miR-103/TRAF3 in mouse adipose tissue. Mol. Ther. Nucleic Acids 14, 192–203. 10.1016/j.omtn.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Shang Q., Pan Z., Bai Y., Li Z., Zhang H., et al. (2018). Exosomes from adipose-derived stem cells attenuate adipose inflammation and obesity through polarizing M2 macrophages and beiging in white adipose tissue. Diabetes 67, 235–247. 10.2337/db17-0356 [DOI] [PubMed] [Google Scholar]
- Zhao X. Y., Li S., Wang G. X., Yu Q., Lin J. D. (2014). A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell. 55, 372–382. 10.1016/j.molcel.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi F., Ding Y., Wang R., Yang Y., Luo K., Hua F. (2021). Exosomal hsa_circ_0006859 is a potential biomarker for postmenopausal osteoporosis and enhances adipogenic versus osteogenic differentiation in human bone marrow mesenchymal stem cells by sponging miR-431-5p. Stem Cell Res. Ther. 12, 157. 10.1186/s13287-021-02214-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu E., Zhang J., Li Y., Yuan H., Zhou J., Wang B. (2019). Long noncoding RNA Plnc1 controls adipocyte differentiation by regulating peroxisome proliferator‐activated receptor γ. FASEB J. 33, 2396–2408. 10.1096/fj.201800739RRR [DOI] [PubMed] [Google Scholar]
- Zhu L., Chen L., Shi C. M., Xu G. F., Xu L. L., Zhu L. L., et al. (2014). MiR-335, an adipogenesis-related microRNA, is involved in adipose tissue inflammation. Cell biochem. Biophys. 68, 283–290. 10.1007/s12013-013-9708-3 [DOI] [PubMed] [Google Scholar]
- Zhu X. X., Yan Y. W., Chen D., Ai C. Z., Lu X., Xu S. S., et al. (2016). Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 7, 63561–63570. 10.18632/oncotarget.11538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Gui W., Lin X., Li H. (2020). Knock-down of circular RNA H19 induces human adipose-derived stem cells adipogenic differentiation via a mechanism involving the polypyrimidine tract-binding protein 1. Exp. Cell Res. 387, 111753. 10.1016/j.yexcr.2019.111753 [DOI] [PubMed] [Google Scholar]