Visual Abstract
Key Words: hypertrophy, post-transcriptional gene regulation, RNA binding protein
Highlights
-
•
YTHDF2 protein is elevated in failing hearts.
-
•
YTHDF2 is essential for maintenance of cardiac homeostasis.
-
•
Loss of YTHDF2 drives cardiac hypertrophy, fibrosis, and dysfunction in mice.
-
•
The proteome of YTHDF2-null cardiomyocytes is remodeled.
-
•
YTHDF2 binds m6A-modified Myzap mRNA and controls its stability.
-
•
Loss of YTHDF2 leads to pathological accumulation of MYZAP protein.
Summary
How post-transcriptional regulation of gene expression, such as through N6-methyladenosine (m6A) messenger RNA methylation, impacts heart function is not well understood. We found that loss of the m6A binding protein YTHDF2 in cardiomyocytes of adult mice drove cardiac dysfunction. By proteomics, we found myocardial zonula adherens protein (MYZAP) within the top up-regulated proteins in knockout cardiomyocytes. We further demonstrated that YTHDF2 binds m6A-modified Myzap messenger RNA and controls its stability. Cardiac overexpression of MYZAP has been associated with cardiomyopathy. Thus, our findings provide an important new mechanism for the YTHDF2-dependent regulation of this target and therein its novel role in the maintenance of cardiac homeostasis.
Maintenance of the heart’s homeostasis is meticulously executed by various cardiac cell types at the level of gene expression.1 Based on robust studies from the last decades, it is now understood that different transcriptional programs are activated in failing cardiomyocytes compared with healthy ones.2, 3, 4 However, cardiac protein levels do not always correlate with the abundance of their mRNA transcripts, hinting at the previously overlooked importance of post-transcriptional pathways in cardiomyocytes.5,6 RNA-binding proteins (RBPs) are the leading modulators of post-transcriptional events and control all aspects of RNA fate, including splicing, localization, stability, and translation. In the heart, several RBPs (ie, RBM20, MBNL1, RBFOX) have already emerged as key regulators of cardiac physiology and disease.7, 8, 9 These studies raise intriguing questions of whether post-transcriptional regulators of RNA biology, such as RBPs, can provide us with new hopes for the understanding and treatment of heart disease.
The YTHDF1, YTHDF2, and YTHDF3 proteins represent a unique class of RBPs, as they only recognize a select subset of mRNAs.10,11 With their conserved C-terminal YTH domains, they preferentially bind to N6-methyladenosines (m6As), on mRNA transcripts.12 m6A formation is accomplished by a methyltransferase complex composed of METTL3, METTL14, and WTAP,13,14 and its removal is achieved by m6A demethylases, such as ALKBH5 or FTO.15,16 Our laboratory and several other groups showed that strict regulation of m6A levels by these enzymes is required for the heart’s ability to maintain homeostasis.17, 18, 19 For example, genetic perturbation of METTL3 or FTO leads to abnormal stress responses in various animal and cell models.17,18 m6A levels are also altered in human heart failure, highlighting the clinical relevance of this pathway.20,21 However, the exact contribution of specific RBPs to the m6A-driven phenotypes remains an important topic to dissect.
Most studies conform to the originally proposed model that each YTHDF protein has a distinct role in the regulation of the fate of m6A mRNA, with YTHDF1 promoting translation,22 YTHDF2 facilitating mRNA decay,23 and YTHDF3 playing a more elusive role.24 However, other works suggest apparent redundancy or functional compensation among YTHDFs depending on the cellular context or modeling methods.25, 26, 27 Recent studies have begun to explore the regulatory changes in YTHDF2 with cardiomyopathy, suggesting a key role for this YTHDF member in the heart.28, 29, 30
Here, we used genetically engineered mice with cardiomyocyte-specific loss of Ythdf2 (YTHDF2 cardiac knockout [cKO]), which resulted in cardiac dysfunction, hypertrophy, and fibrosis. Mechanistically, we found up-regulation of the intercalated disc myocardial zonula adherens protein (MYZAP) (also known as MYOZAP) in cardiomyocytes lacking YTHDF2. We further demonstrated the ability of YTHDF2 to bind m6A-modified Myzap mRNA and facilitate its decay. Overall, we present a novel post-transcriptional mechanism positioning YTHDF2 at the center of m6A RNA regulation and essential for maintenance of cardiac structure and function.
Methods
Animal generation and treatment
Adult male and female C57BL6N mice between 2 and 4 months of age were used in this study. Mice were housed at 72 °F under a 12-hour light/dark cycle and maintained on a standard chow diet. Mice had ad libitum access to food and water. The generation of cardiac-specific YTHDF2 deletion mice was achieved in the following manner. Ythdf2 LoxP-targeted (flox/flox; exon 4) mice (Ythdf2fl/fl) were crossed with mice expressing tamoxifen-inducible Cre recombinase gene under the control of the alpha myosin heavy chain promoter (αMHC-MerCreMer) mice, to obtain cardiomyocyte-restricted Ythdf2 knockout (Y2-cKO) in adult mice.
To induce deletion, tamoxifen (MilliporeSigma) was mixed in sesame oil at 10 mg/mL. 2-month-old mice were given doses of tamoxifen at 40 mg/kg body weight/day for 5 consecutive days by intraperitoneal injection. Mice with wild-type levels of Ythdf2, either expressing Cre recombinase only or the Ythdf2 flox/flox–only littermates (“Ctrl”), were used as control animals. All mice received tamoxifen injections. All in vivo experiments and analyses were performed at 1 month after the first tamoxifen injection. The use of animals was approved by the Institutional Animal Care and Use Committee at The Ohio State University.
Cardiomyocyte isolations and immunostaining
Adult mouse cardiomyocytes were isolated using a previously described Langendorff method.31 For cell length and width measurements, cardiomyocytes were imaged in suspension using bright-field microscopy (magnification ×20) on an EVOS FL Auto II microscope (Thermo Fisher Scientific). At least 50 myocytes were measured for each mouse.
For immunostaining, isolated cardiomyocytes were plated on laminin-coated glass coverslips kept in 24-well dishes, fixed and permeabilized at room temperature in 4% paraformaldehyde in phosphate-buffered saline for 20 minutes and blocked in 5% bovine growth serum in phosphate-buffered saline (0.01% sodium azide, 0.1% Triton X) for 1 hour. Cells were stained with the following primary antibodies overnight in blocking solution: YTHDF2 (1:100; Proteintech; rabbit; #24744-1-AP), MYZAP (1:100; Sigma-Aldrich; rabbit; #ABT407), or N-cadherin (1:200; Life Technologies; mouse; #33-3900). Cells were stained with DAPI (1:1000) and fluorescent secondary antibodies (1:500) in blocking solution for 2 hours at room temperature. Secondary antibodies included Alexa-conjugated donkey anti-mouse 568 and donkey anti-rabbit 488. Images were obtained with Nikon A1R Galvano scanning inverted confocal microscope at the OSU Campus Microscopy and Imaging Facility.
Echocardiography and electrocardiography
Echocardiographic measurements were taken using a VisualSonics Vevo3100 system and MS-550D transducer. The mice were lightly anesthetized (1.5% isoflurane), and the ejection fraction, fractional shortening, and ventricular chamber dimensions were determined in the M-mode using the parasternal short-axis view at the level of the papillary muscles. Measurements were calculated from the average of at least 3 consecutive cardiac cycles using the VevoLAB program (VisualSonics). All echocardiographic measurements are reported in Supplemental Table 1.
Surface electrocardiograms (ECGs) were carried out in anesthetized mice (1.5% isoflurane) placed prone on a heated pad. Needle electrodes were placed under the skin to record in Lead-I configuration using a PowerLab system (ADInstruments). The baseline recording was performed for 5 minutes, and the standard parameters (heart rates; P duration; PR, QRS, QT, JT, QTc, JTc, and RR intervals; ST height; and all wave amplitudes) were calculated using LabChart Pro ECG Analysis v2.3.2 module (ADInstruments). Manual adjustments were made to ensure that each parameter was marked correctly. After 5 minutes of stable baseline recording, epinephrine (1.5 mg/kg) and caffeine (120 mg/kg) were injected intraperitoneally, and the ECGs were recorded for additional 15 minutes. Animals were monitored for ventricular arrhythmic events using timed interval analysis (every fifth, 10th, and 15th minutes following injection). Arrhythmia scoring, adapted from van der Werf et al,32 was used to assign individual scores to each mouse based on the worst ventricular arrhythmia displayed: 0 = no events; 1 = isolated premature ventricular contractions (<10 per minute); 2 = frequent premature ventricular contractions (≥10 per minute); 3 = ventricular bigeminy; 4 = ventricular tachycardia.32 The Wilcoxon rank sum test was used to compare group means for the nonparametric arrhythmia scores between genotypes. Incidence of arrhythmic events in the 2 groups was compared by Fisher exact test.
Tissue staining and quantification
Tissues were fixed in 10% neutral buffered formalin for 18 hours and processed for paraffin embedding. A Shandon Finesse 325 microtome (Thermo Fisher Scientific) was used to trim the surface of the paraffin blocks until all embedded tissues were exposed to the air. The blocks were then placed face down in ice water overnight to hydrate the tissue. Once hydrated, tissue blocks were cut in 5-μm sections. Paraffin sections were stained with hematoxylin and eosin, Picrosirius red staining, or fluorescent wheat germ agglutin using standard procedures. The total area of fibrosis on Picrosirius red–stained images was measured using the threshold function within ImageJ v1.53t (National Institutes of Health), and this value is divided by the total area of tissue to derive a value for % fibrosis.
For the determination of cardiomyocyte cross-sectional areas, paraffin-embedded sections were deparaffinized, subjected to antigen retrieval for 15 minutes in boiling sodium citrate buffer (10 mM sodium citrate, pH 6.0, 0.05% Tween 20), rinsed in distilled water, incubated for 1 hour at room temperature with blocking buffer (3% bovine growth serum, 0.05% Tween 20), and then incubated for 2 hours at room temperature with wheat germ agglutinin, Alexa Fluor 488 conjugate (50 μg/mL; Invitrogen; #W11261). Slides were then rinsed and mounted in VECTASHIELD (Vector Labs; #H-1000-10) and imaged on the EVOS Imaging System (Invitrogen). All images were imported into ImageJ software for quantification of cell cross-sectional area using built-in particle analysis setting. Measurements from 5 different images per cross-section were used to obtain an average cell area for 1 mouse heart.
Western blotting and quantification
Whole heart protein extracts were generated using RIPA buffer (150 mM NaCl, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA, 10 mM NaF, 50 mM Tris pH 7.4) with protease (cOmplete tablets, EDTA free; Roche) and phosphatase (phosphatase inhibitor cocktail sets I + II; Millipore) inhibitors. Briefly, the hearts were snap-frozen in liquid nitrogen and cryopulverized (Cole Parmer Tissue Pulverizer; #40355). The lysates were sonicated for 10 minutes in the water bath, centrifuged (4 °C × 21,000 g × 20 minutes), and quantified using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; # 23225). Standard Western blotting analysis was performed using 10% SDS-polyacrylamide gel electrophoresis gels with YTHDF2 (1:1000; Abcam; #ab220163) or MYZAP (1:1000, Sigma-Aldrich, ABT407) primary antibodies. Secondary antibody incubations were done at room temperature for 60 minutes using horseradish peroxidase–conjugated antibodies (1:10,000) and then imaged using a ChemiDoc system (Bio-Rad). Individual band intensity was quantified with ImageJ software using Ponceau-stained total proteins as a loading control. The normalized signals were calculated by dividing the signal intensities of each target protein (YTHDF2 or MYZAP) by the total loading factor (Ponceau).
Mass spectrometry–based proteomics analysis
Isolated adult cardiomyocytes from control or Y2-cKO mice (3 hearts per genotype) were lysed in TMT-MS lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.5% Na deoxycholate, 1% SDS, 1 mM EDTA without protease inhibitors), sonicated, and cleared at 20,000 g for 20 minutes at 4 °C. Supernatant concentrations were measured with BCA assay, and 100 μg of extracted proteins were submitted to The Ohio State University Campus Chemical Instrument Center Mass Spectrometry and Proteomics Facility. Tandem-mass tag analysis was performed by labeling all samples with reactive isobaric tags, mixed and analyzed in a single liquid chromatography tandem mass spectrometry experiment as previously described.33
RNA immunoprecipitations
Fresh mouse heart tissues were excised, homogenized with handheld homogenizer, and sonicated in buffer A (100 mM KCL, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% NP-40, 1 mM dithiothreitol, protease inhibitors). Sonicated samples were clarified by centrifugation at 4 °C for 15 minutes. Pulldown was performed by incubating 2 mg of protein extract with 5 μg of antibody (anti-YTHDF2, #24744-1-AP [Proteintech]; anti-rabbit immunoglobulin G [IgG], #12-370 [EMD Millipore]) at 4 °C for 4 hours, followed by the addition of 40 μL of protein A/G magnetic beads (#88803; Pierce) to the rotation overnight. The following day, samples were washed 5 times in buffer B (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), subjected to DNase I (#AM2238; Invitrogen) digest at 37 °C for 5 minutes, and then eluted in buffer B, supplemented with 1 mg/mL Proteinase K (#E195-5ML; VWR) and 0.1% SDS, at 55 °C for 30 minutes. Following elution, RNA from input and immunoprecipitation samples was isolated using standard phenol-chloroform extraction followed by reverse transcription and quantitative polymerase chain reaction (qPCR) with gene-specific primers. The experiments were performed in 3 independent technical replicates.
m6A immunoprecipitations
Total RNA was extracted from wild-type mouse heart tissues (n = 3 biological replicates) using TRIzol (Life Technologies). A total of 0.5 μg was reserved for input and subjected to reverse-transcription qPCR. A total of 10 μg of each purified RNA extract was incubated with 5 μg of m6A polyclonal antibody (Synaptic Systems) in ice-cold IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM TrisHCl, pH 7.4, and 200 U/mL RNase inhibitor) for 2 hours at 4 °C. Rabbit IgG antibody was used as a nonspecific control. Following the 2 hours incubation, prewashed Pierce A/G magnetic beads (#88803) were added into RNA-m6A immunoprecipitations and subjected to incubation for additional 2 hours at 4 °C. The beads were then washed 5 times with 1 mL ice-cold IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM TrisHCl, pH 7.4). The immunoprecipitation complexes were digested with proteinase K (in the presence of 0.1% SDS) at 55 °C for 1 hours, and the RNA was isolated using phenol-chloroform extraction followed by reverse transcription and qPCR with gene-specific primers.
mRNA analysis by real-time polymerase chain reaction
RNA was extracted using TRIzol and then reverse-transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Selected gene expression differences were analyzed by qPCR using SsoAdvanced SYBR Green Supermix (Bio-Rad) in a 96-well format and the ddCT method of analysis. Quantified mRNA levels were normalized to the housekeeping gene Rpl7, and expression is presented relative to control levels. Primers used were: mouse Ythdf2 5′-TGGTTCTGTGCATCAAAAGGA-3′ and 5′-CACCTCCAGTAGACCAAGCA-3′; mouse Myzap 5′-GCAGTTAGAGGCTGCTCAGATG-3′ and 5′-GCTTTCTCGTCATCTCCCGCAT-3′; mouse Fbn1 5′- GCTGTGAATGCGACATGGGCTT-3′ and 5′-TCTCACACTCGCAACGGAAGAG-3′; mouse Myh7 5-CCTGCGGAAGTCTGAGAAGG-3′ and 5′-CTCGGGACACGATCTTGGC-3′; mouse Esr1 5′-TCTGCCAAGGAGACTCGCTACT-3′ and 5′-GGTGCATTGGTTTGTAGCTGGAC-3′; mouse My6b 5′-GCTTCGTGTGTTTGACAAAGAGG-3′ and 5′-CTGCCAGAACAGTCTCTACCTC-3′.
Nuclear RNA extraction
Whole heart tissues from control or Y2-cKO mice (n = 4 per genotype) were pulverized and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.6, 50 mM NaCl, 5 mM MgCl2, 0.1% IGEPAL, 1 mM β-mercaptoethanol, with fresh protease and RNase inhibitors). After homogenization with a handheld glass-glass Dounce homogenizer (30 strokes), the samples were incubated on ice for 30 minutes and centrifuged at 2,000 g for 3 minutes. The pellets containing the nuclei were washed 2 times with lysis buffer, then resuspended in TRIzol, and nuclear RNA was extracted according to standard manufacturer’s instructions.
RNA and protein stability assays
mRNA and protein stability assays were performed in H9C2 rat cardiomyoblast cells transfected with either control nontargeting small interfering RNA (siRNA) or a pool of 3 rat-specific siRNAs targeting Ythdf2 gene (Integrated DNA technologies). For RNA stability, experiments were performed 48 hours after siRNA transfection with Lipofectamine RNAiMax (Thermo Fisher Scientific; #13778150) using 10 μg/mL Actinomycin D (#A1410; Sigma-Aldrich) as a transcription inhibitor. Cells were treated with actinomycin D for 0, 1, 2, 4, or 6 hours (n = 4 wells per condition). RNA was extracted by TRIzol, and the Myzap RNA levels were measured by qPCR using the following primers: rat Myzap 5′-TGCAGCAAACCTATGAAGCG-3′ and 5′-AGCTTTCTCGTCATCTCCCG-3′. The experiments were performed in 3 technical replicates.
For protein stability assays, cells transfected with either si-Ctrl or si-Y2 were treated with 50-μg/mL cycloheximide (#C4859; Sigma-Aldrich) as a translation elongation inhibitor. Cells were then collected in RIPA lysis buffer (150 mM NaCl, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 10 mM NaF, 50 mM Tris pH 7.4) with protease and phosphatase inhibitors at different time points (t = 0, 4, 6, 8, 10, 14, and 24 hours). The proteins were isolated, separated on a Western blot, and analyzed using the method described above (refer to Western Blotting and Quantification).
Statistical analysis
All results were presented as mean ± SEM, with dots indicating individual biological samples within a group. The Shapiro-Wilk method was used to determine the distribution of data. Normally distributed data were compared using the unpaired 2-tailed t test with Welch's correction for unequal variances or Wilcoxon rank sum test for skewed data. For the analysis of mass spectrometry, gene ontology over-representation analysis was performed using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit)34 with post hoc adjusted false discovery rate based on Bonferroni test. For RNA immunoprecipitation analysis, paired 1-tailed t test was used to assess for enrichment greater than in the IgG control. The best-fit values for mRNA and protein decay rates (k) and half-lives were calculated using nonlinear least-squares regression curve fitting (1 phase decay). A P value of ≤0.05 was considered statistically significant, and analyses were performed using GraphPad Prism 9 software (GraphPad Software).
Results
YTHDF2 regulation in stressed hearts and generation of mice lacking YTHDF2 in cardiomyocytes
To understand whether YTHDF2 is responsive to cardiac stress, we compared protein samples from murine hearts subjected to either sham operation (nonfailing) or pressure overload injury (failing) (Figures 1A and 1B). We found an increased YTHDF2 protein level in failing hearts compared with uninjured control hearts (Figures 1A and 1B), suggesting a crucial role for this protein in regulating cardiac homeostasis. To assess its physiological relevance, we generated a cardiomyocyte-specific and inducible loss-of-function mouse model for YTHDF2 (Y2-cKO) (Figure 1C). We achieved significant depletion of YTHDF2 levels from total hearts both at the mRNA and protein levels, suggesting a primary contribution of cardiomyocytes to the cardiac expression profile of YTHDF2 (Figures 1D to 1F). We then further validated the efficiency of our model on isolated cardiomyocytes, in which we achieved a near complete deletion of YTHDF2 (Figures 1G and 1H). Upon examining the mRNA levels of the other 2 YTH-domain containing paralogs in the heart, we observed that YTHDF1 but not YTHDF3 was up-regulated in the absence of YTHDF2 (Supplemental Figures 1A and 1B). Our myocyte-specific knockdown was further supported by considerable loss of YTHDF2 signal (green) from immunostained heart sections (Figure 1I). Interestingly, we also noticed enhanced YTHDF2 localization at the areas of cardiomyocyte cell-cell contacts reminiscent of intercalated discs, implying a potential involvement of YTHDF2 in the regulation of these specialized structures (Figure 1I, white arrowheads). In comparison, YTHDF1 and YTHDF3 showed distinct localization patterns (Supplemental Figures 1C and 1D), hinting at their differences in distribution and potentially divergent roles in the heart.
Figure 1.
YTHDF2 Regulation in Stressed Hearts and Generation of Mice Lacking YTHDF2 in Cardiomyocytes
(A) Western blot of YTHDF2 protein expression and total protein by Ponceau stain in nonfailing and failing mouse hearts. Heart failure samples were obtained from mice subjected to transverse aortic constriction, and nonfailing controls have undergone sham surgeries. (B) Quantification of YTHDF2 intensity using total protein by Ponceau stain as a loading control. (C) Schematic of cardiomyocyte-specific tamoxifen-inducible YTHDF2 cardiac knockout (Y2-cKO) mouse generation. (D) Quantitative polymerase chain reaction analysis of Ythdf2 messenger RNA (mRNA) expression in the whole hearts of control and Y2-cKO mice. (E) Western blot analysis of YTHDF2 protein expression in control and Y2-cKO whole hearts. (F) Quantification of YTHDF2 intensity on Western blot using total protein by Ponceau stain as a loading control. (G) Validation of YTHDF2 deletion by Western blot in control and Y2-cKO isolated cardiomyocytes. (H) Quantification of relative expression of YTHDF2/total protein by Ponceau stain. (I) Immunofluorescence analysis of YTHDF2 expression (green) in cardiac cross-sections from control and Y2-cKO mice. White arrowheads indicate YTHDF2 localization at the areas of cell-cell contacts between cardiomyocytes. DAPI = blue. Scale bar = 100 μm. Normally distributed data were compared using the unpaired 2-tailed t test with Welch's correction for unequal variances or Wilcoxon rank sum test for skewed data: ∗P ≤ 0.05, ∗∗∗P ≤ 0.001. Data are presented as mean ± SEM with the individual biological samples shown. Group sizes are listed in order of control and Y2-cKO mice: (A, B) n = 5, 5; (D) n = 8, 12; (E, F) n = 4, 4; (G, H) n = 3, 3. αMHC = alpha myosin heavy chain; ctrl = control; KO = knockout.
Loss of YTHDF2 leads to pathological cardiac remodeling and dysfunction
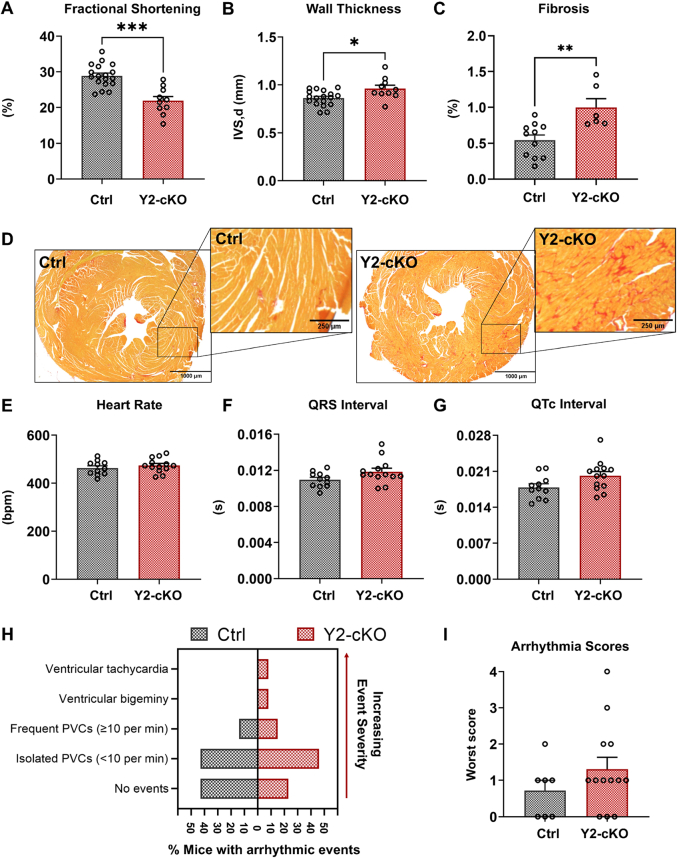
Loss of YTHDF2 from cardiomyocytes resulted in cardiac dysfunction, as illustrated by reduced fractional shortening after 1 month of deletion (Figure 2A, Supplemental Table 1). Echocardiographic analysis also showed increased left ventricular anterior wall thickness, suggesting the development of hypertrophy in mice lacking cardiac YTHDF2 (Figure 2B and Supplemental Table 1). The observed pathological changes were accompanied by increased fibrosis (Figures 2C and 2D), with no significant genotype-dependent contribution from the perivascular area (Supplemental Figure 2A). Using ECG monitoring, we detected mild changes in resting parameters with a trend toward longer QRS (P = 0.07) and QTc (P = 0.05) intervals in Y2-cKO mice, independent of their heart rates (Figures 2E and 2G, Supplemental Table 2). These mice also presented with longer P-wave duration, negative ST-segment heights, and inverted T-wave direction (Supplemental Figures 1B to 1G, Supplemental Table 2). The observed conduction abnormalities are consistent with the presence of hypertrophy and fibrosis but can also indicate a potential for arrhythmia susceptibility. Therefore, we performed caffeine and epinephrine challenge ECG in control and Y2-cKO animals. We found that 16% of our knockout mice developed ventricular arrhythmogenic events (bigeminy and sustained ventricular tachycardia), while none of the control animals displayed arrhythmia of comparable severity during the examined time points (Figure 2H, Supplemental Figure 2H). However, arrythmia scoring showed no significant changes (Figure 2I), and no deaths were observed in either group of mice.
Figure 2.
Loss of YTHDF2 Causes Cardiac Dysfunction and Fibrosis
(A) Fractional shortening (%) by echocardiography of the littermate control animals and Y2-cKO mice. (B) M-mode echocardiographic measurement of interventricular septal thickness in diastole (IVS,d) as a marker of left ventricular anterior wall thickness. (C) Quantification of fibrosis by Picrosirius red stain. (D) Representative images for Picrosirius red stain of heart cross-sections (fibrosis [red]). Scale bar = 1,000 μm for the full-sized images and 250 μm for the callouts. (E to G) Electrocardiography parameters recorded during baseline monitoring of the littermate controls and Y2-cKO mice. The values represent means calculated using LabChart Pro ECG Analysis throughout the 5-minute period of recording: (E) heart rates (beats/min), (F) QRS intervals (seconds), and (G) QTc intervals (seconds) adjusted to heart rates. (H, I) Results of the electrocardiogram (ECG) monitoring 15 minutes after epinephrine and caffeine injections. (H) Percent of mice of each genotype which developed either no events or ventricular arrhythmic events in order of increasing event severity (y-axis): isolated premature ventricular contractions (PVCs), frequent PVCs, bigeminy, or ventricular tachycardia. (I) Assigned worst arrhythmia scores to each mouse were based on the worst ventricular arrhythmia displayed and adapted from van der Werf et al32: 0 = no events; 1 = isolated PVCs (<10 per minute); 2 = frequent PVCs (≥10 per minute); 3 = ventricular bigeminy; 4 = ventricular tachycardia. Normally distributed data were compared using the unpaired 2-tailed t test with Welch's correction for unequal variances or Wilcoxon rank sum test for skewed data: ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗ P ≤ 0.001. Incidence of arrhythmic events in the 2 groups was compared by Fisher exact test. Data are presented as the mean ± SEM with the individual data points shown. Group sizes are listed in order of control and Y2-cKO mice: (A, B) n = 18, 10; (C) n = 11, 6; (E to G) n = 11, 13: (I) n = 7, 13. Abbreviations as in Figure 1.
Upon gravimetric examination, we further observed signs of heart failure such as lung congestion (Figure 3A) and increased heart size in Y2-cKO mice (Figure 3B). This finding correlated with an up-regulation of hypertrophic markers such as Nppa and Nppb in the absence of YTHDF2 (Figures 3C and 3D), altogether linking the lack of YTHDF2 to the development of cardiac pathology. To further characterize the observed hypertrophic remodeling, we quantified cardiomyocyte cross-sectional areas and found their increase in YTHDF2-deficient mice (Figures 3E and 3F). Using isolated cardiomyocytes, we could appreciate a more pronounced increase in length of Y2-cKO cells (Figure 3G) and geometrical changes with increased length/width ratios (Figures 3G to 3J), a morphological hallmark of eccentric pathologic hypertrophy.
Figure 3.
Loss of YTHDF2 Causes Cardiomyocyte Hypertrophic Remodeling
(A) Lung mass and (B) heart mass normalized to body mass after 1 month of YTHDF2 deletion. Quantitative polymerase chain reaction assessment of hypertrophic markers in control and Y2-cKO whole hearts: (C) Nppa and (D) Nppb; fold change in expression was normalized to housekeeping Rpl7 mRNA. (E) Measurements of cardiomyocyte cross-sectional areas by wheat germ agglutinin (WGA) staining. Individual data points represent the means taken from ≥4 images per animal with ≥100 cell areas/image measured. (F) Representative WGA-stained cardiac cross-sections (green). Scale bar = 50 μm. Measurements of individual cardiomyocyte (G) length, (H) width, and (I) length/width ratios (all from an average of n ≥ 50 cells per animal). (J) Representative bright-field images of isolated myocytes from the control and Y2-cKO mice. Scale bar = 50 μm. Normally distributed data were compared using the unpaired 2-tailed t test with Welch's correction for unequal variances or Wilcoxon rank sum test for skewed data: ∗P ≤ 0.05, ∗∗∗P ≤ 0.001. Data are presented as the mean ± SEM with the individual data points shown. Group sizes are listed in order of control and Y2-cKO mice: (A, B) n = 16, 10; (C, D) n = 8, 12; (E) n = 7, 5; (G to I) n = 3, 4. BW = body weight; HW = heart weight; LW = lung weight; other abbreviations as in Figure 1.
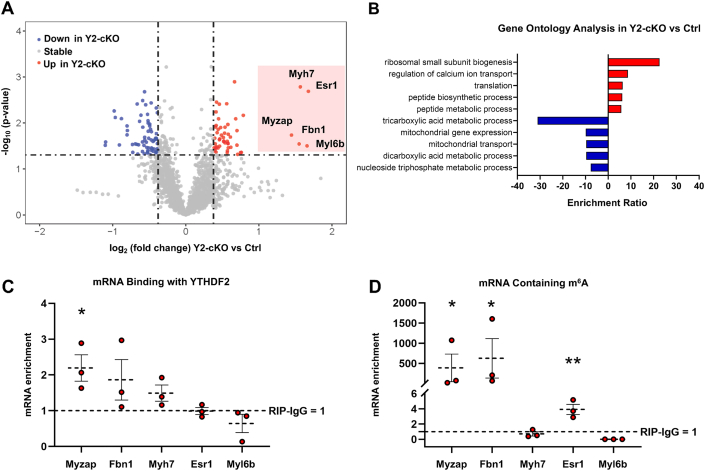
YTHDF2 deletion leads to proteomic reprogramming and up-regulation of m6A-modified Myzap
To dissect the molecular mechanism behind the observed cardiomyopathy in YTHDF2 null mice, we compared the proteomic profiles between control and Y2-cKO cardiomyocytes by mass spectrometry. We found 81 up-regulated and 122 down-regulated proteins in the knockout cardiomyocytes, accounting for 12% of the total detected proteome (Figure 4A, Supplemental Table 3). Gene Ontology analysis of enriched biological processes revealed down-regulation of metabolic pathways and up-regulation of ribosomal biogenesis and calcium signaling (Figure 4B). Notably, 5 proteins showed a >2.5-fold change, all of which were up-regulated in the absence of YTHDF2 (Figure 4A). To screen for the direct targets within these top affected candidates, we examined YTHDF2 binding to the correspondent transcripts by RNA immunoprecipitations from mouse hearts using YTHDF2-specific antibody compared with the nonspecific IgG control antibody. We found Myzap as the most enriched mRNA in complex with YTHDF2, followed by a trend in Fbn1 and Myh7 enrichment, and no binding was observed with Esr1 or Myl6b (Figure 4C). Using m6A immunoprecipitation assays, we then confirmed the presence of m6A-modified sites on Myzap mRNA, establishing it as a genuine m6A-modified and YTHDF2-binding transcript in the heart (Figure 4D). Interestingly, a previous report demonstrated that increased MYZAP expression at the intercalated disc leads to aggregate-associated cardiomyopathy and induction of pathological pro-hypertrophic signaling via serum response factor.35
Figure 4.
YTHDF2 Silencing Leads to Proteomic Reprogramming
(A) Tandem-mass-tag spectrometry to assess proteomic differences between control and Y2-cKO cardiomyocytes. Volcano plot of proteomic results, dashed lines mark fold change Y2-cKO/control = 1.3 and P value cutoff of 0.05. Up-regulated proteins in Y2-cKO are represented as red dots and down-regulated proteins as blue. (B) Gene Ontology over-representation analysis (biological process) was performed on all significantly up-regulated proteins (red) or down-regulated proteins (blue) from the mass spectrometry data using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit). The significance level was set to a false discovery rate ≤0.05 with a post hoc adjusted false discovery rate based on Bonferroni test. (C) RNA immunoprecipitation followed by quantitative polymerase chain reaction from mouse hearts using YTHDF2 antibody compared with control immunoglobulin G (IgG); fold enrichment with IgG was set as 1 (dashed line). (D) RNA enrichment with N6-methyladenosine (m6A) antibody as compared with the same mRNAs immunoprecipitated with IgG control antibody set as 1 (dashed line). Statistics were calculated using a paired 1-tailed t test to assess for enrichment greater than in the IgG control antibody: ∗P ≤ 0.05, ∗∗P ≤ 0.01. Data are presented as the mean ± SEM with the individual data points representing 3 independent replicates. Abbreviations as in Figure 1.
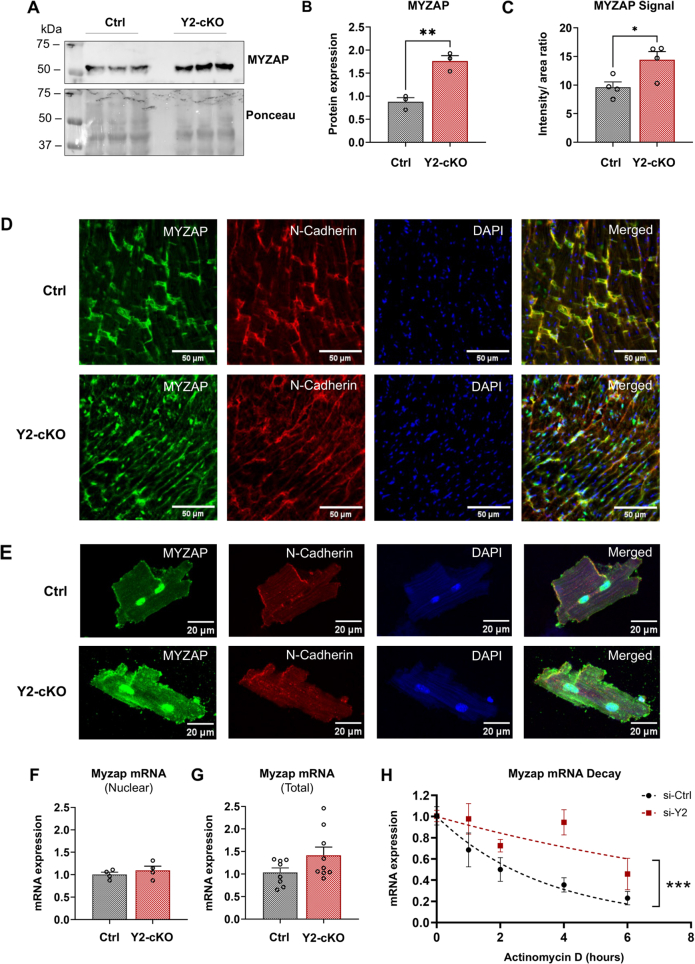
To assess the regulatory effect of YTHDF2 on MYZAP expression, we analyzed our Y2-cKO and control mouse hearts by Western blot and found significant increase in MYZAP protein levels in the absence of YTHDF2 (Figures 5A and 5B). To gain further insight about how the loss of YTHDF2 alters MYZAP in the heart, we performed immunofluorescence analyses in cardiac cryosections (Figures 5C and 5D) and in isolated cardiomyocytes (Figure 5E) from control and Y2-cKO mice. We observed a measurable increase in MYZAP fluorescence signal in Y2-cKO hearts, both at the intercalated discs and beyond (Figures 5C to 5E). Moreover, YTHDF2 deletion was accompanied by a less organized localization of another intercalated disc protein, N-cadherin (Figure 5D), hinting at a role for YTHDF2 in architectural maintenance of the intercalated disc. The observed increase in MYZAP upon YTHDF2 deletion cannot be explained by an effect on transcription, as Myzap mRNA levels were unchanged in the nucleus (Figure 5F) and were only mildly, not significantly, elevated in the total fraction (P = 0.098) (Figure 5G). Given that YTHDF2 has been implicated in controlling mRNA decay,23 we performed mRNA stability assays using H9C2 cardiomyoblasts transfected with YTHDF2-targeting siRNAs or the negative siRNA control cardiomyoblasts (Figure 5H). Loss of YTHDF2 prolonged the half-life of Myzap mRNA (Figure 5H) and slowed its predicted decay rate (si-Ctrl = 0.29 per hour vs si-Y2 = 0.09 per hour). In contrast, analysis of protein stability showed that the estimated rates of MYZAP decay did not differ significantly between the control and si-Y2 H9C2 cells (Supplemental Figures 3A and 3B). These findings are consistent with the notion that YTHDF2 regulates Myzap post-transcriptionally, via its control of mRNA and not protein stability.
Figure 5.
YTHDF2 Controls the Expression of MYZAP by Regulating Its mRNA Stability
(A) Western blot analysis of myocardial zonula adherens protein (MYZAP) expression in control and Y2-cKO whole hearts from 3 mice. (B) Quantification of MYZAP intensity on Western blot using total protein by Ponceau stain as a loading control condition. (C) Quantification of MYZAP fluorescence intensity in the cardiac cryosections taken from control vs Y2-cKO mice; ratios were calculated relative to cross-sectional area. (D) Immunofluorescence analysis of MYZAP expression (green) in cardiac cross-sections from control and Y2-cKO mice. N-cadherin (red) was used as a marker of intercalated disc, while DAPI (blue) marks the nuclei. Scale bar = 50 μm. (E) Representative images of adult cardiomyocytes isolated from control and Y2-cKO mice and immunostained for MYZAP (green), marker of intercalated disc N-cadherin (red), and DAPI (blue). Scale bar = 20 μm. (F) Quantitative polymerase chain reaction analysis of Myzap mRNA expression on nuclear RNA extracted from the hearts of control and Y2-cKO mice. (G) Quantitative polymerase chain reaction analysis of Myzap mRNA expression in the total heart lysates of control and Y2-cKO mice. (H) RNA stability assays using H9C2 cells (n = 4 wells), transfected with negative control cells transfected with nontargeting small interfering RNA (si-Ctrl) or a pool of cells transfected with small interfering RNAs targeting Ythdf2 gene (si-Y2). Myzap mRNA abundance at each time point is normalized to t = 0. The best-fit values for Myzap half-life and k decay rate were calculated using least-squares regression analysis: ∗∗∗P ≤ 0.001. Normally distributed data were compared using the unpaired 2-tailed t test with Welch's correction for unequal variances or Wilcoxon rank sum test for skewed data: ∗P ≤ 0.05, ∗∗P ≤ 0.01. Data are presented as the mean ± SEM with the individual biological samples shown. Group sizes are listed in order of control and Y2-cKO mice: (A, B) n = 3, 3; (C, F) n = 4, 4; (G) n =8, 9. Abbreviations as in Figure 1.
Taken together, our study highlighted a new YTHDF2-dependent post-transcriptional mechanism for regulation of an important intercalated disc protein, MYZAP, and the overall detrimental effect of YTHDF2 loss to the cardiomyocyte homeostasis and cardiac function.
Discussion
Post-transcriptional events, such as regulation of cardiac mRNA fates by RBPs, open exciting new pathways to manipulate pathological gene switches in the heart. Because RBPs coordinate every step of mRNA life, they add another layer of complexity to the orchestration of cardiac gene expression. In this study, we identify an essential role of the cardiac YTHDF2, which binds and regulates mRNAs with m6A modification.
Our study provides important contributions to the recent debate on the functions of the YTHDF m6A binding proteins. Two opposing models exist: in one, they each bind to distinct populations of mRNAs and perform different functions,22, 23, 24 and in another, they bind identical mRNA targets and perform redundant functions to enable mRNA degradation.25, 26, 27 Our findings reveal that YTHDF2 is an indispensable cardiac RBP, and its deletion alone is sufficient to disrupt cardiac homeostasis. Our results align well with the study published by Xu et al,28 who used viral tools to manipulate YTHDF2 in the heart and found a cardioprotective role for YTHDF2 in the setting of injury. In our study, YTHDF1 but not YTHDF3 was up-regulated in the absence of YTHDF2, but this was not sufficient to rescue the physiological deficiency displayed by Y2-cKO animals. Moreover, we found that YTHDF2 had a unique localization profile as compared with its family members YTHDF1 and YTHDF3, further supporting the model in which they take on distinct roles. Taken together, the function of YTHDF2 in the heart could not be compensated by the remaining members of its family, at least in the tested conditions.
Our proteomics screen revealed that 12% of the detectable proteins were significantly altered in the cardiomyocytes lacking YTHDF2. Among these, we identified 5 proteins that were up-regulated more than 2.5-fold in YTHDF2 null cardiomyocytes compared with the control cells, while none of the down-regulated hits were affected to a comparable degree. One logical explanation is that the most direct effects of YTHDF2 silencing in cardiomyocytes lead to increased mRNA stability and the resultant up-regulation of its targets. This aligns with a model in which YTHDF2 is the primary regulator of mRNA degradation in its family, displaying greater efficiency in this role compared with YTHDF1 and YTHDF3.36,37 Indeed, our RNA-YTHDF2 immunoprecipitation assays from mouse hearts showed some degree of enrichment with 3 out of these 5 mRNAs (Myzap, Fbn1, and Myh7). One of these targets is also consistent with a previous publication, in which YTHDF2 was shown to bind and regulate Myh7 mRNA.28
While our mass spectrometry approach allowed us to detect global proteomic changes in response to YTHDF2 deletion, it also presented challenges in differentiating between direct vs indirect effects. For example, we observed down-regulation of metabolic regulators. Because these processes are frequently altered in the failing cardiomyocytes, these findings likely represent indirect effects of YTHDF2 deletion causing cardiomyopathy in our animals. Our YTHDF2-cKO cardiomyocytes also showed higher levels of ribosomal protein biogenesis and calcium ion transport. The former may also be a secondary consequence of the underlying cardiomyocyte hypertrophy. As for the increase in calcium ion transport, this raises interesting future questions about the potential of YTHDF2 to regulate calcium signaling. Aligned with a previous observation that loss of m6A demethylation by FTO results in increased stability of certain calcium-handling mRNAs, our YTHDF2-deficient model may function in a similar fashion by conferring increased stability for calcium ion transporter transcripts.17
Our study highlights one key mRNA coding for the intercalated disc protein MYZAP as a particularly interesting target of YTHDF2 undergoing post-transcriptional regulation. We show that YTHDF2 is enriched at the intercalated disc, can directly bind the m6A-modified mRNA sequence of Myzap and trigger its decay. In response to YTHDF2 deletion in adult mouse hearts, we show aberrantly increased MYZAP expression, both at the intercalated disc and beyond. On the other hand, the increase in total Myzap mRNA levels did not reach significance in the absence of YTHDF2; this could be explained by the post-transcriptional nature of the described gene regulatory mechanism in which the contribution of transcription could mask mRNA stability effects during total mRNA analyses.
The cardiac importance of MYZAP has been well established, and the tight control of its expression and activity is essential for maintenance of contractility and appropriate responses to stress.38, 39, 40 Specifically, the cardiac-restricted overexpression of MYZAP has been shown to result in the activation of the prohypertrophic genes under the control of the serum response transcription factor and MYZAP aggregate-driven cardiomyopathy at intercalated disc.35 Recent data indicate that the intercalated disc serves as a distinct functional unit, controlling not only electromechanical transmission, but also molecular signal transmission between and within the cardiomyocytes.41,42 Given the importance of the intercalated disc in the electrical conduction, it is not surprising that our Y2-cKO mice showed several ECG abnormalities at baseline, such as trending increases in QRS and QTc intervals. These trends are consistent with slowing of conduction and an intercalated disc pathology. However, only 16% of our knockout mice developed severe ventricular arrhythmic events after caffeine and epinephrine stimulation (which did not reach statical significance) and no deaths were observed between the 2 genotypes, implying that YTHDF2 deletion may not be sufficient to fully alter the electrical coupling. Instead, its damaging effect may be more pronounced in regard to the dysregulation of molecular signals relayed via the intercalated disc, primarily by causing adverse hypertrophic switch.42
In our mouse heart failure model induced by pressure overload, we found that YTHDF2 protein expression was increased. This finding is consistent with previous reports in human and rodent heart failure.28,43 Uniquely from previous reports, we also discovered that YTHDF2 is specifically enriched at the intercalated disc in adult cardiomyocytes. This further endorses the novel potential of YTHDF2 to regulate intercalated disc biology and function. Recent studies highlight that localized translation at the intercalated disc can facilitate protein complex assembly, allowing for efficient and timely synthesis of proteins from locally available mRNAs.44 Regulation of mRNA turnover is needed as a failproof checkpoint mechanism to prevent translation of the transcripts that have not passed quality control or that have been improperly localized. It is, therefore, possible that an underappreciated role of YTHDF2 in cardiomyocytes involves the quality control of mRNA-to-protein synthesis by the means of localized mRNA decay. Future studies examining the local control of mRNA fate at the intercalated disc by YTHDF proteins would be key to understanding the m6A-mediated mechanisms of mRNA turnover and quality control in cardiomyocytes.
Study Limitation
While Myzap was critically and directly regulated at the post-transcriptional level by YTHDF2, we cannot currently exclude a potential contribution of additional pathways to our observed cardiac phenotypes. Future transcriptomewide analyses in cardiomyocytes, such as RNA immunoprecipitations followed by sequencing, could uncover a more comprehensive list of YTHDF2 targets in the heart.
Conclusions
Altogether, our work highlights the role of YTHDF2 in regulating the stability of important transcripts in cardiomyocytes, such as Myzap, and the overall necessity of YTHDF2 for maintenance of cardiac homeostasis.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: The preclinical study presented here highlights a new molecular mechanism underlying pathologic cardiac remodeling and heart failure. We demonstrated the importance of preserving YTHDF2 function and maintaining normal levels of its targets (ie, MYZAP) for preservation of cardiac function. We discovered how regulation of mRNA stability by YTHDF2 is critical for controlling gene expression in cardiomyocytes and preventing their hypertrophic remodeling.
TRANSLATIONAL OUTLOOK: YTHDF2 is an RBP that regulates m6A-modified mRNAs. Us and others have established the importance of controlling m6A levels for preservation of cardiac homeostasis. m6A content increases in human heart failure samples independent of the disease etiology. By dissecting this clinically relevant pathway, our study may open the way to the development of new therapeutic strategies for heart disease patients.
Funding Support and Author Disclosures
This work was supported by the National Institutes of Health under grants R01 HL136951 and R01 HL154001 to Dr Accornero and R01 HL142588 to Dr Terentyev; grant F30 HL160104 to Dr Golubeva; and US-Israel Binational Science Foundation grant number 2017094 to Dr Accornero. Resources from the Campus Chemical Instrument Center Mass Spectrometry and Proteomics Facility and the Campus Microscopy and Imaging Facility at the Ohio State University were supported by National Institutes of Health support grants S10 OD025008 and P30CA016058. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors are thankful for the resources from the Campus Chemical Instrument Center Mass Spectrometry and Proteomics Facility and the Campus Microscopy and Imaging Facility at the Ohio State University.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental figures and tables, please see the online version of this paper.
Appendix
References
- 1.Haque Z.K., Wang D.Z. How cardiomyocytes sense pathophysiological stresses for cardiac remodeling. Cell Mol Life Sci. 2017;74:983–1000. doi: 10.1007/s00018-016-2373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molkentin J.D., Lu J.R., Antos C.L., et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang Q., De Windt L.J., Witt S.A., Kimball T.R., Markham B.E., Molkentin J.D. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J Biol Chem. 2001;276:30245–30253. doi: 10.1074/jbc.M102174200. [DOI] [PubMed] [Google Scholar]
- 4.Tham Y.K., Bernardo B.C., Ooi J.Y., Weeks K.L., McMullen J.R. Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch Toxicol. 2015;89:1401–1438. doi: 10.1007/s00204-015-1477-x. [DOI] [PubMed] [Google Scholar]
- 5.Doroudgar S., Hofmann C., Boileau E., et al. Monitoring cell-type-specific gene expression using ribosome profiling in vivo during cardiac hemodynamic stress. Circ Res. 2019;125:431–448. doi: 10.1161/CIRCRESAHA.119.314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Heesch S., Witte F., Schneider-Lunitz V., et al. The translational landscape of the human heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Li D., Morales A., Gonzalez-Quintana J., et al. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3:90–97. doi: 10.1111/j.1752-8062.2010.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis J., Salomonis N., Ghearing N., et al. MBNL1-mediated regulation of differentiation RNAs promotes myofibroblast transformation and the fibrotic response. Nat Commun. 2015;6 doi: 10.1038/ncomms10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher T.L., Arribere J.A., Geurts P.A., et al. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol. 2011;359:251–261. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoilov P., Rafalska I., Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem Sci. 2002;27:495–497. doi: 10.1016/s0968-0004(02)02189-8. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Theler D., Kaminska K.H., et al. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285:14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Ming J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. 2015;290:24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Yue Y., Han D., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia G., Fu Y., Zhao X., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng G., Dahl J.A., Niu Y., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathiyalagan P., Adamiak M., Mayourian J., et al. FTO-dependent N(6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139:518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorn L.E., Lasman L., Chen J., et al. The N(6)-methyladenosine mrna methylase mettl3 controls Cardiac Homeostasis and Hypertrophy. Circulation. 2019;139:533–545. doi: 10.1161/CIRCULATIONAHA.118.036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kmietczyk V., Riechert E., Kalinski L., et al. m(6)A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci Alliance. 2019;2 doi: 10.26508/lsa.201800233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berulava T., Buchholz E., Elerdashvili V., et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. 2020;22:54–66. doi: 10.1002/ejhf.1672. [DOI] [PubMed] [Google Scholar]
- 21.Hinger S.A., Wei J., Dorn L.E., et al. Remodeling of the m(6)A landscape in the heart reveals few conserved post-transcriptional events underlying cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2021;151:46–55. doi: 10.1016/j.yjmcc.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Zhao B.S., Roundtree I.A., et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Lu Z., Gomez A., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H., Wang X., Lu Z., et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaccara S., Jaffrey S.R. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020;181:1582–1595.e18. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasman L., Krupalnik V., Viukov S., et al. Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev. 2020;34:1373–1391. doi: 10.1101/gad.340695.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Bedi R.K., Moroz-Omori E.V., Caflisch A. Structural and dynamic insights into redundant function of YTHDF proteins. J Chem Inf Model. 2020;60:5932–5935. doi: 10.1021/acs.jcim.0c01029. [DOI] [PubMed] [Google Scholar]
- 28.Xu H., Wang Z., Chen M., et al. YTHDF2 alleviates cardiac hypertrophy via regulating Myh7 mRNA decoy. Cell Biosci. 2021;11:132. doi: 10.1186/s13578-021-00649-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y., Mbikyo M., Zhang J., Zhang Y., Zhang N., Li Z. The lncRNA MIAT regulates CPT-1a mediated cardiac hypertrophy through m(6)A RNA methylation reading protein Ythdf2. Cell Death Discov. 2022;8:167. doi: 10.1038/s41420-022-00977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., Yu P., Wang J., et al. Downregulation of circ-ZNF609 promotes heart repair by modulating RNA N(6)-methyladenosine-modified Yap expression. Research (Wash D C) 2022;2022 doi: 10.34133/2022/9825916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louch W.E., Sheehan K.A., Wolska B.M. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol. 2011;51:288–298. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Werf C., Kannankeril P., Sacher F., et al. Flecainide therapy reduces exercise-induced ventricular arrhythmias in patients with catecholaminergic polymorphic ventricular tachycardia. J Am Coll Cardiol. 2011;57:2244–2254. doi: 10.1016/j.jacc.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moliva J.I., Duncan M.A., Olmo-Fontánez A., et al. The lung mucosa environment in the elderly increases host susceptibility to mycobacterium tuberculosis infection. J Infect Dis. 2019;220:514–523. doi: 10.1093/infdis/jiz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank D., Rangrez A.Y., Poyanmehr R., et al. Mice with cardiac-restricted overexpression of Myozap are sensitized to biomechanical stress and develop a protein-aggregate-associated cardiomyopathy. J Mol Cell Cardiol. 2014;72:196–207. doi: 10.1016/j.yjmcc.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y., Choe J., Park O.H., Kim Y.K. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 2020;36:177–188. doi: 10.1016/j.tig.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Flamand M.N., Ke K., Tamming R., Meyer K.D. Single-molecule identification of the target RNAs of different RNA binding proteins simultaneously in cells. Genes Dev. 2022;36:1002–1015. doi: 10.1101/gad.349983.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seeger T.S., Frank D., Rohr C., et al. Myozap, a novel intercalated disc protein, activates serum response factor-dependent signaling and is required to maintain cardiac function in vivo. Circ Res. 2010;106:880–890. doi: 10.1161/CIRCRESAHA.109.213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maver A., Zigman T., Rangrez A.Y., et al. A biallelic loss-of-function variant in MYZAP is associated with a recessive form of severe dilated cardiomyopathy. Cold Spring Harb Mol Case Stud. 2022;8:a006221. doi: 10.1101/mcs.a006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rangrez A.Y., Eden M., Poyanmehr R., et al. Myozap Deficiency promotes adverse cardiac remodeling via differential regulation of mitogen-activated protein kinase/serum-response factor and beta-catenin/GSK-3beta Protein signaling. J Biol Chem. 2016;291:4128–4143. doi: 10.1074/jbc.M115.689620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delmar M. The intercalated disk as a single functional unit. Heart Rhythm. 2004;1:12–13. doi: 10.1016/j.hrthm.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Manring H.R., Dorn L.E., Ex-Willey A., Accornero F., Ackermann M.A. At the heart of inter- and intracellular signaling: the intercalated disc. Biophys Rev. 2018;10:961–971. doi: 10.1007/s12551-018-0430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B., Xu Y., Cui X., et al. Alteration of m6A RNA methylation in heart failure with preserved ejection fraction. Front Cardiovasc Med. 2021;8 doi: 10.3389/fcvm.2021.647806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis Y.E., Moskovitz A., Mutlak M., Heineke J., Caspi L.H., Kehat I. Localization of transcripts, translation, and degradation for spatiotemporal sarcomere maintenance. J Mol Cell Cardiol. 2018;116:16–28. doi: 10.1016/j.yjmcc.2018.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.