Abstract
Force production in muscle is achieved through the interaction of myosin and actin. Strong binding states in active muscle are associated with Mg·ADP bound to the active site; release of Mg·ADP allows rebinding of ATP and dissociation from actin. Thus, Mg·ADP binding is positioned for adaptation as a force sensor. Mechanical loads on the lever arm can affect the ability of myosin to release Mg·ADP but exactly how this is done is poorly defined. Here we use F-actin decorated with double-headed smooth muscle myosin fragments in the presence of Mg·ADP to visualize the effect of internally supplied tension on the paired lever arms using cryoEM. The interaction of the paired heads with two adjacent actin subunits is predicted to place one lever arm under positive and the other under negative strain. The converter domain is believed to be the most flexible domain within myosin head. Our results, instead, point to the segment of heavy chain between the essential and regulatory light chains as the location of the largest structural change. Moreover, our results suggest no large changes in the myosin coiled coil tail as the locus of strain relief when both heads bind F-actin. The method would be adaptable to double-headed members of the myosin family. We anticipate that the study of actin- myosin interaction using double-headed fragments enables visualization of domains that are typically noisy in decoration with single-headed fragments.
Graphical Abstract
1. Introduction
A feature of muscle contraction is its adaptation to changing load requirements through the release of myosin heads from ordered structures organized on the thick filament backbone which minimize ATP consumption in the absence of a work requirement, to disordered myosin heads that can interact with the thin filament to produce tension and work. Once myosin heads bind the thin filament, they undergo conformational changes coupled to: the state of the bound nucleotide, affinity of nucleotide for the myosin head and affinity of the myosin head for actin (Geeves and Holmes, 2005). States that produce tension, must bind the thin filament strongly; those states have either Mg•ADP or no nucleotide bound in the active site. With no nucleotide in the binding pocket, the high concentration and affinity of ATP for myosin will lead to binding and myosin head detachment from the thin filament. ADP-bound states not only can produce tension, but resist exchange with ATP. Thus, ADP is uniquely suited for acting as a tension sensor in muscle. However, the mechanism by which this function is performed is not well understood.
Another feature of myosin•ADP binding to the thin filament is the effect of the nucleotide on the structure. Two experiments first described the effect ADP at saturating levels, which alters the position of the lever arm relative to the nucleotide-free (apo) form (Jontes et al., 1995; Whittaker et al., 1995). More recently an ADP effect on the lever arm position has been observed in Myosin V (Pospich et al., 2021; Wulf et al., 2016) and porcine cardiac muscle S1 (Doran et al., 2023). Subsequent to the initial observation, it was shown that the free energy change between ± ADP is quite small (Cremo and Geeves, 1998) and the tension change when ADP is added to rigor smooth muscle is negligible (Dantzig et al., 1999). Consequently, the change in lever arm position is unlikely to produce much work and instead possibly plays a role in mechanical sensing (Mentes et al., 2018). Myosin heads remain strongly bound to the thin filament until Mg•ADP is exchanged for Mg•ATP.
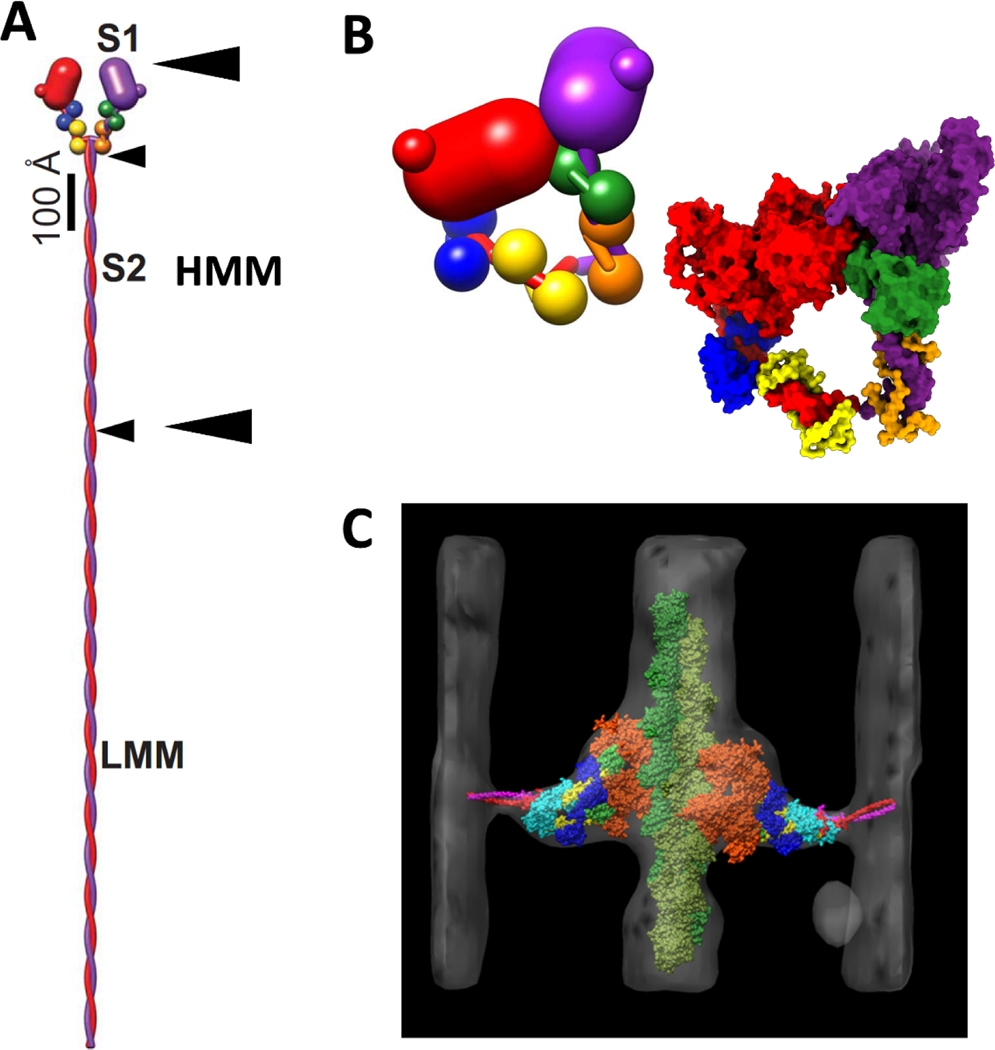
Myosin II, the object of this report, is the only myosin known to form filaments (Chantler et al., 2010; Sellers, 2000). Myosin IIs consist of two heavy chains of ~230 kDa each, and two pairs of light chains (Sellers, 2000), a 17 kDa Essential Light Chain (ELC) and a 20 kDa Regulatory Light Chain (RLC) (Fig. 1A,B). The heavy chains are identical in sequence as are the light chain pairs. Proteolysis of myosin (Lowey et al., 1969; Mornet et al., 1981) followed later by crystallography (Rayment et al., 1993b) and 3-D image reconstruction has defined the separate domains. The N-terminal ~850 residues and the two light chains form the head, also known as subfragment 1 or S1 (Fig. 1A,B), a highly conserved molecular motor (Emerson and Bernstein, 1987; McLachlan and Karn, 1982; Warrick and Spudich, 1987). The head itself consists of a motor domain containing the actin binding and ATPase activities followed by a small folded domain dubbed the converter (Houdusse and Cohen, 1996; Xie et al., 1994) and a long α-helix to which the light chains bind and terminating two residues after the invariant proline (Brown et al., 2008), e.g. residue 849 in chicken smooth muscle myosin, after which the tail begins. The myosin tail forms a long α-helical coiled-coil (McLachlan, 1983) that can be subdivided by controlled proteolysis into two peptides, subfragment 2 (S2) and light meromyosin (LMM) (Lowey et al., 1969). S2 alone forms a stable, soluble dimer at physiological ionic strength and together with the two S1 heads forms a molecule dubbed heavy meromyosin (HMM) (Fig. 1A). LMM is insoluble under the same conditions and is required to form filaments.
Figure 1.
Myosin II structure and conformations. (A) Myosin hexamer with two heavy chains, red and purple, and two pairs of light chains, ELC, blue and green, and RLC yellow and orange. Arrow heads mark the boundaries for the S1, S2, HMM and LMM subfragments. The small arrow heads mark the length of S2 and LMM. The pair of large arrowheads mark the boundaries of HMM. (B) The interacting heads motif in which one head, dubbed the blocked head, red, binds the second head, dubbed the free head, purple. Insert at the lower right is a space-filling rendition using PDB 1i84. (C) Atomic model of a 2-headed, rigor myosin cross-bridge from electron tomograms of swollen Lethocerus flight muscle from Liu et al. (2006) showing convergence toward the head-tail junction. In this orientation, the Z-line would be at the bottom and the M-line at the top. (A,B) Adapted from Hu et al. (2016).
Although myosin IIs have a pair of heads with motor properties, dimerization is not required for the motor function itself. Individual myosin heads have the ability to move actin filaments (Toyoshima et al., 1989). However, some myosin functions require the presence of both heads. For instance, smooth muscle myosin is regulated by RLC phosphorylation (Lowey and Trybus, 2010), but regulation is only observed in two-headed species such as HMM or whole myosin; single headed species show no regulation (Cremo et al., 1995). Inhibition was structurally illustrated by cryoEM of 2-D arrays of dephosphorylated smooth muscle HMM (Wendt et al., 2001) and whole myosin (Liu et al., 2003) which revealed an asymmetric arrangement of the paired heads (Fig. 1B). More recently, near atomic resolution structures have been reported for the soluble form of dephosphorylated smooth muscle myosin (Heissler et al., 2021; Scarff et al., 2020; Yang et al., 2020). A highly similar mechanism of inhibition is found in vertebrate skeletal and cardiac myosin (Jung et al., 2008a; Jung et al., 2008b), which are not intrinsically regulated by RLC phosphorylation.
In addition to the regulation requirement, two-headed constructs also show increased mechanical performance compared to their single-headed forms (Tyska et al., 1999). However, the active 2-headed interaction with actin has not been fully described. A better characterized 2-headed myosin structure is found in rigor muscle where myosin affinity for F-actin is highest and where available actin subunits exceed myosin heads to bind them. Generally, in rigor muscle in the overlap regions with thin filaments, nearly all myosin heads bind actin (Lovell et al., 1981; Taylor et al., 1984; Thomas et al., 1983). In rigor Lethocerus flight muscle fibers, the actin-bound, 2-headed myosin structure has been investigated by electron tomography of thin sections of plastic embedded tissue (Fig. 1C) (Liu et al., 2006; Liu et al., 2004; Taylor et al., 1993) and recently at a much higher resolution using Focused Ion Beam (FIB) milling of frozen-hydrated striated muscle (Wang et al., 2021; Wang et al., 2022). In situ, in fast-frozen, isometrically contracting muscle visualized by electron tomography and subtomogram averaging, two-headed cross-bridges were frequently found (Wu et al., 2010). Following a quick release, two-headed cross-bridges decreased in number, while following a quick stretch, their numbers increased (Wu et al., 2012). A similar increase in two-headed cross-bridges following a stretch was deduced from X-ray fiber diffraction (Brunello et al., 2007).
To distinguish between the two heads of HMM in the following discussion, we use the convention Leading and Trailing as defined by single molecule motility assays which defines the first head to attach as the Trailing Head because it trails the later attaching Leading Head which binds further along the actin filament in the direction of movement. In striated muscle sarcomeres the Trailing Head would be on the M-ward side (closer to the M-band, located at the center of sarcomere) and the Leading Head would be on the Z-ward side (closer to the Z-disk, located at the axial borders of sarcomere).
When cross-bridges are observed in situ in 3-D in plastic sections of rigor muscle by ET (Liu et al., 2006; Liu et al., 2004; Taylor et al., 1989), the pair of heads generally appear different with the Trailing Head having a more steeply angled lever arm than the Leading Head and with the two heads converging toward a common vertex (Fig. 1C). Although, these 3-D images are of low resolution, they do suggest that a higher resolution structure determination must account for some conformational differences between the paired heads similar to what has been seen in ET of FIB milled skeletal muscle (Wang et al., 2021; Wang et al., 2022).
Much of our early knowledge of the manner of myosin binding to actin has come from image reconstructions with imposed helical symmetry of actin filaments decorated with myosin S1, the first of which appeared in 1970 (Moore et al., 1970) and last, in 2008 (Littlefield et al., 2008). Imposition of helical symmetry enforces an identical structure on all the actin-myosin pairs. The technique does not lend itself readily to the visualization of conformational variability within the sample, which when present causes features to become blurred when symmetry is enforced. More recent reconstructions based on a single particle philosophy have yielded higher resolution reconstructions, within regions of myosin heads that are conformationally homogeneous (Behrmann et al., 2012; Fujii and Namba, 2017; Holmes et al., 2003; Mentes et al., 2018; Pospich et al., 2021; von der Ecken et al., 2016; Wulf et al., 2016).
The HMM fragment has been less popular as a specimen for image reconstruction because the binding of two heads originating from a common point to end on adjacent actin subunits violates the actin helical symmetry. Other problems arise from the possibility that some HMM molecules will be unable to bind the same filament with both heads, the so-called “parking problem” (Greene and Eisenberg, 1980). The parking problem introduces a discontinuity in the decoration and removes the registration necessary to visualize the head-tail junction in any spatial averages when helical symmetry is imposed.
In this report, application of single particle cryoEM of HMM-decorated F-actin with focused classification produced a resolution of about 15 A. The resulting reconstructions are very similar to the lower resolution structures produced from plastic sections and as well as the higher resolution reconstructions made from FIB milled frozen rigor muscle. The paired myosin heads have different lever arm angles converging to a common vertex. Lever arm compliance at the interface between the ELC and RLC was suggested in cryoEM structures of several acto-S1 structures (Littlefield et al., 2008), by X-ray crystallography within the RLC itself (Brown et al., 2011), and more recently by cryoET (Wang et al., 2022). Here we are asking the question as to whether this change in lever arm orientation from Mg•ADP-like to rigor-like can be observed by single particle cryoEM when both myosin heads bind actin and the paired heads likely adjust to the differential strain in the lever arms at various sites from the head-tail junction to the nucleotide binding site.
2. Materials and Methods
2.1. Protein preparation
HMM was expressed in Sf9 cells using a baculovirus expression vector and purified as previously described (Wendt et al., 1999). The HMM was stored in 50% glycerol, 50 mM NaCl, 5.7 mM Hepes, 15 mM EGTA, and 1.7 mM ATPγS at a protein concentration of 0.95 mg/ml (stored at −20 °C until used).
Actin was purified from chicken pectoralis muscle according to Spudich and Watt (Spudich and Watt, 1971). G-actin at 1.1 mg/ml was prepared by overnight depolymerization in Ca•ATP followed by clarification using high speed centrifugation (Hampton et al., 2007). Before polymerization, residual ATP in the supernatant was removed using a desalting column. G-actin aliquots (50 μ1) were stored at −80 °C until used.
2.2. Lipid monolayer sample recovery
In addition to conformational heterogeneity, another source of heterogeneity involves diffusion of proteins to the air-water interface and is perhaps the greatest experimental roadblock for cryoEM single particle analysis of biomolecules (Taylor and Glaeser, 2008). The use of lipid monolayers has the potential to prevent this effect. We used two lipid monolayers; one containing a positively charged surfactant; the other a Ni-NTA lipid combined with HMM expressed with a C-terminal His tag (Kornberg and Darst, 1991). Both approaches produced F-actin decorated at close to saturation with myosin heads.
All monolayer samples were prepared on a Teflon block with 1 mm x 5 mm diameter wells, volume ~7 μl. All monolayer samples were recovered using reticulated carbon films on 200 mesh R3/1 Quantifoil copper grids. Monolayers were recovered from the well surface by placing the grid with the grid bar side down, lifting the grid, mounting on the plunge freezer, blotting on the grid-bar side for ~4 sec using Whatman 542 filter paper and immediately plunge frozen (Taylor et al., 2007). The grids were kept in liquid nitrogen (−210°C-−196°C) until used.
2.3. cryoEM sample preparation
The lipid consisted of 1 μl of a mixture consisting of 20% DMPC-Ni NTA (1 mg/ml in chloroform), 80% DPPC lipid (1 mg/ml in chloroform) diluted to 0.5 mg/ml with chloroform. HMM at 0.22 mg/ml was clarified by centrifugation at 85,000 x g for 25 minutes at 4 °C. The HMM (~10 μg) and G-actin (11 μg) were premixed at room temperature for 3 min., followed by dilution in 330 μl of polymerization buffer (50 mM KCl, 2 mM MgCl2, 10 mM Na2PO4, 1 mM ADP, pH 7.6) and kept at room temperature for 1–1.5 hours, and subsequently moved into a °C cold room. At 4 °C this choice of lipids is expected to be in a gel phase, inhibiting HMM diffusion in the plane of the monolayer.
2.4. Single Particle Data Collection and Preprocessing
We used a Titan Krios (FEI Inc.) electron microscope operated at 300 kV with a DE-64 direct electron detector (Direct Electron Ltd) operated in integration mode and Volta phase plate (VPP). Images were collected at 29,000x nominal magnification. The calibrated pixel size of 1.27 Å was used for processing. Movies were collected using Leginon (Suloway et al., 2005) at a dose rate of 6.44 e−/Å2/s with a total exposure of 4.6 seconds and an accumulated dose of 29.6 e/Å2. Intermediate frames were recorded every 0.20 seconds for a total of 23 frames per micrograph. A total of 1,460 images were collected at a nominal defocus range of 0.2 – 2.0 μm.
The movies were corrected for beam-induced motion and dose-weighted using MotionCor2 (Zheng et al., 2017). The non-dose-weighted and dose-weighted micrographs were imported in cisTEM (Grant et al., 2018). CTF and phase shift estimation was performed using CTFFIND4 (Rohou and Grigorieff, 2015) in cisTEM and the result was exported using a simple script. A histogram of phase shift determined by CTFFIND4 is plotted (Supplemental Figure 1). The metadata file was imported in Relion and micrographs with resolution estimates between 2–20Å were selected, resulting in 1,413 micrographs.
2.5. F-actin-HMM Single Particle Image processing
The reconstruction strategy was to first obtain the best alignment of actin subunits and attached myosin heads by imposing helical symmetry. Using 3D-classification without alignment, the decorated segments with better definition were selected, followed by 3D alignment and helical reconstruction. Finally, we would use focused 3-D classification to find the largest number of paired myosin heads attached to successive actin subunits (Supplemental Figure 2).
HMM decorated actin filaments were manually picked from the motion corrected and dose weighted micrographs. The filament segments were extracted into boxes 420 pixels on edge (pixel size 1.27Å) and binned by 2 (pixel size 2.54Å) using Relion 2.1 (Zivanov et al., 2018). These segments were shifted axially in 28 Å steps along the 59Å genetic helix which resulted in 304,437 segments (Table 1 from Supplementary materials). Because the target motif contained two actin subunits, the maximum possible acto-HMM motifs is half that number. The dataset was imported to cisTEM, and 3D refined using Autorefine. An undecorated F-actin density map, from an unpublished result from our lab, was lowpass filtered to 60Å and then was used as the initial model. The result of 3D refinement (resolved at ~14A) clearly showed the decoration with myosin heads. After performing a round of reference free 2D classification in cisTEM, 159,057 segments were selected from the best-looking class averages. Another round of 3D Autorefine in cisTEM resulted in our best reconstruction map. This map was then sharpened in cisTEM.
Separately, the following steps used the Iterative Helical Real Space Reconstruction (IHRSR) implementation in Relion 2.1 (He and Scheres, 2017). The segments (304,437 in total) were helically 3D refined starting from the same actin filament initial model used in cisTEM; lowpass filtered to 60Å. We allowed for local search for helical parameters. Refinement resulted in a map with 18Å global resolution (FSC 0.143) after postprocessing using a soft-edge mask. The result of IHRSR was used for 3D classification without modifying the alignments into four classes (Supplemental Figure 2), with the regularization parameter changed to eight (T=8). Segments from three out of the four classes with slightly more detailed averages were selected (50,203 segments) for further refinement against a lowpass filtered (35Å) map from Class #4 using IHRSR with local searches for symmetry (Supplemental Figure 2). The local resolution of the map was computed using MonoRes (Vilas et al., 2018) varying from ~13–30Å. This resolution is inferior to what would be currently expected from this number of segments. We think this may be the result of the Volta Phase Plate, which in our hands has not produced higher quality reconstructions and the DE-64, which at the time could only produce integrated images and not electron counting images which are today’s standard.
The result of 3D refinement in Relion before local resolution determination was used for masked (focused) 3D classification without alignment modification into 20 classes, using the same regularization parameter (T=8). The soft-edge mask used for focused classification was produced in Relion. Particles from those classes with similar features were merged and reconstructed into four major classes. In cases that we were unable to recognize which group the particles belonged to, we reconstructed the class to see the adjacent motor domains interacting with F-actin for accurate assignment to the corresponding group. Then Groups 1 and 2 were refined separately without imposing helical symmetry in Relion while groups 3 and 4 were only reconstructed without modification of the prior alignments.
2.6. Model Building/Fitting
The structure of full-length smooth muscle S1 interacting with actin has not been resolved to atomic resolution, but some parts of the molecule are available, in particular the motor domain-ELC construct (Dominguez et al., 1998) and the nucleotide-free motor domain alone bound to actin (Banerjee et al., 2017). Hence, we joined various models with a minimum degree of modification. The near rigor state crystal structure of scallop striated muscle myosin subfragment 1 (PDB ID:1KK7) (Himmel et al., 2002) showed an almost perfect fit of the motor domain and ELC into the Trailing Head density thereby providing a template on which to construct a smooth muscle myosin atomic model with only minor adjustments to fit the RLC portion.
Using SWISS model (Waterhouse et al., 2018), we built a homology model starting from the now modified scallop S1 crystal structure and the target sequence of chicken smooth muscle myosin (MYH11_CHICK with primary accession number: P10587). The scallop motor domain (residues 1–780) was replaced with the chicken smooth muscle motor domain and actin subunits from the cryoEM structure of the rigor chicken smooth muscle myosin motor domain bound to actin (PDB ID: 6BIH) (Banerjee et al., 2017), without further modification. Using UCSF Chimera (Pettersen et al., 2004), the lever arm (780–851) was rotated around residue 777 to improve the fit of the density. Finally, coordinates of the RLC and ELC from the inhibited state cryoEM structure of chicken smooth muscle myosin II (PDB ID: 6XE9) (Yang et al., 2020) were fitted as a rigid body into the map.
The fit of the homology model to the Leading Head, except for the motor domain, was poor. Some of this could be improved upon by rotation of the lever arm around residue 777 immediately after the converter domain followed by a rotation around residue 811, which is located in the region between the ELC and RLC for the best fitting of the lever arm and the LCs. The smooth muscle light chains were then rigidly fitted into the density map of the Leading Head.
Finally, Namdinator (Kidmose et al., 2019) was used for molecular dynamic flexible fitting (MDFF) and real space refinement (RSR) followed by validation in Phenix (Afonine et al., 2018) (Table 1 from Supplementary materials). In order to show the conformational changes within the lever arm after MDFF and RSR, we computed the Root Mean Squared Deviation (RMSD) between the models before and after flexible fitting (Supplemental Figure 3).
3. Results
3.1. Single particle analysis of HMM decorated actin
The conditions used for making specimens of HMM decorated actin in the single particle analysis were chosen not to decorate actin with HMM•ADP, but were chosen to see if the asymmetric head-head interaction characteristic of relaxed smooth muscle myosin, otherwise known as the interacting heads motif, could be observed binding to F-actin as suggested biochemically (Sellers et al., 1982) and structurally (Wendt et al., 2001). Single particle processing of free HMM molecules clearly removed from F-actin supported the interpretation that the conditions produced the head-head interaction (Supplemental Figure 4) but provided no new information on the structure of the complex. However, when actin bound the conformation was clearly either rigor-like or ADP-like. The result indicated that if the interacting heads motif were to be trapped binding to actin, a different strategy would be needed.
Decoration of F-actin with myosin heads is generally done in one of two ways. Either F-actin is applied to the grid first followed by washing with a solution of nucleotide free myosin heads (Banerjee et al., 2017; Moore et al., 1970; Seymour and O’Brien, 1985), or F-actin and an excess of myosin heads needed to saturate the filaments are mixed and then applied to the grid (Holmes et al., 2003; Mentes et al., 2018; Schröder et al., 1993). Here we have used a novel method of applying a lipid monolayer doped with Ni-NTA lipid over a solution of G-actin polymerizing in the presence of HMM, ADP and inorganic phosphate. The lipid monolayer was chosen to prevent HMM diffusion to the air-water interface. We obtained near saturation of F-actin with myosin heads consistent with HMM in excess over Ni-NTA binding sites in the monolayer (Fig. 2A).
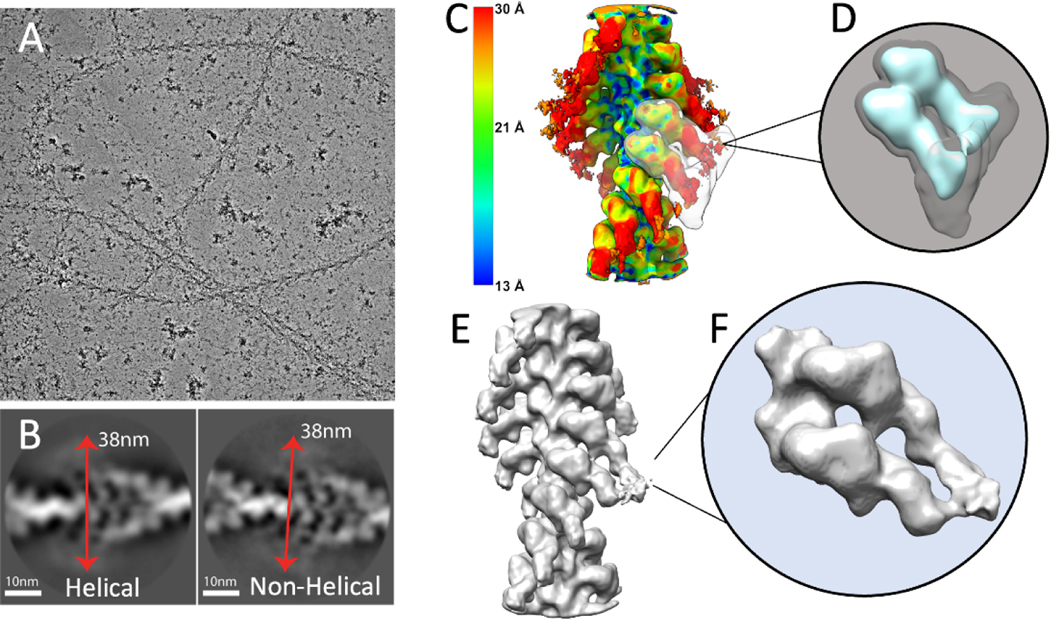
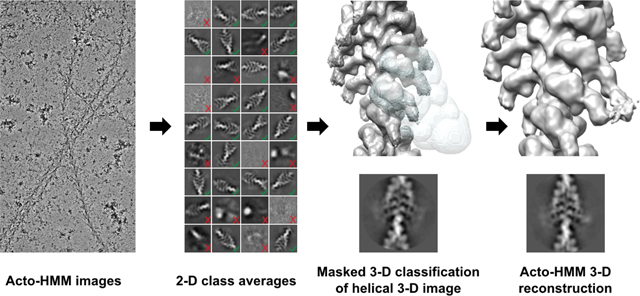
Figure 2.
Cryo-EM image processing of F-actin decorated with smooth muscle HMM. (A) An electron micrograph showing F-actin decorated with HMM. (B) Showing representative good class averages resulted from 2D classification in Relion performed with and without imposed helical symmetry. Red arrows show an estimate of the diameter of the filaments to be approximately 380Å. (C) Result of local resolution determination using Local MonoRes (Vilas et al., 2018) shown as a heatmap of the refinement after selecting the best classes from unmasked 3D classifications. (D) The 3D soft-edge mask (shown in grey) used for 3D classifications. This mask includes both motor domains and the blurred lever arms (shown in cyan). (E) The best Acto-HMM density map, 3D reconstructed by only including the segments from group 1. (F) Magnified view of the segmented map in E.
F-actin decorated with HMM, produces a more sharply defined arrowhead motif characteristic of the rigor state compared to what is obtained by F-actin decoration with S1 (Katayama and Wakabayashi, 1981). The general appearance of the filaments prepared using the lipid monolayer shows a similar arrowhead pattern with clear definition of the polarity. Higher contrast in the micrographs obtained using the VPP produced images of HMM decorated F-actin easily observable in the lowpass filtered micrographs (Fig. 2A). The diameter of the decorated filaments measured from lowpass filtered micrographs and 2D class averages (Fig. 2B) is close to 38 nm. Segments were helically extracted along the genetic helix of F-actin (Supplemental Table 1), which can be considered an implicit form of helical symmetry imposition. Projection classifications of the segments, with (Fig. 2B, left) and without (Fig. 2B, right) imposition of explicit helical symmetry, suggest high saturation of actin sites with myosin heads and blurred distal lever arms.
3.2. 3D-Reconstruction of acto-HMM
Decoration of F-actin with myosin S1, at close to saturation, can follow helical symmetry at least in the motor domain (MD) and proximal parts of the lever arm. The distal parts of the lever arms are likely to show varying amounts of disorder depending on distance from the filament axis and mobility about various hinge points. In contrast, double-headed binding of HMM to F-actin would, in general, not follow the helical symmetry of F-actin because HMM consists of a pair of heads joined at the head-tail junction; the lever arms are not simply disordered, but are structurally different unless the two heavy chains at the head-tail junction unwind enough α-helix to allow separation by some 85 Å. In addition, if one head binds actin and the other does not or an actin subunit goes unlabeled with myosin heads any symmetry with paired heads bound is broken. The paired motor domains when bound to F-actin could be near identical depending on solution conditions and resolution but are not required to be identical. Therefore, to prevent bias in the result of the reconstruction, we did not start with imposing explicit helical symmetry. However, the protocol of extracting segments spaced every 28 Å can be interpreted as implicitly imposing helical symmetry.
To improve the final alignment of the segments, we used Relion to enforce helical symmetry with the result that the lever arm density, which appeared blurred in 2D class averages and the 3-D reconstruction with implicit helical symmetry, was now better resolved. At this point, MonoRes (Vilas et al., 2018) was used to locally filter the map, visualize the local resolution, and illustrate the domains with potential conformational heterogeneity (Fig. 2C). The resolution steadily decreases with increasing radius, being ~15Å for F-actin, ~20Å for the myosin motor domain and ~30Å for the lever arm. The unmasked 3D classification without alignment produced four classes. The class, which had most of the segments (~250,000), had shorter resolved lever arms (cyan colored in Supplemental Figure 2). The other three classes (~50,000 segments in total) showed the more desirable longer and better resolved lever arms (Supplemental Figure 2). Helical refinement of the combination of the three classes with longer arms, resolved more detailed and clearer lever arm density (grey, yellow, and purple classes in Supplemental Figure 2). On the other hand, a similar refinement of the segments from the class with shorter lever arms (Cyan in Supplemental Figure 2) did not produce a map with a comparable improvement in quality to the other three classes. The reason for a major part of the segments with a lower quality is not clear.
Imposing helical symmetry averaged out the 2-headed interactions (Fig. 2C). To recover the head-tail junction of the HMM molecules, we performed a masked 3-D classification limited to a pair of adjacent S1 heads (Fig. 2D), and without modification of the segment alignments. Twenty classes were produced and combined into four major groups based on similarity of features. Three out of the four groups from the 3-D classification show double-headed binding of HMM to F-actin (Supplemental Figure 5). Group 1 (17,394 segments, 32% of 50,203 segments) included those classes with clear adjacency of the RLC regions and juxtaposition of the head-tail junction within the boundaries of the mask (Fig. 2E,F; Supplemental Figure 5A–C). In Group 1, both heads, included in the mask appear to belong to the same HMM molecule. The other three classes showed variations on Group 1. Because segments are selected along the F-actin genetic helix and successive paired heads are included in the classification mask, it is possible for a single myosin head to appear in more than one class. For example, the Trailing Head of a Class 1 member could be in the Leading Head position of another class, and the Leading Head of a Class 1 member could be in the position of a Trailing Head of yet another class. When this occurs, which must occur many times, such a class is likely to have poor definition, which appears to be the case with the other three classes. Thus, there is a good reason to select the class with the best-defined double-headed shape as being representative of the 2-heads of one myosin binding to actin.
3.3. Conformations of the Leading and Trailing heads
The Trailing Head and the Leading Head were segmented out of the Group 1 (panels A-C from Supplemental Figure 5) density map. Using the segmented map, we made a detailed comparison between the Acto-HMM map and various published models of fragments of myosin. The result of a few of these comparisons are shown in Supplemental Figure 6. In order to dock the models into the density map at this resolution, the motor domain from the myosin crystal or cryoEM structure was used for fitting as a single rigid body (Pettersen et al., 2004). This preliminary rigid body docking guided us in determining a good starting point for building the Leading and Trailing Head atomic models (Supplemental Figure 6).
For the Trailing Head (middle row from Supplemental Figure 6) various models show different levels of fitting. The scallop myosin S1 (PDB: 1KK7) as a single rigid body fits the Trailing Head density well but with part of the RLC lying outside of the envelope (Supplemental Figure 6A). The cryo-EM structure of chicken smooth muscle myosin II motor domain (PDB: 6BIH) is also a near perfect fit but it lacks the lever arm (Supplemental Figure 6B). The cryo-EM structure of chicken skeletal S1 also fits the Trailing Head density but with part of the RLC lying outside the density. Note that the parts of the RLC that do not fit in the two cases (scallop vs. skeletal) lay on opposite sides of the RLC region of the reconstruction envelope (Supplemental Figure 6A,C, lower part of top panels).
The density map for the Leading Head indicated a conformation of the RLC segment of the lever arm different from any previous high resolution atomic model. Rigid body alignments of the atomic models (Supplemental Figure 6A,C) achieved similar fitting in the motor domain and ELC positions. However, the RLC domain fell completely outside of the density. Axially, both models with lever arms placed the RLC domain below the Leading Head, but azimuthally they fell to either side of the RLC density indicating that major changes occur in the RLC positions, but they are not identical for all myosin head structures.
To produce the final atomic model of acto-HMM the individual domains, motor, converter, ELC and RLC were fit separately into the density, with the motor domain fit using the chicken smooth muscle structure (PDB: 6BIH). For the lever arm, we produced a homology model for the entire myosin head (PDB: 1KK7) and aligned it to the chicken smooth muscle motor domain on actin. To fit the lever arm into the density, pivot residues were 777 for the ELC and 811 for the RLC. The converter domain fit with minimal adjustment. The ELC domain of the Trailing Head required minimal movement with somewhat more for the Leading Head ELC. The RLC and its heavy chain segment of the Leading Head is unlike any of the existing in vitro models and more like the recent in situ reconstructions (Wang et al., 2021). The density map resolution at the RLC is lower for the Leading Head making the fit more ambiguous. While keeping heavy chain residues 1–810 in place, the rest of the heavy chain and its bound RLC was rotated as a rigid body around residue 811 toward the Trailing Head. Eventually, Molecular Dynamic Flexible Fitting (MDFF) coupled with Real Space Refinement (RSR) were used to generate the final model (Kidmose et al., 2019). Considering the resolution of our map, we cautiously set the values for parameters in MDFF and RSR, following the manual in Namdinator.
The resulting HMM atomic model has some novel features, particularly at the RLC position (Fig. 3A–D; Supplemental Movie 1). The lever arm positions of Leading and Trailing Heads show significant variation. The invariant proline P849 is a good reference for differences at the head-tail junction. The P849 positions for the Leading and Trailing heads differ by 9Å axially and 20Å azimuthally (Fig. 3E,F). The atomic model as constructed had some side chain clashes between the two RLC N-terminal domains, so it is conceivable that the invariant prolines are slightly further separated.
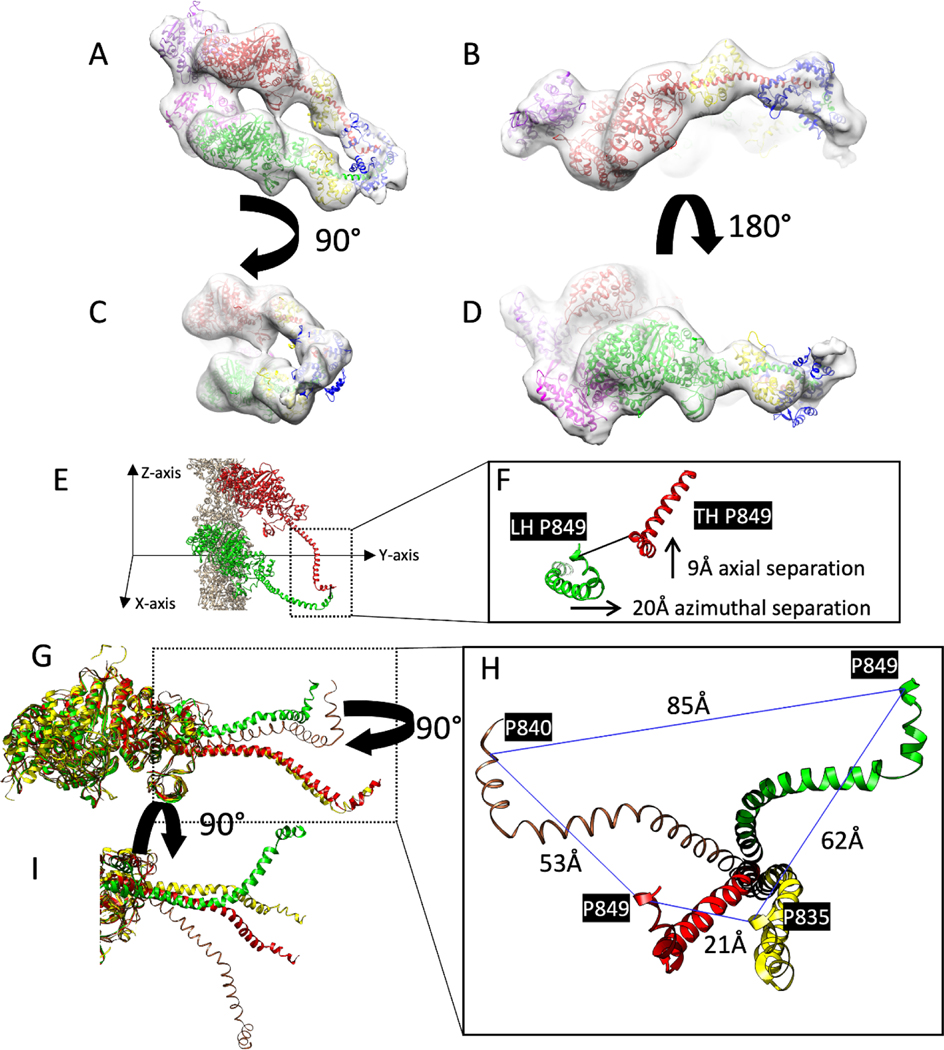
Figure 3.
Comparison of atomic models of HMM decorated F-actin with closely similar structures. These are rigid body fits of the S1 atomic structure into the reconstruction. (A-D) Different views of the acto-HMM atomic model in the density map of Leading and Trailing heads. The two adjacent actin subunits are shown in purple and magenta, heavy chains of the Leading and Trailing heads are shown in green and red; the ELCs and RLCs are shown in blue and yellow, respectively. (E,F) Showing the distance between the conserved pro849 residues from the Leading Head, LH, and the Trailing Head TH, Z-axis showing the direction along the axis of the filament. (G,H) different views of the comparison of the heavy chains from the Trailing Head (red) and Leading Head (green), the crystal structure of scallop myosin S1 in the near rigor conformation (PDB ID: 1KK7) (yellow) and the cryo-EM structure of chicken skeletal S1 in the rigor state (brown) after the alignment of the motor domains. The distances are measured between the invariant proline residues. (I) Axial view showing the conformational difference between Trailing Head (red), Leading Head (green) and the starting model 1KK7 (yellow) after the alignment of the ELC binding segment of the lever arm.
Variation of the lever arm conformations were examined by aligning two structures, the scallop near-rigor conformation and the rigor chicken skeletal model (This model is available as Supplementary Material within the publication (Holmes et al., 2003)) using the motor domains alone (Fig. 3G–I). The difference in conformation between the lever arms of the Trailing Head and scallop myosin S1 are small with only a 21 Å separation at the invariant prolines (Fig. 3I) but are much larger (53Å) for the chicken skeletal structure and to the opposite side of our acto-HMM atomic model. Comparison with the Leading Head atomic model shows good overlap between the lever arm positions azimuthally up to residue 811, which is just after the ELC binding domain, but to opposite sides again (Fig. 3G). The invariant prolines of these three models are quite far apart, 53 Å, 62 Å and 85 Å, and to the same side of the Leading Head (Fig. 3H).
The nearly perfect rigid body fit of the nucleotide-free atomic models of the chicken smooth muscle myosin motor domain (Banerjee et al., 2017) and scallop smooth muscle myosin S1 near rigor structure, PDB 1KK7, (Himmel et al., 2002) to the Trailing and Leading motor domain density provided strong support for a near nucleotide free conformation and also suggests relatively modest changes if any in the motor domain but the resolution here is lower following classification due to the limited data set size. Other published nucleotide free acto-myosin II structures available from public data bases also are good fits to the motor domain density (Fujii and Namba, 2017; Holmes et al., 2004; Holmes et al., 2003).
Comparison of the motor-domain aligned atomic models (Fig. 3E–I) shows that the difference in conformation of the lever arms is not limited to displacement along the axis of the filament (Z-axis); azimuthal differences are present as well, but the chicken skeletal S1 seems to have a different lever arm conformation compared with that of the Trailing Head and the Leading Head as well as the scallop near rigor structure, suggesting species and muscle type differences.
4. Discussion
The in vitro structure of acto-HMM has been investigated far fewer times than has the structure of acto-S1. The literature contains three attempts before the atomic structure of the myosin head had been determined (Kajiyama, 1988; Katayama and Wakabayashi, 1981; Seymour and O’Brien, 1985). The first of these (Katayama and Wakabayashi, 1981) differed from previous reconstructions of acto-S1 in showing density that could plausibly be ascribed to the RLC, referred to at the time as the DTNB light chain. A subsequent reconstruction (Kajiyama, 1988) showed several domains in the region of the lever arm, which were unidentified at the time, but now would probably be assigned to the ELC and RLC. Seymour and O’Brien’s acto-HMM reconstruction was from a single, averaged filament and comparatively noisy.
Distinct from these efforts are several structures obtained of 2-headed myosin contacts in situ in rigor muscle, but these reconstructions are of lower resolution, come from fixed, embedded and sectioned Lethocerus flight muscle, and possess additional sources of heterogeneity due to the effects of the filament lattice (Chen et al., 2001; Chen et al., 2002; Liu et al., 2006; Liu et al., 2004). However, they were determined after the S1 crystal structure (Rayment et al., 1993b) was solved which enabled pseudoatomic models to be built into the density envelopes. In those rigor muscle structures, the 2-headed cross-bridge, dubbed the lead bridge, the shape of the head was distinctly triangular with one edge of the triangle on the thin filament, the opposite point of the triangle positioned at the thick filament surface, the edge of the triangle on the Z-disk side was perpendicular to the filament axis, and the edge of the triangle on the M-line side angled at the classical 45° rigor angle. That shape corresponds well to the higher resolution shape of actin-bound HMM shown here. More recent studies (Wang et al., 2021; Wang et al., 2022) using cryoET of frozen hydrated samples prepared by Focused Ion Beam (FIB) milling avoided all the limitations of the earlier work and showed a distinct improvement in resolution and head definition. Various domains of skeletal muscle myosin bound to actin filaments are clearly resolved, including the lever arms and the light chains. The more recent study also identified two closely related conformations in the lever arm. This plasticity in the structure of the lever arm could be assigned, at least partially, to a location of compliance between the two light chain binding domains.
HMM decorated actin filaments are not strictly speaking helically ordered. Hence, a spatial averaging method such as helical reconstruction is weakly founded for this situation and would average over all possible heterogeneities. However, focused classification scheme proved to be suited for separation of these discrete conformational states. Similar focused classification approaches did not prove successful in separating conformational states of the lever arm in published structures for Acto-S1, resulting in noisy density for at least parts of myosin lever arm.
4.1. Myosin Decorated F-Actin
The wealth of publications on actin filaments decorated with S1 from myosin II provides abundant opportunity to compare our results obtained by cryoEM. Excluding earlier publications based on negative stain or published prior to the availability of the S1 crystal structure, there are 16 papers reporting structures of myosin decorated actin. Some of these use complete S1 subfragments (Jontes and Milligan, 1997; Jontes et al., 1995; Rayment et al., 1993a; Veigel et al., 1999; Whittaker et al., 1995), others contain motor domains alone, or motor domains with the ELC (Banerjee et al., 2017; Behrmann et al., 2012; Fujii and Namba, 2017; Holmes et al., 2003; Littlefield et al., 2008; Mentes et al., 2018; Pospich et al., 2021; Volkmann et al., 2000; Volkmann et al., 2003; Volkmann et al., 2005; von der Ecken et al., 2016).
Given the limited size and quality of the single particle projection data sets, the challenge was less a question of resolution but whether classification and averaging would efficiently find 2-headed HMM attachments to actin. However, at the resolution achieved, a number of observations can be made from the RLC definition which exceeds that typically seen in reconstructions of acto-S1.
The chief takeaway from the actin-HMM reconstruction is that most of the physical change in the myosin head resulting from the two headed binding is accommodated by changes in the orientation of the segment of the lever arm between the ELC and RLC rather than the entire lever arm. Most of the conformational change in the lever arm is accommodated by the Leading Head, as the conformation of the lever arm in the Trailing Head is highly similar to that from selected S1 crystal structures with complete lever arms. Thus, the myosin heads in HMM accommodate two-headed F-actin binding asymmetrically. Consistent with this is little difference between the two motor domains and the ELC when superimposed. There is much more difference in the following portion of the lever arm that contains the RLC. The region of the lever arm between the ELC and the RLC, a region previously suggested to be variable in structure (Littlefield et al., 2008), is the thinnest part of the lever arm and all other things being equal, would be expected to be the most compliant. This contrasts with the recent structure of smooth muscle myosin in the 10S conformation in which only a small change is needed in the same location to close the gap between the head-tail junctions of the two heads (Heissler et al., 2021; Scarff et al., 2020; Yang et al., 2020). However, the affinity of myosin heads for actin when nucleotide-free or with bound ADP is probably a stronger effector to the structure of the heads than is the affinity of the two heads for each other in the 10S conformation.
The structure of acto-HMM provides an opportunity to investigate how the myosin head adjusts its structure in response to strain imposed by the heads binding to sites separated by 55 Å axially and 28° azimuthally. Strain could affect the actin subunit structure and position within the filament, the motor domain orientation on actin, or the relative position of any of the myosin head domains from the converter to the initial segment of the coiled-coil tail. Small changes are likely to be visible at low radius where the resolution is high; larger changes would likely occur at high radius but at a lower resolution. Changes in the position of the RLC domain are both axial and azimuthal and may potentially cause its N-terminal domain to rotate in order to point towards the S2 coiled- coil. Changes axially would be necessary to close the 5.5 nm axial separation; changes azimuthally are ~50% larger to bring the head-tail junctions close enough to form an α-helical coiled coil. Based on that observation alone, one would expect that lever arm stiffness had an orientation dependence. When the muscle is stretched in rigor to evaluate its stiffness it is the axial stiffness that is being measured. Moreover, the muscle’s function is to generate axial force making the expectation that the lever arm is stiff in that direction. Azimuthal stiffness should be minimal as it would reduce the energy available for axial force generation, which comes from the increased actin affinity when the myosin head transitions from weak to strong binding on actin.
It has previously been suggested that the change from weak to strong binding in Lethocerus flight muscle, involves some azimuthal movement of the motor domain on actin (Arakelian et al., 2015). Such a movement in situ with the myosin heads attached to the thick filament would be inhibited by azimuthal stiffness in both the lever arm and the S2 link to the thick filament, which would argue that there is a difference between the lever arm azimuthal and axial stiffness. This is also the conclusion reached recently in detailed analysis of the rigor stiffness in muscle (Caremani and Reconditi, 2022).
The acto-HMM map is in agreement with models obtained from electron tomography of plastic embedded and sectioned muscle (Liu et al., 2006; Liu et al., 2004) with regard to the orientation differences between lever arms of the Trailing and Leading Heads bringing the head-tail junctions to a similar axial position as if there were no alterations to the S2 caused by the 2-headed binding to actin. The more recent tomograms from rigor fibers of rat skeletal muscle are also consistent with this picture (Wang et al., 2021; Wang et al., 2022). Averages of 2-headed rigor cross-bridges were highly similar at the motor domain and ELC but differed primarily at the RLC segment. The Leading Head RLC was rotated M-ward and azimuthally toward the Trailing Head RLC so that their head-tail junctions would converge.
4.2. Effect of ADP on the reconstruction
Myosin in partnership with actin, performs a range of mechanical functions through conformational changes coupled to hydrolysis of ATP (Geeves and Holmes, 2005). In our study, we polymerized desalted G-actin•ATP (no free ATP) in the presence of smooth muscle HMM in a buffer containing ADP and phosphate. On binding actin, the phosphate is almost certainly released leaving the possibility that the HMM may have bound ADP. CryoEM has shown that vertebrate smooth muscle myosin II undergoes a small change in the orientation of the lever arm on ADP release (Whittaker et al., 1995) which might introduce heterogeneity into the structure. Could the acto-HMM structure observed be influenced by the presence of ADP?
Several sources have measured the dissociation constant of ADP for actin-bound smooth muscle myosin II (S1), which range from 1–5 μM (summarized in (Dantzig et al., 1999)). Since our ADP concentration was 0.5–1.0 mM, we expect between 99.9 and 99.5% of the actin bound HMM will have heads with ADP bound. We believe that this percentage is high enough to affect the acto-HMM reconstructions, which predominately contain ADP•HMM. Unclear is whether the lever arm of HMM decorated actin will show the same change in angle observed with the S1 fragment when saturated with ADP (Mentes et al., 2018; Whittaker et al., 1995) given that the paired heads of HMM converge toward a common origin.
Nyitrai and Geeves (Nyitrai and Geeves, 2004) have discussed the effect of strain on the release of ADP from various isoforms of myosin and is largely the source for the following discussion. Smooth muscle myosin II is a kinetically slow myosin that shows a significant conformational change in the lever arm orientation with/without bound ADP (Whittaker et al., 1995). Fast striated muscle myosins do not show this conformational change on ADP release but it has been observed in porcine cardiac muscle myosin, a slow myosin (Doran et al., 2023). Kinetically, smooth muscle myosin II with ADP is almost as tightly bound to actin as nucleotide-free myosin II. Consequently, the myosin in the presence of saturating levels of ADP can remain tightly bound to actin when under strain. Smooth muscle under strain can thus sustain tension with relatively low ATP turnover, which requires release of ADP. The opposite condition, in which the myosin is negatively strained should accelerate ADP release. We suggest that HMM bound to actin with both heads puts the Leading Head under positive strain (capable of performing work) and the Trailing Head under negative strain (having had work performed on it) because the paired lever arms must move in opposite directions to meet at the head-tail junction.
Although the reconstructions lack the resolution to visualize ADP, structurally, the Trailing Head was better fit by a nucleotide-free, scallop myosin head, a completely different species. The rigor like lever arm orientation could be taken as evidence of an apo conformation. The clearest difference between the Trailing Head and the scallop near rigor structure is in the bending of the RLC segment of the lever arm toward the Leading Head, suggesting the Trailing Head is under negative strain (compare red and yellow helices in Figure 3G, which is an axial view). The only published myosin head structure that came close to a fit of the Leading Head was the in situ structure (Wang et al., 2021) which required some small adjustments at the ELC position consistent with ADP binding in our acto-HMM reconstruction. Whether these are due to ADP binding or the connection with the Trailing Head is difficult to distinguish.
In situ structure of two-headed acto-myosin is consistent with this picture, but less dramatically which might be a species difference (Wang et al., 2021). The in situ two-headed rigor crossbridge is arguably an apo form for both heads. Viewed axially, the Trailing Head structures are nearly superimposable (Fig. 4A–C), but the Leading Head structure has the ELC position oriented more toward the F-actin (+) end (Fig. 4A,D) consistent with further lever arm movement toward the end of the power stroke on ADP release. Larger changes in the RLC position were observed for the Leading head (Fig. 4E) which may be due to our attempts to avoid clashes with the paired RLCs.
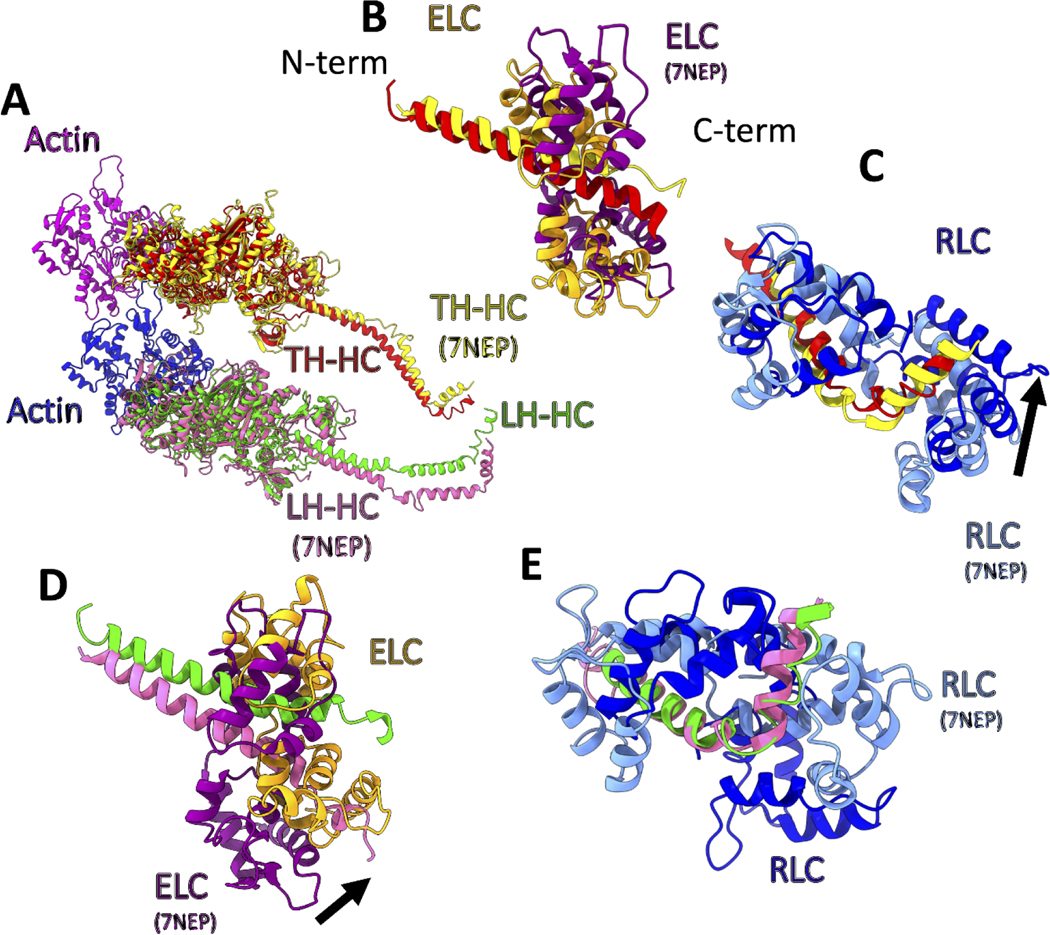
Figure 4.
Comparison of the smooth muscle acto-HMM atomic model with that for skeletal muscle myosin II (PDB ID: 7NEP) obtained by cryoET (Wang et al., 2021). The heavy chains from the two models have been aligned at the motor domains. (A) shows the alignment of the heavy chains from the two models and the actin subunits (magenta and medium blue). Heavy chains of Trailing head (red and yellow for smooth and skeletal muscle myosin, respectively) and Leading heads (green and hot pink for smooth and skeletal muscle myosin, respectively). (B) Comparison of ELCs and the ELC binding domains of the Trailing Head. Skeletal ELC and heavy chain (purple and yellow respectively) and smooth ELC and heavy chain (orange and red respectively). (C) Comparison of the RLCs and the RLC binding domains of the Trailing Head. Skeletal and smooth RLCs are shown in cornflower blue and blue, respectively. The black arrow shows the direction of the movement needed for alignment of the skeletal RLC to the smooth RLC model. (D) Similar to (B), showing ELCs for the Leading Head. The difference in the conformation of the two heads is more significant in the Leading Head. (E) Similar to (C), showing RLCs for the Leading Head, after the alignment of the heavy chain residues in the RLC binding domains.
4.3. Stability of the coiled-coil at the head-tail junction
An important characteristic of the myosin molecule is its pair of heavy chains, which are necessary to produce a coiled-coil tail. The initial segment of the tail, the S2 domain, provides a tether from which myosin heads containing the molecular motor can search for actin subunits. However, S2 may function in ways other than simply as a tether. The S2 coiled coil has been described as unstable in a number of studies (Blankenfeldt et al., 2006; Knight, 1996; Li et al., 2003). What unstable means in these studies is not always precisely defined, but we take it to mean that the α-helices are formed, but the coiled coil is not or is variable (dynamic) for some undefined number of residues, but possibly 14 based on studies of smooth muscle myosin regulation with different tail segments (Trybus et al., 1997).
Optical trapping studies comparing the flexibility of smooth muscle HMM constructs with 15-heptads of coiled-coil versus a construct with a leucine zipper at the head-tail junction, referred to as a 0-hep-zip construct, showed a reduction in unitary displacement from d=9.7±0.6 nm to d=0.1±0.3 nm (Lauzon et al., 2001), illustrating the significance of some coiled-coil instability at the head-tail junction for maximal motor function. Shortly after this study, some similar studies had controversial (Chakrabarty et al., 2003) and in some cases contradicting results (Wahlstrom et al., 2003; Wu et al., 1999). One study (Chakrabarty et al., 2003), could not conclude the necessity of coiled-coil uncoiling as a part of the regulation and mechanical performance. The clearest structural evidence for structural changes at the head-tail junction with myosin heads present comes from the high-resolution structure of the Lethocerus flight muscle myosin tail in relaxed thick filaments which showed that changes in the coiled coil at the head-tail junction consisted of both α-helix unfolding for one head and changes in α-helix winding (pitch) for both heads (Rahmani et al., 2021). Similar changes might be observed in acto-HMM if sufficient resolution could be obtained at the head-tail junction.
4.4. Utility for Myosin II Motor Structure
Typically, atomic structures of myosin motors have utilized X-ray crystallography of expressed constructs (Coureux et al., 2004; Dominguez et al., 1998; Smith and Rayment, 1995). Generally, these constructs lack all or part of the lever arm, which is perceived as an obstacle to obtaining crystals diffracting to high resolution. Acto-S1 or acto-MD specimens solved using cryoEM usually elucidate the interaction of the myosin motor domain with actin (Banerjee et al., 2017; Behrmann et al., 2012; Fujii and Namba, 2017; Holmes et al., 2004; Littlefield et al., 2008). Generally, when present the lever arms of acto-S1 reconstructions have lower resolution than the motor domain because they are more mobile than the motor domain when bound to actin strongly, generally arguing in favor of expressed constructs with the lever arm removed. However, the structure of the lever arm when bound to actin in the rigor state can be quite variable (Fig. 3I, Supplemental Figure 6). This dynamism is quite likely to be of functional importance as a source of compliance in contracting muscle or in limiting or facilitating accessibility to actin during contraction in different muscles. We think a faster route to a near atomic resolution reconstruction of complete myosin heads might be through acto-HMM, which would have a couple of extra benefits: (1) with the lever arms anchored by S2, they would be less dynamic and consequently better ordered. (2) The RLCs of the paired heads in particular are likely to have different structures (Brown et al., 2011) with, as shown here and previously, significant differences in lever arm bending between the ELC and RLC and elsewhere along the lever arm (Littlefield et al., 2008). These differences might be determinable at subnanometer resolution thereby clarifying the source of the flexibility. (3) There is also an opportunity to visualize portions of the S2 at the head-S2 junction which might clarify how the S2 responds to 2-headed actin binding.
HMM is typically obtained by proteolysis from tissue derived myosin in high salt where the myosin is monomeric. This process potentially produces some heterogeneity through clipping at unintended sites but nevertheless circumvents the time intensive process of developing an expression system. Although we used recombinant HMM in this procedure, we think that specimens prepared by trypsin proteolysis in high salt may work as well and be more accessible, especially for striated muscle specimens from many invertebrates, for which expression systems will be difficult to construct. The myosin superfamily of molecular motors contains about 35 classes of myosin and 149 unclassified myosins, of which 18 classes are predicted to contain coiled-coil sequences associated with dimerization (Odronitz and Kollmar, 2007). Structural analysis of these might also be feasible using the same techniques described here for smooth muscle HMM.
5. Conclusion
Here we have used single particle cryoelectron microscopy to determine the structure of smooth muscle heavy meromyosin when bound to actin in the presence of above saturating levels of ADP. Focused classification proved successful in identifying the heterogeneity in the conformations of the lever arms from the 2D projections. Hence, desired two-headed motifs were clearly resolved. Binding of both heads of HMM to successive actins causes structural distortions in the lever arm, which may echo effects of tension on the lever arm. We conclude that the most compliant locus in the lever arm occurs in the myosin heavy chain between the portions that bind the ELC and the RLC. This compliance, which is shown to be asymmetric between the paired heads, has two components, axial and azimuthal. The axial compliance may store elastic energy during isometric contractions, whereas the azimuthal compliance may facilitate myosin-actin attachments.
Supplementary Material
Highlights.
An unanswered question in muscle biology is how some muscles, e.g. smooth muscle, can maintain tension for long periods of time with minimal ATP consumption.
The question could be addressed in vitro using cryoEM by developing a system in which strain was produced internally within the myosin molecule. Such a system could be F- actin in complex the soluble, 2-headed fragment of myosin called heavy meromyosin.
Here we have used a Ni-NTA lipid monolayer systems to prevent the protein components from contacting the air-water interface combined with single particle cryoEM, to visualize changes in the structure of myosin heads when both bind simultaneously to F- actin.
The paired myosin lever arms differ slightly in structure at the converter domain and essential light chain but differ much more in structure at the regulatory light chain consistent with flexibility in the myosin heavy chain between the two light chains.
Importantly, the lever arm changes were in opposite directions with one head appearing positively strained and the other head negatively strained.
6. Acknowledgements
This research supported by NIH grants R01 AR47421, R01 GM30598 and R35 GM139616 to KAT and R35 GM136288 to KMT. The Titan was partially funded from NIH grant S10 RR025080 with funds for the DE-64 and Volta Phase Plate provided by U24 GM116788 and S10 OD018142.
Footnotes
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. Data availability
The raw data are deposited under the accession code EMPIAR-11555. The 3D density map is available in EMDB under the accession code EMD-29646 and the intermediate density maps are available upon request. The molecular model has been deposited in PDB under the accession code 8SYF.
8. Bibliography
- Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, Urzhumtsev A, 2018. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakelian C, Warrington A, Winkler H, Perz-Edwards RJ, Reedy MK, Taylor KA, 2015. Myosin S2 origins track evolution of strong binding on actin by azimuthal rolling of motor domain. Biophys J 108, 1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee C, Hu Z, Huang Z, Warrington JA, Taylor DW, Trybus KM, Lowey S, Taylor KA, 2017. The structure of the actin-smooth muscle myosin motor domain complex in the rigor state. J Struct Biol 200, 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann E, Muller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S, 2012. Structure of the rigor actin-tropomyosin-myosin complex. Cell 150, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W, Thoma NH, Wray JS, Gautel M, Schlichting I, 2006. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci U S A 103, 17713–17717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Kumar VS, O’Neall-Hennessey E, Reshetnikova L, Robinson H, Nguyen-McCarty M, Szent-Gyorgyi AG, Cohen C, 2011. Visualizing key hinges and a potential major source of compliance in the lever arm of myosin. Proc Natl Acad Sci U S A 108, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Yang Y, Reshetnikova L, Gourinath S, Suveges D, Kardos J, Hobor F, Reutzel R, Nyitray L, Cohen C, 2008. An unstable head-rod junction may promote folding into the compact off-state conformation of regulated myosins. J Mol Biol 375, 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunello E, Reconditi M, Elangovan R, Linari M, Sun YB, Narayanan T, Panine P, Piazzesi G, Irving M, Lombardi V, 2007. Skeletal muscle resists stretch by rapid binding of the second motor domain of myosin to actin. Proc Natl Acad Sci U S A 104, 20114–20119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caremani M, Reconditi M, 2022. Anisotropic Elasticity of the Myosin Motor in Muscle. Int J Mol Sci 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty T, Yengo C, Baldacchino C, Chen LQ, Sweeney HL, Selvin PR, 2003. Does the S2 rod of myosin II uncoil upon two-headed binding to actin? A leucine-zippered HMM study. Biochemistry 42, 12886–12892. [DOI] [PubMed] [Google Scholar]
- Chantler PD, Wylie SR, Wheeler-Jones CP, McGonnell IM, 2010. Conventional myosins - unconventional functions. Biophys Rev 2, 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Blanc E, Chapman MS, Taylor KA, 2001. Real space refinement of acto-myosin structures from sectioned muscle. J Struct Biol 133, 221–232. [DOI] [PubMed] [Google Scholar]
- Chen LF, Winkler H, Reedy MK, Reedy MC, Taylor KA, 2002. Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle. J Struct Biol 138, 92–104. [DOI] [PubMed] [Google Scholar]
- Coureux PD, Sweeney HL, Houdusse A, 2004. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J 23, 4527–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremo CR, Geeves MA, 1998. Interaction of actin and ADP with the head domain of smooth muscle myosin: implications for strain-dependent ADP release in smooth muscle. Biochemistry 37, 1969–1978. [DOI] [PubMed] [Google Scholar]
- Cremo CR, Sellers JR, Facemyer KC, 1995. Two heads are required for phosphorylation-dependent regulation of smooth muscle myosin. J Biol Chem 270, 2171–2175. [DOI] [PubMed] [Google Scholar]
- Dantzig JA, Barsotti RJ, Manz S, Sweeney HL, Goldman YE, 1999. The ADP release step of the smooth muscle cross-bridge cycle is not directly associated with force generation. Biophys J 77, 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R, Freyzon Y, Trybus KM, Cohen C, 1998. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell 94, 559–571. [DOI] [PubMed] [Google Scholar]
- Doran MH, Rynkiewicz MJ, Rassici D, Bodt SML, Barry ME, Bullitt E, Yengo CM, Moore JR, Lehman W, 2023. Conformational changes linked to ADP release from human cardiac myosin bound to actin-tropomyosin. Journal of General Physiology 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson CP Jr., Bernstein SI, 1987. Molecular genetics of myosin. Annual Review of Biochemistry 56, 695–726. [DOI] [PubMed] [Google Scholar]
- Fujii T, Namba K, 2017. Structure of actomyosin rigour complex at 5.2 A resolution and insights into the ATPase cycle mechanism. Nat Commun 8, 13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC, 2005. The molecular mechanism of muscle contraction. Adv Protein Chem 71, 161–193. [DOI] [PubMed] [Google Scholar]
- Grant T, Rohou A, Grigorieff N, 2018. cisTEM, user-friendly software for single-particle image processing. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LE, Eisenberg E, 1980. The binding of heavy meromyosin to F-actin. J Biol Chem 255, 549–554. [PubMed] [Google Scholar]
- Hampton CM, Taylor DW, Taylor KA, 2007. Novel structures for alpha-actinin:F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J Mol Biol 368, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Scheres SHW, 2017. Helical reconstruction in RELION. J Struct Biol 198, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissler SM, Arora AS, Billington N, Sellers JR, Chinthalapudi K, 2021. Cryo-EM structure of the autoinhibited state of myosin-2. Sci Adv 7, eabk3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C, 2002. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A 99, 12645–12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Schroder RR, Sweeney HL, Houdusse A, 2004. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci 359, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR, 2003. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425, 423–427. [DOI] [PubMed] [Google Scholar]
- Houdusse A, Cohen C, 1996. Structure of the regulatory domain of scallop myosin at 2 A resolution: implications for regulation. Structure 4, 21–32. [DOI] [PubMed] [Google Scholar]
- Jontes JD, Milligan RA, 1997. Brush border myosin-I structure and ADP-dependent conformational changes revealed by cryoelectron microscopy and image analysis. J. Cell Biol. 139, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jontes JD, Wilson-Kubalek EM, Milligan RA, 1995. A 32 degree tail swing in brush border myosin I on ADP release. Nature 378, 751–753. [DOI] [PubMed] [Google Scholar]
- Jung HS, Komatsu S, Ikebe M, Craig R, 2008a. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell 19, 3234–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ, 2008b. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci U S A 105, 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiyama H, 1988. Shape of the myosin head in the rigor complex. Three-dimensional image reconstruction of the actin-tropomyosin-heavy meromyosin complex. J Mol Biol 204, 639–652. [DOI] [PubMed] [Google Scholar]
- Katayama E, Wakabayashi T, 1981. Three-dimensional image analysis of the complex of thin filaments and myosin molecules from skeletal muscle. III. The multi-domain structure of actin-heavy meromyosin complex. J Biochem 90, 703–714. [DOI] [PubMed] [Google Scholar]
- Kidmose RT, Juhl J, Nissen P, Boesen T, Karlsen JL, Pedersen BP, 2019. Namdinator - automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PJ, 1996. Dynamic behaviour of the head-tail junction of myosin. J Mol Biol 255, 269–274. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Darst SA, 1991. Two-dimensional crystals of proteins on lipid layers. Curr Op Struct Biol 1, 642–646. [Google Scholar]
- Lauzon AM, Fagnant PM, Warshaw DM, Trybus KM, 2001. Coiled-coil unwinding at the smooth muscle myosin head-rod junction is required for optimal mechanical performance. Biophys J 80, 1900–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Brown JH, Reshetnikova L, Blazsek A, Farkas L, Nyitray L, Cohen C, 2003. Visualization of an unstable coiled coil from the scallop myosin rod. Nature 424, 341–345. [DOI] [PubMed] [Google Scholar]
- Littlefield KP, Ward AB, Chappie JS, Reedy MK, Bernstein SI, Milligan RA, Reedy MC, 2008. Similarities and differences between frozen-hydrated, rigor acto-S1 complexes of insect flight and chicken skeletal muscles. J Mol Biol 381, 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wendt T, Taylor DW, Taylor KA, 2003. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J Mol Biol 329, 963–972. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu S, Reedy MC, Winkler H, Lucaveche C, Cheng Y, Reedy MK, Taylor KA, 2006. Electron tomography of swollen rigor fibers of insect flight muscle reveals a short and variably angled S2 domain. J Mol Biol 362, 844–860. [DOI] [PubMed] [Google Scholar]
- Liu J, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, Tregear RT, Lucaveche C, Winkler H, Baumann BA, Squire JM, Irving TC, Reedy MK, Taylor KA, 2004. Electron tomography of fast frozen, stretched rigor fibers reveals elastic distortions in the myosin crossbridges. J Struct Biol 147, 268–282. [DOI] [PubMed] [Google Scholar]
- Lovell SJ, Knight PJ, Harrington WF, 1981. Fraction of myosin heads bound to thin filaments in rigor fibrils from insect flight and vertebrate muscles. Nature 293, 664–666. [DOI] [PubMed] [Google Scholar]
- Lowey S, Trybus KM, 2010. Common structural motifs for the regulation of divergent class II myosins. J Biol Chem 285, 16403–16407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S, Slayter HS, Weeds AG, Baker H, 1969. Substructure of the myosin molecule. I. Subfragments of myosin by enzymic degradation. J Mol Biol 42, 1–29. [DOI] [PubMed] [Google Scholar]
- McLachlan AD, 1983. Analysis of gene duplication repeats in the myosin rod. J Mol Biol 169, 15–30. [DOI] [PubMed] [Google Scholar]
- McLachlan AD, Karn J, 1982. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature 299, 226–231. [DOI] [PubMed] [Google Scholar]
- Mentes A, Huehn A, Liu X, Zwolak A, Dominguez R, Shuman H, Ostap EM, Sindelar CV, 2018. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc Natl Acad Sci U S A 115, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PB, Huxley HE, DeRosier DJ, 1970. Three-dimensional reconstruction of F-actin, thin filaments and decorated thin filaments. J Mol Biol 50, 279–295. [DOI] [PubMed] [Google Scholar]
- Mornet D, Bertrand RU, Pantel P, Audemard E, Kassab R, 1981. Proteolytic approach to structure and function of actin recognition site in myosin heads. Biochemistry 20, 2110–2120. [DOI] [PubMed] [Google Scholar]
- Nyitrai M, Geeves MA, 2004. Adenosine diphosphate and strain sensitivity in myosin motors. Philos Trans R Soc Lond B Biol Sci 359, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M, 2007. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pospich S, Sweeney HL, Houdusse A, Raunser S, 2021. High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani H, Ma W, Hu Z, Daneshparvar N, Taylor DW, McCammon JA, Irving TC, Edwards RJ, Taylor KA, 2021. The myosin II coiled-coil domain atomic structure in its native environment. Proc Natl Acad Sci U S A 118, e202415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA, 1993a. Structure of the actin-myosin complex and its implications for muscle contraction. Science 261, 58–65. [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM, 1993b. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261, 50–58. [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N, 2015. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarff CA, Carrington G, Casas-Mao D, Chalovich JM, Knight PJ, Ranson NA, Peckham M, 2020. Structure of the shutdown state of myosin-2. Nature 588, 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder RR, Manstein DJ, Jahn W, Holden H, Rayment I, Holmes KC, Spudich JA, 1993. Three-dimensional atomic model of F-actin decorated with Dictyostelium myosin S1. Nature 364, 171–174. [DOI] [PubMed] [Google Scholar]
- Sellers JR, 2000. Myosins: a diverse superfamily. Biochim Biophys Acta 1496, 3–22. [DOI] [PubMed] [Google Scholar]
- Sellers JR, Eisenberg E, Adelstein RS, 1982. The binding of smooth muscle heavy meromyosin to actin in the presence of ATP. Effect of phosphorylation. J Biol Chem 257, 13880–13883. [PubMed] [Google Scholar]
- Seymour J, O’Brien EJ, 1985. Structure of myosin decorated actin filaments and natural thin filaments. J Muscle Res Cell Motil 6, 725–755. [DOI] [PubMed] [Google Scholar]
- Smith CA, Rayment I, 1995. X-ray structure of the magnesium(II)-pyrophosphate complex of the truncated head of Dictyostelium discoideum myosin to 2.7 Å resolution. Biochemistry 34, 8973–8981. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S, 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem 246, 4866–4871. [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B, 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Kelly DF, Cheng A, Taylor KA, 2007. On the freezing and identification of lipid monolayer 2-D arrays for cryoelectron microscopy. J Struct Biol 160, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM, 2008. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J Struct Biol 163, 214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Cordova L, Reedy MK, 1984. Three-dimensional reconstruction of rigor insect flight muscle from tilted thin sections. Nature 310, 285–291. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Cordova L, Reedy MK, 1989. Three-dimensional image reconstruction of insect flight muscle. I. The rigor myac layer. J. Cell Biol. 109, 1085–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Reedy MK, Crowther RA, 1993. Crossbridges in the complete unit cell of rigor insect flight muscle imaged by three-dimensional reconstruction from oblique sections. J Mol Biol 233, 86–108. [DOI] [PubMed] [Google Scholar]
- Thomas DD, Cooke R, Barnett VA, 1983. Orientation and rotational mobility of spinlabelled myosin heads in insect flight muscle in rigor. J Muscle Res Cell Motil 4, 367–378. [DOI] [PubMed] [Google Scholar]
- Toyoshima YY, Toyoshima C, Spudich JA, 1989. Bidirectional movement of actin filaments along tracks of myosin heads. Nature 341, 154–156. [DOI] [PubMed] [Google Scholar]
- Trybus KM, Freyzon Y, Faust LZ, Sweeney HL, 1997. Spare the rod, spoil the regulation: necessity for a myosin rod. Proc Natl Acad Sci U S A 94, 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Dupuis DE, Guilford WH, Patlak JB, Waller GS, Trybus KM, Warshaw DM, Lowey S, 1999. Two heads of myosin are better than one for generating force and motion. Proc Natl Acad Sci U S A 96, 4402–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veigel C, Coluccio LM, Jontes JD, Sparrow JC, Milligan RA, Molloy JE, 1999. The motor protein myosin-I produces its working stroke in two steps. Nature 398, 530–533. [DOI] [PubMed] [Google Scholar]
- Vilas JL, Gomez-Blanco J, Conesa P, Melero R, Miguel de la Rosa-Trevin J, Oton J, Cuenca J, Marabini R, Carazo JM, Vargas J, Sorzano COS, 2018. MonoRes: Automatic and Accurate Estimation of Local Resolution for Electron Microscopy Maps. Structure 26, 337–344 e334. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D, Ouyang G, Trybus KM, DeRosier DJ, Lowey S, 2000. Evidence for cleft closure in actomyosin upon ADP release. Nat Struct Biol 7, 1147–1155. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Ouyang G, Trybus KM, DeRosier DJ, Lowey S, Hanein D, 2003. Myosin isoforms show unique conformations in the actin-bound state. Proc Natl Acad Sci U S A 100, 3227–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N, Liu H, Hazelwood L, Krementsova EB, Lowey S, Trybus KM, Hanein D, 2005. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell 19, 595–605. [DOI] [PubMed] [Google Scholar]
- von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S, 2016. CryoEM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 534, 724–728. [DOI] [PubMed] [Google Scholar]
- Wahlstrom JL, Randall MA Jr., Lawson JD, Lyons DE, Siems WF, Crouch GJ, Barr R, Facemyer KC, Cremo CR, 2003. Structural model of the regulatory domain of smooth muscle heavy meromyosin. J Biol Chem 278, 5123–5131. [DOI] [PubMed] [Google Scholar]
- Wang Z, Grange M, Wagner T, Kho AL, Gautel M, Raunser S, 2021. The molecular basis for sarcomere organization in vertebrate skeletal muscle. Cell 184, 2135–2150 e2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Grange M, Pospich S, Wagner T, Kho AL, Gautel M, Raunser S, 2022. Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin. Science 375, abn1934. [DOI] [PubMed] [Google Scholar]
- Warrick HM, Spudich JA, 1987. Myosin structure and function in cell motility. Annu.Rev.Cell Biol 3, 379–421. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T, 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, Taylor K, 2001. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A 98, 4361–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA, 1999. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J. Cell Biol. 147, 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M, Wilson-Kubalek EM, Smith JE, Faust L, Milligan RA, Sweeney HL, 1995. A 35-A movement of smooth muscle myosin on ADP release. Nature 378, 748–751. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu J, Reedy MC, Tregear RT, Winkler H, Franzini-Armstrong C, Sasaki H, Lucaveche C, Goldman YE, Reedy MK, Taylor KA, 2010. Electron tomography of cryofixed, isometrically contracting insect flight muscle reveals novel actin-myosin interactions. PLoS One 5, e12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Liu J, Reedy MC, Perz-Edwards RJ, Tregear RT, Winkler H, Franzini-Armstrong C, Sasaki H, Lucaveche C, Goldman YE, Reedy MK, Taylor KA, 2012. Structural changes in isometrically contracting insect flight muscle trapped following a mechanical perturbation. PLoS One 7, e39422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Clack BA, Zhi G, Stull JT, Cremo CR, 1999. Phosphorylation-dependent structural changes in the regulatory light chain domain of smooth muscle heavy meromyosin. J Biol Chem 274, 20328–20335. [DOI] [PubMed] [Google Scholar]
- Wulf SF, Ropars V, Fujita-Becker S, Oster M, Hofhaus G, Trabuco LG, Pylypenko O, Sweeney HL, Houdusse AM, Schroder RR, 2016. Force-producing ADP state of myosin bound to actin. Proc Natl Acad Sci U S A 113, E1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Harrison DH, Schlichting I, Sweet RM, Kalabokis VN, Szent-Gyorgyi AG, Cohen C, 1994. Structure of the regulatory domain of scallop myosin at 2.8 A resolution. Nature 368, 306–312. [DOI] [PubMed] [Google Scholar]
- Yang S, Tiwari P, Lee KH, Sato O, Ikebe M, Padron R, Craig R, 2020. Cryo-EM structure of the inhibited (10S) form of myosin II. Nature 588, 521–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA, 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH, 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data are deposited under the accession code EMPIAR-11555. The 3D density map is available in EMDB under the accession code EMD-29646 and the intermediate density maps are available upon request. The molecular model has been deposited in PDB under the accession code 8SYF.