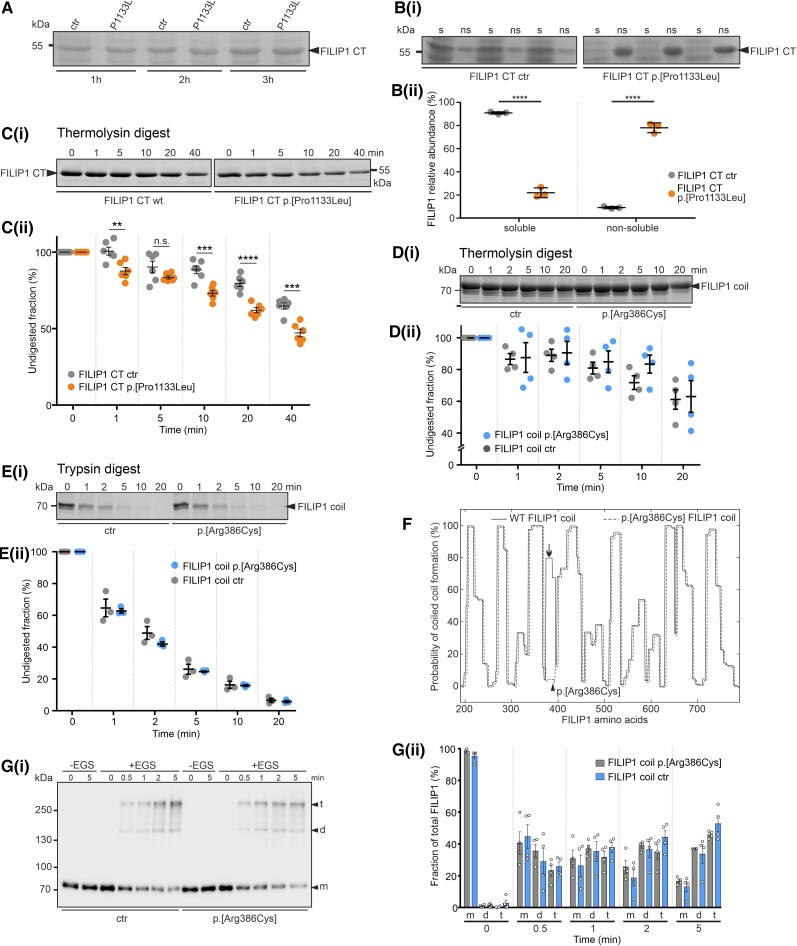
Figure 4.
In vitro studies addressing stability and solubility of recombinant FILIP1 CT and FILIP1 coil constructs. (A) SDS-PAGE analysis of total extracts of bacteria 1, 2 or 3 h after induction of wild-type (wt) or mutant (p.[Pro1133Leu]; mut) protein expression [carboxyterminus (CT), amino acids 776–1177]. Note that equal levels of protein (arrowhead) are expressed. [B(i and ii)] Investigation of the solubility of wt and mutant FILIP1 CT proteins (arrowhead). Note that the mutant protein is found almost exclusively in the non-soluble fraction (ns), whereas approximately 80% of the wt protein is soluble (s). [C(i and ii)] Investigation of protease susceptibility by native, limited thermolysin digestion: wt and mut FILIP1 CT fragments were treated with the protease thermolysin for 0–40 min, as indicated above each lane. Samples were analysed by SDS-PAGE. Note that significantly higher levels of the p.[Pro1133Leu] variant are digested after almost all incubation times, indicating decreased stability of the mutant protein. (D and E) Investigation of protease susceptibility of both FILIP1 coil variants by native, limited thermolysin [D(i and ii)] or trypsin [E(i and ii)] digestion. Recombinantly expressed FILIP1 coil fragments were treated with either thermolysin or trypsin for 0–20 min, as indicated above each lane. Samples were analysed by SDS-PAGE. Both variants do not show any significant differences in protease susceptibility. (F) Prediction of coiled coil formation in the central part of FILIP1. Amino acids 189-786 of both, FILIP1 and the p.[Arg386Cys] variant, are predicted to form extended coiled coil secondary structures when submitted to the COILS Server at embnet.vital-it.ch. Note however, the predicted total loss of coiled coil formation propensity of the region affected by the variation (arrow). The position of Arg386 is indicated by an arrowhead. The graph of the mutant protein was deliberately shifted a few pixels, to be able to distinguish both graphs. [G(i and ii)] Chemical cross-linking experiments using wt FILIP1 coil and the p.[Arg386Cys] variant. Assays were performed in the absence (−EGS) or presence (+EGS) of EGS. Reaction mixtures were analysed by western blotting using specific antibodies against the EEF-immunotag of the constructs. Without the cross-linker EGS, only monomers (m) of the constructs were detected. In the presence of EGS, within 0.5 min both variants were also found as dimers (d) or tetramers (t). With extension of the incubation time, the relative amount of protein found as tetramers increased to up to 45–65% of the total amount of protein. No significant differences were observed between both variants. Panels A–E and G contain cropped gels or blots. For full-size versions, see Supplementary Fig. 4. All data are the result of three to six independent experiments. All statistics were performed using GraphPad Prism and paired t-test. Data are shown as mean ± SEM. **P < 0.01; ***P < 0.001; ****P < 0.0001.