Abstract
Text in electronic health records is organized into sections, and classifying those sections into section categories is useful for downstream tasks. In this work, we attempt to improve the transferability of section classification models by combining the dataset-specific knowledge in supervised learning models with the world knowledge inside large language models (LLMs). Surprisingly, we find that zero-shot LLMs out-perform supervised BERT-based models applied to out-of-domain data. We also find that their strengths are synergistic, so that a simple ensemble technique leads to additional performance gains.
1. Introduction
The text in electronic health record notes is typically organized into multiple sections. Correctly understanding what parts of a note correspond to different section categories has been shown to be useful for a variety of downstream tasks – including abbreviation resolution (Zweigenbaum et al., 2013), cohort retrieval (Edinger et al., 2017), and named entity recognition (Lei et al., 2014). However, documentation of sections is not consistently done across health systems, so building systems to robustly classify clinical text into sections is not trivial. Prior work on text classification has shown that systems trained on a dataset from one source perform quite poorly on different sources (Tepper et al., 2012a).
In this work, we extend recent work on section classification (Zhou et al., 2023) that uses the SOAP ("Subjective", "Objective", "Assessment", "Plan") framework (Podder et al., 2022; Wright et al., 2014). Our previous work (Zhou et al., 2023) mapped heterogeneous section types across three datasets onto SOAP categories (plus "Other") in order to facilitate cross-domain adaptation. However, despite showing improvements, that work showed that the problem was still challenging for a supervised approach that fine tuned pre-trained BERT-style encoder methods.
The insight of this current work is that supervised transformers, while powerful, may overfit to source domain training data. Zero-shot methods, on the other hand, have recently gained attention for their sometimes surprising ability to make accurate classification decisions without supervision. In general, for zero-shot classification to work, (1) the pre-training data must contain enough information about the kind of questions it will be asked, and (2) the prompt must be able to precisely represent the meaning of the classification labels. To work on section classification, then, we explore different base models since it is hard to know a priori which models will satisfy (1), and we explore variations in prompts that inject knowledge about the classification task to satisfy (2).
Therefore, we investigate the following research questions related to the ability of large language models (LLMs) to do SOAP section classification:
RQ1: How do different LLMs perform on the section classification task in zero-shot and few-shot experiments?
RQ2: How do LLMs in the zero-shot setting compare against supervised BERT-based models applied across domains in their ability to classify SOAP sections?
RQ3: Are the strengths of LLMs and BERT-based models complementary so that ensemble methods may be synergistic?
2. Methods
2.1. Datasets
In this study we used three datasets, discharge, thyme and progress, containing 1372, 4223, and 13367 sections, respectively. The discharge dataset consists of discharge summaries from the i2b2 2010 challenge (Tepper et al., 2012b). The thyme dataset consists of colorectal clinical notes from the THYME (Temporal History of Your Medical Events) corpus (Styler IV et al., 2014). The progress dataset consists of progress notes from MIMIC-III (Gao et al., 2022). Although these datasets are common in that they are all medical notes, they differ in both the health care institutions they are coming from and the specialties who wrote them. Following Zhou et al. (2023), the section names in these datasets were mapped to SOAP categories (“Subjective”, “Objective”, “Assessment” and “Plan”). For sections that did not fit into the SOAP categories, the "Others" label was assigned. Therefore, these datasets are tasks that classify a section into one of the 5 categories. We followed the same train/test split as in Zhou et al. (2023).
2.2. Prompt design
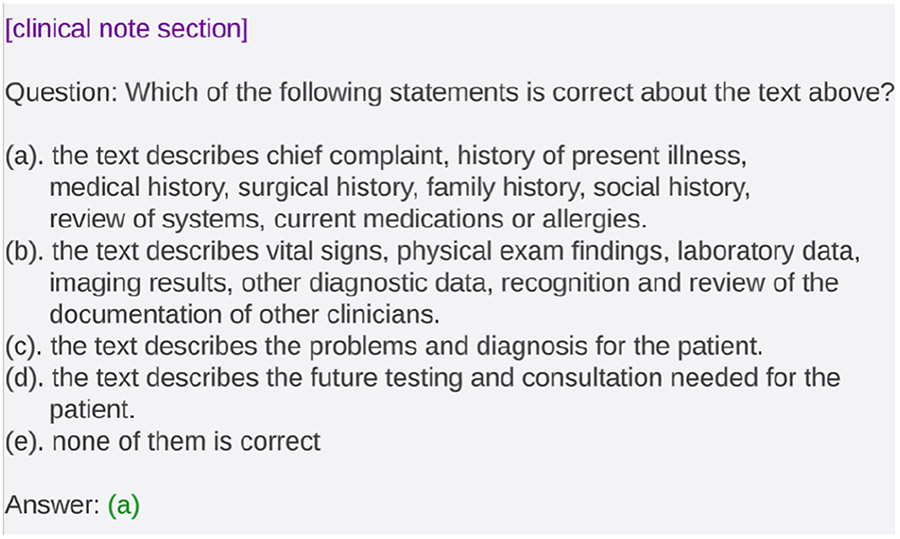
Performing classification with generative LLMs requires the creation of an input prompt that cues the model to generate output that can be deterministically mapped to a classifier output. We design our prompt to be a clinical note section followed by a multiple-choice question. The multiple-choice question begins with "Which of the following statements is correct about the text above" and is followed by statements describing the 5 categories in the SOAP section classification task. The prompt then lists the possible multiple choice answers as categories of SOAP, describing them based on the original definitions (Podder et al., 2022) instead of their labels, to attempt to inject more knowledge into the prompt. We also include the fifth possible answer of "none of them is correct", meaning that the section does not belong to any one of the SOAP categories. Figure 1 shows an example of a prompt with an answer.
Figure 1:
Example of a prompt with the answer provided. It consists of a clinical note section text and a multiple choice question. The options are for "Subjective", "Objective", "Assessment", "Plan" and "Others" respectively.
For few-shot classification, we randomly sample a few examples from the training set with answers, formatted as in Figure 1, and concatenate them together, followed by the query section text with the answer left blank. For zero-shot classification, the prompt contains only the query text with the answer left blank.
Prior work has shown LLMs prefer an option at a specific location for multiple-choice questions, such as always choosing the first or the last option (Singhal et al., 2022). To control for this source of variation, we shuffle the options every time before feeding the prompt into the model such that, for example, the option "Subjective" can be in any one of the five options’ locations. For the very rare cases that a model generates outputs not belonging to one of the 5 options, we consider that to be the "Others" category.
2.3. LLM experiments
To understand the performance of LLMs on section classification, we performed experiments to compare different LLMs and across different number of shots. In this study, we chose to experiment with FLAN-T5 (Chung et al., 2022), BioMedLM (Venigalla et al., 2022) and Galactica (Taylor et al., 2022). 1 We chose these models because, during preliminary work, they performed well with seemingly fewer hallucinations (Ji et al., 2023) than other models we explored.
BioMedLM has 2.7 billion parameters and is trained on biomedical abstracts and papers. FLAN-T5 is trained on the web crawl C4 dataset (Raffel et al., 2020) and additionally more than 1000 tasks, and we used the XXL version which contains 11 billion parameters. Galactica is trained on a large corpus containing scientific literature, and we used the standard version which contains 6.7 billion parameters. For each model we selected the largest variant that could fit in the memory of our GPU.
The maximum input token size is 512 for FLAN-T5, 1024 for BioMedLM, and 2048 for Galactica, which limits the maximum number of shots (input examples in the prompt) to 0, 5, and 10, respectively. Following Zhou et al. (2023), we report the micro-F1 scores. These experiments were done on a 40 GB NVIDIA A40 GPU. The best LLM will be used in the following ensemble model experiments.
2.4. Ensemble of BERT and LLMs
We experiment with improving the performance of cross-domain section classification by ensembling BERT (Vaswani et al., 2017) and LLMs. At a high level, the ensemble model will weight the two models’ prediction by their confidence and choose the one with the highest confidence. Confidence is measured by a model’s prediction probability of a category. For a pair of source and target domain, we first train a BERT model on the source domain and apply it to the target domain. For the target domain, we will obtain the model’s prediction () along with the prediction probability () of that class by applying a softmax function on the model’s output logits. Second, we apply an LLM to the target domain as well. To obtain confidence estimates from LLMs, we introduce a “black-box” method for estimating confidence of an LLM based on bootstrapping. We use this method for maximum generalizability – it could be applied even to black box models like ChatGPT that do not allow access to underlying probability distributions. To estimate confidence values, we make predictions for the same section ten times and vary the order of the five options across the runs. Because the prompt becomes different, the model sometimes makes different option choices. Probabilities are obtained by simply dividing option counts by the number of predictions (ten). We define the LLMs prediction () to be the one with the highest probability (). When ensembling, for each instance, we compare the prediction probabilities (, ) from both models and use the prediction with the highest probability:
As an example, if BERT predicts a section to be "Subjective" with a probability of 0.55 and the LLM predicts it to be "Objective" with a probability of 0.7, the ensemble model will use the LLM’s "Objective" prediction because it has a higher prediction probability. We use BioClinicalBERT (Alsentzer et al., 2019) for the BERT model and the training of BERT follows the same hyperparameter settings as described in Zhou et al. (2023).
3. Results
3.1. Comparing LLMs
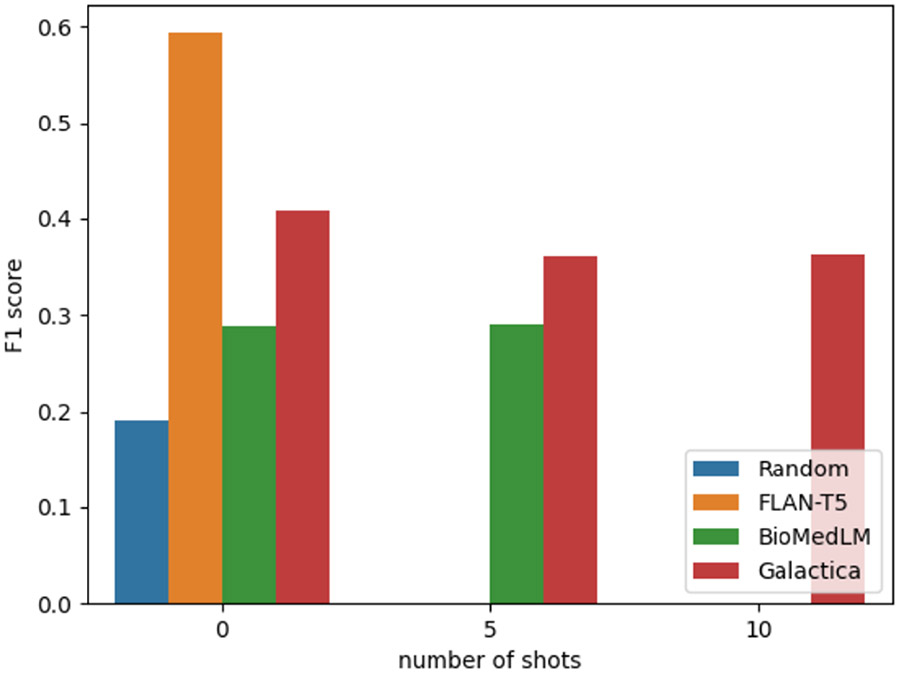
Figure 2 shows the results of running Random (random guess), FLAN-T5, Galactica and BioMedLM with 0-, 5-, and 10-shot experiments, averaged across datasets. Because of the input token size limit, the maximum number of shots for the three models are 0-, 5- and 10-shots respectively. We observe that the best performing LLM is FLAN-T5 at 0-shot (RQ1). We will use FLAN-T5 in the ensembling model development.
Figure 2:
Dataset averaged F1 score of Random, FLAN-T5, BioMedLM and Galactica models using 0-, 5- and 10-shot. Due to different prompt-length restrictions, not all settings could be run with all models.
3.2. Ensemble of BERT and LLMs
Table 1 shows the cross-domain F1 score for BERT, 0-shot FLAN-T5, and their ensemble for each pair of source and target domains. After averaging, we observe that FLAN-T5 is competitive against BERT (RQ2), and the ensemble model that combines both achieves the best performance.
Table 1:
F1 scores of BioClinicalBERT, FLAN-T5 and the ensemble when trained on the source domain and tested on the target domain.
| Source domain |
Target domain |
BioClinicalBERT | FLAN-T5 | Ensemble |
|---|---|---|---|---|
| thyme | discharge | 0.622 | 0.495 | 0.651 |
| progress | 0.465 | 0.495 | 0.491 | |
| discharge | thyme | 0.499 | 0.542 | 0.58 |
| progress | 0.652 | 0.542 | 0.593 | |
| discharge | progress | 0.741 | 0.795 | 0.821 |
| thyme | 0.625 | 0.795 | 0.817 | |
| Average | 0.601 | 0.611 | 0.659 | |
To understand the performance gain of the ensemble method, in Table 2, we show the dataset averaged F1 scores of BERT and FLAN-T5 by SOAP categories. We observe that FLAN-T5 is outperforming BERT on the "Assessment" and "Plan" categories by a large margin, is slightly better on the "Subjective" category, but is under performing on the "Objective" category. Because "Assessment" and "Plan" are less prevalent categories in the datasets, and the "Objective" category is more prevalent, FLAN-T5 achieves a competitive performance against BERT on average. This observation is also indicative that BERT and FLAN-T5 capture different aspects of the task and therefore their ensemble achieves the best performance (RQ3).
Table 2:
The F1 scores of BioClinicalBERT and FLAN-T5 broken down by prediction categories. The rows are the categories and the columns are the models.
| BioClinicalBERT | FLAN-T5 | |
|---|---|---|
| Subjective | 0.676 | 0.691 |
| Objective | 0.696 | 0.613 |
| Assessment | 0.164 | 0.46 |
| Plan | 0.127 | 0.29 |
| Others | 0.166 | 0.16 |
4. Discussion
Our results related to RQ1 were quite surprising. The best-performing LLM, FLAN-T5-XXL, while being the largest model, has the least overlap with our data genre and was unable to fit any example instances into its prompt. The success of FLAN-T5-XXL could be attributed to it both being larger in parameter size and having instruction tuning that other models don’t have. Future work should explore smaller versions of FLAN-T5 to learn whether the model size or fine tuning is more important, but one interesting hypothesis is that explicit fine tuning on tasks with multiple choice setups may have benefited FLAN-T5.
Despite the BioMedLM (2.7b) having fewer than half the parameters of the Galactica (6.7b) models, performance is not as degraded as we might expect. This could be an indicator that incorporating medical knowledge helps LLMs recognize medical texts better and thus performs closer to models that are larger when doing section classification. Here again, it would be valuable to isolate the model size variable from the pre-training genre variable, but the closest Galactica model in size to BioMedLM has 1.3 billion parameters – a closer model size but still not a perfect comparison. Neither model was seemingly able to take advantage of seeing labeled instances in their prompts. One possible explanation is that, because the output space has five unique labels, and the categories are quite heterogeneous, it is just not able to see enough diversity of each category type to meaningfully generalize.
Clinical-T5 (Lehman and Johnson, 2023; Goldberger et al., 2000), which is trained on MIMIC (Johnson et al., 2016, 2020), can be explored in the future too, to examine the effect of pre-training on a more highly aligned domain. However, we note that the pre-training data for Clinical-T5 overlaps with the progress dataset we evaluate on here, which makes it difficult to obtain fair zeroshot comparisons.
Finally, the pace of new releases of LLMs is quite fast, and models released after this work are potentially quite powerful (e.g. Alpaca (Taori et al., 2023) and Vicuna (Team, 2023)). Future work can also include assessing those models’ capability for section classification.
The ensemble model was found to be the best, and a hypothesis can be that LLMs learn better for the rarer categories and supervised learning learns better on prevalent categories. One explanation for this is that the supervised learner implicitly learns a distribution over label frequency, which may bias it towards frequent categories, while the zero-shot learner only has access to the textual evidence to make its decisions. If this same dynamic holds more generally (as seen in other recent work (Yuan et al., 2023)), LLMs may serve as an important supplement to supervised learning in terms of predicting rare categories.
This study estimated the prediction probability for LLM by repeating the experiments, and future work can explore additional methods for obtaining the prediction probability.
5. Conclusion
This paper demonstrates the use of LLMs for section classification and an ensemble method for improving the transferability of section classification models. The supervised learning model and LLMs are competitive, and when ensembled based on the prediction probabilities, we observed a higher performance. In analyzing the prediction performance by categories, we found LLMs complemented the supervised learning by performing better on the rare categories, and the supervised method performed better for the most prevalent category. Future studies can extend to updated LLMs and the use of LLMs for section classification is promising.
6. Limitations
A limitation in this study is we only used open-source models. We were unable to evaluate Chat-GPT, for example, because the data use agreements under which these datasets are made available forbid sending the data to outside APIs. Other models are frequently being released and we did not exhaustively test all publicly available language models. However, the focus of the paper is not to find the best LLMs but instead providing insights into using LLMs to improve transferability.
Footnotes
We were unable to experiment with models like ChatGPT due to the terms of the data use agreements of our datasets.
Contributor Information
Weipeng Zhou, Department of Biomedical, Informatics and Medical Education, School of Medicine, University of Washington.
Dmitriy Dligach, Department of Computer Science, Loyola University Chicago.
Majid Afshar, Department of Medicine, School of Medicine and Public Health, University of Wisconsin.
Yanjun Gao, Department of Medicine, School of Medicine and Public Health, University of Wisconsin.
Timothy A. Miller, Computational Health Informatics Program, Boston Children’s Hospital, Harvard Medical School
References
- Alsentzer Emily, Murphy John R, Boag Willie, Weng Wei-Hung, Jin Di, Naumann Tristan, and McDermott Matthew. 2019. Publicly available clinical bert embeddings. [Google Scholar]
- Chung Hyung Won, Hou Le, Longpre Shayne, Zoph Barret, Tay Yi, Fedus William, Li Eric, Wang Xuezhi, Dehghani Mostafa, Brahma Siddhartha, et al. 2022. Scaling instruction-finetuned language models. [Google Scholar]
- Edinger Tracy, Demner-Fushman Dina, Cohen Aaron M, Bedrick Steven, and Hersh William. 2017. Evaluation of clinical text segmentation to facilitate cohort retrieval. In AMIA Annual Symposium Proceedings, volume 2017, page 660. American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- Gao Yanjun, Dligach Dmitriy, Miller Timothy, Tesch Samuel, Laffin Ryan, Churpek Matthew M., and Afshar Majid. 2022. Hierarchical annotation for building a suite of clinical natural language processing tasks: Progress note understanding. In Proceedings of the Thirteenth Language Resources and Evaluation Conference, pages 5484–5493, Marseille, France. European Language Resources Association. [PMC free article] [PubMed] [Google Scholar]
- Goldberger Ary L, Amaral Luis AN, Glass Leon, Hausdorff Jeffrey M, Ivanov Plamen Ch, Mark Roger G, Mietus Joseph E, Moody George B, Peng Chung-Kang, and Stanley H Eugene. 2000. Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation, 101(23):e215–e220. [DOI] [PubMed] [Google Scholar]
- Ji Ziwei, Lee Nayeon, Frieske Rita, Yu Tiezheng, Su Dan, Xu Yan, Ishii Etsuko, Bang Ye Jin, Madotto Andrea, and Fung Pascale. 2023. Survey of hallucination in natural language generation. ACM Computing Surveys, 55(12):1–38. [Google Scholar]
- Johnson Alistair, Bulgarelli Lucas, Pollard Tom, Horng Steven, Celi Leo Anthony, and Mark Roger. 2020. Mimic-iv. [Google Scholar]
- Johnson Alistair EW, Pollard Tom J, Shen Lu, Lehman Li-wei H, Feng Mengling, Ghassemi Mohammad, Moody Benjamin, Szolovits Peter, Celi Leo Anthony, and Mark Roger G. 2016. Mimic-iii, a freely accessible critical care database. Scientific data, 3(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman Eric and Johnson Alistair. 2023. Clinical-t5: Large language models built using mimic clinical text. https://physionet.org/content/clinical-t5/1.0.0/. [Google Scholar]
- Lei Jianbo, Tang Buzhou, Lu Xueqin, Gao Kaihua, Jiang Min, and Xu Hua. 2014. A comprehensive study of named entity recognition in chinese clinical text. Journal of the American Medical Informatics Association, 21(5):808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podder Vivek, Lew Valerie, and Ghassemzadeh Sassan. 2022. Soap notes. In StatPearls [Internet]. Stat-Pearls Publishing. [PubMed] [Google Scholar]
- Raffel Colin, Shazeer Noam, Roberts Adam, Lee Katherine, Narang Sharan, Matena Michael, Zhou Yanqi, Li Wei, and Liu Peter J.. 2020. Exploring the limits of transfer learning with a unified text-to-text transformer. Journal of Machine Learning Research, 21(140):1–67.34305477 [Google Scholar]
- Singhal Karan, Azizi Shekoofeh, Tu Tao, Mahdavi S Sara, Wei Jason, Chung Hyung Won, Scales Nathan, Tanwani Ajay, Cole-Lewis Heather, Pfohl Stephen, et al. 2022. Large language models encode clinical knowledge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styler William F IV, Bethard Steven, Finan Sean, Palmer Martha, Pradhan Sameer, de Groen Piet C, Erickson Brad, Miller Timothy, Lin Chen, Savova Guergana, et al. 2014. Temporal annotation in the clinical domain. Trans. Assoc. Comput. Linguist, 2:143–154. [PMC free article] [PubMed] [Google Scholar]
- Taori Rohan, Gulrajani Ishaan, Zhang Tianyi, Dubois Yann, Li Xuechen, Guestrin Carlos, Liang Percy, and Hashimoto Tatsunori B.. 2023. Stanford alpaca: An instruction-following llama model. https://github.com/tatsu-lab/stanford_alpaca. [Google Scholar]
- Taylor Ross, Kardas Marcin, Cucurull Guillem, Scialom Thomas, Hartshorn Anthony, Saravia Elvis, Poulton Andrew, Kerkez Viktor, and Stojnic Robert. 2022. Galactica: A large language model for science. arXiv preprint arXiv:2211.09085. [Google Scholar]
- The Vicuna Team. 2023. Vicuna: An open-source chatbot impressing gpt-4 with 90% chatgpt quality. https://lmsys.org/blog/2023-03-30-vicuna/.
- Tepper Michael, Capurro Daniel, Xia Fei, Vanderwende Lucy, and Yetisgen-Yildiz Meliha. 2012a. Statistical section segmentation in free-text clinical records. In Proceedings of the Eighth International Conference on Language Resources and Evaluation (LREC’12), pages 2001–2008, Istanbul, Turkey. European Language Resources Association (ELRA). [Google Scholar]
- Tepper Michael, Capurro Daniel, Xia Fei, Vanderwende Lucy, and Yetisgen-Yildiz Meliha. 2012b. Statistical section segmentation in free-text clinical records. In Lrec, pages 2001–2008. [Google Scholar]
- Vaswani Ashish, Shazeer Noam, Parmar Niki, Uszkoreit Jakob, Jones Llion, Gomez Aidan N, Kaiser Łukasz, and Polosukhin Illia. 2017. Attention is all you need. Advances in neural information processing systems, 30. [Google Scholar]
- Venigalla Abhinav, Frankle Jonathan, and Carbin Michael. 2022. Biomedlm: A domain-specific large language model for biomedical text. [Google Scholar]
- Wright Adam, Sittig Dean F, McGowan Julie, Ash Joan S, and Weed Lawrence L. 2014. Bringing science to medicine: an interview with larry weed, inventor of the problem-oriented medical record. Journal of the American Medical Informatics Association, 21(6):964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Chenhan, Xie Qianqian, and Ananiadou Sophia. 2023. Zero-shot temporal relation extraction with chatgpt. [Google Scholar]
- Zhou Weipeng, Yetisgen Meliha, Afshar Majid, Gao Yanjun, Savova Guergana, and Miller Timothy A.. 2023. Improving model transferability for clinical note section classification models using continued pretraining. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweigenbaum Pierre, Deléger Louise, Lavergne Thomas, Névéol Aurélie, and Bodnari Andreea. 2013. A supervised abbreviation resolution system for medical text. In CLEF (Working Notes). Citeseer. [Google Scholar]