Abstract
There has been a resurgence of interest in bioactive peptides as therapeutic agents. This is particularly interesting for tyrosinase, which can be inhibited by thiol-containing peptides. This work demonstrates that an N-terminal cysteine-containing tetrapeptide can be rationally designed to inhibit tyrosinase activity in vitro and in cells. The tetrapeptide cysteine (C), arginine (R), asparagine (N) and leucine (L) or CRNL is a potent inhibitor of tyrosinase activity with an IC50 value of 39.62 ± 6.21 μM, which is comparable to currently used tyrosinase inhibitors. Through structure-activity studies and computational modeling, we demonstrate the peptide interacts with the enzyme via electrostatic (R with E322), hydrogen bonding (N with N260) and hydrophobic (L with V248) intermolecular interactions and that a combination of these is required for potent activity. Moreover, copper chelating activity might be one of the mechanisms of tyrosinase inhibition by CRNL. Kinetic studies show that tetrapeptide is a competitive inhibitor with two-step irreversible inhibition. In addition, CRNL had no toxicity and could reduce melanin levels in the murine melanoma cell line (B16F1). Overall, CRNL is a very promising candidate for hyperpigmentation treatment.
Keywords: Depigmentation, Enzyme kinetics, Modified peptide, Molecular docking, Tyrosinase inhibitor, Tetrapeptide
Graphical abstract

Highlights
-
•
All designed N-terminal cysteine-containing tetrapeptides exhibited potent tyrosinase inhibitory activity.
-
•
Electrostatic, hydrogen bonding, and hydrophobic interactions were essential for CRNL tetrapeptide's tyrosinase inhibition.
-
•
Kinetic studies showed that the CRNL tetrapeptide was a competitive inhibitor of tyrosinase.
-
•
The CRNL tetrapeptide was not toxic and could reduce melanin levels in B16F1 cells.
1. Introduction
Melanin is a pigment produced in plants, microorganisms, and animals (Alghamdi et al., 2023). In skin, its role is to protect the skin from ultraviolet (UV) radiation damage (Shen et al., 2019; Nazir et al., 2022). However, the overproduction and abnormal accumulation of melanin can result in dark skin, melasma, freckles, lentigo and Riehl melanosis (Nazir et al., 2022). Melanin synthesis is also associated with enzymic browning, which turns food brown and can affect food quality (Moon et al., 2020). Hence, the food and cosmetic industries have tried to control melanin synthesis to prevent food discoloration and skin pigmentation (Shin et al., 2023).
Tyrosinase is a key rate-limiting enzyme in melanogenesis (Joompang et al., 2022; Nazir et al., 2022), the process that creates melanin. As a consequence, inhibitors of this enzyme are key for hyperpigmentation treatment (Lee et al., 2015; Joompang et al., 2020, 2022; Nazir et al., 2022). Known tyrosinase inhibitors such as kojic acid and α-arbutin are widely used in cosmetics, however they can cause skin allergies, inflammation, and other adverse side effects (Curto et al., 1999; DeCaprio, 1999; Hermanns et al., 2000; Parvez et al., 2006; Lee et al., 2015). For example, kojic acid has shown tumorigenic potential in the liver of CBA mice (Takizawa et al., 2003), while arbutin can be metabolized to yield hydroquinone which is potentially carcinogenic (Blaut et al., 2006). Moreover, genetic variation in the human population means the potency of these inhibitors varies from individual to individual (Bachtiar and Lee, 2013). Therefore, the identification of new tyrosinase inhibitors is crucial to improve hyperpigmentation therapy.
Peptides have attracted interest due to their potency and more importantly their minimal adverse side effects (Ubeid et al., 2009, 2012). As a result, tyrosinase inhibitor peptides from both natural peptides (Ateş et al., 2001; Kubglomsong et al., 2018) and artificial peptide mimetics (Ubeid et al., 2009; Hsiao et al., 2014; Ochiai et al., 2016) have been reported and characterized mechanistically. They bind to the catalytic copper active site and interact with nearby residues (Ubeid et al., 2012; Lee et al., 2015; Ochiai et al., 2016). While some information is known on how the amino acid sequence and composition affects tyrosinase inhibition, the studies are not comprehensive and more information is vital to improve peptide inhibitors of tyrosinase.
Herein, we rationally design a tyrosinase inhibiting tetrapeptide (CRNL) that binds to the tyrosinase active site. The potency and mechanism of action are evaluated by biochemical, computational and cellular studies. Our study shows that the peptide is a strong inhibitor of tyrosinase through electrostatic and hydrophobic interactions, and has comparable activity to kojic acid. Most importantly, the peptide is non-toxic in cells and can reduce melanin levels suggesting it could be used in hyperpigmentation treatments. Our studies also provide a rational approach to designing other peptides that strongly inhibit enzymes.
2. Materials and methods
2.1. Materials and reagents
3,4-Dihydroxy-L-phenylalanine, dimethyl sulfoxide (DMSO), mushroom tyrosinase, and pyrocatechol violet were purchased from Sigma-Aldrich (St. Louis, MO, USA). Kojic acid was purchased from TCI (Tokyo, Japan). 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) was purchased from Sigma-Aldrich (Eugene, OR, USA). Dulbecco's modified Eagle's medium (DMEM), Fetal bovine serum (FBS), penicillin/streptomycin and L-glutamine were purchased from Lonza (Walkersville, MD, USA). Trypsin-EDTA was purchased from Corning Inc. (Manassas, VA, USA).
2.2. Tetrapeptide design
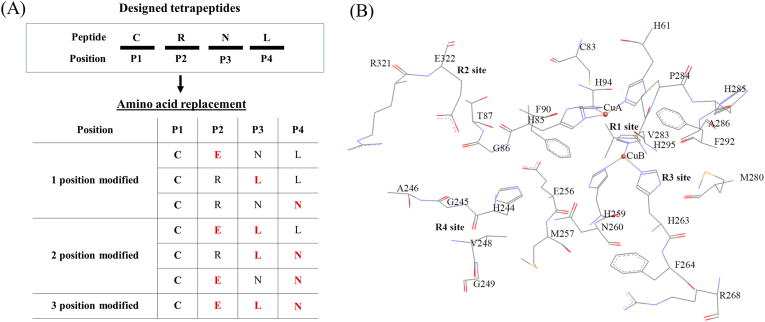
Our designed peptide is expected to interact with amino acids/catalytic copper atoms in the active site of tyrosinase, which has four key regions in close proximity (Fig. 1): R1 (a dinuclear copper complex that interacts with six histidine residues); R2 (contains negatively charged residues); R3 (contains polar residues) and R4 (contains hydrophobic residues). As a consequence, we decided to design a peptide with four amino acids. We refer to each position of the tetrapeptide from N-terminus to C-terminus as P1, P2, P3 and P4 and assigned amino acids to these positions that form complementary interactions with the four positions of the enzyme active site – cysteine (C, P1, metal co-ordination interactions with R1), arginine (R, P2, electrostatic interactions with R2), asparagine (N, P3, polar interactions with R3) and leucine (L, P4, hydrophobic interactions with R4). To study structure-activity relationships, amino acids at positions P2, P3 and P4 were replaced with glutamic acid (E), leucine (L) or asparagine (N) respectively. Various combinations of these substitutions were tested (7 peptides in total, Fig. 1A, Table 1). All peptides were synthesized by GL Biochem Ltd. (Shanghai, China) using Fmoc solid-phase synthesis.
Fig. 1.
Tyrosinase active site and peptide design. (A) Schematic of rationally designed tetrapeptides with a single, double and triple position modifications. (B) Mushroom tyrosinase (PDB: 2Y9X) active site with catalytic amino acids shown. In this research the tyrosinase actvity site is divided into four regions: R1, R2, R3 and R4. The figure was constructed from Discovery Studio 2017 R2 Client.
Table 1.
The ability of CRNL peptide and its variants to inhibit tyrosinase processing of L-DOPA.
| Peptide sequence | Relative tyrosinase activity (%) |
IC50 (μM) | |||||
|---|---|---|---|---|---|---|---|
| 0 μM | 12.50 μM | 25 μM | 50 μM | 100 μM | 200 μM | ||
| CRNL (504.6110 Da) | 100.00 ± 4.86a | 78.38 ± 4.32b | 71.89 ± 5.62b | 46.85 ± 8.10c | 6.74 ± 3.70d | 3.69 ± 1.99d | 39.62 ± 6.21b |
| CENL | 100.00 ± 0.77a | 90.49 ± 3.37ab | 84.15 ± 7.35b | 81.23 ± 6.68b | 65.81 ± 7 .20c | 37.96 ± 6.10d | 150.67 ± 7.37d |
| CRLL | 100.00 ± 0.75a | 85.71 ± 2.01b | 81.95 ± 0.25c | 84.21 ± 4.01bc | 70.43 ± 0.75d | 31.91 ± 1.26e | 143.50 ± 4.82d |
| CRNN | 100.00 ± 2.62a | 85.33 ± 4.71b | 84.69 ± 2.48b | 72.89 ± 3.50c | 51.60 ± 0.87d | 12.90 ± 0.64e | 87.23 ± 1.64c |
| CELL | 100.00 ± 3.21a | 83.60 ± 3.22b | 80.39 ± 4.01b | 76.74 ± 2.94b | 59.54 ± 6.33c | 40.64 ± 1.93d | 148.60 ± 16.13d |
| CRLN | 100.00 ± 1.39a | 88.64 ± 0.55b | 86.70 ± 2.49b | 86.15 ± 0.83b | 79.50 ± 0.83c | 49.31 ± 1.11d | 234.27 ± 8.98e |
| CENN | 100.00 ± 0.65a | 86.36 ± 2.60b | 80.84 ± 0.97b | 79.55 ± 1.62b | 63.30 ± 2.20d | 37.03 ± 3.03e | 144.20 ± 8.02d |
| CELN | 100.00 ± 0.26a | 90.32 ± 5.91b | 77.63 ± 4.63cd | 81.49 ± 0.26c | 72.75 ± 0.77d | 47.81 ± 0.51e | 232.70 ± 4.95e |
| Kojic acid (KA) | 100.00 ± 5.45a | 54.74 ± 1.32b | 49.73 ± 3.27c | 38.77 ± 0.88d | 25.85 ± 1.30e | 13.80 ± 1.62f | 20.80 ± 2.10a |
Values are the mean ± the standard deviation. The different superscript letters are used to indicate the significant difference at p < 0.05.
2.3. Tyrosinase inhibition assay and inhibition mode
The inhibition assay was performed as described by Joompang et al. (2020). Briefly, L-DOPA (3.60 mM, 50 μL in 100 mM phosphate buffer pH 6.8) was mixed with the desired peptide (0 – 400 μM, 47.50 μL) at room temperature before the addition of tyrosinase (400 U/mL, 2.50 μL) to initiate the enzymatic reaction. The absorbance at 475 nm was monitored every 30 s using a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA, USA). The initial rate (slope) of tyrosinase activity was obtained by plotting the absorbance at 475 nm vs time. Kojic acid was used as a positive control. The tyrosinase inhibitory activity and tyrosinase activity were calculated using the following equation:
| (1) |
| (2) |
where ΔA475control and ΔA475test represent the initial rate (slope) of the reaction without the inhibitor and the reaction with the inhibitor, respectively. GraphPad Prism7 (GraphPad Software Inc., California, USA) was used to calculate IC50 values using a non-linear regression model.
To determine the mode of inhibition, the tyrosinase activity assay was performed with the different concentrations of L-DOPA (0 – 0.80 mM) and peptide (0 – 50 μM). The changes of Km and Vmax from the Lineweaver-Burk plot were used to infer the type of inhibition.
The reversible or irreversible inhibition of the peptide was examined from plotting tyrosinase concentration (0 – 10 μg/mL) and tyrosinase activity using different concentrations of the peptides (0 – 50 μM).
2.4. Kinetics of irreversible inhibition
The kinetics of irreversible inhibition was evaluated by pre-incubation of peptide (0 – 50 μM, 47.50 μL) with tyrosinase (400 U/mL, 2.50 μL) for 0, 5, 10, 15, 20, 25, 30, 45 and 60 min at room temperature. Then L-DOPA (3.6 mM, 50 μL in 100 mM phosphate buffer, pH 6.8) was added to initiate the reaction. Tyrosinase activity was detected according to section 2.3. The plot of Ln (remaining tyrosinase activity (%)) vs peptide concentration was used to determine the observed inactivation rate (kobs). The reciprocal plot of 1/kobs vs 1/peptide concentration was used to estimate KI and Kinact.
2.5. Molecular docking
Molecular docking was performed using GOLD Suite 5.5 (Cambridge Crystallographic Data Center, Cambridge, UK) (Jones et al., 1997). Three-dimensional (3D) structures of peptides were constructed using Discovery Studio 2017 R2 Client (Dassault Systèmes BIOVIA, BIOVIA Workbook, Release, 2017; BIOVIA Pipeline Pilot, Release, 2017; San Diego: Dassault Systèmes). The crystal structure of mushroom tyrosinase (PDB: 2Y9X) was selected as the target receptor, and water was removed from the structure before docking. The automated molecular docking was performed with the addition of hydrogen atoms. Then, the tropolone molecule was removed, and the binding site of peptides was set as the tropolone binding site with the radius of 10 Å. Goldscore scoring function was used with 1000 GA running. The search efficiency was flexible and set at 200%. The most feasible complex with the highest gold score was selected to represent the interaction between the peptide and tyrosinase. Discovery Studio 2017 R2 Client was used to detect the modes of interactions.
2.6. Peptide copper chelation assay
Peptide copper chelation activity was determined using a modification of the protocol reported by Fu et al. (2014). Briefly, phosphate buffer (500 mM, 10 μL, pH 6.8) was mixed with peptides (0 – 200 μM, 50 μL) and CuSO4 (aq., 0.69 mM, 20 μL). Subsequently, pyrocatechol violet (PV, 0.69 mM, 20 μL) was added to the mixture and incubated for 20 min. Then the absorbance at 632 nm was recorded. The reaction without the peptide and CuSO4 was performed as a “blank”. The copper chelating activity was calculated using the equation below:
| (3) |
where A632test and A632control represent the absorbance of the reaction with and without the peptide respectively. A632blank is the absorbance of the reaction without the peptide and CuSO4. GraphPad Prism7 (GraphPad Software Inc., California, USA) was used to calculate IC50 values using a non-linear regression model.
2.7. Cell viability assay
Cell viability was determined by a modification of protocol reported by Phosri et al. (2014). B16F1 cells (1.5 x 105 cells/well) were seeded and grown overnight on a 12-well plate, followed by the addition of different concentrations of the CRNL peptide (0 – 200 μM) and incubation at 37 °C, 5% CO2 with 95% relative humidity for 48 h. Next, the culture medium was removed and MTT (0.5 mg/mL) was added to the cells, followed by the incubation at 37 °C for 2 h. The absorbance was then detected at 570 nm using a microplate reader. The percentage of cell viability was calculated using the equation below:
| (4) |
where A570control is the absorbance of untreated cells and A570test is the absorbance of cells treated with the peptide.
2.8. Total melanin content investigation
B16F1 cells (1.5 x 105 cells/well) were seeded and grown overnight on a 12-well plate, followed by the addition of different concentrations of CRNL (0 – 200 μM) at 37 °C, 5% CO2 with 90% relative humidity. After 48 h, the culture medium was removed, and the cells were washed twice with PBS buffer (pH 7.4). Then a DMSO/NaOH (1 M) mixture (1:9, 200 μL) was added and the reaction was incubated at 80 °C for 1 h. The absorbance at 490 nm of 100 μL of supernatant was detected using a microplate reader. The relative melanin content was calculated using the equation below:
| (5) |
where Mtest is the absorbance of the cells treated with the peptide and Mcontrol is the absorbance of the untreated cells.
2.9. Statistical analysis
All experiments were conducted in triplicate (n = 3). The data are presented as the mean ± the standard deviation. The ANOVA test according to Duncan (p < 0.05) was carried out for comparison.
3. Results and discussion
3.1. Peptide inhibition of tyrosinase activity
The tyrosinase active site can be divided into four regions: R1 (a dinuclear copper complex that interacts with six histidine residues; copper atom CuA interacts with H85, H94 and H61; copper atom CuB interacts with H259, H263 and H296); R2 (negatively charged residues such as E256 and E322); R3 (N260 and R268) and R4 (F264, V283, P284, V248, H244 and A246) (Fig. 1B). To target the active site, a tetrapeptide was rationally designed. Previously, cysteine-containing peptides were reported to serve as potent tyrosinase inhibitors (Lee et al., 2015; Joompang et al., 2020). As a consequence, at position 1 (P1), a cysteine (C) was used in order to favor coordinate bond formation between the sulfhydryl group and the catalytic copper atom in R1. At position 2 (P2), arginine (R) was selected to electrostatically interact with the guanidinium and E322 in R2. At position 3 (P3), the very polar asparagine (N) amino acid was selected in order to interact with the polar amino acids of the R3 site. At position 4 (P4), leucine (L) was chosen to facilitate hydrophobic interactions with R4 (Fig. 1A and B). The selection of leucine ensures the rationally designed peptide contains one of the two amino acids that have been reported to give good tyrosinase inhibitory peptides (the other being arginine) (Schurink et al., 2007). To validate the rationale behind the design, the amino acids at the position 2, 3 and 4 were substituted by those with opposite properties (E, L and N respectively), while the C terminal amino acid at P1 was kept constant. Different combinations of these amino acid modifications yield seven versions of the CRNL parent peptide (Fig. 1A and Table 1).
To evaluate their potency as an inhibitor, L-DOPA and tyrosinase were incubated with varying concentrations of the peptides (0 – 200 μM) and the IC50 value was determined with kojic acid as a positive control (IC50 = 20.80 ± 2.10 μM). Promisingly, the rationally designed peptide CRNL gave an IC50 value of 39.62 ± 6.21 μM, which is close to that of the control (Table 1). On the other hand, the other peptide variants had higher IC50 values ranging from 143.50 to 232.70 μM. CRNN was notably better with an IC50 value of 87.23 μM. Overall, these results demonstrate our rational design approach was reasonable.
3.2. Molecular docking
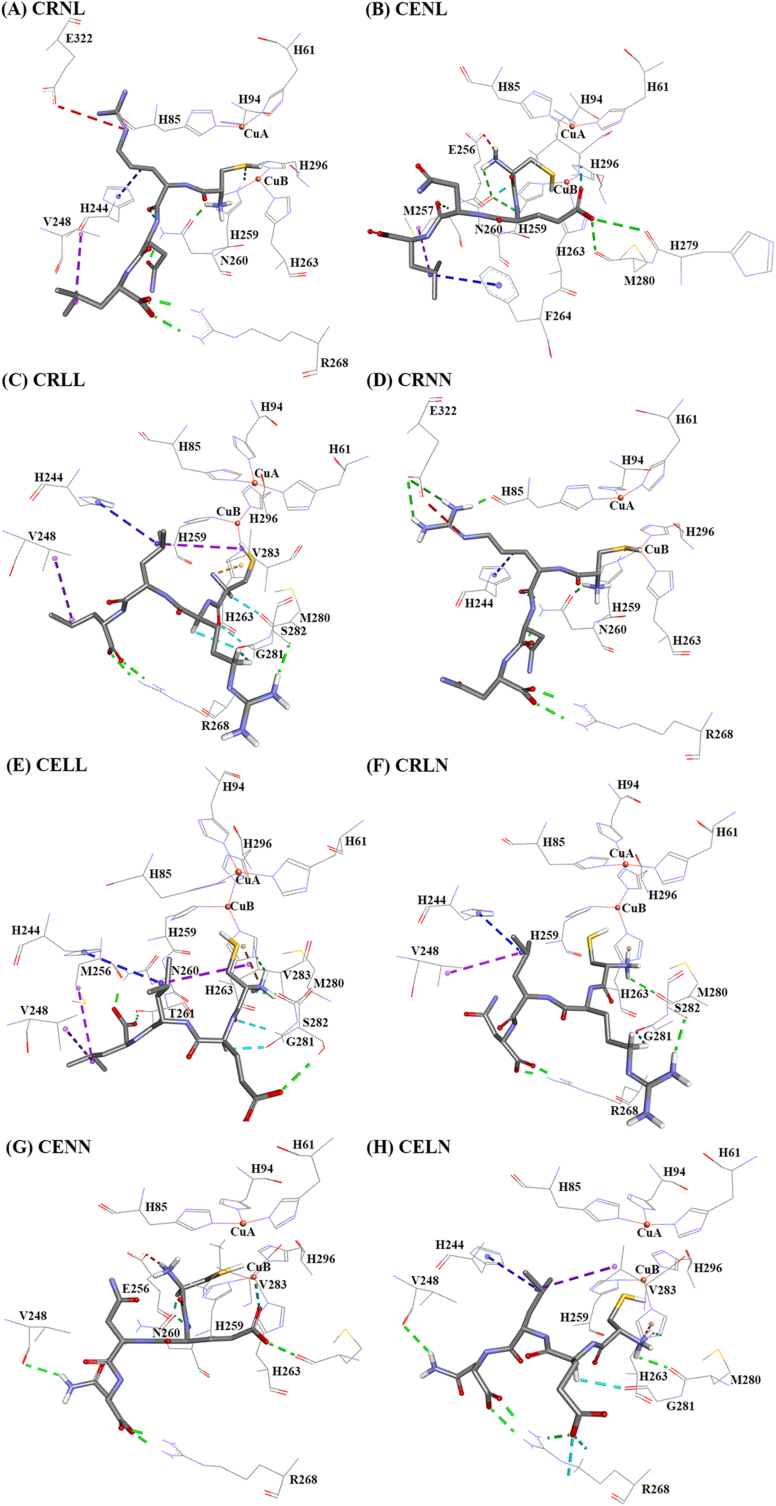
To further support our structure-activity inhibition assay, molecular docking simulations were performed with the CRNL and its variants. All peptides were able to interact with the active site of tyrosinase (Fig. 2A–H and Tables S1–S8). In all cases, the sulfhydryl group of cysteine was predicted to be orientated towards the catalytic copper atoms enabling coordination bonds between the sulfur and copper atoms. This is consistent with previous literature reports (Hsiao et al., 2014; Lee et al., 2015). CRNL formed five hydrogen bonds, one electrostatic interaction (R at P2–E322), and two hydrophobic interactions (R at P2–H244 and L at P4–V248) with the active site. These interactions are consistent with our hypothesis.
Fig. 2.
Tyrosinase complexed with (A) CRNL, (B) CENL, (C) CRLL, (D) CRNN, (E) CELL, (F) CRLN, (G) CENN and (H) CELN. The conventional hydrogen bond and carbon hydrogen bond are represented by the green and cyan dash lines, respectively. The electrostatic interaction, Pi-cation, and Pi-hydrogen bond donor are represented by red, brown, and orange dash lines, respectively. The hydrophobic interactions of alkyl-alkyl and Pi-alkyl are represented by the blue and violet dash lines, respectively. The figure was constructed from Discovery Studio 2017 R2 Client.
From our tyrosinase inhibition assays, all CRNL variants (except CRNN) had similar IC50 values in the range of 143.50 to 232.70 μM. The corresponding molecular docking studies demonstrate that these variants either disrupted electrostatic interactions (CENL and CRLL; IC50 = 150.67 and 143.50 μM) or had more drastic effects on peptide–enzyme interactions (CRLN and CELN; IC50 = 234.27 and 232.70 μM). The CRNL ⟶ CENL variant caused the loss of the electrostatic interaction between the peptide and E322; the CRNL ⟶ CRLL variant disrupted electrostatic interactions (R at P2–E322) and two hydrogen bonds (N at P3 with N260), but did promote hydrophobic interactions. This change does appear to decrease the inhibition of tyrosinase. Finally, CRNL ⟶ CRLN and CRNL ⟶ CELN modifications dramatically altered tyrosinase interactions losing both electrostatic interactions between R at P2 and E322 and hydrophobic interactions between L at P4–V248, which naturally results in the highest IC50 and least potency. This finding suggests the disrupting such both electrostatic and hydrophobic interactions is severely detrimental to tyrosinase inhibition.
Interestingly, for the CRNL ⟶ CRNN variant, docking confirmed that hydrophobic interaction (L at P4–V248) was disrupted. However, given the potency of the CRNN variant was still good (IC50 = 87.23 ± 1.64 μM cf. CRNL IC50 of 39.62 ± 6.21 μM), it appears this hydrophobic interaction is less important than electrostatic interactions.
In combination, these in silico molecular docking studies corroborated our hypothesis and experimental results. They provide interesting extra mechanistic information on how peptides interact with the active site of tyrosinase.
3.3. Peptide copper chelation activity
Compounds with copper chelating functional groups, especially thiol group (-SH) are known to decrease the tyrosinase activity by sequestering the copper atoms of tyrosinase (Pillaiyar et al., 2017; Kubglomsong et al., 2018). The tetrapeptides tested herein all contain an N-terminal cysteine, which is an excellent copper chelator. To evaluate their respective affinity for copper, the peptides were mixed with copper before the addition of pyrocatechol violet. Stronger binders prevent pyrocatechol violet from interacting with copper and therefore changing the absorbance of the dye. The difference in signal from a control (no peptide) and a test sample (with a peptide) gave an indicator of copper binding of the peptide. These results showed that none of the peptides strongly bind to copper unless the concentration of the peptide is higher than 100 μM. At 200 μM, CRNN, CENN and CRNL showed some affinity for copper (41.12, 53.72 and 69.63%; lower values are better chelators; Table 2). However, this does not correlate with their tyrosinase inhibitory activity (IC50 = 87.23, 144.20 and 39.62 μM respectively, Table 1). While this suggests copper chelation is not the primary mechanism by which these peptides inhibit tyrosinase, it must be noted that this assay is binding of the peptide to free copper. When the peptide binds to copper in tyrosinase, other intermolecular interactions may favor copper interactions (see section 3.5 and irreversible inhibition studies). Considering the relation of copper chelating activity and tyrosinase inhibitory activity, it was found that the inhibitory activity of most peptides tends to intensify regarding the increase of the copper chelating activity. However, such relation was not applied to some peptides. These might be due to the different peptide structures and sequences, resulting in the different modes of inhibition towards tyrosinase, consequently affecting the inhibitory activity.
Table 2.
Free copper chelating activity of CRNL and its variants using a pyrocatechol violet assay.
| Peptide | Relative copper (%) |
IC50 (μM) | |||||
|---|---|---|---|---|---|---|---|
| 0 μM | 12.5 0 μM | 25 μM | 50 μM | 100 μM | 200 μM | ||
| CRNL | 100.00 ± 8.85a | 107.70 ± 1.25ab | 102.13 ± 6.92ab | 103.89 ± 2.54ab | 95.01 ± 2.11b | 69.63 ± 3.00c | >200 |
| CENL | 100.00 ± 8.85a | 102.13 ± 6.74a | 100.95 ± 8.05a | 102.13 ± 7.57a | 95.16 ± 4.06a | 73.15 ± 1.43b | >200 |
| CRLL | 100.00 ± 10.46a | 95.08 ± 5.99ab | 93.10 ± 4.47abc | 91.24 ± 2.38abc | 87.82 ± 2.56bc | 83.31 ± 0.45d | >200 |
| CRNN | 100.00 ± 10.46a | 96.28 ± 3.61a | 92.92 ± 2.56a | 91.18 ± 1.00a | 77.61 ± 4.79b | 41.12 ± 2.18c | 171.97 ± 7.70 |
| CELL | 100.00 ± 8.85a | 106.60 ± 3.21a | 106.68 ± 1.47a | 104.11 ± 3.42a | 99.34 ± 4.33a | 84.15 ± 0.46b | >200 |
| CRLN | 100.00 ± 8.85a | 109.17 ± 4.08ab | 103.23 ± 3.24ab | 101.69 ± 3.24ab | 97.29 ± 5.86b | 87.45 ± 3.00c | >200 |
| CENN | 100.00 ± 10.46a | 94.42 ± 1.88ab | 94.96 ± 2.98ab | 90.28 ± 2.52b | 79.11 ± 0.95c | 53.72 ± 2.55d | >200 |
| CELN | 100.00 ± 10.46a | 97.42 ± 4.15ab | 93.16 ± 3.72ab | 93.94 ± 0.28ab | 88.90 ± 2.45c | 72.81 ± 3.26d | >200 |
Values are the mean ± the standard deviation. The different superscript letters are used to indicate the significant difference at p < 0.05.
3.4. Type of inhibition
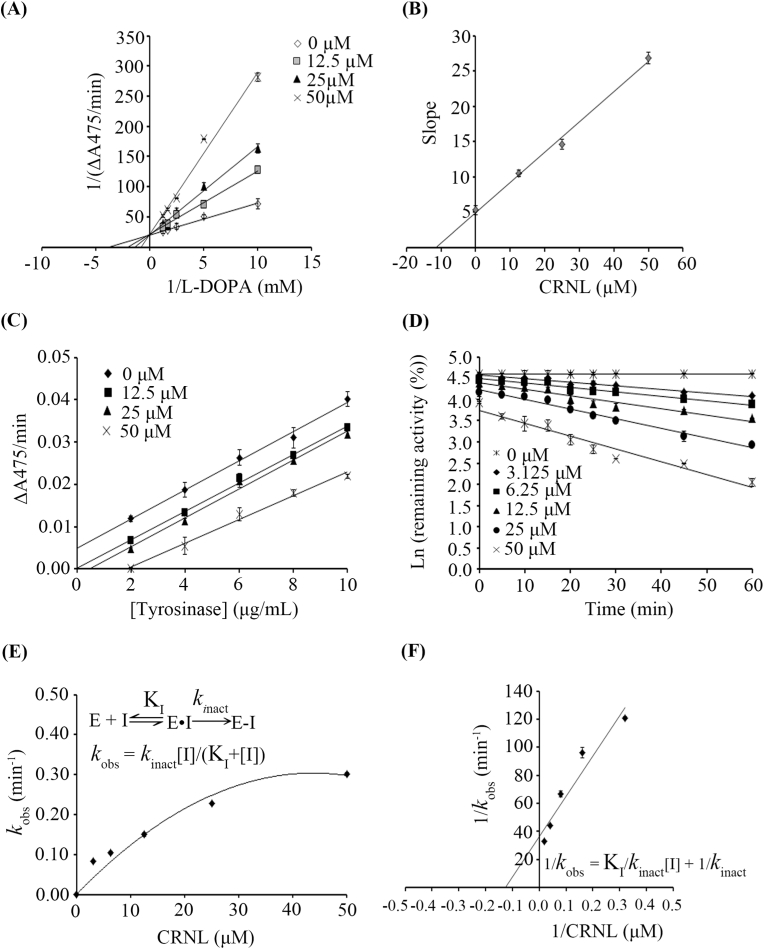
In order to determine the type of inhibition that CRNL causes Lineweaver-Burk plots were made to determine Km and Vmax values. The result indicated that the increase in the concentration of peptides caused an increase of Km, while Vmax did not change. This suggests that CRNL is a competitive inhibitor with Ki of 11.49 ± 1.65 μM (Fig. 3A and B and Table 3). This is consistent with previous reports that a cysteine-containing peptide inhibits tyrosinase a competitive inhibitor (Hsiao et al., 2014; Lee et al., 2015; Tseng et al., 2015).
Fig. 3.
Kinetic studies. (A) Lineweaver-Burk plot of tyrosinase kinetic when L-DOPA was used as a substrate and in the presence of the CRNL peptide from 0 to 50 μM. (B) Secondary plot of Lineweaver-Burk plot in (A). (C) An irreversible inhibition of tyrosinase by the CRNL peptide. (D) The plot of Ln (% remaining tyrosinase activity) vs pre-incubation time at different CRNL concentrations. (E) The plot of kobs (min-1) vs CRNL concentration. (F) Kitz–Wilson re-plot. CRNL at concentration of 0 – 50 μM was used.
Table 3.
Kinetic constants of tyrosinase.
| Peptide name | Concentration (μM) | Km or K'm (μM) | Vmax or V'max (△A475/min) | Mode of inhibition |
|---|---|---|---|---|
| CRNL | 0 | 256.71 ± 21.18a | 0.049 ± 0.0022a | competitive |
| 12.50 | 495.35 ± 30.97b | 0.047 ± 0.0013a | ||
| 20.00 | 715.67 ± 53.43c | 0.049 ± 0.0038a | ||
| 50.00 | 1279.65 ± 143.95d | 0.048 ± 0.0040a |
Values are the mean ± the standard deviation. The different superscript letters are used to indicate the significant difference at p < 0.05.
The thiol group of cysteine could coordinate to the catalytic copper atoms resulting in the irreversible inhibition of tyrosinase. While CRNL peptide does not seem to bind free copper, it may do so once the other intermolecular interactions it forms with the enzyme orientate it for interaction with the copper atoms. As a result, this was evaluated by looking at the linear relationship between each concentration of tyrosinase and its activity in the presence and absence of CRNL (Fig. 3C). The parallel straight lines denoted the occurrence of the irreversible inactivation of these peptides. Based on the irreversible inhibition, the kinetics of peptides then studied for understanding the inactivation mechanism. The plot of natural logarithm (Ln) of the remaining tyrosinase activity vs the pre-incubation times was shown in Fig. 3D. This plot was used to estimate the observed rate constant (kobs). Increasing peptide concentration was found to increase the observed rate constant as follow hyperbolic characteristic (kobs, Fig. 3E), suggesting a two-step irreversible inhibition mechanism, which is consistent with previously reported cysteine-containing peptide, TILI-2 (Joompang et al., 2020). After CRNL reversibly binds to tyrosinase (E•I), tyrosinase is inactivated through coordination bond formation (E-I). The following equation was used to express the two-step inactivation:
| kobs = kinact[I]/(KI+I) |
where kinact is the maximum rate of the enzyme inactivation (min-1), and KI is an inhibition constant (μM) of the reversible binding. In order to obtain the estimated kinact and KI, Kitz–Wilson re-plot (1/kobs = KI/kinact[I] + 1/kinact) was performed (Fig. 3F). The results indicated that CRNL exhibited KI and kinact of 7.87 ± 0.28 μM and 0.027 ± 0.0007 min-1, respectively. In addition, CRNL was observed to have the constant rate of covalent bond formation (kinact/KI) of 0.0035 ± 0.00004 μM-1 min-1.
3.5. Cell viability and melanin content
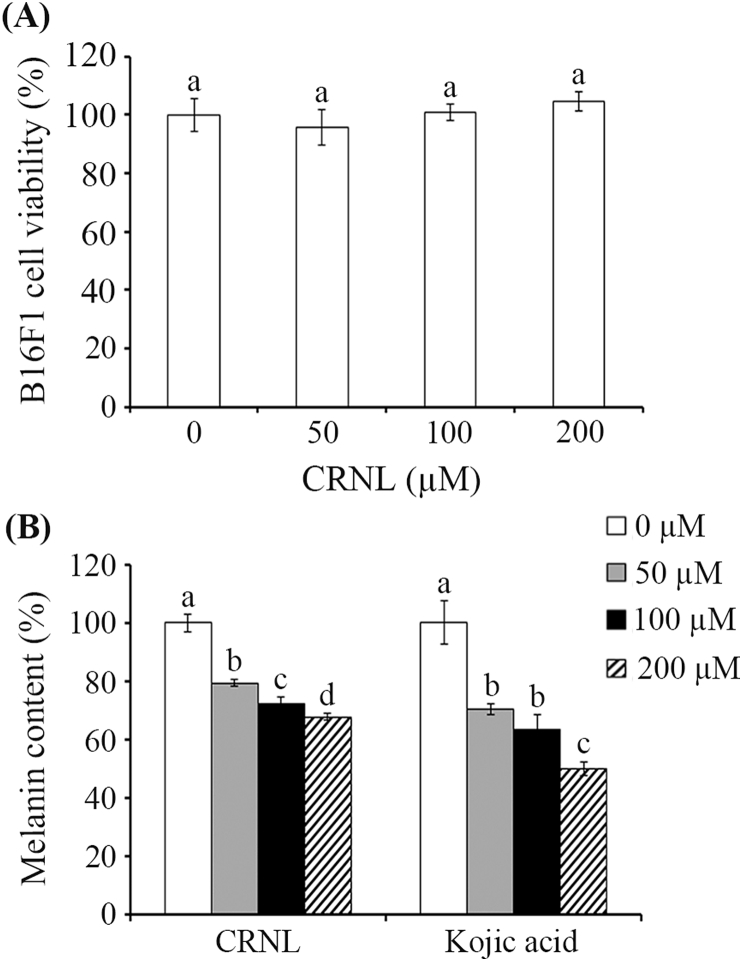
The cytotoxicity and melanin inhibition of CRNL (50 – 200 μM) was investigated in B16F1 cells. Promisingly, no toxicity was observed at the tested concentration (Fig. 4A), while melanin decreased in a dose-dependent manner. At the highest concentration used (200 μM), the melanin content was reduced by 32.13 ± 1.25% (n = 6, Fig. 4B). These results indicated that CRNL could decrease melanin without any toxicity towards B16F1, suggesting the possibility it could be used in hyperpigmentation treatments.
Fig. 4.
The effect of tyrosinase inhibitors on (A) cell viability and (B) melanin content in B16F1 cells. Values are the mean ± the standard deviation. The different superscript letters are used to indicate the significant difference at p < 0.05.
4. Conclusions
A potent tyrosinase inhibitor peptide, CRNL, was developed. Evaluation of structural analogues along with molecular docking suggested that the electrostatic interactions between the peptide's P2 R and the tyrosinase's E322 amino acids, and hydrophobic interactions between the peptide's P4 L and the tyrosinase's V248 amino acids were important for CRNL's strong inhibition of tyrosinase. In addition, all modified tetrapeptides were copper chelators with CRNN being the most potent chelator. Interestingly, there was a good correlation between peptide's ability to chelate copper and its ability to inhibit tyrosinase. Kinetic studies indicated that the CRNL peptide was a competitive inhibitor of tyrosinase through a two-step inactivation mechanism. Moreover, CRNL was not cytotoxic and could also decrease melanin levels in B16F1 cells. Collectively, these results suggest that CRNL is a tyrosinase inhibitor that could be used for hyperpigmentation treatment or as a copper chelator.
CRediT authorship contribution statement
Anupong Joompang: Conceptualization, Investigation, Data curation, Methodology, Formal analysis, Writing – original draft. Preeyanan Anwised: Investigation, Formal analysis. Sompong Klaynongsruang: Conceptualization, Resources, Methodology, Supervision. Lapatrada Taemaitree: Data curation, Visualization, Writing – review & editing. Anuwat Wanthong: Data curation, Visualization. Kiattawee Choowongkomon: Methodology, Supervision. Sakda Daduang: Resources, Methodology, Supervision. Somporn Katekaew: Supervision, Visualization, Writing – review & editing. Nisachon Jangpromma: Conceptualization, Resources, Methodology, Supervision, Data curation, Visualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the Fundamental Fund of Khon Kaen University, The National Science, Research and Innovation Fund (NSRF), and the Protein and Proteomics Research Center for Commercial and Industrial Purposes (ProCCI), Faculty of Science, Khon Kaen University.
Handling Editor: Professor Aiqian Ye
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2023.100598.
Contributor Information
Somporn Katekaew, Email: somkat@kku.ac.th.
Nisachon Jangpromma, Email: nisaja@kku.ac.th.
Abbreviations
- R1
the tyrosinase active site region 1
- R2
the tyrosinase active site region 2
- R3
the tyrosinase active site region 3
- R4
the tyrosinase active site region 4
- P1
position 1 from the N-terminus to the C-terminus of the designed tetrapeptide
- P2
position 2 from the N-terminus to the C-terminus of the designed tetrapeptide
- P3
position 3 from the N-terminus to the C-terminus of the designed tetrapeptide
- P4
position 4 from the N-terminus to the C-terminus of the designed tetrapeptide
- C
cysteine
- R
arginine
- N
asparagine
- L
leucine
- E
glutamic acid
- V
valine
- F
phenylalanine
- P
proline
- H
histidine
- A
alanine
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
The data that has been used is confidential.
References
- Alghamdi K., Alehaideb Z., Kumar A., Al-Eidi H., Alghamdi S.S., Suliman R., Ali R., Almourfi F., Alghamdi S.M., Boudjelal M., Matou-Nasri S. Stimulatory effects of Lycium shawii on human melanocyte proliferation, migration, and melanogenesis: In vitro and in silico studies. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1169812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ateş S., Pekyardimci S., Cokmus C. Partial characterization of a peptide from honey that inhibits mushroom polyphenol oxidase. J. Food Biochem. 2001;25:127–137. [Google Scholar]
- Bachtiar M., Lee C.G.L. Genetics of population differences in drug response. Curr. Genet. Med. Rep. 2013;1(3):162–170. [Google Scholar]
- Blaut M., Braune A., Wunderlich S., Sauer P., Schneider H., Glatt H. Mutagenicity of arbutin in mammalian cells after activation by human intestinal bacteria. Food Chem. Toxicol. 2006;44(11):1940–1947. doi: 10.1016/j.fct.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Curto E.V., Kwong C., Hermersdorfer H., Glatt H., Santis C., Virador V., Hearing V.J., Jr., Dooley T.P. Inhibitors of mammalian melanocyte tyrosinase: in vitro comparisons of alkyl esters of gentisic acid with other putative inhibitors. Biochem. Pharmacol. 1999;57(6):663–672. doi: 10.1016/s0006-2952(98)00340-2. [DOI] [PubMed] [Google Scholar]
- DeCaprio A.P. The toxicology of hydroquinone-relevance to occupational and environmental exposure. Crit. Rev. Toxicol. 1999;29(3):283–330. doi: 10.1080/10408449991349221. [DOI] [PubMed] [Google Scholar]
- Fu R., Zhang Y., Guo Y., Chen F. Antioxidant and tyrosinase inhibition activities of the ethanol-insoluble fraction of water extract of Sapium sebiferum (L.) Roxb. leaves. South Afr. J. Bot. 2014;93:98–104. [Google Scholar]
- Hermanns J.F., Pierard-Franchimont C., Pierard G.E. Skin colour assessment in safety testing of cosmetics. An overview. Int. J. Cosmet. Sci. 2000;22(1):67–71. doi: 10.1046/j.1467-2494.2000.00021.x. [DOI] [PubMed] [Google Scholar]
- Hsiao N.W., Tseng T.S., Lee Y.C., Chen W.C., Lin H.H., Chen Y.R., Wang Y.T., Hsu H.J., Tsai K.C. Serendipitous discovery of short peptides from natural products as tyrosinase inhibitors. J. Chem. Inf. Model. 2014;54(11):3099–3111. doi: 10.1021/ci500370x. [DOI] [PubMed] [Google Scholar]
- Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Joompang A., Anwised P., Klaynongsruang S., Roytrakul S., Taemaitree L., Jangpromma N. Evaluation of TILI-2 as an anti-tyrosinase, anti-oxidative agent and its role in preventing melanogenesis using a proteomics approach. Molecules. 2022;27(10) doi: 10.3390/molecules27103228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joompang A., Jangpromma N., Choowongkomon K., Payoungkiattikun W., Tankrathok A., Viyoch J., Luangpraditkun K., Klaynongsruang S. Evaluation of tyrosinase inhibitory activity and mechanism of leucrocin I and its modified peptides. J. Biosci. Bioeng. 2020;130(3):239–246. doi: 10.1016/j.jbiosc.2020.04.002. [DOI] [PubMed] [Google Scholar]
- Kubglomsong S., Theerakulkait C., Reed R.L., Yang L., Maier C.S., Stevens J.F. Isolation and identification of tyrosinase-inhibitory and copper-chelating peptides from hydrolyzed rice-bran-derived albumin. J. Agric. Food Chem. 2018;66(31):8346–8354. doi: 10.1021/acs.jafc.8b01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.C., Hsiao N.W., Tseng T.S., Chen W.C., Lin H.H., Leu S.J., Yang E.W., Tsai K.C. Phage display-mediated discovery of novel tyrosinase-targeting tetrapeptide inhibitors reveals the significance of N-terminal preference of cysteine residues and their functional sulfur atom. Mol. Pharmacol. 2015;87(2):218–230. doi: 10.1124/mol.114.094185. [DOI] [PubMed] [Google Scholar]
- Moon K.M., Kwon E.B., Lee B., Kim C.Y. Recent trends in controlling the enzymatic browning of fruit and vegetable products. Molecules. 2020;25(12):2754. doi: 10.3390/molecules25122754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir Y., Rafique H., Roshan S., Shamas S., Ashraf Z., Rafiq M., Tahir T., Qureshi Z.U., Aslam A., Asad M. Molecular docking, synthesis, and tyrosinase inhibition activity of acetophenone amide: potential inhibitor of melanogenesis. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/1040693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai A., Tanaka S., Imai Y., Yoshida H., Kanaoka T., Tanaka T., Taniguchi M. New tyrosinase inhibitory decapeptide: molecular insights into the role of tyrosine residues. J. Biosci. Bioeng. 2016;121(6):607–613. doi: 10.1016/j.jbiosc.2015.10.010. [DOI] [PubMed] [Google Scholar]
- Parvez S., Kang M., Chung H.S., Cho C., Hong M.C., Shin M.K., Bae H. Survey and mechanism of skin depigmenting and lightning agents. Phytother Res. 2006;20(11):921–934. doi: 10.1002/ptr.1954. [DOI] [PubMed] [Google Scholar]
- Phosri S., Mahakunakorn P., Lueangsakulthai J., Jangpromma N., Swatsitang P., Daduang S., Dhiravisit A., Thammasirirak S. An investigation of antioxidant and anti-inflammatory activities from blood components of crocodile (Crocodylus siamensis) Protein J. 2014;33(5):484–492. doi: 10.1007/s10930-014-9581-y. [DOI] [PubMed] [Google Scholar]
- Pillaiyar T., Manickam M., Namasivayam V. Skin whitening agents: medicinal chemistry perspective of tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2017;32(1):403–425. doi: 10.1080/14756366.2016.1256882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurink M., van Berkel W.J.H., Wichers H.J., Boeriu C.G. Novel peptides with tyrosinase inhibitory activity. Peptides. 2007;28(3):485–495. doi: 10.1016/j.peptides.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Shen Z., Wang Y., Guo Z., Tan T., Zhang Y. Novel tyrosinase inhibitory peptide with free radical scavenging ability. J. Enzym. Inhib. Med. Chem. 2019;34(1):1633–1640. doi: 10.1080/14756366.2019.1661401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M., Truong V.L., Lee M., Kim D., Kim M.S., Cho H., Jung Y.H., Yang J., Jeong W.S., Kim Y. Investigation of phenyllactic acid as a potent tyrosinase inhibitor produced by probiotics. Curr. Res. Food Sci. 2023;6(2023) doi: 10.1016/j.crfs.2022.100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T., Mitsumori K., Tamura T., Nasu M., Ueda M., Imai T., Hirose M. Hepatocellular tumor induction in heterozygous p53-deficient CBA mice by a 26-week dietary administration of kojic acid. Toxicol. Sci. 2003;73(2):287–293. doi: 10.1093/toxsci/kfg094. [DOI] [PubMed] [Google Scholar]
- Tseng T.S., Tsai K.C., Chen W.C., Wang Y.T., Lee Y.C., Lu C.K., Don M.J., Chang C.Y., Lee C.H., Lin H.H., Hsu H.J., Hsiao N.W. Discovery of potent cysteine-containing dipeptide inhibitors against tyrosinase: a comprehensive investigation of 20 x 20 dipeptides in inhibiting dopachrome formation. J. Agric. Food Chem. 2015;63(27):6181–6188. doi: 10.1021/acs.jafc.5b01026. [DOI] [PubMed] [Google Scholar]
- Ubeid A.A., Do S., Nye C., Hantash B.M. Potent low toxicity inhibition of human melanogenesis by novel indole-containing octapeptides. Biochim. Biophys. Acta. 2012;1820(10):1481–1489. doi: 10.1016/j.bbagen.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Ubeid A.A., Zhao L., Wang Y., Hantash B.M. Short-sequence oligopeptides with inhibitory activity against mushroom and human tyrosinase. J. Invest. Dermatol. 2009;129(9):2242–2249. doi: 10.1038/jid.2009.124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.