Abstract
The mexCD-oprJ and mexAB-oprM operons encode components of two distinct multidrug efflux pumps in Pseudomonas aeruginosa. To assess the contribution of individual components to antibiotic resistance and substrate specificity, these operons and their component genes were cloned and expressed in Escherichia coli. Western immunoblotting confirmed expression of the P. aeruginosa efflux pump components in E. coli strains expressing and deficient in the endogenous multidrug efflux system (AcrAB), although only the ΔacrAB strain, KZM120, demonstrated increased resistance to antibiotics in the presence of the P. aeruginosa efflux genes. E. coli KZM120 expressing MexAB-OprM showed increased resistance to quinolones, chloramphenicol, erythromycin, azithromycin, sodium dodecyl sulfate (SDS), crystal violet, novobiocin, and, significantly, several β-lactams, which is reminiscent of the operation of this pump in P. aeruginosa. This confirmed previous suggestions that MexAB-OprM provides a direct contribution to β-lactam resistance via the efflux of this group of antibiotics. An increase in antibiotic resistance, however, was not observed when MexAB or OprM alone was expressed in KZM120. Thus, despite the fact that β-lactams act within the periplasm, OprM alone is insufficient to provide resistance to these agents. E. coli KZM120 expressing MexCD-OprJ also showed increased resistance to quinolones, chloramphenicol, macrolides, SDS, and crystal violet, though not to most β-lactams or novobiocin, again somewhat reminiscent of the antibiotic resistance profile of MexCD-OprJ-expressing strains of P. aeruginosa. Surprisingly, E. coli KZM120 expressing MexCD alone also showed an increase in resistance to these agents, while an OprJ-expressing KZM120 failed to demonstrate any increase in antibiotic resistance. MexCD-mediated resistance, however, was absent in a tolC mutant of KZM120, indicating that MexCD functions in KZM120 in conjunction with TolC, the previously identified outer membrane component of the AcrAB-TolC efflux system. These data confirm that a tripartite efflux pump is necessary for the efflux of all substrate antibiotics and that the P. aeruginosa multidrug efflux pumps are functional and retain their substrate specificity in E. coli.
Pseudomonas aeruginosa is an opportunistic human pathogen characterized by an intrinsic resistance to a variety of antimicrobial agents. Previously attributed to a highly impermeable outer membrane (24), this property is now recognized to result from the synergy between broadly specific drug efflux pumps and low outer membrane permeability (22). One such efflux system, encoded by the mexAB-oprM operon (8, 32, 33), acts on a range of antibiotics, including tetracycline, chloramphenicol, quinolones, novobiocin, macrolides, trimethoprim, and, apparently, β-lactams and β-lactamase inhibitors (8, 16–18, 33). The β-lactam–β-lactamase inhibitor group of antibiotics are somewhat unique in that their cellular targets are within the periplasm. In contrast to the remaining antibiotic substrates, which act within the cytoplasm and are thus expected to access the pump at the cytoplasmic face of the inner membrane, the β-lactam–β-lactamase inhibitor compounds, if truly eliminated by MexAB-OprM, must interact differently with pump components. Possibilities include interaction with inner membrane constituents at the periplasmic face of the inner membrane or efflux via the outer membrane OprM alone. Still, it is not yet clear that MexAB-OprM and/or the nalB mutation associated with MexAB-OprM overexpression (see below) does not influence β-lactam resistance via an indirect effect on another resistance determinant within P. aeruginosa.
Expressed constitutively in wild-type cells, where it contributes to intrinsic drug resistance (4, 17, 33), the mexAB-oprM operon is hyperexpressed in nalB mutants (34), producing elevated levels of resistance to substrate antibiotics (8, 16, 17, 33). The MexAB-OprM efflux system is highly conserved in serotype, clinical, and environmental strains (2), indicating that it plays an important role in the intrinsic resistance of all examples of this organism. Homologous efflux systems encoded by the mexCD-oprJ (31) and mexEF-oprN (15) operons have also been described. Apparently not expressed during growth under normal laboratory conditions, these systems are expressed in nfxB (31) and nfxC (15) multidrug-resistant mutants, respectively. nfxB strains are resistant to chloramphenicol, tetracycline, quinolones, macrolides, novobiocin, and “fourth generation” cephalosporins (such as cefepime and cefpirome) but display hypersusceptibility to most β-lactam antibiotics (10). nfxC strains elicit resistant to chloramphenicol, trimethoprim, quinolones, and carbapenems, including imipenem, although the last results from the loss of the porin protein OprD in these mutants and not from overexpression of MexEF-OprN (6, 15).
The tripartite efflux pumps consist of an inner membrane component (e.g., MexB, MexD, or MexF) which functions as an RND family H+ antiport exporter (23, 35), an outer membrane, presumably channel-forming component (e.g., OprM, OprJ, or OprN) (22, 25), and a so-called membrane fusion protein predicted to link the membrane-associated efflux components (e.g., MexA, MexC, or MexE) (22, 25). Recent data suggest that the operation of MexAB-OprM (and by analogy the remaining efflux systems) is at least partially dependent upon the TonB energy-coupling protein implicated in the opening of outer membrane gated channels responsible for iron-siderophore uptake across the P. aeruginosa outer membrane (45). Thus, the outer membrane components of these efflux pumps may be gated channels.
Related efflux systems have been described in Escherichia coli (acrAB [20, 22] and acrEF [22], formerly called envCD [14]) and Neisseria gonorrhoeae (mtrCDE [9]). The AcrAB efflux components appear to function in conjunction with the TolC outer membrane protein (5), a pore-forming protein (1) previously implicated in hemolysin (42) and colicin V (44) export across the outer membrane. The acrAB-tolC-encoded efflux system is the primary known efflux system contributing to intrinsic resistance in E. coli, and acrAB deletion strains are markedly susceptible to a variety of antimicrobial agents (21, 30).
In an effort to assess the role of individual efflux system components in resistance and substrate specificity and to demonstrate that antibiotic resistance attributed to the multidrug efflux pumps in P. aeruginosa is a true indication of their efflux by these pumps, and in particular that β-lactams are truly substrates for MexAB-OprM, the mexAB-oprM and mexCD-oprJ operons were expressed in a multidrug-sensitive ΔacrAB E. coli strain. We report here that these systems were indeed expressed in E. coli, where they contributed the expected resistance to dyes, detergents, and antibiotics, including β-lactams.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are described in Table 1. For the selection of nfxB mutants hyperexpressing MexCD-OprJ (e.g., K1111), overnight cultures of P. aeruginosa ML5087 were plated on Luria broth-NaCl (LB [see “Media and culture conditions” below]) plates containing 0.4 μg of ciprofloxacin/ml. MexAB-OprM-hyperexpressing nalB mutants of this strain (e.g., K1112) were selected on 0.2 μg of ciprofloxacin/ml and 8 μg of cefoperazone/ml. The strains were confirmed as nfxB and nalB by antimicrobial susceptibility testing and by Western immunoblotting of cell envelopes with antibodies specific to OprJ and OprM, respectively (see Fig. 1). The mexCD-oprJ operon was previously cloned into plasmid pAK1900 on a ca. 10-kb BamHI fragment to yield pKMJ002. Restriction analysis of pKMJ002 revealed that mexCD-oprJ was in an orientation opposite to that of the lac promoter (plac) of this vector. To facilitate mexCD-oprJ expression in E. coli, the 10-kb BamHI fragment containing mexCD-oprJ was recloned into two other vectors, pRK415 (yielding pRSP15) and pAK1900 (yielding pRSP45), such that the operon was in the same orientation as plac. The mexCD genes were also cloned into pRK415 in the same orientation as plac (yielding pRSP25) on an 8.7-kb BamHI fragment derived from pRSP23, a pKMJ002 derivative in which oprJ had been eliminated by the deletion of two internal EcoRV fragments (0.7 and 0.8 kb). This same BamHI fragment was cloned into pAK1900, again with the mexCD genes in the same orientation as plac, to yield pRSP46. Finally, oprJ was cloned into pRK415 in the same orientation as plac on a 2.5-kb KpnI fragment to yield pRSP06. An 8.5-kb HindIII fragment containing the mexAB-oprM operon of pPV1 was previously cloned into pAK1900, and the resulting plasmid was designated pPV20. Restriction analysis of pPV20 revealed that mexAB-oprM was also transcribed in the opposite direction to the lac promoter of this vector. To facilitate mexAB-oprM expression in E. coli, the 8.5-kb HindIII fragment of pPV20 was recloned into pAK1900 (to yield pRSP01) and pRK415 (to yield pRSP17) in the same orientation as plac. pPV6, a pAK1900 derivative carrying mexAB on a ca. 5-kb HindIII fragment, was initially obtained by cloning a mexAB-containing 9-kb XhoI fragment from pPV1 into the unique SalI site of pT7-6 (to yield pRSK01) followed by its recovery on a ca. 5-kb HindIII fragment (one HindIII site upstream of mexA and a second in the multiple cloning site [MCS], downstream of the SalI-XhoI hybrid site). The 5-kb mexAB-containing HindIII fragment was subsequently cloned from pPV6 into pRK415 in the same orientation as plac to yield pRSP19. pRSP08 is a pRK415 derivative carrying the oprM gene on a 4.2-kb PstI fragment of pRSP01 in the same orientation as plac.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| ML5087 | ilv-220 thr-9001 leu-9001 met-9011 pur-67 aphA | 29 |
| K1111 | ML5087 nfxB | This study |
| K1112 | ML5087 nalB | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 acrAB+ | Bethesda Research Laboratories |
| KZM120b | ΔacrAB::kan | 21 |
| LBB1201 | KZM120b tolC::Tn10 | 5 |
| BL21(DE3) | F−ompT rB−mB−; DE3 is a lambda derivative carrying lacI and T7 RNA polymerase genes under placUV5 control | 40 |
| K113 | BL21(DE3) carrying the phage T7 lysozyme vector pLysS; Cmr | 39 |
| Plasmids | ||
| pAK1900 | E. coli-P. aeruginosa shuttle cloning vector carrying plac upstream of MCS; Apr Cbr | R. Sharp, Queen’s University |
| pKMJ002 | pAK1900::nfxB mexCD-oprJ; mexCD-oprJ in orientation opposite to plac | 31 |
| pADD214 | Mini-D replicon derived from phage D3112; Tcr | 3 |
| pPV1 | pADD214 derivative carrying a P. aeruginosa chromosomal fragment which includes mexAB-oprM | 32 |
| pPV20 | pAK1900::mexAB-oprM mexAB-oprM in orientation opposite to plac | 32 |
| pT7-6 | pBR322-based expression vector carrying MCS downstream of the strong gene 10 promoter of phage T7; Apr | S. Tabor |
| pRSK01 | pT7-6::mexAB | This study |
| pPV6 | pAK1900::mexAB; mexAB in orientation opposite to plac | 32 |
| pRSP01 | pAK1900::mexAB-OprM | This study |
| pRK415 | Low-copy-number, broad-host-range cloning vector carrying MCS downstream of plac; Tcr | 13 |
| pRSP06 | pRK415::oprJ | This study |
| pRSP08 | pRK415::oprM | This study |
| pRSP15 | pRK415::nfxB mexCD-oprJ | This study |
| pRSP17 | pRK415::mexAB-oprM | This study |
| pRSP19 | pRK415::mexAB | This study |
| pRSP23 | pAK1900::mexCD; mexCD in orientation opposite to plac | This study |
| pRSP25 | pRK415::mexCD | This study |
| pRSP45 | pAK1900::nfxB mexCD-oprJ | This study |
| pRSP46 | pAK1900::mexCD | This study |
| pSL1180 | Superlinker phagemid cloning vector | Pharmacia |
| pET21-d(+) | pBR322-derived C-terminal His-Tag vector carrying a T7lac promoter upstream of a MCS; lacI Apr | Novagen |
| pTK15 | pET21-d(+)::mexB | This study |
MCS, multiple cloning site; Apr, ampicillin resistant; Cbr, carbenicillin resistant; Tcr, tetracycline resistant; T7lac, phage T7 promoter followed by a lac operator. Unless otherwise indicated, all genes were cloned in the same orientation as plac of pAK1900 or pRK415.
KZM120 is the same strain as HN818.
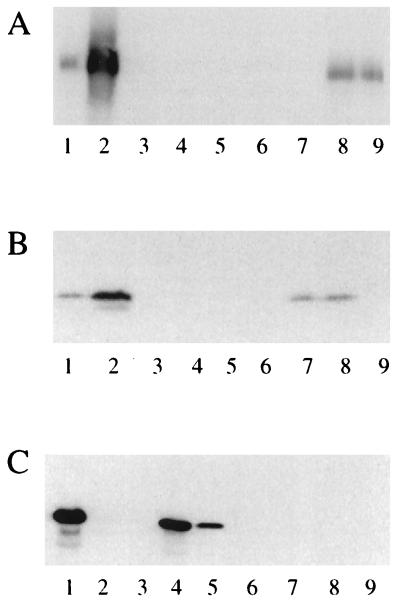
FIG. 1.
Western immunoblots of bacterial cell envelopes (10 μg of protein) developed with antibodies raised against MexB (A), OprM (B), and OprJ (C). Lanes 1, K1111 (nfxB); lanes 2, K1112 (nalB); lanes 3, KZM120 carrying pRK415; lanes 4, KZM120 carrying pRSP06 (oprJ); lanes 5, KZM120 carrying pRSP15 (nfxB mexCD-oprJ); lanes 6, KZM120 carrying pRSP25 (mexCD); lanes 7, KZM120 carrying pRSP08 (oprM); lanes 8, KZM120 carrying pRSP17 (mexAB-oprM); lanes 9, KZM120 carrying pRSP19 (mexAB). (The relevant efflux phenotype or genes expressed are indicated in parentheses.)
Media and culture conditions.
Liquid media for routine culturing of E. coli and P. aeruginosa strains were prepared by dissolving 15.5 g of Miller’s Luria broth base (Difco) and 2 g of NaCl per liter of H2O (LB). For the selection of chromosomal or plasmid antibiotic resistance markers in E. coli, ampicillin at 100 μg per ml, kanamycin at 50 μg per ml, and tetracycline at 10 μg per ml were used in the growth media. Solid media were obtained by the addition of Bacto Agar (1.5% [wt/vol]; Difco).
Molecular biological techniques.
Restriction endonuclease digestions, ligations, and transformations were performed as described by Sambrook et al. (36). E. coli DH5α was used as the host for large-scale isolation of plasmid DNA with a plasmid maxi kit (Qiagen). DNA fragments used in cloning were extracted from agarose gels with Prep-A-Gene (Bio-Rad) according to the manufacturer’s instructions. E. coli cells were made competent by the CaCl2 method (36) or, when highly competent E. coli were required, by the method of Inoue et al. (12).
Isolation of cell envelopes.
Twenty milliliters of bacterial culture was grown in LB to an absorbance at 600 nm (A600) of 1.00, harvested by centrifugation, and stored at −20°C. Cells were later thawed on ice, resuspended in 1 ml of phosphate-buffered saline (1.67 mM NaH2PO4 · H2O–8.09 mM Na2HPO4–150 mM NaCl [pH 7.4]), and sonicated until the cell suspension was clear. Following centrifugation (8,000 × g; 10 min) to pellet unlysed cells and debris, the resulting supernatant was centrifuged (300,000 × g; 15 min) and the cell envelope pellet was resuspended in 100 μl of H2O.
Purification of MexB and generation of rabbit polyclonal antiserum.
To generate antibodies to MexB, the protein was purified as polyhistidine-tagged MexB (MexB-His) following the cloning and expression of the gene in plasmid pET21-d(+) (Pharmacia). To clone mexB into this vector so that the 3′ end of the gene was in frame with the polyhistidine-encoding sequences of pET21-d(+), the following approach was taken. First, the bulk of mexB was released from plasmid pPV6 by digestion with EcoRI and KpnI, with EcoRI cutting upstream of the gene within mexA and KpnI cutting 133 bp upstream of the mexB terminus. The 133 bp at the 3′ end of mexB were then amplified with Vent polymerase (New England Biolabs) and primers TK-1 (5′-GATCGGTACCGGCGTGATCGGCGGCATGGTCACTGCGACCGTCCTGGCGATCTTCTGGGTGCC-3′) and TK-2 (5′-AATTCTCGAGTTGCCCCTTTTCGACGGACG-3′). TK-1 eliminates a KpnI site within the 3′ region of mexB as a result of an A-G change (highlighted in bold italics) which does not alter the amino acid sequence of MexB but does permit subsequent digestion of the PCR product with KpnI and XhoI so that it can be cloned together with the aforementioned mexB-containing EcoRI-KpnI fragment to regenerate an intact mexB gene. PCR mixtures (100 μl) contained 20 ng of pPV6, 1 μM each primer, 200 μM each deoxynucleoside triphosphate, 4 mM MgSO4, 10% (vol/vol) dimethylsulfoxide, and 1 U of Vent polymerase in 1× reaction buffer. Mixtures were heated at 94°C for 2 min followed by 30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min a 72°C before finishing with 10 min at 72°C. The PCR product was then purified with the Qiaquick PCR purification kit (Qiagen), digested with KpnI and XhoI, and cloned into KpnI-XhoI-restricted pSL1180. The insert was sequenced to ensure that the complete 133-bp 3′ region had been cloned and, except for the elimination of the KpnI site, that no alterations in the mexB sequences had been introduced during PCR. The mexB-containing 3.8-kb EcoRI-KpnI fragment was then cloned into EcoRI-KpnI-restricted pSL1180 carrying the cloned PCR product, and the restored mexB gene was then recovered on a ca. 3.9-kb EcoRI-XhoI fragment. Following the cloning of this fragment into EcoRI-XhoI-restricted pET21-d(+), the resultant vector, pTK15, was introduced into E. coli BL21(DE3) carrying the pLysS plasmid (strain K113) and expression of mexB was induced with IPTG (isopropyl-β-d-thiogalactopyranoside). Briefly, overnight cultures of pTK15-carrying K113 in LB containing appropriate antibiotics were diluted 1:49 in the same medium (500 ml) and incubated for 4 h, at which time IPTG was added (0.2 mM final concentration). Two hours later, cells were harvested by centrifugation (8,000 × g) and cell envelopes were prepared (as above) and solubilized in 2 ml of 20 mM Tris-HCl (pH 8.0)–100 mM NaCl–2% (vol/vol) Triton X-100. The MexB-containing Triton X-100-soluble material (supernatant fraction) was then recovered, following centrifugation (300,000 × g), and applied to a 1.5-ml TALON (Clonetech Laboratories, Inc., Palo Alto, Calif.) column equilibrated with 20 mM Tris-HCl (pH 8.0)–100 mM NaCl–0.5% (vol/vol) Triton X-100 (column buffer). The column was washed with 15 ml of column buffer, and bound protein was eluted with column buffer containing 100 mM imidazole at a flow rate of 0.1 ml/min. Three hundred-microliter fractions were collected, and MexB-containing fractions, identified following analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were pooled and dialyzed against 2 liters of 20 mM Tris-HCl (pH 8.0)–0.5% (vol/vol) Triton X-100. Antibodies to purified MexB-His were subsequently raised in rabbits by L. Mutharia, Department of Microbiology, University of Guelph.
SDS-PAGE and immunoblotting.
SDS-PAGE was performed as described previously (19) with 10% (wt/vol) acrylamide in the running gel. Proteins resolved in gels were Coomassie blue stained or transferred electrophoretically to nitrocellulose membranes (BA85; Schleicher & Schuell). Electrophoretic transfer of proteins was as described previously (41) except that SDS (0.1% [wt/vol]) was included in the blotting buffer and transfer was carried out for 16 h at 4°C and 25 mA constant current. Blotted membranes were subsequently incubated in phosphate-buffered saline containing 0.1% (vol/vol) Tween 20 (BDH) (PBST) and 10% (wt/vol) skim milk (Difco) for 60 min. Following two 5-min washes with PBST, the membranes were incubated for 60 min with primary antibody in PBST containing 1% (wt/vol) bovine serum albumin (Sigma Chemical Co., St. Louis, Mo.) and then washed four times for 10 min each time with PBST. Secondary antibody conjugated to horseradish peroxidase (HRP) in PBST containing 1% bovine serum albumin was then added to the membranes, which were subsequently washed four times for 10 min each time with PBST. Substrates for HRP were from the ECL Western blotting detection kit (Amersham) and were used according to the manufacturer’s instructions. The enzymatic activity of HRP was detected as the emission of chemiluminescence after the exposure of blots to XAR 5 film (Kodak). All incubations and washings in the immunoblot procedure were carried out at room temperature with agitation. The primary antibodies anti-OprJ (11) and anti-OprM (7) were previously described mouse monoclonals, and anti-MexB was a rabbit polyclonal antiserum (see above). HRP-conjugated anti-mouse and anti-rabbit secondary antibodies were purchased from Jackson Laboratories and Amersham, respectively.
Antimicrobial susceptibility testing.
The susceptibilities of E. coli strains to a number of antimicrobial agents were tested by inoculating 1 ml of LB containing serial twofold dilutions of each antimicrobial agent with 105 organisms as described previously (17). Bacterial inocula were derived from stocks prepared from overnight cultures grown in LB which had been harvested and resuspended in an equal volume of 10 mM MgSO4.
Antimicrobial agents.
Ampicillin, penicillin G, ticarcillin, cefotaxime, cefoperazone, cefsulodin, cephaloridine, ceftriaxone, ciprofloxacin, norfloxacin, novobiocin, chloramphenicol, and tetracycline were purchased from Sigma. SDS and crystal violet were purchased from ICN Biochemicals, Inc., and Difco, respectively. Cefepime was provided by Bristol-Myers Squibb. Azithromycin was a gift from Pfizer. Sparfloxacin (Rhône-Poulenc Rorer) and pefloxacin (Laboratoire Roger Bellon) were gifts from Microcide Pharmaceuticals Inc., Mountain View, Calif. Cefpirome (Roussel Uclaf) and ceftazidime (Glaxo) were also gifts. Erythromycin (Abbott) and imipenem (Merck Sharpe & Dohme) were purchased from the pharmacy of the Kingston General Hospital.
RESULTS AND DISCUSSION
Expression of MexAB-OprM in E. coli.
To assess the operation of MexAB-OprM in E. coli, it was necessary to express the P. aeruginosa multidrug efflux system in this heterologous host. Using antibodies specific to MexB and OprM (Fig. 1A and B, lanes 2), therefore, we examined the production of pump components in E. coli. Initially, mexAB-oprM was cloned into the low-copy-number, broad-host-range vector pRK415 (to yield pRSP17) in the same orientation as the lac promoter of this vector in case it was necessary to induce expression of the operon in order to obtain detectable expression of the pump components. Introduction of pRSP17 into E. coli DH5α (AcrAB+) (data not shown) or KZM120 (AcrAB−) provided for substantial expression of MexAB-OprM without IPTG induction, as evidenced by the production of MexB (Fig. 1A, lane 8) and OprM (Fig. 1B, lane 8) in cell envelopes of these strains. Similarly, E. coli KZM120 carrying the mexAB vector pRSP19 or the oprM vector pRSP08 produced substantial levels of cell envelope-associated MexB (Fig. 1A, lane 9) and OprM (Fig. 1A, lane 7), respectively, without IPTG induction. Thus, E. coli carrying mexAB-oprM (or mexCD-oprJ [see below]) or its components was assessed for antibiotic susceptibility in the absence of IPTG.
Activity of MexAB-OprM in E. coli.
Despite the expression of MexAB-OprM in E. coli DH5α, the presence of pRSP17 failed to enhance the antibiotic resistance of this strain to any of the tested agents (data not shown) (Tables 2 and 3 list the agents tested). In contrast, the acrAB deletion strain KZM120 carrying pRSP17 exhibited elevated resistance to a variety of agents, including the more hydrophobic quinolones, novobiocin, macrolides, chloramphenicol, detergents, dyes, and the penicillin subgroup of the β-lactam antibiotics (Tables 2 and 3). Although the influence of pRSP17 on tetracycline resistance could not be assessed (the vector carries a tet gene), KZM120 carrying pRSP01, a bla vector with the mexAB-oprM operon, did show a twofold increase in resistance to tetracycline (data not shown).
TABLE 2.
Antimicrobial susceptibility of E. coli expressing components of the P. aeruginosa multidrug efflux pumpsa
| Strain | Plasmid | Efflux genes | MIC (μg/ml)b,c
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | AZI | CAM | SDS | CV | CIP | NOR | SPAR | PEF | NOV | |||
| KZM120 | pRK415 | —d | 16 | 1 | 1 | 32 | 2 | 0.008 | 0.016 | 0.008 | 0.032 | 8 |
| KZM120 | pRSP17 | mexAB-oprM | 64 | 2 | 2 | >256 | 8 | 0.008 | 0.016 | 0.016 | 0.064 | 32 |
| KZM120 | pRSP15 | mexCD-oprJ | >256 | 8 | 2 | 256 | >16 | 0.016 | 0.064 | 0.016 | 0.064 | 8 |
| KZM120 | pRSP25 | mexCD | >256 | 8 | 2 | 256 | >16 | 0.016 | 0.064 | 0.016 | 0.064 | 8 |
| KZM120 | pAK1900 | —; TolC+ | 4 | 0.5 | NDe | 128 | 0.25 | ND | 0.016 | ND | ND | ND |
| KZM120 | pRSP45 | mexCD-oprJ; TolC+ | 64 | 4 | ND | >1,024 | >16 | ND | 0.032 | ND | ND | ND |
| KZM120 | pRSP46 | mexCD; TolC+ | 64 | 4 | ND | >1,024 | >16 | ND | 0.032 | ND | ND | ND |
| LBB1201 | pAK1900 | —; TolC− | 4 | 0.5 | ND | 16 | 0.25 | ND | 0.016 | ND | ND | ND |
| LBB1201 | pRSP45 | mexCD-oprJ; TolC− | 256 | 16 | ND | >1,024 | >16 | ND | 0.128 | ND | ND | ND |
The susceptibility of E. coli KZM120 (ΔacrAB::kan) and LBB1201 (KZM120 tolC::Tn10) carrying P. aeruginosa multidrug efflux genes on the indicated plasmids to various antimicrobial agents was assayed as described in Materials and Methods.
ERY, erythromycin; AZI, azithromycin; CAM, chloramphenicol; SDS, sodium dodecyl sulfate; CV, crystal violet; CIP, ciprofloxacin; NOR, norfloxacin; SPAR, sparfloxacin; PEF, pefloxacin; NOV, novobiocin.
E. coli KZM120 carrying plasmids pRSP08 (oprM), pRSP19 (mexAB), or pRSP06 (oprJ) showed the same pattern of resistance as exhibited by KZM120 carrying the pRK415 vector control. E. coli LBB1201 carrying pRSP46 (mexCD) showed the same pattern of resistance as LBB1201 carrying the pAK1900 vector control.
—, no plasmid-borne efflux genes.
ND, not determined.
TABLE 3.
β-lactam susceptibility of E. coli strains expressing components of the P. aeruginosa multidrug efflux pumpsa
| Plasmid | Efflux genes | MIC (μg/ml)b,c
|
||||
|---|---|---|---|---|---|---|
| AMP | PEN | TIC | CPZ | TAX | ||
| pRK415 | —d | 16 | 32 | 4 | 0.128 | 0.064 |
| pRSP17 | mexAB-oprM | 128 | 128 | 32 | 0.512 | 0.128 |
| pRSP15 | mexCD-oprJ | 32 | 32 | 4 | 0.256 | 0.064 |
| pRSP25 | mexCD | 32 | 32 | 4 | 0.256 | 0.064 |
The susceptibility of E. coli KZM120 (ΔacrAB::kan) carrying P. aeruginosa multidrug efflux genes on the indicated plasmids to various antimicrobial agents was assayed as described in Materials and Methods.
AMP, ampicillin; PEN, penicillin G; TIC, ticarcillin; CPZ, cefoperazone; TAX, cefotaxime.
E. coli KZM120 carrying pRSP08 (oprM), pRSP19 (mexAB) and pRSP06 (oprJ) showed the same resistance pattern as that exhibited by KZM120 carrying pRK415. β-lactams for which no change in susceptibility was observed for KZM120 with or without plasmids containing efflux genes included cephaloridine, cefsulodin, cefepime, cefpirome, imipenem, ceftriaxone, and ceftazidime.
—, no plasmid-borne efflux genes.
This pattern of resistance is less broad than that attributed to MexAB-OprM in P. aeruginosa, where resistance to a variety of cephalosporins, including cefotaxime, ceftazidime, cefsulodin, ceftriaxone, cefepime, and cefpirome, and to the quinolones ciprofloxacin and norfloxacin is also afforded by this efflux system (17, 37, 38). Nonetheless, many of the agents to which MexAB-OprM provided resistance in E. coli are also accommodated by the E. coli acrAB-encoded efflux system, which has been shown to facilitate resistance to chloramphenicol, tetracycline, penicillins (penicillin G and ampicillin), nalidixic acid, novobiocin, erythromycin, SDS, and crystal violet (20, 21, 30). In light of the high degree of homology between MexAB and AcrAB (MexA-AcrA, 57.7% identity; MexB-AcrB, 69.8% identity), it is perhaps not surprising that MexAB-OprM is functional in E. coli and that it accommodates a similar range of agents. Still, even for those agents for which resistance is afforded by MexAB-OprM in E. coli, resistance levels attributed to this pump are markedly lower in this organism than in P. aeruginosa (17, 33, 38). This reduced range of antibiotic substrates and these lower resistance levels likely reflect the markedly higher outer membrane permeability of E. coli compared to that of P. aeruginosa (26, 27), which increases drug influx and probably compromises the contribution of MexAB-OprM-mediated drug efflux to resistance. Indeed, it has been proposed that significant intrinsic antibiotic resistance results from a synergy between reduced influx (i.e., low outer membrane permeability) and active efflux and that both determinants are indispensable (22). Certainly, the higher outer membrane permeability of E. coli has been used to explain the organism’s generally greater susceptibility to antimicrobial agents compared to that of P. aeruginosa, despite the presence of multidrug efflux systems in both (22).
The increased outer membrane permeability of E. coli reflects an increased permeability to hydrophilic agents, which typically use the porin pathway for uptake across the outer membrane. It is not unexpected, therefore, that resistance afforded by MexAB-OprM (and MexCD-OprJ [see below]) in E. coli is generally confined to the more hydrophobic agents, whose uptake will still be somewhat restricted. The contribution of MexAB-OprM to β-lactam resistance in E. coli, for example, was generally limited to the penicillins (e.g., ampicillin, penicillin G, and ticarcillin), whose nuclei are more hydrophobic than those of the cephalosporins (43). Moreover, although MexAB-OprM-mediated resistance to the cephalosporins cefotaxime, ceftazidime, ceftriaxone, cefoperazone, cefpirome, and cefepime was minimal or lacking in E. coli, resistance to these cephalosporins is readily provided by this efflux system in P. aeruginosa (17, 37, 38), indicating that the lack of cephalosporin resistance in MexAB-OprM-expressing E. coli is not due to a deficiency of the pump. The observation that MexAB-OprM-expressing E. coli is less susceptible (though still not resistant) to cefoperazone is likely explained by the presence of an exceptionally bulky side chain (43), which is expected to impede the drug’s passage across the outer membrane, thereby enhancing the contribution of efflux to resistance to this agent.
Arguably, the most significant observation was that β-lactam (penicillin) resistance was afforded by MexAB-OprM in E. coli KZM120, since this confirmed that this efflux system can indeed accommodate β-lactams and that it is directly responsible for the β-lactam resistance attributed to it in, e.g., nalB strains. Interestingly, however, and despite the periplasmic location of their targets, KZM120 expressing oprM alone did not exhibit enhanced resistance to β-lactams (or any other agent) (Tables 2 and 3), indicating that the inner membrane-associated components are essential features of a β-lactam-extruding pump and that OprM alone is unable to facilitate β-lactam efflux and, thus, resistance. Similarly, MexAB alone (pRSP19) also failed to influence antibiotic resistance in the ΔacrAB strain (Tables 2 and 3), demonstrating that a tripartite pump is the only efflux-competent entity.
Although not previously reported for P. aeruginosa, the observation that MexAB-OprM expression afforded resistance to crystal violet and SDS in E. coli indicated that, as with AcrAB, the MexAB-OprM efflux pump can accommodate dyes and detergents. Interestingly, recent comparisons of MexB-OprM+ and MexAB-OprM− strains of P. aeruginosa failed to reveal any differences in susceptibility to either of these agents (data not shown). The MexAB-OprM− strain was, in fact, very resistant to both SDS and crystal violet (data not shown), presumably due to the presence of additional efflux systems in this strain which are capable of exporting these compounds.
Expression of MexCD-OprJ in E. coli.
pRSP15 carrying mexCD-oprJ was also introduced into E. coli DH5α and KZM120, and expression of the efflux system was assessed by immunoblotting with an available anti-OprJ antiserum. As above, E. coli DH5α (data not shown) and KZM120 carrying this vector demonstrated OprJ production (Fig. 1C, lane 5) consistent with the expression of the mexCD-oprJ operon in this strain. Interestingly, introduction of pKMJ002 (the original mexCD-oprJ vector, which carries the efflux genes in an orientation opposite to that of the lac promoter on this plasmid) into KZM120 failed to elicit any antibiotic resistance, indicating that, while IPTG induction may not have been necessary for expression of efflux components, expression from the lac promoter was probably critical. KZM120 carrying oprJ alone on pRSP06 also demonstrated substantial OprJ production (Fig. 1C, lane 4); in fact, levels were markedly higher than those seen for KZM120 carrying pRSP15. This difference in OprJ expression may be due to the presence of the nfxB gene, encoding a repressor of mexCD-oprJ expression (28, 31), on pRSP15, which might have limited mexCD-oprJ (and hence OprJ) expression from this vector. The ability of the nfxB gene to repress expression of a mexC-lacZ fusion in E. coli has been demonstrated (31). In contrast, the mexAB-oprM-encoding plasmid pRSP17 lacks the mexR repressor gene implicated in regulation of mexAB-oprM (34), and accordingly, the levels of OprM detected in KZM120 carrying oprM alone (Fig. 1B, lane 7) or mexAB-oprM (Fig. 1B, lane 8) were similar.
Activity of MexCD-OprJ in E. coli.
E. coli KZM120 carrying mexCD-oprJ on pRSP15 demonstrated elevated resistance to a variety of agents, including macrolides, quinolones, chloramphenicol, SDS, and crystal violet (Table 2). As was seen for MexCD-OprJ in P. aeruginosa, this efflux system did not facilitate resistance to novobiocin (Table 2) or most β-lactams (Table 3) in E. coli. The decreased susceptibility of MexCD-OprJ-expressing E. coli to cefoperazone, however, was consistent with previous observations that nfxB strains of P. aeruginosa show slightly reduced cefoperazone susceptibility (38). Interestingly, although MexCD-OprJ-expressing strains of P. aeruginosa display resistance to the fourth generation cephalosporins cefepime and cefpirome (17, 38), this efflux system failed to alter the susceptibility of E. coli KZM120 to these agents (Table 3), again highlighting the point that the P. aeruginosa efflux systems are not as effective in providing resistance to substrate antibiotics in E. coli. The expression of OprJ alone on pRSP06 (Fig. 1C, lane 4) did not increase the resistance of KZM120 to any tested antimicrobial agent (Table 2), although KZM120 expressing MexCD from pRSP25 exhibited the same profile of resistances as the MexCD-OprJ-expressing KZM120(pRSP15) (Table 2). The dispensability of OprJ suggests that some E. coli outer membrane protein substitutes for OprJ in the operation of this efflux system.
Role of TolC in MexCD-mediated antibiotic resistance in E. coli.
Previous studies have shown that deletion of oprJ in a mexCD-oprJ-hyperexpressing nfxB strain compromised the resistance afforded by the MexCD-OprJ system (38), indicating that the MexCD components are not efflux competent, at least in P. aeruginosa, in the absence of an outer membrane pump constituent. The TolC outer membrane protein, implicated as the outer membrane component of the AcrAB efflux system (5), is one possible candidate for association with MexCD in the reconstitution of an efflux system in E. coli. Precedents exist for the functioning of such so-called chimeric efflux systems, since functional MexCD-OprM and MexAB-OprJ pumps have been successfully constructed in P. aeruginosa (38). To assess the possible involvement of TolC in OprJ-independent, MexCD-mediated multidrug resistance, the mexCD genes were introduced into E. coli LBB1201, a KZM120 derivative carrying a tolC::Tn10 mutation. Because of the Tn10-encoded tetracycline resistance of this strain, however, it was necessary to introduce these genes on the pAK1900-based bla (β-lactam resistance) plasmid pRSP46 rather than on the aforementioned tetracycline resistance vector, pRSP25. The pRSP46 vector failed to increase the resistance of LBB1201 to several agents, including erythromycin, azithromycin, norfloxacin, crystal violet, and SDS, although as expected, it did increase the resistance of the TolC+ strain KZM120 to these agents (Table 2), indicating that the observed MexCD-mediated multidrug resistance in ΔacrAB strains of E. coli requires TolC. Thus, the latter protein likely functions as the outer membrane efflux system component of a MexCD-TolC pump. Introduction of mexCD-oprJ on the pAK1900-based vector pRSP45 also facilitated resistance to erythromycin, azithromycin, norfloxacin, crystal violet, and SDS in both KZM120 and LBB1201 (Table 2), consistent with the native MexCD-OprJ efflux system being operational in E. coli, independent of the AcrAB-TolC components, and with strain LBB1201 being capable of reconstituting a functional MexCD-OprJ efflux system. Interestingly, the MexCD-OprJ system appeared to function better in the TolC− strain LBB1201 than in the TolC+ strain KZM120 (Table 2), suggesting that by virtue of its apparent ability to associate with MexCD, TolC may interfere with the proper association and activity of the MexCD-OprJ components.
These data indicate that the context in which an efflux system operates is important for the expression of resistance and that, limitations in expression of efflux components notwithstanding, MexAB-OprM and MexCD-OprJ in E. coli do not facilitate resistance to as many agents or to levels as high as in P. aeruginosa. Attempts at reconstituting AcrAB in P. aeruginosa (using genes cloned into pRK415 from pUC15C [20]) to see if this system is more active in the less permeable P. aeruginosa have so far failed, apparently because of the lethality of these genes on multicopy vectors in this organism (37). The demonstrated ability to reconstitute MexCD-OprJ and MexAB-OprM activity in E. coli should nonetheless facilitate intended studies of antibiotic efflux by using everted vesicles, a technology which is better developed for this organism. Such studies of the P. aeruginosa pumps should provide novel insights into the molecular and biochemical events that lead to drug efflux.
ACKNOWLEDGMENTS
We are grateful to H. Nikaido and J. Fralick for providing strains.
This work was supported by a grant from the Canadian Cystic Fibrosis Foundation to K.P. R.S. was the recipient of a postdoctoral fellowship from the Natural Sciences and Engineering Research Council (NSERC). K.P. is an NSERC University Research Fellow.
REFERENCES
- 1.Benz R, Maier E, Gentshev I. TolC of Escherichia coli functions as an outer membrane channel. J Bacteriol. 1993;178:5803–5805. doi: 10.1016/s0934-8840(11)80836-4. [DOI] [PubMed] [Google Scholar]
- 2.Bianco N, Neshat S, Poole K. Conservation of the multidrug resistance efflux gene oprM in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:853–856. doi: 10.1128/aac.41.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darzins A, Casadaban M J. In vivo cloning of Pseudomonas aeruginosa genes with mini-D3112 transposable bacteriophage. J Bacteriol. 1989;171:3917–3925. doi: 10.1128/jb.171.7.3917-3925.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans, K., and K. Poole. Unpublished data.
- 5.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda H, Hosaka M, Hirai K, Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1990;34:1757–1761. doi: 10.1128/aac.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotoh N, Itoh N, Tsujimoto H, Yamagishi J-I, Oyamada Y, Nishino T. Isolation of OprM-deficient mutants of Pseudomonas aeruginosa by transposon insertion mutagenesis: evidence of involvement in multiple antibiotic resistance. FEMS Microbiol Lett. 1994;122:267–274. doi: 10.1111/j.1574-6968.1994.tb07179.x. [DOI] [PubMed] [Google Scholar]
- 8.Gotoh N, Tsujimoto H, Poole K, Yamagishi J-I, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 10.Hirai K, Suzue S, Irikura T, Iyobe S, Mitsuhashi S. Mutations producing resistance to norfloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:582–586. doi: 10.1128/aac.31.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosaka M, Gotoh N, Nishino T. Purification of a 54-kilodalton protein (OprJ) produced in NfxB mutants of Pseudomonas aeruginosa and production of a monoclonal antibody specific to OprJ. Antimicrob Agents Chemother. 1995;39:1731–1735. doi: 10.1128/aac.39.8.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1991;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 13.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 14.Klein J R, Henrich B, Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991;230:230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- 15.Koehler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 16.Köhler T, Kok M, Michea-Hamzehpour M, Plesiat P, Gotoh N, Nishino T, Curty L K, Pechere J-C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X.-Z., L. Zhang, R. Srikumar, and K. Poole. β-lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 19.Lugtenberg B, Mrijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the major outer membrane protein of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 20.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 22.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in Gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 23.Nies D. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 26.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido H, Hancock R E W. Outer membrane permeability of Pseudomonas aeruginosa. In: Sokatch J R, editor. The bacteria. Vol. 10. Orlando, Fla: Academic Press, Inc.; 1986. pp. 145–193. [Google Scholar]
- 28.Okazaki T, Hirai K. Cloning and nucleotide sequence of the Pseudomonas aeruginosa nfxB gene, conferring resistance to new quinolones. FEMS Microbiol Lett. 1992;97:197–202. doi: 10.1016/0378-1097(92)90386-3. [DOI] [PubMed] [Google Scholar]
- 29.Okii M, Iyobe S, Mitsuhashi S. Mapping of the gene specifying aminoglycoside 3′-phosphotransferase II on the Pseudomonas aeruginosa chromosome. J Bacteriol. 1983;155:643–649. doi: 10.1128/jb.155.2.643-649.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdin. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 33.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saier M H, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Srikumar, R. 1997. Unpublished data.
- 38.Srikumar R, Li X-Z, Poole K. The inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 40.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandersman C, Delepelaire P. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci USA. 1990;87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimura F, Nikaido H. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L H, Fath M J, Mahanty H K, Tai P C, Kolter R. Genetic analysis of the colicin V secretion pathway. Genetics. 1995;141:25–32. doi: 10.1093/genetics/141.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, Q., X.-Z. Li, A. Mistry, R. Srikumar, O. Lomovskaya, and K. Poole. Unpublished data. [DOI] [PMC free article] [PubMed]