Abstract
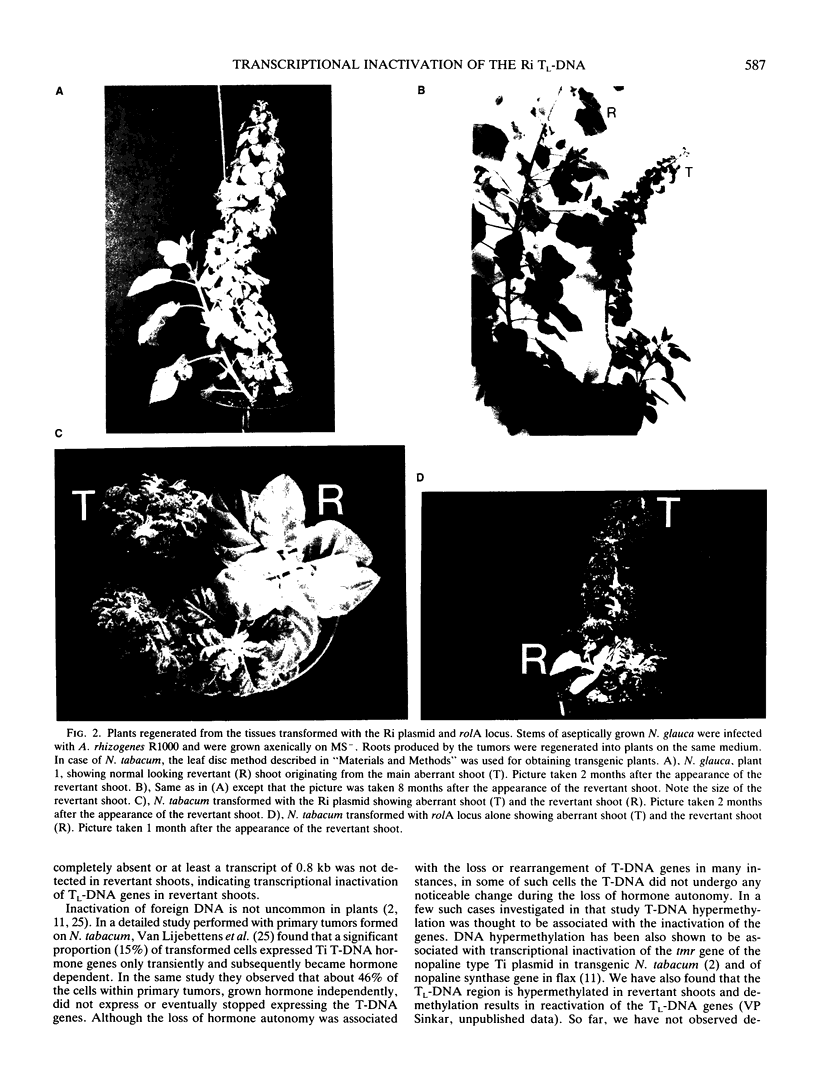
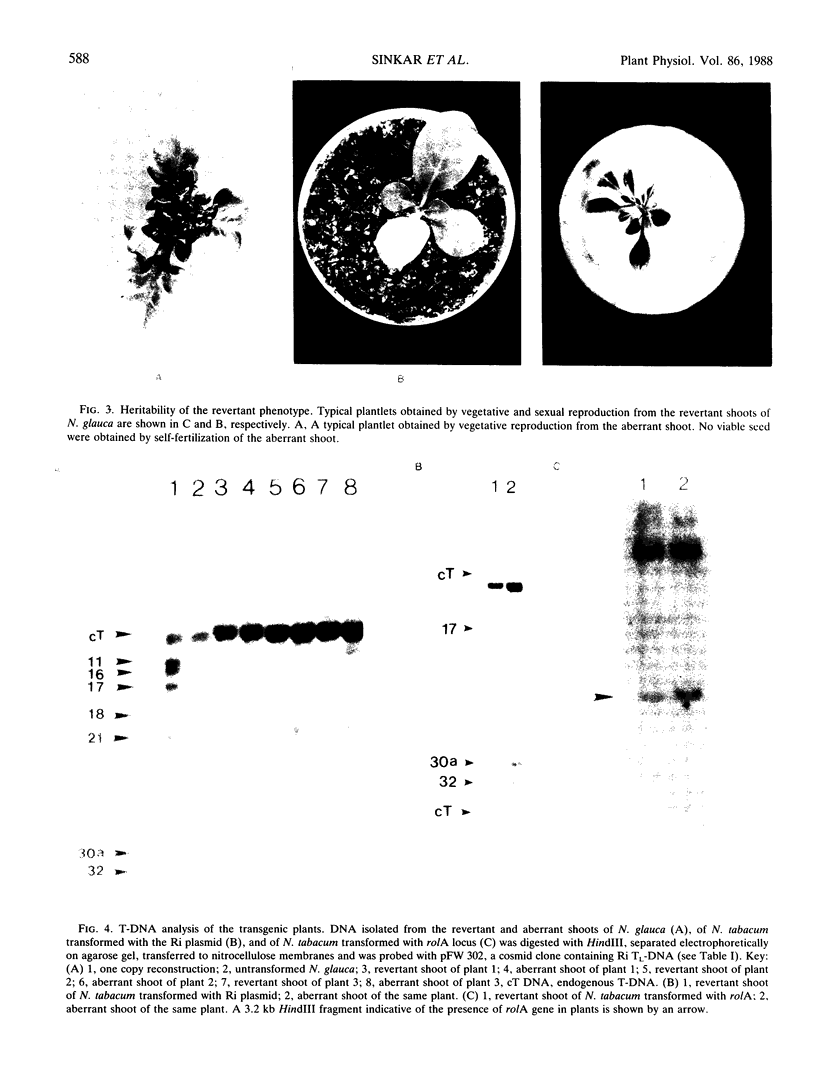
Transgenic plants harboring the left transfer DNA (TL-DNA) of the root inducing plasmid of Agrobacterium rhizogenes show many developmental abnormalities. We observed frequent appearance of normal looking lateral (revertant) shoots from such aberrant plants. Unlike aberrant shoots of the plant, revertant shoots exhibited a very high growth rate and set viable seeds. Sexual and vegetative reproduction studies showed inheritance of the revertant phenotype. Southern hybridization experiments demonstrated that the T-DNA pattern was identical in aberrant and revertant shoots, indicating that the revertant phenotype was not due to deletion or rearrangement of the T-DNA genes. Specific T-DNA transcripts were not expressed in revertant shoots. Thus, the revertant phenotype appears to result from the transcriptional inactivation of T-DNA genes. We propose that similar events in the past may have mediated horizontal acquisition of TL-DNA genes by ancestors of the genus Nicotiana, which are still found as silent endogenous T-DNA in present day untransformed Nicotiana species.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amasino R. M., Powell A. L., Gordon M. P. Changes in T-DNA methylation and expression are associated with phenotypic variation and plant regeneration in a crown gall tumor line. Mol Gen Genet. 1984;197(3):437–446. doi: 10.1007/BF00329940. [DOI] [PubMed] [Google Scholar]
- Costantino P., Spanò L., Pomponi M., Benvenuto E., Ancora G. The T-DNA of Agrobacterium rhizogenes is transmitted through meiosis to the progeny of hairy root plants. J Mol Appl Genet. 1984;2(5):465–470. [PubMed] [Google Scholar]
- Durand-Tardif M., Broglie R., Slightom J., Tepfer D. Structure and expression of Ri T-DNA from Agrobacterium rhizogenes in Nicotiana tabacum. Organ and phenotypic specificity. J Mol Biol. 1985 Dec 5;186(3):557–564. doi: 10.1016/0022-2836(85)90130-5. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Hepburn A. G., Clarke L. E., Pearson L., White J. The role of cytosine methylation in the control of nopaline synthase gene expression in a plant tumor. J Mol Appl Genet. 1983;2(3):315–329. [PubMed] [Google Scholar]
- Huffman G. A., White F. F., Gordon M. P., Nester E. W. Hairy-root-inducing plasmid: physical map and homology to tumor-inducing plasmids. J Bacteriol. 1984 Jan;157(1):269–276. doi: 10.1128/jb.157.1.269-276.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. P., Jr, Fridovich I. Evidence for a natural gene transfer from the ponyfish to its bioluminescent bacterial symbiont Photobacter leiognathi. The close relationship between bacteriocuprein and the copper-zinc superoxide dismutase of teleost fishes. J Biol Chem. 1981 Jun 25;256(12):6080–6089. [PubMed] [Google Scholar]
- Offringa I. A., Melchers L. S., Regensburg-Tuink A. J., Costantino P., Schilperoort R. A., Hooykaas P. J. Complementation of Agrobacterium tumefaciens tumor-inducing aux mutants by genes from the T(R)-region of the Ri plasmid of Agrobacterium rhizogenes. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6935–6939. doi: 10.1073/pnas.83.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G., Waffenschmidt S., Weiler E. W., Schröder J. The T-region of Ti plasmids codes for an enzyme synthesizing indole-3-acetic acid. Eur J Biochem. 1984 Jan 16;138(2):387–391. doi: 10.1111/j.1432-1033.1984.tb07927.x. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Durand-Tardif M., Jouanin L., Tepfer D. Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid. Identification of open reading frames. J Biol Chem. 1986 Jan 5;261(1):108–121. [PubMed] [Google Scholar]
- Tepfer D. Transformation of several species of higher plants by Agrobacterium rhizogenes: sexual transmission of the transformed genotype and phenotype. Cell. 1984 Jul;37(3):959–967. doi: 10.1016/0092-8674(84)90430-6. [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M., Inzé D., Schell J., Van Montagu M. Transformed cell clones as a tool to study T-DNA integration mediated by Agrobacterium tumefaciens. J Mol Biol. 1986 Mar 20;188(2):129–145. doi: 10.1016/0022-2836(86)90299-8. [DOI] [PubMed] [Google Scholar]
- White F. F., Ghidossi G., Gordon M. P., Nester E. W. Tumor induction by Agrobacterium rhizogenes involves the transfer of plasmid DNA to the plant genome. Proc Natl Acad Sci U S A. 1982 May;79(10):3193–3197. doi: 10.1073/pnas.79.10.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White F. F., Taylor B. H., Huffman G. A., Gordon M. P., Nester E. W. Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J Bacteriol. 1985 Oct;164(1):33–44. doi: 10.1128/jb.164.1.33-44.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]