Summary
Adherens junctions (AJs) provide adhesive properties through cadherins and associated cytoplasmic catenins and participate in morphogenetic processes. We examined AJs formed between ISL1+ cardiovascular progenitor cells during differentiation of embryonic stem cells (ESCs) in vitro and in mouse embryogenesis in vivo. We found that, in addition to N-CADHERIN, a percentage of ISL1+ cells transiently formed vascular endothelial (VE)-CADHERIN-mediated AJs during in vitro differentiation on days 4 and 5, and the same pattern was observed in vivo. Fluorescence-activated cell sorting (FACS) analysis extended morphological data showing that VE-CADHERIN+/ISL1+ cells constitute a significant percentage of cardiac progenitors on days 4 and 5. The VE-CADHERIN+/ISL1+ cell population represented one-third of the emerging FLK1+/PDGFRa+ cardiac progenitor cells (CPCs) for a restricted time window (days 4–6). Ablation of VE-CADHERIN during ESC differentiation results in severe inhibition of cardiac differentiation. Disruption of all classic cadherins in the VE-CADHERIN+ population via a cadherin dominant-negative mutant’s expression resulted in a dramatic decrease in the ISL1+ population and inhibition of cardiac differentiation.
Keywords: Adherens Junctions, Cadherins, Cardiac progenitors, ISL1, VE-CADHERIN, N-CADHERIN, MEF2C, FLK1, Embryonic Stem Cells
Highlights
-
•
Early ISL+ cells form AJs mediated by N-, E-, and transient VE-cadherins
-
•
VE-CADHERIN is transiently expressed in ISL1+/FLK1+/PDGFRa+ cardiac progenitors
-
•
VE-CADHERIN ablation results in severe inhibition of cardiac differentiation
Kouklis and colleagues examined adherens junctions mediated by N-, VE-, and E-cadherins in cardiac progenitors during early mesodermal morphogenesis. Using mouse embryonic stem cells and mouse embryos, they identified transient VE-cadherin expression in one-third of ISL1+/FLK1+/PDGFRa+ cardiac progenitor cells. VE-CADHERIN ablation by CRISPR led to cardiac differentiation inhibition because of a reduction in early cardiac progenitors.
Introduction
Among the morphological changes taking place during post-implantation embryogenesis, cell-cell adhesion structures with different properties form to serve structural and functional requirements of emerging and differentiating cell populations. Adherens junctions (AJs) are cell-cell adhesion structures pivotal for morphogenetic processes, lineage specification, and proliferation. They provide mechanical support to forming tissues while also being involved in segregation of various cell types emerging during differentiation (Harris and Tepass, 2010). Classic cadherins and associated catenins are AJ components providing strong adhesion and facilitate signaling cues in proliferation and migration by direct or indirect association with the actin cytoskeleton and signaling receptors and their effectors. E-, N-, and VE-cadherins are typical members of the classic cadherins subfamily, predominantly expressed in epithelial, neuronal, and endothelial tissues, respectively (Liang and Yap, 2015). Differentiation of epiblast into mesodermal cells during gastrulation onset is an EMT (epithelial-to-mesenchymal transition) process characterized by an E- to N-CADHERIN switch (Acloque et al., 2009; Bardot and Hadjantonakis, 2020). N-CADHERIN is the first cadherin expressed in mesoderm after epiblast cells undergo EMT during gastrulation (Acloque et al., 2009), and it has been shown to be essential for cardiovascular system formation (Kostetskii et al., 2005; Radice et al., 1997). The heart is formed from two distinct progenitor populations, called the first and second heart field (FHF and SHF, respectively), derived from a common ancestor and characterized by expression of lineage-specific transcription factors (Meilhac and Buckingham, 2018). FHF progenitors are derived from the anterior splanchnic mesoderm and form the cardiac crescent, which later contributes to the left ventricle and inflow tract (IFT). SHF progenitors are derived from the pharyngeal mesoderm and contribute mainly to the right ventricle and the outflow tract (OFT). Of interest, ISL1 is a hallmark of SHF emergence, with ISL1+ cells differentiating further into cardiomyocytes and endothelial, endocardial, and smooth muscle cells.
There is little information about the nature and the role of AJs between cardiovascular progenitors before their specification into mature phenotypes. SHF cells, however, display epithelial characteristics and form an atypical, apicobasally polarized, epithelium-like cell monolayer in the dorsal pericardial wall, expressing E-CADHERIN on embryonic day 9.5 (E9.5) (Francou et al., 2014). In other studies, it was found that N-CADHERIN is involved in cardiomyocyte specification. Disruption of N-CADHERIN expression in MEF2C+ anterior heart field (AHF) cardiac progenitor cells (CPCs) results in low proliferation and premature differentiation of the ISL1+/MEF2C+ SHF subset, causing embryonic lethality around E9.5–E10.5, and this phenotype can be partially reversed by Wnt pathway activation (Soh et al., 2014). N-CADHERIN AJs have also been detected in cardiomyocyte progenitors during embryonic stem cell (ESC) differentiation, and they can be detected in ISL1 progenitors during mouse heart development on E9.5 (Honda et al., 2006). VE-CADHERIN is an endothelial AJ molecule and plays a crucial role in vasculogenesis and angiogenesis (Szymborska and Gerhardt, 2018). It is well established that VE-CADHERIN is indispensable for endothelial cell-cell adhesion regulation and vascular endothelial barrier function in the adult (Komarova et al., 2017). It also participates in signaling pathways by association and activation of intracellular partners (Maltabe and Kouklis, 2022). At the onset of endothelial differentiation, VE-CADHERIN is expressed in a subset of the early mesodermal FLK1+ cell population that differentiates further into hematopoietic and endothelial lineages (Nikolova-Krstevski et al., 2008). VE-CADHERIN ablation is lethal in the mouse embryo on E9.5 because of defects in endothelial cell survival and vascular remodeling. Interestingly, abnormal cardiac development has been observed in VE-CADHERIN mutant mice, possibly because of endocardial cell disorganization (Carmeliet et al., 1999).
Here we studied, side by side, the expression pattern of VE-, N-, and E-cadherins at the onset of cardiac differentiation in the ISL1+ cell populations using an ESC differentiation system. We report that the majority of ISL1+ cells formed extensive N-CADHERIN AJs at the onset of their emergence during ESC differentiation on day 4. However, E- and VE-CADHERIN AJs were detected in a fraction of N-CADHERIN+/ISL1+ cells on days 4 and 5, and surprisingly distinct VE- and E-CADHERIN AJs formed in the same ISL1+ cell. Fluorescence-activated cell sorting (FACS) analysis demonstrated that a significant percentage of PDGFRa+/FLK1+/ISL1+ SHF cardiac progenitors are VE-CADHERIN+ at that time window. Ablation of VE-CADHERIN in ESCs by CRISPR negatively affected cardiac progenitor specification and terminal differentiation. Expression of an AJ’s dominant-negative mutant under the ve-cadherin promoter resulted in ISL1+ population elimination during ESC differentiation in vitro. Consistent with this, VE-CADHERIN AJs formed transiently in cardiac progenitors during mouse cardiac differentiation on E8.5.
Results
Early ISL1+ cells form AJs mediated by N-, E-, and transient VE-cadherins during ESC differentiation
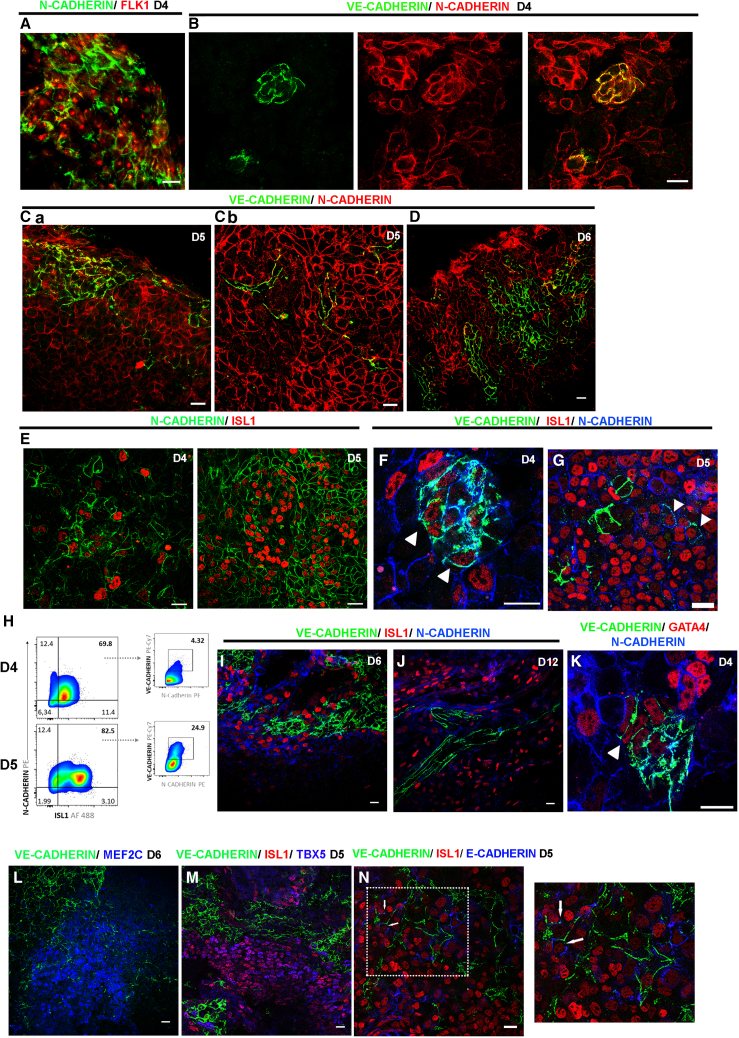
We examined, side by side, the expression patterns of N- and VE-CADHERIN between day 4 and day 6 during ESCs differentiation. N-CADHERIN was expressed in FLK1+cells on day 4 (Figure 1A), and at the same time, VE-CADHERIN+ cells seem to emerge in a subset of the N-CADHERIN+ population co-expressing both cadherins (Figure 1B). At this stage, N-CADHERIN was uniformly distributed, whereas VE-CADHERIN formed distinct punctate AJs. On day 5, the VE-CADHERIN+ population increased (Figure 1Ca); however, co-localization between VE- and N-CADHERIN was observed rarely in cells, surrounded by N-CADHERIN+/VE-CADHERIN− cells (Figure 1Cb). VE-CADHERIN+ population expanded further on day 6, but from this time on, N- and VE-cadherins were expressed in a mutually exclusive manner (Figure 1D).
Figure 1.
N- and VE-CADHERIN AJs during early mesodermal differentiation
(A) N-CADHERIN AJs between FLK1+ cells in EBs on day 4.
(B–D) VE- and N-CADHERIN AJs on days 4, 5, and 6 show co-expression of these cadherins in small cell clusters predominantly on days 4 and 5 (yellow).
(E–J) AJs between ISL1+ cells are mediated by N-cadherin and transient VE-CADHERIN. (E) The majority of ISL1+ cells form N-CADHERIN AJs on days 4 and 5. (F) Small ISL1+ cell clusters form VE- in addition to N-CADHERIN AJs on day 4 (arrowheads). (G and I) VE-CADHERIN AJs in ISL1+ cells form on rare occasions on day 5 (arrowhead) and day 6. Note that high VE-CADHERIN expressors did not express ISL1 (arrow). (H) Flow cytometry analysis (pseudocolor dot plots) for expression of N-CADHERIN and ISL1 in the total population of EBs (left panel) and analysis of VE-CADHERIN in N-CADHERIN+/ISL1+ cells (right panel) on days 4 and 5. Numbers in dot plots represent the percentage of frequencies of cell populations in the quadrants. Blue to red colors represent a low to high cell frequency, respectively. (I and J) VE-CADHERIN AJs in ISL1+ cells on days 6 and 12.
(K–M) VE-CADHERIN AJs in CPCs. (K) Small GATA4+ cell clusters form N- and VE-CADHERIN AJs on day 4 (arrowhead). (L) VE-CADHERIN AJs were formed in MEF2c+ cells. (M) VE-CADHERIN AJs were detected on rare occasions in TBX5+/ISL1+ cells (arrowhead).
(N) E-CADHERIN AJs were formed in some ISL1+cells. On rare occasions, VE- and E-CADHERIN were detected in the same ISL1+ cells, but these cadherins were not co-localized (magnification inset).
See also Figure S1 for separate immunostaining for (I)–(K), (M), and (N). In (B), (Cb), (E)–(G), and (K), EBs were dissociated by mild trypsinization on day 3, and cells were attached on fibronectin and grown as a monolayer. Scale bars, 20 μm. All experiments were performed at least three times, and representative data are shown.
Next, we examined N- and VE-CADHERIN expression in ISL1+ cardiac progenitors at the onset of their appearance. ISL1+cells expressed predominantly N-CADHERIN on days 4 and 5 (Figure 1E), but, surprisingly, a subset of them was VE-CADHERIN+ as well (Figures 1F and 1G). Triple staining of ISL1 and N- and VE-CADHERIN on days 4 and 5 captured a developmental stage where an ISL1+ cell subpopulation formed AJs mediated by both cadherins in the same adhesion structure (Figures 1F and 1G). We quantified the co-expression of N-CADHERIN, VE-CADHERIN, and ISL1 in embryoid bodies (EBs) on days 4 and 5 with flow cytometry. This analysis revealed that the percentages of VE-CADHERIN+ cells were 4.3% (day 4) and 24.9% (day 5) within N-CADHERIN+/ISL1+ cells (Figure 1H). VE-CADHERIN/ISL1 co-expressing cells, although limited, were also found on day 6 (Figure 1I) but not on day 12 (Figure 1J). VE-CADHERIN was also detected in a subset of GATA4+ progenitors on day 4 (Figure 1K) as well as in the MEF2C+ cell population on day 6 (Figure 1L). Next, we examined VE-CADHERIN AJs in the FHF TBX5+ cell population on day 5 and found that they formed in very rare cases (Figure 1M). Because it has been reported that SHF cells display epithelial characteristics, we examined the possibility of E-CADHERIN AJs in ISL1+ cells on day 5. Unexpectedly, we found that distinct E- and VE-CADHERIN AJs formed in the same ISL1+ cells (Figure 1N).
VE-CADHERIN expression in CPCs
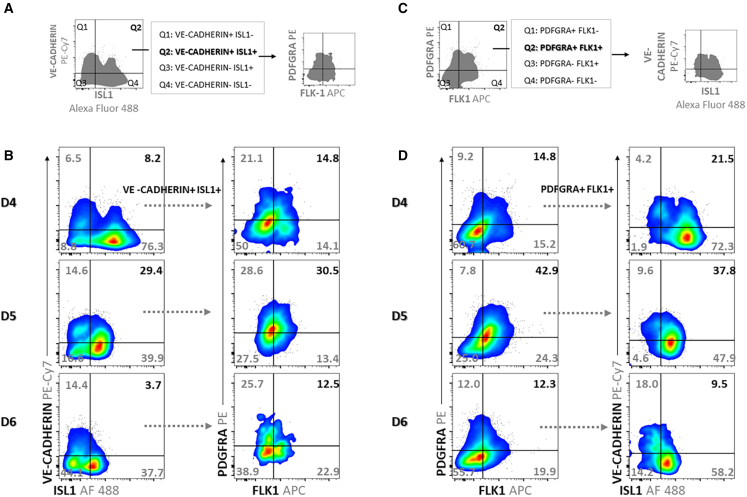
Immunostaining demonstrated that VE-CADHERIN AJs formed in a limited number of cells. However, VE-CADHERIN could be undetectable in more cells by immunofluorescence, in the absence of mature AJ formation. To assess progenitor cell populations where VE-CADHERIN was expressed during early cardiac differentiation in vitro, we employed two surface markers known to tag CPCs, PDGFRa and FLK1, along with the SHF marker ISL1 in FACS analysis. Cells expressing ISL1 and VE-CADHERIN were detected on day 4 up to day 6, peaking on day 5 before showing a vast decrease on day 6 (Figure 2). In more detail, VE-CADHERIN+/ISL1+ cells were 8.2% on day 4, 29.4% on day 5, and 3.7% on day 6. We then quantified CPCs (annotated as FLK1+/PDGFRa+; Figure 2A) and found that their respective percentages were 14.8%, 30.5%, and 12.5%. We also performed an alternative analysis where we measured the percentages of VE-CADHERIN+ cells within the ISL1+/FLK1+/PDGFRa+ CPC population on days 4–6 (Figures 2C and 2D). We found that VE-CADHERIN was expressed in 21.5%, 37.8%, and 9.5%, respectively.
Figure 2.
Flow cytometry analysis of EBs on days 4, 5, and 6
(A and C) Schematic of the gating strategy.
(B) Dot plots displaying expression of VE-CADHERIN and ISL1 in total cells (left panel) and expression of PDGFRa and FLK1 in VE-CADHERIN+/ISL1+ cells (quadrant 2 [Q2] in A, right panel) on days 4, 5, and 6. (B) Density dot plots displaying expression of PDGFRa and FLK1 in total cells (left panel) and expression of VE-CADHERIN and ISL1 in PDGFRa+/FLK1+ cells (Q2 in C, right panel) on days 4, 5, and 6. Numbers in dot plots represent the percentage of frequencies of cell populations in the quadrants (Q1–Q4).
Blue to red color in dot plots (pseudocolor) represents low to high cell frequency, respectively. Representative data from three independent experiments are presented.
Ablation of VE-CADHERIN in ESCs results in cardiac deficiency during in vitro differentiation
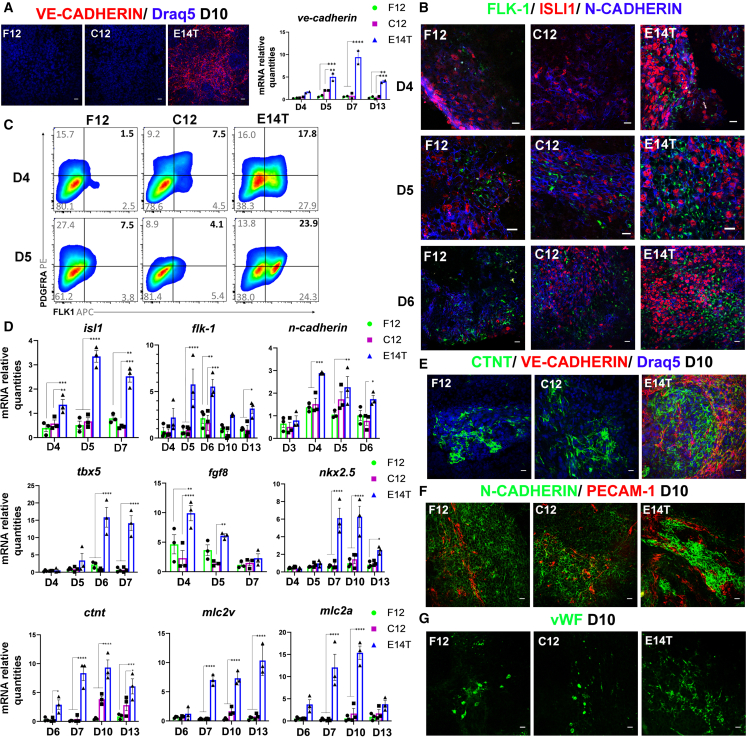
To investigate the role of VE-CADHERIN AJs upon early ESC differentiation, the CRISPR-Cas9 system was used to generate a functional knockout-causing deletion in E14T cells. Ninety clones were isolated, and 30 clones were tested for VE-CADHERIN expression on day 7 by immunofluorescence. Genomic DNA was isolated from 14 clones with no VE-CADHERIN staining, the genomic locus disruption was verified by sequencing (Figure S3A), and homozygous clones C12 and F12 (Figure S3B) were used for further analysis. Embryoid bodies from these clones formed normally (data not shown), and VE-CADHERIN was undetectable in knockout (KO) cells on day 10 by immunofluorescence and by qPCR on days 4, 5, 7, and 13 (Figure 3A). The differentiation potential of the C12 and F12 clones was tested further in vitro by immunofluorescence, qPCR, and FACS analysis. Interestingly, the ISL1+ and FLK1+ populations seemed reduced at the onset of their emergence, on day 4, as well as on days 5 and 6 (Figure 3B). qPCR analysis showed that isl1 and flk1 expression was decreased approximately 3-fold on day 5 (Figure 3D). At earlier stages of differentiation, brachyury and n-cadherin expression was only slightly affected compared with the control (Figure S3C and S3D), but mesp-1 expression was unexpectedly reduced 9-fold in the C12 KO clone and 13-fold in the F12 KO clone compared with the control (Figure S3C). Further qPCR analysis for cardiac developmental markers revealed that tdx5 mRNA expression levels were reduced approximately 8-fold on days 6 and 7, and fgf8, a marker of anterior SHF (aSHF), and nkx2.5 were moderately affected (Figure 3D). MEF2c+, TBX5+, as well as GATA4+ cells were also reduced on days 5 and 6 in KO clones, as indicated by immunofluorescent analysis (Figure S3D–S3F). Relevant to the VE-CADHERIN expression pattern (Figure 2), the PDGFRa+/FLK1+population was substantially reduced in KO clones on days 4 and 5, as shown by FACS analysis (Figure 3C). Cardiac differentiation at later stages was affected in KO clones, as evident by the absence or severe reduction of beating EBs on day 13 (0% in clone C12, 30% in clone F12), in contrast to E14T EBs (90%) under the same conditions (Figure S3G). Also, cTNT+ cells were significantly decreased on day 10 (Figure 3E). In accordance cTNT expression levels were clearly reduced, whereas MLC2v and MLC2a expression was almost lost (Figure 3D). Formation of the endothelium was assessed in KO clones on day 10, when extensive endothelial networks form, using PECAM-1 and von Willebrand factor (vWF) as endothelial markers. Interestingly, we found that, although both proteins were expressed, PECAM-1 was mostly cytoplasmic, and vWF was localized in large aggregates instead of typical Palade bodies (Figures 3F and 3G).
Figure 3.
Impaired differentiation of VE-cadherin-KO ESCs towards cardiac and endothelial cells
(A) VE-CADHERIN AJs in KO and E14T clones on day 10 shown by immunofluorescence and qPCR on days 4–13. Note the absence of VE-CADHERIN in KO EBs.
(B) Immunofluorescence staining in KO and E14T EBs showing downregulation of ISL1 and FLK1 but not N-CADHERIN.
(C) Flow cytometry analysis (pseudocolor dot plots) for expression of PDGFRa and FLK1 in VE-CADHERIN KO EBs on days 4 and 5 compared with control EBs.
(D) isl1, flk1, n-cadherin, tbx5, fgf8, nkx2.5, ctnt, mlc2v, and mlc2a mRNA expression levels in KO and control EBs quantified by real-time qPCR during the indicated differentiation days (three independent biological experiments, n = 3). Data represent mean ± SEM. The statistical significance of difference was determined by two-way ANOVA with Dunnett’s multiple comparisons as post hoc analysis.
(E) cTNT and VE-CADHERIN expression in KO and control EBs on day 10.
(F and G) Immunostaining for the endothelial markers PECAM-1 and vWF on day 10 in KO EBs compared with the control.
Scale bars, 20 μm. All experiments were performed at least three times, and representative data are shown.
Expression of a pan-cadherin dominant-negative mutant in the Pvec+ population inhibits ISL1+ cell specification
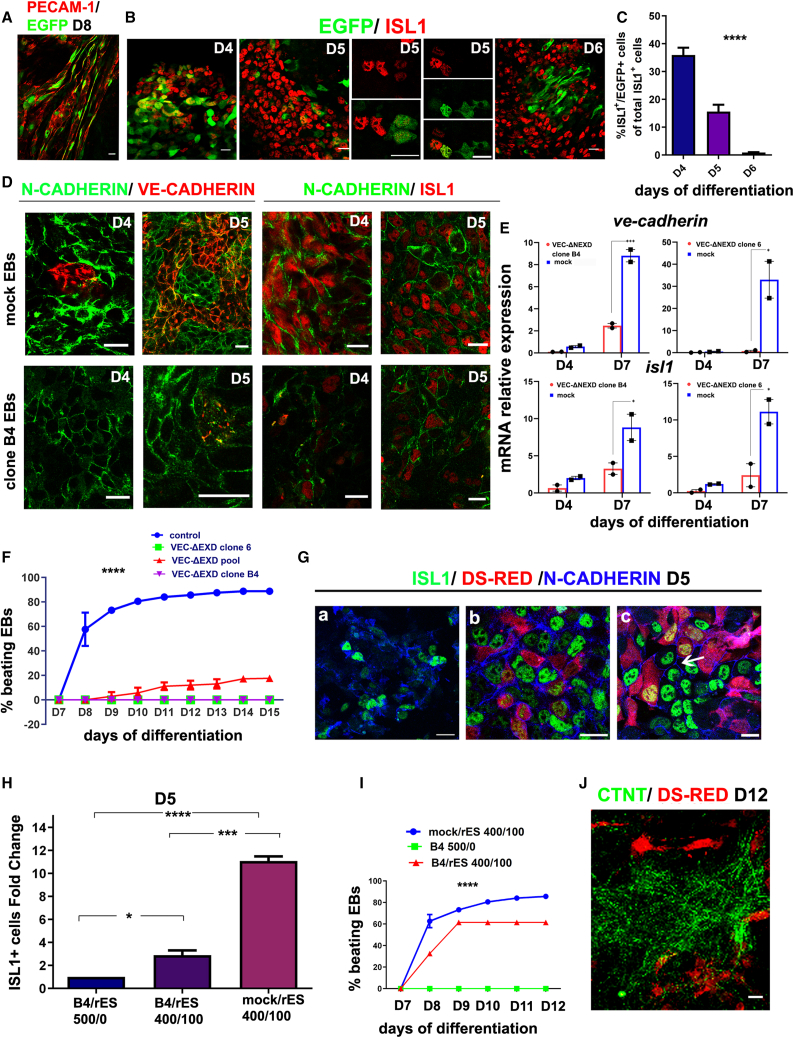
We found that VE-, N-, and E-CADHERIN AJs formed in ISL1+ cells (Figure 1). To eliminate the possibility that N-cadherin, E-cadherin, or other, still unidentified classic cadherins might compensate for VE-CADHERIN loss of function in CPCs, we stably expressed a well-characterized pan-classical cadherin dominant-negative mutant (ΔEXD-VEC; Figure S4A) under a 2.5-kb ve-cadherin promoter (Pvec) (episomal construct pPvec-ΔEXD-VEC; Figure S4B) during ESCs cardiovascular differentiation. Pvec activity was detected in PECAM-1+ and ISL1+ cells by EGFP expression (Figures 4A and 4B). On day 4, 36% of ISL1+ cells co-expressed EGFP, whereas the percentage decreased to 15% on day 5 and to less than 1% by day 6 (Figure 4C). The VE-CADHERIN mutant was expressed stably under Pvec in ESCs, and two mutant clones (6 and B4), a pool of mutant clones (pool), and a mock-transfected clone (mock) were used for further analysis.
Figure 4.
Inhibition of classical cadherins in Pvec+ cells severely affected cardiac differentiation of ESCs
(A) Pvec activity is restricted to endothelial cells on day 8, as indicated by PECAM-1/EGFP co-expression.
(B) ISL1+ cells express EGFP in EBs on days 4–6. Note that Pvec activity was high in low ISL1 expressors and, conversely, on day 5, implying that Pvec is activated transiently during ISL1+ cell specification.
(C) Statistical analysis of ISL1+/EGFP+ cells on days 4–6. Approximately 1,000 ISL1+ cells were counted per experiment. Three independent experiments were performed. ∗∗∗∗p < 0.0001.
(D) VE- and N-CADHERIN AJs in clone B4 and mock EBs on days 4 and 5. Note the absence or disruption of VE-CADHERIN, partial disruption of N-CADHERIN, and significant decrease of ISL1+ cells in clone B4 EBs compared with mock.
(E) Endogenous ve-cadherin and isl1 mRNA levels in clones B4 and 6 and mock quantified by real-time qPCR during the indicated differentiation days (n = 2).
(F) Statistical analysis of beating activity in clones B4, 6, pool, and mock during differentiation days 7–15 (an EB was considered beating when it contained one or more beating areas). The numbers of EBs counted were as follows: mock, 260; clone B4, 185; clone 6, 210; pool, 217 (three independent biological experiments, n = 3). ∗∗∗∗p < 0.0001.
(G) N-CADHERIN, dsRed, and ISL1 triple staining in EBs from clone B4 (a), mock/rES (b), and B4/rES(c) on day 5. EBs were dissociated under mild trypsinization on day 3, and cells were left to attach on fibronectin and grow as a monolayer. Note that the number of ISL1+ cells in hybrid B4/rES EBs are substantially increased compared with B4 and form extensive AJs comparable with mock/rES (arrow).
(H) ISL1+ cell fold change between mutant B4 and hybrid B4/rES and mock/rES. The number of ISL1+ cells counted in each case was derived specifically from mutant and mock ESCs and not from rES. For this experiment, 10 random fields were photographed, and approximately 400 cells per field were counted in three independent experiments. Statistical analysis shows the fold change of ISL1+ cells compared with B4 values set to 1. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(I) Statistical analysis of beating activity in hybrid EBs during differentiation days 7–12 (three independent biological experiments, n = 3). ∗∗∗p < 0.001.
(J) Rescue of cTNT expression in hybrid EBs (B4/rES) on day 12 (maximum projection of confocal image series).
Scale bars, 20 μm. In (C), the clone B4 EB day 5 N/VE-CADHERIN scale bar represents 40 μm. Statistical analysis details are described in the supplemental information. All experiments were performed at least three times, and representative data are shown.
VE-CADHERIN and N-CADHERIN AJs were severely impaired in mutant EBs on days 4 and 5, as evident by N-CADHERIN AJ disruption (Figure 4D). On day 4 and onward, the growth of mutant EBs was inhibited compared with mock EBs (Figure S4C). At the same time, VE- and N-CADHERIN AJs were significantly reduced between ISL1+ cells in mutant EBs (Figure 4D). qPCR analysis showed that ve-cadherin and isl1 levels were dramatically reduced in mutant clones on days 4–7 (Figure 4E). BRACHYURY+ cells were comparable between the mutant and mock EBs on day 4 (Figure S4D), implying that survival of BRACHYURY+ cells was not affected. MEF2c+ cells were absent in mutant EBs on day 7, whereas GATA4 was detected, possibly in non-cardiac cell types (Figure S4E). At later stages of differentiation (days 7–15), beating activity was abolished in mutant EBs, whereas it reached only 20% in pool EBs compared with around 90% in mock EBs (Figure 4F). In agreement, cTNT+ cells were absent in EBs from clones 6 and B4 and significantly reduced in pool compared with mock EBs (Figures S4F and S4G).
AJ rescue was attempted in a hybrid EB system formed between a wild-type ESC line (dsRed.MST, referred to as rES; Vintersten et al., 2004) and B4 mutant ESCs. EBs from rES, distinguished by red staining, showed beating activity similar to mock. B4-ESCs and rESCs were mixed at various cell ratios, and AJ formation was monitored. When hybrid ESCs consisting of 20% rES+ and 80% clone B4 ESCs were used for differentiation, AJ morphology was partially restored in mutant-derived ISL1+ cells on day 5 (Figure 4G). Also, the number of mutant-derived ISL1+ cells increased ∼2.2-fold in B4/rES compared with B4 EBs (Figure 4H). At later stages, cTNT+ mutant-derived cells were detected in extended areas in B4/rES EBs (Figure 4J), and beating activity of hybrid EBs was observed in 10%–30% on days 8 and 9 and ∼60% on days 10–12 compared with 0% in mutant EBs at the same time points (Figure 4I).
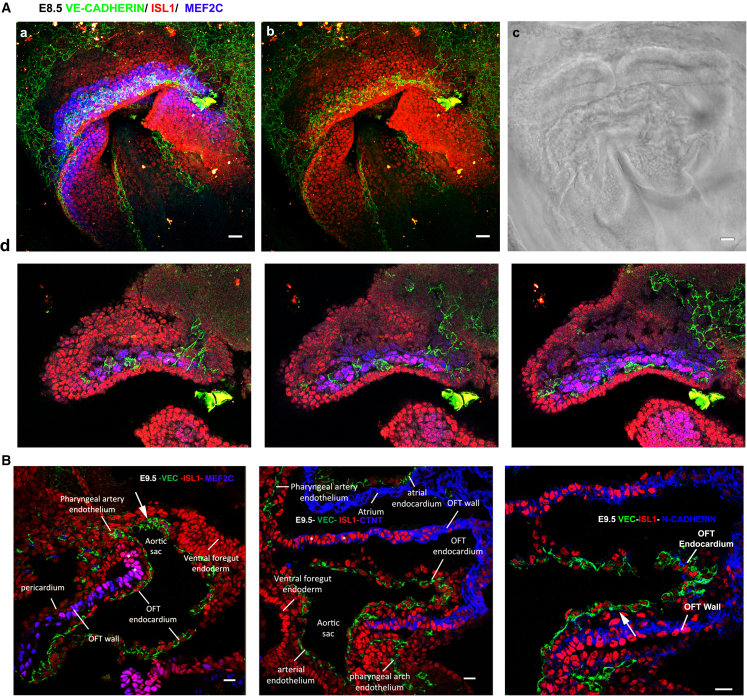
ISL1+/MEF2c+cell population forms VE-CADHERIN AJs during embryogenesis
The unexpected finding that VE-CADHERIN AJs formed transiently in ISL1+cells could mark a previously unidentified cardiac developmental stage. We performed, for the first time, a morphological analysis of ISL1+cardiovascular progenitors in mouse embryos during the developmental stages of the linear heart tube (E8.5), cardiac looping (E9.5), and after chamber formation (E11.5) with respect to VE-CADHERIN AJ formation. Confocal images taken from whole-mount embryos on E8.5 showed VE-CADHERIN junctions formation in a subset of ISL1+ cells, although AJs were punctate (Figures 5A and S6). Interestingly, a percentage of VE-CADHERIN+/ISL1+cells was also MEF2c+ (Figures 5Aa and 5Ad). Quantification in two whole-mount immunostained embryos showed that approximately 20% of MEF2c+ cells were VE-CADHERIN+ on E8.5 (details in Figures S5A). In addition, a distinct VE-CADHERIN+ cell layer emerged between the ISL1+ and MEF2c+ cell layers on E8.5 (Figure S5A; Video S1). On E9.5, we observed that ISL1+ endocardial cells formed extensive VE-CADHERIN AJs (Figure 5B, right panel, arrow) and that, on rare occasions, VE-CADHERIN AJs formed between MEF2c+ cells (Figure 5B, left panel, arrow). Interestingly, it seems that N- and VE-CADHERIN AJs formed in a mutually exclusive manner in ISL1+ cells. On E11.5, the ISL1+/VE-CADHERIN+ population diminished, and the ISL1+/N-CADHERIN+ population was found predominantly (Figure S5B). These findings demonstrate that VE-CADHERIN AJs form transiently in the ISL1+ population on E8.5 and that they represent adhesive structures of the AHF specifically on E9.5.
Figure 5.
VE-CADHERIN is expressed in a subpopulation of SHF cells during murine heart embryonic development on Ε8.5 and E9.5
(A) Stacked confocal microphotographs of a representative whole-mount embryo on E8.5 (somite stage 5) (a and b) phase-contrast image (c), and magnified areas from sections z2–z4 (c). All stacks are presented in Figure S5A and Video S1.
(B) Cryosections of heart development on E9.5, indicative of VE-CADHERIN (green) being expressed in SHF cell populations (ISL1+, red) destined to form future endocardial and pharyngeal endothelial structures. VE-CADHERIN+ cells were shown to co-localize with a subpopulation with MEF2c+ cells (left panel, blue) but not with cTNT+ (blue, center panel) cardiomyocytes or N-CADHERIN (blue, right panel). Arrows depict ISL1+/VE-CADHERIN+ cells. Scale bars, 20 μm.
Discussion
Differentiating cells are equipped with a repertoire of cadherins that determine their associations between the same or different cell types. AJs mediated by classical cadherins associate with the actin cytoskeleton via β- and α-catenin complexes to regulate cell shape as well as cytoskeletal and mechanical properties (Mege and Ishiyama, 2017). These complexes form platforms at cell-cell junctions that regulate actin cytoskeleton rearrangement and migration through activation of Rho GTPases (Braga, 2018). AJs have roles beyond just cell-type-specific glue because they are involved in cell fate decisions through interactions with signaling pathways. The E- to N-CADHERIN switch occurs in early embryogenesis and is a hallmark of EMT. Similarly, the N- to VE-CADHERIN switch takes place during endothelial-to-mesenchymal transition (EndMT) during cardiac valve formation (Alvandi and Bischoff, 2021). Surprisingly, we observed formation of distinct VE- and E-CADHERIN AJs in the same ISL1+ cell. This implies that a similar cadherin switch process might be taking place in a fraction of ISL1+/E-CADHERIN+ SHF progenitors.
In mouse embryos, ve-cadherin transcripts have been detected at the earliest stages of vascular development (E7.5) in mesodermal cells of the yolk sac mesenchyme, and on E9.5, expression of VE-CADHERIN is restricted to the peripheral cell layer of blood islands that gives rise to endothelial cells (Breier and Risau, 1996). Co-expression of VE-CADHERIN with ISL1 has been described in cells found in the lower portion of the OFT septum in human fetal hearts at gestation week 9 (Lui et al., 2013), and ISL1-derived cells contribute to the endothelium of the dorsal aorta and cardinal vein of mouse embryos on E11.5 (Keenan et al., 2012).
In our ESC differentiation system, VE-CADHERIN was transiently co-expressed in a subset of ISL1+ cells when they first emerged on day 4 and formed AJs in small cell clusters mixed with N-CADHERIN. FACS analysis quantified morphological data showing that VE-CADHERIN+/ISL1+ cells constitute a significant percentage of CPCs on days 4 and 5. The double-positive VE-CADHERIN/ISL1 cell population represented one-third of the emerging FLK1+/PDGFRa+ CPCs and for a restricted time window (days 4–6). These relatively high percentages cannot be compared with fluorescence-based morphological data that depict formation of mature AJs only. In agreement, VE-CADHERIN AJs appeared in small numbers of ISL1+ and MEF2c+ cells in mouse embryos on E8.5. The transient presence of that VE-CADHERIN/ISL1 cell population in the SHF on E8.5 demonstrates the emergence of an early SHF CPC subpopulation in vivo. Of interest, transient VE-CADHERIN expression has also been detected in hematopoietic progenitor populations, the “hemogenic endothelium,” indicative of a potential function of this SHF CPC subpopulation in priming formation of the cardiac endothelium (Chen et al., 2009; Lancrin et al., 2009; Oberlin et al., 2010).
VE-CADHERIN is expressed in endocardial cells lining the myocardium (Neri et al., 2019; Zhang et al., 2018), and the ISL1+ SHF population can differentiate to endocardium and OFT endothelium (Meilhac et al., 2015) This could imply that the ISL1+/VE-CADHERIN+ population gives rise to endocardial and endothelial but not myocardial cells. However, in our previous studies, we isolated and characterized the ISL1+ subpopulation where the ve-cadherin promoter (Pvec) is active. This population differentiated into cells with endothelial and cardiac phenotypes (Maltabe et al., 2016).
Ablation of VE-CADHERIN in ESCs by CRISPR-Cas9 technology caused profound changes in myocardial differentiation. Most importantly, beating activity was severely affected because of a dramatic decrease in expression of cardiomyocyte-specific proteins. We postulate that this is a result of a progenitor population decrease, characterized by major cardiac transcription factor expression, at the onset of emergence and not due to endothelial disruption. There is evidence that the endothelium positively affects cardiac differentiation; however, this has been shown to occur at later time points (Chen et al., 2010; Kado et al., 2008). In addition, in our wild-type EBs, endothelial structures were barely detected on day 4.
Lack of VE-CADHERIN in KO mice is lethal because of cardiac and endothelial maturation defects. Specifically, although the primary vascular plexus forms initially, inter-endothelial structures are maintained, possibly through compensatory adhesion molecules, in early stages, and VE-CADHERIN is required later for VEGF signaling and vascular remodeling (Carmeliet et al., 1999; Crosby et al., 2005). Interestingly, cardiac differentiation is also affected in VE-CADHERIN KO mice because of endocardial detachment occluding the ventricular lumen in the OFT on E8.75, leading to myocardial disorganization (Carmeliet et al., 1999). N-CADHERIN expression was not addressed in the developing heart of VE-CADHERIN KO mice. In our ESCs differentiation system, it seems that N-CADHERIN AJs formed, although their expression was affected slightly.
Ablation of N-CADHERIN has been shown to cause embryonic lethality because of cardiovascular defects caused by cardiomyocytes’ inability to aggregate by gestation day 10 (Radice et al., 1997). Heart-specific KO of N-CADHERIN in the adult heart was lethal not because of cardiomyocyte formation failure but because of intercalated disc disruption (Kostetskii et al., 2005), and normal cardiac looping morphogenesis could be partially reversed when E-CADHERIN substituted N-CADHERIN (Luo et al., 2001). This demonstrates that cadherins can function redundantly when expressed ectopically. As a matter of fact, the role of N- and VE-CADHERIN-mediated AJs was studied in the developing embryo by knocking out these molecules individually or in a tissue-specific manner in the adult. It seems that both cadherins were not indispensable for initial specification of progenitor cells but required later during embryonic tissue formation (Carmeliet et al., 1999; Luo and Radice, 2005; Radice et al., 1997). In line with this, we found, quite unexpectedly, that VE- and E-CADHERIN formed distinct AJs in an ISL1+ subpopulation at the same time.
To address cadherin redundancy in VE-CADHERIN+ cells, we used a well-established approach and took advantage of the temporally strict activation pattern of the ve-cadherin promoter (Pvec) characterized previously (Maltabe et al., 2016). Pvec drove the expression of a disruptive VE-CADHERIN mutant (ΔEXD-VEC) lacking the extracellular adhesive domain (Kouklis et al., 2003). Numerous studies have established that such mutants inhibit all classic cadherin-mediated AJs even when expressed ectopically. It is suggested that their expression “poisons” endogenous classic cadherins, possibly through competition for association for β-catenin and p120 catenin, necessary for AJ function (Hermiston and Gordon, 1995; Horikawa and Takeichi, 2001; Kintner, 1992). ΔEXD-VEC expression in Pvec+ cells disrupted AJs mediated by VE-CADHERIN, N-CADHERIN, and presumably other, still unidentified classic cadherins expressed on days 4 and 5. Remarkably, the ISL1+ population’s dramatic decrease in mutant EBs as early as day 4 could be the result of AJ dysfunction caused by the mutant’s expression. These results are in agreement with a recent study demonstrating that ISL1 mutant embryos fail to form mesoderm because of reduced BMP4 signaling from the amnion (Yang et al., 2021). In other studies, BMP4 activation of p38 mitogen-activated protein kinase (MAPK) signaling controls ISL1 protein function (Jing et al., 2021).
We saw VE-CADHERIN AJs first appear in PDGFRα+/FLK1+/ISL1+ CPCs, and their presence could indicate participation in signaling events or represent a beacon for cells originating from different sources to incorporate in the developing heart or regulate cell migration. VE-CADHERIN participates in density-dependent inhibition of endothelial growth by maintaining vascular endothelial growth factor receptor 2 (VEGFR2) at the plasma membrane, resulting in MAPK negative regulation (Grazia Lampugnani et al., 2003; Lampugnani et al., 2006) and, on the other hand, promoting antiapoptotic signaling by positive regulation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway via β-catenin (Carmeliet et al., 1999). In endothelial cells, VE-CADHERIN has been found to be associated with ALK2/BMPRII, resulting in stabilization of the BMP receptor complex and, therefore, support of BMP6-mediated signaling (Benn et al., 2016). Homophilic VE-CADHERIN adhesion and the actomyosin cytoskeleton play central roles in endothelial mechanotransduction properties linked to signaling (Barry et al., 2015). VE-CADHERIN AJs control YAP transcriptional activity/or subcellular localization, phosphorylation, and activity through VE-CADHERIN/PI3K/Akt activation (Choi and Kwon, 2015). VE-CADHERIN forms junctional mechanosensory complexes with PECAM-1, VEGFR2, and VEGFR3, which sense and transmit mechanical forces modulating signaling pathways in adult endothelial cells (Conway et al., 2013; Coon et al., 2015; Dejana and Vestweber, 2013; Tzima et al., 2005). It remains to be seen whether VE-CADHERIN could also be involved in actomyosin-induced tension mechanical properties linked to the epithelial phenotype of SHF cells (Francou et al., 2017).
VE-CADHERIN is involved also in cell shape and polarity, studied in endothelial cells (Iden et al., 2006), and in cell migration processes through VEGFR2 and Rho-family GTPase activation (Abu Taha and Schnittler, 2014; Cao et al., 2017). Its cytoplasmic domain can activate regulators of actin cytoskeleton dynamics in endothelial cells (Broman et al., 2006; Kouklis et al., 2003). Studies in zebrafish revealed that myocardial precursors form a polarized epithelium, which migrates as a coherent population (Trinh and Stainier, 2004). Knockdown of CADHERIN-5 (VE-CADHERIN ortholog) in zebrafish results in early cardiovascular defects, reminiscent of delayed cardiac migration (Mitchell et al., 2010; Montero-Balaguer et al., 2009). Future studies will investigate VE-CADHERIN’s involvement in signaling pathways during the earliest stage of mammalian cardiovascular differentiation by lineage tracing.
Experimental procedures
Resource availability
Corresponding authors
Panos Kouklis is the corresponding author for this paper.
Materials availability
Cell lines and plasmids are available from the lead contact upon reasonable request.
Cell culture
E14T ESCs were kindly provided by Prof. A. Smith and Dr. I. Chambers (MRC Center for Regenerative Medicine, Edinburgh, UK). E14T cells were propagated on gelatin (0.1% swine skin) in high-glucose Glasgow modified Eagle’s medium (Sigma) supplemented with LIF conditioned medium, 15% fetal bovine serum (FBS; Biochrom), 1 mM sodium pyruvate (Invitrogen), 2 mM L-glutamine (Invitrogen), 0.1 mΜ non-essential amino acids (Invitrogen), 0.05 mM β-mercaptoethanol (Sigma), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Invitrogen).
ESC differentiation by “hanging drops” was performed as follows. ESCs were seeded at 500 cells/20-μL drop in IMDM containing 50 ng/mL human VEGF, 100 ng/mL human bFGF, and 10 ng/mL murine interleukin-6 (IL-6; Immunotools, Germany), 2 U/mL human erythropoietin (Life Technologies), and 10 μg/mL human insulin (Sigma) and cultured in hanging drops for 2 days. EBs were then collected and plated on bacterial Petri dishes for further differentiation.
Flow cytometry
EBs were harvested on days 4, 5, and 6. The immunostaining procedure for flow cytometry is described in the supplemental information. Stained cells were acquired on a FACSAria III flow cytometer (BD Life Sciences, Franklin Lakes, NJ, USA). Flow cytometry analysis was performed using FlowJo v.10.8 software (BD Life Sciences). Data clean-up analysis was performed prior to analysis of marker expression and included gating for doublet exclusion and live-dead discrimination (see the gating strategy for flow cytometry analysis in Figure S2). All FACs experiments were performed at least three times, and representative data are shown. Detailed experimental methodology is provided in the supplemental experimental procedures.
CRISPR-Cas9-mediated gene deletion
The CRISPR-double nickase Cas9 system was used for VE-CADHERIN KO clone creation. Specifically, E14T cells were transfected with two plasmids carrying the double Cas9 nickase and the ve-cadherin sgRNAs (sc-419596-NIC-2, Santa Cruz Biotechnology). The gRNAs used to create the ve-cadherin KO are shown in Figure S3A. Transfections were carried out using the UltraCruz transfection reagent (sc-395739, Santa Cruz Biotechnology). 24 h after transfection, 2 μg/mL puromycin was added to the medium for 72 h. The cells were left to recover and to form new colonies for approximately 2 weeks. Newly formed single colonies were picked, transferred into separate wells, and expanded. The screening for positive clones was first performed by VE-CADHERIN staining during clones’ differentiation on day 7. Genomic DNA was isolated from clones with no VE-CADHERIN staining according to a standard protocol. The region of interest was amplified with Kapa HiFi Hot Start (Roche, KK2601) and the following primers: 5ʹ-GCAAGATCTTCGGGGTCGAT-3ʹ (forward) and 5ʹ-AGGATCAAGGCCCAGGCTAT-3ʹ (reverse). PCR products were sequenced, and the results are shown in Figure S3B.
Additional detailed experimental methods are provided in the supplemental information.
Acknowledgments
We thank Dr. Robert Kelly for help with mouse heart embryogenesis, Dr. Mairi Tsikitis for help with mouse embryo isolation and helpful discussions, and Anastasia Apostolidou in the FACS Facility and Vania Tsata for 3D visualization of confocal stacks. The CT3 (cTNT), 39.4D5 (ISL1), and 40.2D6 monoclonal antibodies developed by J.J.-C. Lin, T.M. Jessell, and S. Brennere-Morton, respectively, were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at Department of Biology, The University of Iowa, Iowa City, IA, USA. The dsRed.MST ESC line was a generous gift from Dr. Jenny Nichols of the Wellcome Trust Stem Cell Institute, Cambridge, UK. This research is co-financed by Greece and the European Union (European Social Fund- ESF) through the operational program “Human Resources Development, Education and Lifelong Learning” in the context of the project “Reinforcement of Postdoctoral Researchers – 2nd Cycle” (MIS-5033021), implemented by the State Scholarships Foundation (ΙΚΥ). The project is co-financed by Greece and the European Union - European Regional Development Fund (ERDF) under the operational program “Competitiveness Entrepreneurship Innovation” (EPAnEK), NSRF 2014-2020 (MIS 5047236).
Author contributions
Conceptualization, V.A.M., A.N.M., and P.K.; methodology, V.A.M., A.N.M., D.B., I.K., N.P., and P.K.; validation, V.A.M., A.N.M., and P.K.; formal analysis, V.A.M.; investigation, V.A.M., A.N.M., and N.P.; resources, V.A.M. and A.N.M.; writing – original draft, V.A.M., A.N.M., and P.K.; writing – review & editing, V.A.M., N.P., and P.K.; supervision, P.K.; funding acquisition, V.A.M. and P.K.
Conflict of interests
The authors declare no competing interests.
Published: August 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.07.002.
Supplemental information
Data and code availability
Complementary data used in this publication can be found in the supplemental information.
References
- Abu Taha A., Schnittler H.J. Dynamics between actin and the VE-CADHERIN/catenin complex: novel aspects of the ARP2/3 complex in regulation of endothelial junctions. Cell Adhes. Migrat. 2014;8:125–135. doi: 10.4161/cam.28243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acloque H., Adams M.S., Fishwick K., Bronner-Fraser M., Nieto M.A. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J. Clin. Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvandi Z., Bischoff J. Endothelial-Mesenchymal Transition in Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol. 2021;41:2357–2369. doi: 10.1161/ATVBAHA.121.313788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot E.S., Hadjantonakis A.K. Mouse gastrulation: Coordination of tissue patterning, specification and diversification of cell fate. Mech. Dev. 2020;163 doi: 10.1016/j.mod.2020.103617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry A.K., Wang N., Leckband D.E. Local VE-CADHERIN mechanotransduction triggers long-ranged remodeling of endothelial monolayers. J. Cell Sci. 2015;128:1341–1351. doi: 10.1242/jcs.159954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn A., Bredow C., Casanova I., Vukičević S., Knaus P. VE-CADHERIN facilitates BMP-induced endothelial cell permeability and signaling. J. Cell Sci. 2016;129:206–218. doi: 10.1242/jcs.179960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga V. Signaling by Small GTPases at Cell-Cell Junctions: Protein Interactions Building Control and Networks. Cold Spring Harbor Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier G., Risau W. The role of vascular endothelial growth factor in blood vessel formation. Trends Cell Biol. 1996;6:454–456. doi: 10.1016/0962-8924(96)84935-x. [DOI] [PubMed] [Google Scholar]
- Broman M.T., Kouklis P., Gao X., Ramchandran R., Neamu R.F., Minshall R.D., Malik A.B. Cdc42 regulates adherens junction stability and endothelial permeability by inducing alpha-catenin interaction with the vascular endothelial CADHERIN complex. Circ. Res. 2006;98:73–80. doi: 10.1161/01.RES.0000198387.44395.e9. [DOI] [PubMed] [Google Scholar]
- Cao J., Ehling M., März S., Seebach J., Tarbashevich K., Sixta T., Pitulescu M.E., Werner A.C., Flach B., Montanez E., et al. Polarized actin and VE-CADHERIN dynamics regulate junctional remodelling and cell migration during sprouting angiogenesis. Nat. Commun. 2017;8:2210. doi: 10.1038/s41467-017-02373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Lampugnani M.G., Moons L., Breviario F., Compernolle V., Bono F., Balconi G., Spagnuolo R., Oosthuyse B., Dewerchin M., et al. Targeted deficiency or cytosolic truncation of the VE-CADHERIN gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Chen K., Bai H., Arzigian M., Gao Y.X., Bao J., Wu W.S., Shen W.F., Wu L., Wang Z.Z. Endothelial cells regulate cardiomyocyte development from embryonic stem cells. J. Cell. Biochem. 2010;111:29–39. doi: 10.1002/jcb.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., Speck N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.J., Kwon Y.G. Roles of YAP in mediating endothelial cell junctional stability and vascular remodeling. BMB Rep. 2015;48:429–430. doi: 10.5483/bmbrep.2015.48.8.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway D.E., Breckenridge M.T., Hinde E., Gratton E., Chen C.S., Schwartz M.A. Fluid shear stress on endothelial cells modulates mechanical tension across VE-CADHERIN and PECAM-1. Curr. Biol. 2013;23:1024–1030. doi: 10.1016/j.cub.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon B.G., Baeyens N., Han J., Budatha M., Ross T.D., Fang J.S., Yun S., Thomas J.L., Schwartz M.A. Intramembrane binding of VE-CADHERIN to VEGFR2 and VEGFR3 assembles the endothelial mechanosensory complex. J. Cell Biol. 2015;208:975–986. doi: 10.1083/jcb.201408103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby C.V., Fleming P.A., Argraves W.S., Corada M., Zanetta L., Dejana E., Drake C.J. VE-CADHERIN is not required for the formation of nascent blood vessels but acts to prevent their disassembly. Blood. 2005;105:2771–2776. doi: 10.1182/blood-2004-06-2244. [DOI] [PubMed] [Google Scholar]
- Dejana E., Vestweber D. The role of VE-CADHERIN in vascular morphogenesis and permeability control. Prog. Mol. Biol. Transl. Sci. 2013;116:119–144. doi: 10.1016/B978-0-12-394311-8.00006-6. [DOI] [PubMed] [Google Scholar]
- Francou A., De Bono C., Kelly R.G. Epithelial tension in the second heart field promotes mouse heart tube elongation. Nat. Commun. 2017;8 doi: 10.1038/ncomms14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francou A., Saint-Michel E., Mesbah K., Kelly R.G. TBX1 regulates epithelial polarity and dynamic basal filopodia in the second heart field. Development. 2014;141:4320–4331. doi: 10.1242/dev.115022. [DOI] [PubMed] [Google Scholar]
- Grazia Lampugnani M., Zanetti A., Corada M., Takahashi T., Balconi G., Breviario F., Orsenigo F., Cattelino A., Kemler R., Daniel T.O., Dejana E. Contact inhibition of VEGF-induced proliferation requires vascular endothelial CADHERIN, beta-catenin, and the phosphatase DEP-1/CD148. J. Cell Biol. 2003;161:793–804. doi: 10.1083/jcb.200209019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J.C., Tepass U. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- Hermiston M.L., Gordon J.I. In vivo analysis of CADHERIN function in the mouse intestinal epithelium: essential roles in adhesion, maintenance of differentiation, and regulation of programmed cell death. J. Cell Biol. 1995;129:489–506. doi: 10.1083/jcb.129.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M., Kurisaki A., Ohnuma K., Okochi H., Hamazaki T.S., Asashima M. N-CADHERIN is a useful marker for the progenitor of cardiomyocytes differentiated from mouse ES cells in serum-free condition. Biochem. Biophys. Res. Commun. 2006;351:877–882. doi: 10.1016/j.bbrc.2006.10.126. [DOI] [PubMed] [Google Scholar]
- Horikawa K., Takeichi M. Requirement of the juxtamembrane domain of the CADHERIN cytoplasmic tail for morphogenetic cell rearrangement during myotome development. J. Cell Biol. 2001;155:1297–1306. doi: 10.1083/jcb.200108156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iden S., Rehder D., August B., Suzuki A., Wolburg-Buchholz K., Wolburg H., Ohno S., Behrens J., Vestweber D., Ebnet K. A distinct PAR complex associates physically with VE-CADHERIN in vertebrate endothelial cells. EMBO Rep. 2006;7:1239–1246. doi: 10.1038/sj.embor.7400819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Ren Y., Witzel H.R., Dobreva G. A BMP4-p38 MAPK signaling axis controls ISL1 protein stability and activity during cardiogenesis. Stem Cell Rep. 2021;16:1894–1905. doi: 10.1016/j.stemcr.2021.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado M., Lee J.K., Hidaka K., Miwa K., Murohara T., Kasai K., Saga S., Morisaki T., Ueda Y., Kodama I. Paracrine factors of vascular endothelial cells facilitate cardiomyocyte differentiation of mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2008;377:413–418. doi: 10.1016/j.bbrc.2008.09.160. [DOI] [PubMed] [Google Scholar]
- Keenan I.D., Rhee H.J., Chaudhry B., Henderson D.J. Origin of non-cardiac endothelial cells from an ISL1+ lineage. FEBS Lett. 2012;586:1790–1794. doi: 10.1016/j.febslet.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Kintner C. Regulation of embryonic cell adhesion by the CADHERIN cytoplasmic domain. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- Komarova Y.A., Kruse K., Mehta D., Malik A.B. Protein Interactions at Endothelial Junctions and Signaling Mechanisms Regulating Endothelial Permeability. Circ. Res. 2017;120:179–206. doi: 10.1161/CIRCRESAHA.116.306534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostetskii I., Li J., Xiong Y., Zhou R., Ferrari V.A., Patel V.V., Molkentin J.D., Radice G.L. Induced deletion of the N-CADHERIN gene in the heart leads to dissolution of the intercalated disc structure. Circ. Res. 2005;96:346–354. doi: 10.1161/01.RES.0000156274.72390.2c. [DOI] [PubMed] [Google Scholar]
- Kouklis P., Konstantoulaki M., Malik A.B. VE-CADHERIN-induced Cdc42 signaling regulates formation of membrane protrusions in endothelial cells. J. Biol. Chem. 2003;278:16230–16236. doi: 10.1074/jbc.M212591200. [DOI] [PubMed] [Google Scholar]
- Lampugnani M.G., Orsenigo F., Gagliani M.C., Tacchetti C., Dejana E. Vascular endothelial CADHERIN controls VEGFR-2 internalization and signaling from intracellular compartments. J. Cell Biol. 2006;174:593–604. doi: 10.1083/jcb.200602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin C., Sroczynska P., Stephenson C., Allen T., Kouskoff V., Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap A., Liang X., Gomez G. Current perspectives on CADHERIN-cytoskeleton interactions and dynamics. Cell Health Cytoskeleton. 2015;7:11. doi: 10.2147/CHC.S76107. [DOI] [Google Scholar]
- Lui K.O., Zangi L., Silva E.A., Bu L., Sahara M., Li R.A., Mooney D.J., Chien K.R. Driving vascular endothelial cell fate of human multipotent ISL1+ heart progenitors with VEGF modified mRNA. Cell Res. 2013;23:1172–1186. doi: 10.1038/cr.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Ferreira-Cornwell M., Baldwin H., Kostetskii I., Lenox J., Lieberman M., Radice G. Rescuing the N-CADHERIN knockout by cardiac-specific expression of N- or E-CADHERIN. Development. 2001;128:459–469. doi: 10.1242/dev.128.4.459. [DOI] [PubMed] [Google Scholar]
- Luo Y., Radice G.L. N-CADHERIN acts upstream of VE-CADHERIN in controlling vascular morphogenesis. J. Cell Biol. 2005;169:29–34. doi: 10.1083/jcb.200411127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltabe V., Kouklis P. Vascular Endothelial (VE)-CADHERIN-mediated adherens junctions involvement in cardiovascular progenitor cell specification. Int. J. Dev. Biol. 2022;66:77–83. doi: 10.1387/ijdb.210167pk. [DOI] [PubMed] [Google Scholar]
- Maltabe V.A., Barka E., Kontonika M., Florou D., Kouvara-Pritsouli M., Roumpi M., Agathopoulos S., Kolettis T.M., Kouklis P. Isolation of an ES-Derived Cardiovascular Multipotent Cell Population Based on VE-CADHERIN Promoter Activity. Stem Cell. Int. 2016;2016 doi: 10.1155/2016/8305624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mège R.M., Ishiyama N. Integration of CADHERIN Adhesion and Cytoskeleton at Adherens Junctions. Cold Spring Harbor Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac S.M., Buckingham M.E. The deployment of cell lineages that form the mammalian heart. Nat. Rev. Cardiol. 2018;15:705–724. doi: 10.1038/s41569-018-0086-9. [DOI] [PubMed] [Google Scholar]
- Meilhac S.M., Lescroart F., Blanpain C., Buckingham M.E. Cardiac cell lineages that form the heart. Cold Spring Harb. Perspect. Med. 2015;5:a026344. doi: 10.1101/cshperspect.a026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell I.C., Brown T.S., Terada L.S., Amatruda J.F., Nwariaku F.E. Effect of vascular CADHERIN knockdown on zebrafish vasculature during development. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Balaguer M., Swirsding K., Orsenigo F., Cotelli F., Mione M., Dejana E. Stable vascular connections and remodeling require full expression of VE-CADHERIN in zebrafish embryos. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri T., Hiriart E., van Vliet P.P., Faure E., Norris R.A., Farhat B., Jagla B., Lefrancois J., Sugi Y., Moore-Morris T., et al. Human pre-valvular endocardial cells derived from pluripotent stem cells recapitulate cardiac pathophysiological valvulogenesis. Nat. Commun. 2019;10:1929. doi: 10.1038/s41467-019-09459-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova-Krstevski V., Bhasin M., Otu H.H., Libermann T., Oettgen P. Gene expression analysis of embryonic stem cells expressing VE-CADHERIN (CD144) during endothelial differentiation. BMC Genom. 2008;9:240. doi: 10.1186/1471-2164-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin E., Fleury M., Clay D., Petit-Cocault L., Candelier J.J., Mennesson B., Jaffredo T., Souyri M. VE-CADHERIN expression allows identification of a new class of hematopoietic stem cells within human embryonic liver. Blood. 2010;116:4444–4455. doi: 10.1182/blood-2010-03-272625. [DOI] [PubMed] [Google Scholar]
- Radice G.L., Rayburn H., Matsunami H., Knudsen K.A., Takeichi M., Hynes R.O. Developmental defects in mouse embryos lacking N-CADHERIN. Dev. Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- Soh B.S., Buac K., Xu H., Li E., Ng S.Y., Wu H., Chmielowiec J., Jiang X., Bu L., Li R.A., et al. N-CADHERIN prevents the premature differentiation of anterior heart field progenitors in the pharyngeal mesodermal microenvironment. Cell Res. 2014;24:1420–1432. doi: 10.1038/cr.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymborska A., Gerhardt H. Hold Me, but Not Too Tight-Endothelial Cell-Cell Junctions in Angiogenesis. Cold Spring Harbor Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a029223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh L.A., Stainier D.Y.R. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev. Cell. 2004;6:371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Tzima E., Irani-Tehrani M., Kiosses W.B., Dejana E., Schultz D.A., Engelhardt B., Cao G., DeLisser H., Schwartz M.A. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- Vintersten K., Monetti C., Gertsenstein M., Zhang P., Laszlo L., Biechele S., Nagy A. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- Yang R., Goedel A., Kang Y., Si C., Chu C., Zheng Y., Chen Z., Gruber P.J., Xiao Y., Zhou C., et al. Amnion signals are essential for mesoderm formation in primates. Nat. Commun. 2021;12:5126. doi: 10.1038/s41467-021-25186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lui K.O., Zhou B. Endocardial Cell Plasticity in Cardiac Development, Diseases and Regeneration. Circ. Res. 2018;122:774–789. doi: 10.1161/CIRCRESAHA.117.312136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complementary data used in this publication can be found in the supplemental information.