Abstract
Astrocytes have essential roles in central nervous system (CNS) health and disease. During development, immature astrocytes show complex interactions with neurons, endothelial cells, and other glial cell types. Our work and that of others have shown that these interactions are important for astrocytic maturation. However, whether and how these cells work together to control this process remains poorly understood. Here, we test the hypothesis that cooperative interactions of astrocytes with neurons and endothelial cells promote astrocytic maturation. Astrocytes were cultured alone, with neurons, endothelial cells, or a combination of both. This was followed by astrocyte sorting, RNA sequencing, and bioinformatic analysis to detect transcriptional changes. Across culture configurations, 7,302 genes were differentially expressed by 4 or more-fold and organized into eight groups that demonstrate cooperative and antagonist effects of neurons and endothelia on astrocytes. We also discovered that neurons and endothelial cells caused splicing of 200 and 781 mRNAs, respectively. Changes in gene expression were validated using quantitative PCR, Western blot, and immunofluorescence analysis. We found that the transcriptomic data from the three-culture configurations correlated with protein expression of three representative targets (FAM107A, GAT3, and GLT1) in vivo. Alternative splicing results also correlated with cortical tissue isoform representation of a target (Fibronectin 1) at different developmental stages. By comparing our results to published transcriptomes of immature and mature astrocytes, we found that neurons or endothelia shift the astrocytic transcriptome toward a mature state and that the presence of both cell types has a greater effect on maturation than either cell alone. These results increase our understanding of cellular interactions/pathways that contribute to astrocytic maturation. They also provide insight into how alterations to neurons and/or endothelial cells may alter astrocytes with implications for astrocytic changes in CNS disorders and diseases.
Keywords: astrocytes, astrocyte-neuron interaction, astrocyte-blood vessel interaction, transcriptomics, astrocyte maturation, neurovascular coupling
Graphical Abstract
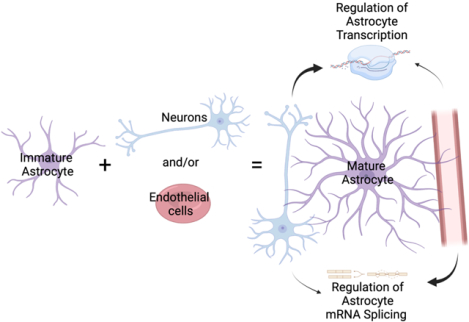
Astrocytes interact with neighboring cells, including neurons and endothelia. Neurons induce maturation of the astrocyte transcriptome and endothelia induce expression of a few markers of mature astrocytes. However, it is not known if and how neurons and endothelia interact to regulate the astrocyte transcriptome. Here we cultured astrocytes alone, with neurons and/or endothelia, and determined how these interactions affect the astrocytic transcriptome. We found that neurons and endothelia have cooperative and antagonist effects on the transcriptome and on mRNA splicing. We also found that neurons and endothelia cooperatively induce a shift in the astrocyte transcriptome reflecting maturation of astrocytes.
INTRODUCTION
Astrocytes have diverse functions in the central nervous system (CNS) including regulation of water balance, buffering of ions, uptake of neurotransmitters, and intercellular trafficking of brain energy substrates (for reviews, see (Jackson and Robinson, 2018; Santello et al., 2019; Souza et al., 2019; Tan et al., 2021; Schober et al., 2022)). They express cell surface receptors and secrete ligands that participate in the formation, maintenance, and refinement of synapses (Reviewed in (Shan et al., 2021; Schober et al., 2022)). Perisynaptic astrocytic processes (PAPs) enwrap synapses and dendrites with each astrocyte contacting approximately 100,000 synapses in mouse cortex (Bushong et al., 2002). Astrocytes also control the formation of tight junctions between brain endothelia and maintain the blood-brain barrier (Janzer and Raff, 1987; Abbott, 2002; Guerit et al., 2021; Heithoff et al., 2021; Pivoriunas and Verkhratsky, 2021). Each astrocyte contacts at least one blood vessel (Bushong et al., 2002; Hosli et al., 2022) with a specialized endfoot that surrounds the brain vasculature (Kacem et al., 1998; Mathiisen et al., 2010). The unique positioning of astrocytes between neurons and blood vessels/endothelia allows them to support energy demands, glutamate recycling, and control of blood flow that are unique to the brain (for reviews see (Petzold and Murthy, 2011; Stackhouse and Mishra, 2021).
Most cortical astrocytes are formed from radial glia that migrate into the cortex prior to birth in the mouse. After a brief period of proliferation, they undergo morphological and functional maturation (for reviews see (Molofsky and Deneen, 2015; Tabata, 2015)). Various markers have been used to study astrocyte maturation in vivo and in vitro, including the cytoskeletal protein glial fibrillary acidic protein (GFAP), the glutamate transporter subtype 1 (GLT1, also called EAAT2 in humans, gene name Slc1a2), the water channel aquaporin 4 (AQP4), and the inward-rectifying K+ channel, Kir4.1 (gene name Kcnj10) (Baba et al., 1997; Gegelashvili et al., 1997; Swanson et al., 1997; Schlag et al., 1998; Camassa et al., 2015; Hasel et al., 2017; Hill et al., 2019; Sakers et al., 2021). GLT1 expression and function increases during synaptogenesis in both mice and humans (Chaudhry et al., 1995; Shibata et al., 1996; Bar-Peled et al., 1997; Furuta et al., 1997; Ullensvang et al., 1997; Sims and Robinson, 1999). Interestingly, astrocytes express little or no GLT1 when cultured alone. However, co-culturing astrocytes with neurons induces expression of GLT1 (Gegelashvili et al., 1997; Swanson et al., 1997; Schlag et al., 1998; Yang et al., 2009; Hasel et al., 2017). This is consistent with the ability of neurons to regulate the astrocytic transcriptome and induce expression of molecules associated with functional astrocytic maturation including Kir4.1, AQP4, and the gap junction protein connexin 43 (Farmer et al., 2016; Hasel et al., 2017; Hill et al., 2019; Farhy-Tselnicker et al., 2021).
We recently demonstrated that brain endothelia also induce expression of GLT1 (Lee et al., 2017; Martinez-Lozada and Robinson, 2020). However, it remains unknown if or how endothelia more broadly affect the astrocytic transcriptome. Similarly, it is unknown if neurons and endothelia work together to regulate GLT1 and, more generally, astrocytic maturation. To address these knowledge gaps, we performed a series of experiments to test if co-culturing astrocytes with neurons, endothelial cells or both caused differential effects on the astrocytic transcriptome. Astrocytes were isolated using fluorescence-activated cell sorting (FACS), and RNA sequencing (RNA-seq) was performed followed by bioinformatic analysis. We discovered that neurons and endothelial cells impacted the astrocytic transcriptome distinctly. Neurons and endothelia cooperatively and, in some cases competitively, regulated the astrocyte transcriptome and had differential effects on mRNA splicing. However, astrocytes grown with neurons and endothelial cells in tandem, showed greater maturation with respect to their morphology and transcriptomic profile. These results demonstrate how signals from neighboring neurons and endothelial cells configure the molecular properties of astrocytes to promote their maturation. They also increase our knowledge of the importance of non-cell autonomous interactions that impact the astrocytic transcriptome with implications for understanding CNS disorders and diseases.
Materials and Methods
Animals.
BAC-GLT1-eGFP reporter mice on a C57/BL6 background (Regan et al., 2007) were maintained at the animal facility of the Children’s Hospital of Philadelphia (CHOP). Aldh1L1-Cre/ERT2 BAC (Srinivasan et al., 2016) (RRID: IMSR_JAX:029655) crossed with Ai9 (Madisen et al., 2010) (RRID: IMSR_JAX:007909) mice were maintained at the Montreal General Hospital. All studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the Children’s Hospital of Philadelphia (animal protocol 030) and followed the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals or by the Montreal General Hospital Facility Animal Care Committee and followed the guidelines of the Canadian Council on Animal Care. Up to five animals were housed per cage in standard controlled temperature, humidity, and light, and had ad libitum access to food and water. These colonies of mice were monitored every other day for any evidence of pain or distress. On the rare occasion that there was an issue, we consulted with the attending veterinarian. Mice were either medically treated or euthanized to minimize pain and distress. One animal from the colony was euthanized in an 18-month period.
Primary Enriched Astrocyte Cultures.
BAC-GLT1-eGFP mice of both sexes were used to prepare primary astrocyte cultures as previously described (Zelenaia and Robinson, 2000). In these mice, the expression of GFP completely overlaps with that of GLT1 (Regan et al., 2007). In addition, the expression of GLT1 and GFP is upregulated in astrocytes when astrocytes are cultured in the presence of neurons or endothelia (Yang et al., 2009; Ghosh et al., 2016; Lee et al., 2017). This allowed us to use GFP as an additional control in the present analyses. A total of 13 BAC-GLT1-eGFP mice were used: 2–3 mice per culture for 5 independent experiments. Briefly, mice 1–3 days of age were decapitated without anesthesia. Mouse brain cortices were dissected, the meninges were removed, the cortices were dissociated into a suspension using trypsin and gentle trituration, and the cells were plated at a density of approximately 2.5 × 105 cells/mL in 75 cm2 flasks (15 mL/flask). Astrocytes were maintained in Dulbecco’s modified eagle’s media (DMEM) (Gibco #11960–014), supplemented with 10% Ham’s F12 (Gibco #11765–047), 10% defined heat-inactivated fetal bovine serum (FBS) (HyClone #SH30070.03), and 0.24% penicillin/streptomycin (Gibco #15140–122), called ‘astrocyte’ media. A complete media exchange was performed every 3–4 days. After 7–10 days (when cell confluency was ~90%), A2B5 positive cells were eliminated using A2B5 hybridoma supernatant (1:50) (Dr. Judith Grinspan Lab, CHOP #anti-A2B5+, RRID: AB_2827951) and Low Tox-M rabbit complement (1:10 Cedarlane #CL3005). After 2–3 days of recovery, the astrocytes were split (1 to 1.5; surface area to surface area) into 24-well culture plates with glass coverslips for immunocytochemical analyses or into 10 cm culture dishes for RNA extraction. Greater than 95% of these cells are astrocytes as defined by using GFAP-immunoreactivity.
Co-cultures.
To study astrocyte/endothelial cell interactions, the mouse brain endothelioma cell line bEND.3 was used (American Type Culture Collection #CRL-2299, RRID: CVCL_0170). We previously showed that this cell line mimics the effects of primary brain endothelia on GLT1 expression (Lee et al., 2017), and this cell line has been used to study the mechanisms that control tight junction formation (Stamatovic et al., 2009; Cibelli et al., 2021; Sasson et al., 2021; Pu et al., 2022). In addition, this cell line has the advantage of being a homogeneous population of cells. These cells were maintained in DMEM supplemented with 4.5 g/L D-glucose (Gibco #11960–014), 10% defined heat-inactivated FBS (HyClone #SH30070.3), 4 mM L-glutamine (Gibco #25030–081), and 1 mM sterile filtered sodium pyruvate, called ‘endothelial’ media, at 37°C and 5% CO2. This media was exchanged every 3–4 days. Cells were never used past passage 30 to limit genetic drift. Two different lots of bEND.3 cells were used for these studies.
In astrocyte/endothelia (AE) co-cultures, astrocytes (A) were cultured on top of endothelial cells. From a confluent dish, bEND.3 cells were split 1:3 (surface area to surface area) into either 10 cm dishes or 24 well plates. Two days later when bEND.3 were ~60–70% confluent, astrocytes were replated directly onto empty wells or on top of endothelia. One third of the astrocyte media was replaced every three days until the time of harvesting.
To study astrocyte-neuron (AN) interactions, cortical cell suspensions that contain neurons and astrocytes were obtained from E17–19 Sprague-Dawley rats (Charles River) prepared by the Neurons R Us service center at the Penn Medicine Translational Neuroscience Center (RRID: SCR_022421) (Calabrese et al., 2022; Gallagher and Holzbaur, 2023). Briefly, five pregnant rats were anesthetized with CO2 and sacrificed by cervical dislocation. The uterine horn was removed, and E17–19 embryos were removed and decapitated one at a time; the heads were placed in cold HBSS. Rat brain cortices were dissected, the meninges were removed, and the cortices were dissociated using trypsin and gentle trituration into a suspension. In these cultures, neurons assume a position on top of astrocytes with some neuronal cell bodies grouping as small clusters and most remaining dispersed. These preparations have been used for decades to study synaptic physiology (Dichter, 1978; Kriegstein and Dichter, 1983; Kriegstein and Dichter, 1984), to study the effects of neurons on GLT1 expression (Schlag et al., 1998; Ghosh et al., 2011; Ghosh et al., 2016), and the role of glutamate transport in glutamate toxicity (Blitzblau et al., 1996). For simplicity, these neuron-enriched suspensions will be referred to as neurons throughout the paper. Three to four days after the astrocytes were plated into either 10 cm dishes or 24 well plates when they were ~60–70% confluent, neurons were added at a density of 6.67 × 105 cells/mL on empty poly-D-lysine-coated wells (N), on top of astrocytes to make double cultures (AN), or on top of astrocytes that also contained endothelia to produce triple cultures (AEN). We arbitrarily plated neurons onto astrocytes or astrocytes with endothelia to form the different culture configurations. The cultures were maintained for 10 days in astrocyte media; one-third of the media was replaced with fresh media every 3–4 days (See Figure S1A).
Immunofluorescence.
Cells were cultured on sterile glass coverslips coated with 5 μg/mL poly-D-lysine as described above. The cells were washed once with PBS 1X and then fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at room temperature. Cells were rinsed three times with PBS 1X, 0.4% Triton X-100, 10 min each wash, and incubated in blocking buffer (5% goat serum in PBS 1X, 0.4% Triton X-100) for 1h at room temperature. This was followed by an overnight incubation at 4°C with a mixture of the following primary antibodies in blocking solution: chicken anti-glial fibrillary acidic protein (1:250, Millipore #AB5541, RRID: AB_177521), rat anti-pecam1/CD31 (1:100, BD Pharmingen #550274, RRID: AB_393571), and mouse anti-NeuN (1:100, Millipore #MAB377, RRID: AB_2298772). Cells were rinsed with three 10 min washes with PBS 1X containing 0.4% Triton X-100, followed by incubation for 1h at room temperature with the following secondary antibodies in blocking buffer: goat anti-chicken Alexa Fluor 633 (1:500, Thermo Fisher Scientific #A21103, RRID: AB_2535756), goat anti-rat Alexa Fluor 594 (1:500, Thermo Fisher Scientific #A11007, RRID: AB_10561522), and goat anti-mouse Alexa Fluor 488 (1:500, Thermo Fisher Scientific #A11029, RRID: AB_2534088). Nuclei were counterstained using a mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories Inc #H-1200, RRID: AB_2336790). Incubations without primary antibodies were included to confirm the specificity of the signal. Pictures were taken with a DMi8 SP8 Leica confocal microscope equipped with a 40X objective (Leica Microsystems, RRID: SCR_018169) using the 405, 488, 594, and 633 nm laser lines. The images were taken using the sequential mode to avoid spillover between fluorophores. For each independent experiment, at least three fields were imaged per culture configuration. Fields were arbitrarily chosen on the 405 (DAPI) channel. Three independent experiments were conducted. The analysis of astrocyte morphology was performed in a blinded fashion using GFAP as a marker for astrocytes.
Cell Dissociation and Flow Cytometry.
Cells were washed twice with Hank’s Balanced Salt Solution and dissociated with 0.05% trypsin (Gibco #25200–056). Cells were then incubated with anti-astrocyte cell surface antigen (ACSA)-2 antibodies coupled to R-Phycoerythrin (PE) to a final concentration of 3 ng/μL (Miltenyi Biotec #130–123-284, RRID: AB_2811488) as previously described (Batiuk et al., 2017; Kantzer et al., 2017) and anti-mouse Pecam1 antibodies coupled to allophycocyanin (APC) to a final concentration of 2.5 ng/μL (Bio Legend #102410, RRID: AB_312905) to separate endothelial cells as previously described (Pal et al., 2017). After incubating cells for 20 min at 4°C, they were washed twice in sorting buffer (PBS 1X with 4% FBS) with centrifugation at 350xg for 5 min, followed by homogenization in ice-cold sorting buffer and filtering through a 35 μm nylon mesh. Fluorescence-activated cell sorting (FACS) was performed in the Flow Cytometry Core Laboratory at CHOP using an electrostatic droplet FACS Jazz sorter (RRID: SCR_019875) with Isoflow sheath fluid (Beckman Coulter #8547008) and a 100 μm nozzle. We excluded debris and aggregates using the pulse width of the triggering parameter before gating populations based on ACSA-2-PE fluorescence (using the yellow-green laser Ex 561nm, Em 585/29 nm) and Pecam1-APC fluorescence (using the red laser Ex 640nm, Em 660/20 nm). Unstained and single fluorophore-labeled cells were used to verify the gates (See Figure 1C and S1A). Cells were purified using a 1.0 Drop Pure sort mode and collected into sorting buffer (at least 400,000 ACSA-2-PE-positive cells, astrocytes, were collected per sample) and concentrated by centrifugation at 350xg for 5 min. Sample size was based on an earlier study that used 3 independent samples to examine the effects of neurons on the astrocyte transcriptome (Hasel et al., 2017). We used an n of 5 to ensure that there were enough independent samples after RNA quality control. For each independent experiment, astrocytes were collected from four different specimens (A, AE, AN, and AEN, see above). Quantification of the percentage of each cell type per culture configuration as estimated using FACS is shown in Table 1.
Figure 1. Co-culture configurations modify the morphology of astrocytes.
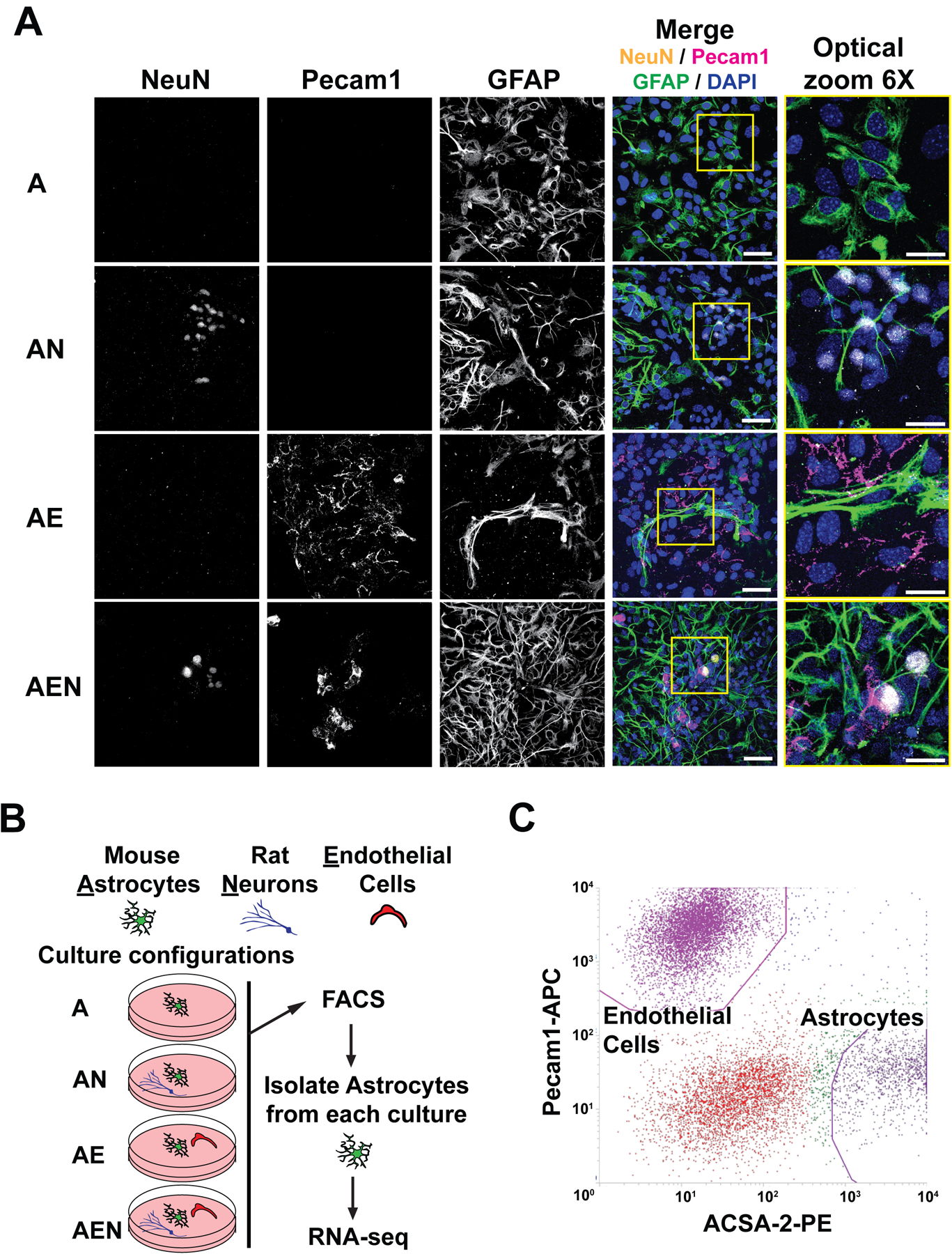
A. Representative images of co-cultures using confocal imaging. Anti-NeuN, anti-Pecam1, and anti-GFAP antibodies were used as markers of neurons, endothelia, and astrocytes, respectively. Nuclei were counter-stained with 4-,6-diamidino-2-phenylindole (DAPI). No staining was observed when the primary antibodies were omitted (data not shown). The magnification is the same in the first four columns (scale bar 50μm). The fifth column is a 6X optical zoom of the yellow-outlined portion of the fourth column (scale bar 25μm). Neurons consistently changed astrocytes to a more stellate/bushy morphology, while endothelial cells induced an elongated morphology, in triple cultures astrocytes have a combination of neuron-induced and endothelia-induced morphology. Fields were chosen using the DAPI channel to blind the analyzer. Data are representative of three independent experiments. B. Outline of experiments: Mouse cortical astrocytes were cultured by themselves (A), with rat cortical neurons (AN), with endothelial cells (AE), or in tricultures with neurons and endothelial cells (AEN). After 10 days, astrocytes were dissociated and labeled using antibodies anti-astrocyte cell surface antigen 2 (ACSA-2) coupled to PE and isolated using fluorescence-activated cell sorting (FACS). RNA was extracted, reverse transcribed, and sequenced. C. A representative dot-plot showing the PE fluorescence (coupled to anti-ACSA-2 antibodies, astrocyte marker) on the X axis and the APC fluorescence (coupled to anti-Pecam1 antibodies, endothelial marker) on the Y axis. Note the separation between the gates selected to sort the cells.
Table 1.
Proportion of astrocytes (ACSA-2-PE+ cells), endothelia (Pecam1-APC+ cells), and negative (ACSA-2-PE−/ Pecam1-APC−) cells as determined by FACS. Cultured cells were dissociated and labeled with anti-ACSA-2 antibodies coupled to the fluorophore PE and with anti-Pecam1 antibodies coupled to APC and subjected to FACS (see methods section for details). Cells were collected from a gate that excluded debris and aggregates. 20,000 events were collected per experiment, and the percentage of cells was calculated as follows: astrocytes all PE+ cells, endothelia all APC+ cells, and negative cells (negative cells = total cells - PE+ - APC+). The mean and standard deviation of five experiments (six samples per experiment) are shown.
| A (STDEV) | E (STDEV) | Neg (STDEV) | |
|---|---|---|---|
| A | 4.4 | 0.0 | 4.4 |
| EA | 12.2 | 11.3 | 1.7 |
| AN | 11.1 | 0.0 | 11.1 |
| EAN | 8.6 | 5.7 | 6.2 |
| E | 0.0 | 1.4 | 1.4 |
| N | 0.2 | 0.1 | 0.3 |
Total RNA Extraction.
Total RNA was extracted from sorted astrocytes or endothelia using the RNeasy Mini Kit according to manufacturer instructions (Qiagen #74104). Three different aliquots were prepared: one for the determination of RNA quality, one for RNA sequencing, and one for qPCR. RNA purity and concentration were measured using the Agilent RNA ScreenTape Assay (Agilent #5067–5576) on the 4200 TapeStation (RRID: SCR_018435).
RNA Sequencing.
Initial RNA quality control, library preparation, and sequencing were performed at the Centre d’Expertise et de Services at Génome Québec. 19 of 20 RNA samples passed initial quality control as assessed using a Bioanalyzer (Agilent) with a mean RNA integrity number of 8.7 ± 0.6. Astrocytes from one of the AEN triple culture experiments did not pass this step (RIN: 0, concentration: 9.9 ng/μL) and were excluded from further analyses. The data for the effects of astrocytes, neurons, or their combination on the endothelial transcriptome will be analyzed and validated in a future study. Libraries were synthesized using the NEBNext library Prep kit (New England BioLabs #E7645) and sequenced on a single lane of Illumina NovaSeq 6000 (RRID: SCR_016387) to generate 3.0×109 100bp paired-end, stranded reads with a mean of 8.9 × 107 ± 8.6 × 106 reads per sample. All libraries passed an initial quality control step using the FASTQC (RRID: SCR_014583). Adapters and low-quality stretches were removed using Trimmomatic (Bolger et al., 2014) (RRID: SCR_011848).
Sargasso Alignment and Counts.
As indicated above, neuronal cell suspensions introduce both rat neurons and astrocytes onto mouse astrocytes. We filter contaminating rat RNAs using Sargasso v2.0.2 (Qiu et al., 2018) (RRID: SCR_023489) set to default parameters. Briefly, sequences were aligned using STAR 2.7.7a (Dobin et al., 2013) (RRID: SRC_004463) against both the mouse (GRCm38 with the addition of the eGFP gene sequence to reflect the experimental design) and the rat (Rnor 6.0) genomes. The resulting filtered sequences that corresponded to mouse were used for all subsequent analyses. The filtered sequences were counted at the gene level with HTseq-count (Putri et al., 2022) (RRID: SRC_011867) using the GRCm38 annotation containing an additional entry for the eGFP transgene. See pipeline in Figure S2A.
Estimation of Endothelial Contamination and Differential Gene Expression Analysis.
DESeq2 (Love et al., 2014) (RRID: SCR_015687) was used to estimate size factors, estimate dispersion, and fit the count data of all samples to a negative binomial generalized linear model initially using only culture configuration as a factor. Due to the presence of endothelial cell-specific transcripts in the sequences from astrocytes isolated from mixed cultures (e.g. Tie2), the unmix function of DESeq2 was used to estimate endothelial contamination by comparing them to pure cultures of endothelial cells and astrocytes. DESeq2 unmix calculated the average endothelial contamination of 12 ± 1.5% in the AE samples and 6 ± 1.7% in the AEN samples (Figure S2B). Following the estimation of endothelial contamination for each astrocyte sample, size factors and dispersion were re-estimated for the astrocyte samples alone. To control for the presence of endothelial cell mRNA in the astrocyte samples during differential gene expression testing and log-fold change estimation, the estimated endothelial contamination was used as a factor in the DESeq2 model.
All differential expression analyses were performed using DESeq2. Shrinkage of log2-fold changes were applied using DESeq2 and shrunken estimates were used throughout the analysis. Genes were annotated using AnnotationDbi (Pagès, 2023) (RRID: SCR_023487) and org.Mm.eg.db (Carlson, 2019) (RRID: SCR_023488). Normalized and variance-stabilized transformed data were used for all heatmaps.
The likelihood ratio test (LRT) was used to test for differentially expressed genes across all culture conditions while controlling for endothelial contamination (reduced model). The 7,302 differentially expressed genes (p adjusted value < 0.05 and log2-fold change ≥ 2; i.e. ≥ 4-fold change) were clustered using the clusterProfiler function (Wu et al., 2021) (RRID: SCR_016884), DEGpatterns, on the variance stabilized transformed counts to reveal the patterns of gene expression across conditions. A cooperative effect of neurons and endothelial cells on astrocytic transcription was attributed to clusters in which neurons or endothelia change astrocyte gene expression in the same direction and where the combination of these cells causes an even greater effect. Clusters in which the changes went in opposing directions were considered antagonistic. Three additional clusters were identified based on changes caused by neurons or endothelial cells with no cooperative or antagonistic effects and were named redundant.
To estimate the shrunken fold changes between each condition, the Adaptive Shrinkage in R (ashr) algorithm (Stephens, 2017) (RRID: SCR_023486) was applied to each possible combination of pairwise comparisons. The reduced model was used to control for endothelial contamination.
Quantitative PCR (qPCR).
cDNA synthesis and qPCR were performed by the Center for Applied Genomics at CHOP. Briefly, complementary DNA (cDNA) was prepared in triplicates using Reverse Transcription Master Mix (Standard BioTools, previously known as Fluidigm #100–6299), and preamplified using Preamp Master Mix (Standard BioTools #100–5580). qPCR was performed using SsoFast EvaGreen (BioRad #172–5211) and DELTAgene Assays (Standard BioTools #ASY-GE, see Table 2 for list of qPCR primers) in 96 by 96 Dynamic Array integrated fluidic circuit (Standard BioTools #BMK-M-96.96 and 85000802). The analyses were conducted using the Juno and Biomark equipment (Standard BioTools). Hars2 (an aminoacyl-tRNA synthetase), Srp68 (a subunit of the signal recognition particle), and Srbd1 (a structural component of the ribosome) were used as internal controls due to their low coefficient of variance between the different culture configurations as determined using the DESeq2 analyses. We confirmed that these controls did not change between culture configurations in this PCR analysis (Table 3). The ddCt method was used to calculate relative gene expression in R and correlated to log-fold change estimates from DESeq2.
Table 2.
Sequences of the primers used for qPCR. Delta gene Assays (Project ID: 20842_FDGP_21, Standar BioTools, previously known as Fluidigm) were used for qPCR. The table includes target gene name, assay ID, and forward and reverse primer sequences.
| Target | Assay ID | Forward Primer | Rerverse Primer |
|---|---|---|---|
| ADCY1 | GEP00118848 | TGTGGAGATGGGACTTGACA | CACACGCATGTTCAGGTCTA |
| ALDOC | GEA00042712 | GACGGAGACCATGACCTCAA | GGTCACTCAGGGCCTTGTATA |
| APPL2 | GEP00118842 | CCCTCACAGATTACACCAACCA | AGTGGCCAGGCACATTTCA |
| AQP4 | GEP00118862 | GGCATCCTCTACCTGGTCAC | CCAGCGGTGAGGTTTCCA |
| ATP1B1 | GEP00118845 | CCAAATGTCCTGCCTGTTCA | TATCCGCCCATCCCAAAGTA |
| CCND1 | GEA00007896 | TGCCGAGAAGTTGTGCATCTA | TGTTCACCAGAAGCAGTTCCA |
| CCND2 | GEA00011726 | GGAGAAGCTGTCCCTGATCC | CGGGTACATGGCAAACTTGAA |
| CDK1 | GEA00011734 | AAGTACCTGGACTCCATCCC | TCCCTGGAGGATTTGGTGTAA |
| CLCN5 | GEP00118830 | GGCCCAGCTCATCATCAATACA | AAGAGCCCAGAGGACGTACA |
| CLDN10 | GEP00118836 | CAAAGTCGGAGGCTCAGATCAA | AATACAATCCCGGCCAAGCA |
| DAPP1 | GEP00118841 | CAGAGTGCTCAGCTGTTCAA | CCCGTCTTTGCACAGAGATAA |
| EMC7 | GEP00118852 | GCTGGACAGACTTTCTGATGAA | CACCACTTTAGGCAGAAGCA |
| EPHB1 | GEP00118826 | TCATGGAGAACGGCGCTTTA | CATCCCCACAAGCTGGATCA |
| EPHB2 | GEP00118861 | CAACGGTGTGATCCTGGACTA | TATGGCCGTGGCGTTGTA |
| FABP7 | GEP00118829 | AACCTGGAAGCTGACAGACA | TCACGTTTCCCACTTGCCTA |
| FAM43A | GEP00118853 | GGGACATCACTTCCTCTGTCA | GGTCGCCCAGGTCACTAATA |
| eGFP | GEP00118549 | ACTTCAAGATCCGCCACAAC | ATCGGGGTGTTCTGCTGATA |
| FZD1 | GEA00012045 | CACGGTGCTCACGTACCTA | TGTAACAGCCGGACAGGAAA |
| FZD10 | GEP00118825 | TTGAAGCCAACAGCAGCTAC | CTGCGCATCACCAAGATCAA |
| GJA1 | GEA00022912 | TCAGCCTCCAAGGAGTTCCA | ACCTTGTCCAGCAGCTTCC |
| GLI1 | GEA00012054 | CAGAATCGGACCCACTCCAA | GCGAGCTGGGATCTGTGTA |
| GLUL | GEA00041926 | GACCTGTTCACCCATCCATCA | GAGGTGGCCATGGTGGAA |
| GPC4 | GEP00118863 | GTTTGGCCAACCAAGGAGAC | CTCTCTGCCACCATCAGCATA |
| GSN | GEA00048001 | TCATCTCCAAGATGCAGTACCC | TGCTTAAAGAGAGGGGTCTCAC |
| HARS2 | GEP00118858 | CCGAACCATCTGCTCCTCAA | GCCACCATCTCATGCCTCA |
| HES5 | GEP00118828 | AAGAGCCTGCACCAGGACTA | GTGCAGGGTCAGGAACTGTAC |
| HEY1 | GEP00057170 | CGAGACCATCGAGGTGGAAA | ATGTCGTTGGGGACATGGAA |
| HEY2 | GEP00118838 | GTGGGGAGCGAGAACAATTAC | TGTCGGTGAATTGGACCTCA |
| KALRN | GEP00118827 | GACTCTTCAGGACACACGAA | CTTCTACATGCTCGGTCACA |
| KCNJ10 | GEP00118851 | TATCAGAGCAGCCACTTCAC | CGTATTCCTGGGGCCACTA |
| MKI67 | GEP00055177 | GAGACATACCTGAGCCCATCA | GCTTTGCTGCATTCCGAGTA |
| NFIA | GEP00118843 | CAGCCAAGTGAAGCTGACA | GTGACAAAGCTGTCCTGGAA |
| NRP2 | GEA00031939 | GTGGATCAGCAGCGCTAAC | GCCATCACTCTGCAGTTTCAA |
| OLIG2 | GEP00118831 | CTGGCGCGAAACTACATCC | CCCCGTAGATCTCGCTCAC |
| PDLIM1 | GEP00118847 | AGACAACATGACGCTCACAGTA | TTTCCCCTCCTCGGTCACTA |
| PLD2 | GEP00118840 | CCTCAACCGCCTCCTGAC | TGGAGCCAAGGTCTGGGATA |
| S100B | GEA00012109 | ACAACGAGCTCTCTCACTTCC | ATCTTCGTCCAGCGTCTCC |
| S1PR1 | GEP00062246 | CGGTGTAGACCCAGAGTCC | GAGAGGCCTCCGAGAAACA |
| SEMA3C | GEP00118850 | CTGGAAACTGACAATCCAAGGAC | ACACACTGCCGATCCCTTAA |
| SHH | GEP00118846 | AGGAAAACACGGGAGCAGAC | ACAGAGATGGCCAAGGCATTTA |
| SLC1A2 | GEP00118832 | AAGAAGGGCCTGGAGTTCAA | ATGGCAATGCCGAAAGCAA |
| SLC1A3 | GEP00118834 | AATGCCTTCGTTCTGCTCAC | TTATACGGTCGGAGGGCAAA |
| SLC1A4 | GEP00118857 | CTCCGGTCTCCAAAGAGACA | CGGCAACCACAAGATTGGAA |
| SLC6A11 | GEP00118855 | TGGGCCACCTTGTTCTTCA | GGCTGTCACAAGACTCTCCA |
| SLC7A11 | GEP00118833 | TTGTTCGAGTCTGGGTGGAA | TCCAGGATGTAGCGTCCAAA |
| SMIM12 | GEP00118854 | CCTGTTGCTCCCGGCTA | AAAGCACAGGCCACATGAC |
| SOX9 | GEP00118835 | AGTACCCGCATCTGCACAA | GTCTCTTCTCGCTCTCGTTCA |
| SPARC | GEA00045025 | GAAACCGTGGTGGAGGAGAC | TGCACCGTCCTCAAATTCTCC |
| SPARCL1 | GEP00118856 | GTGTTTGCCAAGATCCAGAGAC | TGGCGTAGGTTTGGTTGTCA |
| SRBD1 | GEP00118860 | CAGCCCAAAACCTTTGACATCC | TGCAGGTCTTCCAGACACAC |
| SRP68 | GEP00118859 | GAGAGTCTGGGAAGGTGTCAAA | TTCGCCTGATTGCTGTTGAC |
| SYT11 | GEP00118839 | TGACAGTGGTGGTCCTCAAA | TTCTGCCGTAGTAGACGTTCA |
| THBS1 | GEA00023044 | CCCCAGAAGACATTCTCAGGAA | CGTTCACCACGTTGTTGTCA |
| TLN2 | GEP00118849 | GGCTGTGTCTGATTTGCTGAA | TTCCGGCAGCAGTCAAAAC |
| TNF | GEP00055421 | CAAATGGCCTCCCTCTCATCA | GCTACAGGCTTGTCACTCGAA |
| TNFRSF19 | GEP00118837 | GAAACTGTGTCCTCTGCAAACA | CACTGTGCATCCTCCCCATA |
| VIM | GEA00009549 | GATTTCTCTGCCTCTGCCAAC | CAACCAGAGGAAGTGACTCCA |
| ZBTB20 | GEP00118844 | ACAAACTCTCACGCTCACAC | CGAGCACGGAATTGCTGAA |
Table 3.
qPCR results with full statistical report. The table includes raw Ct values for fifty-five target genes and for three housekeeping genes (Hars2, Srbd1, and Srp68, highlighted in yellow columns), calculation of △△CT values and fold changes. Mean fold changes, standard deviations (SD), and standard error of mean (SEM) are highlighted in green rows. n=5 independent experiments, each with 4 samples (A, AN, AE, AEN).
| housekeeping genes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Triplicate mean Ct | HARS2 | SRBD1 | SRP68 | ADCY1 | ALDOC | APPL2 | AQP4 | ATP1B1 | CCND1 | CCND2 | CDK1 | CLCN5 | CLDN10 | DAPP1 | eGFP | EMC7 | EPHB1 | EPHB2 | FABP7 | FAM43A | FZD1 | FZD10 | GJA1 | GLI1 | GLUL | GPC4 | GSN | HES5 | HEY1 | HEY2 | KALRN | KCNJ10 | MKI67 | NFIA | NRP2 | OLIG2 | PDLIM1 | PLD2 | S100B | S1PR1 | SEMA3C | SHH | SLC1A2 | SLC1A3 | SLC1A4 | SLC6A11 | SLC7A11 | SMIM12 | SOX9 | SPARC | SPARCL1 | SYT11 | THBS1 | TLN2 | TNF | TNFRSF19 | VIM | ZBTB20 |
| A_G1 | 11.579 | 12.689533 | 10.632867 | 12.912 | 8.4878333 | 11.6816 | 8.4048 | 7.8249 | 7.4518333 | 7.8992 | 10.800433 | 13.607033 | 15.3486 | 11.807367 | 19.9026 | 9.4475667 | 15.281267 | 9.8918667 | 5.8906667 | 11.1743 | 13.121 | 15.4508 | 9.4114667 | 13.694867 | 10.1352 | 7.1642667 | 9.9924333 | 14.675833 | 11.054167 | 14.1181 | 15.336133 | 7.6148667 | 11.7028 | 9.2911 | 13.331967 | 11.564067 | 11.9488 | 11.0105 | 12.946133 | 11.009967 | 10.910667 | 11.2075 | 10.932767 | 7.4944 | 11.617733 | 15.7903 | 16.5152333 | 19.354167 | 8.6986667 | 5.4468333 | 9.5356 | 8.1963667 | 7.5312333 | 10.665867 | 10.6779 | 13.561933 | 4.7531 | 10.509467 |
| A_G2 | 11.4825 | 12.4694 | 10.3724 | 12.9756 | 8.1165667 | 11.645267 | 8.2525 | 7.6151 | 7.5714667 | 8.0035333 | 10.4604 | 13.731367 | 14.7203 | 11.388133 | 18.966033 | 9.2051 | 14.8293 | 9.7568667 | 5.7257667 | 11.033033 | 12.744633 | 15.3936 | 8.9161667 | 13.551733 | 9.9743333 | 6.9075333 | 9.8411667 | 15.666567 | 10.7587 | 13.943533 | 15.4075 | 7.5178667 | 11.7191 | 9.0542333 | 13.676433 | 11.404833 | 11.634067 | 10.866467 | 12.557267 | 10.773 | 10.803533 | 11.069467 | 10.8708 | 7.1223667 | 11.261033 | 15.283433 | 16.767 | 19.580933 | 8.5718667 | 5.3054 | 9.3577 | 7.7308 | 7.4609 | 10.6466 | 10.991633 | 13.386867 | 4.4952333 | 10.577167 |
| A_G3 | 11.657567 | 12.498433 | 10.496 | 13.220133 | 8.1112 | 11.613433 | 8.2024667 | 7.6092667 | 7.4972667 | 7.841 | 10.8158 | 13.780467 | 14.782267 | 11.444633 | 18.720433 | 9.1362667 | 14.9517 | 9.7789 | 5.6789333 | 11.2085 | 12.922 | 15.5271 | 8.9906 | 13.521967 | 9.9291333 | 6.9905 | 9.6957667 | 17.2793 | 10.7577 | 14.074967 | 15.511533 | 7.3093333 | 12.017733 | 9.0275 | 13.0834 | 11.327067 | 11.868567 | 10.825867 | 12.395767 | 10.682067 | 10.675667 | 11.415867 | 10.4744 | 7.154 | 11.265467 | 15.079733 | 16.4091333 | 20.007367 | 8.4933333 | 5.1854667 | 9.2735333 | 7.7983667 | 7.477 | 10.6222 | 11.703367 | 13.375333 | 4.6026333 | 10.495533 |
| A_G4 | 11.543567 | 12.585867 | 10.344633 | 13.294667 | 8.0132 | 11.603467 | 8.0499333 | 7.5199333 | 7.3771 | 7.9236333 | 10.47 | 13.891167 | 15.165 | 11.471367 | 19.916033 | 9.0639 | 14.957867 | 9.6661667 | 6.0262333 | 10.881733 | 12.8114 | 15.8638 | 8.9933 | 13.345767 | 9.8886333 | 6.757 | 9.5652667 | 19.007267 | 10.699 | 14.4345 | 15.500033 | 7.3453333 | 12.149233 | 9.1577333 | 13.352433 | 12.049133 | 11.7129 | 10.829433 | 13.113933 | 10.7646 | 10.554467 | 11.374 | 11.045533 | 7.3381 | 11.350633 | 15.766633 | 16.4478667 | 19.685533 | 8.3974333 | 5.1451333 | 9.4956667 | 7.8941 | 7.2432333 | 10.628067 | 11.960633 | 13.522767 | 4.5749333 | 10.809467 |
| A_G5 | 12.102767 | 13.132133 | 11.035033 | 13.503667 | 8.3408 | 12.208133 | 8.7431333 | 8.4738667 | 8.2982333 | 8.6158333 | 11.3041 | 14.623633 | 14.8948 | 12.362667 | 18.9057 | 9.8087333 | 14.736133 | 10.468433 | 6.4991667 | 11.806133 | 13.0922 | 15.601533 | 9.8186333 | 14.206133 | 10.514933 | 7.5655333 | 10.657433 | 16.353633 | 11.327833 | 14.887467 | 15.776233 | 8.1488333 | 12.4349 | 10.033333 | 14.069467 | 11.9734 | 12.3345 | 11.561033 | 13.903733 | 11.8927 | 11.5754 | 13.435633 | 10.9352 | 7.8528667 | 11.775833 | 16.440267 | 17.6587 | 21.102733 | 9.4282667 | 5.8915667 | 9.9346333 | 8.4866 | 8.0540667 | 11.204533 | 13.0706 | 14.199133 | 4.9620333 | 11.223267 |
| Mean | 11.67308 | 12.675073 | 10.576187 | 13.181213 | 8.21392 | 11.75038 | 8.3305667 | 7.8086133 | 7.63918 | 8.05664 | 10.770147 | 13.926733 | 14.982193 | 11.694833 | 19.28216 | 9.3323133 | 14.951253 | 9.9124467 | 5.9641533 | 11.22074 | 12.938247 | 15.567367 | 9.2260333 | 13.664093 | 10.088447 | 7.0769667 | 9.9504133 | 16.59652 | 10.91948 | 14.291713 | 15.506287 | 7.5872467 | 12.004753 | 9.31278 | 13.50274 | 11.6637 | 11.899767 | 11.01866 | 12.983367 | 11.024467 | 10.903947 | 11.700493 | 10.85174 | 7.3923467 | 11.45414 | 15.672073 | 16.7595867 | 19.946147 | 8.7179133 | 5.39488 | 9.5194267 | 8.0212467 | 7.5532867 | 10.753453 | 11.680827 | 13.609207 | 4.6775867 | 10.72298 |
| SD | 0.2484033 | 0.2695246 | 0.2807707 | 0.24141 | 0.1945794 | 0.2577089 | 0.2631449 | 0.388378 | 0.3751201 | 0.3180001 | 0.3442886 | 0.4027111 | 0.2663966 | 0.4078196 | 0.5796413 | 0.3028707 | 0.2062171 | 0.321029 | 0.3292689 | 0.3518349 | 0.1665387 | 0.1833331 | 0.3844383 | 0.3274565 | 0.2561273 | 0.3101941 | 0.4262513 | 1.6500555 | 0.2671738 | 0.3787018 | 0.1670694 | 0.3379031 | 0.307751 | 0.4159139 | 0.3804439 | 0.3296717 | 0.2728949 | 0.3123997 | 0.5899881 | 0.5005106 | 0.3985052 | 0.9797395 | 0.2201338 | 0.2979658 | 0.23122 | 0.5279555 | 0.52157339 | 0.6879646 | 0.4121424 | 0.301509 | 0.2548734 | 0.315207 | 0.3007304 | 0.2527407 | 0.9342268 | 0.3397984 | 0.1844049 | 0.3067361 |
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| SEM | 0.1110893 | 0.1205351 | 0.1255645 | 0.1079619 | 0.0870186 | 0.1152509 | 0.117682 | 0.1736879 | 0.1677588 | 0.142214 | 0.1539706 | 0.1800979 | 0.1191362 | 0.1823825 | 0.2592235 | 0.1354479 | 0.0922231 | 0.1435685 | 0.1472535 | 0.1573453 | 0.0744784 | 0.0819891 | 0.171926 | 0.146443 | 0.1145436 | 0.138723 | 0.1906254 | 0.7379272 | 0.1194838 | 0.1693606 | 0.0747157 | 0.1511148 | 0.1376304 | 0.1860024 | 0.1701397 | 0.1474337 | 0.1220423 | 0.1397094 | 0.2638507 | 0.2238351 | 0.1782169 | 0.4381528 | 0.0984468 | 0.1332544 | 0.1034047 | 0.2361089 | 0.23325471 | 0.3076671 | 0.1843157 | 0.1348389 | 0.1139828 | 0.1409648 | 0.1344907 | 0.1130291 | 0.4177989 | 0.1519625 | 0.0824684 | 0.1371766 |
| AN_G1 | 11.340067 | 12.2161 | 10.3718 | 13.739267 | 6.7452333 | 10.8474 | 7.0579333 | 7.036 | 8.4655667 | 8.2855667 | 11.8638 | 14.416867 | 12.937633 | 10.693967 | 18.2123 | 8.7978333 | 12.527667 | 10.171967 | 5.6588667 | 11.457433 | 11.354567 | 13.137067 | 7.6547 | 12.649467 | 8.4943333 | 7.5134333 | 9.5910667 | 12.057633 | 9.9583667 | 12.0371 | 13.8229 | 6.8085333 | 13.039233 | 8.7416 | 13.1477 | 9.6403667 | 11.9162 | 9.956 | 9.8854 | 9.7110333 | 11.2121 | 13.454667 | 8.8794333 | 6.2241667 | 10.551567 | 10.987867 | 14.9864 | 19.7624 | 8.0228 | 5.1556 | 7.5377333 | 7.1497 | 7.8466333 | 10.804267 | 12.528833 | 11.019 | 4.8791 | 9.2164 |
| AN_G2 | 12.162067 | 13.088767 | 11.063733 | 14.786533 | 7.5789333 | 11.864133 | 8.1242333 | 7.8736667 | 9.1505333 | 9.2722 | 11.852033 | 15.026333 | 14.017267 | 11.7556 | 18.5142 | 9.8033 | 14.845033 | 11.0714 | 6.7333 | 12.677067 | 12.565667 | 14.5732 | 8.4046667 | 14.000033 | 10.202767 | 8.1797667 | 10.260267 | 15.499 | 10.863067 | 13.397467 | 15.9004 | 7.8457 | 13.4624 | 9.6559667 | 14.318567 | 11.373367 | 12.897267 | 11.0812 | 11.347867 | 10.573 | 12.090033 | 13.8577 | 10.321667 | 7.2153667 | 11.362633 | 13.639633 | 16.9781 | 20.5364 | 9.0707333 | 5.8747667 | 8.8898667 | 8.1569 | 8.8791667 | 11.828433 | 13.694533 | 12.193133 | 5.3674333 | 10.779867 |
| AN_G3 | 11.258367 | 12.0614 | 10.244533 | 13.7933 | 6.9007 | 10.9379 | 7.1492667 | 6.8803333 | 8.1633 | 8.0562333 | 11.228633 | 14.2723 | 13.213233 | 10.756767 | 18.0089 | 8.8273333 | 13.6522 | 9.9274667 | 5.8455 | 11.863867 | 11.4955 | 13.374367 | 7.5735333 | 12.835 | 9.2956 | 7.1236 | 9.3748333 | 14.793133 | 9.8510333 | 12.795533 | 14.754033 | 6.5841 | 12.4915 | 8.5865333 | 13.1071 | 10.877167 | 11.774867 | 10.2062 | 10.6308 | 9.5782 | 10.732467 | 12.790233 | 9.1161667 | 6.2067 | 10.3691 | 12.659467 | 15.5735333 | 18.6643 | 8.0274 | 4.9283333 | 7.9401 | 7.2259 | 7.6284 | 10.666567 | 12.703933 | 11.201633 | 4.6071333 | 9.2124333 |
| AN_G4 | 12.2048 | 13.192433 | 11.144867 | 14.728133 | 7.4118333 | 11.937533 | 8.0448333 | 7.6165333 | 8.9448 | 8.8109667 | 11.933733 | 15.3149 | 14.253833 | 11.6509 | 19.310667 | 9.6116667 | 14.2204 | 10.6792 | 6.6986667 | 12.349733 | 12.4837 | 14.4528 | 8.4475 | 13.447267 | 9.6330667 | 7.7644667 | 9.9861667 | 15.8969 | 10.781233 | 13.828633 | 15.524833 | 7.3348333 | 13.862633 | 9.3511 | 14.118133 | 12.327633 | 12.865533 | 11.0035 | 12.076967 | 10.392233 | 11.424733 | 14.035933 | 10.330433 | 7.2529333 | 11.509267 | 14.2528 | 16.3472333 | 20.272833 | 8.6962667 | 5.4715333 | 8.8478667 | 8.1894 | 8.4596333 | 11.369167 | 13.459867 | 12.051033 | 5.4052333 | 10.652167 |
| AN_G5 | 12.2657 | 13.598067 | 11.529067 | 14.604233 | 8.0345 | 12.5001 | 8.5523333 | 8.2469333 | 9.2451 | 9.1388667 | 11.969233 | 15.624267 | 14.279667 | 12.3493 | 18.556767 | 10.083867 | 14.204767 | 11.0658 | 6.8025667 | 12.2157 | 12.712467 | 14.673933 | 9.1596 | 13.964267 | 10.258433 | 7.8097333 | 10.6567 | 16.883267 | 11.334067 | 14.3792 | 15.887267 | 7.8253 | 13.319067 | 9.8361 | 14.946633 | 12.5171 | 12.757433 | 11.4603 | 12.9053 | 11.2644 | 11.747667 | 14.833767 | 10.836233 | 7.7477333 | 11.929867 | 15.356733 | 17.5731 | 20.494233 | 9.3629667 | 5.9451333 | 9.6369333 | 8.6565333 | 8.6009667 | 11.641567 | 15.138867 | 12.8902 | 5.6949667 | 11.455267 |
| Mean | 11.8462 | 12.831353 | 10.8708 | 14.330293 | 7.33424 | 11.617413 | 7.78572 | 7.5306933 | 8.79386 | 8.7127667 | 11.769487 | 14.930933 | 13.740327 | 11.441307 | 18.520567 | 9.4248 | 13.890013 | 10.583167 | 6.34778 | 12.11276 | 12.12238 | 14.042273 | 8.248 | 13.379207 | 9.57684 | 7.6782 | 9.9738067 | 15.025987 | 10.557553 | 13.287587 | 15.177887 | 7.2796933 | 13.234967 | 9.23426 | 13.927627 | 11.347127 | 12.44226 | 10.74144 | 11.369267 | 10.303773 | 11.4414 | 13.79446 | 9.8967867 | 6.92938 | 11.144487 | 13.3793 | 16.2916733 | 19.946033 | 8.6360333 | 5.4750733 | 8.5705 | 7.8756867 | 8.28296 | 11.262 | 13.505207 | 11.871 | 5.1907733 | 10.263227 |
| SD | 0.5015139 | 0.6625433 | 0.5447159 | 0.5194105 | 0.5222895 | 0.7066014 | 0.6527199 | 0.5713337 | 0.4634109 | 0.5286081 | 0.3062299 | 0.577777 | 0.6231806 | 0.7060938 | 0.4955906 | 0.5836531 | 0.8707514 | 0.5195087 | 0.5489683 | 0.4681335 | 0.6437727 | 0.7271362 | 0.6523552 | 0.6247068 | 0.7263663 | 0.3908457 | 0.5132773 | 1.8232569 | 0.633352 | 0.9086058 | 0.8889963 | 0.5759628 | 0.5108372 | 0.5513536 | 0.7919992 | 1.1688468 | 0.5494722 | 0.6332989 | 1.1840491 | 0.6857312 | 0.5171961 | 0.752728 | 0.8508096 | 0.6847937 | 0.6614856 | 1.6566632 | 1.04112755 | 0.7798102 | 0.6057049 | 0.4420723 | 0.8336994 | 0.6588812 | 0.5259833 | 0.5100154 | 1.0370739 | 0.7663071 | 0.438517 | 1.0048835 |
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| SEM | 0.2242839 | 0.2962984 | 0.2436044 | 0.2322874 | 0.233575 | 0.3160018 | 0.2919052 | 0.2555082 | 0.2072437 | 0.2364007 | 0.1369502 | 0.2583897 | 0.2786949 | 0.3157747 | 0.2216348 | 0.2610176 | 0.3894119 | 0.2323313 | 0.2455061 | 0.2093557 | 0.2879039 | 0.3251852 | 0.2917421 | 0.2793774 | 0.3248409 | 0.1747915 | 0.2295446 | 0.8153853 | 0.2832436 | 0.4063409 | 0.3975713 | 0.2575784 | 0.2284533 | 0.2465728 | 0.3541928 | 0.5227242 | 0.2457314 | 0.2832199 | 0.5295228 | 0.3066683 | 0.2312971 | 0.3366302 | 0.3804936 | 0.306249 | 0.2958254 | 0.7408823 | 0.4656064 | 0.3487417 | 0.2708795 | 0.1977007 | 0.3728417 | 0.2946606 | 0.2352269 | 0.2280858 | 0.4637935 | 0.342703 | 0.1961108 | 0.4493976 |
| AE_G1 | 11.9896 | 12.9426 | 10.796333 | 14.8048 | 6.6187 | 11.694933 | 7.0299 | 8.0402333 | 9.0651 | 8.3194667 | 11.1695 | 13.727967 | 13.354933 | 11.599033 | 15.319133 | 9.4110667 | 11.728833 | 12.4612 | 4.4984 | 11.8527 | 13.669333 | 13.459633 | 9.1156333 | 19.113267 | 8.8897333 | 9.7416667 | 9.5018333 | 12.288233 | 10.797833 | 12.1481 | 13.287033 | 8.6962333 | 11.908167 | 9.5903333 | 12.565267 | 12.0053 | 12.655167 | 10.747833 | 12.447733 | 9.6479333 | 14.3682 | 18.3739 | 8.6503667 | 6.5448667 | 10.995867 | 11.107533 | 11.3877 | 20.551433 | 8.8584 | 6.2179 | 7.5306667 | 7.2485333 | 11.0152 | 12.180367 | 11.400067 | 13.052933 | 5.2466 | 11.225233 |
| AE_G2 | 11.079033 | 11.938333 | 10.084867 | 13.831767 | 6.0221667 | 11.0711 | 6.3990333 | 7.4522333 | 8.1575 | 7.4113 | 10.152467 | 12.979133 | 12.949167 | 10.721167 | 14.159367 | 8.5902667 | 10.940833 | 11.5908 | 3.7367 | 11.077633 | 13.008367 | 12.8108 | 8.2455333 | 17.692133 | 8.2778667 | 8.7997333 | 8.6964 | 12.082367 | 9.9956333 | 11.3796 | 12.433 | 7.9723 | 10.835233 | 8.6379333 | 11.7518 | 10.6804 | 12.2856 | 9.9703 | 11.605467 | 8.9658 | 13.3903 | 16.857867 | 8.3298667 | 5.8122667 | 10.2442 | 10.374467 | 11.2126 | 19.3108 | 8.1303333 | 5.3026 | 7.0818333 | 6.4491667 | 9.7763 | 11.3124 | 12.041367 | 12.189467 | 4.6530667 | 10.314267 |
| AE_G3 | 11.495 | 12.354233 | 10.3557 | 14.387733 | 5.9564 | 11.172233 | 6.5798333 | 7.8843667 | 8.8964 | 7.9738 | 11.1064 | 13.2162 | 12.595533 | 11.032733 | 13.805033 | 8.9012667 | 11.1699 | 12.420733 | 4.2388667 | 11.377367 | 13.1563 | 12.810267 | 8.6413333 | 19.941967 | 8.4963333 | 9.8204667 | 9.1460667 | 12.505067 | 10.219067 | 11.785833 | 12.798233 | 8.4824 | 11.6773 | 9.3969 | 11.8334 | 11.1459 | 11.646767 | 10.286933 | 12.0825 | 9.3529333 | 14.312333 | 19.534 | 7.7451667 | 5.9411 | 10.4665 | 10.419067 | 10.9179 | 19.8303 | 8.6951667 | 5.8778 | 6.9361333 | 6.6995333 | 11.280533 | 12.162833 | 12.892667 | 12.494133 | 4.7721 | 10.6009 |
| AE_G4 | 11.851133 | 12.760933 | 10.6809 | 15.4119 | 6.6416333 | 11.668433 | 7.1106 | 8.2684333 | 9.2771333 | 8.4904667 | 11.443467 | 13.6594 | 13.2646 | 11.2117 | 15.1631 | 9.3853 | 11.765533 | 12.510767 | 4.5943 | 11.5183 | 13.6342 | 13.324967 | 8.9342667 | 18.314733 | 8.9143333 | 9.8460333 | 9.5150333 | 13.561667 | 10.725867 | 12.3613 | 13.770933 | 8.9401 | 12.663167 | 9.7421333 | 12.174967 | 12.201567 | 12.073167 | 10.750667 | 13.309967 | 9.853 | 14.092367 | 18.287267 | 8.8086 | 6.4244 | 11.249333 | 11.971467 | 11.7382667 | 20.155567 | 8.9456 | 6.0528667 | 7.894 | 7.2716333 | 11.156867 | 12.750933 | 12.6712 | 13.223433 | 4.9471667 | 11.246 |
| AE_G5 | 11.3918 | 12.742633 | 11.303 | 14.9016 | 6.3007667 | 11.5315 | 7.2375667 | 8.2883667 | 9.1991667 | 8.7099333 | 11.2252 | 13.522833 | 12.077367 | 11.541667 | 14.527133 | 9.4835333 | 11.2858 | 12.709733 | 4.8201 | 10.861667 | 12.765067 | 13.320033 | 9.0215667 | 20.440533 | 8.8374333 | 10.2157 | 9.4014667 | 12.693467 | 10.8599 | 12.768167 | 13.3508 | 8.8381 | 11.557067 | 9.7083 | 11.980533 | 11.076067 | 11.6938 | 10.668767 | 11.997367 | 9.8931333 | 14.591933 | 20.244533 | 8.2175667 | 6.4292333 | 11.198033 | 10.870233 | 11.0552333 | 19.686 | 8.8745333 | 6.2750667 | 7.4566 | 7.2193 | 11.499167 | 12.4472 | 13.018733 | 12.846567 | 5.2481333 | 11.189667 |
| Mean | 11.561313 | 12.547747 | 10.64416 | 14.66756 | 6.3079333 | 11.42764 | 6.8713867 | 7.9867267 | 8.91906 | 8.1809933 | 11.019407 | 13.421107 | 12.84832 | 11.22126 | 14.594753 | 9.1542867 | 11.37818 | 12.338647 | 4.3776733 | 11.337533 | 13.246653 | 13.14514 | 8.7916667 | 19.100527 | 8.68314 | 9.68472 | 9.25216 | 12.62616 | 10.51966 | 12.0886 | 13.128 | 8.5858267 | 11.728187 | 9.41512 | 12.061193 | 11.421847 | 12.0709 | 10.4849 | 12.288607 | 9.54256 | 14.151027 | 18.659513 | 8.3503133 | 6.2303733 | 10.830787 | 10.948553 | 11.26234 | 19.90682 | 8.7008067 | 5.9452467 | 7.3798467 | 6.9776333 | 10.945613 | 12.170747 | 12.404807 | 12.761307 | 4.9734133 | 10.915213 |
| SD | 0.3650861 | 0.4025452 | 0.4622215 | 0.5925591 | 0.3214114 | 0.2883433 | 0.362132 | 0.3425525 | 0.4495807 | 0.507465 | 0.5009637 | 0.3156827 | 0.5240196 | 0.3643831 | 0.6452782 | 0.3904626 | 0.3592308 | 0.4325398 | 0.4145351 | 0.3852457 | 0.3955126 | 0.3105458 | 0.3532014 | 1.1296951 | 0.2824362 | 0.5274546 | 0.3441862 | 0.5710237 | 0.3875 | 0.5324625 | 0.5196311 | 0.3835535 | 0.6587521 | 0.4549569 | 0.324482 | 0.650737 | 0.4211776 | 0.3455346 | 0.6446857 | 0.3869718 | 0.4609037 | 1.2983729 | 0.4136124 | 0.3296172 | 0.4511351 | 0.649556 | 0.31867988 | 0.4709718 | 0.3318177 | 0.391231 | 0.3803567 | 0.3790917 | 0.6773767 | 0.5361803 | 0.6759197 | 0.4196007 | 0.2710812 | 0.4303448 |
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| SEM | 0.1632715 | 0.1800237 | 0.2067117 | 0.2650005 | 0.1437396 | 0.128951 | 0.1619503 | 0.1531942 | 0.2010586 | 0.2269453 | 0.2240378 | 0.1411776 | 0.2343487 | 0.1629571 | 0.2885772 | 0.1746202 | 0.1606529 | 0.1934377 | 0.1853857 | 0.1722871 | 0.1768786 | 0.1388803 | 0.1579565 | 0.505215 | 0.1263093 | 0.2358848 | 0.1539248 | 0.2553695 | 0.1732953 | 0.2381244 | 0.2323861 | 0.1715303 | 0.2946029 | 0.2034629 | 0.1451128 | 0.2910184 | 0.1883563 | 0.1545278 | 0.2883122 | 0.173059 | 0.2061224 | 0.58065 | 0.1849731 | 0.1474093 | 0.2017538 | 0.2904903 | 0.14251798 | 0.210625 | 0.1483934 | 0.1749638 | 0.1701007 | 0.169535 | 0.3029321 | 0.2397871 | 0.3022805 | 0.1876511 | 0.1212312 | 0.192456 |
| AEN_G1 | 12.989867 | 14.1618 | 12.184833 | 17.6474 | 7.4669667 | 11.753067 | 8.8193667 | 9.6138333 | 12.113067 | 11.462633 | 15.256433 | 15.387667 | 12.024633 | 12.056533 | 15.098233 | 11.022067 | 13.147833 | 15.239667 | 7.4937333 | 12.9409 | 13.691533 | 14.836933 | 8.9574 | 15.339967 | 9.1646333 | 12.000767 | 10.156967 | 12.880767 | 12.5596 | 13.579733 | 15.381533 | 9.9716333 | 17.0373 | 10.8421 | 13.6881 | 12.508533 | 14.108 | 11.089033 | 12.095533 | 10.476367 | 16.342567 | 23.7607 | 8.2019667 | 7.3051667 | 11.688767 | 11.243533 | 11.6517667 | 21.277733 | 9.5414333 | 7.1863 | 7.7601667 | 8.1919333 | 12.7568 | 14.626333 | 13.9658 | 12.286533 | 7.1744667 | 11.1961 |
| AEN_G2 | 11.9364 | 12.7655 | 10.840733 | 16.006467 | 5.9798 | 10.910967 | 7.0704333 | 7.8112 | 10.1082 | 9.3573667 | 12.907467 | 14.199867 | 11.4423 | 10.827067 | 13.1792 | 9.4247333 | 11.5698 | 13.245767 | 5.4605667 | 11.894667 | 12.9256 | 13.596567 | 7.6389333 | 14.4656 | 8.2069667 | 9.9540667 | 8.7218667 | 12.232833 | 10.820367 | 11.8826 | 13.737467 | 8.2468667 | 13.912 | 9.0464 | 12.5236 | 11.860567 | 13.587567 | 10.044133 | 11.117867 | 8.9000667 | 14.368 | 20.754733 | 7.4005667 | 5.9837667 | 10.329733 | 10.165133 | 10.6250333 | 19.781367 | 8.1109667 | 5.6159333 | 6.7194667 | 6.7516 | 11.301267 | 12.8117 | 13.1425 | 11.214333 | 5.5886667 | 10.455933 |
| AEN_G3 | 11.8241 | 13.245767 | 10.7493 | 15.4111 | 5.5614667 | 11.000233 | 6.5519 | 7.5416333 | 9.9750667 | 8.805 | 12.980233 | 14.016633 | 11.172 | 10.903 | 12.676733 | 9.0898667 | 11.187267 | 13.039433 | 4.6716 | 12.0489 | 12.8023 | 12.6417 | 7.6545 | 15.143367 | 8.0288 | 9.8229 | 8.6932 | 12.153333 | 10.3221 | 11.5785 | 13.520833 | 7.8905333 | 14.6998 | 9.0371 | 12.369133 | 11.7463 | 12.946467 | 9.9424 | 11.354567 | 8.7419333 | 14.516833 | 21.6864 | 6.7447 | 5.6467 | 10.065033 | 9.7771 | 9.80996667 | 19.8751 | 8.1184 | 5.5747 | 6.2166 | 6.4746667 | 11.5793 | 12.772067 | 13.051367 | 11.056333 | 5.2675667 | 10.1847 |
| AEN_G4 | 12.133333 | 13.095667 | 11.068933 | 16.5516 | 6.1255333 | 11.335633 | 7.6346 | 8.1608667 | 10.5775 | 9.5750667 | 13.394033 | 14.339233 | 11.475233 | 11.0686 | 13.345567 | 9.5602 | 11.742 | 13.182267 | 5.7083667 | 11.759333 | 13.136767 | 13.752433 | 7.9951333 | 14.6934 | 8.373 | 10.320067 | 8.9708 | 12.822233 | 11.246 | 12.504133 | 14.264967 | 8.4399333 | 14.921333 | 9.3453667 | 12.757633 | 11.911733 | 13.276 | 10.334333 | 12.027433 | 9.1154667 | 14.188333 | 22.7563 | 7.2922 | 6.2458 | 10.847967 | 10.973833 | 10.4874667 | 20.702333 | 8.331 | 5.7574667 | 7.2205333 | 6.9475 | 11.975433 | 13.4258 | 12.812467 | 11.249033 | 5.7717667 | 10.5385 |
| AEN_G5 | 12.274367 | 13.7451 | 11.957833 | 16.813 | 6.4232667 | 11.9489 | 8.4101667 | 8.7572667 | 11.111833 | 10.229933 | 14.505467 | 15.235933 | 11.480167 | 12.086667 | 13.6528 | 10.112767 | 12.027333 | 13.918467 | 6.0122 | 12.471833 | 13.037233 | 13.885767 | 8.7718 | 14.638267 | 9.1423667 | 11.0585 | 9.8768 | 13.582867 | 11.936867 | 13.313533 | 14.753567 | 8.5327333 | 15.0294 | 9.9743 | 14.047133 | 11.975367 | 13.243433 | 10.976333 | 12.007167 | 9.872 | 15.1784 | 22.827867 | 7.6742333 | 6.6136333 | 11.395667 | 10.742933 | 10.7366667 | 21.229 | 9.0657 | 6.3557 | 7.4862 | 7.5163333 | 12.695333 | 13.716667 | 14.371833 | 11.7717 | 6.5975333 | 11.083267 |
| Mean | 12.231613 | 13.402767 | 11.360327 | 16.485913 | 6.3114067 | 11.38976 | 7.6972933 | 8.37696 | 10.777133 | 9.886 | 13.808727 | 14.635867 | 11.518867 | 11.388373 | 13.590507 | 9.8419267 | 11.934847 | 13.72512 | 5.8692933 | 12.223127 | 13.118687 | 13.74268 | 8.2035533 | 14.85612 | 8.5831533 | 10.63126 | 9.2839267 | 12.734407 | 11.376987 | 12.5717 | 14.331673 | 8.61634 | 15.119967 | 9.6490533 | 13.07712 | 12.0005 | 13.432293 | 10.477247 | 11.720513 | 9.4211667 | 14.918827 | 22.3572 | 7.4627333 | 6.3590133 | 10.865433 | 10.580507 | 10.66218 | 20.573107 | 8.6335 | 6.09802 | 7.0805933 | 7.1764067 | 12.061627 | 13.470513 | 13.468793 | 11.515587 | 6.08 | 10.6917 |
| SD | 0.4581629 | 0.551904 | 0.664279 | 0.8434223 | 0.7167206 | 0.455119 | 0.932208 | 0.8275023 | 0.8699681 | 1.0184248 | 1.031012 | 0.6298453 | 0.3103951 | 0.6298728 | 0.9139571 | 0.7558272 | 0.7431714 | 0.9118484 | 1.0352116 | 0.4822859 | 0.3436924 | 0.7828578 | 0.6234748 | 0.3684637 | 0.5347492 | 0.9037368 | 0.6849481 | 0.5784119 | 0.8876668 | 0.8706776 | 0.7574947 | 0.796592 | 1.1575349 | 0.7679598 | 0.7456413 | 0.2961453 | 0.4406849 | 0.5285458 | 0.4511402 | 0.7319586 | 0.8793525 | 1.1585003 | 0.5337875 | 0.6366033 | 0.6868306 | 0.599727 | 0.66011405 | 0.7171924 | 0.6405106 | 0.684288 | 0.6170123 | 0.6841313 | 0.6525078 | 0.7616386 | 0.6659531 | 0.508102 | 0.7845639 | 0.4312334 |
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| SEM | 0.2048967 | 0.246819 | 0.2970746 | 0.3771899 | 0.3205272 | 0.2035354 | 0.4168961 | 0.3700703 | 0.3890616 | 0.4554534 | 0.4610826 | 0.2816754 | 0.1388129 | 0.2816877 | 0.4087341 | 0.3380162 | 0.3323564 | 0.407791 | 0.4629607 | 0.2156848 | 0.1537039 | 0.3501047 | 0.2788264 | 0.164782 | 0.2391471 | 0.4041634 | 0.3063181 | 0.2586737 | 0.3969766 | 0.3893789 | 0.3387619 | 0.3562468 | 0.5176653 | 0.3434421 | 0.3334609 | 0.1324402 | 0.1970803 | 0.2363729 | 0.201756 | 0.3273418 | 0.3932584 | 0.5180971 | 0.238717 | 0.2846977 | 0.30716 | 0.2682061 | 0.29521198 | 0.3207382 | 0.286445 | 0.3060229 | 0.2759363 | 0.3059528 | 0.2918104 | 0.3406151 | 0.2978233 | 0.2272301 | 0.3508677 | 0.1928534 |
| △CT= CT gene of interest - Ct house keeping gene (ave of 3) | Ave HARS2-SRBD1-SRP68 | ADCY1 | ALDOC | APPL2 | AQP4 | ATP1B1 | CCND1 | CCND2 | CDK1 | CLCN5 | CLDN10 | DAPP1 | eGFP | EMC7 | EPHB1 | EPHB2 | FABP7 | FAM43A | FZD1 | FZD10 | GJA1 | GLI1 | GLUL | GPC4 | GSN | HES5 | HEY1 | HEY2 | KALRN | KCNJ10 | MKI67 | NFIA | NRP2 | OLIG2 | PDLIM1 | PLD2 | S100B | S1PR1 | SEMA3C | SHH | SLC1A2 | SLC1A3 | SLC1A4 | SLC6A11 | SLC7A11 | SMIM12 | SOX9 | SPARC | SPARCL1 | SYT11 | THBS1 | TLN2 | TNF | TNFRSF19 | VIM | ZBTB20 | ||
| A_G1 | 11.6338 | 1.2782 | −3.145967 | 0.0478 | −3.229 | −3.8089 | −4.181967 | −3.7346 | −0.833367 | 1.9732333 | 3.7148 | 0.1735667 | 8.2688 | −2.186233 | 3.6474667 | −1.741933 | −5.743133 | −0.4595 | 1.4872 | 3.817 | −2.222333 | 2.0610667 | −1.4986 | −4.469533 | −1.641367 | 3.0420333 | −0.579633 | 2.4843 | 3.7023333 | −4.018933 | 0.069 | −2.3427 | 1.6981667 | −0.069733 | 0.315 | −0.6233 | 1.3123333 | −0.623833 | −0.723133 | −0.4263 | −0.701033 | −4.1394 | −0.016067 | 4.1565 | 4.88143333 | 7.7203667 | −2.935133 | −6.186967 | −2.0982 | −3.437433 | −4.102567 | −0.967933 | −0.9559 | 1.9281333 | −6.8807 | −1.124333 | ||
| A_G2 | 11.441433 | 1.5341667 | −3.324867 | 0.2038333 | −3.188933 | −3.826333 | −3.869967 | −3.4379 | −0.981033 | 2.2899333 | 3.2788667 | −0.0533 | 7.5246 | −2.236333 | 3.3878667 | −1.684567 | −5.715667 | −0.4084 | 1.3032 | 3.9521667 | −2.525267 | 2.1103 | −1.4671 | −4.5339 | −1.600267 | 4.2251333 | −0.682733 | 2.5021 | 3.9660667 | −3.923567 | 0.2776667 | −2.3872 | 2.235 | −0.0366 | 0.1926333 | −0.574967 | 1.1158333 | −0.668433 | −0.6379 | −0.371967 | −0.570633 | −4.319067 | −0.1804 | 3.842 | 5.32556667 | 8.1395 | −2.869567 | −6.136033 | −2.083733 | −3.710633 | −3.980533 | −0.794833 | −0.4498 | 1.9454333 | −6.9462 | −0.864267 | ||
| A_G3 | 11.550667 | 1.6694667 | −3.439467 | 0.0627667 | −3.3482 | −3.9414 | −4.0534 | −3.709667 | −0.734867 | 2.2298 | 3.2316 | −0.106033 | 7.1697667 | −2.4144 | 3.4010333 | −1.771767 | −5.871733 | −0.342167 | 1.3713333 | 3.9764333 | −2.560067 | 1.9713 | −1.621533 | −4.560167 | −1.8549 | 5.7286333 | −0.792967 | 2.5243 | 3.9608667 | −4.241333 | 0.4670667 | −2.523167 | 1.5327333 | −0.2236 | 0.3179 | −0.7248 | 0.8451 | −0.8686 | −0.875 | −0.1348 | −1.076267 | −4.396667 | −0.2852 | 3.5290667 | 4.85846667 | 8.4567 | −3.057333 | −6.3652 | −2.277133 | −3.7523 | −4.073667 | −0.928467 | 0.1527 | 1.8246667 | −6.948033 | −1.055133 | ||
| A_G4 | 11.491356 | 1.8033111 | −3.478156 | 0.1121111 | −3.441422 | −3.971422 | −4.114256 | −3.567722 | −1.021356 | 2.3998111 | 3.6736444 | −0.019989 | 8.4246778 | −2.427456 | 3.4665111 | −1.825189 | −5.465122 | −0.609622 | 1.3200444 | 4.3724444 | −2.498056 | 1.8544111 | −1.602722 | −4.734356 | −1.926089 | 7.5159111 | −0.792356 | 2.9431444 | 4.0086778 | −4.146022 | 0.6578778 | −2.333622 | 1.8610778 | 0.5577778 | 0.2215444 | −0.661922 | 1.6225778 | −0.726756 | −0.936889 | −0.117356 | −0.445822 | −4.153256 | −0.140722 | 4.2752778 | 4.95651111 | 8.1941778 | −3.093922 | −6.346222 | −1.995689 | −3.597256 | −4.248122 | −0.863289 | 0.4692778 | 2.0314111 | −6.916422 | −0.681889 | ||
| A_G5 | 12.0900 | 1.4137 | −3.7492 | 0.1182 | −3.3468 | −3.6161 | −3.7917 | −3.4741 | −0.7859 | 2.5337 | 2.8048 | 0.2727 | 6.8157 | −2.2812 | 2.6462 | −1.6215 | −5.5908 | −0.2838 | 1.0022 | 3.5116 | −2.2713 | 2.1162 | −1.5750 | −4.5244 | −1.4325 | 4.2637 | −0.7621 | 2.7975 | 3.6863 | −3.9411 | 0.3449 | −2.0566 | 1.9795 | −0.1166 | 0.2445 | −0.5289 | 1.8138 | −0.1973 | −0.5146 | 1.3457 | −1.1548 | −4.2371 | −0.3141 | 4.3503 | 5.5687 | 9.0128 | −2.6617 | −6.1984 | −2.1553 | −3.6034 | −4.0359 | −0.8854 | 0.9806 | 2.1092 | −7.1279 | −0.8667 | ||
| Mean | 11.641447 | 1.5397667 | -3.4275267 | 0.1089333 | -3.31088 | -3.8328333 | -4.0022667 | -3.5848067 | -0.8713 | 2.2852867 | 3.3407467 | 0.0533867 | 7.6407133 | -2.3091333 | 3.3098067 | -1.729 | -5.6772933 | -0.4207067 | 1.2968 | 3.92592 | -2.4154133 | 2.0226467 | -1.553 | -4.56448 | -1.6910333 | 4.9550733 | -0.7219667 | 2.6502667 | 3.86484 | -4.0542 | 0.3633067 | -2.3286667 | 1.8612933 | 0.0222533 | 0.25832 | -0.6227867 | 1.34192 | -0.61698 | -0.7375 | 0.0590467 | -0.7897067 | -4.2491 | -0.1873067 | 4.0306267 | 5.11814 | 8.3047 | -2.9235333 | -6.2465667 | -2.12202 | -3.6202 | -4.08816 | -0.8879933 | 0.03938 | 1.96776 | -6.96386 | -0.9184667 | ||
| SD | 0.260774 | 0.206532 | 0.2213484 | 0.0611756 | 0.1016155 | 0.1401581 | 0.1652809 | 0.1343016 | 0.1244066 | 0.2093326 | 0.3720531 | 0.1618004 | 0.6937176 | 0.1075445 | 0.3851685 | 0.0786918 | 0.1550081 | 0.124745 | 0.1796975 | 0.3107193 | 0.1564112 | 0.110502 | 0.0670695 | 0.100535 | 0.1997657 | 1.7195114 | 0.0913891 | 0.2078567 | 0.1568902 | 0.1365106 | 0.2189902 | 0.1699073 | 0.2684161 | 0.3075832 | 0.0561693 | 0.075881 | 0.3873688 | 0.2520951 | 0.1721068 | 0.7323576 | 0.3120521 | 0.1095847 | 0.119595 | 0.3409636 | 0.31449735 | 0.4757621 | 0.1721429 | 0.1025839 | 0.1038791 | 0.1223128 | 0.1003979 | 0.0658105 | 0.7605047 | 0.107922 | 0.0957236 | 0.175103 | ||
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||
| SEM | 0.1166217 | 0.0923639 | 0.09899 | 0.0273585 | 0.0454438 | 0.0626806 | 0.0739159 | 0.0600615 | 0.0556363 | 0.0936164 | 0.1663872 | 0.0723593 | 0.3102399 | 0.0480954 | 0.1722526 | 0.035192 | 0.0693217 | 0.0557877 | 0.0803631 | 0.1389579 | 0.0699492 | 0.049418 | 0.0299944 | 0.0449606 | 0.0893379 | 0.7689889 | 0.0408705 | 0.0929564 | 0.0701634 | 0.0610494 | 0.0979354 | 0.0759849 | 0.1200393 | 0.1375554 | 0.0251197 | 0.033935 | 0.1732366 | 0.1127403 | 0.0769685 | 0.3275203 | 0.139554 | 0.0490078 | 0.0534845 | 0.1524835 | 0.14064749 | 0.2127673 | 0.0769846 | 0.0458769 | 0.0464561 | 0.0546999 | 0.0448993 | 0.0294313 | 0.3401081 | 0.0482642 | 0.0428089 | 0.0783084 | ||
| AN_G1 | 11.309322 | 2.4299444 | −4.564089 | −0.461922 | −4.251389 | −4.273322 | −2.843756 | −3.023756 | 0.5544778 | 3.1075444 | 1.6283111 | −0.615356 | 6.9029778 | −2.511489 | 1.2183444 | −1.137356 | −5.650456 | 0.1481111 | 0.0452444 | 1.8277444 | −3.654622 | 1.3401444 | −2.814989 | −3.795889 | −1.718256 | 0.7483111 | −1.350956 | 0.7277778 | 2.5135778 | −4.500789 | 1.7299111 | −2.567722 | 1.8383778 | −1.668956 | 0.6068778 | −1.353322 | −1.423922 | −1.598289 | −0.097222 | 2.1453444 | −2.429889 | −5.085156 | −0.757756 | −0.321456 | 3.67707778 | 8.4530778 | −3.286522 | −6.153722 | −3.771589 | −4.159622 | −3.462689 | −0.505056 | 1.2195111 | −0.290322 | −6.430222 | −2.092922 | ||
| AN_G2 | 12.104856 | 2.6816778 | −4.525922 | −0.240722 | −3.980622 | −4.231189 | −2.954322 | −2.832656 | −0.252822 | 2.9214778 | 1.9124111 | −0.349256 | 6.4093444 | −2.301556 | 2.7401778 | −1.033456 | −5.371556 | 0.5722111 | 0.4608111 | 2.4683444 | −3.700189 | 1.8951778 | −1.902089 | −3.925089 | −1.844589 | 3.3941444 | −1.241789 | 1.2926111 | 3.7955444 | −4.259156 | 1.3575444 | −2.448889 | 2.2137111 | −0.731489 | 0.7924111 | −1.023656 | −0.756989 | −1.531856 | −0.014822 | 1.7528444 | −1.783189 | −4.889489 | −0.742222 | 1.5347778 | 4.87324444 | 8.4315444 | −3.034122 | −6.230089 | −3.214989 | −3.947956 | −3.225689 | −0.276422 | 1.5896778 | 0.0882778 | −6.737422 | −1.324989 | ||
| AN_G3 | 11.1881 | 2.6052 | −4.2874 | −0.2502 | −4.038833 | −4.307767 | −3.0248 | −3.131867 | 0.0405333 | 3.0842 | 2.0251333 | −0.431333 | 6.8208 | −2.360767 | 2.4641 | −1.260633 | −5.3426 | 0.6757667 | 0.3074 | 2.1862667 | −3.614567 | 1.6469 | −1.8925 | −4.0645 | −1.813267 | 3.6050333 | −1.337067 | 1.6074333 | 3.5659333 | −4.604 | 1.3034 | −2.601567 | 1.919 | −0.310933 | 0.5867667 | −0.9819 | −0.5573 | −1.6099 | −0.455633 | 1.6021333 | −2.071933 | −4.9814 | −0.819 | 1.4713667 | 4.38543333 | 7.4762 | −3.1607 | −6.259767 | −3.248 | −3.9622 | −3.5597 | −0.521533 | 1.5158333 | 0.0135333 | −6.580967 | −1.975667 | ||
| AN_G4 | 12.1807 | 2.5474333 | −4.768867 | −0.243167 | −4.135867 | −4.564167 | −3.2359 | −3.369733 | −0.246967 | 3.1342 | 2.0731333 | −0.5298 | 7.1299667 | −2.569033 | 2.0397 | −1.5015 | −5.482033 | 0.1690333 | 0.303 | 2.2721 | −3.7332 | 1.2665667 | −2.547633 | −4.416233 | −2.194533 | 3.7162 | −1.399467 | 1.6479333 | 3.3441333 | −4.845867 | 1.6819333 | −2.8296 | 1.9374333 | 0.1469333 | 0.6848333 | −1.1772 | −0.103733 | −1.788467 | −0.755967 | 1.8552333 | −1.850267 | −4.927767 | −0.671433 | 2.0721 | 4.16653333 | 8.0921333 | −3.484433 | −6.709167 | −3.332833 | −3.9913 | −3.721067 | −0.811533 | 1.2791667 | −0.129667 | −6.775467 | −1.528533 | ||
| AN_G5 | 12.464278 | 2.1399556 | −4.429778 | 0.0358222 | −3.911944 | −4.217344 | −3.219178 | −3.325411 | −0.495044 | 3.1599889 | 1.8153889 | −0.114978 | 6.0924889 | −2.380411 | 1.7404889 | −1.398478 | −5.661711 | −0.248578 | 0.2481889 | 2.2096556 | −3.304678 | 1.4999889 | −2.205844 | −4.654544 | −1.807578 | 4.4189889 | −1.130211 | 1.9149222 | 3.4229889 | −4.638978 | 0.8547889 | −2.628178 | 2.4823556 | 0.0528222 | 0.2931556 | −1.003978 | 0.4410222 | −1.199878 | −0.716611 | 2.3694889 | −1.628044 | −4.716544 | −0.534411 | 2.8924556 | 5.10882222 | 8.0299556 | −3.101311 | −6.519144 | −2.827344 | −3.807744 | −3.863311 | −0.822711 | 2.6745889 | 0.4259222 | −6.769311 | −1.009011 | ||
| Mean | 11.849451 | 2.4808422 | -4.5152111 | -0.2320378 | -4.0637311 | -4.3187578 | -3.0555911 | -3.1366844 | -0.0799644 | 3.0814822 | 1.8908756 | -0.4081444 | 6.6711156 | -2.4246511 | 2.0405622 | -1.2662844 | -5.5016711 | 0.2633089 | 0.2729289 | 2.1928222 | -3.6014511 | 1.5297556 | -2.2726111 | -4.1712511 | -1.8756444 | 3.1765356 | -1.2918978 | 1.4381356 | 3.3284356 | -4.5697578 | 1.3855156 | -2.6151911 | 2.0781756 | -0.5023244 | 0.5928089 | -1.1080111 | -0.4801844 | -1.5456778 | -0.4080511 | 1.9450089 | -1.9526644 | -4.9200711 | -0.7049644 | 1.5298489 | 4.44222222 | 8.0965822 | -3.2134178 | -6.3743778 | -3.2789511 | -3.9737644 | -3.5664911 | -0.5874511 | 1.6557556 | 0.0215489 | -6.6586778 | -1.5862244 | ||
| SD | 0.5661479 | 0.2115588 | 0.1775117 | 0.176865 | 0.1332152 | 0.1417425 | 0.1698158 | 0.2208728 | 0.4022119 | 0.093849 | 0.1777253 | 0.1921515 | 0.4153441 | 0.1113305 | 0.5989434 | 0.189516 | 0.1503071 | 0.3707183 | 0.1498704 | 0.2322744 | 0.171887 | 0.2516759 | 0.4049814 | 0.355836 | 0.1843569 | 1.4108655 | 0.1069493 | 0.4543644 | 0.4867689 | 0.2141439 | 0.3520184 | 0.1380536 | 0.2666363 | 0.738224 | 0.1859678 | 0.1571683 | 0.7006329 | 0.215411 | 0.342696 | 0.3093817 | 0.3108672 | 0.1354914 | 0.1088484 | 1.1812605 | 0.56894883 | 0.3964284 | 0.1776849 | 0.2322094 | 0.3371176 | 0.1257111 | 0.2444192 | 0.2309942 | 0.5903588 | 0.2684048 | 0.150259 | 0.4508756 | ||
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||
| SEM | 0.253189 | 0.094612 | 0.0793857 | 0.0790965 | 0.0595756 | 0.0633892 | 0.075944 | 0.0987773 | 0.1798746 | 0.0419705 | 0.0794812 | 0.0859328 | 0.1857475 | 0.0497885 | 0.2678557 | 0.0847542 | 0.0672194 | 0.1657903 | 0.0670241 | 0.1038763 | 0.0768702 | 0.1125529 | 0.1811132 | 0.1591347 | 0.0824469 | 0.6309582 | 0.0478292 | 0.2031979 | 0.2176897 | 0.0957681 | 0.1574274 | 0.0617394 | 0.1192434 | 0.3301438 | 0.0831673 | 0.0702878 | 0.3133325 | 0.0963347 | 0.1532583 | 0.1383597 | 0.139024 | 0.0605936 | 0.0486785 | 0.5282758 | 0.25444165 | 0.1772882 | 0.0794631 | 0.1038472 | 0.1507636 | 0.0562197 | 0.1093076 | 0.1033037 | 0.2640165 | 0.1200343 | 0.0671979 | 0.2016377 | ||
| AE_G1 | 11.909511 | 2.8952889 | −5.290811 | −0.214578 | −4.879611 | −3.869278 | −2.844411 | −3.590044 | −0.740011 | 1.8184556 | 1.4454222 | −0.310478 | 3.4096222 | −2.498444 | −0.180678 | 0.5516889 | −7.411111 | −0.056811 | 1.7598222 | 1.5501222 | −2.793878 | 7.2037556 | −3.019778 | −2.167844 | −2.407678 | 0.3787222 | −1.111678 | 0.2385889 | 1.3775222 | −3.213278 | −0.001344 | −2.319178 | 0.6557556 | 0.0957889 | 0.7456556 | −1.161678 | 0.5382222 | −2.261578 | 2.4586889 | 6.4643889 | −3.259144 | −5.364644 | −0.913644 | −0.801978 | −0.5218111 | 8.6419222 | −3.051111 | −5.691611 | −4.378844 | −4.660978 | −0.894311 | 0.2708556 | −0.509444 | 1.1434222 | −6.662911 | −0.684278 | ||
| AE_G2 | 11.034078 | 2.7976889 | −5.011911 | 0.0370222 | −4.635044 | −3.581844 | −2.876578 | −3.622778 | −0.881611 | 1.9450556 | 1.9150889 | −0.312911 | 3.1252889 | −2.443811 | −0.093244 | 0.5567222 | −7.297378 | 0.0435556 | 1.9742889 | 1.7767222 | −2.788544 | 6.6580556 | −2.756211 | −2.234344 | −2.337678 | 1.0482889 | −1.038444 | 0.3455222 | 1.3989222 | −3.061778 | −0.198844 | −2.396144 | 0.7177222 | −0.353678 | 1.2515222 | −1.063778 | 0.5713889 | −2.068278 | 2.3562222 | 5.8237889 | −2.704211 | −5.221811 | −0.789878 | −0.659611 | 0.17852222 | 8.2767222 | −2.903744 | −5.731478 | −3.952244 | −4.584911 | −1.257778 | 0.2783222 | 1.0072889 | 1.1553889 | −6.381011 | −0.719811 | ||
| AE_G3 | 11.401644 | 2.9860889 | −5.445244 | −0.229411 | −4.821811 | −3.517278 | −2.505244 | −3.427844 | −0.295244 | 1.8145556 | 1.1938889 | −0.368911 | 2.4033889 | −2.500378 | −0.231744 | 1.0190889 | −7.162778 | −0.024278 | 1.7546556 | 1.4086222 | −2.760311 | 8.5403222 | −2.905311 | −1.581178 | −2.255578 | 1.1034222 | −1.182578 | 0.3841889 | 1.3965889 | −2.919244 | 0.2756556 | −2.004744 | 0.4317556 | −0.255744 | 0.2451222 | −1.114711 | 0.6808556 | −2.048711 | 2.9106889 | 8.1323556 | −3.656478 | −5.460544 | −0.935144 | −0.982578 | −0.4837444 | 8.4286556 | −2.706478 | −5.523844 | −4.465511 | −4.702111 | −0.121111 | 0.7611889 | 1.4910222 | 1.0924889 | −6.629544 | −0.800744 | ||
| AE_G4 | 11.764322 | 3.6475778 | −5.122689 | −0.095889 | −4.653722 | −3.495889 | −2.487189 | −3.273856 | −0.320856 | 1.8950778 | 1.5002778 | −0.552622 | 3.3987778 | −2.379022 | 0.0012111 | 0.7464444 | −7.170022 | −0.246022 | 1.8698778 | 1.5606444 | −2.830056 | 6.5504111 | −2.849989 | −1.918289 | −2.249289 | 1.7973444 | −1.038456 | 0.5969778 | 2.0066111 | −2.824222 | 0.8988444 | −2.022189 | 0.4106444 | 0.4372444 | 0.3088444 | −1.013656 | 1.5456444 | −1.911322 | 2.3280444 | 6.5229444 | −2.955722 | −5.339922 | −0.514989 | 0.2071444 | −0.0260556 | 8.3912444 | −2.818722 | −5.711456 | −3.870322 | −4.492689 | −0.607456 | 0.9866111 | 0.9068778 | 1.4591111 | −6.817156 | −0.518322 | ||
| AE_G5 | 11.812478 | 3.0891222 | −5.511711 | −0.280978 | −4.574911 | −3.524111 | −2.613311 | −3.102544 | −0.587278 | 1.7103556 | 0.2648889 | −0.270811 | 2.7146556 | −2.328944 | −0.526678 | 0.8972556 | −6.992378 | −0.950811 | 0.9525889 | 1.5075556 | −2.790911 | 8.6280556 | −2.975044 | −1.596778 | −2.411011 | 0.8809889 | −0.952578 | 0.9556889 | 1.5383222 | −2.974378 | −0.255411 | −2.104178 | 0.1680556 | −0.736411 | −0.118678 | −1.143711 | 0.1848889 | −1.919344 | 2.7794556 | 8.4320556 | −3.594911 | −5.383244 | −0.614444 | −0.942244 | −0.7572444 | 7.8735222 | −2.937944 | −5.537411 | −4.355878 | −4.593178 | −0.313311 | 0.6347222 | 1.2062556 | 1.0340889 | −6.564344 | −0.622811 | ||
| Mean | 11.584407 | 3.0831533 | -5.2764733 | -0.1567667 | -4.71302 | -3.59768 | -2.6653467 | -3.4034133 | -0.565 | 1.8367 | 1.2639133 | -0.3631467 | 3.0103467 | -2.43012 | -0.2062267 | 0.75424 | -7.2067333 | -0.2468733 | 1.6622467 | 1.5607333 | -2.79274 | 7.51612 | -2.9012667 | -1.8996867 | -2.3322467 | 1.0417533 | -1.0647467 | 0.5041933 | 1.5435933 | -2.99858 | 0.14378 | -2.1692867 | 0.4767867 | -0.16256 | 0.4864933 | -1.0995067 | 0.7042 | -2.0418467 | 2.56662 | 7.0751067 | -3.2340933 | -5.3540333 | -0.75362 | -0.6358533 | -0.3220667 | 8.3224133 | -2.8836 | -5.63916 | -4.20456 | -4.6067733 | -0.6387933 | 0.58634 | 0.8204 | 1.1769 | -6.6109933 | -0.6691933 | ||
| SD | 0.3627475 | 0.3334723 | 0.2107539 | 0.1277771 | 0.1306317 | 0.1551245 | 0.1849076 | 0.2184354 | 0.2567793 | 0.0893519 | 0.6155884 | 0.1115322 | 0.4414019 | 0.0752318 | 0.1998676 | 0.2065789 | 0.1574919 | 0.4079064 | 0.4068647 | 0.134842 | 0.0248311 | 1.006458 | 0.1038929 | 0.3071857 | 0.0785546 | 0.5099557 | 0.086677 | 0.2839809 | 0.2667081 | 0.1477918 | 0.4704571 | 0.1781031 | 0.2189733 | 0.4474997 | 0.5264232 | 0.0606165 | 0.5058273 | 0.1423648 | 0.2629244 | 1.1404759 | 0.4085007 | 0.0865915 | 0.1845184 | 0.4880831 | 0.38523663 | 0.2835939 | 0.1294223 | 0.1001884 | 0.2723718 | 0.0801646 | 0.4536923 | 0.3112617 | 0.7761299 | 0.1648839 | 0.1585957 | 0.1060764 | ||
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||
| SEM | 0.1622256 | 0.1491333 | 0.094252 | 0.0571437 | 0.0584203 | 0.0693738 | 0.0826932 | 0.0976873 | 0.1148352 | 0.0399594 | 0.2752995 | 0.0498787 | 0.1974009 | 0.0336447 | 0.0893835 | 0.0923849 | 0.0704325 | 0.1824213 | 0.1819554 | 0.0603032 | 0.0111048 | 0.4501017 | 0.0464623 | 0.1373776 | 0.0351307 | 0.2280591 | 0.0387631 | 0.1270001 | 0.1192755 | 0.0660945 | 0.2103948 | 0.0796501 | 0.0979279 | 0.200128 | 0.2354236 | 0.0271085 | 0.2262129 | 0.0636675 | 0.1175834 | 0.5100363 | 0.1826871 | 0.0387249 | 0.0825192 | 0.2182774 | 0.17228306 | 0.126827 | 0.0578794 | 0.0448056 | 0.1218084 | 0.0358507 | 0.2028974 | 0.1392005 | 0.3470959 | 0.0737383 | 0.0709262 | 0.0474388 | ||
| AEN_G1 | 13.112167 | 4.5352333 | −5.6452 | −1.3591 | −4.2928 | −3.498333 | −0.9991 | −1.649533 | 2.1442667 | 2.2755 | −1.087533 | −1.055633 | 1.9860667 | −2.0901 | 0.0356667 | 2.1275 | −5.618433 | −0.171267 | 0.5793667 | 1.7247667 | −4.154767 | 2.2278 | −3.947533 | −1.1114 | −2.9552 | −0.2314 | −0.552567 | 0.4675667 | 2.2693667 | −3.140533 | 3.9251333 | −2.270067 | 0.5759333 | −0.603633 | 0.9958333 | −2.023133 | −1.016633 | −2.6358 | 3.2304 | 10.648533 | −4.9102 | −5.807 | −1.4234 | −1.868633 | −1.4604 | 8.1655667 | −3.570733 | −5.925867 | −5.352 | −4.920233 | −0.355367 | 1.5141667 | 0.8536333 | −0.825633 | −5.9377 | −1.916067 | ||
| AEN_G2 | 11.847544 | 4.1589222 | −5.867744 | −0.936578 | −4.777111 | −4.036344 | −1.739344 | −2.490178 | 1.0599222 | 2.3523222 | −0.405244 | −1.020478 | 1.3316556 | −2.422811 | −0.277744 | 1.3982222 | −6.386978 | 0.0471222 | 1.0780556 | 1.7490222 | −4.208611 | 2.6180556 | −3.640578 | −1.893478 | −3.125678 | 0.3852889 | −1.027178 | 0.0350556 | 1.8899222 | −3.600678 | 2.0644556 | −2.801144 | 0.6760556 | 0.0130222 | 1.7400222 | −1.803411 | −0.729678 | −2.947478 | 2.5204556 | 8.9071889 | −4.446978 | −5.863778 | −1.517811 | −1.682411 | −1.2225111 | 7.9338222 | −3.736578 | −6.231611 | −5.128078 | −5.095944 | −0.546278 | 0.9641556 | 1.2949556 | −0.633211 | −6.258878 | −1.391611 | ||
| AEN_G3 | 11.939722 | 3.4713778 | −6.378256 | −0.939489 | −5.387822 | −4.398089 | −1.964656 | −3.134722 | 1.0405111 | 2.0769111 | −0.767722 | −1.036722 | 0.7370111 | −2.849856 | −0.752456 | 1.0997111 | −7.268122 | 0.1091778 | 0.8625778 | 0.7019778 | −4.285222 | 3.2036444 | −3.910922 | −2.116822 | −3.246522 | 0.2136111 | −1.617622 | −0.361222 | 1.5811111 | −4.049189 | 2.7600778 | −2.902622 | 0.4294111 | −0.193422 | 1.0067444 | −1.997322 | −0.585156 | −3.197789 | 2.5771111 | 9.7466778 | −5.195022 | −6.293022 | −1.874689 | −2.162622 | −2.1297556 | 7.9353778 | −3.821322 | −6.365022 | −5.723122 | −5.465056 | −0.360422 | 0.8323444 | 1.1116444 | −0.883389 | −6.672156 | −1.755022 | ||
| AEN_G4 | 12.099311 | 4.4522889 | −5.973778 | −0.763678 | −4.464711 | −3.938444 | −1.521811 | −2.524244 | 1.2947222 | 2.2399222 | −0.624078 | −1.030711 | 1.2462556 | −2.539111 | −0.357311 | 1.0829556 | −6.390944 | −0.339978 | 1.0374556 | 1.6531222 | −4.104178 | 2.5940889 | −3.726311 | −1.779244 | −3.128511 | 0.7229222 | −0.853311 | 0.4048222 | 2.1656556 | −3.659378 | 2.8220222 | −2.753944 | 0.6583222 | −0.187578 | 1.1766889 | −1.764978 | −0.071878 | −2.983844 | 2.0890222 | 10.656989 | −4.807111 | −5.853511 | −1.251344 | −1.125478 | −1.6118444 | 8.6030222 | −3.768311 | −6.341844 | −4.878778 | −5.151811 | −0.123878 | 1.3264889 | 0.7131556 | −0.850278 | −6.327544 | −1.560811 | ||
| AEN_G5 | 12.6591 | 4.1539 | −6.235833 | −0.7102 | −4.248933 | −3.901833 | −1.547267 | −2.429167 | 1.8463667 | 2.5768333 | −1.178933 | −0.572433 | 0.9937 | −2.546333 | −0.631767 | 1.2593667 | −6.6469 | −0.187267 | 0.3781333 | 1.2266667 | −3.8873 | 1.9791667 | −3.516733 | −1.6006 | −2.7823 | 0.9237667 | −0.722233 | 0.6544333 | 2.0944667 | −4.126367 | 2.3703 | −2.6848 | 1.3880333 | −0.683733 | 0.5843333 | −1.682767 | −0.651933 | −2.7871 | 2.5193 | 10.168767 | −4.984867 | −6.045467 | −1.263433 | −1.916167 | −1.9224333 | 8.5699 | −3.5934 | −6.3034 | −5.1729 | −5.142767 | 0.0362333 | 1.0575667 | 1.7127333 | −0.8874 | −6.061567 | −1.575833 | ||
| Mean | 12.331569 | 4.1543444 | -6.0201622 | -0.9418089 | -4.6342756 | -3.9546089 | -1.5544356 | -2.4455689 | 1.4771578 | 2.3042978 | -0.8127022 | -0.9431956 | 1.2589378 | -2.4896422 | -0.3967222 | 1.3935511 | -6.4622756 | -0.1084422 | 0.7871178 | 1.4111111 | -4.1280156 | 2.5245511 | -3.7484156 | -1.7003089 | -3.0476422 | 0.4028378 | -0.9545822 | 0.2401311 | 2.0001044 | -3.7152289 | 2.7883978 | -2.6825156 | 0.7455511 | -0.3310689 | 1.1007244 | -1.8543222 | -0.6110556 | -2.9104022 | 2.5872578 | 10.025631 | -4.8688356 | -5.9725556 | -1.4661356 | -1.7510622 | -1.6693889 | 8.2415378 | -3.6980689 | -6.2335489 | -5.2509756 | -5.1551622 | -0.2699422 | 1.1389444 | 1.1372244 | -0.8159822 | -6.2515689 | -1.6398689 | ||
| SD | 0.5381395 | 0.4184232 | 0.2918579 | 0.2547244 | 0.4695958 | 0.321943 | 0.3577657 | 0.5284538 | 0.4948176 | 0.1825241 | 0.3214349 | 0.2076569 | 0.468221 | 0.2736399 | 0.3100247 | 0.4299767 | 0.5929382 | 0.1839346 | 0.3014286 | 0.4494505 | 0.1503641 | 0.4637488 | 0.1815562 | 0.3787426 | 0.1810626 | 0.4506088 | 0.4094968 | 0.4044064 | 0.2722632 | 0.3959419 | 0.705868 | 0.2437935 | 0.3721346 | 0.2985794 | 0.4186187 | 0.149124 | 0.3433726 | 0.2121139 | 0.4095895 | 0.7307249 | 0.2753842 | 0.2008904 | 0.2542813 | 0.3894011 | 0.3614749 | 0.3288948 | 0.1104289 | 0.1792794 | 0.3133924 | 0.1967848 | 0.2274443 | 0.2771018 | 0.3927475 | 0.1052601 | 0.2816618 | 0.2009528 | ||
| N | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||
| SEM | 0.2406633 | 0.1871245 | 0.1305228 | 0.1139162 | 0.2100096 | 0.1439773 | 0.1599977 | 0.2363317 | 0.2212892 | 0.0816273 | 0.1437501 | 0.092867 | 0.2093948 | 0.1223755 | 0.1386472 | 0.1922914 | 0.26517 | 0.082258 | 0.134803 | 0.2010004 | 0.0672449 | 0.2073948 | 0.0811944 | 0.1693789 | 0.0809736 | 0.2015184 | 0.1831325 | 0.1808561 | 0.1217598 | 0.1770706 | 0.3156738 | 0.1090278 | 0.1664236 | 0.1335288 | 0.187212 | 0.0666903 | 0.1535609 | 0.0948602 | 0.183174 | 0.3267901 | 0.1231556 | 0.0898409 | 0.1137181 | 0.1741455 | 0.16165649 | 0.1470862 | 0.0493853 | 0.0801762 | 0.1401534 | 0.0880049 | 0.1017162 | 0.1239237 | 0.175642 | 0.0470738 | 0.125963 | 0.0898688 | ||
| △△CT= △CT co-culture - △CT monoculture | ADCY1 | ALDOC | APPL2 | AQP4 | ATP1B1 | CCND1 | CCND2 | CDK1 | CLCN5 | CLDN10 | DAPP1 | eGFP | EMC7 | EPHB1 | EPHB2 | FABP7 | FAM43A | FZD1 | FZD10 | GJA1 | GLI1 | GLUL | GPC4 | GSN | HES5 | HEY1 | HEY2 | KALRN | KCNJ10 | MKI67 | NFIA | NRP2 | OLIG2 | PDLIM1 | PLD2 | S100B | S1PR1 | SEMA3C | SHH | SLC1A2 | SLC1A3 | SLC1A4 | SLC6A11 | SLC7A11 | SMIM12 | SOX9 | SPARC | SPARCL1 | SYT11 | THBS1 | TLN2 | TNF | TNFRSF19 | VIM | ZBTB20 | |||
| A_G1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| AN_G1 | 1.1517444 | −1.418122 | −0.509722 | −1.022389 | −0.464422 | 1.3382111 | 0.7108444 | 1.3878444 | 1.1343111 | −2.086489 | −0.788922 | −1.365822 | −0.325256 | −2.429122 | 0.6045778 | 0.0926778 | 0.6076111 | −1.441956 | −1.989256 | −1.432289 | −0.720922 | −1.316389 | 0.6736444 | −0.076889 | −2.293722 | −0.771322 | −1.756522 | −1.188756 | −0.481856 | 1.6609111 | −0.225022 | 0.1402111 | −1.599222 | 0.2918778 | −0.730022 | −2.736256 | −0.974456 | 0.6259111 | 2.5716444 | −1.728856 | −0.945756 | −0.741689 | −4.477956 | −1.2043556 | 0.7327111 | −0.351389 | 0.0332444 | −1.673389 | −0.722189 | 0.6398778 | 0.4628778 | 2.1754111 | −2.218456 | 0.4504778 | −0.968589 | |||
| AE_G1 | 1.6170889 | −2.144844 | −0.262378 | −1.650611 | −0.060378 | 1.3375556 | 0.1445556 | 0.0933556 | −0.154778 | −2.269378 | −0.484044 | −4.859178 | −0.312211 | −3.828144 | 2.2936222 | −1.667978 | 0.4026889 | 0.2726222 | −2.266878 | −0.571544 | 5.1426889 | −1.521178 | 2.3016889 | −0.766311 | −2.663311 | −0.532044 | −2.245711 | −2.324811 | 0.8056556 | −0.070344 | 0.0235222 | −1.042411 | 0.1655222 | 0.4306556 | −0.538378 | −0.774111 | −1.637744 | 3.1818222 | 6.8906889 | −2.558111 | −1.225244 | −0.897578 | −4.958478 | −5.4032444 | 0.9215556 | −0.115978 | 0.4953556 | −2.280644 | −1.223544 | 3.2082556 | 1.2387889 | 0.4464556 | −0.784711 | 0.2177889 | 0.4400556 | |||
| AEN_G1 | 3.2570333 | −2.499233 | −1.4069 | −1.0638 | 0.3105667 | 3.1828667 | 2.0850667 | 2.9776333 | 0.3022667 | −4.802333 | −1.2292 | −6.282733 | 0.0961333 | −3.6118 | 3.8694333 | 0.1247 | 0.2882333 | −0.907833 | −2.092233 | −1.932433 | 0.1667333 | −2.448933 | 3.3581333 | −1.313833 | −3.273433 | 0.0270667 | −2.016733 | −1.432967 | 0.8784 | 3.8561333 | 0.0726333 | −1.122233 | −0.5339 | 0.6808333 | −1.399833 | −2.328967 | −2.011967 | 3.9535333 | 11.074833 | −4.209167 | −1.6676 | −1.407333 | −6.025133 | −6.3418333 | 0.4452 | −0.6356 | 0.2611 | −3.2538 | −1.4828 | 3.7472 | 2.4821 | 1.8095333 | −2.753767 | 0.943 | −0.791733 | |||
| A_G2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| AN_G2 | 1.1475111 | −1.201056 | −0.444556 | −0.791689 | −0.404856 | 0.9156444 | 0.6052444 | 0.7282111 | 0.6315444 | −1.366456 | −0.295956 | −1.115256 | −0.065222 | −0.647689 | 0.6511111 | 0.3441111 | 0.9806111 | −0.842389 | −1.483822 | −1.174922 | −0.215122 | −0.434989 | 0.6088111 | −0.244322 | −0.830989 | −0.559056 | −1.209489 | −0.170522 | −0.335589 | 1.0798778 | −0.061689 | −0.021289 | −0.694889 | 0.5997778 | −0.448689 | −1.872822 | −0.863422 | 0.6230778 | 2.1248111 | −1.212556 | −0.570422 | −0.561822 | −2.307222 | −0.4523222 | 0.2920444 | −0.164556 | −0.094056 | −1.131256 | −0.237322 | 0.7548444 | 0.5184111 | 2.0394778 | −1.857156 | 0.2087778 | −0.460722 | |||
| AE_G2 | 1.2635222 | −1.687044 | −0.166811 | −1.446111 | 0.2444889 | 0.9933889 | −0.184878 | 0.0994222 | −0.344878 | −1.363778 | −0.259611 | −4.399311 | −0.207478 | −3.481111 | 2.2412889 | −1.581711 | 0.4519556 | 0.6710889 | −2.175444 | −0.263278 | 4.5477556 | −1.289111 | 2.2995556 | −0.737411 | −3.176844 | −0.355711 | −2.156578 | −2.567144 | 0.8617889 | −0.476511 | −0.008944 | −1.517278 | −0.317078 | 1.0588889 | −0.488811 | −0.544444 | −1.399844 | 2.9941222 | 6.1957556 | −2.133578 | −0.902744 | −0.609478 | −4.501611 | −5.1470444 | 0.1372222 | −0.034178 | 0.4045556 | −1.868511 | −0.874278 | 2.7227556 | 1.0731556 | 1.4570889 | −0.790044 | 0.5651889 | 0.1444556 | |||
| AEN_G2 | 2.6247556 | −2.542878 | −1.140411 | −1.588178 | −0.210011 | 2.1306222 | 0.9477222 | 2.0409556 | 0.0623889 | −3.684111 | −0.967178 | −6.192944 | −0.186478 | −3.665611 | 3.0827889 | −0.671311 | 0.4555222 | −0.225144 | −2.203144 | −1.683344 | 0.5077556 | −2.173478 | 2.6404222 | −1.525411 | −3.839844 | −0.344444 | −2.467044 | −2.076144 | 0.3228889 | 1.7867889 | −0.413944 | −1.558944 | 0.0496222 | 1.5473889 | −1.228444 | −1.845511 | −2.279044 | 3.1583556 | 9.2791556 | −3.876344 | −1.544711 | −1.337411 | −5.524411 | −6.5480778 | −0.205678 | −0.867011 | −0.095578 | −3.044344 | −1.385311 | 3.4342556 | 1.7589889 | 1.7447556 | −2.578644 | 0.6873222 | −0.527344 | |||
| A_G3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| AN_G3 | 0.9357333 | −0.847933 | −0.312967 | −0.690633 | −0.366367 | 1.0286 | 0.5778 | 0.7754 | 0.8544 | −1.206467 | −0.3253 | −0.348967 | 0.0536333 | −0.936933 | 0.5111333 | 0.5291333 | 1.0179333 | −1.063933 | −1.790167 | −1.0545 | −0.3244 | −0.270967 | 0.4956667 | 0.0416333 | −2.1236 | −0.5441 | −0.916867 | −0.394933 | −0.362667 | 0.8363333 | −0.0784 | 0.3862667 | −0.087333 | 0.2688667 | −0.2571 | −1.4024 | −0.7413 | 0.4193667 | 1.7369333 | −0.995667 | −0.584733 | −0.5338 | −2.0577 | −0.4730333 | −0.9805 | −0.103367 | 0.1054333 | −0.970867 | −0.2099 | 0.5139667 | 0.4069333 | 1.3631333 | −1.811133 | 0.3670667 | −0.920533 | |||
| AE_G3 | 1.3166222 | −2.005778 | −0.292178 | −1.473611 | 0.4241222 | 1.5481556 | 0.2818222 | 0.4396222 | −0.415244 | −2.037711 | −0.262878 | −4.766378 | −0.085978 | −3.632778 | 2.7908556 | −1.291044 | 0.3178889 | 0.3833222 | −2.567811 | −0.200244 | 6.5690222 | −1.283778 | 2.9789889 | −0.400678 | −4.625211 | −0.389611 | −2.140111 | −2.564278 | 1.3220889 | −0.191411 | 0.5184222 | −1.100978 | −0.032144 | −0.072778 | −0.389911 | −0.164244 | −1.180111 | 3.7856889 | 8.2671556 | −2.580211 | −1.063878 | −0.649944 | −4.511644 | −5.3422111 | −0.028044 | 0.3508556 | 0.8413556 | −2.188378 | −0.949811 | 3.9525556 | 1.6896556 | 1.3383222 | −0.732178 | 0.3184889 | 0.2543889 | |||
| AEN_G3 | 1.8019111 | −2.938789 | −1.002256 | −2.039622 | −0.456689 | 2.0887444 | 0.5749444 | 1.7753778 | −0.152889 | −3.999322 | −0.930689 | −6.432756 | −0.435456 | −4.153489 | 2.8714778 | −1.396389 | 0.4513444 | −0.508756 | −3.274456 | −1.725156 | 1.2323444 | −2.289389 | 2.4433444 | −1.391622 | −5.515022 | −0.824656 | −2.885522 | −2.379756 | 0.1921444 | 2.2930111 | −0.379456 | −1.103322 | 0.0301778 | 0.6888444 | −1.272522 | −1.430256 | −2.329189 | 3.4521111 | 9.8814778 | −4.118756 | −1.896356 | −1.589489 | −5.691689 | −6.9882222 | −0.521322 | −0.763989 | 0.0001778 | −3.445989 | −1.712756 | 3.7132444 | 1.7608111 | 0.9589444 | −2.708056 | 0.2758778 | −0.699889 | |||
| A_G4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| AN_G4 | 0.7441 | −1.2907 | −0.3553 | −0.6944 | −0.5927 | 0.8784 | 0.1980 | 0.7744 | 0.7344 | −1.6005 | −0.5098 | −1.2947 | −0.1416 | −1.4268 | 0.3237 | −0.0169 | 0.7787 | −1.0170 | −2.1003 | −1.2351 | −0.5878 | −0.9449 | 0.3181 | −0.2684 | −3.7997 | −0.6071 | −1.2952 | −0.6645 | −0.6998 | 1.0241 | −0.4960 | 0.0764 | −0.4108 | 0.4633 | −0.5153 | −1.7263 | −1.0617 | 0.1809 | 1.9726 | −1.4044 | −0.7745 | −0.5307 | −2.2032 | −0.7900 | −0.1020 | −0.3905 | −0.3629 | −1.3371 | −0.3940 | 0.5271 | 0.0518 | 0.8099 | −2.1611 | 0.1410 | −0.8466 | |||
| AE_G4 | 1.8442667 | −1.644533 | −0.208 | −1.2123 | 0.4755333 | 1.6270667 | 0.2938667 | 0.7005 | −0.504733 | −2.173367 | −0.532633 | −5.0259 | 0.0484333 | −3.4653 | 2.5716333 | −1.7049 | 0.3636 | 0.5498333 | −2.8118 | −0.332 | 4.696 | −1.247267 | 2.8160667 | −0.3232 | −5.718567 | −0.2461 | −2.346167 | −2.002067 | 1.3218 | 0.2409667 | 0.3114333 | −1.450433 | −0.120533 | 0.0873 | −0.351733 | −0.076933 | −1.184567 | 3.2649333 | 6.6403 | −2.5099 | −1.186667 | −0.374267 | −4.068133 | −4.9825667 | 0.1970667 | 0.2752 | 0.6347667 | −1.874633 | −0.895433 | 3.6406667 | 1.8499 | 0.4376 | −0.5723 | 0.0992667 | 0.1635667 | |||
| AEN_G4 | 2.6489778 | −2.495622 | −0.875789 | −1.023289 | 0.0329778 | 2.5924444 | 1.0434778 | 2.3160778 | −0.159889 | −4.297722 | −1.010722 | −7.178422 | −0.111656 | −3.823822 | 2.9081444 | −0.925822 | 0.2696444 | −0.282589 | −2.719322 | −1.606122 | 0.7396778 | −2.123589 | 2.9551111 | −1.202422 | −6.792989 | −0.060956 | −2.538322 | −1.843022 | 0.4866444 | 2.1641444 | −0.420322 | −1.202756 | −0.745356 | 0.9551444 | −1.103056 | −1.694456 | −2.257089 | 3.0259111 | 10.774344 | −4.361289 | −1.700256 | −1.110622 | −5.400756 | −6.5683556 | 0.4088444 | −0.674389 | 0.0043778 | −2.883089 | −1.554556 | 4.1242444 | 2.1897778 | 0.2438778 | −2.881689 | 0.5888778 | −0.878922 | |||
| A_G5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| AN_G5 | 0.7263 | −0.6806 | −0.0823 | −0.5651 | −0.6012 | 0.5726 | 0.1487 | 0.2908 | 0.6263 | −0.9894 | −0.3877 | −0.7232 | −0.0992 | −0.9057 | 0.2231 | −0.0709 | 0.0353 | −0.7540 | −1.3019 | −1.0333 | −0.6162 | −0.6308 | −0.1301 | −0.3750 | 0.1553 | −0.3681 | −0.8826 | −0.2633 | −0.6978 | 0.5099 | −0.5715 | 0.5029 | 0.1694 | 0.0486 | −0.4750 | −1.3727 | −1.0026 | −0.2020 | 1.0238 | −0.4733 | −0.4794 | −0.2203 | −1.4578 | −0.4599 | −0.9828 | −0.4396 | −0.3207 | −0.6720 | −0.2044 | 0.1726 | 0.0627 | 1.6940 | −1.6832 | 0.3586 | −0.1423 | |||
| AE_G5 | 1.6754 | −1.7625 | −0.3991 | −1.2281 | 0.0920 | 1.1784 | 0.3716 | 0.1986 | −0.8233 | −2.5399 | −0.5435 | −4.1011 | −0.0477 | −3.1728 | 2.5188 | −1.4016 | −0.6670 | −0.0496 | −2.0040 | −0.5196 | 6.5119 | −1.4000 | 2.9277 | −0.9785 | −3.3827 | −0.1904 | −1.8418 | −2.1479 | 0.9668 | −0.6003 | −0.0475 | −1.8114 | −0.6198 | −0.3632 | −0.6148 | −1.6289 | −1.7221 | 3.2940 | 7.0864 | −2.4401 | −1.1461 | −0.3003 | −5.2925 | −6.3260 | −1.1392 | −0.2762 | 0.6610 | −2.2005 | −0.9898 | 3.7226 | 1.5202 | 0.2256 | −1.0751 | 0.5636 | 0.2439 | |||
| AEN_G5 | 2.7402 | −2.4867 | −0.8284 | −0.9021 | −0.2857 | 2.2445 | 1.0450 | 2.6322 | 0.0432 | −3.9838 | −0.8451 | −5.8220 | −0.2651 | −3.2779 | 2.8809 | −1.0561 | 0.0966 | −0.6241 | −2.2849 | −1.6160 | −0.1370 | −1.9417 | 2.9238 | −1.3498 | −3.3399 | 0.0399 | −2.1431 | −1.5918 | −0.1852 | 2.0254 | −0.6282 | −0.5915 | −0.5672 | 0.3398 | −1.1538 | −2.4657 | −2.5898 | 3.0339 | 8.8231 | −3.8301 | −1.8084 | −0.9493 | −6.2665 | −7.4912 | −0.4429 | −0.9317 | −0.1050 | −3.0176 | −1.5394 | 4.0721 | 1.9430 | 0.7321 | −2.9966 | 1.0664 | −0.7091 | |||
| Fold Change | ADCY1 | ALDOC | APPL2 | AQP4 | ATP1B1 | CCND1 | CCND2 | CDK1 | CLCN5 | CLDN10 | DAPP1 | eGFP | EMC7 | EPHB1 | EPHB2 | FABP7 | FAM43A | FZD1 | FZD10 | GJA1 | GLI1 | GLUL | GPC4 | GSN | HES5 | HEY1 | HEY2 | KALRN | KCNJ10 | MKI67 | NFIA | NRP2 | OLIG2 | PDLIM1 | PLD2 | S100B | S1PR1 | SEMA3C | SHH | SLC1A2 | SLC1A3 | SLC1A4 | SLC6A11 | SLC7A11 | SMIM12 | SOX9 | SPARC | SPARCL1 | SYT11 | THBS1 | TLN2 | TNF | TNFRSF19 | VIM | ZBTB20 | |||
| A_G1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| AN_G1 | 0.4500807 | 2.6723745 | 1.423776 | 2.0312797 | 1.3797647 | 0.3955108 | 0.6109624 | 0.3821353 | 0.4555524 | 4.2471318 | 1.7277832 | 2.5772317 | 1.2528864 | 5.3856565 | 0.6576638 | 0.9377805 | 0.6562825 | 2.7168889 | 3.9703207 | 2.6987454 | 1.6482353 | 2.4904197 | 0.626921 | 1.0547411 | 4.9031953 | 1.7068334 | 3.3788264 | 2.2795603 | 1.3965387 | 0.3162394 | 1.1687953 | 0.9073864 | 3.0297993 | 0.8168382 | 1.6586646 | 6.6633864 | 1.9648995 | 0.6480104 | 0.1682124 | 3.3146477 | 1.9261974 | 1.6721322 | 22.284297 | 2.30434313 | 0.601772 | 1.2757882 | 0.9772202 | 3.1896296 | 1.6496831 | 0.6417673 | 0.7255376 | 0.2213788 | 4.6539495 | 0.7318005 | 1.9569256 | |||
| AE_G1 | 0.3259926 | 4.4224457 | 1.199454 | 3.139666 | 1.0427388 | 0.3956905 | 0.904658 | 0.9373401 | 1.1132501 | 4.8211515 | 1.3986592 | 29.024067 | 1.2416092 | 14.203203 | 0.2039628 | 3.1776887 | 0.7564471 | 0.8278136 | 4.8128044 | 1.4861136 | 0.0283072 | 2.8702527 | 0.2028255 | 1.7009151 | 6.3348529 | 1.4459768 | 4.7427082 | 5.0100018 | 0.5721021 | 1.0499673 | 0.9838278 | 2.059667 | 0.8916057 | 0.7419246 | 1.4523385 | 1.7101361 | 3.1117894 | 0.1101986 | 0.0084274 | 5.889361 | 2.3379506 | 1.8629356 | 31.092135 | 42.3193172 | 0.5279395 | 1.0837093 | 0.7093868 | 4.8589495 | 2.3351973 | 0.1081979 | 0.4237282 | 0.7338436 | 1.7227473 | 0.8598823 | 0.7371062 | |||
| AEN_G1 | 0.1046009 | 5.6538489 | 2.6516677 | 2.0904304 | 0.806325 | 0.1101188 | 0.2356852 | 0.126953 | 0.8109772 | 27.90271 | 2.3443695 | 77.855839 | 0.935537 | 12.225317 | 0.0684202 | 0.9171947 | 0.8189042 | 1.8762256 | 4.2640765 | 3.8169845 | 0.8908576 | 5.4601226 | 0.0975217 | 2.4860121 | 9.6694467 | 0.9814137 | 4.0466648 | 2.7000136 | 0.5439704 | 0.0690539 | 0.9509007 | 2.1768369 | 1.4478378 | 0.6238048 | 2.638711 | 5.0244534 | 4.0333166 | 0.0645458 | 0.0004636 | 18.496324 | 3.1768567 | 2.6524643 | 65.124719 | 81.1114305 | 0.7344825 | 1.5535837 | 0.8344514 | 9.5387485 | 2.7949065 | 0.0744698 | 0.1789837 | 0.2852832 | 6.7447579 | 0.5201501 | 1.7311531 | |||
| A_G2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| AN_G2 | 0.4514033 | 2.2990782 | 1.3608948 | 1.7310998 | 1.3239563 | 0.530107 | 0.65736 | 0.603652 | 0.645485 | 2.5783633 | 1.2276979 | 2.1663338 | 1.0462461 | 1.5666565 | 0.6367897 | 0.7877932 | 0.506765 | 1.7930167 | 2.7968875 | 2.2578071 | 1.1608023 | 1.3519004 | 0.6557369 | 1.1845361 | 1.7789043 | 1.4733044 | 2.3125569 | 1.1254658 | 1.2618924 | 0.4730689 | 1.0436868 | 1.0148657 | 1.6187598 | 0.6598556 | 1.3647994 | 3.6624834 | 1.8193489 | 0.6492843 | 0.229281 | 2.3174779 | 1.4849581 | 1.4761325 | 4.9492922 | 1.36824086 | 0.8167438 | 1.1208207 | 1.0673664 | 2.1904929 | 1.1788027 | 0.5926103 | 0.6981403 | 0.2432518 | 3.6229265 | 0.86527 | 1.3762306 | |||
| AE_G2 | 0.4165258 | 3.2199637 | 1.1225744 | 2.7247259 | 0.8441148 | 0.5022965 | 1.1367207 | 0.9334067 | 1.2700434 | 2.573582 | 1.197156 | 21.102048 | 1.1546677 | 11.166546 | 0.2114973 | 2.9932465 | 0.7310512 | 0.6280325 | 4.517249 | 1.2002024 | 0.0427552 | 2.4437744 | 0.2031257 | 1.6671814 | 9.0432694 | 1.2796162 | 4.4585598 | 5.9263525 | 0.5502698 | 1.3913748 | 1.0062191 | 2.8625041 | 1.2458046 | 0.4800016 | 1.403288 | 1.4584586 | 2.6387313 | 0.1255103 | 0.0136424 | 4.3880433 | 1.8696192 | 1.5257068 | 22.6527 | 35.4335583 | 0.9092682 | 1.0239731 | 0.755469 | 3.6515554 | 1.8330902 | 0.1514847 | 0.4752783 | 0.3642273 | 1.7291277 | 0.6758669 | 0.9047207 | |||
| AEN_G2 | 0.1621324 | 5.8275027 | 2.2044383 | 3.0066934 | 1.1566971 | 0.2283594 | 0.5184504 | 0.2430027 | 0.957677 | 12.853694 | 1.9550124 | 73.158036 | 1.137982 | 12.68992 | 0.1180288 | 1.5925196 | 0.7292462 | 1.1688943 | 4.604819 | 3.2117163 | 0.7033158 | 4.5110953 | 0.1603813 | 2.8786874 | 14.318857 | 1.269662 | 5.5290992 | 4.2167879 | 0.7994674 | 0.2898164 | 1.3323235 | 2.9463819 | 0.9661893 | 0.3421287 | 2.3431421 | 3.5938025 | 4.8535638 | 0.1120057 | 0.0016095 | 14.685744 | 2.9174564 | 2.5269745 | 46.027083 | 93.5767214 | 1.153228 | 1.8238804 | 1.0684932 | 8.2497159 | 2.6122828 | 0.0925094 | 0.2954552 | 0.2983845 | 5.9737814 | 0.6210054 | 1.4412738 | |||
| A_G3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| AN_G3 | 0.5227767 | 1.7999207 | 1.2422596 | 1.6139919 | 1.2891022 | 0.4901856 | 0.6699847 | 0.5842266 | 0.5530953 | 2.3077176 | 1.2529249 | 1.273648 | 0.9635067 | 1.9144544 | 0.701671 | 0.6929709 | 0.4938232 | 2.0906236 | 3.4585484 | 2.0769982 | 1.2521436 | 1.206616 | 0.7092339 | 0.9715544 | 4.3578 | 1.4581104 | 1.8880103 | 1.314882 | 1.2858004 | 0.5600652 | 1.0558464 | 0.765107 | 1.0624046 | 0.8299713 | 1.195074 | 2.6434096 | 1.6716815 | 0.7477528 | 0.3000067 | 1.9940017 | 1.4997617 | 1.4477375 | 4.1632206 | 1.38802479 | 1.9731491 | 1.0742775 | 0.9295257 | 1.9600177 | 1.156608 | 0.7002943 | 0.7542249 | 0.3887371 | 3.5091785 | 0.7753574 | 1.8928149 | |||
| AE_G3 | 0.4014738 | 4.0160515 | 1.2244873 | 2.7771616 | 0.7452921 | 0.341947 | 0.8225514 | 0.7373277 | 1.3335246 | 4.1059359 | 1.1998697 | 27.215899 | 1.0614069 | 12.40438 | 0.1445003 | 2.4470515 | 0.8022429 | 0.7666701 | 5.9290917 | 1.148893 | 0.0105324 | 2.434757 | 0.1268338 | 1.320128 | 24.678984 | 1.3100402 | 4.4079599 | 5.9145884 | 0.3999554 | 1.1418801 | 0.6981349 | 2.1450002 | 1.0225309 | 1.0517398 | 1.3103127 | 1.1205791 | 2.2659423 | 0.0725094 | 0.0032459 | 5.980272 | 2.0905431 | 1.5691078 | 22.810789 | 40.5663364 | 1.0196291 | 0.784119 | 0.5581189 | 4.5579269 | 1.9316197 | 0.0645895 | 0.3100009 | 0.3954803 | 1.6611447 | 0.8019094 | 0.8383422 | |||
| AEN_G3 | 0.2867944 | 7.6676734 | 2.0031293 | 4.1113786 | 1.3723884 | 0.2350852 | 0.6713121 | 0.2921178 | 1.1117935 | 15.992485 | 1.906186 | 86.387793 | 1.3523378 | 17.796096 | 0.1366467 | 2.6324185 | 0.731361 | 1.4228224 | 9.6763004 | 3.3061577 | 0.4256252 | 4.88849 | 0.1838569 | 2.6237354 | 45.728517 | 1.7711121 | 7.3897329 | 5.2044855 | 0.8753037 | 0.2040492 | 1.3008508 | 2.1484887 | 0.9792996 | 0.6203505 | 2.4158355 | 2.6949445 | 5.0252274 | 0.0913716 | 0.0010602 | 17.372766 | 3.722716 | 3.0094271 | 51.685543 | 126.959296 | 1.4352701 | 1.6981794 | 0.9998768 | 10.89798 | 3.277863 | 0.0762434 | 0.2950822 | 0.5144332 | 6.5344036 | 0.8259476 | 1.6243797 | |||
| A_G4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| AN_G4 | 0.597031 | 2.4464861 | 1.2792319 | 1.6182612 | 1.5081129 | 0.5439871 | 0.871765 | 0.5846362 | 0.6010726 | 3.0325073 | 1.4238638 | 2.4532787 | 1.1031109 | 2.688518 | 0.7990242 | 1.0117909 | 0.5829098 | 2.0237687 | 4.2881175 | 2.3540492 | 1.5029994 | 1.9250703 | 0.8021132 | 1.2045084 | 13.92602 | 1.523206 | 2.454129 | 1.5850677 | 1.6243296 | 0.4917321 | 1.4102762 | 0.9484505 | 1.3294638 | 0.7253308 | 1.4292693 | 3.3088069 | 2.0874058 | 0.8821389 | 0.2547954 | 2.6471583 | 1.7106103 | 1.4446411 | 4.6049254 | 1.72904783 | 1.0732934 | 1.3108577 | 1.286048 | 2.5265075 | 1.3140721 | 0.6939696 | 0.9647616 | 0.5704258 | 4.4724885 | 0.9069183 | 1.7983134 | |||
| AE_G4 | 0.2784969 | 3.1264671 | 1.1550858 | 2.3170674 | 0.7192009 | 0.3237458 | 0.8157129 | 0.6153589 | 1.4188611 | 4.5107479 | 1.4465672 | 32.579668 | 0.9669858 | 11.044835 | 0.1682136 | 3.2600634 | 0.7772227 | 0.683099 | 7.0216009 | 1.2587572 | 0.0385801 | 2.3739123 | 0.1419971 | 1.2511025 | 52.657483 | 1.1859967 | 5.0847141 | 4.0057341 | 0.4000355 | 0.8461781 | 0.8058407 | 2.7329013 | 1.0871367 | 0.9412827 | 1.2760929 | 1.0547736 | 2.2729511 | 0.1040296 | 0.0100247 | 5.695806 | 2.2762621 | 1.2961805 | 16.77375 | 31.6156432 | 0.8723224 | 0.8263358 | 0.644045 | 3.6670841 | 1.8601685 | 0.0801771 | 0.2774116 | 0.7383619 | 1.4868921 | 0.9335074 | 0.8928151 | |||
| AEN_G4 | 0.159433 | 5.6397149 | 1.8350112 | 2.0325472 | 0.9774008 | 0.1658046 | 0.4851565 | 0.2008127 | 1.1172011 | 19.667235 | 2.0149195 | 144.85064 | 1.0804674 | 14.160715 | 0.1332175 | 1.8997667 | 0.829524 | 1.2163757 | 6.5856335 | 3.0443247 | 0.5988731 | 4.3577665 | 0.1289505 | 2.3012572 | 110.89026 | 1.0431565 | 5.8091304 | 3.5876079 | 0.7136831 | 0.2231144 | 1.3382264 | 2.3017889 | 1.6763874 | 0.5157899 | 2.1480917 | 3.2365472 | 4.7802593 | 0.122775 | 0.000571 | 20.553168 | 3.2495852 | 2.1593876 | 42.246372 | 94.9012744 | 0.7532264 | 1.5959206 | 0.9969702 | 7.3772795 | 2.9374322 | 0.0573428 | 0.2191852 | 0.8444724 | 7.370124 | 0.6648599 | 1.8390009 | |||
| A_G5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| AN_G5 | 0.6044661 | 1.6028062 | 1.058729 | 1.4794901 | 1.5170129 | 0.6724194 | 0.9020421 | 0.8174298 | 0.6478208 | 1.985405 | 1.3082758 | 1.6508778 | 1.0711546 | 1.87341 | 0.8567424 | 1.0503717 | 0.9758514 | 1.6865012 | 2.4655337 | 2.0467478 | 1.532797 | 1.5484234 | 1.0943696 | 1.2968695 | 0.8979249 | 1.2906221 | 1.8436524 | 1.2001932 | 1.6220669 | 0.7022873 | 1.4861022 | 0.7057031 | 0.8892124 | 0.9668518 | 1.3899503 | 2.5896073 | 2.0036076 | 1.1503185 | 0.4918078 | 1.3882493 | 1.3941959 | 1.1649489 | 2.7469551 | 1.37544648 | 1.9762973 | 1.3562282 | 1.2489652 | 1.5932802 | 1.1521804 | 0.8872423 | 0.9574484 | 0.309076 | 3.2114689 | 0.779903 | 1.1036632 | |||
| AE_G5 | 0.3130721 | 3.3929339 | 1.3187155 | 2.3425286 | 0.9382212 | 0.441831 | 0.7729248 | 0.8713958 | 1.7694488 | 5.8156213 | 1.4575042 | 17.161059 | 1.0336158 | 9.0181614 | 0.174488 | 2.6418832 | 1.5877312 | 1.0350018 | 4.0111057 | 1.4335246 | 0.0109578 | 2.6390158 | 0.131427 | 1.9703701 | 10.429996 | 1.1411064 | 3.5845698 | 4.4319246 | 0.5116515 | 1.5160668 | 1.0334964 | 3.5099083 | 1.5366976 | 1.2862758 | 1.5313103 | 3.0926995 | 3.2990866 | 0.1019523 | 0.0073584 | 5.4269188 | 2.2131992 | 1.2314004 | 39.193251 | 80.2242597 | 2.2026394 | 1.2110289 | 0.6324398 | 4.5964923 | 1.9859097 | 0.0757505 | 0.3486456 | 0.8552195 | 2.1068194 | 0.6766117 | 0.8444594 | |||
| AEN_G5 | 0.1496629 | 5.6047715 | 1.7756602 | 1.8687698 | 1.2190204 | 0.2110303 | 0.4846524 | 0.161293 | 0.9705149 | 15.820854 | 1.7964169 | 56.572233 | 1.2017101 | 9.6995796 | 0.1357561 | 2.079287 | 0.9352489 | 1.5412372 | 4.8732657 | 3.0651455 | 1.0996077 | 3.841551 | 0.1317756 | 2.5486894 | 10.125273 | 0.9727149 | 4.4169655 | 3.0142287 | 1.1369921 | 0.2456408 | 1.5455877 | 1.5067662 | 1.4815995 | 0.7901448 | 2.225026 | 5.5239065 | 6.0202451 | 0.1220989 | 0.0022079 | 14.222359 | 3.5024284 | 1.9309207 | 76.982337 | 179.912991 | 1.3592921 | 1.9075077 | 1.0754861 | 8.0979434 | 2.9067135 | 0.0594514 | 0.2600731 | 0.6020223 | 7.9809227 | 0.4775164 | 1.6348091 | |||
| Mean Fold Change | ADCY1 | ALDOC | APPL2 | AQP4 | ATP1B1 | CCND1 | CCND2 | CDK1 | CLCN5 | CLDN10 | DAPP1 | eGFP | EMC7 | EPHB1 | EPHB2 | FABP7 | FAM43A | FZD1 | FZD10 | GJA1 | GLI1 | GLUL | GPC4 | GSN | HES5 | HEY1 | HEY2 | KALRN | KCNJ10 | MKI67 | NFIA | NRP2 | OLIG2 | PDLIM1 | PLD2 | S100B | S1PR1 | SEMA3C | SHH | SLC1A2 | SLC1A3 | SLC1A4 | SLC6A11 | SLC7A11 | SMIM12 | SOX9 | SPARC | SPARCL1 | SYT11 | THBS1 | TLN2 | TNF | TNFRSF19 | VIM | ZBTB20 | |||
| Mean (A) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| SD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| n | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |||
| SEM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Mean (AN) | 0.525 | 2.164 | 1.273 | 1.695 | 1.404 | 0.526 | 0.742 | 0.594 | 0.581 | 2.830 | 1.388 | 2.024 | 1.087 | 2.686 | 0.730 | 0.896 | 0.643 | 2.062 | 3.396 | 2.287 | 1.419 | 1.704 | 0.778 | 1.142 | 5.173 | 1.490 | 2.375 | 1.501 | 1.438 | 0.509 | 1.233 | 0.868 | 1.586 | 0.800 | 1.408 | 3.774 | 1.909 | 0.816 | 0.289 | 2.332 | 1.603 | 1.441 | 7.750 | 1.633 | 1.288 | 1.228 | 1.102 | 2.292 | 1.290 | 0.703 | 0.820 | 0.347 | 3.894 | 0.812 | 1.626 | |||
| SD | 0.075 | 0.448 | 0.139 | 0.208 | 0.105 | 0.100 | 0.134 | 0.154 | 0.080 | 0.880 | 0.204 | 0.551 | 0.106 | 1.565 | 0.094 | 0.152 | 0.197 | 0.401 | 0.767 | 0.263 | 0.204 | 0.515 | 0.189 | 0.129 | 5.176 | 0.149 | 0.620 | 0.469 | 0.176 | 0.140 | 0.204 | 0.129 | 0.853 | 0.116 | 0.166 | 1.678 | 0.165 | 0.210 | 0.123 | 0.719 | 0.215 | 0.181 | 8.168 | 0.405 | 0.649 | 0.123 | 0.160 | 0.606 | 0.212 | 0.112 | 0.130 | 0.141 | 0.632 | 0.072 | 0.369 | |||
| n | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |||
| SEM | 0.034 | 0.200 | 0.062 | 0.093 | 0.047 | 0.045 | 0.060 | 0.069 | 0.036 | 0.394 | 0.091 | 0.246 | 0.047 | 0.700 | 0.042 | 0.068 | 0.088 | 0.180 | 0.343 | 0.118 | 0.091 | 0.231 | 0.085 | 0.058 | 2.315 | 0.067 | 0.277 | 0.210 | 0.079 | 0.063 | 0.091 | 0.058 | 0.382 | 0.052 | 0.074 | 0.750 | 0.074 | 0.094 | 0.055 | 0.322 | 0.096 | 0.081 | 3.653 | 0.181 | 0.290 | 0.055 | 0.071 | 0.271 | 0.095 | 0.050 | 0.058 | 0.063 | 0.283 | 0.032 | 0.165 | |||
| Mean (AE) | 0.347 | 3.636 | 1.204 | 2.660 | 0.858 | 0.401 | 0.891 | 0.819 | 1.381 | 4.365 | 1.340 | 25.417 | 1.092 | 11.567 | 0.181 | 2.904 | 0.931 | 0.788 | 5.258 | 1.305 | 0.026 | 2.552 | 0.161 | 1.582 | 20.629 | 1.273 | 4.456 | 5.058 | 0.487 | 1.189 | 0.906 | 2.662 | 1.157 | 0.900 | 1.395 | 1.687 | 2.718 | 0.103 | 0.009 | 5.476 | 2.158 | 1.497 | 26.505 | 46.032 | 1.106 | 0.986 | 0.660 | 4.266 | 1.989 | 0.096 | 0.367 | 0.617 | 1.741 | 0.790 | 0.843 | |||
| SD | 0.059 | 0.560 | 0.075 | 0.341 | 0.135 | 0.073 | 0.146 | 0.140 | 0.244 | 1.184 | 0.131 | 6.211 | 0.108 | 1.909 | 0.027 | 0.349 | 0.368 | 0.158 | 1.211 | 0.147 | 0.015 | 0.204 | 0.038 | 0.296 | 19.267 | 0.119 | 0.557 | 0.864 | 0.082 | 0.268 | 0.146 | 0.590 | 0.248 | 0.306 | 0.104 | 0.829 | 0.475 | 0.019 | 0.004 | 0.644 | 0.185 | 0.250 | 8.735 | 19.576 | 0.640 | 0.179 | 0.076 | 0.566 | 0.203 | 0.035 | 0.081 | 0.223 | 0.227 | 0.113 | 0.066 | |||
| n | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |||
| SEM | 0.027 | 0.250 | 0.034 | 0.153 | 0.060 | 0.033 | 0.065 | 0.062 | 0.109 | 0.530 | 0.059 | 2.778 | 0.048 | 0.854 | 0.012 | 0.156 | 0.165 | 0.071 | 0.541 | 0.066 | 0.007 | 0.091 | 0.017 | 0.132 | 8.616 | 0.053 | 0.249 | 0.387 | 0.037 | 0.120 | 0.065 | 0.264 | 0.111 | 0.137 | 0.047 | 0.371 | 0.212 | 0.009 | 0.002 | 0.288 | 0.083 | 0.112 | 3.907 | 8.755 | 0.286 | 0.080 | 0.034 | 0.253 | 0.091 | 0.016 | 0.036 | 0.100 | 0.101 | 0.051 | 0.030 | |||
| Mean (AEN) | 0.173 | 6.079 | 2.094 | 2.622 | 1.106 | 0.190 | 0.479 | 0.205 | 0.994 | 18.447 | 2.003 | 87.765 | 1.142 | 13.314 | 0.118 | 1.824 | 0.809 | 1.445 | 6.001 | 3.289 | 0.744 | 4.612 | 0.140 | 2.568 | 38.146 | 1.208 | 5.438 | 3.745 | 0.814 | 0.206 | 1.294 | 2.216 | 1.310 | 0.578 | 2.354 | 4.015 | 4.943 | 0.103 | 0.001 | 17.066 | 3.314 | 2.456 | 56.413 | 115.292 | 1.087 | 1.716 | 0.995 | 8.832 | 2.906 | 0.072 | 0.250 | 0.509 | 6.921 | 0.622 | 1.654 | |||
| SD | 0.068 | 0.892 | 0.354 | 0.944 | 0.219 | 0.052 | 0.156 | 0.065 | 0.127 | 5.812 | 0.207 | 33.709 | 0.153 | 2.977 | 0.029 | 0.633 | 0.085 | 0.285 | 2.241 | 0.314 | 0.261 | 0.605 | 0.033 | 0.211 | 43.339 | 0.337 | 1.317 | 1.000 | 0.219 | 0.083 | 0.215 | 0.512 | 0.320 | 0.165 | 0.190 | 1.206 | 0.712 | 0.025 | 0.001 | 2.648 | 0.309 | 0.422 | 14.404 | 39.899 | 0.330 | 0.150 | 0.097 | 1.393 | 0.244 | 0.014 | 0.050 | 0.232 | 0.775 | 0.137 | 0.147 | |||
| n | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |||
| SEM | 0.030 | 0.399 | 0.158 | 0.422 | 0.098 | 0.023 | 0.070 | 0.029 | 0.057 | 2.599 | 0.092 | 15.075 | 0.069 | 1.331 | 0.013 | 0.283 | 0.038 | 0.127 | 1.002 | 0.141 | 0.117 | 0.271 | 0.015 | 0.094 | 19.382 | 0.151 | 0.589 | 0.447 | 0.098 | 0.037 | 0.096 | 0.229 | 0.143 | 0.074 | 0.085 | 0.540 | 0.319 | 0.011 | 0.000 | 1.184 | 0.138 | 0.189 | 6.442 | 17.843 | 0.148 | 0.067 | 0.043 | 0.623 | 0.109 | 0.006 | 0.023 | 0.104 | 0.347 | 0.061 | 0.066 |
Gene Ontology Analysis.
The gene ontology (GO) functions (enrichGO and simplify) of clusterProfiler (Wu et al., 2021) (RRID: SCR_016884) were used to determine the molecular function and biological processes associated with each cluster and to determine biological processes associated with differential splicing of mRNAs.
Gene Set Enrichment Analyses.
To determine if the gene set associated with astrocytic maturation from Zhang et al. (Zhang et al., 2016), as well as the gene sets associated with immature and mature astrocytes from Lattke et al. (Lattke et al., 2021) were enriched in any of the co-culture configurations, gene set enrichment analysis was performed using the R package fgsea (Korotkevich, 2019) (RRID: SCR_020938). Gene expression was ranked using the shrunken log2-fold changes from the comparisons between the A, AN, AE, or AEN configurations as estimated by DESeq2. The human gene symbols used in Zhang et. al, 2016 were converted to the orthologous mouse gene symbols using biomaRt (Durinck et al., 2009) (RRID: SCR_019214), and outdated gene symbols were replaced with current symbols manually.
Analyses of Alternative mRNA Splicing.
To identify differential splicing between the culture configurations, the Sargasso-filtered sequences were tested using rMATS-turbo v4.1.2 (Shen et al., 2014) (RRID: SCR_023485). The resulting data were visualized using the R package maser v1.12.1 (Veiga, 2022) (RRID: SCR_023484). The differential splice events with false discovery rate < 0.05 and a difference in proportion of inclusion > 0.1 were considered significant. Sashimi plots we generated with the Integrated Genome Viewer (Thorvaldsdottir et al., 2013) (RRID: SCR_011793) using a representative Sargasso filtered alignment for each displayed condition. Spliced exons were mapped to protein location using ensembldb (Rainer et al., 2019) (RRID: SCR_019103) with EnsDb.Mmusculus.v79 (Rainer, 2017). Protein structures were drawn using UniprotR (Soudy et al., 2020) (RRID: SCR_023483) with the mapped exons as added features.
Data and code availability.
All code used for data processing and for generating figures can be found on GitHub (https://github.com/murailab/Astrocyte_Triple_Co-Culture). The trimmed sequence reads can be found on the NCBI SRA (www.ncbi.nlm.nih.gov/bioproject/PRJNA961849).
Tamoxifen administration.
Aldh1L1-Cre/ERT2:Ai9 mice were used to visualize astrocytes. Both males and females were used for experiments and littermate pairs were used whenever possible. The stock solution for in vivo activation of Cre recombinase was prepared by dissolving tamoxifen (MilliporeSigma #T5648) in ¼ the final volume with 100% EtOH by vortexing. Once the tamoxifen was in solution, corn oil (MilliporeSigma #C8267) was added for the remaining ¾ final volume to make a final concentration of 10 mg/ml. This stock solution was stored at 4°C for a maximum of 2 days. The stock solution was diluted to 1 mg/ml with corn oil directly before injections. Mice at postnatal day 0 (P0) received one injection of 50 μl of 1 mg/ml solution once per day for a total of 2 days. Mice were collected for immunohistochemistry analysis at P4, P7, and P28 to assess protein expression of markers at key developmental time points for astrocytes.
Immunohistochemistry Immunofluorescence.
Mice were briefly anesthetized with 5% isoflurane/oxygen in an induction box. Once anesthetized (as assessed by absence of toe pinch reflex), mice were fitted with a nose cone delivering constant 5% isoflurane/oxygen. The mice were then transcardially perfused with ice-cold heparinized PBS followed by ice-cold 4% PFA in PBS. Brains were extracted and post-fixed in 4% PFA overnight at 4°C then equilibrated in 30% sucrose at 4°C, before embedding in OCT Compound (Somagen #SAK4583). A cryostat (Leica CM1950) was used to slice the tissue into 30 μm thick sections that were then stored in Dulbecco’s Phosphate-Buffered Saline (DPBS, Thermo Fisher #14190250) until use. For labeling, sections underwent antigen retrieval in a solution of 0.05% Tween/10mM Citrate/pH 6.0 for 15 mins at 70°C. Sections were then permeabilized with 1% Triton X-100 in DPBS at RT for 20 min with agitation. Next, sections were blocked with 0.2% Triton X-100 and Mouse-on-Mouse blocking reagent (Thermo Fisher #R37621) in DPBS for 30 min at RT to aid in specificity of binding by the mouse antibody NeuN (Millipore # MAB377, RRID: AB_2298772). After washing for 5 min in DPBS, sections were blocked with 10% normal donkey serum (NDS, Jackson ImmunoResearch Labs #017–000-121, RRID: AB_2337258) and 0.2% Triton X-100 in DPBS for 1 hour at RT with agitation. Sections were then incubated with primary antibodies and 5% NDS/0.1% Triton X-100 at 4°C with agitation for 3 nights. The following antibodies were used: guinea pig anti-GLT1 (1:250, Millipore #AB1783, RRID: AB_90949), rabbit anti-SLC6A11 (1:200, Proteintech #139201AP, RRID: AB_2877988), rabbit anti-FAM107A (1:50, Thermo Fisher #PA563200, RRID: AB_2641212), and mouse anti-NeuN (1:500, Millipore # MAB377, RRID: AB_2298772). After washing sections 3 times for 5 min in DPBS, fluorescent secondary antibodies were added with 5% NDS/0.1% Triton X-100 for 1 hour at RT with agitation. The following secondaries were used: donkey anti-guinea pig Alexa Fluor 647 (1:500, Jackson ImmunoResearch Labs #706605148, RRID: AB_2340476), donkey anti-rabbit Alexa Fluor 647 (1:500, Jackson ImmunoResearch Labs #711605152, RRID: AB_2492288), and donkey anti-mouse Alexa Fluor 405 (1:500, Jackson ImmunoResearch Labs #715475150, RRID:AB_2340839). Sections were washed twice for 5 min in DPBS, mounted on Superfrost Plus slides (Fisher Scientific #1255015), and cover slipped with Slowfade Gold anti-fade mounting medium (Thermo Fisher #S3696). Mosaic images of cortical areas were obtained using an LSM880 laser scanning confocal microscope (Zeiss, RRID: SCR_020925) with a 20X objective and stitched automatically within the ZEN Blue 2.1 software (RRID: SCR_013672). Higher magnification images were obtained using an FV-1000 laser scanning confocal microscope (Olympus, RRID: SCR_020337) with a 60X objective and 1.6 optical zoom. Images used for quantification of expression at developmental time points were obtained using a 20X objective on the FV-1000 LSM under identical acquisition parameters. Images were taken from 2 slices from each brain, one upper and one lower cortex for a total of 8 images per mouse and 4 mice (2 males, 2 females) for each time point (total of 12 Aldh1L1-Cre/ERT2:Ai9 mice). Images were analyzed using the ImageJ software (RRID: SCR_003070) by performing background subtraction and quantifying average fluorescence intensity per visual field. One-way ANOVA with Tukey post-hoc was performed on the mean fluorescence intensity values of each animal versus developmental time point (n = 4 per Timepoint) in R using the rstatix package (RRID: SCR_021240) (See Figure S1C).
Western Blot Analysis.
Fibronectin expression was examined in cortical tissue from C57BL/6 mice (Jackson laboratory stock #000664, RRID: IMSR_JAX:000664) at embryonic day 18 (E18) and P28; 3 mice from each age group (total of 6 C57BL/6 mice). Briefly, pregnant mice (for E18) were decapitated without anesthesia beforehand in accordance with IACUC. The uterine horn was removed and E18 embryos were removed and decapitated one at a time. P28 mice were decapitated without anesthesia. The heads were placed in cold HBSS and mouse brain cortices were dissected and solubilized by mechanical homogenization in 20 volumes of 50mM Tris-HCl, 150mM NaCl, 5mM EDTA, 1% NP-40, 1% sodium dodecyl sulfate pH 7.4 (Dunlop et al., 2003) with protease inhibitors (1μg/ml leupeptin, 250μM phenylmethylsulfonyl fluoride, 1μg/ml aprotinin, 1mM iodoacetamide) then centrifuged at 14,000xg for 10 min at 4°C. Protein concentration was determined using a Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific, #23225) according to kit instructions. 60μg of each sample was resolved on a 6% sodium dodecyl sulfate-polyacrylamide gel by electrophoresis then transferred for 22 hours at 30 volts at 4°C to an Immobilon FL membrane (Millipore #IPFL00010) using a Bio-Rad transblot apparatus (Bio-Rad Laboratories). After incubation for 1 hour at RT in blocking solution (TBS-T: 50mM Tris, 200mM NaCl, 0.1% Tween 20 with 5% nonfat dry milk), immunoblots were probed with anti-Fibronectin antibody at 1:1000 dilution (Sigma #ab1954, RRID: AB_11213226) overnight at 4°C with gentile shaking. Immunoblots were washed with TBS-T three times for 10 min each then incubated with secondary antibodies for 1 hour at room temperature in blocking solution containing anti-rabbit IRDye 800 antibody (1:10,000; LI-COR Biosciences #926–32213, RRID: AB_621848), washed with TBS-T three times for 10 min each and visualized using an Odyssey Infrared Imaging system (LI-COR Biosciences) (See Figure S1B).
RESULTS
Neurons and/or endothelial cells shift the morphology of astrocytes to a more ‘mature’ phenotype.
Astrocytes in vivo develop a complex stellate or bushy morphology with fine processes that contact synapses and larger processes that extend to, and envelop, the vasculature with endfeet (Bushong et al., 2002; Hosli et al., 2022). In culture, astrocytes are polygonal-shaped or fibroblast-like and have few processes (Raff et al., 1983). Several factors, such as dibutyryl-cyclic AMP, fibroblast growth factor, and others cause a shift in astrocytic morphology to a more in vivo-like phenotype (Fahrig and Sommermeyer, 1993; Schlag et al., 1998; Stork et al., 2014). Co-culturing astrocytes with neurons also induces a shift in astrocytic morphology (Matsutani and Yamamoto, 1997; Schlag et al., 1998; Hasel et al., 2017). We cultured mouse cortical astrocytes by themselves (A), or in direct contact with rat cortical neurons (AN), mouse brain endothelial cells (AE), or a combination of both (AEN) for ten days (Figure 1A). To evaluate the effects of the different co-culture configurations on astrocytic morphology, we stained astrocytes with antibodies against glial fibrillary acidic protein (GFAP), which allowed us to visualize the cytoarchitecture of astrocytes. We found that, as reported earlier (Raff et al., 1983), astrocytes grown alone display a polygonal (fibroblast-like) shape (Figure 1A, top row, 3rd and 5th columns). After co-culturing with neurons, which were visualized using the anti-NeuN antibody, astrocytes acquired a more stellate morphology that has been reported elsewhere (Matsutani and Yamamoto, 1997; Hasel et al., 2017) (Figure 1A, second row, 3rd and 5th columns). In the presence of endothelial cells, which were visualized using the anti-pecam1 antibody, astrocytes stretch to form an elongated cellular phenotype, described before as “elongated multicellular columns” (Yoder, 2002) (Figure 1A, third row, 3rd and 5th columns). In the tri-cultures, we observed astrocytes bearing both neuron-induced stellate and endothelial cell-induced elongated morphologies (Figure 1A, bottom row, 3rd column). These results replicate previous studies demonstrating that neurons and endothelia shape astrocytic morphology and indicate that the culture configurations can be used to evaluate non-cell autonomous changes to astrocytes with possible effects on astrocytic maturation.
Neurons and/or endothelial cells modify the transcriptome of astrocytes.
To test the hypothesis that neurons and/or endothelial cells modify the astrocytic transcriptome, astrocytes from the four different culture configurations (astrocytes alone, A; astrocytes + neurons, AN; astrocytes + endothelia, AE; astrocytes + neurons + endothelial cells, AEN) were isolated using FACS. mRNA was then isolated and subjected to high-throughput sequencing (Figure 1B). Anti-ACSA-2 antibodies recognize an epitope on the ATPase Na+/K+ transporting subunit beta 2 (ATP1ß2) and allow for the isolation of astrocytes (Batiuk et al., 2017). These antibodies coupled to the PE fluorophore were used to isolate astrocytes (Kantzer et al., 2017) (X-axis Figure 1C) and could be distinguished from endothelial cells isolated using anti-Pecam1 antibodies coupled to the APC fluorophore (Y-axis Figure 1C). mRNA was extracted from 5 independent biological replicates. One triple culture (AEN) sample did not pass quality control and was discarded. The remainder of the samples were further processed and sequenced. These analyses generated 3.0×109 100bp paired-end, stranded sequences with a mean of 8.9 × 107 ± 8.6 × 106 sequences per sample. 15,711 genes remained after filtering out all of the genes that did not have at least 20 counts in each of the pure astrocyte samples. To evaluate changes to the astrocytic transcriptome under the different co-culture configurations, we performed a principal component analysis (Figure 2A). This revealed four clusters corresponding to the different co-culture configurations and showed low variance between independent biological replicates. Thus, variations in the astrocytic transcriptome were impacted by the presence of neurons and endothelial cells with the largest variation due to the presence/absence of endothelial cells and a combination of neurons and endothelial cells (Figure 2A). We tested for differential gene expression using DESeq2 (Love et al., 2014) with a likelihood-ratio test (LRT) to identify significant differences in the various culture configurations followed by pair-wise estimation of log2-fold differences. Co-culture configurations robustly altered the transcriptome of the astrocytes (7,302 differentially expressed genes (DEG) with an adjusted p-value of <0.05 and a log2-fold change >=2 or <=−2).
Figure 2. Co-culture configurations modify the transcriptome of astrocytes.
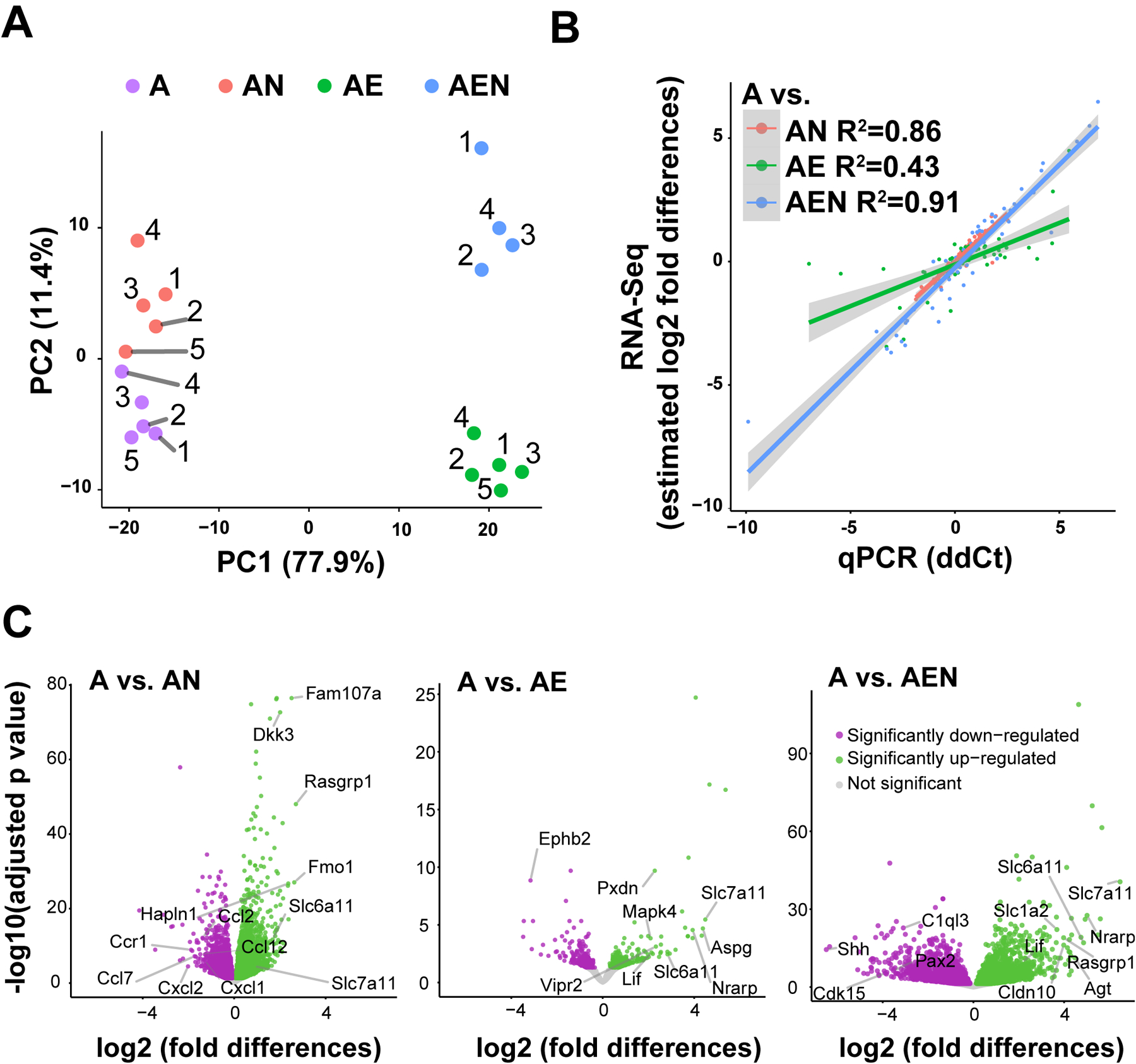
A. Principal component analysis of RNAseq data shows that samples cluster based on culture configurations, that the main driver of clustering is the presence/absence of endothelial cells, and the low variance between our independent biological replicates. B. Quantitative PCR was performed using microfluidics to validate the RNAseq. The average between Hars2, Srbd1, and Srp68 was used as normalized control due to their low variance between culture configurations. The ddCt method was used to calculate relative gene expression. See complete data set and description of statistics in Table 3. ddCt (x-axis) was compared to estimated log2-fold differences calculated by DESeq2 (y-axis). C. Volcano plots show all genes identified and highlight differentially expressed genes (DEG) in astrocyte monocultures (A) compared to AN, AE, and AEN respectively. Genes differentially up- or down-regulated are shown in green and magenta respectively, the rest of the genes that were identified by sequencing but with non-significant differences between culture configurations (p>0.05) are shown in gray. Some genes mentioned in the results/discussion sections are highlighted. Solute carrier family 6 member 11 (Slc6a11, gene that encodes the GABA transporter 3 (GAT3)) and solute carrier family 7 member 11 (Slc7a11, gene that encodes the cystine/glutamate transporter) are shown as examples of genes that are modified by all co-culture configurations. Note that while the X-axis shows log2-fold differences between A and co-culture configurations using the same scale in all volcano plots, the Y-axis shows adjusted p-value on a different scale. The list with all DEG with statistical reports is found in Table S1.
To validate our RNA sequencing data and DESeq2 estimated differences, we performed qPCR using microfluidics technology with Delta Gene assays (see Table 2). We selected 55 genes, based on the following criteria: large differences between co-culture configurations (14 genes with > 3-fold change), some of the lowest adjusted p values and therefore unlikely to be a false discovery, or high abundance transcripts (>300 sequences per fragment). We chose some genes regulated by neurons but not endothelial cells and vice versa. We also included some genes that are highly enriched/almost exclusively expressed by astrocytes (i.e., aquaporin 4 (Aqp4), connexin 43 (Gja1), glutamine synthetase (Glul), nuclear factor I A (Nfia), glutamate transporter 1 (Slc1a2), and SRY-box transcription factor 9 (Sox9)). We selected histidyl-tRNA synthetase 2 (Hars2), S1 RNA binding domain 1 (Srbd1), and signal recognition particle 68 (Srp68) as controls for the qPCR since they have the lowest coefficient of variance between culture configurations and are required for basic cellular functions (Subramanian, 1983; Freist et al., 1999; Harada et al., 2001). The results of these analyses can be found in Table 3. We found significant correlations between our qPCR data and changes estimated from RNA-seq analyses when comparing the effects of neurons (AN) (r2= 0.862, p < 2.2 X 1016), endothelial cells (AE) (r2= 0.431, p < 2.2 X 108), or the tri-culture (AEN) (r2= 0.906, p < 2.2 X 1016) on the transcriptome of astrocytes (Figure 2B). These analyses were conducted using the data that were corrected for endothelial contamination (see methods, Figure S2B). To determine if the correction might be influencing the correlations observed in the co-cultures containing endothelia, we tested the correlation of the uncorrected estimated changes versus the qPCR data. In the triple culture (AEN), there was less contamination with endothelia-’specific’ genes (~6%) and the resulting correlation was essentially the same (0.98 instead of 0.91). In the astrocyte-endothelia (AE) co-cultures, the estimated contamination was 12% and the correlation dramatically improved from 0.43 to 0.99 (Figure S2C). These data suggest that the correction for contamination is modestly blunting the predicted effects of endothelia.
Volcano plots were used to facilitate the visualization of all genes that were identified and all genes that were differentially regulated in response to the culture configurations. Several previously reported genes regulated by neurons or endothelia were identified in the current analysis in addition to newly identified genes. Figure 2C (left graph) shows all genes that were identified and the subset of genes that are differentially expressed when astrocytes were cultured by themselves compared to when astrocytes were cultured with neurons. A total of 3,098 and 2,904 genes were significantly (p<0.05) up- and downregulated respectively (representing 19.7% and 18.5% of the total genes identified) (Table S1). In contrast to the effect of neurons, endothelial cells regulated the expression of far fewer astrocytic genes with 377 up- and 346 down-regulated respectively (representing 2.4% and 2.2% of the total genes identified) (Figure 2C, middle graph and Table S1).
The combination of neurons and endothelial cells upregulated 2,875 and downregulated 2,707 astrocytic genes (representing 18.3% and 17.2% of the total gene count) (Figure 2C, right graph, and Table S1). This indicates that neurons and endothelia cooperatively regulate astrocytic genes, however, they also have some antagonistic or competitive effects on expression of other genes. To further evaluate the effects of neurons and/or endothelial cells on the astrocytic transcriptome, we generated a heatmap showing eighty DEGs with the smallest adjusted p-values from LRT analysis (Figure 3A).
Figure 3. Neurons and endothelial cells induce competitive and cooperative changes on the astrocyte transcriptome.
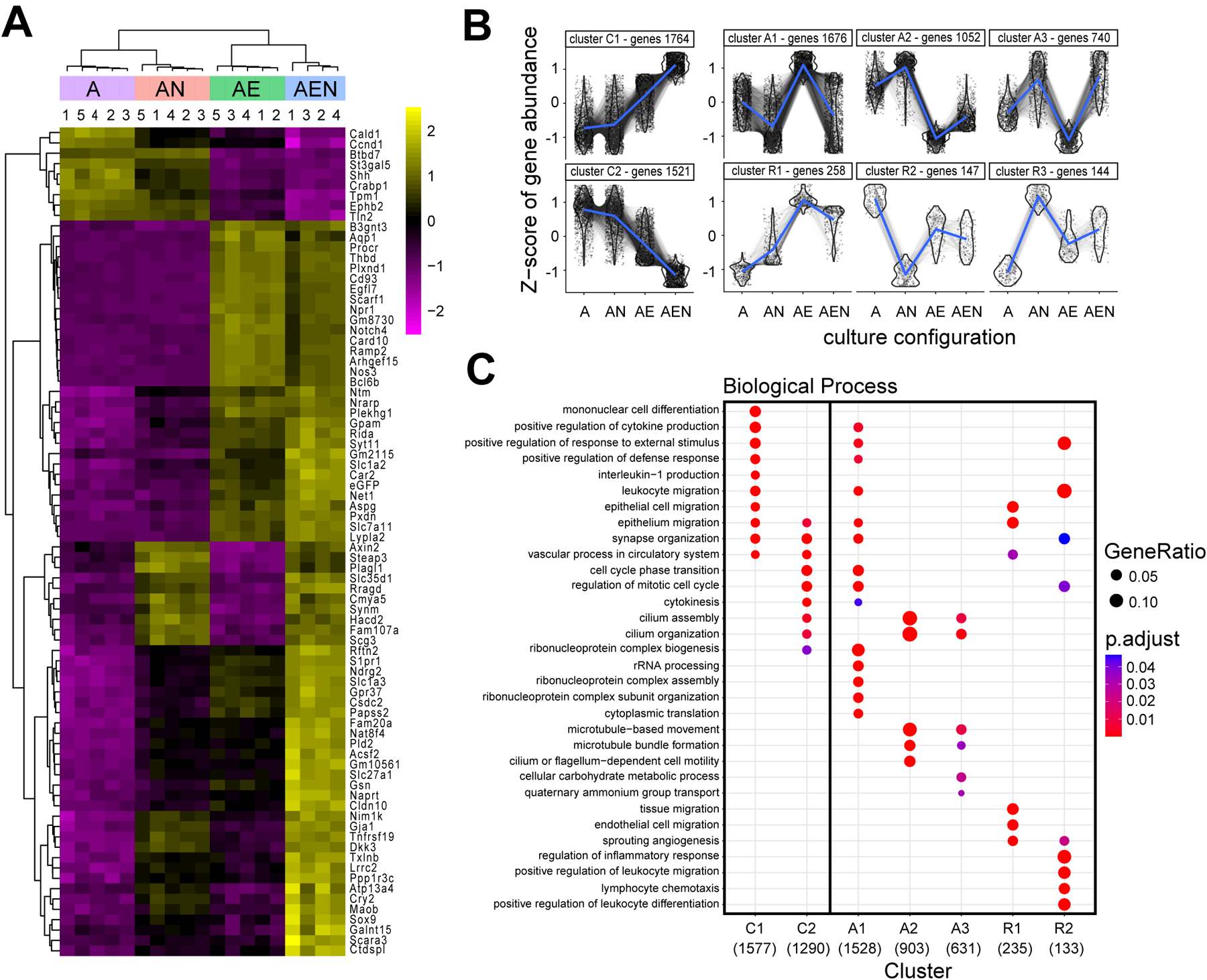
A. Unsupervised hierarchical clustering heat map of 80 significantly DEGs between purified astrocytes from the different co-culture configurations: A, AN, AE, and AEN. Each row represents a gene (named on the right), the numbers at the top of the columns represent biological replicates, and the color code represents the relative expression level (magenta for lower expression and yellow for higher expression). B. Unsupervised clustering of differentially expressed genes. Y-axis shows Z-score gene abundance and the X-axis co-culture configurations. Clusters C1 and C2 (cooperative), contain astrocytic genes that are cooperatively regulated by neurons and endothelial cells. Clusters A1-A3 (antagonist) contain astrocytic genes on which neurons and endothelial cells have competitive regulation. Clusters R1-R3 (redundant) contain astrocytic genes that are regulated in the same direction by neurons and endothelial cells but without additive effects. Each dot represents a gene. C. Gene ontology biological process enrichment analysis of differentially expressed genes in the clusters assigned in Figure 3B. The color code represents the adjusted p value. The size of the circles represents the gene ratio (percentage of total DEGs in the gene ontology term). The list with all gene ontology biological processes, the gene ratios, adjusted p values, and gene lists, are found in Table S2.
The clustering of astrocytic DEGs on the heat map indicated that subsets of genes are differentially regulated by neurons, endothelial cells, and the combination of both. To gain additional insight into the way that the different culture conditions impacted astrocyte gene expression, all of the DEGs that were significantly different by more than 4-fold (7,302 genes) were clustered based on the pattern of responsiveness to culture configurations. From this, eight clusters were identified (Figure 3B). In the first two clusters, neurons or endothelia changed astrocytic expression in the same direction with the combination of both causing a greater change (cooperative clusters, C1 and C2). A total of 3,285 genes were in these two clusters, cluster C1 contained 1,764 cooperatively up-regulated genes and cluster C2 1,521 cooperatively down-regulated genes. We used gene ontology enrichment analysis to better understand the potential biological implications of cooperative up- or down-regulation (Figure 3C). We found that neurons and endothelial cells cooperatively increase the expression of genes involved in cell differentiation, cytokine production, migration, synapse organization, anion transport, response to oxidative stress, and calcium ion transport, among others (cluster C1, a full list of GO terms is given in Table S2). Neurons and endothelial cells cooperatively downregulate molecules involved in cell cycle phase transition, regulation of mitotic cell cycle, cytokinesis, and axonogenesis (cluster C2 and Table S2). Based on this gene ontology, neurons and endothelia are shifting the astrocyte transcriptome from a proliferative phase toward a transcriptome that supports neuronal and immune cell signaling.
We also found that endothelial cells and neurons could have opposite effects on the astrocytic transcriptome with one cell population dominating the astrocytic transcriptome in the triple culture configuration (antagonistic clusters, A1, A2, and A3) (Figure 3B). A total of 3,468 genes were in these three clusters. Interestingly, we observed that endothelial cells, but not neurons, regulate genes involved in biogenesis, assembly, and organization of RNA-protein complexes, DNA repair, cell cycle transition, and mitochondrial transmembrane transport (cluster A1, 1,676 genes) (Figure 3C and Table S2). On the other hand, neurons but not endothelial cells, activated the following biological processes in astrocytes: microtubule-based movement and microtubule bundle formation, carbohydrate metabolic process, ammonium group transport, smoothened signaling pathway, cell-cell signaling by Wnt, extracellular transport, and lysosomal transport (clusters A2 and A3, 1,052 and 740 genes, respectively).
In the remaining three clusters, the effects of neurons or endothelia were in the same direction, but their effects were not additive in tri-cultures (redundant clusters, R1, R2, and R3) (Figure 3B). In cluster R1, we identified 258 genes upregulated by both neurons and endothelial cells, but without cooperative effects. These genes belong to the following GO terms: axonogenesis, RNA splicing, positive regulation of kinase activity, and cell junction assembly. In cluster R2 (147 genes), both neurons and endothelial cells down-regulate genes associated with leukocyte migration, regulation of the inflammatory response, and positive regulation of cell adhesion. In cluster R3, there were only 144 differentially expressed genes and they were not significantly enriched in any gene ontology terms.
We also performed gene ontology for molecular functions (Figure S3A) and found that neurons and endothelial cells cooperatively induce genes involved in phospholipid binding, immune receptor activity, GTPase activity, actin-binding, and cytokine activity in astrocytes. Endothelial cells induce the expression of genes that participate in rRNA and mRNA binding, while neurons induce the transport of intracellular cargo through the regulation of microtubules and dynein. The association between these molecular functions can be observed in Figure S3B. These data demonstrate that neurons and endothelia have both cooperative and competitive effects on the astrocyte transcriptome.
Neurons and endothelial cells cooperatively induce astrocyte maturation.
It has been previously reported that astrocytes in culture have an ‘immature’ transcriptome that differs from that observed in mature astrocytes in vivo (Cahoy et al., 2008). Furthermore, it was demonstrated that neurons can shift the astrocytic transcriptome to a more mature state (Hasel et al., 2017). To test if neurons and endothelial cells cooperatively promote astrocytic maturation, we evaluated if the culture configurations modified the expression of genes previously identified as enriched in mature astrocytes in a coordinated fashion by using Gene Set Enrichment Analysis (GSEA). GSEA allows for the comparison of differences in expression of specific genes observed in different studies (Subramanian et al., 2005). As recently discussed, it is not appropriate to compare relative expression of different genes in different studies because of technical issues ranging from RNA preparation and library preparation to normalization (for a recent review, see (Zhao et al., 2020)); therefore, we did not directly compare the transcriptome of astrocytes observed in the present study to previous publications. Instead, we performed GSEA using a gene list of twenty genes that were previously shown to have the highest difference between fetal and adult human astrocytes (Zhang et al., 2016). Of these twenty genes, two genes (alanine-glyoxylate aminotransferase 2-like 1 (Agxt2l1) and hydroxysteroid 17-beta dehydrogenase 6 (Hsd17b6)) were not present in our data set. Of the remaining eighteen genes, sixteen were found to be significantly differentially expressed by the LRT while eleven were found to be significantly upregulated by co-culturing with neurons and/or endothelia by pair-wise tests (Figure S4A). We also performed GSEA using the list of genes identified as enriched in immature astrocytes (678 genes) isolated from postnatal day 4 mice or mature astrocytes (359 genes) isolated from mice 6–10 weeks of age (Lattke et al., 2021). We found that neurons, significantly decreased the expression of genes enriched in immature astrocytes (Figure S3B, left column). Interestingly, endothelia did not significantly decrease expression of genes enriched in immature astrocytes. We also found that both neurons or endothelia significantly increase the expression of genes enriched in mature astrocytes (Figure S4B, right column). We found that the combination of neurons and endothelia significantly decreased expression of genes enriched in immature astrocytes and significantly increased expression of genes enriched in mature astrocytes (Figure 4, top row). A major goal of this study was to test the prediction that the combination of neurons and endothelia would cause a bigger shift toward a mature astrocytic transcriptome than either cell alone. Consistent with this prediction, we found that the combination of neurons and endothelial cells had a significantly greater effect on downregulation of genes associated with the immature state and had a significantly greater effect on genes associated with the mature state than either neurons or endothelia alone (Figure 4, bottom two rows). Thus, neurons and endothelial cells cooperatively shift the transcriptome of astrocytes toward a mature state.
Figure 4. The combination of neurons and endothelial cells have a larger effect in decreasing expression of genes enriched in immature astrocytes and increasing expression of mature astrocyte markers than either cell type alone.
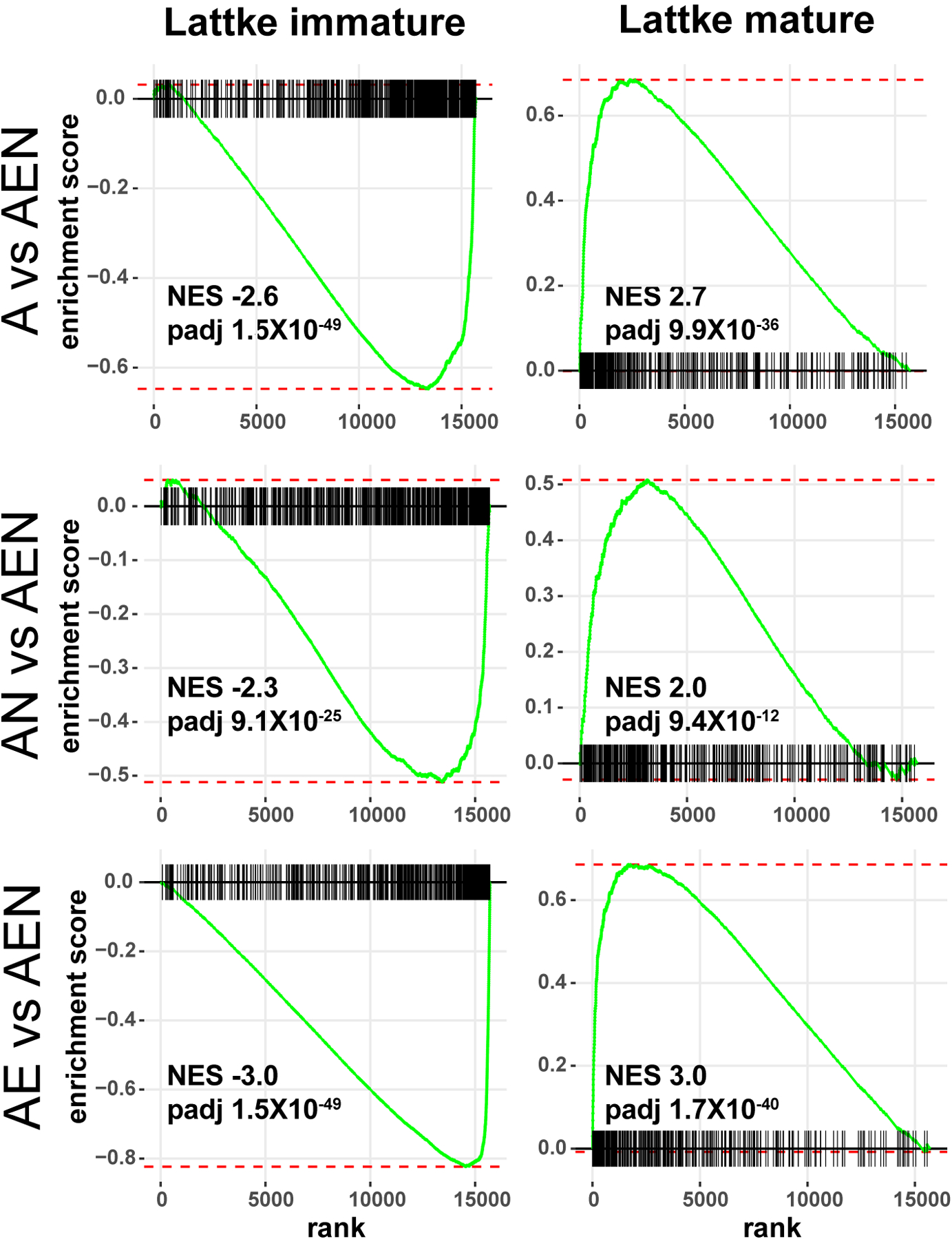
Gene set enrichment analysis of genes identified as enriched in immature astrocytes (left graphs) or genes enriched in mature astrocytes (right graphs) (Lattke et al., 2021) in astrocytes isolated from the different co-culture configuration. The normalized enrichment score (NES) and adjusted p values are shown. Each vertical black line in the X-axis represents a gene and indicates its position in the studied gene set. Genes on the far left are enriched in the gene set analyzed (immature or mature markers), while genes on the far right are underrepresented. The Y-axis represents the enrichment score.
Neurons and endothelial cells regulate mRNA splicing.
Alternative splicing diversifies the set of proteins in a cell and/or developmental-specific context, with the highest rates of alternative mRNA splicing occurring in the brain (Mills and Janitz, 2012). The high sequencing depth of our samples (8.9 × 107 reads per sample) permitted the analysis of low-abundance transcripts and analysis of mRNA splicing. We found that neurons changed 200 splicing events (Figure 5A, 1st pie chart). Gene ontology analysis of these alternatively spliced transcripts identified the following biological processes: Ion transport, dendrite and cell morphogenesis, and postsynaptic organization (Figure S5A and Table S3).
Figure 5. Neurons and endothelial cells modify the splicing of mRNAs in astrocytes.
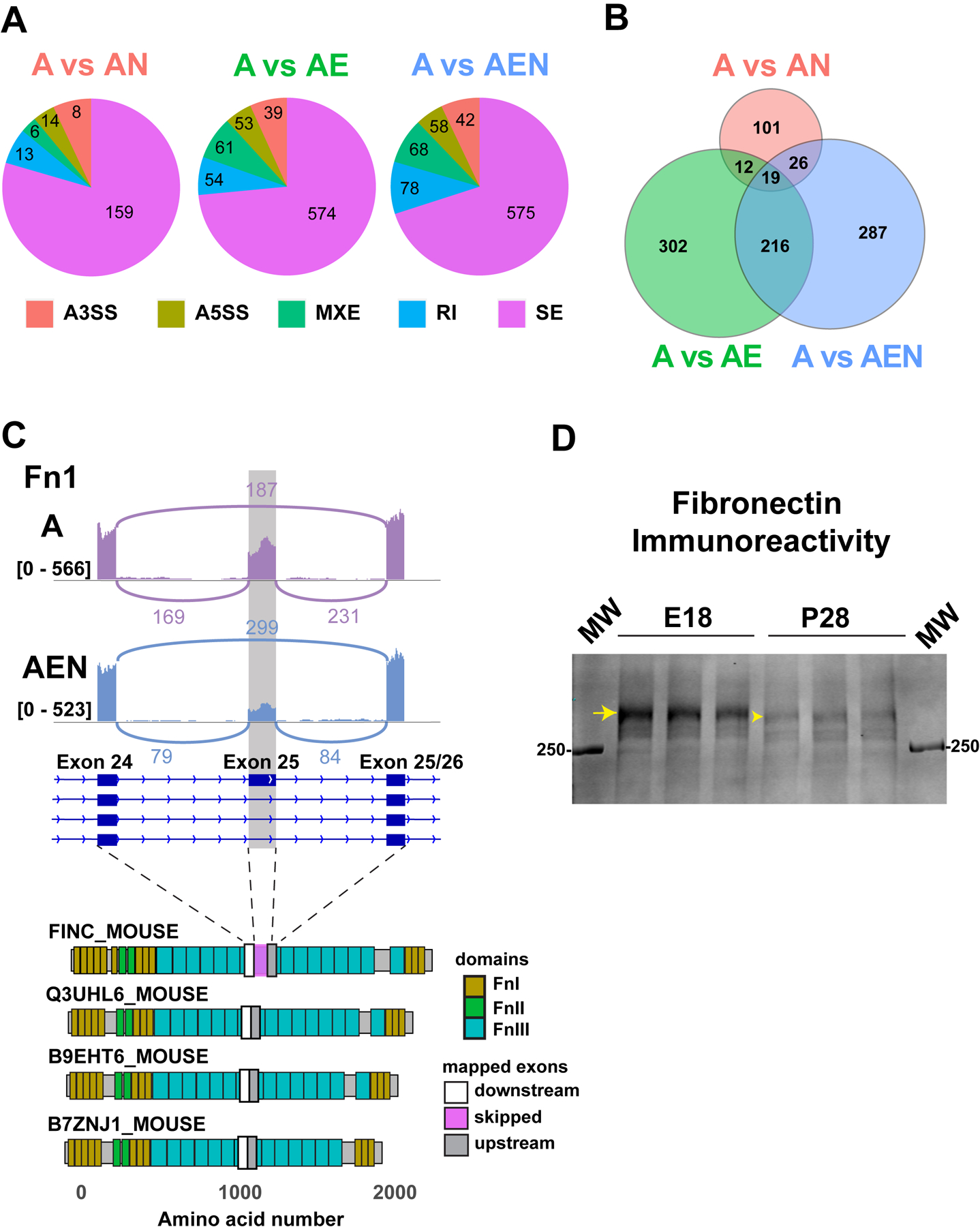
A. Frequencies of the five types of alternative splicing differentially detected in astrocytes monocultures versus astrocytes culture with neurons (A vs AN), endothelial cells (A vs AE), or with neurons and endothelial cells (A vs AEN). Five different types of alternative splicing were identified including alternative 3’ splice site (A3SS), alternative 5’ splice site (A5SS), mutually exclusive exons (MXEs), retained introns (RI), and skipped exons (SE). The numbers indicate the number of genes with differentially alternative events in the culture configurations indicated. Skipped exons were the most abundant alternative event. B. Scale vent diagram showing the unique versus shared skipped exons events in the different culture configurations. Only 19 skipped exon events were present in all co-culture configurations but absent in astrocyte monocultures. C. Fibronectin 1 (Fn1) is an example of a gene differentially regulated by neurons and endothelial cells through alternative splicing, specifically by exon skipping. The purple and blue traces represent raw data of the number of reads mapped to the Fn1 gene in astrocytes grown as monocultures and of astrocytes grown in the presence of neurons and endothelial cells respectively. The height of the purple and blue columns represents the number of reads. The gray column highlights exon 25. The transcript model of the Fn1 gene in dark blue is from the genome browser Ensembl (ENSMUST00000055226), boxes represent exons. D. Western blot analysis of mouse embryonic (E18) and adult (P28) brain cortex (tissue from three different animals each). Fibronectin bands were detected. The arrow highlights Fn1 long isoform (containing exon 25), while the arrowhead highlights Fn1 short isoform (lacking exon 25).
Consistent with the effects of endothelia on transcripts that encode RNA processing enzymes and mRNA binding proteins (Figure S3, cluster A1), both of which control splicing (Wang et al., 2015), we found that endothelia cause far more alternative splicing events (781) than those observed with neurons (200) (Figure 5A, 2nd pie chart). Skipped exons (SE) (also known as cassette exon events) were the most abundant form of alternative splicing in our dataset and represent greater than 70% of all events (Figure 5A). Although there were comparable numbers of alternatively spliced transcripts in the presence of endothelia or endothelia with neurons (Figure 5A, 2nd and 3rd pie charts), approximately 60% of the alternative spliced transcripts were unique to either endothelia or neurons (Figure 5B). Endothelial cells induced alternative splicing of astrocytic transcripts associated with the regulation of neuronal morphology and functions, including synapse organization, axonogenesis, and regulation of neurogenesis, in addition to cell junction assembly, cell-matrix adhesion, regulation of cell morphogenesis, and small GTPase mediated signal transduction (Figure S5A and Table S3). Although the majority of differentially spliced mRNAs were different in astrocyte-endothelia co-cultures, the same biologic processes were linked to the alternative splicing observed in the tri-culture, suggesting that the effects of endothelia dominate even in the presence of neurons.
To better understand the extent of the splicing events occurring when astrocytes were co-cultured with neurons and endothelial cells, we focused on two targets, fibronectin 1 (Fn1) and prominin 1 (Prom1) (Figure 5C and S5C). We chose Fn1 because its splicing has been studied, and it is known that at least 20 different proteins can be generated by alternative mRNA splicing in three regions: extra domain A (EDA, also known as EIIIA), extra domain B (EDB, also known as EIIIB), and the variable region (V, also known as type III connecting segment or IIICS) ((Schwarzbauer et al., 1987) for a review, see (White et al., 2008)). We found that 57.64 ± 0.03% of Fn1 transcripts in astrocytes grown by themselves retained exon 25 (EDB), however, only 22.75 ± 0.03% retained it in the triple cultures (Figure 5C). We validated this by Western blot for FN1 in embryonic (E18) and adult (P28) mouse brain cortex. We found a ~10 kDa switch in the migration of FN1, consistent with this alternative splicing (Figure 5D, arrow vs arrowhead). Accordingly this alternate splicing of Fn1 was previously reported in vivo using PCR with high levels of EDB transcripts observed during embryogenesis and low levels of this transcript observed in adult mice (Bencharit et al., 2007). This agrees with our Western blot results and further validates our transcriptomics data.
We also detected differences in the alternative splicing of prominin 1 mRNA (Prom1) (also known as CD133) and chose to follow up on Prom1 as it is associated with glioma recurrence and cancer therapy resistance (Abdoli Shadbad et al., 2021). We found that this gene undergoes three alternative splicing events that are differentially present in the culture configurations: one in the extracellular domain (EC), and two in the intracellular region (IC1 and IC2) (Figure S5 B&C). We found that the presence of endothelia increased the splicing of EC from 40% to 64%, while neurons did not modify the splicing in this region. In the presence of both endothelia and neurons (AEN), the effect of endothelia dominated with 67% of transcripts retaining the EC spliced. (Figure S5 B&C). Meanwhile neurons and endothelia cooperatively down regulated splicing in the intracellular domains. In astrocytes grown on their own, 30% of transcripts retained IC1, in astrocytes grown in the presence of neurons or endothelia, this decreased to 22%, and in the presence of the three cell types, IC1 splicing decreased even further to 19%. IC2 was spliced 94% in astrocytes monocultures and 89, 70, and 60% in astrocytes grown in the presence of neurons, endothelia, or their combination, respectively. This shows that even in the same gene there are exclusive and cooperative effects of neurons and endothelia on the splicing of astrocytic mRNAs. The relevance of these Prom1 splicing events in vivo is still unknown. As shown by these two examples, our data provide a valuable resource for glial biologists interested in understanding alternative RNA splicing in astrocytes.
Astrocytic transcripts regulated by neurons and endothelial cells in co-culture are differentially expressed in vivo during brain development.
To better understand how differentially expressed mRNAs from co-culturing astrocytes with neurons and/or endothelial cells are reflective of changes in protein expression in vivo, we performed immunolabeling on mouse coronal sections at different developmental stages (Figure 6A). We evaluated the expression of proteins whose transcripts responded differently to the culture configurations: FAM107A (actin-associated protein, DRR1) induced by neurons but not endothelial cells, and GAT3 (GABA transporter 3, Slc6a11) and GLT1 induced by both neurons and endothelial cells. We first verified that these proteins were expressed by astrocytes, as reported before (Lehre et al., 1995; Minelli et al., 1996; Regan et al., 2007; Cahoy et al., 2008; Batiuk et al., 2020). To do this we used Aldh1L1-Cre/ERT2 mice crossed with Ai9 TdTomato (Tom) reporter mice to visualize astrocytes. Mice were given tamoxifen to induce Tom at P0 and collected at either P4, P7, or P28, corresponding to time points after which neural progenitors have switched from production of neurons to glial progenitors (P4), when astrocytes continue to divide, migrate, and develop in cortex (P7), or when they have stabilized and reached a more mature state (P28) (Schober et al., 2022). Expression of each target was investigated in the somatosensory cortex.
Figure 6. GAT3, FAM107A, and GLT1 are regulated through development in the mouse somatosensory cortex.
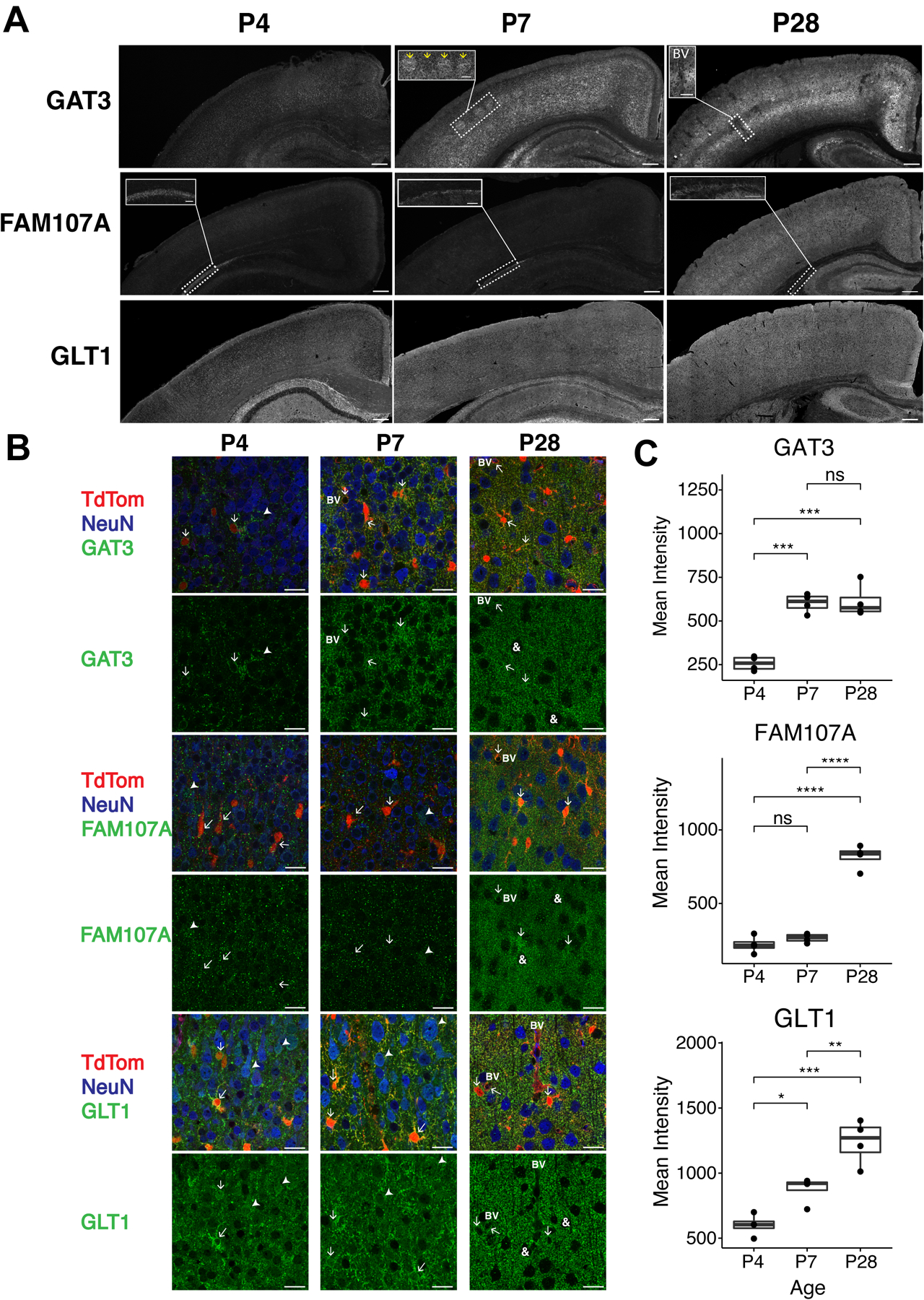
A. Mosaic images taken from P4, P7, and P28 mouse somatosensory cortex for GAT3 (encoded by Slc6a11) (top row), FAM107A (middle row), and GLT1 (encoded by Slc1a2) (bottom row). Outsets in GAT3 show presence of barrels in the barrel cortex (P7) and heterogeneity around blood vessels (P28). Outsets in FAM107A show a distinct region of the hippocampal CA1 where expression is higher. Scale bars = 250 μm and 100 μm for the outsets. Images acquired with LSM880 at 20X. B. Higher magnification images of P4, P7, and P28 mouse somatosensory cortex for GAT3 (top 2 rows), FAM107A (middle 2 rows), and GLT1 (bottom 2 rows). TdTom = tdTomato signal in astrocytes, NeuN = neuronal cell bodies, arrows = astrocytes, arrow heads = neurons, BV = blood vessel, &= highlight that neurons lack immunoreactivity for these targets, scale bars = 20 μm. Images acquired with Olympus FV1000 LSM at 60X with 1.6x optical zoom. C. Quantification of expression of GAT3 (top), FAM107A (middle), and GLT1 (bottom) in mouse somatosensory cortex at P4, P7, and P28. Graphs show data points for each individual animal. One-way ANOVA with Tukey post-hoc was performed on the mean fluorescence intensity values per visual field of each animal versus developmental time point (n = 4 per Timepoint). * p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant.
At P4, GAT3, FAM107A, and GLT1 were present throughout the cortex (Figure 6A) in a punctate pattern that was largely astrocytic (Figure 6B, arrows), with some neuronal expression (Figure 6B, arrowheads). Some heterogeneity was seen with FAM107A with higher expression in layers II/III and IV as well as a small area near the dorsal aspect of hippocampal areas CA3 and CA1 (Figure 6A, outset). At P7, there was a noticeable difference in the expression of all targets, with more clearly defined astrocytic domains (Figure 6A). Expression of GAT3 and GLT1 increased at P7 by ~57% (p < 0.001) and ~31% (p < 0.01), respectively, while FAM107A remained the same (Figure 6C). Expression of GAT3 was more specific to astrocytes at P7 and was observed in endfeet surrounding blood vessels (Figure 6B, BV), as well as processes that were adjacent to neurons (Figure 6B). GAT3 at P7 displayed both regional (low levels in layer I and II/III, low levels in CA1) and local heterogeneity (variable expression within layers) (Figure 6A). Notably, GAT3 expression at P7 showed cortical barrel expression, which to our knowledge has not been reported (Figure 6A, yellow arrows in outset). At P7, GLT1 expression was more evident in the fine processes of astrocytes, although it was still found in some neurons (Figure 6B) and appeared to be more locally heterogeneous (Figure 6A). At P7, expression of FAM107A was similar to the expression at P4 (Figure 6B) but began to show heterogeneous patterns especially in layer IV and V astrocytes (Figure 6A). Interestingly, the increased expression of FAM107A within a stream of cells at the dorsal boundary of hippocampal CA3 and CA1 seen at P4 was still present at P7, albeit at lower levels (Figure 6A, outset).
By P28, all targets appeared specific to astrocytes in the cortex (Figure 6B, & symbol highlights lack of immunoreactivity in neurons). From P7 levels, FAM107A expression at P28 increased dramatically by ~68% (p < 0.001), GLT1 increased by ~29% (p < 0.01), and GAT3 levels stayed the same (Figure 6C). GAT3 at P28 showed a wide range of local and regional heterogeneity and was seen on astrocyte membranes including endfeet (Figure 6A and 6B). Of note, GAT3 expression adjacent to some blood vessels (but not all) appeared to be dramatically reduced (Figure 6A, BV in outset). FAM107A expression in the cortex at P28 was specific to astrocytes and was seen uniformly on astrocyte membranes, including in endfeet (Figure 6A and 6B). Heterogeneous expression of FAM107A was more apparent by P28, with notably lower expression specifically in layer V astrocytes (Figure 6A). The high levels of FAM107A seen in hippocampal CA1 from P4 and P7 appeared to be similar to adjacent regions by P28 (Figure 6A, outset). GLT1 expression by P28 had become specific to astrocytes with no remaining detectable neuronal expression (Figure 6B). GLT1 at P28 was expressed on astrocytic membranes including at endfeet (Figure 6B, BV), and displayed some local and regional heterogeneity (Figure 6A). Overall, age had a significant effect on GAT3, GLT1, and FAM107A expression levels (p < 0.001 for all) and reached peak expression by P28 as compared to expression at P4 (Figure 6C). This aligns with the in vitro transcriptomics data (A vs AEN) presented (Figure 2C and Table S1). As shown by these three examples, the maturation pattern of gene expression correlates with that observed in vivo (P4 vs P28).
DISCUSSION
In the present study, we demonstrated that neurons and endothelia modify the astrocytic transcriptome and work together to establish a more ‘mature’ gene expression profile of astrocytes. We also found that neurons regulate more genes than endothelial cells in astrocytes, neurons and endothelial cells control different patterns of astrocytic gene regulation, and that endothelial cells cause a substantial number of mRNA alternative splicing events in astrocytes.
We found that astrocytes are highly adaptive cells at the molecular level, having more than 15,000 differentially expressed genes that relate to specific culture configurations. Among the genes that were upregulated in astrocytes by neurons was the family with sequence similarity 107 member A gene (Fam107a), also known as DRR1 and TU3A (Figure 2C and Table S1). Fam107a is predicted to be a stress-induced actin-binding protein (Schmidt et al., 2011) and was previously identified as a gene that is restricted to astrocytes in vivo and transcriptionally activated during astrocytic maturation (Cahoy et al., 2008). Upon overexpression of Fam107a, mice display increased sociability (Masana et al., 2014). We found low levels of FAM107A immunoreactivity in the somatosensory cortex at P1 and P4, with significant increased expression by P28 (Figure 6). Neurons also caused a robust down-regulation of several chemokine-binding proteins that have been implicated in inflammation (Charo and Ransohoff, 2006), including the c-c motif chemokine ligands 2, 7, and 12 (Ccl2, Ccl7, and Ccl12), c-c motif chemokine receptor 1 (Ccr1), and c-x-c motif chemokine 2 ligands 1 and 2 (Cxcl1 and Cxcl2) (Figure 2C and Table S1).
In contrast, endothelia caused a 20-fold increase in Slc7a11 expression in astrocytes compared to a relatively modest increase caused by neurons (1.8-fold) (Figure 2C). Slc7a11 encodes the functional subunit of the system Xc- (xCT) that exchanges cystine and glutamate to support glutathione synthesis (Bannai and Kitamura, 1980; Sato et al., 1998). However, the regulation of expression of this transporter remains poorly understood (Lewerenz et al., 2013). Endothelia also caused a 19-fold increase in the expression of the gene that encodes asparaginase (Aspg), an enzyme that converts asparagine to aspartate and that is targeted in the treatment of acute lymphoblastic leukemia and lymphoblastic lymphoma (Dinndorf et al., 2007). Among the astrocytic genes downregulated by endothelial cells, we found the ephrin receptor B2 (Ephb2) (10-fold reduction; Figure 2C and Table S1). EphB2 expression increases in reactive astrocytes (Bundesen et al., 2003), and deletion of EphB2 improves outcomes in an animal model of stroke (Ernst et al., 2019) and synaptic function in an animal model of Alzheimer’s disease (APP/PS1 model) (Qi et al., 2019).
Within the group of upregulated genes, we found some genes that were upregulated in all three co-culture configurations (AN, AE, and AEN; i.e. GABA transporter 3/GAT3, (Slc6a11) and Slc7a11), while others were regulated by neurons but not endothelial cells (i.e Fam107a and Ras guanyl releasing protein 1 (Rasgrp1)), or vice versa (i.e. Aquaporin 1 (Aqp1) and leukemia inhibitory factor (Lif)) (Figure 2C and Table S1). We also found that the combination of neurons and endothelia induced larger changes than either one alone to GLT1 (Slc1a2), angiotensinogen (Agt), claudin 10 (Cldn10), and others.
Our results indicate that neurons and endothelia shift the astrocyte transcriptome towards a more mature phenotype likely by using overlapping and distinct signaling pathways. Several different signals have been implicated in astrocyte maturation. For example, components of the Notch signaling pathway are highly enriched in astrocytes compared to other cell types in vivo (Cahoy et al., 2008). The increased expression of GLT1 induced by neurons or endothelia is blocked by pharmacologic and/or genetic inhibition of this pathway (Hasel et al., 2017; Lee et al., 2017; Martinez-Lozada and Robinson, 2020). As was previously demonstrated, we found that endothelia or neurons both increase expression of Slc1a2 (GLT1). We found that the combination of neurons and endothelia had a greater effect on Slc1a2 transcription than either cell alone revealing a cooperative interaction (Figure 3A and 3B–C1). We also found that neurons, endothelia, or the combination of both cell types induced expression of one of the transcription factors that is downstream of Notch signaling, Hes5 (Table S1), consistent with the known contribution of Notch to both neuron- and endothelia-induced astrocyte maturation (Hasel et al., 2017; Lee et al., 2017; Martinez-Lozada and Robinson, 2020). In addition to Notch, other signaling pathways have been implicated in astrocyte specification and maturation, including activation of JAK/STAT by ciliary neurotrophic factor (CNTF) (Hughes et al., 1988) or LIF (Bonni et al., 1997; Mi et al., 2001), activation of Smad by BMP (Gross et al., 1996; Scholze et al., 2014), activation of ß-catenin by Wnt (Bejoy et al., 2020), and activation of Gli by sonic hedgehog (Shh) (Farmer et al., 2016; Hill et al., 2021; Xie et al., 2022). We found that endothelial cells, but not neurons, induced expression of Lif and its receptor (Lifr) in astrocytes. On the other hand, neurons, but not endothelia, increased expression of several components of Shh signaling (Gli, Ptch, Fgf8, Smo, Fgfr2, and Runx2) and of the Wnt signaling pathway (Fzd1, Wnt2, Wnt2b, Dkk3, and Fzd10). Exogenous expression of four transcription factors, Rorb, Dbx2, Lhx2, and Fezf2, in cultured astrocytes shifted the astrocyte transcriptome to a more ‘mature’ transcriptome (Lattke et al., 2021). As was observed by Lattke et al., we observed low levels of transcripts for these factors when astrocytes were cultured on their own. We found that Rorb and Dbx2 were induced by neurons, endothelia, or their combination in a cooperative fashion (C1), and Fezf2 was induced by neurons, but not endothelia. Lhx2 was not detected in any of the culture configurations.
Attributing the activation and suppression of distinct signaling pathways to specific cell types and non-cell autonomous cellular interactions will lead to a better understanding of how cells cooperate to support astrocyte maturation. Most astrocytes migrate into the cortex after the vascular bed has started to form, and when neurons are present, but the majority of synapses have not been established (Mi et al., 2001; Allen and Eroglu, 2017; Tan et al., 2021). This suggests that the proportion of signals received from neurons and endothelia likely evolves during maturation.
Astrocytes display inter- and intra-regional diversity across the CNS (Schober et al., 2022), but how this diversity is generated is still poorly understood. Astrocyte diversity is likely driven by different lineages of precursor cells and further sculpted by different environmental signals. However, the extent to which diversity is pre-determined or due to environmental signals requires further investigation (For a discussion on this topic see (Chierzi et al., 2023)). In our immunolabeling analysis, we investigated three targets, one of which was influenced by neurons alone (FAM107A) and two of which were influenced by both neurons and endothelia (Slc6a11/GAT3 and Slc1a2/GLT1). All three displayed different patterns of expression throughout development: FAM107A had low expression at P4 and P7 before peaking at P28, GAT3 peaked at P7, and GLT1 increased expression across all ages (Figure 6). These patterns of expression throughout development are consistent with prior findings for FAM107A (Lu et al., 2021), GLT1 (Furuta et al., 1997), and GAT3 (Minelli et al., 2003). Although all targets displayed differences in expression patterns, all three increased their expression over time with highest levels at P28, indicating that these are markers of astrocyte maturation. Exactly how neurons and endothelia contribute to the expression of FAM107A, GAT3, and GLT1 in vivo will need to be further investigated. It is also possible that the antagonistic effect of neurons and endothelial cells identified here contributes to the generation of astrocyte diversity. Astrocytes in different brain regions coordinate their transcriptome/proteome to region-specific neuronal functions. For example, we previously showed that neuronal signaling, through Sonic hedgehog (Shh), diversifies cerebellar, cortical, and hippocampal astrocytes (Farmer et al., 2016), while Hill and colleagues demonstrated that Shh induces expression of Kir4.1 in astrocytes found in cortical layers IV and V (Hill et al., 2019). Consistent with this, we found that Gli1, Ptch1 (Shh targets), and Kcnj10 (Kir4.1 gene) are induced by neurons, but not by endothelial cells. A subpopulation of cortical astrocytes enriched in cortical layer V has a unique molecular repertoire, including expression of Norrin and Leucine-rich repeat-containing G-protein-coupled receptor 6 (LGR6) (Miller et al., 2019). Remarkably, we found that neurons induce expression of Lgr6 in astrocytes, while endothelia decrease it. Astrocytes from different cortical layers have distinct gene signatures and morphology; these characteristics are dependent upon discrete neuronal layering (Lanjakornsiripan et al., 2018). This can be seen by the heterogeneous expression patterns we observed for GAT3, GLT1, and FAM107A, all of which are influenced by neurons, throughout the different layers in the somatosensory cortex (Figure 6A). The contributions of endothelial cells to astrocyte diversity have not yet been explored.
The results from the present study have implications for the studies in which brain development and neuropathologies are being modeled using multi-cellular configurations like in organoid cultures involving human induced pluripotent stem cells (hIPSCs). One of the current limitations of brain organoids is that they lack blood vessels. This had been associated with a lack of oxygenation and nutrient supply (Di Lullo and Kriegstein, 2017; Qian et al., 2019). However, based on the data presented here, the lack of endothelial cells in brain organoid cultures will also affect the maturation of astrocytes and thus must be considered. Accordingly, the field is now developing organoids with ‘blood vessels’. One approach is to express transcription factors to induce the formation of endothelial cells and vascular-like structures in brain organoids (Cakir et al., 2019). Another method is to independently generate brain and blood vessel organoids and combine them (Sun et al., 2022). Using the latter, the Luo group showed that brain organoids with blood vessels have more neural progenitors in agreement with the idea that endothelial cells regulate neural development (Sun et al., 2022). The authors of this study did not examine astrocytes. Based on our data it is tempting to speculate that the astrocytes present in the brain-blood vessel-fused organoids would more closely mimic the environment observed in vivo.
Alternative mRNA splicing helps to diversify the repertoire of proteins in cells (Mills and Janitz, 2012) and is known to regulate all stages of CNS development including cell specification and synaptogenesis (Su et al., 2018). Alternative splicing is important for neuronal differentiation, migration, synapse maturation, and regulation (Li et al., 2021). Errors in splicing contribute to several neurodegenerative diseases including Parkinson’s disease, Alzheimer’s disease, spinal muscular atrophy, and inherited frontotemporal dementia (Dredge et al., 2001; Scotti and Swanson, 2016). Alternative splicing changes during development and is more prevalent in the brain than in other body organs (Mazin et al., 2021). Zhang and collaborators identified 2,500 astrocyte-specific alternative splicing events (Zhang et al., 2014). Here we found that neurons or endothelia change mRNA splicing and that endothelia caused alternate splicing of ~4 times more astrocytic transcripts than neurons. Although it is not surprising that alternative splicing will be regulated by the cellular environment as it occurs in a cell- and development-specific context, it is intriguing to observe that alternative splicing in astrocytes is highly regulated by cell-cell interactions.
Recently Larionova and colleagues demonstrated that alternative splicing of Rpl22L1 contributes to the diversity of glioblastoma cell populations (Larionova et al., 2022), and that this alternative splicing is regulated by the microenvironment. Blood vessels are key components of the glioma microenvironment; they are not only required for oxygen and nutrient supply, but they also promote glioblastoma proliferation and migration (McCoy et al., 2019). We demonstrated that endothelia have a much greater effect on splicing of astrocytic mRNAs than neurons and that the transcripts spliced are known participants in cell growth and cell-matrix adhesion (Figure S5A). Interestingly, increased expression of the cystine/glutamate antiporter (Slc7a11, system Xc-) is observed in glioblastoma and the magnitude of the increase predicts outcome, including the occurrence of seizures (Lyons et al., 2007). It is thought that this transporter operates in the reverse direction to increase extracellular glutamate and excitability (Robert et al., 2015; Sorensen et al., 2018). Although we found that neurons or endothelia induced expression of Slc7a11, the effect of neurons was small (1.8-fold) compared to those of endothelia (20-fold). Our data suggest that endothelial cells normally induce/maintain astrocyte maturation and that neovascularization in glioblastoma favors glioma growth, migration, and progression of the disease.
Previously, we and others have shown that the effects of neurons on the astrocytic expression of GLT1 are reversible with GLT1 decreasing after neuronal death in culture or in vivo (Schlag et al., 1998; Yang et al., 2009). In the same way, the loss of Shh signaling from neurons reduces expression of Bergmann glia markers and of Kir4.1 in cortical astrocytes (Farmer et al., 2016). This suggests that neuronal signals are required to maintain the expression of proteins associated with the mature astrocytic phenotype. In response to injury, a fraction of astrocytes proliferate (Pekny and Pekna, 2014; Escartin et al., 2021). This ‘stem cell response’ only occurs in astrocytes that are associated with blood vessels (Bardehle et al., 2013). In agreement with this, in our experiments, neurons decreased the expression of immature ‘stem cell-like’ genes, while endothelia did not. This suggests that a loss of neurons, combined with an increase in endothelia, may shift astrocytes to a proliferative state. After stroke, the expression of GLT1 decreases, while the expression of inflammatory chemokines increases (Ma et al., 2022). In our experiments, neurons induced GLT1 expression and inhibited the expression of inflammatory chemokines. Therefore, the stroke-induced changes may reflect the loss of signals from neurons. Further experiments aimed at testing these hypotheses may increase our understanding of the molecular mechanisms involved in cell damage after stroke.
It is important to note that our study has some limitations. 1) For our analysis, we are purely relying on astrocytic mRNA levels and not considering astrocytic protein levels which may be locally regulated for example in astrocytic perisynaptic processes and endfeet (Boulay et al., 2017; Sakers et al., 2017; Mazare et al., 2020). Thus, astrocytic mRNAs regulated by neurons and/or endothelial cells may not be reflective of differences in levels of astrocytic proteins. 2) We used co-culture systems to dissect the signals from neurons and endothelial cells that induce astrocyte maturation. These culture configurations represent ‘simplified’ systems that cannot fully recapitulate the complexity of extrinsic/environmental factors that may contribute to astrocyte maturation in vivo (for a discussion on the topic see (Lattke et al., 2021) discussion section). 3) We cultured astrocytes in the presence of FBS, which has been shown to modify the transcriptome of astrocytes (Foo et al., 2011). However, it is important to note that FBS was included in all culture configurations and therefore was a controlled variable.
In conclusion, our study identified neuron- and endothelial-dependent regulation of the astrocytic transcriptome. Our findings demonstrate that the maturation of astrocytes is orchestrated by a mixture of signals from their neighboring cells. Interestingly, we also demonstrate that neurons and endothelia competitively regulate the astrocytic transcriptome and presumably astrocytic functions. The dataset presented here provides a valuable resource for future studies interested in the exploration of signaling mechanisms involved in astrocyte-neuron and astrocyte-endothelial interactions.
Supplementary Material
Figure S1. Graphical Timeline. A. Mouse astrocytes were cultured by themselves, in presence of neurons, endothelia, or their combination. After 10 days in culture cells were harvested for immunocytochemistry immunofluorescence (ICC-IF) or dissociated and labelled with anti-astrocyte cell surface antigen 2 (ACSA-2) antibodies or anti-Pecam1 antibodies and separated using fluorescence activated cell sorting (FACS). RNA was then isolated from the sorted astrocytes. RNA was analyzed by qPCR or sequenced. B. Cortical tissue was isolated from E19 and P28 mice, resolved in a western blot (WB) and probed with anti-Fibronectin antibodies. C. Coronal brain sections were obtained from P4, P7, and P28 mice and immunohistochemistry immunofluorescence (IHC-IF) was performed for GAT3, FAM107A, and GLT1. See methods sections for details.
Figure S2. A. Pipeline schematic of RNA sequencing analysis. See methods for steps description. All code that processed the data can be found at GitHub (https://github.com/murailab/Astrocyte_Triple_Co-Culture). B. To estimate the percent composition of endothelia in each sample we used the average read counts for each gene in pure astrocytes and pure endothelial samples. As expected, A and AN do not contain any endothelial-specific sequences. Although we set conservative gates (see Figure 1C), we estimated 12% and 6% of endothelial content in the astrocytes isolated from the AE and the AEN co-cultures respectively. C. Correlation of RNAseq uncorrected log2-fold differences (Y-axis) versus quantitative PCR ddCt data (X-axis).
Figure S3. A. Gene ontology molecular function enrichment analysis of differentially expressed genes in the clusters assigned in Figure 3B. The color code represents the adjusted p value. The size of the circle represents the gene ratio. B. Associated networks for the molecular function enrichment analysis shown in panel A. Each node represents an enrichment term, their size represents the number of genes enriched within that molecular function. The color code represents each gene cluster.
Figure S4. A. Gene set enrichment plots for astrocyte mature markers derived from a custom gene set composed from 18 genes (Zhang et al., 2016). This set of genes was significantly enriched in the co-culture configuration AEN, but not in AN or AE, compared to astrocytes grown in monocultures. Of this gene set we found 10, 0, and 9 genes that were significantly upregulated in the AN, AE, AEN co-culture configurations when compared to astrocytes grown in monoculture. B. GSEA of the 678 genes identified as enriched in immature astrocytes (left graphs) or the 359 genes enriched in mature astrocytes (right graphs) (Lattke et al., 2021) in astrocytes grown on their own versus astrocytes grown in the presence of neurons (AN) or endothelia (AE). The normalized enrichment score (NES) and adjusted p values are shown. Each vertical black line in the X-axis represents a gene and indicates its position in the studied gene set. Genes on the far left are enriched in the gene set analyzed, while genes on the far right are underrepresented. The Y-axis represent the enrichment score.
Figure S5. A. Gene ontology biological process enrichment analysis of differentially spliced mRNAs in astrocytes grown as monocultures versus those of astrocytes grown in the presence of neurons and endothelial cells. The color code represents the adjusted p value. The size of the circles represents the gene ratio. The list with all gene ontology biological processes, the gene ratios, adjusted p values, and gene lists, are found in Table S3. B. Alternative splicing in three regions of the prominin 1 (Prom1) gene are shown from astrocytes isolated from monocultures (in purple), astrocytes grown in the presence of neurons (in pink), or endothelia (in green), or their combination (in blue). C. Sashimi plots representing the raw data of the number of reads mapped to the Prom1 gene in astrocytes grown in the four different culture configurations, same color code that panel B. The height of the traces represents the number of reads. The gray column highlights differentially spliced exons. The transcript model of the Prom1 gene in dark blue is from the genome browser Ensembl, boxes represent exons. EC= extracellular, IC1= intracellular region 1, IC2= intracellular region 2 are used to identify the areas in the Prom 1 protein affected by the alternative splicing events.
Table S1. List of all differentially expressed genes in the different culture configurations with full statistical report. Here we report 15,711 genes differentially regulated in the culture configurations. Results of the likelihood-ratio test (LRT) including base mean (column B), log2 fold changes (column C), and adjusted p values (column D) are shown. DESeq2 estimated differences between all culture configurations (columns E to P) are listed.
Table S2. Complete list of the gene ontology biological processes associated with the clusters identified in Figure 3B. Each cluster (cooperative: C1-C2, antagonistic: A1-A3, and redundant: R1-R2) is listed in a different sheet. Cluster, description of gene ontology biological process term, gene ratio (number of genes associated with the term divided by the total number of genes in that cluster), adjusted p-value, and the list of all genes associated with each biological process (gene ID) are listed.
Table S3. Complete list of the gene ontology biological processes and molecular functions associated with the differentially spliced mRNAs. Cluster, identifier, description of gene ontology biological process (BP sheet) or molecular functions (MF sheet) term, gene ratio (number of genes associated with the term over the total number of genes in that cluster), Bg ratio (number of genes included in the gene set that belong to the gene ontology term over the total number of genes identified in the gene set), p-value (raw), adjusted p-value, q value (false discovery rate adjusted p-value), the list of all genes associated with each biological process (gene ID), and count (number of genes differentially spliced associated in that gene ontology category) are listed.
SIGNIFICANCE STATEMENT.
Astrocytes have complex functions in central nervous system (CNS) health and disease. However, how astrocytes mature within the CNS environment remains poorly understood. We tested the hypothesis that neurons and endothelial cells cooperatively drive astrocytic maturation. By culturing astrocytes alone, with neurons, endothelial cells, or a combination of both, we found that these cells individually and together shifted the astrocyte transcriptome towards a mature state observed in vivo. Importantly, the combination of both cells had a larger effect on astrocytic maturation than either cell alone. Neurons and endothelial cells also antagonistically regulated expression of select astrocytic genes and had differential effects on mRNA splicing. These results demonstrate that extrinsic signals from neighboring cells help configure the molecular properties of astrocytes.
ACKNOWLEDGEMENTS:
This work was supported by NIH (R01 NS92067, R01 NS106693 to M.B.R.), Canadian Institutes of Health Research (PJT148569, 156247, and 180573 to K.K.M.), Natural Science and Engineering Research Council of Canada (69404 and RGPIN-2022–03395 to K.K.M.), a Joint Canada-Israel Research Program Award from IDRC/ISF/CIHR/Azrieli Foundation, The Children’s Hospital of Philadelphia (Bridge to Faculty fellowship to Z.M.L.) and Fonds de recherche du Québec - Santé (postdoctoral fellowship to A.S.). We would like to thank Dr. Judy Grinspan of the Preclinical Models Core of the Institutional Intellectual and Developmental Research Center at CHOP/Penn (U54 HD086984) for providing the A2B5 hybridoma supernatant, Sarah Woidill from the laboratory of Dr. Adeline Vanderver for help with the analysis of RNA samples in TapeStation, Renata Pellegrino Da Silva from the Center for Applied Genomics at CHOP and the Genomics and Data Integration Core of the Intellectual and Developmental Research Center (IDDRC) at CHOP/Penn (P50 105354) for her help with the qPCR using microfluidics, and Prof. Kai Wang of the same IDDRC core for his advice on statistical tests. We would also like to thank the staff from the Research Institute of the McGill University Health Centre Molecular Imaging Platform, Genome Quebec Centre D’expertise et de Services, and Canadian Centre for Computational Genomics.
Acknowledgments:
This work was funded by Fonds de recherche du Québec - Santé, (Grant / Award Number: ‘postdoctoral fellowship ‘)
Joint Canada-Israel Research Program Award from IDRC/ISF/CIHR/Azrieli Foundation, (Grant / Award Number:)
Natural Science and Engineering Research Council of Canada, (Grant / Award Number: ‘69404’,’RGPIN-2022–03395’)
Canadian Institutes of Health Research, (Grant / Award Number: ‘156247’,’180573’,’PJT148569’)
The Children’s Hospital of Philadelphia, (Grant / Award Number: ‘Bridge to Faculty Fellowship’)
NIH, (Grant / Award Number: ‘R01 NS106693 ‘,’R01 NS92067’) (grant number): This information is usually included already, but please add to the Acknowledgments if not.
Footnotes
if ‘none’, insert “The authors have no conflict of interest to declare.” else insert info unless it is already included
Conflict of Interest Statement: The authors declare no competing financial interests.
Human subjects: Involves human subjects:
If yes: Informed consent & ethics approval achieved:
if yes, please ensure that the info “Informed consent was achieved for all subjects, and the experiments were approved by the local ethics committee.” is included in the Methods.
Data availability.
Original code and data sets are publicly available as of the date of publication at GitHub (https://github.com/murailab/Astrocyte_Triple_Co-Culture) and at NCBI SRA (project number PRJNA961849).
References
- Abbott NJ (2002) Astrocyte-endothelial interactions and blood-brain barrier permeability. J Anat 200:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdoli Shadbad M, Hosseinkhani N, Asadzadeh Z, Brunetti O, Silvestris N, Baradaran B (2021) The Prognostic Value of CD133 in Predicting the Relapse and Recurrence Pattern of High-Grade Gliomas on MRI: A Meta-Analysis. Front Oncol 11:722833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NJ, Eroglu C (2017) Cell Biology of Astrocyte-Synapse Interactions. Neuron 96:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba H, Nakahira K, Morita N, Tanaka F, Akita H, Ikenaka K (1997) GFAP gene expression during development of astrocyte. Dev Neurosci 19:49–57. [DOI] [PubMed] [Google Scholar]
- Bannai S, Kitamura E (1980) Transport interaction of L-cystine and L-glutamate in human diploid fibroblasts in culture. J Biol Chem 255:2372–2376. [PubMed] [Google Scholar]
- Bar-Peled O, Ben-Hur H, Biegon A, Groner Y, Dewhurst S, Furuta A, Rothstein JD (1997) Distribution of glutamate transporter subtypes during human brain development. J Neurochem 69:2571–2580. [DOI] [PubMed] [Google Scholar]
- Bardehle S, Kruger M, Buggenthin F, Schwausch J, Ninkovic J, Clevers H, Snippert HJ, Theis FJ, Meyer-Luehmann M, Bechmann I, Dimou L, Gotz M (2013) Live imaging of astrocyte responses to acute injury reveals selective juxtavascular proliferation. Nat Neurosci 16:580–586. [DOI] [PubMed] [Google Scholar]
- Batiuk MY, de Vin F, Duque SI, Li C, Saito T, Saido T, Fiers M, Belgard TG, Holt MG (2017) An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J Biol Chem 292:8874–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiuk MY, Martirosyan A, Wahis J, de Vin F, Marneffe C, Kusserow C, Koeppen J, Viana JF, Oliveira JF, Voet T, Ponting CP, Belgard TG, Holt MG (2020) Identification of region-specific astrocyte subtypes at single cell resolution. Nat Commun 11:1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejoy J, Bijonowski B, Marzano M, Jeske R, Ma T, Li Y (2020) Wnt-Notch Signaling Interactions During Neural and Astroglial Patterning of Human Stem Cells. Tissue Eng Part A 26:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencharit S, Cui CB, Siddiqui A, Howard-Williams EL, Sondek J, Zuobi-Hasona K, Aukhil I (2007) Structural insights into fibronectin type III domain-mediated signaling. J Mol Biol 367:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzblau R, Gupta S, Djali S, Robinson MB, Rosenberg PA (1996) The glutamate transport inhibitor L-trans-pyrrolidine-2,4-dicarboxylate indirectly evokes NMDA receptor mediated neurotoxicity in rat cortical cultures. Eur J Neurosci 8:1840–1852. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME (1997) Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278:477–483. [DOI] [PubMed] [Google Scholar]
- Boulay AC, Saubamea B, Adam N, Chasseigneaux S, Mazare N, Gilbert A, Bahin M, Bastianelli L, Blugeon C, Perrin S, Pouch J, Ducos B, Le Crom S, Genovesio A, Chretien F, Decleves X, Laplanche JL, Cohen-Salmon M (2017) Translation in astrocyte distal processes sets molecular heterogeneity at the gliovascular interface. Cell Discov 3:17005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundesen LQ, Scheel TA, Bregman BS, Kromer LF (2003) Ephrin-B2 and EphB2 regulation of astrocyte-meningeal fibroblast interactions in response to spinal cord lesions in adult rats. J Neurosci 23:7789–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH (2002) Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci 22:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir B et al. (2019) Engineering of human brain organoids with a functional vascular-like system. Nat Methods 16:1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese B, Jones SL, Shiraishi-Yamaguchi Y, Lingelbach M, Manor U, Svitkina TM, Higgs HN, Shih AY, Halpain S (2022) INF2-mediated actin filament reorganization confers intrinsic resilience to neuronal ischemic injury. Nat Commun 13:6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camassa LMA, Lunde LK, Hoddevik EH, Stensland M, Boldt HB, De Souza GA, Ottersen OP, Amiry-Moghaddam M (2015) Mechanisms underlying AQP4 accumulation in astrocyte endfeet. Glia 63:2073–2091. [DOI] [PubMed] [Google Scholar]
- Carlson M (2019) org.Mm.eg.db: Genome wide annotation for Mouse. In: R package, 3.8.2 Edition, pp Genome wide annotation for Mouse, primarily based on mapping using Entrez Gene identifiers. [Google Scholar]
- Charo IF, Ransohoff RM (2006) The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med 354:610–621. [DOI] [PubMed] [Google Scholar]
- Chaudhry FA, Lehre KP, van Lookeren Campagne M, Ottersen OP, Danbolt NC, Storm-Mathisen J (1995) Glutamate transporters in glial plasma membranes: highly differentiated localizations revealed by quantitative ultrastructural immunocytochemistry. Neuron 15:711–720. [DOI] [PubMed] [Google Scholar]
- Chierzi S, Kacerovsky JB, Fok AH, Lahaie S, Shibi-Rosen A, Farmer WT, Murai KK (2023) Astrocytes transplanted during early postnatal development integrate, mature, and survive long-term in mouse cortex. J Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibelli A, Veronica Lopez-Quintero S, McCutcheon S, Scemes E, Spray DC, Stout RF Jr., Suadicani SO, Thi MM, Urban-Maldonado M (2021) Generation and Characterization of Immortalized Mouse Cortical Astrocytes From Wildtype and Connexin43 Knockout Mice. Front Cell Neurosci 15:647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E, Kriegstein AR (2017) The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci 18:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter MA (1978) Rat cortical neurons in cell culture: culture methods, cell morphology, electrophysiology, and synapse formation. Brain Res 149:279–293. [DOI] [PubMed] [Google Scholar]
- Dinndorf PA, Gootenberg J, Cohen MH, Keegan P, Pazdur R (2007) FDA drug approval summary: pegaspargase (oncaspar) for the first-line treatment of children with acute lymphoblastic leukemia (ALL). Oncologist 12:991–998. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Polydorides AD, Darnell RB (2001) The splice of life: alternative splicing and neurological disease. Nat Rev Neurosci 2:43–50. [DOI] [PubMed] [Google Scholar]
- Dunlop J, Beal McIlvain H, She Y, Howland DS (2003) Impaired spinal cord glutamate transport capacity and reduced sensitivity to riluzole in a transgenic superoxide dismutase mutant rat model of amyotrophic lateral sclerosis. J Neurosci 23:1688–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W (2009) Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst D, Westerbergh J, Sogkas G, Jablonka A, Ahrenstorf G, Schmidt RE, Heidecke H, Wallentin L, Riemekasten G, Witte T (2019) Lowered anti-beta1 adrenergic receptor antibody concentrations may have prognostic significance in acute coronary syndrome. Sci Rep 9:14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escartin C et al. (2021) Reactive astrocyte nomenclature, definitions, and future directions. Nat Neurosci 24:312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrig T, Sommermeyer H (1993) Dibutyryl cyclic AMP-induced morphological differentiation of rat brain astrocytes increases alpha 1-adrenoceptor induced phosphoinositide breakdown by a mechanism involving protein synthesis. Brain Res 602:318–324. [DOI] [PubMed] [Google Scholar]
- Farhy-Tselnicker I, Boisvert MM, Liu H, Dowling C, Erikson GA, Blanco-Suarez E, Farhy C, Shokhirev MN, Ecker JR, Allen NJ (2021) Activity-dependent modulation of synapse-regulating genes in astrocytes. Elife 10:e70514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WT, Abrahamsson T, Chierzi S, Lui C, Zaelzer C, Jones EV, Bally BP, Chen GG, Theroux JF, Peng J, Bourque CW, Charron F, Ernst C, Sjostrom PJ, Murai KK (2016) Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science 351:849–854. [DOI] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, Cahoy JD, Daneman R, Zong H, Ellisman MH, Barres BA (2011) Development of a method for the purification and culture of rodent astrocytes. Neuron 71:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freist W, Verhey JF, Ruhlmann A, Gauss DH, Arnez JG (1999) Histidyl-tRNA synthetase. Biol Chem 380:623–646. [DOI] [PubMed] [Google Scholar]
- Furuta A, Rothstein JD, Martin LJ (1997) Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J Neurosci 17:8363–8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher ER, Holzbaur ELF (2023) The selective autophagy adaptor p62/SQSTM1 forms phase condensates regulated by HSP27 that facilitate the clearance of damaged lysosomes via lysophagy. Cell Rep 42:112037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegelashvili G, Danbolt NC, Schousboe A (1997) Neuronal soluble factors differentially regulate the expression of the GLT1 and GLAST glutamate transporters in cultured astroglia. J Neurochem 69:2612–2615. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Yang Y, Rothstein JD, Robinson MB (2011) Nuclear factor-kappaB contributes to neuron-dependent induction of glutamate transporter-1 expression in astrocytes. J Neurosci 31:9159–9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Lane M, Krizman E, Sattler R, Rothstein JD, Robinson MB (2016) The transcription factor Pax6 contributes to the induction of GLT-1 expression in astrocytes through an interaction with a distal enhancer element. J Neurochem 136:262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross RE, Mehler MF, Mabie PC, Zang Z, Santschi L, Kessler JA (1996) Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron 17:595–606. [DOI] [PubMed] [Google Scholar]
- Guerit S, Fidan E, Macas J, Czupalla CJ, Figueiredo R, Vijikumar A, Yalcin BH, Thom S, Winter P, Gerhardt H, Devraj K, Liebner S (2021) Astrocyte-derived Wnt growth factors are required for endothelial blood-brain barrier maintenance. Prog Neurobiol 199:101937. [DOI] [PubMed] [Google Scholar]
- Harada H, Nagai H, Mine N, Terada Y, Fujiwara H, Mikami I, Tsuneizumi M, Yabe A, Miyazaki K, Yokota T, Imoto I, Inazawa J, Emi M (2001) Molecular cloning, tissue expression, and chromosomal assignment of a novel gene encoding a subunit of the human signal-recognition particle. J Hum Genet 46:70–75. [DOI] [PubMed] [Google Scholar]
- Hasel P, Dando O, Jiwaji Z, Baxter P, Todd AC, Heron S, Markus NM, McQueen J, Hampton DW, Torvell M, Tiwari SS, McKay S, Eraso-Pichot A, Zorzano A, Masgrau R, Galea E, Chandran S, Wyllie DJA, Simpson TI, Hardingham GE (2017) Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nat Commun 8:15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff BP, George KK, Phares AN, Zuidhoek IA, Munoz-Ballester C, Robel S (2021) Astrocytes are necessary for blood-brain barrier maintenance in the adult mouse brain. Glia 69:436–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SA, Fu M, Garcia ADR (2021) Sonic hedgehog signaling in astrocytes. Cell Mol Life Sci 78:e45545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SA, Blaeser AS, Coley AA, Xie Y, Shepard KA, Harwell CC, Gao WJ, Garcia ADR (2019) Sonic hedgehog signaling in astrocytes mediates cell type-specific synaptic organization. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosli L, Zuend M, Bredell G, Zanker HS, Porto de Oliveira CE, Saab AS, Weber B (2022) Direct vascular contact is a hallmark of cerebral astrocytes. Cell Rep 39:110599. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Lillien LE, Raff MC, Rohrer H, Sendtner M (1988) Ciliary neurotrophic factor induces type-2 astrocyte differentiation in culture. Nature 335:70–73. [DOI] [PubMed] [Google Scholar]
- Jackson JG, Robinson MB (2018) Regulation of mitochondrial dynamics in astrocytes: Mechanisms, consequences, and unknowns. Glia 66:1213–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzer RC, Raff MC (1987) Astrocytes induce blood-brain barrier properties in endothelial cells. Nature 325:253–257. [DOI] [PubMed] [Google Scholar]
- Kacem K, Lacombe P, Seylaz J, Bonvento G (1998) Structural organization of the perivascular astrocyte endfeet and their relationship with the endothelial glucose transporter: a confocal microscopy study. Glia 23:1–10. [PubMed] [Google Scholar]
- Kantzer CG, Boutin C, Herzig ID, Wittwer C, Reiss S, Tiveron MC, Drewes J, Rockel TD, Ohlig S, Ninkovic J, Cremer H, Pennartz S, Jungblut M, Bosio A (2017) Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia 65:990–1004. [DOI] [PubMed] [Google Scholar]
- Korotkevich GS V; Budin N; Shpak B; Artyomov MN; Sergushichev A (2019) Fast gene set enrichment analysis. bioRxiv. [Google Scholar]
- Kriegstein A, Dichter MA (1984) Neuron generation in dissociated cell cultures from fetal rat cerebral cortex. Brain Res 295:184–189. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Dichter MA (1983) Morphological classification of rat cortical neurons in cell culture. J Neurosci 3:1634–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjakornsiripan D, Pior BJ, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, Suzuki Y, Fukazawa Y, Gotoh Y (2018) Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat Commun 9:1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionova TD et al. (2022) Alternative RNA splicing modulates ribosomal composition and determines the spatial phenotype of glioblastoma cells. Nat Cell Biol 24:1541–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattke M, Goldstone R, Ellis JK, Boeing S, Jurado-Arjona J, Marichal N, MacRae JI, Berninger B, Guillemot F (2021) Extensive transcriptional and chromatin changes underlie astrocyte maturation in vivo and in culture. Nat Commun 12:4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ML, Martinez-Lozada Z, Krizman EN, Robinson MB (2017) Brain endothelial cells induce astrocytic expression of the glutamate transporter GLT-1 by a Notch-dependent mechanism. J Neurochem 143:489–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehre KP, Levy LM, Ottersen OP, Storm-Mathisen J, Danbolt NC (1995) Differential expression of two glial glutamate transporters in the rat brain: quantitative and immunocytochemical observations. J Neurosci 15:1835–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M, Smith SB, Ganapathy V, Maher P (2013) The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal 18:522–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, McIntosh CS, Mastaglia FL, Wilton SD, Aung-Htut MT (2021) Neurodegenerative diseases: a hotbed for splicing defects and the potential therapies. Transl Neurodegener 10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Zhu L, Mu B, Jia X, Wang J, Mu P (2021) Down-regulated in renal cell carcinoma 1 (DRR1) regulates axon outgrowth during hippocampal neuron development. Biochem Biophys Res Commun 558:36–43. [DOI] [PubMed] [Google Scholar]
- Lyons SA, Chung WJ, Weaver AK, Ogunrinu T, Sontheimer H (2007) Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res 67:9463–9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhou Y, Li Z, Zhu L, Li H, Zhang G, Wang J, Gong H, Xu D, Hua W, Liu P, Zhang X, Zhang Y, Zhang L, Hong B, Zhou W, Yang P, Liu J (2022) Single-Cell RNA-Sequencing Analyses Revealed Heterogeneity and Dynamic Changes of Metabolic Pathways in Astrocytes at the Acute Phase of Ischemic Stroke. Oxid Med Cell Longev 2022:1817721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lozada Z, Robinson MB (2020) Reciprocal communication between astrocytes and endothelial cells is required for astrocytic glutamate transporter 1 (GLT-1) expression. Neurochem Int 139:104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masana M, Su YA, Liebl C, Wang XD, Jansen L, Westerholz S, Wagner KV, Labermaier C, Scharf SH, Santarelli S, Hartmann J, Schmidt MV, Rein T, Muller MB (2014) The stress-inducible actin-interacting protein DRR1 shapes social behavior. Psychoneuroendocrinology 48:98–110. [DOI] [PubMed] [Google Scholar]
- Mathiisen TM, Lehre KP, Danbolt NC, Ottersen OP (2010) The perivascular astroglial sheath provides a complete covering of the brain microvessels: an electron microscopic 3D reconstruction. Glia 58:1094–1103. [DOI] [PubMed] [Google Scholar]
- Matsutani S, Yamamoto N (1997) Neuronal regulation of astrocyte morphology in vitro is mediated by GABAergic signaling. Glia 20:1–9. [DOI] [PubMed] [Google Scholar]
- Mazare N, Oudart M, Moulard J, Cheung G, Tortuyaux R, Mailly P, Mazaud D, Bemelmans AP, Boulay AC, Blugeon C, Jourdren L, Le Crom S, Rouach N, Cohen-Salmon M (2020) Local Translation in Perisynaptic Astrocytic Processes Is Specific and Changes after Fear Conditioning. Cell Rep 32:108076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin PV, Khaitovich P, Cardoso-Moreira M, Kaessmann H (2021) Alternative splicing during mammalian organ development. Nat Genet 53:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MG, Nyanyo D, Hung CK, Goerger JP, W RZ, Williams RM, Nishimura N, Fischbach C (2019) Endothelial cells promote 3D invasion of GBM by IL-8-dependent induction of cancer stem cell properties. Sci Rep 9:9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Haeberle H, Barres BA (2001) Induction of astrocyte differentiation by endothelial cells. J Neurosci 21:1538–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SJ et al. (2019) Molecularly defined cortical astroglia subpopulation modulates neurons via secretion of Norrin. Nat Neurosci 22:741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JD, Janitz M (2012) Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol Aging 33:1012 e1011–1024. [DOI] [PubMed] [Google Scholar]
- Minelli A, Barbaresi P, Conti F (2003) Postnatal development of high-affinity plasma membrane GABA transporters GAT-2 and GAT-3 in the rat cerebral cortex. Brain Res Dev Brain Res 142:7–18. [DOI] [PubMed] [Google Scholar]
- Minelli A, DeBiasi S, Brecha NC, Zuccarello LV, Conti F (1996) GAT-3, a high-affinity GABA plasma membrane transporter, is localized to astrocytic processes, and it is not confined to the vicinity of GABAergic synapses in the cerebral cortex. J Neurosci 16:6255–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Deneen B (2015) Astrocyte development: A Guide for the Perplexed. Glia 63:1320–1329. [DOI] [PubMed] [Google Scholar]
- Pagès HC M; Falcon S; Li N (2023) AnnotationDbi: Manipulation of SQLite-based annotations in Bioconductor. In: R package 1.60.2 Edition. [Google Scholar]
- Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, Wilcox S, Fu N, Liu KH, Jackling FC, Davis MJ, Lindeman GJ, Smyth GK, Visvader JE (2017) Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat Commun 8:1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Pekna M (2014) Astrocyte reactivity and reactive astrogliosis: costs and benefits. Physiol Rev 94:1077–1098. [DOI] [PubMed] [Google Scholar]
- Petzold GC, Murthy VN (2011) Role of astrocytes in neurovascular coupling. Neuron 71:782–797. [DOI] [PubMed] [Google Scholar]
- Pivoriunas A, Verkhratsky A (2021) Astrocyte-Endotheliocyte Axis in the Regulation of the Blood-Brain Barrier. Neurochem Res 46:2538–2550. [DOI] [PubMed] [Google Scholar]
- Pu Y, Zhao L, Xi Y, Xia Y, Qian Y (2022) The protective effects of Mirtazapine against lipopolysaccharide (LPS)-induced brain vascular hyperpermeability. Bioengineered 13:3680–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F (2022) Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 38:2943–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Cui EH, Ji CM, Zhang XB, Wang ZA, Sun YZ, Xu JC, Zhai XF, Chen ZJ, Li J, Zheng JY, Yu RT (2019) Specific knockdown of hippocampal astroglial EphB2 improves synaptic function via inhibition of D-serine secretion in APP/PS1 mice. Am J Transl Res 11:1073–1083. [PMC free article] [PubMed] [Google Scholar]
- Qian X, Song H, Ming GL (2019) Brain organoids: advances, applications and challenges. Development 146:dev166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Dando O, Baxter PS, Hasel P, Heron S, Simpson TI, Hardingham GE (2018) Mixed-species RNA-seq for elucidation of non-cell-autonomous control of gene transcription. Nat Protoc 13:2176–2199. [DOI] [PubMed] [Google Scholar]
- Raff MC, Abney ER, Cohen J, Lindsay R, Noble M (1983) Two types of astrocytes in cultures of developing rat white matter: differences in morphology, surface gangliosides, and growth characteristics. J Neurosci 3:1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainer J (2017) EnsDb.Mmusculus.v79: Ensembl based annotation package R package version 2990. [Google Scholar]
- Rainer J, Gatto L, Weichenberger CX (2019) ensembldb: an R package to create and use Ensembl-based annotation resources. Bioinformatics 35:3151–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD (2007) Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci 27:6607–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert SM, Buckingham SC, Campbell SL, Robel S, Holt KT, Ogunrinu-Babarinde T, Warren PP, White DM, Reid MA, Eschbacher JM, Berens ME, Lahti AC, Nabors LB, Sontheimer H (2015) SLC7A11 expression is associated with seizures and predicts poor survival in patients with malignant glioma. Sci Transl Med 7:289ra286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakers K, Lake AM, Khazanchi R, Ouwenga R, Vasek MJ, Dani A, Dougherty JD (2017) Astrocytes locally translate transcripts in their peripheral processes. Proc Natl Acad Sci U S A 114:E3830–E3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakers K, Liu Y, Llaci L, Lee SM, Vasek MJ, Rieger MA, Brophy S, Tycksen E, Lewis R, Maloney SE, Dougherty JD (2021) Loss of Quaking RNA binding protein disrupts the expression of genes associated with astrocyte maturation in mouse brain. Nat Commun 12:1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santello M, Toni N, Volterra A (2019) Astrocyte function from information processing to cognition and cognitive impairment. Nat Neurosci 22:154–166. [DOI] [PubMed] [Google Scholar]
- Sasson E, Anzi S, Bell B, Yakovian O, Zorsky M, Deutsch U, Engelhardt B, Sherman E, Vatine G, Dzikowski R, Ben-Zvi A (2021) Nano-scale architecture of blood-brain barrier tight-junctions. Elife 10:e63253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Kuriyama-Matsumura K, Siow RC, Ishii T, Bannai S, Mann GE (1998) Induction of cystine transport via system x-c and maintenance of intracellular glutathione levels in pancreatic acinar and islet cell lines. Biochim Biophys Acta 1414:85–94. [DOI] [PubMed] [Google Scholar]
- Schlag BD, Vondrasek JR, Munir M, Kalandadze A, Zelenaia OA, Rothstein JD, Robinson MB (1998) Regulation of the glial Na+-dependent glutamate transporters by cyclic AMP analogs and neurons. Mol Pharmacol 53:355–369. [DOI] [PubMed] [Google Scholar]
- Schmidt MV, Schulke JP, Liebl C, Stiess M, Avrabos C, Bock J, Wochnik GM, Davies HA, Zimmermann N, Scharf SH, Trumbach D, Wurst W, Zieglgansberger W, Turck C, Holsboer F, Stewart MG, Bradke F, Eder M, Muller MB, Rein T (2011) Tumor suppressor down-regulated in renal cell carcinoma 1 (DRR1) is a stress-induced actin bundling factor that modulates synaptic efficacy and cognition. Proc Natl Acad Sci U S A 108:17213–17218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober AL, Wicki-Stordeur LE, Murai KK, Swayne LA (2022) Foundations and implications of astrocyte heterogeneity during brain development and disease. Trends Neurosci 45:692–703. [DOI] [PubMed] [Google Scholar]
- Scholze AR, Foo LC, Mulinyawe S, Barres BA (2014) BMP signaling in astrocytes downregulates EGFR to modulate survival and maturation. PLoS One 9:e110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer JE, Patel RS, Fonda D, Hynes RO (1987) Multiple sites of alternative splicing of the rat fibronectin gene transcript. EMBO J 6:2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS (2016) RNA mis-splicing in disease. Nat Rev Genet 17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Zhang T, Fan K, Cai W, Liu H (2021) Astrocyte-Neuron Signaling in Synaptogenesis. Front Cell Dev Biol 9:680301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 111:E5593–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Watanabe M, Tanaka K, Wada K, Inoue Y (1996) Dynamic changes in expression of glutamate transporter mRNAs in developing brain. Neuroreport 7:705–709. [DOI] [PubMed] [Google Scholar]
- Sims KD, Robinson MB (1999) Expression patterns and regulation of glutamate transporters in the developing and adult nervous system. Crit Rev Neurobiol 13:169–197. [DOI] [PubMed] [Google Scholar]
- Sorensen MF, Heimisdottir SB, Sorensen MD, Mellegaard CS, Wohlleben H, Kristensen BW, Beier CP (2018) High expression of cystine-glutamate antiporter xCT (SLC7A11) is an independent biomarker for epileptic seizures at diagnosis in glioma. J Neurooncol 138:49–53. [DOI] [PubMed] [Google Scholar]
- Soudy M, Anwar AM, Ahmed EA, Osama A, Ezzeldin S, Mahgoub S, Magdeldin S (2020) UniprotR: Retrieving and visualizing protein sequence and functional information from Universal Protein Resource (UniProt knowledgebase). J Proteomics 213:103613. [DOI] [PubMed] [Google Scholar]
- Souza DG, Almeida RF, Souza DO, Zimmer ER (2019) The astrocyte biochemistry. Semin Cell Dev Biol 95:142–150. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, Khakh BS (2016) New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron 92:1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackhouse TL, Mishra A (2021) Neurovascular Coupling in Development and Disease: Focus on Astrocytes. Front Cell Dev Biol 9:702832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatovic SM, Keep RF, Wang MM, Jankovic I, Andjelkovic AV (2009) Caveolae-mediated internalization of occludin and claudin-5 during CCL2-induced tight junction remodeling in brain endothelial cells. J Biol Chem 284:19053–19066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M (2017) False discovery rates: a new deal. Biostatistics 18:275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Sheehan A, Tasdemir-Yilmaz OE, Freeman MR (2014) Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83:388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CH, D D, Tarn WY (2018) Alternative Splicing in Neurogenesis and Brain Development. Front Mol Biosci 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian AR (1983) Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol 28:101–142. [DOI] [PubMed] [Google Scholar]
- Sun XY, Ju XC, Li Y, Zeng PM, Wu J, Zhou YY, Shen LB, Dong J, Chen YJ, Luo ZG (2022) Generation of vascularized brain organoids to study neurovascular interactions. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RA, Liu J, Miller JW, Rothstein JD, Farrell K, Stein BA, Longuemare MC (1997) Neuronal regulation of glutamate transporter subtype expression in astrocytes. J Neurosci 17:932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H (2015) Diverse subtypes of astrocytes and their development during corticogenesis. Front Neurosci 9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CX, Burrus Lane CJ, Eroglu C (2021) Role of astrocytes in synapse formation and maturation. Curr Top Dev Biol 142:371–407. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullensvang K, Lehre KP, Storm-Mathisen J, Danbolt NC (1997) Differential developmental expression of the two rat brain glutamate transporter proteins GLAST and GLT. Eur J Neurosci 9:1646–1655. [DOI] [PubMed] [Google Scholar]
- Veiga DFT (2022) maser: Mapping Alternative Splicing Events to pRoteins. . R package version 1121. [Google Scholar]
- Wang Y, Liu J, Huang BO, Xu YM, Li J, Huang LF, Lin J, Zhang J, Min QH, Yang WM, Wang XZ (2015) Mechanism of alternative splicing and its regulation. Biomed Rep 3:152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White ES, Baralle FE, Muro AF (2008) New insights into form and function of fibronectin splice variants. J Pathol 216:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G (2021) clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kuan AT, Wang W, Herbert ZT, Mosto O, Olukoya O, Adam M, Vu S, Kim M, Tran D, Gomez N, Charpentier C, Sorour I, Lacey TE, Tolstorukov MY, Sabatini BL, Lee WA, Harwell CC (2022) Astrocyte-neuron crosstalk through Hedgehog signaling mediates cortical synapse development. Cell Rep 38:110416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gozen O, Watkins A, Lorenzini I, Lepore A, Gao Y, Vidensky S, Brennan J, Poulsen D, Won Park J, Li Jeon N, Robinson MB, Rothstein JD (2009) Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron 61:880–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder EJ (2002) Modifications in astrocyte morphology and calcium signaling induced by a brain capillary endothelial cell line. Glia 38:137–145. [DOI] [PubMed] [Google Scholar]
- Zelenaia OA, Robinson MB (2000) Degradation of glial glutamate transporter mRNAs is selectively blocked by inhibition of cellular transcription. J Neurochem 75:2252–2258. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA 3rd, Cheshier SH, Shuer LM, Chang EF, Grant GA, Gephart MG, Barres BA (2016) Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Ye Z, Stanton R (2020) Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 26:903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Graphical Timeline. A. Mouse astrocytes were cultured by themselves, in presence of neurons, endothelia, or their combination. After 10 days in culture cells were harvested for immunocytochemistry immunofluorescence (ICC-IF) or dissociated and labelled with anti-astrocyte cell surface antigen 2 (ACSA-2) antibodies or anti-Pecam1 antibodies and separated using fluorescence activated cell sorting (FACS). RNA was then isolated from the sorted astrocytes. RNA was analyzed by qPCR or sequenced. B. Cortical tissue was isolated from E19 and P28 mice, resolved in a western blot (WB) and probed with anti-Fibronectin antibodies. C. Coronal brain sections were obtained from P4, P7, and P28 mice and immunohistochemistry immunofluorescence (IHC-IF) was performed for GAT3, FAM107A, and GLT1. See methods sections for details.
Figure S2. A. Pipeline schematic of RNA sequencing analysis. See methods for steps description. All code that processed the data can be found at GitHub (https://github.com/murailab/Astrocyte_Triple_Co-Culture). B. To estimate the percent composition of endothelia in each sample we used the average read counts for each gene in pure astrocytes and pure endothelial samples. As expected, A and AN do not contain any endothelial-specific sequences. Although we set conservative gates (see Figure 1C), we estimated 12% and 6% of endothelial content in the astrocytes isolated from the AE and the AEN co-cultures respectively. C. Correlation of RNAseq uncorrected log2-fold differences (Y-axis) versus quantitative PCR ddCt data (X-axis).
Figure S3. A. Gene ontology molecular function enrichment analysis of differentially expressed genes in the clusters assigned in Figure 3B. The color code represents the adjusted p value. The size of the circle represents the gene ratio. B. Associated networks for the molecular function enrichment analysis shown in panel A. Each node represents an enrichment term, their size represents the number of genes enriched within that molecular function. The color code represents each gene cluster.
Figure S4. A. Gene set enrichment plots for astrocyte mature markers derived from a custom gene set composed from 18 genes (Zhang et al., 2016). This set of genes was significantly enriched in the co-culture configuration AEN, but not in AN or AE, compared to astrocytes grown in monocultures. Of this gene set we found 10, 0, and 9 genes that were significantly upregulated in the AN, AE, AEN co-culture configurations when compared to astrocytes grown in monoculture. B. GSEA of the 678 genes identified as enriched in immature astrocytes (left graphs) or the 359 genes enriched in mature astrocytes (right graphs) (Lattke et al., 2021) in astrocytes grown on their own versus astrocytes grown in the presence of neurons (AN) or endothelia (AE). The normalized enrichment score (NES) and adjusted p values are shown. Each vertical black line in the X-axis represents a gene and indicates its position in the studied gene set. Genes on the far left are enriched in the gene set analyzed, while genes on the far right are underrepresented. The Y-axis represent the enrichment score.
Figure S5. A. Gene ontology biological process enrichment analysis of differentially spliced mRNAs in astrocytes grown as monocultures versus those of astrocytes grown in the presence of neurons and endothelial cells. The color code represents the adjusted p value. The size of the circles represents the gene ratio. The list with all gene ontology biological processes, the gene ratios, adjusted p values, and gene lists, are found in Table S3. B. Alternative splicing in three regions of the prominin 1 (Prom1) gene are shown from astrocytes isolated from monocultures (in purple), astrocytes grown in the presence of neurons (in pink), or endothelia (in green), or their combination (in blue). C. Sashimi plots representing the raw data of the number of reads mapped to the Prom1 gene in astrocytes grown in the four different culture configurations, same color code that panel B. The height of the traces represents the number of reads. The gray column highlights differentially spliced exons. The transcript model of the Prom1 gene in dark blue is from the genome browser Ensembl, boxes represent exons. EC= extracellular, IC1= intracellular region 1, IC2= intracellular region 2 are used to identify the areas in the Prom 1 protein affected by the alternative splicing events.
Table S1. List of all differentially expressed genes in the different culture configurations with full statistical report. Here we report 15,711 genes differentially regulated in the culture configurations. Results of the likelihood-ratio test (LRT) including base mean (column B), log2 fold changes (column C), and adjusted p values (column D) are shown. DESeq2 estimated differences between all culture configurations (columns E to P) are listed.
Table S2. Complete list of the gene ontology biological processes associated with the clusters identified in Figure 3B. Each cluster (cooperative: C1-C2, antagonistic: A1-A3, and redundant: R1-R2) is listed in a different sheet. Cluster, description of gene ontology biological process term, gene ratio (number of genes associated with the term divided by the total number of genes in that cluster), adjusted p-value, and the list of all genes associated with each biological process (gene ID) are listed.
Table S3. Complete list of the gene ontology biological processes and molecular functions associated with the differentially spliced mRNAs. Cluster, identifier, description of gene ontology biological process (BP sheet) or molecular functions (MF sheet) term, gene ratio (number of genes associated with the term over the total number of genes in that cluster), Bg ratio (number of genes included in the gene set that belong to the gene ontology term over the total number of genes identified in the gene set), p-value (raw), adjusted p-value, q value (false discovery rate adjusted p-value), the list of all genes associated with each biological process (gene ID), and count (number of genes differentially spliced associated in that gene ontology category) are listed.
Data Availability Statement
All code used for data processing and for generating figures can be found on GitHub (https://github.com/murailab/Astrocyte_Triple_Co-Culture). The trimmed sequence reads can be found on the NCBI SRA (www.ncbi.nlm.nih.gov/bioproject/PRJNA961849).
Original code and data sets are publicly available as of the date of publication at GitHub (https://github.com/murailab/Astrocyte_Triple_Co-Culture) and at NCBI SRA (project number PRJNA961849).


