The mechanistically designed, biomarker-matched strategy of combining alpelisib and tipifarnib is efficacious in PIK3CA- and HRAS-dysregulated head and neck squamous carcinoma and could improve outcomes for many patients with recurrent, metastatic disease.
Abstract
Outcomes for patients with recurrent/metastatic (R/M) head and neck squamous cell carcinoma (HNSCC) are poor, with median overall survival (OS) ranging from 6 to 18 months. For those who progress on standard-of-care (chemo)immunotherapy, treatment options are limited, necessitating the development of rational therapeutic strategies. Toward this end, we targeted the key HNSCC drivers PI3K–mTOR and HRAS via the combination of tipifarnib, a farnesyltransferase (FTase) inhibitor, and alpelisib, a PI3Kα inhibitor, in multiple molecularly defined subsets of HNSCC. Tipifarnib synergized with alpelisib at the level of mTOR in PI3Kα- or HRAS-dependent HNSCCs, leading to marked cytotoxicity in vitro and tumor regression in vivo. On the basis of these findings, the KURRENT-HN trial was launched to evaluate the effectiveness of this combination in PIK3CA-mutant/amplified and/or HRAS-overexpressing R/M HNSCC. Preliminary evidence supports the clinical activity of this molecular biomarker-driven combination therapy. Combined alpelisib and tipifarnib has potential to benefit >45% of patients with R/M HNSCC. By blocking feedback reactivation of mTORC1, tipifarnib may prevent adaptive resistance to additional targeted therapies, enhancing their clinical utility.
Significance:
The mechanistically designed, biomarker-matched strategy of combining alpelisib and tipifarnib is efficacious in PIK3CA- and HRAS-dysregulated head and neck squamous carcinoma and could improve outcomes for many patients with recurrent, metastatic disease.
Introduction
With nearly 900,000 new cases diagnosed each year, head and neck squamous cell carcinoma (HNSCC) is the sixth most prevalent cancer worldwide, and its incidence is increasing (1). HNSCC is a heterogeneous disease with tumors arising from epithelial linings of the oral cavity, pharynx, and larynx. Regardless of the subsite, locoregional recurrences, and distant metastases are not uncommon despite curative-intent treatment with surgery, radiation, and/or chemotherapy. Recurrent/metastatic (R/M) HNSCC is a challenging disease to treat, and patient prognosis is poor, with a dismal median overall survival (OS) rate of about 12 months (2). In 2019, the immune checkpoint inhibitor pembrolizumab targeting programmed cell death protein 1 (PD-1) was approved as first-line therapy for patients with R/M HNSCC based on the findings of the KEYNOTE-048 trial (3). Patients treated with pembrolizumab plus chemotherapy (platinum + 5-fluorouracil, 5-FU) had an improved median OS of 13.0 months compared with 10.7 months for patients receiving cetuximab plus chemotherapy [HR, 0.77; confidence interval (CI) 95%, 0.63–0.93; P = 0.0034]. While pembrolizumab improved the duration of response in the monotherapy arm, the overall response rates (ORR) in this study were the same as achieved by historical standard-of-care chemotherapy (platinum +5-FU +cetuximab) since 2008 (36%). Few options currently exist for patients who progress on (chemo)immunotherapy and to date, no biomarker-selected therapies targeted to molecular subtypes of head and neck cancers have been approved. Therefore, it is of critical importance to identify new therapies tailored to the specific molecular characteristics of HNSCCs.
In head and neck cancers, the PI3K–AKT–mTOR pathway is the most frequently dysregulated signaling cascade. Activation of this oncogenic signaling pathway is commonly achieved via amplification or gain-of-function mutation of PIK3CA, the gene encoding the α isoform of the PI3K p110 catalytic subunit, making PI3Kα an attractive therapeutic target in this setting (4, 5). Although the PI3Kα inhibitor alpelisib (Novartis) has shown some activity in HNSCC in early clinical trials (6), its efficacy as a single agent is limited by feedback reactivation of PI3K (7–10) or compensatory parallel pathways such as RAS-MAPK (11–13), necessitating the development of rational combination strategies. Although combined EGFR and PI3K inhibition targets the signaling pathways of interest in HNSCC, significant toxicity was observed when alpelisib was combined with cetuximab (14). Thus, the mechanistic strategy must be weighed carefully against the toxicity profile of potential combination drugs. For example, though the combination of alpelisib with a rapalog or TOR kinase inhibitor would sustainably block pathway signaling, overlapping toxicities primarily attributed to mTORC2 inhibition make the combination hard to justify from a tolerability standpoint. An ideal combination partner would target the feedback reactivated pathway(s) to synergize mechanistically with alpelisib, while maintaining acceptable tolerability.
Tipifarnib, a potent and selective FTase inhibitor (FTI), can inhibit several proteins involved in oncogenic growth factor signaling, including HRAS and RHEB, which depend on farnesylation for their activation and/or cellular localization (15, 16). Tipifarnib's ability to inhibit both HRAS and RHEB makes it a promising candidate partner drug for alpelisib in HNSCC. HRAS is the dominant RAS isoform in squamous cell carcinomas (SCC; ref. 17) and although all RAS isoforms regulate PI3K (18), HRAS preferentially signals through this route (19). As RHEB is a nonredundant TORC1 activator (20), tipifarnib simultaneously acts as an inhibitor of mTOR, which has been shown to be an important driver of both tumor growth and PI3K inhibitor (PI3Ki) resistance in HNSCC (21–23). Indeed, sustained inhibition of mTORC1 activity is required for PI3Ki efficacy (24, 25). By blocking both HRAS-MAPK and PI3K–AKT–mTOR signaling, tipifarnib can potentially blunt the feedback (re)activation of the two major pathways driving PI3Ki resistance and greatly enhance the efficacy of alpelisib in head and neck cancers. Importantly, tipifarnib is well tolerated in humans as evidenced by over two decades of safety and tolerability data and has shown encouraging clinical activity in HRAS-mutant SCC (26, 27).
Here, we demonstrate that combined alpelisib and tipifarnib is efficacious in preclinical models of HNSCC and provide mechanistic rationale for advancing this combination into the clinic. Via concurrent inhibition of HRAS and RHEB, tipifarnib blocks the feedback reactivation of MAPK and mTOR following alpelisib exposure, leading to durable pathway inhibition and tumor cell death in both PI3Kα and HRAS-dysregulated models. The KURRENT-HN trial (NCT04997902) is underway to evaluate this combination in recurrent/metastatic HNSCCs and encouraging preliminary results have been observed.
Materials and Methods
Cell lines and reagents
Cell lines were obtained from ATCC (SCC25, SCC9, FaDu, and Detroit 562), JCRB (SAS, HSC2, HSC3), DSMZ (CAL33), or Sigma (PECAP15J) and maintained in a humidified atmosphere with 5% CO2 at 37°C. Cells were maintained in DMEM (Detroit 562, FaDu, HSC2, HSC3, CAL33, PECAP15J) or DMEM/F12 (SCC25, SCC9, SAS) supplemented with 10% FBS and penicillin/streptomycin. All lines tested negative for Mycoplasma by the Lonza MycoAlert and/or IDEXX STAT-Myco RT-PCR prior to manuscript submission. Alpelisib was purchased from MedChemExpress. Tipifarnib was manufactured by STA Pharmaceutical Co., Ltd., a WuXi AppTec Company (WuXi STA). Dharmacon ON-TARGETplus siRNA SMARTPools against RHEB and HRAS were obtained from Horizon Discovery and transfected using Lipofectamine RNAiMAX (Thermo Fisher Scientific).
Tumor spheroid growth assays
Cells were resuspended in 4% Matrigel and seeded in 96-well ultra-low attachment plates at a density of 1 to 1.5000 cells/well. The next day, spheroids were treated with alpelisib and/or tipifarnib (dissolved in DMSO for in vitro studies) and baseline growth measured using 3D Cell Titer Glo reagent (Promega). Spheroids were incubated with drug for 7 days and a final cardiotocography (CTG) reading taken. Percentage growth was calculated by [(Ti-Tz)/(C-Tz)] × 100 for concentrations for which Ti >/ = Tz and [(Ti-Tz)/Tz] × 100 for concentrations for which Ti < Tz, where Tz = time zero, C = control growth, and Ti = test growth at each drug concentration. Calculation of combination sensitivity/synergy scores was performed using the SynergyFinder R package (28). Loewe synergy score of less than -10 indicates antagonistic combination activity, -10 to 10 an additive effect, and greater than +10 a synergistic effect. Half maximal inhibitory concentrations (IC50) were determined by nonlinear regression analysis of plots of percentage growth inhibition versus log inhibitor concentration in GraphPad Prism.
Xenograft models and in vivo experiments
Cell line–derived xenograft experiments were performed at the University of California, San Diego (San Diego, CA), under protocol ASP # S15195, and approved by the Institutional Animal Care and Use Committee (IACUC). Briefly, 2 million cells were implanted into the flanks of female athymic (Charles River Laboratories). When tumors reached approximately 150 to 200 mm3, mice were randomized and treated orally with tipifarnib twice a day (in 20% w/v hydroxypropyl-β-cyclodextrin, pH 2.5) and/or alpelisib every day (in 1% methylcellulose/0.5% Tween 80).
Patient-derived xenograft (PDX) experiments were conducted at Crown Bioscience. The protocol and any amendment(s) or procedures involving the care and use of animals were approved by the IACUC of Crown Bioscience prior to initiation. During the study, the care and use of animals was conducted in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care. Tumor fragments were implanted into flanks of female athymic (nu/nu) mice (Vital River Laboratories). When tumors reached approximately 300 mm3, mice were randomized into groups and treated for 28 days.
Immunoblotting
Cell lysates were prepared on ice by washing cells once with PBS, resuspending in 1× cell lysis buffer (Cell Signaling Technology, #9803) or RIPA buffer supplemented with Halt protease inhibitor cocktail (Thermo Fisher Scientific, #78430) and briefly sonicating or vortexing. Tumor lysates were prepared by placing tumor pieces in tissue homogenizing tubes (Thermo Fisher Scientific, #15–340–154) containing RIPA buffer and homogenizing with a bead mill. Lysates were cleared by centrifugation (maximum speed, 10 minutes) and protein concentration determined by BCA assay (Pierce). Twenty to 60 μg of lysate was loaded on to 4%–12% Bis-Tris gels (NuPAGE, Invitrogen) for electrophoresis and immunoblotting. Active RAS was detected using Thermo Fisher Scientific Kit #16117. Membrane and cytosolic fractions were isolated using the Mem-PER Plus Membrane Protein Extraction Kit (Thermo Fisher Scientific kit, #89842).
IHC
IHC was performed as described previously (29). The following antibodies were used: Cleaved PARP (Cell Signaling Technology, #9661; 1:400 dilution) and Ki67 (DAKO, #M7240, 1:50). Percent positive cells in three fields per slide were counted using a microscope.
Clinical trial
The KURRENT-HN trial (NCT04997902) has been approved by the Institutional Review Board or ethics committee at participating institutions and is conducted in accordance with recognized ethical guidelines (U.S. Common Rule). Written informed consent was obtained from all patients enrolled in the study.
Data availability
PIK3CA mutation rates and HRAS expression data were obtained from The Cancer Genome Atlas (TCGA) PanCancer Atlas via http://cbioportal.org. All other data are available from the corresponding author upon reasonable request.
Results
The combination of alpelisib and tipifarnib inhibits spheroid growth in PIK3CA- and HRAS-dysregulated HNSCC cell line models
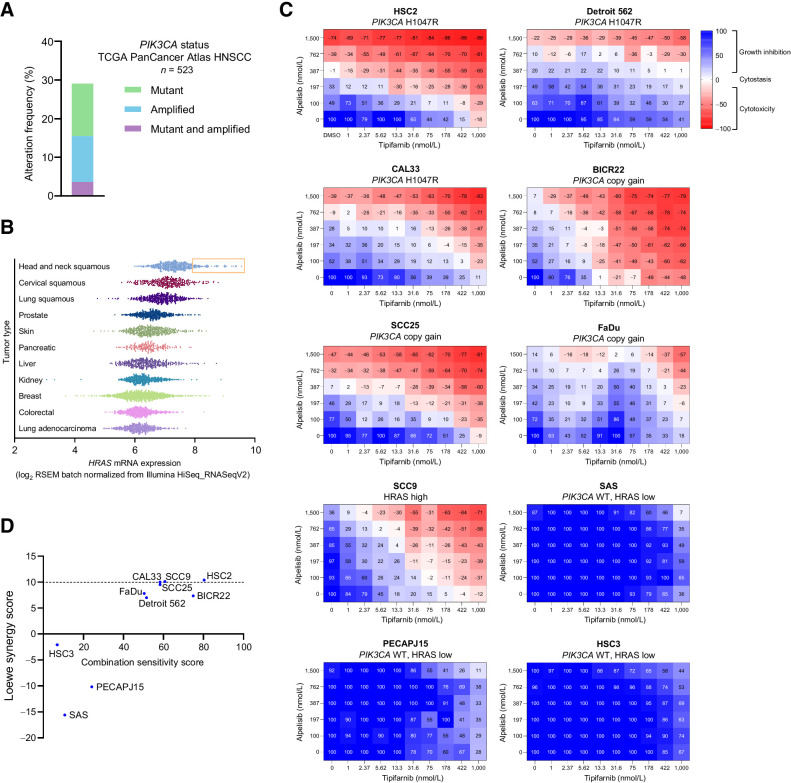
HRAS-MAPK and PI3K-AKT–mTOR are highly interdependent pathways in SCC. HRAS requires PIK3α to transform squamous epithelial cells (30), helical domain PIK3CA mutants must bind RAS for transformation (31), and overexpression of mutant or wild-type (WT) HRAS in a PIK3CA-mutant HNSCC cell line drives resistance to alpelisib (22). On the basis of such close cross-talk between HRAS and PI3Kα, we hypothesized that the combination of tipifarnib and alpelisib would be effective in head and neck tumors with high HRAS expression, PIK3CA amplification, or PIK3CA mutations. To ascertain what proportion of head and neck cancers might benefit from this combination, we evaluated rates of mutation and amplification of PIK3CA and HRAS in TCGA dataset. Approximately 30% of head and neck tumors harbored mutations or amplification of PIK3CA (Fig. 1A). As has been previously described (26), HRAS mutations are relatively rare, occurring in 4% to 8% of HNSCCs. However, the fact that HRAS mutations are restricted to only a handful of cancer subtypes, including HNSCC, implies that HRAS may play an important role in these tumors (32). Consistent with this notion, SCCs expressed elevated levels of HRAS mRNA compared with other tumor types in the TCGA PanCancer Atlas dataset, with up to 15% of head and neck tumors exhibiting high expression (defined as HRAS expression of one SD above the mean of HNSCC tumors; Fig. 1B). Taken together, we hypothesize that approximately 45% of all patients with HNSCC could potentially benefit from the simultaneous inhibition of HRAS and PI3K–mTOR signaling by tipifarnib and alpelisib.
Figure 1.
PIK3CA mutation/amplification and HRAS overexpression are common in HNSCC and cell lines harboring those alterations that are sensitive to combined tipifarnib and alpelisib. A, Frequency of PIK3CA gain-of-function mutations and amplifications in the TCGA HNSCC dataset (n = 523). B,HRAS mRNA expression in various tumor types in the TCGA PanCancer Atlas. Subset of HNSCCs with HRAS expression exceeding one SD from the mean is highlighted by orange box. C, HNSCC cell lines of the indicated PIK3CA/HRAS statuses were cultured as 3D tumor spheroids and treated with increasing concentrations of tipifarnib and/or alpelisib for 7 days. Heat maps indicate the degree of growth inhibition or cytotoxicity induced by the compounds. Data are representative of three biological replicates. D, Combination sensitivity and synergy scores of cell lines treated with alpelisib and tipifarnib in C. Scores were calculated from raw luminescence values (three biological replicates per cell line) using the SynergyFinder R package. Loewe synergy score of less than −10 indicates antagonistic combination activity, −10 to 10 an additive effect, and greater than +10 a synergistic effect.
To explore the therapeutic potential of this combination, we first assessed the sensitivity of a panel of PIK3CA- or HRAS-dysregulated HNSCC cell lines to tipifarnib and alpelisib. Cell lines with greater than 2.5 copies of PIK3CA were defined as copy gain and a cell line with the highest ratio of active (GTP-bound) to total HRAS was selected as the HRAS-high model (Supplementary Fig. S1A and S1B). When cultured as 3D tumor spheroids and treated in a checkerboard dose–response fashion, cell lines with gain-of-function PIK3CA mutations, PIK3CA copy gain, or high HRAS activity were sensitive to combined tipifarnib and alpelisib (Fig. 1C). Additive or synergistic inhibitory effects on cellular viability were observed in these lines with induction or enhancement of cytotoxicity by the combination (Fig. 1D; Supplementary Fig. S1C). In contrast, cell lines lacking both PIK3CA mutation/amplification and HRAS overexpression did not respond to either single agent or the combination (Fig. 1C and D). These results demonstrate that tipifarnib potentiates the antiproliferative effects of alpelisib in in vitro models of PI3Kα/HRAS-dysregulated HNSCC.
Combined tipifarnib and alpelisib treatment inhibits mTOR and RSK more potently and durably than either agent alone
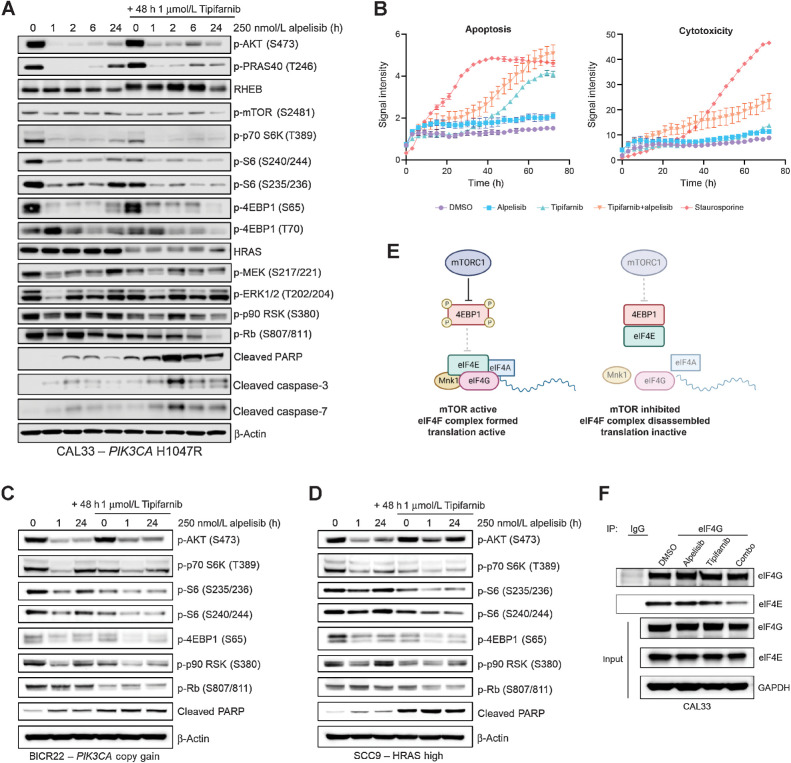
We next interrogated the molecular mechanism underlying the enhanced antiproliferative and cytotoxic activity of the combination compared with the single agents in our in vitro HNSCC models. As the single-agent efficacy of PI3K pathway inhibitors has historically been limited by feedback reactivation of PI3K–AKT–mTOR and RAS–MAPK signaling (7–13), we examined the effect of addition of tipifarnib on the kinetics of alpelisib's inhibition of these pathways in cell lines representative of each genetic subtype. In PIK3CA H1047R–mutant CAL33 cells, alpelisib rapidly inhibited the activity of AKT, mTOR, and ERK, as indicated by dephosphorylation of their substrates PRAS40, S6K, and RSK, respectively, after 1 hour of treatment (Fig. 2A). However, by 24 hours, phosphorylation of these proteins partially or completely rebounded. In contrast, when cells were treated with tipifarnib (causing HRAS and RHEB to become defarnesylated, as indicated by mobility shift) 24 hours prior to addition of alpelisib, basal phosphorylation of S6K, S6, and RSK was reduced, the initial inhibition by alpelisib was deeper, and the subsequent rebound was blocked. This potent and durable inhibition of mTOR and RSK by the tipifarnib–alpelisib combination corresponded with cell-cycle arrest (dephosphorylation of Rb) and induction of apoptosis (PARP, caspase-3, and caspase-7 cleavage; Fig. 2A). Live cell imaging experiments corroborated these findings; combination treatment more potently induced cytotoxicity in CAL33 cells than tipifarnib or alpelisib alone (Fig. 2B). In PIK3CA WT, HRAS low HSC3 cells, neither PARP cleavage (Supplementary Fig. S2A) nor Annexin V/cytotoxicity (Supplementary Fig. S2B) were significantly induced by the combination.
Figure 2.
Tipifarnib blunts mTOR and RSK reactivation following alpelisib treatment and induces cell-cycle arrest and apoptosis in PIK3CA-mutant, PIK3CA-amplified, and HRAS-high cell lines. A, Immunoblot of the indicated signaling proteins in PIK3CA-mutant CAL33 cells treated with 250 nmol/L alpelisib for 0, 1, 2, 6, or 24 hours in the presence or absence of 1 μmol/L tipifarnib. Cells were treated with tipifarnib for 48 hours. Images are representative of three biological replicates. B, Apoptosis (Annexin V) and cytotoxicity (loss of membrane integrity, nuclei exposure) over time in CAL33 cells treated with DMSO, 250 nmol/L alpelisib, 1 μmol/L tipifarnib, or the combination for 72 hours measured via Incucyte live cell imaging. Staurosporine (100 nmol/L) was used as a positive control. Data are means ± SD of three biological replicates. C and D, Immunoblots of indicated proteins in PIK3CA copy gain BICR22 cells (C) or HRAS-high SCC9 cells (D) treated with 250 nmol/L alpelisib for 0, 1, or 24 hours in the presence of absence of 1 μmol/L tipifarnib (48-hour treatment). E, Graphical overview of role of mTORC1 in regulating assembly of the eIF4F translation initiation complex. mTORC1 phosphorylates 4EBP1, impeding its binding to eIF4E, allowing for complex assembly. F, eIF4G (or IgG control) was immunoprecipitated from CAL33 cells treated with DMSO, 250 nmol/L alpelisib (24 hours), 1 μmol/L tipifarnib (48 hours), or the combination. Immunoblots indicate the levels of eIF4G and eIF4E coimmunoprecipitated and in the input cell lysate. Images are representative of two biological replicates.
In PIK3CA copy gain BICR22 cells and HRAS-high SCC9 cells, combined tipifarnib–alpelisib treatment had a similar impact on signaling. Phosphorylation of S6K, S6, 4EBP1, and RSK was transiently reduced by alpelisib, but rebounded by 24 hours (Fig. 2C and D). Addition of tipifarnib reduced baseline phosphorylation of these proteins, deepened their inhibition by alpelisib, and blunted their rebound after 24 hours of alpelisib exposure. More potent TOR and RSK blockade correlated with diminished Rb phosphorylation and induction of PARP cleavage, indicating that the combination inhibited cell cycling and induced cell death, as in PIK3CA-mutant cells.
As we observed a synergistic inhibition of mTOR substrate phosphorylation by combined treatment with alpelisib and tipifarnib in our cell line models, we examined the effect these agents had on eIF4F complex formation. mTORC1 phosphorylates 4EBP1, preventing its binding to eIF4E, allowing eIF4E:eIF4G interaction and initiation of translation (Fig. 2E). In CAL33 cells, the interaction of eIF4E with eIF4G was markedly reduced by combination treatment, as measured by eIF4G immunoprecipitation (Fig. 2F). Thus, tipifarnib and alpelisib may decrease protein translation via mTOR inhibition. Taken together, these findings demonstrate that addition of tipifarnib increases the depth and duration of mTOR inhibition compared with alpelisib alone, leading to cell death in PIK3CA/HRAS-dysregulated HNSCC.
The combination of tipifarnib and alpelisib inhibits mTORC1 activity and induces tumor regression in a PIK3CA-mutant cell line–derived xenograft model
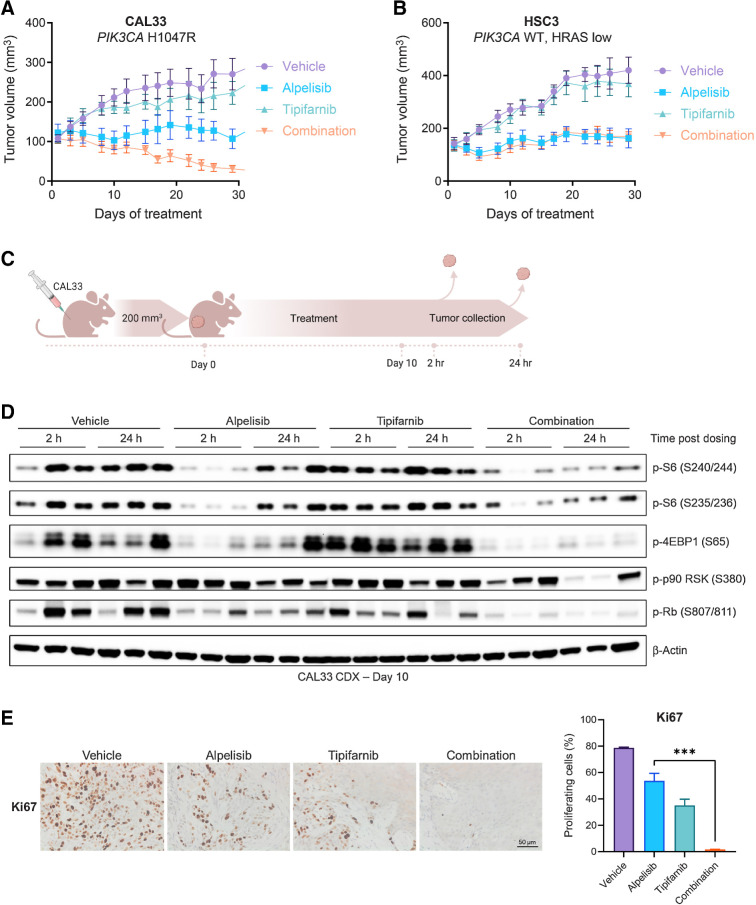
To determine whether enhanced sensitivity to the alpelisib-tipifarnib doublet also occurred in vivo, we implanted PIK3CA-mutant (CAL33) and PIK3CA WT/HRAS-low (HSC3) xenografts and treated the tumors with alpelisib, tipifarnib, or the combination. The combination blocked the growth of the PIK3CA-mutant tumors and induced tumor regression better than either agent alone (Fig. 3A). No additive effect was observed in the control PIK3CA WT/HRAS-low tumors (Fig. 3B).
Figure 3.
The tipifarnib and alpelisib doublet inhibits mTOR signaling and induces apoptosis and tumor regression in PIK3CA-mutant cell line–derived xenograft tumors. A and B, Growth of CAL33 (A) and HSC3 (B) cell line–derived xenograft tumors treated with vehicle, tipifarnib (60 mg/kg twice a day), alpelisib (40 mg/kg every day), or the combination. Data represent means ± SEM; n = 10 mice per group. C, Overview of in vivo pharmacodynamic study. CAL33 cells were implanted in the flanks of athymic mice and allowed to reach 200 mm3, at which time, treatment with vehicle, tipifarnib (60 mg/kg twice a day), alpelisib (40 mg/kg every day), or the combination was initiated and continued for 10 days. On day 10, tumors were collected 2 hours and 24 hours after alpelisib dosing for analysis by IHC and immunoblot. D, Immunoblots of indicated signaling proteins in CAL33 tumors treated with tipifarnib, alpelisib, or the combination for 10 days and collected at 2 and 24 hours post-alpelisib dosing. Tumors from three animals per treatment group/collection time point are shown. E, Representative IHC analysis (left) and quantification (right) of Ki67 in CAL33 tumors from C collected after 10 days of treatment, 24 hours after final alpelisib dose. Data are means ± SEM; n = 3 fields per condition. ***, P < 0.01 by two-tailed Student t test.
We next asked whether the mechanism underlying this tumor regression was consistent with that observed in vitro. We treated PIK3CA-mutant CAL33 tumors with tipifarnib, alpelisib, or the combination for 10 days and collected tumors 2 and 24 hours after the final dose of alpelisib for IHC and immunoblot analysis (Fig. 3C). Tipifarnib treatment resulted in defarnesylation of HRAS and RHEB (Supplementary Fig. S3A). Alpelisib alone transiently inhibited mTOR activity (4EBP1, S6 phosphorylation at 2 hours), but by 24 hours, a significant rebound was observed (Fig. 3D). Addition of tipifarnib blocked this rebound and promoted cell-cycle arrest as indicated by decreased Rb phosphorylation. IHC analyses revealed the tipifarnib–alpelisib doublet inhibited tumor proliferation (Ki67 staining; Fig. 3E) and enhanced induction of apoptosis (PARP cleavage; Supplementary Fig. S3B) compared to monotherapy. Thus, the combination of alpelisib and tipifarnib durably inhibits mTORC1 activity in vivo, leading to cell death and tumor regression.
Genetic depletion of RHEB and HRAS phenocopies tipifarnib treatment, implicating them as key FTase targets in HNSCC
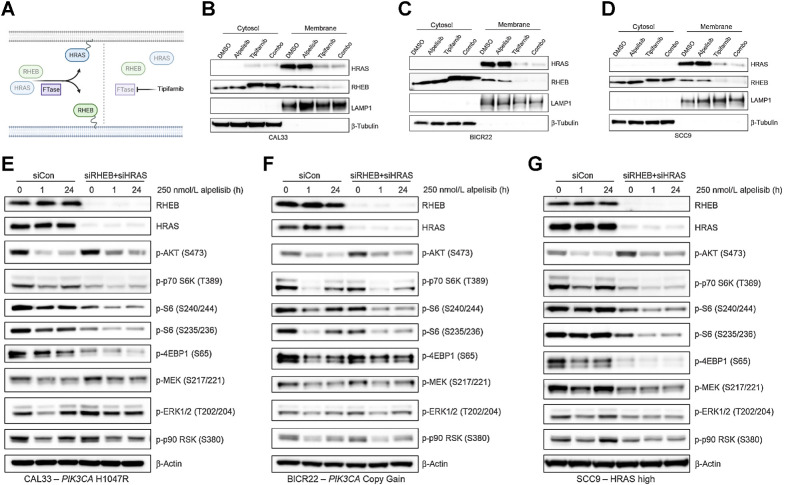
Although our mechanistic data point to HRAS and RHEB being the most important farnesylated targets in PI3Kα/HRAS-dysregulated HNSCCs, a number of signaling proteins are obligately farnesylated (16) and could contribute to tipifarnib sensitivity. Upon FTase inhibition, these proteins are defarnesylated and lose their membrane localization and activity (Fig. 4A). Using subcellular fractionation, we confirmed that tipifarnib treatment resulted in the membrane delocalization of the majority of both HRAS and RHEB in CAL33, BICR22, and SCC9 cells (Fig. 4B–D). This result suggests that these proteins were sufficiently defarnesylated to impair their activity. Tipifarnib also displaced RHEB from lysosomes, a major site for mTORC1 activation (Supplementary Fig. S4).
Figure 4.
Depletion of the FTase targets RHEB and HRAS phenocopies tipifarnib treatment in PIK3CA/HRAS–dysregulated HNSCC cell lines. A, Graphical overview of the effect of tipifarnib on the membrane localization of HRAS and RHEB. Inhibition of FTase activity leads to defarnesylation of HRAS and RHEB and loss of membrane localization and activity. B–D, Immunoblots of HRAS, RHEB, LAMP1, and β-tubulin in the cytosol and membrane fractions of CAL33 (B), BICR22 (C), and SCC9 (D) cells treated with DMSO, 250 nmol/L alpelisib (24 hours), 1 μmol/L tipifarnib (48 hours), or the combination. E–G, Immunoblots of indicated signaling proteins in CAL33 (E), BICR22 (F), and SCC9 (G) cells treated with siRNAs to knock down RHEB and HRAS expression (vs. control nontargeting pool) for 48 hours prior to addition of 250 nmol/L alpelisib. Cells were collected and lysed after 0, 1, or 24 hours alpelisib treatment and immunoblot analysis performed.
To further clarify the role of RHEB and HRAS in mediating tipifarnib response, we asked whether genetic depletion of these two targets was sufficient to phenocopy the effect of tipifarnib on cell signaling. We transfected cells with siRNAs to deplete RHEB and HRAS expression or a control nontargeting pool and, after allowing 48 hours for knock down, treated the cells with alpelisib. Like tipifarnib treatment, reduction of RHEB and HRAS expression decreased basal phosphorylation of S6K, S6, and 4EBP1 to varying degrees across the PIK3CA-mutant/amplified and high active cell lines (Fig. 4E–G). Inhibition of phospho-S6K, phospho-S6, and phospho-4EBP1 by alpelisib was greater in the double knock-down (siRHEB and siHRAS) cells compared with control-transfected cells, and this inhibition was more durable, exhibiting minimal rebound of phosphorylation after 24 hours. The effect of codepletion of RHEB and HRAS expression on mTOR substrate phosphorylation was greatest in PIK3CA-mutant cells (Fig. 4E), whereas its effect on MAPK activity was largest in HRAS-high cells (Fig. 4G), pointing to a potential hierarchy of target dependence corresponding to genotype. As genetic targeting of HRAS and RHEB closely mimicked tipifarnib treatment in these models, we surmise these are the key farnesylation-dependent proteins in the context of PI3Kα inhibition in HNSCC.
Combined, synchronous tipifarnib–alpelisib treatment robustly inhibits the growth of PDX models harboring PIK3CA alterations or HRAS overexpression
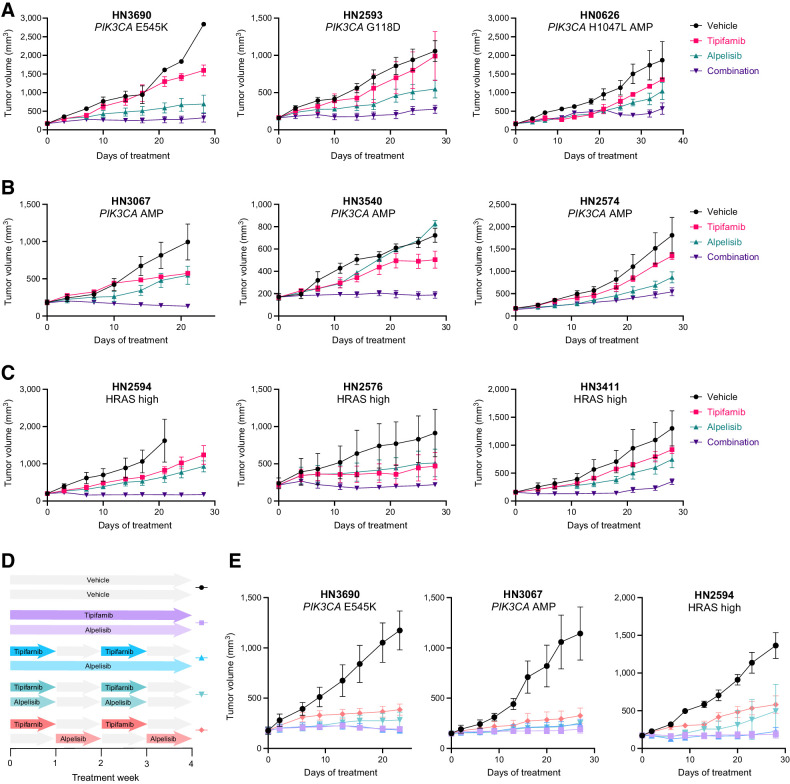
We next assessed the sensitivity of a panel of PIK3CA-mutant, PIK3CA-amplified, or HRAS-overexpressing (Supplementary Fig. S5) PDX models to single-agent tipifarnib/alpelisib or the combination. Combination treatment was well tolerated (Supplementary Fig. S5B), consistently slowed, or blocked tumor growth more effectively than alpelisib or tipifarnib alone, and induced tumor regressions in several models (Fig. 5A–C).
Figure 5.
Combined, synchronous tipifarnib–alpelisib treatment robustly inhibits the growth of PIK3CA- and HRAS-dysregulated HNSCC PDX models. A–C, Growth of HNSCC PDX tumors harboring the indicated PIK3CA mutations (A), PIK3CA amplification (AMP; B), or HRAS overexpression (models in the top 25% of HRAS expression; C) treated with vehicle, tipifarnib (60 mg/kg twice a day), alpelisib (40 mg/kg every day), or the combination on a continuous schedule. Data represent means ± SEM; n = 3 mice per group. D, Schematic overview of the tipifarnib (60 mg/kg twice a day) and alpelisib (40 mg/kg every day) dosing schedules evaluated in select HNSCC PDX models. E, Impact of the dosing schedules outlined in D on the growth of PIK3CA- or HRAS-dysregulated PDX tumors. Data represent means ± SEM; n = 5 mice per group.
The extensive cross-talk between the HRAS-MAPK and PI3K–AKT–mTOR pathways implies that synchronous administration of tipifarnib and alpelisib would have the greatest antitumor effects. However, concomitant blockade of multiple oncogenic signaling pathways can be challenging in the clinic. To explore scheduling options and the role of pathway interdependence in the observed combination activity, we tested various dosing schedules in PIK3CA- and HRAS-dysregulated PDX models (Fig. 5D). Continuous dosing of tipifarnib and alpelisib was most effective in blocking tumor growth but administering tipifarnib on alternate weeks (which is the established clinical regimen in HRAS-mutant HNSCC) while dosing alpelisib continuously was nearly as efficacious (Fig. 5E). Dosing alpelisib every other week slightly reduced the activity of the combination. Nevertheless, the observation that synchronous weekly on/off dosing retains robust activity may provide flexibility to optimize the dose scheduling of the combination in the clinic. It is important to note that nonsynchronous intermittent dosing of alpelisib and tipifarnib was markedly less effective, highlighting the degree of cooperativity between HRAS/RHEB/PI3K and the synergistic potential of combination treatment. Although synchronous and nonsynchronous discontinuous dosing appears similarly effective in the HRAS-high PDX model (Fig. 5E, right), this is due to a single outlier in the synchronous group. In this cohort, 4 of 5 tumors were maintained in full stasis with one rapidly growing breakout tumor, whereas 4 of 5 animals displayed tumor progression in the nonsynchronous cohort.
KURRENT-HN clinical trial
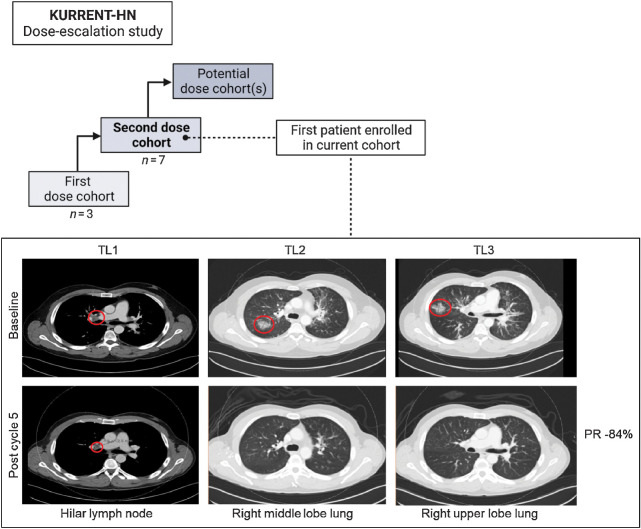
On the basis of these findings, we launched the KURRENT-HN trial (NCT04997902) to assess the safety, tolerability, pharmacokinetics, pharmacodynamics, and preliminary antitumor activity of combined tipifarnib and alpelisib in advanced HNSCC. This study is a phase I/II open-label dose escalation study enrolling patients with HNSCC with PIK3CA mutation/amplification and/or HRAS overexpression that have progressed on at least one prior line of therapy. Although it is early in this clinical trial, preliminary safety and efficacy data have been encouraging. The first patient enrolled to the second dose cohort (Fig. 6) was a 35-year-old male nonsmoker with PIK3CA R88Q mutant, HPV16+ SCC of the tonsil that had progressed following cisplatin/radiation and cemiplimab/ISA101b treatment. The patient was administered tipifarnib and alpelisib and after one 28-day cycle, experienced an 81% reduction and a complete disappearance of the two target lesions in the lung. After three cycles, the complete response of the lung was maintained, and the patient had an 84% reduction in the target lesions (Fig. 6). Since being on treatment, the patient has also experienced improvement in respiratory symptoms. As of the most recent data cut (September 2022), the response was maintained (>27 weeks), and the patient remained on study.
Figure 6.
Overview of ongoing KURRENT-HN tipifarnib–alpelisib dose escalation study and partial response (PR) of first patient enrolled in current dose cohort. CT of a patient with PIK3CA-mutant metastatic HNSCC at baseline and after five cycles of tipifarnib and alpelisib. Target lesions (red circles) are in the hilar lymph node and right upper lobe of the lungs. Patient experienced a partial response and remains on study at time of data cut (September 2022).
Discussion
Improvement in outcomes for patients with R/M HNSCC has been hindered by a lack of targeted therapies. In this study, we provide in vitro and in vivo evidence that the mechanistically designed, biomarker-matched strategy of combining alpelisib and tipifarnib may be efficacious in PIK3CA- and HRAS-dysregulated HNSCCs. Showing encouraging early activity in the clinic and a manageable safety profile (NCT04997902), this combination has potential to benefit approximately half of patients with HNSCC, for whom treatment options are currently limited.
As approximately 30% of head and neck cancers harbor alterations in PIK3CA, pharmacologic targeting of p110α is an appealing therapeutic strategy. Unfortunately, feedback reactivation of PI3K and/or its parallel pathways greatly reduces the single-agent effectiveness of PI3K inhibitors. For example, in estrogen receptor–positive (ER+) breast cancer, where alpelisib is approved in combination with the ER antagonist fulvestrant (33), PI3Kα inhibition induces ER signaling while the latter activates the PI3K pathway as a mechanism of endocrine therapy escape (34–36). When combined, alpelisib and fulvestrant effectively block the bypass mechanisms tumor cells employ to evade the inhibitory effects of alpelisib monotherapy, leading to superior antitumor activity.
In HNSCC, mTOR and RAS–MAPK are the primary pathways driving adaptive resistance to PI3K inhibitors, but cotargeting PI3K and these escape pathways has proven challenging in the clinic. Direct inhibition of PI3K and mTOR via dual kinase inhibitors demonstrated unacceptable toxicity and/or lacked meaningful clinical activity (37, 38). Although the combination of alpelisib and the allosteric mTOR inhibitor everolimus was tolerable in solid tumors (like breast cancer and renal cell carcinoma), overlapping toxicities resulting from PI3K–mTOR pathway inhibition, such as hyperglycemia and stomatitis, make it difficult to assess the overall benefit-risk ratio (39, 40). Similarly, while backed by a strong mechanistic rationale, the clinical development of therapeutic strategies combining inhibitors of the PI3K and MAPK pathways has been hindered by safety concerns. The combination of PI3K and MEK inhibition showed favorable activity in solid cancers but at the expense of tolerability (41). Thus, further effort is needed to design translatable methods of coinhibiting the PI3K–mTOR and RAS–MAPK pathways.
In this work, we propose a new therapeutic strategy—the combination of the FTI tipifarnib with alpelisib—to target the cross-talk of the PI3K and MAPK pathways in HNSCCs and limit their feedback re-activation upon PI3K inhibition. We demonstrate that tipifarnib achieves both vertical and lateral inhibition of PI3K signaling via depletion of RHEB and HRAS. Treatment of PI3Kα- and HRAS-dysregulated HNSCC cells with alpelisib exposes a dependency on mTOR that is effectively blocked by tipifarnib, thereby greatly enhancing sensitivity to PI3Kα inhibition.
Although RHEB and HRAS are the two obligately farnesylated proteins most closely linked to PI3K–MAPK signaling, FTase inhibition undoubtedly perturbs the function of other farnesylated targets that may contribute (or detract) from the observed phenotype. Profiling farnesylation changes in an unbiased manner (via mass spectrometry) would help confirm the mechanism we propose in this study. Moreover, questions remain regarding the durability of the antitumor effect we describe. Even with combination therapy regimens, development of resistance is common, and the signaling networks governing HNSCC tumor growth are complex. Analysis of xenograft tumors exposed to prolonged combined tipifarnib and alpelisib would be helpful in identifying potential mechanisms of resistance. As our clinical trial is in the early-stages of dose escalation, we were able to provide only one example of a sustained PR in a PIK3CA-mutant patient in this report. We will gather more information regarding the durability of this particular combination strategy as more patients are enrolled and we move towards an optimal biologically active dose.
Importantly, by blocking farnesylation of RHEB, tipifarnib is likely to inhibit mTORC1, the driver of feedback-mediated resistance to alpelisib, while sparing mTORC2, the primary source of TOR inhibitor-associated toxicity (42, 43). Tipifarnib blocks localization of RHEB to all cellular membranes, including the lysosomes, a key site for RHEB's regulation of mTORC1 activity. Recently, a small pool of RHEB was reported to be functional in the nucleus and found to be refractory to farnesyltransferase inhibition (44). While the exact role of this nuclear form of RHEB is unclear, exclusive inhibition of membrane bound RHEB by tipifarnib may be a desirable property from a tolerability perspective when combining with a PI3K inhibitor.
Although RHEB has been nominated, albeit infrequently, as an oncogenic driver and therapeutic target in cancer (45–48), this work is the first to demonstrate that RHEB inhibition may prevent adaptive resistance to a targeted therapy. We describe how RHEB–mTORC1 blockade via tipifarnib potentiates the antitumor effects of PI3K inhibition in HNSCC, but sufficient TORC1 suppression is a common requirement for sensitivity to numerous other targeted agents, including inhibitors of RAF (49) and KRASG12C (50). Positioned downstream of the majority of oncogenic drivers, mTORC1 is a critical mediator of tumor growth, survival, and therapy escape. We posit that by blocking feedback reactivation of mTORC1, tipifarnib may enhance the efficacy and expand the utility of other targeted therapies approved or currently in development, broadly benefiting patients with advanced malignancies.
Supplementary Material
Supplementary figures related to figures 1-5.
Acknowledgments
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
A.E. Smith reports patents for US 17/488,194, US 18/300,924, and PCT/US2023/65417 pending to Kura Oncology; in addition, A.E. Smith is an employee and stockholder of Kura Oncology. S. Chan is an employee and stockholder of Kura Oncology. Q. Reilly is an employee and stockholder of Kura Oncology. J.Z. Wang is a former employee of Kura Oncology, Inc. L. Kessler reports personal fees from Kura Oncology during the conduct of the study and personal fees from Kura Oncology outside the submitted work. A.L. Dayoub is a stakeholder at Kura Oncology. V.M. Villaflor reports other support from Johnson & Johnson and AstraZeneca and grants from Takeda outside the submitted work. D.R. Adkins reports grants and other support from Kura Oncology during the conduct of the study; grants and other support from Merck, Cue Biopharma, Blueprint Medicine, Exelixis, Vaccinex, Boeringer Ingelheim, Gilead Sciences, Immunitas, TargImmune Therapeutics, twoXAR, Xilio Therapeutics, Eisai Europe, Coherus Biosciences, Jazz Pharmaceuticals, Pfizer, Eli Lilly, Bristol Myers Squibb/Celgene, Novartis, AstraZeneca, CoFactor Genomics, Debiopharm, ISA Pharmaceuticals, Beigene, Roche, Immutep, Hookipa Biotech, Epizyme, Adlai Nortye, BioAtla, Calliditas Therapeutics, Genmab, Tizona Therapeutics, and NATCO Pharma outside the submitted work. J.Y. Bruce reports other support from Kura during the conduct of the study. A.L. Ho reports grants from Kura Oncology during the conduct of the study; grants and personal fees from Eisai, Kura Oncology, Merck, and Ayala; other support from Remix Therapeutics and Klus Pharma; personal fees from Affyimmune, Coherus, Exelixis, Rgenta, Elevar Therapeutics, and Prelude Therapeutics; nonfinancial support from Cellestia and Inxmed; grants from BMS, Bayer, Genentech, Astellas, Novartis, Bioatla, Posieda Therapeutics, TILT biotherapeutics, and grants from Verastem outside the submitted work; in addition, A.L. Ho has patent 63/193700 for “Lesional Dosimetry Methods For Tailoring Targeted Radiotherapy” issued. C.A. Perez reports grants from Kura Oncology, Accutar Biotech, Artios Pharma, Dracen Pharmaceuticals, Elpiscience Biopharmaceuticals, Elucida Oncology, Genentech Inc., Hyamab Inc., Jazz Pharmaceuticals, Kinnate Biopharma, Mirati Pharmaceuticals, Relay Therapeutics, Ribbon Therapeutics, Seagen Inc., Tallac Inc., Xilio Therapeutics, and Zhuhai Yufan Biotechnologies Co. outside the submitted work. G.J. Hanna reports grants and nonfinancial support from Kura Oncology outside the submitted work. A. Saunders holds stock options in Kura Oncology. J. Gutkind reports other support from Kura Oncology during the conduct of the study and personal fees from Domain Therapeutics, io9, and Pangea outside the submitted work; in addition, J. Gutkind is a founder of Kadima Pharmaceuticals. F. Burrows is an employee and stockholder of Kura Oncology. S. Malik reports patents for US 17/488,194, US 18/300,924, and PCT/US2023/65417 pending to Kura Oncology; in addition, S. Malik is an employee and stockholder of Kura Oncology. No disclosures were reported by the other authors.
Authors' Contributions
A.E. Smith: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Chan: Conceptualization, data curation, formal analysis, investigation, visualization, and methodology. Z. Wang: Resources, investigation, visualization, methodology, writing–review and editing. A. McCloskey: Conceptualization, formal analysis, investigation, visualization, and methodology. Q. Reilly: Investigation and visualization. J.Z. Wang: Investigation and visualization. H.V. Patel: Investigation and methodology. K. Koshizuka: Resources, investigation, visualization, methodology, writing–review and editing. H.S. Soifer: Conceptualization, data curation, writing–review and editing. L. Kessler: Conceptualization, writing–review and editing. A. Dayoub: Data curation, project administration, writing–review and editing. V. Villaflor: Investigation, writing–review and editing. D.R. Adkins: Investigation, writing–review and editing. J.Y. Bruce: Investigation, writing–review and editing. A.L. Ho: Investigation, writing–review and editing. C.A. Perez: Investigation, writing–review and editing. G.J. Hanna: Investigation, writing–review and editing. A. Gascó Hernández: Conceptualization, project administration, writing–review and editing. A. Saunders: Conceptualization, project administration, writing–review and editing. S. Dale: Conceptualization, resources, supervision, project administration, writing–review and editing. J. Gutkind: Conceptualization, resources, data curation, formal analysis, supervision, methodology, project administration, writing–review and editing. F. Burrows: Conceptualization, resources, data curation, formal analysis, supervision, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Malik: Conceptualization, data curation, formal analysis, supervision, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers 2020;6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ionna F, Bossi P, Guida A, Alberti A, Muto P, Salzano G, et al. Recurrent/metastatic squamous cell carcinoma of the head and neck: a big and intriguing challenge which may be resolved by integrated treatments combining locoregional and systemic therapies. Cancers (Basel) 2021;13:2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet North Am Ed 2019;394:1915–28. [DOI] [PubMed] [Google Scholar]
- 4. Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov 2013;3:761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Juric D, Rodon J, Tabernero J, Janku F, Burris HA, Schellens JHM, et al. Phosphatidylinositol 3-Kinase α-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: results from the first-in-human study. J Clin Oncol 2018;36:1291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2011;19:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 2006;66:1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haruta T, Uno T, Kawahara J, Takano A, Egawa K, Sharma PM, et al. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol Endocrinol 2000;14:783–94. [DOI] [PubMed] [Google Scholar]
- 10. Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof Raphael A, Brook S, et al. AXL mediates resistance to PI3Kα inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell 2015;27:533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest 2008;118:3065–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011;332:1322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 2011;30:2547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunn LA, Riaz N, Fury MG, McBride SM, Michel L, Lee NY, et al. A phase 1b study of cetuximab and BYL719 (Alpelisib) concurrent with intensity modulated radiation therapy in stage III-IVB head and neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2020;106:564–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun 2006;344:869–80. [DOI] [PubMed] [Google Scholar]
- 16. Storck EM, Morales-Sanfrutos J, Serwa RA, Panyain N, Lanyon-Hogg T, Tolmachova T, et al. Dual chemical probes enable quantitative system-wide analysis of protein prenylation and prenylation dynamics. Nat Chem 2019;11:552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci 2016;129:1287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Hee W, Shin M-G, Lee S, Kim J-R, Park Wei S, Cho K-H, et al. Cooperative Activation of PI3K by Ras and Rho Family Small GTPases. Mol Cell 2012;47:281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-Kinase. J Biol Chem 1998;273:24052–6. [DOI] [PubMed] [Google Scholar]
- 20. Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol 2005;15:702–13. [DOI] [PubMed] [Google Scholar]
- 21. Wang Z, Feng X, Molinolo AA, Martin D, Vitale-Cross L, Nohata N, et al. 4E-BP1 is a tumor suppressor protein reactivated by mTOR inhibition in head and neck cancer. Cancer Res 2019;79:1438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruicci KM, Pinto N, Khan MI, Yoo J, Fung K, MacNeil D, et al. ERK-TSC2 signalling in constitutively-active HRAS mutant HNSCC cells promotes resistance to PI3K inhibition. Oral Oncol 2018;84:95–103. [DOI] [PubMed] [Google Scholar]
- 23. Li Z, Yang Z, Passaniti A, Lapidus RG, Liu X, Cullen KJ, et al. A positive feedback loop involving EGFR/Akt/mTORC1 and IKK/NF-kB regulates head and neck squamous cell carcinoma proliferation. Oncotarget 2016;7:31892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, et al. mTORC1 inhibition is required for sensitivity to PI3K p110α inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med 2013;5:196ra99–ra99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ, et al. PDK1-SGK1 signaling sustains AKT-independent mTORC1 activation and confers resistance to PI3Kα inhibition. Cancer Cell 2016;30:229–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gilardi M, Wang Z, Proietto M, Chillà A, Calleja-Valera JL, Goto Y, et al. Tipifarnib as a precision therapy for HRAS-mutant head and neck squamous cell carcinomas. Mol Cancer Ther 2020;19:1784–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ho AL, Brana I, Haddad R, Bauman J, Bible K, Oosting S, et al. Tipifarnib in head and neck squamous cell carcinoma with HRAS mutations. J Clin Oncol 2021;39:1856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng S, Wang W, Aldahdooh J, Malyutina A, Shadbahr T, Tanoli Z, et al. SynergyFinder plus: toward better interpretation and annotation of drug combination screening datasets. Genomics Proteomics Bioinformatics 2022;20:587–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z, Goto Y, Allevato MM, Wu VH, Saddawi-Konefka R, Gilardi M, et al. Disruption of the HER3-PI3K-mTOR oncogenic signaling axis and PD-1 blockade as a multimodal precision immunotherapy in head and neck cancer. Nat Commun 2021;12:2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, et al. Binding of Ras to phosphoinositide 3-Kinase p110α is required for Ras- driven tumorigenesis in mice. Cell 2007;129:957–68. [DOI] [PubMed] [Google Scholar]
- 31. Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110α of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 2008;105:2652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kessler L, Malik S, Leoni M, Burrows F. Potential of farnesyl transferase inhibitors in combination regimens in squamous cell carcinomas. Cancers 2021;13:5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narayan P, Prowell TM, Gao JJ, Fernandes LL, Li E, Jiang X, et al. FDA approval summary: alpelisib plus fulvestrant for patients with HR-positive, HER2-negative, PIK3CA-mutated, advanced or metastatic breast cancer. Clin Cancer Res 2021;27:1842–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller TW, Hennessy BT, González-Angulo AM, Fox EM, Mills GB, Chen H, et al. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest 2010;120:2406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor–positive breast cancer. Sci Transl Med 2015;7:283ra51–ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res 2010;12:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carlo MI, Molina AM, Lakhman Y, Patil S, Woo K, DeLuca J, et al. A phase Ib study of BEZ235, a dual inhibitor of phosphatidylinositol 3-Kinase (PI3K) and mammalian target of rapamycin (mTOR), in patients with advanced renal cell carcinoma. Oncologist 2016;21:787–8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tarantelli C, Lupia A, Stathis A, Bertoni F. Is there a role for dual PI3K/mTOR inhibitors for patients affected with lymphoma? Int J Mol Sci 2020;21:1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Curigliano G, Martin M, Jhaveri K, Beck JT, Tortora G, Fazio N, et al. Alpelisib in combination with everolimus ± exemestane in solid tumours: Phase Ib randomised, open-label, multicentre study. Eur J Cancer 2021;151:49–62. [DOI] [PubMed] [Google Scholar]
- 40. Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 2018;15:273–91. [DOI] [PubMed] [Google Scholar]
- 41. Shimizu T, Tolcher AW, Papadopoulos KP, Beeram M, Rasco DW, Smith LS, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res 2012;18:2316–25. [DOI] [PubMed] [Google Scholar]
- 42. Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 2012;335:1638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soefje SA, Karnad A, Brenner AJ. Common toxicities of mammalian target of rapamycin inhibitors. Targeted Oncology 2011;6:125–9. [DOI] [PubMed] [Google Scholar]
- 44. Zhong Y, Zhou X, Guan KL, Zhang J. Rheb regulates nuclear mTORC1 activity independent of farnesylation. Cell Chem Biol 2022;29:1037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Basso AD, Mirza A, Liu G, Long BJ, Bishop WR, Kirschmeier P. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits rheb farnesylation and mTOR signaling: role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J Biol Chem 2005;280:31101–8. [DOI] [PubMed] [Google Scholar]
- 46. Mavrakis KJ, Zhu H, Silva RL, Mills JR, Teruya-Feldstein J, Lowe SW, et al. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev 2008;22:2178–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ding H, McDonald JS, Yun S, Schneider PA, Peterson KL, Flatten KS, et al. Farnesyltransferase inhibitor tipifarnib inhibits Rheb prenylation and stabilizes Bax in acute myelogenous leukemia cells. Haematologica 2014;99:60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nardella C, Chen Z, Salmena L, Carracedo A, Alimonti A, Egia A, et al. Aberrant Rheb-mediated mTORC1 activation and Pten haploinsufficiency are cooperative oncogenic events. Genes Dev 2008;22:2172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Corcoran RB, Rothenberg SM, Hata AN, Faber AC, Piris A, Nazarian RM, et al. TORC1 suppression predicts responsiveness to RAF and MEK inhibition in BRAF-mutant melanoma. Sci Transl Med 2013;5:196ra98–ra98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures related to figures 1-5.
Data Availability Statement
PIK3CA mutation rates and HRAS expression data were obtained from The Cancer Genome Atlas (TCGA) PanCancer Atlas via http://cbioportal.org. All other data are available from the corresponding author upon reasonable request.