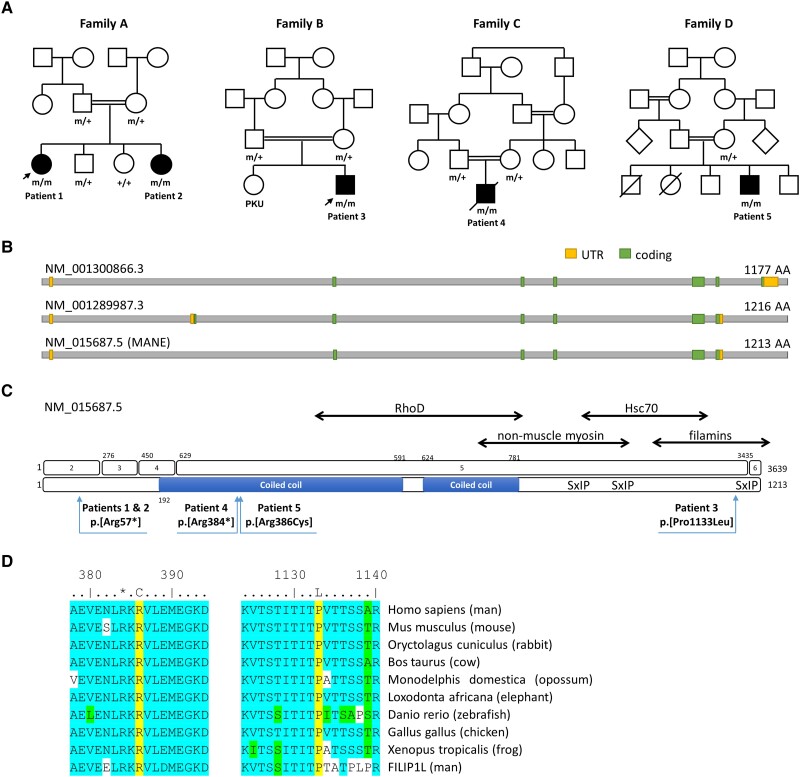
Figure 2.
Pedigrees of patients of our FILIP1-cohort and schematic representation of FILIP1 gene and protein sequence along with distribution and conservation of identified variants. (A) Pedigrees of the consanguineous families of our four patients with identified FILIP1-variants. +/+ = wild-type, +/m = heterozygous, m/m = homozygous mutant. Arrows indicate patients for which muscle biopsies were studied. (B) Schematic representation of the FILIP1 protein encoded by different transcript isoforms including the MANE transcript NM_015687.5. (C) Schematic representation of the gene and protein sequence of FILIP1/FILIP1. Exons are numbered 1–6 (top). Pathogenic variants on protein level refer to the canonical/MANE transcript NM_015687.5 (bottom). The predicted coiled coil region is shown as a blue bar. Known binding partners are shown at the top. SxIP indicates the position of three putative EB1 and EB3 binding sites. Further amino acid regions of FILIP1 mediating interactions with RhoD, non-muscle myosin IIb, HSC70 and filamins are indicated by double-headed arrows. (D) Sequence alignments showing that Arg386 and Pro1133 (both yellow) are conserved between species and between FILIP1 and FILIP1L. *Arg384. Many of the amino acids flanking the variant are highly conserved (blue) or similar (green). UTR = untranslated region.