Abstract
Background
Idiopathic pulmonary fibrosis (IPF) has an unknown aetiology and limited treatment options. A recent meta-analysis identified three novel causal variants in the TERT, SPDL1, and KIF15 genes. This observational study aimed to investigate whether the aforementioned variants cause clinical phenotypes in a well-characterised IPF cohort.
Methods
The study consisted of 138 patients with IPF who were diagnosed and treated at the Helsinki University Hospital and genotyped in the FinnGen FinnIPF study.
Data on > 25 clinical parameters were collected by two pulmonologists who were blinded to the genetic data for patients with TERT loss of function and missense variants, SPDL1 and KIF15 missense variants, and a MUC5B variant commonly present in patients with IPF, or no variants were separately analysed.
Results
The KIF15 missense variant is associated with the early onset of the disease, leading to progression to early-age transplantation or death. In patients with the KIF15 variant, the median age at diagnosis was 54.0 years (36.5–69.5 years) compared with 72.0 years (65.8–75.3 years) in the other patients (P = 0.023). The proportion of KIF15 variant carriers was 9- or 3.6-fold higher in patients aged < 55 or 65 years, respectively. The variants for TERT and MUC5B had similar effects on the patient’s clinical course, as previously described. No distinct phenotypes were observed in patients with the SPDL1 variant.
Conclusions
Our study indicated the potential of KIF15 to be used in the genetic diagnostics of IPF. Further studies are needed to elucidate the biological mechanisms of KIF15 in IPF.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12931-023-02540-0.
Keywords: Idiopathic pulmonary fibrosis, KIF15, TERT, SPDL1, Missense variant
Background
Idiopathic pulmonary fibrosis (IPF) is a progressive lung disease that is accompanied by respiratory symptoms, primarily dyspnoea, and a poor quality of life. The impacts of current antifibrotic therapies are limited, and the prognosis has not improved sufficiently [1, 2].
Studies are slowly unravelling the aetiology of IPF with evidence of genetic susceptibility combined with external risk factors and possible exposure. More than 25 different genetic regions and numerous variants have been reported to be involved in IPF [3–7]. Although data on genetic variants are abundant, little is known about their clinical significance. The clinical aspects have been best described in variants in the telomerase and promoter regions of MUC5B. A common variant in MUC5B has the largest effect on IPF risk, but rare variants, such as TERT, are considered more important in disease pathogenicity [5, 7, 8]. Variants in non-telomerase-related genes, such as SDPL1, KIF15, and surfactant-related genes, have recently been suggested to be of importance in IPF [5, 7, 9–11]. These single studies focused on genetic data with limited clinical features. There is a lack of studies where detailed clinical data are presented along with the genetic variants.
A recent large-scale meta-analysis of IPF genetics suggested causal coding variants at three loci –TERT, SPDL1, and KIF15, in the FinnGen IPF (FinnIPF) population [12]. The unique genetic background of an isolated population in Finland has provided a special basis for several genetic research studies [13, 14]. Using data from the national biobank and large GWAS studies, we aimed to correlate this genetic data with a well-defined clinical cohort of 138 patients with IPF.
We studied the effects of the predicted rare causal variants of TERT, SPDL1, and KIF15 and the common variant of MUC5B on the clinical phenotype and disease course of patients with IPF.
Methods
The FinnishIPF study is a nationwide epidemiological registry study [15]. The inclusion criteria are written informed consent and a diagnosis of IPF according to the ATS/ERS criteria [16]. FinnGen (https://www.finngen.fi/en) is a large biobank study in which diagnoses are based on ICD codes. FinnGen samples were genotyped using multiple Illumina and Affymetrix arrays (Illumina, San Diego, CA, USA and Thermo Fisher Scientific, Santa Clara, CA, USA, respectively), filtered, and imputed with a population-specific reference panel as previously described [15].
In 2017, a subgroup of patients in the FinnishIPF study was contacted to collect DNA and serum samples and was included in the FinnGen analysis (FinnIPF cohort, N = 235). Our study cohort consisted of FinnishIPF study patients who were followed up at the Helsinki University Hospital (N = 138).
In the present study, causal variants predicted by fine-mapping in the FinnGen study, two TERT loss of function (Lof) and missense variants (rs770066110 and rs776981958) and SPDL1 (rs116483731) and KIF15 missense (rs138043992) variants, were included in the analyses [12]. We also analysed the most well-known genetic variant for IPF, MUC5B (rs35705950) [4, 9, 17]. Due to the extremely high prevalence (97/138), the patients with the MUC5B variant were investigated separately.
Using electronic medical records and CT scans at the Helsinki University Hospital, we collected pre-specified clinical characteristics of our patient cohort (N = 138) in July 2022. Two pulmonologists specialising in interstitial lung diseases re-evaluated these clinical characteristics. The genetic data were blinded to the pulmonologists and added to the analysis after the clinical evaluation was completed by unblinded researchers. Before the unblinding and analysis, three patients (2.1%, 3/138) were excluded from the final cohort: one did not have IPF, and two were immigrants with a genetically remote ethnicity. None of the excluded patients possessed any of the genetic variants under investigation.
Statistical analysis
All patients with either KIF15, TERT, or SPDL1 missense variants were grouped according to variant status, and the patients without any of the studied variants were grouped as “no Qv”. One of the patients (1/135) harboured two of the studied variants (TERT and SPDL1 missense) and was excluded from the analyses that simultaneously included both the TERT and SPDL1 missense groups to maintain the independence of the groups necessary for statistical testing.
We used a Kruskal–Wallis or Fisher's exact test (with a Freeman–Halton extension for contingency tables larger than 2 × 2) to evaluate the differences between variant groups, depending on the variable type. Furthermore, the variant groups were individually compared with the no Qv group using a Mann–Whitney U or Fisher’s exact test. As the differences between the KIF15 and no Qv groups were consistent, the KIF15 group was further compared with a group consisting of all other patients. We also conducted Kaplan–Meier one-minus survival and survival analyses using the Mantel–Cox log-rank test to estimate disease occurrence and progression. In these analyses, the exact timing and occurrence of events (diagnosis, death, or transplantation) were known without any missing data or dropouts. As a result, only the patients who survived to the end of the study period without death or transplantation were censored (no patients were censored in the disease occurrence analysis, as all patients were diagnosed with IPF).
All data are presented as median (IQR) or % (n/N), and a two-tailed P < 0.05 was considered significant. Statistical analyses were performed using IBM SPSS Statistics (version 25, SPSS Inc., Chicago, IL, USA).
Results
The patient characteristics of the variant groups are shown in Table 1. Most patients were male and smokers with no family history of pulmonary fibrosis. Comorbidities included hypertension, hyperlipidaemia, coronary artery disease, obesity, type 2 diabetes mellitus, and hypothyroidism, and 33 patients had cancer: 12 lung, 11 prostate, 6 gastrointestinal tract, 4 bladder, 3 breast, 1 cervix, and 1 renal cancer. The variant groups differed, especially in variables relating to treatment and prognosis, such as the probability of lung transplantation, age at diagnosis, or death (Table 1).
Table 1.
Patient characteristics of all patients and by variant groups, and the statistical comparison between variant groups, KIF15 missense, SPDL1 missense, TERT, and No Qv (patients without any of these variants)
| All patients (N = 135) | KIF15 missense (N = 5) | SPDL1 missense (N = 21) | TERT (N = 8) | No Qv (N = 100) | Sig | |
|---|---|---|---|---|---|---|
| Background and comorbidities | ||||||
| Female sex | 43 (32%) | 0 (0%) | 7 (33%) | 1 (13%) | 35 (35%) | 0.313 |
| Male sex | 92 (68%) | 5 (100%) | 14 (67%) | 7 (88%) | 65 (65%) | 0.313 |
| Family history of pulmonary fibrosis | 8 (6%) | 0 (0%) | 1 (5%) | 2 (25%) | 5 (5%) | 0.173 |
| Smoking history | 86 (64%) | 4 (80%) | 17 (81%) | 5 (63%) | 59 (59%) | 0.250 |
| Cancers | 33 (24%) | 1 (20%) | 6 (29%) | 1 (13%) | 25 (25%) | 0.899 |
| BMI, kg/m2 | 28.0 (25.0–30.0) | 34.0 (28.1–36.0) | 28.0 (25.4–31.1) | 27.4 (25.2–32.8) | 27.3 (24.9–30.0) | 0.131 |
| Comorbidities, N | 4.0 (2.0–6.0) | 5.0 (2.0–7.5) | 3.0 (1.5–7.0) | 3.5 (0.5–4.8) | 4.0 (2.0–6.0) | 0.450 |
| Treatment and prognosis | ||||||
| Antifibrotic treatment | 79 (59%) | 4 (80%) | 10 (48%) | 4 (50%) | 60 (60%) | 0.519 |
| Lung transplantation | 15 (11%) | 2 (40%) | 1 (5%) | 3 (38%) | 8 (8%) | 0.013* |
| Oxygen therapy | 43 (32%) | 4 (80%) | 7 (33%) | 3 (38%) | 29 (29%) | 0.126 |
| Acute exacerbations | 34 (25%) | 1 (20%) | 5 (24%) | 1 (13%) | 27 (27%) | 0.925 |
| Deceased | 62 (46%) | 1 (20%) | 12 (57%) | 2 (25%) | 46 (46%) | 0.318 |
| Age at diagnosis, years | 71.0 (65.0–75.0) | 54.0 (36.5–69.5) | 68.0 (65.0–76.0) | 67.0 (59.3–70.3) | 72.0 (66.0–76.0) | 0.016* |
| Age at death, years | 78.8 (74.1–83.0) | 61.2 (61.2–61.2) | 77.5 (69.9–80.2) | 74.1 (71.3–74.1) | 79.6 (74.7–84.2) | 0.075 |
| Age at transplant, years | 61.1 (49.8–65.0) | 40.2 (30.9–40.2) | 63.5 (63.5–63.5) | 68.0 (63.7–68.0) | 57.4 (50.7–61.6) | 0.033* |
| Age at death or transplant, years | 76.5 (68.5–81.4) | 49.4 (30.9–49.4) | 76.3 (68.7–80.0) | 68.7 (64.8–74.9) | 78.6 (72.3–83.8) | 0.013* |
| Symptoms at diagnosis | ||||||
| Dyspnoea | 68 (50%) | 3 (60%) | 13 (62%) | 3 (38%) | 49 (49%) | 0.637 |
| Cough | 69 (51%) | 1 (20%) | 8 (38%) | 4 (50%) | 55 (55%) | 0.269 |
| Laboratory findings at diagnosis | ||||||
| Macrocytosis | 30 (22%) | 1 (20%) | 5 (24%) | 2 (25%) | 21 (21%) | 0.942 |
| Thrombocytopenia | 18 (13%) | 1 (20%) | 2 (10%) | 1 (13%) | 13 (13%) | 0.895 |
| Radiological findings at diagnosis | ||||||
| Traction bronchiectasis | 132 (98%) | 5 (100%) | 21 (100%) | 8 (100%) | 97 (97%) | 1.000 |
| Honeycombing | 108 (80%) | 4 (80%) | 15 (71%) | 4 (50%) | 85 (85%) | 0.050* |
| Ground-glass opacity | 25 (19%) | 2 (40%) | 4 (19%) | 0 (0%) | 18 (18%) | 0.324 |
| Right ventricular strain | 17 (13%) | 1 (20%) | 3 (14%) | 0 (0%) | 13 (13%) | 0.697 |
| Emphysema | 32 (24%) | 1 (20%) | 6 (29%) | 1 (13%) | 24 (24%) | 0.896 |
| Pulmonary function at diagnosis | ||||||
| FVC, L | 2.96 (2.42–3.70) | 3.35 (2.77–3.58) | 2.97 (2.52–3.48) | 3.41 (2.71–4.22) | 2.90 (2.29–3.76) | 0.468 |
| FVC, % predicted | 81.0 (68.0–93.0) | 59.0 (51.5–90.0) | 79.0 (69.5–89.5) | 73.0 (64.3–97.5) | 84.5 (70.3–94.8) | 0.327 |
| FEV1, L | 2.37 (2.00–3.00) | 2.73 (2.27–2.76) | 2.21 (2.08–2.72) | 2.55 (2.20–3.57) | 2.34 (1.91–3.04) | 0.587 |
| FEV1, % predicted | 80.0 (72.8–94.3) | 66.0 (55.0–84.0) | 77.0 (72.5–90.5) | 77.0 (62.0–103.0) | 83.0 (74.0–95.8) | 0.275 |
| DLCO, % predicted | 59.0 (50.0–69.3) | 52.0 (37.0–64.5) | 56.0 (45.5–70.5) | 62.5 (47.5–75.5) | 60.0 (52.0–69.0) | 0.423 |
Bold values are statistically significant p < 0.05
KIF15 missense variant
Patients with the KIF15 missense variant were the youngest at the time of diagnosis, transplantation, death, or both endpoints combined (Table 1).
They were diagnosed at an earlier age (median 54.0 years, 36.5–69.5, n1 = 5) compared with those without any of the studied variants (72.0 years, 66.0–76.0, n2 = 100, U = 99.5, P = 0.023). They also died at a younger age (61.2 years, n1 = 1) than the patients in the no Qv group (72.0 years, 66.0–76.0, n2 = 100, U = 99.5, P = 0.023). When both endpoints (transplantation and death) were combined, the patients with the KIF15 missense variant were noticeably younger (49.4 years, 30.9–49.4, n1 = 3) compared with those in the no Qv group (78.6 years, 72.3–83.8, n2 = 53, U = 7.0, P = 0.002).
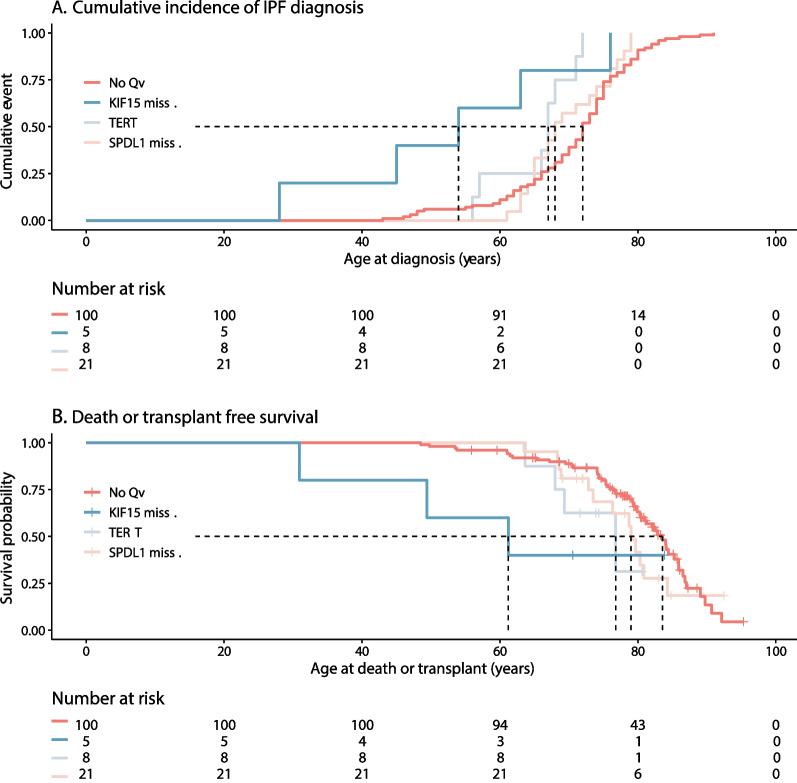
According to the Kaplan–Meier one-minus survival analysis, the IPF occurred significantly earlier among the patients with the KIF15 missense variant compared with those in the no Qv group (χ2(1) = 6.53, P = 0.011, Fig. 1A). In addition, the death or transplant-free survival was significantly weaker in the KIF15 missense group compared with that in the no Qv group (χ2(1) = 3.96, P = 0.047, Fig. 1B). The KIF15 missense group also clearly stood out from others in terms of death and transplant-free survival (Fig. 1B).
Fig. 1.
A, B Disease progression in the KIF15 missense, TERT, SPDL1 missense, and No Qv groups ("No Qv", patients without studied TERT, SPDL1, and KIF15 missense variants)
Patients with the KIF15 missense variant had a higher median body mass index (BMI) of 34.0 (28.1–36.0, n1 = 5) than those in the no Qv group (27.3, n2 = 100, U = 398.5, P = 0.025). Most patients with the KIF15 missense variant, 80% (4/5), were started on supplemental oxygen therapy, relative to the 29% (29/100) of the no Qv patients (P = 0.033).
As the differences between the patients in the KIF15 missense and no Qv groups were consistent, we compared the KIF15 group to those without KIF15 variant carriers (“Others”). The results were even more pronounced when patients with other variants were included in the analyses (Table 2). The patients in the KIF15 group were diagnosed (U = 129.5, P = 0.023), died (U = 0.0, P = 0.032), and either received the transplant or died (U = 7.0, P = 0.001) at a significantly younger age than those of the others. In addition, the patients with the KIF15 variant needed oxygen therapy more often than the other patients (P = 0.036).
Table 2.
Patient characteristics and the statistical comparison between patients with the KIF15 missense variant and all other patients
| KIF15-missense (N = 5) | Others (N = 130) | Sig | |
|---|---|---|---|
| Background and comorbidities | |||
| Female sex | 0 (0%) | 43 (33%) | 0.177 |
| Male sex | 5 (100%) | 87 (67%) | 0.177 |
| Family history of pulmonary fibrosis | 0 (0%) | 8 (6%) | 1.000 |
| Smoking history | 4 (80%) | 82 (63%) | 0.653 |
| Cancers | 1 (20%) | 32 (25%) | 1.000 |
| BMI, kg/m2 | 34.0 (28.1–36.0) | 27.8 (25.0–30.0) | 0.028* |
| Comorbidities, N | 5.0 (2.0–7.5) | 4.0 (2.0–6.0) | 0.432 |
| Treatment and prognosis | |||
| Oxygen therapy | 4 (80%) | 39 (30%) | 0.036* |
| Antifibrotic treatment | 4 (80%) | 75 (58%) | 0.402 |
| Lung transplantation | 2 (40%) | 13 (10%) | 0.095 |
| Deceased | 1 (20%) | 61 (47%) | 0.374 |
| Age at diagnosis, years | 54.0 (36.5–69.5) | 72.0 (65.8–75.3) | 0.023* |
| Age at death, years | 61.2 (61.2–61.2) | 79.0 (74.1–83.2) | 0.032* |
| Age at transplant, years | 40.2 (30.9–40.2) | 61.8 (53.6–65.2) | 0.038* |
| Age at death or transplant, years | 49.4 (30.9–49.4) | 76.8 (69.3–82.0) | < 0.001* |
| Symptoms | |||
| Dyspnoea | 3 (60%) | 65 (50%) | 1.000 |
| Cough | 1 (20%) | 68 (52%) | 0.202 |
| Acute exacerbations | 1 (20%) | 33 (25%) | 1.000 |
| Laboratory findings | |||
| Macrocytosis | 1 (20%) | 29 (23%) | 1.000 |
| Thrombocytopenia | 1 (20%) | 17 (13%) | 0.520 |
| Radiological findings | |||
| Right ventricular strain | 1 (20%) | 16 (12%) | 0.495 |
| Emphysema | 1 (20%) | 31 (24%) | 1.000 |
| Honeycombing | 4 (80%) | 104 (80%) | 1.000 |
| Traction bronchiectasis | 5 (100%) | 127 (98%) | 1.000 |
| Ground-glass opacity | 2 (40%) | 23 (18%) | 0.231 |
| Pulmonary function test findings | |||
| FVC, L | 3.35 (2.77–3.58) | 2.95 (2.40–3.71) | 0.578 |
| FVC, % predicted | 59.0 (51.5–90.0) | 81.0 (68.0–93.0) | 0.128 |
| FEV1, L | 2.73 (2.27–2.76) | 2.36 (1.96–3.04) | 0.562 |
| FEV1, % predicted | 66.0 (55.0–84.0) | 82.0 (73.0–95.0) | 0.086 |
| DLCO, % predicted | 52.0 (37.0–64.5) | 59.0 (50.5–69.5) | 0.188 |
Bold values are statistically significant p < 0.05
Considering the timing of disease onset, the proportion of KIF15 missense carriers was noticeably high among all the patients with early diagnosis: 50.0% (1/2) of patients with diagnosis before 45 years, 33.3% (3/9) of patients with diagnosis before 55 years, and 13.3% (4/30) of patients with diagnosis before 65 years. The total proportion of KIF15 missense variant carriers in the study population was 3.7% (5/135).
Other variants
The patients with the SPDL1 variant died at a younger age (76.3 years, 68.7–80.0, n1 = 13) than those in the no Qv group (79.6 years, 74.7–84.2, n2 = 46, U = 412.0, P = 0.039).
Four and five patients possessed a TERT Lof and a TERT missense variant, respectively. These patients were pooled for statistical analysis. The proportion of patients reporting dyspnoea at the time of diagnosis (75.0% for Lof and 0.0% for missense) was the only parameter that differed among the groups (P = 0.048).
The patients with the TERT variants were younger (67.0 years, 58.5–69.5, n1 = 9) at the time of diagnosis than the no Qv patients (72.0 years, 66.0–76.0, n2 = 100, U = 678.5, P = 0.012). In addition, the cumulative incidence of IPF diagnosis differed from that among the no Qv patients (χ2(1) = 11.62, P = 0.001). Patients with TERT variants received a lung transplant more often (44.4%, 4/9) compared with the 8.0% of patients with no Qv (8/100, P = 0.008). They were also younger at the time of death or transplantation (68.0 years, 64.3–73.1, N1 = 5) compared with the no Qv patients (78.6 years, 72.3–83.8, N2 = 53, U = 61.0, P = 0.047). Patients with the TERT variants had lesser honeycombing in the HRCT (44.4%, 4/9) than no Qv patients (85.0%, 85/100, P = 0.010).
The MUC5B variant was found in 70% (97/138) of the patients. Due to its high prevalence in the total sample and the variant groups, it was not included in the variant group comparisons. Additional file 1: Table S1 shows the patient characteristics of the MUC5B variant group and other patients; the only significant difference was that the MUC5B carriers were significantly older at the time of transplantation than other patients. These groups did not differ regarding the prevalence of the other studied variants (p = 0.548 for KIF15 missense, p = 0.537 for SPDL1 missense, and p = 0.720 for TERT variants). The exclusion of patients with any of the other variants (KIF15 missense, SPDL1 missense, TERT variants) did not significantly change the results in any of the studied.
Discussion
We investigated the possibility of defining a clinical IPF phenotype based on distinct genetic variants associated with IPF susceptibility. To the best of our knowledge, this is one of the first studies in which systematically collected clinical characteristics were compared with a detailed genetic evaluation.
This study showed that the KIF15 missense variant (rs138043992) was associated with markedly early disease onset, a high need for supplemental oxygen, and disease progression to transplantation or death at an early age.
The studied KIF15 missense variant causes a nucleotide substitution from arginine to leucine at the 501st amino acid (p.Arg501Leu). Hence, it could affect the stability and function of the KIF15 protein. KIF15 is a motor protein involved in mitotic spindle assembly and affects cell proliferation. It is upregulated in various cancers, including breast and gastric cancer. Overexpression of KIF15 promotes lung carcinogenesis and cancer cell proliferation and is associated with a poor prognosis in non-small cell lung carcinoma [18–20]. Several KIF15 variants (missense, Lof, and intron) have also been associated with IPF risk, but their impact on disease course and phenotype remains unclear [4, 10, 11].
The KIF15 variant in this study was suggested to be causal in the fine-mapping of the FinnGen IPF population [12]. It was recently observed that variants in the Finnish population that were enriched by more than twofold were 1.7-fold more likely to be associated with a phenotype expected by chance [14]. Allelic frequency (AF) in our IPF cohort was 0.018, which showed a twofold increase compared with that of the reference Finnish population (AF 0.009), over fourfold compared with that of the non-Finnish Europeans (AF 0.004), and sixfold compared with that of ALL individuals (AF 0.003) based on the whole genome variant set version 3.1.2 of the Genome Aggregation Database (GnomAD). This shows that the KIF15 missense variant allele is enriched in the Finnish population and could be a possible causal variant for IPF, as shown earlier [12].
Our main finding was that patients with the KIF15 missense variant were diagnosed at a substantially younger age than other patients. The difference in the median diagnosis age between the KIF15 variant carriers and others was large (over 10 years) and not explained by family history of IPF or statistical outliers. The proportion of KIF15 variant carriers was ninefold higher in patients aged < 55 years. Our findings imply a potential link between the early onset of disease and the KIF15 missense variant. Among the three earlier studies reporting on KIF15 and IPF, only Zhang et al. reported the median age of the patients [4, 10, 11]. In their study, the median age of patients with KIF15 variants (stop gain, missense, splice acceptor, and frameshift) was 64 years (N = 12), and that of all patients was 67 years. The study included both patients with IPF and other interstitial lung diseases [11]. It is important to note that the comprehensive KIF15 mutation variant list in the Zhang data doesn’t include our KIF15 variant found in an all-Finnish population.
Our KIF15 missense variant patients received transplantation or died at a younger age than other patients. This was probably explained by the early onset of the disease and increased morbidity. Patients diagnosed at a younger age are more likely to meet the requirements for lung transplantation, and human lifespan is limited and influenced by many factors other than the variants (such as the presence and severity of comorbid conditions, smoking, and exposures). Hence, the large difference in the timing of disease onset has the potential to mask the effects of the variant on overall survival. Some results might suggest that the KIF15 variant impacts the disease course. Patients with the KIF15 variant needed oxygen therapy markedly more often than the others. Previously, Allen et al. reported an association between lower pulmonary function and the KIF15 variant, rs78238620 [4]. In our study, the KIF15 missense group performed the worst in pulmonary function tests, but probably due to the sample size, the results did not reach statistical significance. This might also obscure the finding that none of the patients with the KIF15 missense variant had a known family history of pulmonary fibrosis despite the early onset of the disease. More studies on the possible effects of KIF15 variants on disease course, mortality, and familial forms of the disease are needed.
Considering the notable effect on disease onset and the possible effect on disease progression, our findings support the view of Moss and Rosas, who highlighted that KIF15 has the potential to uncover disease mechanisms and even contribute to drug discovery [7]. Many centres employ genetic panels for clinical diagnosis, and the identified mutations influence clinical decisions [21, 22]. Our findings suggest that KIF15 could be included in genetic screening panels.
The findings on the other, better-described variants (in TERT, SPDL1, and MUC5B) were in line with earlier studies. Telomerase reverse transcriptase (TERT) maintains telomere length by adding nucleotides to the ends of telomeres and plays a role in cellular senescence and cancer development. Shorter telomere lengths have been associated with IPF and poor survival [23–25]. The TERT variant was also associated with worse outcomes in our study. SPDL1 variants are reportedly upregulated in the lung tissue of patients with IPF [9]. We did not find a distinct clinical phenotype for the SPDL1 variant. Previously, Dhindsa et al. reported a summary of the clinical features of 26 IPF patients with the SPLD1 variant without a clear or significant difference from other study patients. These results suggest that the SPDL1 variants do not have any major impact on the clinical phenotype of IPF. The best-known single genetic risk factor for IPF is a variant in MUC5B, which is present in more than half of the patients with IPF and has been associated with susceptibility to IPF and a mild disease course [26–29]. In our study, 70% of the patients carried the MUC5B variant, and these patients needed lung transplantation at an older age than other patients. The other variants did not affect this result.
Our study has several strengths. Repeated genetic bottlenecks and isolation from other countries create a distinct genetic profile for the Finnish people, providing a unique setting for genetic studies. Patient samples were genotyped as part of a large FinnGen IPF study, and all patients had IPF and met the ATS/ERS criteria for the disease. We had access to detailed clinical histories of all patients, and all data were reviewed by two pulmonologists specialising in IPF care. The number of patients was limited, but the statistical assumptions were met, and there were no distinct outliers in any of the variant groups. The number of censored patients in the survival analyses was minimal.
Our study also has limitations. Our patient population was relatively small and from a single university hospital. Only four variants were analysed; therefore, the significance of other potentially significant variants remains unknown. The small number of patients in some variant groups limits the possibilities of adjusting the analyses with confounding factors, such as disease severity, sex, age, and comorbidities, that are known to influence IPF survival. It was not possible to analyse the association between genetic variants and pulmonary function over time. The observatory and exploratory nature of the study limit final conclusions regarding distinct phenotypes. Larger confirmatory studies within multi-ancestry genetic populations and multiple cohorts are needed to confirm these results and detect possible milder effects caused by other variants.
Conclusions
Our study highlights the importance of connecting clinical characteristics to genetic data. We found that the KIF15 missense variant is associated with a remarkably early onset of the disease, leading to transplantation and death at an early age. Our findings suggest the potential of KIF15 to be used in diagnostic procedures, patient monitoring, and screening, especially among young patients. We also confirmed the previously identified clinical phenotypes of TERT variants and that many patients with IPF are genetically susceptible to the disease. In the future, larger studies on KIF15 focusing on clinical aspects, especially disease course and survival, are needed to confirm its significance.
Supplementary Information
Additional file 1. Table S1. Characteristics of the MUC5B positive patients.
Acknowledgements
We thank Professor Mark Daly, the participants and investigators of the FinnGen study, and the study participants for their generous participation in the THL and Helsinki Biobanks. We thank Helsinki University Hospital, the University of Helsinki, the Finnish TB Foundation, the Finnish Research Foundation of Pulmonary Diseases, Boehringer Ingelheim, and Nummela Sanatorium for funding.
Abbreviations
- IPF
Idiopathic pulmonary fibrosis
- KIF15
Kinesin family member 15
- MUC5B
Mucin 5b
- SPDL1
Spindle apparatus coiled-coil protein 1
- TERT
Telomerase reverse transcriptase
Author contributions
MM and MH designed the study and collected the data. JP, JK and MA performed the genetic variant analyses. AL and MH performed the statistical analyses. MH, AL, MM, MA, and JP drafted the manuscript. MH, AL, MA, JP, JK, MM, ES and RK reviewed and edited the manuscript. MH, MM and AL were responsible for the decision to submit the manuscript. MH, MM and MA verified the data. All authors had access to the underlying blinded data, and they read and approved the final text. ES and MM are the project administrators and managers of resources. MM acquired funding.
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital. This study was funded by the University of Helsinki, Helsinki University Hospital Funds, Nummela Sanatorium, the Finnish TB-Foundation, the Finnish Research Foundation of the Pulmonary Diseases, Academy of Finland, the doctoral program in population health, university of Helsinki, and Boehringer Ingelheim. Funding was used for data acquisition and to assist staff. The researchers in this study did not receive additional salaries or payments, apart from those from the research institutions where they were employed. The funding sources played no role in the study design, data collection, analysis, and interpretation, or writing of the manuscript. The corresponding author has access to all the data in the study and has the final responsibility for the decision to submit the manuscript. The data presented are owned by two organisations, the University of Helsinki and the Helsinki University Hospital, and access is granted with research permission from these organisations.
Availability of data and materials
A large portion of the data can be found in a meta-analysis by Partanen et al. [12]. The data can be shared upon request by the corresponding author. Our data is from a small population with a rare disease and patients in distinct groups of causal variants (KIF15 and TERT). Hence, variants and patient demographics are identity attributes, and according to legislation on data protection, it is not possible to share de-identified patient data.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS) (HUS/2550/2017) and a research permit (HUS/8/2022). All participants provided written informed consent to participate in the Finnish IPF study and separate consent and blood samples from any of the Finnish biobanks.
Consent for publication
Not applicable.
Competing interests
RK reports grants for the clinical group from the Tampere Tuberculosis Foundation, the Jalmari and Rauha Ahokas Foundation, the Research Foundation of the Pulmonary Diseases, the Finnish Anti-Tuberculosis Association Foundation, and the State subsidy of Oulu University Hospital; honoraria for lectures, consultancies, and virtual congress fees from Boehringer-Ingelheim, MSD, Novartis, and Roche; and leadership or fiduciary role as a President of the Finnish Respiratory Society (2017–2020), a member of the board of the Finnish Medical Foundation (1/2020–12/2022), and a member of the board of the Finnish Lung Health Association (1/2021–onwards). MH reports honoraria for lectures from Boehringer-Ingelheim, MSD, and AstraZeneca; participation on an advisory board for Boehringer-Ingelheim; travel support from MSD; and owns stock in Eli Lilly. Reported interests are all outside the submitted work. All other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Raghu G, Chen SY, Yeh WS, Maroni B, Li Q, Lee YC, et al. Idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older: incidence, prevalence, and survival, 2001–11. Lancet Respir Med. 2014;2:566–572. doi: 10.1016/S2213-2600(14)70101-8. [DOI] [PubMed] [Google Scholar]
- 2.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 3.Kaur A, Mathai SK, Schwartz DA. Genetics in idiopathic pulmonary fibrosis pathogenesis, prognosis, and treatment. Front Med (Lausanne) 2017;4:154. doi: 10.3389/fmed.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen RJ, Guillen-Guio B, Oldham JM, Ma SF, Dressen A, Paynton ML, et al. Genome-wide association study of susceptibility to idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2020;201:564–574. doi: 10.1164/rccm.201905-1017OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peljto AL, Blumhagen RZ, Walts AD, Cardwell J, Powers J, Corte TJ, et al. Idiopathic pulmonary fibrosis is associated with common genetic variants and limited rare variants. Am J Respir Crit Care Med. 2023;207:1194–1202. doi: 10.1164/rccm.202207-1331OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M, Zhang Y, Adams T, Ji D, Jiang W, Wain LV, et al. Integrative analyses for the identification of idiopathic pulmonary fibrosis-associated genes and shared loci with other diseases. Thorax. 2022 doi: 10.1136/thorax-2021-217703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss BJ, Rosas IO. Defining the genetic landscape of idiopathic pulmonary fibrosis: role of common and rare variants. Am J Respir Crit Care Med. 2023;207:1118–1120. doi: 10.1164/rccm.202301-0177ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton CA, Oldham JM, Applegate C, Carmichael N, Powell K, Dilling D, et al. The role of genetic testing in pulmonary fibrosis: a perspective from the pulmonary fibrosis foundation genetic testing work group. Chest. 2022;162:394–405. doi: 10.1016/j.chest.2022.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhindsa RS, Mattsson J, Nag A, Wang Q, Wain LV, Allen R, et al. Identification of a missense variant in SPDL1 associated with idiopathic pulmonary fibrosis. Commun Biol. 2021;4:392. doi: 10.1038/s42003-021-01910-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D, Povysil G, Kobeissy PH, Li Q, Wang B, Amelotte M, et al. Rare and common variants in KIF15 contribute to genetic risk of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2022;206:56–69. doi: 10.1164/rccm.202110-2439OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Newton CA, Wang B, Povysil G, Noth I, Martinez FJ, et al. Utility of whole genome sequencing in assessing risk and clinically relevant outcomes for pulmonary fibrosis. Eur Respir J. 2022;60:2200577. doi: 10.1183/13993003.00577-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partanen JJ, Häppölä P, Zhou W, Lehisto AA, Ainola M, Sutinen E, et al. Leveraging global multi-ancestry meta-analysis in the study of idiopathic pulmonary fibrosis genetics. Cell Genom. 2022;2:100181. doi: 10.1016/j.xgen.2022.100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locke AE, Steinberg KM, Chiang CWK, Service SK, Havulinna AS, Stell L, et al. Exome sequencing of Finnish isolates enhances rare-variant association power. Nature. 2019;572:323–328. doi: 10.1038/s41586-019-1457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaunisto J, Kelloniemi K, Sutinen E, Hodgson U, Piilonen A, Kaarteenaho R, et al. Re-evaluation of diagnostic parameters is crucial for obtaining accurate data on idiopathic pulmonary fibrosis. BMC Pulm Med. 2015;15:92. doi: 10.1186/s12890-015-0074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghu G, Remy-Jardin M, Myers JL, Richeldi L, Ryerson CJ, Lederer DJ, et al. Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Crit Care Med. 2018;198:e44–e68. doi: 10.1164/rccm.201807-1255ST. [DOI] [PubMed] [Google Scholar]
- 17.Allen RJ, Stockwell A, Oldham JM, Guillen-Guio B, Schwartz DA, Maher TM, et al. Genome-wide association study across five cohorts identifies five novel loci associated with idiopathic pulmonary fibrosis. Thorax. 2022;77:829–833. doi: 10.1136/thoraxjnl-2021-218577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding L, Li B, Yu X, Li Z, Li X, Dang S, et al. KIF15 facilitates gastric cancer via enhancing proliferation, inhibiting apoptosis, and predict poor prognosis. Cancer Cell Int. 2020;20:125. doi: 10.1186/s12935-020-01199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao Y, Chen J, Ma C, Liu Y, Li P, Wang Y, et al. Increased KIF15 expression predicts a poor prognosis in patients with lung adenocarcinoma. Cell Physiol Biochem. 2018;51:1–10. doi: 10.1159/000495155. [DOI] [PubMed] [Google Scholar]
- 20.Bidkhori G, Narimani Z, Hosseini Ashtiani SH, Moeini A, Nowzari-Dalini A, Masoudi-Nejad A. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS ONE. 2013;8:e67552. doi: 10.1371/journal.pone.0067552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borie R, Kannengiesser C, Sicre de Fontbrune F, Gouya L, Nathan N, Crestani B. Management of suspected monogenic lung fibrosis in a specialised centre. Eur Respir Rev. 2017;26:160122. doi: 10.1183/16000617.0122-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terwiel M, Grutters JC, van Moorsel CHM. Clustering of lung diseases in the family of interstitial lung disease patients. BMC Pulm Med. 2022;22:134. doi: 10.1186/s12890-022-01927-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molina-Molina M, Borie R. Clinical implications of telomere dysfunction in lung fibrosis. Curr Opin Pulm Med. 2018;24:440–444. doi: 10.1097/MCP.0000000000000506. [DOI] [PubMed] [Google Scholar]
- 24.Snetselaar R, van Batenburg AA, van Oosterhout MFM, Kazemier KM, Roothaan SM, Peeters T, et al. Short telomere length in IPF lung associates with fibrotic lesions and predicts survival. PLoS ONE. 2017;12:e0189467. doi: 10.1371/journal.pone.0189467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dressen A, Abbas AR, Cabanski C, Reeder J, Ramalingam TR, Neighbors M, et al. Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study. Lancet Respir Med. 2018;6:603–614. doi: 10.1016/S2213-2600(18)30135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stock CJ, Sato H, Fonseca C, Banya WA, Molyneaux PL, Adamali H, et al. Mucin 5B promoter polymorphism is associated with idiopathic pulmonary fibrosis but not with development of lung fibrosis in systemic sclerosis or sarcoidosis. Thorax. 2013;68:436–441. doi: 10.1136/thoraxjnl-2012-201786. [DOI] [PubMed] [Google Scholar]
- 27.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peljto AL, Zhang Y, Fingerlin TE, Ma SF, Garcia JG, Richards TJ, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA. 2013;309:2232–2239. doi: 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei R, Li C, Zhang M, Jones-Hall YL, Myers JL, Noth I, et al. Association between MUC5B and tert polymorphisms and different interstitial lung disease phenotypes. Transl Res. 2014;163:494–502. doi: 10.1016/j.trsl.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Characteristics of the MUC5B positive patients.
Data Availability Statement
A large portion of the data can be found in a meta-analysis by Partanen et al. [12]. The data can be shared upon request by the corresponding author. Our data is from a small population with a rare disease and patients in distinct groups of causal variants (KIF15 and TERT). Hence, variants and patient demographics are identity attributes, and according to legislation on data protection, it is not possible to share de-identified patient data.