Abstract
Intraneuronal accumulation of misfolded α-synuclein is the pathological hallmark of Parkinson’s disease and dementia with Lewy bodies, often co-occurring with variable degrees of Alzheimer’s disease related neuropathology. Genetic association studies have successfully identified common variants associated with disease risk and phenotypic traits in Lewy body disease, yet little is known about the genetic contribution to neuropathological heterogeneity.
Using summary statistics from Parkinson’s disease and Alzheimer’s disease genome-wide association studies, we calculated polygenic risk scores and investigated the relationship with Lewy, amyloid-β and tau pathology. Associations were nominated in neuropathologically defined samples with Lewy body disease from the Netherlands Brain Bank (n = 217) and followed up in an independent sample series from the Mayo Clinic Brain Bank (n = 394). We also generated stratified polygenic risk scores based on single-nucleotide polymorphisms annotated to eight functional pathways or cell types previously implicated in Parkinson’s disease and assessed for association with Lewy pathology in subgroups with and without significant Alzheimer’s disease co-pathology.
In an ordinal logistic regression model, the Alzheimer’s disease polygenic risk score was associated with concomitant amyloid-β and tau pathology in both cohorts. Moreover, both cohorts showed a significant association between lysosomal pathway polygenic risk and Lewy pathology, which was more consistent than the association with a general Parkinson’s disease risk score and specific to the subset of samples without significant concomitant Alzheimer’s disease related neuropathology.
Our findings provide proof of principle that the specific risk alleles a patient carries for Parkinson’s and Alzheimer’s disease also influence key aspects of the underlying neuropathology in Lewy body disease. The interrelations between genetic architecture and neuropathology are complex, as our results implicate lysosomal risk loci specifically in the subset of samples without Alzheimer’s disease co-pathology. Our findings hold promise that genetic profiling may help predict the vulnerability to specific neuropathologies in Lewy body disease, with potential relevance for the further development of precision medicine in these disorders.
Keywords: Lewy body disease, Parkinson’s disease, genetics, neuropathology, lysosomal pathway
Tunold et al. studied genetics and neuropathology in 611 samples from individuals with Parkinson’s disease or dementia with Lewy bodies. Genetic risk of Alzheimer’s disease was associated with amyloid and tau accumulation whereas Lewy body pathology increased with a higher burden of Parkinson’s disease risk variants affecting the lysosomal pathway.
Introduction
Lewy body disease (LBD) represents a continuum of closely related neurodegenerative diseases with overlapping clinical characteristics, genetic risk factors and neuropathological features. The most common manifestations of LBD are Parkinson’s disease (PD) and dementia with Lewy bodies (DLB). A defining neuropathological hallmark of LBD is the deposition of α-synuclein (α-syn) rich intra-neuronal inclusions called Lewy bodies and Lewy neurites, collectively referred to as Lewy pathology.1 In addition to Lewy pathology, varying degrees of Alzheimer’s disease (AD) co-pathology, including amyloid-β plaques and tau positive neurofibrillary tangles (NFT), are often present.2 Elucidating biological mechanisms that underlie the heterogenous neuropathological substrates of LBD is crucial for understanding disease aetiology and progression, with the ultimate aim to discover novel therapeutic avenues for disease modification.
Genome-wide association studies (GWAS) have provided insights into the complex polygenic architecture of clinical LBD phenotypes, and successfully identified common genetic risk variants in PD3 and to a lesser extent in DLB.4,5 Each GWAS locus explains only a small proportion of disease susceptibility, yet the cumulative effect of many risk variants can be estimated as a polygenic risk score (PRS). A PRS is calculated as the weighted sum of the number of risk alleles an individual carries. In PD, PRSs have been applied successfully to a number of traits, including age at onset, disease status, motor progression and cognitive decline.6-9 In DLB, the PRS has been linked to disease risk.10
The general PRS approach includes all independent single-nucleotide polymorphisms (SNPs) with associated P-values below a specified threshold in summary statistics from GWAS. However, stratified PRSs may be generated from subsets of SNPs that are annotated to specific pathways, thus helping to nominate mechanisms that contribute to disease development.11-13 Pathway-specific PRS studies have provided further support for a number of biological pathways and mechanisms previously implicated in PD, including mitochondrial dysfunction, lysosomal mediated autophagy/lysosomal dysfunction, endocytic membrane trafficking, α-syn misfolding and neuroinflammation.11,13,14
A major challenge in understanding how genetic risk influences LBD relates to the clinical and neuropathological heterogeneity. While LBD is defined by the accumulation of Lewy bodies, comorbid AD pathology is common and found more frequently in DLB and PD with dementia than in non-demented PD.2,15-18 The level of AD co-pathology shows association with the severity of Lewy pathology,19 which makes it challenging to determine the causal relationships underlying genetic associations. Two recent publications have shown that risk variants in the β-glucocerebrosidase (GBA) gene are primarily associated with ‘pure’ DLB, while the APOE ε4 allele is a risk factor for DLB with AD co-pathology,10,20 suggesting the existence of distinct genetic architectures within the LBD continuum.
To assess how common genetic risk variants associated with PD and AD influence the multifaceted neuropathologies of LBD, we generated PD- and AD-susceptibility PRSs as well as stratified PD-PRSs and explored their relationship to key neuropathological markers in two post-mortem cohorts, using the Netherlands Brain Bank (n = 217) for discovery and the Mayo Clinic Brain Bank (n = 394) for replication. Based on the hypothesis that distinct genetic profiles associate with Lewy pathology depending on the presence or absence of AD co-pathology, we divided the LBD samples into two subgroups based on the level of AD co-pathology.
Materials and methods
Subjects
Cases of LBD with available data from neuropathological assessment of Lewy pathology and AD pathology, as well as genotype data, were considered for inclusion. From the Netherlands Brain Bank (NBB, www.brainbank.nl), donors enrolled from 1989 to 2017 (n = 3853) were assessed, and 222 subjects with a neuropathologically confirmed diagnosis of PD or DLB were included. In addition, neurologically healthy controls (n = 82) and samples with a neuropathological diagnosis of AD (n = 64) were included to evaluate the discriminative ability of AD- and PD-PRSs. Written, informed consent for the use of clinical information and tissue samples for research purposes, was collected from the donors or their next of kin.
Brain dissection was performed according to international guidelines of Brain Net Europe II (BNE) consortium (www.brainnet-europe.org) and the National Institute on Aging-Alzheimer’s Association (NIA-AA)21 by an experienced neuropathologist (A.R.). Formalin-fixed paraffin-embedded 6-μm thick sections were immunostained with antibodies against p-tau (clone AT8, 1:500, Thermo Fisher Scientific), amyloid-β (clone 6F/3D, 1:500, Dako) and α-syn (clone KM51, 1:500, Monosan Xtra), or stained with haematoxylin and eosin (H&E) or Congo red according to current diagnostic guidelines of BrainNet Europe.22,23
To assign a Braak NFT stage (Braak NFT 0–VI), NFTs were scored in association cortices (medial frontal gyrus, medial temporal and superior parietal cortex), primary cortices (primary visual cortex and pre/postcentral gyrus), hippocampus (CA1, CA4 and subiculum) and adjacent (trans)entorhinal and fusiform cortex, amygdala, caudate-putamen and cerebellum (if available), as previously described.22 Thal amyloid-β phases (0–4) were scored according to Thal et al.24 on the medial temporal lobe. For the majority of the cases, a distinction between Thal phase 4 and 5 could not be made, as the cerebellum was not available. Pathological staging for neuritic plaques in the above-described cortical brain regions was based on the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) score.25
Braak Lewy pathology stages, ranging from 3 to 6 in LBD cases, were based on α-syn immunostaining in the neocortices (medial frontal, medial temporal, superior parietal, primary visual and motor cortex), anterior cingulate gyrus, hippocampus (CA1, CA2), (trans)entorhinal cortex, amygdala, basal forebrain, midbrain (substantia nigra), tegmentum (locus coeruleus) and medulla oblongata (dorsal motor nucleus of the vagal nerve), according to the protocol described by Alafuzoff et al.23 Owing to the low number of samples with Braak Lewy pathology stage 3, stages 3 and 4 were collapsed into a single group for the statistical analyses.
Clinical information was extracted from the medical records provided by the NBB. PD was diagnosed based on the combination of UK Parkinson’s Disease Society Brain Bank criteria26 and moderate to severe loss of neurons in the substantia nigra with concurrent Lewy pathology in at least the brainstem.27 Criteria for DLB were a clinical diagnosis of probable DLB according to the consensus criteria of the DLB Consortium,28 combined with presence of limbic-transitional or diffuse-neocortical Lewy pathology upon autopsy. Dementia was diagnosed prior to death by a neurologist or geriatrician, or retrospectively based on neuropsychological test results29 or a Mini-Mental State Examination (MMSE) score <20.
From the Mayo Clinic Jacksonville Brain Bank for Neurodegenerative Disorders, we included a total of 402 autopsy-confirmed LBD cases, characterized by a single neuropathologist (D.W.D.). All subjects were Caucasian, non-Hispanic and unrelated, with written, informed consent for the use of clinical information and tissue samples for research purposes collected from the donors or their next of kin.
Paraffin-embedded 5-μm thick sections mounted on glass slides were stained with thioflavin S. To assign a Braak NFT stage (0–VI) and Thal amyloid-β phase (0–5), NFTs and senile plaques were quantified using thioflavin S fluorescence microscopy in association cortices (frontal, temporal and parietal), primary cortices (visual and motor), hippocampus (CA1, CA4 and subiculum) and adjacent cortex, amygdala, basal ganglia and cerebellum, as previously described.21,30
Lewy pathology was assessed in the neocortices (frontal, temporal, parietal, visual and motor), cingulate gyrus, transentorhinal cortex, amygdala, basal forebrain, midbrain, pons and medulla using α-syn immunohistochemistry (NACP, 1:3000 rabbit polyclonal, Mayo Clinic antibody).31 Lewy pathology was staged as brainstem, transitional or diffuse LBD according to Kosaka et al.32
Clinical information was extracted from the medical records by three investigators (S.K., H.S. and N.B.M.) to identify the clinical diagnosis and determine the age at onset of either motor symptoms or dementia.33 Donors with an ante-mortem diagnosis of either PD or DLB were included in the study.26,28
We used an adaption of the NIA-AA criteria, where the combination of Thal phase and Braak NFT stage was used to calculate a composite AD-score.34 Samples with Thal phase 0 or Braak NFT 0 were classified as ‘no’, Thal phase 1–2 and Braak NFT I-VI or Thal phase 3–5 and Braak NFT I–II as ‘low’, Thal phase 3 and Braak NFT III–VI or Thal 4–5 and Braak III–IV as ‘intermediate’ and Thal phase 4–5 and Braak NFT V–VI as ‘high’. The LBD samples were divided into two subgroups by severity of AD co-pathology. LBD − ADpath was defined as ‘no’ or ‘low’ AD-score and LBD + ADpath as ‘intermediate’ or ‘high’ AD-score.
Genotyping
Genotyping of NBB samples was carried out on the Infinium NeuroChip Consortium Array (Illumina).35 Mayo Clinic brain bank samples were genotyped on the Infinium OmniExpress-24 (version 1.3) array (Illumina). Standard quality control and filtering were performed and variants imputed using reference data from the Haplotype Reference Consortium as reported previously in detail.36
Polygenic risk scores
For each individual, we generated AD-PRS and PD-PRS based on summary statistics from Jansen et al.37 and Nalls et al.3 (including 23andMe, Inc.), respectively, using PRSice2 with standard linkage disequilibrium clumping thresholds (clumping SNPs within a 250 kb window and r2 > 0.1).38 To improve the linkage disequilibrium estimation for clumping, the 1000 Genomes European samples (n = 503) were used as an external reference panel, as is recommended for small datasets in particular.38 In each of the GWAS summary statistics (the base datasets), duplicated and ambiguous SNPs (C/G or A/T SNPs) were removed as is standard practice.38 Only variants with a minor allele frequency > 1% were included.
Based on previously published studies of stratified PD-PRS, we selected six pathways (adaptive immune system, α-syn, endocytic membrane trafficking, innate immune system, lysosomal and mitochondrial pathways) and two cell types (microglia and monocytes) of interest for which a significant enrichment of PD risk has been reported.11-14 One study reported as many as 46 partly overlapping gene sets,11 and from these we prioritized only a few corresponding to widely studied disease pathways in order to limit multiple testing. We used the same lists of genes or genomic coordinates as these previously published studies to generate pathway-specific PD-PRS, applying the PRSet function in PRSice2. Pathway gene lists were selected by using The Molecular Signatures Database (MsigDB)39 as well as curated lists of mitochondrial and endocytic membrane trafficking genes applied in previous reports.13,14 SNPs were mapped to genes using the physical gene boundaries. Cell-type annotations for monocytes and microglia were based on publicly available data on open chromatin regions mapped by assay for transposase-accessible chromatin with sequencing (ATACseq).40,41
The PRS algorithm includes SNPs with P-values below a user-specified threshold in the original GWAS, which could in theory be less stringent than the threshold for genome-wide significance. To test different thresholds, we evaluated the ability of susceptibility PD-PRS and AD-PRS to discriminate PD and AD samples, respectively, from controls without neurological disease, estimating the area under the receiver operator curve (AUC). For both PD-PRS and AD-PRS, a genome-wide threshold of P < 5 × 10−8 was superior to P < 1 × 10−5 and P < 0.05. We therefore chose to use the genome-wide threshold in subsequent analyses, although we acknowledge that assessing susceptibility PRS as predictors for quantitative neuropathologic outcomes in a case-only analysis is principally different from the standard approach differentiating cases from controls. Each PRS was standardized to have a mean of 0 and standard deviation (SD) of 1. The number of SNPs as well as lists of SNPs used to build each PRS are provided in Supplementary Tables 1–25. Genotype imputation ensures that most common SNPs are present in both the NBB and Mayo Clinic datasets, despite not being directly genotyped. Nevertheless, minor differences in the specific SNPs included from the PRSice algorithm were seen for a few of the PRS.
Statistical analyses
All statistical analyses were performed in R version 4.2.1 (www.r-project.org). Demographic data were compared between groups using Pearson’s chi-square test for categorical variables, t-tests or the Wilcoxon rank sum test for continuous variables and ordinal variables, as appropriate.
Associations between neuropathology scores (Braak Lewy pathology stage or Kosaka’s stage, CERAD neuritic plaque score, Thal amyloid-β phase and Braak NFT stage) and standardized PRS were tested with proportional odds (PO) ordinal logistic regression models to account for the ordered nature of the outcome measure using the vglm() function in the R package ‘VGAM’.42 To assess the PO assumption, we fitted a partial proportional odds (PPO) model where the PO assumption was relaxed for the explanatory variable (i.e. PRS). When comparing PO with PPO models, the likelihood ratio test P-values were non-significant, indicating the PO assumption to be reasonable. Due to the small number of LBD + ADpath individuals in the NBB cohort with Braak Lewy pathology stage <5, stages 3–5 were collapsed, and associations with Lewy pathology were tested with logistic regression using the R package ‘rms’.43 The models included sex, age at death and first five principal components (PC1–5) as covariates. The odds ratio estimates corresponded to the effect size per 1 SD increase in PRS.
All statistical tests were two-sided. We applied a two-stage design where association signals passing a threshold of P < 0.05 in the NBB discovery cohort (NBB) were nominated for independent replication in the Mayo Clinic cohort. We interpreted signals replicating at P < 0.05 with a consistent direction of effect across both stages as positive findings.
To further explore the power of PRS to predict neuropathology, we generated an AD co-pathology risk score using coefficients from the ordinal logistic regression in the NBB dataset and assessed the performance of the score in the independent Mayo Clinic dataset. The model included AD-PRS, age at onset and sex. We evaluated the ability of the score to differentiate between LBD − ADpath and LBD + ADpath samples estimating the AUC from the R package ‘pROC’. We also calculated a Lewy pathology risk score in the Mayo Clinic samples using coefficients from the ordinal logistic regression in the NBB cohort, where the model included the lysosomal PD-PRS, age at death, sex and dichotomized AD-score. The AUC was used to assess the power to predict DLBD.
To investigate if the highlighted PRS also influence dementia onset, we conducted survival analysis using the R package ‘survival’. Cox proportional hazards regression models were employed to assess the relationship between PRS and time to dementia for the NBB samples. The presence of dementia was used as the event variable. The time variable was the interval between symptom onset and dementia diagnosis for cases who developed dementia prior to death and disease duration at death for non-demented cases. Age at onset, sex and the first five genetic principal components were used as covariates. To assess the proportional hazards assumption, a combined plotting and testing approach was employed.
Data availability
Data on NBB donors that support the findings of this study can be obtained from the Netherlands Neurogenetics Database (https://www.brainbank.nl/nnd-project/). Mayo Clinic data are available from the authors on request. Analysis code used in this manuscript is available on GitHub at https://github.com/lpihlstrom/projects.
Results
After filtering extreme age outliers (n = 1) and cases with an atypical distribution of Lewy pathology that prevented the assignment of a Braak Lewy pathology stage (n = 4), 217 cases with LBD were included from the NBB in the final analyses. Overall, 161 cases (74%) were classified as LBD − ADpath and 56 (26%) as LBD + ADpath. The clinical and demographic details split across the LBD − ADpath and LBD + ADpath groups for NBB cases are summarized in Table 1. Gender distribution and age at death were comparable among the two subgroups of LBD. LBD + ADpath subjects were older at disease onset (70.6 versus 64.2 years) and had a shorter disease duration (8.1 versus 13.2 years) but a similar age at death as LBD − ADpath cases (77.5 versus 78.8). For 211 NBB cases, dementia status was also available. A larger proportion of LBD + ADpath cases had developed dementia prior to death compared to LBD − ADpath cases [49/54 (90.7%) versus 95/159 (59.7%)], and LBD + ADpath cases had a shorter interval between disease onset and onset of dementia (3.6 years versus 10.1 years). Braak Lewy pathology stage and, as expected, all measures of AD neuropathology were significantly higher in the LBD + ADpath subgroup.
Table 1.
Demographics for NBB samples
| LBD − ADpath (n = 161) |
LBD + ADpath (n = 56) |
|
|---|---|---|
| Sex, n (%) | ||
| Female | 56 (34.8) | 25 (44.6) |
| Male | 105 (65.2) | 31 (55.4) |
| Age at onset, mean (SD) | 64.2 (11.9) | 70.6 (8.8) |
| Age at death, mean (SD) | 77.5 (8.0) | 78.8 (7.8) |
| Disease duration, mean (SD) | 13.2 (7.5) | 8.1 (5.1) |
| Time to dementia (SD) | 10.1 (8.2) | 3.6 (5.9) |
| Braak Lewy pathology stage, n (%) | ||
| 3–4 | 14 (8.7) | 2 (3.6) |
| 5 | 60 (37.3) | 5 (8.9) |
| 6 | 87 (54.0) | 49 (87.5) |
| CERAD, median (Q1, Q3) | 0 (0, 1) | 1 (1, 2) |
| Thal phase, median (Q1, Q3) | 1 (0, 3) | 3 (3, 4) |
| Braak NFT stage, median (Q1, Q3) | I (I, II) | IV (III, IV) |
ADpath = Alzheimer's disease co-pathology; CERAD = Consortium to Establish a Registry for Alzheimer’s Disease; LBD = Lewy body disease; NFT = neurofibrillary tangle; Q1 = 1st quartile; Q3 = 3rd quartile; SD = standard deviation.
In the Mayo Clinic dataset, extreme age outliers (n = 8) were excluded and a total of 394 LBD cases included in the final analysis. Of these, 196 cases (50%) were categorized as LBD − ADpath and 198 cases (50%) as LBD + ADpath. A larger male predominance was seen in the LBD − ADpath than in the LBD + ADpath subgroup [71.9% (141/196) versus 66.2% (131/198)]. LBD + ADpath cases had a higher age at onset and age at death (70.4 versus 65.9 and 78.3 versus 76.0 years, respectively) and a shorter disease duration (7.9 versus 10.0 years). The clinical and demographic variables are summarized in Table 2.
Table 2.
Demographic table for Mayo Clinic samples
| LBD − ADpath (n = 196) |
LBD + ADpath (n = 198) |
|
|---|---|---|
| Sex, n (%) | ||
| Female | 55 (28.1) | 67 (33.8) |
| Male | 141 (71.9) | 131 (66.2) |
| Age at onset, mean (SD) | 65.9 (10.8) | 70.4 (8.3) |
| Age at death, mean (SD) | 76.0 (8.6) | 78.3 (6.6) |
| Disease duration, mean (SD) | 10.0 (7.4) | 7.9 (5.5) |
| LBD type/Kosaka, n (%) | ||
| BLBD | 54 (27.6) | 6 (3.0) |
| TLBD | 84 (42.9) | 34 (17.2) |
| DLBD | 58 (29.6) | 158 (79.8) |
| Thal phase, median (Q1, Q3) | 1 (0, 2) | 4 (3, 5) |
| Braak NFT stage, median (Q1, Q3) | II (II, III) | IV (III, V) |
ADpath = Alzheimer's disease co-pathology; BLBD = brainstem Lewy body disease; DLBD = diffuse Lewy body disease; NFT = neurofibrillary tangle; Q1 = 1st quartile; Q3 = 3rd quartile; SD = standard deviation; TLBD = transitional Lewy body disease.
AD-PRS is associated with the level of AD co-pathology in LBD
As expected, a higher genetic risk for AD was strongly associated with all measures of AD pathology in the NBB cohort. These included Thal phase, Braak NFT stage and CERAD score as well as the dichotomized AD-score (Table 3). The associations between AD-PRS and measures of AD pathology were replicated in the Mayo Clinic cohort (Table 3). In the Mayo Clinic cohort, these associations were also significant when removing the APOE component from the AD-PRS [Mayo Clinic cohort: Thal phase P = 0.044, 95% confidence interval (CI) of odds ratio (OR) = 1.0–1.44; Braak NFT stage P = 0.0095, 95% CI of OR = 1.06–1.52; AD-score P = 0.032, 95% CI of OR = 1.02–1.55].
Table 3.
Association between polygenic risk for AD and measures of AD pathology in samples with LBD, regardless of level of concomitant AD pathology
| Outcome | PRS model | OR | 95% CI | P-value |
|---|---|---|---|---|
| NBB discovery cohort (n = 217) | ||||
| CERAD score | AD-PRS | 2.14 | 1.61–2.85 | 1.5 × 10−7* |
| Thal phase | AD-PRS | 2.08 | 1.60–2.71 | 5.5 × 10−8* |
| Braak NFT stage | AD-PRS | 1.39 | 1.08–1.78 | 0.010* |
| AD-pathology score | AD-PRS | 1.84 | 1.33–2.60 | 3.2 × 10−4* |
| Mayo Clinic replication cohort (n = 394) | ||||
| Thal phase | AD-PRS | 2.07 | 1.70–2.52 | 3.5 × 10−13* |
| Braak NFT stage | AD-PRS | 1.75 | 1.45–2.11 | 5.1 × 10−9* |
| AD-pathology score | AD-PRS | 2.04 | 1.62–2.62 | 5.7 × 10−9* |
Associations were assessed in proportional odds ordinal logistic regression models. AD = Alzheimers' disease; NBB = Netherlands Brain Bank; NFT = neurofibrillary tangle; OR = odds ratio; PRS = polygenic risk score. *P < 0.05.
A risk score predicts AD co-pathology from the AD-PRS, age at onset and sex
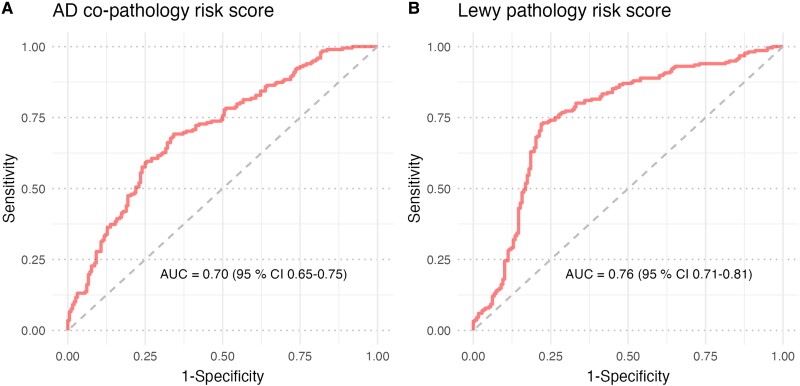
To investigate the power to distinguish LBD − ADpath from LBD + ADpath based on genetics and basic demographic variables, we generated an AD co-pathology risk score for each Mayo Clinic donor based on coefficients from ordinal logistic regression in the NBB data, where the model included AD-PRS, age at onset and sex. The AUC for this score was 0.70 (95% CI 0.65–0.75) (Fig. 1A).
Figure 1.
Performance of prediction models for AD co-pathology and Lewy pathology. (A) The AD) co-pathology risk score was calculated in Mayo Clinic samples (n = 394) based on coefficient weights for AD-PRS, sex and age at onset from ordinal logistic regression in Netherlands Brain Bank (NBB) samples (n = 213). (B) The Lewy pathology risk score was calculated in Mayo Clinic samples (n = 394) based on coefficient weights for lysosomal PD-PRS, AD-pathology, sex and age at onset from proportional odds ordinal logistic regression in NBB samples (n = 217). AD-PRS = Alzheimer's disease polygenic risk score; PD-PRS = Parkinson's disease polygenic risk score.
Lysosomal PRS is associated with Lewy pathology in LBD without AD co-pathology
In a sample set including donors both with and without LBD, by definition, diagnosis alone would drive an association between PD-PRS and Lewy pathology. However, there is also an interesting variation ‘within’ the LBD group, where some have more widespread Lewy pathology than others. This difference is potentially relevant for clinical trials but is not currently well captured by any available in vivo biomarker. We hypothesized that the way genetic burden is distributed across specific disease pathways partly determines the extent of Lewy pathology in the individual LBD patient. Several recent reports have indicated that the genetic architecture of ‘pure’ LBD may be different from cases where Lewy pathology co-exists with changes associated with AD. To further investigate this hypothesis in a neuropathological context and identify genetic drivers of Lewy pathology, we split each cohort into two subgroups based on the level of AD co-pathology and assessed ordinal logistic regression models separately for each subgroup. Results from association analyses of Lewy pathology are presented in Table 4. Based on previous reports, we expected genetic risk factors for PD to have the strongest effect on the Lewy pathology stage in the subgroup of cases without AD co-pathology.20,44,45 In line with this hypothesis, a general PD-PRS was associated with Lewy pathology stage in LBD − ADpath samples in the NBB cohort. However, this signal did not replicate in the Mayo Clinic cohort. In contrast, LBD + ADpath subgroups showed no clear trend towards association between the Lewy pathology stage and PD-PRS in either cohort.
Table 4.
Associations between PD polygenic risk scores and Lewy pathology, stratified by level of AD co-pathology
| PRS model | LBD − ADpath | LBD + ADpath | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value | |
| NBB discovery cohort (LBD − AD, n = 161, LBD + AD, n = 56) | ||||||
| Full PD-PRS | 1.62 | 1.15–2.27 | 0.0055* | 0.70 | 0.24–2.07 | 0.52 |
| Adaptive immunity | 1.15 | 0.85–1.57 | 0.37 | 0.44 | 0.16–1.20 | 0.11 |
| α-Synuclein | 1.15 | 0.84–1.58 | 0.39 | 0.62 | 0.23–1.66 | 0.34 |
| Endocytic membrane trafficking | 0.92 | 0.67–1.25 | 0.58 | 1.16 | 0.38–3.54 | 0.80 |
| Innate immunity | 1.04 | 0.76–1.42 | 0.81 | 0.52 | 0.18–1.53 | 0.24 |
| Lysosomal | 1.48 | 1.04–2.09 | 0.027* | 1.62 | 0.44–5.94 | 0.47 |
| Lysosomal excluding GBA | 1.25 | 0.90–1.73 | 0.18 | 2.32 | 0.74–7.25 | 0.15 |
| Microglia | 1.21 | 0.87–1.67 | 0.25 | 1.21 | 0.43–3.39 | 0.72 |
| Mitochondria | 0.93 | 0.68–1.28 | 0.66 | 2.90 | 0.81–10.4 | 0.10 |
| Monocytes | 1.08 | 0.78–1.49 | 0.64 | 0.77 | 0.24–2.53 | 0.67 |
| Mayo Clinic replication cohort (LBD − AD, n = 196) | ||||||
| Full PD-PRS | 0.98 | 0.75–1.28 | 0.86 | – | – | – |
| Lysosomal | 1.46 | 1.11–1.92 | 0.0070* | – | – | – |
| Lysosomal excluding GBA | 1.42 | 1.08–1.86 | 0.011* | – | – | – |
Proportional odds (PO) ordinal logistic regression with Braak Lewy pathology stage (NBB) or Kosaka’s LBD type (Mayo Clinic) as outcome. Binary rather than ordinal logistic regression was used in the NBB LBD + ADpath analysis (see ‘Materials and methods’ section). ADpath = Alzheimer's disease co-pathology; CI = confidence interval; LBD = Lewy body disease; NBB = Netherlands Brain Bank; OR = odds ratio; PD-PRS = Parkinson's disease polygenic risk score. *P < 0.05.
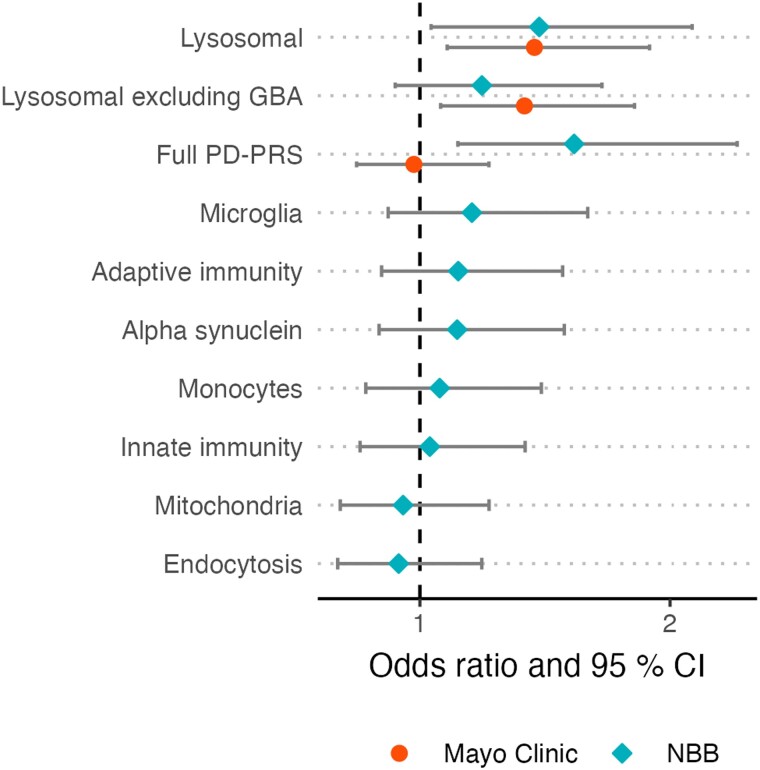
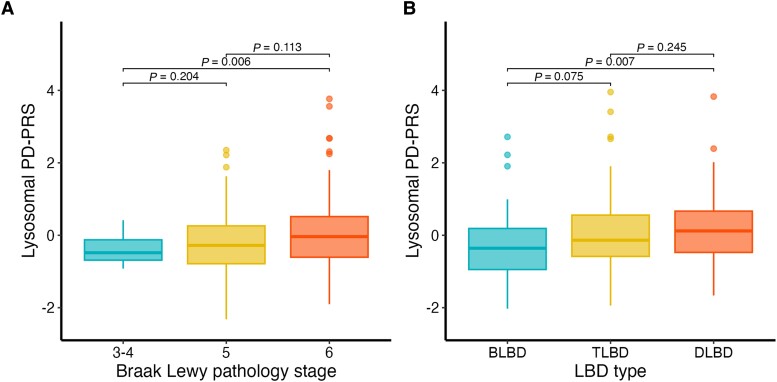
We then moved on to investigate the association with eight stratified PD-PRS capturing common genetic risk variants annotated to specific pathways and cell types (Fig. 2). One of these risk scores, the lysosomal PD-PRS, was associated with Lewy pathology in the LBD − ADpath group in the NBB cohort. This association was replicated in the Mayo clinic cohort (Table 4). We note that the Lewy pathology was classified differently across the two cohorts, although both analyses included three stages. Notwithstanding this methodological difference, the effect size was similar, with 1 SD increase in lysosomal PD-PRS corresponding to an odds ratio of 1.48 (NBB) or 1.46 (Mayo Clinic), respectively, for a higher Lewy pathology stage (Fig. 3). As GBA variants are known strong risk factors for both PD and DLB, and have previously been linked to Lewy pathology, we also generated lysosomal PD-PRS excluding the GBA region. The association with Lewy pathology stage in LBD − ADpath samples remained significant in the Mayo Clinic dataset and showed a similar trend in the NBB dataset, indicating that other lysosomal genes also contribute to Lewy pathology burden in LBD − ADpath.
Figure 2.
Associations between Lewy pathology and PRS in the subgroup without AD co-pathology. The figure shows the effect size and confidence intervals of the association between different polygenic risk scores (PD-PRS) and Lewy pathology stage in the subgroup without Alzheimer’s disease co-pathology (LBD − ADpath). Hypothesis testing was performed using proportional odds ordinal logistic regression models and associations passing P < 0.05 in the Netherlands Brain Bank (NBB) cohort (n = 161) were followed-up in the Mayo Clinic cohort (n = 196). ADpath = Alzheimer's disease co-pathology; LBD = Lewy body disease; PD-PRS = Parkinson's disease polygenic risk score.
Figure 3.
Box plots illustrating increasing lysosomal PD-PRS with higher Lewy pathology stages in LBD − ADpath samples. The figure shows box plots of lysosomal PD-PRS for different stages of Lewy pathology in the Netherlands Brain Bank (NBB) (A) and Mayo Clinic (B) data for the subgroup without AD co-pathology (n = 161 and 196, respectively). Plots are created with the default R parameters where the box ranges from the first (Q1) to the third (Q3) quartile and whiskers extend to the most extreme observation that is less than 1.5 times the Q1–Q3 distance from the box. Mean lysosomal PD-PRS between the samples with different levels of Lewy pathology were compared with t-tests. AD = Alzheimer's disease; BLBD = brainstem Lewy body disease; DLBD = diffuse Lewy body disease; PD-PRS = Parkinson's disease polygenic risk score; TLBD = transitional Lewy body disease.
No significant association between stratified PD-PRS were observed in the LBD + ADpath subgroup; however, less variation in Lewy pathology was observed with a majority of donors in the highest stages, thus statistical power was limited.
A joint model of AD co-pathology and lysosomal PRS predicts Lewy pathology stage
The neuropathological data indicate that LBD donors with intermediate or high AD co-pathology are likely also to have the most advanced stage of Lewy pathology. In ‘pure’ LBD, without AD co-pathology, we identified a higher lysosomal genetic burden as a risk factor for higher Lewy pathology stage. From a clinical perspective, the presence of AD co-pathology can be assessed by amyloid PET imaging or CSF biomarkers in living patients. To assess how well we could predict the Lewy pathology stage from genetics combined with data on AD co-pathology in LBD patients, we performed ordinal regression in the full NBB cohort with both dichotomized AD co-pathology status and lysosomal PD-PRS included in the same model. The lysosomal PD-PRS remained independently significant, with only marginally weaker effect size (odds ratio 1.40, 95% CI 1.03–1.89, P = 0.030). We used the coefficients from the NBB ordinal logistic regression to generate a joint risk score for neocortical Lewy pathology in the Mayo Clinic dataset. The score had an AUC of 0.76 (95% CI 0.71–0.81) for predicting DLBD (Fig. 1B).
AD-PRS and lysosomal PD-PRS are associated with dementia onset
To further investigate if the highlighted PRS also associate with disease progression we performed survival analysis for AD-PRS and lysosomal PD-PRS and time to dementia in NBB donors. In a Cox proportional hazards model, AD-PRS was associated with a shorter time between disease onset and dementia diagnosis (hazard ratio 1.36 per SD increase in PRS, 95% CI 1.15–1.61, P = 0.00040) when all NBB samples were analysed together. When the samples were split based on the level of AD co-pathology the lysosomal PD-PRS was also associated with a shorter time to dementia in the LBD − ADpath samples (hazard ratio 1.31 per SD increase in PRS, 95% CI 1.07–1.62, P = 0.010), but not in LBD + ADpath samples (hazard ratio 0.81 per SD increase in PRS, 95% CI 0.54–1.22, P = 0.32).
Discussion
An increasing number of genetic variants are recognized as risk factors for LBD, but the specific genetic architecture of the underlying neuropathological substrate of disease remains undetermined. To address this knowledge gap, we explored the association between AD-PRS, PD-PRS, and pathway-stratified PD-PRSs with key neuropathological measures in thoroughly characterized LBD samples from two independent brain bank cohorts. The relationship between neuropathological outcomes and PRS were assessed using ordinal logistic regression models that take into account the ordered fashion of the neuropathological stages. Firstly, we showed that AD-PRS was associated with co-morbid amyloid-β and tau pathology in samples with LBD. Secondly, we provide evidence that a stratified PD-PRS reflecting the genetic burden on the lysosomal pathway is associated with Lewy pathology in LBD, specifically in the subgroup without AD co-pathology. Interestingly, this pathway-stratified PRS showed a stronger and more consistent association with Lewy pathology than the overall PD-PRS, which includes a larger number of risk variants reflecting several biological pathways. Thirdly, our data suggests that both the AD-PRS and lysosomal PD-PRS are associated with an accelerated onset of dementia, the latter specifically in donors without AD co-pathology. Our findings provide novel insights into the complex relationships between neuropathology and genetics and indicate a future potential for the use of multiple, specific PRSs in clinical patient stratification.
As expected, we observed a strong association between AD-PRS and AD co-pathology, including measures of both amyloid-β and tau pathology in both datasets. AD-PRS also remained strongly associated with AD pathology in the larger Mayo Clinic dataset after removing the APOE component, indicating that genetic risk variants beyond APOE influence amyloid-β and tau pathology. Previously, PRS based on AD GWAS have also been shown to be associated with AD pathology in brain bank cohorts of AD patients.46-48 In samples from LBD patients, the APOE ε4 allele has in several reports been associated with higher likelihood of both amyloid-β pathology and tau NFT co-pathology,2,36,49-51 including studies from the NBB36 and Mayo Clinic51 based on sample sets overlapping those of our present study. Moreover, a ‘clinical-genetic risk score’ based on APOE ε4 alleles, two additional risk SNPs (BIN1 and SORL1) and age at onset, was reported to predict the presence of intermediate or high AD co-pathology in samples with LBD.52 Our study adds to prior findings by showing association between a full AD-PRS and the two key measures of AD-pathology in post-mortem LBD samples, also demonstrating significance of polygenic burden when the strong APOE effect is excluded.
Genetic association studies of Lewy pathology have thus far focused primarily on targeted variants in the SNCA, MAPT, GBA, and APOE loci.20,44,49,51,53 In a previous Mayo Clinic study, Heckman and colleagues did not find an association between a PD genetic risk score and Lewy body count or LBD subtype, yet the analysis did not stratify by the presence of AD co-pathology.54 In the present study, the full PD-PRS was associated with Lewy pathology only in the LBD − ADpath subgroup of the NBB cohort, but did not replicate in the Mayo Clinic cohort. The absence of a consistent strong signal for the general PD-PRS indicates that not all PD risk factors act through mechanisms that increase Lewy pathology, underlining the rationale for a pathway-stratified PRS approach. In principle, genetic variants not associated with Lewy pathology could increase PD risk through mechanisms that primarily cause neuronal loss.
To our knowledge, there are no previous reports examining associations between pathway-stratified PRS and neuropathological outcomes in samples with LBD. Testing eight selected PRS stratified by specific pathways or cell-types we found that the lysosomal PD-PRS was associated with Lewy pathology in both the discovery and replication cohort. This result is highly plausible considering previous research. Genetic studies have provided a link between lysosomal function and LBD risk. Heterozygous mutations in the GBA gene, which in the biallelic state are known to cause the lysosomal storage disorder Gaucher’s disease, are major genetic risk factors for both PD55,56 and DLB,57 conferring a more than 5-fold increase of PD risk,55 6-fold increase in risk of PD with dementia57 and more than 8-fold increase in DLB risk.57 Moreover, low-frequency variants in the GBA locus have consistently shown significant association with both PD and DLB in GWAS.3-5,58,59 Neuropathological studies of patients carrying GBA variants have demonstrated that these patients tend to have severe Lewy pathology,44,45 although a small neuropathological study found no significant association comparing to sporadic PD.60
In the current study, we suspected variation in the GBA locus to be a strong driver of the lysosomal PD-PRS signal. However, even after removing the GBA component, the lysosomal PD-PRS remained significantly associated with Lewy pathology in the LBD − ADpath samples from the Mayo Clinic, indicating that lysosomal variants beyond GBA are involved. Substantial evidence suggests a wider contribution of lysosomal mechanisms in LBD liability and pathogenesis. In addition to GBA, other genes involved in lysosomal functioning have been nominated by both PD and DLB GWAS, including SCARB2, TMEM175, CTSB, ATP6V0A1, GALC, GUSB, GRN and NEU1.3,4,58,59,61 Moreover, an excessive burden of lysosomal storage disorder gene variants has been found in PD.56 Our findings add to these insights by showing that the cumulative effect of multiple gene loci converging on the lysosomal pathway increases the Lewy pathology burden in LBD.
Interestingly, the association between lysosomal PD-PRS and Lewy pathology was specific to the subgroup of donors with no or low AD co-pathology. In agreement with this result, GBA has been proposed to be associated with ‘pure’ LBD with extensive Lewy pathology and less severe AD co-pathology, supported by several autopsy studies.20,44,45 Data from neuropathological post-mortem studies are corroborated by a recent study where CSF biomarkers were used as an in vivo proxy of AD co-pathology. Here, van der Lee and colleagues found the GBA p.E365K variant to be more strongly associated with ‘pure’ DLB than DLB with AD co-pathology.10 In a large neuropathological study of different forms of dementia, a DLB-PRS was associated with Lewy pathology stage only if the APOE component was excluded.48 The reason why genetic association signals differ depending on the degree of AD co-pathology is currently unclear. One possible explanation could be that AD-related changes make the brain more susceptible to additional neuropathologies, creating a vulnerable environment where genetic risk factors specific to LBD are relatively less important.
Genetics and neuropathology probably also contribute to shaping the clinical progression of disease. LBD + ADpath donors had a later disease onset and more rapid progression to dementia than LBD − ADpath donors. Further, we showed that the AD-PRS was associated with a more rapid progression to dementia, in line with longitudinal and cross-sectional studies that have shown that the APOE E4 allele is associated with an increased risk of cognitive decline and dementia in PD.36,62-64 Notably, the lysosomal PD-PRS was also associated with a faster development of dementia, exclusively in the LBD − ADpath samples. Several studies have shown that GBA variants (both pathogenic and non-pathogenic) adversely affects the prognosis of PD, including increasing the risk of dementia in PD patients.65-67
A strength of our study is the relatively large sample size, including a total of more than 600 neuropathologically characterized LBD cases. However, the NBB and Mayo Clinic cohorts differ in several important ways, the latter being larger and having a much larger proportion of donors positive for AD co-pathology. Furthermore, there are differences in neuropathological assessment between the two brain banks. In particular, the protocols for defining the Lewy pathology stage and Thal phases were not identical. However, both staging schemes for Lewy pathology are based on the same assumption of a generally caudal to rostral progression Lewy pathology within the CNS,23 and a high correlation between the two protocols for determining Thal phases used in this study has previously been reported.68 Differences in neuropathological methodology are likely to make signals less reproducible across the cohorts. This represents a clear limitation to our study design, yet with respect to the findings that we do highlight as consistent across both datasets, similar results across heterogeneous independent cohorts are arguably also an indication of methodologically robust signals.
We acknowledge that post-mortem datasets are skewed towards advanced disease stages and will not be representative of living patients the same age, limiting the relevance for e.g. patient stratification in clinical trials. Furthermore, we chose to emphasize the neuropathology of LBD as a common group, with the caveat that there might be relevant differences between PD and DLB that are not captured by our design. Limited statistical power led us to select eight pathways and cell types for stratified PD-PRS based on previous literature, although a hypothesis-free approach would have been preferable. The analysis of time to dementia was performed only in 211 donors with available data in the NBB dataset and should therefore be regarded as exploratory and interpreted with particular caution. Finally, our analysis was restricted to donors of European ancestry, a reminder that efforts to extend PD research to underrepresented populations should also involve brain bank donor programs.
Our study holds promise that PRS may be useful as an enrichment marker of Lewy pathology and AD co-pathology in future clinical trials assessing the effect of therapeutics targeting α-syn, amyloid-β or tau. Future work should also aim to further characterize the relationship between the correlations studied here and clinical outcomes in LBD. A number of large clinico-genetic studies have recently started to shed light on the genetics basis of clinical variability in PD, yet the positive associations with established risk variants from GWAS have been few and inconsistent.63,67,69 Our findings suggest that endophenotypes such as neuropathology could capture relevant pathological processes with higher precision than clinical symptoms, thereby providing a promising path for further genetic association studies.
Conclusion
In this study on neuropathologically defined samples from two independent cohorts we show that genetic variants known to be associated with the risk of AD and PD also influence key neuropathological measures in LBD donors. We extend the current knowledge about the influence of AD risk variants on AD co-pathology. Furthermore, we provide novel evidence that genetic variants linked to the lysosomal pathway are associated with higher Lewy pathology stage in ‘pure’ LBD. Our findings hold promise that larger genetic association studies of neuropathology and other endophenotypes will provide further insights into the pathogenic mechanisms of LBD. With further refinements, it is also our hope that a more fine-grained understanding of polygenic risk will make stratified PRS a useful tool for patient stratification in a precision medicine context.
Supplementary Material
Acknowledgements
The PD GWAS summary statistics used in this study were generated in a meta-analysis including data from 23andMe, Inc. We would like to thank the research participants and employees of 23andMe for making this work possible. The full GWAS summary statistics for the 23andMe discovery data set will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/dataset-access/for more information and to apply to access the data. The authors are grateful to the Netherlands Brain Bank and its funders for providing the samples that made the study possible.
Contributor Information
Jon-Anders Tunold, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway; Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, 0372 Oslo, Norway.
Manuela M X Tan, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway.
Shunsuke Koga, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Hanneke Geut, Department of Anatomy and Neurosciences, Amsterdam UMC, Vrije Universiteit, 1081 HV Amsterdam, The Netherlands.
Annemieke J M Rozemuller, Department of Anatomy and Neurosciences, Amsterdam UMC, Vrije Universiteit, 1081 HV Amsterdam, The Netherlands; Department of Pathology, Amsterdam UMC, Vrije Universiteit, 1081 HV Amsterdam, The Netherlands; Program Neurodegeneration, Amsterdam Neuroscience, 1081 HV Amsterdam, The Netherlands.
Rebecca Valentino, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Hiroaki Sekiya, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Nicholas B Martin, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Michael G Heckman, Division of Clinical Trials and Biostatistics, Mayo Clinic, Jacksonville, FL 32224, USA.
Jose Bras, Department of Neurodegenerative Science, Van Andel Institute, Grand Rapids, MI 49503, USA; Division of Psychiatry and Behavioral Medicine, Michigan State University College of Human Medicine, Grand Rapids, MI 49503, USA.
Rita Guerreiro, Department of Neurodegenerative Science, Van Andel Institute, Grand Rapids, MI 49503, USA; Division of Psychiatry and Behavioral Medicine, Michigan State University College of Human Medicine, Grand Rapids, MI 49503, USA.
Dennis W Dickson, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Mathias Toft, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway; Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, 0372 Oslo, Norway.
Wilma D J van de Berg, Department of Anatomy and Neurosciences, Amsterdam UMC, Vrije Universiteit, 1081 HV Amsterdam, The Netherlands; Program Neurodegeneration, Amsterdam Neuroscience, 1081 HV Amsterdam, The Netherlands.
Owen A Ross, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Lasse Pihlstrøm, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway.
Funding
J.-A.T. and L.P. were funded by grants from the South-Eastern Norway Regional Health Authority (Helse Sør-Øst RHF, Norway). W.B. received funding from the Dutch Parkinson association, Health Holland, and Rotary Club Aalsmeer-Uithoorn. O.A.R. is supported by NIH (RF1 NS085070; U54-NS100693; U01 NS100620; R01 AG056366; U19 AG071754), DOD (W81XWH-17-1-0249), The Michael J. Fox Foundation, The Little Family Foundation, the Mayo Clinic Dorothy and Harry T. Mangurian Jr. Lewy Body Dementia Program at Mayo Clinic, Ted Turner and family with the Functional Genomics of LBD Program and the Mayo Clinic LBD Center without Walls (U54-NS110435). Mayo Clinic (O.A.R.) was a recipient of the inaugural Cure One, Cure Many Award from the American Brain Foundation for the study of Lewy body dementia. Mayo Clinic is an American Parkinson Disease Association (APDA) Mayo Clinic Information and Referral Center, an APDA Center for Advanced Research, and a Lewy Body Dementia Association (LBDA) Research Center of Excellence.
Competing interests
The authors declare no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. [DOI] [PubMed] [Google Scholar]
- 2. Robinson JL, Lee EB, Xie SX, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain. 2018;141:2181–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chia R, Sabir MS, Bandres-Ciga S, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 2021;53:294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guerreiro R, Ross OA, Kun-Rodrigues C, et al. Investigating the genetic architecture of dementia with Lewy bodies: A two-stage genome-wide association study. Lancet Neurol. 2018;17:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Escott-Price V, Nalls MA, et al. Polygenic risk of Parkinson disease is correlated with disease age at onset. Ann Neurol. 2015; 77:582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ibanez L, Dube U, Saef B, et al. Parkinson Disease polygenic risk score is associated with Parkinson disease status and age at onset but not with alpha-synuclein cerebrospinal fluid levels. BMC Neurol. 2017;17:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pihlstrom L, Morset KR, Grimstad E, Vitelli V, Toft M. A cumulative genetic risk score predicts progression in Parkinson's disease. Mov Disord. 2016;31:487–490. [DOI] [PubMed] [Google Scholar]
- 9. Paul KC, Schulz J, Bronstein JM, Lill CM, Ritz BR. Association of polygenic risk score with cognitive decline and motor progression in Parkinson disease. JAMA Neurol. 2018;75:360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van der Lee SJ, van Steenoven I, van de Beek M, et al. Genetics contributes to concomitant pathology and clinical presentation in dementia with Lewy bodies. J Alzheimers Dis. 2021;83:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bandres-Ciga S, Saez-Atienzar S, Kim JJ, et al. Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease. Acta Neuropathol. 2020;140:341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersen MS, Bandres-Ciga S, Reynolds RH, et al. Heritability enrichment implicates microglia in Parkinson's disease pathogenesis. Ann Neurol. 2021;89:942–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Billingsley KJ, Barbosa IA, Bandres-Ciga S, et al. Mitochondria function associated genes contribute to Parkinson's disease risk and later age at onset. NPJ Parkinsons Dis. 2019;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bandres-Ciga S, Saez-Atienzar S, Bonet-Ponce L, et al. The endocytic membrane trafficking pathway plays a major role in the risk of Parkinson's disease. Mov Disord. 2019;34:460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamilton RL. Lewy Bodies in Alzheimer's disease: A neuropathological review of 145 cases using alpha-synuclein immunohistochemistry. Brain Pathol. 2000;10:378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Irwin DJ, White MT, Toledo JB, et al. Neuropathologic substrates of Parkinson disease dementia. Ann Neurol. 2012;72:587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dugger BN, Adler CH, Shill HA, et al. Concomitant pathologies among a spectrum of parkinsonian disorders. Parkinsonism Relat Disord. 2014;20:525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walker L, Stefanis L, Attems J. Clinical and neuropathological differences between Parkinson's disease, Parkinson's disease dementia and dementia with Lewy bodies—current issues and future directions. J Neurochem. 2019;150:467–474. [DOI] [PubMed] [Google Scholar]
- 19. Irwin DJ, Grossman M, Weintraub D, et al. Neuropathological and genetic correlates of survival and dementia onset in synucleinopathies: a retrospective analysis. Lancet Neurol. 2017;16:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaivola K, Shah Z, Chia R, International LBDGC, Scholz SW. Genetic evaluation of dementia with Lewy bodies implicates distinct disease subgroups. Brain. 2022;145:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montine TJ, Phelps CH, Beach TG, et al. National institute on aging-Alzheimer's association guidelines for the neuropathologic assessment of Alzheimer's disease: a practical approach. Acta Neuropathol. 2012;123:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alafuzoff I, Arzberger T, Al-Sarraj S, et al. Staging of neurofibrillary pathology in Alzheimer's disease: A study of the BrainNet Europe consortium. Brain Pathol. 2008;18:484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alafuzoff I, Ince PG, Arzberger T, et al. Staging/typing of Lewy body related alpha-synuclein pathology: a study of the BrainNet Europe consortium. Acta Neuropathol. 2009;117:635–652. [DOI] [PubMed] [Google Scholar]
- 24. Thal DR, Rub U, Schultz C, et al. Sequence of Abeta-protein deposition in the human medial temporal lobe. J Neuropathol Exp Neurol. 2000;59:733–748. [DOI] [PubMed] [Google Scholar]
- 25. Mirra SS, Heyman A, McKeel D, et al. The consortium to establish a registry for Alzheimer's disease (CERAD). part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. [DOI] [PubMed] [Google Scholar]
- 26. Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. J Neurol Neurosurg Psychiatry. 1988;51:745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dickson DW, Braak H, Duda JE, et al. Neuropathological assessment of Parkinson's disease: Refining the diagnostic criteria. Lancet Neurol. 2009;8:1150–1157. [DOI] [PubMed] [Google Scholar]
- 28. McKeith IG, Boeve BF, Dickson DW, et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB consortium. Neurology. 2017;89:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emre M, Aarsland D, Brown R, et al. Clinical diagnostic criteria for dementia associated with Parkinson's disease. Mov Disord. 2007;22:1689–1707. [DOI] [PubMed] [Google Scholar]
- 30. Koga S, Zhou X, Dickson DW. Machine learning-based decision tree classifier for the diagnosis of progressive supranuclear palsy and corticobasal degeneration. Neuropathol Appl Neurobiol. 2021;47:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dickson DW, Liu W, Hardy J, et al. Widespread alterations of alpha-synuclein in multiple system atrophy. Am J Pathol. 1999;155:1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kosaka K, Yoshimura M, Ikeda K, Budka H. Diffuse type of Lewy body disease: Progressive dementia with abundant cortical Lewy bodies and senile changes of varying degree–a new disease? Clin Neuropathol. 1984;3:185–192. [PubMed] [Google Scholar]
- 33. Koga S, Sekiya H, Kondru N, Ross OA, Dickson DW. Neuropathology and molecular diagnosis of synucleinopathies. Mol Neurodegener. 2021;16:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hyman BT, Phelps CH, Beach TG, et al. National Institute on Aging-Alzheimer's association guidelines for the neuropathologic assessment of Alzheimer's disease. Alzheimers Dement. 2012;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blauwendraat C, Faghri F, Pihlstrom L, et al. Neurochip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol Aging. 2017;57:247 e249-247 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tunold JA, Geut H, Rozemuller JMA, et al. APOE And MAPT are associated with dementia in neuropathologically confirmed Parkinson's disease. Front Neurol. 2021;12:631145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet. 2019;51:404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi SW, O'Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8:giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Corces MR, Buenrostro JD, Wu B, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nott A, Holtman IR, Coufal NG, et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 2019;366:1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. VGAM: Vector Generalized Linear and Additive Models. Version R package version 1.1–7. 2022. https://CRAN.R-project.org/package=rms
- 43. rms: Regression Modeling Strategies. Version R package version 6.3–0. 2022. https://CRAN.R-project.org/package=rms
- 44. Clark LN, Kartsaklis LA, Wolf Gilbert R, et al. Association of glucocerebrosidase mutations with dementia with Lewy bodies. Arch Neurol. 2009;66:578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsuang D, Leverenz JB, Lopez OL, et al. GBA Mutations increase risk for Lewy body disease with and without Alzheimer disease pathology. Neurology. 2012;79:1944–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan CH, Bonham LW, Fan CC, et al. Polygenic hazard score, amyloid deposition and Alzheimer's neurodegeneration. Brain. 2019;142:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hannon E, Shireby GL, Brookes K, et al. Genetic risk for Alzheimer's disease influences neuropathology via multiple biological pathways. Brain Commun. 2020;2:fcaa167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spencer BE, Jennings RG, Fan CC, Brewer JB. Assessment of genetic risk for improved clinical-neuropathological correlations. Acta Neuropathol Commun. 2020;8:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Compta Y, Parkkinen L, O'Sullivan SS, et al. Lewy- and Alzheimer-type pathologies in Parkinson's disease dementia: Which is more important? Brain. 2011;134(Pt 5):1493–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lashley T, Holton JL, Gray E, et al. Cortical alpha-synuclein load is associated with amyloid-beta plaque burden in a subset of Parkinson's disease patients. Acta Neuropathol. 2008;115:417–425. [DOI] [PubMed] [Google Scholar]
- 51. Dickson DW, Heckman MG, Murray ME, et al. APOE Epsilon4 is associated with severity of Lewy body pathology independent of Alzheimer pathology. Neurology. 2018;91:1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dai DL, Tropea TF, Robinson JL, et al. ADNC-RS, a clinical-genetic risk score, predicts Alzheimer's pathology in autopsy-confirmed Parkinson's disease and dementia with Lewy bodies. Acta Neuropathol. 2020;140:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Colom-Cadena M, Gelpi E, Marti MJ, et al. MAPT H1 haplotype is associated with enhanced alpha-synuclein deposition in dementia with Lewy bodies. Neurobiol Aging. 2013;34:936–942. [DOI] [PubMed] [Google Scholar]
- 54. Heckman MG, Kasanuki K, Diehl NN, et al. Parkinson's disease susceptibility variants and severity of Lewy body pathology. Parkinsonism Relat Disord. 2017;44:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sidransky E, Nalls MA, Aasly JO, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med. 2009;361:1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robak LA, Jansen IE, van Rooij J, et al. Excessive burden of lysosomal storage disorder gene variants in Parkinson's disease. Brain. 2017;140:3191–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nalls MA, Duran R, Lopez G, et al. A multicenter study of glucocerebrosidase mutations in dementia with Lewy bodies. JAMA Neurol. 2013;70:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014;46:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang D, Nalls MA, Hallgrimsdottir IB, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet. 2017;49:1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Parkkinen L, Neumann J, O'Sullivan SS, et al. Glucocerebrosidase mutations do not cause increased Lewy body pathology in Parkinson's disease. Mol Genet Metabol. 2011;103:410–412. [DOI] [PubMed] [Google Scholar]
- 61. Do CB, Tung JY, Dorfman E, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet. 2011;7:e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huertas I, Jesus S, Garcia-Gomez FJ, et al. Genetic factors influencing frontostriatal dysfunction and the development of dementia in Parkinson's disease. PLoS One. 2017;12:e0175560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan MMX, Lawton MA, Jabbari E, et al. Genome-Wide association studies of cognitive and motor progression in Parkinson's disease. Mov Disord. 2021;36:424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu G, Peng J, Liao Z, et al. Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson’s disease. Nat Genet. 2021;53:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stoker TB, Camacho M, Winder-Rhodes S, et al. Impact of GBA1 variants on long-term clinical progression and mortality in incident Parkinson's disease. J Neurol Neurosurg Psychiatry. 2020; 91:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Winder-Rhodes SE, Evans JR, Ban M, et al. Glucocerebrosidase mutations influence the natural history of Parkinson's disease in a community-based incident cohort. Brain. 2013;136(Pt 2):392–399. [DOI] [PubMed] [Google Scholar]
- 67. Iwaki H, Blauwendraat C, Leonard HL, et al. Genetic risk of Parkinson disease and progression: An analysis of 13 longitudinal cohorts. Neurol Genet. 2019;5:e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Thal DR, Ronisz A, Tousseyn T, et al. Different aspects of Alzheimer's disease-related amyloid β-peptide pathology and their relationship to amyloid positron emission tomography imaging and dementia. Acta Neuropathol Commun. 2019;7:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Iwaki H, Blauwendraat C, Leonard HL, et al. Genomewide association study of Parkinson's disease clinical biomarkers in 12 longitudinal patients’ cohorts. Mov Disord. 2019;34:1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data on NBB donors that support the findings of this study can be obtained from the Netherlands Neurogenetics Database (https://www.brainbank.nl/nnd-project/). Mayo Clinic data are available from the authors on request. Analysis code used in this manuscript is available on GitHub at https://github.com/lpihlstrom/projects.