Summary
Sporadic amyotrophic lateral sclerosis (sALS) is the majority of ALS, and the lack of appropriate disease models has hindered its research. Induced pluripotent stem cell (iPSC) technology now permits derivation of iPSCs from somatic cells of sALS patients to investigate disease phenotypes and mechanisms. Most existing differentiation protocols are time-consuming or low efficient in generating motor neurons (MNs). Here we report a rapid and simple protocol to differentiate MNs in monolayer culture using small molecules, which led to nearly pure neural stem cells in 6 days, robust OLIG2+ pMNs (73%–91%) in 12 days, enriched CHAT+ cervical spinal MNs (sMNs) (88%–97%) in 18 days, and functionally mature sMNs in 28 days. This simple and reproducible protocol permitted the identification of hyperexcitability phenotypes in our sALS iPSC-derived sMNs, and its application in neurodegenerative diseases should facilitate in vitro disease modeling, drug screening, and the development of cell therapy.
Keywords: induced Pluripotent Stem Cells, Spinal Motor Neuron Differentiation, Amyotrophic Lateral Sclerosis, Notch pathway, Whole-Cell Patch-Clamp, Multielectrode array, Calcium imaging
Highlights
-
•
A protocol is developed to differentiate human iPSCs into cervical sMNs
-
•
Notch inhibition is used to accelerate MN maturation
-
•
Hyper-excitability is detected in iPSC-derived MNs from sporadic ALS
In this article, Shen et al. describe a novel strategy to generate homogeneous cervical motor neurons from human iPSCs under a monolayer condition. The maturity of the derived neurons was accelerated with Compound E to inhibit γ-secretase and Notch signaling. The application of the protocol may facilitate investigation of motor neuron diseases and advance cell replacement therapy.
Introduction
Since the establishment of human induced pluripotent stem cell (iPSC) technology in 2007 (Takahashi et al., 2007), iPSCs have been widely used in regenerative medicine and disease modeling. iPSCs can self-renew and differentiate into almost any cell types in the body, including disease-relevant ones. Motor neuron diseases (MNDs) include amyotrophic lateral sclerosis (ALS), spinal muscular atrophy, and spinal and bulbar muscular atrophy and are fatal diseases characterized by degeneration of MNs in the CNS. There is no effective treatment for MNDs, which is thought, in part, to be caused by the lack of suitable human disease models (Hardiman et al., 2017). The development of a robust protocol for derivation of functional MNs is, therefore, crucial for understanding disease mechanisms and developing novel therapies for MNDs.
Spinal MNs (sMNs) are responsible for transmitting commands from the CNS to muscles to control motor activity appropriately. Signaling pathways and transcription factors contributing to the sMN development have been unveiled (Jessell, 2000; Muñoz-Sanjuán and Brivanlou, 2002; Shirasaki and Pfaff, 2002; Stifani, 2014). For example, neuroectoderm is activated in the fibroblast growth factor (FGF) and WNT pathways and/or inhibited by bone morphogenetic protein (BMP) and transforming growth factor-β (TGF-β) signalings (De Robertis, 2006; Nordström et al., 2002). Retinoid acid (RA) and sonic hedgehog (SHH) serve as morphogens for the rostrocaudal and dorsoventral axes along the spinal cord (Muhr et al., 1999; Patani et al., 2011; Roelink et al., 1995), which define MN progenitor (pMN) domain and repress other progenitor domains, by co-expressing transcription factors of PAX6 (neuroectodermal markers), NKX6.1 (ventral progenitor marker), and OLIG2 (a pMN-specific marker) (Sagner and Briscoe, 2019). OLIG2 promotes MN development by directly repressing HES expression, which in turn induces NGN2 (Sagner et al., 2018). NGN2 is required for cell cycle exit and induction of terminal MN transcription factors including MNX1, ISL1, and LHX3, to establish MN identity (Lee et al., 2005; Novitch et al., 2001). MNX1+ MNs further differentiate into mature MNs expressing choline acetyltransferase (CHAT), which synthesize acetylcholine for signal conduction at neuromuscular junctions (Stifani, 2014). The stage-specific MN genes and their expression patterns during development can be used as indicators for in vitro differentiation.
Previously, several protocols for sMN differentiation have been developed by mimicking in vivo neurogenesis using extrinsic signaling molecules (Figure S1 and Table S1). Different doses, timing, and delivery procedures of signaling molecules among different protocols led to substantial variations in the duration (1–2 months) and efficiency (30%–90%) of MN differentiation. In addition, the derived MNs were often immature, and a lengthy maturation period was required before the detection of electrophysiological activity (Figure S1). As most MNDs including ALS are late-onset diseases, these deficiencies limit the value of the iPSC-derived MNs in revealing disease characteristics.
In this study, we developed an efficient strategy to differentiate iPSCs into cervical sMNs under a monolayer-adherent condition. This protocol can shorten the duration required to detect electrophysiological phenotypes in ALS cell models, by generating nearly pure neural stem cells (NSCs) in 6 days, robust pMNs in 12 days, enriched CHAT+ sMNs in 18 days, and functional sMNs with extensive network activities in 28 days. This novel MN differentiation protocol, therefore, facilitates the production of mature sMNs for MND research, high-throughput drug screening, and potential cell transplantation therapy.
Results
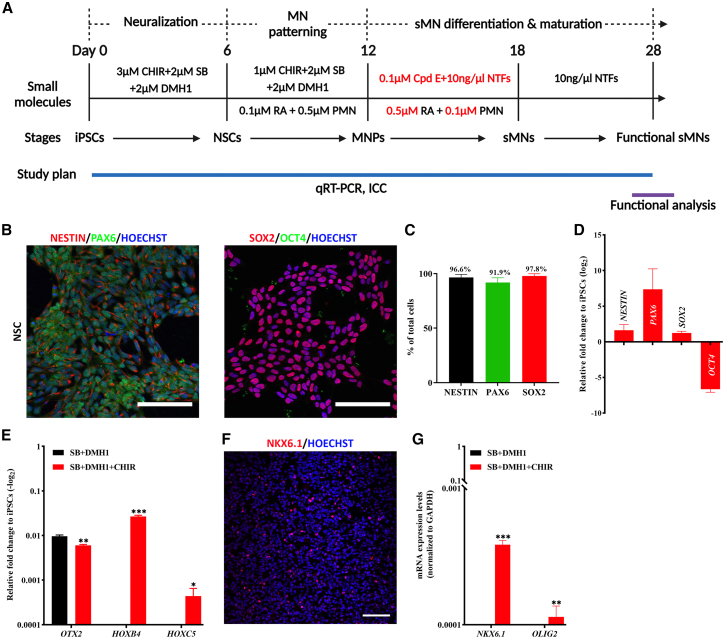
Homogeneous caudal and ventral NSCs are generated in 6 days
Dual SMAD inhibition of BMP and TGF-β signaling is widely used in neural induction to generate rostral NSCs (Chambers et al., 2009) expressing OTX2, a rostral marker for the fore- and midbrain. However, WNT activation is required for generating NSCs with a caudal fate expressing HOX genes (Chambers et al., 2009; Nordström et al., 2002). We therefore in this study first neuralized human iPSCs for 6 days with 2 μM SB431542 (SB) to inhibit TGF-β signaling, 2 μM DMH1 to inhibit BMP pathway, and 3 μM CHIR99021 (CHIR) to activate WNT signaling (Figure 1A) (days 0–6). This resulted in the production of homogeneous NSCs, which were positive for NSC markers of NESTIN (96.6% ± 2.7%), PAX6 (91.9% ± 4.3%), and SOX2 (97.8% ± 2.1%), but negative for pluripotency marker of OCT4 (Figures 1B and 1C). The consistent efficiency of NSC derivation was validated in three control and four sALS iPSC lines (Figure S2A). Reverse transcrtipase qPCR (RT-qPCR) results further confirmed high expression of NESTIN, PAX6, and SOX2, but repression of OCT4 mRNA (Figure 1D).
Figure 1.
Combination of dual SMAD inhibition with WNT activation converts human iPSCs into caudal/ventral NSCs in 6 days
(A) Schematic diagram of the MN differentiation protocol from iPSCs.
(B) Representative images of NESTIN+/PAX6+ and SOX2+/OCT4– NSCs generated after 6 days of SB + DMH1 + CHIR treatment.
(C) The proportion of NESTIN+, PAX6+, and SOX2+ cells on day 6 of differentiation, N = 7 cell lines, n = 3 replicates.
(D) RT-qPCR analysis of NESTIN, PAX6, SOX2 and OCT4 mRNA expression after 6 days of SB + DMH1+CHIR treatment, N = 2, n = 3.
(E) CHIR-induced WNT activation conferred a caudal identity of the NSCs expressing fore/midbrain marker OTX2 and hindbrain/spinal markers of HOXB4 and HOXC5. N = 4, n = 3.
(F) The NSCs were positively immunostained for a ventral marker NKX6.1 after 6 days of SB + DMH1+CHIR treatment.
(G) RT-qPCR analysis showed activation of NKX6.1 and OLIG2 mRNA by addition of CHIR, N = 1, n = 3. All representative images were from the control line of ALSH84C5, Scale bar, 100 μM.
See also Figure S2.
In addition, this triple treatment was found to significantly caudalize the NSCs, and this was supported by marked activation of HOXB4 and HOXC5, and decreased expression of OTX2 compared with conventional dual SMAD inhibition (SB + DMH1) (Figures 1E and 1F). The derived NSCs also displayed ventral feature, with induced expression of NKX6.1 (a ventral progenitor marker) and OLIG2 (an MNP-specific gene) (Figures 1G and S2B). Thus, the combination of dual SMAD inhibition with WNT activation effectively converted human iPSCs into homogeneous caudal and ventral NSCs in 6 days.
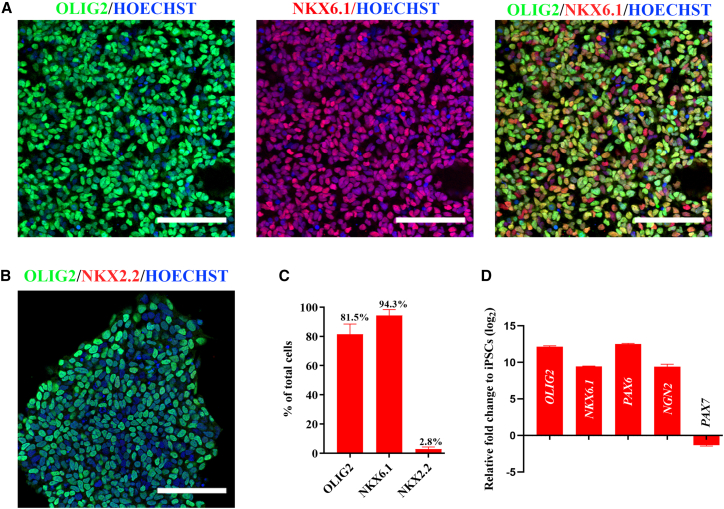
Robust MNPs are derived in 12 days of iPSC differentiation
To convert the NSCs into MNPs, 0.1 μM RA and 0.5 μM purmorphamine (an SHH agonist) were applied to the 6-day NSCs for the subsequent 6 days in addition to 1 μM CHIR + 2 μM SB + 2 μM DMH1 (Figure 1A) (days 6–12) (Du et al., 2015). The cell cultures was evaluated on day 12 for the expression of OLIG2 (an MNP marker), NKX6.1 (a ventral progenitor marker), PAX6, NKX2.2 (an interneuron progenitor marker), and PAX7 (a dorsal progenitor marker) to define the pMN domain according to spinal cord development (Sagner and Briscoe, 2019) (Figure S2C).
In the day 12 cultures, 81.5% ± 6.9% cells were positive for OLIG2+ (up to 91.4% ± 1.4%) and 94.3% ± 0.7% positive for NKX6.1+ (up to 99.0 ± 0.5%), whereas only 2.8% ± 1.5% of cells were positive for NKX2.2 (Figures 2A–2C). The high efficiency of ventral MNP production was consistently validated in seven individual iPSC lines from six donors (Figure S2B). Additionally, RT-qPCR data confirmed the elevated expression of MNP markers of OLIG2, NKX6.1, and PAX6 as well as repression of a dorsal marker PAX7. Enhanced expression of NGN2, a transcription factor critical for post-mitotic MN induction (Lee et al., 2005; Novitch et al., 2001), was also observed in the differentiated cells (Figure 2D). Together, these data showed robust derivation of MNPs in 12 days of iPSC differentiation, by treating monolayer iPSCs with 6 days of CHIR + SB + DMH1 and subsequent 6 days of CHIR + SB + DMH1 + RA + purmorphamine.
Figure 2.
Robust MNPs are derived in 12 days of iPSC differentiation
(A) Representative images of OLIG2+/NKX6.1+ MNPs generated in 12 days of differentiation.
(B) Representative images of OLIG2+/NKX2.2– MNPs generated in 12 days of differentiation.
(C) The proportion of OLIG2+, NKX6.1+, and NKX2.2+ cells.
(D) RT-qPCR data showing relative expression of OLIG2, NKX6.1, PAX6, NGN2, and PAX7 genes in 12-day culture compared to iPSCs. N = 2 cell lines, n = 3 replicates. All representative images were from the control line ALSH84C5, Scale bar, 100 μM. See also Figure S3.
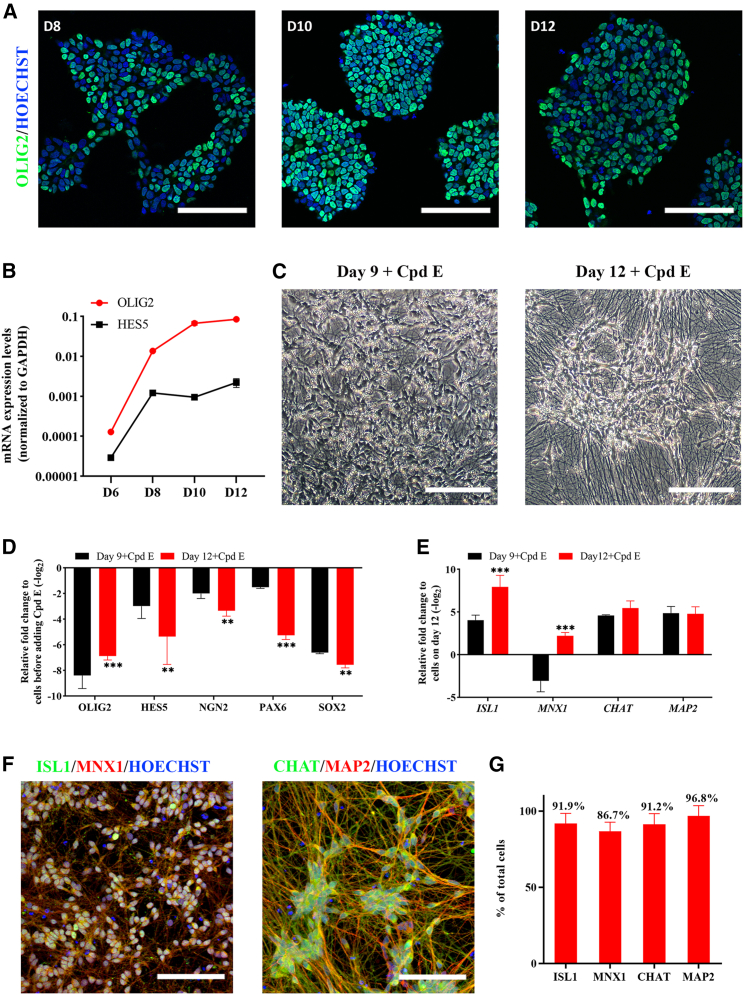
Compound E effectively promotes the conversion of MNPs into sMNs
In the pMN domain, OLIG2-expressing MNPs can become MNs or oligodendrocytes (Lu et al., 2002; Zhou and Anderson, 2002). Therefore, a timely conversion of OLIG2+ MNPs into MNs is vital to increase MN productions in vitro. Notch activity is known to promote MNP proliferation via γ-secretase cleavage, which releases the Notch intracellular domain and activates expression of genes including HES5, a repressor of proneural genes (Ben-Shushan et al., 2015). We therefore used a γ-secretase inhibitor, Compound E, to inhibit HES5 expression and accelerate MN differentiation.
We first examined the time course of OLIG2 expression to determine the optimal time for Compound E treatment, and found OLIG2 expression was peaked at day 10 (83.3% ± 6.9%) and 12 (80.2% ± 7.7%) at both mRNA and protein levels (Figures 3A, 3B, and S3A), and this was also accompanied by increased HES5 expression (Figure 3B). Therefore, we chose day 9 (before the peak) and day 12 (at the plateau) to test the effect of Compound E (0.1 μM). We also increased RA to 0.5 μM as a high concentration of RA was shown to induce neuronal differentiation, and reduced purmorphamine to 0.1 μM since MNP differentiation required a low level of SHH (Saade et al., 2013). Meanwhile, CHIR, SB, and DMH1 were withdrawn from the medium because they were used to maintain MNP proliferation (Kalani et al., 2008).
Figure 3.
Compound E effectively represses the expression of MNP genes and promotes the generation of sMNs
(A) Representative images of OLIG2+ MNPs on days 8, 10, and 12 of differentiation.
(B) A time course induction of OLIG2 and HES5 expression from day 6 to day 12 of differentiation. N = 3 cell lines, n = 3 replicates.
(C) Representative morphology of cells after 6 days of Compound E (Cpd E) treatment from day 9 or 12 of differentiation.
(D) Repression of MNP gene expression after 6 days of Cpd E treatment.
(E) The mRNA expression of MN-specific genes ISL1, MNX1, mature MN gene CHAT, and mature neuronal marker MAP2 after 6 days of Cpd E treatment (MN stage) over that in their respective MNPs at day 9 or day 12. N = 3, n = 3.
(F) Representative images of ISL1+/MNX1+ and CHAT+/MAP2+ cells on day 18 under Cpd E treatment.
(G) The proportion of ISL1+, MNX1+, CHAT+, and MAP2+ cells among Hoechst-stained cells on day 18. N = 7, n = 2–3. All representative images were from the control line ALSH84C5, Scale bar, 100 μM. See also Figure S3.
Following the next 6 days of exposure to 0.5 μM RA + 0.1 μM purmorphamine + 0.1 μM Compound E + Neurotrophic factors (NTFs) (10 ng/mL each), cells exhibited typical neuronal morphology with long neurites and high expression of mature neuronal marker MAP2 under both conditions (Figures 3D and 3E). The expression of MNP markers OLIG2, HES5, NGN2, PAX6, and SOX2 was markedly repressed under both conditions, with greater reductions by adding Compound E from day 12, with the exception of OLIG2 (Figure 3D). Surprisingly, only when Compound E was added from day 12, both MN-specific genes of ISL1 and MNX1 were induced; the addition of Compound E to day 9 MNPs induced ISL1 but not MNX1 expression (Figure 3E). Therefore, the timing of Compound E addition plays a crucial role in promoting MN differentiation.
We subsequently quantified the proportion of MNs on day 18 and consistently showed high efficiency of MN differentiation across 7 iPSC lines (Figure S3B). On day 18, 91.9% ± 6.7% of ISL+, 86.7% ± 6.1% of HB9+, 91.2% ± 7.0% of CHAT+, and 96.8% ± 6.8% of MAP2+ cells were identified (Figures 3F and 3G). Meanwhile, Ki67+ proliferating cells (1.28% ± 0.95%) or OLIG2+ MNPs (3.37% ± 2.87%) were rarely detected, and GFAP+ glia was absent (Figures S3C and S3D). In addition, high levels of mRNA expression were detected for synaptic excitatory markers (SYN1, SHANK1, and PSD-95), vesicular glutamate transporter (VGlut2), and glutamate receptor markers including ionotropic AMPA (GRIA1 and GRIA2), NMDA (GRIN1 and GRIN2A), and kainate (GRIK1) receptors (Figure S3E). Together, these data demonstrated that our protocol produced a high purity of sMNs with excitatory and glutamatergic properties on day 18 of iPSC differentiation.
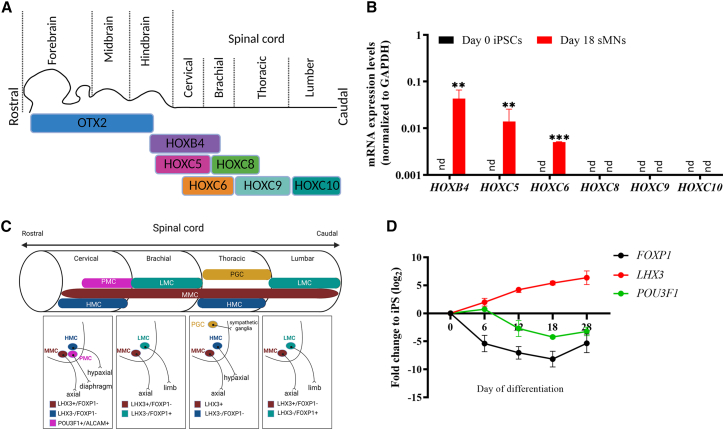
The sMNs display a cervical MMC MN identity
During spinal cord patterning, sMNs acquire a diverse of positional and columnar characteristics, resulting in various subtypes of sMNs innervating different muscle groups in the body (Peljto and Wichterle, 2011; Philippidou and Dasen, 2013; Sagner and Briscoe, 2019). To construct clinically relevant in vitro disease models, it is crucial to determine the spatial identity of MNs because there is wide variation in illness onset sites among ALS patients.
During embryonic development, HOX genes are expressed in an overlapping rostrocaudal pattern from brain to the cervical, brachial, thoracic, and lumbar spinal cord (Figure 4A). Compared with iPSCs, HOXB4, HOXC,5 and HOXC6 expression were significantly activated on day 18 sMNs, but not HOXC8, HOXC9, or HOXC10 genes (Figure 4B). This HOX expression pattern confined the derived sMNs to the cervical region of the spinal cord, which was consistent with previous findings that RA and purmorphamine enabled cells to acquire a cervical character (Peljto et al., 2010; Wichterle et al., 2002).
Figure 4.
The iPSC-derived sMNs resemble cervical MMC in the spinal cord
(A) Schematic diagram showing differential expression of OTX2 and HOX genes in the hindbrain and spinal cord. Created with BioRender.com and adapted from Philippidou and Dasen (2013) and Sagner and Briscoe (2019).
(B) Expression of spinal cord positional markers of HOXB4, HOXC5, HOCX6, HOXC8, HOXC9, and HOXC10 in day 0 iPSCs and day 18 iPSC-derived sMNs. N = 3, n = 3.
(C) Schematic diagram presenting MN columnar pattern, cell types, positions, general muscle targets, and specific marker genes. MMC neurons innervate axial muscles, HMC cells innervate hypaxial muscles, PMC neurons innervate diaphragm, LMC neurons innervate limb muscles, and preganglionic motor column (PGC) neurons innervate sympathetic ganglia. Adapted from (Peljto and Wichterle, 2011; Philippidou and Dasen, 2013) and created with BioRender.com.
(D) The expression of FOXP1, LHX3 and POU3F1 mRNA during the course of differentiation. N = 3 cell lines, n = 3 replicates. nd, not detected, as the values were too small to be visible on the graph.
To assess the columnar nature of iPSC-derived sMNs, we examined the expression of FOXP1, LHX3, and POU3F1, which are markers for hypaxial (HMC), medial (MMC), and phrenic motor column (PMC), respectively (Figure 4C). FOXP1 and POU3F1 expression were repressed during the course of MN differentiation, indicating the absence of HMC or PMC sMNs. Meanwhile, LHX3 expression was steadily increased during the 4 weeks of differentiation in comparison with that in day 0 iPSCs (Figure 4D). This suggests that Compound E made no changes to the neuronal identity and the iPSC-derived sMNs belongs to the MMC subtype that innervate axial musculature.
Compound E accelerates functional maturation of iPSC-derived cervical sMNs
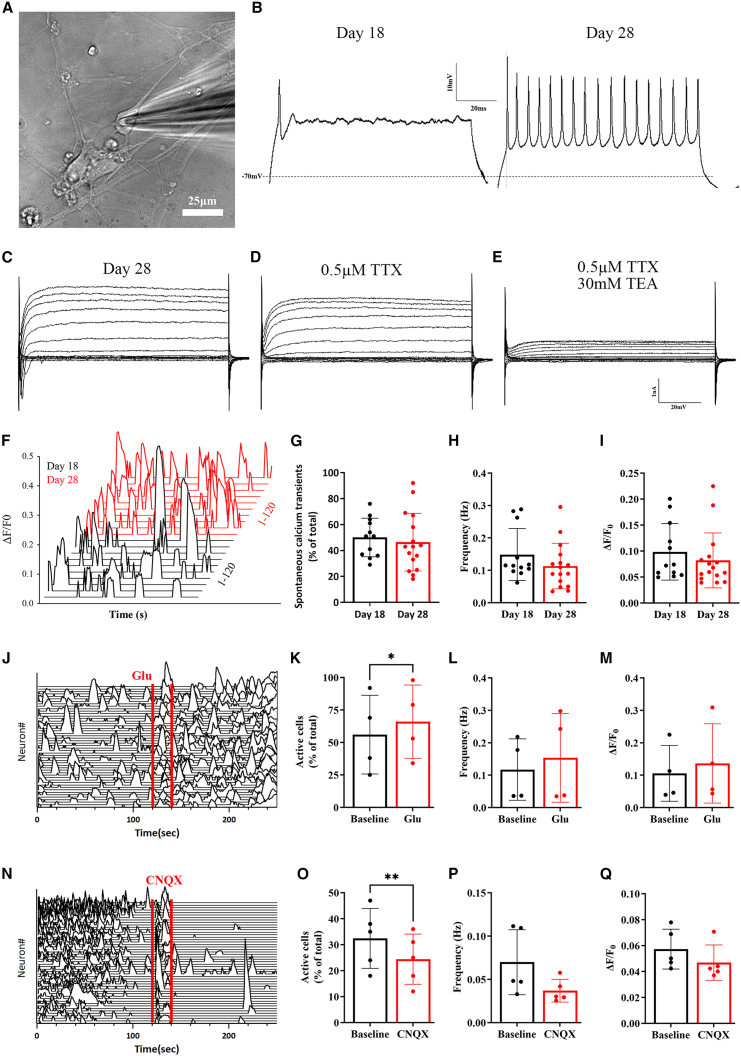
Variations in sMN differentiation protocols may lead to incomparable results among different studies (Du et al., 2015; Fujimori et al., 2018; Maury et al., 2015) (Figure S1). However, a standardized criterion can compensate for this deficiency if neuronal maturation is equivalent before phenotype comparison. Electrical activities and functional networks are essential characteristics of adult neurons. Therefore, we performed whole-cell patch clamp recordings on iPSC-derived sMNs on days 18 and 28 of differentiation. The cells on day 28 showed significantly greater electrophysiological maturity than those on day 18, as repeated action potentials (AP) were recorded in day 28 sMNs, but only single AP events were detected on day 18 (Figures 5A and 5B). Day 28 sMNs displayed input resistance of 546 ± 62 MΩ, Rheobase current of 10.7 ± 2.3 pA, AP threshold of −33.5 ± 1.1 mV, and AP Amplitude of 29.0 ± 2.5 mV, which appeared to be comparable with 18-week human fetal sMNs (Tadros et al., 2015) (Table S3). Moreover, they exhibited a voltage-dependent current, which was abolished by 0.5 μM of the Na+ channel blocker tetrodotoxin (TTX) or 30 mM of the K+ channel blocker tetraethylammonium (TEA) (Figures 5C and 5D). The functional maturity of the sMNs were further supported by calcium imaging experiments (Figures 5F–5Q). The iPSC-sMNs displayed spontaneous Ca2+ transients on both day 18 and 28 of differentiation (Figures 5F–5I). Additionally, day 28 sMNs were shown to be responsive to glutamate activation and CNQX blockade (Figures 5J–5Q).
Figure 5.
The iPSC-derived sMNs on day 28 show greater functional maturity
(A) Representative image of a patched sMN on day 28 of differentiation. Scale bar, 25 μm.
(B) Representative current-clamp recordings of iPSC-derived sMNs on day 18 (n = 18 neurons) and 28 (n = 26 neurons) of differentiation from the control ALSH47C1 line.
(C) Representative voltage-clamp recordings of a cell held at −70mV.
(D) Representative traces of the currents from the same cell as in (C) after the application of 0.5 μM Na+ channel blocker TTX.
(E) Representative traces of the currents from the same cell as in (D) with 30 mM K+ channel blocker TEA after TTX treatment.
(F–I) (F) Representative waterfall traces of spontaneous Ca2+ transients on day 18 (black) and 28 (red) from a 120-s recording. No significant difference in proportion (%) (G), frequency (Hz) (H), or the amplitude (ΔF/F0) (I) was shown on iPSC-derived sMNs between day 18 (N = 4 cell lines, n = 12 recordings/8 coverslips, 6,868 cells examined) and day 28 (N = 4, n = 16/8, 7,977 cells examined).
(J–Q) Representative waterfall traces and statistical results demonstrating the spontaneous Ca2+ transients and their responses to glutamate (Glu, 10 mM, [J–M], N = 2, n = 4/4, 2,143 cells examined) or CNQX (50 μM, [N–Q], N = 2, n = 5/4, 1,622 cells examined) on day 28.
Sporadic ALS-derived sMNs exhibit enhanced spontaneous intrinsic excitability
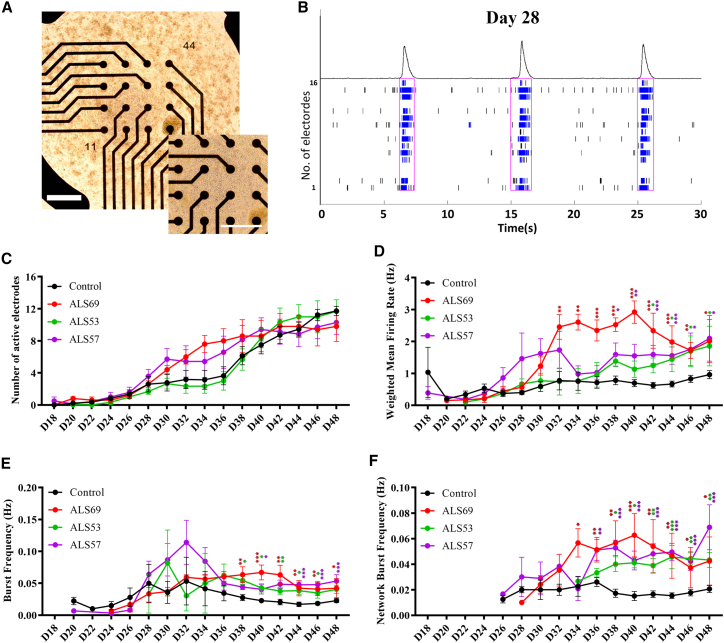
Altered excitability of MNs has previously been described in ALS patients (Kanai et al., 2006; Vucic et al., 2008), familial ALS-derived sMNs with known mutations (Devlin et al., 2015; Naujock et al., 2016; Wainger et al., 2014), and transgenic animal models (Kuo et al., 2004; Pieri et al., 2003; van Zundert et al., 2008). Although the affected CNS region varies with the progression of ALS, the detection of network and population electrophysiological properties in sALS iPSC-derived sMNs may provide insights into the underlying mechanism. We therefore replated day 12 MNPs onto the 48-well multi-electrode array (MEA) plates with 16 micro-electrodes/well. MEA recording was carried out every other day, before medium change, from day 18 to day 48 of differentiation (Figure 6A). Consistent with the patch-clamp and Ca2+ imaging data, MEA recording detected significant spontaneous electrical activities on sMNs from day 20 or after, with extensive network activities on day 28 of differentiation, indicating formation of functional neuronal networks with extensive neurotransmission (Figure 6B).
Figure 6.
The iPSC-derived sMNs from sALS patients exhibit an increased spontaneous firing
(A) Representative images of iPSC-derived day 28 sMNs on a 48-well MEA plate with 16 microelectrodes/well. Scale bar, 100 μM.
(B) The MEA recording plot showing typical network firing patterns detected from iPSC-derived sMNs on day 28.
(C) The MEA recordings were carried out on iPSC-derived sMNs from day 18 to day 48. The control data were averaged from three lines of ALSH84C5, ALSH47C1, and LQTH002C4 of three independent donors, and patient data from four lines (ALS69C4, ALS53C5, ALS57C1, and ALS57C4) of three sALS patients.
(D–F) sMNs generated from patients ALS53, ALS57, and ALS69 had a significantly higher AP rate (weighted mean firing rate) (D), burst frequency (E), and network burst frequency (F) at the late stages of differentiation than the sMNs from the control group. The total MEA wells examined were n = 22 wells from N = 3 control iPSC lines, and n = 15 wells from N = 4 patient iPSC lines. The data were presented as mean ± SEM. See also Table S3.
During the time course of MEA recording, there was no significant difference in the number of active electrodes between the sALS (ALS69C4, ALS53C5, ALS57C1, and ALS57C4 lines) and the control group (ALSH84C5, ALSH47C1, and LQTH002C4) (Figure 6C), suggesting that sALS patient-derived iPSCs did not exhibit significant defects in MN differentiation or maturation. The sMNs from the patient group, however, fired significantly more spontaneous spikes, bursts, and network bursts than that in the control group, with an increased weighted mean firing rate (Figure 6D), burst frequency (Figure 6E), and network burst frequency (Figure 6F) from day 32 to day 48 of differentiation. The raw data for each donor are included in Table S3. These results indicate that the sALS samples exhibited elevated intrinsic excitability. Moreover, this hyperexcitability phenotype was phasic over time and became even more prominent in the late phases of sMN differentiation and during MN aging in vitro.
Discussion
Numerous efforts have been made to differentiate human pluripotent stem cells into MNs for modeling MNDs. The research has, however, been hindered by the lengthy differentiation process, low efficiency, and extended maturation time to display functional properties (Figure S1). Here we have developed a novel monolayer MN induction protocol that enables derivation of nearly pure NSCs in 6 days, MNPs (73%–91%) in 12 days, cervical sMNs (88%–97%) in 18 days, and functionally mature sMNs in 28 days. These data were supported by comprehensive characterization and functional assays. This protocol also enables phenotyping of pre-symptomatic changes of the disease in a relatively short duration, and we have identified hyper-excitability of sMNs derived from sALS iPSC lines via MEA recording after 32 days of differentiation.
One feature of our protocol is the monolayer-adherent culture. Serum-free floating embryoid body (EB) culture has been a default stage for neuronal differentiation in most previous MN differentiation protocols, as this was thought to mimic the three-dimensional environment in vivo (Table S1). However, the EB method can be accompanied with challenges including difficulty in observing cell morphology, variability in EB size, low permeability of morphogens to the innermost parts, and low cell viability after EB dissociation. The monolayer differentiation was explored previously, but ended up with a low efficiency of MNs or a lengthy maturation time (Bianchi et al., 2018; Qu et al., 2014) (Table S1). In this study, we demonstrate that monolayer culture together with timely addition of different molecules can result in up to 97% of CHAT+ mature sMNs in 18 days of iPSC differentiation.
The MN development is governed by dorsal-ventral and anterior-posterior morphogens. We aimed to generate ventral sMNs and, therefore, used dual SMAD inhibition to repress dorsal pathways. Consequently, no expression of PAX7 (a dorsal spinal marker) was detected. This was consistent with previous data that activation of the dorsal signaling by BMP4 was required to generate dorsal spinal interneurons from ESCs or iPSCs (Gupta et al., 2018; Ogura et al., 2018). Meanwhile, the iPSC-sMNs derived in this study showed cervical identity with high expression of HOXB4, HOXC5, and HOXC6, but no expression of HOXC8, HOXC9, or HOXC10. This is likely to result from the use of 0.1 μM RA during NSC-MNP transition. RA is essential to activate class I ventral transcription factors of PAX6, DBX2, DBX1, and IRX3 (Patani, 2016). The endogenous RA at the cervical region is relatively low, as a two-tailed RA gradient is established with high concentration at the trunk mesoderm, which gradually declines anteriorly toward the hindbrain, and posteriorly toward the caudal progenitor zone where FGF8 is highly expressed (Cunningham and Duester, 2015). A previous study also showed that 0.1 μM RA could induce neural tube explants to express Hoxc5 (cervical), but not Hoxc8/9 (thoracic) or Hoxc10 (lumber) (Liu et al., 2001), whereas generation of lumbosacral sMNs requires addition of FGF8 in the absence of RA (Xu et al., 2023).
An SHH agonist, purmorphamine, is deployed in this protocol for ventralization and activation of class II transcription factors of OLIG2, NKX6.1, NKX6.2, and NKX2.2 (Patani, 2016). During MN differentiation, LHX3 interacts with ISL1 and determines MN identity; however, in mature MNs, LHX3 expression is retained exclusively in the MMC (Lee et al., 2012). The sMNs from this study exhibit MMC identity, as they increasingly express LHX3 but not FOXP1 or POU3F1. This is likely to result from the use of a medium-high concentration (0.5 μM) of purmorphamine, as the pMN domain is anatomically the second domain most close to the floor plate, where SHH is most abundant, among the p0, p1, p2, pMN, and p3 domains (Andrews et al., 2019).
MMC sMNs innervate axial muscles in the neck, trunk, and hips, which are often deteriorated at the late stages of ALS progression. This may result in difficulties in head and neck control, balance, and stability, and decreased mobility (Duleep and Shefner, 2013). Cervical MNs at C2–C7 innervate respiratory muscles and axial muscle weakness is also linked to diaphragmatic denervation and disease progression, and consequently most ALS patients die of respiratory impairment (Cykowski et al., 2018; Niedermeyer et al., 2019; Portaro et al., 2018). Atrophy of the cervical spinal cord is therefore used as a predictor of respiratory dysfunction (Cohen et al., 2017; Grolez et al., 2018; Patzig et al., 2019).
An ability to regenerate cervical sMNs is, therefore, vital for ALS, and we show here that the conversion of MNPs into cervical MMC MNs can be expedited by addition of 0.1 μM Compound E, a γ-secretase and Notch inhibitor. Blocking of Notch signaling was shown to accelerate neuronal differentiation by delaying the G1/S phase transition (Borghese et al., 2010; Crawford and Roelink, 2007), but previous efforts were associated with low differentiation efficiency (<74% ISL1+/HB9+ cells) or a long duration (28–31 days) of CHAT+ MN generation (Du et al., 2015; Maury et al., 2015). We demonstrate that the timing of Compound E addition is crucial, and inhibition of γ-secretase on day 12 (not day 9) at the plateau stage of OLIG2 expression not only resulted in high CHAT+ sMN production (88%–97%) in 18 days, but also accelerated MN maturation, with multiple APs, extensive network activities, and strong calcium transients detected in day 28 sMNs.
Whereas the bona fide electrophysiological data of the adult human MNs is unavailable, our day 28 sMNs appeared to be comparable with 18-week human fetal MNs (Tadros et al., 2015) or iPSC-MNs in some other protocols (Table S3). Although reprogramming can be influenced by epigenetic memory and prolonged passaging (P20) may improve pluripotency (Koehler et al., 2011). Our seven iPSC lines at P20–30 showed similar capacity to generate NSCs (92%–98%), MNPs (73%–91%), sMNs (88%–97%), and comparable maturity at day 28. Replication of the protocol to derive sMNs from iPSCs of other ALS patients will provide independent cell models for investigating pathogenic mechanisms, particularly those associated with respiratory failure, as well as donor cells for subsequent preclinical and clinical trials, which may require a quantity of MNs. This may be done by expansion of MNPs as it was reported that OLIG2+ MNPs could be expanded for more than five passages without significantly losing MNP identity or capacity to generate sMNs (Du et al., 2015). Therefore, our monolayer sMN differentiation protocol, in combination with MNP expansion, may provide large quantities of cervical sMNs with consistency and high maturity for large-scale studies, such as disease phenotyping, high-throughput drug screening, and future cell transplantation trials.
Experimental procedures
Resource availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors, Sanbing Shen (sanbing.shen@universityofgalway.ie).
Materials availability
This study did not generate new unique reagents.
Cell culture and expansion
The iPSC lines used in this study are listed in Table S2 and have been characterized in previous studies (de la Cruz et al., 2020; Ge et al., 2020; Yang et al., 2019, 2020). All iPSC lines were maintained on Geltrex (A1413302, Gibco)-coated six-well plates in Essential 8 Flex Medium (A2858501, Gibco).
MN differentiation
The iPSCs at passages 20–30 were dissociated with Accutase (A6964, Sigma) and seeded at 30,000 cells/cm2 on Geltrex-coated six-well plates in E8 with 10 μM Y-27632 (72304, STEMCELL) on day −1. On day 0, E8 medium was switched to neuronal induction medium (NIM) consisting of 1:1 DMEM/F12 (BE-12-719F, Lonza) and neurobasal medium (21103049), 1% P/S (15140122), 0.5× N2 (17502048), 0.5× B27 (17504044), 0.1 mM ascorbic acid (AA, A4403, Sigma), and 1% GlutaMAX (35050061) from Gibco. The medium was renewed every other day with fresh 3 μM CHIR (HY-10182, MCE), 2 μM SB (HY-10431, MCE), and 2 μM DMH1 (HY-12273, MCE) for 6 days. Cells were then split 1:3 onto Geltrex-coated plates in NIM with 1 μM CHIR, 2 μM SB, 2 μM DMH1, 0.1 μM RA (HY-14649, MCE), and 0.5 μM purmorphamine (HY-15108, MCE) for the subsequent 6 days with medium changed every 2 days. On day 9 or 12, the patterned MNPs were re-seeded at 50,000 cells/cm2 on 0.01% polyethyleneimine (PEI, P3143, Sigma)-coated plates for terminal differentiation. We added 0.5 μM RA, 0.1 μM purmorphamine, and 0.1 μM Compound E (HY-14176, MCE) freshly for the subsequent 6 days into the NMM medium, consisting of Neurobasal plus medium (A358290, Gibco), 1% P/S, 0.5× N2, 0.5× B-27 Plus (A3582801, Gibco), 0.1 mM AA, and 1% GlutaMAX. The medium was half-changed every other day for 6 days, and small molecules were withdrawn from the culture medium from day 15 or day 18, respectively. NTFs of BDNF (450-02, PeproTech), IGF-1(450-13, PeproTech), and CTNF (450-10, PeproTech) were added at 10 ng/mL each, from day 9 or 12 onwards.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (30525-89-4, Santa Cruz), permeabilized and blocked simultaneously in 1% BSA (A2153, Sigma) with 0.3% Triton X-100 (T8787, Sigma), and incubated with primary antibodies (Table S4) at 4°C overnight. They were then washed, incubated with secondary antibodies and Hoechst 33258, imaged using a confocal microscope (Olympus FluoView 1000 system) or the Operetta High Content Imaging System (PerkinElmer), and quantified using ImageJ or the Operetta High Content Imaging System with Harmony software (PerkinElmer).
RNA extraction for RT-qPCR
Total RNA was extracted using RNeasy Plus Mini Kit (74134, Qiagen), and reverse transcribed to cDNA using SensiFAST cDNA Synthesis Kit (BIO-65053, Bioline). RT-qPCR was performed on a Step One Plus Real-Time PCR System (4376600, Applied Biosystems) using Fast SYBR Green Master Mix (4385612, Applied Biosystems). Primers are listed in Table S4.
Whole-cell patch clamp
Patch clamp was performed as described previously with some modification (Avazzadeh et al., 2021). MNPs were seeded on 13-mm glass coverslips on day 12. Whole-cell patch clamp recordings were performed on day 18 (n = 12 neurons) and 28 (n = 26 neurons) in a warm extracellular bath solution (18°C–20°C), containing 135 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES sodium salt, and 10 mM glucose at pH 7.4 (Wainger et al., 2014). Patch pipettes were filled with intracellular solution containing 150 mM KCl, 6 mM MgCl2, 10 mM HEPES, 1 mM EGTA (E3889, Sigma), 4 mM ATP (HY-B2176, MCE), 0.3 mM Na2GTP (HY-12695, MCE), and 10 mM Na2PhosCr (HY-D0885B, MCE) at pH 7.4 with a resistance of 2–3 MΩ. Images were captured using Zeiss Axiovert 200 (original magnification ×40).
Recordings were performed using an EPC10 patch-clamp amplifier from HEKA. Intrinsic firing properties of neurons were recorded in current-clamp mode, with step current injections from −40 to +100 pA in 10-pA increments. Voltage-dependent Na+ and K+ currents were recorded in voltage-clamp mode from a holding potential of −70 mV and step depolarization from −100 to +40 mV in 10-mV increments for 200 ms. Additional recordings were performed in the presence of 1 μM TTX (T-550, Alomone Labs) or 30 mM TEA (140023, Sigma) to block Na+ and K+ currents, respectively. All data were recorded as unfiltered voltage and current clamp recording at 50.0 kHz and 20.0 kHz, respectively.
Multi-electrode array assay
Approximately 50,000 MNPs in 8 μL NMM were plated on 0.1% PEI-coated CytoView 48-well MEA plate (M768-tMEA-48W, AXION) on day 12 of differentiation. Neuronal activities were recorded for 300 s every other day before medium change, from day 18 to day 48. In short, the 48-well MEA plate was docked into the Maestro MEA recording amplifier with a heater to maintain at 37°C (Axion Biosystems). Signals were sampled at 12.5 kHz, digitized and analyzed using Axion Integrated Studio Navigator (AxIS) 2.5.2 with a 200-Hz high-pass and 3-kHz low-pass filter. An adaptive spike detection threshold was set at 5.5 times of the standard deviation for each electrode with 1-s binning (Wainger et al., 2014).
The exported spike files (.spk) were batch-processed using Neural Metric Tool (Axion Biosystems). All data reflect well-wide averages from active electrodes, with the number of wells per condition represented by n values. Briefly, the active electrode was defined as having ≥5 spikes/min. The weighted Mean Firing Rate (Hz) was calculated as the total number of spikes divided by the number of active electrodes over a recording duration (300 s). A single electrode burst detector was set to detect the inter-spike interval (ISI) threshold with ≥5 spikes in a maximum of 100 ms ISI. Burst frequency was calculated as the total number of single-electrode bursts divided by the recording duration (300 s). The network burst detector was set to >50 spikes with a maximum of 100 ms of ISI, with >35% of active electrodes involved in bursting. The network burst rate was calculated as the total number of network bursts divided by the recording duration (300 s). The processed data were exported as a comma format.csv file. For comparison (i.e., time-course, control-patient pair), the.csv file was uploaded to the Axion Metric Plotting Tool. The summarized.csv file with parameters was exported for statistical analysis.
Calcium imaging
Calcium images was performed and analyzed as described previously (Avazzadeh et al., 2019).
Statistical analyses
Statistical analyses were performed with GraphPad Prism version 9.3.1 using paired or unpaired Student’s t test with a level of significance set at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. N represents the total number of cell lines; n is the number of independent experiments. Data are presented as mean ± SD or mean ± SEM.
Author contributions
Conceptualization: S.S., O.H., T.O., M.Y., and D.C.H.; experimental investigation: S.S., M.Y., and M.L.; data analysis: M.Y., M.L.G.Y., Y.L., and G.L.; Whole-cell patch-clamp and calcium imaging: Y.S., S.A., and L.Q.; original draft: M.Y. and M.L.; revision: S.S., O.H., T.O., D.C.H., G.Y., Y.L., G.L., and L.Q.
Acknowledgments
We acknowledge volunteers for participating in this study. This research was supported by SFI Investigator award (13/IA/1787) and Center Grant Number 16/RC/3948, which was co-funded under the European Regional Development Fund and by FutureNeuro industry partners, Galway University Foundation, and the One Hundred Person Project of Hebei Province (E2019050021). This research was supported by the HRB-Clinical Research Facility Galway, a unit of the University of Galway and Saolta University Health Care Group, and with scientific and technical assistance of the NCBES Genomics Facility and the Center for Microscopy & Imaging, which are funded by the University of Galway and the Irish Government's Programme for Research in Third Level Institutions, Cycles 4/5, National Development Plan 2007–2013. The study was approved by the Clinical Research Ethics Committee of Galway University Hospitals (C.A.750).
Conflict of interests
The authors declare no competing interests.
Published: August 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.07.004.
Contributor Information
David C. Henshall, Email: dhenshall@rcsi.ie.
Orla Hardiman, Email: hardimao@tcd.ie.
Sanbing Shen, Email: sanbing.shen@universityofgalway.ie.
Supplemental information
Data and code availability
No large datasets were generated in this study. No original code was generated in this study.
References
- Andrews M.G., Kong J., Novitch B.G., Butler S.J. New perspectives on the mechanisms establishing the dorsal-ventral axis of the spinal cord. Curr. Top. Dev. Biol. 2019;132:417–450. doi: 10.1016/bs.ctdb.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avazzadeh S., McDonagh K., Reilly J., Wang Y., Boomkamp S.D., McInerney V., Krawczyk J., Fitzgerald J., Feerick N., O'Sullivan M., et al. Increased Ca(2+) signaling in NRXN1α (+/-) neurons derived from ASD induced pluripotent stem cells. Mol. Autism. 2019;10:52. doi: 10.1186/s13229-019-0303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avazzadeh S., Quinlan L.R., Reilly J., McDonagh K., Jalali A., Wang Y., McInerney V., Krawczyk J., Ding Y., Fitzgerald J., et al. NRXN1α(+/-) is associated with increased excitability in ASD iPSC-derived neurons. BMC Neurosci. 2021;22:56. doi: 10.1186/s12868-021-00661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shushan E., Feldman E., Reubinoff B.E. Notch signaling regulates motor neuron differentiation of human embryonic stem cells. Stem Cells. 2015;33:403–415. doi: 10.1002/stem.1873. [DOI] [PubMed] [Google Scholar]
- Bianchi F., Malboubi M., Li Y., George J.H., Jerusalem A., Szele F., Thompson M.S., Ye H. Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling. Stem Cell Res. 2018;32:126–134. doi: 10.1016/j.scr.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Borghese L., Dolezalova D., Opitz T., Haupt S., Leinhaas A., Steinfarz B., Koch P., Edenhofer F., Hampl A., Brüstle O. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cell s. 2010;28:955–964. doi: 10.1002/stem.408. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y., Anaby D., Morozov D. Diffusion MRI of the spinal cord: from structural studies to pathology. NMR Biomed. 2017;30:e3592. doi: 10.1002/nbm.3592. [DOI] [PubMed] [Google Scholar]
- Crawford T.Q., Roelink H. The notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev. Dynam. 2007;236:886–892. doi: 10.1002/dvdy.21083. [DOI] [PubMed] [Google Scholar]
- Cunningham T.J., Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015;16:110–123. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cykowski M.D., Powell S.Z., Appel J.W., Arumanayagam A.S., Rivera A.L., Appel S.H. Phosphorylated TDP-43 (pTDP-43) aggregates in the axial skeletal muscle of patients with sporadic and familial amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 2018;6:28. doi: 10.1186/s40478-018-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz B.M., Ding Y., McInerney V., Krawczyk J., Lu Y., Yang G., Qian X., Li W., Howard L., Allen N.M., et al. Derivation of two iPSC lines from a sporadic ASD patient (NUIGi033-A) and a paternal control (NUIGi034-A) Stem Cell Res. 2020;44 doi: 10.1016/j.scr.2020.101722. [DOI] [PubMed] [Google Scholar]
- De Robertis E.M. Spemann's organizer and self-regulation in amphibian embryos. Nat. Rev. Mol. Cell Biol. 2006;7:296–302. doi: 10.1038/nrm1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin A.C., Burr K., Borooah S., Foster J.D., Cleary E.M., Geti I., Vallier L., Shaw C.E., Chandran S., Miles G.B. Human iPSC-derived motoneurons harbouring TARDBP or C9ORF72 ALS mutations are dysfunctional despite maintaining viability. Nat. Commun. 2015;6:5999. doi: 10.1038/ncomms6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.W., Chen H., Liu H., Lu J., Qian K., Huang C.L., Zhong X., Fan F., Zhang S.C. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 2015;6:6626. doi: 10.1038/ncomms7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleep A., Shefner J. Electrodiagnosis of motor neuron disease. Phys. Med. Rehabil. Clin. N. Am. 2013;24:139–151. doi: 10.1016/j.pmr.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Fujimori K., Ishikawa M., Otomo A., Atsuta N., Nakamura R., Akiyama T., Hadano S., Aoki M., Saya H., Sobue G., Okano H. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 2018;24:1579–1589. doi: 10.1038/s41591-018-0140-5. [DOI] [PubMed] [Google Scholar]
- Ge N., Liu M., Zhu X., Krawczyk J., McInerney V., Shen S., O'Brien T., Prendiville T. Generation and characterization of two induced pluripotent stem cell lines (NUIGi038-A, NUIGi038-B) from dermal fibroblasts of a healthy individual. Stem Cell Res. 2020;49 doi: 10.1016/j.scr.2020.101996. [DOI] [PubMed] [Google Scholar]
- Grolez G., Kyheng M., Lopes R., Moreau C., Timmerman K., Auger F., Kuchcinski G., Duhamel A., Jissendi-Tchofo P., Besson P., et al. MRI of the cervical spinal cord predicts respiratory dysfunction in ALS. Sci. Rep. 2018;8:1828. doi: 10.1038/s41598-018-19938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Sivalingam D., Hain S., Makkar C., Sosa E., Clark A., Butler S.J. Deriving Dorsal Spinal Sensory Interneurons from Human Pluripotent Stem Cells. Stem Cell Rep. 2018;10:390–405. doi: 10.1016/j.stemcr.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman O., Al-Chalabi A., Chio A., Corr E.M., Logroscino G., Robberecht W., Shaw P.J., Simmons Z., van den Berg L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers. 2017;3 [Google Scholar]
- Jessell T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kalani M.Y.S., Cheshier S.H., Cord B.J., Bababeygy S.R., Vogel H., Weissman I.L., Palmer T.D., Nusse R. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc. Natl. Acad. Sci. USA. 2008;105:16970–16975. doi: 10.1073/pnas.0808616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai K., Kuwabara S., Misawa S., Tamura N., Ogawara K., Nakata M., Sawai S., Hattori T., Bostock H. Altered axonal excitability properties in amyotrophic lateral sclerosis: impaired potassium channel function related to disease stage. Brain. 2006;129:953–962. doi: 10.1093/brain/awl024. [DOI] [PubMed] [Google Scholar]
- Koehler K.R., Tropel P., Theile J.W., Kondo T., Cummins T.R., Viville S., Hashino E. Extended passaging increases the efficiency of neural differentiation from induced pluripotent stem cells. BMC Neurosci. 2011;12:82. doi: 10.1186/1471-2202-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J.J., Schonewille M., Siddique T., Schults A.N.A., Fu R., Bär P.R., Anelli R., Heckman C.J., Kroese A.B.A. Hyperexcitability of Cultured Spinal Motoneurons From Presymptomatic ALS Mice. J. Neurophysiol. 2004;91:571–575. doi: 10.1152/jn.00665.2003. [DOI] [PubMed] [Google Scholar]
- Lee S., Cuvillier J.M., Lee B., Shen R., Lee J.W., Lee S.K. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc. Natl. Acad. Sci. USA. 2012;109:3383–3388. doi: 10.1073/pnas.1114515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Lee B., Ruiz E.C., Pfaff S.L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q.R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C.D., Rowitch D.H. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Maury Y., Côme J., Piskorowski R.A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C., Nedelec S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015;33:89–96. doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- Muhr J., Graziano E., Wilson S., Jessell T.M., Edlund T. Convergent inductive signals specify midbrain, hindbrain, and spinal cord identity in gastrula stage chick embryos. Neuron. 1999;23:689–702. doi: 10.1016/s0896-6273(01)80028-3. [DOI] [PubMed] [Google Scholar]
- Muñoz-Sanjuán I., Brivanlou A.H. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- Naujock M., Stanslowsky N., Bufler S., Naumann M., Reinhardt P., Sterneckert J., Kefalakes E., Kassebaum C., Bursch F., Lojewski X., et al. 4-Aminopyridine Induced Activity Rescues Hypoexcitable Motor Neurons from Amyotrophic Lateral Sclerosis Patient-Derived Induced Pluripotent Stem Cells. Stem cells (Dayton, Ohio) 2016;34:1563–1575. doi: 10.1002/stem.2354. [DOI] [PubMed] [Google Scholar]
- Niedermeyer S., Murn M., Choi P.J. Respiratory Failure in Amyotrophic Lateral Sclerosis. Chest. 2019;155:401–408. doi: 10.1016/j.chest.2018.06.035. [DOI] [PubMed] [Google Scholar]
- Nordström U., Jessell T.M., Edlund T. Progressive induction of caudal neural character by graded Wnt signaling. Nat. Neurosci. 2002;5:525–532. doi: 10.1038/nn0602-854. [DOI] [PubMed] [Google Scholar]
- Novitch B.G., Chen A.I., Jessell T.M. Coordinate Regulation of Motor Neuron Subtype Identity and Pan-Neuronal Properties by the bHLH Repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ogura T., Sakaguchi H., Miyamoto S., Takahashi J. Three-dimensional induction of dorsal, intermediate and ventral spinal cord tissues from human pluripotent stem cells. Development. 2018;145:dev162214. doi: 10.1242/dev.162214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patani R. Generating Diverse Spinal Motor Neuron Subtypes from Human Pluripotent Stem Cells. Stem Cells Int. 2016;2016:1036974. doi: 10.1155/2016/1036974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patani R., Hollins A.J., Wishart T.M., Puddifoot C.A., Alvarez S., de Lera A.R., Wyllie D.J.A., Compston D.A.S., Pedersen R.A., Gillingwater T.H., et al. Retinoid-independent motor neurogenesis from human embryonic stem cells reveals a medial columnar ground state. Nat. Commun. 2011;2:214. doi: 10.1038/ncomms1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzig M., Bochmann K., Lutz J., Stahl R., Küpper C., Liebig T., Reilich P., Dieterich M. Measurement of structural integrity of the spinal cord in patients with amyotrophic lateral sclerosis using diffusion tensor magnetic resonance imaging. PLoS One. 2019;14 doi: 10.1371/journal.pone.0224078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peljto M., Dasen J.S., Mazzoni E.O., Jessell T.M., Wichterle H. Functional Diversity of ESC-Derived Motor Neuron Subtypes Revealed through Intraspinal Transplantation. Cell Stem Cell. 2010;7:355–366. doi: 10.1016/j.stem.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peljto M., Wichterle H. Programming embryonic stem cells to neuronal subtypes. Curr. Opin. Neurobiol. 2011;21:43–51. doi: 10.1016/j.conb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Dasen J.S. Hox Genes: Choreographers in Neural Development, Architects of Circuit Organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieri M., Albo F., Gaetti C., Spalloni A., Bengtson C.P., Longone P., Cavalcanti S., Zona C. Altered excitability of motor neurons in a transgenic mouse model of familial amyotrophic lateral sclerosis. Neurosci. Lett. 2003;351:153–156. doi: 10.1016/j.neulet.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Portaro S., Morini E., Santoro M.E., Accorinti M., Marzullo P., Naro A., Calabrò R.S. Breathlessness in amyotrophic lateral sclerosis: A case report on the role of osteoporosis in the worsening of respiratory failure. Medicine. 2018;97 doi: 10.1097/MD.0000000000013026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q., Li D., Louis K.R., Li X., Yang H., Sun Q., Crandall S.R., Tsang S., Zhou J., Cox C.L., et al. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat. Commun. 2014;5:3449. doi: 10.1038/ncomms4449. [DOI] [PubMed] [Google Scholar]
- Roelink H., Porter J.A., Chiang C., Tanabe Y., Chang D.T., Beachy P.A., Jessell T.M. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- Saade M., Gutiérrez-Vallejo I., Le Dréau G., Rabadán M.A., Miguez D.G., Buceta J., Martí E. Sonic Hedgehog Signaling Switches the Mode of Division in the Developing Nervous System. Cell Rep. 2013;4:492–503. doi: 10.1016/j.celrep.2013.06.038. [DOI] [PubMed] [Google Scholar]
- Sagner A., Briscoe J. Establishing neuronal diversity in the spinal cord: a time and a place. Development. 2019;146:dev182154. doi: 10.1242/dev.182154. [DOI] [PubMed] [Google Scholar]
- Sagner A., Gaber Z.B., Delile J., Kong J.H., Rousso D.L., Pearson C.A., Weicksel S.E., Melchionda M., Mousavy Gharavy S.N., Briscoe J., Novitch B.G. Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. PLoS Biol. 2018;16:e2003127. doi: 10.1371/journal.pbio.2003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R., Pfaff S.L. Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Stifani N. Motor neurons and the generation of spinal motor neuron diversity. Front. Cell. Neurosci. 2014;8:293. doi: 10.3389/fncel.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros M.A., Lim R., Hughes D.I., Brichta A.M., Callister R.J. Electrical maturation of spinal neurons in the human fetus: comparison of ventral and dorsal horn. J. Neurophysiol. 2015;114:2661–2671. doi: 10.1152/jn.00682.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- van Zundert B., Peuscher M.H., Hynynen M., Chen A., Neve R.L., Brown R.H., Constantine-Paton M., Bellingham M.C. Neonatal Neuronal Circuitry Shows Hyperexcitable Disturbance in a Mouse Model of the Adult-Onset Neurodegenerative Disease Amyotrophic Lateral Sclerosis. J. Neurosci. 2008;28:10864–10874. doi: 10.1523/JNEUROSCI.1340-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic S., Nicholson G.A., Kiernan M.C. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain. 2008;131:1540–1550. doi: 10.1093/brain/awn071. [DOI] [PubMed] [Google Scholar]
- Wainger B.J., Kiskinis E., Mellin C., Wiskow O., Han S.S.W., Sandoe J., Perez N.P., Williams L.A., Lee S., Boulting G., et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Xu H.J., Yao Y., Yao F., Chen J., Li M., Yang X., Li S., Lu F., Hu P., He S., et al. Generation of functional posterior spinal motor neurons from hPSCs-derived human spinal cord neural progenitor cells. Cell Regen. 2023;12:15. doi: 10.1186/s13619-023-00159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Liu M., Ding Y., Vajda A., Ma J., Cui H., O'Brien T., Henshall D., Hardiman O., Shen S. Generation of twelve induced pluripotent stem cell lines from two healthy controls and two patients with sporadic amyotrophic lateral sclerosis. Stem Cell Res. 2020;44:101752. doi: 10.1016/j.scr.2020.101752. [DOI] [PubMed] [Google Scholar]
- Yang M., Liu M., Vajda A., O'Brien T., Henshall D., Hardiman O., Shen S. Generation of six induced pluripotent stem cell (iPSC) lines from two patients with amyotrophic lateral sclerosis (NUIGi043-A, NUIGi043-B, NUIGi043-C, NUIGi044-A, NUIGi044-B, NUIGi044-C) Stem Cell Res. 2019;40:101558. doi: 10.1016/j.scr.2019.101558. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Anderson D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No large datasets were generated in this study. No original code was generated in this study.