Abstract
This study is aimed to explore the performance of texture-based machine learning and image-based deep-learning for enhancing detection of Transitional-zone prostate cancer (TZPCa) in the background of benign prostatic hyperplasia (BPH), using a one-to-one correlation between prostatectomy-based pathologically proven lesion and MRI. Seventy patients confirmed as TZPCa and twenty-nine patients confirmed as BPH without TZPCa by radical prostatectomy. For texture analysis, a radiologist drew the region of interest (ROI) for the pathologically correlated TZPCa and the surrounding BPH on T2WI. Significant features were selected using Least Absolute Shrinkage and Selection Operator (LASSO), trained by 3 types of machine learning algorithms (logistic regression [LR], support vector machine [SVM], and random forest [RF]) and validated by the leave-one-out method. For image-based machine learning, both TZPCa and BPH without TZPCa images were trained using convolutional neural network (CNN) and underwent 10-fold cross validation. Sensitivity, specificity, positive and negative predictive values were presented for each method. The diagnostic performances presented and compared using an ROC curve and AUC value. All the 3 Texture-based machine learning algorithms showed similar AUC (0.854–0.861)among them with generally high specificity (0.710–0.775). The Image-based deep learning showed high sensitivity (0.946) with good AUC (0.802) and moderate specificity (0.643). Texture -based machine learning can be expected to serve as a support tool for diagnosis of human-suspected TZ lesions with high AUC values. Image-based deep learning could serve as a screening tool for detecting suspicious TZ lesions in the context of clinically suspected TZPCa, on the basis of the high sensitivity.
Keywords: artificial intelligence, diagnostic performance, texture analysis, transitional zone prostate cancer
1. Introduction
Transitional zone prostate cancer (TZPCa) accounts for 20% to 30% of all prostate cancers.[1] However, TZPCa is not easily detected in digital rectal examinations because of its location.[2] Moreover, since the biopsy specimen in the standard prostate biopsy technique does not fully cover the transitional zone (TZ), the rate of detection of prostate cancer (PCa) in the initial biopsy is very low (2%–4%).[3] Therefore, detection and localization of TZPCa through imaging is important for the development of an appropriate treatment plan.
Magnetic resonance imaging (MRI) is widely used for detection and localization of PCa because of its high soft-tissue contrast and the availability of function-based imaging techniques, such as diffusion-weighted imaging (DWI) or dynamic contrast-enhanced (DCE) imaging. The Prostate Imaging Reporting and Data System (PI-RADS), which was introduced in 2012 and revised to version 2.1 in 2019, aims to reduce variability in MRI interpretation and improve detection, localization, and characterization of PCa lesions. Although T2-weighted imaging (T2WI) is generally accepted as the most important technique in the detection of TZPCa and is therefore designated as the primary sequence in PI-RADS for TZ lesions, accurate visualization of TZPCa in T2WI is often difficult, especially when TZPCa coexists with benign prostatic hyperplasia (BPH).[4] Since BPH shows a heterogeneous signal intensity (SI) with lower SI components, similar to PCa, it is often misdiagnosed as TZPCa on T2WI.[5] Several studies have reported the added value of DWI for detection of TZPCa; however, its final accuracy varies among the studies and observers (68%–98%).[6,7]
Artificial intelligence (AI) is expected to compensate for human errors in diagnosis, such as misdiagnosis and inter- and intraobserver variability. Therefore, a series of studies have been conducted using AI for the detection and localization of PCa on MRI. Many of these studies extracted features merely from functional MR sequences (e.g., Ktrans from DCE imaging or ADC from DWI) or from first-order (histogram) analysis.[8] Sidhu et al used first-order texture analysis for TZPCa detection, reporting an overall good diagnostic performance with an area under the receiver operating characteristic (ROC) curve (AUC) value of 0.86.[9] However, with the evolution of computing power, second-order parameters can now be used for texture analysis.[10] Niu et al applied texture analysis for detection of PCa in both the peripheral zone (PZ) and TZ using the dominant sequence of each zone.[11] However, this approach showed limitations such as a lack of precise lesion allocation (random transrectal ultrasonography guided with cognitive target biopsy) and the availability of only 6 texture parameters arbitrarily selected by the authors. Wang et al[12] evaluated the incremental value of radiomics features for improving the diagnostic performance of PI-RADS, although their comparison was not completely zone-specific (e.g., all PCa lesions vs normal PZ or normal TZ), and only 8 first-order parameters were included in their analysis. Similarly, image-based deep learning algorithms such as convolutional neural networks (CNNs) have been recently applied to the detection of PCa, although the source images did not meet the conditions set in PI-RADS, the criteria of ground truth were not definite, and the study was not zone-specific.[13]
Considering these issues, in the present study, we attempted to explore the performance of texture feature-based machine learning and image-based deep learning for enhancing the detection of TZPCa in the background of BPH by using a one-to-one correlation between prostatectomy-based pathologically proven lesions and MRI.
2. Materials and methods
This retrospective study was approved by the Institutional Review Board, which waived the requirement for informed consent.
2.1. Study population
The study population in this study includes the patients who underwent radical prostatectomy and subsequently diagnosed as pathologically proven PCa. From July 2014 to October 2018, 364 patients underwent radical prostatectomy at our institute. Three of these patients had no PCa detected in their postoperative specimens. Thus, 361 patients were included in our study population at the beginning of the study.
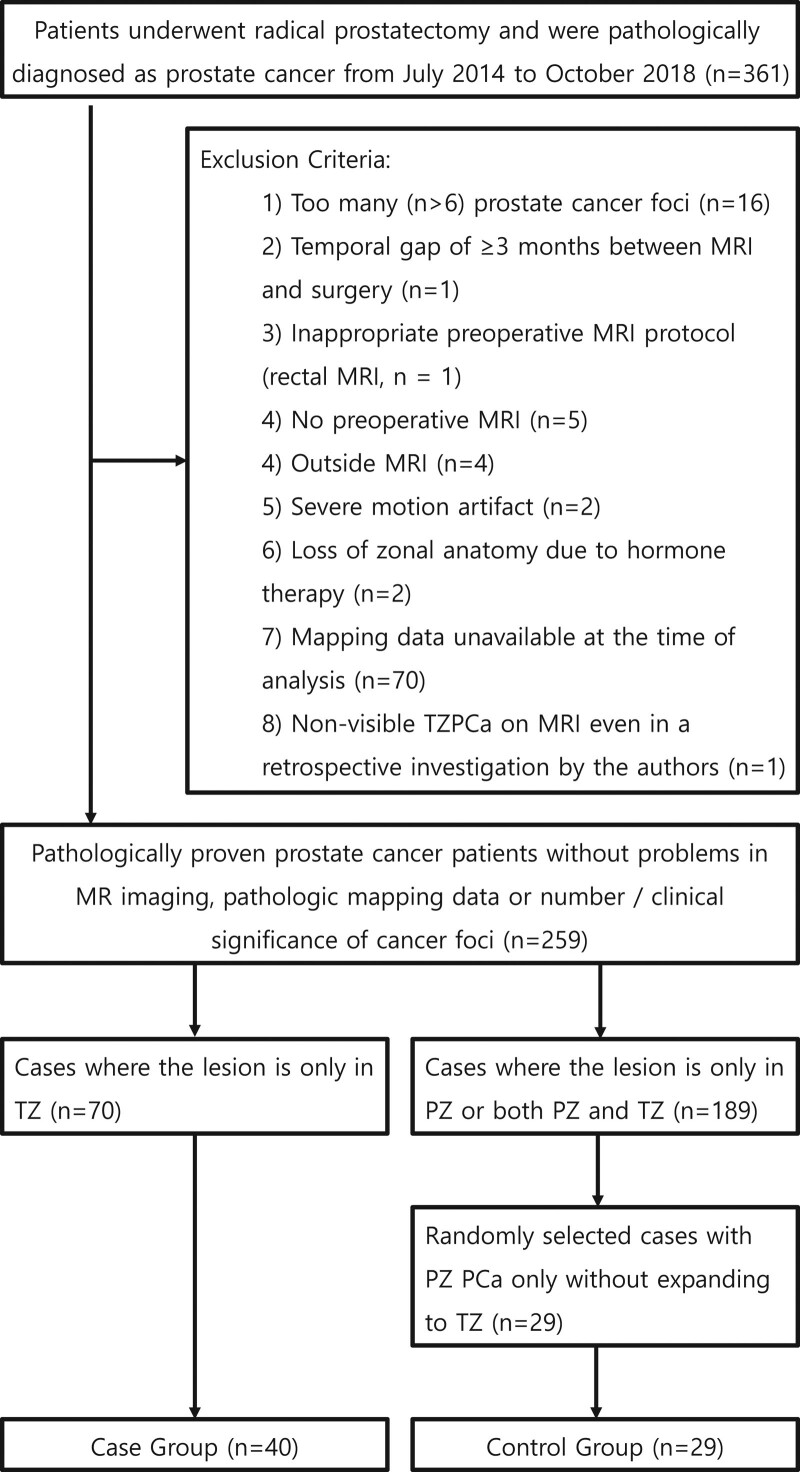
The TZPCa patient group consisted of a total of 70 patients. From aforementioned 361 of initial study population, 291 patients were excluded for the following reasons: too many (>6) PCA lesions in the prostate gland (n = 16), a temporal gap of ≥3 months between MRI and surgery (n = 1), inappropriate preoperative MRI protocol (rectal MRI, n = 1), no preoperative MRI (n = 5), MRI performed in an outside hospital (n = 4), severe motion artifact (n = 2), loss of zonal anatomy due to hormone therapy before prostatectomy (n = 2), and pathological mapping data of the prostatectomy specimen unavailable at the time of analysis (n = 70). A total of 189 patients who had PZ cancer or a cancer lesion spanning both the TZ and PZ were also excluded, since this study concerns the detection and localization of TZPCa from a non-cancerous TZ background so the authors decided to include TZPCa confined to the TZ. One patient was confirmed to have TZPCa; however, the lesion could not be visualized on MRI even in a retrospective investigation by the authors; this case was also excluded since the purpose of this study was to evaluate the diagnostic value of texture analysis and deep learning in facilitating assessments of suspicious lesions detected by radiologists.
The final study population consisted of 99 patients, since the authors randomly selected 29 patients as a control group from the excluded patients with peripheral zone prostate cancer (PZPCa) only, for extracting an image of benign hyperplasia of the TZ without a cancer focus (Fig. 1).
Figure 1.
Flow chart depicting the patient section process with exclusion criteria.
2.2. MRI protocol
MRI was performed with a 3-Tesla MR scanner (Achieva 3T, Philips Healthcare, Best, The Netherlands). Before MRI acquisition, 20 mg of butyl scopolamine (Buscopan; Boehringer-Ingelheim, Ingelheim, Germany) was injected intravenously for suppression of bowel peristalsis. Our prostate MRI protocol generally meets the recommendation of PI-RADS v2[1]: multiplanar T2-weighted image (T2WI) in 4 planes (axial, coronal, sagittal, and oblique axial); oblique axial T1-weighted image (T1WI); oblique axial, fat-saturated, single-shot echo-planar DWI with b values of 0 and 1000 s/mm2, with generation of apparent diffusion coefficient maps on a voxel-wise basis; dynamic contrast-enhanced MRI (DCE-MRI) after intravenous injection of 0.1 mmol/kg of Dotarem (Guerbet, Villepinte, France) at a rate of 2 mL/s with an automatic injector (Spectris Solaris EP; Medrad, Warrendale, PA) in the oblique axial plane using a 3-dimensional, T1-weighted, spoiled gradient-echo sequence. We used an axial oblique reference plane perpendicular to the rectal surface of the prostate, similar to the sectioning plane of prostatectomy specimens.[14] Additional DWI with an ADC map using an ultra-high b-value of 1400 s/mm2 has been included in our institutional protocol since August 2017. Although PI-RADS recommended not to have a slice gap in the axial sequences, but our institute applied a 1mm interslice gap to reduce exam time and image noise. Detailed MRI parameters of each sequence are described in Table 1.
Table 1.
MR imaging sequences and parameters.
| Sequence | TR/TE (mSec) | Matrix (mm) | FoV (mm) | NEX | ST/ gap(mm) | FA (degree) |
|---|---|---|---|---|---|---|
| True FISP Localizer | ||||||
| T2 TSE Sagittal Pelvis | 3457/100 | 316 × 310 | 222 × 222 | 2.0 | 3.0/ 0.3 | 90 |
| T2 TSE Oblique Axial | 3457/100 | 300 × 291 | 200 × 200 | 3.0 | 3.0/ 1.0 | 90 |
| T2 TSE Coronal | 3457/100 | 316 × 310 | 220 × 220 | 2.0 | 3.0/ 0.3 | 90 |
| T1 TSE Oblique Axial | 497.4/10 | 284 × 279 | 200 × 200 | 2.0 | 3.0/ 1.0 | 90 |
| Diffusion Weighted Echo Planar 2D Axial (b = 0, 1000, 1400) and ADC Map |
4495.4/62.8 | 168 × 168 | 250 × 250 | 4.0 | 3.0/ 1.0 | 90 |
| Dynamic contrast T1 FS VIBE axial (25 phases) |
7.0/4.5 | 244 × 246 | 220 × 220 | 2.0 | 4.0/ no gap | 10 |
| Subtraction Image | 1st phase was digitally subtracted from the other phases based on dynamic contrast image. | |||||
| T1 FS SPIR Oblique Axial | 713.9/10 | 284 × 275 | 200 × 200 | 2.0 | 3.0/ 1.0 | 90 |
| T2 TSE True Axial | 3320/90 | 312 × 300 | 250 × 250 | 2.0 | 5.0/ 1.0 | 90 |
FA = flip angle, FoV = field of view, FS = fat suppression, NEX = number of excitation, SPIR = spectral presaturation with inversion recovery, ST = slice thickness, TE = echo time, TR = relaxation time, True FISP = true fast imaging with steady state free precession, TSE = turbo spin echo, VIBE = volume interpolated breath hold examination.
2.3. Creation of a pathologic mapping sheet
For pathologic evaluation, the prostate specimen was coated with India ink and fixed in 4% buffered formalin. After the distal 5-mm portion of the apex was amputated and coned, the prostate was sliced from the base to the apex along the longitudinal axis at 4-mm intervals, followed by paraffin embedding. Subsequently, microslices were placed on glass slides and stained with hematoxylin-eosin. All slices, including cancer foci, were transferred to a pathologic mapping sheet, and a genitourinary pathologist (J.H.P with 11 years of experience in pathology) recorded the Gleason score (GS) and presence of extraprostatic extension for every detectable PCa lesion on the mapping sheet. Cases with 6 or more PCa lesions in 1 prostate specimen were excluded from the pathologic analysis, as mentioned above.
2.4. Pathology-matched image segmentation
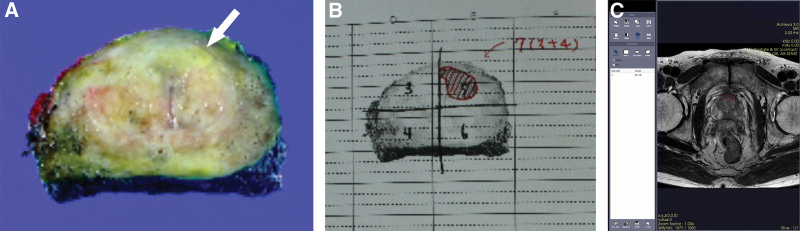
Two genitourinary subspecialized radiologists (M.S.L with 13 years and M.H.M with 25 years of experience in radiology) and the genitourinary pathologist mentioned above performed one-to-one correlation of each TZPCa on the pathologic map with TZ lesions in MR oblique axial T2WI, since the dominant sequence for the TZ in PI-RADS is T2WI.[15] One of the radiologists (M.S.L) manually drew the region of interest (ROI) for the pathologically correlated TZPCa lesion and the surrounding BPH on oblique axial T2WI (Fig. 2). One hundred 7 ROIs of TZPCa were extracted from the 70 TZPCa patients.
Figure 2.
Example ROI drawing in a 57-year-old patient with transitional zone prostate cancer. A yellowish focal lesion suggestive of cancer is present in the left transitional zone (A). After prostatectomy, a genitourinary pathologist draws the boundary of the cancer in the pathologic map (B). Two genitourinary radiologists and the pathologist arrive at a consensus for the cancerous part on the MR image on the basis of the pathologic map, and one of the radiologists subsequently draws the ROI on the T2-weighted oblique axial image using an in-house program (C). ROI = region of interest.
Due to its limited throughput in daily practice, prostate adenoma (also known as a benign prostatic hyperplasia “nodule”) could not be delineated pathologically, and thus, a one-to-one correlation could not be performed. Thus, the authors decided to draw the ROI upon lesions that showed circumscribed hypointense or heterogeneous encapsulated features on T2WI, as mentioned in the MRI features of BPH nodules in PI-RADS version 2[1] (Fig. 3). Three or less ROIs of BPH nodules were extracted from each patient with TZPCa. In addition, in order to minimize the effects of tiny nests of cancer cells inside the BPH nodule that remained undetected despite pathological confirmation and image-pathology correlation, the authors selected an additional 29 patients without TZPCa and extracted the ROIs of normal transitional zones and BPH nodules. Finally, 177 ROIs of usual BPH were extracted from patients with and without TZPCa.
Figure 3.
Representative ROI drawings of benign prostate hyperplasia according to the MRI features of benign prostate hyperplasia nodules in PI-RADS version 2; a circumscribed hypointense nodule (A) or heterogeneous encapsulated nodule (B). PI-RADS = Prostate Imaging Reporting and Data System, ROI = region of interest.
2.5. MR texture analysis
Texture feature-based machine learning was performed using the previously mentioned 107 ROIs of TZPCa and 177 ROIs of usual BPH drawn on MR T2W images. After normalizing the MR images using min-max scales, a total of 75 features were extracted using statistics-based first-order (histogram) and second-order (gray level co-occurrence matrix [GLCM] and gray level run length matrix) texture parameters with an in-house program. First-order features were extracted from both original images and 8-bit (256 gray color) converted images on the basis of the window width and length described in DICOM data of the original images (marked as _uc). The initially extracted features are listed in Table 2.
Table 2.
The 75 initially extracted texture features.
| Histogram | Volume | Min | Max | Mean | Stdev |
| Kurtosis | Energy | Entropy | Min_uc | Max_uc | |
| Variance_uc | Skewness_uc | Kurtosis_uc | Energy_uc | Entropy_uc | |
| GLCM | GLCM_Contrast_0 | GLCM_Dissimilarity_0 | GLCM_Homogeneity_0 | GLCM_ASM_0 | GLCM_Energy_0 |
| GLCM_Probability_max_0 | GLCM_Entropy_0 | GLCM_Correlation_0 | GLCM_Contrast_45 | GLCM_Dissimilarity_45 | |
| GLCM_Homogeneity_45 | GLCM_ASM_45 | GLCM_Energy_45 | GLCM_Probability_max_45 | GLCM_Entropy_45 | |
| GLCM_Correlation_45 | GLCM_Contrast_90 | GLCM_Dissimilarity_90 | GLCM_Homogeneity_90 | GLCM_ASM_90 | |
| GLCM_Energy_90 | GLCM_Probability_max_90 | GLCM_Entropy_90 | GLCM_Correlation_90 | GLCM_Contrast_135 | |
| GLCM_Dissimilarity_135 | GLCM_Homogeneity_135 | GLCM_ASM_135 | GLCM_Energy_135 | GLCM_Probability_max_135 | |
| GLCM_Entropy_135 | GLCM_Correlation_135 | ||||
| GLRLM | GLRLM_LRE_0 | GLRLM_GLN_0 | GLRLM_RLN_0 | GLRLM_RP_0 | GLRLM_LGRE_0 |
| GLRLM_HGRE_0 | GLRLM_LRE_45 | GLRLM_GLN_45 | GLRLM_RLN_45 | GLRLM_RP_45 | |
| GLRLM_LGRE_45 | GLRLM_HGRE_45 | GLRLM_LRE_90 | GLRLM_GLN_90 | GLRLM_RLN_90 | |
| GLRLM_RP_90 | GLRLM_LGRE_90 | GLRLM_HGRE_90 | GLRLM_LRE_135 | GLRLM_GLN_135 | |
| GLRLM_RLN_135 | GLRLM_RP_135 | GLRLM_LGRE_135 | GLRLM_HGRE_135 | GLRLM_RLN_135 |
ASM = angular second moment, GLCM = gray level co-occurrence matrix, GLN = gray level nonuniformity, GLRLM = gray level run length matrix, HGRE = high gray level run emphasis, LGRE = low gray level run emphasis, LRE = long runs emphasis, RLN = run length nonuniformity, RP = run percentage, uc = unsigned char.
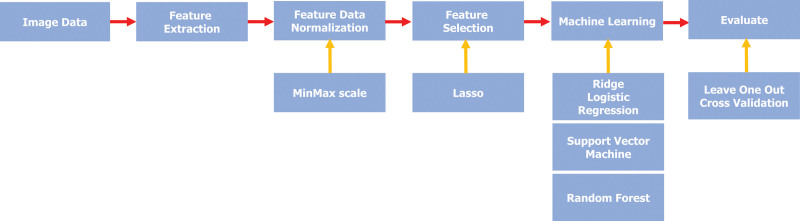
Significant features were selected using Least Absolute Shrinkage and Selection Operator (LASSO), and subsequently trained by 3 types of machine learning algorithms: logistic regression (LR), support vector machine (SVM), and random forest (RF) (Fig. 4). The final model was validated by the leave-one-out method.
Figure 4.
Flowchart of texture feature-based machine learning.
2.6. Image-based deep learning
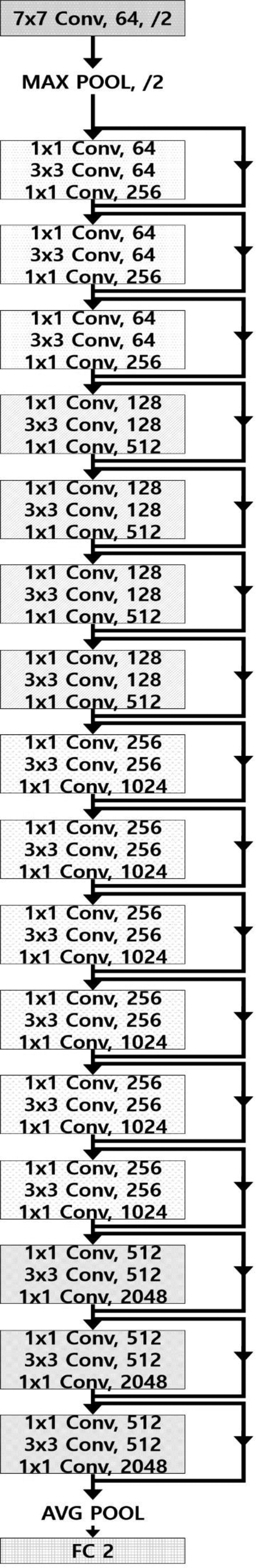
A total of 93 cancer-containing T2W image slices were selected from the aforementioned 70 TZPCa patients, and 42 slices of usual BPH were selected from the aforementioned 29 patients without TZPCa. Image-based deep learning was performed using these 135 images. A total of 123 images (86 and 37 images for cancer and BPH, respectively) were assigned to the training set, and the remaining 12 images (7 and 5 images for cancer and BPH, respectively) were assigned to the test set. In the training set, the number of images was augmented to 5477 (2748 and 2729 images for cancer and BPH, respectively) based on rotation, zoom, and translation to compensate for the small number of images. Then, the images were resampled to 224 × 224 pixels, and 12-bit pixel values were discretized into 8-bit pixel values. Deep learning was performed using an application programming interface based on Tensorflow in an Ubuntu system. The learning model was based on the ResNet-50 architecture and was trained for the classification of cancer and BPH with 35 batch sizes and 70 epochs (Fig. 5). The trained deep-learning model was validated by 10-fold cross validation, which maintained the best compromise between computational cost and reliable estimates.[16]
Figure 5.
Diagram of ResNet-50 architecture for image-based deep learning.
2.7. Statistical analysis
Sensitivity, specificity, and positive and negative predictive values were calculated for each machine learning algorithm for texture analysis and image-based deep learning. The diagnostic performance of each texture feature-based machine learning algorithm and image-based deep learning was presented using an ROC curve and the AUC value. Comparisons between the AUC of each machine learning algorithm for texture analysis and the AUC of deep learning were performed using a previously reported method.[17]
3. Results
3.1. Patient demographics
The mean age of the TZPCa patients was 71.0 years. The median PSA and PSA densities were 10.0 ng/dL and 0.30 ng/dL/mL. Among the aforementioned 107 TZPCa lesions, 17 lesions showed a GS of 6; 41 showed a GS of 7 (3 + 4); 25 showed a GS of 7 (4 + 3); and 24 showed GS of 8 or higher. The detailed data are provided in Table 3.
Table 3.
Demographic data of included transitional zone prostate cancer patients.
| Variables | Median (IQR) or n (%) |
|---|---|
| Patient based analysis (n = 70) | |
| Age, yr | 71.0 (64.0–74.5) |
| PSA, ng/dL | 10.0 (6.9–22.3) |
| PSAD, ng/dL/cm3 | 0.30 (0.19–0.73) |
| Prostate volume (cm3) | 32.8 (25.4–42.9) |
| TZ volume (cm3) | 16.7 (11.2–20.4) |
| Lesion based analysis (n = 107) | |
| Gleason score (GS) | |
| 6 | 17 (15.9%) |
| 7(3 + 4) | 41 (38.3%) |
| 7(4 + 3) | 25 (23.4%) |
| 8 or more | 24 (22.4%) |
IQR = interquartile range, PSA = prostate-specific antigen, PSAD = PSA density, TZ = transitional zone.
3.2. Diagnostic performances of texture feature-based machine learning
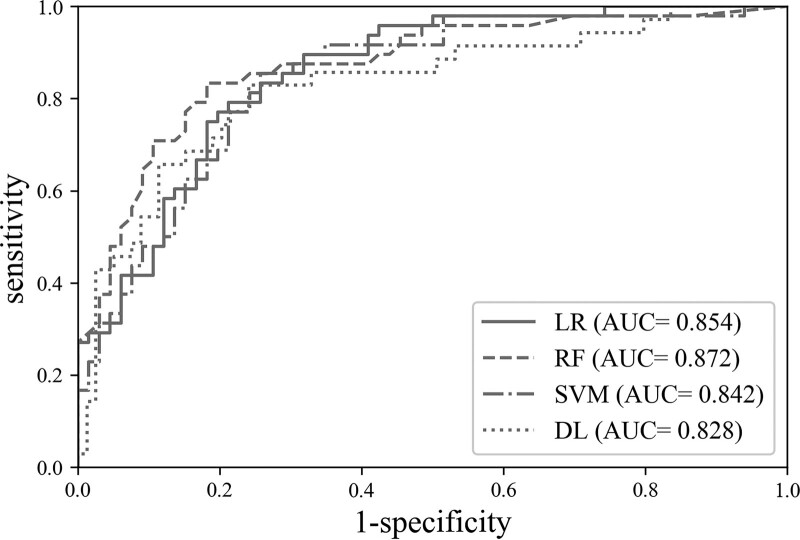
Four first-order texture features (“Entropy,” “Mean,” “Min,” “Stdev”) and 2 second-order texture features (“GLCM_Entropy_0” and “GLCM_Homogeneity_135”) were finally selected through LASSO. With these features, all 3 machine learning algorithms (LR, SVM, and RF) showed similar accuracy (0.782–0.789) and diagnostic performance (AUC = 0.854–0.861) for differentiating the selected ROI as a cancerous lesion or not. As a diagnostic decision support tool for human-detected suspicious cancerous lesions, the positive likelihood ratios (LR+) of the methods ranged from 3.813 to 4.335 (Table 4) (Fig. 6).
Table 4.
Performance of cross-validation by each machine learning algorithm.
| TP | FN | FP | TN | Sensitivity | Specificity | Accuracy | AUC | PPV | NPV | LR+ | LR− | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LR | 76 | 31 | 29 | 148 | 0.710 | 0.836 | 0.789 | 0.854 | 0.724 | 0.827 | 4.335 | 0.346 |
| SVM | 79 | 28 | 34 | 143 | 0.738 | 0.808 | 0.782 | 0.842 | 0.699 | 0.836 | 3.843 | 0.324 |
| RF | 83 | 24 | 36 | 141 | 0.775 | 0.797 | 0.789 | 0.872 | 0.697 | 0.854 | 3.814 | 0.282 |
AUC = area under receiver-operating curve, FN = false negative cases, FP = false positive cases, LR = logistic regression, LR− = negative likelihood ratio, LR+ = positive likelihood ratio, NPV = negative predictive value, PPV = positive predictive value, RF = random forest, SVM = support vector machine, TN = true negative cases, TP = true positive cases.
Figure 6.
ROC curves with AUC values of each machine learning algorithm for texture analysis and the image-based deep learning algorithm. AUC = area under the ROC curve, DL = image-based deep learning, LR = logistic regression, RF = random forest, ROC = receiver operating characteristic, SVM = support vector machine.
3.3. Diagnostic performance of image-based deep learning
The image-based deep learning model showed accuracy and AUC values of 0.852 and 0.802, respectively for detecting image containing TZPCa (Table 5) (Fig. 6).
Table 5.
Performance of image-based machine learning after 10-fold cross validation.
| TP | FN | FP | TN | Sensitivity | Specificity | Accuracy | AUC | PPV | NPV | LR+ | LR− |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 88 | 5 | 15 | 27 | 0.946 | 0.643 | 0.852 | 0.828 | 0.854 | 0.843 | 2.649 | 0.084 |
AUC = area under receiver-operating curve, FN = false negative cases, FP = false positive cases, LR− = negative likelihood ratio, LR+ = positive likelihood ratio, NPV = negative predictive value, PPV = positive predictive value, TN = true negative cases, TP = true positive cases.
4. Discussion
In this study, we present the diagnostic performance of texture feature-based machine learning with 3 different algorithms as a classifier of TZPCa or BPH for lesions suspected by humans. We also evaluated the performance of image-based deep learning powered by CNN for detecting TZPCa in the background of BPH as well as for classifying non-cancerous hypertrophied TZ on a non-cancerous image. The 2 approaches mentioned above differ in that the former is based on a human-drawn lesion ROI, while the latter uses images with and without lesions as sources of analysis. Therefore, although the same image of the same patient is used, the 2 algorithms have slightly different concern: the texture-based machine learning determines whether the lesion where the observer placed the ROI is cancer or BPH, whereas deep learning determines whether the given image contains cancer. Moreover, the 2 approaches use different number of data for analysis, and thus there is a difference in the prevalence between the 2 approaches. Thus, one-to-one comparison of the positive and negative predictive values of the 2 analyses is difficult. For radiologists, the former approach simulates a situation in which the radiologist diagnoses a lesion as a TZPCa- or BPH-related change when the lesion is found in the TZ, while the latter simulates a situation in which radiologists look for TZ lesions suspected to be TZPCa that show heterogenicity due to BPH-related changes.
In studies before the advent of PI-RADS, MR T2WI showed sensitivity and specificity of 64% to 80% and 44% to 87% and AUC values of 0.61 to 0.75 from the perspective of per-patient-base identification of TZPCa.[4,6] Using PI-RADS version 2 with T2WI as the primary sequence, the sensitivity and specificity were 68.5% to 68.8% and 77.8.90.2%, with an AUC value of 0.786 to 0.788[18–20] for detecting TZPCa. The PI-RADS approach is a system in which the detection and differentiation of lesions are performed at the same time, and it is thought that a simple comparison with the 2 algorithms of our study will be meaningful to some extent. Our results for the 3 texture-based and image-based algorithms showed comparable specificity and slightly better AUCs and sensitivity in comparison with the T2WI-based findings. Thus, we can conclude that AI-based algorithms that use texture feature-based machine learning and image-based deep learning using the CNN architecture for detecting TZPCa could improve diagnostic performance and sensitivity, which is important for screening serious diseases such as cancer.
Previous studies using texture-based machine learning for detecting TZPCa also showed similar results as our studies: sensitivity of 83.6%, specificity of 81.3%, and AUC value of 0.83 to 0.87 for LR[9,11]; sensitivity of 88.8%, specificity of 86.4%, and an AUC value of 0.955 for SVM[12]; and sensitivity of 92%, specificity of 53%, and AUC value of 0.88 on a per-lesion basis for RF as a learning algorithm.[21] Our results showed slightly lower sensitivity and specificity, possibly due to differences in the types and selection methods of the hyperparameters from initially extracted texture features. However, the accuracies and AUC values in our results were comparable to those of previous studies.[9,11,12,21] On the basis of these findings, texture-based machine learning can be considered to show excellent diagnostic performance (AUC value > 0.8) without any significant difference between the learning algorithms, so this approach should provide incremental value for MRI-based classification of TZPCa and the BPH background.
Both texture feature-based machine learning (LR, SVM, and RF) and image-based deep learning showed excellent AUC values. Characteristically, the texture-based machine learning algorithm seems to focus on improving specificity relatively, while the image-based deep learning is thought to aim to achieve relatively high sensitivity. The major application of deep learning represented by CNN in radiology is focused on the detection and classification of cancer or suspected cancer lesions in the breast, lung, or prostate.[22] Therefore, most architectures based on CNN are presumed to be designed to increase diagnostic sensitivity, and the high sensitivity and comparable AUC value of image-based deep learning in our result could also be attributed to the CNN-based architecture. In the same vein, texture analysis has evolved to characterize tumors primarily in the oncology domain, taking over some of the role of biopsy.[23] The relatively high specificity of texture-based machine learning is thought to be due to this circumstance.
Development of high-quality machine learning algorithms requires tens of thousands of image data (rubbish in, rubbish out).[24] However, for uncommon diseases such as cancer and infrequently obtained images such as MR images, acquisition of such a large amount of image data may be difficult. Therefore, with a proper augmentation technique, it is essential to ensure that the image data are of good quality.[25,26] The prostate MR data at our institute matched the pathological findings 1:1; thus, our imaging database could be considered to contain pathologically verified information, which can improve the reliability of the aforementioned results.
In summary, texture feature-based machine learning would be suitable for application in diagnostic support tools that add confidence to assessments showing whether a human-found transitional zone lesion is a cancer or not. Image-based deep learning with a CNN architecture may be appropriate for screening of TZPCa suspected on the basis of clinical examinations or other modalities (e.g., ultrasonography) before radiologists’ interpretation, reducing the risk of missed lesions by radiologists and improving their diagnostic performance. To improve this approach, more studies on radiomics and deep learning based on larger patient populations and better-quality images are needed.
Our study has several limitations. First, in comparison with plain radiography or CT, fewer patients undergo MRI, making it very difficult to collect sufficient high-quality MR data for use in machine learning. In particular, in this study, it was difficult to avoid the limitation of the small sample size because we used data in which the imaging and pathologic findings were paired 1:1 based on reviews by the radiologist and the pathologist. This approach can lead to overfitting.[27] We also conducted temporal validation using 23 TZPCa patients and 10 PZPCa patients without TZPCa (see Supplemental digital content including Supplemental text S1, http://links.lww.com/MD/J738 Supplemental Table S1–S3, http://links.lww.com/MD/J739, http://links.lww.com/MD/J742, http://links.lww.com/MD/J744 and Supplemental Figure S1, http://links.lww.com/MD/J746 which briefly describing the process and result of temporal validation). For LR and image-based deep learning, the temporal validation showed markedly decreased sensitivity with highly increased specificity in comparison with the internal validation result, suggestive of overfitting. However, the temporal validation results for SVM and RF showed a simultaneous decrease in sensitivity and specificity with a relatively even distribution of true/false-positive and negative results, indicating effective machine learning without excessive overfitting. Our results suggest that machine learning using SVM or RF may be more effective in the classification of cancer when the sample size is small, such as in the case of prostate MR.
Second, this was a single-institution, retrospective study. Further multicenter, prospective studies are needed to validate our results. However, the findings may show reproducibility problems due to the lack of external validation and the manual drawing of ROIs by a consensus of 2 radiologists in the same institute. As mentioned previously, we observed a difference in prevalence between texture feature-based machine learning and image-based deep learning due to differences in the preparation of the analysis material. Thus, direct comparison of positive and negative predictive values was not possible.
Because many centers used true axial planes rather than oblique axial plane (perpendicular to prostate long axis), and not provided the detailed pathologic map including location and Gleason score/grade of each PCa focus, we cannot collect sufficient outside data for external validation. The authors used temporal validation as an alternative, but it can be a limitation of the study in that it can be a cause of the overfitting problem as data from the same institution. The authors manually outlined a single-slice TZ ROI; thus, volume data were not obtained. In addition, data regarding the shape and contour of lesions could not be included in the analysis because of the aforementioned reasons. In the image-based deep learning process, data on the shape or contour of the lesion may have been considered, which may have affected the overall diagnostic performance.
It might be not appropriate to directly compare the diagnostic performance of manual ROI-based texture analysis and image-based deep learning because the former is regarding the discrimination of TZPCa and BPH, whereas the latter is about the given image contains TZPCa in its background transitional zone or not. In general, the prostatic transitional zone of the elderly is hyperplastic and often contains a lot of BPH nodules, making it difficult to detect TZPCa, and even if the lesion is specified, it is difficult to distinguish whether it is cancer or not due to the imaging similarity between TZPCa and BPH nodule. The usability assessment, such as comparing performance enhancement of users (i.e., Radiologists) with and without each learning methods may provide important clues as to whether radiologists feel it more difficult to find lesions in the BPH background prostatic transitional zone, or to determine whether or not they are cancerous. Performing the usability assessment as a follow-up study will be the best way to complement the methodological pitfall and enforce the strength of our study. In conclusion, both texture feature-based machine learning using LR, SVM, or RF as learning algorithms and image-based machine learning powered by the CNN architecture provide excellent diagnostic performance (AUC value > 0.8) with comparable sensitivity, specificity, and accuracy for determining TZPCa in a BPH background on T2WI, which shows heterogeneous signals that make it difficult to distinguish between areas with and without cancer in the TZ. Texture feature-based machine learning can be expected to serve as a support tool for diagnosis of human-suspected TZ lesions with high AUC values, while image-based deep learning could serve as a screening tool for detecting suspicious TZ lesions in the context of clinically suspected TZPCa, on the basis of the high sensitivity it showed in our study. We are currently developing an AI diagnostic support tool that shows whether an ROI drawn by a radiologist on an indiscernible lesion is closer to a TZPCa or BPH. This study aims to develop and evaluate algorithms to meet this aim.
Author contributions
Conceptualization: Myoung Seok Lee, Min Hoan Moon, Kwang Gi Kim.
Data curation: Myoung Seok Lee, Young Jae Kim, Jeong Hwan Park, Chang Kyu Sung, Hyeon Jeong, Hwancheol Son.
Formal analysis: Myoung Seok Lee, Young Jae Kim.
Funding acquisition: Myoung Seok Lee.
Investigation: Myoung Seok Lee, Young Jae Kim, Min Hoan Moon, Kwang Gi Kim.
Methodology: Myoung Seok Lee, Young Jae Kim, Min Hoan Moon, Kwang Gi Kim.
Resources: Young Jae Kim, Kwang Gi Kim.
Software: Young Jae Kim, Kwang Gi Kim.
Supervision: Min Hoan Moon, Kwang Gi Kim.
Validation: Myoung Seok Lee, Young Jae Kim, Min Hoan Moon, Kwang Gi Kim.
Visualization: Myoung Seok Lee, Young Jae Kim, Min Hoan Moon, Jeong Hwan Park, Chang Kyu Sung, Hyeon Jeong, Hwancheol Son.
Writing – original draft: Myoung Seok Lee, Min Hoan Moon.
Writing – review & editing: Myoung Seok Lee, Min Hoan Moon, Kwang Gi Kim.
Supplementary Material
Abbreviations:
- AI
- artificial intelligence
- BPH
- benign prostatic hyperplasia
- CNN
- convolutional neural network
- DWI
- diffusion-weighted imaging
- GLCM
- gray level co-occurrence matrix
- LASSO
- Least Absolute Shrinkage and Selection Operator
- LR
- logistic regression
- PCa
- prostate cancer
- PZ
- peripheral zone
- SVM
- support vector machine
- T2WI
- T2-weighted imaging
- TZ
- transitional zone
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant number: 2016R1D1A1B03936175).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental Digital Content is available for this article.
This retrospective, single-institution study was approved by our Institutional Review Board and Ethics Committee. Informed consent was waved because of the retrospective nature of the study.
How to cite this article: Lee MS, Kim YJ, Moon MH, Kim KG, Park JH, Sung CK, Jeong H, Son H. Transitional zone prostate cancer: Performance of texture-based machine learning and image-based deep learning. Medicine 2023;102:39(e35039).
Contributor Information
Myoung Seok Lee, Email: achieva1004@gmail.com.
Young Jae Kim, Email: kimkg@gachon.ac.kr.
Kwang Gi Kim, Email: kimkg@gachon.ac.kr.
Jeong Hwan Park, Email: hopemd@hanmail.net.
Chang Kyu Sung, Email: sckmd@snu.ac.kr.
Hyeon Jeong, Email: drjeongh@gmail.com.
Hwancheol Son, Email: volley@snu.ac.kr.
References
- [1].Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS prostate imaging - reporting and data system: 2015, Version 2. Eur Urol. 2016;69:16–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Augustin H, Erbersdobler A, Hammerer PG, et al. Prostate cancers in the transition zone: Part 2; clinical aspects. BJU Int 2004;94:1226–9. [DOI] [PubMed] [Google Scholar]
- [3].Erbersdobler A, Hammerer P, Huland H, et al. Numerical chromosomal aberrations in transition-zone carcinomas of the prostate. J Urol. 1997;158:1594–8. [PubMed] [Google Scholar]
- [4].Akin O, Sala E, Moskowitz CS, et al. Transition zone prostate cancers: features, detection, localization, and staging at endorectal MR imaging. Radiology. 2006;239:784–92. [DOI] [PubMed] [Google Scholar]
- [5].Hoeks MA, Hambrock T, Yakar D, et al. Transition zone prostate cancer: detection and localization with 3-T multiparametric MR imaging. Radiology. 2013;266:207–17. [DOI] [PubMed] [Google Scholar]
- [6].Jung S, Donati OF, Vargas HA, et al. Transition zone prostate cancer: incremental value of diffusion-weighted endorectal MR imaging in tumor detection and assessment of aggressiveness. Radiology. 2013;269:493–503. [DOI] [PubMed] [Google Scholar]
- [7].Rosenkrantz AB, Kim S, Campbell N, et al. Transition zone prostate cancer: revisiting the role of multiparametric MRI at 3 T. Am J Roentgenol 2015;204:W266–72. [DOI] [PubMed] [Google Scholar]
- [8].Wang S, Burtt K, Turkbey B, et al. Computer aided-diagnosis of prostate cancer on multiparametric MRI: a technical review of current research. Biomed Res Int. 2014;2014:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sidhu HS, Benigno S, Ganeshan B, et al. “Textural analysis of multiparametric MRI detects transition zone prostate cancer.”. Eur Radiol. 2017;27:2348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Varghese BA, Cen SY, Hwang DH, et al. Texture analysis of imaging: what radiologists need to know. Am J Roentgenol. 2019;212:520–8. [DOI] [PubMed] [Google Scholar]
- [11].Niu X, Chen Z, Chen L, et al. Clinical application of biparametric MRI texture analysis for detection and evaluation of high-grade prostate cancer in zone-specific regions. Am J Roentgenol. 2018;210:549–56. [DOI] [PubMed] [Google Scholar]
- [12].Wang J, Wu CJ, Bao ML, et al. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur Radiol. 2017;27:4082–90. [DOI] [PubMed] [Google Scholar]
- [13].Ishioka J, Matsuoka Y, Uehara S, et al. Computer-aided diagnosis of prostate cancer on magnetic resonance imaging using a convolutional neural network algorithm. BJU Int. 2018;122:411–7. [DOI] [PubMed] [Google Scholar]
- [14].Villers A, Puech P, Mouton D, et al. Dynamic contrast enhanced, pelvic phased array magnetic resonance imaging of localized prostate cancer for predicting tumor volume: correlation with radical prostatectomy findings. J Urol. 2006;176:2432–7. [DOI] [PubMed] [Google Scholar]
- [15].Lee MS, Moon MH, Kim YA, et al. Is prostate imaging reporting and data system version 2 sufficiently discovering clinically significant prostate cancer? Per-Lesion Radiology-Pathology Correlation Study. Am J Roentgenol. 2018;211:114–20. [DOI] [PubMed] [Google Scholar]
- [16].Larroza A, Moratal D, Paredes-Sánchez A, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging. 2015;42:1362–8. [DOI] [PubMed] [Google Scholar]
- [17].Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. [DOI] [PubMed] [Google Scholar]
- [18].Thai JN, Narayanan HA, George AK, et al. Validation of PI-RADS version 2 in transition zone lesions for the detection of prostate cancer. Radiology. 2018;288:485–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang X, Bao J, Ping X, et al. The diagnostic value of PI-RADS V1 and V2 using multiparametric MRI in transition zone prostate clinical cancer. Oncol Lett. 2018;16:3201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tamada T, Kido A, Takeuchi M, et al. Comparison of PI-RADS version 2 and PI-RADS version 2.1 for the detection of transition zone prostate cancer. Eur J Radiol. 2019;121:108704. [DOI] [PubMed] [Google Scholar]
- [21].Bonekamp D, Kohl S, Wiesenfarth M, et al. Radiomic machine learning for characterization of prostate lesions with MRI: comparison to ADC values. Radiology. 2018;289:128–37. [DOI] [PubMed] [Google Scholar]
- [22].Gao J, Jiang Q, Zhou B, et al. Convolutional neural networks for computer-aided detection or diagnosis in medical image analysis: an overview. Math Biosci Eng. 2019;6536:6561. [DOI] [PubMed] [Google Scholar]
- [23].Corrias G, Micheletti G, Barberini L, et al. Texture analysis imaging “what a clinical radiologist needs to know.”. Eur J Radiol. 2022;146:110055. [DOI] [PubMed] [Google Scholar]
- [24].Machine learning collaborations accelerate materials discovery – Physics World [https://physicsworld.com/a/machine-learning-collaborations-accelerate-materials-discovery/]
- [25].Do S, Song KD, Chung JW. Basics of deep learning: a radiologist’s guide to understanding published radiology articles on deep learning. Korean J Radiol. 2020;21:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mazurowski MA, Buda M, Saha A, et al. Deep learning in radiology: an overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging. 2019;49:939–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee D, Lee J, Ko J, et al. Deep learning in MR image processing. Investig Magn Reson Imaging 2019;23:81–99. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.