Abstract
Genetic modification of cells using viral vectors has shown huge therapeutic benefit in multiple diseases. However, inefficient transduction contributes to the high cost of these therapies. Several transduction-enhancing small molecules have previously been identified; however, some may be toxic to the cells or patient, otherwise alter cellular characteristics, or further increase manufacturing complexity. In this study, we aimed to identify molecules capable of enhancing lentiviral transduction of T cells from available small-molecule libraries. We conducted a high-throughput flow-cytometry-based screen of 27,892 compounds, which subsequently was narrowed down to six transduction-enhancing small molecules for further testing with two therapeutic lentiviral vectors used to manufacture GSK’s clinical T cell therapy products. We demonstrate enhanced transduction without a negative impact on other product attributes. Furthermore, we present results of transcriptomic analysis, suggesting alteration of ribosome biogenesis, resulting in reduced interferon response, as a potential mechanism of action for the transduction-enhancing activity of the lead compound. Finally, we demonstrate the ability of the lead transduction enhancer to produce a comparable T cell product using a 3-fold reduction in vector volume in our clinical manufacturing process, resulting in a predicted 15% reduction in the overall cost of goods.
Keywords: transduction enhancer, lentivirus, lentiviral transduction, T cell, T cell therapy, cancer immunotherapy, cell therapy, gene therapy, cell and gene therapy
Graphical abstract

Malach and colleagues identify novel compounds capable of enhancing lentiviral transduction of human T cells using a high-throughput screening approach. Verification studies identified a transduction enhancer capable of producing a comparable T cell product using a 3-fold reduction in vector volume in a large-scale manufacturing process.
Introduction
Genetic modification of cells using viral vectors for transduction forms the keystone of many cellular therapies, including the treatment of rare genetic diseases using modified hematopoietic stem cells (HSCs) or cancer treatment using adoptive T cell therapy. In many cases, vesicular stomatitis virus G (VSV-G)-pseudotyped lentiviral vectors are used, as they are able to transduce a wide range of both dividing and non-dividing cells, resulting in long-term transgene expression. However, the low efficiency of transduction with lentiviral vectors carrying large therapeutic cassettes poses challenges to production of cell therapies,1 particularly where high levels of transgene expression are needed for therapeutic effect, for example, where production of the therapeutic protein by the transduced cell provides a wider therapeutic effect through cross-correction or where a high number of transduced cells is required. For these reasons, large volumes of lentiviral vector are often required in the production of these therapies, which in turn increases the cost of manufacturing.2
The use of small molecules to enhance transduction has the potential to address this challenge. Several transduction enhancers (TEs) have been previously identified and are reviewed by Kaygisiz and Synatschke.3 Different TEs act at different stages of the viral replication cycle, suggesting the existence of multiple innate barriers to gene transfer. Entry enhancers, including RetroNectin,4,5 LentiBOOST,6 protamine sulfate,7 Vectofusin-1,8 polybrene,9 DEAE-dextran,10 and staurosporine,11 act by reducing the electrostatic repulsion between the two negatively charged surfaces of the cell membrane and the viral envelope, thereby increasing co-localization of the cell and vector particle, or by increasing fusion of the viral particles with the cell. Post-entry enhancers, including prostaglandin E2 (PGE2),12,13 cyclosporine H,14,15 and rapamycin,16 act on intracellular processes. Several TEs have been shown to work synergistically.17 While much of the focus has been on TEs for lentiviral transduction of HSCs, their use has also been reported in other cell types, including T cells,14,18,19,20,21 mesenchymal stem cells (MSCs),10,22 and dendritic cells,23 and with other types of vectors including adenovirus23 and α-retrovirus.17
Aside from their transduction-enhancing activity, there are other considerations for introduction of a TE into cell therapy manufacturing. Although polybrene has been widely used because of its low cost, ease of use, and efficacy in many cell types, it is also toxic to cells at higher concentrations and has been shown to alter cellular characteristics, including differentiation and proliferation of MSCs22 and T cells.18 RetroNectin resulted in a change in the proportion of T cell memory subtypes,18 PGE2 reduced CD90+ long-term repopulating HSCs,17 and rapamycin reduced cell-cycle progression and proliferation in HSCs.16 Not all the TEs described are compatible with good manufacturing practice (GMP). In addition, some could pose safety concerns, which render them less suitable for clinical use: for example, rapamycin is an immunosuppressant.16 Finally, the use of TEs should not increase the complexity of the manufacturing process, as is the case for RetroNectin, which must be applied as a coating to cell-culture vessels used during transduction.4 The most widely used TE is LentiBOOST, a mixture of poloxamer synperonic F108 and polybrene, which has been shown to be effective in increasing transduction in several different cell types including HSCs and T cells.24 While some commercially available TEs have shown to be beneficial in enhancing transduction, the cost associated with their use may be prohibitive due to restrictions placed on intellectual property and license requirements for clinical and commercial use. GSK has access to a large library of small molecules; to find an enhancer suitable for use in the manufacture of cell therapies, a library of biologically annotated compounds was selected for screening.
In this article, we describe the results of the screen for 27,892 compounds through primary, confirmatory, and full dose-response curve experiments to identify hit compounds for further testing and, finally, identification of a lead molecule. We verify that these compounds enhance transduction with two different therapeutic vectors currently used to manufacture clinical T cell therapy products (registered clinical trial NCT04526509), encoding a T cell receptor targeting the NY-ESO-1 antigen together with either the cluster of differentiation 8 α (CD8α) cell-surface receptor or the dominant-negative transforming growth factor β receptor type II (dnTGF-β RII) cell-surface receptor. We show no negative impact of compound treatment on other product attributes including viability, expansion, phenotype, and cytotoxicity. We present results of transcriptomic and integration site analysis which suggest a potential mechanism of action for the transduction-enhancing activity of the lead compound. Finally, we demonstrate production of a comparable T cell product using a 3-fold reduction in vector volume in a large-scale, automated manufacturing process suitable for clinical and commercial production of autologous cell therapies.
Results
Screening of a small-molecule library identifies compounds capable of enhancing lentiviral transduction of T cells
A high-throughput screen of 27,892 compounds from GSK’s small-molecule library was conducted on enriched CD4+/CD8+ T cells from a single healthy donor at 10 mM concentration with GFP vector, as described in materials and methods. In total, 347 compounds were identified that met the QC criteria and which showed an increase in transduction of ≥3 standard deviations over that of the vector-only control as determined by flow cytometry for GFP (Figure 1A) while not decreasing viability. The proportion of active molecules identified from the primary screen was 1.2%, which confirmed that our selection of compounds annotated as biologically active led to a significant increase in active molecules. These compounds were screened again in enriched CD4+/CD8+ T cells from two additional healthy donors; 160 compounds which showed a consistent increase in transduction in both donors were then identified (Figure 1B).
Figure 1.
High-throughput screening identifies compounds capable of enhancing transduction in CD3+ T cells using GFP vector
(A) Initial screen of 27,892 compounds in CD3+ T cells from a single donor, showing %GFP+ cells (gray dots indicate compounds not resulting in increased transduction above the vector-only control, blue dots indicate compounds resulting in increased transduction of 3 standard deviations above the vector-only control, red dots indicate the lead compound identified in subsequent experiments). (B) Plot showing increase in %GFP+ cells in T cells from three different healthy donors: primary, confirmation 2 (CON 2), and confirmation 3 (CON 3); darker blue markers represent increasing cLogP, marker size increasing with molecular weight. (C) Representative example of full-curve results for a compound showing a dose-dependent increase in GFP expression in CD3+, CD4+, and CD8+ T cell subsets. (D) Representative example of full-curve results for a compound showing a dose-dependent increase in GFP expression in CD3+ and CD8+ T cell subsets, with minimal increase in CD4+ GFP expression.
These 160 compounds were taken forward into full-curve screening, where they were tested in an 11-point dose-response series to produce a dose-response curve for transduction efficiency, using enriched CD4+/CD8+ T cells from two healthy donors. Transduction efficiency was measured in CD3+, CD4+, and CD8+ T cell populations and viability. Compounds that produced a clear dose-response curve in all three populations (Figure 1C), as well as compounds which initially showed an increase in GFP expression in the CD3+ cell population and a selective effect in either the CD4+ or CD8+ populations (Figure 1D), were selected for further study to evaluate their impact on other T cell product attributes.
Small-scale experiments identify compounds capable of enhancing transduction of T cells with therapeutic vectors
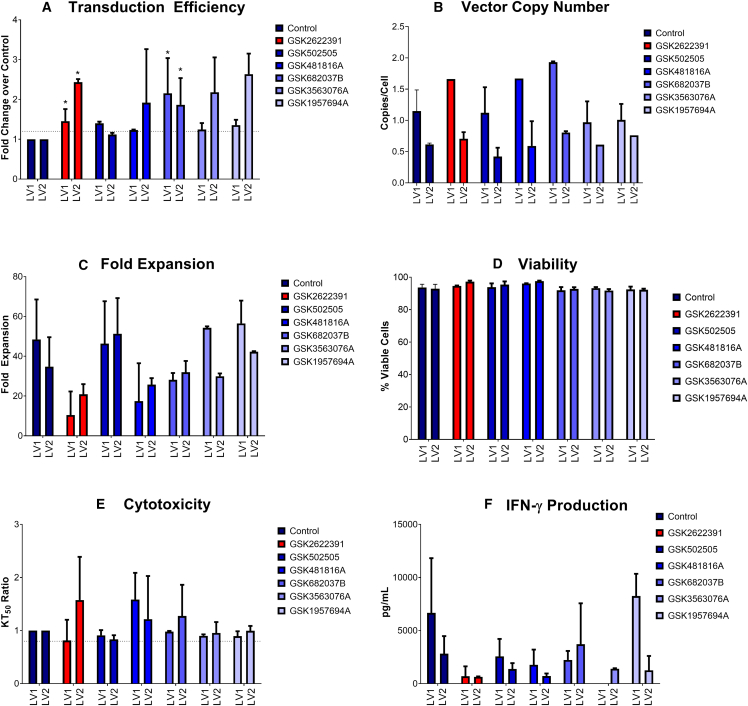
To confirm that the results obtained during the primary and full-curve screen with GFP vector was reproducible with vectors encoding therapeutic genes, 60 compounds were evaluated in small-scale experiments for their ability to increase transduction efficiency without negatively impacting other product attributes, such as viability and fold expansion (data not shown). From these experiments, the top-performing six compounds were selected and further evaluated for their impact on transduction efficiency, viability, fold expansion, vector copy number, antigen-specific cell killing, and cytokine production (Figure 2).
Figure 2.
Impact of compound treatment on transduction, vector copy number, expansion, viability, cytotoxicity, and cytokine production following transduction with two vectors encoding different TCRs (LV1 and LV2)
(A) Fold change in transduction efficiency as measured using a flow-cytometry assay to detect a TCR-specific dextramer reagent in compound-treated samples compared to untreated control; dotted line indicates 1.2× transduction compared to control. (B) Vector copy number in compound-treated samples compared to untreated control. (C) Fold expansion over 11 days in compound-treated samples compared to untreated control. (D) Viability in compound-treated samples compared to untreated control. (E) Ratio of time taken to reach 50% killing of antigen-expressing target cells (KT50) in compound-treated samples compared to untreated control; dotted line indicates target of 0.8× vs. control sample. (F) Levels of interferon-γ (IFN-γ) 48 h post antigen exposure. Graphs show mean and standard deviation, n = 2 for compound-treated samples, n = 6 for untreated controls. Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005. Lead compound highlighted in red.
Transduction efficiency was determined in CD3+ T cells using flow cytometry to measure a T cell receptor (TCR)-specific dextramer. Results were normalized to the untreated control sample. A target increase of 1.2× the transduction efficiency seen in the control sample (shown by the dotted line) was achieved for all six compounds with lentivirus 1 (LV1) and five of six compounds with lentivirus 2 (LV2) (Figure 2A). A statistically significant increase in transduction efficiency was seen in samples treated with GSK2622391 (p = 0.0144 for LV1 and p = 0.0345 for LV2) and GSK682037B (p = 0.0139 for LV1 and p = 0.0155 for LV2). A slight, although non-significant, increase in vector copy number was seen for several compounds in combination with both LV1 and LV2 (Figure 2B), but in all cases the vector copy number (VCN) remained below the Food and Drug Administration-recommended limit of 5 copies per cell.
Calculation of fold expansion was based on the number of viable cells after 10 days of expansion in small-scale culture compared to the number of cells seeded at the start of culture. There was no significant difference in fold expansion for any of the compound-treated samples in comparison with the untreated control (Figure 2C). Viability of cells throughout culture was monitored and was above 90% in all samples at all time points (Figure 2D).
Antigen-specific cell killing was tested on cells after 10 days of culture using the xCELLigence system. Compound-treated and untreated cells were incubated with a cell line expressing the antigen recognized by the TCR construct. The transduction efficiency was normalized by mixing transduced and untransduced cells from the same donor to eliminate differences in cytotoxicity resulting from different numbers of transduced cells present in the assay. The time taken for 50% of the target cells to be killed (KT50) was evaluated for each sample and expressed as a ratio of the untreated control to eliminate assay variability. A criterion of ≥80% of the control KT50 was set, as shown by the dotted line, which was met by all compounds (Figure 2E). There was no significant difference in antigen-specific cell killing as a result of treatment with any compounds compared to the untreated control.
To evaluate cytokine production after antigen exposure, supernatant samples were removed from the xCELLigence co-culture experiments after either 24 h or 48 h and analyzed via Meso Scale Discovery (MSD) for interferon-γ (IFN-γ), a commonly used surrogate of T cell-mediated killing. Higher levels of IFN-γ were produced after 48 h compared to 24 h (data not shown). Figure 2F shows levels of IFN-γ at 48 h post antigen exposure. There was no statistically significant difference in IFN-γ levels in any compound-treated population compared to the untreated control population, although all compound-treated samples had slightly lower levels of IFN-γ.
Additional flow-cytometry analysis for markers of memory subpopulations was performed on cells after 10 days of culture and is shown in Figures S3–S6. Some variability in memory phenotype was observed, with GSK2622391 resulting in a lower proportion of effector memory T cells (TEM cells) in the CD4+ T cell population transduced with LV1 (mean for control = 36.95%, mean for GSK2622391 = 15.05%, p = 0.0017) and LV2 (mean for control = 44.00%, mean for GSK2622391 = 13.40%, p < 0.0001), a higher proportion of central memory T cells (TCM cells) in the CD4+ population transduced with LV2 (mean for control = 22.95%, mean for compound = 31.15%, p = 0.0438), and a higher proportion of TCM cells in the CD4+ population transduced with LV2 (mean for control = 26.28%, mean for GSK2622391 = 43.15%, p = 0.0410). GSK481816A showed a similar phenotype, also resulting in a lower proportion of TEM cells in the CD4+ population transduced with LV1 (mean for control = 36.95%, mean for GSK481816A = 23.55%, p = 0.0303) and LV2 (mean for control = 44.00%, mean for GSK481816A = 20.05%, p = 0.0006), and a higher proportion of TCM cells in the CD4+ population transduced with LV2 (mean for control = 26.28%, mean for GSK481816A = 49.20%, p = 0.0059). GSK1957694A resulted in a lower proportion of TEM cells in the CD4+ population transduced with LV2 only (mean for control = 44.00%, mean for GSK2622391 = 27.50%, p = 0.0376). No differences in the proportions of other memory phenotypes were observed.
Analysis of markers of activation (CD25 and CD69) and exhaustion (PD-1, TIM3, and LAG3) was performed on cells after 10 days of culture and is shown in Figures S7 and S8. The number of TIM3+ cells was variable across different compound treatments, different T cell populations, and different vectors used, with GSK2622391 and GSK481816A most frequently resulting in an increased proportion of TIM3+ cells. Increased levels of TIM3+ cells were seen in CD4+ cells treated with GSK2622391, GSK481816A, and GSK3563076A transduced with LV1 (mean for control = 70.83%, mean for GSK2622391 = 86.9% [p = 0.0161], mean for GSK481816A = 87.3 [p = 0.0138], mean for GSK3563076A = 83.3% [p = 0.0231]), and with compounds GSK2622391, GSK481816A, GSK3563076A, and GSK1957694A transduced with LV2 (mean for control = 68.92%, mean for GSK2622391 = 91.85% [p < 0.0001], mean for GSK481816A = 87.9% [p < 0.0001], mean for GSK3563076A = 83.70% [p = 0.0002], mean for GSK1957694A = 77.85% [p = 0.0045]). Increased levels of TIM3 were also seen in CD8+ cells transduced with LV2 for GSK2622391 and GSK481816A (mean for control = 94.13%, mean for GSK2622391 = 98.6% [p = 0.0255], mean for GSK481816A = 98.4% [p = 0.0438]). Reduced levels of TIM3+ cells were seen in CD8+ cells treated with GSK682037B and transduced with LV2 (mean for control = 68.92%, mean for GSK682037B = 60.1%, p = 0.0035) and in CD8+ cells treated with GSK1957694A and transduced with LV2 (mean for control = 94.13%, mean for GSK1957694A = 91.95%, p = 0.008). The differences, although statistically significant, are small, and therefore it is difficult to determine the biological impact (if any) on the T cell product. In addition, no significant differences in the expression of the exhaustion markers PD-1 or LAG3 were observed. No significant differences in the expression of the activation markers CD25 and CD69 were observed.
Identification of lead compounds for further testing
Subsequent hit validation of compounds GSK2622391, GSK502505, GSK481816A, GSK682037B, GSK3563076A, and GSK1957694A identified compounds GSK2622391 and GSK502505 as genuine screening hits. GSK2622391 and GSK502505 are structurally distinct neutral organic compounds with favorable drug-like properties, such as molecular weight <500, hydrogen bond donor count = 1, hydrogen bond acceptor count ≤3, aromatic ring count = 3, rotatable bond count <10, and total polar surface area <100 Å.25
To assess the suitability of these compounds for use in clinical studies, the physicochemical properties and toxicology (in silico flags and in vitro genetic toxicity) information available for each compound were reviewed. GSK2622391 has moderate lipophilicity (chromlogD7.4 = 3.7) and moderate kinetic solubility (36 μM), while GSK502505 has high lipophilicity (chromlogD7.4 = 7.6) and low kinetic solubility (1 μM).26 Achieving full compound washout (based on analysis of residual compound at the end of culture) was more challenging with GSK502505, which is consistent with increased lipophilicity driving increased non-specific binding to cellular compartments. GSK502505 also had lower kinetic solubility, making it more challenging to integrate into the large-scale manufacturing process. Both molecules were profiled against an in-house panel of known liability targets, and no significant activity was identified. GSK2622391 and GSK502505 were also found to be Ames27 and mouse lymphoma assay28 negative, indicating low potential for mutagenicity. Based on performance in small-scale experiments and its superior physicochemical properties, GSK2622391 was identified as the lead compound for further studies, and GSK502505 was identified as a potential backup.
Compound treatment has no impact on reverse transcription kinetics
Two steps in the retroviral life cycle, reverse transcription (RT) and integration, are essential for the function of lentiviral vectors,25 and changes in these, or in gene expression, as a result of transduction enhancer treatment may explain a possible mechanism of action.
The progression of RT can be mapped by measuring the quantity and accumulation of DNA products that correspond to the early, intermediate, and late stages of the reaction. Each RT product was measured by digital droplet PCR (ddPCR), and Figures 3A–3C show the quantity of early, mid, and late RT products, respectively, plotted against time for samples treated with either GSK2622391 (red) or GSK502505 (blue) compared to untreated controls (gray). GSK502505 treatment leads to increases in early RT and late RT products from the 10-h time point. The effect of GSK2622391 on RT is less pronounced, with a slight increase seen in early and late RT products. However, the variability between replicates is largely due to the low levels of the product detected, and the differences are not consistent between donors; therefore, the differences between treated and untreated controls may not be significant and the compounds may be enhancing transduction by other mechanisms.
Figure 3.
Quantity of DNA products during reverse transcription
Early (A), intermediate (B), and late (C) stages of reverse transcription (RT). GSK502505 treatment leads to increases in early RT and late RT products from the 10-h time point. The effect of GSK2622391 on reverse transcription is less pronounced, with a slight increase seen in early and late RT products. Graph shows mean + Poisson error, n ∼ 40,000 droplets.
Compound treatment has no impact on vector integration profile
To assess the impact of compound addition on the integration profile of the lentiviral vector, samples for gene integration analysis were collected at day 7 after compound treatment. Integration site analysis was performed using the kernel convolution framework,29 using a scale parameter of 100 kb. The results are shown in Table 1. There was a slightly higher total number of insertion sites identified within the GSK2622391-treated sample for both donors compared to the GSK502505-treated and untreated samples.
Table 1.
Number of insertion sites and peaks identified using kernel convolution framework with 100 kb kernel scale parameters
| Donor | Condition | Total number of insertions | Number of peaks | Percentage of total insertion sites |
|---|---|---|---|---|
| 1 | GSK2622391 treated | 33,891 | 615 | 1.8 |
| GSK502505 treated | 24,467 | 574 | 2.3 | |
| Untreated | 21,847 | 543 | 2.4 | |
| 2 | GSK2622391 treated | 24,713 | 631 | 2.5 |
| GSK502505 treated | 24,574 | 582 | 2.4 | |
| untreated | 24,771 | 528 | 2.1 | |
| Polyclonal HEK cell control | 18,957 | 468 | 2.4 | |
The number of peaks identified within the 100-kb kernel analysis were analyzed using an R script to identify common insertion site (CIS) locations with gene annotations and enable the plotting of UpSet graphs to display the number of gene name overlaps between the samples (Figure 4). A comparison of the overlapping gene names between untreated, GSK502505-treated, and GSK2622391-treated demonstrates 93 and 99 overlapping CIS locations for the two respective donors. Overall, CIS locations were more frequently shared between T cell samples from different donors than between T cell samples and the HEK polyclonal control sample.
Figure 4.
Intersection of annotated gene occurrences displayed as UpSet graphs
UpSet Graphs displaying the overlap of common insertion site (CIS) locations for compound-treated and untreated samples for two donors (A and B). The top section displays the number of overlapping genes within each comparison with comparisons performed shown below. The bar graph to the lower left of each plot demonstrates the size of each CIS set.
In addition to the kernel convolution method of analysis, insertion site tables were also generated within an R analysis package, RIPAT (RIPAT_1.0.0), which identifies the genes and transcription start sites closest to the identified insertion sites. The number of occurrences of annotated gene names within each sample enables the identification of common insertion sites. The top ten occurrences of genes identified closest to insertion sites through RIPAT analysis are shown in Table 2. PACS1, NPLOC4, and PPP6R2 were all genes identified as T cell CISs in the literature30 and were also identified within the top ten CISs in our samples. Similarity between CIS sets associated with T cells and T cells treated with either GSK2622391 or GSK502505 suggests that treatment with either compound does not alter vector integration at the level of CIS.
Table 2.
Top ten genes closest to common insertion sites for each cell population using RIPAT analysis for two donors transduced with LV2 alone (untreated) or in combination with GSK2622391 or compound B, compared to a polyclonal HEK cell line
| Donor 1 |
Donor 2 |
Polyclonal HEK | ||||
|---|---|---|---|---|---|---|
| Compound A | Compound B | Untreated | Compound A | Compound B | Untreated | |
| PACS1 | NPLOC4 | NPLOC4 | KDM2A | PACS1 | HSF1 | RP11-953B20.2 |
| NPLOC4 | PACS1 | BOP1 | BOP1 | JPT2 | NPLOC4 | TBC1D22A |
| KDM2A | SBF1 | MAPK8IP3 | SCX | POLR2E | NOSIP | CDC42BPG |
| BAG6 | MAPK8IP3 | ZNF34 | PACS1 | NOSIP | PRRG2 | NPLOC4 |
| MAPK8IP3 | ABCA7 | POLR2A | NPLOC4 | PRRG2 | PACS1 | TSPAN10 |
| BOP1 | MROH1 | MSH5 | JPT2 | NPLOC4 | KDM2A | MAPK8IP3 |
| SBNO2 | LTA | MSH5-SAPCD1 | SBNO2 | KDM2A | TNRC6C | TRAF2 |
| TRAF2 | TRAF7 | KDM2A | ALYREF | BOP1 | ARHGAP45 | ZGPAT |
| PPP6R2 | VWA7 | SBF1 | PPP1R2P1 | PRRC2A | MAPK8IP3 | RP4-583P15.15 |
| PSMB9 | TNRC6C | RP11-953B20.2 | SBF1 | SBF1 | IL32 | TNFSF12 |
Transcriptomic analysis of compound-treated samples shows enrichment of gene sets associated with innate immune response
To assess the impact of compound treatment on the target cell transcriptome, samples from T cells treated with GSK2622391 and untreated controls were collected at 6, 10, and 24 hours post transduction from three donors. Analysis of RNA expression at each individual time point, as well as the analysis of the combined time points, showed that a large proportion of the total RNA was differentially expressed (6,860–7,958 [20%–23%] of 33,694 transcripts analyzed) in compound-treated samples vs. untreated control samples. As so many individual genes were differentially expressed, further analysis focused on gene set enrichment analysis (GSEA) of biological processes, which evaluates entire datasets rather than a selected set of differentially expressed genes.
Cell transduction with lentiviral vectors activates pattern recognition receptors and induces the cellular inflammatory response.31 Previous analysis by our group showed enrichment of genes corresponding to gene ontology GO:0060337 “Cellular response to type I interferon” following transduction (data not shown) in transduced cells compared to untransduced cells, while in the present study we observed enrichment of gene ontology GO:0002758 “Innate immune response-activating signaling pathway” (Figure 5). A comparison of the two gene ontologies using the open-source software Revigo shows a connection between these GO annotations, suggesting that the enrichment of genes related to the immune response detected in the present study may be attributed to the presence of the lentiviral vector rather than to the compound treatment.
Figure 5.
Gene set enrichment analysis of gene ontology biological process
Gene set enrichment analysis of gene ontology biological process in compound-treated compared to untreated control samples at 6 h (A), 10 h (B), and 24 h (C) post transduction. Treatment with GSK2622391 shows an upregulation of genes related to RNA processing and ribosome biogenesis at each time point.
Treatment with GSK2622391 showed a signature related to RNA processing and ribosome biogenesis for each time point (Figure 5). These findings suggest that GSK2622391 may act as an inhibitor of MDN1, a member of the AAA ATPase family, thereby affecting ribosome assembly. Kawashima et al.32 showed that a series of compounds known as ribozinoindoles are potent inhibitors of ribosome assembly by interfering with the MDN1 ATPases. Bianco and Mohr33 showed that inactivation of ribosome activity resulted in impaired induction of type I IFN production, which allowed human cytomegalovirus (HCMV) replication. As a result of type I IFN response the chromatin-associated protein HMGB2 is expressed, which facilitates double-stranded DNA (dsDNA) sensing by protein cGAS, which in turn stimulates IFNB1 mRNA accumulation. Interfering with ribosome RNA accumulation therefore reduces the antiviral IFN response and reduces dsDNA sensing. Similarly, upon infection with HIV-1, CD4+ T cells showed a reduction in expression of genes associated with ribosome biogenesis.34 A later study observed similar effects after cells were infected with HIV-1 encoded protein Nef or a Nef-deleted (Δ-nef) version of HIV-1. In these experiments, Nef was shown to downregulate genes related to ribosome biogenesis and IFN response, suggesting that mechanisms similar to those observed in HCMV infection might be active after HIV infection.35 Together, this suggests that GSK2622391 may enhance transduction by altering ribosome biogenesis, resulting in a reduced IFN response.
Large-scale experiments demonstrate compatibility of lead compound with clinical manufacturing process for T cell products
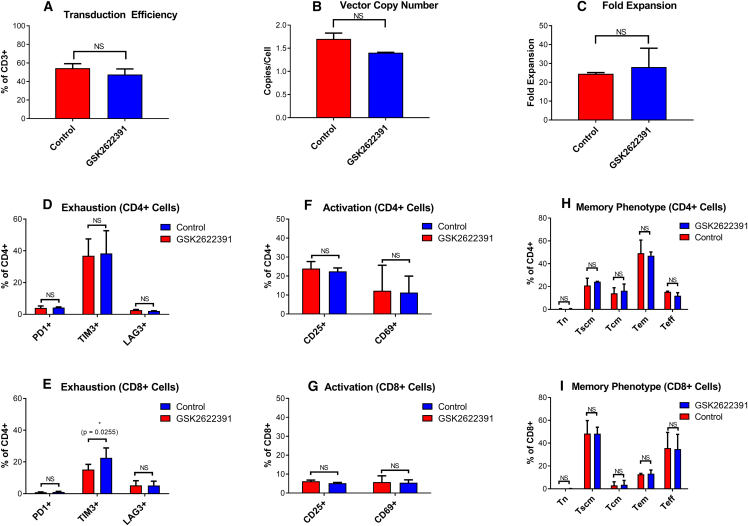
At-scale experiments were performed to demonstrate the potential of the TE to generate a comparable T cell drug product using GSK’s large-scale clinical manufacturing process and 3-fold less lentiviral vector than the current manufacturing process. Initial experiments were performed to assess compatibility of the compound with the filters, accessories, and tubing used on the Prodigy device, and analysis of compound concentrations before and after transfer into the Prodigy chamber showed minimal loss in the tubing set or filter (data not shown). In addition, a design-of-experiment approach was used to define the optimal conditions for compound addition, including compound concentration, timing of compound addition in relation to T cell activation and vector addition, and timing of washout (data not shown). Based on the results of this study, the compound was added 24 h after activation and 2 h prior to transduction, and washed out after 36 h in the large-scale experiments.
Figure 6 shows that use of GSK2622391 resulted in a comparable T cell therapy product to the standard condition while using a 3-fold reduction in lentiviral vector. Both test and control arms met all previously defined acceptance criteria set for product manufacturing for clinical studies. There was no significant difference in transduction efficiency (Figure 6A), VCN (Figure 6B), or fold expansion (Figure 6C). Viability remained above 70% at all time points throughout the process (data not shown). Extended characterization of activation, exhaustion, and memory markers also showed comparability between the two conditions, suggesting that the differences in phenotype observed during small-scale experiments may have been a result of donor variability rather than an effect of compound treatment. There was a slight (although statistically significant) difference in TIM-3 expression between GSK2622391-treated and untreated samples (mean for GSK2622391-treated = 15.2%, mean for untreated = 22.6%, p = 0.026), but this was seen only in the CD8+ T cell population. This is in contrast to previous small-scale studies, which showed an increase in TIM-3 with GSK2622391 treatment. The relationship between these in vitro markers of exhaustion and the efficacy of the final product post infusion is not well understood.
Figure 6.
Comparison of T cell therapy products manufactured with transduction enhancer and an MOI of 1 (red) or without transduction enhancer and an MOI of 3 (blue)
(A) Transduction efficiency measured using a TCR-specific detection reagent. (B) Vector copy number measured using ddPCR. (C) Fold expansion calculation based on cell numbers at the end of the process compared to the number of cells seeded. Exhaustion markers PD1, TIM3, and LAG2 measured by flow cytometry in CD4+ (D) and CD8+ (E) T cells. Activation markers CD25 and CD69 measured by flow cytometry in CD4+ (F) and CD8+ (G) T cells. Memory subpopulations measured by flow cytometry in CD4+ (H) and CD8+ (I) T cells. Graphs show mean + SD, n = 2. Paired t test was used to determine differences between test and control conditions. Statistical significance is indicated as follows: NS (not significant), ∗p ≤ 0.05), ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.005.
GSK2622391 concentration in the culture medium was measured using liquid chromatography-mass spectrometry (LC-MS) on days 0, 1, 5, and 12 of the process. We were able to confirm the expected concentration (100 μM) of GSK2622391 on day 1 of the experiment, shortly after the compound was added to the cell culture, demonstrating the feasibility of adding GSK2622391 to the Prodigy system in a closed and automated way. Following medium exchanges on day 5, designed to wash out residual vector and TransAct, the concentration of GSK2622391 was reduced to 4–6 μM. The concentration of GSK2622391 was reduced further by day 12 to 0.05 μM in the two autologous drug substances. Additional washing and formulation steps included in the current clinical manufacturing process were not carried out in these studies but would be expected to further reduce residual compound levels in the final drug product.
Discussion
Recent years have demonstrated the enormous potential of T cell immunotherapy, notably in treating cancer but in other diseases as well. In addition, genetic modification of other types of immune cells such as B cells and natural killer cells are also being investigated (reviewed in Finck et al.36). However, the manufacturing process for autologous cell therapies is complex and expensive, resulting in high prices for these medicines and, therefore, limited patient access.2 Viral vector production costs contribute significantly to the cost of manufacturing.37 In addition, later generations of therapies may use multiple constructs to enhance the function of the modified cells,38 and studies have previously demonstrated reduced transduction with increased construct sizes.1 For these reasons, identifying methods to improve transduction efficiency, and therefore reduce vector requirements, is important.
In this study, we report the results of a high-throughput screening (HTS) approach to identify a small molecule from the GSK small-molecule library, which can enhance lentiviral transduction in T cells without negatively impacting other product attributes important for the safety and efficacy of the product, including viability, expansion, VCN, antigen-specific cell killing, and cytokine production. We observed enhanced transduction with three different viral vectors, including GFP and two different second-generation TCR vectors, each including a second genetic element to enhance the function of the transduced cells,39 currently used in GSK’s early-stage clinical studies. In addition, we demonstrated consistent results across several donors. While this method focused on lentiviral transduction of T cells, further studies could also evaluate whether the enhancer molecule could also be applied to other cell and/or vector types or even to a biological effect different from transduction efficiency, depending on the therapeutic need: for example, modulation of the number of corrected cells vs. modulation of the number of copies per cell, as has been shown for other TEs.
Finally, we demonstrated the compatibility of this TE with GMP requirements in GSK’s clinical manufacturing process using a closed and automated system and showed that production of a comparable T cell product is possible while achieving a 3-fold reduction in vector requirements. Internal cost-of-goods modeling shows that this would result in approximately 15% reduction in cost of goods (even when the cost of manufacturing the compound is considered) and reduce the number of vector batches required to supply pivotal studies from four to one (depending on the number of patients in any given trial design). Further modeling analysis suggests that using the transduction enhancer with a higher multiplicity of infection (MOI) to increase the number of transduced cells earlier in the manufacturing process could potentially reduce the vein-to-vein time for each product from 12 days to 5 days, which would reduce the cost of goods by approximately 25% and reduce the time taken to complete patient treatment in a planned pivotal study from 4 years to 2 years. However, this option would require further process development, in particular to determine whether adequate vector and compound washout could be achieved with a shorter process.
Testing of residual levels of GSK2622391 in the representative drug product from the large-scale studies demonstrated very low levels of compound remaining at the drug substance stage, which together with the one-time dosing strategy for these products means it is likely that the levels of compound infused into the patient would be below the threshold of toxicological concern, as per ICH M7 guidance.40 In addition, in vitro and in silico safety screening of the small molecules alone did not raise any concerns. A key safety concern for genetically modified T cell therapies is off-target cell killing, and while no differences in on- or off-target killing was observed between compound-treated and untreated samples in the cytotoxicity assay, which includes both antigen-positive and antigen-negative cell lines, further assessment of this using in vitro or in vivo studies may be required prior to use of the compound in product manufacturing for human clinical studies.
We suggested a potential mechanism of action for the transduction enhancer, based on analysis of gene expression using RNA sequencing, which showed an upregulation of genes related to RNA processing and ribosome biogenesis. GSK2622391 may potentially act as a post-entry enhancer by inhibiting MDN1, thereby affecting ribosome assembly, which has previously been shown to increase infection with HIV-1.32,33,34,35 Further mechanism-of-action studies are warranted. No impact on RT kinetics or lentiviral integration profile was identified in this study. Several studies have demonstrated the applicability of previously described TEs to other cell and vector types,10,14,17,18,19,20,21,22,23 suggesting some commonalities in their mechanisms of action. It would be of interest to conduct further studies to determine whether GSK2622391 is also able to enhance transduction in other cell or vector types.
In summary, we presented a technology to screen a large number of small molecules for their ability to enhance lentiviral transduction of T cells, which allowed us to identify a number of compounds that enhance transduction without negatively impacting other product attributes. Finally, we demonstrated compatibility of the lead compound with GSK’s existing clinical manufacturing platform, resulting in a comparable drug product while significantly reducing vector requirements and, therefore, the cost of manufacturing.
Materials and methods
Source of T cells
All experiments were performed at GSK (Medicines Research Center, Stevenage, UK or Upper Providence, Collegeville, PA, USA). Whole blood, buffy coats, and leukapheresis were obtained from Research Donors (London, UK), Hemacare (Los Angeles, CA, USA), or BioIVT (Cambridge, UK). The human biological samples were sourced ethically, and their research use was in accordance with the terms of the informed consents under an Institutional Review Board/Ethics Committee-approved protocol. T cells were isolated from leukapheresis using MACS CD4 and CD8 beads and columns (Miltenyi Biotec, Bergisch Gladbach, Germany). Purified T cells were cryopreserved in 5% DMSO.
Source of lentiviral vectors
Third-generation self-inactivating lentiviral vectors based on HIV-1 pseudotyped with the envelope glycoprotein from VSV-G were manufactured using transient transfection of suspension culture-adapted HEK293T cells. The transgene was under the control of the human phosphoglycerate kinase promoter and a woodchuck hepatitis virus post-transcriptional regulatory element. Screening was performed using a vector encoding GFP manufactured at GSK, while verification was performed with two different therapeutic vectors, each encoding an engineered TCR against the NY-ESO-1 antigen as well as either CD8α (LV1) or dominant-negative TGF-β receptor II (LV2) as a second construct, manufactured at AGC Biologics (Bresso, Italy). Following transfection, the supernatant containing the viral particles was purified, concentrated, and stored at −80°C. The infectious titer of each batch was determined by flow cytometry for GFP or a TCR-specific peptide complex.
Small-molecule high-throughput screening
Because of time and cost factors, it was not feasible to screen the entire GSK library, containing around 1.8 million compounds. Instead, we ran approximately 30,000 compounds taken from internal compound sets with known biological activity, where annotations suggested the potential for a return of a higher percentage of true active compounds relative to the overall HTS set. In normal HTS, we would expect a hit rate of roughly 0.1%, yielding slightly less than 20,000 compounds at the end of the primary screen. With this set enriched for biologically active compounds, we hoped to achieve a similar yield of hits but with much lower numbers of plates run.
T cells from one donor were thawed and resuspended in TexMACS medium (Miltenyi Biotec) containing 5% human AB serum (Sigma, Darmstadt, Germany) + 1% penicillin-streptomycin (Corning, Corning, NY, USA) + 50 U/mL interleukin-2 (IL-2) (Miltenyi Biotec). Cells were activated using 10 μL per mL of medium TransACT (Miltenyi Biotec). Cells were incubated in an Erlenmeyer flask (Sigma) overnight at 37°C and 5% CO2 with shaking.
The following day (day 1), expression of activation markers was evaluated using flow cytometry for CD69 and CD25 to confirm activation (antibody details are provided in supplemental information). T cells were seeded into 384-well plates (Costar, Washington, DC, USA) containing the compound library. Poloxamer Synperonic F108 (0.1 mg/mL; Sigma) was included as a positive control, while DMSO only was included as a negative control. Cells were incubated with the compounds for 2 h prior to transduction with the GFP lentiviral vector at an MOI of 2.
The plates were sealed using Breathe-right seals (Diversified Biotech, Dedham, MA, USA) and incubated overnight at 37°C and 5% CO2. On day 2, old medium was aspirated out and fresh medium was added. On day 4, transduction was measured using flow cytometry for CD3 and GFP. The gating strategy used is described in supplemental information. The total count of cells, percentage of CD3+/GFP+ cells, median fluorescence intensity of GFP+ cells, and the percentage of viable cells were analyzed for each well. For quality control purposes, the control wells must have a coefficient of variation (%CV) of <20% for CD3+/GFP+, and any wells with <85% viability were excluded. Compounds which increased the percentage of GFP+ cell in the CD3+ population by more than 3 standard deviations relative to the vehicle control of the assay were designated as hits.
For the full-curve screens, T cells from two donors were prepared as described above and again plated into 384-well plates containing the compounds with one donor used for each replicate. Compounds identified in the initial screen were tested at 11 different concentrations from 10−9 M to 10−4 M. On day 4, the percentage of GFP+ cells and the percent viability of cells were analyzed in CD3+, CD4+, and CD8+ populations.
Small-scale tests with therapeutic vectors
Hit compounds identified from HTS were tested at small scale with two lentivectors, each encoding a therapeutic TCR transgene. Enriched CD4+/CD8+ T cells from three donors were thawed and resuspended in complete TexMACS medium (Miltenyi Biotec) supplemented with 100 IU/mL IL-2 (Miltenyi Biotec) and 5% human antibody serum (Access Biologics, Vista, CA, USA). Cell count and viability was determined using the NC-250 NucleoCounter with solution 18 (both ChemoMetec, Hovedstaden, Denmark). Cells were seeded into 24-well flat-bottomed cell-culture plates (Greiner Bio-One, Kremsmünster, Austria) and activated using TransACT. Cells were incubated overnight at 37°C and 5% CO2.
On day 1, expression of the activation markers CD69 and CD25 was evaluated using flow cytometry as described. Cells were seeded into a 48-well flat-bottomed cell-culture plate (Greiner Bio-One). The appropriate dose of compound, 0.1 mg/mL Poloxamer Synperonic F108 (positive control) (Sigma) or DMSO alone (negative control), was added to each well. The optimal concentration of each compound was determined based on the results of the full-curve screen, typically within the range 10–100 μM. The cells were incubated at 37°C and 5% CO2 for 2 h, followed by addition of therapeutic lentivector at an MOI of 8 for NY-ESO-1 dnTGFβRII and MOI of 3 for NY-ESO-1 CD8α. Cells were incubated overnight at 37°C and 5% CO2.
On day 2, cells were washed with complete TexMACS, resuspended in complete TexMACS, and transferred into a 24-well G-Rex plate (Wilson Wolf, Saint Paul, MN, USA). Medium change or supplementation took place on days 4, 6, and 9 of culture. Samples for cell count and viability, and flow-cytometry analysis (for transduction efficiency, memory phenotype, activation, and exhaustion), were collected on days 0, 1, 4, 7, and 11 of the cell-culture process. On day 11, samples were harvested for VCN and co-culture with antigen-expressing cell targets.
GraphPad Prism v7.03 (GraphPad Software, San Diego, CA, USA) was used for generation of graphs. Statistical analysis was performed in SAS v3.81. Data for cells treated with test compounds were compared to data from donor-matched untreated controls. The estimated least-square mean differences for each compound-treated sample against the untreated were calculated for each of the vectors. The mean differences were tested for significance and the corresponding p values reported; significant values were those with p < 0.05. Dunnett’s adjustment was used to allow for multiple comparisons against the control. Donor variability has been excluded from the estimated differences by treating each donor sample as a separate subject.
Verification in large-scale manufacturing process
One lead compound (GSK2622391) identified in the small-scale experiments was tested in the full-scale process for T cell therapy products using the cliniMACS Prodigy (Miltenyi Biotec). The cliniMACS Prodigy is an automated cell-processing platform capable of performing all manufacturing operations including T cell selection, activation, transduction, expansion, and formulation in an automated closed system. A customized version of the T cell transduction process, developed in collaboration with Miltenyi Biotec, was used.
Apheresis from two healthy donors was obtained as described and enriched for CD4+/CD8+ T cells. Enriched T cells from each of the two donors were split into two experimental arms: one untreated control, transduced at the standard MOI of 3 for the therapeutic vector, and one treated with the lead transduction enhancer and an MOI of 1. Cells were activated by addition of MACS GMP T cell TransAct—Large Scale (Miltenyi Biotec).
GSK2622391 was dissolved first in DMSO to create a stock solution and then in TexMACS to create a working solution. The working solution was sterile filtered into a transfer bag (Terumo BCT, Lakewood, CO, USA). Separate studies were conducted to demonstrate no loss of the compound in the filter and tubing (data not shown). The cells were treated with the enhancer for 2 h prior to transduction with lentiviral vector. The following day a culture wash was performed to remove compound and lentivirus. Separate studies were conducted to evaluate the optimal timing of compound and vector addition and washout (data not shown). The cells were then expanded for 12 days in TexMACS medium supplemented with 100 IU/mL IL-2 and 5% AB serum for the first 5 days, and TexMACS and 100 IU/mL IL-2 only for the remaining days. On day 12, samples were taken for analysis of immunophenotype, VCN, and cytotoxicity as described.
Samples of supernatant and cell pellets were taken at defined time points throughout the process: prior to compound addition (negative control), immediately after compound addition, after compound washout, and at the end of the manufacturing process. Cell pellets were washed twice, and supernatants and cell pellets were frozen at −80°C before analysis. Residual compound concentration in the sample was evaluated using an LC-MS method developed at GSK.
GraphPad Prism v7.03 was used for generation of graphs and statistical analysis: paired t test was used to compare compound-treated vs. untreated for each of the two donors, with statistical significance determined by a p value of ≤0.05.
Flow-cytometry analysis
Flow-cytometry analysis was performed using a 14-color panel on the Fortessa X-20 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Antibodies used are listed in supplemental information. Cells were transferred into a 96-well round-bottomed plate (Corning) and resuspended in fluorescence-activated cell sorting (FACS) buffer (D-PBS + 0.5% human serum albumin (Irvine Scientific, Santa Ana, CA, USA). Zombie NIR dye (Biolegend, San Diego, CA, USA) was added and cells were incubated for 15 min. Cells were washed and resuspended in FACS buffer, followed by addition of dextramer reagent for detection of the engineered TCR (Immudex, Copenhagen, Denmark). Cells were incubated for 25 min, washed, and resuspended in FACS buffer, and all other antibodies (except CD45RA and CD45RO) were added as a pre-prepared master mix. Cells were incubated for 30 min, washed, and resuspended in CytoFix fixation buffer (BD Biosciences). Cells were incubated for 30 min, washed, and resuspended in FACS buffer. Cells were incubated for 30 min with CD45RA and CD45RO, then washed and resuspended in FACS buffer. Data acquisition was performed using the Fortessa flow cytometer. The gating strategy applied is described in supplemental information. Appropriate fluorescence-minus-one and compensation controls were prepared. Data analysis was performed using FlowJo software version 10 (FlowJo, Ashland, OR, USA).
Vector copy number analysis
The average number of copies of integrated lentivirus per cell was determined using a ddPCR method on the QX200 auto droplet digital PCR system (Bio-Rad, Hercules, CA, USA). The method is performed with two sets of primers and probes: one specific for quantification of LV (LTR region and gag) and one specific for quantification of the cellular genome (RPLP0). VCN is determined by the ratio of LV copies to cell genome.
Cell pellets were frozen at −80°C and thawed for genomic DNA extraction using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). DNA digestion was performed using the Fast Digest MIuI kit (Thermo Fisher, Waltham, MA, USA). The PCR reaction mixture was prepared using ddPCR Supermix for probes and ddPCR copy number assay kit (both Bio-Rad). Primers and probe mixes were supplied by Invitrogen (Waltham, MA, USA) or Thermo Fisher. Droplet generation was performed using an automated droplet generator (Bio-Rad). PCR amplification was performed using the QX200 system thermocycler (Bio-Rad). Droplet reading was performed using the QX200 droplet reader (Bio-Rad). Data analysis was performed using the QX manager software (Bio-Rad).
Cytotoxicity analysis and cytokine release
Analysis of antigen-specific cell killing of transduced T cells was performed using the xCELLigence system (Agilent, Santa Clara, CA, USA), which measures killing of target cells via changes in impedance of electrical current between gold electrodes in a microtiter plate. Cells were cryopreserved prior to analysis and thawed for co-culture with cells expressing the peptide recognized by the TCR construct. A background equilibration step was performed by adding 50 μL of pre-warmed xCELLigence medium (RPMI 1640 + 10% [v/v] fetal bovine serum + 1% [v/v] Glutamax I [all from Thermo Fisher]) to each well of the xCELLigence E-plate before placing the plates into the cradles. Antigen-positive and antigen-negative cell lines were resuspended in xCELLigence medium, and 2 × 104 target cells were added to each well. Plates were left at room temperature for 45 min to allow cells to evenly settle across the well before being placed into the xCELLigence cradles for initiation of assay reading.
Transduced and untransduced T cells from compound-treated and untreated samples were thawed on the same day as target cell seeding. Each transduced T cell population was normalized by addition of untransduced T cell populations treated with the same compound to within 5% of the transduction efficiency of the relevant untreated transduced control population for each donor to ensure that any differences in antigen-specific cell killing observed were as a result of compound treatment rather than variability in the number of transduced cells. T cells were seeded into 24-well G-REX plates and incubated for 24 h at 37°C and 5% CO2.
After 24 h, T cell populations were harvested, counted, and resuspended to a concentration of 2 × 105 cells/mL in xCELLigence medium. The xCELLigence assay was paused, and 2 × 104 T cells were added to each well of the xCELLigence plate. One hundred microliters of medium only or 0.1% Triton X-100 solution were added to no-killing wells and 100% lysis control wells, respectively. xCELLigence plates were returned to the xCELLigence cradles and plates were incubated for a further 48 h, with assay measurements performed every hour. Impact of the compound treatment on T cell cytotoxicity was assessed by comparing the time to achieve 50% killing (KT50) between compound-treated and untreated transduced T cells.
Supernatant samples were collected from the xCELLigence co-culture experiments after either 24 h or 48 h and analyzed using the MSD Sector 600 Imager (Meso-Scale Discovery, Rockville, MD, USA) for IFN-γ production. Supernatants were stored at −80°C and thawed at room temperature for analysis. Samples and calibrators were prepared as per manufacturer’s instructions, and 25 μL of each was added to the MSD plates.
Lentiviral reverse transcription analysis
Lentiviral RT analysis was performed on compound-treated cells and matched controls. A ddPCR method on the QX200 auto ddPCR system was used to evaluate the impact of compound addition on the progression of RT by measuring the quantity and accumulation of DNA products which correspond to the early, intermediate, and late stages of RT. Primer/probe sequences are listed in supplemental information. Samples were collected 2, 4, 6, 8, 10, 24, and 48 h post transduction. Sample preparation, genomic DNA extraction, and digestion were performed using the same method as for VCN analysis. The ddPCR reaction was performed using a set of primers and probes designed to detect viral DNA products from different stages of RT. Primer and probe mixes were supplied by Invitrogen or Thermo Fisher. Droplet generation and PCR amplification were performed as described previously.
Insertion site analysis
Samples for insertion site analysis were collected at day 7 post transduction and analyzed using a site-directed, ligase-independent mutagenesis (SLiM) PCR assay.41 DNA extraction and purification was performed using the Maxwell RSC 48 (Promega, Madison, WI, USA). DNA concentration was determined using the Qubit dsDNA HS assay kit (Thermo Fisher). DNA was sheared to a target peak of 1,000 base pairs using the Biorupter Pico Sonicator (Diagenode, Denville, NJ, USA). End repair and adenylation of the sheared DNA fragments was performed using the NEBNext Ultra DNA library prep kit for Illumina (New England Biolabs, Ipswich, MA, USA). Linker cassettes were generated from single-strand oligos the day before use and ligated to each sample of DNA using the NEBNext Ultra DNA library prep kit (New England Biolabs). Agencourt Ampure XP beads (Beckman Coulter, Brea, CA, USA) was used to remove any unligated linkers and enzymes less than 150 base pairs in length. The purified ligation reaction product was amplified by PCR using Taq DNA polymerase (Qiagen). A second clean-up step to remove PCR primers, DNA polymerase, and fragments of genomic DNA was performed using Ampure XP beads. A second PCR reaction was performed using a unique LTR Illumina P5 primer for each replicate, which adds a unique barcode to the DNA sequences for each sample and adds the Illumina adapter to enable sequencing to occur. A third clean-up was performed using Ampure XP beads. Final libraries were analyzed by Tapestation (Agilent), and DNA was quantified using the Illumina Library Quantification kit (Kapa Biosystems, Wilmington, MA, USA) before being diluted or concentrated as required to a concentration of 4 nM. Sequencing was carried out on the Illumina MiSeq, using the MiSeq reagent kit v3 (Illumina, San Diego, CA, USA).
Raw sequencing data in the format of FASTQ files were uploaded and analyzed using an internally developed analysis pipeline to generate, determine, and quantify integration sites. The abundance of each integration site was calculated by dividing the number of unique barcodes detected for that site by the total number of unique barcodes detected in that replicate and multiplying by 100 to give a percentage. CIS analysis was performed using the kernel convolution framework,29 using a scale parameter of 100 kb (i.e., insertion sites within 100 kb of one another are smoothed into a single insertion site reading).
In addition to the kernel convolution method of analysis, insertion site tables were also generated within the R analysis package RIPAT (RIPAT_1.0.0), which identifies the genes and transcription start sites closest to the identified insertion site. The number of occurrences of annotated gene names within each sample enables the identification of CISs.
RNA-sequencing analysis
Samples were harvested for RNA expression analysis at 0, 6, 10, and 24 h post transduction from both compound-treated and untreated transduced cells from three healthy donors to assess the impact of compound addition on the target cell transcriptome. Cell pellets were frozen at −80°C. RNA extraction and sequencing were performed by GeneWiz (Leipzig, Germany). FASTQ files were analyzed using the DNAnexus native STAR mapping app (https://platform.dnanexus.com/app/app-G5By8b804zV5201j5KVfq18P) to generate gene count tables for each sample, which were further analyzed using Rstudio. Comparison of compound-treated and untreated samples was performed at each time point separately and across all time points in combination with the time factor included in the design. As a large number of genes were differentially expressed, GSEA of biological processes, which evaluates entire datasets rather than a selected set of differentially expressed genes, was performed. Differential gene expression was determined using DESeq242 in Bioconductor. GSEA was performed using the gseGO function from the Bioconductor clusterProfiler package.43
Acknowledgments
The authors gratefully acknowledge the contributions of the following, without whom this publication would not have been possible: Amy Quinn, Gemma Smith, Wilson Li, Georgia Flawn-Thomas, Ryan Roberts, Ashkenaz Richard, Ceara Rea, Natasha Sanjrani, Simran Judge, and Rochishnu Ghosh.
Author contributions
Conceptualization and methodology, N.F., C.M.-E., and M.R.d.C.; data curation, P.M., C.K., J.W., N.F., F.P., and B.D.; formal analysis, N.F., C.M.-E., M.B., F.P., and B.D.; investigation, F.P., B.D., B.J., C.T., P.M., C.K., J.W., M.B., C.M.-E., and N.F.; writing, N.F.
Declaration of interests
All authors are current or past employees and/or shareholders of GSK.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2023.101113.
Supplemental information
Data and code availability
The data that support the findings of this study are available from the corresponding author on reasonable request, subject to the relevant legal agreements being in place.
References
- 1.Sweeney N.P., Vink C.A. The impact of lentiviral vector genome size and producer cell genomic to gag-pol mRNA ratios on packaging efficiency and titre. Mol. Ther. Methods Clin. Dev. 2021;21:574–584. doi: 10.1016/j.omtm.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishna D., Rittié L., Tran H., Zheng X., Chen-Rogers C.E., McGillivray A., Clay T., Ketkar A., Tarnowski J. Short Time to Market and Forward Planning Will Enable Cell Therapies to Deliver R&D Pipeline Value. Hum. Gene Ther. 2021;32:433–445. doi: 10.1089/hum.2020.212. [DOI] [PubMed] [Google Scholar]
- 3.Kaygisiz K., Synatschke C.V. Materials promoting viral gene delivery. Biomater. Sci. 2020;8:6113–6156. doi: 10.1039/d0bm01367f. [DOI] [PubMed] [Google Scholar]
- 4.Hanenberg H., Xiao X.L., Dilloo D., Hashino K., Kato I., Williams D.A. Colocalization of retrovirus and target cells on specific fibronectin fragments increases genetic transduction of mammalian cells. Nat. Med. 1996;2:876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
- 5.Lee H.J., Lee Y.S., Kim H.S., Kim Y.K., Kim J.H., Jeon S.H., Lee H.W., Kim S., Miyoshi H., Chung H.M., Kim D.K. Retronectin enhances lentivirus-mediated gene delivery into hematopoietic progenitor cells. Biologicals. 2009;37:203–209. doi: 10.1016/j.biologicals.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Hauber I., Beschorner N., Schrödel S., Chemnitz J., Kröger N., Hauber J., Thirion C. Improving Lentiviral Transduction of CD34(+) Hematopoietic Stem and Progenitor Cells. Hum. Gene Ther. Methods. 2018;29:104–113. doi: 10.1089/hgtb.2017.085. [DOI] [PubMed] [Google Scholar]
- 7.Cornetta K., Anderson W.F. Protamine sulfate as an effective alternative to polybrene in retroviral-mediated gene-transfer: implications for human gene therapy. J. Virol. Methods. 1989;23:187–194. doi: 10.1016/0166-0934(89)90132-8. [DOI] [PubMed] [Google Scholar]
- 8.Fenard D., Ingrao D., Seye A., Buisset J., Genries S., Martin S., Kichler A., Galy A. Vectofusin-1, a new viral entry enhancer, strongly promotes lentiviral transduction of human hematopoietic stem cells. Mol. Ther. Nucleic Acids. 2013;2:e90. doi: 10.1038/mtna.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konopka K., Stamatatos L., Larsen C.E., Davis B.R., Düzgüneş N. Enhancement of human immunodeficiency virus type 1 infection by cationic liposomes: the role of CD4, serum and liposome-cell interactions. J. Gen. Virol. 1991;72:2685–2696. doi: 10.1099/0022-1317-72-11-2685. [DOI] [PubMed] [Google Scholar]
- 10.Amadeo F., Hanson V., Murray P., Taylor A. DEAE-Dextran Enhances the Lentiviral Transduction of Primary Human Mesenchymal Stromal Cells from All Major Tissue Sources Without Affecting Their Proliferation and Phenotype. Mol. Biotechnol. 2023;65:544–555. doi: 10.1007/s12033-022-00549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis G., Christiansen L., McKenzie J., Luo M., Pasackow E., Smurnyy Y., Harrington S., Gregory P., Veres G., Negre O., Bonner M. Staurosporine Increases Lentiviral Vector Transduction Efficiency of Human Hematopoietic Stem and Progenitor Cells. Mol. Ther. Methods Clin. Dev. 2018;9:313–322. doi: 10.1016/j.omtm.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heffner G.C., Bonner M., Christiansen L., Pierciey F.J., Campbell D., Smurnyy Y., Zhang W., Hamel A., Shaw S., Lewis G., et al. Prostaglandin E(2) Increases Lentiviral Vector Transduction Efficiency of Adult Human Hematopoietic Stem and Progenitor Cells. Mol. Ther. 2018;26:320–328. doi: 10.1016/j.ymthe.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masiuk K.E., Zhang R., Osborne K., Hollis R.P., Campo-Fernandez B., Kohn D.B. PGE2 and Poloxamer Synperonic F108 Enhance Transduction of Human HSPCs with a beta-Globin Lentiviral Vector. Mol. Ther. Methods Clin. Dev. 2019;13:390–398. doi: 10.1016/j.omtm.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrillo C., Thorne L.G., Unali G., Schiroli G., Giordano A.M.S., Piras F., Cuccovillo I., Petit S.J., Ahsan F., Noursadeghi M., et al. Cyclosporine H Overcomes Innate Immune Restrictions to Improve Lentiviral Transduction and Gene Editing In Human Hematopoietic Stem Cells. Cell Stem Cell. 2018;23:820–832.e9. doi: 10.1016/j.stem.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang Y., Kim Y.S., Wielgosz M.M., Ferrara F., Ma Z., Condori J., Palmer L.E., Zhao X., Kang G., Rawlings D.J., et al. Optimizing lentiviral vector transduction of hematopoietic stem cells for gene therapy. Gene Ther. 2020;27:545–556. doi: 10.1038/s41434-020-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C.X., Sather B.D., Wang X., Adair J., Khan I., Singh S., Lang S., Adams A., Curinga G., Kiem H.P., et al. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124:913–923. doi: 10.1182/blood-2013-12-546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schott J.W., León-Rico D., Ferreira C.B., Buckland K.F., Santilli G., Armant M.A., Schambach A., Cavazza A., Thrasher A.J. Enhancing Lentiviral and Alpharetroviral Transduction of Human Hematopoietic Stem Cells for Clinical Application. Mol. Ther. Methods Clin. Dev. 2019;14:134–147. doi: 10.1016/j.omtm.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasiri F., Muhammadnejad S., Rahbarizadeh F. Effects of polybrene and retronectin as transduction enhancers on the development and phenotypic characteristics of VHH-based CD19-redirected CAR T cells: a comparative investigation. Clin. Exp. Med. 2022 doi: 10.1007/s10238-022-00928-8. [DOI] [PubMed] [Google Scholar]
- 19.Lo Presti V., Cornel A.M., Plantinga M., Dünnebach E., Kuball J., Boelens J.J., Nierkens S., van Til N.P. Efficient lentiviral transduction method to gene modify cord blood CD8(+) T cells for cancer therapy applications. Mol. Ther. Methods Clin. Dev. 2021;21:357–368. doi: 10.1016/j.omtm.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim-Hoehamer Y.I., Riberdy J.M., Zheng F., Park J.J., Shang N., Métais J.Y., Lockey T., Willis C., Akel S., Moore J., et al. Development of a cGMP-compliant process to manufacture donor-derived, CD45RA-depleted memory CD19-CAR T cells. Gene Ther. 2023;30:222–231. doi: 10.1038/s41434-021-00307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon B., Harrer D.C., Thirion C., Schuler-Thurner B., Schuler G., Uslu U. Enhancing lentiviral transduction to generate melanoma-specific human T cells for cancer immunotherapy. J. Immunol. Methods. 2019;472:55–64. doi: 10.1016/j.jim.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Collon K., Gallo M.C., Bell J.A., Chang S.W., Rodman J.C.S., Sugiyama O., Kohn D.B., Lieberman J.R. Improving Lentiviral Transduction of Human Adipose-Derived Mesenchymal Stem Cells. Hum. Gene Ther. 2022;33:1260–1268. doi: 10.1089/hum.2022.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strack A., Deinzer A., Thirion C., Schrodel S., Dorrie J., Sauerer T., Steinkasserer A., Knippertz I. Breaking Entry-And Species Barriers: LentiBOOST((R)) Plus Polybrene Enhances Transduction Efficacy of Dendritic Cells and Monocytes by Adenovirus 5. Viruses. 2022;14 doi: 10.3390/v14010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Höfig I., Atkinson M.J., Mall S., Krackhardt A.M., Thirion C., Anastasov N. Poloxamer synperonic F108 improves cellular transduction with lentiviral vectors. J. Gene Med. 2012;14:549–560. doi: 10.1002/jgm.2653. [DOI] [PubMed] [Google Scholar]
- 25.Rishton G.M. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov. Today. 2003;8:86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- 26.Bunally S.B., Luscombe C.N., Young R.J. Using Physicochemical Measurements to Influence Better Compound Design. SLAS Discov. 2019;24:791–801. doi: 10.1177/2472555219859845. [DOI] [PubMed] [Google Scholar]
- 27.Zeiger E. The test that changed the world: The Ames test and the regulation of chemicals. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019;841:43–48. doi: 10.1016/j.mrgentox.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd M., Kidd D. The mouse lymphoma assay. Methods Mol. Biol. 2012;817:35–54. doi: 10.1007/978-1-61779-421-6_3. [DOI] [PubMed] [Google Scholar]
- 29.de Ridder J., Uren A., Kool J., Reinders M., Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput. Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biffi R., Orsi F., Pozzi S., Maldifassi A., Radice D., Rotmensz N., Zampino M.G., Fazio N., Peruzzotti G., Didier F. No impact of central venous insertion site on oncology patients' quality of life and psychological distress. A randomized three-arm trial. Support. Care Cancer. 2011;19:1573–1580. doi: 10.1007/s00520-010-0984-9. [DOI] [PubMed] [Google Scholar]
- 31.Annoni A., Gregori S., Naldini L., Cantore A. Modulation of immune responses in lentiviral vector-mediated gene transfer. Cell. Immunol. 2019;342 doi: 10.1016/j.cellimm.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawashima S.A., Chen Z., Aoi Y., Patgiri A., Kobayashi Y., Nurse P., Kapoor T.M. Potent, Reversible, and Specific Chemical Inhibitors of Eukaryotic Ribosome Biogenesis. Cell. 2016;167:512–524.e14. doi: 10.1016/j.cell.2016.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianco C., Mohr I. Ribosome biogenesis restricts innate immune responses to virus infection and DNA. Elife. 2019;8 doi: 10.7554/eLife.49551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinman C.L., Doria M., Orecchini E., Giuliani E., Galardi S., De Jay N., Michienzi A. HIV-1 infection causes a down-regulation of genes involved in ribosome biogenesis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furler R.L., Ali A., Yang O.O., Nixon D.F. Nef-induced differential gene expression in primary CD4+ T cells following infection with HIV-1 isolates. Virus Gene. 2019;55:541–544. doi: 10.1007/s11262-019-01670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finck A.V., Blanchard T., Roselle C.P., Golinelli G., June C.H. Engineered cellular immunotherapies in cancer and beyond. Nat. Med. 2022;28:678–689. doi: 10.1038/s41591-022-01765-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capra E G.A., Loche A., Temps C. Viral-vector therapies at scale: Today’s challenges and future opportunities. 2022. https://www.mckinsey.com/industries/life-sciences/our-insights/viral-vector-therapies-at-scale-todays-challenges-and-future-opportunities
- 38.Young R.M., Engel N.W., Uslu U., Wellhausen N., June C.H. Next-Generation CAR T-cell Therapies. Cancer Discov. 2022;12:1625–1633. doi: 10.1158/2159-8290.CD-21-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silk J.D., Abbott R.J.M., Adams K.J., Bennett A.D., Brett S., Cornforth T.V., Crossland K.L., Figueroa D.J., Jing J., O'Connor C., et al. Engineering Cancer Antigen-Specific T Cells to Overcome the Immunosuppressive Effects of TGF-beta. J. Immunol. 2022;208:169–180. doi: 10.4049/jimmunol.2001357. [DOI] [PubMed] [Google Scholar]
- 40.Agency E.M. ICH Guideline M7 on Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk. 2021. https://www.ema.europa.eu/en/documents/scientific-guideline/draft-ich-guideline-m7-assessment-control-dna-reactive-mutagenic-impurities-pharmaceuticals-limit_en.pdf
- 41.Cesana D., Calabria A., Rudilosso L., Gallina P., Benedicenti F., Spinozzi G., Schiroli G., Magnani A., Acquati S., Fumagalli F., et al. Retrieval of vector integration sites from cell-free DNA. Nat. Med. 2021;27:1458–1470. doi: 10.1038/s41591-021-01389-4. [DOI] [PubMed] [Google Scholar]
- 42.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request, subject to the relevant legal agreements being in place.