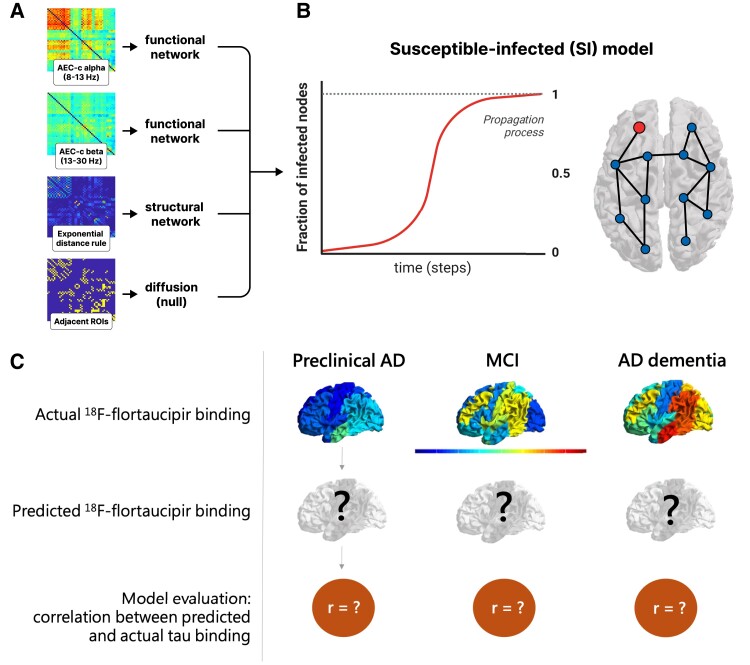
Abstract
Recent studies on Alzheimer’s disease (AD) suggest that tau proteins spread through the brain following neuronal connections. Several mechanisms could be involved in this process: spreading between brain regions that interact strongly (functional connectivity); through the pattern of anatomical connections (structural connectivity); or simple diffusion. Using magnetoencephalography (MEG), we investigated which spreading pathways influence tau protein spreading by modelling the tau propagation process using an epidemic spreading model. We compared the modelled tau depositions with 18F-flortaucipir PET binding potentials at several stages of the AD continuum.
In this cross-sectional study, we analysed source-reconstructed MEG data and dynamic 100-min 18F-flortaucipir PET from 57 subjects positive for amyloid-β pathology [preclinical AD (n = 16), mild cognitive impairment (MCI) due to AD (n = 16) and AD dementia (n = 25)]. Cognitively healthy subjects without amyloid-β pathology were included as controls (n = 25). Tau propagation was modelled as an epidemic process (susceptible-infected model) on MEG-based functional networks [in alpha (8–13 Hz) and beta (13–30 Hz) bands], a structural or diffusion network, starting from the middle and inferior temporal lobe. The group-level network of the control group was used as input for the model to predict tau deposition in three stages of the AD continuum. To assess performance, model output was compared to the group-specific tau deposition patterns as measured with 18F-flortaucipir PET. We repeated the analysis by using networks of the preceding disease stage and/or using regions with most observed tau deposition during the preceding stage as seeds.
In the preclinical AD stage, the functional networks predicted most of the modelled tau-PET binding potential, with best correlations between model and tau-PET [corrected amplitude envelope correlation (AEC-c) alpha C = 0.584; AEC-c beta C = 0.569], followed by the structural network (C = 0.451) and simple diffusion (C = 0.451). Prediction accuracy declined for the MCI and AD dementia stages, although the correlation between modelled tau and tau-PET binding remained highest for the functional networks (C = 0.384; C = 0.376). Replacing the control-network with the network from the preceding disease stage and/or alternative seeds improved prediction accuracy in MCI but not in the dementia stage.
These results suggest that in addition to structural connections, functional connections play an important role in tau spread, and highlight that neuronal dynamics play a key role in promoting this pathological process. Aberrant neuronal communication patterns should be taken into account when identifying targets for future therapy. Our results also suggest that this process is more important in earlier disease stages (preclinical AD/MCI); possibly, in later stages, other processes may be influential.
Keywords: Alzheimer’s disease, tau protein, spreading model, magnetoencephalography, functional connectivity
Schoonhoven et al. combine MEG data with 18F-flortaucipir PET to construct epidemic spreading models that mimic the propagation of tau tangles throughout the brain. The results show that the functional architecture of the brain plays a key role in determining the pattern by which tau pathology spreads in Alzheimer’s disease.
Introduction
Tau protein is a microtubule-associated protein that accumulates in the Alzheimer’s disease (AD) brain as neurofibrillary tangles. In vitro studies have suggested that tau proteins may propagate through the brain in a prion-like manner, and that this spreading is enhanced by neuronal activity.1-3 This has led to the ‘transneuronal spread’ hypothesis, in which a toxic agent (tau) propagates along connected neurons, driven by aberrant neuronal activity.4 However, evidence regarding the role of functional dynamics in AD remains incomplete.
Discovery of tau-binding PET radiotracers has made it possible to not only quantify the amount of aggregated tau proteins in the brain, but also to assess the spatial patterns of tau protein distribution in vivo. Currently, 18F-flortaucipir is the most widely used tracer to image tau pathology in vivo.5,6 Importantly, in vivo18F-flortaucipir PET studies have shown agreement with Braak-staging regarding the distribution of tau tangles throughout the brain.7,8 Owing to the availability of this tau-PET tracer, we can investigate the contribution of functional and structural network characteristics and the propagation of tau non-invasively.
Previous studies have combined functional MRI (fMRI) and tau-PET to investigate if, and how, tau deposition is related to functional connectivity and brain networks. These studies have provided evidence that tau topographies show spatial overlap with functional brain networks.9-12 Additionally, studies have shown that strongly connected regions accrue more tau pathology,13,14 yet the strength of the connections decreases with increasing tau pathology.13 These findings suggest that there is a close correlation between functional connectivity and tau levels shared by functionally connected brain regions. In another paper, Franzmeier and colleagues15 were able to improve tau spreading predictions over Braak staging methods by using functional connectivity networks. They found that tau accumulation rates correlated with connectivity strength to patient-specific tau epicentres. Interestingly, by using atlases based on either anatomical or fMRI-based functional connectivity to simulate tau spread, Vogel and colleagues16 found that a model based on anatomical connectivity performed better than the one based on functional connectivity, predicting 70% of the variance of the observed tau PET pattern while the functional connectivity model explained 58% of the observed variance.
In contrast to fMRI, magnetoencephalography (MEG) has the capability to directly assess neuronal activity of functional brain networks with high temporal resolution.17,18 Beamforming approaches make it possible to reconstruct functional networks of oscillatory activity at source level, enabling a direct comparison with 18F-flortaucipir PET. In a recent study, Ranasinghe and colleagues19 evaluated the association between 18F-flortaucipir PET and an MEG measure of functional connectivity between neuronal populations (imaginary coherence), and observed that 18F-flortaucipir uptake strongly co-localized with alpha band hypo-synchrony and delta-theta band hyper-synchrony. In a small early AD sample, another MEG study found that higher tau burden was related to a significant decline in both local and global efficiency of information transfer, as well as loss of functional connectivity and global level communication in higher frequency bands, whereas functional connectivity in the lower frequency bands increased.20 However, these studies were correlational, comparing tau-PET signal with brain-wide functional connectivity, and did not model spread over anatomical pathways or via functional connections. Additionally, both studies focused on functional connectivity, but did not study structural connectivity, which was found to be the best predictor of tau deposition by Vogel and colleagues.16
The main objective of the present pseudo-longitudinal study was to investigate the contribution of different types of network connectivity to the stereotypical spread of tau pathology (18F-flortaucipir retention) in the different stages of AD, i.e. preclinical AD, mild cognitive impairment due to AD, and dementia due to AD. Based on previous studies, we hypothesized that several mechanisms could be involved in this process: spreading between brain regions that interact strongly (functional connectivity); spreading through the pattern of anatomical/axonal connections (structural connectivity); or, as null hypothesis, simple diffusion through the extracellular space to spatially adjacent regions. We investigated this by modelling the tau propagation process on these different networks using an epidemic spreading model (ESM), a mathematical model that simulates the propagation of an agent from some given location to other connected areas.21 Such models have been used previously to study the spreading of pathological proteins on brain networks16,22 or propagation of seizure activity.23,24 We subsequently compared the modelled tau depositions with in vivo tau depositions at several stages of the AD continuum as measured with 18F-flortaucipir PET. Based on the strong correlates between tau and functional connectivity, as well as neuronal activity, we expected that tau propagates over anatomical connections, but that this is propagation is strongest for connections that are most used functionally. We therefore expected a higher correlation between the simulated tau load and tau-PET binding potentials for the functional network model than for the structural or diffusion network models.
Materials and methods
Subjects
Subjects were recruited from the Amsterdam Dementia Cohort, consisting of patients who visited the memory clinic at Alzheimer Centre Amsterdam, Amsterdam UMC. All subjects received a standardized diagnostic work-up including medical history taking, neurological and neuropsychological examination, laboratory testing, MRI, EEG or MEG and, if possible, a lumbar puncture to collect CSF.25 Cognitive function was assessed through a battery of neuropsychological tests, including the Mini-Mental State Examination (MMSE26) and Montreal Cognitive Assessment (MoCA27).
Diagnoses were generated during a multidisciplinary consensus meeting, according to recent international guidelines. Subjects were diagnosed with subjective cognitive decline (SCD) when reported cognitive complaints remained undetected through formal neuropsychological testing. Patients with mild cognitive impairment (MCI) due to AD exhibited objectively detectable cognitive deficits with preserved functional independence. A diagnosis of ‘probable AD dementia’ was assigned according to criteria of the National Institute on Aging-Alzheimer’s Association.28 The presence or absence of AD pathology was confirmed using CSF and/or amyloid-PET. To classify as amyloid-positive, subjects had to have either positive CSF amyloid-β1–42 and/or positive amyloid-PET. For CSF amyloid-β1–42, drift-corrected values were used; the cut-off was set at 813 pg/ml.29 When both amyloid-PET and CSF data were available, amyloid-PET was decisive.
To be eligible to participate in this study, subjects needed to fulfil the diagnostic criteria for SCD, MCI due to AD or probable AD dementia; be at least 50 years of age; and have known amyloid status (in the case of MCI and probable AD dementia, amyloid-positivity was required). Subjects were excluded when they met any of the following criteria: having a cardiac pacemaker, implantable cardioverter-defibrillator (ICD) or other intra-corporal devices interfering with MEG signals; severe claustrophobia; evidence of structural abnormality on MRI scan; females of childbearing potential who were using reliable methods for contraception; relevant history of severe drug allergy or hypersensitivity; participation in an experimental study with a tau or amyloid targeting agent; history of moderate or severe traumatic brain injury; and/or specific exclusion criteria for MRI scanning.
For the purpose of the study, subjects were finally split into four groups: amyloid-negative SCD (henceforth referred to as controls); amyloid-positive SCD (henceforth referred to as preclinical AD); MCI due to Ad; and AD dementia. As a result of a shift in diagnoses at the Alzheimer Center Amsterdam (‘MCI due to AD’ was often labelled as ‘early phase probable AD dementia’), MCI due to AD and probable AD dementia were taken together and subsequently divided into MCI and dementia using a median split based on MMSE. The local Institutional Review Board of the VUmc (Amsterdam, The Netherlands) approved the study protocol (2018.070). Subjects’ consent was obtained according to the Declaration of Helsinki and all subjects provided written informed consent prior to participating in the study.
In total, 85 subjects received both an MEG measurement and 18F-flortaucipir PET in the context of the study. Three 18F-flortaucipir PET scans did not meet scan quality criteria for tracer quantification, leaving a final study population of 82 subjects.
MRI acquisition procedure
Three-dimensional T1-weighted MRI scans were acquired for all patients on a 3.0 T Ingenuity TF PET/MR system (Philips Medical Systems) in the context of the memory clinic screening, within a maximum of 6 months from the 18F-flortaucipir PET scan. If the MRI scan and MEG measurement took place on the same day, the MRI was performed after the MEG measurement, to prevent artefacts arising from magnetization of, for instance, dental implants. If an MRI scan had been performed more than 6 months before the 18F-flortaucipir PET scan, or had not been performed on the PET/MR Ingenuity system, the MRI scan was repeated for this study.
18F-flortaucipir PET acquisition procedure
Subjects underwent a dual time point dynamic 18F-flortaucipir PET scan starting immediately after injection of 229 ± 12 MBq 18F-flortaucipir and including at least the 0–30 min and 80–100 min post-injection time interval as previously described.30,31 Scans were acquired on an Ingenuity TF PET/CT scanner (Philips Medical Systems), preceded by a low-dose CT scan for attenuation correction purposes. All PET data were 3D reconstructed using the vendor provided image reconstruction method (RAMLA) with a matrix size of 128 × 128 × 90 and a final voxel size of 2 × 2 × 2 m3. All standard corrections for attenuation, scatter, randoms, decay and dead time were performed.
MEG acquisition procedure
Subjects underwent MEG measurement within 6 months from the 18F-flortaucipir PET scan [median: 135 days, interquartile range (IQR): 81–159 days]. MEG data were acquired with a 306-channel whole-head MEG-system (Elekta Neuromag Oy), while subjects were in supine position inside a magnetically shielded room (VacuumSchmelze GmbH). For each subject, two 5-min eyes-closed resting state recordings were made. Subjects were instructed to open and close their eyes several times during the recordings to prevent drowsiness. Magnetic fields were recorded at a sample frequency of 1250 Hz, with an anti-aliasing filter of 410 Hz and a high-pass filter of 0.1 Hz. The subject’s head position in relation to the MEG sensors was recorded using signals from five head-localization coils.
PET and MEG analyses
Structural 3D T1-weighted magnetic resonance images were co-registered to the PET images using Vinci software (Max Planck Institute, Cologne, Germany). For each subject, regions of interest (ROIs) were defined on the co-registered MRI scan with the Hammers template,32 which is incorporated in PVElab, a software programme that uses a probability map of delineated (grey matter) ROIs that has been validated previously.33 These ROIs were then superimposed onto the dynamic PET scans to extract regional time activity curves (TACs). For 18F-flortaucipir, receptor parametric mapping (RPM)-derived binding potential (BPND), with grey matter cerebellum as reference region, were obtained, which is a validated method to quantify tau load.30,31 A partial volume correction method that combines iterative deconvolution methods with highly constrained back projection (HYPR) was applied to the 18F-flortaucipir BPND data for a better quantification of the dynamic PET signal.34
Raw MEG data were visually inspected (A.N. and D.N.S.) for malfunctioning and noisy channels, which were subsequently removed, after which the temporal extension of Signal Space Separation (tSSS) in MaxFilter software (Elekta Neuromag Oy, version 2.2.12)35 was applied to remove artefacts from the data.36 Data were subsequently filtered in the 0.5–48 Hz band using MaxFilter software. Each subject’s MEG data were co-registered with the PET co-registered MRI using surface matching, after which the same transformation was applied to the subject-space Hammers template. In order to reconstruct neuronal activity at source level, an atlas-based beamforming approach was applied to each 5-min resting state recording,37 using the centroids38 of 50 parcels in an adjusted version of the Hammers template (Supplementary Table 1). For each of these centroid voxels, time series of neuronal activity (so-called virtual electrodes) were reconstructed by projecting the sensor signals to source space. Broadband data (0.5–48 Hz) were used for the estimation of the beamformer weights, to avoid overestimation of covariance between channels,39 as well as a unity noise covariance matrix, a spherical head model (fitted to the scalp surface as extracted from the co-registered MRI), and an equivalent current dipole as source model. On average 302 s of data (range: 297–394 s) were used for the estimation of the data covariance matrix, which was regularized using singular value truncation with the default setting of 1 × 10−6 times the maximum singular value. The optimum orientation of the equivalent current dipole was found using singular value decomposition.40 The broadband sensor-level data were subsequently and sequentially projected through the normalized beamformer weights41 resulting in a time series for each ROI. The time series for these ROIs were subsequently used for further analysis.
For each subject, 10 non-overlapping, artefact-free, drowsiness-free, eyes-closed, down-sampled (factor 4) epochs of 4096 samples (13.1072 s) were selected (D.N.S. and A.A.G.), based on careful visual inspection. Inspection, cutting of epochs and calculation of the functional connectivity measure (see below) was done using in-house software Brainwave (version 0.9.152.12.26, available from http://home.kpn.nl/stam7883/brainwave.html). Further analyses were done in MATLAB version R2018b.
Network construction
Functional network
Functional connectivity was estimated for each epoch using the corrected amplitude envelope correlation (AEC-c) in the alpha (8–13 Hz) and beta (13–30 Hz) frequency bands, based on previous work that identified these two bands as most robust, for the AEC-c, for AD.42,43 The amplitude envelope correlation44 is an amplitude-based metric that estimates the coupling between two time series by computing the Pearson correlation between the amplitude envelopes of these time series. The AEC-c overcomes the effects of spatial leakage by using pair-wise orthogonalization prior to the AEC estimation for each pair of band-pass filtered time series. The correction is performed in two directions by means of linear regression, meaning that time series x is regressed out from time series y, and time series y is regressed out from time series x and the AEC values for both directions are computed and averaged.45 A value of 1 was added to all AEC-c values, which were then divided by two in order to obtain values in the range (0–1). AEC-c values were averaged over epochs for each subject.
For each band separately, AEC-c values were computed for all pair-wise combinations between ROIs, resulting in a symmetric 50 × 50 connectivity matrix, and subsequently averaged over rows (i.e. resulting in one connectivity value for each ROI, denoting the connectivity strength of that ROI with the rest of the brain). Finally, the matrices were averaged over all epochs and all subjects in each group, to create one group-level functional connectivity matrix for each band.
Structural network
Since diffusion tensor imaging (DTI) data were not available for our patient cohort, we used the well validated Exponential Distance Rule (EDR) as an approximation of structural connectivity. While structural connectivity is usually obtained from DTI tractography, tract tracing studies in non-human primates have shown that the core of anatomical structural brain connectivity can be fairly well described by a simple rule, the EDR. Based on animal studies, the EDR specifies that the weights of structural connections in the brain, wij, decay exponentially with distance dij,46-48 i.e. wij ∼ exp(−αdij), where α is the decay exponent. Recent studies have repeatedly shown that the EDR also reproduces human DTI data well.49,50 Rather than the 0.188 exponent used in Deco and Kringelbach’s work, we used an exponent of 0.052 as validated by Millán and colleagues24 for an atlas with a comparable resolution to the Hammers atlas (whereas Deco and Kringelbach used a brain parcelation with 1000 ROIs). Similar to the functional networks, a group-level symmetric 50 × 50 connectivity matrix was used in the analyses, using the ROI centroids for the computation of dij. Additionally, in order to further validate our choice of structural network proxy, we compared EDR-MEG networks to DTI networks in an external dataset of n = 18 epilepsy subjects, obtaining a correlation of r = 0.61 (Supplementary material).
Diffusion network
To simulate tau diffusion to nearby ROIs, we defined the diffusion network such that two ROIs were connected if they were adjacent in the atlas. This network was defined as binary: two ROIs were either adjacent (and therefore connected) or not.
Analysis I: functional connectivity differences between groups
To investigate functional connectivity differences between clinical stages along the AD continuum, the AEC-c in the alpha and beta band for each ROI was compared between patient groups using the ANOVA test with Bonferroni post hoc testing for pairwise comparisons between groups. Results were corrected for multiple comparisons using the false discovery rate (FDR).51
Analysis II: epidemic spreading model
To simulate the spreading of tau protein on brain networks we used the susceptible-infected (SI) model.21 Each brain region (nodes in the network) can be in one of two states. Initially, all regions are in the susceptible state S (inactive), except for a (set of) seed region(s) in the infected state I (active). Each infected region j can then propagate the infected state to each of its susceptible neighbours i with independent probability βij, where βij is the local spreading rate. This model does not include any deactivation or recovery mechanisms (once a region becomes infected, it stays infected). Eventually all connected regions will be in the infected state and the spreading ends.
Here we considered two factors in the local spreading rate, βij = βwij, where β is the global spreading rate characterizing the speed and temporal structure of the spreading process and wij is the coupling strength between ROIs i and j, and it characterizes the spatial structure of the spreading process. In this manner wij couples tau spreading with the underlying network structure, so that different connectivity profiles will lead to different spreading profiles. In order to reduce the effect of spurious or weak connections, the functional connectivity matrices were rescaled such that , with ν ≥ 1. In this manner the contribution of weak connections became negligible, but the functional networks always remained fully connected. The EDR weights already decay exponentially so more scaling is not needed, and the diffusion network is binary so again no re-scaling is needed.
To simulate the SI dynamics we made use of Monte Carlo algorithms and custom-made MATLAB codes (available from Github).
We defined the simulated tau load mi(t) for each model configuration by iterating 104 times and measuring the fraction of iterations during which node i was infected at step t. The simulated tau load was compared with the corresponding group level tau binding pattern, as observed in the 18F-flortaucipir PET scans in each of the three disease stages of the AD continuum: preclinical AD, MCI due to AD, or AD dementia in order to fit the model parameters to the data. In particular, for each model configuration and group-level tau pattern, we identified the values of the control parameters ν, β and t (range: until all connected nodes were infected) that best reproduced the observed tau pattern. For this, we used a range of ν and β values, with ν = 1, 2, …, 10, and β = 10−x, x = − 2 + 2i/9, with i = 1, 2, …, 10. Then, we simulated the SI dynamics for each parameter configuration and measured the goodness of fit of the model for each simulation step as:
| (1) |
where τ is the measured PET-tau load, C(x, y) is the Pearson’s correlation coefficient, d(x, y) is the L1 distance between x and y, and bold symbols indicate vectors. In this manner we consider both the total tau load (through the distance) and the spatial pattern of the tau load (through the correlation). Since the correlation is sensitive to small changes, to avoid the correlation from dominating the goodness of fit metric the distance has to be weighted. The exponent of 3 was included to weigh the different contributions based on preliminary analyses indicating most robust results for an exponent equal to 3. The optimal fit was defined as the set of model parameters (β, ν, t) leading to the maximum value of r, r*. We also measured the corresponding correlation, C*. The significance of the correlation was computed by transforming the correlation to create a t-statistic having n − 2 degrees of freedom, where n is the number of ROIs. Each corresponding P-value is the probability of getting a correlation as large as the observed value by random chance, when the true correlation is zero.
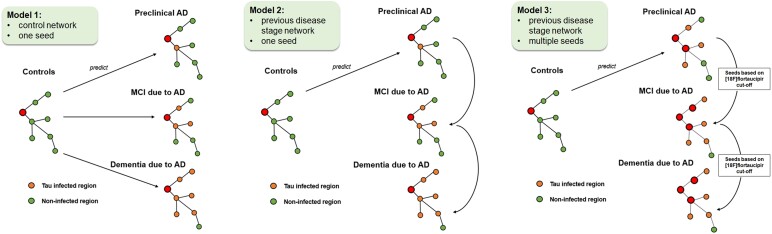
Model 1: Control network and one seed region
We first ran the susceptible-infected model over the previously described networks (functional in two bands, one structural, one diffusion) using the group-level network of the control group (Figs 1 and 2). Tau propagation was modelled to start from one a priori defined seed region (left middle and inferior temporal gyri), which was selected based on the reliability to accurately measure tau deposition at that location and its proximity to the hypothesized origin of tau pathology, the entorhinal cortex.32,52
Figure 1.
Workflow visualizing the prediction model. (A) The group level connectivity between 50 regions of interest (ROIs) of the adjusted Hammers atlas serves as the basis for the functional (AEC-c in alpha and beta band), structural (EDR) and diffusion (adjacent ROIs) networks. (B) The networks are used as the backbone for the epidemic spreading model (SI model), where the fraction of newly infected nodes (red line), is represented as a function of time. (C) For each network and each disease stage, the spreading dynamics were simulated and the propagation pattern was constructed across all 50 ROIs, and compared with the actual 18F-flortaucipir pattern. AD = Alzheimer’s disease; AEC-c = corrected amplitude envelope correlation; MCI = mild cognitive impairment.
Figure 2.
Visual representation of the prediction models of tau spread. Model 1: starting from the seed region (red), in the control group (amyloid-negative SCD) tau spread is modelled to the different stages of the Alzheimer’s disease (AD) continuum: preclinical AD (amyloid-positive SCD), MCI due to AD, and dementia due to AD. Orange nodes represent tau-infected areas, whereas green nodes represent non-infected areas. Model 2: to account for neurodegeneration in later disease stages, models were repeated with magnetoencephalography networks of the preceding disease stage. Model 3: alternative seed regions were added in the preclinical and mild cognitive impairment (MCI) stage, based on a 18F-flortaucipir cut-off, in order to model tau seeding from previously infected regions. Regions above this cut-off were considered tau-positive and were entered as seeds into the model. SCD = subjective cognitive decline.
Model 2: previous disease stage network
We expected that in the course of the disease the network would deteriorate as the result of neurodegeneration. We therefore investigated, as a second model, whether using the network of the previous disease stage (i.e. preclinical AD to predict MCI, and MCI to predict AD dementia) would give a better prediction than using the network of the controls to predict all subsequent stages (Fig. 2).
Model 3: previous disease stage network and alternative seeds
Finally, it can be assumed that as tau spreads through the brain, more regions become saturated with tau and thus begin seeding tau as well. In the third model, in addition to using the previous disease stage networks, we therefore added alternative seed regions based on a 18F-flortaucipir cut-off (Fig. 2). Regions above this cut-off were considered tau-infected and were entered as seeds into the model. To determine the cut-offs, we used the actual 18F-flortaucipir data of the control group (for preclinical AD) and preclinical AD (for MCI). We averaged the tau-binding across each ROI, and selected all ROIs with tau binding cut-off of 1.96 standard deviation (SD) (P = 0.05/95% confidence level) above the mean as ‘infected’. To clarify, in order to predict tau binding in the MCI stage, we used the preclinical AD network, and as seeds we used the ‘infected’ ROIs from this disease stage. The tau binding cut-off for preclinical AD was based on the 18F-flortaucipir from the control group. To predict the AD dementia stage we used the MCI network, and defined the ‘infected’ ROIs in the MCI stage based on the 18F-flortaucipir data of the preclinical AD stage.
Data availability
Due to the clinical nature of the data, the data that support the findings of this study are not freely available but can be made available by the corresponding author, upon reasonable request. A formal data sharing agreement is needed before any data can be shared. MATLAB codes for the ESM model are available on Github.
Results
Demographic characteristics
Table 1 presents the demographic characteristics of the four diagnosis groups. Functional connectivity differences between groups (Analysis I) are shown in the Supplementary material.
Table 1.
Baseline characteristics
| Controls (n = 25) | Preclinical AD (n = 16) | MCI (n = 16) | AD dementia (n = 25) | |
|---|---|---|---|---|
| Age in years, mean (SD)a | 64.7 (7.0) | 70.5 (6.9) | 65.8 (7.2) | 66.7 (7.8) |
| Sex, female (%) | 12 (48.0%) | 7 (43.8%) | 10 (62.5%) | 10 (39.1%) |
| Education level, median (IQR)b | 6 (5–7) | 6 (5–6) | 6 (5–7) | 5 (4–6) |
| MMSE score, median (IQR) | 30 (28–30) | 29 (27–30) | 26 (25–27) | 22 (19–23) |
| Global 18F-flortaucipir BPND, mean (SD)c | 0.034 (0.085) | 0.082 (0.130) | 0.260 (0.283) | 0.307 (0.349) |
Depicted are mean values (standard deviation, SD) or median with the interquartile range (IQR) where appropriate. AD = Alzheimer’s disease; MCI = mild cognitive impairment; MMSE = Mini-Mental State Examination.
Age at time of MEG measurement.
Education level according to Verhage score.
Volume-weighted mean cortical 18F-flortaucipir BPND over 50 regions of interest.
Analysis II: epidemic spreading model
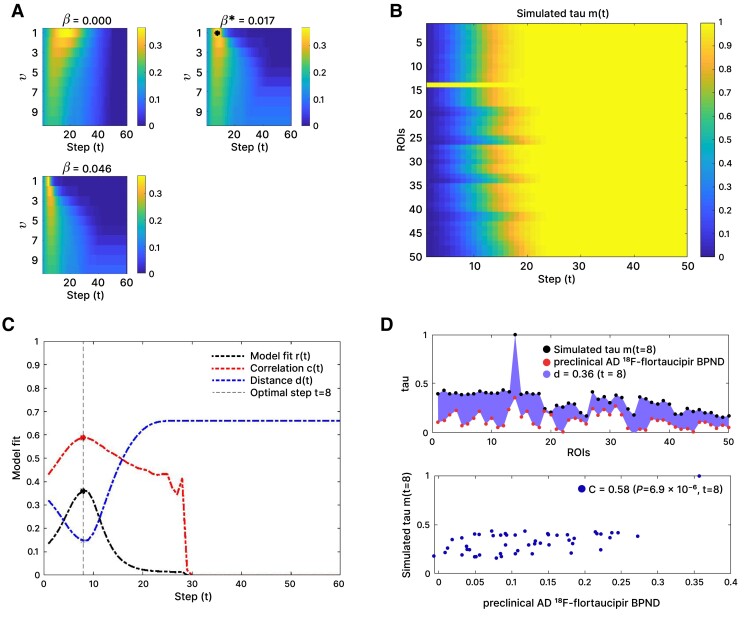
We constructed a tau propagation pattern using the ESM for each network and disease stage. An exemplary susceptible-infected propagation process for the alpha band AEC-c network, predicting preclinical AD, can be found in Fig. 3.
Figure 3.
Exemplary susceptible-infected propagation process for the functional network (AEC-c, alpha band) predicting tau spread in the preclinical Alzheimer’s disease stage using the control network. (A) Correlation between actual and predicted tau deposition for different spreading rates (β) and different weightings (v) of the functional connections. The optimum fit is indicated with an asterisk. (B) Model propagation pattern showing the probability pi(t), that a given region of interest (ROI) i (y-axis) is infected at simulation step t (x-axis). The seed (‘left middle and inferior temporal gyri’, ROI 14) is by definition infected at simulation Step 0. (C) Goodness of model fit, considering the total tau load [distance d(t)], the spatial pattern of the tau load [correlation C(t)], and final model fit [r(t)] for each simulation step (t). The dashed vertical line indicates the simulation step with the optimum model fit, in this case t = 8. (D) Simulated tau spread and observed tau spread by ROI (top), and the predicted tau spread versus observed tau spread for the optimal time point (t = 8), with the corresponding correlation and P-value (bottom).
Model 1: control network and one seed region
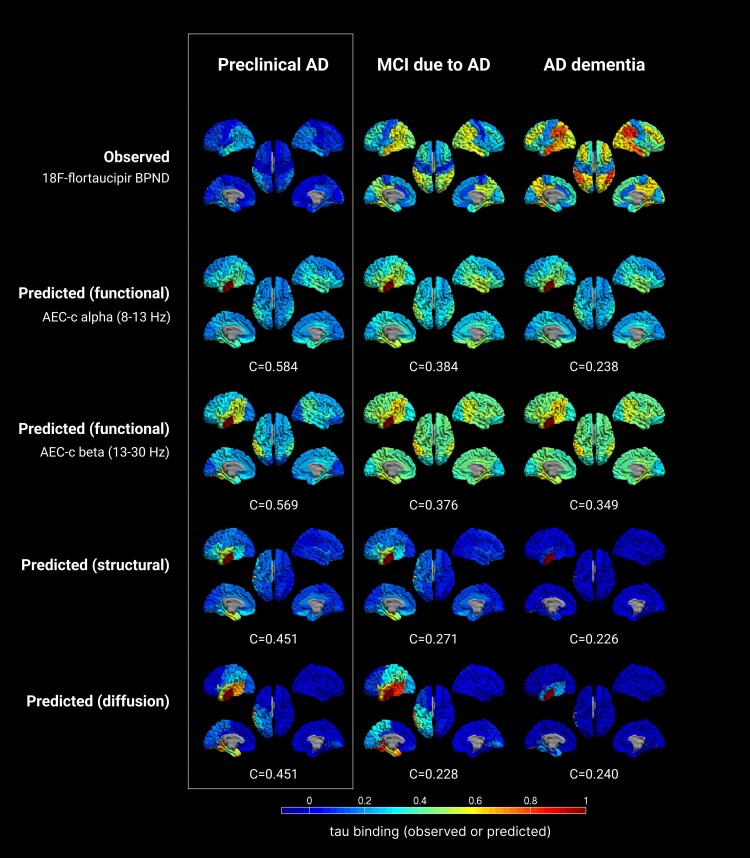
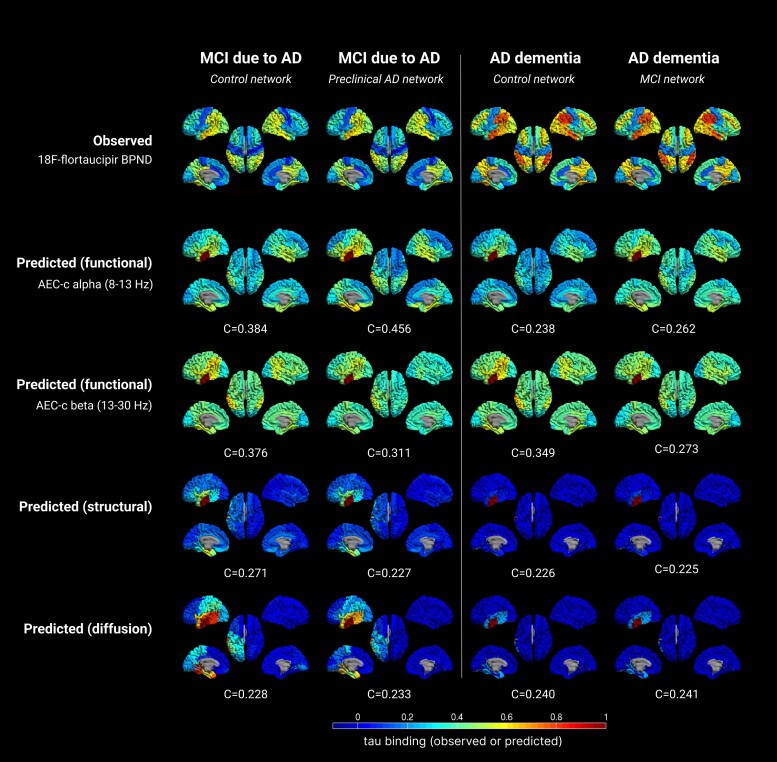
When using the control network as backbone for the susceptible-infected model, and the left middle and inferior temporal gyri as seed, we found that the best reproduction of the tau propagation patterns was for the alpha and beta band AEC-c functional network models (see Table 2 and Fig. 4 for the spreading patterns and Supplementary Table 6 for specific model details). When predicting tau deposition in the preclinical AD stage, the alpha band AEC-c network model showed an optimal correlation of C = 0.584, and for the beta band C = 0.569. For the structural network model, the correlation was C = 0.451; the diffusion model also showed an optimal correlation of C = 0.451. These model fits were all statistically significant (P < 0.001, P < 0.001, P = 0.001 and P = 0.001, respectively). The functional networks also spread to the contralateral temporal lobe, in contrast to the structural and diffusion networks.
Table 2.
Model performance
| Model | Disease stage | Network | Correlation, C (mean ± SD) |
Model fit, r (mean ± SD) |
P-value |
|---|---|---|---|---|---|
| Model 1 | Preclinical AD | AEC-c alpha | 0.584 ± 0.004 | 0.360 ± 0.003 | <0.001* |
| AEC-c beta | 0.569 ± 0.003 | 0.354 ± 0.002 | <0.001* | ||
| EDR | 0.451 ± 0.002 | 0.243 ± 0.002 | 0.001* | ||
| Diffusion | 0.451 ± 0.002 | 0.196 ± 0.001 | 0.001* | ||
| MCI due to AD | AEC-c alpha | 0.384 ± 0.004 | 0.212 ± 0.002 | 0.005* | |
| AEC-c beta | 0.376 ± 0.008 | 0.212 ± 0.004 | 0.007* | ||
| EDR | 0.271 ± 0.003 | 0.080 ± 0.001 | 0.130 | ||
| Diffusion | 0.228 ± 0.001 | 0.064 ± 0.001 | 0.110 | ||
| Dementia due to AD | AEC-c alpha | 0.238 ± 0.004 | 0.113 ± 0.002 | 0.090 | |
| AEC-c beta | 0.349 ± 0.009 | 0.185 ± 0.005 | 0.012* | ||
| EDR | 0.226 ± 0.001 | 0.033 ± 0.001 | 0.115 | ||
| Diffusion | 0.240 ± 0.001 | 0.035 ± 0.002 | 0.090 | ||
| Model 2 | MCI due to AD | AEC-c alpha | 0.456 ± 0.003 | 0.257 ± 0.002 | <0.001* |
| AEC-c beta | 0.311 ± 0.006 | 0.168 ± 0.004 | 0.030* | ||
| EDR | 0.227 ± 0.003 | 0.077 ± 0.001 | 0.112 | ||
| Diffusion | 0.233 ± 0.002 | 0.058 ± 0.001 | 0.103 | ||
| Dementia due to AD | AEC-c alpha | 0.262 ± 0.005 | 0.135 ± 0.003 | 0.070 | |
| AEC-c beta | 0.273 ± 0.007 | 0.142 ± 0.003 | 0.060 | ||
| EDR | 0.225 ± 0.001 | 0.033 ± 0.001 | 0.117 | ||
| Diffusion | 0.241 ± 0.001 | 0.035 ± 0.001 | 0.090 | ||
| Model 3 | MCI due to AD | AEC-c alpha | 0.469 ± 0.003 | 0.248 ± 0.002 | <0.001* |
| AEC-c beta | 0.329 ± 0.007 | 0.170 ± 0.002 | 0.020* | ||
| EDR | 0.297 ± 0.002 | 0.111 ± 0.001 | 0.036* | ||
| Diffusion | 0.290 ± 0.001 | 0.075 ± 0.002 | 0.041* | ||
| Dementia due to AD | AEC-c alpha | 0.368 ± 0.002 | 0.131 ± 0.001 | 0.008* | |
| AEC-c beta | 0.316 ± 0.001 | 0.096 ± 0.000 | 0.025* | ||
| EDR | 0.206 ± 0.001 | 0.075 ± 0.001 | 0.150 | ||
| Diffusion | 0.256 ± 0.002 | 0.095 ± 0.001 | 0.072 |
For each group, the correlations and optimal model fit between the clinical and modelled 18F-flortaucipir PET patterns, and the corresponding P-values, are given for each network type separately. Model 1 used the baseline control network and one a priori defined seed region (from the control group). Model 2 used the network of the previous disease stage and one a priori defined seed region (from the control group). Since the pattern for the preclinical Alzheimer’s disease (AD) stage remained unaltered these results are not included. Model 3 used the network of the previous disease stage and alternative seed regions. Since the pattern for the preclinical AD stage remained unaltered these results are not included. AEC-c = corrected amplitude envelope correlation; EDR = exponential distance rule; MCI = mild cognitive impairment; SD = standard deviation.
Indicates a significant correlation (P < 0.05).
Figure 4.
Observed versus predicted patterns of tau spreading in several stages of the Alzheimer’s disease continuum. The top row displays the observed clinical 18F-flortaucipir-PET BPND for each group. The other rows display the predicted spreading patterns for the different (functional, structural, diffusion) networks, based on the group level control network and a single seed region, with the corresponding optimum correlation coefficient evaluating model performance. Warmer colours represent a higher proportion of regional tau-binding, or higher probability of infection. Note that in the predicted data, the seed region (ROI 14) is always infected with maximum tau binding (by definition). AD = Alzheimer’s disease; AEC-c = corrected amplitude envelope correlation; MCI = mild cognitive impairment.
In the MCI stage, the optimal correlation was lower compared to the preclinical AD stage (C = 0.384; P = 0.005 and C = 0.376; P = 0.007 for the alpha and beta band AEC-c network models, respectively), but remained statistically significant. For the structural network model, the correlation dropped to C = 0.271; P = 0.130, and for the diffusion network model the correlation declined to C = 0.228; P = 0.110. Both the structural and diffusion models lost statistical significance.
In the AD dementia stage, the correlation between modelled and observed tau deposition declined further for the alpha band AEC-c, to C = 0.238, whereas for the beta band, interestingly, it declined only slightly to C = 0.349. For the structural and diffusion network model the correlation remained approximately equal to the MCI stage. Only the correlation for beta band AEC-c reached statistical significance (P = 0.012). Thus, for all stages, the models with tau propagation via the functional network were the best-fitting models, with the best prediction of tau deposition in the preclinical AD stage on the basis of the functional network of the healthy group.
Model 2: previous disease stage network
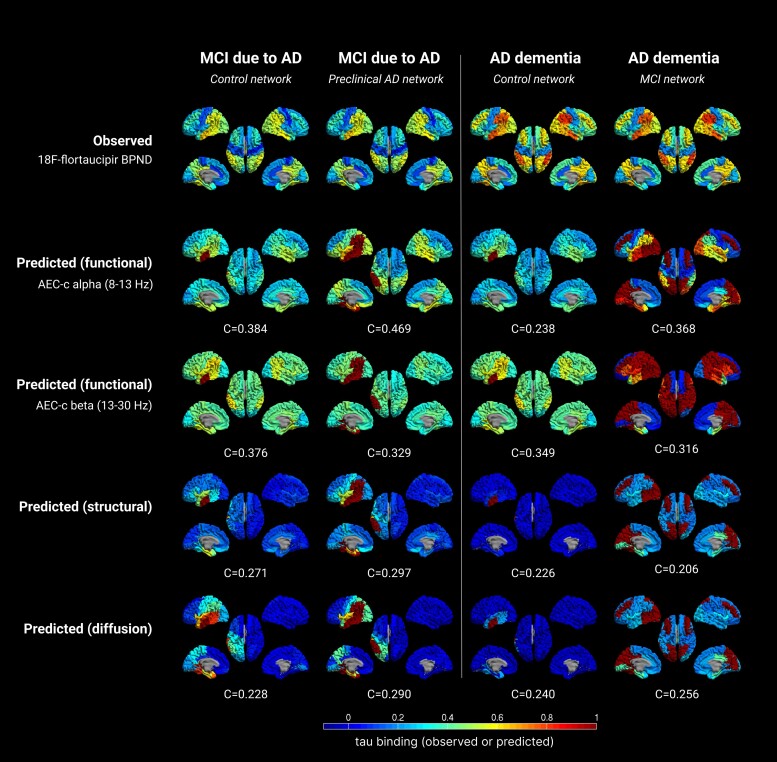
Next, we constructed a tau propagation pattern using the susceptible-infected model on the network of the previous disease stage instead of the control group, to account for neurodegeneration in later disease stages (Figs 2 and 5 and Table 2). For specific model details see Supplementary Table 7. In the MCI stage, the results for the alpha band AEC-c network model improved from C = 0.384 to C = 0.456; P < 0.001. The optimal correlation for the beta band AEC-c network-model declined, from C = 0.376 to C = 0.311; P = 0.030. The structural network model declined as well, to C = 0.227, and the diffusion model remained the same. The models based on the functional networks remained the best predictors and were the only ones to reach statistical significance. For the AD dementia stage, using the networks from the MCI stage did not improve the model performance for any of the networks except the alpha band AEC-c (from C = 0.238 to C = 0.262), and the correlations between modelled and measured tau patterns remained low. The correlations did not reach significance for any of the networks.
Figure 5.
Observed versus predicted patterns of tau spreading in several stages of the Alzheimer’s disease continuum, for the second model. The first and third columns show the original results from the first model, using the control network as backbone, with the second and fourth columns showing the new results, based on the networks of the preceding disease stage, with the corresponding fit evaluating model performance. The rows display the observed clinical 18F-flortaucipir-PET BPND, and the predicted spreading patterns for the different (functional, structural, diffusion) networks. Warmer colours represent higher proportion of regional tau-binding. Note that in the predicted data, the seed region (ROI 14) is always infected with maximum tau binding. AD = Alzheimer’s disease; AEC-c = corrected amplitude envelope correlation; MCI = mild cognitive impairment.
Model 3: previous disease stage network and alternative seeds
Finally, in the third model we used alternative seed regions in the MCI and AD dementia stage, based on cut-off that was based on the observed 18F-flortaucipir tau deposition from the clinical stage before the stage on which the networks were constructed. Regions above this cut-off were entered as seeds into the model (Fig. 2). The calculation of the tau cut-offs and resulting seeds can be found in Supplementary Tables 4 and 5. Table 2 and Fig. 6 show the resultant tau propagation patterns (the pattern for the preclinical AD stage remains unaltered). For specific model details, see Supplementary Table 8. For the MCI stage, the optimal correlation for the functional alpha band AEC-c network-model improved compared to Model 1, but not compared to Model 2, from C = 0.384 and C = 0.456 to C = 0.469; P < 0.001, respectively. The correlation for the beta band AEC-c network model was, similar to Model 2, not improved compared to Model 1. The structural network model showed slight improvement compared to Models 1 and 2, from C = 0.271 and C = 0.227, to C = 0.297; P = 0.036, respectively as well as the diffusion network model, with C = 0.228 and C = 0.233 for the previous models, to C = 0.290; P = 0.041. The correlations reached statistical significance for all networks. For the AD dementia stage, adding the alternative seeds improved the model performance when using the alpha band AEC-c as functional network, from C = 0.238 (Model 1) and C = 0.262 (Model 2) to C = 0.368; P = 0.008. For the beta band AEC-c, model performance improved compared to Model 2 (from C = 0.273, to C = 0.316; P = 0.025) but not to Model 1. For the other network models prediction accuracy remained poor.
Figure 6.
Observed versus predicted patterns of tau spreading in several stages of the Alzheimer’s disease continuum, for the third model. The first and third columns show the original results from the first model, using the control network as backbone, with the second and fourth columns showing the new results, based on the networks of the preceding disease stage and alternative seed regions (based on tau cut-offs), with the corresponding correlation evaluating model performance. The rows again display the observed clinical 18F-flortaucipir-PET BPND, and the predicted spreading patterns for the different (functional, structural, diffusion) networks. Warmer colours represent higher proportion of regional tau-binding. Note that in the predicted data, the seed regions are always infected with maximum tau binding. AD = Alzheimer’s disease; AEC-c = corrected amplitude envelope correlation; MCI = mild cognitive impairment.
Discussion
Using the brain network topology of subjects without AD pathology we were able to predict tau propagation in all pre-dementia stages of the AD continuum. The tau deposition patterns predicted by the spreading model that was based on functional networks showed highest correlations with observed 18F-flortaucipir tau depositions, followed by those based on the structural network or the diffusion process. When taking into account the ongoing neurodegeneration and tau spread in later disease stages, namely by using the previous disease network and its regions where tau binding was most prominent, the model showed slight improvement for the functional network based on the alpha band AEC-c, but no or only marginal improvement for the other networks. The models based on the functional networks remained the best predictors of tau spreading. These results suggest that in addition to structural connections, functional connections play an important role in tau spread, and highlight that neuronal dynamics play a key role in promoting this pathological process. Aberrant neuronal communication patterns should be taken into account when identifying targets for future therapy that halt disease progression.
The role of functional connectivity in tau spread
Because of the current availability of specific tracers for PET imaging of tau deposition, enabling the visualization of tau in living humans,5,6 we can investigate the relationships between network connectivity and the spread of tau throughout the brain. Multiple previous 18F-flortaucipir versus fMRI studies have provided support for the possibility of a close relationship between tau deposition and functional connectivity.9-14,53 Interestingly, structural connectivity was found to be the best predictor of tau deposition in recent work by Vogel and colleagues, comparing fMRI-based functional networks to DTI-based structural networks.16 MEG holds the possibility to expand upon fMRI-based functional connectivity research since it is a direct measure of neuronal activity with high temporal and spatial resolution. To our knowledge, only three studies have investigated MEG in relation to tau-PET, two of which opted to explore covariance between tau patterns and brain networks of functional connectivity.19,20 Both studies showed tau-related increases in functional connectivity in the lower frequencies (delta and theta bands) combined with a loss of functional connectivity in the higher frequency bands (alpha and beta). This is in agreement with previous studies showing decreases in number and within-network connections of functional subnetworks for beta and gamma bands, but paradoxical increases in the delta and theta bands for MCI and AD patients.54 A third study from our group combined MEG and tau-PET with 11C-UCB-J-PET and showed that both higher 18F-flortaucipir and lower 11C-UCB-J uptake were associated with altered synaptic function, indicative of slowing of oscillatory activity, indicating that in AD, tau pathology is closely associated with reduced synaptic density and synaptic dysfunction.55
Our study expands upon these previous works by introducing an MEG-based epidemic spreading model, a mathematical model that simulates the propagation of an agent from some given location to other connected areas, in this case designed to mimic the hypothetical spreading of tau. In this model, we evaluated both functional and structural networks, as well as a diffusion network as null hypothesis, in order to investigate the role of different network types in tau spread. In our spreading model, functional networks in two frequency bands were the best predictors of tau spread, outperforming the structural and diffusion networks. In other words, the spread of tau may be strongly determined by the way neurons are interacting, and strong functional interactions with neurons at the initial seeding site may be more important than simple anatomical connections. While structural networks are believed to shape and provide constraints for the dynamics of functional connectivity56-58 (and to an extent, functional networks can be predicted from the underlying structural connectome),59-62 functional networks are not merely a one-to-one reflection of the underlying structural network.63 Interestingly, multiple sclerosis patients with cognitive problems may show functional network changes without severe structural damage.64 Functional imaging may therefore provide another layer of information by identifying the role of aberrant neuronal dynamics in the pathogenesis of AD and subsequent tau spread through the brain.
While our model could capture the early stages of tau spreading accurately, later disease stages were modelled less precisely. Possible explanations for this include the initial use of the control network as a backbone to predict all subsequent stages of the AD continuum, which does not take into account the decline of connectivity or network degradation due to neurodegeneration in later disease stages. However, when adding the previous disease stage network to account for this, only the prediction for the alpha band functional network improved, and the prediction for the beta band network and the structural network did not. A possible explanation (Supplementary Fig. 1) is that, as a result of neurodegenerative loss of connectivity, the networks of subsequent disease stages lose ‘structure’, i.e. display less hierarchy in connectivity values, which makes it harder for the model to predict which connections are important for tau spread. The beta band functional network, even in the control and preclinical AD stage, shows less structure than the alpha band, which may explain why prediction accuracy did not improve for this band when the previous disease stage networks were used, as it did for the alpha band. A similar explanation may explain the results for the structural network. The diffusion network, as a network of adjacent ROIs, is most likely little affected by neurodegeneration, which may be why prediction accuracy in later stages was largely unaltered when using the previous disease stage network. Another relevant explanation for the lack of accuracy in later stages is the use of a relatively low-resolution atlas, which may not capture the subtleties in network degeneration as the disease progresses and allows fewer parameters to be fit to the data, resulting in less accurate predictions.
When adding additional tau-infected regions at starting point into the model in order to model tau seeding from previously infected regions, model performance showed some improvement for most networks in the MCI stage, and only for the alpha band in the dementia stage. Possible explanations for the limited improvements are that by adding more seeds to the model, a larger percentage of the total number of ROIs is used as seed (for instance, in the AD dementia stage 14 ROIs were entered as seeds, which is 28% of the total number of ROIs), which the model compensates for by adjusting the spreading rate, resulting in either over- or underestimates of tau spread.
This is not the first study that uses a spreading model in an attempt to explain tau pathology in AD. Vogel and colleagues16 provided valuable insights in a recent paper that used ESM to model the spread of tau tangles across the brain using atlases based on either anatomical (DTI) or functional connectivity (fMRI) maps, and then compared the resulting patterns with actual data from tau-PET scans of 312 people. Both maps fit the data, but, contrary to our results, the model based on structural connectivity performed best, explaining 70% of the observed pattern of tangles within the brain. A possible explanation for these discrepant results is the measures used to estimate connectivity. In fMRI, a change in brain activity is inferred via measurement of local changes in haemodynamics, which necessarily assumes a relationship between neuronal activity and the haemodynamic response [blood oxygen level-dependent (BOLD) signal]. The dependence of the BOLD signal on haemodynamic and neurovascular coupling parameters means that it must be viewed with some caution as a quantitative metric of brain activity. A change in the BOLD signal could arise from a change in neuronal activity, neurovascular coupling, haemodynamic state or a combination of these.65 In contrast, MEG measurements of electrical current in the brain are inferred directly via measurement of extracranial magnetic fields. Since DTI data were not available for our cohort, we used a well validated surrogate model for structural connectivity: the EDR, which has repeatedly been shown to reproduce human DTI data well.49,50,66 Future work may expand upon the current model by adding long-range connections.66 Using EDR-MEG networks to define the backbone for the dynamical model allows for more versatility, as well as the ability to use our approach in patients for whom DTI data are not available, reducing the burden associated with the use of computational models in clinical practice.
Our understanding of the pathological mechanisms underlying AD has grown over the past years. Tau pathology has long been considered to appear late in the disease course, but it may actually be one of the earliest signs of the disease. Pathological studies indicate that early stage tau deposits can present locally in the medial temporal lobe before the first amyloid plaques appear.67,68 Contrary to amyloid-β studies, previous tau studies have shown that tau load correlates strongly with cognitive decline, preferentially accumulates in functionally relevant brain areas and co-localizes with hypometabolic regions and atrophy.11,69-72 These relationships indicate that tau may well be a very important factor in the development of AD. Preclinical studies in several brain diseases indicate that tau proteins transfer trans-synaptically from neuron to neuron.73-75 Studies in transgenic mice expressing human tau in the entorhinal cortex found age-dependent tau accumulation in synaptically downstream regions,73 not only locally, but also in distant regions with strong synaptic connections to the initial site.76 These studies suggest that the spread of tau is determined by the way neurons are interconnected. Our current findings are in line with these studies, and propose that tau pathology spreads through neuronal connections, principally those that are heavily used functionally. Interestingly, recent work found evidence that from Braak stage III onward, local replication rather than spreading between brain regions, is the main process controlling the overall rate of accumulation of tau in neocortical regions, but which specific factors influence this process remains speculative.77 Possibly, the process of neurodegeneration, i.e. neurons that degenerate and show tau deposition, is mediated by local pathological neuronal activity that makes use of functional connections to influence downstream activity of other neurons, which in turn will start to exhibit pathological (hyper)activity and enhance tau deposition. This then gives rise to a self-perpetuating loop of further damage to inhibitory interneurons and neuronal hyperactivity in (sub)networks involved in memory encoding, independently of amyloid. To which regions tau is spread is determined by trans-synaptic propagation of tau proteins. In later stages, disinhibition and tau deposition lead to massive loss of synapses and cell death, reflected by hypoconnectivity of large scale networks. The present results may contribute to increased understanding of pathophysiological mechanisms in AD, as well as identify novel targets for future therapy that halt disease progression, for instance by manipulating the neuronal activity levels of target regions in early stages.
Strengths
An important strength of the current study is the inclusion of subjects from the full scale of the Alzheimer’s continuum, all with biomarker confirmation of AD pathology. All subjects received an elaborate work-up and diagnoses were generated during a multidisciplinary consensus meeting, according to recent international guidelines. For all subjects, we acquired data from different modalities: MEG, MRI and 18F-flortaucipir PET, and integrated them into a predictive model, with the simplicity of the model being a main advantage. ESMs do not intend to capture the details of the underlying biological basis of tau spreading, only the stereotypical patterns of its propagation. This simplicity allows for faster calculations and fewer free parameters, and also comes with a large body of theoretical and computational studies that can be used to design the study and interpret the results.
Limitations
Our study comes with a number of limitations. First, due to atlas restrictions, we were not able to model the initial spread of tau pathology from the entorhinal cortex, which is generally accepted as the starting point of tau spread through the brain. However, while the entorhinal cortex plays a central role in the early appearance of tau, recent work has shown that it may be the inferior temporal cortex that is the critical region for rapid tau accumulation in preclinical AD,52 which was used as the seed in the current study. In a recent paper, Insel and colleagues52 used three large cohorts to identify regions with high rates of tau accumulation, showing that the inferior temporal cortex, fusiform gyrus and middle temporal cortex had the largest effect sizes of tau accumulation longitudinally. They postulate that while the entorhinal cortex is the initial site to show abnormal levels of tau with age, this does not appear to be the result of a high rate of accumulation in the short term, but possibly a more moderate rate occurring early as a result of ageing. The inferior temporal cortex in particular was shown to be a site of both amyloid-β deposition as well as rapid tau accumulation; it could thus subsequently act as a central hub for rapid, widespread tau propagation due to local amyloid-β–tau interaction. Additionally, the limited resolution of the current atlas entails that fewer subtleties of the parameters of the dynamical model can be reliably fit to the data. Another possible limitation is the validation of the results: in order to assess model performance, we compared the model prediction with 18F-flortaucipir binding. However, it must be noted that the 18F-flortaucipir tracer provides a measure for neurofibrillary tangle pathology, while most preclinical studies have shown oligomeric tau as the pathological tau isoform at the synapse. Finally, it must be noted that participants in the study were relatively young as the result of drawing from a tertiary memory clinic population, as well as selection bias due to the fact that participants had to be in sufficient condition to undergo 80–100 min of 18F-flortaucipir PET. Possibly, a single spreading model with a single epicentre may not be sufficiently adequate to describe patterns of tau spread, which have been shown to conform to several different patterns.78 One positive aspect of this young group is that the AD pathology may be less affected by other problems such as vascular changes. Future studies could improve upon our work and possibly improve correlations by testing multiple tau epicentres as original seeds.
Future studies
Future models may be able to improve upon the current results by using higher resolution networks, larger samples and incorporating more complex network features, such as centrality, in order to identify which network characteristics play an important role in tau spread. Additionally, the role of neuronal (hyper)activity in promoting tau spread requires further examination. Finally, we investigated only the alpha and beta bands, while other frequency bands could capture different aspects of network pathology in AD. Given previous work showing the importance of theta and gamma waves for cognitive processes and the development of AD pathology,79 it would be interesting for future studies to investigate the role of these bands in tau spread.
Conclusion
In conclusion, our data support the concept that tau pathology spreads through neuronal connections, principally via functional connections. These results suggest that in addition to structural connections, neuronal dynamics play a key role in promoting tau spread. We hypothesize that aberrant neuronal activity, especially in highly functionally connected regions, could drive this process. These results may contribute to the identification of vulnerability factors for disease progression and identify potential targets for future therapy.
Supplementary Material
Acknowledgements
The authors would like to thank all participants of the MANTA study for their contribution. The authors thank technicians P. J. Ris, C. H. Plugge, N. Sijsma, N. C. Akemann, N. Zwagerman, T. Oorebeek and M. C. Alting Siberg for acquisition of the MEG data. The authors thank E. Scheijbeler for her contribution to preprocessing of the MEG data.
Contributor Information
Deborah N Schoonhoven, Department of Clinical Neurophysiology and MEG Center, Neurology, Amsterdam UMC location Vrije Universiteit Amsterdam, 1081 HZ Amsterdam, The Netherlands; Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands.
Emma M Coomans, Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands; Radiology and Nuclear Medicine, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands.
Ana P Millán, Department of Clinical Neurophysiology and MEG Center, Neurology, Amsterdam UMC location Vrije Universiteit Amsterdam, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands.
Anne M van Nifterick, Department of Clinical Neurophysiology and MEG Center, Neurology, Amsterdam UMC location Vrije Universiteit Amsterdam, 1081 HZ Amsterdam, The Netherlands; Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands.
Denise Visser, Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands; Radiology and Nuclear Medicine, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands.
Rik Ossenkoppele, Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands; Radiology and Nuclear Medicine, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands; Clinical Memory Research Unit, Lund University, 221 00 Lund, Sweden.
Hayel Tuncel, Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands; Radiology and Nuclear Medicine, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands.
Wiesje M van der Flier, Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands.
Sandeep S V Golla, Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands; Radiology and Nuclear Medicine, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands.
Philip Scheltens, Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands.
Arjan Hillebrand, Department of Clinical Neurophysiology and MEG Center, Neurology, Amsterdam UMC location Vrije Universiteit Amsterdam, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands; Amsterdam Neuroscience, Systems and Network Neuroscience, 1081 HV Amsterdam, The Netherlands.
Bart N M van Berckel, Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands; Radiology and Nuclear Medicine, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Brain Imaging, 1081 HV Amsterdam, The Netherlands.
Cornelis J Stam, Department of Clinical Neurophysiology and MEG Center, Neurology, Amsterdam UMC location Vrije Universiteit Amsterdam, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands.
Alida A Gouw, Department of Clinical Neurophysiology and MEG Center, Neurology, Amsterdam UMC location Vrije Universiteit Amsterdam, 1081 HZ Amsterdam, The Netherlands; Alzheimer Center Amsterdam, Neurology, Vrije Universiteit Amsterdam, Amsterdam UMC location VUmc, 1081 HZ Amsterdam, The Netherlands; Amsterdam Neuroscience, Neurodegeneration, 1081 HV Amsterdam, The Netherlands.
Funding
Research of Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting VUmc fonds. The chair of Wiesje van der Flier is supported by the Pasman Stichting. Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting VUmc fonds. This study was made possible by ZonMW Memorabel, Dioraphte, Avid Radiopharmaceuticals, and Janssen Pharmaceuticals. This work was specifically supported by ZonMW (Grant No: 733050812), and ZonMW/Dutch Epilepsy Foundation (Grant No: 95105006).
Competing interests
Research programs of W.v.d.F. have been funded by ZonMW, NWO, EU-FP7, EU-JPND, Alzheimer Nederland, Hersenstichting CardioVascular Onderzoek Nederland, Health∼Holland, Topsector Life Sciences & Health, stichting Dioraphte, Gieskes-Strijbis fonds, stichting Equilibrio, Edwin Bouw fonds, Pasman stichting, stichting Alzheimer & Neuropsychiatrie Foundation, Philips, Biogen MA Inc, Novartis-NL, Life-MI, AVID, Roche BV, Fujifilm, Combinostics. W.v.d.F. holds the Pasman chair. W.v.d.F. is recipient of ABOARD, which is a public-private partnership receiving funding from ZonMW (#73305095007) and Health∼Holland, Topsector Life Sciences & Health (PPP-allowance; #LSHM20106). All funding is paid to her institution. P.S. is a full time employee of EQT Life Sciences (formerly LSP). He has received consultancy fees (paid to the university) from Alzheon, Brainstorm Cell and Green Valley. Within his university affiliation he is global PI of the phase 1b study of AC Immune, Phase 2b study with FUJI-film/Toyama and phase 2 study of UCB. He is past chair of the EU steering committee of the phase 2b program of Vivoryon and the phase 2b study of Novartis Cardiology and presently co-chair of the phase 3 study with NOVO-Nordisk. All other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Wu JW, Hussaini SA, Bastille IM, et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat Neurosci. 2016;19:1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pooler AM, Phillips EC, Lau DH, Noble W, Hanger DP. Physiological release of endogenous tau is stimulated by neuronal activity. EMBO Rep. 2013;14:389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamada K, Holth JK, Liao F, et al. Neuronal activity regulates extracellular tau in vivo. J Exp Med. 2014;211:387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia CF, Arteaga J, Chen G, et al. [(18)F]T807, a novel tau positron emission tomography imaging agent for Alzheimer's disease. Alzheimers Dement. 2013;9:666–676. [DOI] [PubMed] [Google Scholar]
- 6. Chien DT, Bahri S, Szardenings AK, et al. Early clinical PET imaging results with the novel PHF-tau radioligand [F-18]-T807. J Alzheimers Dis. 2013;34:457–468. [DOI] [PubMed] [Google Scholar]
- 7. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. [DOI] [PubMed] [Google Scholar]
- 8. Schöll M, Lockhart SN, Schonhaut DR, et al. PET Imaging of tau deposition in the aging human brain. Neuron. 2016;89:971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansson O, Grothe MJ, Strandberg TO, et al. Tau pathology distribution in Alzheimer's disease corresponds differentially to cognition-relevant functional brain networks. Front Neurosci. 2017;11:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoenig MC, Bischof GN, Seemiller J, et al. Networks of tau distribution in Alzheimer's disease. Brain. 2018;141:568–581. [DOI] [PubMed] [Google Scholar]
- 11. Jones DT, Graff-Radford J, Lowe VJ, et al. Tau, amyloid, and cascading network failure across the Alzheimer's disease spectrum. Cortex. 2017;97:143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schultz AP, Chhatwal JP, Hedden T, et al. Phases of hyperconnectivity and hypoconnectivity in the default mode and salience networks track with amyloid and tau in clinically normal individuals. J Neurosci. 2017;37:4323–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cope TE, Rittman T, Borchert RJ, et al. Tau burden and the functional connectome in Alzheimer's disease and progressive supranuclear palsy. Brain. 2018;141:550–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Franzmeier N, Rubinski A, Neitzel J, et al. Functional connectivity associated with tau levels in ageing, Alzheimer's, and small vessel disease. Brain. 2019;142:1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franzmeier N, Dewenter A, Frontzkowski L, et al. Patient-centered connectivity-based prediction of tau pathology spread in Alzheimer's disease. Sci Adv. 2020;6:eabd1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vogel JW, Iturria-Medina Y, Strandberg OT, et al. Spread of pathological tau proteins through communicating neurons in human Alzheimer's disease. Nat Commun. 2020;11:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hari R, Baillet S, Barnes G, et al. IFCN-endorsed practical guidelines for clinical magnetoencephalography (MEG). Clin Neurophysiol. 2018;129:1720–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lopes da Silva F. EEG And MEG: Relevance to neuroscience. Neuron. 2013;80:1112–1128. [DOI] [PubMed] [Google Scholar]
- 19. Ranasinghe KG, Cha J, Iaccarino L, et al. Neurophysiological signatures in Alzheimer's disease are distinctly associated with TAU, amyloid-beta accumulation, and cognitive decline. Sci Transl Med. 2020;12:eaaz4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kocagoncu E, Quinn A, Firouzian A, et al. Tau pathology in early Alzheimer's disease is linked to selective disruptions in neurophysiological network dynamics. Neurobiol Aging. 2020;92:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pastor-Satorras R, Castellano C, Van Mieghem P, Vespignani A. Epidemic processes in complex networks. Rev Mod Phys. 2015;87:925–979. [Google Scholar]
- 22. Peraza LR, Díaz-Parra A, Kennion O, et al. Structural connectivity centrality changes mark the path toward Alzheimer's disease. Alzheimers Dement (Amst). 2019;11:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nissen IA, Millan AP, Stam CJ, et al. Optimization of epilepsy surgery through virtual resections on individual structural brain networks. Sci Rep. 2021;11:19025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Millan AP, van Straaten ECW, Stam CJ, et al. Epidemic models characterize seizure propagation and the effects of epilepsy surgery in individualized brain networks based on MEG and invasive EEG recordings. Sci Rep. 2022;12:4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Flier WM, Scheltens P. Amsterdam Dementia cohort: Performing research to optimize care. J Alzheimers Dis. 2018;62:1091–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Folstein MF, Folstein SE, McHugh PR. Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- 27. Nasreddine ZS, Phillips NA, Bedirian V, et al. The Montreal cognitive assessment, MoCA: A brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53:695–699. [DOI] [PubMed] [Google Scholar]
- 28. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tijms BM, Willemse EAJ, Zwan MD, et al. Unbiased approach to counteract upward drift in cerebrospinal fluid amyloid-β 1–42 analysis results. Clin Chem. 2018;64:576–585. [DOI] [PubMed] [Google Scholar]
- 30. Golla SSV, Timmers T, Ossenkoppele R, et al. Quantification of tau load using [F-18]AV1451 PET. Mol Imaging Biol. 2017;19:963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tuncel H, Visser D, Yaqub M, et al. Effect of shortening the scan duration on quantitative accuracy of [(18)F-flortaucipir studies. Mol Imaging Biol. 2021;23:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hammers A, Allom R, Koepp MJ, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19:224–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Svarer C, Madsen K, Hasselbalch SG, et al. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage. 2005;24:969–979. [DOI] [PubMed] [Google Scholar]
- 34. Golla SVS, Lubberink M, Lammertsma AA, Boellaard R. Partial volume correction of brain pet studies using iterative deconvolution in combination with Hypr denoising. J Cerebr Blood F Met. 2016;36:690–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taulu S, Simola J. Spatiotemporal signal space separation method for rejecting nearby interference in MEG measurements. Phys Med Biol. 2006;51:1759–1768. [DOI] [PubMed] [Google Scholar]
- 36. Hillebrand A, Fazio P, de Munck JC, van Dijk BW. Feasibility of clinical magnetoencephalography (MEG) functional mapping in the presence of dental artefacts. Clin Neurophysiol. 2013;124:107–113. [DOI] [PubMed] [Google Scholar]
- 37. Hillebrand A, Barnes GR, Bosboom JL, Berendse HW, Stam CJ. Frequency-dependent functional connectivity within resting-state networks: An atlas-based MEG beamformer solution. Neuroimage. 2012;59:3909–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hillebrand A, Tewarie P, van Dellen E, et al. Direction of information flow in large-scale resting-state networks is frequency-dependent. Proc Natl Acad Sci U S A. 2016;113:3867–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hillebrand A, Barnes GR. Beamformer analysis of MEG data. Int Rev Neurobiol. 2005;68:149–171. [DOI] [PubMed] [Google Scholar]
- 40. Sekihara K, Nagarajan SS, Poeppel D, Marantz A. Asymptotic SNR of scalar and vector minimum-variance beamformers for neuromagnetic source reconstruction. IEEE Trans Biomed Eng. 2004;51:1726–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheyne D, Bostan AC, Gaetz W, Pang EW. Event-related beamforming: A robust method for presurgical functional mapping using MEG. Clin Neurophysiol. 2007;118:1691–1704. [DOI] [PubMed] [Google Scholar]
- 42. Schoonhoven DN, Briels CT, Hillebrand A, Scheltens P, Stam CJ, Gouw AA. Sensitive and reproducible MEG resting-state metrics of functional connectivity in Alzheimer's disease. Alzheimers Res Ther. 2022;14:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Briels CT, Schoonhoven DN, Stam CJ, de Waal H, Scheltens P, Gouw AA. Reproducibility of EEG functional connectivity in Alzheimer's disease. Alzheimers Res Ther. 2020;12:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bruns A, Eckhorn R, Jokeit H, Ebner A. Amplitude envelope correlation detects coupling among incoherent brain signals. Neuroreport. 2000;11:1509–1514. [PubMed] [Google Scholar]
- 45. Hipp JF, Hawellek DJ, Corbetta M, Siegel M, Engel AK. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat Neurosci. 2012;15:884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ercsey-Ravasz M, Markov NT, Lamy C, et al. A predictive network model of cerebral cortical connectivity based on a distance rule. Neuron. 2013;80:184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gămănuţ R, Kennedy H, Toroczkai Z, et al. The mouse cortical connectome, characterized by an ultra-dense cortical graph, maintains specificity by distinct connectivity profiles. Neuron. 2018;97:698–715.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Theodoni P, Majka P, Reser DH, Wójcik DK, Rosa MGP, Wang XJ. Structural attributes and principles of the neocortical connectome in the marmoset monkey. Cerebral Cortex. 2022;32:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deco G, Kringelbach ML. Turbulent-like dynamics in the human brain. Cell Rep. 2020;33:108471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roberts JA, Perry A, Roberts G, Mitchell PB, Breakspear M. Consistency-based thresholding of the human connectome. Neuroimage. 2017;145:118–129. [DOI] [PubMed] [Google Scholar]
- 51. Benjamini Y, Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 52. Insel PS, Young CB, Aisen PS, et al. Tau positron emission tomography in preclinical Alzheimer's disease. Brain. 2022;146:700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ossenkoppele R, Iaccarino L, Schonhaut DR, et al. Tau covariance patterns in Alzheimer's disease patients match intrinsic connectivity networks in the healthy brain. Neuroimage Clin. 2019;23:101848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Haan W, van der Flier WM, Koene T, Smits LL, Scheltens P, Stam CJ. Disrupted modular brain dynamics reflect cognitive dysfunction in Alzheimer's disease. Neuroimage. 2012;59:3085–3093. [DOI] [PubMed] [Google Scholar]
- 55. Coomans EM, Schoonhoven DN, Tuncel H, et al. In vivo tau pathology is associated with synaptic loss and altered synaptic function. Alzheimers Res Ther. 2021;13:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O'Neill GC, Tewarie P, Vidaurre D, Liuzzi L, Woolrich MW, Brookes MJ. Dynamics of large-scale electrophysiological networks: A technical review. Neuroimage. 2018;180:559–576. [DOI] [PubMed] [Google Scholar]
- 57. Tewarie P, Prasse B, Meier JM, et al. Mapping functional brain networks from the structural connectome: Relating the series expansion and eigenmode approaches. Neuroimage. 2020:216:116805. [DOI] [PubMed] [Google Scholar]
- 58. Tewarie P, Prasse B, Meier J, et al. Predicting time-resolved electrophysiological brain networks from structural eigenmodes. Hum Brain Mapp. 2022;43:4475–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mišić B, Betzel RF, de Reus MA, et al. Network-level structure-function relationships in human neocortex. Cerebral Cortex. 2016;26:3285–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Honey CJ, Kötter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc Natl Acad Sci U S A. 2007;104:10240–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tewarie P, Abeysuriya R, Byrne A, et al. How do spatially distinct frequency specific MEG networks emerge from one underlying structural connectome? The role of the structural eigenmodes. Neuroimage. 2019;186:211–220. [DOI] [PubMed] [Google Scholar]
- 62. Meier J, Tewarie P, Hillebrand A, et al. A mapping between structural and functional brain networks. Brain Connect. 2016;6:298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Honey CJ, Sporns O, Cammoun L, et al. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eijlers AJC, Meijer KA, van Geest Q, Geurts JJG, Schoonheim MM. Determinants of cognitive impairment in patients with multiple sclerosis with and without atrophy. Radiology. 2018;288:544–551. [DOI] [PubMed] [Google Scholar]
- 65. Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31:1536–1548. [DOI] [PubMed] [Google Scholar]
- 66. Deco G, Perl YS, Vuust P, Tagliazucchi E, Kennedy H, Kringelbach ML. Rare long-range cortical connections enhance human information processing. Curr Biol. 2021;31:4436–4448.e5. [DOI] [PubMed] [Google Scholar]
- 67. Braak H, Thal DR, Ghebremedhin E, Del Tredici K. Stages of the pathologic process in Alzheimer disease: Age categories from 1 to 100 years. J Neuropathol Exp Neurol. 2011;70:960–969. [DOI] [PubMed] [Google Scholar]
- 68. Braak H, Zetterberg H, Del Tredici K, Blennow K. Intraneuronal tau aggregation precedes diffuse plaque deposition, but amyloid-β changes occur before increases of tau in cerebrospinal fluid. Acta Neuropathol. 2013;126:631–641. [DOI] [PubMed] [Google Scholar]
- 69. Hanseeuw BJ, Betensky RA, Schultz AP, et al. Fluorodeoxyglucose metabolism associated with tau-amyloid interaction predicts memory decline. Ann Neurol. 2017;81:583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ossenkoppele R, Schonhaut DR, Schöll M, et al. Tau PET patterns mirror clinical and neuroanatomical variability in Alzheimer's disease. Brain. 2016;139:1551–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sepulcre J, Schultz AP, Sabuncu M, et al. In vivo tau, amyloid, and gray matter profiles in the aging brain. J Neurosci. 2016;36:7364–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johnson KA, Schultz A, Betensky RA, et al. Tau positron emission tomographic imaging in aging and early Alzheimer disease. Ann Neurol. 2016;79:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. de Calignon A, Polydoro M, Suárez-Calvet M, et al. Propagation of tau pathology in a model of early Alzheimer's disease. Neuron. 2012;73:685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Calafate S, Buist A, Miskiewicz K, et al. Synaptic contacts enhance cell-to-cell tau pathology propagation. Cell Rep. 2015;11:1176–1183. [DOI] [PubMed] [Google Scholar]
- 75. Goedert M, Masuda-Suzukake M, Falcon B. Like prions: The propagation of aggregated tau and α-synuclein in neurodegeneration. Brain. 2017;140:266–278. [DOI] [PubMed] [Google Scholar]
- 76. Ahmed Z, Cooper J, Murray TK, et al. A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: The pattern of spread is determined by connectivity, not proximity. Acta Neuropathol. 2014;127:667–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Meisl G, Hidari E, Allinson K, et al. In vivo rate-determining steps of tau seed accumulation in Alzheimer's disease. Sci Adv. 2021;7:eabh1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vogel JW, Young AL, Oxtoby NP, et al. Four distinct trajectories of tau deposition identified in Alzheimer's disease. Nat Med. 2021;27:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Martorell AJ, Paulson AL, Suk HJ, et al. Multi-sensory gamma stimulation ameliorates Alzheimer's-associated pathology and improves cognition. Cell. 2019;177:256–271.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to the clinical nature of the data, the data that support the findings of this study are not freely available but can be made available by the corresponding author, upon reasonable request. A formal data sharing agreement is needed before any data can be shared. MATLAB codes for the ESM model are available on Github.