Abstract
The dynamic addition of O-GlcNAc to target proteins is now recognized as a major signaling paradigm impacting phosphorylation, protein turnover, gene expression, and other posttranslational modifications influencing epigenetics. Here we describe the production of and methods for assay of the recombinant enzymes of O-GlcNAc cycling: O-linked GlcNAc Transferase (OGT) and O-GlcNAcase (OGA).
Keywords: Recombinant, Fluorogenic substrates, Immunoblots, Bioorthogonal chemistry, O-GlcNAc
1. Introduction
The O-GlcNAc modification is an abundant and highly dynamic nucleocytoplasmic posttranslational modification of protein Ser and Thr residues [1, 2]. The enzymes of O-GlcNAc cycling play critical roles in development, signaling, gene expression, and are emerging as important players in epigenetic regulation [1, 3, 4]. The ability to produce recombinant forms of O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) has facilitated both highly informative structural studies and led to inhibitor and small molecule inhibitors of these key enzymes.
Human O-GlcNAc transferase was first produced in recombinant form to demonstrate the identity of this transferase as the enzyme catalyzing O-GlcNAc transfer [5]. Subsequent work led to identification of the 3–12 tetratricopeptide repeats (TPR) as important determinants of target specificity [6, 7]. The C-terminal domain contains a glycosyltransferase domain belonging to the GT41 family in the CAZY database and uses UDP-GlcNAc as a glycosyl donor [8]. Recombinant expression allowed us to determine the structure of the human OGT TPR domain [9]. Subsequent recombinant production of a 4 TPR version of OGT allowed crystallization of the human OGT catalytic domain [10]. These studies have revealed that OGT exhibits enzymatic features consistent with an ordered bi–bi reaction mechanism involving initial tight binding of sugar nucleotide followed by target peptide binding. The reaction mechanism also involves a proposed conformational entrapment by the TPR domain of the peptide linked to catalysis [10]. The strategy for the detection of OGT activity described here takes advantage of several detection strategies including radiochemical and bioorthogonal chemical approaches.
The human O-GlcNAcase was originally identified as hexosaminidase C and is a CAZY GH84 family member with a TIM barrel structure similar to the CAZY GH20 members of hexosaminidase A and B. When expressed in E. coli, the human O-GlcNAcase shows little activity against either GalNAc or capping GlcNAc residues and exhibits a pH optimum near pH 7. It exhibits rather high specificity for O-GlcNAc residues and has been shown to accommodate extension of the N-acetyl to longer acyl groups including N-pentanoyl. Such substrate flexibility is not exhibited by the CAZY 20 family members of hexosaminidases A and B. There is some evidence that the protein sequence surrounding the O-GlcNAc modification is also an important determinant of O-GlcNAcase specificity [11]. The methods we describe here take advantage of a highly sensitive fluorogenic substrate [12] which can be tailored to measuring enzyme activity in rather crude extracts.
2. Materials
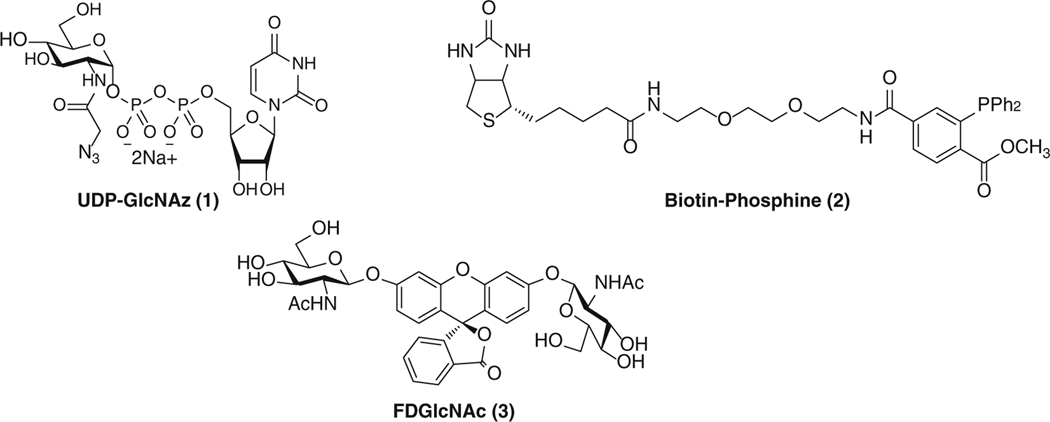
Prepare all solutions using ultrapure water and analytical grade reagents unless indicated otherwise. Structures of some compounds are presented in Fig. 1.
Fig. 1.
Chemical structures of UDP-GlcNAz, FDGlcNAc, and Biotin-Phosphine
2.1. Components of OGT Expression in E. coli
Cloned plasmids of human ncOGT and mOGT inserting the coding regions in pET43.1 Ek/LIC expression vector (Novagen, San Diego, CA, USA) (see Note 1).
BL21(DE3) chemically competent E. coli (Invitrogen, Carlsbad, CA, USA).
Luria–Bertani (LB) broth.
Ampicillin solution (100 mg/mL): Weigh 0.5 g Ampicillin sodium salt (Sigma-Aldrich, St. Louis, MO, USA) and prepared in 5 mL volume by adding water to a total volume of 5 mL. Mix and filter-sterilize with a 0.22 μm filter (EMD Millipore, Billerica, MA, USA). Aliquot in 1 mL volume and store at −20 °C.
LB Agar plates with 50 μg/mL Ampicillin.
Incubator for a microbiological culture.
OGT lysis buffer (see Note 2): 20 mM Tris–HCl, pH 7.5, 2 mM EDTA, 1 mg/mL of lysozyme (Sigma), complete mini EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA). Prepare OGT lysis buffer right before use and store at 4 °C.
Lysozyme solution (100 mg/mL): Weigh 0.5 g lysozyme (Sigma) and prepare 5 mL solution by adding water to a total volume of 5 mL. Mix and filter-sterilize with a sterilized 0.22 μm filter (EMD Millipore). Aliquot in 1 mL volume and store at −20 °C.
Triton X-100™ (Sigma) 10 % solution: Add 1 mL of Triton X-100 to 9 mL of distilled water.
A centrifuge with temperature control: Sorvall Stratos Centrifuge (Thermo Scientific, Rockford, IL, USA).
A microcentrifuge with temperature control: Eppendorf® Refrigerated Microcentrifuge (Eppendorf, Hauppauge, NY, USA).
2.2. Components of OGT Purification
S-Protein Agarose (Novagen, San Diego, CA, USA).
Dithiothreitol (DTT) solution (1 M): Dissolve 0.787 g DTT (Sigma) in distilled water to give a total volume of 5 mL. Mix and filter-sterilize with a 0.22 μm filter device (EMD Millipore). Aliquot in 1 mL volume and store at −30 °C.
OGT Assay buffer (see Note 3): 50 mM Tris–HCl, pH 7.5, 1 mM DTT, 12.5 mM MgCl2.
An orbital shaker.
A microcentrifuge with temperature control: Eppendorf® Refrigerated Microcentrifuge.
2.3. Components of OGT Enzymatic Activity Assay
2.3.1. Components of Radiometric OGT Activity Assay (Fig. 2)
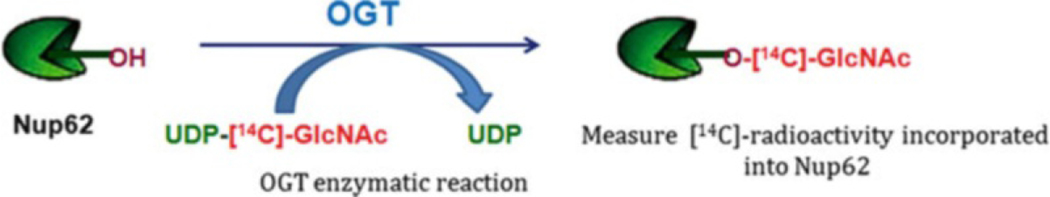
Fig. 2.
Radiometric OGT activity assay. This method uses a radiolabeled sugar donor substrate and the activity is measured by quantitating radiolabeled GlcNAc incorporation into a protein such as Nup62
Uridine diphosphate N-acetyl [1-14C]D-glucosamine (UDP-[14C]-GlcNAc): UDP-[14C]-GlcNAc, 0.1 mCi/mL, 40–60 mCi/mmol (American Radiolabeled Chemicals, St. Louis, MO, USA).
Recombinant, purified Nup62 (Bioclone, Inc., San Diego, CA, USA): Concentration of Nup62 is 1 μg/μL in 20 mM sodium phosphate, pH 7.5, 0.5 M NaCl, 1 M imidazole. Recombinant and purified Nup62 can also be prepared as described previously [13].
Incubator for microbiological culture.
SDS-PAGE sample buffer: 4× NuPAGE® LDS sample buffer (Invitrogen).
SDS-PAGE gels: 10 % or 4–12 % NuPAGE® Bis-Tris gels (Invitrogen).
SDS running buffer: 20× NuPage® MOPS SDS Running Buffer (Invitrogen). Dilute it to 1× SDS Running Buffer with distilled water for the gel electrophoresis.
A low speed orbital shaker.
Simply Blue Safestain (Invitrogen).
En3Hance (Perkin Elmer, Wellesley, MA, USA).
PEG solution: 10 % PEG solution (MW 8000).
OGT assay buffer: 50 mM Tris–HCl, pH 7.5, 1 mM DTT, 12.5 mM MgCl2.
A Gel Dryer equipped with a vacuum pump (Bio-Rad, Hercules, CA, USA).
A phosphor screen: A BAS-IIIs imaging plate (Fuji Film Co., Tokyo, Japan).
A phosphorimager: BAS-1500 phosphor imager (Fuji Film Co.).
Albumin (bovine serum) [methyl-14C] methylated: [methyl-14C]-BSA, 0.01 mCi/mL, 3−30 μCi/mg (American Radiolabeled Chemicals).
2.3.2. Components of Chemoenzymatic OGT Activity Assay (Fig. 3)
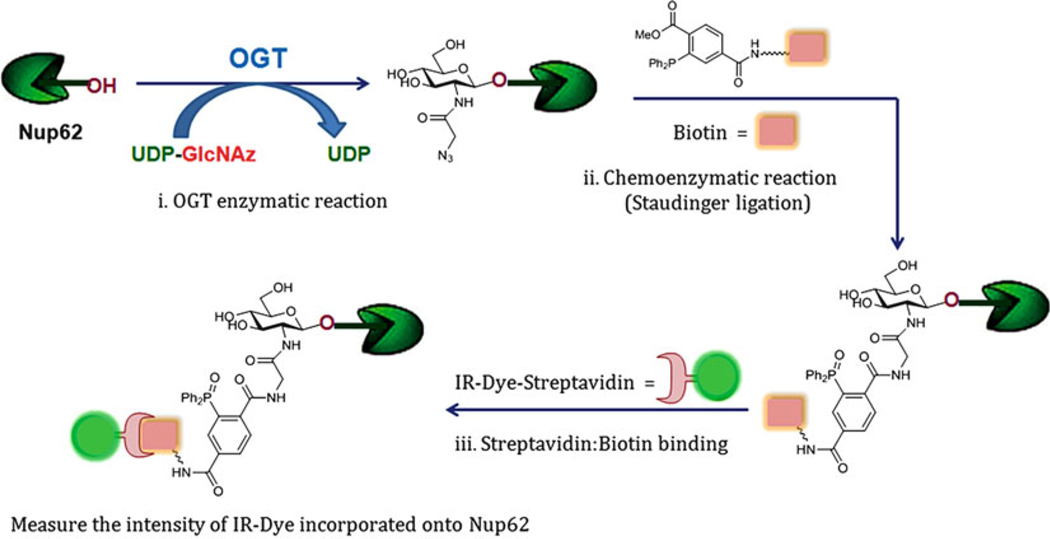
Fig. 3.
Chemoenzymatic OGT activity assay. This method involves azide labeling using UDP-GlcNAz, followed by chemoenzymatic reaction either Staudinger ligation with a phosphine reagent as shown in this section, or Click reaction with a terminal alkyne reagent and Cu+1 ion (or strained cycloalkyne without Copper ion catalyst; not provided here)
Uridine 5′-diphospho-2-azidoacetamido-2-deoxy-α-D-glucopyranose disodium salt (UDP-GlcNAz 1, 2 mM in distilled water): UDP-GlcNAz is synthesized as described previously [14]. In order to prepare 2 mM UDP-GlcNAz, first, prepare 10 mM of UDP-GlcNAz by dissolving 6.92 mg in distilled water to give a final volume of 1 mL. Transfer 100 μL of 10 mM UDP-GlcNAz solution into a new vial and add 400 μL of distilled water to make a final concentration of 2 mM solution. Store both 10 and 2 mM solutions of UDP-GlcNAz at −30 °C.
Recombinant, purified Nup62 (Bioclone, Inc.): Concentration of Nup62 is 1 μg/μL in 20 mM sodium phosphate, pH 7.5, 0.5 M NaCl, 1 M imidazole. Recombinant and purified Nup62 can also be prepared as described previously [13].
Purified OGT bound to S-protein Agarose.
OGT assay buffer: 50 mM Tris–HCl, pH 7.5, 1 mM DTT, 12.5 mM MgCl2.
Incubator for microbiological culture.
A microcentrifuge with temperature control: Eppendorf® Refrigerated Microcentrifuge.
Microcon YM-30 filter device (EMD Millipore).
Dimethyl sulfoxide (DMSO) (Sigma).
A Biotin-Phosphine reagent 2, a Staudinger reaction reagent (10 mM in DMSO): Biotin-phosphine reagent is prepared as described previously [15]. Phosphine-PEG3-Biotin (Thermo Fisher Scientific) is also commercially available. To prepare 10 mM Biotin-Phosphine (2), dissolve 7.21 mg of 2 in DMSO to give a final volume of 1 mL.
SDS-PAGE sample buffer: 4× NuPAGE® LDS sample buffer (Invitrogen).
Precast SDS-PAGE gels: 10 % or 4–12 % NuPAGE® Bis–Tris gels (Invitrogen).
SDS running buffer: 20× NuPage® MOPS SDS Running Buffer (Invitrogen). Dilute it to 1× SDS Running Buffer with distilled water for the gel electrophoresis.
A low speed orbital shaker.
Nitrocellulose membrane (Invitrogen).
IR-Dye conjugated streptavidin: IRDye800CW-Streptavidin (Li-COR Biosciences, Lincoln, NE, USA).
An Odyssey® Infrared Imaging System (Li-COR Biosciences).
2.4. Components of OGA Expression in E. coli
Cloned plasmids of human OGA were used to generate inserts spanning the ORF of O-GlcNAcase which was ligated into pBADHisA expression vector (Novagen) (see Note 4).
BL21(DE3) chemically competent E. coli (Invitrogen).
Luria–Bertani (LB) broth.
Ampicillin solution (100 mg/mL), prepared as described above.
LB Agar plate with 50 μg/mL Ampicillin.
L-Arabinose solution (10 %): Weigh 0.5 g L-Arabinose (Sigma) and prepare 4.5 mL solution by adding water in a small falcon tube. Mix and filter-sterilize with a 0.22 μm filter (EMD Millipore). Aliquot in 1 mL volume and store at −20 °C.
Lysozyme solution (100 mg/mL): Weigh 0.5 g lysozyme (Sigma) and prepare 5 mL solution by adding water to a total volume of 5 mL. Mix and filter-sterilize with a sterilized 0.22 μm filter (EMD Millipore). Aliquot in 1 mL volume and store at −20 °C.
OGA lysis buffer (see Note 5): 20 mM Tris–HCl, pH 7.5, 0.1 mg/mL of lysozyme, and Complete mini EDTA-free protease inhibitor cocktail (Roche Applied Science). Place on ice until it is used.
Triton X-100™ (Sigma) 10 % solution.
A centrifuge: Sorvall_RC 5C Plus centrifuge (DuPont, Delaware City, DE, USA)
A sonicator: Misonix Sonicator® S-4000 Ultrasonic Processor (Cole-Parmer, Vernon Hills, IL, USA).
2.5. Components of OGA Purifi cation
HisTrap HP column (GE Healthcare Biosciences, Pittsburgh, PA, USA): Column is pre-charged with Ni2+.
8× Phosphate buffer stock solution, pH 7.4 (GE Healthcare Biosciences).
2 M imidazole, pH 7.4 (GE Healthcare Biosciences).
A 5-mL syringe.
A 0.45 μm filter (EMD Millipore).
A UV–vis Spectrophotometer: NanoDrop 2000 (Thermo Scientific).
A fluorescence microplate reader: Victor 2 Microplate Reader (Perkin-Elmer Life Sciences, Waltham, MA, USA).
2.6. Components of OGA Enzymatic Assay
2.6.1. Components of Standard OGA Activity Assay (Fig. 4)
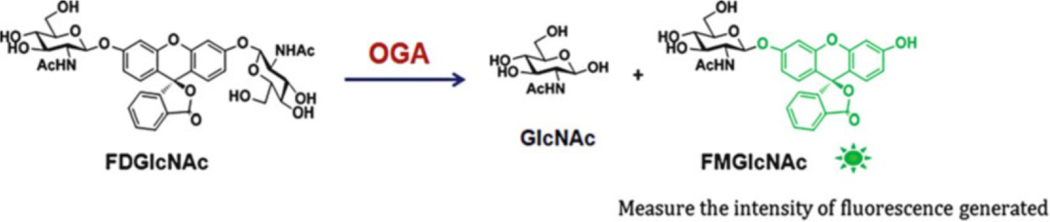
Fig. 4.
Fluorogenic OGA activity assay. This method uses nonfluorescent FDGlcNAc as a OGA substrate. Upon enzymatic cleavage of FDGlcNAc by OGA, fluorescent FMGlcNAc is generated
A fluorogenic substrate (FDGlcNAc 3): FDGlcNAc is synthesized as described previously [12]. In order to prepare 1 mM FDGlcNAc, first, prepare 10 mM of FDGlcNAc by dissolving 7.38 mg in distilled water to give a final volume of 1 mL. Then transfer 50 μL from 10 mM FDGlcNAc solution into a new vial and add 450 μL of distilled water to make a final concentration of 1 mM solution. Store both 10 and 1 mM solutions of FDGlcNAc at −30 °C.
OGA assay buffer: 0.5 M citrate–phosphate buffer, pH 6.5 (see Note 6).
N-Acetyl-D-galactosamine (GalNAc) solution (0.1 M): Dissolve 22.6 mg of GalNAc (Sigma) in distilled water to a final volume of 1 mL. Store at −30 °C.
Na2CO3 solution (0.5 M): Dissolve 5.30 g of sodium carbonate (Sigma) in distilled water to a final volume of 100 mL. Filter-sterilize with a sterilized 0.22 μm filter (EMD Millipore). Store at room temperature.
A fluorescence microplate reader: Victor 2 Microplate Reader (Perkin-Elmer Life Sciences).
2.6.2. Components of OGA Activity Assay in a 96-Well Plate Format
A fluorogenic substrate (FDGlcNAc 3): FDGlcNAc is synthesized as described above in Subheading 2.6.1.
OGA assay buffer, 0.1 M GalNAc and 0.5 M Na2CO3 solutions: as described in Subheading 2.6.1.
A 96-well plate (BD Biosciences, Bedford, MA, USA).
A fluorescence microplate reader: Victor 2 Microplate Reader (Perkin-Elmer Life Sciences).
3. Methods
3.1. OGT Expression in E. coli
pET43.1 Ek/LIC/ncOGT or mOGT is transformed into competent BL21(DE3) cells according to the manufacturer’s instructions. Grow overnight on a LB Agar plate with 50 μg/mL Ampicillin (LB Agar/Amp plate) at 37 °C in the incubator.
Inoculate a single colony on the LB Agar/Amp plate in a 20 mL culture of LB broth supplemented with Ampicillin (50 μg/mL).
Grow overnight at room temperature (see Note 7).
Centrifuge the cells at 2,500 × g at 4 °C for 10 min.
Remove the supernatant and resuspend the pellets in 0.99 mL of OGT lysis buffer containing 20 mM Tris–HCl, pH 7.5, 2 mM EDTA, 1 mg/mL of lysozyme, and complete mini EDTA-free protease inhibitor cocktail.
Incubate at room temperature for 5 min to perform the lysozyme digestion.
Add 10 μL of 10 % Triton X-100 to give 0.1 % final concentration of Triton X-100 and vortex.
Sonicate the lysate for 15 s on ice with 30 s pause in between each until DNA is completely sheared (see Note 8).
Centrifuge lysate at 14,000 × g for 10 min and aliquot supernatant (~200 μL) in a clean tube and store at −80 °C.
Determine the OGT expression level in the supernatant (see Note 9).
3.2. Purification of Expressed OGT in Lysate
Perform the OGT purification procedure in the cold room (~4 °C).
Gently suspend S-protein Agarose by inversion and transfer 240 μL of the slurry (equivalent to 120 μL settled resin) to a clean tube (see Note 10).
Add 600 μL of OGT assay buffer to the resin and gently mix by inversion to wash the resin. Centrifuge at 500 × g for 5 min and carefully discard the supernatant.
Repeat step 2.
Add 120 μL of E. coli lysate containing OGT to the pre-washed S-protein Agarose.
Mix thoroughly and incubate at 4 °C on an orbital shaker for 1–2 h.
Centrifuge the entire volume at 500 × g at 4 °C for 5 min and carefully decant supernatant.
Resuspend the S-protein Agarose, which now contains bound S-tag fusion OGT enzyme, in 600 μL OGT assay buffer. Mix thoroughly by inverting the tube 5−7 times.
Repeat steps 6 and 7 twice more to wash away unbound proteins.
Centrifuge at 500 × g at 4 °C for 5 min and carefully remove the final supernatant.
Resuspend OGT bound to S-protein Agarose in 240 μL of OGT assay buffer and use 10−15 μL for each reaction.
3.3. OGT Enzymatic Activity Assay
3.3.1. Radiometric OGT Activity Assay
Keep UDP-[14C]-GlcNAc, E. coli lysate containing active OGT enzyme, and Nup62 on ice at all times during the experiment. Experiment has been performed at room temperature unless indicated otherwise.
Calculate the volume of OGT assay buffer to make a total reaction volume be 40 μL: If there are UDP-[14C]-GlcNAc 0.4 μL (20 μM), E. coli lysate containing active OGT enzyme 8 μL, and Nup62 1 μL (1 μg), then amount of OGT assay buffer needed is 30.6 μL.
Place appropriate amount of OGT assay buffer calculated from the step 1 in a clean tube and add 10–20 μM of UDP-[14C]-GlcNAc and 1 μg of recombinant, purified Nup62.
Add 8 μL of the lysate containing OGT enzyme into reaction mixture and vortex briefly.
Incubate reactions at 37 °C and rotate at 220 rpm for 1–2 h.
Stop the enzymatic reaction by adding 20 μL of 4× NuPAGE® LDS sample buffer and boil samples for 3 min.
Load samples onto a SDS-PAGE gel (10 % or 4–12 % NuPAGE® Bis-Tris gel) and run with 1× NuPage® MOPS SDS Running Buffer for 50 min at 200 V.
After finishing gel electrophoresis, rinse the gel with distilled water by transferring the gel into a tray containing distilled water and incubating on a shaker for 5 min.
Carefully pour off distilled water from the tray. Repeat the step 7 twice more.
Carefully pour off distilled water and pour stain solution, e.g., Simply Blue Safestain solution to cover the gel in the tray, and incubate the gel in the stain solution on a shaker for 1 h.
Carefully pour off the stain solution and replace with distilled water to de-stain the gel and incubate the gel in distilled water to de-stain on a shaker for 1 h.
Pour off distilled water and soak the gel in En3Hance in a fume hood and incubate on a shaker for 1 h.
Carefully pour off the En3Hance solution and rinse the gel with a 10 % PEG (MW8000) solution for 30 min.
After rinsing, dry the gel using a gel dryer (see Note 11).
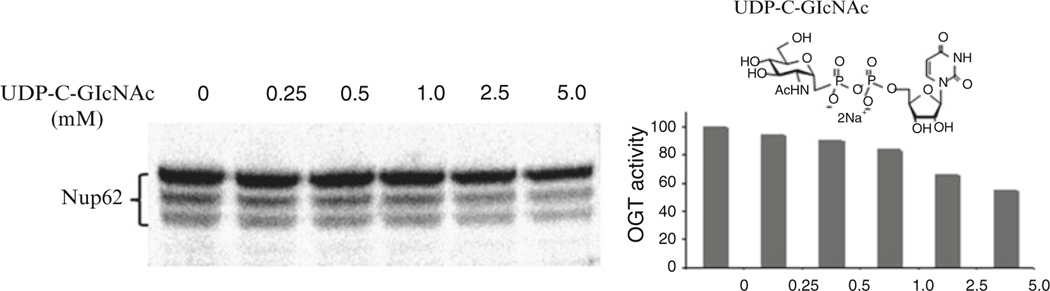
Expose the gel to a phosphor screen. Scan and analyze the imaging plate on the Fujifilm BAS-1500 phosphorimager (Fig. 5, see Note 12). Densitometry of the phosphoimage data is performed with Image Gauge 3.0 software (see Note 13).
Fig. 5.
Radiometric ncOGT activity assay in the absence and presence of UDP-C-GlcNAc. Inhibition of ncOGT by UDP-C-GlcNAc can be evaluated using Nup62 and UDP-[14C]-GlcNAc substrate [16]
3.3.2. Chemoenzymatic OGT Activity Assay
Keep UDP-GlcNAz, purified OGT bound to S-protein Agarose, and Nup62 on the ice all the time during the experiment. Experiment has been performed at room temperature unless indicated otherwise.
Calculate the volume of OGT assay buffer to make a total reaction volume of 40 μL: If UDP-GlcNAz solution is required, add 1 μL (50 μM), Purified OGT bound to S-protein Agarose 15 μL, and Nup62 2 μL (2 μg), then OGT assay buffer needed is 22 μL.
Place appropriate amount of OGT assay buffer calculated from the step 1 in a clean tube and add 10−50 μM of UDP-GlcNAz and 2 μg of recombinant, purified Nup62.
Add 10−15 μL of purified OGT bound to S-protein Agarose into reaction mixture and vortex briefly.
Incubate reactions at 37 °C with frequent mixing for 1–2 h.
Centrifuge at 500 × g for 5 min and carefully transfer the supernatant to the new clean tube.
Wash the resin with 50 μL of OGT assay buffer. Centrifuge at 500 × g for 5 min and combine the wash solution with the supernatant in the tube obtained from step 5.
Remove excess, unreacted UDP-GlcNAz and buffer-exchange into 1× phosphate buffer using a Microcon YM-30 from the combined solution (see Note 14).
To the buffer-exchanged filtrate (~40 μL) containing Nup62, add 1 μL of Biotin-Phosphine reagent (10 mM stock solution) and vortex.
Perform the Staudinger ligation by incubating reaction mixture at 37 °C at 200 rpm for 2 h (see Note 15).
Add 20 μL of 4× NuPAGE® LDS sample buffer and boil samples for 3 min.
Load samples onto a SDS-PAGE gel (10 % or 4–12 % NuPAGE® Bis-Tris gel) and run with 1× NuPage® MOPS SDS Running Buffer for 50 min at 200 V.
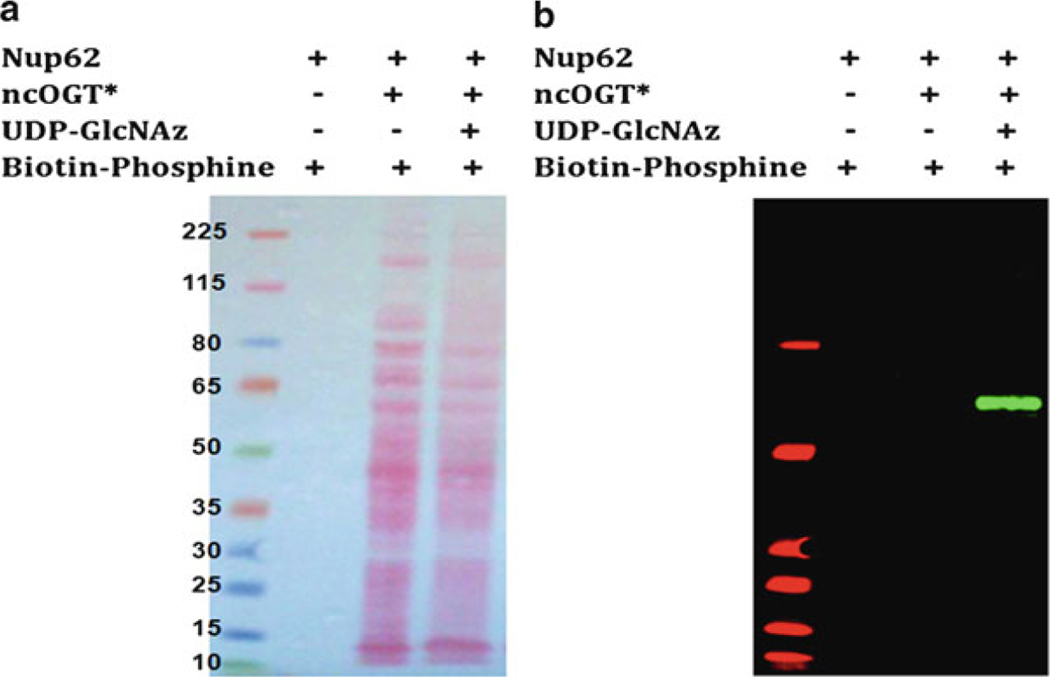
Electrophoretically transfer the protein from the gel onto the nitrocellulose membrane and probe the membrane with IR-Dye conjugated Streptavidin (Fig. 6).
Fig. 6.
Ponce S. Stained (a) and Western blot (b) of Nup62. ncOGT utilizes UDP-GlcNAz to label Nup62 giving a distinctive signal after chemoselective reaction with Biotin-Phosphine. Nitrocellulose membrane was probed with IRDye800CW-Streptavidin. Asterisk (*) represents ncOGT in a crude extract of E. coli
3.4. OGA Expression in E. coli
Carry out all procedures at room temperature unless otherwise specified.
pBADHisA/MEGE5 plasmid is transformed into competent BL21(DE3) cells according to the manufacturer’s instruction. Grow overnight on a LB Agar plate with 50 μg/mL Ampicillin (LB Agar/Amp plate) at 37 °C in the incubator.
Pick a single colony on the LB Agar/Amp plate in 5 mL LB medium containing Ampicillin (50 μg/mL).
Grow overnight at 37 °C at 200 rpm.
Inoculate 1 mL of overnight culture into 99 mL of fresh LB medium supplemented with Ampicillin (100 μg/mL) and cultivate at 37° at 200 rpm until they reach mid-log phase (OD600 ~ 0.5; 2.5−3.5 h).
Induce the culture by adding 200 μL of 10 % arabinose solution to a final concentration of 0.02 % and culture for an additional 3−4 h at 30 °C at 200 rpm.
Harvest cells by centrifugation at 3,000 × g for 10 min. Discard the supernatant. Store the cell pellet at −80 °C until the cells are lysed.
Freeze-thaw the cell pellet prepared in the previous step and suspend in 1.8 mL OGA lysis buffer (add 0.9 mL OGA breaking buffer for 50 mL culture volume) containing 20 mM Tris–HCl, pH 7.5, 100 μg/mL of lysozyme, and complete mini EDTA-free protease inhibitor cocktail.
Add 18 μL of 10 % Triton X-100™ solution and vortex.
Incubate at room temperature for 15 min and sonicate lysate for 15 s on ice with 30 s pause in between each until DNA is completely sheared.
Centrifuge lysate at 14,000 × g for 10 min and aliquot supernatant (~200 μL) in a clean tube and store at −80 °C.
Determine the OGA expression level of the supernatant (see Note 16).
3.5. Purification of His-Tag Fused OGA
To avoid clogging of the column it is recommended to filter lysate containing recombinant OGA through a 0.45 μm filter. Carry out the purification procedure in the cold room (4 °C). For 2 mL volume of lysate, 1-mL size of HisTrap HP column can be used. Use a buffer containing 1× Phosphate buffer (20 mM sodium phosphate, 500 mM NaCl, pH 7.4) and 40 mM imidazole as binding and wash buffer and a flow rate of 1 mL/min.
Prepare the column for His-tagged OGA’s purification (see Note 17) at room temperature. But perform the purification in a cold room.
Thaw the lysate (2 mL) and add 40 μL of 2 M imidazole solution to a final concentration of 40 mM (see Note 18).
Apply the lysate containing 40 mM imidazole to the column using a syringe with a flow rate of 1 mL/min.
Collect the flowthrough fraction in a falcon tube.
Wash with 20 mL binding buffer.
Start elution with 5 mL of 1× phosphate buffer containing 100 mM imidazole and collect the eluate in five 1-mL fractions.
Next add 5 mL of 1× phosphate buffer containing 300 mM imidazole and collect the eluate in five 1-mL fractions (see Note 19).
Finally add 3 mL of 1× phosphate buffer containing 500 mM imidazole and collect the eluate in three 1-mL fractions (see Note 20).
Check the different fractions for protein by measuring the absorbance of eluate at 280 nm (A280nm) for protein assays (see Note 21), and by performing an OGA activity assay using a protocol described below in this chapter.
Pool the fractions that exhibit OGA activity. Perform buffer exchange into a buffer containing 20 mM Tris–HCl, pH 7.5 using a Microcon YM-100.
Aliquot purified OGA and store at −80 °C.
Concentration of purified OGA can be determined using a BCA assay and the purity of OGA can be determined by Western blot analysis using anti-His-tag antibody.
3.6. OGA Activity Assay
3.6.1. Standard OGA Activity Assay
First, calculate distilled water required to make the total volume of assay be 100 μL. (If there are enzyme volume 2 μL, FDGlcNAc 4 μL, OGA assay buffer 20 μL, GalNAc solution 10 μL, then volume of distilled water needed is 72 μL).
Place distilled water that is calculated from step 1 in a clean tube.
Add 20 μL of OGA assay buffer.
Add 10 μL of 0.1 M GalNAc solution and briefly vortex.
Add 10 μL of 1 mM FDGlcNAc solution and briefly vortex.
Carefully, add 2 μL of OGA overexpressed lysate and briefly vortex. Upon addition of the lysate, OGA enzymatic reaction starts.
Incubate assay solution at 37 °C for 20 min.
Terminate the enzymatic reaction by adding 900 μL of 0.5 M Na2CO3 solution and vortex.
Measure the fluorescence generated from the enzymatic reaction on a fluorescence spectrofluorometer or on a fluorescence microplate reader with the excitation wavelength of 485 nm and the emission wavelength of 535 nm. For fluorescence measurement on a fluorescence microplate reader, transfer 200 μL of OGA assay reaction solution to a 96-well plate and read the fluorescence at the excitation wavelength of 485 nm and the emission wavelength of 535 nm.
3.6.2. OGA Activity Assay in a 96-Well Plate Format
First, calculate distilled water required to make the total volume of assay be 50 μL (If there is enzyme volume 1 μL, FDGlcNAc 2 μL, OGA assay buffer 10 μL, GalNAc solution 5 μL, then volume of distilled water needed is 32 μL).
Place distilled water that is calculated from step 1 in a clean tube.
Add 10 μL of OGA assay buffer.
Add 5 μL of 0.1 M GalNAc solution and briefly vortex.
Add 2 μL of 1 mM FDGlcNAc solution and briefly vortex.
Carefully, add 1 μL of OGA overexpressed lysate and briefly vortex. Upon addition of the lysate, OGA enzymatic reaction starts.
Incubate assay solution at 37 °C for 20 min.
Terminate the enzymatic reaction by adding 200 μL of 0.75 M Na2CO3 solution.
Read the fluorescence on a fluorescence microplate reader at the excitation wavelength of 485 nm and the emission wavelength of 535 nm.
4. Notes
Recombinant human ncOGT and mOGT have both His-Tag and S-Tag.
To prepare a10 mL volume of OGT lysis buffer, place 200 μL of 1 M Tris–HCl, pH 7.5, 40 μL of 0.5 M EDTA, 100 μL of 100 mg/mL lysozyme stock solution, and a complete mini EDTA-free protease inhibitor cocktail tablet and add water to give a final volume of 10 mL. Mix thoroughly and place the lysis buffer on ice while cells are lysed.
To prepare a 10 mL volume of OGT assay buffer, place 500 μL of 1 M Tris–HCl, pH 7.5, 125 μL of 1 M MgCl2, and 10 μL of 1 M DTT and add water to give a final volume of 10 mL.Mix thoroughly and store at 4 °C.
Recombinant human OGA has a His-tag.
To prepare a 10 mL volume of OGA lysis buffer, place 200 μL of 1 M Tris–HCl, pH 7.5, 10 μL of 100 mg/mL lysozyme stock solution, and a complete mini EDTA-free protease inhibitor cocktail tablet and add water to give a final 10 mL volume. Mix thoroughly and place the OGA breaking buffer on ice while cells are lysed.
To make 0.5 M citrate–phosphate buffer, first dissolve 15.0 g of anhydrous sodium dihydrogen phosphate (NaH2PO4) in 200 mL of distilled water. Prepare 0.5 M citric acid solution by dissolving 7 mL (10.51 g) of citric acid monohydrate in distilled water to a final volume of 100 mL. Adjust sodium phosphate buffer’s pH to be 6.5 by adding 0.5 M citric acid solution. When pH of phosphate buffer gets 6.5, add distilled water to give a final volume of 250 mL and filter-sterilize with a 0.22 μm filter.
Temperature control is critical to obtain active OGT enzyme. No not allow the temperature of cell culturing over 30 °C or an OGT enzymatic activity will be dramatically decreased.
Perform sonication of the lysate on ice all the time because heat generated during sonication can cause the loss of an OGT activity.
Total protein level in the supernatant can be determined using the Pierce® bicinchoninic acid (BCA) protein assay protocol as described by the manufacturer (Thermo Fisher Scientific). OGT expression can be determined by detecting His-tag fusion protein or S-tag fusion protein by immunoblotting with anti-His-tag antibody (Abcam, Cambridge, MA, USA) or anti-S-tag antibody (Abcam).
The resin is most conveniently transferred with a 1 mL wide-mouth pipet tip.
For gel drying experiment, empty gel dryer cold trap flask before turning on the dryer to prevent solvent accumulation and potential vacuum cutoff. Prechill gel dryer cold trap. Place gel onto a Whatman 3 MM filter paper and cover with plastic wrap. Put gel sandwich (paper, gel, and plastic wrap) on dryer. Cover with flexible plastic membrane and turn on vacuum. Then turn on heat for 2 h at 75 °C. When the gel is completely dried, turn off the vacuum and trap chiller and remove gel sandwich. Remove plastic wrap.
For phosphorimaging of dried gel, first, blank phosphor screen (BAS III) on light source for about 20 min. Put the gel in the phorphor cassette (BAS cassette 2040, Fuji Film Co.) and carefully place the erased screen phosphor side down on the gel. Close cassette and leave to expose at room temperature for 16−24 h. When ready to read, take the screen to the phosphorimager.
Stoichiometry of O-GlcNAc modified Nup62 can be determined using phosphorimager quantitation against [methyl-14C]-BSA internal control.
Insert Microcon sample reservoir into vial. Pipette the combined solution into sample reservoir without touching the filter membrane with the pipette tip. Seal with attached cap and centrifuge at 14,000 × g at 4 °C. Place 100 μL of 1× PBS and centrifuge at 14,000 × g at 4 °C. Add 100 μL of 1× PBS and centrifuge at 14,000 × g at 4 °C one more time. Separate vial from sample reservoir and place sample reservoir upside down in a new vial, then spin 3 min at 1,000 × g at 4 °C to transfer concentrate to vial. Adjust the concentrate volume to be about 40 μL.
Azide can be selectively reacted with phosphine reagent by Staudinger ligation or with terminal alkyne and copper catalyst by Click chemistry. In this protocol, we demonstrated the Staudinger ligation method.
Total protein level in the supernatant can be determined using the Pierce® bicinchoninic acid (BCA) protein assay protocol as described by the manufacturer. OGA expression can be determined by detecting His-tag fusion protein by immunoblotting with anti-His-tag antibody (Abcam).
Preparation of HisTrap HP column for His-tagged protein’s purification can be performed at room temperature. Remove the snap-off end of the column and wash the column with 5 mL distilled water at a rate of 1 mL/min. Equilibrate the column with 10 mL binding buffer (20 mM sodium phosphate, 500 mM NaCl, pH 7.4, and 40 mM imidazole) using the syringe.
To minimize nonspecific binding of host cell proteins, lysate to be purified is adjusted to have the same concentration of imidazole as the binding and wash buffer (40 mM) before it is applied to the column.
The purified OGA is most likely found in the fractions eluted with a 1× Phosphate buffer containing 300 mM imidazole.
After the OGA protein has been eluted, the column can be re-equilibrated with 10 mL of binding buffer. The column is ready for a new purification.
For A280 measurement, use the elution buffer as a blank.
Acknowledgments
This work was supported by NIDDK intramural funds (NIH) and the National Research Foundation of Korea (2011–0027257).
References
- 1.Hanover JA, Krause MW, Love DC (2010) The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta 1800:80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butkinaree C, Park K, Hart GW (2010) O-linked beta-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta 1800:96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem 80:825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Love DC, Krause MW, Hanover JA (2010) O-GlcNAc cycling: emerging roles in development and epigenetics. Semin Cell Dev Biol 21:646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubas WA, Frank DW, Krause M, Hanover JA (1997) O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem 272:9316–9324 [DOI] [PubMed] [Google Scholar]
- 6.Kreppel LK, Hart GW (1999) Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem 274:32015–32022 [DOI] [PubMed] [Google Scholar]
- 7.Lubas WA, Hanover JA (2000) Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem 275:10983–10988 [DOI] [PubMed] [Google Scholar]
- 8.Coutinho P, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328:307–317 [DOI] [PubMed] [Google Scholar]
- 9.Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E (2004) The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol 11:1001–1007 [DOI] [PubMed] [Google Scholar]
- 10.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469:564–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schimpl M, Borodkin VS, Gray LJ, van Aalten DM (2012) Chem Biol 19:173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim EJ, Kang DO, Love DC, Hanover JA (2006) Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr Res 341:971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubas WA, Smith M, Starr CM, Hanover JA (1995) Analysis of nuclear pore protein p62 glycosylation. Biochemistry 34: 1686–1694 [DOI] [PubMed] [Google Scholar]
- 14.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR (2003) A chemical approach for identifying O-GlcNAc modified proteins in cells. Proc Natl Acad Sci USA 100: 9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxon E, Bertozzi C (2000) Cell surface engineering by a modified Staudinger reaction. Science 287:2007–2010 [DOI] [PubMed] [Google Scholar]
- 16.Hajduch J, Nam G, Kim EJ, Fröhlich R, Hanover JA, Kirk KL (2008) A convenient synthesis of the C-1-phosphonate analogue of UDP-GlcNAc and its evalution as an inhibitor of O-linked GlcNAc transferase (OGT). Carbohydr Res 343:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]