Abstract
Advances in multiagent chemotherapy have led to recent improvements in survival for patients with acute lymphoblastic leukemia (ALL); however, a significant fraction do not respond to frontline chemotherapy or later relapse with recurrent disease, after which long-term survival rates remain low. To develop new, effective treatment options for these patients, we conducted a series of high-throughput combination drug screens to identify chemotherapies that synergize in a lineage-specific manner with MRX-2843, a small molecule dual MERTK and FLT3 kinase inhibitor currently in clinical testing for treatment of relapsed/refractory leukemias and solid tumors. Using experimental and computational approaches, we found that MRX-2843 synergized strongly—and in a ratio-dependent manner—with vincristine to inhibit both B-ALL and T-ALL cell line expansion. Based on these findings, we developed multiagent lipid nanoparticle formulations of these drugs that not only delivered defined drug ratios intracellularly in T-ALL, but also improved anti-leukemia activity following drug encapsulation. Synergistic and additive interactions were recapitulated in primary T-ALL patient samples treated with MRX-2843 and vincristine nanoparticle formulations, suggesting their clinical relevance. Moreover, the nanoparticle formulations reduced disease burden and prolonged survival in an orthotopic murine xenograft model of early thymic precursor T-ALL (ETP-ALL), with both agents contributing to therapeutic activity in a dose-dependent manner. In contrast, nanoparticles containing MRX-2843 alone were ineffective in this model. Thus, MRX-2843 increased the sensitivity of ETP-ALL cells to vincristine in vivo. In this context, the additive particles, containing a higher dose of MRX-2843, provided more effective disease control than the synergistic particles. In contrast, particles containing an even higher, antagonistic ratio of MRX-2843 and vincristine were less effective. Thus, both the drug dose and the ratio-dependent interaction between MRX-2843 and vincristine significantly impacted therapeutic activity in vivo. Together, these findings present a systematic approach to high-throughput combination drug screening and multiagent drug delivery that maximizes the therapeutic potential of combined MRX-2843 and vincristine in T-ALL and describe a novel translational agent that could be used to enhance therapeutic responses to vincristine in patients with T-ALL. This broadly generalizable approach could also be applied to develop other constitutively synergistic combination products for the treatment of cancer and other diseases.
Keywords: combination therapy, drug delivery, nanotechnology, synergy, lipid nanoparticle, drug screening
INTRODUCTION
Combination approaches to cancer therapy have greatly improved treatment outcomes since their inception in the mid-1950s [1–3] and today, a majority of cancer patients receive some form of multiagent therapy [4] that can encompass a wide range of drug types including small molecules, proteins, nucleic acids, viral vectors, or engineered cells. In addition to diversity of size and structure, these agents also vary widely in their route or frequency of administration and, as a result, large time-dependent fluctuations in both tissue and plasma drug concentration—and therefore drug ratio—are commonly observed following the administration of combination therapies [5–7].
Drug synergy, which is the observation of combined drug effect that exceeds the expected sum of component drug effects, is a common basis for the use of multiagent therapies in cancer, infectious diseases, and neurological disorders. Yet, despite the strong therapeutic potential of synergistic multiagent approaches, prior large-scale screening studies suggest that the identification of synergistic drug pairs is in fact rare (4–10%) [8], while others indicate that synergistic effects, when they do occur, are highly ratio-dependent, whereby some ratios of particular drugs may be supra-additive (i.e. synergistic) while others are additive or sub-additive (i.e. antagonistic) [9–11].
Recent approaches to maximize the therapeutic potential of multiagent cocktails have aimed to co-deliver therapeutics in a manner that constitutively maintains drug synergy [12]. Such approaches include nanometer-scale drug carriers that deliver agents in a time-staggered [13, 14] or ratiometric [15–19] fashion, as well as those that harness synthetic lethal gene interactions [20] or target adaptive drug resistance mechanisms a priori [21, 22]. Liposomal cytarabine and daunorubicin (CPX-351, Vyxeos®) is one such example and was originally discovered through in vitro screens that identified synergistic cell growth inhibition at a 5:1 mole ratio of cytarabine:daunorubicin [23, 24]. By constitutively maintaining ratiometric drug synergy between these two agents via lipid nanoparticle formulation, CPX-351 improved treatment outcomes relative to free combination chemotherapy in both mouse models and clinical trials, and is now FDA-approved for the treatment of both adult and pediatric patients with leukemia [25–27].
The receptor tyrosine kinase, MERTK, is ectopically expressed in a majority of acute myeloid leukemias (AML) and approximately 50% of T-cell acute lymphoblastic leukemias (T-ALL) and we have described key roles for this protein in leukemia cell survival and leukemogenesis [28–31]. More recently, we developed small molecule inhibitors that target MERTK and FLT3, a clinically-validated therapeutic target in AML [32]. These agents improve survival in murine ALL and AML models [33–5] and the lead compound, MRX-2843, is currently being tested in Phase 1b clinical trials (NCT03510104, NCT04762199, NCT04872478, NCT04946890). We hypothesized that the therapeutic potential of combined MERTK/FLT3 inhibition and chemotherapy for leukemia could be maximized via ratiometric drug screening and formulation. To this end, here we describe a novel and systematic approach to high-throughput combination drug screening and nanoscale drug delivery that maximizes drug synergy between MRX-2843 and cytotoxic chemotherapy. This novel, broadly generalizable approach to formulation discovery could lead to the development of constitutively synergistic combination products for the treatment of cancer and other diseases.
RESULTS
Guided by prior reports indicating the potential for synergy between MERTK inhibition and methotrexate or vincristine [28, 31, 33], we developed a high-throughput drug combination screen (HTS) in which we sought to identify ratiometric synergy between these compounds following cell exposure to drug interaction matrices comprising concentration gradients of MRX-2843, vincristine and methotrexate (Fig 1a). As an in vitro model, we selected a genotypically and phenotypically diverse set of leukemia cell lines spanning B- and pro-B ALL, as well as T- and early T-cell precursor (ETP-) ALL, exposing each of these cell lines to drug interaction matrices comprising >530 discrete ratiometric drug combinations, and their equivalent single-agents, in 384-well plates using high-throughput liquid handling robotics (Fig 1b). Cell density was measured in quadruplicate via luminescent cell viability assay using Z’≥0.5 as a cutoff for data quality.
Figure 1. Combined MRX-2843 and vincristine synergize to inhibit T-ALL cell growth in high-throughput combination drug screens.
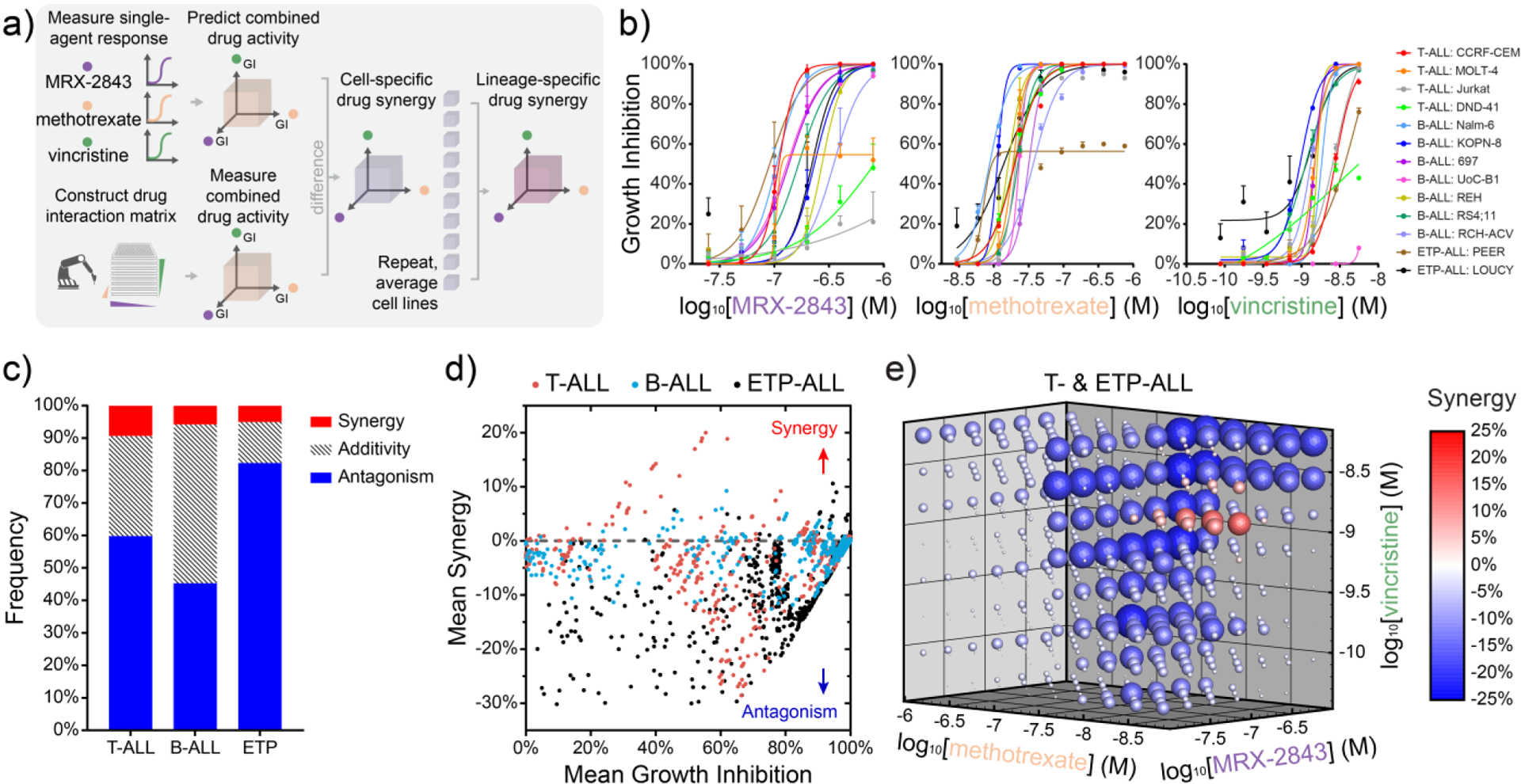
a) Illustration of a ratiometric drug screen combining the dual MERTK/FLT3 inhibitor, MRX-2843, with methotrexate and/or vincristine in a panel of 13 B- and T-ALL cell lines as measured by luminescent viability assay (72 h, Z’≥0.5). b) Single-agent dose response curves and (c) lineage-specific frequencies of combined drug synergy, additivity, or antagonism for 537 unique pairwise or triplet drug combinations as measured in parallel and assessed via the Bliss Independence model. d) Scatter plots of mean drug synergy as a function of mean growth inhibition grouped by cell lineage. Ratiometric drug synergy is conserved across T- and ETP-ALL cell lines with synergy among distinct molar ratios of MRX-2843 and vincristine. e) Drug synergy and antagonism appear at distinct doses and molar ratios in cells from the T- and ETP-ALL lineage. (c-e) Synergy represents percent reduction in cell density greater or lesser than that predicted by the Bliss Independence model with synergy (>1%) and antagonism (<−1%). (e) Sphere size and saturation correspond to the mean magnitude of synergy observed across T- and ETP-ALL cell lines (n=6) for each pairwise and triplet drug combination among 537 tested.
Consistent with other large scale screening studies [9, 36, 37], we observed that synergy among these ratiometric combinations was rare (<6.7% of combinations overall) as assessed using the Bliss Independence model of drug synergy (Fig 1c,d) [38]. Interestingly, drug synergy was largely absent when averaged over B lineage ALL cells (Fig S1); however, we observed strong synergy and antagonism that was ratio-dependent across T and ETP lineage ALL cell lines (Fig 1e). The consistency of these trends in synergy was confirmed using the Response Additivity and Zero Interaction Potency synergy models [38] (Fig S2a). HTS results were reproduced in a validation study using an alternative 96-well assay format and new drug and cell stocks (Fig S2b,c).
To guide the selection of a drug combination with maximum potency in T-ALL, we next determined whether synergy observed in our prior screen was principally attributable to either higher order (3-drug) or lower order (2-drug) effects. By comparing concordance between measured cell viabilities and those predicted from synergy models [39], we found that synergy observed in 3-drug combinations via HTS was predominantly attributable to lower order (2-drug), rather than higher order (3-drug), interactions (Fig 2a, S3a-c). To confirm which among the three pairwise drug combinations was most synergistic, we conducted a microwell assay in which hydrogel-embedded Jurkat (T-ALL) cells were exposed to continuous gradients of each drug pair and assessed for cell viability. Comparing viability data to corresponding additivity isoboles, we found that the combination of MRX-2843 and vincristine was consistently synergistic and most effective among these pairwise combinations (Fig 2b, S2d,e).
Figure 2. In silico and in vitromodels prioritize pairwise drug synergy between MRX-2843 and vincristine among screened combinations.
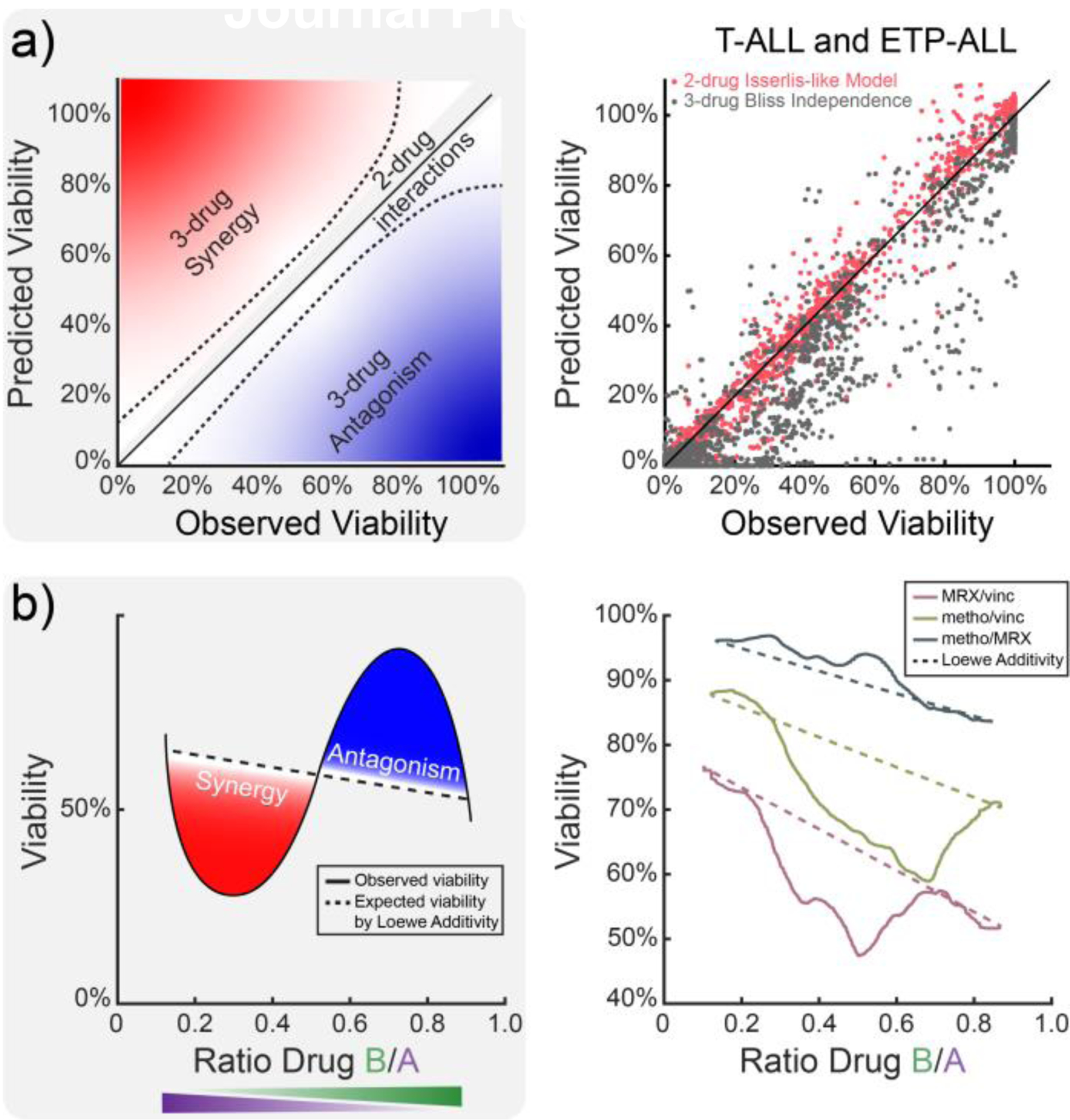
a) Comparison of higher order (3-drug) and lower order (2-drug) synergy models to high-throughput combination drug screening data indicates that experimentally observed drug synergy is predominantly attributable to pairwise drug interactions. b) Comparison of expected (dashed) and observed (solid) Jurkat (T-ALL) cell viability following exposure to continuous pairwise drug gradients demonstrates that MRX-2843 and vincristine are most consistently synergistic. (a) Data points show 378 triplet drug combination responses measured in T- and ETP-ALL cell lines (n=6) via HTS. Data in (b) represent mean cell viability (72 h) as measured by nuclear dye exclusion.
To better define the biological underpinnings of synergy between MRX-2843 and vincristine in T-cell ALL, we performed transcriptomic profiling on Jurkat (T-ALL) cells treated with MRX-2843:vincristine or their respective single-agent and vehicle controls (Fig S4a). Although the set of differentially expressed genes (DEGs) associated with a synergistic combination was unique, it was also limited in mapping to enriched ontologies after adjusting for multiple comparisons (Fig S4b,c). However, the union of DEGs in cells treated with MRX-2843 and vincristine monotherapies mapped with high significance to a network of similar and interconnected shared biological processes (GO, Reactome, KEGG and WikiPathways) (Fig S4d-g) associated with respiratory electron transport, RNA metabolism, ribosomal complex biogenesis, and anaphase initiation via the degradation of securin (Fig S4d,e). Insight into these ontological relationships may lead to development of novel drug combinations targeting shared mechanisms of action.
Having determined that MRX-2843 and vincristine act in a ratio-dependent manner to decrease T-ALL cell density, we next sought to develop drug formulations that both deliver and constitutively maintain ratiometric synergy between these compounds. Using a cheminformatics-guided approach, we determined pH-dependent physicochemical properties associated with MRX-2843 and vincristine via ACD/Labs software (see Cheminformatics Computation, Methods), and then devised a pH-gradient based method of drug loading (Fig 3a,b) in which the compounds were simultaneously co-encapsulated within lipid nanoparticles composed of DSPC (1,2-distearoyl-sn-glycero-3-phosphocoline), DSPG (1,2-distearoyl-sn-glycero-3-phosphoglycerol, sodium salt) and cholesterol lipids. Using this approach, we synthesized drug formulations of MRX-2843 and vincristine that recapitulated drug ratios which we had identified via HTS as synergistic (8.9 – 90 mol:mol and 4.7 – 48 mg:mg MRX-2843:vincristine), additive (90 – 160 mol:mol; 48 – 85 mg:mg), or antagonistic (≥160 mol:mol; ≥85 mg:mg) in their inhibition of T-ALL and ETP-ALL cell expansion (Fig 3c), referred to hereafter as Syn, Add and Antag nanoparticles, respectively. Characterization of dual-agent lipid nanoparticle formulations by dynamic light scattering, transmission electron microscopy, and liquid chromatography-mass spectrometry (LC-MS) indicated that pH-gradient co-loading achieved size uniformity with low polydispersity indices (PDI 0.11 – 0.14), efficient drug loading (6 wt% for dual-agent nanoparticles) and precise ratiometric encapsulation using these methods (Fig 3d,e, S5a,b, see Methods for details). In addition, we evaluated nanoparticle stability following lyophilization. Dual- and single-agent nanoformulations demonstrated zeta-potentials <−30 mV (Fig S5c) and lost <3% of total drug following storage (Fig S5d). A dual-agent nanoformulation released <7.5% of MRX-2843 over 72 hours of dialysis in PBS at 37°C without any apparent loss in vincristine, demonstrating stable ratiometric maintenance, whereas both drugs were rapidly released and at varying rates in acidic dialysis buffer (pH=5.5) (Fig S5e). Dual-agent nanoformulations maintained potent activity in cell-based assays for up to 1 year (Syn range: 1.36 – 5.30 nM (vincristine); Add range: 0.96 – 1.70 nM (vincristine)) (Fig S5f). Suggesting further formulation stability, nanoparticles showed minimal changes in size and PDI following rehydration or in solution with serum (Fig S6a,b).
Figure 3. Lipid nanoparticles co-encapsulating MRX-2843 and vincristine constitutively maintain defined drug ratios following co-formulation and intracellular delivery.
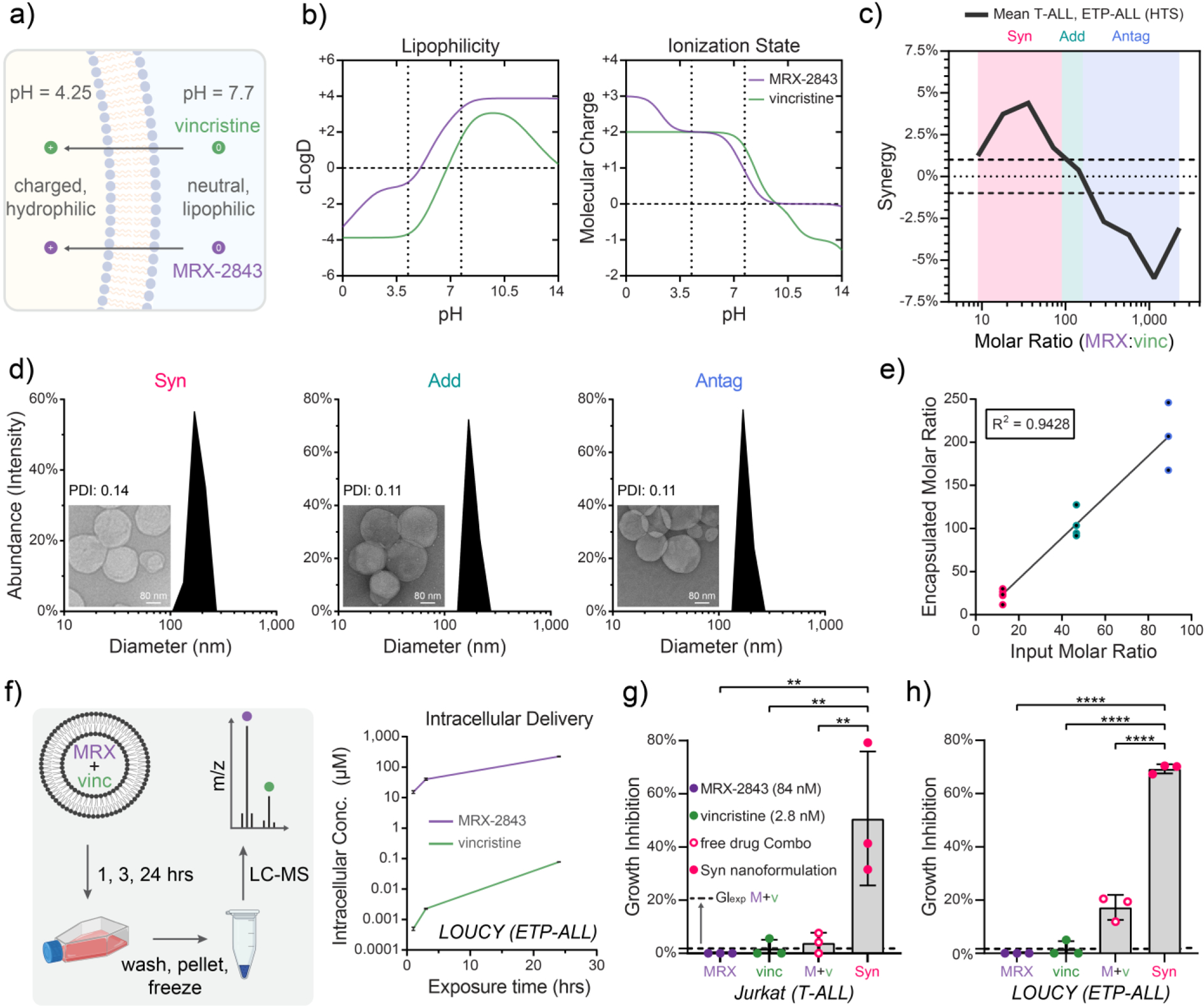
a) Illustration of a pH gradient-based method of drug co-loading that relies upon (b) pH-dependent MRX-2843 and vincristine lipophilicity and charge. c) A plot of mean drug synergy versus MRX-2843:vincristine mole ratio defines synergistic (8.9 – 90), additive (90 – 160), and antagonistic (≥160) drug ratios. d) T-ALL tailored combination drug formulations encapsulating synergistic (Syn), additive (Add), and antagonistic (Antag) drug ratios shown at nanometer-scale size and uniformity as measured by dynamic light scattering and (inset) transmission electron microscopy e) Drug co-loading as measured by LC-MS. f) Intracellular drug delivery kinetics following treatment of LOUCY (ETP-ALL) cells with Antag nanoparticles over 24 h as measured by LC-MS. g) Comparison of growth inhibition following treatment with Syn nanoparticles, as well as combined and free MRX-2843/vincristine demonstrates that nanoparticle encapsulation further enhances in vitro drug synergy as measured by luminescent cell viability assay in Jurkat (T-ALL) cells (72 h) and (h) LOUCY (ETP-ALL) (72 h). (c) The line traces the trend in mean response between tested ratios, and hashes delineate the range of +/− 1% synergy, which we define as additivity. (d) Scale bars measure 80 nm, and PDI stands for polydispersity index. (e) Data points represent drug ratios from distinct nanoparticles batches as measured by LC-MS. Best fit by linear regression is shown with coefficient of linearity. (f) Vertices are mean values ± SD of intracellular drug concentrations of 3 replicates per time point as measured by LC-MS. (g,h) Individual values (dots) and mean values ± SD of 3 replicates from a single experiment are shown, where the dashed line represents the expected effect for an additive interaction (GIexp). Mean differences were evaluated by one-way ANOVA with Tukey’s correction for multiple comparisons, where * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
To measure the ratiometric delivery of MRX-2843 and vincristine, we exposed LOUCY (ETP-ALL) cells to drug-loaded lipid nanoparticles for 24 hours and measured intracellular drug delivery via LC-MS (Fig 3f). We found that drug-loaded nanoparticles not only achieved highly efficient delivery of MRX-2843 to T-lineage ALL cells, but that they also co-delivered MRX-2843 and vincristine at concentrations that monotonically increased in parallel over time. To further characterize the therapeutic performance of dual-agent ratiometric nanoparticles, we examined their impact on LOUCY and Jurkat cell density via luminescent viability assay. Strikingly, drug synergy was even further enhanced in both cell lines following nanoparticle encapsulation, whereby growth inhibition far exceeded that of the already synergistic combination of free MRX-2843 and vincristine (Jurkat, P=0.0090; LOUCY, P<0.0001) (Fig 3g,h, S5g). Syn, Add and Antag dual-agent nanoformulations demonstrated higher potency than both the expected effect for an additive interaction and the observed effect in cultures treated with the free drug combination (Fig S5g).
We next investigated the clinical relevance of these combination drug formulations using primary pediatric T-ALL patient samples collected at time of diagnosis. Mononuclear cells from bone marrow or blood were treated with liposomal MRX-2843, free vincristine, vehicle lipid nanoparticles, or Syn or Add nanoparticles that reflect optimal synergy or additivity in LOUCY (ETP-ALL) cells (Fig 4a). GI50 values of cell density, measured via luminescent viability assay, demonstrated growth inhibition mediated by both Syn and Add nanoparticles (Fig 4b), and both formulations recapitulated their expected synergism or additivity in patient samples compared to expectation based on the effects of liposomal MRX-2843 and free drug vincristine measured in parallel (Fig 4c,d).
Figure 4. Comparative potency of ratiometric drug formulations in primary T-ALL samples ex vivo.
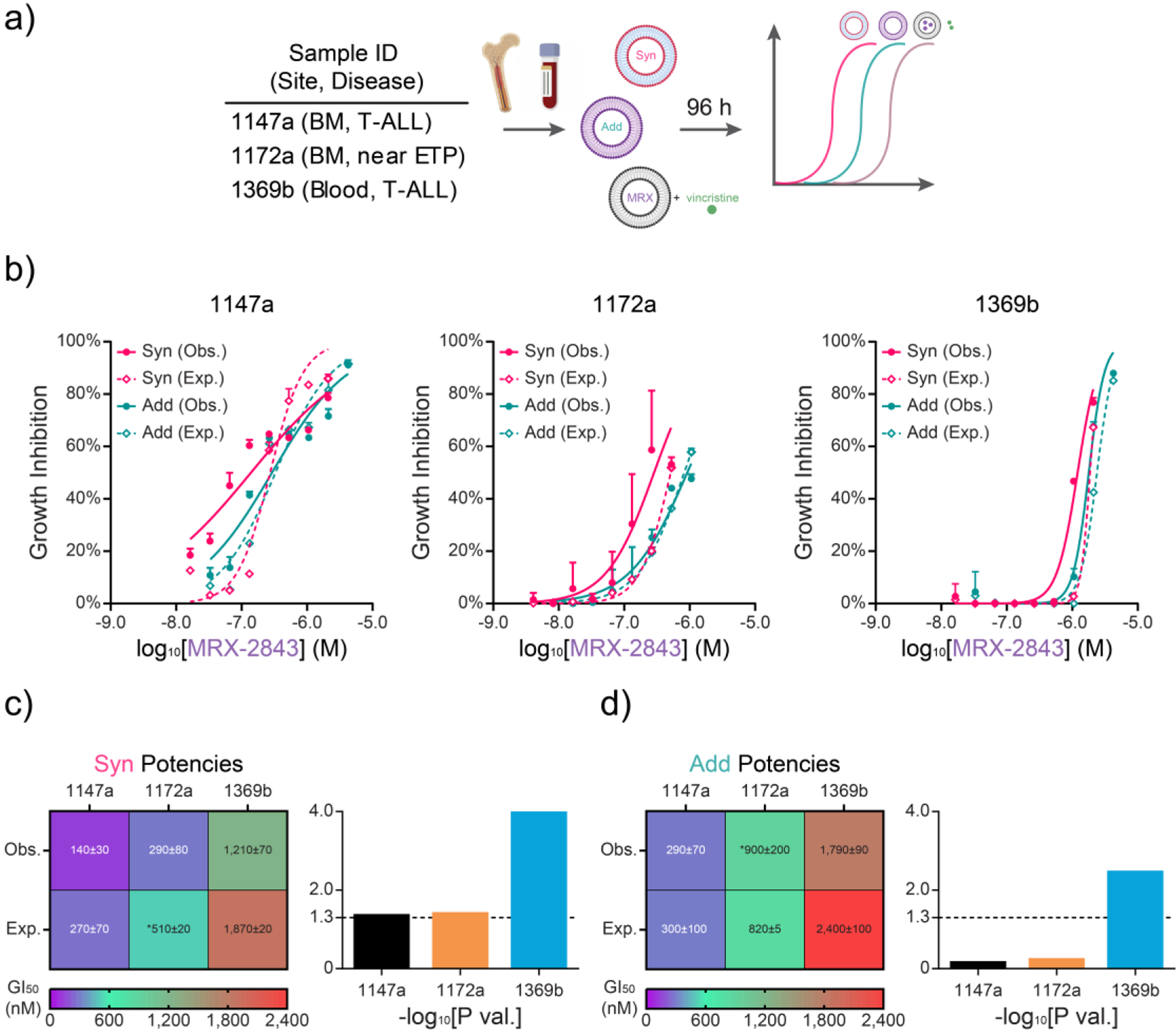
a) Approach to testing ratiometric nanoformulations in T-ALL or near ETP-ALL samples obtained from patient blood or bone marrow (BM) at initial diagnosis. b) Expected (Exp.; dashed) and observed (Obs.; solid) dose-dependent growth inhibition from synergistic and additive ratiometric drug formulations as measured by luminescent viability assay (96 h). c) Heatmap of GI50s and P values from (c) Syn and (d) Add drug formulations shown in (b). (b) Mean values ± SD of growth inhibition scaled against the concomitant MRX-2843 doses in the nanoparticles. Exp. dose response curves in (b) were calculated with the Bliss Independence model of drug synergy between single-agent liposomal MRX and vincristine free drug dose responses. (c,d) Heatmaps show GI50 ± standard error (nM) of Syn and Add potencies scaled by MRX-2843 dose. Bar charts show associated log transformed P values of unpaired parametric t tests. Asterisks (*) denote extrapolated GI50 values. All Obs. dose responses were run in triplicate, while Exp. dose responses were run either in duplicate (1172a) or triplicate (1147a, 1369b) in a single experiment.
Next ratiometric lipid nanoparticles were compared in a murine ETP-ALL xenograft model. For these studies, a luciferase-expressing derivative of the LOUCY cell line (LOUCY-luc) [40] was injected intravenously into NRG mice to establish orthotopic xenografts, and mice were treated with nanoparticles containing Syn, Add and Antag formulations of MRX-2843 and vincristine, vincristine (Vinc) or MRX-2843 (MRX) monotherapies, or empty liposomes (vehicle, Veh) once weekly for four weeks to mimic the schedule of administration for vincristine in patients with leukemia (Fig S7a,b). In an initial study various formulations were administered to span a range of MRX-2843 and vincristine doses and Vinc only particles contained the same dose as the Syn formulation. Mice treated with Syn and Vinc only nanoparticles delivering a dose of 1.152 mg/kg vincristine were removed from the study by 80 days after initiation of treatment due to poor health, despite a lack of significant disease burden (Fig S7c-e). Add particles delivering a dose of 0.57 mg/kg vincristine were well-tolerated and bioluminescence intensity increased coincident with removal of mice from study, suggesting leukemia as a cause of death. Thus, the maximum tolerated dose of vincristine nanoparticles in this model is between 0.57–1.152 mg/kg. When treatment was initiated with minimal disease burden, bioluminescence intensity was significantly decreased in mice treated with the Add formulation compared to mice treated with Antag nanoparticles (p=0.0159) (Fig S7c) and median survival was significantly prolonged (74 versus 121 days after tumor inoculation, p=0.0059) (Fig S7d). When initiation of treatment was delayed until the mice had developed higher disease burden, the Add nanoparticles were no longer sufficient to provide therapeutic activity, while the Syn particles still prevented disease progression and significantly reduced disease burden compared to treatment with Add particles (p=0.0317) (Fig S7f). Thus, both toxicity and therapeutic efficacy varied with vincristine dose. In contrast, MRX nanoparticles were well tolerated at doses of up to 60 mg/kg twice weekly, but did not have therapeutic activity in mice with LOUCY-luc xenografts (Fig S8a-g).
To determine whether MRX-2843 contributes to the therapeutic activity mediated by the combination nanoparticles in this model, Syn and Add nanoparticle formulations containing vincristine at a fixed, tolerated and effective dose (0.713 mg/kg) were directly compared in mice with LOUCY-luc xenografts (Fig 5a,b). Both formulations significantly decreased disease burden compared to treatment with empty liposomes (p=0.0106 and p=0.0042, respectively) (Fig 5c). Leukemia was also significantly decreased in mice treated with Add compared to Syn particles (p=0.0204). Similarly, median survival was significantly prolonged in mice treated with Add nanoparticles (85 days post-treatment) compared to Veh-treated mice (41 days; p = 0.0287) and survival was prolonged but the difference was not significant in mice treated with Syn particles (71 days) compared to Veh (p=0.1269) (Fig 5d). Thus, both Add and Syn nanoparticle formulations of MRX-2843 and vincristine were therapeutically effective in mice with LOUCY-luc xenografts, but the Add particles, which deliver a higher dose of MRX-2843, provided greater therapeutic benefit.
Figure 5. Nanoparticles containing MRX-2843 and vincristine at an additive ratio mediate superior antileukemia activity in a murine ETP-ALL xenograft model.
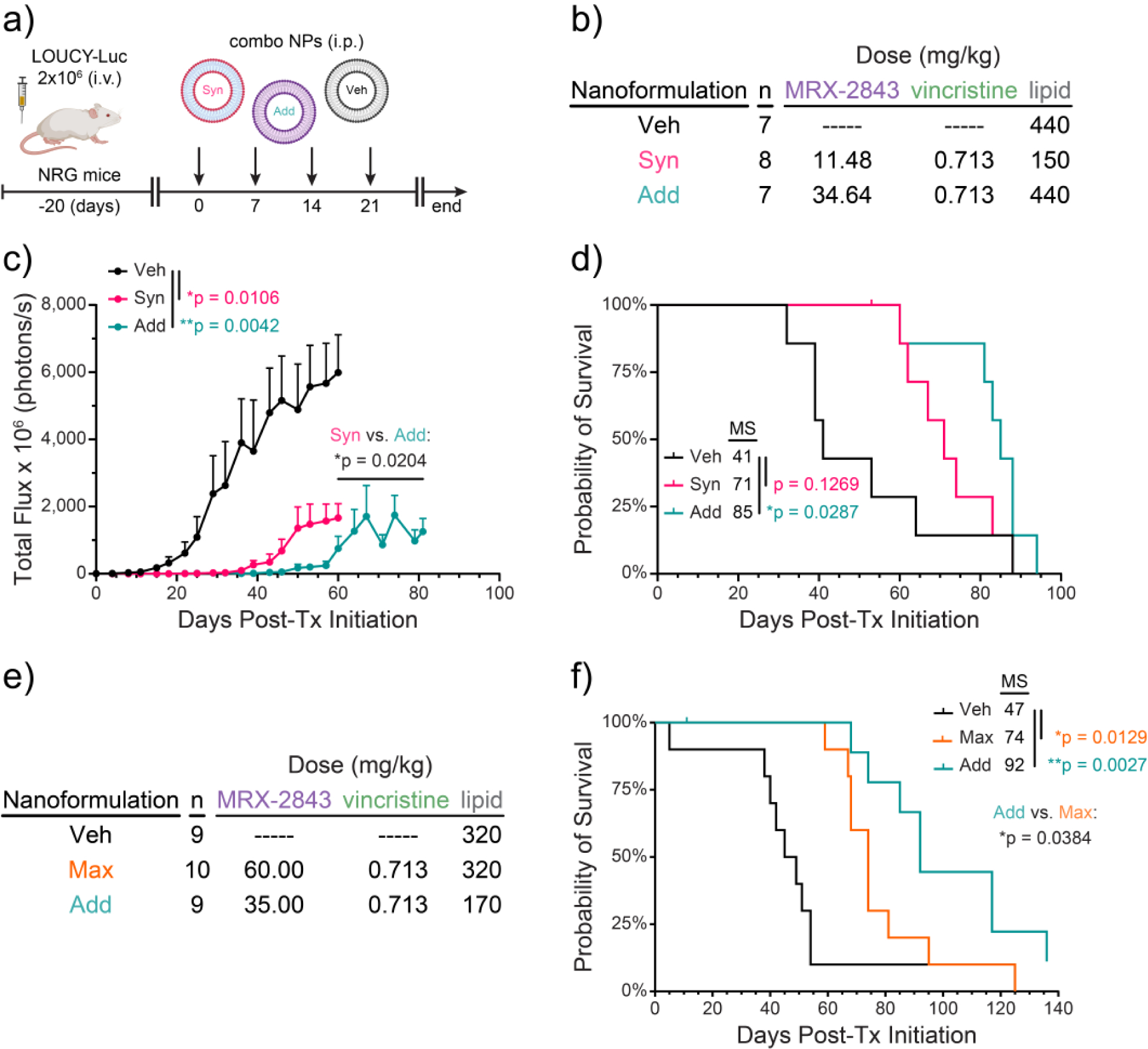
a,b) NRG mice were inoculated with a luciferase-expressing ETP-ALL cell line (LOUCY-luc; 2 million cells). Mice were randomized to groups with equal starting disease burden (n=7–8) and treatment with synergistic (Syn) or additive (Add) liposomal formulations of MRX-2843 and vincristine or vehicle nanoparticles without drug (Veh) was initiated 20 days later. Arrows indicate treatment administration. c) Disease burden was determined at intervals by bioluminescence imaging. Data are reported as mean ± SEM. Differences were determined by two-way ANOVA. d) Differences in median survival (MS) were calculated by the log-rank method. e,f) NRG mice were inoculated intravenously with 3 million LOUCY-luc cells, then randomized to groups (n=9–10) and treatment with the indicated liposomal formulations of MRX-2843 and vincristine or Veh was initiated 20 days later. f) Differences in median survival were calculated by the log-rank method.
In contrast, nanoparticles containing an even higher dose of MRX-2843 (Max, MRX-2843:vincrstine = 159 mol:mol) provided decreased therapeutic efficacy compared to the Add nanoparticles in this same model (Fig 5e,f). Both Add and Max nanoformulations significantly prolonged survival compared to vehicle (p=0.0027 and p=0.0129, respectively) and median survival was also significantly decreased from 92 days in mice treated with the Add nanoformulation to 74 days in mice treated with Max nanoparticles (p=0.0384). Of note, any changes in body weight in mice treated with nanoparticles containing 0.713 mg/kg vincirstine in combination with MRX-2843 were coincident with advanced disease (Fig S9). Thus, among dual-agent nanoformulations of MRX-2843 and vincristine, the Add nanoparticles provided optimal therapeutic activity in vivo.
DISCUSSION
Here we address the urgent and unmet need to develop new, more potent therapies for patients with relapsed/refractory T-ALL. While advancements in chemotherapy dose-intensification and patient risk-stratification have led to gradual improvements in patient survival for T-ALL in recent years, a substantial proportion of patients have treatment refractory disease or later relapse. As a result, mortality rates for pediatric and adult T-ALL patients are 20% and >50%, respectively [41]. Patients diagnosed with ETP-ALL experience particularly high rates of induction therapy failure, necessitating intensified treatments associated with increased toxicity and increasing the likelihood for bone marrow transplant [42–46]. To address the challenge of improving outcomes in these patients, we devised a systematic approach to maximize drug synergy between small molecule anticancer drug combinations and to tailor these effects to a specific disease indication, in this case TALL.
Using a novel, combinatorial high-throughput drug screen, we found that combined treatment with the tyrosine kinase inhibitor, MRX-2843, and the cytotoxic chemotherapy, vincristine, synergized in a ratio-dependent manner to inhibit T-ALL cell expansion, a finding that is significant because (i) vincristine is a mainstay of frontline induction and consolidation therapies for T-ALL [47] and (ii) both MRX-2843 targets (MERTK/FLT3) are frequently actionable in T-ALL. For example, in our previous studies, approximately 50% of pediatric T-ALL patient samples ectopically expressed MERTK [28] and 50% of both pediatric and adult samples were sensitive to treatment with a MERTK/FLT3 inhibitor [48]. Similarly, approximately 15% and 35% of pediatric and adult ETP-ALLs, respectively, carry activating FLT3 mutations [49, 50].
Given the dual targets of MRX-2843 and the multifarious effects of MERTK on oncogenic signaling pathways [51], the mechanism(s) by which MRX-2843 synergizes with vincristine in T-ALL are speculative; however, pharmacologic inhibitors of several pathways and proteins that are known to be downstream targets of MERTK signaling also synergize with vincristine, suggesting their potential as mediators of synergy. MERTK promotes expression of the anti-apoptotic protein BCL-XL in ALL and other cancer cells and can inhibit expression of the related protein, BCL-2, in some circumstances [31, 52]. Gossypol, a BCL-2 and BCL-XL inhibitor, synergized with vincristine in non-Hodgkin’s lymphoma cells [53]. Likewise, inhibition of BCR/ABL whose downstream signaling, like MERTK, converges on the MAPK pathway, synergized with vincristine against ALL cells [54]. Lastly, inhibitors of PI3K – positively regulated by MERTK signaling – synergized with vincristine in T-ALL cell culture and murine models [55].
Based on the findings from our screen and subsequent analyses indicating ratiometric drug synergy between MRX-2843 and vincristine, we developed multiagent lipid nanoparticles closely related in lipid composition to FDA-approved products such as AmBisome and Vyxeos, both of which are comprised of phosphatidylcholine, phosphatidylglycerol and cholesterol [56]. To further enhance the feasibility of clinical-scale manufacturing, we also devised a method to co-load defined ratios of MRX-2843 and vincristine using a gradient-based method which is notable in its typically high encapsulation efficiency, scalability and reproducibility, as well as its utilization in the manufacture of FDA-approved products, such as Onivyde and Doxil [57, 58]. In addition, we demonstrated a scalable method of freeze-dried storage that stabilizes drug activity for at least 1 year, retains encapsulated drug upon reconstitution, and maintains nanoparticle size and dispersion. Given the safe and effective clinical use of these lipid excipients, together with a commercially viable method of manufacturing and storage, these findings further support the translational potential of liposomal MRX-2843 and vincristine.
Having developed lipid nanoparticle formulations co-loading these compounds, we further examined the extent to which combination formulations deliver synergistic and antagonistic drug ratios intracellularly. Indeed, the Antag formulation delivered an antagonistic drug ratio of MRX-2843 and vincristine in vitro that varied by only 11.43-fold over 24 hours. These findings support the success and importance of the pH gradient-based loading method described here. Drugs contained within the lumen of lipid vesicles are typically released less rapidly than those that reside in the lipid bilayer [14], where lipophilic compounds often reside following passive drug loading or incomplete gradient-based loading [57, 58]. The comparable intracellular drug release profiles between MRX-2843 (clogD7.4=3.07) and vincristine (clogD7.4=1.14) observed here thus suggest that our loading method was not only successful in luminal drug entrapment, but that it was also critical to maintaining ratiometric drug synergy following cell delivery. Further, the intracellular drug concentrations achieved were well in-excess (3x – 100x) of their corresponding GI50, 72h values in LOUCY (ETP-ALL) cells. Together, these findings support both the therapeutic utility and clinical relevance of our described approach to co-formulating MRX-2843 and vincristine.
In addition to ratiometric drug delivery, the synergistic anti-leukemia activity observed in cell cultures treated with the free drug combination was further enhanced following nanoparticle co-encapsulation, a phenomenon that we and others have previously observed with other drug pairs and carriers [15]. While studies to elucidate the mechanisms by which drug co-encapsulation further improves synergy between MRX-2843 and vincristine are ongoing, such effects may be attributable in part to nanoparticle-accelerated intracellular drug transport, augmented target engagement whereby the intracellular kinase domains of both MERTK and FLT3 (enzymatic IC50~ 1.3 and 0.64 nM [34]) are exposed to drug concentrations in excess of those achieved via passive drug diffusion, or differential plasma/serum protein binding. Further, in addition to prominent cell-surface expression, MERTK has also been reported to co-localize with early endosomes [59] where lipid nanoparticles frequently accumulate following cellular uptake [60]. Thus, nanoparticle-mediated drug delivery may augment the magnitude, duration, and/or location of protein inhibition in order to further enhance drug synergy.
Nanoparticles containing MRX-2843 and vincristine were also therapeutically effective against T-ALL cells in blood and bone marrow cultures derived from diagnostic T-ALL patient samples. Both drug synergy and additivity were maintained by Syn and Add drug formulations, respectively, following ex vivo treatment. These findings are significant in that the delivery of drug carriers and transfection reagents to primary T lymphocytes is notoriously difficult [61].
Dual-agent MRX-2843 and vincristine nanoformulations were also therapeutically effective in a murine ETPALL model. While vincristine was the primary driver of efficacy in the studies described here, when the vincristine dose was fixed to allow a direct comparison of particles containing synergistic or additive ratios of MRX-2843 and vincristine, mice treated with Add particles had decreased disease burden and prolonged survival compared to mice treated with Syn particles. Thus, therapeutic efficacy increased with increasing MRX-2843. However, when MRX-2843 was increased even further, therapeutic efficacy decreased relative to the Add particles, consistent with the antagonistic drug interaction observed in cell-based assays at higher concentrations of MRX-2843. These observations have several implications.
First, both vincristine and MRX-2843 contribute to the therapeutic activity mediated by the nanoparticles and the activity of both drugs is dose dependent. In Syn and Add nanoparticles, ratiometric dosing limits exposure to MRX-2843. Thus, the Syn formulation, with its lower dose of MRX-2843, does not attain maximum efficacy, despite a more favorable interaction between drugs. These studies demonstrate that in at least some cases, increasing drug exposure should be prioritized over maintenance of ratiometric dosing to optimize therapeutic benefit for translational applications, even if the interaction between drugs may be less synergistic under these conditions. This is likely to be particularly relevant in cases where toxicity is driven primarily by one drug in a combination. In contrast, interactions between drugs may be more important when the drugs are synergistic at ratios where both can be administered at or near their maximum tolerated or maximally effective doses. In addition, because nanoparticles containing only MRX-2843 do not have therapeutic activity in vivo, but MRX-2843 still provided enhanced efficacy when combined with vincristine, the interaction between MRX-2843 and vincristine is synergistic in vivo, even when the ratiometric dose of drugs provides an additive response in cell-based assays.
In addition, while Add nanoparticles were more effective than Syn nanoparticles, increasing the concentration of MRX-2843 to a molar ratio that is predicted to be more antagonistic reduced therapeutic efficacy, consistent with previous reports comparing ratiometric nanoformulations of other drugs [6, 24, 62, 63]. Thus, the interaction between drugs can drive therapeutic efficacy in vivo in some circumstances and ratiometric dosing is critical to maintain optimal anti-leukemia activity. Ultimately, in vitro modeling alone is insufficient to predict the optimal dose and ratio for application in animal models a priori. Development of computer models that incorporate and interface data describing drug interactions and relative therapeutic activity in cell-based assays as well as toxicity and dosing information for the single agents in mice and humans will be necessary to accurately predict optimal therapeutic doses and ratios for combination therapies.
Taken together, these data demonstrate both the safety and efficacy of nanoparticle formulations of MRX-2843 and vincristine and demonstrate activity mediated by both drugs in this context, with higher doses of either MRX-2843 or vincristine providing greater therapeutic benefit in Syn or Add particles and Add particles providing optimal therapeutic efficacy. Moreover, co-administration of MRX-2843 enhanced sensitivity to vincristine without evidence of increased toxicity. These data suggest that MRX-2843 could be added to current therapeutic regimens utilizing vincristine to improve outcomes for patients with newly diagnosed or relapsed T-ALL. Given the potent anti-leukemia activity and synergistic interaction mediated by MRX-2843 and vincristine and the efficient formulation and delivery of the combination described here, this work may serve to guide future clinical studies of these or related drug formulations incorporating MRX-2843 and/or other anticancer agents for the treatment of ALL.
MATERIALS AND METHODS
Cell Lines and Culture
CCRF-CEM, Jurkat, MOLT-4 and LOUCY were obtained from the American Type Culture Collection (Manassa, VA). 697, PEER and DND-41 were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH (Braunschweig, Germany). Nalm-6, KOPN-8 and UoC-B1 were provided by Dr. Christopher Porter (Emory University). REH, RCH-ACV, and RS4;11 were from Dr. Lia Gore (University of Colorado Denver). Cells were cultured in RPMI 1640 with L-Glutamine supplemented with 10–20% heat-inactivated fetal bovine serum or as recommended by the original supplier (ATCC, DSMZ). All cells were cultured in a 5% CO2 humidified atmosphere and were tested for mycoplasma every four months.
High-Throughput Combination Drug Screening
MRX-2843 (Meryx, Inc.; Chapel Hill, NC) and methotrexate (Sigma-Aldrich; Burlington, MA) were reconstituted in dry DMSO and stored in checkerboard array within 384-well polypropylene plates (VWR; Radnor, PA) covered in sterile adhesive plate film prior to use. Vincristine sulfate (Cayman Chemical; Ann Arbor, MI) was solvated in molecular biology grade water (VWR) on the day of experimentation. 15,000 viable cells per well were dispensed into white polystyrene 384-well microplates (Greiner Bio-One) using a Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific; Waltham, MA). Vincristine was subsequently dispensed onto these cell-containing microplates. MRX-2843 and methotrexate were then transferred to these cell + vincristine microplates using a Beckman NX Liquid Handler (Beckman Coulter; Brea, CA) at the Emory Chemical Biology Discovery Center. Cell- and drug-containing plates were then centrifuged for 5 minutes at 135 x g (Eppendorf 5810R; Eppendorf, Hamburg, Germany), then incubated for 72 hours at 37°C in 5% CO2 humidified atmosphere with gas permeable plate sealing film (VWR). All wells had a DMSO concentration of 0.5% v/v with drugs MRX-2843 (25 – 800 nM), methotrexate (2.9375 – 752 nM), and vincristine (0.0875 – 5.6 nM). After 72 hours, viable cell numbers were assessed using CellTiter-Glo 2.0 Assay (Promega; Madison, WI) and the Integra Viaflo Assist Pipetting Platform (Integra Biosciences; Hudson, NH) according to manufacturer’s instructions. Luminescence was measured using a SpectraMax iD3 plate reader (Molecular Devices; San Jose, CA) and viability data were background corrected to empty wells containing DMSO/water/media and normalized to positive control cells treated with DMSO. All experiments were performed in quadruplicate using the following metrics as criteria for acceptability: CV<20% (positive and negative controls), SNR≥5 (positive controls), Z’≥0.5 (per plate). Drug synergy was calculated using the Bliss Independence model, reported as GI% beyond the expected additive value, and nominally classified using >1% and <−1% synergy as cutoffs for synergistic and antagonistic effects, respectively. Alternative calculations of drug synergy were performed using the Response Additivity [64] and Zero Interaction Potency (ZIP) [65] models.
Screening Validation
High-throughput screening (HTS) results were validated using an alternate assay plate format and new drug stocks and cell lots. Briefly, MRX-2843 and vincristine sulfate were dissolved in molecular biology grade water (VWR) on the day of experimentation, then serially diluted in cell media to obtain 0.01% v/v of water in all media dilutions. 45,000 viable LOUCY or Jurkat cells were then dispensed into drug/media-containing white polystyrene 96-well μClear microplates and subsequently handled as described for HTS experiments.
2- and 3-drug Effect Models
To determine whether synergy emerged from 2-drug or 3-drug interactions, we calculated pairwise and triplet combination synergy and antagonism using both lower order and higher order predictive models. For the lower order model, an Isserlis-like formula developed by Wood et al. [39] was used,
where gi, gij, and gijk refer to cell growth rates when exposed to single, pairwise, or triplet drug combinations, respectively, of drugs i, j, and k. When comparing predictions with measured effects, the Isserlis-like formula may indicate whether drug interactions can be explained by single or pairwise drug responses. For the higher order model, Bliss Independence was similarly used to assess correlations between measured and predicted values.
Microwell Assay
Jurkat cells were embedded in 3 mg/mL collagen hydrogels (Corning; Corning, NY) and exposed concurrently to overlapping concentration gradients of free drug or solvent controls for 72 hours. Following drug exposure, cell viability was assessed using viability dyes Calcein AM (Thermo Fisher) and Propidium Iodide (Thermo Fisher), and nuclei were stained using Hoechst 33342. Images were acquired using a Keyence BZ-X800 Fluorescence Microscope. Cell segmentation was performed using CellProfiler (v3.1, Broad Institute; Cambridge, MA), and cells were classified as live or dead based on mean per cell intensity measurements done in MATLAB (MATLAB 2021a; Mathworks, Natick, MA). A rolling ball averaging method was then used to determine average viability at each drug combination dose ratio. Expected viabilities were computed based on Loewe Additivity isobolograms of single drugs at 10% of the respective maximum doses whereby the line of additivity is assumed to be a linear combination between the maximum effect at the highest concentrations of drug 1 and drug 2. Deviation from the line of additivity towards lower viability indicates synergy.
RNA sequencing (RNAseq)
Three separate Jurkat (T-ALL) cultures were divided and exposed for 72 hours to either a free drug combination comprising MRX-2843 (100 nM) and vincristine (1.4 nM, water), its component monotherapies, or vehicle (DMSO, 0.5% v/v) in separate cell culture flasks (0.6e6 live cells/mL) at 37°C in 5% CO2. At harvest, cells were centrifuged (200 × g, 5 min) and aspirated three times to remove dead cells. Isolated cell pellets were flash frozen on dry ice and stored at −80°C. RNA was extracted from frozen cell pellets using the miRNEasy mini kit (Qiagen, Cat. No. 217004; Maryland, USA). RNA quantification was determined using the Infinite M200Pro (Tecan; Zurich, Switzerland). RNA quality was assessed using the Bioanalyzer RNA 6000 Nano or Pico reagents (Agilent). RNA extracts were sequenced and analyzed by Novogene. RNA libraries were prepared using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina®. Libraries were then read [66] with the NovaSeq 6000 using PE150 sequencing.
Downstream analysis, including read alignments, reference genome mapping, gene expression quantification and differential expression, was performed using a combination of programs [67–73] including STAR, HTseq, Cufflinks [74] and Novogene wrapped scripts. Alignments were parsed using TopHat program. [72, 73] Reference genome and gene model annotation files were downloaded from genome website browsers (NCBI/UCSC/Ensembl) directly. Indexes of the reference genome were built using STAR and paired-end clean reads were aligned to the reference genome using STAR (v2.5). STAR used the method of Maximal Mappable Prefix (MMP), which can generate a precise mapping result for junction reads. HTSeq v0.6.1 was used to count the read numbers mapped for each gene. FPKM (Fragments Per Kilobase of transcript sequence per Million base pairs sequenced) [75] of each gene was calculated based on the length of the gene and read counts mapped to the same gene, where the threshold for expression was set to FPKM>1. [74, 76] Differential expression analysis was performed using the DESeq2 R package (2_1.6.3). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using Benjamini and Hochberg’s approach which controls for the False Discovery Rate (FDR). Genes with an adjusted P value <0.05 found by DESeq2 were assigned as differentially expressed genes (DEGs). TFCat [74] and the COSMIC database were used to annotate differentially expressed genes. TFCat is a curated catalog of mouse and human transcription factors based on a reliable core collection of annotations obtained by expert review of the scientific literature.
Differential Gene Expression Analyses
DEGs identified by RNAseq were organized by hierarchical clustering (Morpheus, https://software.broadinstitute.org/morpheus). DEG expression levels (FPKM) were used as input values, which were then organized using the Euclidean distance of average linkages between clusters. MRX-2843 and vincristine-induced DEGs were analyzed for functional enrichment using the Metascape portal (https://metascape.org). As described by Zhou et al., [77] DEGs were converted into Gene IDs and then queried for membership across multiple databases, including GO, KEGG, Reactome and WikiPathways. DEGs were analyzed for functional enrichment in biological processes using the hypergeometric distribution. Functional terms with P values <0.01 were then hierarchically clustered into trees based on Kappa similarity scores. [78] Functional terms were considered to be similar with gene membership similarity score ≥0.3. [77] Functional terms with high similarity were grouped as clusters, and representative terms within each cluster were selected to eliminate intra-cluster term redundancy while also establishing differences between clusters. Ontology clustering was visualized using Cytoscape (v 3.7.2).
Cheminformatics Computation
All calculations of MRX-2843 and vincristine physicochemical properties (molecular charge, lipophilicity) were performed using ACD/Labs software and SMILES representations of chemical structures. In brief, ionizable microspecies were identified within the parent molecular structures and partial charge distributions were predicted based on orbital electronegativities of constituent atoms. Total molecular charge was then computed as the weighted sum of net charge for each microspecies based on their relative distributions per unit pH (step size: 0.1) and pKa. Lipophilicity was calculated as the logarithm of the Distribution coefficient (cLogD), which is the concentration ratio of all microspecies in octanol compared to water. The partition coefficient (octanol:water) of each microspecies was computed using pre-defined values of structural fragments [79–81].
Lipid Nanoparticle Formulation
DSPC (NOF Corporation; Shanghai, China), DSPG (NOF), and cholesterol (Sigma-Aldrich) were combined at a 7:2:1 mole ratio from lipid stocks solvated to 25 mg/mL in chloroform (DSPC, cholesterol) or 5 mg/mL chloroform:methanol (5:1 v/v, DSPG). Solvent from the lipid mixture was removed via rotary evaporation under vacuum at 30°C (Rotavapor R-100; Buchi, New Castle, DE) and the resulting lipid cake was further dried under house vacuum overnight. Lipid films were rehydrated via rapid addition of ammonium sulfate buffer (pH 4.25, 500 – 600 mM) followed by high power cup horn sonication at 60° C (Q700; Qsonica, Newton, CT). Multilamellar vesicles were then extruded ten times at (60°C) through 0.08 μm polycarbonate membranes (Nuclepore; Whatman) supported by polyester drain discs (230600; Whatman) using a high pressure N2 extruder (Liposofast LF-50; Avestin, Ottawa, ON, Canada). Ammonium sulfate buffer was then exchanged for phosphate buffer (pH 7.7) via centrifugal diafiltration (100kDa Amicon; EMD Millipore, Burlington, MA) or tangential flow filtration (Krosflo KR2i; Repligen, Waltham, MA) and unilamellar vesicles were then sterile filtered through 0.45 μm cellulose acetate membranes (VWR).
To prepare drug-loaded lipid nanoparticles, MRX-2843 (10.0 mg/mL in molecular biology grade water) and vincristine (sulfate) (1.0 mg/mL in molecular biology grade water) were mixed at various molar ratios with freshly prepared, concentrated gradient liposomes. Drug and lipid nanoparticle mixtures were then agitated at 700 rpm (60° C) for 1 hour (Mixer HC; USA Scientific, Ocala, FL). Drug-loaded liposomes were dialyzed twice in phosphate buffered saline using 100 kDa MWCO dialysis bags (Float-A-Lyzer G2; Spectrum Laboratories).
UPLC-MS Analysis of Liposomal Drug Concentrations
Concentrations of MRX-2843, vincristine (sulfate), and DSPC were measured by LC-MS using a Vanquish Horizon UPLC (Thermo Fisher Scientific) fitted with a Waters Corporation ACQUITY UPLC BEH C18 column (2.1×100 mm, 1.7 μm particle size) coupled to a high-resolution accurate mass Orbitrap ID-X Tribrid mass spectrometer (Thermo Fisher Scientific). The chromatographic method for sample analysis involved elution with 20:80 MeCN:water + 10 mM ammonium formate + 0.1% formic acid (mobile phase A) and 10:90 MeCN:IPA + 10 mM ammonium formate + 0.1% formic acid (mobile phase B) with flow rate at 0.4 mL/min and column temperature set to 50°C. In a separate iteration of drug quantification, drug analytes were measured using a Waters Corporation Cortecs UPLC T3 column (2.1×150mm, 1.6 μm particle size). The chromatographic gradient for MRX-2843 and vincristine (sulfate) involved elution with water + 0.1% formic acid (mobile phase A) and MeCN + 0.1% formic acid (mobile phase B) with flow rate at 0.6 mL/min and column temperature set to 40°C. The chromatographic method for DSPC involved elution with water + 0.1% formic acid (mobile phase A) and 90:10 IPA:MeCN + 0.1% formic acid + 10mM ammonium formate (mobile phase B) with flow rate at 0.25 mL/min and column temperature set to 40°C. UPLC-MS2 experiments were performed by acquiring mass spectra with targeted MS/MS (tMS2) acquisition. Data processing was performed with Thermo Scientific Xcalibur Version 4.3.73.11. The MSMS transitions were integrated, and the data was exported to excel. The transitions for each adduct were summed and quantified from standard calibration curves.
Alternatively, drug concentrations were measured by LC-MS using an Agilent 6120 mass spectrometer (Agilent Technologies; Santa Clara, CA) with an Agilent 1220 Infinity liquid chromatography inlet. The chromatographic method involved a linear gradient of 5% acetonitrile in water (0.1% formic acid, mobile phase A) and 80% acetonitrile in water (0.1% formic acid, mobile phase B). Elution occurred over 2 minutes an Agilent Zorbax SB-C18 column (1.8 μm particle size, 2.1 mm × 50 mm) with a flow rate of 0.650 mL/min at ambient temperature. Drug concentration was quantified by integrating UV absorbance at 254 nm and normalized against an internal standard (ketoconazole) peak area.
Characterization of Lipid Formulations
For downstream in vitro and ex vivo assays of growth inhibitory synergy, dual-agent MRX-2843:vincristine nanoparticles were mixed at defined ratios that recapitulated observed synergy, additivity, and antagonism in vitro. To determine these molar ratios, combination responses observed via HTS were averaged for all doses tested. Ratiometric responses were analyzed for mean synergistic responses across T-ALL and ETP-ALL cell lines by filtering doses of MRX-2843 (>200 nM) and vincristine (>2.8 nM) near or slightly above the mean lineage GI50s of 208 nM and 2.6 nM, respectively.
Hydrodynamic size and polydispersity were obtained via dynamic light scattering (DLS) (DynaPro, Plate Reader III; Wyatt Technologies, Santa Barbara, CA). Nanoparticles were syringe filtered through 0.45 μm cellulose acetate membranes (VWR) and briefly sonicated prior to size measurements at fixed temperature (25°C) in black polystyrene microwell plates (Greiner Bio-One). Routine observations of all dual-agent, single-agent, and non-drug loaded formulations stored at 4°C over a 120-day period revealed no diameter changes >11% from baseline. Size and polydispersity stability were evaluated for nanoparticles incubating in heat-inactivated fetal bovine serum (0%, 10%, 50% v/v) and at 37°C for up to 72 hours via number abundance.
Nanoparticle morphology was imaged via transmission electron microscopy (TEM) at 80 kV using a Hitachi HT-7700 instrument. Liposomes were diluted in ultrapure water and applied to charge coated formvar/carbon coated copper grids (400 mesh, Electron Microscopy Sciences; Hatfield, PA) for 15 minutes followed by washing for 2 seconds with ultrapure water. Samples were then negatively stained with phosphotungstic acid for 20 seconds and washed again for 2 seconds with ultrapure water.
Lipid nanoparticles were lyophilized to test for the effects of freeze-dried storage on drug activity and drug retention. Formulations were lyophilized by mixing in 99.3 mg/mL Trehalose (Sigma) in PBS and snap freezing in liquid nitrogen. Formulations were then lyophilized under lower pressure (<0.05 mBar) and temperature (−50°C) (Labconco FreeZone; Kansas City, MO), and subsequently stored in −80°C. Long-term storage effects on drug activity were evaluated for Syn and Add formulations by measuring growth inhibitory dose responses in LOUCY cells as described for the Screening Validation. Drug retention was evaluated by comparing drug concentrations of reconstituted dual-agent and single-agent lyophilisates to spin filtered filtrates (100kDa Amicon Ultracel; EMD Millipore) via LC-MS.
Zeta-potential of nanoparticles dispersed in deionized water were measured with a Zetasizer Nano ZS (Malvern Panalytical Ltd; Malvern, UK) using the Smoluchowski model.
Intracellular Drug Delivery
LOUCY cells were exposed to Antag nanoparticles (10x HTS dose) under normal culture conditions. Cells were washed 3x in PBS and counted prior to snap-freezing on dry ice after 1, 3 and 24 hours of nanoparticle exposure. Frozen cell pellets were prepared for LC-MS analysis by mixing in 500 μL IPA and 50 μL of 500 μm glass beads. Samples were then homogenized in a Tissuelyzer II (QIAGEN) at 30 Hz for 5 minutes and the supernatant was transferred to separate microcentrifuge vials. The original samples in glass beads were washed twice with 100 μL of 80% MeOH, with each wash additive to the IPA supernatant. The cellular extractions were then dried by vacuum centrifugation and reconstituted in 250 μL of 80% MeOH. Reconstitution was assisted by sonication for 5 minutes. The samples were centrifuged at 21,100 x g prior to being transferred to LC vials for LC-MS analysis as described above. LOUCY cells not exposed to nanoparticles were processed as the negative control at 0 hours of exposure.
Drug Release Kinetics
Nanoparticles (1 mL) were suspended in 300 kDa MWCO dialysis bags (Float-A-Lyzer G2; Spectrum Laboratories) in 1 liter of either PBS (pH=7.4) or citrate buffer (pH=5.5) at 37°C under continuous stirring. Aliquots from diluent buffer (filtrates) were withdrawn for drug quantitation (LC-MS) at various time intervals up to 72 hours. Samples were diluted in MeOH and centrifuged at 21,100 x g for 5 minutes prior to LC-MS analysis. MRX-2843 lower limit of detection (LLOD) =0.19 ng/mL< or 0.023% of starting sample mass released into diluent buffer; vincristine LLOD=1.20 ng/mL, or 5.52% of starting sample mass released into dialysis buffer.
In vitro Syn Nanoformulation Dose Responses
LOUCY and Jurkat cells were treated with dose response series of Syn, Add, or Antag nanoparticles, their constituent free drug monotherapies, and free drug combinations matched for vincristine doses. MRX-2843 was dosed in multiples of 30 nM to mimic the dose effects of Syn, Add, and Antag molar ratios (MRX-2843:vincristine) at 30.13, 127.59, and 246.06. Free drug and nanoparticles were dissolved in PBS vehicle (0.05% v/v) in media on white 96-well polystyrene μClear Microplates. 45,000 viable cells (Countess IIl Invitrogen) were added per well and cultured for 72 hours. Cells were then treated with CellTiter-Glo 2.0 (Promega) and analyzed for luminescence with a SpectraMax iD3 (Molecular Devices). Data represent triplicate measurements from a single experiment.
Primary Sample Responses to Ratiometric Nanoformulations
Viably frozen, de-identified blood and bone marrow samples collected from patients with T-ALL at initial diagnosis were obtained from the Aflac Cancer and Blood Disorders Center Leukemia and Lymphoma Biorepository at Children’s Healthcare of Atlanta. Cells (12,500 per 384-well) were cultured in cytokine-sera, consisting of 20 ng/mL human recombinant IL-7, IGF-1, and SCF (StemCell Technologies; Vancouver, BC, Canada) in 90% RPMI + 10% heat inactivated FBS + 0.1 mM beta-mercaptoethanol (Sigma) [82–84] and exposed in triplicate to Syn or Add nanoparticles for 96 hours. Expected GI values for an additive interaction were interpolated from dose response curves (GraphPad Prism) of free vincristine and liposomal MRX-2843 using the Bliss Independence model of drug synergy. Each well contained 0.5% v/v PBS/media. Positive controls were cells treated with PBS/media, and negative controls contained PBS/media but no cells. Following incubation, cultures were treated with CellTiter-Glo 2.0 and relative viable cell numbers were measured as described above.
Animal Studies
A luciferase-expressing ALL cell line (LOUCY-luc) was injected into the tail vein of NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ (NRG) or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (2×106 or 3×106 cells/mouse). Leukemia engraftment and disease burden were monitored by bioluminescence imaging as previously described [85]. Briefly, mice were anesthetized with isoflurane and then sterile filtered D-luciferin in PBS (150 mg/kg) was injected intraperitoneally (i.p.) at a dose of 10 mL/kg. Bioluminescence images were captured on an IVIS Lumina Series 3 (PerkinElmer; Waltham, MA) and quantitated with Living Image software. Mice were randomized to treatment groups with statistically equal starting disease burden (data not shown) and nanoparticles were administered by i.p. injection at a dose of 20 mL/kg unless otherwise noted. Health status was evaluated daily using a standardized scoring system and mice with >20% weight loss or meeting other standardized criteria [33] were removed from study and euthanized. All animal protocols have been reviewed and approved by the Emory University Institutional Animal Care and Use Committee (PROTO201700276).
Statistical Analyses and Software
All dose response series were modeled as nonlinear 4-parameter logistic functions using GraphPad Prism. Multidimensional synergy plots were produced in Tecplot 360. Differences between lipid nanoparticle and free drug dose responses in cell lines were measured by one-way ANOVA with Tukey’s multiple comparisons test. Observed and expected dose responses (GI50) for primary samples were compared by unpaired parametric t test. Heat maps for primary sample potencies were rendered in GraphPad Prism. Experimental diagrams were drawn using BioRender software. Survival comparisons in mice were made with the Mantel-Cox log-rank method. Disease burden was compared by two-way ANOVA. Bioluminescence data were collected until fewer than half of mice in that group remained on study. Maximum bioluminescence and body mass values were carried forward for each subject after removal.
Supplementary Material
Fig. S1: B-ALL lineage-specific drug synergy
Fig. S2: Experimental validations of high-throughput screening
Fig. S3: Computational assessment of lower- and higher-order drug synergy per cell line
Fig. S4: RNA sequencing analysis of synergistic MRX-2843 and vincristine ontologies
Fig. S5: Characterization of lipid nanoparticle formulations
Fig. S6: Nanoparticle size stability post-lyophilization and in serum
Fig. S7: Therapeutic efficacy and toxicity in vivo
Fig. S8: Activity and safety of MRX-2843 single-agent formulations in vivo
Fig. S9: In vivo body mass following treatment
Highlights.
MRX-2843 and vincristine mediate ratio-dependent synergistic anti-leukemia activity
Liposomes deliver defined mole ratios of MRX-2843 and vincristine to cells
Ratiometric nanoparticles constitutively synergize against primary T-ALL cells
MRX-2843 and vincristine nanoformulations are therapeutically effective in vivo
In vivo efficacy of the nanoformulations is dependent on drug ratio and drug dose
ACKNOWLEDGEMENTS
This work was supported in part by research grants from CURE Childhood Cancer (DD), No More Kids With Cancer (DKG, DD), the National Institutes of Health Research Training Program in Immunoengineering (T32EB021962), and the Coulter Department of Biomedical Engineering. We gratefully acknowledge Peter Van Vlierberghe for providing the luciferase-expressing LOUCY cell line. The RNAseq study was supported in part by the Emory Integrated Genomics Core (EIGC), subsidized by the Emory University School of Medicine. We are also grateful for assistance from the Pediatric General Equipment Core, the Robert P. Apkarian Integrated Electron Microscopy Core, Georgia Institute of Technology’s Systems Mass Spectrometry Core Facility, and the Emory Chemical Biology Discovery Center. Patient samples were provided by the Aflac Cancer and Blood Disorders Leukemia and Lymphoma Biorepository at Children’s Healthcare of Atlanta; other investigators may have received specimens from the same subjects. The content here is solely the responsibility of the authors and does not necessarily represent the official views of the organizations acknowledged herein.
Footnotes
COMPETING INTERESTS
D.K.G. is a founder and serves on the Board of Directors of Meryx, Inc. X.W., D.K.G., and D.D. are equity holders in Meryx, Inc. X.W. is inventor on a patent for MRX-2843. J.M.K., J.J., D.K.G., D.D., and E.C.D. are inventors on a patent related to this work describing combination drug screening, formulation, and treatment.
CRediT Author Statement
James M Kelvin: Conceptualization, Methodology, Validation, Investigation, Writing – Original Draft, Writing – Review and Editing, Visualization
Madison L Chimenti: Investigation, Validation
Dan Y Zhang: Software, Formal analysis
Evelyn K Williams: Investigation, Validation
Samuel G Moore: Investigation, Validation
Gabrielle M Humber: Investigation
Travon A Baxter: Investigation
Lacey A Birnbaum: Investigation
Min Qui: Investigation
Henry Zecca: Investigation
Aashis Thapa: Methodology
Juhi Jain: Investigation
Nathan T Jui: Conceptualization, Resources
Xiaodong Wang: Conceptualization, Resources
Haian Fu: Conceptualization, Resources
Yuhong Du: Conceptualization, Resources, Writing – Review and Editing
Melissa L Kemp: Conceptualization, Resources, Writing – Review and Editing
Wilbur A Lam: Conceptualization, Resources, Writing – Review and Editing
Douglas K Graham: Conceptualization, Resources, Writing – Review and Editing, Funding acquisition
Deborah DeRyckere: Conceptualization, Methodology, Formal Analysis, Resources, Writing – Original Draft, Writing – Review and Editing, Supervision, Funding acquisition
Erik C Dreaden: Conceptualization, Methodology, Resources, Writing – Original Draft, Writing – Review and Editing, Visualization, Supervision, Funding acquisition
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Frei III E, Karon M, Levin RH, Freireich EJ, Taylor RJ, Hananian J, Selawry O, Holland JF, Hoogstraten B, Wolman IJ, The effectiveness of combinations of antileukemic agents in inducing and maintaining remission in children with acute leukemia, Blood, 26 (1965) 642–656. [PubMed] [Google Scholar]
- [2].Moxley JH, De Vita VT, Brace K, Frei E, Intensive Combination Chemotherapy and X-irradiation in Hodgkin’s Disease, Cancer Res, 27 (1967) 1258–1263. [PubMed] [Google Scholar]
- [3].Devita V, Serpick A, Carbone P, Combination Chemotherapy in the Treatment of Advanced Hodgkin’s Disease, Ann. Intern. Med, 73 (1970) 881–895. [DOI] [PubMed] [Google Scholar]
- [4].Palmer AC, Chidley C, Sorger PK, A curative combination cancer therapy achieves high fractional cell killing through low cross-resistance and drug additivity, eLife, 8 (2019) e50036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bayne WF, Mayer LD, Swenson CE, Pharmacokinetics of CPX-351 (cytarabine/daunorubicin HCl) liposome injection in the mouse, Journal of pharmaceutical sciences, 98 (2009) 2540–2548. [DOI] [PubMed] [Google Scholar]
- [6].Harasym TO, Liboiron BD, Mayer LD, Drug ratio-dependent antagonism: a new category of multidrug resistance and strategies for its circumvention, Methods in molecular biology (Clifton, N.J.), 596 (2010) 291–323. [DOI] [PubMed] [Google Scholar]
- [7].Lim WS, Tardi PG, Dos Santos N, Xie X, Fan M, Liboiron BD, Huang X, Harasym TO, Bermudes D, Mayer LD, Leukemia-selective uptake and cytotoxicity of CPX-351, a synergistic fixed-ratio cytarabine:daunorubicin formulation, in bone marrow xenografts, Leukemia research, 34 (2010) 1214–1223. [DOI] [PubMed] [Google Scholar]
- [8].Cokol M, Chua HN, Tasan M, Mutlu B, Weinstein ZB, Suzuki Y, Nergiz ME, Costanzo M, Baryshnikova A, Giaever G, Nislow C, Myers CL, Andrews BJ, Boone C, Roth FP, Systematic exploration of synergistic drug pairs, Mol. Syst. Biol, 7 (2011) 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehár J, Price ER, Serbedzija G, Zimmermann GR, Foley MA, Stockwell BR, Keith CT, Systematic discovery of multicomponent therapeutics, Proc. Natl. Acad. Sci. U.S.A, 100 (2003) 7977–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Eetezadi S, Evans JC, Shen Y-T, De Souza R, Piquette-Miller M, Allen C, Ratio-Dependent Synergism of a Doxorubicin and Olaparib Combination in 2D and Spheroid Models of Ovarian Cancer, Mol. Pharmaceutics, 15 (2018) 472–485. [DOI] [PubMed] [Google Scholar]
- [11].Shen YT, Evans JC, Zafarana G, Allen C, Piquette-Miller M, BRCA Status Does Not Predict Synergism of a Carboplatin and Olaparib Combination in High-Grade Serous Ovarian Cancer Cell Lines, Mol. Pharmaceutics, 15 (2018) 2742–2753. [DOI] [PubMed] [Google Scholar]
- [12].Ma L, Kohli M, Smith A, Nanoparticles for Combination Drug Therapy, ACS Nano, 7 (2013) 9518–9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sengupta S, Eavarone D, Capila I, Zhao G, Watson N, Kiziltepe T, Sasisekharan R, Temporal targeting of tumour cells and neovasculature with a nanoscale delivery system, Nature, 436 (2005) 568–572. [DOI] [PubMed] [Google Scholar]
- [14].Morton S, Lee M, Deng Z, Dreaden E, Siouve E, Shopsowitz K, Shah N, Yaffe M, Hammond P, A Nanoparticle-Based Combination Chemotherapy Delivery System for Enhanced Tumor Killing by Dynamic Rewiring of Signaling Pathways, Sci. Signal, 7 (2014) ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dreaden EC, Kong YW, Morton SW, Correa S, Choi KY, Shopsowitz KE, Renggli K, Drapkin R, Yaffe MB, Hammond PT, Tumor-Targeted Synergistic Blockade of MAPK and PI3K from a Layer-by-Layer Nanoparticle, Clin. Cancer Res, 21 (2015) 4410–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Houdaihed L, Evans JC, Allen C, In Vivo Evaluation of Dual-Targeted Nanoparticles Encapsulating Paclitaxel and Everolimus, Cancers (Basel: ), 11 (2019) 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Goldman A, Kulkarni A, Kohandel M, Pandey P, Rao P, Natarajan SK, Sabbisetti V, Sengupta S, Rationally Designed 2-in-1 Nanoparticles Can Overcome Adaptive Resistance in Cancer, ACS Nano, 10 (2016) 5823–5834. [DOI] [PubMed] [Google Scholar]
- [18].Mikhail AS, Eetezadi S, Allen C, Multicellular Tumor Spheroids for Evaluation of Cytotoxicity and Tumor Growth Inhibitory Effects of Nanomedicines In Vitro: A Comparison of Docetaxel-Loaded Block Copolymer Micelles and Taxotere®, PLOS ONE, 8 (2013) e62630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Do P, Perdue LA, Chyong A, Hunter R, Dougan J, Henry CJ, Porter CC, Dreaden EC, Rapid Assembly and Screening of Multivalent Immune Cell-Redirecting Therapies for Leukemia, ACS Comb. Sci, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kong YW, Dreaden EC, Morandell S, Zhou W, Dhara SS, Sriram G, Lam FC, Patterson JC, Quadir M, Dinh A, Shopsowitz KE, Varmeh S, Yilmaz ÖH, Lippard SJ, Reinhardt HC, Hemann MT, Hammond PT, Yaffe MB, Enhancing chemotherapy response through augmented synthetic lethality by co-targeting nucleotide excision repair and cell-cycle checkpoints, Nat. Commun, 11 (2020) 4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Choi KY, Correa S, Min J, Li J, Roy S, Laccetti KH, Dreaden E, Kong S, Heo R, Roh YH, Lawson EC, Palmer PA, Hammond PT, Binary Targeting of siRNA to Hematologic Cancer Cells In Vivo Using Layer-by-Layer Nanoparticles, Adv. Funct. Mater, 29 (2019) 1900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT, Layer-by-Layer Nanoparticles for Systemic Codelivery of an Anticancer Drug and siRNA for Potential Triple-Negative Breast Cancer Treatment, ACS Nano, 7 (2013) 9571–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Alfayez M, Kantarjian H, Kadia T, Ravandi-Kashani F, Daver N, CPX-351 (vyxeos) in AML, Leukemia & lymphoma, 61 (2020) 288–297. [DOI] [PubMed] [Google Scholar]
- [24].Tardi P, Johnstone S, Harasym N, Xie S, Harasym T, Zisman N, Harvie P, Bermudes D, Mayer L, In vivo maintenance of synergistic cytarabine:daunorubicin ratios greatly enhances therapeutic efficacy, Leukemia research, 33 (2009) 129–139. [DOI] [PubMed] [Google Scholar]
- [25].Hu Y, Caldwell KJ, Onciu M, Federico SM, Salek M, Lewis S, Lei S, Zhang J, Nichols KE, Takemoto CM, Triplett BM, Farrar JE, Rubnitz JE, Ribeiro RC, Wlodarski MW, CPX-351 induces remission in newly diagnosed pediatric secondary myeloid malignancies, Blood Adv, 6 (2022) 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Krauss AC, Gao X, Li L, Manning ML, Patel P, Fu W, Janoria KG, Gieser G, Bateman DA, Przepiorka D, Shen YL, Shord SS, Sheth CM, Banerjee A, Liu J, Goldberg KB, Farrell AT, Blumenthal GM, Pazdur R, FDA Approval Summary: (Daunorubicin and Cytarabine) Liposome for Injection for the Treatment of Adults with High-Risk Acute Myeloid Leukemia, Clinical cancer research : an official journal of the American Association for Cancer Research, 25 (2019) 2685–2690. [DOI] [PubMed] [Google Scholar]
- [27].Lancet JE, Uy GL, Cortes JE, Newell LF, Lin TL, Ritchie EK, Stuart RK, Strickland SA, Hogge D, Solomon SR, Stone RM, Bixby DL, Kolitz JE, Schiller GJ, Wieduwilt MJ, Ryan DH, Hoering A, Chiarella M, Louie AC, Medeiros BC, Final results of a phase III randomized trial of CPX-351 versus 7+3 in older patients with newly diagnosed high risk (secondary) AML, Journal of Clinical Oncology, 34 (2016) 7000–7000. [Google Scholar]
- [28].Brandao LN, Winges A, Christoph S, Sather S, Migdall-Wilson J, Schlegel J, McGranahan A, Gao D, Liang X, DeRyckere D, Graham DK, Inhibition of MerTK increases chemosensitivity and decreases oncogenic potential in T-cell acute lymphoblastic leukemia, Blood Cancer Journal, 3 (2013) e101–e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Keating AK, Salzberg DB, Sather S, Liang X, Nickoloff S, Anwar A, Deryckere D, Hill K, Joung D, Sawczyn KK, Park J, Curran-Everett D, McGavran L, Meltesen L, Gore L, Johnson GL, Graham DK, Lymphoblastic leukemia/lymphoma in mice overexpressing the Mer (MerTK) receptor tyrosine kinase, Oncogene, 25 (2006) 6092–6100. [DOI] [PubMed] [Google Scholar]
- [30].Lee-Sherick AB, Eisenman KM, Sather S, McGranahan A, Armistead PM, McGary CS, Hunsucker SA, Schlegel J, Martinson H, Cannon C, Keating AK, Earp HS, Liang X, DeRyckere D, Graham DK, Aberrant Mer receptor tyrosine kinase expression contributes to leukemogenesis in acute myeloid leukemia, Oncogene, 32 (2013) 5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Linger RMA, Lee-Sherick AB, DeRyckere D, Cohen RA, Jacobsen KM, McGranahan A, Brandão LN, Winges A, Sawczyn KK, Liang X, Keating AK, Tan AC, Earp HS, Graham DK, Mer receptor tyrosine kinase is a therapeutic target in pre–B-cell acute lymphoblastic leukemia, Blood, 122 (2013) 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Daver N, Schlenk RF, Russell NH, Levis MJ, Targeting FLT3 mutations in AML: review of current knowledge and evidence, Leukemia, 33 (2019) 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].DeRyckere D, Lee-Sherick AB, Huey MG, Hill AA, Tyner JW, Jacobsen KM, Page LS, Kirkpatrick GG, Eryildiz F, Montgomery SA, Zhang W, Wang X, Frye SV, Earp HS, Graham DK, UNC2025, a MERTK Small-Molecule Inhibitor, Is Therapeutically Effective Alone and in Combination with Methotrexate in Leukemia Models, Clinical cancer research : an official journal of the American Association for Cancer Research, 23 (2017) 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Minson KA, Smith CC, DeRyckere D, Libbrecht C, Lee-Sherick AB, Huey MG, Lasater EA, Kirkpatrick GD, Stashko MA, Zhang W, Jordan CT, Kireev D, Wang X, Frye SV, Earp HS, Shah NP, Graham DK, The MERTK/FLT3 inhibitor MRX-2843 overcomes resistance-conferring FLT3 mutations in acute myeloid leukemia, JCI insight, 1 (2016) e85630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang W, DeRyckere D, Hunter D, Liu J, Stashko MA, Minson KA, Cummings CT, Lee M, Glaros TG, Newton DL, Sather S, Zhang D, Kireev D, Janzen WP, Earp HS, Graham DK, Frye SV, Wang X, UNC2025, a potent and orally bioavailable MER/FLT3 dual inhibitor, Journal of medicinal chemistry, 57 (2014) 7031–7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang L, Yan K, Zhang Y, Huang R, Bian J, Zheng C, Sun H, Chen Z, Sun N, An R, Min F, Zhao W, Zhuo Y, You J, Song Y, Yu Z, Liu Z, Yang K, Gao H, Dai H, Zhang X, Wang J, Fu C, Pei G, Liu J, Zhang S, Goodfellow M, Jiang Y, Kuai J, Zhou G, Chen X, High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections, Proc. Natl. Acad. Sci. U.S.A, 104 (2007) 4606–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Farha MA, Brown ED, Chemical Probes of Escherichia coli Uncovered through Chemical-Chemical Interaction Profiling with Compounds of Known Biological Activity, Chem. Biol, 17 (2010) 852–862. [DOI] [PubMed] [Google Scholar]
- [38].Lederer S, Dijkstra TMH, Heskes T, Additive Dose Response Models: Defining Synergy, Front. Pharmacol, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wood K, Nishida S, Sontag ED, Cluzel P, Mechanism-independent method for predicting response to multidrug combinations in bacteria, Proceedings of the National Academy of Sciences of the United States of America, 109 (2012) 12254–12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Peirs S, Matthijssens F, Goossens S, Van de Walle I, Ruggero K, de Bock CE, Degryse S, Canté-Barrett K, Briot D, Clappier E, Lammens T, De Moerloose B, Benoit Y, Poppe B, Meijerink JP, Cools J, Soulier J, Rabbitts TH, Taghon T, Speleman F, Van Vlierberghe P, ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia, Blood, 124 (2014) 3738–3747. [DOI] [PubMed] [Google Scholar]
- [41].Cordo’ V, van der Zwet JCG, Canté-Barrett K, Pieters R, Meijerink JPP, T-cell Acute Lymphoblastic Leukemia: A Roadmap to Targeted Therapies, Blood Cancer Discov., 2 (2021) 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Conter V, Valsecchi MG, Buldini B, Parasole R, Locatelli F, Colombini A, Rizzari C, Putti MC, Barisone E, Nigro LL, Santoro N, Ziino O, Pession A, Testi AM, Micalizzi C, Casale F, Pierani P, Cesaro S, Cellini M, Silvestri D, Cazzaniga G, Biondi A, Basso G, Early T-cell precursor acute lymphoblastic leukaemia in children treated in AIEOP centres with AIEOP-BFM protocols: a retrospective analysis, Lancet Haematol, 3 (2016) e80–e86. [DOI] [PubMed] [Google Scholar]
- [43].Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, Biondi A, Pui CH, Downing JR, Campana D, Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia, Lancet Oncol, 10 (2009) 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Haydu JE, Ferrando AA, Early T-cell precursor acute lymphoblastic leukaemia, Curr. Opin. Hematol, 20 (2013) 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Patrick K, Wade R, Goulden N, Mitchell C, Moorman AV, Rowntree C, Jenkinson S, Hough R, Vora A, Outcome for children and young people with Early T-cell precursor acute lymphoblastic leukaemia treated on a contemporary protocol, UKALL 2003, Br. J. Haematol, 166 (2014) 421–424. [DOI] [PubMed] [Google Scholar]
- [46].Wood B, Winter S, Dunsmore K, Raetz E, Borowitz MJ, Devidas M, Winick NJ, Carroll WL, Hunger SP, Loh ML, Patients with Early T-Cell Precursor (ETP) Acute Lymphoblastic Leukemia (ALL) Have High Levels of Minimal Residual Disease (MRD) at the End of induction—A Children’s Oncology Group (COG) Study, Blood, 114 (2009) 9–9. [Google Scholar]
- [47].Stephenson R, Singh A, Drug discovery and therapeutic delivery for the treatment of B and T cell tumors, Advanced drug delivery reviews, 114 (2017) 285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Summers RJ, Jain J, Vasileiadi E, Smith B, Stout M, Kelvin J, Wang X, Frye SV, Earp HS, Tyner JW, Dreaden E, DeRyckere D, Graham DK, Therapeutic Targeting of Mertk and BCL-2 in T-Cell and Early T-Precursor Acute Lymphoblastic Leukemia, Blood, 138 (2021) 1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Neumann M, Coskun E, Fransecky L, Mochmann LH, Bartram I, Sartangi NF, Heesch S, Gökbuget N, Schwartz S, Brandts C, Schlee C, Haas R, Dührsen U, Griesshammer M, Döhner H, Ehninger G, Burmeister T, Blau O, Thiel E, Hoelzer D, Hofmann WK, Baldus CD, FLT3 mutations in early T-cell precursor ALL characterize a stem cell like leukemia and imply the clinical use of tyrosine kinase inhibitors, PloS one, 8 (2013) e53190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen S-C, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui C-H, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG, The genetic basis of early T-cell precursor acute lymphoblastic leukaemia, Nature, 481 (2012) 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Huelse JM, Fridlyand DM, Earp S, DeRyckere D, Graham DK, MERTK in cancer therapy: Targeting the receptor tyrosine kinase in tumor cells and the immune system, Pharmacol. Ther, 213 (2020) 107577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Linger RMA, Cohen RA, Cummings CT, Sather S, Migdall-Wilson J, Middleton DHG, Lu X, Barón AE, Franklin WA, Merrick DT, Jedlicka P, DeRyckere D, Heasley LE, Graham DK, Mer or Axl receptor tyrosine kinase inhibition promotes apoptosis, blocks growth and enhances chemosensitivity of human non-small cell lung cancer, Oncogene, 32 (2013) 3420–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li Z-M, Jiang W-Q, Zhu Z-Y, Zhu X-F, Zhou J-M, Liu Z-C, Yang D-J, Guan Z-Z, Synergistic cytotoxicity of Bcl-xL inhibitor, gossypol and chemotherapeutic agents in non-Hodgkin’s lymphoma cells, Cancer Biol. Ther, 7 (2008) 51–60. [DOI] [PubMed] [Google Scholar]
- [54].Kano Y, Akutsu M, Tsunoda S, Mano H, Sato Y, Honma Y, Furukawa Y, In vitro cytotoxic effects of a tyrosine kinase inhibitor STI571 in combination with commonly used antileukemic agents, Blood, 97 (2001) 1999–2007. [DOI] [PubMed] [Google Scholar]
- [55].Mues M, Karra L, Romero-Moya D, Wandler A, Hangauer MJ, Ksionda O, Thus Y, Lindenbergh M, Shannon K, McManus MT, Roose JP, High-Complexity shRNA Libraries and PI3 Kinase Inhibition in Cancer: High-Fidelity Synthetic Lethality Predictions, Cell Rep., 27 (2019) 631–647.e635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].van Hoogevest P, Tiemessen H, Metselaar JM, Drescher S, Fahr A, The Use of Phospholipids to Make Pharmaceutical Form Line Extensions, Eur. J. Lipid Sci. Technol, 123 (2021) 2000297. [Google Scholar]
- [57].Large DE, Abdelmessih RG, Fink EA, Auguste DT, Liposome composition in drug delivery design, synthesis, characterization, and clinical application, Adv. Drug Deliv. Rev, 176 (2021) 113851. [DOI] [PubMed] [Google Scholar]
- [58].Cullis P, Bally M, Madden T, Mayer L, Hope M, pH gradients and membrane transport in liposomal systems, Trends Biotechnol, 9 (1991) 268–272. [DOI] [PubMed] [Google Scholar]
- [59].Shelby SJ, Colwill K, Dhe-Paganon S, Pawson T, Thompson DA, MERTK Interactions with SH2-Domain Proteins in the Retinal Pigment Epithelium, PLOS ONE, 8 (2013) e53964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Saei AA, Yazdani M, Lohse SE, Bakhtiary Z, Serpooshan V, Ghavami M, Asadian M, Mashaghi S, Dreaden EC, Mashaghi A, Nanoparticle Surface Functionality Dictates Cellular and Systemic Toxicity, Chem. Mater, 29 (2017) 6578–6595. [Google Scholar]
- [61].McKinlay CJ, Benner NL, Haabeth OA, Waymouth RM, Wender PA, Enhanced mRNA delivery into lymphocytes enabled by lipid-varied libraries of charge-altering releasable transporters, Proc. Natl. Acad. Sci. U.S.A, 115 (2018) E5859–E5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Detappe A, Nguyen HVT, Jiang Y, Agius MP, Wang W, Mathieu C, Su NK, Kristufek SL, Lundberg DJ, Bhagchandani S, Ghobrial IM, Ghoroghchian PP, Johnson JA, Molecular bottlebrush prodrugs as mono- and triplex combination therapies for multiple myeloma, Nature Nanotechnology, 18 (2023) 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mayer LD, Harasym TO, Tardi PG, Harasym NL, Shew CR, Johnstone SA, Ramsay EC, Bally MB, Janoff AS, Ratiometric dosing of anticancer drug combinations: controlling drug ratios after systemic administration regulates therapeutic activity in tumor-bearing mice, Molecular cancer therapeutics, 5 (2006) 1854–1863. [DOI] [PubMed] [Google Scholar]
- [64].Foucquier J, Guedj M, Analysis of drug combinations: current methodological landscape, Pharmacology research & perspectives, 3 (2015) e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yadav B, Wennerberg K, Aittokallio T, Tang J, Searching for Drug Synergy in Complex Dose-Response Landscapes Using an Interaction Potency Model, Computational and structural biotechnology journal, 13 (2015) 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jiang L, Schlesinger F, Davis CA, Zhang Y, Li R, Salit M, Gingeras TR, Oliver B, Synthetic spike-in standards for RNA-seq experiments, Genome research, 21 (2011) 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic acids research, 25 (1997) 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM, The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants, Nucleic acids research, 38 (2009) 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloë D, Le Gall C, Schaëffer B, Le Crom S, Guedj M, Jaffrézic F, A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis, Briefings in bioinformatics, 14 (2013) 671–683. [DOI] [PubMed] [Google Scholar]
- [70].Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ, Alta-Cyclic: a self-optimizing base caller for next-generation sequencing, Nature methods, 5 (2008) 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A, The Pfam protein families database: towards a more sustainable future, Nucleic acids research, 44 (2016) D279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data, Genome research, 20 (2010) 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome research, 13 (2003) 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nature Biotechnology, 28 (2010) 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B, Mapping and quantifying mammalian transcriptomes by RNA-Seq, Nature methods, 5 (2008) 621–628. [DOI] [PubMed] [Google Scholar]
- [76].Yu G, Wang LG, Han Y, He QY, clusterProfiler: an R package for comparing biological themes among gene clusters, Omics : a journal of integrative biology, 16 (2012) 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets, Nature communications, 10 (2019) 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Cohen J, A Coefficient of Agreement for Nominal Scales, Educational and Psychological Measurement, 20 (1960) 37–46. [Google Scholar]
- [79].Csizmadia F, Tsantili-Kakoulidou A, Panderi I, Darvas F, Prediction of Distribution Coefficient from Structure. 1. Estimation Method, Journal of Pharmaceutical Sciences, 86 (1997) 865–871. [DOI] [PubMed] [Google Scholar]
- [80].Klopman G, Li J-Y, Wang S, Dimayuga M, Computer Automated log P Calculations Based on an Extended Group Contribution Approach, Journal of Chemical Information and Computer Sciences, 34 (1994) 752–781. [Google Scholar]
- [81].Viswanadhan VN, Ghose AK, Revankar GR, Robins RK, Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics, Journal of Chemical Information and Computer Sciences, 29 (1989) 163–172. [Google Scholar]
- [82].Delgado-Martin C, Meyer LK, Huang BJ, Shimano KA, Zinter MS, Nguyen JV, Smith GA, Taunton J, Winter SS, Roderick JR, Kelliher MA, Horton TM, Wood BL, Teachey DT, Hermiston ML, JAK/STAT pathway inhibition overcomes IL7-induced glucocorticoid resistance in a subset of human T-cell acute lymphoblastic leukemias, Leukemia, 31 (2017) 2568–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Armstrong F, Brunet P Grange de la, Gerby B, Rouyez MC, Calvo J, Fontenay M, Boissel N, Dombret H, Baruchel A, Landman-Parker J, Roméo PH, Ballerini P, Pflumio F, NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity, Blood, 113 (2009) 1730–1740. [DOI] [PubMed] [Google Scholar]
- [84].Yost AJ, Shevchuk OO, Gooch R, Gusscott S, You MJ, Ince TA, Aster JC, Weng AP, Defined, serum-free conditions for in vitro culture of primary human T-ALL blasts, Leukemia, 27 (2013) 1437–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Christoph S, Lee-Sherick AB, Sather S, DeRyckere D, Graham DK, Pre-clinical evaluation of tyrosine kinase inhibitors for treatment of acute leukemia, Journal of visualized experiments : JoVE, (2013) e50720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: B-ALL lineage-specific drug synergy
Fig. S2: Experimental validations of high-throughput screening
Fig. S3: Computational assessment of lower- and higher-order drug synergy per cell line
Fig. S4: RNA sequencing analysis of synergistic MRX-2843 and vincristine ontologies
Fig. S5: Characterization of lipid nanoparticle formulations
Fig. S6: Nanoparticle size stability post-lyophilization and in serum
Fig. S7: Therapeutic efficacy and toxicity in vivo
Fig. S8: Activity and safety of MRX-2843 single-agent formulations in vivo
Fig. S9: In vivo body mass following treatment


