Abstract
The staphylococcal multidrug efflux pump QacA mediates resistance to a broad spectrum of monovalent and divalent antimicrobial cations. Resistance toward various classes of these compounds identified features of the substrate that may be important for interaction with QacA. Analysis of combinations of two substrates suggested that the same mechanism is used for the extrusion of different classes of compounds.
The plasmid-encoded multidrug resistance gene qacA from Staphylococcus aureus mediates resistance to a number of classes of antimicrobial organic cations, including intercalating dyes, quaternary ammonium compounds, diamidines, and biguanidines (3, 10). qacA has been shown to encode a protein, QacA, with 14 transmembrane segments (6, 9) that confers resistance via export of the compound energized by the proton motive force (3). QacA is a member of the major facilitator superfamily of transport proteins, which are involved in the uniport, symport, and antiport of a wide range of substances across the cell membrane (4, 7). A closely related protein, QacB, also from clinical isolates of S. aureus, characteristically differs from QacA in that it mediates significantly reduced levels of resistance to divalent cationic drugs, such as diamidines and biguanidines (3, 6). Random and site-directed mutagenesis showed that the difference in substrate specificity between QacA and QacB is due to a single amino acid substitution at position 323, where the presence of an acidic residue in QacA is essential for high levels of resistance to diamidines and biguanidines (6).
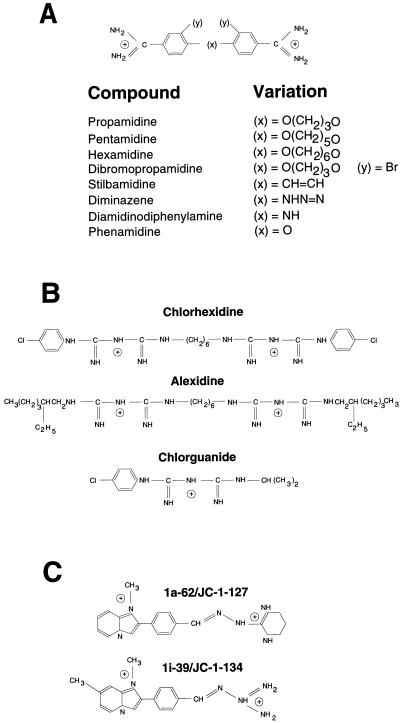
A series of diamidine variants was used to establish if the difference in substrate specificity between QacA and QacB exists for a wide range of structures. The various diamidine structures enabled the effects of altered interamidine linkage (x) and the addition of side chains (y) to be examined (Fig. 1A). The diamidine amicarbalide differs from those shown in Fig. 1A by the position of the amidine groups attached to the aromatic rings. MIC analysis demonstrated that QacA conferred significantly higher levels of resistance than did QacB for all of the diamidines tested, irrespective of the interamidine linkage, the addition of side chains, or the position of the amidine group on the aromatic ring (Table 1). These results are consistent with the hypothesis (6) that the negative charge of the acidic amino acid at position 323 in transmembrane segment 10 of QacA interacts directly with one of the positively charged moieties of the divalent cation and, as such, may be involved in substrate binding or recognition, resulting in increased QacA-mediated export of these compounds.
FIG. 1.
Chemical structures of diamidines, biguanidines, and guanylhydrazones used in this study. (A) Diamidines: structural variations at x (interamidine linkage) and y (side chains). (B) Biguanidines: chlorhexidine (digluconate and dihydrochloride) and alexidine represent aromatic and aliphatic biguanidines, respectively. Chlorguanide is a monovalent derivative of chlorhexidine. (C) Guanylhydrazones: 1a-62, JC-1-127, 1i-39, and JC-1-134 are divalent aromatic guanylhydrazones. 1a-62 and JC-1-127 have the same structure but represent dibromide and dichloride salts, respectively (11). 1i-39 and JC-1-134 vary in the position of a methyl side chain in that 1i-39 is 1,7-dimethyl (shown) and JC-1-134 is 1,6-dimethyl; they also represent dibromide and dichloride salts, respectively (11). Structures were taken from information provided by suppliers.
TABLE 1.
Resistance to diamidines, biguanidines, and guanylhydrazones
| Compounda | MIC (μg/ml)b
|
||
|---|---|---|---|
| QacA | QacB | Control | |
| Biguanidines | |||
| Alexidine | 6 | 4 | 4 |
| Chlorguanide | 250 | 250 | 250 |
| Chlorhexidinec | 12 | 6 | 1 |
| Diamidines | |||
| Amicarbalide | 1,200 | 400 | 200 |
| Diamidinodiphenylamine | 250 | 50 | 50 |
| Dibromopropamidine | 10 | 1 | 1 |
| Diminazene | 400 | 200 | 200 |
| Hexamidine | 300 | 200 | 100 |
| Pentamidine | 350 | 200 | 100 |
| Phenamidine | 1,800 | 200 | 200 |
| Propamidine | 300 | 100 | 100 |
| Stilbamidine | 400 | 200 | 100 |
| Guanylhydrazones | |||
| 1i-39/JC-1-134d | >2,000 | 1,600 | 100 |
| 1a-62/JC-1-127d | 1,600 | 1,600 | 500 |
| Methylglyoxal-bisguanylhydrazone | 1,200 | 1,200 | 1,200 |
Amicarbalide, chlorguanide, diamidinodiphenylamine, dibromopropamidine, phenamidine, propamidine, and stilbamidine were obtained from Rhône Poulenc Rorer (Dagenham, United Kingdom); hexamidine was obtained from Chauvin Laboratoire (Montpellier, France); and the guanylhydrazones, JC-1-127, JC-1-134, 1a-62, and 1i-39 (11), were obtained from Richard Sundberg (University of Virginia, Charlottesville).
MICs were determined in triplicate with Escherichia coli K-12 strain BHB2600 (2) carrying plasmids pSK4219 (qacA) and pSK4270 (qacB) and the vector pBluescript (control) (6).
Chlorhexidine dihydrochloride and digluconate gave equivalent results.
Similar results were obtained for the different salts (see Fig. 1 legend) of these compounds.
The biguanidines and guanylhydrazones are structurally related to the diamidines (Fig. 1). These compounds represent different chemical classes that all have the common feature of amidine groups attached to aliphatic or aromatic structures. The guanylhydrazones contain the amidine (guanyl) group attached to the hydrazone group, and the biguanidines possess the guanido in place of the amidine group. QacA and QacB have previously been shown to differ in their substrate specificity for the biguanidine chlorhexidine (3). However, phenotypic analysis showed that neither QacA nor QacB conferred resistance to alexidine, which represents an aliphatic derivative of chlorhexidine, or to chlorguanide, a monovalent derivative of chlorhexidine (Table 1). MIC analysis revealed that both QacA and QacB conferred resistance to divalent aromatic guanylhydrazones but not to the divalent aliphatic guanylhydrazone methylglyoxal-bisguanylhydrazone (Table 1) or to trivalent guanylhydrazones (data not shown). Notably, the characteristic difference in substrate specificity of QacA and QacB for the diamidines was absent for the divalent aromatic guanylhydrazones, indicating that at least one amino acid common to both proteins is involved in conferring resistance to these compounds. Phenotypic analysis of different salts of the guanylhydrazones and chlorhexidine confirmed that the nature of the anionic component of the salt is relatively unimportant (Table 1), reiterating the view that the Qac proteins interact with a solubilized cationic substrate. In addition, the cationic amidine moiety may not in itself be sufficient for interaction with the Qac proteins but may need to be attached to or contained within an aromatic ring to facilitate recognition and transport of these compounds. Unlike some other multidrug efflux proteins in the major facilitator superfamily which can recognize both anionic and cationic substrates (7), neither QacA nor QacB conferred resistance to anionic substances such as hydrophilic (enoxacin and norfloxacin) or hydrophobic (nalidixic acid) quinolones (data not shown).
To further examine the hypothesis that QacA-mediated resistance to structurally dissimilar compounds is conferred via a common mechanism, fractional inhibitory concentration (FIC) analysis of various combinations of two QacA substrates was performed. Monovalent-divalent combinations were represented by ethidium-propamidine and benzalkonium-propamidine. Divalent-divalent combinations were represented by propamidine-pentamidine and propamidine-chlorhexidine. Increasing increments of each compound were combined by the checkerboard procedure (8) as follows: 200 to 800 μg/ml for ethidium, 20 to 80 μg/ml for benzalkonium, 50 to 300 μg/ml for pentamidine and propamidine, and 1 to 12 μg/ml for chlorhexidine. Microtiter plates and 20-ml agar plates were incubated for 48 h at 37°C. FIC index = x/MIC(x) + y/MIC(y), where x and y represent the lowest concentration of each compound (in combination) at which there is no growth and MIC(x) and MIC(y) represent the individual MIC of each compound. FIC indices have been defined as follows: ≤0.50, synergy; 0.51 to 2.00, additivity; 2.01 to 4.00, indifference; and >4, antagonism (1, 8). In this study, we have adapted the FIC index based on the assumption that if QacA extrudes two different compounds via the same mechanism, then an additive result would be expected, whereas if distinct mechanisms for different chemicals operate independently, then the result would be one of indifference. The results for all combinations tested fell within a range of FIC indices from 0.67 to 1.33, consistent with an additive effect.
The data presented in this report, together with those from previous studies (3, 5, 10), demonstrate that the multidrug efflux protein QacA is able to confer resistance to more than 30 cationic lipophilic antimicrobial compounds that belong to 11 distinct chemical classes. No resistance was observed for trivalent cationic substances or anionic substances, indicating that the resistance specificity of QacA is restricted to monovalent and divalent cationic substrates. Furthermore, FIC analysis of combinations of two substrates from various chemical classes showed an additive result, suggesting that QacA operates via a single antiport mechanism for the export of structurally dissimilar lipophilic cationic substrates.
Acknowledgments
We thank R. Sundberg, Rhône-Poulenc Rorer and Chauvin Laboratoire, for the generous supply of chemicals used in this study.
This work was supported by a project grant from the National Health and Medical Research Council (Australia). B.A.M. was a recipient of an Australian Postgraduate Award.
REFERENCES
- 1.Eliopoulos G M, Moellering R C., Jr . Antimicrobial combinations. In: Lorian V, editor. Antibiotics in laboratory medicine. 3rd ed. Baltimore, Md: Williams & Wilkins; 1991. pp. 432–492. [Google Scholar]
- 2.Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- 3.Littlejohn T G, Paulsen I T, Gillespie M T, Tennent J M, Midgley M, Jones I G, Purewal A S, Skurray R A. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1992;95:259–266. doi: 10.1016/0378-1097(92)90439-u. [DOI] [PubMed] [Google Scholar]
- 4.Marger M D, Saier M H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 5.Paulsen I T. Ph.D. thesis. Victoria, Australia: Monash University; 1994. [Google Scholar]
- 6.Paulsen I T, Brown M H, Littlejohn T G, Mitchell B A, Skurray R A. Molecular characterization of the multidrug resistance proteins QacA and QacB: membrane topology and identification of residues involved in specificity for divalent cations. Proc Natl Acad Sci USA. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pendland S L, Piscitelli S C, Schreckenberger P C, Danziger L H. In vitro activities of metronidazole and its hydroxy metabolite against Bacteroides spp. Antimicrob Agents Chemother. 1994;38:2106–2110. doi: 10.1128/aac.38.9.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouch D A, Cram D S, DiBerardino D, Littlejohn T G, Skurray R A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 10.Tennent J M, Lyon B R, Midgley M, Jones I G, Purewal A S, Skurray R A. Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. J Gen Microbiol. 1989;135:1–10. doi: 10.1099/00221287-135-1-1. [DOI] [PubMed] [Google Scholar]
- 11.Walzer P D, Foy J, Runck J, Steele P, White M, Klein R S, Otter B A, Sundberg R J. Guanylhydrazones in therapy of Pneumocystis carinii pneumonia in immunosuppressed rats. Antimicrob Agents Chemother. 1994;38:2572–2576. doi: 10.1128/aac.38.11.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]