Abstract
Positron emission tomography (PET) with a reduced injection dose, i.e., low-dose PET, is an efficient way to reduce radiation dose. However, low-dose PET reconstruction suffers from a low signal-to-noise ratio (SNR), affecting diagnosis and other PET-related applications. Recently, deep learning-based PET denoising methods have demonstrated superior performance in generating high-quality reconstruction. However, these methods require a large amount of representative data for training, which can be difficult to collect and share due to medical data privacy regulations. Moreover, low-dose PET data at different institutions may use different low-dose protocols, leading to non-identical data distribution. While previous federated learning (FL) algorithms enable multi-institution collaborative training without the need of aggregating local data, it is challenging for previous methods to address the large domain shift caused by different low-dose PET settings, and the application of FL to PET is still under-explored. In this work, we propose a federated transfer learning (FTL) framework for low-dose PET denoising using heterogeneous low-dose data. Our experimental results on simulated multi-institutional data demonstrate that our method can efficiently utilize heterogeneous low-dose data without compromising data privacy for achieving superior low-dose PET denoising performance for different institutions with different low-dose settings, as compared to previous FL methods.
Keywords: Low-dose PET, Denoising, Federated learning, Transfer learning
I. Introduction
Positron emission tomography (PET) is a functional imaging modality that measures molecular-level activities inside tissues and has wide applications in oncology, cardiology, neurology, and biomedical research. To obtain high-quality PET images for diagnostic purposes, a given dose of radioactive tracer is injected into patients, which inevitably introduces radiation exposure to both patients and healthcare providers. According to the principle of As Low As Reasonably Achievable (ALARA) [1], reducing the administered injection dose is of great interest to the patients, in particular for oncology applications where serial scans are performed to measure response to therapy. However, compared to full-dose PET, low-dose PET with a reduced injection dose often results in low signal-to-noise ratio (SNR) and potential image artifacts in the reconstructed image, leading to negative impacts on the downstream clinical tasks.
To reduce the injection dose and to maintain the PET image quality, deep learning-based PET denoising methods have been developed [2]–[13] to generate full-dose PET image from low-dose PET image, demonstrating superior reconstruction performance when compared with conventional methods [14]–[16]. However, the superior denoising performance of deep learning-based methods often relies on a large amount of representative paired data, which could be prohibitively expensive to collect. One way to alleviate this issue is through building a centralized dataset with multi-institutional data, but the patient data privacy concern poses a major challenge to such a solution [17], [18].
Recently, federated learning (FL) addresses the data privacy issue by building a client-to-cloud platform for different local clients to learn collaboratively using their local data and computing resources without sharing any private local data. Specifically, the cloud server periodically communicates with local clients to collect local models for aggregating a global model, and re-distributes to local clients for local updates. Instead of directly transferring local data for global training, the FL method only involves the communication of model parameters or gradients, thus solving the data privacy concerns.
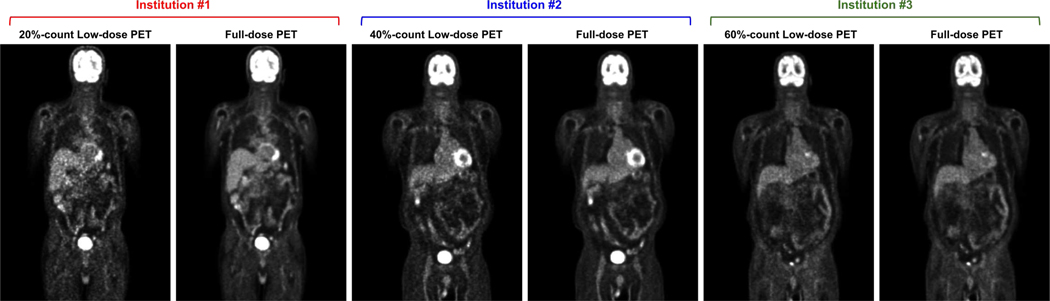
In the context of low-dose PET imaging, the different low-dose settings with various injection doses of different institutions present data heterogeneity, leading to domain shifts between different institutions. Examples of PET data with different low-dose settings, simulating multi-institution scenarios, are illustrated in Figure 1. The noise level of reconstruction gradually increases as the dose level decreases across different institution’s settings, leading to potential domain shifts. Unfortunately, the generalization capability of the global model trained with classical FL algorithm, such as FedAvg [19], may be non-ideal under these conditions. Previous works have attempted to address the domain shift issues, but only focuses on image classification task and only consider small domain shift scenarios [20]–[23]. In the application of MR reconstruction using FL, Guo et al. [24] tried to address the domain shift issue by iteratively aligning the latent feature of UNet [25] between target and other client sites. However, their cross-site strategy requires the target client to share both the latent feature and the network parameter with other client sites in each communication rounds, which could result in data privacy concerns [26], [27]. Moreover, the cross-site strategy requires communications between local clients with their local data which may contradict the purpose of FL. The communication process during the training will also be tedious and expensive when the number of clients is large, requiring high-frequency communication of both model parameters and latent features. Similarly, Feng et al. [28] tried to address the domain shift issue by using a UNet [25] with a globally shared encoder for generalized representation learning and a client-specific decoder for domain-specific reconstruction. While achieving promising performance for the MR reconstruction task, the network architecture is limited to UNet or its variants due to the constraint in their FL algorithm designs. Moreover, instead of simple encoder-decoder structures, state-of-the-art deep learning-based image restoration networks often involve advanced designs, such as original resolution restoration [29], recurrent restoration [30], and multi-stage restoration [31]. Therefore, designing an FL algorithm that is not limited to specific network architecture while addressing the domain shift issue is an important task for medical imaging restoration. Furthermore, none of the aforementioned FL methods is for low-dose PET denoising and the application of federated learning for low-dose PET denoising is still highly under-explored.
Fig. 1.
Illustration of PET patient data with three unique low-dose settings, where 20%-count, 40%-count, 60%-count low-dose settings were utilized to simulate the multi-institutional data, respectively. The corresponding full-dose PET is shown on the right of the low-dose PET. The lower the dose level, the higher the noise level in the reconstructed image, thus causing a non-identical data distribution.
To address these issues, we propose a federated transfer learning (FTL) framework for low-dose PET denoising using heterogeneous low-dose data. The general idea is to first obtain a global pre-trained model from collaborative training from multi-institution datasets. With a global model trained with large-scale and diverse data, it is then transferred to local institutions for fine-tuning, such that the domain shift issue can be mitigated with local data only training and better generalized to the local data domain. Thus, our FTL consists of two major steps. First, we employ a standard federated averaging method where the models are independently trained at each institution and iteratively sent over to the central server for aggregation. After convergence, the pre-trained global model is downloaded by each institution for transfer learning on local low-dose data distribution. The proposed FL framework can be flexibly adapted to different denoising network architectures. From our experimental results on simulated multi-institutional data, we found our FTL, a simple yet efficient solution, can generate superior PET denoising results at different institutions with different low-dose settings, as compared to previous baseline methods.
II. Related Works
Low-dose PET Denoising.
Prior works on low-dose PET denoising can be categorized into conventional image post-processing methods [14]–[16] and deep learning-based methods [2]–[13]. Conventional methods, such as Gaussian filtering, are standard post-processing procedures for PET denoising. However, with amplified noise levels under low-dose PET settings, these conventional methods are prone to over-smoothing the image and have challenges to preserve local structures. With recent advances in deep learning for natural image restoration [29]–[31], deep learning-based denoising methods have been developed for low-dose PET and have achieved promising performance. Kaplan et al. [5] used a 2D GAN [32] with UNet [25] as a generator to predict full-dose PET images from low-dose PET images. Wang et al. [3] further advanced the idea and used a 3D conditional GAN [33] to directly translate 3D low-dose PET images to full-dose PET images. Similarly, Gong et al. [7] proposed to use a 3D Wasserstein GAN [34] which also achieved promising low-dose PET denoising performance. Based on previous GAN designs, Ouyang et al. [9] proposed to reinforce the denoising performance by incorporating patient-specific information. In addition to approaches using only the low-dose PET images as network input, low-dose PET denoising facilitated by other imaging modalities has also been investigated. For example, Xiang et al. [2] developed an auto-context CNN using both low-dose PET images and T1 MR images as inputs for full-dose PET generation. Similarly, Chen et al. [10] proposed to input low-dose PET images along with multi-contrast MR images into a UNet [25] for ultra-low-dose PET denoising. While deep learning-based methods have achieved promising denoising performance on low-dose PET, these previous works only investigated low-dose PET under single-institution with single-low-dose-level settings, where different institutions usually do not share the same low-dose protocol. Investigation on how to use multi-institutional low-dose data with different low-dose protocols while considering the data privacy issue to train models that can be generalized to different institutions is an important research topic and is still under-explored.
Federated Learning.
FL with a decentralized learning framework enables multiple local institutions to collaborate in training shared models, while maintaining their local data privacy [35]. Even though a classical FL algorithm, such as FedAvg [19], allows collaborative training without sharing data, it does not address the domain shift issue caused by the data distribution difference between different clients. In the classification task, recent studies mitigate this issue by keeping parts of the network parameters local, with solutions such as local batch normalization [20] and keeping the last output layer local [21], [22]. However, these methods are limited to small domain shift, e.g. covariance shift, and may have sub-optimal performance when domain shift is large. To protect patient data privacy while enabling collaborative training, FL has also been applied to medical imaging tasks and demonstrated promising performance [24], [28], [36]–[39]. For example, Li et al. [38] applied the FedAvg [19] for the brain MR tumor segmentation task. Similarly, Feki el al. [37] investigated the use of FedAvg [19] for the chest X-ray COVID-19 classification task. However, none of these works consider the domain shift caused by the data heterogeneity across different institutions. Recently, for the fMRI classification task, Li et al. [39] investigated using federated adversarial domain alignment [40] to reduce the domain shift for improved classification performance. Guo et al. [24] proposed federated learning with cross-site modeling (FLCM) to address the domain shift issues in the application of MR reconstruction by iteratively aligning the latent feature distribution between the clients. However, their method requiring frequent communication between clients increases the communication cost and the risk of potential privacy leakage. As an alternative, Feng et al. [28] proposed to use a client-specific decoder to reduce the domain shift when reconstructing MR images. However, these methods require specific types of network architecture, i.e. the encoder-decoder structure with a latent feature, and cannot be generalized to applications requiring other network architectures for optimal performance. For example, in the application of medical image reconstruction and restoration, cascaded/recurrent network design with fully-complete backbone network architectures have demonstrated superior performance as compared to encoder-decoder type networks [30], [41]–[44]. Therefore, it is desirable to develop a federated learning framework that can be adapted to networks with flexible architecture while addressing the domain shift issue. Furthermore, federated learning for low-dose PET denoising using heterogeneous low-dose data has not been well studied in previous works.
Algorithm 1:
Personalized Federated Learning with Transfer Learning for PET Denoising.
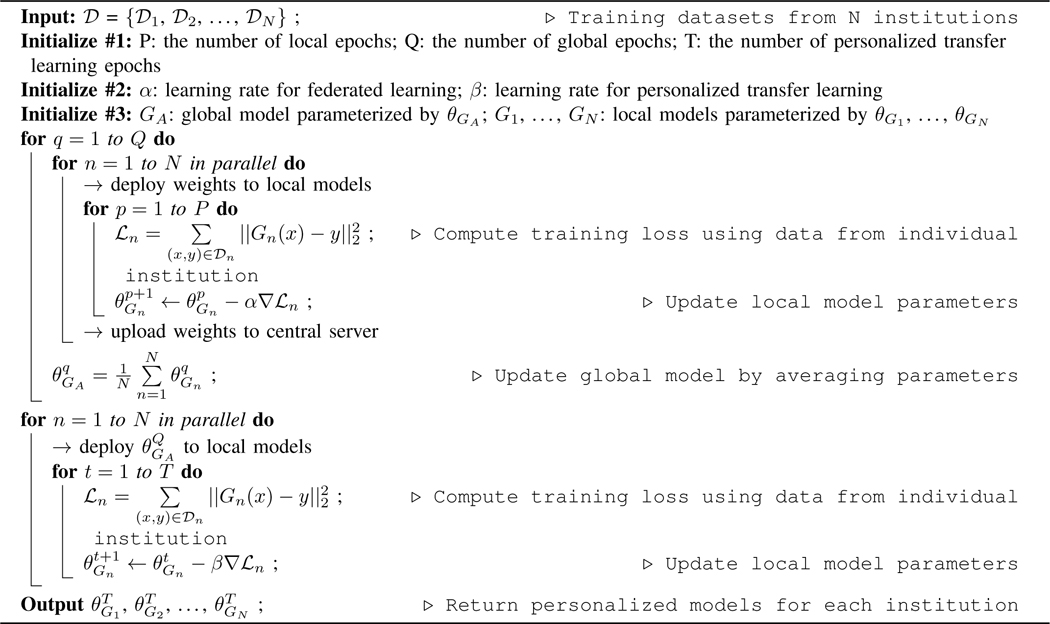
|
III. Methods
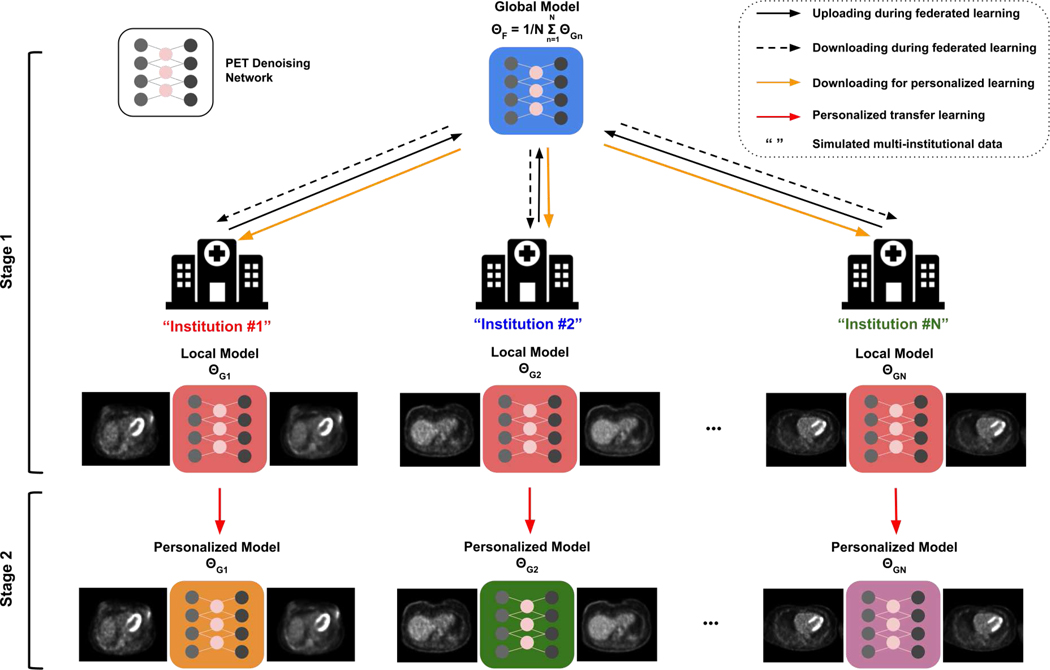
The general pipeline of our federated transfer learning (FTL) is illustrated in Figure 2. The FTL is a two-stage framework, consisting of collaborative training of a global model using all institution data with federated learning, and then using transfer learning to train individual models with local data only, such that the data privacy is preserved.
Fig. 2.
The framework of the Federated Transfer Learning (FTL). During the first stage, with iterative communication between central server and local models in individual institutions, a global model collaboratively trained by all the institution data is obtained in a data-preserving manner. During the second stage, the global model is downloaded by individual institution and further fine-tuned by the dataset hold by each institution.
A. Federated Transfer Learning Framework
Denoting as low-dose PET datasets from N different institutions. Each local dataset contains pairs of full-dose and low-dose PET 3D images, but with different low-dose settings. Within each institution, the local denoising model can be trained using the local data by iteratively optimizing:
| (1) |
where Gn is the local denoising model at the nth institution, and parameterized by . x and y form a low-dose and full-dose PET image pair from. The L2 loss is computed between the predicted full-dose image and the ground truth full dose image y. Since we assume the data collected at each institution uses different low-dose settings, non-identical data distribution is expected among. Therefore, a locally trained denoising model may not generalize well to another institution’s low-dose PET data. One potential way to alleviate this issue is to train the denoising model using a combined dataset from N institutions, i.e.,. However, it is infeasible to directly combine multi-institution data due to data privacy concerns that might prevent data sharing across institutions.
To address this issue and allow multiple institutions to collaboratively train personalized PET denoising models that perform well on their own data distribution, we use a two-stage framework. In stage 1, we employ a federated averaging method to obtain a global model. Assuming there are Q global training epochs and P local training epochs at each institution, the iterative optimization of local model parameter can be written as:
| (2) |
where is the denoising loss function defined in Eq. (1), and α is the learning rate in stage 1. At the end of each global training epoch, the local model parameters at each institution are uploaded to a central server for aggregating the local model updates. The central server updates the global model with local model updates by averaging the updated parameters from all local models:
| (3) |
where q denotes the q-th global epoch. After Q rounds of communication between local institutions and the central server, we can obtain a collaboratively-trained global model GA with parameter by utilizing multi-institutional datasets while without directly accessing them.
In stage 2, the trained global model with is downloaded by each institution for personalized fine-tuning. Assuming there are T fine-tuning training epochs at each institution, the iterative fine-tuning of local model can be written as:
| (4) |
where t denotes the t-th fine-tuning epoch, and β is the learning rate in stage 2. After T epochs of training using the local dataset, we obtain the personalized models G1,...,GN with parameters of . The algorithm is summarized in Algorithm 1.
B. Network Architecture and Implementation Details
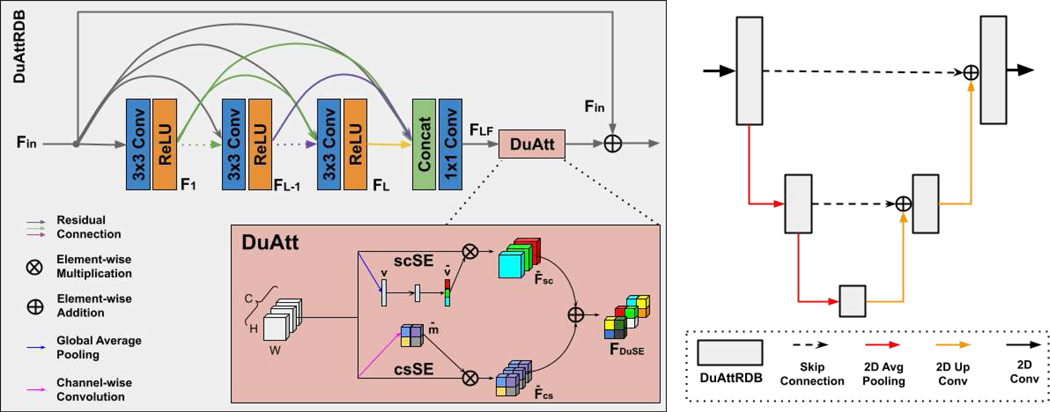
In our experiment, we use a dual attention residual dense U-Net (DuAttRDUNet) as our PET denoising network [45]. The architecture details are illustrated in Figure 3. The DuAttRDUNet is a U-shape network with dual attention residual dense blocks (DuAttRDB) embedded at different resolution levels. The DuAttRDB consists of a densely connected module [46], [47], a dual attention module [48], and a residual connection. Here, we set the number of input feature of each DuAttRDB to 64. Since PET is 3D imaging data and its FOV varies in the z direction, we implement a 2.5D DuAttRDUNet that takes five continuous axial slices as inputs and predicts the denoised central slice. We implemented our FTL in Python with Pytorch1. The Adam solver [49] was used to optimize the denoising network in both stages of the FTL. During the first stage training, we used a constant learning rate of 1e−4 and trained for 90 global epochs. The number of local epoch is set to 2. During the second stage training, we used a reduced constant learning rate of 2e − 5 and trained the local model using local dataset for 10 epochs. The batch size is set to 8 in both stages.
Fig. 3.
The network architecture of Dual Attention Residual Dense U-Net (DuAttRDUNet) used in the FTL framework (Figure 2).
C. Heterogeneous Low-dose Human Data
To simulate a multi-institutional dataset with heterogeneous low-dose settings for our study, we collected 175 subjects in our experiments. The subjects were injected with a 18F-FDG tracer and the whole-body protocol with continuous-bedmotion scanning was used. All data were acquired using a Siemens Biograph mCT PET/CT system. The 175 subjects were split into 120 subjects for training and 55 subjects for evaluation. The 120 subjects were further split into 3 groups with 40 subjects in each group and with no patient overlapping between groups. Each group was treated as one individual institutional data. To create heterogeneous low-dose data that vary between groups, we used uniform down-sampling of the patient list-mode data with down-sampling ratios of 20%, 40%, and 60% for the three groups, respectively. For the 55 subjects in the evaluation set, we also used uniform down-sampling of the patient list-mode data with down-sampling ratios of 20%, 40%, and 60%, such that each subject has three low-dose images for evaluation. In the evaluation set, 10 subjects with lung lesions were annotated on tumor regions at voxel-level which were used for the evaluation of tumor quantification. For both the low-dose and full-dose images, they were reconstructed using the ordered-subsets expectation maximization (OSEM) algorithm with 2 iterations and 21 subsets, provided by the vendor. A post-reconstruction Gaussian filter with 5mm full width at half maximum (FWHM) was used. The voxel size of the reconstructed image was 4.07 × 4.07 × 3mm3. The image size was 200 × 200 in the transverse plane and varied in the axial direction depending on patient height.
D. Evaluation Metrics and Baselines
For denoising evaluation, we computed the Peak Signal-to-Noise Ratio (PSNR), Structural Similarity Index (SSIM), and Normalized Mean Square Errors (NMSE) between the predicted full-dose image and the ground truth full-dose image. RMSE and PSNR stress the evaluation of intensity profile recovery, while SSIM focuses on the evaluation of structural recovery. To evaluate the denoising performance on important pathological regions, such as lung tumor, we also calculated the lesion region’s bias using:
| (5) |
where R is the lesion region. and are the predicted and ground-truth full dose image, respectively. For comparative study, we compared our method against four previous medical image restoration with FL methods, including FedAvg [19], FLCM [24], FedPer [21], [28], and FedSP [28] without the contrastive regularization. For a fair comparison, we used the DuAttRDUNet as the denoising network in all the compared methods. In FedPer, the last output layer was set as the personalized layer and is local.
IV. Results
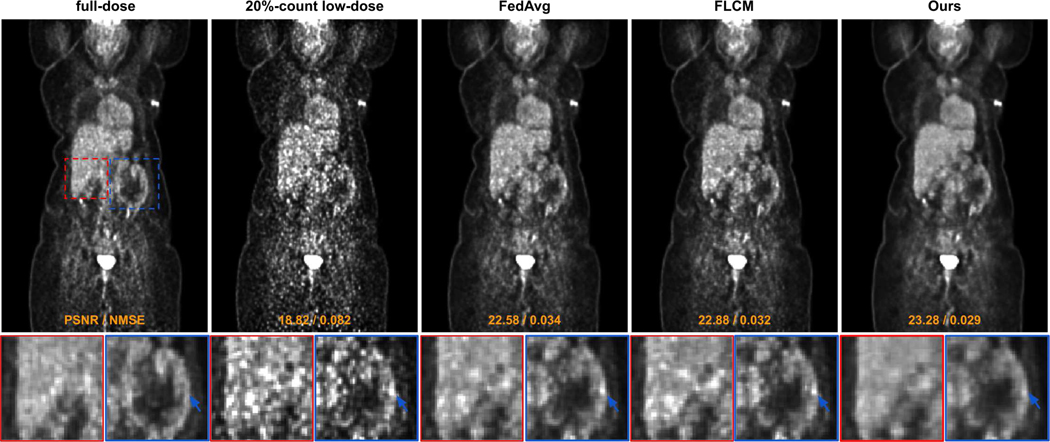
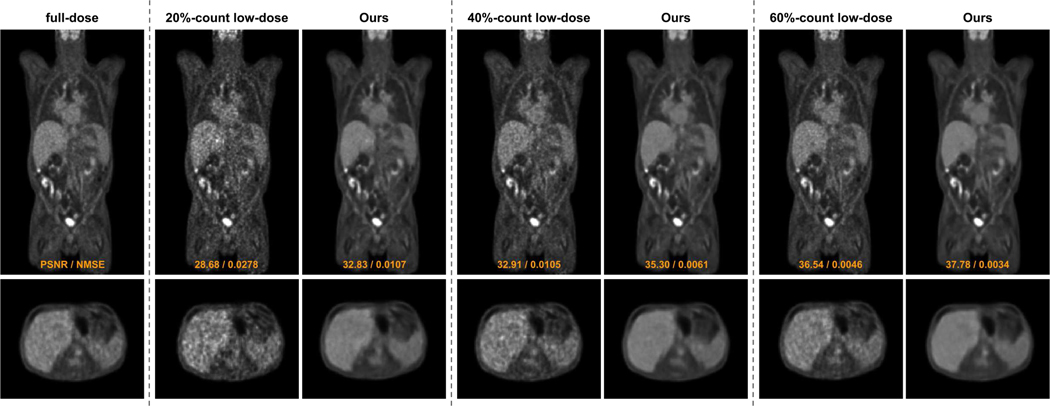
The visual comparisons between our FTL and previous FL methods under 20%-count low-dose setting are shown in Figure 4. With the coronal view visualization, we can observe that the 20%-count low-dose image suffers from a high noise level due to insufficient counts in the acquisition. Comparing the full-dose to the low-dose image, we can observe that the noise causes artefactual hot spots across the chest, abdominal, and pelvic regions. While FedAvg can reduce the global noise, the artefactual hot spots remain in the liver and intestine regions (zoom-in regions in Figure 4). Similar observations can be found for the FLCM where the false positive hot spots presented in the original low-dose image still remain in the denoised image. As compared to previous methods, our FTL can generate reconstruction with intensity and structure best matched with the full-dose ground truth image, where the artefactual hot spots are eliminated in the liver, intestine, and cardiac regions by our method. In Figure 5, we show an additional visual comparison of our denoising performance under three different low-dose settings corresponding to three different institutional settings. As we can see, original reconstruction with a lower dose level leads to a higher noise level. The personalized denoising model from the FTL can consistently generate high-quality denoised images across various low-dose levels of different institutional settings.
Fig. 4.
Qualitative comparison of low-dose reconstructions using different federated learning methods under the 20%-count low-dose setting (institution #1).
Fig. 5.
Qualitative comparison of FTL reconstructions on multi-institution dataset with 20%-count, 40%-count, and 60%-count low-dose settings.
The quantitative evaluation of different FL methods under different low-dose settings from different institutions is shown in Table I. In the 20%-count low-dose experiment, compared to the original low-dose reconstruction, our method can improve the image quality with NMSE decreased from 0.0292 to 0.0124 and PSNR increased from 25.86 to 29.24 on average, outperforming the previous baseline methods. Similarly, in the 40%-count low-dose experiment, our method achieves the best performance as compared to previous baseline methods, maintaining the NMSE lower than 0.0065 and PSNR over 32.30. Comparing the experiments from 20%-count to 40%-count low-dose settings, the reconstruction improvement of our method is mainly due to the increase in dose level, where the PSNR already increased from 25.86 to 30.24 in the original reconstruction. Similar observations can be found in the 60%-count low-dose experiment. The evaluation of lesion region’s bias is summarized in Table II. In the 20%-count low-dose experiment, compared to the original low-dose reconstruction, our method can reduce the bias from 0.007 ± 0.023 to 0.004 ± 0.015, better than the previous baseline methods. Similar observations can be found in both the 40%-count and 60%-count low-dose experiments.
TABLE I.
Quantitative comparison of reconstructions using different federated learning methods. We evaluated on three different institution data with three different low-dose settings. Best results are marked in bold.
| Evaluation | 20%-count data | 40%-count data | 60%-count data | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SSIM | NMSE | PSNR | SSIM | NMSE | PSNR | SSIM | NMSE | PSNR | |
| LDPET | .964 | 0.0292 | 25.86 | .982 | 0.0106 | 30.24 | .991 | 0.0047 | 33.77 |
| FedAvg [19] | .977 | 0.0138 | 28.83 | .985 | 0.0070 | 31.76 | .986 | 0.0042 | 34.03 |
| FLCM [24] | .978 | 0.0132 | 29.01 | .986 | 0.0065 | 32.13 | .991 | 0.0042 | 34.09 |
| FedPer [21] | .977 | 0.0131 | 29.02 | .985 | 0.0072 | 31.73 | .992 | 0.0038 | 34.48 |
| FedSP [28] | .978 | 0.0129 | 29.09 | .986 | 0.0066 | 32.09 | .991 | 0.0039 | 34.37 |
| Ours | .978†∗ | 0.0124 †‡∗⋆ | 29.24 †‡∗⋆ | .987†‡∗⋆ | 0.0064 †‡∗⋆ | 32.38 †‡∗⋆ | .993†‡∗⋆ | 0.0037 †‡∗⋆ | 34.67 †‡∗⋆ |
TABLE II.
Comparison of lesion region’s bias from different federated learning reconstruction methods. We evaluated on three different institution data with three different low-dose settings. Standard deviations were reported in (). Best results are marked in bold.
| 20%-count | 40%-count | 60%-count | |
|---|---|---|---|
| LDPET | 0.007(0.023) | 0.006(0.019) | 0.002(0.010) |
| FedAvg [19] | 0.006(0.021) | 0.005(0.017) | 0.001(0.009) |
| FLCM [24] | 0.005(0.019) | 0.004(0.015) | 0.001(0.008) |
| FedPer [21] | 0.005(0.018) | 0.004(0.016) | 0.001(0.008) |
| FedSP [28] | 0.005(0.019) | 0.004(0.014) | 0.001(0.007) |
| Ours | 0.004(0.015) | 0.003(0.012) | 0.001(0.004) |
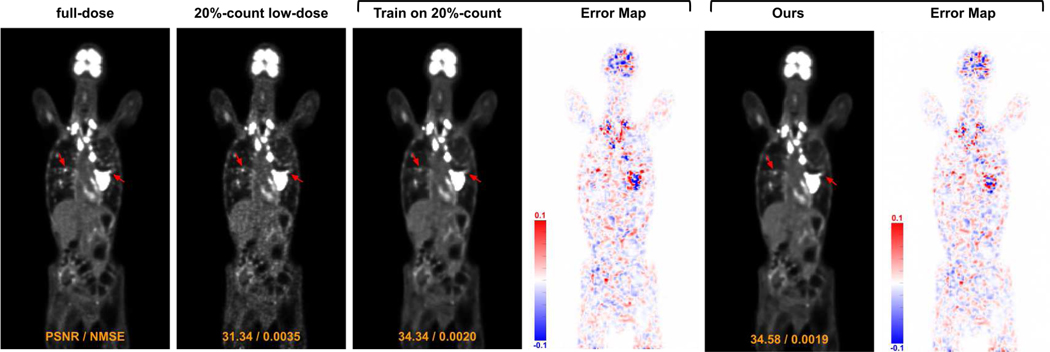
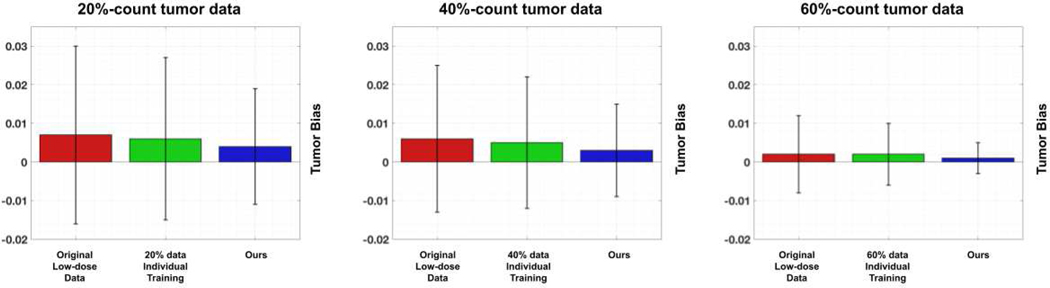
For ablative studies, we first compared the performance of denoising models trained from scratch using only local data v.s trained using our FTL framework. In Figure 6, we demonstrate the visual comparison of low-dose reconstructions from denoising models trained from a single institution 20%count low-dose paired data and trained from our FTL. As we can see, while the denoising model trained with only local data can reduce the global noise level, it also reduces the hypermetabolic hot spot signals presented in the full-dose images. On the other hand, our FTL not only reduces the global noise level but also maintains the hot spot signals in the final reconstructions. The corresponding quantitative evaluation is summarized in Table III. When the denoising model is trained from scratch using only 20%-count low-dose paired data (1st row), it yields PSNR of 28.56 and NMSE of 0.0144 when evaluating on the same institutional data. Due to the domain shift caused by different low-dose settings, when applying the denoising model trained from one institutional data to another institutional data, it yields inferior denoising performance. For example, when applying the model trained with 20%-count data to those 40%-count data (1st row and 2nd column), it only yields PSNR of 30.63 and NMSE of 0.0089 which is significantly lower than those of the model trained with 40%-count data and applied on the 40%-count data (2nd row and 2nd column). While training and applying the same institutional local data mitigates the domain shift issue, our FTL (last row) with diverse multi-institutional data collaborative training still yields the best performance with PSNR boost from 28.56 to 29.24 for the 20%-count experiment for example. The corresponding tumor region’s bias is reported in Figure 7. In the 20%-count experiment, the bias can be reduced by using the denoising models trained from scratch using only local data. Our FTL method provides further reduced mean bias and reduced standard deviation. Similar observations can be seen in both the 40% and 60% experiments.
Fig. 6.
Visual comparison of low-dose reconstruction from denoising model trained from single institution data and from our FTL.
TABLE III.
Quantitative comparison of reconstructions when training only on individual institution dataset. The results with training and testing on same institution data are marked with underline. Best results are marked in bold.
| Evaluation | 20%-count data | 40%-count data | 60%-count data | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SSIM | NMSE | PSNR | SSIM | NMSE | PSNR | SSIM | NMSE | PSNR | |
| 20%-count data | .977 | 0.0144 | 28.56 | .982 | 0.0089 | 30.63 | .985 | 0.0069 | 31.65 |
| 40%-count data | .975 | 0.0160 | 28.22 | .986 | 0.0076 | 31.54 | .988 | 0.0051 | 33.11 |
| 60%-count data | .974 | 0.0182 | 27.71 | .985 | 0.0078 | 31.33 | .991 | 0.0041 | 34.20 |
| Ours | .978 | 0.0124 | 29.24 | .987 | 0.0064 | 32.38 | .993 | 0.0037 | 34.67 |
Fig. 7.
Comparison of lesion region’s bias from the original low-dose reconstruction (red bar), the reconstruction using model trained only from its own institutional data (green bar), and our FTL reconstruction (blue bar). We evaluated on three different institution data with 20%-count (left), 40%-count (middle), and 60%-count (right) low-dose settings.
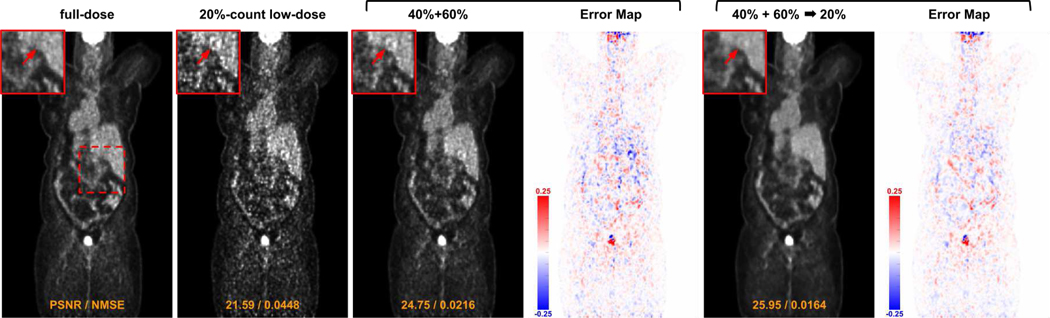
We also studied the denoising performance when using different federated learning settings of FTL. A visual comparison example is shown in Figure 8. As we can observe, when training a denoising model using FL with multi-institutional data consisting of 40%-count and 60%-count data, directly applying it to the 20%-count data can reduce the global noise level but with noise-induced hot spots remain in the liver region. When the model was further fine-tuned with 20%count local data, the denoising performance further improves with these hot spots being eliminated. The corresponding quantitative evaluation is summarized in Table IV. A denoising model trained with FL and multi-institutional data consisting of 40%-count and 60%-count data yields PSNR of 28.43 when evaluating on 20%-count data (1st row and 4th column), which is inferior to the performance of the denoising model directly trained on 20%-count from scratch (Table III 1st row and 4th column). However, when the model was further fine-tuned on 20%-count local data (2nd row and 4th column), it boosts the PSNR up to 29.15 which is better than the model trained from scratch. Similar observations can be found for the other FTL setups. The last row reports the performance of our FTL which was first trained using all three institutional local data with FL and then fine-tuned on individual local data, providing the best performance across different setups.
Fig. 8.
Visual comparison of low-dose reconstruction using different federated learning settings of FTL. Results on 20%-count low-dose data are shown here. 40% + 60% means two institution data used during the federated learning stage of FTL. → 20% means transfer learning on target institution data.
TABLE IV.
Quantitative comparison of reconstructions when using different federated learning settings of FTL. Best results are marked in bold. For instance, X + Y → Z means federated learning using X and Y dataset, and then transfer learning to Z dataset.
| Evaluation | 20%-count data | 40%-count data | 60%-count data | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SSIM | NMSE | PSNR | SSIM | NMSE | PSNR | SSIM | NMSE | PSNR | |
| 40% + 60% | .976 | 0.0153 | 28.43 | - | - | - | - | - | - |
| 40% + 60% → 20% | .978 | 0.0128 | 29.15 | - | - | - | - | - | - |
| 20% + 60% | - | - | - | .986 | 0.0067 | 31.97 | - | - | - |
| 20% + 60% → 40% | - | - | - | .987 | 0.0065 | 32.16 | - | - | - |
| 20% + 40% | - | - | - | - | - | - | .988 | 0.0051 | 33.16 |
| 20% + 40% → 60% | - | - | - | - | - | - | .992 | 0.0038 | 34.49 |
| Ours | .978 | 0.0124 | 29.24 | .987 | 0.0064 | 32.38 | .993 | 0.0037 | 34.67 |
V. Discussion
In this work, we developed a federated learning framework, called FTL, for low-dose PET denoising using simulated heterogeneous multi-institutional low-dose data. Specifically, the proposed FTL consists of two major steps. In the first step, a standard federated averaging is used, such that the denoising models are independently trained at individual institutions and iteratively sent over to the central server for model aggregation. During this process, no local data is shared between institutions. After the convergence of federated training, the global model is re-distributed to each institution for transfer learning on local data only, generating a personalized denoising model for each institution such that the domain shift can be mitigated. Unlike previous works of FLCM [24] and FedPer [21], [28] that are bound to specific network architecture, our FTL framework allows flexible network architecture to be integrated.
From our experimental results, we demonstrated the feasibility of using our FTL for low-dose PET denoising with three different low-dose settings corresponding to three different simulated institutions. First of all, as we can observe from Table I and Figure 4, our method generating three personalized denoising models for each individual institution can consistently outperform previous methods, including FLCM, FedPer, and FedSP that also generate three personalized models, one for each institution, as well as FedAvg that only generates one global model for all institutions. The lesion bias analysis, as shown in Table II, also demonstrated the FTL provides less bias quantification for lung lesions as compared to previous methods. Please note the lesion regions usually have higher SNR as compared to other regions, due to the higher amount of tracer concentration in these regions. Thus, the bias in the lesion region is already very low from the original low-dose reconstruction, e.g. 0.007±0.023 from 20%-count data. Even under this condition, our method further reduced the bias to a lower level with reduced standard deviation as well. Second, one of the goals of our FTL is to address the domain shift issue. As we can observe from Table III, Figure 6, and Figure 7, while training a denoising model from scratch using local data with limited diversity can avoid domain shift issues and provide reasonable denoising performance for each institution, our method yields a superior performance as compared to them since we utilize all institutional data with diverse distribution for the federated pre-training. Similar observations can be found in Table IV where the best performance is given when all the institutional data is used in the first step of FTL, and then fine-tune using local data. Even though our FTL achieves superior performance as compared to models trained from local data only, we think increasing the amount of local data and diversity may further boost our performance and may also potentially close the gap between the model trained from local data only and federated learning-based methods.
The presented work also has limitations with several potential improvements that are the subjects of our future studies. First, our work considers a three-institution scenario with three different low-dose settings, i.e. 20%-count, 40%-count, and 60%-count. However, the number of institutions could be more than three with more diverse low-dose settings. For example, Kaplan et al. [5], Ladefoged et al. [50], and Zhou et al. [51] used a 10%-count low-dose setting. Wang et al. [3] and Whiteley et al. [52] deployed a 25%-count low-dose setting, while Ouyang et al. [9] considered a 1%count ultra-low-dose setting. Even though we only consider three different low-dose settings in our work, our proposed framework can be flexibly adapted to a different number of institutions with different low-dose settings. In fact, we believe that expanding the number of institutions with more diverse low-dose settings could potentially further improve our method’s performance. Second, we created our multi-institutional low-dose PET dataset based on simulation which uses data all acquired from the Siemens mCT PET/CT systems and reconstructed with the OSEM reconstruction algorithm incorporated with point-spread-function (PSF) and time-offlight (TOF) information. In real-world applications, low-dose PET data from different institutions could be acquired with different vendor systems, reconstructed with different algorithms, and administered with different tracer types. Our pilot study mainly focuses on low-dose 18F-FDG PET with OSEM reconstruction which is the most common protocol in clinic PET and previous works of low-dose PET. In our future work, we will investigate the performance of our framework with data acquired from different tracer types, vendor systems, and reconstruction protocols from different institutions. We believe our method with transfer learning could potentially be adapted to changing elements in the system and their reconstruction algorithms. Thirdly, for a fair comparison, we performed all our experiments using the same denoising network architecture, i.e. DuAttRDUNet (Figure 3). While our framework is technically simple, it can be flexibly adapted with different advanced denoising networks that do not use U-shape design. For instance, the residual dense network based on the residual dense block without any downsampling operation has demonstrated superior performance in general image restoration tasks [46]. Similarly, SwinIR [53] based on residual Swin Transformer blocks [54] without any downsampling operation also showed significant advantages for image restoration tasks. Combining these networks with dual domain learning has also shown to yield superior medical image reconstruction performance [30], [44], [55]–[57]. Deploying these networks in our FTL could potentially further improve our denoising performance and will be an important direction of our future studies. Lastly, in our current experiment, we assumed a similar amount of data from each institution, thus we used a equally weighted parameter averaging strategy in our first stage’s training. In our future work with a more diverse and potential imbalanced amount of real data, we will investigate using weighed parameter averaging strategies [58] to further improve our method’s performance.
There are also additional challenges and considerations that the current work did not consider which would require attention in the future works on using FL for PET applications [59]–[61]. First, even though FL has demonstrated its effectiveness in protecting patients’ privacy by keeping institutional data locally, there are still privacy and security challenges in FL. For example, the parameter in the deep learning model still contains sensitive information that could allow one to reconstitute the patients’ information in the local training set [62]. Adversarial attacks from malicious parties can also degrade the deep learning model’s performance, thus causing severe consequences when a model is deployed in real clinical scenarios [63]. Additional data safety measurements in FL, such as model encryption, differential privacy, and adversarial defense against malicious clients, should be considered to better protect the data privacy [64], [65]. Second, the FL requires standardized data from different institutions to allow stable training of deep learning models. Different institutions may have different infrastructure and imaging data management protocols, resulting in imaging data stored in different formats. This introduces additional data heterogeneity that may affect the federated training’s convergence which could negatively impact the model’s final performance. Thus, appropriate data preprocessing and curation measurements should be performed to standardize the data before performing federated learning [66], [67]. Lastly, an appropriate system architecture is also required to perform FL. For example, local institutions need private or cloud-based computation resources in order to perform local training before federated training. Federated learning also requires internet connections with high-performance bandwidth between different institutions to enable model parameter passing. Many institutions may still lack these resources to enable the proposed FL methods. Therefore, the design of FL algorithms should also consider different system architectures, especially when these resources are limited.
VI. Conclusion
We developed a federated transfer learning framework (FTL) for low-dose PET denoising using heterogeneous data. Our FTL aims to generate a personalized denoising model for each institution by first collaboratively training a global model using all institutional data while without sharing data during this process, and then fine-tune using each institution’s local data to alleviate the domain shift issue. Our experimental results demonstrate that our method can provide high-quality denoising reconstruction, superior to previous baseline image reconstruction with federated learning methods under simulated multi-institution with diverse low-dose settings.
Acknowledgment
This work was supported by National Institutes of Health (NIH) grant R01EB025468. All authors declare that they have no known conflicts of interest in terms of competing financial interests or personal relationships that could have an influence or are relevant to the work reported in this paper.
Footnotes
This work involved human subjects or animals in its research. The authors confirm that all human/animal subject research procedures and protocols are exempt from review board approval.
Contributor Information
Bo Zhou, Department of Biomedical Engineering, Yale University, New Haven, CT, 06511, USA..
Tianshun Miao, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, 06511, USA..
Niloufar Mirian, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, 06511, USA..
Xiongchao Chen, Department of Biomedical Engineering, Yale University, New Haven, CT, 06511, USA..
Huidong Xie, Department of Biomedical Engineering, Yale University, New Haven, CT, 06511, USA..
Zhicheng Feng, Department of Biomedical Engineering, University of Southern California, Los Angeles, CA, 90007, USA..
Xueqi Guo, Department of Biomedical Engineering, Yale University, New Haven, CT, 06511, USA..
Xiaoxiao Li, Electrical and Computer Engineering Department, University of British Columbia, Vancouver, Canada..
S. Kevin Zhou, School of Biomedical Engineering & Suzhou Institute for Advanced Research, University of Science and Technology of China, Suzhou, China and the Institute of Computing Technology, Chinese Academy of Sciences, Beijing, 100190, China..
James S. Duncan, Department of Biomedical Engineering and the Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, 06511, USA..
Chi Liu, Department of Biomedical Engineering and the Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, 06511, USA..
References
- [1].Strauss KJ and Kaste SC, “The alara (as low as reasonably achievable) concept in pediatric interventional and fluoroscopic imaging: striving to keep radiation doses as low as possible during fluoroscopy of pediatric patientsa white paper executive summary,” Radiology, vol. 240, no. 3, pp. 621–622, 2006. [DOI] [PubMed] [Google Scholar]
- [2].Xiang L, Qiao Y, Nie D, An L, Lin W, Wang Q, and Shen D, “Deep auto-context convolutional neural networks for standard-dose pet image estimation from low-dose pet/mri,” Neurocomputing, vol. 267, pp. 406–416, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, Wu X, Zhou J, Shen D, and Zhou L, “3d conditional generative adversarial networks for high-quality pet image estimation at low dose,” Neuroimage, vol. 174, pp. 550–562, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, and Liu C, “An investigation of quantitative accuracy for deep learning based denoising in oncological pet,” Physics in Medicine & Biology, vol. 64, no. 16, p. 165019, 2019. [DOI] [PubMed] [Google Scholar]
- [5].Kaplan S and Zhu Y-M, “Full-dose pet image estimation from low-dose pet image using deep learning: a pilot study,” Journal of digital imaging, vol. 32, no. 5, pp. 773–778, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hu Z, Xue H, Zhang Q, Gao J, Zhang N, Zou S, Teng Y, Liu X, Yang Y, Liang D et al. , “Dpir-net: Direct pet image reconstruction based on the wasserstein generative adversarial network,” IEEE Transactions on Radiation and Plasma Medical Sciences, 2020. [Google Scholar]
- [7].Gong Y, Shan H, Teng Y, Tu N, Li M, Liang G, Wang G, and Wang S, “Parameter-transferred wasserstein generative adversarial network (pt-wgan) for low-dose pet image denoising,” IEEE Transactions on Radiation and Plasma Medical Sciences, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou L, Schaefferkoetter JD, Tham IW, Huang G, and Yan J, “Supervised learning with cyclegan for low-dose fdg pet image denoising,” Medical Image Analysis, vol. 65, p. 101770, 2020. [DOI] [PubMed] [Google Scholar]
- [9].Ouyang J, Chen KT, Gong E, Pauly J, and Zaharchuk G, “Ultralow-dose pet reconstruction using generative adversarial network with feature matching and task-specific perceptual loss,” Medical physics, vol. 46, no. 8, pp. 3555–3564, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Boumis A, Khalighi M, Poston KL, Sha SJ, Greicius MD, Mormino E et al. , “Ultra–low-dose 18f-florbetaben amyloid pet imaging using deep learning with multi-contrast mri inputs,” Radiology, vol. 290, no. 3, pp. 649–656, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu H, Wu J, Lu W, Onofrey JA, Liu Y-H, and Liu C, “Noise reduction with cross-tracer and cross-protocol deep transfer learning for low-dose pet,” Physics in Medicine & Biology, vol. 65, no. 18, p. 185006, 2020. [DOI] [PubMed] [Google Scholar]
- [12].Liu J, Malekzadeh M, Mirian N, Song T-A, Liu C, and Dutta J, “Artificial intelligence-based image enhancement in pet imaging: Noise reduction and resolution enhancement,” PET clinics, vol. 16, no. 4, pp. 553–576, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Song T-A, Yang F, and Dutta J, “Noise2void: unsupervised denoising of pet images,” Physics in Medicine & Biology, vol. 66, no. 21, p. 214002, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dutta J, Leahy RM, and Li Q, “Non-local means denoising of dynamic pet images,” PloS one, vol. 8, no. 12, p. e81390, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Maggioni M, Katkovnik V, Egiazarian K, and Foi A, “Nonlocal transform-domain filter for volumetric data denoising and reconstruction,” IEEE transactions on image processing, vol. 22, no. 1, pp. 119–133, 2012. [DOI] [PubMed] [Google Scholar]
- [16].Mejia J, Mederos B, Mollineda RA, and Maynez LO, “Noise reduction in small animal pet images using a variational non-convex functional,” IEEE Transactions on Nuclear Science, vol. 63, no. 5, pp. 2577–2585, 2016. [Google Scholar]
- [17].Rieke N, Hancox J, Li W, Milletari F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K et al. , “The future of digital health with federated learning,” NPJ digital medicine, vol. 3, no. 1, pp. 1–7, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Roski J, Bo-Linn GW, and Andrews TA, “Creating value in health care through big data: opportunities and policy implications,” Health affairs, vol. 33, no. 7, pp. 1115–1122, 2014. [DOI] [PubMed] [Google Scholar]
- [19].McMahan B, Moore E, Ramage D, Hampson S, and y Arcas BA, “Communication-efficient learning of deep networks from decentralized data,” in Artificial intelligence and statistics. PMLR, 2017, pp. 1273–1282. [Google Scholar]
- [20].Li X, Jiang M, Zhang X, Kamp M, and Dou Q, “Fedbn: Federated learning on non-iid features via local batch normalization,” arXiv preprint arXiv:2102.07623, 2021. [Google Scholar]
- [21].Arivazhagan MG, Aggarwal V, Singh AK, and Choudhary S, “Federated learning with personalization layers,” arXiv preprint arXiv:1912.00818, 2019. [Google Scholar]
- [22].Collins L, Hassani H, Mokhtari A, and Shakkottai S, “Exploiting shared representations for personalized federated learning,” arXiv preprint arXiv:2102.07078, 2021. [Google Scholar]
- [23].Fallah A, Mokhtari A, and Ozdaglar A, “Personalized federated learning: A meta-learning approach,” arXiv preprint arXiv:2002.07948, 2020. [Google Scholar]
- [24].Guo P, Wang P, Zhou J, Jiang S, and Patel VM, “Multi-institutional collaborations for improving deep learning-based magnetic resonance image reconstruction using federated learning,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2021, pp. 2423–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241. [Google Scholar]
- [26].Lyu L, Yu H, Ma X, Sun L, Zhao J, Yang Q, and Yu PS, “Privacy and robustness in federated learning: Attacks and defenses,” arXiv preprint arXiv:2012.06337, 2020. [DOI] [PubMed] [Google Scholar]
- [27].Huang Y, Gupta S, Song Z, Li K, and Arora S, “Evaluating gradient inversion attacks and defenses in federated learning,” Advances in Neural Information Processing Systems, vol. 34, 2021. [Google Scholar]
- [28].Feng C-M, Yan Y, Fu H, Xu Y, and Shao L, “Specificity-preserving federated learning for mr image reconstruction,” arXiv preprint arXiv:2112.05752, 2021. [DOI] [PubMed] [Google Scholar]
- [29].Zhang Y, Tian Y, Kong Y, Zhong B, and Fu Y, “Residual dense network for image super-resolution,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 2472–2481. [Google Scholar]
- [30].Zhou B and Zhou SK, “Dudornet: Learning a dual-domain recurrent network for fast mri reconstruction with deep t1 prior,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2020, pp. 298–313. [Google Scholar]
- [31].Zamir SW, Arora A, Khan S, Hayat M, Khan FS, Yang M-H, and Shao L, “Multi-stage progressive image restoration,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2021, pp. 14821–14831. [Google Scholar]
- [32].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, “Generative adversarial nets,” in Advances in neural information processing systems, 2014, pp. 2672–2680. [Google Scholar]
- [33].Isola P, Zhu J-Y, Zhou T, and Efros AA, “Image-to-image translation with conditional adversarial networks,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 1125–1134. [Google Scholar]
- [34].Arjovsky M, Chintala S, and Bottou L, “Wasserstein gan,” arXiv preprint arXiv:1701.07875, 2017. [Google Scholar]
- [35].Li T, Sahu AK, Talwalkar A, and Smith V, “Federated learning: Challenges, methods, and future directions,” IEEE Signal Processing Magazine, vol. 37, no. 3, pp. 50–60, 2020. [Google Scholar]
- [36].Sheller MJ, Edwards B, Reina GA, Martin J, Pati S, Kotrotsou A, Milchenko M, Xu W, Marcus D, Colen RR et al. , “Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data,” Scientific reports, vol. 10, no. 1, pp. 1–12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Feki I, Ammar S, Kessentini Y, and Muhammad K, “Federated learning for covid-19 screening from chest x-ray images,” Applied Soft Computing, vol. 106, p. 107330, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li W, Milletarì F, Xu D, Rieke N, Hancox J, Zhu W, Baust M, Cheng Y, Ourselin S, Cardoso MJ et al. , “Privacy-preserving federated brain tumour segmentation,” in International workshop on machine learning in medical imaging. Springer, 2019, pp. 133–141. [Google Scholar]
- [39].Li X, Gu Y, Dvornek N, Staib LH, Ventola P, and Duncan JS, “Multi-site fmri analysis using privacy-preserving federated learning and domain adaptation: Abide results,” Medical Image Analysis, vol. 65, p. 101765, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Peng X, Huang Z, Zhu Y, and Saenko K, “Federated adversarial domain adaptation,” arXiv preprint arXiv:1911.02054, 2019. [Google Scholar]
- [41].Schlemper J, Caballero J, Hajnal JV, Price AN, and Rueckert D, “A deep cascade of convolutional neural networks for dynamic mr image reconstruction,” IEEE transactions on Medical Imaging, vol. 37, no. 2, pp. 491–503, 2017. [DOI] [PubMed] [Google Scholar]
- [42].Guo P, Valanarasu JMJ, Wang P, Zhou J, Jiang S, and Patel VM, “Over-and-under complete convolutional rnn for mri reconstruction,” arXiv preprint arXiv:2106.08886, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhou B, Zhou SK, Duncan JS, and Liu C, “Limited view tomographic reconstruction using a cascaded residual dense spatial-channel attention network with projection data fidelity layer,” IEEE transactions on medical imaging, vol. 40, no. 7, pp. 1792–1804, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhou B, Chen X, Zhou SK, Duncan JS, and Liu C, “Dudodr-net: Dual-domain data consistent recurrent network for simultaneous sparse view and metal artifact reduction in computed tomography,” Medical Image Analysis, vol. 75, p. 102289, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen X, Zhou B, Shi L, Liu H, Pang Y, Wang R, Miller EJ, Sinusas AJ, and Liu C, “Ct-free attenuation correction for dedicated cardiac spect using a 3d dual squeeze-and-excitation residual dense network,” Journal of Nuclear Cardiology, pp. 1–16, 2021. [DOI] [PubMed] [Google Scholar]
- [46].Zhang Y, Tian Y, Kong Y, Zhong B, and Fu Y, “Residual dense network for image restoration,” IEEE Transactions on Pattern Analysis and Machine Intelligence, 2020. [DOI] [PubMed] [Google Scholar]
- [47].Huang G, Liu Z, Van Der Maaten L, and Weinberger KQ, “Densely connected convolutional networks,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 4700–4708. [Google Scholar]
- [48].Roy AG, Navab N, and Wachinger C, “Recalibrating fully convolutional networks with spatial and channel squeeze and excitation blocks,” IEEE transactions on medical imaging, vol. 38, no. 2, pp. 540–549, 2018. [DOI] [PubMed] [Google Scholar]
- [49].Kingma DP and Ba J, “Adam: A method for stochastic optimization,” arXiv preprint arXiv:1412.6980, 2014. [Google Scholar]
- [50].Ladefoged CN, Hasbak P, Hornnes C, Højgaard L, and Andersen FL, “Low-dose pet image noise reduction using deep learning: application to cardiac viability fdg imaging in patients with ischemic heart disease,” Physics in Medicine & Biology, vol. 66, no. 5, p. 054003, 2021. [DOI] [PubMed] [Google Scholar]
- [51].Zhou B, Tsai Y-J, Chen X, Duncan JS, and Liu C, “Mdpet: A unified motion correction and denoising adversarial network for low-dose gated pet,” IEEE Transactions on Medical Imaging, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Whiteley W, Panin V, Zhou C, Cabello J, Bharkhada D, and Gregor J, “Fastpet: near real-time reconstruction of pet histo-image data using a neural network,” IEEE Transactions on Radiation and Plasma Medical Sciences, vol. 5, no. 1, pp. 65–77, 2020. [Google Scholar]
- [53].Liang J, Cao J, Sun G, Zhang K, Van Gool L, and Timofte R, “Swinir: Image restoration using swin transformer,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 2021, pp. 1833–1844. [Google Scholar]
- [54].Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, and Guo B, “Swin transformer: Hierarchical vision transformer using shifted windows,” arXiv preprint arXiv:2103.14030, 2021. [Google Scholar]
- [55].Lin W-A, Liao H, Peng C, Sun X, Zhang J, Luo J, Chellappa R, and Zhou SK, “Dudonet: Dual domain network for ct metal artifact reduction,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2019, pp. 10512–10521. [Google Scholar]
- [56].Zhou B, Zhou SK, Duncan JS, and Liu C, “Limited view tomographic reconstruction using a deep recurrent framework with residual dense spatial-channel attention network and sinogram consistency,” arXiv preprint arXiv:2009.01782, 2020. [Google Scholar]
- [57].Zhou B, Schlemper J, Dey N, Salehi SSM, Liu C, Duncan JS, and Sofka M, “Dsformer: A dual-domain self-supervised transformer for accelerated multi-contrast mri reconstruction,” arXiv preprint arXiv:2201.10776, 2022. [Google Scholar]
- [58].Hong M, Kang S-K, and Lee J-H, “Weighted averaging federated learning based on example forgetting events in label imbalanced non-iid,” Applied Sciences, vol. 12, no. 12, p. 5806, 2022. [Google Scholar]
- [59].Darzidehkalani E, Ghasemi-Rad M, and van Ooijen P, “Federated learning in medical imaging: Part i: Toward multicentral health care ecosystems,” Journal of the American College of Radiology, 2022. [DOI] [PubMed] [Google Scholar]
- [60].Darzidehkalani E, Ghasemi-Rad M, and van Ooijen P, “Federated learning in medical imaging: Part ii: Methods, challenges, and considerations,” Journal of the American College of Radiology, 2022. [DOI] [PubMed] [Google Scholar]
- [61].Kumar Y and Singla R, “Federated learning systems for healthcare: perspective and recent progress,” Federated Learning Systems, pp. 141–156, 2021. [Google Scholar]
- [62].Carlini N, Liu C, Erlingsson U, Kos J, and Song D, “The secret´ sharer: Evaluating and testing unintended memorization in neural networks,” in 28th USENIX Security Symposium (USENIX Security 19), 2019, pp. 267–284. [Google Scholar]
- [63].Zhang J, Chen J, Wu D, Chen B, and Yu S, “Poisoning attack in federated learning using generative adversarial nets,” in 2019 18th IEEE International Conference On Trust, Security And Privacy In Computing And Communications/13th IEEE International Conference On Big Data Science And Engineering (TrustCom/BigDataSE). IEEE, 2019, pp. 374–380. [Google Scholar]
- [64].Abadi M, Chu A, Goodfellow I, McMahan HB, Mironov I, Talwar K, and Zhang L, “Deep learning with differential privacy,” in Proceedings of the 2016 ACM SIGSAC conference on computer and communications security, 2016, pp. 308–318. [Google Scholar]
- [65].Hitaj B, Ateniese G, and Perez-Cruz F, “Deep models under the gan: information leakage from collaborative deep learning,” in Proceedings of the 2017 ACM SIGSAC conference on computer and communications security, 2017, pp. 603–618. [Google Scholar]
- [66].Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, Soussan M, Frouin F, Frouin V, and Buvat I, “A postre-construction harmonization method for multicenter radiomic studies in pet,” Journal of Nuclear Medicine, vol. 59, no. 8, pp. 1321–1328, 2018. [DOI] [PubMed] [Google Scholar]
- [67].Zhou B, Harrison AP, Yao J, Cheng C-T, Xiao J, Liao C-H, and Lu L, “Ct data curation for liver patients: phase recognition in dynamic contrast-enhanced ct,” in Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data. Springer, 2019, pp. 139–147. [Google Scholar]