Abstract
The SARS-CoV-2 pandemic has highlighted the need for vaccines that are effective, but quickly produced. Of note, vaccines with plug-and-play capabilities that co-deliver antigen and adjuvant to the same cell have shown remarkable success. Our approach of utilizing a nitrilotriacetic acid (NTA) histidine (His)-tag chemistry with viral adjuvants incorporates both of these characteristics: plug-and-play and co-delivery. We specifically utilize the cowpea mosaic virus (CPMV) and the virus-like particles from bacteriophage Qβ as adjuvants and bind the model antigen ovalbumin (OVA). Successful binding of the antigen to the adjuvant/carrier was verified by SDS-PAGE, western blot, and ELISA. Immunization in C57BL/6J mice demonstrates that with Qβ - but not CPMV - there is an improved antibody response against the target antigen using the Qβ-NiNTA:His-OVA versus a simple admixture of antigen and adjuvant. Antibody isotyping also shows that formulation of the vaccines can alter T helper biases; while the Qβ-NiNTA:His-OVA particle produces a balanced Th1/Th2 bias the admixture was strongly Th2. In a mouse model of B16F10-OVA, we further demonstrate improved survival and slower tumor growth in the vaccine groups compared to controls. The NiNTA:His chemistry demonstrates potential for rapid development of future generation vaccines enabling plug-and-play capabilities with effectiveness boosted by co-delivery to the same cell.
Graphical Abstract
INTRODUCTION
The SARS-CoV-2 pandemic highlights the importance of vaccines that can be produced and scaled quickly.1,2 The first COVID-19 vaccines introduced into the clinic had a modular platform with rapid antigen exchange capabilities, often referred to as plug-and-play.3 For instance, the mRNA vaccines by Moderna and BioNTech/Pfizer were both being studied previously for cancer vaccine applications,4,5 but the encapsulated mRNA encoding the cancer antigens were replaced with mRNA encoding the spike proteins of SARS-CoV-2 – a plug- and-play strategy.6,7 The ability to swap the antigen of choice so rapidly while keeping the core technology the same is a defining feature of these vaccine platforms, and it provided the opportunity to develop their vaccines at a rapid rate. This is most likely the reason why the mRNA and viral vector vaccines (e.g., by Janssen and the University of Oxford/AstraZeneca), which also have plug-and-play capabilities, were the first to be developed and then utilized in the clinic.8 In fact, Moderna started phase I clinical trials for its SARS-CoV-2 vaccine candidate in 10 weeks, a speed that is unparalleled by traditional vaccine efforts.9
There are also efforts to co-deliver the adjuvant and antigen in vaccine applications. While traditional vaccines may inject the antigen and adjuvant as simple admixtures, newer research suggests that co-delivery can boost vaccine effectiveness and reduce side effects.10 This is mainly due to activation of the antigen presenting cell to the actual target antigen and not off-target self-antigens.11 Co-delivered vaccines improve effector B and T cell responses improving therapeutic and prophylactic response in not only infectious disease applications but also in cancer and chronic diseases.11–15
Here, we set out to develop a vaccine platform capable of both plug-and-play and co-delivery. We utilized a nitrilotriacetic acid (NTA) linker conjugated to cowpea mosaic virus (CPMV), a plant virus, or virus-like particles (VLPs) from bacteriophage Qβ, through simple lysine, N-hydroxysuccinimide (NHS) chemistry. Both CPMV and Qβ have shown remarkable efficacy as vaccine adjuvants.16–21 The NTA group complexes with any histidine (His)-tagged protein of interest in the presence of a nickel (Ni) ion.22 We hypothesized that with this method, co-delivery of His-tagged antigen bound to the viral adjuvants (CPMV or Qβ) would be achieved. Furthermore, plug-and-play is achieved as the target antigen can be exchanged if it contains a His-tag. In fact, many recombinant proteins are already His-tagged to aid in the protein purification process.23,24 Assuming the His-tag does not alter the function or immunogenicity of the antigen, post-purification cleavage and additional processing would not be required potentially saving time and lowering costs during vaccine formulation.23,25
As with any vaccine platform, the adjuvant choice is as important as the antigen design itself. In our case, we utilized the viral adjuvants CPMV and Qβ, which has demonstrated efficacy in cancer and infectious disease vaccines.16–21 The virus-based nanotechnologies (a plant virus and VLP) cannot replicate in mammals improving safety but are recognized as pathogens by pathogen associated molecular patterns (PAMPs).26,27 More specifically, they activate pattern recognition receptors such as toll-like receptors thereby instigating innate immune responses.26,28,29 Their size also allows for efficient trafficking to the draining lymph nodes where uptake by antigen presenting cells can lead to priming of the adaptive response.30 Past studies have also demonstrated the safety of both viruses as adjuvants with no reported toxicities.20,31
Traditional protocols for viral nanoparticle vaccine formulations have focused on chemical conjugation. For example, Kentucky Bioprocessing, Inc. creates their SARS-CoV-2 vaccine through chemical conjugation of the receptor binding domain of SARS-CoV-2 onto tobacco mosaic virus.32 However, chemical conjugation can have its fair share of drawbacks. First, conjugation of large protein antigens is difficult, and it must be tailored to the protein of interest, which does not allow for plug-and-play capabilities. Second, chemical conjugation may lead to antigen display in different configurations (i.e., when a protein has multiple conjugation sites). This can lead to batch-to-batch variability and inconsistent immune responses against the target antigen. Lastly, conjugation can lead to epitope masking, aggregation, and disruption of protein structures, which must be experimentally resolved and can take extended periods of time.33 To overcome this, peptide epitopes of the antigen have been used. These vaccine formulations require simplified bioconjugation/genetic display procedures leading to greater yields, better reproducibility, and increased quality control and assurance. However, with peptide vaccination, the breadth of antibody response becomes quite narrowed leading to limited neutralization34 – vaccine ineffectiveness then leads to a complete restart of the vaccine formulation starting from epitope design. The NiNTA:His-tag approach combats these drawbacks: by binding the full-length antigen in a controlled manner through engineered His-tags, we get simple, non-tailored binding with a broad antibody response to the full-length protein. We tested this design using the CPMV and Qβ adjuvants and the common model antigen ovalbumin (OVA). We test these vaccines for improved antibody production compared to simple admixtures of OVA and CPMV/Qβ and demonstrate efficacy in a mouse model of OVA-expressing melanoma (B16F10-OVA) in C57BL/6J mice.
RESULTS AND DISCUSSION
Vaccine Production and Characterization.
CPMV was harvested from black-eyed pea no. 5 plants while Qβ VLPs were expressed and purified from Bl21 (DE3) E. coli as previously reported.19,35 The capsids of CPMV and Qβ both contain external lysines (300 on CPMV,36 720 on Qβ37) – thus, the exterior lysines on CPMV and Qβ were first conjugated to an NTA-PEG2K-NHS linker (Figure 1). The NTA group was then reacted with Ni overnight. Following purification of unbound Ni, we ascertained the presence of bound Ni using dithiothreitol (DTT), which causes a color change of the solution to a light brown color following reduction of Ni ions (Figure S1).38 The NiNTA group then serves as a linker to bind a His-tagged protein of interest. Proteins are commonly genetically engineered with His-tags for purification.23 For our studies, we manually inserted His-tags into native target proteins through bioconjugation (Figure S2). This was accomplished by reacting 2-iminothiolane to solvent-exposed lysines on the model antigen, OVA, therefore introducing a thiol group to couple to maleimide-terminated His6 peptides. The successful addition of the His-tag (His6) by chemical conjugation to OVA was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot (WB) (Figure S2). We acknowledge that chemical tagging of the protein with a His-tag may not provide orientational control of display, but we reasoned it would be highly effective as a proof-of-concept and to demonstrate that several His-tagged proteins could be easily obtained through the chemical coupling of the His-tagged peptides (see Figure S3). This approach demonstrates wide pertinency for a variety of applications due to the plug-and-play nature of the vaccine formulation.
Figure 1.
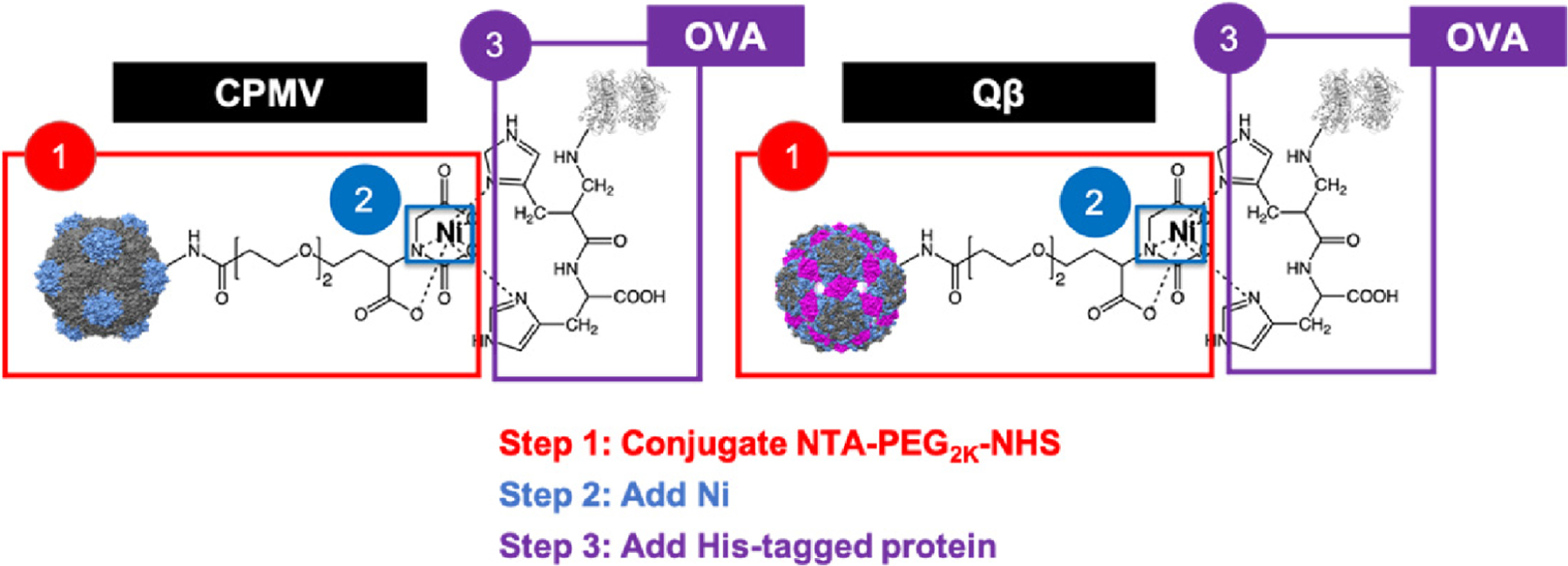
Binding schematic of His-tagged OVA to NiNTA-conjugated CPMV/Qβ. The CPMV, Qβ, and OVA structures were created using Chimera 1.14 (CPMV PDB ID: 1NY7, Qβ PDB ID: 1QBE, OVA PDB ID: 1OVA). The chemical structures were created on ChemDraw 19.0. The small and large CPs of CPMV are shown in gray and blue, respectively. The Qβ CPs down the 5–3-2-fold axis are colored in gray, blue, and magenta, respectively.
To ensure that the NiNTA-conjugated virus-based nanoparticles and the His-tagged OVA (His-OVA) were indeed coupled with the antigen, dot blots (DBs) were carried out (Figure 2a). His-OVA was spotted onto a nitrocellulose membrane and then probed with either CPMV-NiNTA or native CPMV. Binding was visualized using an α-CPMV antibody and a horseradish peroxidase (HRP)-conjugated secondary antibody. Binding to His-OVA was only observed using CPMV-NiNTA (Figure 2a). Next, the complex was formed in solution and purified, followed by characterization using WB and SDS-PAGE (Figure 2b,c).
Figure 2.
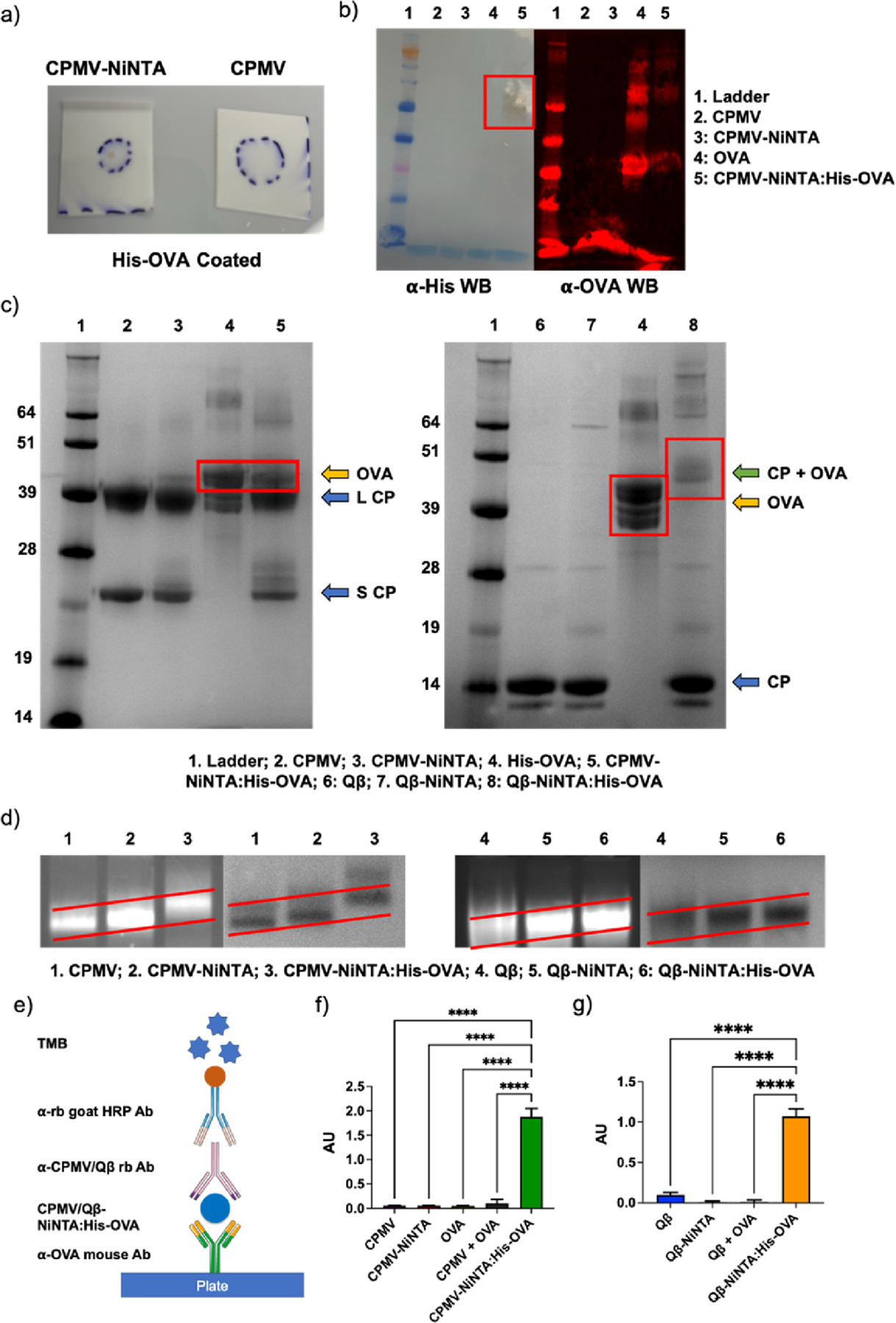
Characterization of NiNTA:His-OVA vaccine formulations. (a) DB of CPMV-NiNTA vs CPMV against His-OVA on a nitrocellulose membrane. (b) WB against His-tag (left) and OVA (right). (c) SDS-PAGE. In CPMV-NiNTA:His-OVA (left), OVA dissociates from the complex (lane 4); however, in Qβ-NiNTA:His-OVA (right), the CP and OVA remain associated (lane 8). (d) Agarose gel electrophoresis of the vaccine formulations. The increasing molecular weight is better demonstrated by the sloped red lines. Left is RNA staining, right is protein staining. (e) Schematic of ELISA. (f) CPMV-NiNTA:His-OVA ELISA and controls. (g) Qβ-NiNTA:His-OVA ELISA and controls. **** = p < 0.0001. The schematic in (e) was created using Biorender.com.
In the WB probed with α-His and α-OVA antibodies, His and OVA were successfully detected in the CPMV-NiNTA:His-OVA samples (Figure 2b, for WB, OVA served as the control). In SDS-PAGE, it should be noted that free His-OVA is observed, not in complex with the CPMV CPs (Figure 2c). However, it is expected that the Ni-NTA:His-OVA complex dissociates under the denaturing conditions of SDS-PAGE. While we cannot rule out entirely the presence of unbound His-OVA, native gels and ELISA further support the successful formation of the CPMV-NiNTA:His-OVA complex (Figure 2d–g). Somewhat contrasting, in the Qβ-NiNTA:His-OVA samples, the His-OVA remains bound to the Qβ coat protein (CP) as evidenced by the upwards shift of the His-OVA protein band by ∼14 kDa, the molecular weight of one Qβ CP.39 While somewhat puzzling, the His-OVA may not release after heating and denaturation from Qβ-NiNTA possibly due to the multivalency and avidity effects because Qβ exhibits a higher density of lysines and hence NiNTA. The amount of NTA bound per particle could not be determined, but there were ∼15 and 13 His-OVA per CPMV and Qβ, respectively. The amount of bound OVA was calculated using densitometry analysis on ImageJ. As mentioned above, CPMV and Qβ contain 300 and 720 addressable lysines on the exterior of the viral capsid.36,37 However based on labeling with fluorophores (small molecules) we expect ∼100 NTA displayed per nanoparticle; thus we estimate an OVA display efficiency at ∼15%. Based on the size of OVA and the available surface area of a 30-nm sized nanoparticle, a densely packed arrangement would equate to ∼39 OVA per nanoparticle.
The CPMV-NiNTA:His-OVA and Qβ-NiNTA:His-OVA vaccines were further characterized by agarose gel electrophoresis (Figure 2d). Changes in molecular weight and charge can influence the electrophoretic mobility. Coupling of the NTA-PEG2K-NHS linker and binding of Ni reduced the mobility of the particles. The binding of His-OVA reduced mobility even further indicating a step-by-step increase in molecular weight. Co-localization of the RNA (genomic RNA for CPMV and host RNA for Qβ) and protein component indicates stable particle formulations.40 Furthermore, transmission electron microscopy (TEM) and circular dichroism (CD) of both native and OVA-bound viruses confirm structural integrity of the vaccine formulations (Figures S4 and S5). TEM shows intact 30 nm-sized nanoparticles and binding of OVA did not change the morphology or structure. Consistent with TEM imaging, the overlayed CD spectra show consistency between the native and OVA-bound viruses with minimal differences in the secondary and tertiary structures which can be explained by the OVA protein loading onto the viral nanoparticles.
Modified ELISA protocols were carried out to validate that the His-OVA was indeed binding to CPMV/Qβ-NiNTA (Figure 2e–g). The plate was first coated with an α-OVA antibody followed by the addition of CPMV/Qβ-NiNTA:His-OVA and controls, and then probed using α-CPMV/Qβ antibodies. By utilizing two specific antibodies (α-OVA and α-CPMV/Qβ), only samples containing bound OVA and CPMV/Qβ would be able to produce a signal (see schematic in Figure 2e). While CPMV-NiNTA:His-OVA produced a strong discernible signal, the controls did not produce a significant signal above background (Figure 2f). Most importantly, there was no indication of binding or association in the CPMV and OVA admixture (CPMV + OVA). We noticed a similar pattern with the Qβ samples (Figure 2g).
Longitudinal studies utilizing the modified ELISA protocols, fast protein liquid chromatography (FPLC), and dynamic light scattering (DLS) were also carried out to investigate the structural and binding properties of the CPMV/Qβ-NiNTA:-His-OVA with respect to time. The ELISAs show that even 4 weeks past the production and assembly of the vaccines, the samples produce multi-fold improvements in absorbance over the controls indicating binding between the CPMV/Qβ and the His-OVA occurs long-term (Figure S6). The DLS data of CPMV-NiNTA:His-OVA shows minor levels of aggregation starting at day 7 most likely due to the decreased stability of the formulations following OVA disassembly (Figure S7a,c). The Qβ-NiNTA:His-OVA displayed slightly different properties in that there was almost immediate aggregation of the particles starting 1 day after the vaccines were generated, however, to a much smaller degree compared to the CPMV-NiNTA:His-OVA (Figure S7b,d). The aggregated particles were in the range of 100–200 nm, and only ∼50% of the particles were aggregated. The constant nature of the aggregation most likely indicates an equilibrium of association/dissociation of the OVA had occurred, which is also represented by the similar ELISA levels seen throughout the longitudinal study (Figure S6). Unlike the DLS data, the FPLC spectra for both the CPMV and Qβ-NiNTA:His-OVA did not show any levels of aggregation and the particles were intact (Figure S8). Additionally, there was no presence of unbound OVA at any of the timepoints – a control experiment with native CPMV and an equimolar ratio of OVA indicated that if 100% of the OVA was unbound, it could be detected by FPLC (Figure S9). This indicates that either (1) the OVA remains bound to the CPMV/Qβ-NiNTA or (2) the dissociated OVA is too low in concentration to be detected by FPLC.
Demonstration of Plug-and-Play Capabilities.
To validate that the vaccine formulation strategy indeed could be utilized as plug-and-play candidates for future vaccine applications, the Qβ-NiNTA was also tested to be complexed with other proteins such as bovine serum albumin (BSA) and carbonic anhydrase (CA). We chemically His-tagged both of these proteins and then bound them to the Qβ-NiNTA. SDS-PAGE characterization of the Qβ-NiNTA:His-CA and Qβ-NiNTA:His-BSA (Figure S3) demonstrates the successful binding of these antigens to Qβ-NiNTA and thereby the modular platforms capability.
SDS-PAGE reveals the presence of His-CA and His-BSA, but the pattern is distinct: His-CA dissociates from the Ni-NTA complex under the SDS-PAGE conditions (Figure S3a). In contrast, His-BSA remains stably bound as was observed with His-OVA (Figures S3b and 2c). Therefore, in addition to avidity effects from multivalent NiNTA display on the Qβ-NiNTA nanoparticles, the charge/hydrophobicity of the target protein may come into play to determine the overall stability of the complex. For the Qβ-NiNTA:His-CA complex that disassembles under SDS-PAGE conditions, we noted that only His-tagged CA and not a mixture of native and His-CA was detectable, which supports that free CA was not present in the complex.
Mice Immunization.
Mice Immunization and Antibody Titers.
The CPMV/Qβ-NiNTA:His-OVA vaccine formulations were then tested in mice to evaluate effectiveness in generating antibodies against the target antigen, OVA. C57BL/6J mice were immunized using a prime and double-boost regimen spaced two weeks apart (Figure 3a). Each vaccine was standardized to the OVA concentration with dosing of 5 μg/mouse subcutaneously (s.c.). To estimate the OVA concentration in CPMV-NiNTA:His-OVA vs Qβ-NiNTA:His-OVA, SDS-PAGE and ImageJ lane analysis was performed. Mice received 41 μg of CPMV or CPMV-NiNTA:His-OVA and 25 μg of Qβ or Qβ-NiNTA:His-OVA. The molecular weight of CPMV is ∼2.25× greater than Qβ, so although similar amounts of OVA were bound to each virus, a greater weight of CPMV was injected. Blood was collected every two weeks until week 6 and antibody titers and subtypes were evaluated using ELISA.
Figure 3.
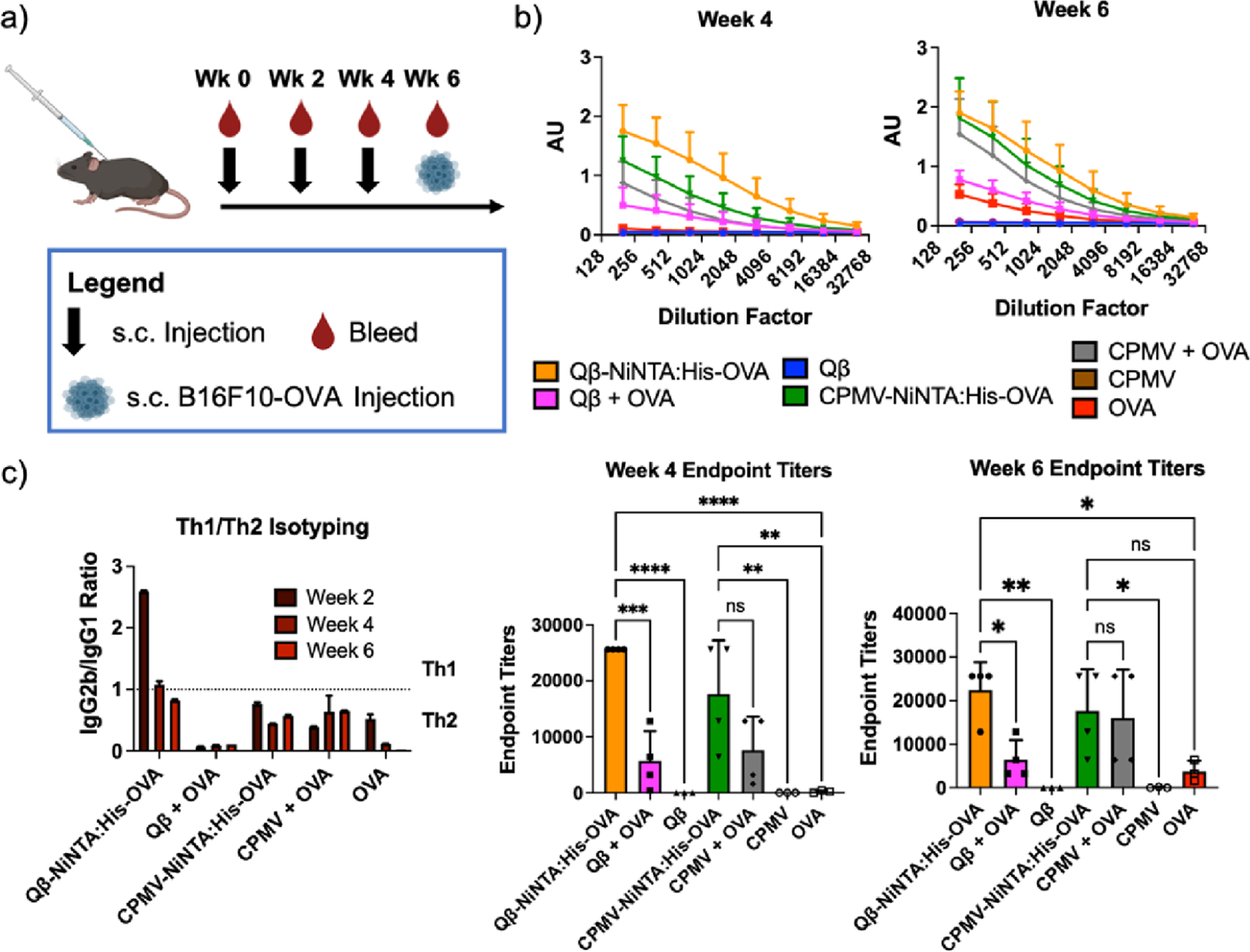
Antibody titers against OVA. (a) Injection and bleeding schedule. (b) Complete ELISAs at weeks 4 and 6 as well as the endpoint titers. The endpoint titer was determined as the dilution at which the absorbance was greater than twice the blank. The week 2 data can be found in Figure S5. (c) IgG isotyping. An IgG2b IgG1−1 ratio < 1 was considered Th2 while a ratio > 1 was considered Th1. The full antibody profile can be found in Figure S5. * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001, ns = not significant. The injection schedule schematic was created on Biorender.com.
At week 2, the titers are low as expected; however, even at week 2, data indicate a 4.4-fold increase in titers for the Qβ-NiNTA:His-OVA vs the Qβ + OVA admixture (Figure S10). By week 4, the Qβ-NiNTA:His-OVA titers were 4.5- and 128-fold that of the admixture (p < 0.001) and OVA only control (p < 0.0001), respectively (Figure 3b). At the last measured timepoint (week 6), the Qβ-NiNTA:His-OVA titers remained 2.3- and 6-fold improved compared to the admixture (p < 0.05) and OVA (p < 0.05), respectively. However, unlike with the Qβ groups, in the CPMV groups, there was no clear difference between the NiNTA vaccine and admixture. At week 4, the CPMV-NiNTA:His-OVA was 2.3- and 88-fold that of the admixture (p > 0.05) and OVA only control (p < 0.01), respectively. By week 6, the titers between CPMV-NiNTA:His-OVA and CPMV + OVA were identical and notably ∼4.7-fold greater than the OVA only control (p < 0.05).
The results demonstrate that with the Qβ formulation, Qβ-NiNTA:His-OVA outperformed the simple admixture of Qβ + OVA in terms of antibody production against the target protein OVA. This may be explained by the fact that OVA is being co-delivered with Qβ in the Qβ-NiNTA:His-OVA formulation, therefore achieving co-delivery of antigen and adjuvant to the same cell.41–43
With CPMV there were no clear differences between the CPMV-NiNTA:His-OVA and CPMV + OVA formulations which may indicate that (i) not all viral adjuvants may require co-delivery to achieve potency, (ii) the complex had dissociated, as observed in the SDS-PAGE (see Figure 2c), or (iii) that complexation in media led to co-delivery even with the admixture. In fact, it was recently shown that proteins may adsorb onto plant viral adjuvants even without integrated complexation chemistry and that some plant viral adjuvants remain efficacious without complexation.44,45 The data demonstrates that whether or not an antigen binds is a function of the protein chemistry of the antigen and viral adjuvant. For future experiments, one could utilize a trivalent NTA as opposed to the monovalent one used here to improve the binding kinetics between CPMV and OVA (monovalent NTA has a Kd of ∼10 μM and trivalent NTA has a Kd of ∼1 nM).46,47 The data also indicates that OVA by itself can elicit titers after 3 total injections, but that an adjuvant like CPMV or Qβ greatly improves titer production.
Lastly, we do concede that conjugation of OVA to the viruses would most likely boost antibody response compared to the NTA:His chemistry as the OVA would not dissociate in vivo – others have indeed showed that conjugation provides the best antibody response.46 However, conjugation can be difficult, and in our own experiments, conjugation of OVA to our virus particles utilizing both EDC/NHS and NHS maleimide chemistry were unsuccessful (data not shown) providing further evidence that for rapid development of vaccine candidates, a non-tailored approach such as with the NTA:His can greatly improve the development speed.
Antibody Isotyping.
The antibodies were further investigated for their IgG isotypes as well as any other Ig subtypes. A ratio of IgG2b IgG1−1 < 1 is seen as a Th2 balance while a ratio > 1 is Th1.48 In the Qβ-NiNTA:His-OVA group, the bias skewed strongly Th1 at week 2 and then moved to a balanced Th1/Th2 bias starting from week 4 and remained balanced at week 6 (Figure 3c). All the other groups (Qβ + OVA, CPMV-NiNTA:His-OVA, CPMV + OVA, and OVA) skewed strongly Th2 starting from week 2 and remained Th2 biased. The difference in bias between the Qβ-NiNTA:His-OVA and Qβ + OVA indicates that even with the same adjuvant/antigen combination, co-delivery can vastly affect the types of antibodies that are produced. When looking at the other antibody isotypes, all the vaccine groups were able to produce small quantities of IgM, as part of the onset of antibody production and IgM-to-IgG switch (Figure S11).49 Other antibody classes were not detected. Of note, IgE, which is known to elicit allergic responses, was not detected indicating the safety of the vaccines.
For cancer vaccines, generally a Th1 bias is desired, as this promotes cytotoxic T cell priming and destruction of cancer cells with increased safety when targeting self-antigens.50 Alternatively, active immunization to generate therapeutic antibodies (which is Th2-mediated) also has shown success, for instance, against HER2-positive cancers.51,52 Our past research with CPMV and peptide epitopes has generally indicated that CPMV vaccination induces a strong Th1 bias.19,53–55 However in complex with OVA, immunization promotes Th2 bias – therefore, it appears that the T helper cell bias is directly affected by the antigen (this data), vaccine formulation (e.g. implant, microneedle, or bolus injection), and the adjuvant, and the bias will have to be experimentally discovered for each antigen/adjuvant combination.19,53–55
Tumor Challenge and Vaccine Efficacy.
The same mice from above were challenged at week 6 post-immunization with 200,000 B16F10-OVA cells s.c. to determine whether α-OVA antibodies exhibited a therapeutic effect. Indeed efficacy was observed, in particular for the Qβ-NiNTA:His-OVA as well as the Qβ + OVA groups, with Qβ-NiNTA:His-OVA being the most potent formulation significantly reducing tumor burden. On day 20, the average tumor volume was 28.48 and 70.54 mm3 for Qβ-NiNTA:His-OVA and Qβ + OVA treated animals (Figure 4a). In comparison, the Qβ- and OVA-treated control groups displayed average tumor volumes of 247.5 and 799 mm3. By day 26, the difference in tumor volume between Qβ-NiNTA:His-OVA and Qβ + OVA became more pronounced as Qβ-NiNTA:His-OVA had a 3-fold smaller tumor volume than Qβ + OVA. In the CPMV vaccine groups, the CPMV-NiNTA:His-OVA and CPMV + OVA mice had tumor volumes of 52.4 and 56 mm3, respectively (Figure 4a). The CPMV only control had a tumor volume of 319.8 mm3, a 6.1- and 5.7-fold difference, respectively. Efficacy data are in good agreement with the antibody titers (see Figure 3): Qβ-NiNTA:His-OVA produced more α-OVA antibodies vs the Qβ + OVA admixture and is the more potent vaccine formulation. In contrast, the CPMV-NiNTA:His-OVA and CPMV + OVA groups demonstrated similar antibody production, which was reflected by the tumor rejection. It is important to note that the CPMV and Qβ experiments were run at the same time, which is why they both show identical OVA tumor volumes. They were split into two groups for ease of viewing. The tumor volume curves for all groups can be found in Figure S12a.
Figure 4.
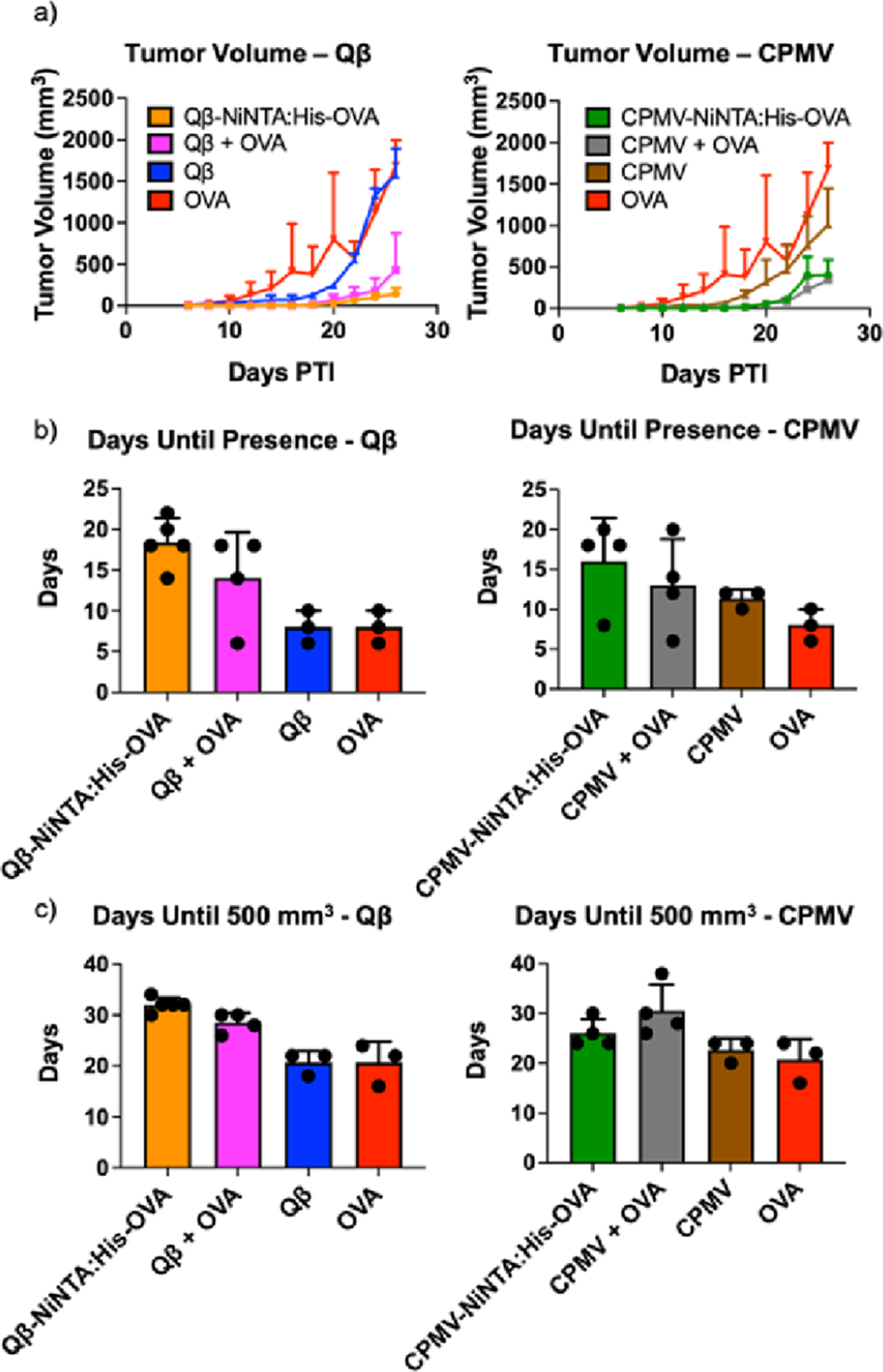
Tumor volume curves and graphs. (a) Tumor volume curves. (b) Bar graph indicating how long it took before the tumors were discernible for measurement. (c) Bar graph indicating how long it took the tumors to reach a volume of 500 mm3. The CPMV and Qβ experiments in a-c were all done at the same time, but were separated into two for ease of viewing. The full graphs can be seen in Figure S6.
Efficacy is not only apparent by reduced tumor burden, but also by delayed onset of tumor growth. Therefore, we also analyzed how many days passed until tumors were palpable and then reached a size of 500 mm3. For Qβ-NiNTA:His-OVA, it was 18.4 days until tumors were palpable (Figure 4b). This was 1.3-, 2.3-, and 2.3-fold slower than Qβ + OVA (p > 0.05), Qβ (p < 0.05), and OVA (p < 0.05), respectively. When measuring the days it took tumors to reach 500 mm3, Qβ-NiNTA:His-OVA took 1.1-, 1.5-, and 1.5-fold greater time than Qβ + OVA (p > 0.05), Qβ (p < 0.01), and OVA (p < 0.01), respectively (Figure 4c). While CPMV-NiNTA:His-OVA fared better than the negative controls, it was in line with the CPMV + OVA group (Figure 4b,c). The bar graph displaying all groups simultaneously can be found in Figure S12b,c.
The mice were also measured for survival and were sacrificed at a tumor volume endpoint of 1500 mm3 (Figure S13). Qβ-NiNTA:His-OVA and Qβ + OVA improved survival compared to Qβ and OVA with a median survival of 34 and 31 days compared to 26 and 26 days, respectively. Survival was not extended with the CPMV vaccine groups compared to the negative controls.
CONCLUSIONS
We developed and validated a modular vaccine platform making use of plant viral and VLP adjuvant nanoparticles displaying NiNTA for binding of His-tagged antigens. We demonstrate the modularity of this platform by binding OVA as well as other model antigens allowing for a plug-and-play approach for the generation of future vaccines. We utilized the OVA vaccine formulations and demonstrated efficacy in a tumor model using OVA-expressing melanoma cells (B16F10-OVA). Antibody titers and efficacy (reduction of tumor burden/delayed onset of tumor growth) were mirrored demonstrating that Qβ-NiNTA:His-OVA was the most potent formulation outperforming the Qβ + OVA admixture. In contrast, α-OVA antibodies and antitumor efficacy were comparable between the CPMV-NiNTA:His-OVA vs CPMV + OVA admixture group. Therefore, the Qβ platform appears to be most suitable for the proposed modular vaccine strategy. Potential exists to further improve efficacy with the use of trivalent NiNTA linkers, and future research should detail whether Th1/2 biases could be defined as a function of antigen, adjuvant, and display strategy.
EXPERIMENTAL METHODS
Materials.
Potassium phosphate monobasic, potassium phosphate dibasic, and Tween-20 were purchased from Fisher Scientific. Phosphate buffered saline (PBS) was purchased from G Biosciences. Dimethyl sulfoxide (DMSO), OVA, bovine serum albumin (BSA), and nickel(II) chloride hexahydrate were purchased from Sigma-Aldrich. 2-iminothiolane, 3-morpholinopropane-1-sulfonic acid (MOPS) buffer, and Tris acetate EDTA (TAE) buffer were purchased from Thermo Fisher Scientific. Dithiothreitol (DTT) was purchased from Gold Biotechnologies. The His6-maleimide peptide was purchased from Genscript.
Cells.
The B16F10-OVA cell line was a generous gift from Dr. Mary Jo Turk at Dartmouth College. The B16F10-OVA was grown and passaged in RPMI-1640 supplemented with 10% (w/v) fetal bovine serum (FBS) + 1% (v/v) penicillin/streptomycin (P/S). RPMI was purchased from Corning, FBS was purchased from R&D Systems, and P/S was purchased from Cytiva. The cells were kept in 5% CO2 and 37 °C.
Preparation of CPMV-NiNTA:His-OVA and Qβ-NiNTA:-His-OVA Vaccines.
CPMV was propagated in black-eyed pea No. 5 plants and purified as previously reported.35 Qβ VLPs were produced in Bl21 E. coli (DE3) (New England BioLabs) and purified as previously reported.19 CPMV was stored in 0.1 M potassium phosphate (KP) buffer pH 7.2 while Qβ was stored in 1× PBS pH 7.2. Both virus nanoparticles were stored at 4 °C until further use.
CPMV and Qβ were resuspended in 10 mM KP by buffer exchange using 100 kDa molecular weight cut off (MWCO) spin filters (EMD Millipore). The viral capsids were modified with NTA through the addition of 3000 mol equivalents (equiv) per virus nanoparticle of NTA-PEG2K-NHS (Nanocs) diluted in DMSO and allowed to react overnight (ON) at 4 °C; the final DMSO concentration was kept to a maximum of 10% by volume. Excess NTA-PEG2K-NHS was removed using Sephadex G-25 columns (Cytiva) according to the manufacturer’s protocols. Ni (5 mM) was added to the solution and incubated ON at 4 °C before removal of Ni through dialysis ON in 10 mM KP. The resulting samples were stored at 4 °C in 10 mM KP until further use. To ensure the presence of bound Ni, the CPMV-NiNTA sample was incubated with 330 mM of DTT; a brown color change indicates the presence of Ni within the solution.
OVA was chemically His-tagged for binding to Qβ and CPMV. The OVA was resuspended to 10 mg mL−1 in water before the addition of 10 mol equiv of 2-iminothiolane (2 mg mL−1 in deionized (DI) water) per OVA. The reaction was run for 2 h followed by removal of excess 2-iminothiolane using 10 kDa MWCO spin filters. 4 mol equiv of a His-tag with an N-terminal maleimide (Genscript, sequence: maleimide-HHHHHHHH or maleimide-His6) was conjugated to the introduced thiol groups and allowed to react ON at 4 °C. Excess His-tag was removed through dialysis using a 12–14 kDa MWCO dialysis membrane, and the OVA-His was stored at 4 °C in DI water until further use.
To create the CPMV-NiNTA:His-OVA or Qβ-NiNTA:His-OVA vaccines, 500 mol equiv of the His-OVA was added per CPMV-NiNTA and Qβ-NiNTA and allowed to react ON at 4 °C. The unbound His-OVA was removed with a 100 kDa MWCO dialysis membrane in 10 mM KP, and the resulting CPMV-NiNTA:His-OVA and Qβ-NiNTA:His-OVA were stored at 4 °C in 10 mM KP until further use. The same procedures were carried out using CA and BSA proteins.
Characterization.
Concentration.
The concentration of CPMV-NiNTA:His-OVA was analyzed using UV–vis (Nano-drop 2000). The absorbance was measured at 260 and 280 nm, and an absorbance ratio of 260 to 280 near 1.8 was used to ascertain unbroken, pure particles. The concentration was measured using Beer’s Law and the absorbance value at 260 nm with a path length of 0.1 cm and extinction coefficient of 8.1 mL mg−1 cm−1. The concentration of Qβ-NiNTA:His-OVA was analyzed using a Pierce BCA assay (Thermo Scientific) according to the manufacturer’s protocols. It is noted that the concentrations determined are estimates because the additional protein displayed will also be measured.
SDS-PAGE.
SDS-PAGE was carried out to ensure successful conjugation of the His-tag to the OVA and binding of His-OVA to CPMV/Qβ-NiNTA. 10 μg of sample was loaded with 4× lithium dodecyl sulfate Sample Buffer (Life Technologies). In samples with Qβ, a 10× sample reducing agent (Invitrogen) was also added. The samples were then heated at 95 °C for 5 min before running on a 12% NuPAGE gel (ThermoFisher Scientific) at 200 V, 120 mA, and 25 W in 1× MOPS buffer. The gel was visualized with GelCode Blue Safe Protein Stain (ThermoFisher Scientific) according to the manufacturer’s instructions. The gel was imaged on an AlphaImager (Protein Simple). The amount of bound OVA was calculated using densitometry analysis on ImageJ.
Western Blot (WB).
To further ensure successful conjugation of the His-tag to OVA, WBs were carried out against the His-tag. Following SDS-PAGE of the His-OVA, the proteins were transferred onto a nitrocellulose paper (VWR) for 1 h at 25 V, 160 mA, and 17 W. The paper was blocked with 5% (w/v) milk (RPI) for 1 h and washed 3× with 1× PBS. An α-His HRP antibody (Biolegend) at 0.5 μg mL−1 in 1× PBS was incubated for 1 h at RT and washed 3× with 1× PBS. A 3,3′-diaminobenzidine (DAB) substrate was incubated for 5 min before washing away 3× with 1× PBS. The nitrocellulose was then read under the AlphaImager System.
The CPMV-NiNTA:His-OVA sample was also assessed through WB. The protocol was unchanged from before except the samples were incubated with either an α-His HRP antibody (0.5 μg mL−1) or an α-OVA mouse antibody (1:1000 dilution, Novus Biologicals). In the samples bound with α-OVA, the nitrocellulose was washed 3× with 1× PBS followed by the addition of an α-mouse goat AF647 antibody (1:1000 dilution, Biolegend) for 1 h at RT. The unbound secondary antibody was washed away 3× with 1× PBS before imaging on the AlphaImager System.
Agarose Gel Electrophoresis.
Electrophoresis was carried out using 10 μg of CPMV-NiNTA:His-OVA and Qβ-NiNTA:His-OVA and a 1.2% (w/v) agarose gel in 1× TAE buffer. 1 μL of GelRed nucleic acid gel stain (Gold Biotechnologies) was added to the gel before running the gel at 30 min at 120 V and 400 mA. The RNA was first visualized using the AlphaImager using a red filter, and then the protein was visualized by incubating the gel in 0.25% (w/v) Coomassie Blue ON followed by imaging on the AlphaImager under white light.
ELISA.
Greiner Bio-One 96-well medium-binding microplates were coated with 100 μL of 10 μg mL−1 of an α-OVA mouse antibody (Novus Biologicals) ON at 4 °C. The plate was washed 3× with 100 μL of PBS + 0.1% (v/v) Tween-20 (PBST). The CPMV/Qβ-NiNTA:His-OVA and control samples were then added to appropriate wells at 50 μg mL−1 and incubated for 1 h at RT. The wells were washed 3× with PBST and incubated with 100 μL of an α-CPMV or α-Qβ rabbit antibody (Pacific Immunology) at 10 μg mL−1 for 1 h at RT. The wells were washed 3× with PBST followed by incubation of an α-rabbit goat HRP antibody (1:5000 dilution, Fisher Scientific) for 1 h at RT. The plate was washed 3× with PBST, and 100 μL of 1-Step Ultra TMB was added to each well. The TMB was reacted for 2 min followed by the addition of 100 μL of 2 N H2SO4. The plate was read on a microplate reader (Tecan) at 450 nm. All samples were run in triplicate. The ELISAs were carried out on samples 7 and 28 days following the generation of the vaccines.
TEM.
TEM was carried out on Formvar carbon film coated TEM supports with 400-mesh hexagonal copper grids (VWR International). The grids were first incubated with the viruses, which were diluted to 0.1 mg mL−1 in DI water, for 2 min followed by staining with 2% uranyl acetate for 2 min. The images were taken on a Joel 1400 TEM at 80 kV.
CD.
CD measurements were carried out on an Aviv model 21D CD spectrometer. OVA was diluted to 0.5 mg mL−1 while the viruses were diluted to 0.3 mg mL−1 in 0.1 M KP buffer. Measurements were taken from 180 to 320 nm at RT at stepwise increments of 1 nm. Readings were taken 2–3 times and averaged.
DLS.
The samples were diluted to 0.1 mg mL−1 in DI water before reading on a Zetasizer Nano ZSP/Zen5600 (Malvern Panalytical). The samples were run at 25 °C with a 20 s equilibration time. The OVA-bound samples were measured on days 3, 7, 14, 21, and 28 following the binding.
FPLC.
FPLC measurements were taken on an Akta pure 25 M1 (Cytiva). Samples were diluted to 1 mg mL−1 in 150 μL of 10 mM KP. The flow rate was set to 0.5 mg mL−1 and an isocratic elution profile was used. Absorbance measurements were taken at 260 and 280 nm. FPLC was run on samples 7, 14, 21, and 28 days following the generation of the vaccines.
Mice Immunization.
All animal experiments were carried out in accordance with the guidelines set out by the IACUC of the University of California, San Diego. All mice were purchased from Jackson Labs and housed at the Moores Cancer Center. The mice were granted unlimited food and water at all times.
C57BL/6J mice were immunized through 3 injections spaced two weeks apart of CPMV-NiNTA:His-OVA, Qβ-NiNTA:His-OVA, CPMV + OVA, Qβ + OVA, CPMV, Qβ, and OVA. The injections were done s.c. and standardized to the OVA concentration (5 μg), which meant that for the CPMV- and Qβ-containing groups, 41 and 25 μg of CPMV/Qβ-NiNTA:His-OVA and control samples were injected, respectively. Mice blood was collected every two weeks through retroorbital (r.o.) bleeding until 6 weeks past the first dose. The sera were isolated through centrifugation of blood at 2000 × g for 10 min at 4 °C and collection of the supernatant. Sera were stored at −80 °C until further use.
Antibody Titer Measurements and Antibody Isotyping.
Antigen-specific antibodies were quantified using ELISA. Greiner Bio-One medium-binding 96-well plates were coated with 100 μL of 10 μg mL−1 of OVA in 50 mM carbonate–bicarbonate buffer pH 9.6 ON at 4 °C. The plates were washed with PBST 3× and blocked with 1× casein blocking buffer with fish gelatin (Bioworld) in 1× PBS for 1 h at RT. Following washing, the sera of the mice were added at a starting dilution of 1:200 followed by serial dilutions of 2. The sera were incubated for 1 h at RT followed by washing. Goat α-mouse HRP IgG secondary antibodies specific to the Fc region were added to the plate and incubated for 1 h at RT. The secondaries were washed 3× with PBST, and 100 μL of 1-Step Ultra TMB was incubated for 2 min followed by the addition of 100 μL of 2 N H2SO4. The plate was then read on a microplate reader at 450 nm. The endpoint titer was considered the dilution at which the absorbance of the samples was greater than twice that of the blanks.
The isotype of the antibodies that were produced was also investigated through ELISA. In this case, the sera within each group were pooled and diluted 1:1000. When adding the secondary antibodies, isotype specific antibodies with conjugated HRP were added (IgGtotal, IgG1, IgG2b, IgG2c, IgA, IgM, and IgE). All secondaries were added at a dilution of 1:5000 except for IgE, which was diluted 1:1000. The ratio of IgG2b IgG1−1 and IgG2c IgG1−1 was calculated, and a value <1 was considered a Th2 response while >1 was considered Th1. All the secondary antibodies were purchased from Biolegend.
Tumor Inoculation.
In the same mice used above for antibody titer measurements, at week 6, 200,000 B16F10-OVA cells were injected s.c. in 200 μL of 1× PBS. The tumors were measured every 2 days, and the survival of the mice was followed. Mice were euthanized when their tumors reached >1500 mm3 with tumor volume measured using the equation: V = l × w2/2.
Statistical Analysis.
ELISA data proving binding between OVA and CPMV/Qβ was analyzed using one-way ANOVA. Endpoint titers were analyzed with two-way ANOVA. Tumor volume curves were analyzed with two-way ANOVA while the delay of tumor onset bar graphs were analyzed with one-way ANOVA. All analyses were done on GraphPad Prism.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by a grant from NIH (R21AI161306) and by NSF through the UC San Diego Materials Research Science and Engineering Center (UCSD MRSEC; DMR-2011924).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.bioconjchem.2c00601.
Reduction of Ni with dithiothreitol; SDS-PAGE and WB of His-OVA; SDS-PAGE of Qβ-NiNTA:His-CA and Qβ-NiNTA:His-BSA; TEM of CPMV, Qβ, CPMV-NiNTA:-His-OVA, and Qβ-NiNTA:His-OVA; CD of CPMV, Qβ, OVA, CPMV-NiNTA:His-OVA, and Qβ-NiNTA:His-OVA, Longitudinal ELISAs of CPMV-NiNTA:His-OVA and Qβ-NiNTA:His-OVA; Longitudinal DLS of CPMV-NiNTA:His-OVA and Qβ-NiNTA:His-OVA; Longitudinal FPLC of CPMV-NiNTA:His-OVA and Qβ-NiNTA:-His-OVA; FPLC of CPMV and unbound OVA; Week 2 titers; Isotyping of vaccines; Tumor volume curves and delayed onset of tumor development graphs for all groups; Survival curves for all groups (PDF)
The authors declare the following competing financial interest(s): Dr. Steinmetz is a co-founder of, has equity in, and has a financial interest with Mosaic ImmunoEnginering Inc. Dr. Steinmetz serves as Director, Board Member, and Acting Chief Scientific Officer, and paid consultant to Mosaic. The other authors declare no potential COI.
Contributor Information
Young Hun Chung, Department of Bioengineering and Moores Cancer Center, University of California, San Diego, La Jolla, California 92093, United States.
Britney A. Volckaert, Department of NanoEngineering, University of California, San Diego, La Jolla, California 92093, United States
Nicole F. Steinmetz, Department of Bioengineering, Moores Cancer Center, Department of NanoEngineering, Department of Radiology, and Center for Nano-ImmunoEngineering, University of California, San Diego, La Jolla, California 92093, United States; Institute for Materials Discovery and Design, University of California, San Diego, La Jolla, California 92093, United States; Center for Engineering in Cancer, Institute for Engineering in Medicine, University of California, San Diego, La Jolla, California 92093, United States
REFERENCES
- (1).Bok K; Sitar S; Graham BS; Mascola JR Accelerated COVID-19 Vaccine Development: Milestones, Lessons, and Prospects. Immunity 2021, 54, 1636–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kashte S; Gulbake A; El-Amin SF III; Gupta A COVID-19 Vaccines: Rapid Development, Implications Challenges and Future Prospects. Hum. Cell 2021, 34, 711–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Shin MD; Shukla S; Chung YH; Beiss V; Chan SK; Ortega-Rivera OA; Wirth DM; Chen A; Sack M; Pokorski JK; Steinmetz NF COVID-19 Vaccine Development and a Potential Nanomaterial Path Forward. Nat. Nanotechnol 2020, 15, 646–655. [DOI] [PubMed] [Google Scholar]
- (4).Cafri G; Gartner JJ; Zaks T; Hopson K; Levin N; Paria BC; Parkhurst MR; Yossef R; Lowery FJ; Jafferji MS; et al. MRNA Vaccine-Induced Neoantigen-Specific T Cell Immunity in Patients with Gastrointestinal Cancer. J. Clin. Invest 2020, 130, 5976–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Beck JD; Reidenbach D; Salomon N; Sahin U; Türeci O; Vormehr M; Kranz LM MRNA Therapeutics in Cancer Immunotherapy. Mol. Cancer 2021, 20, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Baden LR; El Sahly HM; Essink B; Kotloff K; Frey S; Novak R; Diemert D; Spector SA; Rouphael N; Creech CB; et al. COVE Study Group. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med 2021, 384, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Polack FP; Thomas SJ; Kitchin N; Absalon J; Gurtman A; Lockhart S; Perez JL; Pérez Marc G; Moreira ED; Zerbini C; Bailey R; Swanson KA; Roychoudhury S; Koury K; Li P; Kalina WV; Cooper D; Frenck RW Jr; Hammitt LL; Türeci Ö; Nell H; Schaefer A; Ünal S; Tresnan DB; Mather S; Dormitzer PR; Şahin U; Jansen KU; Gruber WC; C4591001 Clinical Trial Group. C4591001 Clinical Trial Group . Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med 2020, 383, 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chung YH; Beiss V; Fiering SN; Steinmetz NF COVID-19 Vaccine Frontrunners and Their Nanotechnology Design. ACS Nano 2020, 14, 12522–12537. [DOI] [PubMed] [Google Scholar]
- (9).Borah P; Deb PK; Al-Shar’i NA; Dahabiyeh LA; Venugopala KN; Singh V; Shinu P; Hussain S; Deka S; Chandrasekaran B; et al. Perspectives on RNA Vaccine Candidates for COVID-19. Front. Mol. Biosci 2021, 8, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wang Z-B; Xu J Better Adjuvants for Better Vaccines: Progress in Adjuvant Delivery Systems, Modifications, and Adjuvant–Antigen Codelivery. Vaccines 2020, 8, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kreutz M; Giquel B; Hu Q; Abuknesha R; Uematsu S; Akira S; Nestle FO; Diebold SS Antibody-Antigen-Adjuvant Conjugates Enable Co-Delivery of Antigen and Adjuvant to Dendritic Cells in Cis but Only Have Partial Targeting Specificity. PLoS One 2012, 7, No. e40208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wortham C; Grinberg L; Kaslow DC; Briles DE; McDaniel LS; Lees A; Flora M; Snapper CM; Mond JJ Enhanced Protective Antibody Responses to PspA after Intranasal or Subcutaneous Injections of PspA Genetically Fused to Granulocyte-Macrophage Colony-Stimulating Factor or Interleukin-2. Infect. Immun 1998, 66, 1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Huang Q; Yu W; Hu T Potent Antigen-Adjuvant Delivery System by Conjugation of Mycobacterium Tuberculosis Ag85B-HspX Fusion Protein with Arabinogalactan-Poly(I:C) Conjugate. Bioconjugate Chem 2016, 27, 1165–1174. [DOI] [PubMed] [Google Scholar]
- (14).Li WM; Dragowska WH; Bally MB; Schutze-Redelmeier MP Effective Induction of CD8+ T-Cell Response Using CpG Oligodeoxynucleotides and HER-2/Neu-Derived Peptide Co-Encapsulated in Liposomes. Vaccine 2003, 21, 3319–3329. [DOI] [PubMed] [Google Scholar]
- (15).Fang R; Qiao S; Liu Y; Meng Q; Chen X; Song B; Hou X; Tian W Sustained Co-Delivery of BIO and IGF-1 by a Novel Hybrid Hydrogel System to Stimulate Endogenous Cardiac Repair in Myocardial Infarcted Rat Hearts. Int. J. Nanomed 2015, 10, 4691–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nkanga CI; Ortega-Rivera OA; Shin MD; Moreno-Gonzalez MA; Steinmetz NF Injectable Slow-Release Hydrogel Formulation of a Plant Virus-Based COVID-19 Vaccine Candidate. Biomacromolecules 2022, 23, 1812–1825. [DOI] [PubMed] [Google Scholar]
- (17).Ray S; Wirth DM; Ortega-Rivera OA; Steinmetz NF; Pokorski JK Dissolving Microneedle Delivery of a Prophylactic HPV Vaccine. Biomacromolecules 2022, 23, 903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ortega-Rivera OA; Shukla S; Shin MD; Chen A; Beiss V; Moreno-Gonzalez MA; Zheng Y; Clark AE; Carlin AF; Pokorski JK; et al. Cowpea Mosaic Virus Nanoparticle Vaccine Candidates Displaying Peptide Epitopes Can Neutralize the Severe Acute Respiratory Syndrome Coronavirus. ACS Infect. Dis 2021, 7, 3096–3110. [DOI] [PubMed] [Google Scholar]
- (19).Ortega-Rivera OA; Shin MD; Chen A; Beiss V; Moreno-Gonzalez MA; Lopez-Ramirez MA; Reynoso M; Wang H; Hurst BL; Wang J; et al. Trivalent Subunit Vaccine Candidates for COVID-19 and Their Delivery Devices. J. Am. Chem. Soc 2021, 143, 14748–14765. [DOI] [PubMed] [Google Scholar]
- (20).Ortega-Rivera OA; Pokorski JK; Steinmetz NF A Single-Dose, Implant-Based, Trivalent Virus-like Particle Vaccine against “Cholesterol Checkpoint” Proteins. Adv. Ther 2021, 4, No. 2100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lizotte PH; Wen AM; Sheen MR; Fields J; Rojanasopondist P; Steinmetz NF; Fiering S In Situ Vaccination with Cowpea Mosaic Virus Nanoparticles Suppresses Metastatic Cancer. Nat. Nanotechnol 2016, 11, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Knecht S; Ricklin D; Eberle AN; Ernst B Oligohis-Tags: Mechanisms of Binding to Ni2+-NTA Surfaces. J. Mol. Recognit 2009, 22, 270–279. [DOI] [PubMed] [Google Scholar]
- (23).Spriestersbach A; Kubicek J; Schäfer F; Block H; Maertens B Purification of His-Tagged Proteins. Methods Enzymol 2015, 559, 1–15. [DOI] [PubMed] [Google Scholar]
- (24).Andersen KR; Leksa NC; Schwartz TU; Optimized E Coli Expression Strain LOBSTR Eliminates Common Contaminants from His-Tag Purification. Proteins 2013, 81, 1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Abdullah N; Chase HA Removal of Poly-Histidine Fusion Tags from Recombinant Proteins Purified by Expanded Bed Adsorption. Biotechnol. Bioeng. 2005, 92, 501–513. [DOI] [PubMed] [Google Scholar]
- (26).Mohsen MO; Augusto G; Bachmann MF The 3Ds in Virus-like Particle Based-Vaccines: “Design, Delivery and Dynamics.”. Immunol. Rev 2020, 296, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chung YH; Church D; Koellhoffer EC; Osota E; Shukla S; Rybicki EP; Pokorski JK; Steinmetz NF Integrating Plant Molecular Farming and Materials Research for Next-Generation Vaccines. Nat. Rev. Mater 2022, 7, 372–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Spohn G; Guler R; Johansen P; Keller I; Jacobs M; Beck M; Rohner F; Bauer M; Dietmeier K; Kündig TM; et al. A Virus-Like Particle-Based Vaccine Selectively Targeting Soluble TNF-α Protects from Arthritis without Inducing Reactivation of Latent Tuberculosis. J. Immunol 2007, 178, 7450–7457. [DOI] [PubMed] [Google Scholar]
- (29).Bachmann MF; Dyer MR Therapeutic Vaccination for Chronic Diseases: A New Class of Drugs in Sight. Nat. Rev. Drug Discov 2004, 3, 81–88. [DOI] [PubMed] [Google Scholar]
- (30).Cubas R; Zhang S; Kwon S; Sevick-Muraca EM; Li M; Chen C; Yao Q Virus-like Particle (VLP) Lymphatic Trafficking and Immune Response Generation after Immunization by Different Routes. J. Immunother 2009, 32, 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Singh P; Prasuhn D; Yeh RM; Destito G; Rae CS; Osborn K; Finn MG; Manchester M Bio-Distribution, Toxicity and Pathology of Cowpea Mosaic Virus Nanoparticles in Vivo. J. Controlled Release 2007, 120, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Maharjan PM; Choe S Plant-Based COVID-19 Vaccines: Current Status, Design, and Development Strategies of Candidate Vaccines. Vaccines 2021, 9, 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Spicer CD; Jumeaux C; Gupta B; Stevens MM Peptide and Protein Nanoparticle Conjugates: Versatile Platforms for Biomedical Applications. Chem. Soc. Rev 2018, 47, 3574–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li W; Joshi MD; Singhania S; Ramsey KH; Murthy AK Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wen AM; Lee KL; Yildiz I; Bruckman MA; Shukla S; Steinmetz NF Viral Nanoparticles for In Vivo Tumor Imaging. J. Visualized Exp 2012, 69, No. e4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Shukla S; Wang C; Beiss V; Cai H; Washington T; Murray AA; Gong X; Zhao Z; Masarapu H; Zlotnick A; Fiering S; Steinmetz NF The Unique Potency of Cowpea Mosaic Virus (CPMV) in Situ Cancer Vaccine. Biomater. Sci 2020, 8, 5489–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Pokorski DJK; Hovlid ML; Finn PMG Cell Targeting with Hybrid Qβ Virus-Like Particles Displaying Epidermal Growth Factor. Chembiochem 2011, 12, 2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Le DHT; Commandeur U; Steinmetz NF Presentation and Delivery of Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand via Elongated Plant Viral Nanoparticle Enhances Antitumor Efficacy. ACS Nano 2019, 13, 2501–2510. [DOI] [PubMed] [Google Scholar]
- (39).Gorzelnik KV; Cui Z; Reed CA; Jakana J; Young R; Zhang J Asymmetric Cryo-EM Structure of the Canonical Allolevivirus Qβ Reveals a Single Maturation Protein and the Genomic SsRNA in Situ. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shahrivarkevishahi A; Luzuriaga MA; Herbert FC; Tumac AC; Brohlin OR; Wijesundara YH; Adlooru AV; Benjamin C; Lee H; Parsamian P; Gadhvi J; et al. PhotothermalPhage: A Virus-Based Photothermal Therapeutic Agent. J. Am. Chem. Soc 2021, 143, 16428–16438. [DOI] [PubMed] [Google Scholar]
- (41).Hou Y; Wang Y; Tang Y; Zhou Z; Tan L; Gong T; Zhang L; Sun X Co-Delivery of Antigen and Dual Adjuvants by Aluminum Hydroxide Nanoparticles for Enhanced Immune Responses. J. Controlled Release 2020, 326, 120–130. [DOI] [PubMed] [Google Scholar]
- (42).Dong X; Liang J; Yang A; Qian Z; Kong D; Lv F A Visible Codelivery Nanovaccine of Antigen and Adjuvant with Self-Carrier for Cancer Immunotherapy. ACS Appl. Mater. Interfaces 2019, 11, 4876–4888. [DOI] [PubMed] [Google Scholar]
- (43).Lim J-W; Na W; Kim H-O; Yeom M; Kang A; Park G; Park C; Ki J; Lee S; Jung B; et al. Co-Delivery of Antigens and Immunostimulants via a Polymersome for Improvement of Antigen-Specific Immune Response. J. Mater. Chem. B 2020, 8, 5620–5626. [DOI] [PubMed] [Google Scholar]
- (44).Evtushenko EA; Ryabchevskaya EM; Nikitin NA; Atabekov JG; Karpova OV Plant Virus Particles with Various Shapes as Potential Adjuvants. Sci. Rep 2020, 10, 10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Acosta-Ramírez E; Pérez-Flores R; Majeau N; Pastelin-Palacios R; Gil-Cruz C; Ramírez-Saldaña M; Manjarrez-Orduño N; Cervantes-Barragán L; Santos-Argumedo L; Flores-Romo L; et al. Translating Innate Response into Long-Lasting Antibody Response by the Intrinsic Antigen-Adjuvant Properties of Papaya Mosaic Virus. Immunology 2008, 124, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Watson DS; Platt VM; Cao L; Venditto VJ; Szoka FC Antibody Response to Polyhistidine-Tagged Peptide and Protein Antigens Attached to Liposomes via Lipid-Linked Nitrilotriacetic Acid in Mice. Clin. Vaccine Immunol 2011, 18, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).van Broekhoven CL; Altin JG The Novel Chelator Lipid 3(Nitrilotriacetic Acid)-Ditetradecylamine (NTA(3)-DTDA) Promotes Stable Binding of His-Tagged Proteins to Liposomal Membranes: Potent Anti-Tumor Responses Induced by Simultaneously Targeting Antigen, Cytokine and Costimulatory Signals to T Cells. Biochim. Biophys. Acta 2005, 1716, 104–116. [DOI] [PubMed] [Google Scholar]
- (48).Nazeri S; Zakeri S; Mehrizi AA; Sardari S; Djadid ND Measuring of IgG2c Isotype Instead of IgG2a in Immunized C57BL/6 Mice with Plasmodium Vivax TRAP as a Subunit Vaccine Candidate in Order to Correct Interpretation of Th1 versus Th2 Immune Response. Exp. Parasitol 2020, 216, No. 107944. [DOI] [PubMed] [Google Scholar]
- (49).Schroeder HW; Cavacini L Structure and Function of Immunoglobulins. J. Allergy Clin. Immunol 2010, 125, S41–S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Tatsumi T; Kierstead LS; Ranieri E; Gesualdo L; Schena FP; Finke JH; Bukowski RM; Mueller-Berghaus J; Kirkwood JM; Kwok WW; et al. Disease-Associated Bias in T Helper Type 1 (Th1)/Th2 CD4+ T Cell Responses Against MAGE-6 in HLA-DRB10401+ Patients With Renal Cell Carcinoma or Melanoma. J. Exp. Med 2002, 196, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Maximiano S; Magalhaes P; Guerreiro MP; Morgado M Trastuzumab in the Treatment of Breast Cancer. BioDrugs 2016, 30, 75–86. [DOI] [PubMed] [Google Scholar]
- (52).Ishii K; Morii N; Yamashiro H Pertuzumab in the Treatment of HER2-Positive Breast Cancer: An Evidence-Based Review of Its Safety, Efficacy, and Place in Therapy. Core Evid 2019, Volume 14, 51–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Shukla S; Myers JT; Woods SE; Gong X; Czapar AE; Commandeur U; Huang AY; Levine AD; Steinmetz NF Plant Viral Nanoparticles-Based HER2 Vaccine: Immune Response Influenced by Differential Transport, Localization and Cellular Interactions of Particulate Carriers. Biomaterials 2017, 121, 15–27. [DOI] [PubMed] [Google Scholar]
- (54).Shukla S; Jandzinski M; Wang C; Gong X; Bonk KW; Keri RA; Steinmetz NF A Viral Nanoparticle Cancer Vaccine Delays Tumor Progression and Prolongs Survival in a HER2+ Tumor Mouse Model. Adv. Ther 2019, 2, No. 1800139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Cai H; Shukla S; Wang C; Masarapu H; Steinmetz NF Heterologous Prime-Boost Enhances the Antitumor Immune Response Elicited by Plant-Virus-Based Cancer Vaccine. J. Am. Chem. Soc 2019, 141, 6509–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


