Abstract
The mammalian gastrointestinal tract (GIT) hosts a diverse and highly active microbiota composed of bacteria, eukaryotes, archaea, and viruses. Studies of the GIT microbiota date back more than a century, although modern techniques, including mouse models, sequencing technology, and novel therapeutics in humans, have been foundational to our understanding of the roles of commensal microbes in health and disease. Here, we review the impacts of the GIT microbiota on viral infection, both within the GIT and systemically. GIT-associated microbes and their metabolites alter the course of viral infection through a variety of mechanisms, including direct interactions with virions, alteration of the GIT landscape, and extensive regulation of innate and adaptive immunity. Mechanistic understanding of the full breadth of interactions between the GIT microbiota and the host is still lacking in many ways but will be vital for the development of novel therapeutics for viral and nonviral diseases alike.
Keywords: microbiota, metabolites, microbiome, virome, interferon, signaling
INTRODUCTION
All multicellular organisms, from plants to fungi to mammals, host communities of microbes within and on their bodies, collectively known as the microbiota. While the microbiota has been increasingly well characterized in humans and manipulated in diverse model organisms such as Caenorhabditis elegans (roundworm), Drosophila melanogaster (fruit fly), and Mus musculus (house mouse), more comprehensive survey efforts are ongoing to develop a fuller understanding of animal-microbiota interactions (1). All mucosal and likely some other body sites are colonized by microbes that mediate numerous local and systemic effects on the host as well as on other members of the microbial community. Here, we predominantly focus upon the microbiota of the mammalian gastrointestinal tract (GIT) as the most rich, complex, and well-studied site. Because it is easily sampled by analysis of fecal material, which can provide an approximation of the GIT microbiota, this site has been thoroughly profiled longitudinally, across many individuals, and in a variety of disease states, including in several Human Microbiome Projects (2).
The GIT microbiota is composed of bacteria, archaea, fungi, other microbial eukaryotes, and viruses (Figure 1). The best-studied component of the GIT microbiota is the bacteria. A large portion of GIT-associated bacteria are cultivable and have been studied for more than a century. The advent of next-generation sequencing has allowed for culture-independent study of the microbiome, the genetic component of the microbiota, primarily through the study of the 16S ribosomal RNA (rRNA) genes of bacteria and archaea, as well as the internal transcribed spacer and 18S rRNA genes of eukaryotes (Figure 2). More recently, metagenomics and viromics, made possible by the decreasing costs of high-throughput sequencing technologies, have emerged as techniques to describe the enormous genomic diversity contained in the GIT microbiome, both within and between individuals (3).
Figure 1.
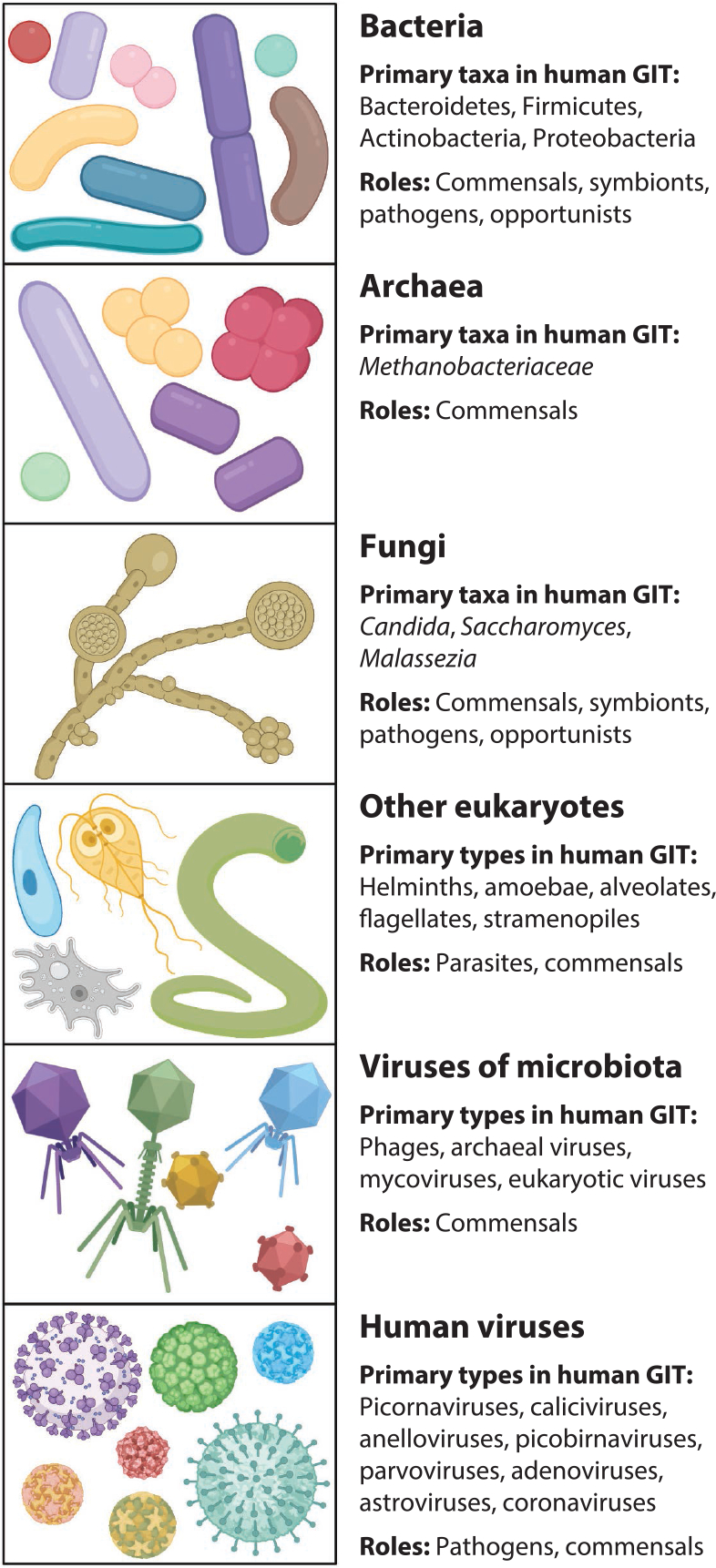
Components of the mammalian gastrointestinal tract (GIT) microbiota. The complex community of the GIT includes bacteria such as major phyla Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria; archaea such as family Methanobacteriaceae; and fungi such as genera Candida, Saccharomyces, and Malassezia. Other eukaryotes such as helminths may also variably be present. Additionally, viruses that target these various microbes as well as viruses that directly target humans are critical components of the microbiota. Figure adapted from images created with BioRender.com.
Figure 2.
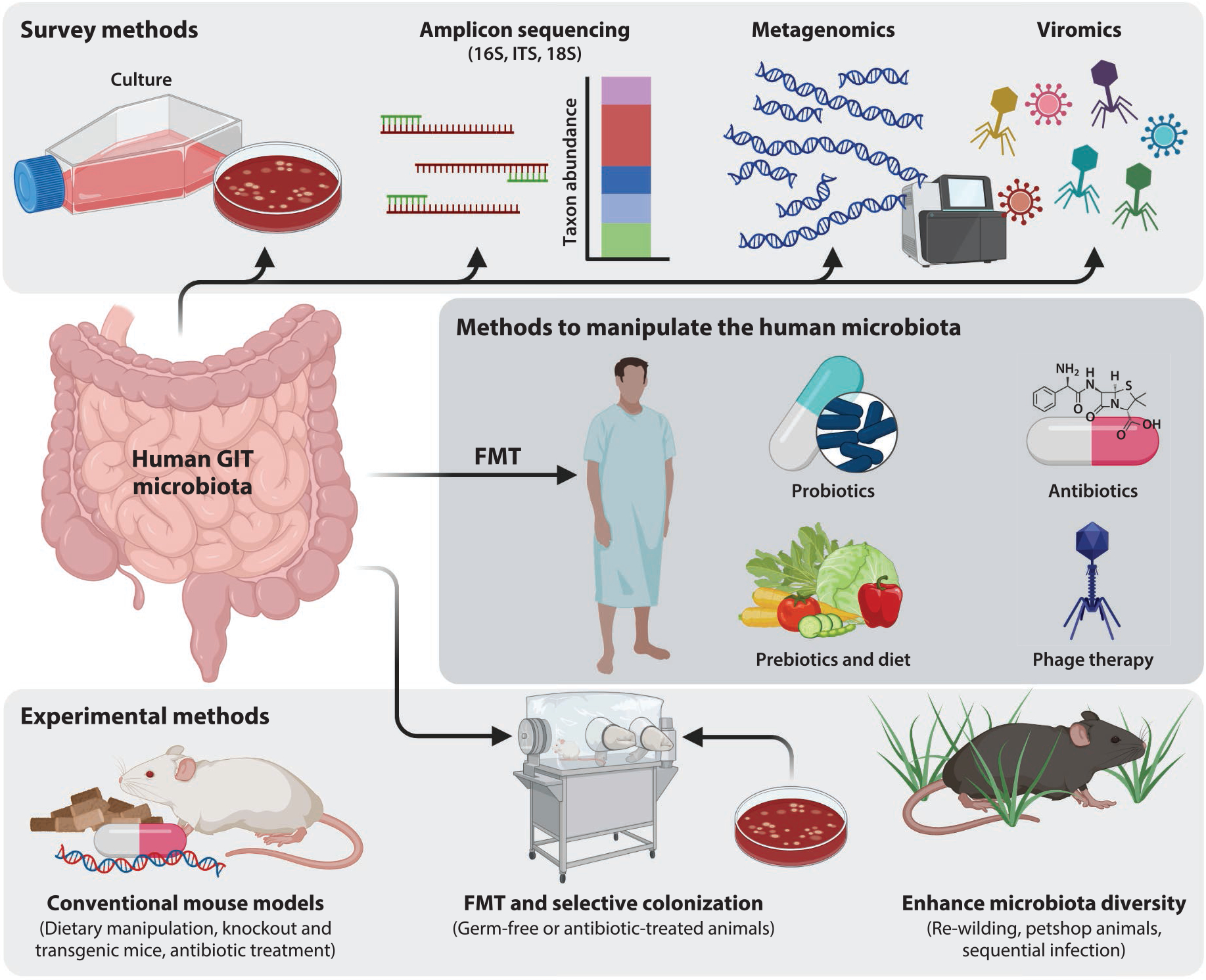
Methods to study and manipulate the gastrointestinal tract (GIT) microbiota. Surveys of the gut microbiota can leverage traditional microbial cultivation or sequence-based analysis, which avoids biases in cultivation. The in situ human GIT microbiota can be manipulated through fecal microbiota transplant (FMT); alterations to the diet; and the administration of probiotics, antibiotics, prebiotics, and phage therapy. Experimental methods to study the human GIT microbiota rely heavily on the use of mouse models, which are readily manipulated in many ways. Figure adapted from images created with BioRender.com.
The GIT microbiota is a vital component of human biology and development, and it is increasingly recognized as a functional organ. The microbiota shapes the gut environment through its diverse metabolic activities, which are often beneficial to human hosts, such as the breakdown of digestion-resistant food components such as fiber, synthesis of vitamin K, exclusion of pathogens, and immune development and modulation. Functional study of the GIT microbiota has been made possible primarily through the manipulation of mouse models (Figure 2). These include methods to reduce or tailor microbial diversity, such as antibiotic treatments and germ-free (GF) models, as well as methods to humanize or alter the microbiota through fecal microbiota transplant (FMT), dietary changes, or addition of microbes or metabolites. Recently, interest has increased in methods to enhance the microbial diversity of animal models raised in clean facilities, such as rewilding, cohousing with pet shop animals, and sequential infection, to better mimic the microbial exposures of humans. With regard to direct microbiota manipulation in humans, FMT and probiotic administration are used as therapeutic avenues for intestinal diseases including Clostridioides difficile infection and inflammatory bowel diseases (IBDs) (4, 5). Prebiotic approaches leveraging dietary manipulations (6) and phage therapies (7) are emerging treatments for numerous human diseases (Figure 2).
Here, we describe what is currently known about the impact of the GIT microbiota on both enteric viruses that infect the intestine directly, such as norovirus and rotavirus (RV), and viruses that infect at distal body sites or systemically, such as influenza virus and flaviviruses. First, we summarize what is known about the effects of the GIT microbiota on initial viral infection and replication, including direct interactions between commensal microbes and viruses, effects of microbial metabolites, and interactions between viral and bacterial pathogens. Then, we describe how the GIT microbiota modulates local and systemic effects of the initial intrinsic host response to viral infection. Finally, we review the roles of the microbiota in the maintenance and regulation of immune cell populations to contribute to adaptive responses to viruses.
EFFECTS OF THE MICROBIOTA ON INITIAL VIRAL INFECTION AND REPLICATION
All enteric viruses, which predominantly infect via the fecal-oral route, have naturally evolved with the microbiota as part of the environment present in the host’s gut lumen at the time of infection. Interactions with the GIT microbiota can be either detrimental or beneficial for a virus. Moreover, these outcomes can occur via direct physical interactions between the microbiota and virus particles or via indirect interactions mediated by microbial metabolites.
Associations Between the Microbiota and Viral Disease
Enteric viral infection causing diarrhea, known as acute viral gastroenteritis (AVG), often leads to bacterial dysbiosis, or an imbalance of bacterial types, of the GIT microbiota. The mechanisms causing dysbiosis, as well as its consequences, following viral infection are poorly understood. While chronic dysbiosis of the GIT microbiota is implicated in several human disease states, including IBD and cardiovascular disease, the microbiota of otherwise healthy individuals generally recovers to a healthy state after alleviation of AVG symptoms (8, 9).
AVG is broadly associated with overall decreased microbial diversity, decreased abundances of Bacteroidetes, and increased abundances of Firmicutes (9–11). Some alterations to the microbiota are specific to the viral pathogen. Human norovirus (HNoV) is a single-stranded RNA virus belonging to the Caliciviridae family, RV is a double-stranded RNA virus in the Reoviridae family, and human astrovirus (HAstV) is a positive-sense RNA virus in the Astroviridae family. These pathogens are transmitted predominantly via the fecal-oral route and are common causes of AVG, with RV causing severe diarrheal disease in children, while HNoV causes epidemic outbreaks across broader age ranges and HAstV is more variably associated with diarrheal illness. Reductions in the abundance of Bifidobacterium are most associated with RV and HNoV, and less pronounced during infection with HAstV (10). RV infection causes especially severe loss of bacterial diversity, at least partly attributable to increased abundances of Gammaproteobacteria (8).
Dysbiosis of the GIT microbiota during AVG can allow for the proliferation of opportunistic pathogens, including Campylobacter, Neisseria, and Enterobacteriaceae, which are linked to disease complications (12). In children with hand, foot, and mouth disease (HFMD), caused by several types of enteroviruses (EVs), increased abundances of Clostridium sp. L2–50 and Bacteroides stercoris are linked to more severe disease symptoms (13). Metagenomic analysis of the same cohort revealed enrichment of genes related to bacterial secretion systems, pathogenicity, and drug resistance in severe HFMD compared to mild cases, suggesting that specific bacterial metabolic activities are linked to disease severity (13). Thus, prevention or minimization of bacterial dysbiosis during AVG through the use of probiotics, prebiotics, or phage therapy may be an important therapeutic avenue in the future.
A major limitation of observational studies examining the GIT microbiota during viral infection is that they are generally unable to establish the preinfection state of an individual’s microbiota. Although broad patterns of dysbiosis subsequent to viral infection have been established, specific taxa and genetic factors of the human GIT microbiota that exacerbate AVG have been difficult to determine. One HNoV human challenge study found that a preinfection GIT microbiota enriched in Bacteroidetes and depleted in Clostridia was associated with asymptomatic HNoV infection (11). Metagenomic analysis of the same cohort identified specific glycan metabolism and cell-cell signaling pathways enriched in the microbiomes of symptomatic individuals prior to infection, implicating both the composition and activities of the GIT microbiota in AVG severity and onset.
Enteric viral infection can further alter the microbiota at distal body sites. During symptomatic HFMD, increased abundances of Streptococcus spp. were found in the oral microbiome and were positively correlated with viral RNA levels in saliva (14). The same study observed an altered salivary virome, including both human viruses and phages, during HFMD compared to healthy controls (14). Further, viral infection outside the GIT has been linked to an altered GIT microbiota, such as in the case of respiratory syncytial virus (RSV) infection (15). This link between respiratory viral infection and GIT microbiota dysbiosis has been investigated in a mouse model of influenza and is mediated by adaptive immune cells (16). Moreover, members of the respiratory tract microbiota can be associated with the onset of viral respiratory diseases (17, 18). Thus, the entirety of the human microbiota, spanning microbial types and body sites local and distal to the site of infection, may affect or be affected by viral infection.
Direct Interactions Between Viruses and the Bacterial Microbiota
Direct interactions between viruses and bacteria are inherently complex and depend on the bacterial surface structures, such as lipopolysaccharide (LPS) and peptidoglycan (PGN), and viral capsid moieties involved in these interactions. In many cases, the direct binding of viral particles to bacterial cells and the effects of virus-bacteria interaction depend on the viral and bacterial strains involved in the interaction (19), although investigation with a broader diversity of bacterial strains is needed to better understand the selectivity of viral capsids for bacterial binding partners and to reflect the immensely diverse human GIT microbiota. The outcomes of bacterial binding likely depend on a given virus-bound bacterium’s affinity to mammalian cells, ability to persist in the gut lumen, and effects on the mammalian host.
A diverse array of enteric viruses, including poliovirus (PV), murine norovirus (MNoV), and coxsackievirus B3 (CVB3), are less infectious and/or pathogenic in GF or bacteria-depleted animal models (20–23), supporting the idea that interactions between the GIT microbiota and invading viruses are broadly important for infection outcomes (Figure 3a). PV, a nonenveloped, single-stranded RNA virus from the Picornaviridae family that can cause central nervous system infections and paralysis in humans, is currently the best-characterized system for studying direct bacteria-virus interactions in the GIT, although mechanistic understanding of most bacteria-virus interactions is still lacking.
Figure 3.
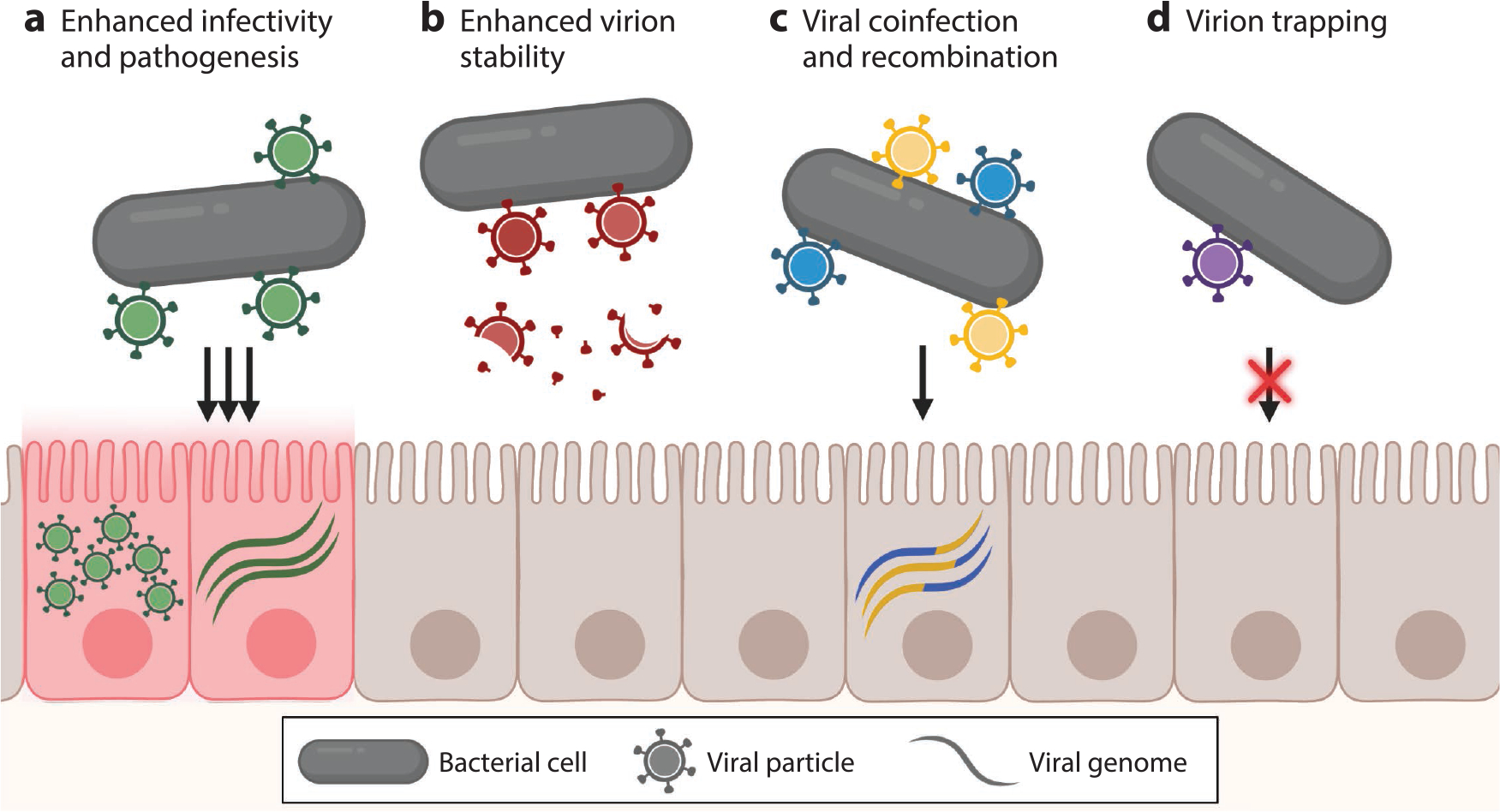
Potential outcomes of direct interactions between enteric viruses and bacteria. (a) Bacterial binding of viral particles can enhance viral infectivity and pathogenesis. (b) Viral particles bound to bacterial cells or bacterial components may be more stable than free viral particles. (c) Binding of multiple viral particles to bacterial cells can lead to viral coinfection of the same cell and recombination of viral genomes. (d) Binding of viral particles to bacterial cells can trap them, preventing or limiting infection of mammalian host cells. Figure adapted from images created with BioRender.com.
Some of the increased infectivity of viruses bound to bacteria or bacterial cell wall components is directly caused by increased viral stability (Figure 3b). PV binds to bacterial LPS and PGN via the viral protein 1 capsid protein, leading to increased stability in the mouse gut and transmissibility between hosts (24). Similarly, other picornaviruses bind to bacterial cells and LPS, stabilizing them to heat treatment and bleach exposure in vitro (19). Binding of specific species of bacteria by CVB3, a cardiopathogenic virus also in the Picornaviridae family, increases viral infectivity and stability in a mouse model and is mediated by O-antigen, a highly polymorphic component of LPS (19, 25). Reovirus, a nonenveloped, double-stranded RNA virus from the Reoviridae family that causes intestinal inflammation and tissue damage in immunocompromised mice, directly interacts with LPS and PGN on diverse bacteria, increasing its thermostability in cell culture (26).
Multiple PV virions can bind to single bacterial cells, creating locally high multiplicities of infection that allow for coinfection of individual host cells and for subsequent genomic recombination to occur (27) (Figure 3c). Although PV binds to a broad diversity of bacteria, only specific strains promote viral coinfection, which is likely due to differential affinities of bacterial strains to mammalian cells (28). This coinfection of the same host cell by multiple PV particles allows for genomic recombination, and bacterial binding increases PV recombination rates nearly fivefold (28). In humans, recombination has been observed between PV strains, including vaccine strains (29, 30), as well as recombination of PV with other EVs (31), together suggesting that bacterial binding plays an important role in picornavirus evolution.
Viruses may also interact with bacterial products that resemble mammalian host attachment factors. HNoV and RV bind to histo-blood group antigen-like (HBGA-like) glycoproteins expressed on bacterial cells, which resemble HBGAs, an important attachment factor for these viruses on human intestinal cells (23, 32, 33). HNoV binds to HBGA-like glycoproteins on the surface of diverse GIT-associated bacteria, including Escherichia coli, Enterobacter, and C. difficile, which protect viral particles from heat stress and lead to enhanced attachment to and infection of cultured cells (23, 32, 34). MNoV, a small-animal model virus that shares many characteristics with HNoV, binds to both Gram-negative and Gram-positive bacterial cells, but only binding to Gram-positive bacteria leads to increased thermostability of viral particles and does not lead to increases in infectivity in cell culture (35).
Converse to these proviral effects of enteric bacteria, infection by some enteric viruses is inhibited by specific microbes (Figure 3d). A probiotic strain of Lactobacillus reuteri binds to enterovirus 71 (EV71) and coxsackieviruses (CVs) A6 and A16, causative agents of HFMD, thereby inhibiting viral entry into host cells (36). Segmented filamentous bacteria (SFB), common GIT commensals in mice, provide protection against infection by murine RV (mRV) via multiple effects on the host GIT environment, including altered host gene expression and accelerated epithelial cell turnover, as well as by neutralizing viral particles via direct interactions (37). HAstV presents a peculiar case, wherein LPS, PGN, and individual gut-associated bacteria enhance HAstV thermostability and infectivity in Caco2 cells through direct interactions (38). However, examination of the effects of complex human fecal material on HAstV infection revealed a large degree of variation across individual donors, ranging from increased HAstV infectivity to protection from HAstV infection, suggesting that unknown, variable components of the GIT microbiota can have protective effects against HAstV infection (38). Infection by murine astrovirus (muAstV), a small-animal model for HAstV, however, proceeds uninhibited in GF mice, suggesting a potentially minimal role for the bacterial microbiota in regulating some astrovirus strains (39).
Indirect Interactions Between Enteric Viruses and Bacterial Microbiota
Enteric microbes drastically alter the gut landscape via their diverse metabolic activities, which produce a variety of bioactive compounds that modulate the chemical composition of the lumen, host cellular populations, and immune activities. Further, because many of the small metabolites produced by the GIT microbiota can circulate to trigger systemic signals, the microbiota also affects viral infection even at distal body sites.
Bacterial metabolism in the intestine is dominated by the fermentation of undigested complex carbohydrates from the diet as well as host-derived mucins. Mucin degradation is an important avenue by which the microbiota alters the GIT environment and the course of viral infection. Increases in mucin-digesting Bacteroides and Akkermansia and degradation of mucin during mRV infection reduce the binding capacity of mRV virions for host cells in vitro and in vivo (40). Carbohydrate fermentation by the GIT microbiota produces short-chain fatty acids (SCFAs), primarily acetate, butyrate, and propionate (Figure 4a). SCFAs are readily circulated to other tissues via the bloodstream and alter both local and systemic immune responses to infection. Many SCFAs function as histone deacetylase (HDAC) inhibitors and have broad effects on epigenetic regulation in host cells, such as altering immune responses (41) and activating the latent herpesvirus Epstein-Barr virus (42). Butyrate is an especially potent HDAC inhibitor (43) and causes increased expression of the coxsackie and adenovirus receptor (CAR) in a colon cancer cell line (44). In this way, microbial-derived butyrate may play an indirect role in susceptibility to HFMD caused by CVs and EVs that use CAR for cell entry (45).
Figure 4.
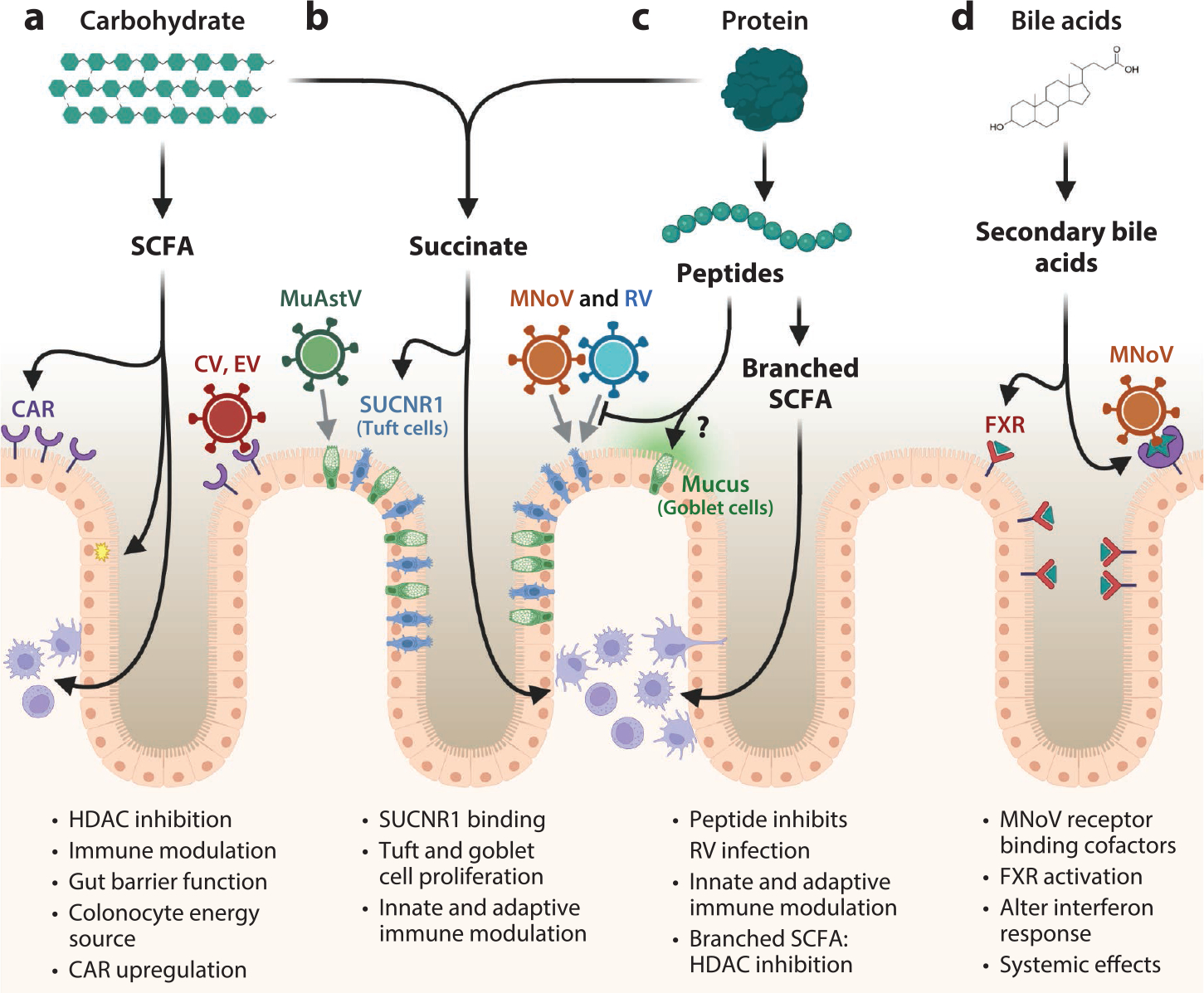
Interactions between microbial metabolites and viral infection. The by-products of microbial metabolism—(a) short chain fatty acid (SCFA), (b) succinate, (c) peptides and branched SCFA, and (d) secondary bile acids—exert strong effects on the gut environment and enteric viral infection. (a) Bacterial fermentation of carbohydrates produces several SCFA species, which are important energy sources for colonocytes and upregulate the coxsackievirus (CV) and adenovirus receptor (CAR) to enhance CV and enterovirus (EV) infection. (b) Succinate is produced from the fermentation of both amino acids and carbohydrates. Succinate binding to succinate receptor 1 (SUCNR1) causes goblet and tuft cell proliferation, which are host cells for murine astrovirus (muAstV), and murine norovirus (MNoV) and rotavirus (RV), respectively. (c) Protein fermentation and decarboxylation produce a variety of by-products that can influence host immune activities. SCFA, succinate, and branched SCFA are all potent histone deacetylase (HDAC) inhibitors and affect host gene expression on broad scales, including in important immune cell populations. (d) Gut-associated bacteria deconjugate and modify bile acids produced by the mammalian host, which serve as cofactors for the binding of MNoV and exert immunomodulatory effects through the farnesoid X receptor (FXR). Figure adapted from images created with BioRender.com.
Another abundant microbial by-product is succinate, which is produced by fermentation of both carbohydrates and proteins (Figure 4b). Succinate is an important cross-feeding metabolite in the intestine, being produced and consumed by different microbes, and its levels normally remain low. However, microbial dysbiosis after antibiotic treatment or during IBD (46) can lead to increased succinate levels. Succinate is sensed by the receptor succinate receptor 1 primarily expressed on tuft cells, a rare epithelial cell type in the gut (47), leading to modulation of mucosal immune responses and tuft cell proliferation (48). As tuft cells are target cells for both MNoV (49) and RV (50), succinate production by the microbiota may facilitate viral infection of these cells in the gut epithelium.
Protein metabolism by the GIT-associated microbiota produces a wide variety of bioactive compounds, including peptides and small metabolites produced from the diversity of amino acid side chains (Figure 4c). In humans, most dietary protein is absorbed as di- and tripeptides in the duodenum and proximal jejunum, although excess unabsorbed peptides and undigestible proteins reach the colon. Many strains of Bifidobacterium, a common probiotic, are protective against RV infection in cell culture, in animal models of disease, and in pediatric patients (51–53), an interaction likely mediated by a small peptide produced from Bifidobacterium metabolism of milk casein (54). Although full mechanistic understanding of this interaction remains elusive, it may be related to proliferation of and increased mucin production by goblet cells (55, 56). Bacterial fermentation of valine, leucine, and isoleucine produces branched SCFAs, which, like butyrate, are potent HDAC inhibitors (57). Products of amino acid metabolism by the microbiota also mediate effects on both innate and adaptive immunity (58). Although relatively little is currently known about the roles of microbially derived amino acid metabolites during viral infection, at least one study linked valine metabolism by GIT-resident Bifidobacterium to decreased disease severity during influenza infection in mice (59). Protein and amino acid metabolites likely constitute a major route of chemical communication between the GIT microbiota and the host immune system that could modulate viral infections.
Bile acids (BAs) are unique molecules synthesized from cholesterol in the liver and secreted into the lumen of the intestine after the ingestion of food to solubilize lipids. BAs are secreted in their primary form, mainly chenodeoxycholic acid (CDCA) in humans, or as amino acid conjugates linked to glycine or taurine: glycochenodeoxycholic acid (GCDCA) and taurochenodeoxycholic acid. A diversity of gut-resident microbes express bile salt hydrolases, enzymes that deconjugate taurine- or glycine-conjugated BAs to their primary forms. Bacterial BA deconjugation has three important effects: (a) enhanced BA tolerance for the microbiota, (b) increased reabsorption and recycling of BAs by the human host, and (c) availability of primary BAs for further modification by the microbiota, producing secondary BAs, such as lithocholic acid (LCA) and deoxycholic acid (DCA) (Figure 4d). Taken together, the microbiota plays a central role in regulating the composition of the luminal BA pool, which can influence virus-human interactions. For example, MNoV uses GCDCA and LCA, a secondary BA produced only through bacterial biotransformation, as cofactors during binding to the viral receptor CD300LF (60). Similarly, porcine enteric calicivirus requires BAs for host cell entry (61), for endosomal escape (62), and to alter antiviral interferon (IFN) responses (discussed in greater detail below) (63). Conversely, RV replication in the intestine is reduced by CDCA and DCA via activation of the farnesoid X receptor (FXR), the primary BA sensor involved in regulation of cholesterol, BA, and lipid homeostasis, potentially via down-regulation of lipid synthesis (64). The intestinal BA pool and its effects via FXR may also mediate a variety of systemic effects on viral infection, as in the case of hepatitis C virus infection (65, 66).
Taken together, the diverse metabolic capabilities of the GIT microbiota clearly have the capacity to drastically alter the gut lumen and participate in viral infection directly as cofactors for cell entry, as well as indirectly by modulating host activities. However, only a small fraction of these interactions have been explored, warranting further research to discover how diverse microbial metabolisms contribute to health and disease.
Synergism Between Bacterial and Viral Pathogens
Bacterial and viral coinfection rates can be high (67, 68), especially in low- and middle-income countries and areas with poor sanitation. The order of infection is often important, as immune effects, gut barrier damage, and altered host gene expression resulting from the primary infecting microbe can lead to increased or decreased susceptibility to subsequent viral and bacterial pathogens alike. For example, preinfection with Salmonella protects macrophages from MNoV infection in cell culture (69), although preinfection of mice with MNoV does not impact Salmonella infection (70).
There exists a long history of studying bacterial-viral synergism in the human lung, where influenza infection leads to increased susceptibility to bacterial pneumonia (71). Similar patterns of bacterial-viral cooperation have been characterized for other respiratory viruses, such as RSV and severe acute respiratory syndrome coronavirus 2 (72, 73). Bacterial-viral interactions during mixed infection of the GIT are likely complicated significantly by the increased bacterial burden, diversity, and effects of the microbiota (68). Synergistic bacterial-viral interactions can be specific to the pathogens at play, although investigation with a broader diversity of bacterial and viral strains is warranted. For example, entry and replication of the bacterial pathogen Listeria monocytogenes in human Caco2 cells is reduced by preinfection with PV but increased by RV (74). RV-infected cells exhibit increased susceptibility to diverse bacterial pathogens, including Yersinia (75), Salmonella, Shigella, and E. coli (76).
Increased susceptibility to bacterial infection following viral infection has long been thought to be mediated by opportunistic bacteria taking advantage of viral-induced damage to the gut barrier. This view, however, is oversimplistic, as recent work has shown the complex nature of some interacting bacterial-viral pairs. For example, transmissible gastroenteritis virus induces an epithelial-mesenchymal transition in intestinal epithelial cells (IECs) that results in increased expression of integrin and fibronectin, which subsequently infecting bacterial pathogens, such as E. coli, can utilize for adhesion and invasion (77). More study is needed to clarify interactions between viral and bacterial pathogens, as the mechanisms underlying most of these clinically relevant interactions remain poorly understood.
Viral Interactions with Nonbacterial Components of the Microbiota
In addition to bacteria, the GIT microbiota hosts a diversity of other microbes: archaea, fungi, parasites, bacteriophages, and other viruses. These components of the microbiome are not as well studied as their bacterial counterparts, and their interactions with enteric viruses largely remain speculative or poorly understood.
Although parasites are a common component of the GIT microbiota, especially in low- and middle-income countries, their roles in viral infection are not well studied because they are excluded from specific pathogen-free animal models of disease. Enteric parasites include helminth worms, flagellates, alveolates, and amoebae, and they can cause a diversity of altered immune states with potential effects on viral infection (78, 79). For example, helminths infecting the GIT produce the metabolic by-product succinate (Figure 4b), which is sensed by tuft cells and alters immune signaling on a systemic level via interleukin (IL-)4/IL-25 signaling, leading to worsened outcomes in a mouse model of West Nile virus (WNV) infection (80). Succinate production by helminths further induces the proliferation of tuft and goblet cells (81), target host cells of MNoV and muAstV, respectively (49, 82). Viral replication of both viruses is thus exacerbated during helminth coinfection (82, 83). Physical damage to host tissues by parasites may also play a role in viral susceptibility, as is the case during helminth-lymphocytic choriomeningitis virus (LCMV) coinfection in the liver (84).
Bacteriophages (phages), viruses that infect bacteria, play a vital role in controlling bacterial cell populations in vivo through lytic infection (85) and potentially modulate bacterial interactions with eukaryotic viruses in this way. Further, temperate phages can integrate as latent prophages, which can modulate bacterial phenotypes in GIT commensals and symbionts. Phage BV01, a prophage in the ubiquitous GIT-associated commensal Phocaeicola vulgatus, abrogates host BA metabolism and alters the expression of amino acid decarboxylases producing bioactive compounds (86), which may in turn affect viral infection. Finally, phages can directly interact with epithelial, endothelial, and immune cells (87). Although the effects of direct phage-mammal interactions on viral infection have yet to be fully characterized, one study found that a phage alters the innate immune response and is antiviral against MNoV in cell culture (88). Thus, diverse elements of the microbiota can contribute to modulation of eukaryotic virus infection.
MICROBIOTA EFFECTS ON INTRINSIC IMMUNE RESPONSES TO VIRAL INFECTIONS
The intestinal epithelium serves as the primary barrier separating underlying lamina propria (LP) and deeper tissues from the commensal microorganisms in the intestinal lumen. IECs and mononuclear phagocytes (MNPs), as well as the gut-associated lymphoid tissues, together maintain this spatial segregation, sense microbes, and regulate immune responses to prevent inflammation (89). Pattern recognition receptors (PRRs), including membrane-bound Toll-like receptors (TLRs) and a variety of other host factors, serve as frontline sensors for microbial signals in the GIT and initiate innate immune defenses as well as the development of antigenspecific adaptive immune responses. PRRs recognize highly conserved microbial-, pathogen-, and damage-associated molecular patterns, triggering an array of canonical antimicrobial immune responses through the induction of various inflammatory cytokines, chemokines, and IFNs that further induce the secretion of antimicrobial peptides and mucus by IECs, as well as recruit and activate intestinal MNPs. PRR sensing in the intestinal mucosa is strictly regulated to induce effective immune responses against invasive pathogens while maintaining immune tolerance to harmless commensals, thus preventing intestinal pathology and maintaining homeostasis.
Among the early immune responses stimulated by PRR-mediated signaling is induction of IFNs, comprising type I, II, and III IFN cytokine families, which activate distinct but overlapping downstream signaling cascades to restrict viral infections. Generally, both local and systemic control of viruses, as well as consequent immune pathology, is mediated by type I IFNs, whereas type III IFNs predominantly provide frontline protection at mucosal barriers (90). Here, we focus on what has been described about how the microbiota influences expression of and signaling by type I and III IFNs as well as several other cytokines critical for viral regulation.
Microbial Modulation of Viral Infection via Type I Interferons
The type I IFN family consists of 13 partially homologous cytokines, of which IFN-α subtypes and IFN-β are the most well characterized. Nearly all cell types in the body can produce type I IFNs, usually in response to sensing of microbial products by PRRs. Type I IFNs bind to their cognate receptor, the heterodimeric interferon-α/β receptor (IFNAR)1/IFNAR2 complex, and via a signaling cascade induce the rapid expression of hundreds of IFN-stimulated genes (ISGs), which can mediate both direct and indirect antiviral effects on viral entry, replication, and dissemination (91). Growing evidence over the last decade has indicated that the GIT microbiota controls low-level constitutive expression of type I IFNs, particularly by dendritic cells (DCs), that is critical for the rapid induction of antiviral activities upon infection (92–94). MNPs in GF mice, including plasmacytoid DCs (pDCs) that are a key source of type I IFNs, exhibit a failure to mount a normal type I IFN response to viral stimuli (92, 94).This defect in type I IFN production is associated with a number of broad immunological defects including diminished natural killer (NK) cell priming (92). Further, classical DCs (cDCs) from GF mice exhibit a severe reduction in histone activation markers on many ISGs (94). Thus, microbiota-mediated tonic type I IFN signaling is key in keeping the immune system poised against incoming pathogens (Figure 5a).
Figure 5.
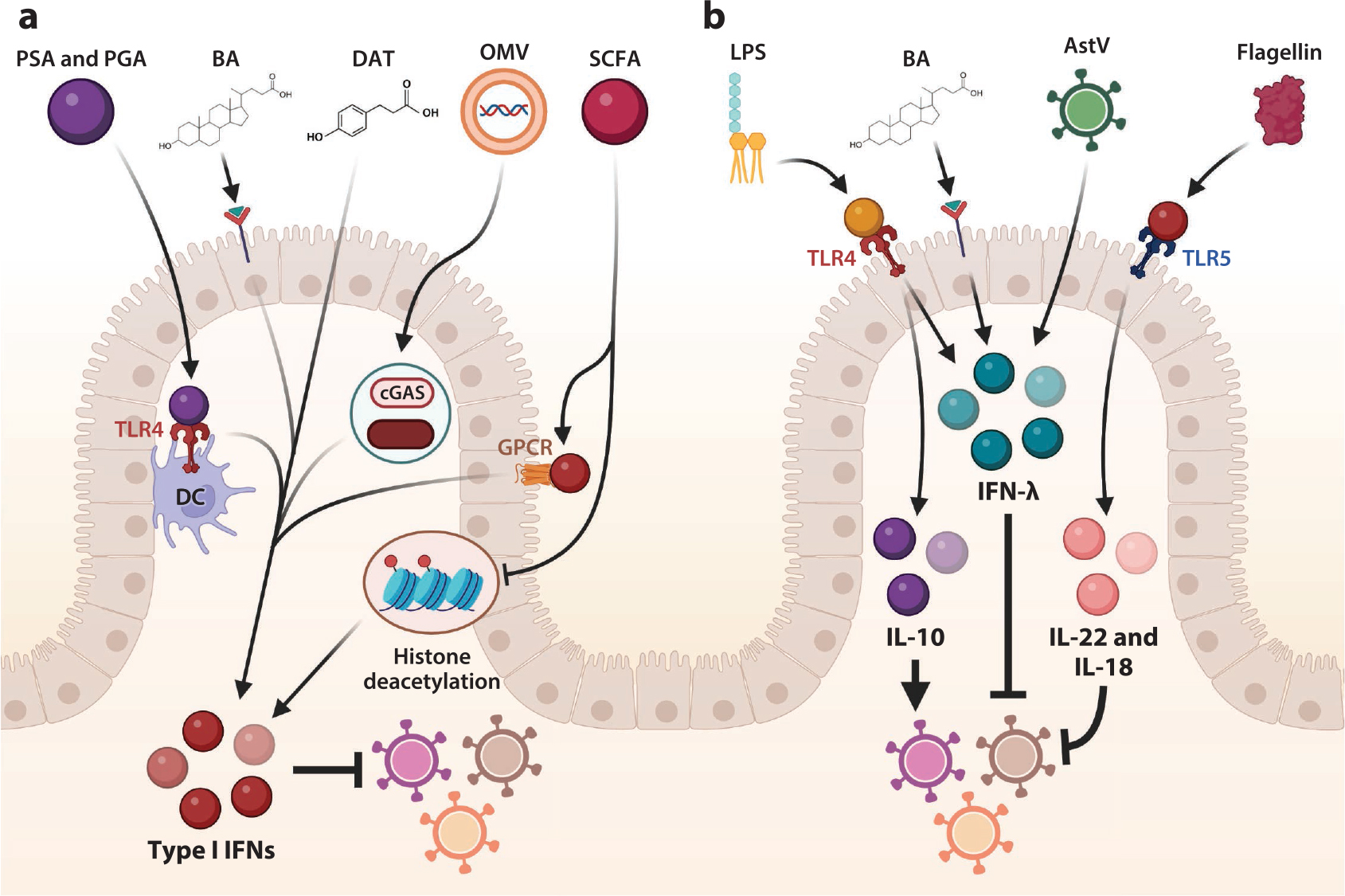
Components of the microbiota alter interferon (IFN) and cytokine signaling. (a) Microbial products such as polysaccharide A (PSA) and poly-γ-glutamic acid (PGA) are sensed by Toll-like receptor (TLR)4 for type I IFN secretion by dendritic cells (DCs), and microbially regulated products desaminotyrosine (DAT) and bile acids (BAs) also induce type I IFNs to inhibit viral infection. Outer membrane vesicles (OMVs) containing bacterial products and DNA activate the cyclic GMP-AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway to induce type I IFNs, and short chain fatty acids (SCFAs) induce histone changes to alter type I IFNs to control viral infection. SCFAs can also enhance type I IFN responses via activation of specific G-protein coupled receptors (GPCR). (b) Lipopolysaccharide (LPS) can bind to TLR4 to induce IFN-λ and/or interleukin (IL-)10, which mediates antiviral or proviral effects in a virus-dependent context. BAs also regulate IFN-λ, as do virome elements such as astrovirus (AstV) strains to drive viral interference. Flagellin is sensed by TLR5 and induces IL-22 and IL-18 to restrict viral infection. Figure adapted from images created with BioRender.com.
Diminished type I IFN signaling in the absence of the microbiota has been shown to have important consequences for numerous viruses. Depletion of the microbiota in mice is associated with enhanced systemic infection by a variety of viral pathogens, including LCMV and encephalomyocarditis virus, due to an inadequate type I IFN response (93, 95). Microbial cells and cell components regulate systemic infection via type I IFN by a variety of mechanisms, including TLR signaling and cyclic GMP-AMP synthase (cGAS)–stimulator of interferon genes (STING) signaling. Sensing of microbial membrane-derived glycolipids, such as polysaccharide A (PSA), via TLR4 on the cell surface of DCs has been shown to enhance the type I IFN response against influenza virus (96). Similarly, poly-γ-glutamic acid, a component of the cell envelope of Bacillus spp. and ligand for TLR4, induces IFN-β, resulting in reduced MNoV infection of immune tissues (97). Several strains of lactic acid bacteria have been shown to induce type I IFN responses in human peripheral blood mononuclear cells and mouse macrophages via STING and mitochondrial antiviral signaling (MAVS) signaling (98). Intriguingly, a recent study showed that microbiota-derived DNA-containing membrane vesicles trigger systemic antiviral type I IFN responses after being sensed by the cGAS-STING pathway to control herpes simplex virus type 1 (HSV-1) and vesicular stomatitis virus (VSV) infection (99).
A diversity of microbial metabolites, including BAs, SCFAs, and flavonoid derivatives, also influence type I IFN signaling. In the context of chikungunya virus infection, type I IFN responses in pDCs are diminished in microbiota-deplete mice, but the secondary BA DCA, produced by microbial modification of primary BAs, can rescue type I IFN responses, implicating BA transformation by the microbiota in the maintenance of type I IFN signaling responses (100). Flavonoids, plantderived polyphenols ubiquitous in fruits and vegetables, largely pass through the upper GIT and are metabolized and modified by the microbiota in the colon. One flavonoid-derived metabolite, desaminotyrosine, produced by Clostridium orbiscindens in the GIT, is protective against influenza infection by amplifying the type I IFN response (101). The SCFA butyrate suppresses type I IFN induction of ISGs via HDAC inhibition, resulting in increased human immunodeficiency virus 1, VSV, and human metapneumovirus infection (102). In contrast, dietary supplementation with the SCFA acetate enhances type I IFN responses in lung epithelial cells against RSV by activation of specific G-protein coupled receptors (103), suggesting that SCFAs may mediate context-specific regulation of IFN activity. Thus, there may be numerous mechanisms by which the GIT microbiota promotes and regulates type I IFN induction to limit viral infection.
Microbial Modulation of Viral Infection via Type III Interferons
The type III IFNs, which include IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4, play important roles in controlling antiviral immune responses at barrier surfaces (90). IFN-λ acts through receptor interferon lambda receptor 1 (IFNLR1), which is highly expressed in epithelial cells in humans and mice, and IFN-λ can potently inhibit viral replication (104). The limited expression of IFNLR1 underlies the localized function of IFN-λ at barrier surfaces, and type III IFNs therefore serve as important cytokines to limit local infection without inducing excessive systemic inflammation (105). As these mucosal sites are also rich in microbiota, interactions between microbes and type III IFNs have been a growing area of interest.
The microbiota influences type III IFN responses via multiple mechanisms (Figure 5b). The GIT microbiota drives baseline expression of tonic IFN-λ signaling in IECs to control early mRV infection, with TLR4 ligand LPS and potentially other microbial ligands implicated as key contributors (106). Microbial cues have also been shown to modulate priming of IFN-λ-dependent antiviral responses upon enteric viral infections. For MNoV, the microbiota promotes viral infection in the distal intestinal tissues of the ileum and colon, but this enhancement of infection is counteracted by IFN-λ signaling (22, 23). In contrast, the microbiota limits MNoV infection in the proximal small intestine via modification of BAs that prime IFN-λ-dependent antiviral responses (107). Beyond regulation of IFN-λ by the bacterial microbiota, chronic muAstV in the intestine of immunocompromised mice as a component of the commensal virome was found to induce IFN-λ to limit infection by other enteric viruses, supporting the conclusion that diverse elements of the microbiota can regulate this signaling pathway (39).
Toll-Like Receptor Ligands from the Microbiota Influence Inflammatory Cytokine Responses
Beyond IFNs, other microbiota-regulated cytokines have been found to regulate viral infections (Figure 5b). Pro-inflammatory cytokines IL-1β and IL-18 are regulated by the microbiota to facilitate development of adaptive immune responses to influenza infection, with diverse TLR ligands contributing to this effect (108). Bacterial flagellin, acting via TLR5, induces IL-22 and IL-18 cytokines to limit RV replication via stimulation of protective gene expression programs in IECs and elimination of infected cells, respectively (109), and it has further been shown that the microbiota as a whole is important for IL-22 induction to limit RV replication (110). Finally, bacterial LPS-bound mouse mammary tumor virus (MMTV) particles engage TLR4 to produce the immunosuppressive cytokine IL-10, which facilitates MMTV persistence (111), a proviral consequence of the microbiota. In contrast, IL-10 secretion from T cells, driven by PSA binding to B cells, helps to protect against virus-mediated pathology by HSV-1 (112). Thus, the complex milieu of microbial ligands and metabolic products mediates diverse effects on numerous cell types and contributes to a number of regulatory effects for viral infections via cytokine signaling.
MICROBIOTA EFFECTS ON CELLULAR AND ADAPTIVE IMMUNE RESPONSES TO VIRAL INFECTIONS
Interactions between animal hosts and their microbiota are vital to train the host immune system, which, in turn, affects the composition and function of the microbiome. Disruption of these mutually beneficial interactions can mediate important effects on disease susceptibility and the capacity to mount an effective host immune response to challenges. In the GIT, beyond the first line of defense mediated by surface IECs, a complex and integrated collection of subepithelial stromal cells, neural structures, enteric glial cells, and immune cells in both the LP and deeper intestinal layers serve as the mucosal barrier and play critical roles in spatial segregation, intestinal microbial sensing, and immunoregulatory responses (89) (Figure 6). A number of cells straddle the boundary between innate and adaptive immunity, including intraepithelial lymphocytes (IELs), lamina propria lymphocytes (LPLs), and innate lymphoid cells (ILCs). These immune cells play essential roles in the maintenance of a delicate balance between intestinal immune tolerance to the continuous stimulation of commensal microbes and the effective immune responses against invading pathogens (113). Disruption or dysregulation of such homeostatic conditions may lead to infection, inflammation, and tissue injury, and conversely, excessive immune responses to innocuous commensal microbiota can contribute to the pathogenesis of human IBD (114). Here, we detail what is known about the role of these cells in regulating viral infections and how the microbiota may modulate the infection response by influencing the abundance and activation status of various immune cells.
Figure 6.
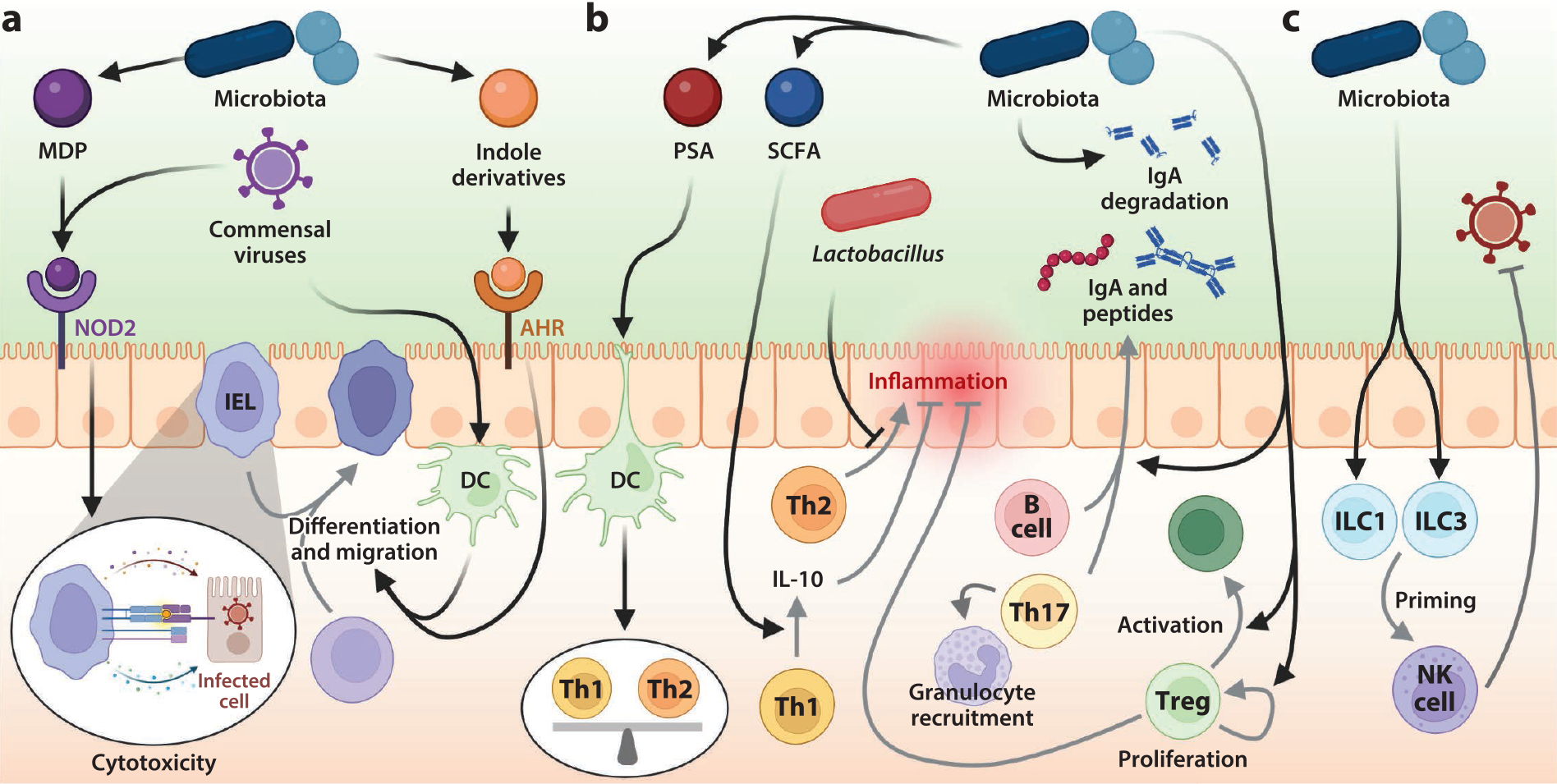
Gut microbes and their metabolites regulate cellular and adaptive immunity during viral infections. (a) The microbiota modulates the activities of intraepithelial lymphocytes (IELs) by several mechanisms. The intestinal epithelium recognizes viral and bacterial signals such as muramyldipeptide (MDP) via pattern recognition receptors (PRRs) such as nucleotide-binding oligomerization domain–containing protein 2 (NOD2). Microbiota and viral antigens captured by dendritic cells (DCs) and macrophages traffic to the lymphoid tissues and lead to the activation, migration, and differentiation of intestinal lymphocytes. Indole derivatives modulated by the microbiota activate the aryl hydrocarbon receptor (AHR) to facilitate this differentiation. Activated cytotoxic T cells migrate to the intestinal tissue (IEL) and elicit antiviral immunity by cytokine secretion or cytotoxic activity. (b) Commensal microbes extensively control the composition and activities of lamina propria (LP) lymphocytes. Presentation of antigens by intestinal DCs leads to the differentiation of commensal-specific regulatory T cells (Tregs); helper T (Th)1, Th2, and Th17 cells; and immunoglobulin A (IgA)-producing B cells in the LP. The gastrointestinal tract (GIT) microbiota and its metabolites can modulate the function of DCs and other innate cells both locally and systemically in a manner that promotes the induction of effector T and B cell responses against pathogens and regulate inflammation. For example, polysaccharide A (PSA)-producing bacteria modulate the balance between Th1 and Th2 cells, and short-chain fatty acids (SCFAs) promote antigen-specific Th1 interleukin (IL)-10 production to maintain intestinal homeostasis. (c) Perturbations to the microbiota alter innate lymphoid cell (ILC) activity during intestinal infections, including priming of the natural killer (NK) cell antiviral response to viral infection. Figure adapted from images created with BioRender.com.
Intraepithelial Lymphocytes
As one of the main branches of the intestinal immune system, IELs provide protective immunity and support the integrity of the intestinal epithelial barrier (Figure 6a). IELs protect IECs from microbial invaders by promoting intraepithelial cell scanning and expression of antimicrobial genes (115). As sentinels for epithelial integrity, IELs can kill IECs under stress from infection, transformation, or invasion by other cells via granzyme or by engagement of cell death molecule Fas, and/or produce a broad spectrum of immunoregulatory cytokines, including IFN-γ, tumor necrosis factor-α, IL-4, IL-10, and IL-17. In the context of viral infection, activated IELs can rapidly secrete type I and III IFNs to upregulate ISGs in the villus epithelium to limit infection (116). Adoptive transfer of antigen-specific IELs can reduce enteric viral burden (117), and IELs also contribute to the recruitment of antiviral immune cells via cytokine and chemokine expression (118).
IEL homeostasis is strongly influenced by the microbiota. GF mice exhibit an overall reduction in IELs, although IEL subsets are differentially affected (119). IEL numbers are also significantly reduced in Nod2−/− mice, suggesting that recognition of muramyl dipeptide, a component of the bacterial cell wall, by the PRR nucleotide-binding oligomerization domain–containing protein 2 (NOD2) is essential for the homeostasis of IELs (120). The mouse microbiota stimulates both the development of IELs (121) and their capacity for cytolytic activity, particularly during the weaning period (122). IEL differentiation correlates with the presence of Lactobacillus reuteri, which produces indole derivatives from tryptophan, activating the aryl hydrocarbon receptor (123). The microbiota also regulates the migration of LP regulatory T cells (Tregs) to the intestinal epithelium and their conversion to IELs (124). Interestingly, commensal viruses and dietary components also maintain the IEL population (125). Depletion of commensal viruses results in loss of IELs, in a process reliant on viral recognition via retinoic acid-inducible gene I/MAVS by intraepithelial antigen-presenting cells (APCs). MNoV infection of GF or antibiotic-treated mice restores IEL function without inducing overt inflammation and disease via type I IFN and IL-22 signaling (126), and indeed different enteric viruses can contribute to distinct immunological programs similar to distinct bacterial taxa of the microbiota (127). Together, these findings suggest important cross talk between viral and bacterial ligands in IEL homeostasis and support the importance of the microbiota in maintaining IEL-dependent protection from infectious challenges.
Lamina Propria Lymphocytes
The LP contains a large and heterogeneous population of immune cells, with an abundance of antigen-experienced memory T and B cells, IgA-secreting plasma cells, and MNPs including macrophages and DCs (Figure 6b). DCs, which play central roles in tolerance and immunity in the LP, uptake and process antigens from the gut lumen and migrate to the T cell areas of intestinal lymphoid tissues where they prime naive T and B cells. Thus, differentiation and maturation of lymphocytes from naive T and B cells to distinct helper T cell (Th) subpopulations and Tregs as well as IgA+ plasma cells are strongly influenced by the gut microbiota (113).
Among Th cell subsets, Th1, Th2, and Th17 CD4+ cells are associated with responses to viral infections, parasitic colonization, and extracellular bacterial and fungal infections, respectively. Colonization with PSA-producing Bacteroides fragilis corrects imbalances between Th1 and Th2 cells in GF mice, a process mediated by PSA binding to APCs that then secrete IL-12 to promote Th1 levels (128). Addition of Lactobacillus to the maternal gut microbiota reduces Th2 cell-mediated immune responses and lung inflammation after RSV infection in offspring, suggesting multiple bacterial taxa can regulate Th1/Th2 skewing (129). Moreover, gut microbiota-derived SCFAs promote antigen-specific Th1 IL-10 production to maintain intestinal homeostasis (130) (Figure 4a). Under the influence of microbiota-induced cytokines and chemokines from DCs and macrophages, Th17 cells differentiate and secrete cytokines that stimulate IEC proliferation and production of antimicrobial proteins, mediate IgA transport, and recruit granulocytes (131). Th17 cells are scarce in the intestinal LP in GF mice, while colonization with microbes that adhere to IECs, including SFB, induces Th17 cells (132).
Tregs maintain immunological tolerance to self-antigens and prevent excessive immune responses, secreting anti-inflammatory cytokines such as IL-10 and inducing IgA production (133). The GIT microbiota promotes Treg levels, inducing the expression of IL-1β from LP macrophages to activate a cytokine signaling cascade through neighboring ILCs and DCs that ultimately induces Tregs (134). A mixture of Clostridia strains was shown to be sufficient to enhance Treg abundance and induce the expression of anti-inflammatory molecules in mice (135), and PSA from B. fragilis can also selectively induce Tregs (136). In addition, the SCFAs acetate, propionate, and butyrate regulate the size and function of the colonic Treg populations (137). Beyond T cell subsets, IgA-expressing B cells in the LP are greatly reduced in GF animals, and the colonization of GF mice with a microbiota quickly triggers the production of IgA (138). Additionally, variation within the microbiota has been shown to dramatically affect secretory IgA levels in the intestinal lumen, as different microbes may variably stimulate or degrade both total and microbe-specific IgA levels (139).
Regulation of enteric viruses by lymphocytes including LPLs has been well established. Mice depleted of B and T cells become chronically infected with mRV (140), and antibody and T cell responses are both critical for MNoV clearance (141). Similarly, muAstV establishes chronic infection in the absence of B and T cell responses (39). While the regulation of LPL populations by the microbiota would suggest an important shift in the capacity to mount an adaptive immune response to enteric viruses, a limited number of studies have directly explored this connection. Antibiotic treatment has been reported to suppress mRV infection via enhancement of mRV-specific IgA-secreting cells in the small intestine (142), suggesting there could be proviral effects of a diverse microbial population for adaptive targeting.
In the regulation of extraintestinal viruses by the GIT microbiota, systemic lymphocyte responses are also critical. Antibiotic treatment leads to increased viral levels and immunopathology after infection by multiple flaviviruses including Zika virus, WNV, and dengue virus, mainly due to impaired development of systemic T cell responses (143). Similarly, impaired T cell responses contribute to enhanced severity of influenza infection after antibiotics treatment (108), and the microbiota has been implicated in regulating LCMV-specific memory CD8+ T cells (144). Further studies are sorely needed to better understand the mechanisms of microbiota-associated T cell–mediated control of both enteric and systemic viruses.
Innate Lymphoid Cells
ILCs, including ILC1–3 and NK cells, are a relatively rare and mucosal tissue–resident family of cells that are thought of as the innate counterparts of adaptive T cells (145) (Figure 6c). They mirror the functions of CD8+ and CD4+ T cells but undergo rapid activation during the early immune responses to infection via cytokine secretion. The cytokines ILCs produce induce innate responses in the LP and IECs against infection by intracellular pathogens (ILC1), helminths (ILC2), and extracellular pathogens (ILC3). Tissue-resident ILC1s provide early protection at the initial site of viral infection in response to local cDC1-derived proinflammatory cytokines (146), and synergism between IFN-λ and IL-22 produced by ILC3s plays an important role in the control of RV infection (147). ILC3s as well as NK cells also serve as important sources of IFN-γ during infection by viral and bacterial pathogens (92).
The microbiota influences the development and function of intestinal ILCs throughout the host’s lifetime. Exposure to aryl hydrocarbon receptor ligands produced by the maternal microbiota plays an important role in establishing the intestinal ILC3 pool during early life (148), while ILC1 cells are shaped by microbiota colonization and maturation after birth (149). During adulthood, human ILC1s can differentiate into ILC3 driven by IL-23 and accelerated by IL-1β and retinoic acid, which are partially regulated by the commensal microbiota (149). Microbiome depletion blunts the expansion and migration of intestinal NK cells (150), and NK cell priming and antiviral responses are also compromised (92). In sum, the microbiota regulation of ILC populations is also a contributing factor to its overall effects upon the response to viral infection.
CONCLUSIONS
Mechanistic understanding of the roles of the microbiota in human health and disease is still lacking in many ways. The complexity of an individual’s microbiome, as well as the diversity of the microbiome observed across human populations, makes these studies inherently difficult. Direct interactions between bacterial cells and human viruses are now being explored with greater depth, which will help to clarify the specificity of these interactions and their outcomes. Further work is also warranted to explore the effects of microbial metabolites directly on invading viral particles as well as on the human innate and adaptive immune responses both locally in the intestine and systemically. While research is increasingly integrating study of the microbiota into analyses of the host immune response to viral infection, we are still only beginning to understand the relationship of endogenous microbial factors with the first-line defense as well as the development of protective memory responses. Greater understanding of these microbiota-human-virus interactions will be vital for the development and clinical utility of novel therapeutics, such as prebiotics, probiotics, and phage therapies, for viral and nonviral diseases alike.
ACKNOWLEDGMENTS
D.E.C. was supported by National Institutes of Health (NIH) grant T32 DK077653-29 and Crohn’s & Colitis Foundation Research Fellowship Award 935619.M.T.B. was supported by NIH grants R01AI139314 and R01AI127552, as well as the Burroughs Wellcome Fund Pathogenesis of Infectious Disease Program. We sincerely apologize to our colleagues for the work we could not include in the present review because of space constraints.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ryu EP, Davenport ER. 2022. Host genetic determinants of the microbiome across animals: from Caenorhabditis elegans to cattle. Annu. Rev. Anim. Biosci. 10:203–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Integr. HMP Res. Netw. Consort. 2019. The Integrative Human Microbiome Project. Nature 569:641–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, et al. 2015. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput. Struct. Biotechnol. J. 13:390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JW, Kuo CH, Kuo FC, Wang YK, Hsu WH, et al. 2019. Fecal microbiota transplantation: review and update. J. Formos. Med. Assoc. 118:S23–31 [DOI] [PubMed] [Google Scholar]
- 5.Weingarden AR, Vaughn BP. 2017. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 8:238–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen RY, Mostafa I, Hibberd MC, Das S, Mahfuz M, et al. 2021. A microbiota-directed food intervention for undernourished children. N. Engl. J. Med. 384:1517–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25:219–32 [DOI] [PubMed] [Google Scholar]
- 8.Dinleyici EC, Martinez-Martinez D, Kara A, Karbuz A, Dalgic N, et al. 2018. Time series analysis of the microbiota of children suffering from acute infectious diarrhea and their recovery after treatment. Front. Microbiol. 9:1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P, Teal TK, Marsh TL, Tiedje JM, Mosci R, et al. 2015. Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma C, Wu X, Nawaz M, Li J, Yu P, et al. 2011. Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr. Microbiol. 63:259–66 [DOI] [PubMed] [Google Scholar]
- 11.Patin NV, Pena-Gonzalez A, Hatt JK, Moe C, Kirby A, Konstantinidis KT. 2020. The role of the gut microbiome in resisting norovirus infection as revealed by a human challenge study. mBio 11(6):e02634–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen SY, Tsai CN, Lee YS, Lin CY, Huang KY, et al. 2017. Intestinal microbiome in children with severe and complicated acute viral gastroenteritis. Sci. Rep. 7:46130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen C, Xu Y, Ji J, Wei J, Jiang Y, et al. 2021. Intestinal microbiota has important effect on severity of hand foot and mouth disease in children. BMC Infect. Dis. 21:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho SX, Min N, Wong EPY, Chong CY, Chu JJH. 2021. Characterization of oral virome and microbiome revealed distinctive microbiome disruptions in paediatric patients with hand, foot and mouth disease. NPJ Biofilms Microbiomes 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groves HT, Higham SL, Moffatt MF, Cox MJ, Tregoning JS. 2020. Respiratory viral infection alters the gut microbiota by inducing inappetence. mBio 11(1):e03236–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Li F, Wei H, Lian ZX, Sun R, Tian Z. 2014. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 211:2397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu R, Lu R, Zhang T, Wu Q, Cai W, et al.2021. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun. Biol. 4:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KH, Gordon A, Shedden K, Kuan G, Ng S, et al. 2019. The respiratory microbiome and susceptibility to influenza virus infection. PLOS ONE 14:e0207898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguilera ER, Nguyen Y, Sasaki J, Pfeiffer JK. 2019. Bacterial stabilization of a panel of picornaviruses. mSphere 4(2):e00183–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson CM, Woods Acevedo MA, McCune BT, Pfeiffer JK. 2019. Related enteric viruses have different requirements for host microbiota in mice. J. Virol. 93(23):e01339–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, et al. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, et al. 2015. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347:266–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, et al. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson CM, Jesudhasan PR, Pfeiffer JK. 2014. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 15:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhalech AH, Fuller TD, Robinson CM. 2021. Specific bacterial cell wall components influence the stability of coxsackievirus B3. J. Virol. 95:e0142421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger AK, Yi H, Kearns DB, Mainou BA. 2017. Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLOS Pathog. 13:e1006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilera ER, Pfeiffer JK. 2019. Strength in numbers: mechanisms of viral co-infection. Virus Res.265:43–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. 2018. Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe 23:77–88.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahourou G, Guillot S, Le Gall O, Crainic R. 2002. Genetic recombination in wild-type poliovirus. J. Gen. Virol. 83:3103–10 [DOI] [PubMed] [Google Scholar]
- 30.Furione M, Guillot S, Otelea D, Balanant J, Candrea A, Crainic R. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199–208 [DOI] [PubMed] [Google Scholar]
- 31.Holmblat B, Jegouic S, Muslin C, Blondel B, Joffret ML, Delpeyroux F. 2014. Nonhomologous recombination between defective poliovirus and coxsackievirus genomes suggests a new model of genetic plasticity for picornaviruses. mBio 5:e01119–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura T, Sano D, Suenaga A, Yoshimura T, Fuzawa M, et al. 2013. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J. Virol. 87:9441–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monedero V, Buesa J, Rodriguez-Diaz J. 2018. The interactions between host glycobiology, bacterial microbiota, and viruses in the gut. Viruses 10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D, Breiman A, le Pendu J, Uyttendaele M. 2015. Binding to histo-blood group antigen-expressing bacteria protects human norovirus from acute heat stress. Front. Microbiol. 6:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budicini MR, Pfeiffer JK. 2022. Stabilization of murine norovirus by bacteria. mSphere 7:e0004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ang LY, Too HK, Tan EL, Chow TK, Shek LP, et al. 2016. Antiviral activity of Lactobacillus reuteri Protectis against Coxsackievirus A and Enterovirus 71 infection in human skeletal muscle and colon cell lines. Virol. J. 13:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Z, Zou J, Zhang Z, Zhao X, Noriega J, et al. 2019. Segmented filamentous bacteria prevent and cure rotavirus infection. Cell 179:644–58.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Rodriguez FJ, Vieille G, Turin L, Yildiz S, Tapparel C, Kaiser L. 2019. Fecal components modulate human astrovirus infectivity in cells and reconstituted intestinal tissues. mSphere 4(6):e00568–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingle H, Lee S, Ai T, Orvedahl A, Rodgers R, et al. 2019. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat. Microbiol. 4:1120–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engevik MA, Banks LD, Engevik KA, Chang-Graham AL, Perry JL, et al. 2020. Rotavirus infection induces glycan availability to promote ileum-specific changes in the microbiome aiding rotavirus virulence. Gut. Microbes 11:1324–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W, Yu T, Huang X, Bilotta AJ, Xu L, et al. 2020. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 11:4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorres KL, Daigle D, Mohanram S, Miller G. 2014. Activation and repression of Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus lytic cycles by short- and medium-chain fatty acids. J. Virol. 88:8028–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Hee B, Wells JM. 2021. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 29:700–12 [DOI] [PubMed] [Google Scholar]
- 44.Kuster K, Grotzinger C, Koschel A, Fischer A, Wiedenmann B, Anders M. 2010. Sodium butyrate increases expression of the coxsackie and adenovirus receptor in colon cancer cells. Cancer Invest. 28:268–74 [DOI] [PubMed] [Google Scholar]
- 45.Guo X, Lan Z, Wen Y, Zheng C, Rong Z, et al. 2021. Synbiotics supplements lower the risk of hand, foot, and mouth disease in children, potentially by providing resistance to gut microbiota dysbiosis. Front. Cell Infect. Microbiol. 11:729756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagao-Kitamoto H, Shreiner AB, Gillilland MG III, Kitamoto S, Ishii C, et al. 2016. Functional characterization of inflammatory bowel disease–associated gut dysbiosis in gnotobiotic mice. Cell. Mol. Gastroenterol. Hepatol. 2:468–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider C, O’Leary CE, von Moltke J, Liang HE, Ang QY, et al. 2018. A metabolite-triggered tuftcell-ILC2 circuit drives small intestinal remodeling. Cell 174:271–84.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei W, Ren W, Ohmoto M, Urban JF Jr., Matsumoto I, et al. 2018. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. PNAS 115:5552–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, et al. 2018. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360:204–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bomidi C, Robertson M, Coarfa C, Estes MK, Blutt SE. 2021. Single-cell sequencing of rotavirus infected intestinal epithelium reveals cell-type specific epithelial repair and tuft cell infection. PNAS 118(45):e2112814118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee DK, Park JE, Kim MJ, Seo JG, Lee JH, Ha NJ. 2015. Probiotic bacteria, B. longum and L. acidophilus inhibit infection by rotavirus in vitro and decrease the duration of diarrhea in pediatric patients. Clin. Res. Hepatol. Gastroenterol. 39:237–44 [DOI] [PubMed] [Google Scholar]
- 52.Duffy LC, Zielezny MA, Riepenhoff-Talty M, Dryja D, Sayahtaheri-Altaie S, et al. 1994. Effectiveness of Bifidobacterium bifidum in mediating the clinical course of murine rotavirus diarrhea. Pediatr.Res.35:690–95 [DOI] [PubMed] [Google Scholar]
- 53.Munoz JA, Chenoll E, Casinos B, Bataller E, Ramon D, et al. 2011. Novel probiotic Bifidobacterium longum subsp. infantis CECT 7210 strain active against rotavirus infections. Appl. Environ. Microbiol. 77:8775–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chenoll E, Casinos B, Bataller E, Buesa J, Ramon D, et al. 2016. Identification of a peptide produced by Bifidobacterium longum CECT 7210 with antirotaviral activity. Front. Microbiol. 7:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen CC, Baylor M, Bass DM. 1993. Murine intestinal mucins inhibit rotavirus infection. Gastroenterology 105:84–92 [DOI] [PubMed] [Google Scholar]
- 56.Kawahara T, Makizaki Y, Oikawa Y, Tanaka Y, Maeda A, et al. 2017. Oral administration of Bifidobacterium bifidum G9–1 alleviates rotavirus gastroenteritis through regulation of intestinal homeostasis by inducing mucosal protective factors. PLOS ONE 12:e0173979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho RH, Chan JCY, Fan H, Kioh DYQ, Lee BW, Chan ECY. 2017. In silico and in vitro interactions between short chain fatty acids and human histone deacetylases. Biochemistry 56:4871–78 [DOI] [PubMed] [Google Scholar]
- 58.Kim CH. 2018. Immune regulation by microbiome metabolites. Immunology 154:220–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Q, Hu J, Feng JW, Hu XT, Wang T, et al. 2020. Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome Biol. 21:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson CA, Wilen CB, Dai YN, Orchard RC, Kim AS, et al. 2018. Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. PNAS 115:E9201–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shivanna V, Kim Y, Chang KO. 2014. The crucial role of bile acids in the entry of porcine enteric calicivirus. Virology 456–457:268–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shivanna V, Kim Y, Chang KO. 2015. Ceramide formation mediated by acid sphingomyelinase facilitates endosomal escape of caliciviruses. Virology 483:218–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang KO, Sosnovtsev SV, Belliot G, Kim Y, Saif LJ, Green KY. 2004. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. PNAS 101:8733–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim Y, Chang KO. 2011. Inhibitory effects of bile acids and synthetic farnesoid X receptor agonists on rotavirus replication. J. Virol. 85:12570–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scholtes C, Diaz O, Icard V, Kaul A, Bartenschlager R, et al. 2008. Enhancement of genotype 1 hepatitis C virus replication by bile acids through FXR. J. Hepatol. 48:192–99 [DOI] [PubMed] [Google Scholar]
- 66.Wu ZY, Li H, Li JR, Lv XQ, Jiang JD, Peng ZG. 2019. Farnesoid X receptor agonist GW4064 indirectly inhibits HCV entry into cells via down-regulating scavenger receptor class B type I. Eur. J. Pharmacol. 853:111–20 [DOI] [PubMed] [Google Scholar]
- 67.Shrivastava AK, Kumar S, Mohakud NK, Suar M, Sahu PS. 2017. Multiple etiologies of infectious diarrhea and concurrent infections in a pediatric outpatient-based screening study in Odisha, India. Gut Pathog. 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathew S, Smatti MK, Al Ansari K, Nasrallah GK, Al Thani AA, Yassine HM. 2019. Mixed viral-bacterial infections and their effects on gut microbiota and clinical illnesses in children. Sci. Rep. 9:865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agnihothram SS, Basco MD, Mullis L, Foley SL, Hart ME, et al. 2015. Infection of murine macrophages by Salmonella enterica serovar Heidelberg blocks murine norovirus infectivity and virus-induced apoptosis. PLOS ONE 10:e0144911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Higgins PD, Johnson LA, Sauder K, Moons D, Blanco L, et al. 2011. Transient or persistent norovirus infection does not alter the pathology of Salmonella typhimurium induced intestinal inflammation and fibrosis in mice. Comp. Immunol. Microbiol. Infect. Dis. 34:247–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW. 2019. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat. Microbiol. 4:1328–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith CM, Sandrini S, Datta S, Freestone P, Shafeeq S, et al. 2014. Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am. J. Respir. Crit. Care Med. 190:196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kruglikov IL, Shah M, Scherer PE. 2020. Obesity and diabetes as comorbidities for COVID-19: underlying mechanisms and the role of viral–bacterial interactions. eLife 9:e61330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Superti F, Petrone G, Pisani S, Morelli R, Ammendolia MG, Seganti L. 1996. Superinfection by Listeria monocytogenes of cultured human enterocyte-like cells infected with poliovirus or rotavirus. Med. Microbiol. Immunol. 185:131–37 [DOI] [PubMed] [Google Scholar]
- 75.Di Biase AM, Petrone G, Conte MP, Seganti L, Ammendolia MG, et al. 2000. Infection of human enterocyte-like cells with rotavirus enhances invasiveness of Yersinia enterocolitica and Y. pseudotuberculosis. J. Med. Microbiol. 49:897–904 [DOI] [PubMed] [Google Scholar]
- 76.Bukholm G 1988. Human rotavirus infection enhances invasiveness of enterobacteria in MA-104 cells. APMIS 96:1118–24 [DOI] [PubMed] [Google Scholar]
- 77.Xia L, Dai L, Yu Q, Yang Q. 2017. Persistent transmissible gastroenteritis virus infection enhances enterotoxigenic Escherichia coli K88 adhesion by promoting epithelial-mesenchymal transition in intestinal epithelial cells. J. Virol. 91(21):e01256–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Desai P, Diamond MS, Thackray LB. 2021. Helminth-virus interactions: determinants of coinfection outcomes. Gut Microbes 13:1961202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakada-Tsukui K, Nozaki T. 2016. Immune response of amebiasis and immune evasion by Entamoeba histolytica. Front. Immunol. 7:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desai P, Janova H, White JP, Reynoso GV, Hickman HD, et al. 2021. Enteric helminth coinfection enhances host susceptibility to neurotropic flaviviruses via a tuft cell-IL-4 receptor signaling axis. Cell 184:1214–31.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.von Moltke J, Ji M, Liang HE, Locksley RM. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature 529:221–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ingle H, Hassan E, Gawron J, Mihi B, Li Y, et al. 2021. Murine astrovirus tropism for goblet cells and enterocytes facilitates an IFN-λ response in vivo and in enteroid cultures. Mucosal Immunol. 14:751–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, et al. 2014. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 345:578–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edwards MJ, Buchatska O, Ashton M, Montoya M, Bickle QD, Borrow P. 2005. Reciprocal immunomodulation in a schistosome and hepatotropic virus coinfection model. J. Immunol. 175:6275–85 [DOI] [PubMed] [Google Scholar]
- 85.Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Lyon L, et al. 2019. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 25:803–14.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campbell DE, Ly LK, Ridlon JM, Hsiao A, Whitaker RJ, Degnan PH. 2020. Infection with bacteroides phage BV01 alters the host transcriptome and bile acid metabolism in a common human gut microbe. Cell Rep. 32:108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carroll-Portillo A, Lin HC. 2019. Bacteriophage and the innate immune system: access and signaling. Microorganisms 7:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang L, Ma C, Liu J, Shahin K, Hou X, et al. 2021. Antiviral effect of a bacteriophage on murine norovirus replication via modulation of the innate immune response. Virus Res. 305:198572. [DOI] [PubMed] [Google Scholar]
- 89.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14:141–53 [DOI] [PubMed] [Google Scholar]
- 90.Ingle H, Peterson ST, Baldridge MT. 2018. Distinct effects of type I and III interferons on enteric viruses. Viruses 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schoggins JW. 2019. Interferon-stimulated genes: What do they all do? Annu. Rev. Virol. 6:567–84 [DOI] [PubMed] [Google Scholar]
- 92.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, et al. 2012. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 37:171–86 [DOI] [PubMed] [Google Scholar]
- 93.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, et al. 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37:158–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schaupp L, Muth S, Rogell L, Kofoed-Branzk M, Melchior F, et al. 2020. Microbiota-induced type I interferons instruct a poised basal state of dendritic cells. Cell 181:1080–96.e19 [DOI] [PubMed] [Google Scholar]
- 95.Yang XL, Wang G, Xie JY, Li H, Chen SX, et al. 2021. The intestinal microbiome primes host innate immunity against enteric virus systemic infection through type I interferon. mBio 12(3):e00366–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stefan KL, Kim MV, Iwasaki A, Kasper DL. 2020. Commensal microbiota modulation of natural resistance to virus infection. Cell 183:1312–24. e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee W, Kim M, Lee SH, Jung HG, Oh JW. 2018. Prophylactic efficacy of orally administered Bacillus poly-γ-glutamic acid, a non-LPS TLR4 ligand, against norovirus infection in mice. Sci. Rep. 8:8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gutierrez-Merino J, Isla B, Combes T, Martinez-Estrada F, Maluquer De Motes C. 2020. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microbes 11:771–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Erttmann SF, Swacha P, Aung KM, Brindefalk B, Jiang H, et al. 2022. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity 55:847–61.e10 [DOI] [PubMed] [Google Scholar]
- 100.Winkler ES, Shrihari S, Hykes BL Jr., Handley SA, Andhey PS, et al. 2020. The intestinal microbiome restricts alphavirus infection and dissemination through a bile acid-type I IFN signaling axis. Cell 182:901–18.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, et al. 2017. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357:498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chemudupati M, Kenney AD, Smith AC, Fillinger RJ, Zhang L, et al. 2020. Butyrate reprograms expression of specific interferon-stimulated genes. J. Virol. 94(16):e00326–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Antunes KH, Fachi JL, de Paula R, da Silva EF, Pral LP, et al. 2019. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 10:3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, et al. 2011. IFN-λ determines the intestinal epithelial antiviral host defense. PNAS 108:7944–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dai J, Zhou P, Li S, Qiu HJ. 2022. New insights into the crosstalk among the interferon and inflammatory signaling pathways in response to viral infections: defense or homeostasis. Viruses 14(12):2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Van Winkle JA, Peterson ST, Kennedy EA, Wheadon MJ, Ingle H, et al. 2022. A homeostatic interferon-lambda response to bacterial microbiota stimulates preemptive antiviral defense within discrete pockets of intestinal epithelium. eLife 11:e74072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grau KR, Zhu S, Peterson ST, Helm EW, Philip D, et al. 2020. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat. Microbiol. 5:84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, et al. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. PNAS 108:5354–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Z, Zou J, Shi Z, Zhang B, Etienne-Mesmin L, et al. 2020. IL-22-induced cell extrusion and IL-18-induced cell death prevent and cure rotavirus infection. Sci. Immunol. 5(52):eabd2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schnepf D, Hernandez P, Mahlakoiv T, Crotta S, Sullender ME, et al. 2021. Rotavirus susceptibility of antibiotic-treated mice ascribed to diminished expression of interleukin-22. PLOS ONE 16:e0247738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, et al. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramakrishna C, Kujawski M, Chu H, Li L, Mazmanian SK, Cantin EM. 2019. Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat. Commun. 10:2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ansaldo E, Farley TK, Belkaid Y. 2021. Control of immunity by the microbiota. Annu. Rev. Immunol. 39:449–79 [DOI] [PubMed] [Google Scholar]
- 114.Alexander KL, Targan SR, Elson CO III. 2014. Microbiota activation and regulation of innate and adaptive immunity. Immunol. Rev. 260:206–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, Mucida D. 2017. Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell 171:783–94.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Swamy M, Abeler-Dorner L, Chettle J, Mahlakoiv T, Goubau D, et al. 2015. Intestinal intraepithelial lymphocyte activation promotes innate antiviral resistance. Nat. Commun. 6:7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dharakul T, Labbe M, Cohen J, Bellamy AR, Street JE, et al. 1991. Immunization with baculovirus-expressed recombinant rotavirus proteins VP1, VP4, VP6, and VP7 induces CD8+ T lymphocytes that mediate clearance of chronic rotavirus infection in SCID mice. J. Virol. 65:5928–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Salazar-Mather TP, Hokeness KL. 2006. Cytokine and chemokine networks: pathways to antiviral defense. Curr. Top. Microbiol. Immunol. 303:29–46 [DOI] [PubMed] [Google Scholar]
- 119.Imaoka A, Matsumoto S, Setoyama H, Okada Y, Umesaki Y. 1996. Proliferative recruitment of intestinal intraepithelial lymphocytes after microbial colonization of germ-free mice. Eur. J. Immunol. 26:945–48 [DOI] [PubMed] [Google Scholar]
- 120.Jiang W, Wang X, Zeng B, Liu L, Tardivel A, et al. 2013. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J. Exp. Med. 210:2465–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jung J, Surh CD, Lee YJ. 2019. Microbial colonization at early life promotes the development of diet-induced CD8αβ intraepithelial T cells. Mol. Cells 42:313–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kawaguchi-Miyashita M, Shimizu K, Nanno M, Shimada S, Watanabe T, et al. 1996. Development and cytolytic function of intestinal intraepithelial T lymphocytes in antigen-minimized mice. Immunology 89:268–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, et al. 2017. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 357:806–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sujino T, London M, Hoytema van Konijnenburg DP, Rendon T, Buch T, et al. 2016. Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science 352:1581–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu L, Gong T, Tao W, Lin B, Li C, et al. 2019. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat. Immunol. 20:1681–91 [DOI] [PubMed] [Google Scholar]
- 126.Neil JA, Matsuzawa-Ishimoto Y, Kernbauer-Holzl E, Schuster SL, Sota S, et al. 2019. IFN-I and IL-22 mediate protective effects of intestinal viral infection. Nat. Microbiol. 4:1737–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dallari S, Heaney T, Rosas-Villegas A, Neil JA, Wong SY, et al. 2021. Enteric viruses evoke broad host immune responses resembling those elicited by the bacterial microbiome. Cell Host Microbe 29:1014–29.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–18 [DOI] [PubMed] [Google Scholar]
- 129.Fonseca W, Malinczak CA, Fujimura K, Li D, McCauley K, et al. 2021. Maternal gut microbiome regulates immunity to RSV infection in offspring. J. Exp. Med. 218(11):e20210235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun M, Wu W, Chen L, Yang W, Huang X, et al. 2018. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 9:3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maynard CL, Elson CO, Hatton RD, Weaver CT. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, et al. 2015. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163:367–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, et al. 2014. Foxp3+ T cells regulate immunoglobulin A selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41:152–65 [DOI] [PubMed] [Google Scholar]
- 134.Kim M, Galan C, Hill AA, Wu WJ, Fehlner-Peach H, et al. 2018. Critical role for the microbiota in CX3CR1+ intestinal mononuclear phagocyte regulation of intestinal T cell responses. Immunity 49:151–63.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, et al.2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500:232–36 [DOI] [PubMed] [Google Scholar]
- 136.Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, et al. 2015. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39+Foxp3+ T cells and Treg function. Gut Microbes 6:234–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, et al. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Smith K, McCoy KD, Macpherson AJ. 2007. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19:59–69 [DOI] [PubMed] [Google Scholar]
- 139.Moon C, Baldridge MT, Wallace MA, Burnham CA, Virgin HW, Stappenbeck TS. 2015. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature 521:90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Franco MA, Greenberg HB. 1995. Role of B cells and cytotoxic T lymphocytes in clearance of and immunity to rotavirus infection in mice. J. Virol. 69:7800–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chachu KA, LoBue AD, Strong DW, Baric RS, Virgin HW. 2008. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLOS Pathog. 4:e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. 2014. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 210:171–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, et al. 2018. Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. 22:3440–53.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goncalves P, El Daker S, Vasseur F, Serafini N, Lim A, et al. 2020. Microbiota stimulation generates LCMV-specific memory CD8+ T cells in SPF mice and determines their TCR repertoire during LCMV infection. Mol. Immunol. 124:125–41 [DOI] [PubMed] [Google Scholar]
- 145.Sonnenberg GF, Artis D. 2015. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat. Med. 21:698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, et al. 2017. ILC1 confer early host protection at initial sites of viral infection. Cell 171:795–808.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, et al. 2015. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 16:698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, et al. 2016. The maternal microbiota drives early postnatal innate immune development. Science 351:1296–302 [DOI] [PubMed] [Google Scholar]
- 149.Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, et al. 2015. Interleukin-12 and −23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43:146–60 [DOI] [PubMed] [Google Scholar]
- 150.Pal S, Perrien DS, Yumoto T, Faccio R, Stoica A, et al. 2022. The microbiome restrains melanoma bone growth by promoting intestinal NK and Th1 cell homing to bone. J. Clin. Invest. 132(12):e157340. [DOI] [PMC free article] [PubMed] [Google Scholar]


