Upregulation of SLC7A11 can be induced by androgen receptor variants to inhibit antiandrogen-induced prostate cancer cell ferroptosis and to drive castration resistance in prostate cancer.
Abstract
Androgen receptor (AR) inhibition by androgen deprivation and/or antiandrogen administration is the mainstay therapy for advanced prostate cancer. However, most prostate cancers ultimately become resistant to these therapies, indicating the importance of identifying mechanisms driving resistance to improve patient outcomes. Here we demonstrated that acute treatment with the antiandrogen enzalutamide (ENZ) decreased glutathione (GSH) production, increased lipid peroxidation, and induced ferroptosis in prostate cancer cells. Consistently, meta-analysis of transcriptomic data linked the androgen-AR axis to metabolism-related biological processes, including lipid metabolism. The cystine transporter gene SLC7A11 was a key AR target, and full-length AR (AR-FL) transactivated SLC7A11 transcription by directly occupying the SLC7A11 promoter and putative enhancer regions. AR variants (AR-V) preferentially bound the SLC7A11 enhancer and upregulated SLC7A11 expression, thereby conferring resistance to ferroptosis induced by ENZ treatment. However, this effect was abolished following downregulation of AR-Vs using the dual CBP/p300 and BET inhibitor NEO2734. These findings reveal ferroptosis induction as an anticancer mechanism of antiandrogens and SLC7A11 as a direct target gene of AR-FL and AR-Vs. AR-V-mediated SLC7A11 expression represents a mechanism coupling ferroptosis resistance to prostate cancer progression.
Significance:
Upregulation of SLC7A11 can be induced by androgen receptor variants to inhibit antiandrogen-induced prostate cancer cell ferroptosis and to drive castration resistance in prostate cancer.
Introduction
Ferroptosis is a unique form of cell death that is biochemically and morphologically distinct from other types of cell death such as apoptosis, necrosis, autophagy, and pyroptosis (1–3). Ferroptosis is known to be caused by lipid peroxidation and intracellular iron-mediated membrane oxidative damage induced by cystine depletion (4–6). Solute carrier family 7 member 11 (SLC7A11) is a subunit of the cystine/glutamate antiporter system Xc− and is the major transporter of extracellular cystine (7–9). Cystine depletion or treatment of chemicals such as erastin, which blocks SLC7A11-mediated cystine uptake, triggers ferroptosis (3). Intercellular cystine is rapidly converted to cysteine, which subsequently serves as the rate-limiting precursor for glutathione synthesis. Glutathione peroxidase 4 (GPX4) utilizes reduced glutathione (GSH) to decrease lipid peroxides to lipid alcohols to protect cells against membrane lipid peroxidation and inhibit ferroptosis (10, 11). It is well established that cell death plays important roles in tumor suppression (12, 13). Ferroptosis can be regulated at epigenetic, transcriptional, posttranscriptional, and posttranslational levels. Activation of ferroptosis results in inhibition of certain cancer types including prostate cancer (14). The roles and regulatory mechanisms of ferroptosis in prostate cancer progression remain poorly understood.
Prostate cancer is the leading cause of cancer-related mortality in men in Western countries. Androgen receptor (AR) is the critical transcription factor that is required for normal prostate development (15–17). AR also plays a pivotal role in prostate cancer development and progression. Androgen signaling pathway inhibition is only effective in AR-positive prostate cancer, and hormone naïve patients with prostate cancer have benefitted largely from androgen deprivation therapies (ADT; refs. 18, 19). However, most prostate cancer cases eventually progress into castration-resistant prostate cancer (CRPC). The underlying mechanisms remain elusive, and it is unclear whether castration resistance is related to deregulation of ferroptosis in prostate cancer.
AR variants (AR-V) have been implicated in the development of CRPC (20, 21). AR-Vs can be derived through mechanisms such as intronic polyadenylation-coupled alternative splicing of the AR gene (22, 23). AR-Vs encode various truncated AR proteins partially or completely lacking a functional ligand-binding domain in the C terminus (20). Increasing evidence indicates that AR-Vs are constitutively active in the absence of androgens or in the present of antiandrogens, thus promoting prostate cancer progression by conferring antiandrogen resistance (21, 24, 25). Previous studies have shown that androgen deprivation or treatment of antiandrogen such as enzalutamide (ENZ) induces AR-V expression in prostate cancer cells in vitro and in vivo (22, 24).
In this study, we identified the SLC7A11 gene as a bona fide transactivation target of AR in prostate cancer cells. Inhibition of the AR by ENZ impaired GSH production and sensitized prostate cancer cells to ferroptosis by repressing SLC7A11 expression. However, sustained expression of AR-Vs in ENZ-treated cells drove SLC7A11 expression and thereby promoted castration-resistance due to ferroptosis inhibition. Cotreatment of CBP/p300 and BET dual inhibitor NEO2734 inhibited AR-V expression and overcame resistance to ferroptosis and ENZ in prostate cancer cells.
Materials and Methods
Cell lines, cell culture, plasmids, and chemicals
LNCaP, 22Rv1, and HEK293T cell lines were purchased from the ATCC. C4–2 cell line was purchased from Uro Corporation. LNCaP, C4–2, and 22Rv1 cells were cultured in RPMI1640 cell culture medium (Corning) containing 10% FBS, together with 100 μg/mL streptomycin and 100 U/mL penicillin. HEK293T cells were cultured in DMEM cell culture medium (Corning) containing 10% FBS, together with 100 μg/mL streptomycin and 100 U/mL penicillin. Cells were cultured in incubator with 37°C and 5% CO2. All cell lines were authenticated by short tandem repeat profiling and tested as Mycoplasma free. FLAG-AR expression vector was generated as described previously (26). To generate HA-SLC7A11 expression vector, SLC7A11 cDNA was amplified by PCR using indicated primers (SLC7A11-F: 5′-CGGTCGACCATGGTCAGAAAGCCTGTTGTGTCCA CC-3′, SLC7A11-R: 5′-CAGCGGCCGCTTATAACTTATCTTCTTCTGGTACAACTTCC AGTAT-3′) and subcloned into the pCMV-HA vector. Ferroptosis inhibitor Ferr-1 and deferoxamine (DFO) were obtained from Cayman Chemical. ENZ and Bavdegalutamide ARV110 were obtained from TargetMOL. dihydrotestosterone (DHT) was purchased from Selleckchem. Z-VAD-FMK was obtained from MedChemExpress. NEO2734 was kindly provided by Epigenetix Inc. The activity of NEO2734 in inhibition of BET as well as CBP/p300 bromodomains was initially confirmed with the BROMOscan platform (DiscoverX/Eurofins; ref. 27).
Prostate cancer patient samples
Prostate cancer specimens used for IHC were obtained from the tissue registry of Mayo Clinic. The studies were approved by the Institute Review Board of Mayo Clinic. All specimens were de-identified from patient information. All participants provided written informed consent. This study was conducted in accordance with the guidelines and tenets of the Declaration of Helsinki.
Transfection and stable cell-line generation
Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) was used to transfect prostate cancer cells with siRNAs. Lentivirus transduction system was utilized to generate stable cell lines with specific gene knockdown. PEI was used to transfect shRNA plasmids together with lentivirus package plasmids (PSPAX2 and PMD2.G) into HEK293T cells. Forty-eight hours after transfection, supernatant containing viruses was collected, filtered, and utilized to infect indicated cells. Polybrene (8 μg/mL) was added to the viral supernatant to increase the infection efficiency. Forty-eight hours after infection, culture medium was replaced with fresh medium, and puromycin (1 μg/mL) was administrated for cell selection. Sequence information of siRNAs and shRNAs is provided in Supplementary Table S1.
Lipid peroxidation assay
Cells were incubated in a 60 mm dish containing 5 μmol/L BODIPY 581/591 C11 dye (Cayman, 27086) for 25 minutes. Cells were washed with PBS, trypsinized and subjected to flow cytometry analysis using flow cytometer as described previously (28).
Western blotting and IHC
Detailed information regarding antibodies used for Western blotting and IHC is provided in Supplementary Table S2. For Western blotting analysis, cells were lysed in modified RIPA buffer (50 mmol/L Tris-HCl pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mmol/L NaCl, 0.1% SDS, and 1 mmol/L EDTA) supplemented with 1% protease inhibitor cocktail. Protein concentration was determined using DC protein assay reagent (Bio-Rad). Cell extracts were supplemented with 10% DTT (Thermo Fisher Scientific) and boiled at 95°C for 3 minutes. Samples were subjected to SDS-PAGE (Bio-Rad) separation, and the gels were further transferred to nitrocellulose (NC) membranes (Thermo Fisher Scientific). After transferring, the NC membranes were blocked in 5% nonfat milk (Bio-Rad) for 1 hour at room temperature and incubated with the indicated primary antibodies at 4°C overnight. Next day, the NC membranes were washed with 1× TBST for 10 minutes three times and incubated with matched secondary antibody for 1 hour at room temperature. The membranes were washed with 1× TBST for 10 minutes three times. Finally, the signals were developed with SuperSignal West Pico Luminal Enhancer Solution (Thermo Fisher Scientific) on autoradiography films (HyBlot CL).
For IHC staining, prostate cancer patient specimens and xenograft tumor samples were fixed by formalin and embedded in paraffin. Four-micrometer-thick sections were cut from the samples and mounted onto slides. Antigen retrieval and immuno-staining were performed as described previously (29). IHC staining was scored based on the “most common” criteria. Staining score = staining intensity × staining positivity. Staining intensity was graded into four categories: 0, 1, 2, and 3. Specifically, 0 = no staining, 1 = weak staining (staining obvious only at ×400), 2 = medium staining (staining obvious at ×100 but not ×40), and 3 = strong staining (staining obvious at ×40). For staining positivity, 0 = no positive cells, 1 = 10% or less of positive cells, 2 = 11% to 50% positive cells, 3 = 51% to 70% positive cells, 4 = 71% or more positive cells.
Biochemical experiments
To assess intracellular GSH concentration, GSH assay (Cayman Chemical) was performed according to manufacturer's specifications. In brief, cell lysate supernatant from indicated treatments were reacted with monochlorobimane to generate a highly fluorescent product that can be measured using excitation and emission wavelengths of 380 and 480 nm, respectively.
To assess cytotoxicity, LDH assay (Eton Bioscience) was performed according to the manufacturer's introductions. In brief, LNCaP and C4–2 cells were cultured in 6-well adherent plates treated with ENZ or AR PROTAC ARV110 (1 μmol/L) for 48 hours. Lactic acid from indicated culture media was oxidized by enzyme reactions to yield color product, which can be measured at 570 nm for colorimetric assay.
Transmission electron microscopy
Indicated prostate cancer cells were fixed with 2.5% glutaraldehyde in phosphoric acid buffer for 2 hours, followed by after fixation in 1% osmium acid for 2 hours. Cell sections were dehydrated in ethanol and embedded in acetone. The sections (50–60 nm) were stained with 3% uranium acetate and lead citrate. TEM images were captured using the transmission electron microscope (JEM-1011).
Quantitative reverse transcription and PCR
Total RNA was extracted from cells using TRIzol reagent (Ambion) and reversely transcribed into cDNA using the GoScript Kit (Promega). The SYBR Green Mix (Bio-Rad) and CFX96 Real-Time System (Bio-Rad) were utilized to conduct the real-time PCR according to manufacturer's instruction. Expression of ACTB housekeeping gene was used as an internal control, and data were present as mean ± SD. Sequence information for primers used for qRT-PCR is provided in Supplementary Table S3.
Quantitative chromatin immunoprecipitation PCR
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (22). In brief, chromatin was cross-linked for 10 minutes at room temperature with 11% formaldehyde/PBS solution added to cell culture medium. Cross-linked chromatin was sonicated, diluted, and immunoprecipitated with Protein G-plus Agarose beads (Bio-Rad) prebound with antibody at 4°C overnight. Precipitated protein–DNA complexes were eluted, and cross-linking was reversed at 65°C for 16 hours. DNA fragments were purified and analyzed by qPCR. qChIP-PCR data were analyzed as % input after normalizing each ChIP DNA fraction's Ct value to the input DNA fraction's Ct value. Sequence information for primers used for qChIP-PCR is provided in Supplementary Table S3.
Bioinformatic analysis
The raw reads of RNA-seq were aligned to the human genome reference (hg19) using STAR 2.7.10. The bam2wig.py in RSeQC was used to transform the bam files to wiggle format, which was transferred to bigwig format to be visualized in the UCSC Genome Browser.
MTS cell proliferation assay
Cell proliferation was measured using MTS assay (Promega) according to manufacturer's instruction. Briefly, cells were seeded in a 96-well plate with a density of 1,500 cells per well. At the indicated time points, 10 μL CellTiter 96R Aqueous One Solution reagent (Promega) was added to cells. After incubating at 37°C incubator for 2 hours, cell growth was measured in a microplate reader with absorbance at 490 nm.
Colony formation assay
Cells were resuspended in fresh culture medium, seeded in triplicate with a density of 2,000 cells/well for 22Rv1, 1,500 cells/well for LNCaP and C4–2 in six-well plates, and cultured under normal growth conditions for 10 to 14 days. Colonies were washed three times in PBS, and fixed with 4% paraformaldehyde for 0.5 hours. The colonies were further stained by 0.5% crystal violet for 1 hour and washed with water twice to remove the crystal violet. The number of colonies in each well was counted using an inverted microscope.
Generation of xenografts and PDX tumors and drug treatment
The animal studies were approved by the Institutional Animal Care and Use Committee at the Mayo Clinic. C4–2 xenograft experiments were performed as described previously (26). Briefly, C4–2 cells (3 × 106) were mixed with Matrigel [in 50 μL of 1 × PBS plus 50 μL of Matrigel (BD Biosciences)] and injected subcutaneously into the right flank of 6-week-old male mice. After xenografts reached a size of ∼200 mm3, animals were randomly divided into two groups for treatment with vehicle or ENZ (10 mg/kg daily). 22Rv1 cells (3 × 106) were mixed with Matrigel [in 50 μL of 1 × PBS plus 50 μL of Matrigel (BD Biosciences)] and injected subcutaneously into the right flank of 6-week-old male mice. After xenografts reached a size of ∼100 mm3 and animals were randomly divided into different groups for treatment daily with vehicle, ENZ (10 mg/kg), ARV110 (5 mg/kg), NEO2734 (10 mg/kg), or combination of ENZ and NEO2734. Treatments were administrated 5 days per week by intraperitoneal injection and tumor volumes were measured using a digital caliper. LuCaP35 PDX was provided by E. Corey from the University of Washington. For the LuCaP35 PDX study, PDX tumors were divided into small pieces (∼1 mm3) and injected subcutaneously into 6-week-old male mice. After xenografts reached a size of ∼200 mm3, animals were randomly divided into two groups. One group was treated with sham castration and the other group was castrated surgically. After 1 week, tumors were harvested and proceeded for Western blot and qRT-PCR.
Statistical analysis
All data are presented as the mean ± SE from three or more independent experiments. Differences between two groups were analyzed using unpaired Student t test. Survival curves were obtained using the Kaplan–Meier method, and the log-rank test was used to test the difference in survival curves. P values <0.05 were considered statistically significant.
Data availability
The next-generation sequencing data from The Cancer Genome Atlas (TCGA), Stand Up To Cancer (SU2C), and Neuroendocrine Prostate Cancer (NEPC) cohort were obtained and analyzed via cBioPortal (https://www.cbioportal.org/). The microarray data from the primary and mCRPC cohort were obtained from the Gene Expression Omnibus database with the accession number GSE32269. All data supporting the findings of this study are available within the article and the Supplementary Information files. The relevant reagents such as plasmids are available from the corresponding authors upon request. All other raw data are available upon request from the corresponding authors.
Results
AR inhibition suppresses GSH production and promotes ferroptosis in prostate cancer
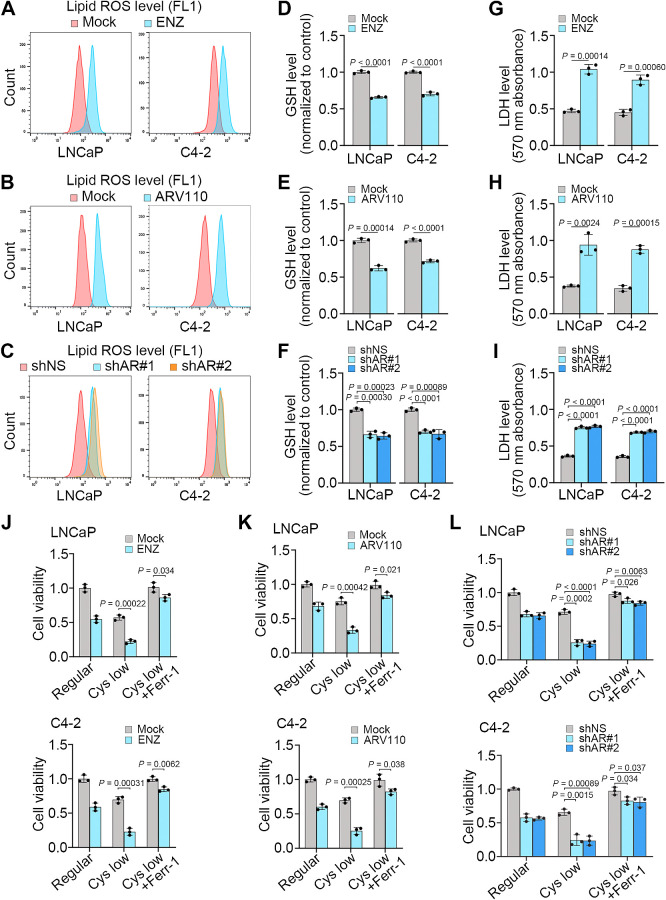
Androgen ablation has long been documented to induce cell death such as apoptosis in the normal prostate and prostate cancer (30). Prostate cancer is a malignancy having a high-rate lipid metabolism (31). We therefore sought to determine whether AR inhibition induces lipid peroxidation and ferroptosis in prostate cancer cells. As expected, inhibition of AR by ENZ decreased expression of KLK3, a well-studied AR target gene in both LNCaP and its castration-resistant derivative C4–2 cells (Supplementary Fig. S1A). Notably, ENZ treatment increased lipid peroxidation in these two cell lines (Fig. 1A). Similar results were obtained by treatment with either ARV110, a degrader of full-length AR (AR-FL; ref. 32) or two independent AR-specific small hairpin RNAs (shRNA; Fig. 1B and C; Supplementary Fig. S1B–S1D). Both pharmacologic inhibition and genetic depletion of AR also decreased GSH but increased soluble enzyme lactate dehydrogenase (LDH) levels, indicating the compromise of cell member integrity in LNCaP and C4–2 cell lines (Fig. 1D–I). Treatment with ferroptosis inhibitors such as ferrostatin (Ferr-1) or iron chelator (DFO) but not apoptosis inhibitor (Z-VAD-FMK) largely blocked ENZ-induced loss of cell viability (Supplementary Fig. S1E–S1G). In contrast, treatment with the ferroptosis inducer erastin largely enhanced cell viability inhibition induced by ENZ, ARV110, or AR shRNAs (Supplementary Fig. S1H–S1J). To further validate these observations, we cultured LNCaP and C4–2 cells with regular or cystine-low medium and measured cell viability. We demonstrated that decrease in cystine concentration reduced viability in LNCaP and C4–2 cells and this effect was largely enhanced by ENZ, ARV110, or AR shRNAs (Fig. 1J–L). Notably, cystine reduction-induced cell viability loss was reversed by Ferr-1 (Fig. 1J–L). Together, these data indicate that AR inhibition induces ferroptosis in prostate cancer cells.
Figure 1.
AR antagonists promote ferroptosis in prostate cancer cells. A–I, LNCaP and C4–2 cells treated with vehicle (mock) or ENZ, vehicle (mock) or ARV110, or transfected with nonspecific shRNA (shNS) or AR-specific shRNAs (shAR#1 and #2). At 48 hours after treatment, cells were subjected to measurement of lipid peroxidation by C11-BODIPY staining and flow cytometry (A–C), and measurement of intracellular levels of GSH (D–F) and LDH (G–I). Experiments were repeated three times independently and similar results were obtained. J–L, Cell viability in LNCaP and C4–2 cells cultured in regular or cystine-low (2 μmol/L) medium in the presence or absence of Ferr-1 and treated with vehicle or ENZ (J), ARV110 (K), or infected with lentivirus expressing nonspecific shRNA (shNS) or AR-specific shRNAs (L) for 72 hours.
Identification of SLC7A11 as an androgen-induced gene
To define the molecular mechanism by which the androgen-AR axis regulates ferroptosis in prostate cancer cells, we performed meta-analysis of RNA-seq data generated from LNCaP cells cultured in androgen-depleted (charcoal-stripped serum or CSS) medium and treated with or without DHT (33). We found that lipid metabolic process genes were among the top groups of genes upregulated by DHT [log2 (fold change) ≥ 1; Fig. 2A and B). Importantly, meta-analysis of RNA-seq data suggests that SLC7A11, a known ferroptosis inhibitory gene (Fig. 2C), is a putative androgen-upregulated gene (Fig. 2A and D). Androgen regulation of SLC7A11 mRNA expression was further validated by qRT-PCR and Western blot analysis in both LNCaP and C4–2 cell lines (Fig. 2E and F). In contrast, RNA-seq analysis showed that SLC7A11 mRNA expression was downregulated by ENZ treatment in LNCaP (Fig. 2G), and this result was further confirmed by qRT-PCR and Western blot analysis in LNCaP and C4–2 cell lines (Fig. 2H and I). Furthermore, we found that knockdown of AR by two independent shRNAs also decreased SLC7A11 expression at both protein and mRNA levels in LNCaP and C4–2 cells (Fig. 2J and K). Although DHT or ENZ treatment had no obvious effect on mRNA expression of GPX4, another key regulator of ferroptosis (Supplementary Fig. S2A–S2C), DHT increased and ENZ decreased GPX4 expression at the protein level (Supplementary Fig. S2D and S2E). These findings are consistent with a previous report that cystine uptake caused by upregulation of SLC7A11 only increases GPX4 protein synthesis, but not GPX4 gene mRNA expression (34), indicating that antiandrogens regulate ferroptosis via modulating expression of both SLC7A11 and GPX4, two key regulators of ferroptosis. Both qRT-PCR and Western blot analyses further showed that SLC7A11 expression was significantly downregulated in hormone-sensitive LuCaP35 patient-derived xenografts (PDX) after castration of mice (Fig. 2L and M; Supplementary Fig. S2F–S2I).
Figure 2.
Genome-wide analyses link AR to metabolism-related biological processes. A, Heatmap showing expression of genes downregulated (left) and upregulated (right) by DHT treatment relative to mock in LNCaP cells cultured in medium containing charcoal-stripped serum (CSS). B, GO enrichment analysis of the 415 upregulated genes [log2 (fold change) > 1] using the tools from the GSEA website (https://www.gsea-msigdb.org/gsea/msigdb/human/annotate.jsp). Top 10 annotation clusters are shown according to their enrichment scores [−log10 (P value)]. C, Diagram showing the role of SLC7A11 in the context of ferroptosis regulation. D, UCSC Genome Browser screenshot of RNA-seq data showing SLC7A11 expression levels in LNCaP (GSM2432771 vs. GSM2432769) and C4–2 cells (GSM2432783 vs. GSM2432781) cultured in CSS medium and treated with or without DHT (10 nmol/L) for 24 hours. E and F, qRT-PCR (E) and Western blot (F) analysis of SLC7A11 expression in LNCaP and C4–2 cells treated as in D. G–I, UCSC screenshot of RNA-seq data (G) showing SLC7A11 expression levels in LNCaP cells (GSM6132392 vs. GSM6132398) treated with or without ENZ (10 μmol/L) for 72 hours and qRT-PCR (H) and WB (I) analysis of SLC7A11 expression in LNCaP and C4–2 cells with the same treatments as in G. J and K, Western blot (J) and qRT-PCR (K) analysis of SLC7A11 protein (J) and mRNA expression (K) in LNCaP and C4–2 cells infected with lentivirus expressing nonspecific shRNA (shNS) or AR-specific shRNAs for 72 hours. L and M, qRT-PCR (L) and Western blot (M) analysis of LuCaP35 xenograft tumors from male mice (n = 5 mice/group) treated with sham castration or castration for 1 week. N, The dot plots showing the comparison of expression levels of AR (left) and SLC7A11 (right) between primary tissues and corresponding metastatic prostate cancer patient samples (GSE32269). O, The scatter plots showing the positive correlation between AR and SLC7A11 mRNA expression in GSE32269 dataset. P and Q, Representative images (P) and quantitative data (Q) for SLC7A11 protein IHC staining in a group of primary tissues (n = 20) and metastatic CRPC samples (n = 20).
AR signaling plays a central role in prostate cancer progression and aberrant AR mRNA expression positively correlates with shorter disease-free survival of TCGA patients (Supplementary Fig. S2J). SLC7A11 expression was higher in prostate tumors compared with matched normal prostatic tissues in the TCGA cohort (Supplementary Fig. S2K). SLC7A11 level was positively associated with AR expression in TCGA prostate cancer specimens and higher SLC7A11 expression correlated with shorter survival of patients (Supplementary Fig. S2L and S2M). SLC7A11 expression was much lower in AR-negative NEPC samples (35) compared with AR-positive CRPC samples (Supplementary Fig. S2N; ref. 36). To investigate SLC7A11 expression in CRPC, we analyzed the microarray data (GSE32269) from paired primary prostate cancer and metastatic CRPC samples (37). We found that expression of both AR and SLC7A11 mRNA was higher in CRPC samples compared with primary tumors (Fig. 2N) and that SLC7A11 also positively correlated with AR expression in this cohort (Fig. 2O). We further performed IHC analysis of SLC7A11 protein expression in patient samples. We confirmed that SLC7A11 level was higher in CRPC samples compared with primary tumors (Fig. 2P and Q). Together, the results from cultured cell lines, PDX tumors and patient samples reveal that SLC7A11 is a target gene transcriptionally upregulated by the androgen signaling.
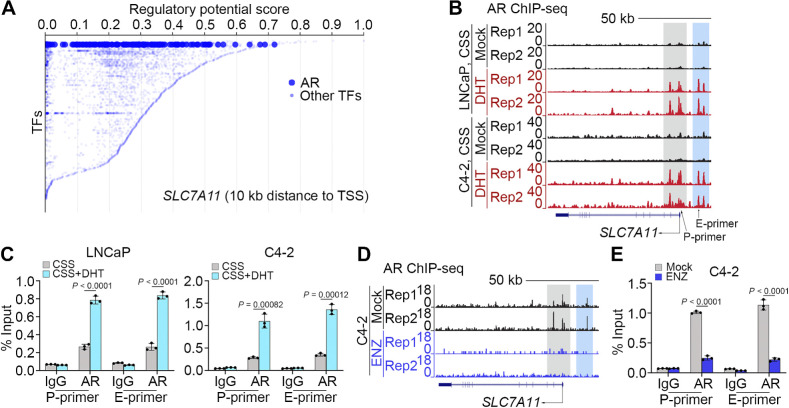
Identification of SLC7A11 as an AR-binding target gene
Consistent with gene expression data, meta-analysis of chromatin immunoprecipitation sequencing (ChIP-seq) data of specific transcription factors, including those highly relevant in prostate cancer such as AR, p300, FOXA1, and FOXA2, revealed that AR was one of the top 20 transcription factors with a strong potential to transactivate expression of the SLC7A11 gene (Fig. 3A), suggesting that upon activation AR might regulate SLC7A11 expression by directly binding to this gene locus. In support of this notion, analysis of the AR ChIP-seq data we generated previously (38) revealed that DHT treatment markedly increased AR occupancy at promoter (P) and non-promoter [e.g., putative enhancer (E)] regions of SLC7A11 locus in both LNCaP and C4–2 cell lines (Fig. 3B). Androgen-induced AR binding in these genomic regions was further confirmed by qChIP-PCR in these two cell lines (Fig. 3C). In contrast, ENZ treatment abolished AR occupancy at the SLC7A11 gene locus (Fig. 3D and E). These data indicate that AR promotes SLC7A11 expression through occupancy at different regions in this gene locus and AR binding can be abrogated by inhibition of the androgen/AR axis.
Figure 3.
AR promotes SLC7A11 expression at transcriptional level. A, Meta-analysis of ChIP data of transcription factors that potentially bind to the SLC7A11 gene locus using the online-based platform Cistrome software (http://dbtoolkit.cistrome.org). B and C, UCSC screenshot of AR ChIP-Seq showing AR occupancy at the SLC7A11 loci in LNCaP and C4–2 cells cultured in CSS medium and treated with or without DHT (B), and qChIP-PCR analysis of AR binding at the SLC7A11 promoter and enhancer regions in LNCaP and C4–2 (C) cells with the same treatment as in B. D and E, AR ChIP-Seq occupancy profiles at the SLC7A11 loci in C4–2 cells treated with or without ENZ (D). qChIP-PCR confirming AR binding at the SLC7A11 promoter and enhancer region was abolished in C4–2 cells (E) with the same treatment as in D.
AR inhibits ferroptosis through upregulation of SLC7A11
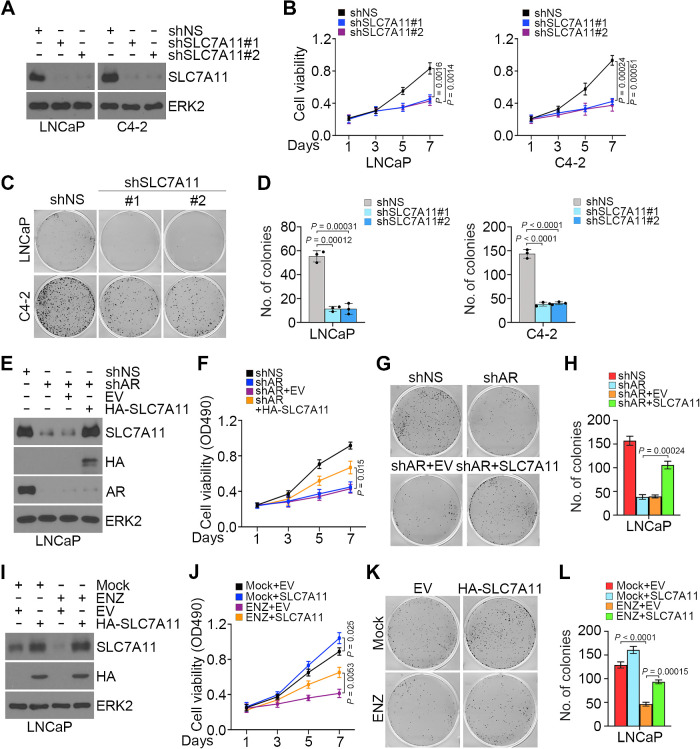
SLC7A11 imports extracellular cystine into cells for glutathione synthesis in the cytoplasm, and GPX4 utilizes glutathione to detoxify lipid peroxide to protect cells from ferroptosis (28). To investigate the impact of SLC7A11 on prostate cancer cell growth and survival, we performed MTS assay and showed that SLC7A11 depletion significantly suppresses cell growth (Fig. 4A and B). In contrast, SLC7A11 overexpression significantly increased cell viability in different prostate cancer cell lines (Supplementary Fig. S3A and S3B). Colony growth assay further demonstrated that SLC7A11 knockdown decreased colony numbers compared with control cells (Fig. 4C and D). Furthermore, we showed that restoration of SLC7A11 in AR-depleted cells largely restored cell viability (Fig. 4E–H). Importantly, forced expression of SLC7A11 overcame, at least in part, ENZ-induced inhibition of cell viability and growth (Fig. 4I–L). AR depletion-induced loss of viability in LNCaP and C4–2 cells cultured in cystine-low medium was reversed by restored expression of either AR or SLC7A11 (Supplementary Fig. S4A–S4D).
Figure 4.
Role of SLC7A11 in AR-mediated prostate cancer cell growth. A–D, LNCaP and C4–2 cells transfected with shNS or shSLC7A11 and stable cell lines were used for Western blot analysis (A), MTS assays (B), and colony formation assays, followed by photographing (C) and quantification (D). Three biological replicates were analyzed. ERK2 was used as a loading control. E–H, LNCaP cells were infected with lentivirus expressing the indicated shRNAs or expression vectors for 72 hours and subjected to Western blot analysis (E), MTS assays (F), and colony formation assays, followed by photographing (G) and quantification (H). Three biological replicates were analyzed. I–L, LNCaP cells infected with lentivirus expressing the indicated plasmids were treated with or without ENZ and subjected to Western blot analysis (I), MTS assays (J), and colony formation assays, followed by photographing (K) and quantification (L). Three biological replicates were analyzed.
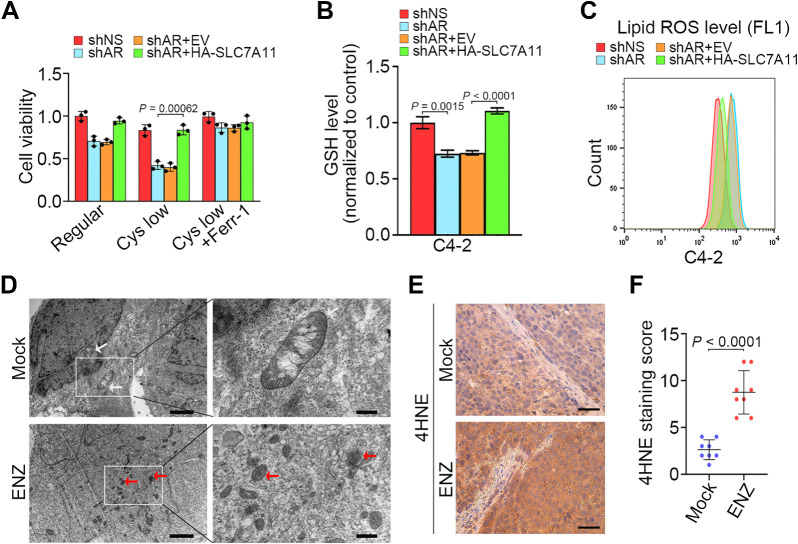
We further demonstrated that AR depletion-induced inhibition of cell viability and reduction in GSH cellular level were reversed by forced expression of SLC7A11 (Fig. 5A–C). Transmission electron microscopy (TEM) analysis revealed that ENZ-treated C4–2 cells exhibited shrunken mitochondria with increased membrane density, a morphologic feature of ferroptosis in comparison to control cells (Fig. 5D; ref. 28). Because ferroptosis is characterized by overproduction of lipid peroxidation, we performed 4-Hydroxynonenal (4HNE) IHC analysis to determine lipid peroxidation levels in C4–2 xenograft tumors treated with or without ENZ. We demonstrated that ENZ treatment largely decreased tumor volume and SLC7A11 expression but induced 4HNE level in C4–2 xenografts (Fig. 5E and F; Supplementary Fig. S5A–S5C). These data suggest that ferroptosis is responsible, at least in part, for AR inhibition-induced cell death in vitro and in vivo.
Figure 5.
SLC7A11 attenuates ferroptosis induced by AR antagonist. A–C, C4–2 cells infected with lentivirus expressing the indicated shRNAs and/or expression vectors and cultured in regular or cystine-low medium (2 μmol/L) for 48 hours and subjected to MTS assays (A), measurement of intracellular GSH levels (B), and detection of lipid peroxidation by flow cytometry after C11-BODIPY staining (C). Three biological replicates were analyzed. D, C4–2 cells treated as indicated were subjected to transmission electron microscopy. White arrows, mitochondria with obvious cristae. Red arrows, shrunken mitochondria. Scale bars, left, 2 μm; right, 500 nm. Experiment was repeated three times independently with similar results. E and F, Representative images (E) and quantitative data (F) for IHC staining of 4HNE in C4–2 xenograft tumors in mice treated with or without ENZ for 20 days.
AR-Vs suppress ferroptosis by inducing SLC7A11 expression
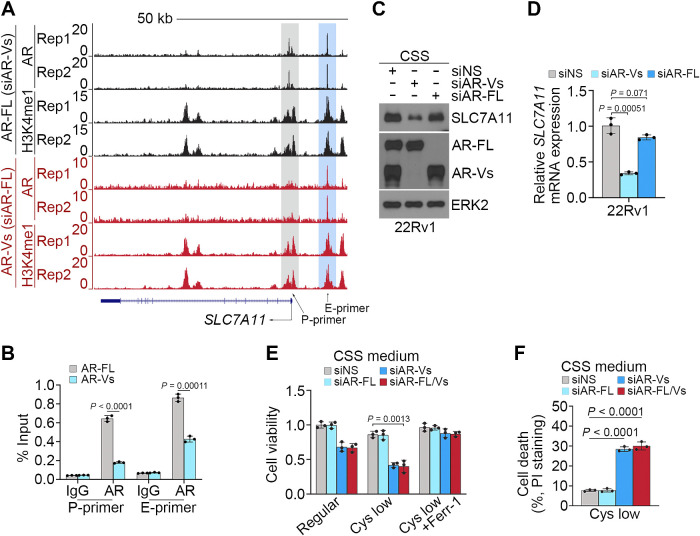
AR-Vs, which either partially or completely lack a functional ligand binding domain, confer constitutive AR activity and AR signaling inhibitor resistance in prostate cancer (20, 39). We have shown previously that AR-Vs bind to and transactivate a unique subset of genes that are related to cell-cycle transition and cancer progression (40). 22Rv1 is a known ENZ-resistant cell line, which expresses AR-Vs (39). We found that ENZ treatment failed to suppress SLC7A11 mRNA expression in 22Rv1 cells (Supplementary Fig. S6A). By analyzing the AR-FL and AR-V ChIP-seq data in 22Rv1 cells we generated previously (40), we found that AR-FL bound to both the promoter and enhancer regions [indicated by high enrichment of histone H3 lysine 4 monomethylation (H3K4me1), an enhancer histone mark] in the SLC7A11 gene locus in 22Rv1 cells; however, AR-Vs preferentially bound to the putative enhancer regions (Fig. 6A). AR-FL and AR-V binding in this locus in 22Rv1 cells was further confirmed by qChIP-PCR (Fig. 6B). Previous studies suggest that expression of AR-Vs, but not AR-FL confers ENZ resistance in 22Rv1 cells (41). In agreement with this report and our finding that ENZ treatment failed to abrogate AR-V occupation and suppress SLC7A11 expression in 22Rv1 cells (Supplementary Fig. S6A and S6B), AR-FL knockdown had little or no effect on SLC7A11 expression in 22Rv1 cells cultured in androgen-depleted medium whereas AR-V knockdown largely decreased SLC7A11 expression at both mRNA and protein levels (Fig. 6C and D). These data suggest that AR-Vs play a pivotal role in upregulating SLC7A11 expression in ENZ-resistant 22Rv1 cells. Importantly, under androgen depletion conditions, AR-V ablation largely sensitized 22Rv1 cells to ferroptosis induced by low concentration of cystine (Fig. 6E and F). These data suggest that AR-Vs promote SLC7A11 expression to suppress ferroptosis, thereby contributing to ENZ resistance in prostate cancer cells.
Figure 6.
AR-Vs promote SLC7A11 expression and suppress ferroptosis. A, UCSC screenshot of ChIP-seq data of AR and H3K4me1 showing AR occupancy and the level of H3K4me1 at the SLC7A11 gene locus in 22Rv1 cells in which endogenous AR-FL and AR-Vs were specifically knocked down individually. B, qChIP-PCR confirming the binding of AR-FL and AR-Vs at the SLC7A11 promoter and enhancer regions in 22Rv1 cells. C and D, 22Rv1 cells transfected with nonspecific siRNA (siNS), siRNA specific for AR-FL (siAR-FL), and siRNAs specific for AR-Vs (siAR-Vs, including siAR-V1, V3, V4, and V7) and cultured in CSS medium were subjected to Western blot (C) and qRT-PCR (D) analysis. ERK2 was used as a loading control. E and F, 22Rv1 cells transfected with indicated siRNAs and cultured in regular or cystine-low CSS medium in the presence or absence of Ferr-1 were subjected to MTS (E) and FACS analysis (F) after cells were stained with propidium iodide (PI).
CBP/p300 and BET dual inhibitor NEO2734 overcomes AR-V–mediated ferroptosis and ENZ resistance in prostate cancer
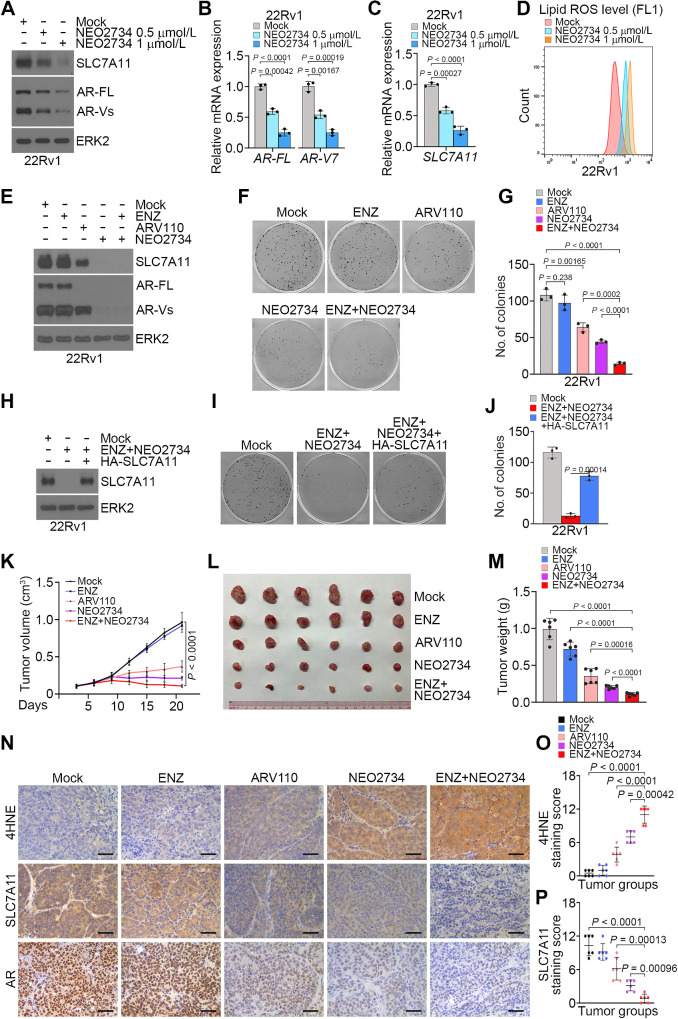
Our previous studies show that NEO2734, a dual inhibitor of CBP/p300 and BET proteins largely decreases AR-FL protein expression (26, 27). Given that AR-FL and AR-Vs share the same regulatory elements [such as the promoter and enhancer(s)] of the AR gene (39), we sought to determine whether NEO2734 also inhibits AR-V expression. We treated 22Rv1 cells with different doses of NEO2734. We demonstrated that NEO2734 inhibited expression of AR-FL and AR-vs. at both mRNA and protein levels (Fig. 7A and B). Different from ENZ treatment (Supplementary Fig. S6A), NEO2734 treatment suppressed SLC7A11 expression in 22Rv1 cells (Fig. 7C). NEO2734 treatment also increased lipid ROS in 22Rv1 cells (Fig. 7D). Co-treatment of NEO2734 and ENZ substantially reduced 22Rv1 cell viability (Fig. 7E–G). Most importantly, the effect of dual treatments was largely reversed by restored expression of HA-SLC7A11 (Fig. 7H–J), suggesting a pivotal role of SLC7A11 downregulation in mediating the cell viability loss induced by treatment with NEO2734 and ENZ. We also treated 22Rv1 xenograft tumors with ENZ, ARV110, NEO2734, and the combination of NEO2734 and ENZ. Compared to each individual treatment, coadministration of NEO2734 and ENZ resulted in much greater inhibition of tumor growth, stronger suppression of SLC7A11 expression, and higher level of the lipid peroxidation indicator 4HNE (Fig. 7K–P). These data indicate that the CBP/p300 and BET dual inhibitor NEO2734 can inhibit AR-V expression and AR-V-mediated resistance to ENZ and ferroptosis in prostate cancer in vitro and in vivo.
Figure 7.
Dual inhibitor NEO2734 cotreatment overcomes AR-V-mediated resistance to ferroptosis and ENZ in prostate cancer. A–C, 22Rv1 cells treated with vehicle or different doses of NEO2734 for 24 hours were subjected to WB (A) and qRT-PCR analysis of AR-FL and AR-V7 (B), and SLC7A11 mRNA expression (C). D, 22Rv1 cells treated with vehicle or different doses of NEO2734 for 36 hours were subjected to the detection of lipid peroxidation by flow cytometry after C11-BODIPY staining. E–G, 22Rv1 cells were treated as indicated and subjected to WB analysis at 48 hours posttreatment (E), colony formation assay after 12 days of treatment followed by colony photographing (F) and quantification (G). H–J, 22Rv1 cells infected with lentivirus expressing empty vector or HA-SLC7A11 and stable cells were subjected to WB analysis (H) and treated with vehicle or ENZ plus NEO2734 followed by colony formation assay for 12 days. Colonies were photographed (I) and quantified (J) at the end of treatment. K–M, Mice with 22Rv1 xenograft tumors were treated with the indicated drugs and tumor volumes were measured at the indicated time points (K) and tumors were photographed (L) and weighted (M) at the end of drug treatment (day 21). Data shown as mean ± SD (n = 6 replicates/group). N–P, Representative images (N) and quantitative data for IHC staining of 4HNE (O) and SLC7A11 (P) proteins in 22Rv1 xenograft tumors.
Discussion
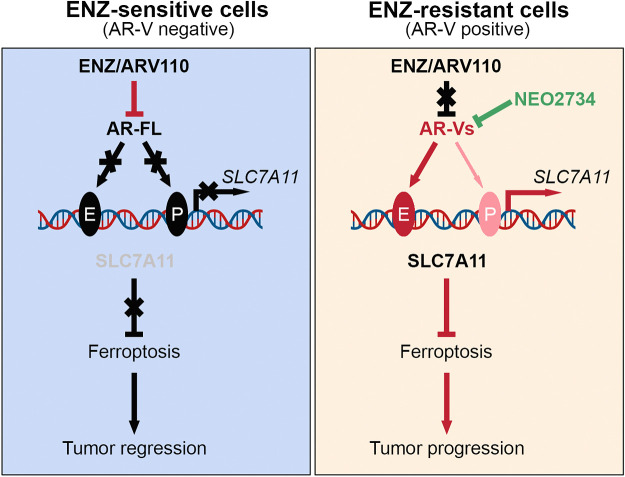
AR-targeted therapy remains the mainstay treatment of advanced prostate cancer in patients. Androgen ablation triggers apoptotic cell death in both normal prostate glandular epithelial cells and androgen-dependent prostate cancer cells (30, 42), and ADT-induced prostate cancer cell apoptosis has been implicated in regression of prostate cancer in patients (43). Ferroptosis is a form of cell death that is distinct from apoptosis and is an iron-dependent cell death induced by lipid peroxidation on cell membrane. Notably, a previous study suggests that ferroptosis induction can be a novel therapeutic strategy for the treatment of advanced prostate cancer (14). In this study, we provide evidence that antiandrogen treatment or AR protein depletion decreased cellular level of GSH and induced ferroptosis of prostate cancer cells. Specifically, we show that AR inhibitory agents suppress expression of SLC7A11, an essential negative regulator of ferroptosis. Our current studies hence reveal ferroptosis as a new form of cell death induced by AR ablation therapies (Fig. 8, left).
Figure 8.
Hypothetical working model. AR-FL transactivates SLC7A11 gene transcription by directly occupying at the promoter and enhancer regions of SLC7A11 gene. Antiandrogen treatment suppresses SLC7A11 expression and induces ferroptosis in prostate cancer cells by inhibiting AR-FL-mediated transactivation of SLC7A11 expression (left). In contrast, AR-Vs preferentially bind to the SLC7A11 gene enhancer and upregulate SLC7A11 expression, thereby conferring resistance to ferroptosis induced by ENZ treatment (right). However, the effect of AR-Vs can be abolished by CBP/p300 and BET dual inhibitor treatment-mediated inhibition of AR-V expression.
Our studies in cell lines, PDX tumors, and patient samples show that SLC7A11 is an androgen-regulated gene. By analyzing the AR ChIP-seq data, we demonstrate that DHT treatment induces AR-FL occupancy at both the promoter and enhancer regions of SLC7A11 gene locus. This observation is corroborated by the finding that ENZ treatment eliminated AR-FL occupancy in prostate cancer cells. Our data reveal that SLC7A11 is a bona fide AR target gene and importantly, our findings also provide a plausible explanation as to why acute androgen ablation or antiandrogen treatment induces ferroptosis. Intriguingly, we provide evidence that AR-Vs also occupy at the SLC7A11 gene locus although it appears that AR-Vs preferentially bind to the enhancer region. Importantly, we show that SLC7A11 expression is primarily regulated by AR-Vs under androgen deprivation conditions in AR-V-expressing prostate cancer cells. We and others have shown that androgen depletion or antiandrogen treatment induces AR-V expression in prostate cancer cells in vitro or xenograft tumors in vivo (22, 24). On the basis of the previous findings and those from the present study, we envisage a model wherein AR targeting therapies induce AR-V expression, which in turn drives SLC7A11 expression in a manner independent of androgens, thereby contributing tumor progression by suppressing ferroptosis (Fig. 8, right). We provide further evidence that the effect of AR-Vs on SLC7A11 expression and ferroptosis resistance can be therapeutically targeted by the CBP/p300 and BET dual inhibitor NEO2734, although such effect of NEO2734 can be mediated, at least partially by other targets in addition to AR since this compound can also inhibit the growth of AR-negative prostate cancer cells.
Intriguingly, it has been reported previously that ENZ treatment increases SLC7A11 expression in LNCaP cells (44). It is well known that growth of LNCaP cells is highly sensitive to androgen levels and the LNCaP cell line can evolve into different cellular states depending on the cell culture conditions. Indeed, a number of androgen refractory LNCaP sublines have been derived when LNCaP cells are cultured in various androgen-depleted medium (45, 46). Given that androgen deprivation induces expression of AR-Vs (22, 24) and that AR-Vs can induce SLC7A11 expression in androgen depletion conditions as we demonstrated in the present study, it is not surprising that ENZ treatment increases SLC7A11 expression when AR-V expression is induced.
In summary, we demonstrate for the first time that antiandrogen therapy decreases GSH level, elevates lipid peroxidation, and induces ferroptosis. We identify the cystine transporter gene SLC7A11 as a bona fide AR target gene. Notably, both AR-FL and AR-Vs can transactivate SLC7A11 gene through direct occupancy at the SLC7A11 promoter and/or enhancer regions. Antiandrogen therapy induces ferroptosis by blocking the regulation of SLC7A11 expression by AR-FL; however, this effect is abolished by subsequent induction of AR-Vs. Most importantly, we show that cotreatment of the CBP/p300 and BET dual inhibitor NEO2734 restores antiandrogen-induced ferroptosis by blocking the undesired expression of AR-Vs. Thus, blocking AR-V-mediated inhibition of ferroptosis represents a new tactic to overcome ENZ resistance in CRPC.
Supplementary Material
SI
Acknowledgments
The authors thank Dr. B.Q. Huang for assistance in transmission electron microscopy analysis. This work was supported by the Mayo Clinic Foundation (to H. Huang) and Fudan University Shanghai Cancer Center (to D. Ye).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No author disclosures were reported.
Authors' Contributions
R. Sun: Conceptualization, data curation, formal analysis, investigation, writing–original draft. B. Yan: Data curation. H. Li: Data curation, formal analysis, investigation. D. Ding: Data curation, formal analysis, investigation. L. Wang: Data curation, supervision, investigation. J. Pang: Supervision, writing–review and editing. D. Ye: Conceptualization, supervision, project administration, writing–review and editing. H. Huang: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Green DR. The coming decade of cell death research: five riddles. Cell 2019;177:1094–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koren E, Fuchs Y. Modes of regulated cell death in cancer. Cancer Discov 2021;11:245–65. [DOI] [PubMed] [Google Scholar]
- 3. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res 2021;31:107–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22:266–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol 2021;18:280–96. [DOI] [PubMed] [Google Scholar]
- 7. Conrad M, Sato H. The oxidative stress-inducible cystine/glutamate antiporter, system x c (-) : cystine supplier and beyond. Amino Acids 2012;42:231–46. [DOI] [PubMed] [Google Scholar]
- 8. Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun (Lond) 2018;38:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim JC, Donaldson PJ. Focus on molecules: the cystine/glutamate exchanger (system x(c)(-)). Exp Eye Res 2011;92:162–3. [DOI] [PubMed] [Google Scholar]
- 10. Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 2014;16:1180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer 2002;2:277–88. [DOI] [PubMed] [Google Scholar]
- 13. Green DR, Evan GI. A matter of life and death. Cancer Cell 2002;1:19–30. [DOI] [PubMed] [Google Scholar]
- 14. Ghoochani A, Hsu EC, Aslan M, Rice MA, Nguyen HM, Brooks JD, et al. Ferroptosis inducers are a novel therapeutic approach for advanced prostate cancer. Cancer Res 2021;81:1583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011;20:457–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grossmann ME, Huang H, Tindall DJ. Androgen receptor signaling in androgen-refractory prostate cancer. J Natl Cancer Inst 2001;93:1687–97. [DOI] [PubMed] [Google Scholar]
- 17. Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015;15:701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agus DB, Cordon-Cardo C, Fox W, Drobnjak M, Koff A, Golde DW, et al. Prostate cancer cell cycle regulators: response to androgen withdrawal and development of androgen independence. J Natl Cancer Inst 1999;91:1869–76. [DOI] [PubMed] [Google Scholar]
- 19. Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev 2004;25:276–308. [DOI] [PubMed] [Google Scholar]
- 20. Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer 2011;18:R183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun R, Wei T, Ding D, Zhang J, Chen S, He HH, et al. CYCLIN K down-regulation induces androgen receptor gene intronic polyadenylation, variant expression and PARP inhibitor vulnerability in castration-resistant prostate cancer. Proc Natl Acad Sci USA 2022;119:e2205509119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Etten JL, Nyquist M, Li Y, Yang R, Ho Y, Johnson R, et al. Targeting a single alternative polyadenylation site coordinately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Cancer Res 2017;77:5228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci USA 2010;107:16759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bohrer LR, Liu P, Zhong J, Pan Y, Angstman J, Brand LJ, et al. FOXO1 binds to the TAU5 motif and inhibits constitutively active androgen receptor splice variants. Prostate 2013;73:1017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He Y, Wei T, Ye Z, Orme JJ, Lin D, Sheng H, et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat Commun 2021;12:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan Y, Ma J, Wang D, Lin D, Pang X, Wang S, et al. The novel BET-CBP/p300 dual inhibitor NEO2734 is active in SPOP mutant and wild-type prostate cancer. EMBO Mol Med 2019;11:e10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol 2018;20:1181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun R, Xie HY, Qian JX, Huang YN, Yang F, Zhang FL, et al. FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res 2018;78:5274–86. [DOI] [PubMed] [Google Scholar]
- 30. Kyprianou N, Isaacs JT. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology 1988;122:552–62. [DOI] [PubMed] [Google Scholar]
- 31. Baron A, Migita T, Tang D, Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem 2004;91:47–53. [DOI] [PubMed] [Google Scholar]
- 32. Proof-of-concept with PROTACs in prostate cancer. Cancer Discov 2020;10:1084. [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y, Pitchiaya S, Cieslik M, Niknafs YS, Tien JC, Hosono Y, et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet 2018;50:814–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y, Swanda RV, Nie L, Liu X, Wang C, Lee H, et al. mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat Commun 2021;12:1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cai C, Wang H, He HH, Chen S, He L, Ma F, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest 2013;123:1109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao Y, Wang L, Ren S, Wang L, Blackburn PR, McNulty MS, et al. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep 2016;15:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 2008;68:5469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He Y, Lu J, Ye Z, Hao S, Wang L, Kohli M, et al. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res 2018;46:1895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013;73:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Denmeade SR, Lin XS, Isaacs JT. Role of programmed (apoptotic) cell death during the progression and therapy for prostate cancer. Prostate 1996;28:251–65. [DOI] [PubMed] [Google Scholar]
- 43. Ohlson N, Bergh A, Nygren K, Stattin P, Wikstrom P. The magnitude of early castration-induced primary tumour regression in prostate cancer does not predict clinical outcome. Eur Urol 2006;49:675–83. [DOI] [PubMed] [Google Scholar]
- 44. Yuan F, Hankey W, Wu D, Wang H, Somarelli J, Armstrong AJ, et al. Molecular determinants for enzalutamide-induced transcription in prostate cancer. Nucleic Acids Res 2019;47:10104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murillo H, Huang H, Schmidt LJ, Smith DI, Tindall DJ. Role of PI3K signaling in survival and progression of LNCaP prostate cancer cells to the androgen refractory state. Endocrinology 2001;142:4795–805. [DOI] [PubMed] [Google Scholar]
- 46. Leung JK, Tam T, Wang J, Sadar MD. Isolation and characterization of castration-resistant prostate cancer LNCaP95 clones. Hum Cell 2021;34:211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI
Data Availability Statement
The next-generation sequencing data from The Cancer Genome Atlas (TCGA), Stand Up To Cancer (SU2C), and Neuroendocrine Prostate Cancer (NEPC) cohort were obtained and analyzed via cBioPortal (https://www.cbioportal.org/). The microarray data from the primary and mCRPC cohort were obtained from the Gene Expression Omnibus database with the accession number GSE32269. All data supporting the findings of this study are available within the article and the Supplementary Information files. The relevant reagents such as plasmids are available from the corresponding authors upon request. All other raw data are available upon request from the corresponding authors.



![Figure 2. Genome-wide analyses link AR to metabolism-related biological processes. A, Heatmap showing expression of genes downregulated (left) and upregulated (right) by DHT treatment relative to mock in LNCaP cells cultured in medium containing charcoal-stripped serum (CSS). B, GO enrichment analysis of the 415 upregulated genes [log2 (fold change) > 1] using the tools from the GSEA website (https://www.gsea-msigdb.org/gsea/msigdb/human/annotate.jsp). Top 10 annotation clusters are shown according to their enrichment scores [−log10 (P value)]. C, Diagram showing the role of SLC7A11 in the context of ferroptosis regulation. D, UCSC Genome Browser screenshot of RNA-seq data showing SLC7A11 expression levels in LNCaP (GSM2432771 vs. GSM2432769) and C4–2 cells (GSM2432783 vs. GSM2432781) cultured in CSS medium and treated with or without DHT (10 nmol/L) for 24 hours. E and F, qRT-PCR (E) and Western blot (F) analysis of SLC7A11 expression in LNCaP and C4–2 cells treated as in D. G–I, UCSC screenshot of RNA-seq data (G) showing SLC7A11 expression levels in LNCaP cells (GSM6132392 vs. GSM6132398) treated with or without ENZ (10 μmol/L) for 72 hours and qRT-PCR (H) and WB (I) analysis of SLC7A11 expression in LNCaP and C4–2 cells with the same treatments as in G. J and K, Western blot (J) and qRT-PCR (K) analysis of SLC7A11 protein (J) and mRNA expression (K) in LNCaP and C4–2 cells infected with lentivirus expressing nonspecific shRNA (shNS) or AR-specific shRNAs for 72 hours. L and M, qRT-PCR (L) and Western blot (M) analysis of LuCaP35 xenograft tumors from male mice (n = 5 mice/group) treated with sham castration or castration for 1 week. N, The dot plots showing the comparison of expression levels of AR (left) and SLC7A11 (right) between primary tissues and corresponding metastatic prostate cancer patient samples (GSE32269). O, The scatter plots showing the positive correlation between AR and SLC7A11 mRNA expression in GSE32269 dataset. P and Q, Representative images (P) and quantitative data (Q) for SLC7A11 protein IHC staining in a group of primary tissues (n = 20) and metastatic CRPC samples (n = 20).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/2434/10543964/ed7a876e6045/3192fig2.jpg)