Abstract
Myotonic dystrophy type 1 is a dominantly inherited multisystemic disease caused by CTG tandem repeat expansions in the DMPK 3′ untranslated region. These expanded repeats are transcribed and produce toxic CUG RNAs that sequester and inhibit activities of the MBNL family of developmental RNA processing factors. Although myotonic dystrophy is classified as a muscular dystrophy, the brain is also severely affected by an unusual cohort of symptoms, including hypersomnia, executive dysfunction, as well as early onsets of tau/MAPT pathology and cerebral atrophy. To address the molecular and cellular events that lead to these pathological outcomes, we recently generated a mouse Dmpk CTG expansion knock-in model and identified choroid plexus epithelial cells as particularly affected by the expression of toxic CUG expansion RNAs.
To determine if toxic CUG RNAs perturb choroid plexus functions, alternative splicing analysis was performed on lateral and hindbrain choroid plexi from Dmpk CTG knock-in mice. Choroid plexus transcriptome-wide changes were evaluated in Mbnl2 knockout mice, a developmental-onset model of myotonic dystrophy brain dysfunction. To determine if transcriptome changes also occurred in the human disease, we obtained post-mortem choroid plexus for RNA-seq from neurologically unaffected (two females, three males; ages 50–70 years) and myotonic dystrophy type 1 (one female, three males; ages 50–70 years) donors. To test that choroid plexus transcriptome alterations resulted in altered CSF composition, we obtained CSF via lumbar puncture from patients with myotonic dystrophy type 1 (five females, five males; ages 35–55 years) and non-myotonic dystrophy patients (three females, four males; ages 26–51 years), and western blot and osmolarity analyses were used to test CSF alterations predicted by choroid plexus transcriptome analysis.
We determined that CUG RNA induced toxicity was more robust in the lateral choroid plexus of Dmpk CTG knock-in mice due to comparatively higher Dmpk and lower Mbnl RNA levels. Impaired transitions to adult splicing patterns during choroid plexus development were identified in Mbnl2 knockout mice, including mis-splicing previously found in Dmpk CTG knock-in mice. Whole transcriptome analysis of myotonic dystrophy type 1 choroid plexus revealed disease-associated RNA expression and mis-splicing events. Based on these RNA changes, predicted alterations in ion homeostasis, secretory output and CSF composition were confirmed by analysis of myotonic dystrophy type 1 CSF. Our results implicate choroid plexus spliceopathy and concomitant alterations in CSF homeostasis as an unappreciated contributor to myotonic dystrophy type 1 CNS pathogenesis.
Keywords: short tandem repeat expansion, RNA disease, alternative splicing, development, neurodegeneration
Myotonic dystrophy type 1 is a neuromuscular disease that also severely affects the brain. Using mouse models and patient samples, Nutter et al. describe how this disease alters the gene expression program of cells that produce CSF and propose that these alterations are linked to disease-associated CNS symptoms.
Introduction
The alternative splicing (AS) of mammalian RNAs plays a key role in the expression and timing of RNA and protein isoforms required at each stage of embryonic and postnatal development. To achieve this precise expression program, AS is regulated by coordinated interactions between nascent RNA sequences/structures, numerous RNA splicing factors and the spliceosome.1–4 The importance of AS regulation during development is exemplified by the autosomal dominant hereditary disease myotonic dystrophy type 1 (DM1), which is caused by CTG short tandem repeat expansions (CTGexp) in the DMPK 3′ untranslated region.5,6 Transcription of the DM1 mutant allele results in CUGexp RNAs that alter the activities of the CELF and MBNL splicing factor families. During development, MBNL1 and MBNL2 promote foetal-to-adult isoform switching by preferential binding to structurally organized compound YGCY (Y = pyrimidines) motifs on its target RNAs.7–10 Since the density of this binding motif is exceptionally high on CUGexp RNAs, MBNL is sequestered by these mutant RNAs in nuclear ribonucleoprotein (RNP) foci, which results in impaired MBNL activity, reversion to foetal isoform patterns for its RNA targets and disease manifestations.11 Inhibition of MBNL activity due to both germline and somatic CTGexp mutations leads to severe deficits in the CNS, including early onset tau neurofibrillary tangles (NFTs), altered sleep regulation, impaired executive function, progressive cerebral atrophy with coordinate ventricle/CSF volume increases and intellectual disability in congenital myotonic dystrophy.12–18 Interestingly, several adult-onset myotonic dystrophy disease symptoms appear to model accelerated brain ageing,19,20 although how the reversion to foetal splicing patterns due to loss of MBNL activity can lead to these CNS manifestations is unclear.
While previous studies on transcriptomic alterations in the DM1 brain have focused on neuronal AS,21,22 we recently generated Dmpk CTGexp knock-in (KI) mouse models for this disease and determined that the initial impact of CUGexp expression on molecular pathology occurs in the choroid plexus (ChP) due to the relatively high level of Dmpk expression in this tissue.23 The ChP, a highly active fenestrated capillary network responsible for producing CSF, is composed of specialized epithelial (ependymal) cells that envelope a capillary and connective tissue core.24,25 The four ChPs are located in the two lateral (telencephalic), third (diencephalic) and fourth (hindbrain) ventricles, where each ChP plays distinct roles through differences in their secretome.26,27 In addition to ChP spatial specificity, their functions are temporally controlled with a major switch from regulating CNS development during embryogenesis and postnatal development to a maintenance function in adults.27–29 ChP-CSF axis functions range from providing nutrients, growth factors, clearing metabolites, circadian rhythm regulation and CNS immune surveillance to the regulation of neural stem cells (NSCs) in the subventricular zone.28–32 Interestingly, mouse models of other late adult-onset short tandem repeat expansion (STRexp) diseases, such as fragile-X tremor/ataxia syndrome and Huntington’s disease, have provided evidence that repeat expansions in the lower pathogenic range can affect neurodevelopment, which may influence disease onset and progression later in life.33,34 It is possible that a similar phenomenon occurs in myotonic dystrophy by affecting neurodevelopment through the ChP-CSF axis.
Overall, the ChP is in a unique position to elicit global changes to the CNS, since it may regulate pronounced shifts from foetal to adult CSF composition. Based on this possibility, we investigated the pathology of the ChP in human patients with DM1 as well as mouse models, to further understand how ChP-CSF axis alterations might impact the brain. Here, we show splicing patterns are most affected in the lateral ventricle ChP (LVChP) and are primarily regulated by MBNL2 in our Dmpk CTGexp mouse model, and a similar set of mis-splicing events occurs in the human DM1 LVChP. Furthermore, RNA-seq analysis of DM1 versus control LVChP revealed alterations in ion channel homeostasis as well as specific neurotrophic and growth factor mis-splicing with coordinate downstream effects on CSF composition. Our results demonstrate that the ChP is a particularly vulnerable tissue in DM1 and implicate ChP mis-splicing in disease-relevant CSF alterations.
Materials and methods
Human choroid plexus and CSF samples
Human ChP tissues (Alzheimer’s disease and neurologically unaffected controls) were obtained from the NIH NeuroBioBank.35 DM1 ChP tissues were obtained both from the University of Florida (UF) Center for NeuroGenetics and the Stanford University Neuromuscular BioBanks. All human ChP tissues used in this study were pre-existing and deidentified. Human CSF samples were obtained (details below) from 10 patients with DM1, who suffered from hypersomnolence, and seven non-myotonic dystrophy patients at the University of Rochester Medical Center, under a protocol approved by the Institutional Review Board (IRB #STUDY00004736). Details of the human patient tissues and CSF with relevant information are included in Supplementary Table 1.
Animal experiments
All animal procedures were conducted in accordance with National Institutes of Health Guidelines and approved by the UF IACUC. Timed matings were performed to obtain wild-type FVB/NJ prenatal time point embryos, and embryonic time points [embryonic Day (E)13.5, E15.5 and E18.5] were collected followed by ChP isolation for RNA analysis (2–4 samples were pooled for sufficient RNA). Postnatal time point [postnatal Day (P)0, P1, P3, P7, P14, P21 and P63] mice were euthanized and the ChP isolated for RNA (P0, P1 and P3 were 2–4 sample pools) or imaging. DmpkCTG480 knock-in (Dmpk CTGexp KI)23 and Mbnl2Δe2/Δe2 knockout (Mbnl2 KO)36 mice were generated previously and bred to obtain congenic (N10) animals on the FVB/NJ background. Genotyping methods are described in the Supplementary material, and the primers are listed in Supplementary Table 2. Embryonic time point E13.5 was obtained as described above and ChP postnatal time points (P1, P14, P63 and P54–P84) were isolated for RNA or imaging. All experiments prior to P21 were composed of equal numbers of males/females, in contrast to P21+, which were all males. Females were needed to generate additional embryonic time points. For ChP isolation, adult wild-type FVB mice were euthanized, and the brain carefully transferred to cold PBS. To isolate the LVChP, the brain was cut coronally approximately one-third from the top to open the lateral ventricles. The lateral ventricles were identified and carefully opened to allow visualization and removal of the LVChP by gentle separation along the base. To isolate the fourth ventricle (4V)ChP, the hindbrain including cerebellum and brainstem was separated from the rest of the brain to open the fourth ventricle. The 4VChP was identified and removed gently but firmly with forceps, carefully breaking the connection to other tissues at the base of the ChP.
CSF collection and western blotting
CSF samples from DM1 and control patients were obtained in a seated position and fluid was processed immediately after collection. One control patient sample was excluded from further analysis due to diagnosis of chronic inflammatory demyelinating polyneuropathy and cerebellar stroke, both of which result in CSF protein alterations, including increased total protein.37,38 Fluid was moved to separate tubes and centrifuged at 2500g for 10 min at 4°C; then, supernatants were removed into aliquots of approximately 1.5 ml and stored at −80°C. Time to freezer on average did not exceed 1 h. Protein concentration in CSF samples was assayed using the Qubit 4 Fluorometer protein assay (Invitrogen) and sample loading was normalized to approximately 10 μg per lysate load. Samples were lysed in protein lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Igepal, 0.25% sodium deoxycholate and protease inhibitors). Protein lysates were separated on 7.5% TGX Stain-Free Criterion gels (Bio-Rad), imaged in a ChemiDoc+ system (Bio-Rad) for total protein loading and transferred to a PVDF membrane (Millipore). Blotting was performed using primary antibodies (Supplementary Table 3) diluted in 4% fat-free milk in PBS. Horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit secondaries were used at 1:10 000. Signals were developed with ProSignal Dura ECL substrate (Prometheus). Images were captured using the Amersham Imager 680 (GE) and signal intensity was calculated using ImageLab software (Bio-Rad).
Immunofluorescence and RNA-fluorescence in situ hybridization
ChPs were isolated intact and fixed in 4% paraformaldehyde (PFA) for 10 min. For fluorescence in situ hybridization (FISH), samples were permeabilized in 70% ethanol overnight at −20°C. Samples were rehydrated in 2× saline-sodium citrate (SSC)/40% formamide for 1 h at room temperature (RT), then blocked with pre-hybridization solution for 1 h at 50°C. The probe used was TYE563-LNA-CAG10 (400 ng/ml), which was denatured in hybridization solution (2× SSC, 40% formamide, 100 mg/ml dextran sulfate, 1 mg/ml yeast tRNA, 0.2 mg/ml BSA and 2 mM ribonucleoside vanadyl complex) before adding to the samples for overnight hybridization at 50°C. The following day, slides were washed twice for 30 min each in 2× SSC/40% formamide at 60°C, counterstained with DAPI and mounted in slides using ProLong Glass anti-fade mountant (Invitrogen). For immunofluorescence processing, samples were permeabilized in 0.2% Triton X-100 and blocked in Background Sniper (Biocare Medical) for 30 min at RT. Primary and secondary antibodies were diluted in 10% Background Sniper/PBS (Supplementary Table 3). Samples were counterstained with DAPI and mounted in slides using ProLong Glass anti-fade mountant (Invitrogen). All images were collected using a Zeiss LSM 880 confocal laser scanning microscope.
RNA-sequencing
Total RNA was isolated as described in the Supplementary material, DNase digested and quality checked using a Fragment Analyzer (Agilent). RNA was depleted of rRNA (rRNA Depletion Kit v2 Human/Mouse/Rat, NEB) and cDNA libraries prepared (Ultra II Directional RNA Library, NEB). Mouse developmental ChP libraries were generated using 168 ng, and human ChP libraries using 500 ng, of total RNA input, and sequenced on Illumina NextSeq500 and NextSeq2000 platforms. Demultiplexed fastq files were inspected using FastQC, aligned to the mouse (GRCm38/mm10) or human (GRCh38/hg38) genomes using STAR (v2.7.10a).39 Alignments were processed using DESeq2 (v1.36.0) for differential expression (DE) and rMATS turbo (v4.1.2) for AS analysis.40 rMATS outputs were filtered using the R package maser, using cut-off criteria of mean reads ≥5, false discovery rate (FDR) < 0.05 and a minimum change in AS events of 10% (|ΔΨ| ≥ 0.1). Multiple sclerosis patient ChP RNA-seq datasets are publicly available online (GSE137619).
Protein structure modelling and prediction
Protein structures for the Saposin-B region of PSAP (1N69) and the central domains of PGF (1FZV) are publicly available on Protein Data Bank (PDB).41 The modelled structures for alterations due to splicing changes were predicted using the UCSF ChimeraX AlphaFold tool with the use of ColabFold, an optimized version of AlphaFold2, with default parameters.42,43 Protein structures modelled with AlphaFold2 are included in Supplementary Table 4.
Osmolarity
CSF was obtained from DM1 and non-myotonic dystrophy patients as described above. The total solute concentration (any charged molecule: ions, metabolites, etc.) was analysed using the Vapro vapour pressure osmometer model 5600 (EliTech Group Biomedical Sciences). Three readings of 10 μl each were made per sample, and the average of these three replicates was used as one data-point. Samples were analysed at random in groups of four, and after every fourth reading, the osmometer was recalibrated to ensure no drift occurred by using 100, 290 and 1000 mOsm standards (EliTech Group Biomedical Sciences).
Statistical analysis
Statistical significance was determined in GraphPad Prism (v9.5.0.525) by ordinary one-way ANOVA with Tukey’s multiple comparisons test. A t-test (two-tailed) was used for either paired or unpaired comparisons of two groups as stated in the figure legends. Standard deviation (SD) was used for graphing of error bars throughout the figures. All statistical analysis was performed using at least three biologically independent samples.
Data availability
All data are openly available at the Gene Expression Omnibus (GEO)44 (https://www.ncbi.nlm.nih.gov/geo/) under the SuperSeries GSE228459. Mbnl2 KO mouse ChP developmental RNA-seq datasets are deposited under the SubSeries GSE228314; human patient ChP RNA-seq datasets for neurologically unaffected controls, DM1 and Alzheimer’s disease are deposited in the GEO under SubSeries GSE228458. All code used to process, analyse and visualize data is openly available at https://github.com/helmutcarter/Nutter_et_al_2023.
Results
Lateral ventricle choroid plexus spliceopathy in Dmpk CTGexp knock-in mice
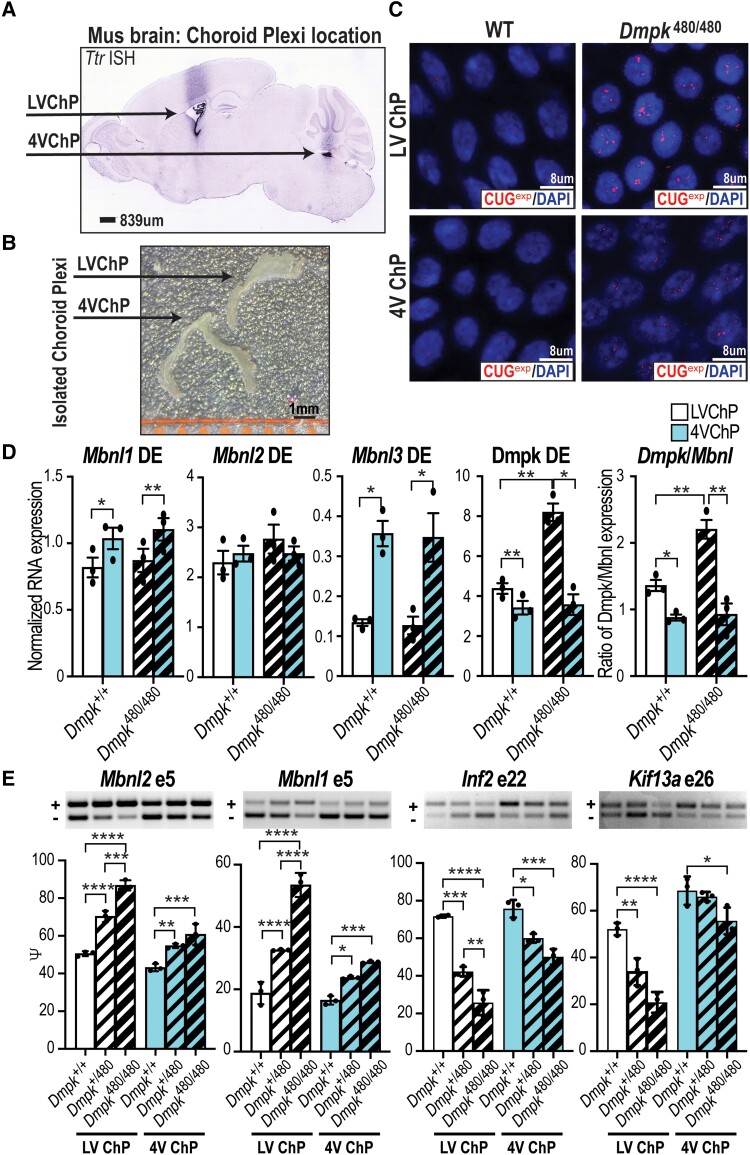
Using a mouse Dmpk CTGexp KI model for DM1, we recently demonstrated that expression of toxic CUGexp RNAs in the brain has the greatest impact on RNA mis-splicing in ChP epithelial cells,23 which produce the majority of CSF.24,45 While these findings suggested that CSF alterations induced by ChP dysfunction play a role in DM1 CNS aetiology,23 this study only evaluated ChP mis-splicing in the adult 4VChP omitting the LVChP (Fig. 1A).46,47 Since progressive cerebral atrophy is a major feature of DM1 brain pathology, the LVChP is of particular interest due to its regulation of neural stem cell proliferation/differentiation in the subventricular zone and the subgranular layer of the hippocampal dentate gyrus.27,48 We first isolated and tested if there are differences in the molecular pathology of toxic RNA foci between 4VChP and LVChP (Fig. 1B). Our analysis of CUGexp RNA revealed larger and more intense RNA foci in the LVChP compared to the 4VChP (Fig. 1C), suggesting either longer repeats or more prevalent Dmpk transcripts containing toxic repeats in the LVChP. Repeat length was not the cause since somatic expansion between tissues or different ChP ventricular regions was not detected (Supplementary Fig. 1A). To test if Dmpk expression was higher, we used reverse transcription quantitative PCR (RT-qPCR) and found that, while wild-type mice exhibited a small increase in Dmpk RNA levels in the LVChP compared to 4VChP, there was a greater increase in the KI mice (Fig. 1D). Furthermore, RNA steady-state levels of Mbnl1 and Mbnl3, but not Mbnl2, were higher in 4VChP versus LVChP in both wild-type and Dmpk CTGexp KI mice (Fig. 1D). This resulted in a Dmpk/Mbnl ratio exhibiting a nearly 2-fold increase in KI mice LVChP compared to KI 4VChP or wild-type LVChP (Fig. 1D). Since this ratio increase was expected to further impair splicing, we compared Dmpk CTGexp KI mis-splicing of several previously identified DM1-relevant genes (Mbnl2, Mbnl1, Inf2, Kif13a) in the LVChP versus the 4VChP (Fig. 1E). We observed that the magnitude of spliceopathy increased approximately 2-fold in LVChP compared to 4VChP (Fig. 1E and Supplementary Fig. 1B), which coincided with the 2-fold increase in Dmpk/Mbnl ratio (Fig. 1D). In conclusion, the LVChP was more severely affected in this mouse model of DM1.
Figure 1.
LVChP spliceopathy in Dmpk CTGexp KI mice. (A) In situ hybridization (dark stain) showing localization of LVChP and 4VChP in a sagittal brain section from an adult C57BL6 mouse using Ttr expression as the ChP marker (Allen Brain Atlas Mouse database). Scale = 839 μm. (B) Morphological comparison of isolated LVChP and 4VChP. Scale = 1 mm. (C) RNA-FISH of CUG repeat expansion (CUGexp) showing RNA foci (red) in the nucleus (blue, DAPI) of adult Dmpk+/+ FVB control versus Dmpk480/480 LVChP and 4VChP. Scale = 8 μm. WT = wild-type. (D) Mbnl1, Mbnl2, Mbnl3, and Dmpk RNA expression by RT-qPCR in LVChP (white) and 4VChP (blue) isolated from wild-type FVB littermate mice and Dmpk480/480 KI (thick crosshatch). Normalized RNA expression is based on the geometric mean of three housekeeping genes (Gorasp1, Psmb4, Rpl38) while the ratio of Dmpk/Mbnl expression was calculated as the normalized expression of Dmpk/(Mbnl1 + Mbnl2 + Mbnl3). (E) RT-PCR gels (representative top) and analysis (graph bottom) of myotonic dystrophy associated AS events (Mbnl2 exon 5, Mbnl1 exon 5, Inf2 exon 22, Kif13a exon 26) from LVChP (white) and 4VChP (blue) isolated from adult littermate Dmpk+/+ wild-type FVB (outlined), Dmpk+/480 KI (thin crosshatch), and Dmpk480/480 KI (thick crosshatch) mice *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA or paired t-test. 4V ChP = fourth ventricle choroid plexus; DE = differential expression; LV ChP = lateral ventricle choroid plexus.
Alternative splicing regulation during choroid plexus development
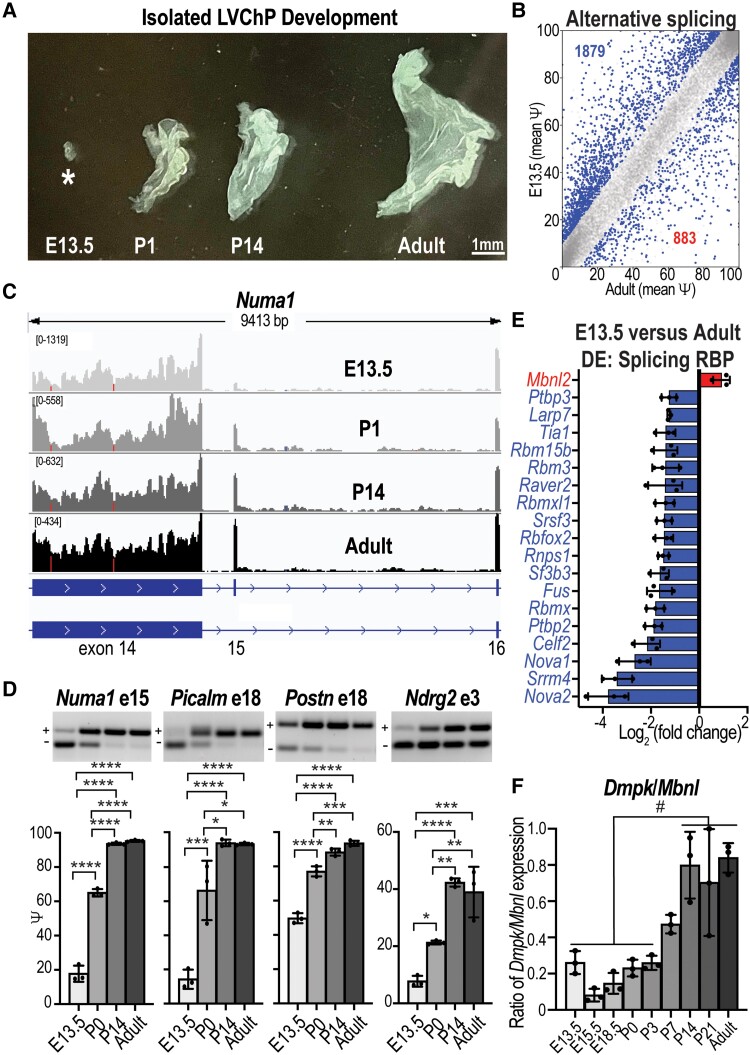
Since a hallmark of DM1 spliceopathy is the reversion of splicing to earlier developmental patterns,49 and the LVChP showed a more severe molecular pathology (Fig. 1), we next investigated LVChP temporal RNA processing. Splicing changes throughout ChP development have not been investigated previously, so we analysed RNA AS and DE during wild-type LVChP development to identify the period when CUGexp RNA toxicity might be the most severe. Using the mouse developmental Allen Brain Atlas collection47,50,51 and expression of ChP-specific genes (Ttr, Aqp1, Kl), we determined that the earliest stage to reproducibly isolate wild-type LVChP based on tissue morphology was E13.5 (Supplementary Fig. 2A). To positively identify the tissue as LVChP, we used morphology52 (Fig. 2A) and expression analysis of the ChP marker Ttr (Supplementary Fig. 2B). Next, we used RNA-sequencing (RNA-seq) (Supplementary material) to determine the splicing of wild-type LVChP isolated from E13.5 versus postnatal (P1, P14) (Supplementary Fig. 2C) and adult (P63) stages (Fig. 2B). The developmental transition that occurred between E13.5 and P1 showed the most splicing changes, in comparison to more modest additive increases with each progressive stage of development (P14 and adult), including considerable overlap between time points (Supplementary Fig. 2D). Developmental splicing changes (Fig. 2C) were validated by RT-PCR, including Numa1 exon 15, Picalm exon 18, Postn exon 18 and Ndrg2 exon 3 (Fig. 2D). We further analysed the RNA-seq to look at the DE changes (Supplementary Fig. 2E and F). Significantly DE genes in E13.5 versus adult LVChPs revealed that nearly all annotated mRNA binding/splicing factors had decreased RNA levels, including Nova1, Srrm4, Celf2 and Ptbp2 (Fig. 2E and Supplementary Table 5). In contrast, Mbnl2 was the only mRNA binding/splicing factor with significantly increased expression in adulthood. RT-qPCR validated that Mbnl2 RNA expression increased approximately 4-fold from E13.5 to P63, with more moderate changes in Mbnl1 and Mbnl3 expression (Supplementary Fig. 2G). Particularly relevant to DM1, Dmpk RNA levels were observed to be dynamically regulated during LVChP development, staying relatively level until P3, then increasing steadily to adult (P63) stages (Supplementary Fig. 2G). The cumulative changes in Mbnl and Dmpk expression resulted in a clear increase in the Dmpk/Mbnl ratio over development, which reached its peak at P14 to adult (Fig. 2F). Since these results indicated that higher MBNL2 levels promote adult specific splicing patterns during LVChP development and that impaired MBNL activity in DM1 may occur as early as P14, we next assessed the effects of MBNL2 loss in ChP development using Mbnl2 KO mice.
Figure 2.
Alternative splicing regulation during ChP development. (A) Developmental stage- (E13.5, P1, P14, adult) specific variations in size and morphology of LVChP. Scale = 1 mm. (B) Plot of AS events (Ψ) in wild-type FVB mice ChP comparing E13.5 to adult per cent spliced in (PSI) with the significant events in blue. (C) Numa1 exon 15 AS RNA-seq reads during development (E13.5, P0, P14, adult, light grey to black gradient). (D) RT-PCR gels (representative) and analysis (bar graph) validations for E13.5, P0, P14 and adult stages of Numa1 exon 15, Picalm exon 18, Postn exon 18 and Ndrg2 exon 3. (E) In contrast to other splicing factors, Mbnl2 expression increases during postnatal development. Relative expression (RNA-seq) is plotted for significant differentially expressed genes during LVChP development. (F) The ratio of Dmpk/Mbnl expression was calculated as the normalized expression of Dmpk/(Mbnl1 + Mbnl2 + Mbnl3) for each developmental stage. #Significance of at least P < 0.05 for all the compared stages, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA. LVChP = lateral ventricle choroid plexus.
MBNL2 promotes adult splicing events during choroid plexus development
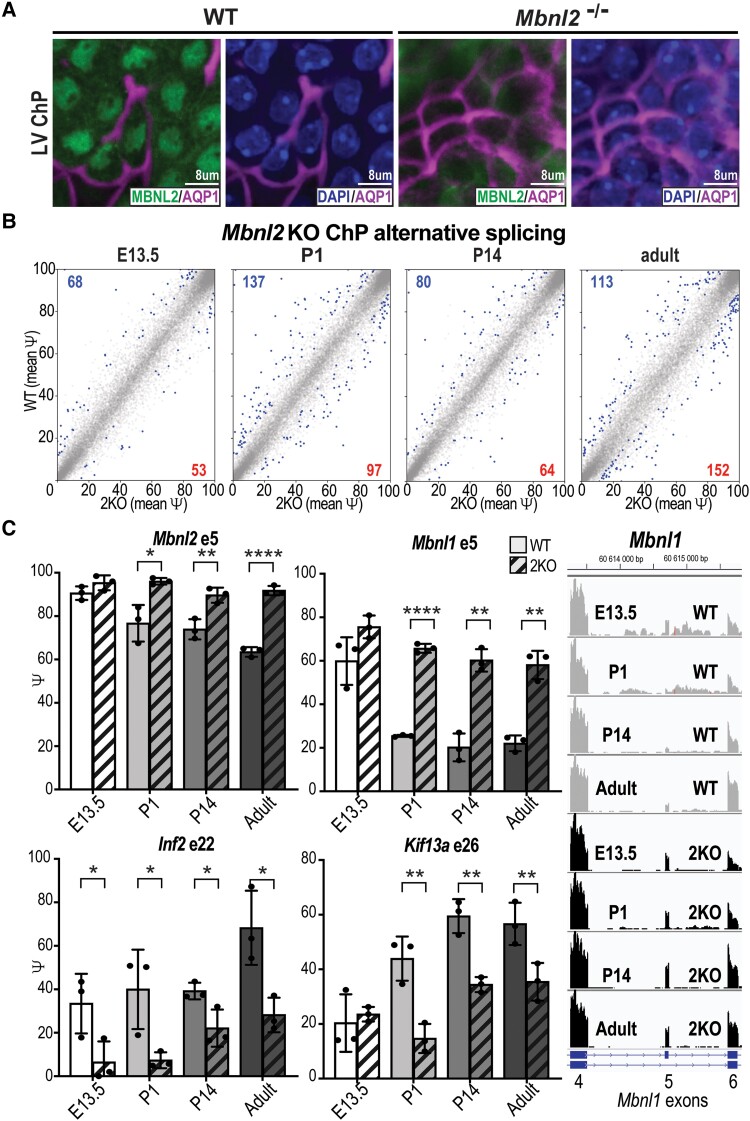
Mbnl2 KO mice have been previously evaluated as a model for CNS symptoms of myotonic dystrophy, since MBNL2 is the major MBNL paralogue in brain and neurons.36 Indeed, Mbnl2 KO mice exhibit sleep dysfunction and cerebral atrophy, similar to symptoms observed in human DM1.19,36,53 Therefore, our goal was to analyse how Mbnl2 KO influences LVChP, especially whether transitions to adult splicing patterns are impaired. Using isolated LVChP tissue, we first determined that MBNL2 protein primarily localized to the nucleus of both LV and 4VChP epithelial cells in wild-type mice, while nuclear MBNL2 was undetectable in Mbnl2 KO mice (Fig. 3A and Supplementary Fig. 3A). RNA-seq was performed on LVChP from both Mbnl2 KO and wild-type littermate mice at developmental stages E13.5, P1, P14 and adult (P63) (Supplementary material). MBNL2 loss did not cause a reduction in the expression of ChP markers, including Ttr (Supplementary Fig. 3B). MBNL2 loss was associated with significant shifts at different developmental stages, for splicing this varied between 121 and 265 splicing events (Fig. 3B), while for DE there were between 286 and 709 expression changes (Supplementary Fig. 3C). Supporting our hypothesis, throughout development Mbnl2 KOs showed an impaired shift to adult splicing patterns for MBNL targets Numa1 exon 15 and Postn exon 18, in contrast to non-MBNL targets Picalm exon 18 and Ndrg2 exon 3 (Supplementary Fig. 3D). Additionally, genes mis-spliced in Dmpk CTGexp KI mice (Fig. 1E) were mis-spliced in Mbnl2 KO mice at almost all stages of development. Interestingly, in three of four events (Mbnl2 exon 5, Mbnl1 exon 5 and Kif13a exon 26) the Mbnl2 KO essentially maintained splicing at E13.5 patterns, while Inf2 exon 22 was further repressed even at E13.5 (Fig. 3C). In adult Mbnl2 KOs, RT-PCR validated these mis-splicing events in both LVChP and 4VChP (Supplementary Fig. 3E). Since MBNL2 is sequestered in DM1, resulting in a loss of splicing activity, this strongly suggested that ChP mis-splicing could revert to embryonic patterns in patients with DM1 or possibly fail to transition to adult patterns in congenital myotonic dystrophy patients.
Figure 3.
MBNL2 promotes adult splicing events during ChP development. (A) MBNL2 is the major MBNL paralogue in ChP. Immunofluorescence of nuclear MBNL2 (green) and the choroid plexus (ChP) apical membrane marker AQP1 (magenta) in the LVChP of wild-type (WT) (left) versus Mbnl2 KO (right) mice. Localization of AQP1 and DAPI (blue, nucleus). Scale = 8 μm. (B) LVChP RNA splicing events (Ψ) in Mbnl2−/− (Mbnl2 KO) compared to wild-type FVB (WT) littermate mice at E13.5, P1, P14 and adult (P63), with significant events in blue. (C) LVChP AS changes by RNA-seq in Mbnl2 KO compared to FVB WT littermate mice at developmental stages E13.5, P1, P14 and P63 (gradient of light to dark grey with crosshatch for Mbnl2KO) for Mbnl2 exon 5, Mbnl1 exon 5, Inf2 exon 22 and Kif13a exon 26. Right: Mbnl1 exon 5 AS RNA-seq reads during development (E13.5, P0, P14, adult) for FVB WT littermates (light grey) compared to Mbnl2 KO (black). *P < 0.05, **P < 0.01, ****P < 0.0001, unpaired t-test.
Choroid plexus spliceopathy in myotonic dystrophy type 1
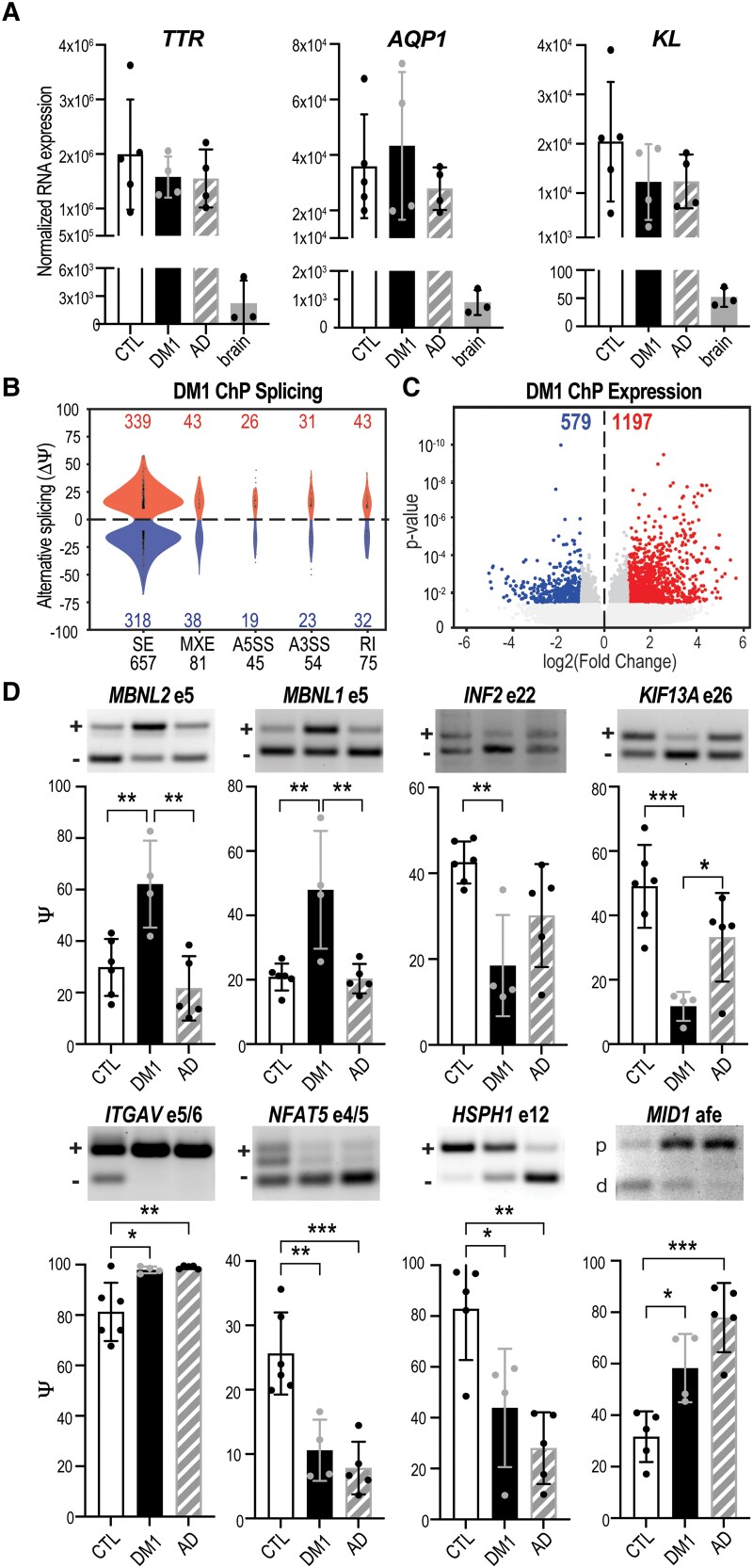
Based on our findings from mouse models that LVChP is particularly susceptible to Dmpk CUG repeat expansions (Fig. 1) and MBNL2 regulates transitions to adult specific splicing patterns (Figs 2 and 3), we hypothesized extensive transcriptomic alterations occur in DM1 LVChP. To test this hypothesis, LVChP was isolated from post-mortem DM1 patients (one female, three males; age 50–70 years), neurologically unaffected patients (two females, three males; age 50–70 years) and disease control patients (Alzheimer’s disease; three females, one male; age 70–85 years) (Supplementary Table 1). To ensure accurate identification of ChP tissues, all samples were evaluated for positive expression of the ChP markers TTR, AQP1 and KL (Fig. 4A) and negative expression of the neuronal markers MAP2, RBFOX3/NEUN and SYN1 (Supplementary Fig. 4A). Furthermore, ChP isolates were positively identified as LVChP by expression of SIX3 (Supplementary Fig. 4B) and excluded as the 4VChP by low expression of EN2 (Supplementary Fig. 4C).26 Using RNA-seq (Supplementary material), we observed that DM1 LVChP had 912 significant splicing changes approximately equally distributed in terms of inclusion versus exclusion (Fig. 4B) in contrast to DE of 1776 genes with a bias toward increased expression (Fig. 4C and Supplementary Fig. 4E). Notably, mis-splicing of MBNL2 exon 5, MBNL1 exon 5, INF2 exon 22 and KIF13A exon 26 identified in the Dmpk CTGexp KI (Fig. 1E) and Mbnl2 KO (Fig. 3C and Supplementary Fig. 3C) mouse models also occurred in DM1 patients’ ChP (Fig. 4D, top row). This subset of AS events was not significantly affected in Alzheimer’s disease or multiple sclerosis patients’ ChP (Fig. 4D, top row and Supplementary Fig. 4F). To further support our hypothesis that the spliceopathy we identified in Dmpk CTGexp KI and Mbnl2 KO mouse models is representative of human myotonic dystrophy ChP, we generated heat maps of the conserved mis-splicing events in DM1 LV ChP (Supplementary Fig. 4G) and Mbnl2 KO mice LV ChP (Supplementary Fig. 4H).
Figure 4.
ChP spliceopathy in DM1. (A) Normalized RNA expression (RNA-seq) for choroid plexus (ChP) markers TTR, AQP1 and KL for neurologically unaffected controls (CTL), myotonic dystrophy type 1 (DM1) and Alzheimer’s disease (AD), together with non-ChP brain tissues (brain). (B) Violin plots of AS events (Ψ) quantified by change in per cent spliced in (ΔPSI). A5SS = alternative 5′ splice site; A3SS = alternative 3′ splice site; MXE = mutually exclusive exon; SE = skipped exon. (C) DE of RNA transcripts showing the log2 transformed fold change plotted against significance (P-value). (D) RT-PCR validations of splicing changes in DM1 ChP compared to CTL and AD for MBNL2 exon 5, MBNL1 exon 5, INF2 exon 22, KIF13A exon 26, ITGAV exons 5 and 6, NFAT5 exons 4 and 5, HSPH1 exon 12 and MID1 alternative first exon (afe). *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA.
Although Alzheimer’s disease samples did not show the same mis-splicing that was concordant between our KI mouse model and human DM1 patients’ ChP (Fig. 4D, top row), recent reports have implicated the ChP in Alzheimer’s disease pathology.54,55 Thus, we further analysed the RNA-seq data of Alzheimer’s disease ChP (Supplementary material). In contrast to myotonic dystrophy, this analysis revealed 1174 AS events, with a bias toward exclusion, and 1520 DE genes in Alzheimer’s disease ChP (Supplementary Fig. 4I). We identified splicing events specific to Alzheimer’s disease ChP (Supplementary Fig. 4J). Unexpectedly, approximately half the splicing events with concordance in the RNA-seq reads and statistics showed similar mis-splicing between Alzheimer’s disease and DM1 patients’ ChP (Supplementary Fig. 4K), including four we validated by RT-PCR (Fig. 4D, bottom row). We subsequently analysed publicly available RNA-seq datasets of multiple sclerosis ChP56 as an additional neurological disease control, which exhibited 1030 splicing changes (Supplementary material and Supplementary Fig. 4L–N); however, the mis-splicing events we validated for DM1 and Alzheimer’s disease ChP (Fig. 4D) were unaffected in multiple sclerosis ChP (Supplementary Fig. 4F and N). These findings indicated there is a specific mis-splicing program of myotonic dystrophy ChP, although some novel events overlap with Alzheimer’s disease.
Choroid plexus mis-splicing alters CSF composition
Choroid plexus transporters and channels
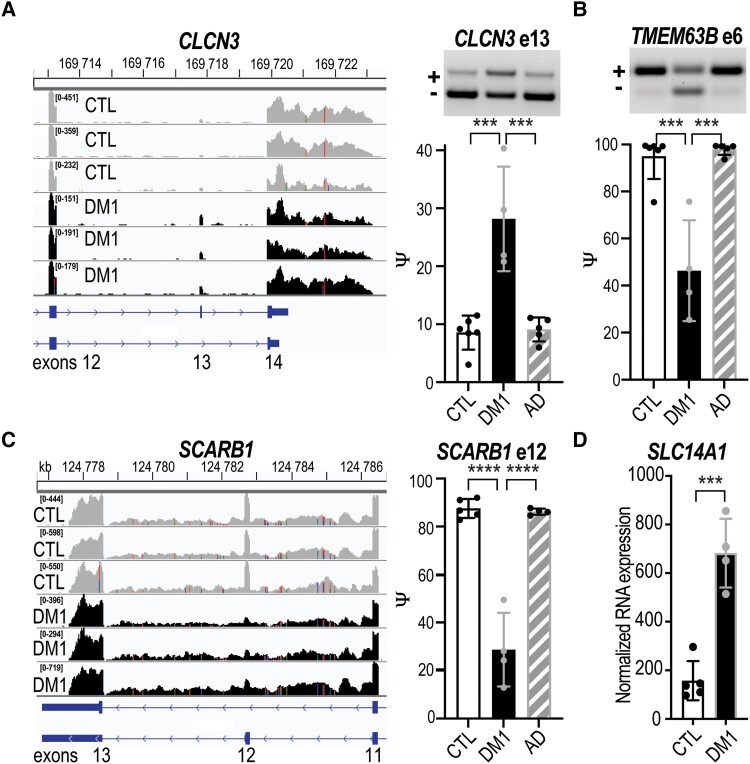
Dysregulation of chloride and calcium ion homeostasis are prominent features of myotonic dystrophy. For example, skeletal muscle myotonia is caused by mis-splicing of the chloride channel CLCN1, resulting in its depletion due to nonsense mediated decay.57–59 Thus, we next investigated if the AS and/or DE of ion channels were impaired in DM1 ChP. We screened the RNA-seq datasets for ‘transporter’ or ‘voltage-gated ion channel’ annotated genes and identified 177 genes with altered RNA AS and/or DE (Supplementary Table 6). Since chloride homeostasis is of special interest in myotonic dystrophy, we further investigated mis-splicing of CLCN3, an outward rectifying chloride-proton exchanger related to CLCN1.60 Interestingly, constitutive Clcn3 KO mice have a severe CNS neurodegenerative phenotype61,62 and CLCN3 variants have been associated with human neurodevelopmental disorders.63 In DM1 ChP, we found increased inclusion of CLCN3 exon 13 (Fig. 5A) in the absence of a change in the RNA level (Supplementary Fig. 5A). Splicing has been shown to influence CLCN3 localization; N-terminal isoforms have been shown to alter endosomal preferences while C-terminal isoforms are implicated in plasma membrane localization.64,65 Splicing of CLCN3 exon 13 causes a frameshift to a longer C-terminal isoform that localizes to the plasma-membrane66 and while maintaining the core structure adds a 48 amino acid tail (Supplementary Fig. 5B). Another mis-spliced (top 1%) ion channel gene was TMEM63B, a calcium permeable osmosensitive ion channel.67 Splicing and co-regulated Q/R RNA editing of TMEM63B regulates the osmosensitivity and calcium permeability of this channel.68 In DM1 ChP, decreased inclusion of TMEM63B exon 6 (Fig. 5B) occurred in the absence a change in the RNA levels (Supplementary Fig. 5A). Structural predictions suggest exon 6 modifies the intracellular regulatory region of TMEM63B, resulting in a hyper-osmosensitive isoform.68
Figure 5.
ChP mis-splicing alters transporters and ion channels. Neurologically unaffected (CTL) versus myotonic dystrophy type 1 (DM1) LVChP transcriptomic analysis of alternative splicing (AS) and differential expression (DE) was screened for either ‘ion channel’ or ‘transporter’ candidates. (A) CLCN3 was identified as a top ion channel candidate by RNA-seq (left) and validated (right) by RT-PCR gel (representative) and analysis (bar graph) of CLCN3 exon 13 (e13) AS in DM1 versus CTL and Alzheimer’s disease (AD) LVChP. (B) TMEM63B was also identified from RNA-seq and validated by RT-PCR for AS of exon 6. (C) SCARB1 was identified as a top transporter candidate and RNA-seq revealed SCARB1 exon 12 AS for DM1 versus CTL and AD LVChP. (D) Another transporter, SLC14A1, exhibited increased DE in DM1 versus CTL patients’ ChP. ***P < 0.001, ****P < 0.0001, one-way ANOVA or unpaired t-test. LVChP = lateral ventricle choroid plexus.
In addition to controlling the production of CSF content, the ChP also plays a role in the clearance of waste and other by-products from the CSF.29 Among the 177 genes screened from DM1 ChP RNA-seq datasets for ‘transporter’ annotation (Supplementary Table 6), we identified several genes with roles in CSF clearance. The #1 mis-splicing event was SCARB1, which encodes the high-density lipoprotein (HDL) scavenger receptor BI (SR-BI) that mediates HDL binding and selective lipid uptake at the cell surface.69SCARB1 has two primary isoforms regulated by exclusion of exon 12, resulting in differential localization and a switch to HDL endocytosis for lipid uptake.70 We identified a shift from RNA encoding SR-BI (exon 12 inclusion) to SR-BII (exon 12 exclusion) (Fig. 5C), without a change in the RNA levels (Supplementary Fig. 5A). Skipping of exon 12 leads to expression of the HDL endocytosis isoform, which is predicted to drastically alter the C-terminal structure of the protein due to a frameshift.71 Another interesting (#15) DE event for annotated ‘transporters’ was SLC14A1, which encodes the primary urea transporter for the brain, UT-B.72 We detected a >3-fold increase in SLC14A1 expression in DM1 patients’ ChP (Fig. 5D). These RNA processing changes suggested an imbalance in ion homeostasis and metabolic molecule trafficking that is consistent with other myotonic dystrophy molecular changes in tissues like skeletal muscle and heart that lead to symptoms such as myotonia, arrythmias and insulin resistance.58,73–76
Choroid plexus secretome
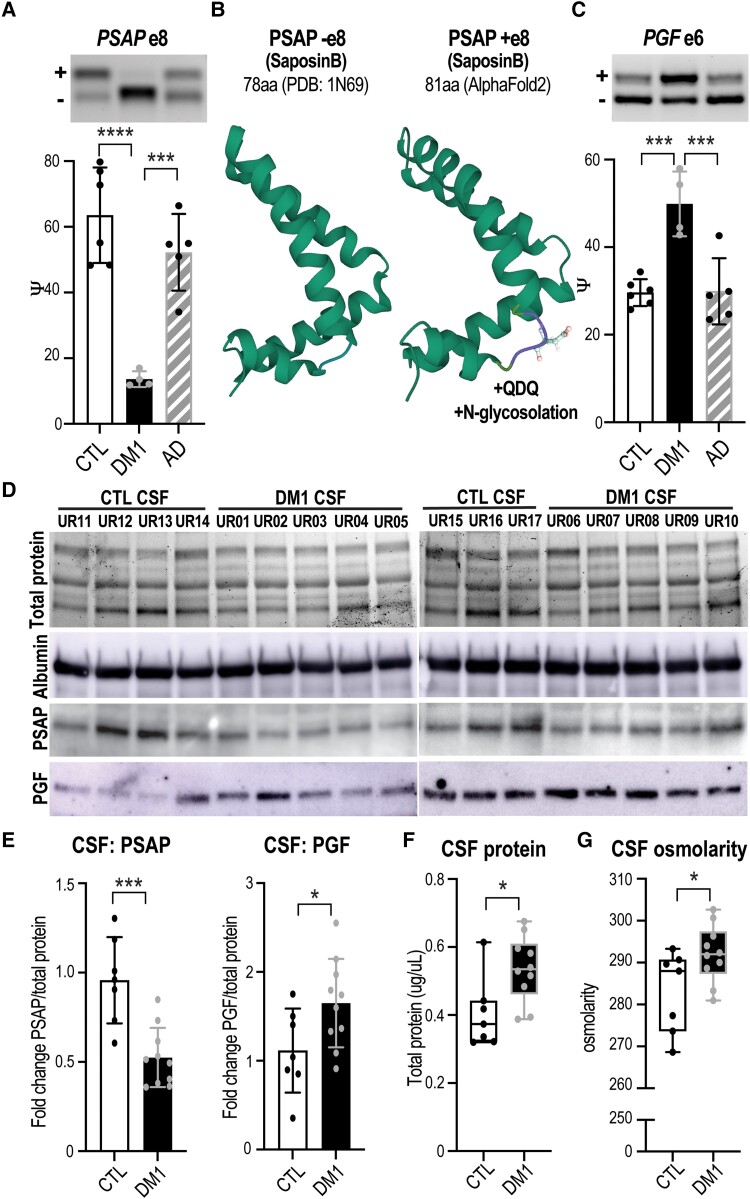
ChP secretion of a variety of factors into the CSF is required to support proper CNS development, function and maintenance.24,45,77 In our transcriptome analysis of DM1 ChP, 373 genes annotated as ‘secreted’ were significantly mis-spliced or showed DE changes (Supplementary Table 7), which could alter secretion levels and/or function of the corresponding proteins. The #1 mis-spliced event among secreted genes was in prosaposin (PSAP), a multifunctional protein that either acts intracellularly as a regulator of lysosomal enzyme activity78–80 or is secreted as a signalling factor to protect neuronal and glial cell health.78,81,82 The neurotrophic effect of secreted PSAP originates from a short 18–21 amino acid peptide sequence that binds to G-protein receptors on neurons82–84 and when mutated results in neurodegeneration in mice.85,86 RT-PCR of DM1 ChP validated exclusion of PSAP exon 8 (Fig. 6A) in the absence of expression changes (Supplementary Fig. 6A). Splicing of PSAP exon 8 is associated with targeting of the protein to either the lysosome (−exon 8) for intracellular use or to vesicles (+exon 8) for secretory export.87–91 The residues encoded by exon 8 include an aspartate positioned on the surface of the saposin-B region of PSAP, which is a substrate for N-glycosylation, facilitating subsequent PSAP oligomerization, which inhibits lysosomal sorting and promotes secretion (PDB: 1N69; Fig. 6B).87,92
Figure 6.
Mis-splicing alters the ChP secretome and CSF composition. Neurologically unaffected (CTL) versus DM1 LVChP transcriptomic analysis of alternative splicing (AS) and differential expression (DE) was screened for ‘secreted’ GO annotation candidates. (A) PSAP was selected as a top secreted candidate and validated by RT-PCR gel (representative, top) and analysis (bar graph, bottom), which showed exclusion of exon 8 in DM1 ChP. (B) Predicted (AlphaFold2) protein structural changes due to PSAP exon 8 AS with the inclusion of a +QDQ (glutamine-aspartate-glutamine) in PSAP region SaposinB (PDB: 1N69). aa = amino acids. (C) Another top secreted candidate was PGF, which was validated by RT-PCR for increased exon 6 inclusion in DM1. (D) To test if ChP changes correlate with CSF composition, we obtained DM1 and non-DM1 (CTL) patient CSF for protein lysates and assayed top candidates, PSAP and PGF. Loading was normalized based on protein quantification, total protein signal and albumin western blot. (E) Western blot of PSAP and PGF proteins in DM1 patients’ CSF compared to CTLs was quantified as fold-change normalized over total protein signal. (F and G) To test if ChP changes also correlate with cumulative measures of CSF composition, CTL versus DM1 patients’ CSF was assayed for (F) total protein as well as (G) total osmolarity. *P < 0.05, ***P < 0.001, ****P < 0.0001, one-way ANOVA or unpaired t-test.
Two additional (#8 and #9 of 373) mis-splicing events in closely related genes annotated as secreted were placental growth factor (PGF) and vascular endothelial growth factor A (VEGFA).93,94 These genes are part of the highly conserved VEGF family of growth factors, which are upregulated in CSF in response to numerous CNS injuries (stroke, trauma) and neurodegenerative diseases (multiple sclerosis, Alzheimer’s disease, Parkinson’s disease).95–99 In addition to cell type-specific effects such as endothelial growth, signalling by these growth factors contributes to vascular permeability and can lead to breakdown of the brain’s protective barriers.99–102 In DM1 ChP, we identified exon 6 mis-splicing of PGF (Fig. 6C) and VEGFA (Supplementary Fig. 6B), with a trend towards increased expression (Supplementary Fig. 6C).103 Analysis of PGF -exon 6 suggests the C-terminal tail is unstructured; however, the +exon 6 tail is much longer, and predictions suggest interactions with the main protein structure (PDB: 1FZV; Supplementary Fig. 6D).
Mis-splicing of secreted proteins in DM1 ChP may alter their CSF levels, so we obtained patient CSF to test this possibility. Since PSAP switches from approximately equal expression of +/−exon 8 isoforms in control patients to primarily excluding exon 8 in DM1 patients, we predicted that PSAP CSF protein levels would decrease. This was confirmed by western blot analysis, which detected an approximately 2-fold decrease in PSAP protein levels in DM1 versus control CSF (Fig. 6D and E). PGF/VEGFA splicing of exon 6 regulates heparin binding and the formation of signalling gradients,94,104 and we predicted this would alter PGF protein levels. Since PGF splicing was more robust than VEGFA, we used an anti-PGF antibody for western blot analysis and tested DM1 patients’ CSF, which revealed an approximately 1.5-fold increase in PGF protein levels (Fig. 6D and E).
Previous fluid biomarker analysis has shown elevation of neurofilament light chain (NEFL) in DM1 blood and changes in Aβ42 and tau levels in DM1 CSF.105 Our prior studies have also shown APP and MAPT AS changes in the hippocampus and temporal cortex.106 For DM1 ChP, we detected decreased exon 7 inclusion (∼20%) of APP (Supplementary Fig. 6E) and increased expression (2.1 log2fold) of MAPT (Supplementary Fig. 6F).
Based on the occurrence of 177 transcriptomic changes in ChP transporters/ion channels involved in CSF production/clearance, as well as 373 genes that encode secreted proteins, we tested if the net levels of protein or solute were altered in DM1 CSF. Total protein levels were found to be increased significantly in DM1 patients (Fig. 6F), which could be a marker of inflammation or neurodegeneration. Since CSF osmolarity (total solutes) is tightly regulated,107 it was noteworthy that osmolarity consistently increased in DM1 CSF compared to control CSF (Fig. 6G). Overall, our results indicated that myotonic dystrophy ChP AS changes predict altered CSF composition and levels of secreted proteins implicated in neurodegeneration and neuronal response to damage.
Discussion
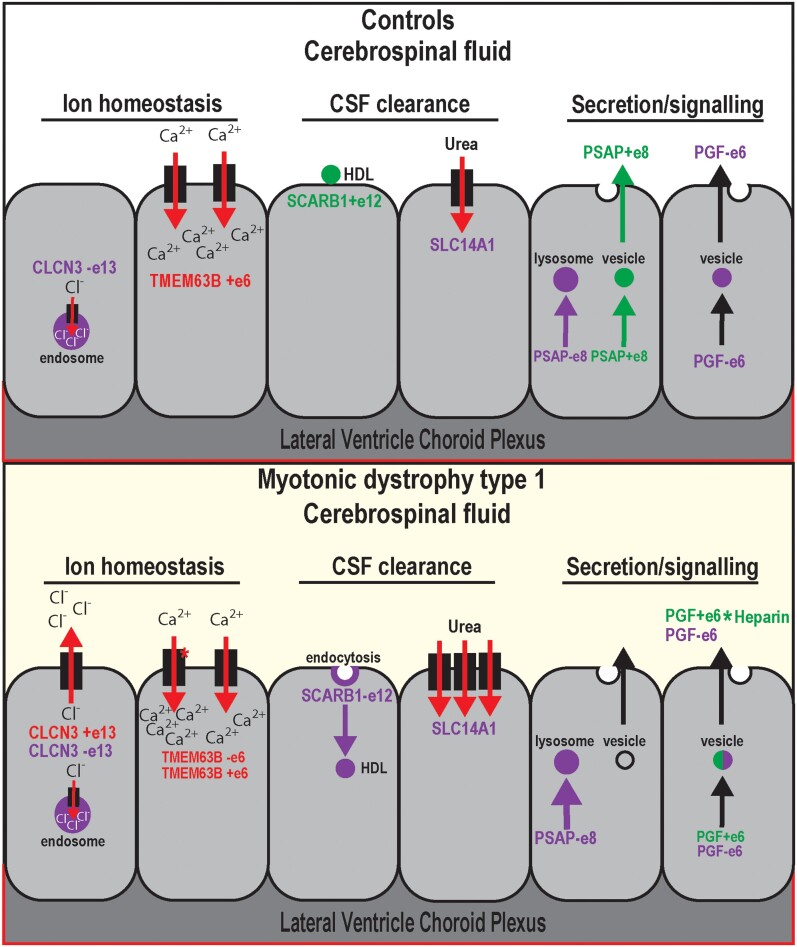
Pathomechanistic studies of neurodegenerative diseases have generally overlooked the choroid plexus in favor of neuronal and other cell types in the brain. This has certainly been the case for DM1 which is characterized by widespread CNS symptoms of uncertain origin,11,14,15 with most prior studies focusing on neuronal spliceopathy and some addressing potential glial origins.21,22,36,108 Our prior development of Dmpk CTGexp mouse models for DM1 enabled the discovery that the ChP is highly susceptible to Dmpk CUG repeat expansions.23 In this study, we first demonstrated that AS in the LVChP is more severely affected compared to 4VChP in this Dmpk CTGexp mouse model. Mis-splicing in the LVChP has important pathomechanistic implications, since this ventricular region has specific functions that impact neural development and the regulation of NSCs in the subventricular zone.24 Second, we showed that loss of MBNL2 function, previously implicated in neuronal AS, also represses LVChP developmental splicing transitions. Developmental splicing regulation by MBNL has important implications for tissue-specific mechanisms, since inappropriate expression of developmental isoforms has been shown as a primary cause of myotonic dystrophy symptoms.49,57–59,73,75,76 Third, we confirmed that the spliceopathy detected in our Dmpk CTGexp KI model also occurs in the human DM1 LVChP. Fourth, we showed that ChP mis-splicing results in downstream alterations in CSF composition and factors that are known to have detrimental impacts on CNS development and health (Fig. 7).
Figure 7.
Choroid plexus mis-splicing correlates with CSF alterations in DM1. Proposed model of non-DM1 (unaffected and disease controls) and DM1 ChP AS of CLCN3 exon 13, TMEM63B exon 6, SCARB1 exon 12, SLC14A1 DE, PSAP exon 8 and PGF exon 6, as well as potential influences on CSF content. Both CLCN3 and TMEM63B are involved in ChP regulation of ion homeostasis. CLCN3 exon 13 is associated with a localization switch from endosomal to plasma membrane, leading to a gain in outward rectifying chloride ion flow. TMEM63B exon 6 loss is associated with a switch to a more osmosensitive isoform of the calcium permeable ion channel. Both SCARB1 and SLC14A1 are involved in clearance of the CSF. SCARB1 exon 12 loss is associated with a switch to isoform SR-BII, which has been shown to promote endocytosis of high-density lipoprotein (HDL) particles. SLC14A1 encodes urea transporter B, the primary transport protein for urea in the CNS, whose increased expression has been reported in neurodegenerative diseases. Both PSAP and PGF are highly expressed secretory proteins produced by the ChP to modulate signalling throughout the CSF. PSAP exon 8 skipping is associated with a switch from trafficking of the protein to vesicles for secretion to primarily lysosomal for non-signalling functions. PGF exon 6 splicing is associated with a change in heparin binding activity and thus altered distribution gradients.
Our study revealed ChP ion channel dysregulation that we hypothesized alters CSF content directly as well as indirectly through dysregulation of ChP secretory cycles, which is supported by the increased osmolarity we identified in patient CSF. Splicing changes in ion channels were also revealed during this study that are predicted to elicit a similar ion dyshomeostasis as seen in myotonic muscle, where mis-splicing of CLCN1 results in myotonia.57,59 The inappropriate inclusion of CLCN3 exon 13, which occurs in DM1 ChP, is predicted to increase chloride export into the CSF,66 which could impair ChP ependymal cell recovery after an initial depolarization signalling event. The TMEM63B splicing shift to a more active and osmosensitive isoform in DM1 ChP suggests increased intracellular calcium storage. In myotonic muscle there is an increase in calcium and decrease in chloride ions which, upon depolarization, leads to prolonged contraction with impaired hyperpolarization necessary for recovery.58,73 Mis-splicing in DM1 ChP of CLCN3, TMEM63B and other ion channels or regulators could have a similarly detrimental effect. Dyshomeostasis of ion channels or regulators in the ChP could impair CSF production, flux and secretory release patterns such as those that occur differentially during day versus night,30 which in turn could be a primary contributor to insomnia and hypersomnia symptoms in myotonic dystrophy patients.
DM1 is associated with white matter abnormalities, altered sleep regulation, impaired executive function, progressive cerebral atrophy with parallel ventricle/CSF volume increases, as well as intellectual disability in congenital myotonic dystrophy.11,14,15,17,18 Since the choroid plexus plays a major role in CSF volume, flow and composition,29 we pursued the hypothesis that ChP spliceopathy alters these critical functions and uncovered mis-splicing events in DM1 patients’ LVChP and CSF that support this possibility. For example, mis-splicing of PSAP (a neurotrophic factor) in ChP correlates with decreased PSAP secretion into the CSF of DM1 patients. Secreted PSAP has been shown to promote the proliferation and survival of a wide range of cells, including in the CNS, where it has important functions for development as well as maintenance/protection.80–83,85,109 We predict that CSF loss of secreted PSAP, in conjunction with changes in other ChP secreted proteins, could be detrimental to the survival and resilience of neurons/glia, resulting in a major contribution to the CNS symptoms of DM1. Further, we suggest a compound effect with symptoms of early-onset ageing such that decreased PSAP in CSF leads to an imbalance of NSC proliferation in the subventricular zone and results in early-onset depletion of NSCs and inability to replenish glial cells and possibly neurons.27,31,48,110
By understanding ChP changes in DM1, we can predict and rationally select CSF biomarkers that are relevant for CNS health. Since previous biomarker analysis has shown consistent elevation of NEFL in DM1 blood,105 it would be interesting to determine if this also occurs in CSF. Our prior studies have also shown APP and MAPT AS changes in the hippocampus and temporal cortex,106 while other studies have indicated decreased Aβ42 and increased total tau levels in DM1 CSF.105 For DM1 ChP, we detected mis-splicing of APP and increased expression of MAPT, which is consistent with the previously reported CSF changes. The ChP transcriptomic alterations we report here in human DM1 and mouse DM1 models may be involved in abnormal levels of these disease biomarkers in CSF, but this possibility requires further study.
Since the choroid plexus is a barrier between blood and the CSF, it is both more accessible to drug treatments and a useful avenue to deliver treatments to the rest of the brain. This is an important consideration for DM1, since this disease has unanticipated effects on ChP functions. Potentially, ChP targeting or systemic treatments for DM1 which correct ChP mis-splicing could have the added benefit of restoring proper CSF composition and thus potentially improve CNS symptoms.
Supplementary Material
Acknowledgements
We would like to thank Ramadan Ajredini, Jodi L. Bubenik, Avery Engelbrecht, Charles A. Thornton, and Marta Trzeciak for technical support and discussion. We would like to acknowledge the patients who donated tissues and cerebrospinal fluid to this study. Additionally, we would like to recognize the NIH NeuroBioBank for providing neurologically unaffected ChP and Alzheimer’s disease ChP tissues.
Contributor Information
Curtis A Nutter, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Benjamin M Kidd, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Helmut A Carter, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Johanna I Hamel, Department of Neurology, University of Rochester, Rochester, NY 14642, USA.
Philip M Mackie, Department of Neuroscience, McKnight Brain Institute, College of Medicine, University of Florida, Gainesville, FL 32610, USA.
Nayha Kumbkarni, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Mackenzie L Davenport, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Dana M Tuyn, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Adithya Gopinath, Department of Neuroscience, McKnight Brain Institute, College of Medicine, University of Florida, Gainesville, FL 32610, USA.
Peter D Creigh, Department of Neurology, University of Rochester, Rochester, NY 14642, USA.
Łukasz J Sznajder, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Eric T Wang, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Laura P W Ranum, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, McKnight Brain Institute and the Fixel Institute for Neurological Diseases, College of Medicine, University of Florida, Gainesville, FL 32610, USA.
Habibeh Khoshbouei, Department of Neuroscience, McKnight Brain Institute, College of Medicine, University of Florida, Gainesville, FL 32610, USA.
John W Day, Department of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford, CA 94304, USA.
Jacinda B Sampson, Department of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford, CA 94304, USA.
Stefan Prokop, Department of Pathology, Immunology, and Laboratory Medicine, Center for Translational Research in Neurodegenerative Disease, McKnight Brain Institute and the Fixel Institute for Neurological Diseases, College of Medicine, University of Florida, Gainesville, FL 32610, USA.
Maurice S Swanson, Department of Molecular Genetics and Microbiology, Center for NeuroGenetics and the Genetics Institute, University of Florida, College of Medicine, Gainesville, FL 32610, USA.
Funding
This study was supported by grants from the NIH (NS048843 and NS103172 to M.S.S.), a Myotonic Dystrophy Foundation postdoctoral fellowship (C.A.N.), Myotonic Dystrophy Foundation and University of Florida McKnight Brain Institute predoctoral fellowships (B.M.K.), a Muscular Dystrophy Association Development Grant (MDA546770 to Ł.J.S.), and a Schmitt Program in Integrative Neuroscience (SPIN) pilot award (J.I.H.).
Supplementary material
Supplementary material is available at Brain online.
Competing interests
M.S.S. is a Scientific Advisory Board member for Kate Therapeutics, Skyhawk Therapeutics and Tacit Therapeutics.
References
- 1. Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17:265–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Manning KS, Cooper TA. The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol. 2017;18:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weskamp K, Olwin BB, Parker R. Post-transcriptional regulation in skeletal muscle development, repair, and disease. Trends Mol Med. 2021;27:469–481. [DOI] [PubMed] [Google Scholar]
- 5. Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. [DOI] [PubMed] [Google Scholar]
- 6. Mahadevan M, Tsilfidis C, Sabourin L, et al. Myotonic dystrophy mutation: An unstable CTG repeat in the 3’ untranslated region of the gene. Science. 1992;255:1253–1255. [DOI] [PubMed] [Google Scholar]
- 7. Taylor K, Sznajder ŁJ, Cywoniuk P, Thomas JD, Swanson MS, Sobczak K. MBNL Splicing activity depends on RNA binding site structural context. Nucleic Acids Res. 2018;46:9119–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas JD, Sznajder ŁJ, Bardhi O, et al. Disrupted prenatal RNA processing and myogenesis in congenital myotonic dystrophy. Genes Dev. 2017;31:1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goers ES, Purcell J, Voelker RB, Gates DP, Berglund JA. MBNL1 Binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010;38:2467–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang C, Lee K-Y, Swanson MS, Darnell RB. Prediction of clustered RNA-binding protein motif sites in the mammalian genome. Nucleic Acids Res. 2013;41:6793–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sznajder ŁJ, Swanson MS. Short tandem repeat expansions and RNA-mediated pathogenesis in myotonic dystrophy. Int J Mol Sci. 2019;20:3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hamilton MJ, Atalaia A, McLean J, et al. Clinical and neuroradiological correlates of sleep in myotonic dystrophy type 1. Neuromuscul Disord. 2022;32:377–389. [DOI] [PubMed] [Google Scholar]
- 13. Langbehn KE, van der Plas E, Moser DJ, Long JD, Gutmann L, Nopoulos PC. Cognitive function and its relationship with brain structure in myotonic dystrophy type 1. J Neurosci Res. 2021;99:190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subramony SH, Wymer JP, Pinto BS, Wang ET. Sleep disorders in myotonic dystrophies. Muscle Nerve. 2020;62:309–320. [DOI] [PubMed] [Google Scholar]
- 15. Miller JN, Kruger A, Moser DJ, et al. Cognitive deficits, apathy, and hypersomnolence represent the core brain symptoms of adult-onset myotonic dystrophy type 1. Front Neurol. 2021;12:700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laberge L, Gallais B, Auclair J, Dauvilliers Y, Mathieu J, Gagnon C. Predicting daytime sleepiness and fatigue: A 9-year prospective study in myotonic dystrophy type 1. J Neurol. 2020;267:461–468. [DOI] [PubMed] [Google Scholar]
- 17. van der Plas E, Hamilton MJ, Miller JN, et al. Brain structural features of myotonic dystrophy type 1 and their relationship with CTG repeats. J Neuromuscul Dis. 2019;6:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Douniol M, Jacquette A, Cohen D, et al. Psychiatric and cognitive phenotype of childhood myotonic dystrophy type 1. Dev Med Child Neurol. 2012;54:905–911. [DOI] [PubMed] [Google Scholar]
- 19. Sta Maria NS, Zhou C, Lee SJ, et al. Mbnl1 and Mbnl2 regulate brain structural integrity in mice. Commun Biol. 2021;4:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okkersen K, Monckton DG, Le N, Tuladhar AM, Raaphorst J, van Engelen BGM. Brain imaging in myotonic dystrophy type 1: A systematic review. Neurology. 2017;89:960–969. [DOI] [PubMed] [Google Scholar]
- 21. Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. [DOI] [PubMed] [Google Scholar]
- 22. Otero BA, Poukalov K, Hildebrandt RP, et al. Transcriptome alterations in myotonic dystrophy frontal cortex. Cell Rep. 2021;34:108634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nutter CA, Bubenik JL, Oliveira R, et al. Cell-type-specific dysregulation of RNA alternative splicing in short tandem repeat mouse knockin models of myotonic dystrophy. Genes Dev. 2019;33:1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lun MP, Monuki ES, Lehtinen MK. Development and functions of the choroid plexus-cerebrospinal fluid system. Nat Rev Neurosci. 2015;16:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwerk C, Tenenbaum T, Kim KS, Schroten H. The choroid plexus-a multi-role player during infectious diseases of the CNS. Front Cell Neurosci. 2015;9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dani N, Herbst RH, McCabe C, et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell. 2021;184:3056–3074.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Silva-Vargas V, Maldonado-Soto AR, Mizrak D, Codega P, Doetsch F. Age-Dependent niche signals from the choroid Plexus regulate adult neural stem cells. Cell Stem Cell. 2016;19:643–652. [DOI] [PubMed] [Google Scholar]
- 28. Lehtinen MK, Walsh CA. Neurogenesis at the brain-cerebrospinal fluid interface. Annu Rev Cell Dev Biol. 2011;27:653–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fame RM, Lehtinen MK. Emergence and developmental roles of the cerebrospinal fluid system. Dev Cell. 2020;52:261–275. [DOI] [PubMed] [Google Scholar]
- 30. Christensen J, Li C, Mychasiuk R. Choroid plexus function in neurological homeostasis and disorders: The awakening of the circadian clocks and orexins. J Cereb Blood Flow Metab. 2022;42:1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lepko T, Pusch M, Müller T, et al. Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain. EMBO J. 2019;38:e100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Myung J, Schmal C, Hong S, et al. The choroid plexus is an important circadian clock component. Nat Commun. 2018;9:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cunningham CL, Martínez Cerdeño V, Navarro Porras E, et al. Premutation CGG-repeat expansion of the Fmr1 gene impairs mouse neocortical development. Hum Mol Genet. 2011;20:64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Molero AE, Arteaga-Bracho EE, Chen CH, et al. Selective expression of mutant huntingtin during development recapitulates characteristic features of Huntington’s disease. Proc Natl Acad Sci USA. 2016;113:5736–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. NIH NeuroBioBank. Accessed 23 December 2022. https://neurobiobank.nih.gov/
- 36. Charizanis K, Lee K-Y, Batra R, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barohn RJ, Kissel JT, Warmolts JR, Mendell JR. Chronic inflammatory demyelinating polyradiculoneuropathy. Clinical characteristics, course, and recommendations for diagnostic criteria. Arch Neurol. 1989;46:878–884. [DOI] [PubMed] [Google Scholar]
- 38. Peltier AC, Donofrio PD. Chronic inflammatory demyelinating polyradiculoneuropathy: From bench to bedside. Semin Neurol. 2012;32:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen Y, Yue F, McCleary DF, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Organization of 3D Structures in the Protein Data Bank. RCSB Protein Data Bank. Accessed 23 December 2022. https://www.rcsb.org/docs/general-help/organization-of-3d-structures-in-the-protein-data-bank
- 42. Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. Colabfold: Making protein folding accessible to all. Nat Methods. 2022;19:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tunyasuvunakool K, Adler J, Wu Z, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gene Expression Omnibus. NCBI. Accessed 23 December 2022. https://www.ncbi.nlm.nih.gov/geo/
- 45. Damkier HH, Brown PD, Praetorius J. Cerebrospinal fluid secretion by the choroid plexus. Physiol Rev. 2013;93:1847–1892. [DOI] [PubMed] [Google Scholar]
- 46. Gene Detail:: Allen Brain Atlas: Mouse Brain. Accessed 23 December 2022. https://mouse.brain-map.org/gene/show/21896
- 47. Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. [DOI] [PubMed] [Google Scholar]
- 48. Bond AM, Ming G-L, Song H. Ontogeny of adult neural stem cells in the mammalian brain. Curr Top Dev Biol. 2021;142:67–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thomas JD, Oliveira R, Sznajder ŁJ, Swanson MS. Myotonic dystrophy and developmental regulation of RNA processing. Compr Physiol. 2018;8:509–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. ISH Data:: Allen Brain Atlas: Developing Mouse Brain. Accessed 23 December 2022. https://developingmouse.brain-map.org/search/index
- 51. Gene Detail:: Allen Brain Atlas: Developing Mouse Brain. Accessed 23 December 2022. https://developingmouse.brain-map.org/gene/show/21896
- 52. Sturrock RR. A morphological study of the development of the mouse choroid plexus. J Anat. 1979;129:777–793. [PMC free article] [PubMed] [Google Scholar]
- 53. Edokpolor KS, Banerjee A, McEachin ZT, et al. Altered behavioral responses show GABA sensitivity in muscleblind-like 2-deficient mice: Implications for CNS symptoms in myotonic dystrophy. eNeuro. 2022;9:ENEURO.0218-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gião T, Teixeira T, Almeida MR, Cardoso I. Choroid Plexus in Alzheimer’s disease-the current state of knowledge. Biomedicines. 2022;10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mesquita SD, Ferreira AC, Sousa JC, et al. Modulation of iron metabolism in aging and in Alzheimer’s disease: Relevance of the choroid plexus. Front Cell Neurosci. 2012;6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rodríguez-Lorenzo S, Ferreira Francisco DM, Vos R, et al. Altered secretory and neuroprotective function of the choroid plexus in progressive multiple sclerosis. Acta Neuropathol Commun. 2020;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Charlet-B N, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. [DOI] [PubMed] [Google Scholar]
- 58. Mankodi A, Takahashi MP, Jiang H, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. [DOI] [PubMed] [Google Scholar]
- 59. Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jentsch TJ, Pusch M. CLC Chloride channels and transporters: Structure, function, physiology, and disease. Physiol Rev. 2018;98:1493–1590. [DOI] [PubMed] [Google Scholar]
- 61. Yoshikawa M, Uchida S, Ezaki J, et al. CLC-3 deficiency leads to phenotypes similar to human neuronal ceroid lipofuscinosis. Genes Cells. 2002;7:597–605. [DOI] [PubMed] [Google Scholar]
- 62. Dickerson LW, Bonthius DJ, Schutte BC, et al. Altered GABAergic function accompanies hippocampal degeneration in mice lacking ClC-3 voltage-gated chloride channels. Brain Res. 2002;958:227–250. [DOI] [PubMed] [Google Scholar]
- 63. Duncan AR, Polovitskaya MM, Gaitán-Peñas H, et al. Unique variants in CLCN3, encoding an endosomal anion/proton exchanger, underlie a spectrum of neurodevelopmental disorders. Am J Hum Genet. 2021;108:1450–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Okada T, Akita T, Sato-Numata K, Islam MR, Okada Y. A newly cloned ClC-3 isoform, ClC-3d, as well as ClC-3a mediates Cd-sensitive outwardly rectifying anion currents. Cell Physiol Biochem. 2014;33:539–556. [DOI] [PubMed] [Google Scholar]
- 65. Ogura T, Furukawa T, Toyozaki T, et al. ClC-3B, a novel ClC-3 splicing variant that interacts with EBP50 and facilitates expression of CFTR-regulated ORCC. FASEB J. 2002;16:863–865. [DOI] [PubMed] [Google Scholar]
- 66. Guzman RE, Miranda-Laferte E, Franzen A, Fahlke C. Neuronal ClC-3 splice variants differ in subcellular localizations, but mediate identical transport functions. J Biol Chem. 2015;290:25851–25862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Du H, Ye C, Wu D, et al. The cation channel TMEM63B is an osmosensor required for hearing. Cell Rep. 2020;31:107596. [DOI] [PubMed] [Google Scholar]
- 68. Wu D, Zang Y-Y, Shi Y-Y, et al. Distant coupling between RNA editing and alternative splicing of the osmosensitive cation channel Tmem63b. J Biol Chem. 2020;295:18199–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mulcahy JV, Riddell DR, Owen JS. Human scavenger receptor class B type II (SR-BII) and cellular cholesterol efflux. Biochem J. 2004;377:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Eckhardt ERM, Cai L, Shetty S, et al. High density lipoprotein endocytosis by scavenger receptor SR-BII is clathrin-dependent and requires a carboxyl-terminal dileucine motif. J Biol Chem. 2006;281:4348–4353. [DOI] [PubMed] [Google Scholar]
- 71. Paterson HAB, Yu S, Artigas N, et al. Liver RBFOX2 regulates cholesterol homeostasis via Scarb1 alternative splicing in mice. Nat Metab. 2022;4:1812–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jones AC, Pinki F, Stewart GS, Costello DA. Inhibition of Urea Transporter (UT)-B modulates LPS-induced inflammatory responses in BV2 microglia and N2a neuroblastoma cells. Neurochem Res. 2021;46:1322–1329. [DOI] [PubMed] [Google Scholar]
- 73. Lueck JD, Mankodi A, Swanson MS, Thornton CA, Dirksen RT. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J Gen Physiol. 2007;129:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rao AN, Campbell HM, Guan X, et al. Reversible cardiac disease features in an inducible CUG repeat RNA-expressing mouse model of myotonic dystrophy. JCI Insight. 2021;6:e143465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Freyermuth F, Rau F, Kokunai Y, et al. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat Commun. 2016;7:11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. [DOI] [PubMed] [Google Scholar]
- 77. Mazucanti CH, Liu Q-R, Lang D, et al. Release of insulin produced by the choroid plexis is regulated by serotonergic signaling. JCI Insight. 2019;4:e131682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mendsaikhan A, Tooyama I, Serrano GE, Beach TG, Walker DG. Loss of lysosomal proteins progranulin and prosaposin associated with increased neurofibrillary tangle development in Alzheimer disease. J Neuropathol Exp Neurol. 2021;80:741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen Y-P, Gu X-J, Ou R-W, et al. Genetic analysis of prosaposin, the lysosomal storage disorder gene in Parkinson’s disease. Mol Neurobiol. 2021;58:1583–1592. [DOI] [PubMed] [Google Scholar]
- 80. Tian R, Abarientos A, Hong J, et al. Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat Neurosci. 2021;24:1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nabeka H. Prosaposin, a neurotrophic factor, protects neurons against kainic acid-induced neurotoxicity. Anat Sci Int. 2021;96:359–369. [DOI] [PubMed] [Google Scholar]
- 82. Liu B, Mosienko V, Vaccari Cardoso B, et al. Glio- and neuro-protection by prosaposin is mediated by orphan G-protein coupled receptors GPR37L1 and GPR37. Glia. 2018;66:2414–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gao HL, Li C, Nabeka H, et al. Attenuation of MPTP/MPP(+) toxicity in vivo and in vitro by an 18-mer peptide derived from prosaposin. Neuroscience. 2013;236:373–393. [DOI] [PubMed] [Google Scholar]
- 84. Kunihiro J, Nabeka H, Wakisaka H, et al. Prosaposin and its receptors GRP37 and GPR37L1 show increased immunoreactivity in the facial nucleus following facial nerve transection. PLoS One. 2020;15:e0241315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sun Y, Qi X, Witte DP, et al. Prosaposin: Threshold rescue and analysis of the “neuritogenic” region in transgenic mice. Mol Genet Metab. 2002;76:271–286. [DOI] [PubMed] [Google Scholar]
- 86. Oya Y, Nakayasu H, Fujita N, Suzuki K, Suzuki K. Pathological study of mice with total deficiency of sphingolipid activator proteins (SAP knockout mice). Acta Neuropathol. 1998;96:29–40. [DOI] [PubMed] [Google Scholar]
- 87. Carvelli L, Libin Y, Morales CR. Prosaposin: A protein with differential sorting and multiple functions. Histol Histopathol. 2015;30:647–660. [DOI] [PubMed] [Google Scholar]
- 88. Saito S, Saito K, Nabeka H, Shimokawa T, Kobayashi N, Matsuda S. Differential expression of the alternatively spliced forms of prosaposin mRNAs in rat choroid plexus. Cell Tissue Res. 2014;356:231–242. [DOI] [PubMed] [Google Scholar]
- 89. Siri L, Rossi A, Lanza F, et al. A novel homozygous splicing mutation in PSAP gene causes metachromatic leukodystrophy in two Moroccan brothers. Neurogenetics. 2014;15:101–106. [DOI] [PubMed] [Google Scholar]
- 90. Cohen T, Ravid L, Altman N, et al. Conservation of expression and alternative splicing in the prosaposin gene. Brain Res Mol Brain Res. 2004;129(1–2):8–19. [DOI] [PubMed] [Google Scholar]
- 91. Yuan L, Morales CR. Prosaposin sorting is mediated by oligomerization. Exp Cell Res. 2011;317:2456–2467. [DOI] [PubMed] [Google Scholar]
- 92. Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Privé GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci USA. 2003;100:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. De Falco S, Gigante B, Persico MG. Structure and function of placental growth factor. Trends Cardiovasc Med. 2002;12:241–246. [DOI] [PubMed] [Google Scholar]
- 94. De Falco S. The discovery of placenta growth factor and its biological activity. Exp Mol Med. 2012;44:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Janelidze S, Lindqvist D, Francardo V, et al. Increased CSF biomarkers of angiogenesis in Parkinson disease. Neurology. 2015;85:1834–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Saul J, Hutchins E, Reiman R, et al. Global alterations to the choroid plexus blood-CSF barrier in amyotrophic lateral sclerosis. Acta Neuropathol Commun. 2020;8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dewerchin M, Carmeliet P. PlGF: A multitasking cytokine with disease-restricted activity. Cold Spring Harb Perspect Med. 2012;2:a011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pikula A, Beiser AS, Chen TC, et al. Serum brain-derived neurotrophic factor and vascular endothelial growth factor levels are associated with risk of stroke and vascular brain injury: Framingham study. Stroke. 2013;44:2768–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. D’Onofrio PM, Thayapararajah M, Lysko MD, et al. Gene therapy for traumatic central nervous system injury and stroke using an engineered zinc finger protein that upregulates VEGF-A. J Neurotrauma. 2011;28:1863–1879. [DOI] [PubMed] [Google Scholar]
- 100. Licht T, Keshet E. Delineating multiple functions of VEGF-A in the adult brain. Cell Mol Life Sci. 2013;70:1727–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zachary I. Neuroprotective role of vascular endothelial growth factor: Signalling mechanisms, biological function, and therapeutic potential. Neurosignals. 2005;14:207–221. [DOI] [PubMed] [Google Scholar]
- 102. Kim HM, Hwang DH, Lee JE, Kim SU, Kim BG. Ex vivo VEGF delivery by neural stem cells enhances proliferation of glial progenitors, angiogenesis, and tissue sparing after spinal cord injury. PLoS One. 2009;4:e4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Iyer S, Leonidas DD, Swaminathan GJ, et al. The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 Å resolution. J Biol Chem. 2001;276:12153–12161. [DOI] [PubMed] [Google Scholar]
- 104. Woolard J, Bevan HS, Harper SJ, Bates DO. Molecular diversity of VEGF-A as a regulator of its biological activity. Microcirculation. 2009;16:572–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rossi S, Silvestri G. Fluid biomarkers of central nervous system (CNS) involvement in myotonic dystrophy type 1 (DM1). Int J Mol Sci. 2023;24:2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Goodwin M, Mohan A, Batra R, et al. MBNL Sequestration by toxic RNAs and RNA misprocessing in the myotonic dystrophy brain. Cell Rep. 2015;12:1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Praetorius J, Damkier HH. Transport across the choroid plexus epithelium. Am J Physiol Cell Physiol. 2017;312:C673–C686. [DOI] [PubMed] [Google Scholar]
- 108. Dincã DM, Lallemant L, González-Barriga A, et al. Myotonic dystrophy RNA toxicity alters morphology, adhesion and migration of mouse and human astrocytes. Nat Commun. 2022;13:3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Motta M, Tatti M, Furlan F, et al. Clinical, biochemical and molecular characterization of prosaposin deficiency. Clin Genet. 2016;90:220–229. [DOI] [PubMed] [Google Scholar]
- 110. Saunders NR, Dziegielewska KM, Fame RM, Lehtinen MK, Liddelow SA. The choroid plexus: A missing link in our understanding of brain development and function. Physiol Rev. 2023;103:919–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are openly available at the Gene Expression Omnibus (GEO)44 (https://www.ncbi.nlm.nih.gov/geo/) under the SuperSeries GSE228459. Mbnl2 KO mouse ChP developmental RNA-seq datasets are deposited under the SubSeries GSE228314; human patient ChP RNA-seq datasets for neurologically unaffected controls, DM1 and Alzheimer’s disease are deposited in the GEO under SubSeries GSE228458. All code used to process, analyse and visualize data is openly available at https://github.com/helmutcarter/Nutter_et_al_2023.