Abstract
A sulfathiazole-resistant dihydropteroate synthase (DHPS) present in two different laboratory strains of Escherichia coli repeatedly selected for sulfathiazole resistance was mapped to folP by P1 transduction. The folP mutation in each of the strains was shown to be identical by nucleotide sequence analysis. A single C→T transition resulted in a Pro→Ser substitution at amino acid position 64. Replacement of the mutant folP alleles with wild-type folP significantly reduced the level of resistance to sulfathiazole but did not abolish it, indicating the presence of an additional mutation(s) that contributes to sulfathiazole resistance in the two strains. Transfer of the mutant folP allele to a wild-type background resulted in a strain with only a low level of resistance to sulfathiazole, suggesting that the presence of the resistant DHPS was not in itself sufficient to account for the overall sulfathiazole resistance in these strains of E. coli. Additional characterization of an amplified secondary resistance determinant, sur, present in one of the strains, identified it as the previously identified bicyclomycin resistance determinant bcr, a member of a family of membrane-bound multidrug resistance antiporters. An additional mutation contributing to sulfathiazole resistance, sux, has also been identified and has been shown to affect the histidine response to adenine sensitivity displayed by these purU strains.
Sulfonamide antimicrobial agents such as sulfanilamide, sulfathiazole, and sulfamethoxazole prevent the biosynthesis of reduced folate compounds by inhibiting the production of dihydropteroate from 6-hydroxymethyl-dihydropterin pyrophosphate and p-aminobenzoate (PABA) (7). Sulfonamide derivatives are structural analogs of PABA and compete with PABA for enzymatic condensation (catalyzed by dihydropteroate synthase or DHPS) with the dihydropterin substrate (7, 29). The dihydropterin-sulfonamide adducts are thought not to be further inhibitory to cellular function and to passively diffuse out of the cell (29). Since DHPS is inhibited from using PABA, the cell becomes depleted for the dihydropterin substrate necessary to produce dihydrofolate and tetrahydrofolate. Growth is thus thought to be inhibited by a lack of the vitamin cofactor necessary for the biosynthesis of macromolecular precursors (2, 13).
Pato and Brown (23) showed that a sulfonamide-resistant DHPS could be recovered from laboratory strains of Escherichia coli selected for resistance to sulfathiazole. Many transmissible R-determinant plasmids confer sulfonamide resistance on E. coli by expressing a sulfonamide-resistant form of DHPS (25, 32, 34), corroborating the role of a resistant DHPS in sulfonamide resistance. The acquisition of such plasmids is, in fact, the clinically significant mechanism by which E. coli becomes resistant to sulfonamide antimetabolites.
DHPS is encoded by folP, and the wild type as well as temperature-sensitive, sulfonamide-resistant variants have been cloned and sequenced (9, 10). Recently, the three-dimensional structure has been determined for the enzymes from E. coli (1) and Staphylococcus aureus (12). The mutant DHPS enzymes were each characterized to have elevated Kis for sulfathiazole (150- and 3,000-fold, respectively, and elevated Kms for PABA (10- and 300-fold, respectively). In addition, the activity of the mutant enzyme characterized by Dallas et al. (9) was only about 1/10 that of the wild-type enzyme, resulting in an auxotrophic requirement for the end products of folate metabolism. Wild-type and sulfonamide-resistant DHPS sequences have also been obtained from a variety of other microorganisms (6, 16, 26, 33, 35, 37).
Nichols and Guay (21) reported that one laboratory strain of E. coli selected for resistance to high levels of sulfathiazole contained a resistant DHPS, as well as a secondary resistance determinant, sur, present on a tandemly amplified segment of the chromosome. The level of sulfathiazole resistance correlated with the copy number of the sur amplification, but resistance was not abolished upon loss of the amplified DNA segment. sur was cloned from the resistant and wild-type strains and was shown to confer only a low level of resistance to sulfathiazole.
We have further characterized two laboratory strains of E. coli selected for resistance to high levels of sulfathiazole. Each contained a mutant folP allele in addition to at least one secondary mutation. We have characterized the contributions of the mutant folP and sur alleles and an additional mutation, sux, to sulfathiazole resistance in each strain and in wild-type backgrounds.
MATERIALS AND METHODS
Microbiological methods.
The bacterial strains used in this study are listed in Table 1. Basal minimal medium for the growth of E. coli consisted of 0.4% (wt/vol) glucose and the inorganic salts recommended by Vogel and Bonner (36). Enriched medium was Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl [17]). Final antibiotic concentrations were as follows: ampicillin, 75 μg/ml; tetracycline, 10 μg/ml; kanamycin, 75 μg/ml; and nalidixic acid, 20 μg/ml. Amino acids were added at 20 μg/ml when necessary. Colorimetric screenings for pKAN1 and recombinants were facilitated by the addition of 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside per ml and 48 μg of isopropyl-β-d-thiogalactopyranoside per ml to LB medium.
TABLE 1.
E. coli strains used in this study
| Strain | Genotype | Reference or derivation |
|---|---|---|
| BN102 | trpA33 cysG (am) purU tyrT Nalr | 21 |
| BN122 | folP122 sur (bcr)amptrpA33 cysG (am) purU tyrT Nalr | 21 |
| BN123 | folP123 sux trpA33 cysG (am) purU tyrT Nalr | Selection |
| BN1038 | folP+ zha-203::Tn10 sur (bcr)amptrpA33 cysG (am) purU tyrT Nalr | BN122 × CAG12072 |
| BN1039 | folP122 zha-203::Tn10 sur (bcr)amptrpA33 cysG (am) purU tyrT Nalr | BN122 × CAG12072 |
| BN1040 | folP+ zha-203::Tn10 sux trpA33 cysG (am) purU tyrT Nalr | BN123 × CAG12072 |
| BN1041 | folP123 zha-203::Tn10 sux trpA33 cysG (am) purU tyrT Nalr | BN123 × CAG12072 |
| BN1042 | folP122 zha-203::Tn10 | MG1655 × BN1039 |
| BN1057 | folP123 sux+ zef-3129::Tn10 trpA33 cysG (am) purU tyrT Nalr | BN123 × CAG12099 |
| BN1066 | folP123 sux zef-3129::Tn10 trpA33 cysG (am) purU tyrT Nalr | BN123 × CAG12099 |
| BN1079 | sux zef-3129::Tn10 | MG1655 × BN1066 |
| MG1655 | Prototroph | 31 |
| CAG12072 | zha-203::Tn10 | 31 |
| CAG12153 | zhc-6::Tn10 | 31 |
| CAG12099 | zef-3129::Tn10 | 31 |
| CAG18451 | zed-3069::Tn10 | 31 |
Selection of spontaneously arising sulfathiazole-resistant E. coli strains by the method of Sköld (32) was described previously (21). Because the growth of E. coli in the presence of sulfathiazole is dependent on the size of the inoculum and sulfathiazole is depleted from the medium over time, we established a standard method for determining the MIC of sulfathiazole for resistant strains. Cultures grown overnight were diluted 10−5 in 0.9% saline, and 0.1 ml was spread onto plates containing minimal medium and different concentrations of sulfathiazole. Plates were incubated at 37°C and were scored for growth after 48 h, although colonies would continue to arise upon additional incubation of the plates. The lowest concentration at which no colonies were visible after 48 h was recorded as the MIC. The concentrations of sulfathiazole typically tested were 0, 0.25, 0.5, 1, 2, 4, 6, 8, 10, 12, 14, and 16 μg/ml.
E. coli strains containing cotransductional markers for folP were constructed by transduction with bacteriophage P1 (18) by using as donors phage lysates derived from several strains containing Tn10 near 70 min (31). E. coli BN122 and BN123 were infected, and approximately 20 Tcr recipients from each transduction experiment were screened for reduced sulfathiazole resistance (MIC, <6 μg/ml). Lysates from CAG12072 (zha-203::Tn10) and CAG12153 (zhc-6::Tn10) yielded recipients with the expected phenotypes. Additional transduction experiments showed that folP, as determined by reduced sulfathiazole resistance, was approximately 70 to 75% cotransducible with tetracycline resistance by using lysates from CAG12072 (zha-203::Tn10), in which the Tn10 resided at 68.75 min, and was approximately 25 to 30% cotransducible with tetracycline resistance by using lysates from CAG12153 (zhc-6::Tn10), in which the Tn10 resided at 70.0 min. These data placed folP close to 69 min on the E. coli chromosome, consistent with the physical map location of folP determined by Dallas et al. (8). These experiments yielded strains that replaced the folP122 and folP123 alleles with wild-type folP (E. coli BN1038 and BN1040) and also produced strains that contained Tn10 near the mutant folP alleles (E. coli BN1039 and BN1066). To produce a strain that contained a mutant folP in isolation from other mutations that affect sulfathiazole resistance, bacteriophage P1 lysates were made from E. coli BN1039 and BN1066 and were used to transduce MG1655 to tetracycline resistance. Several colonies were screened for resistance to 2 μg of sulfathiazole per ml. No colonies appeared after 48 h of incubation at 37°C, but after additional incubation, about 70% of the transductants tested grew in the presence of 2 μg of sulfathiazole per ml. These strains were designated BN1042 and BN1083. No colonies appeared under similar circumstances when using lysates from strains in which tetracycline resistance was unlinked to folP.
A similar approach was used to map and move the sux mutation of E. coli BN123, except that lysates from E. coli CAG18451 (zed-3069::Tn10) and CAG12099 (zef-3129::Tn10) (31) were used as the source of cotransductional markers.
DNA manipulations.
E. coli chromosomal DNA was prepared by established methods (17, 39). Small-scale plasmid samples were prepared either by the rapid alkaline lysis technique of Birnboim and Doly (4) or by the rapid boiling technique (17).
Restriction endonuclease digestions and ligation reactions were performed with commercially available buffer preparations (New England Biolabs, Inc.; Boehringer-Mannheim Biochemicals, Inc.; International Biotechnologies, Inc.; and Bethesda Research Laboratories, Inc.), in accordance with the manufacturers’ recommendations.
Deletion analysis with exonuclease III and S1 nuclease was performed as described by Roberts and Lauer (28).
Plasmids containing a kanamycin resistance cassette within the sur-coding region were constructed from pBNSurB (21). pBN99 (BglII::kan) was constructed by digesting pBNSurB with BglII and ligating it with the isolated BamHI fragment containing the kanamycin resistance determinant isolated from pMB2190. Apr Knr transformants were selected, and the relevant plasmid was identified by restriction analysis. sur::kan derivatives of E. coli were constructed by transforming the strain with pBN99, selecting for growth on kanamycin, and screening for ampicillin sensitivity. Because pBNSurB is unstable, Knr Aps recombinants were readily obtained. Southern hybridization was used in several instances to confirm the chromosomal location of the kanamycin resistance determinant.
DNA containing wild-type or mutant folP was amplified from chromosomal DNA preparations as a 1,065-bp fragment by using a Perkin-Elmer thermocycler and Amplitaq DNA polymerase (Perkin-Elmer). Primers flanking the folP-coding region were selected with the aid of the program Primer, version 2 (Scientific and Educational Software). The primers used for amplification were 5′-GTAGTCCAAAGGCTCCTCGTC-3′ (for the 5′ end of folP) and 5′-GATGCGCCGATCACACCTGAT-3′ (for the 3′ end of folP). Amplification was carried out for 29 cycles with the following specific temperatures and times: denaturation at 94°C for 2.0 min, annealing at 61°C for 1.5 min, and extension at 72°C for 1.5 min. A 10-min extension at 72°C was carried out after 29 cycles of amplification. PCR products were purified with Geneclean (Bio 101) and were analyzed on a 0.8% agarose gel. A single PCR amplification product was obtained by using these conditions.
Direct PCR sequencing was performed according to the protocol supplied with the Sequenase PCR Sequencing kit purchased from United States Biochemicals.
Enzyme assays.
DHPS assays were carried out as described by Richey and Brown (27). 6-Hydroxymethylpterin (Sigma Chemical Co.) was reduced by the method of Blakely (5). 14C-PABA was purchased from Amersham. 4-Amino-4-deoxychorismate synthase and 4-amino-4-deoxychorismate lysate activities were determined as described by Nichols et al. (22).
RESULTS
Identification of folP mutations in two sulfathiazole-resistant E. coli strains.
Spontaneously occurring sulfathiazole-resistant derivatives of E. coli BN102 were obtained by prolonged selection as described previously (21, 32), and two of the most resistant strains were chosen for analysis of the mutations contributing to resistance. The sulfathiazole MICs for E. coli BN122 (previously designated BN102sur-122 [21]) and BN123 were determined to be 10 and 14 μg/ml, respectively. The sulfathiazole MIC for the parent strain, E. coli BN102, was 0.25 μg/ml. Since each resistant strain had more than one mutation that contributed to sulfathiazole resistance (21) (see below), we first sought to study the role of folP mutations both in isolation from and in combination with the endogenous host backgrounds.
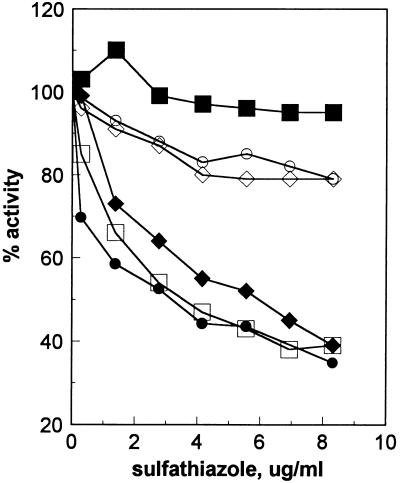
Each resistant strain was shown to express a sulfathiazole-resistant DHPS (21) (Fig. 1). Derivatives of E. coli BN122 and BN123 that replaced the folP122 and folP123 alleles with wild-type folP (e.g., E. coli BN1038) were constructed by transduction with bacteriophage P1. In addition, wild-type backgrounds containing only the mutant folP alleles were constructed. As shown in Fig. 1, strains that received a wild-type folP allele produced a sulfathiazole-sensitive DHPS (50% inhibitory concentration, 2.2 μg/ml), and those that received a mutant allele produced a sulfathiazole-resistant DHPS (50% inhibitory concentration, 18 μg/ml). The phenotypic resistance to high levels of sulfathiazole correlated with the presence of a sulfathiazole-resistant DHPS in the E. coli BN122 and BN123 backgrounds (Table 2), but the presence of the folP122 or folP123 alleles in the E. coli MG1655 wild-type background resulted in only a low level of phenotypic resistance to sulfathiazole, even though a resistant DHPS was expressed.
FIG. 1.
In vitro inhibition of DHPSs by sulfathiazole. Symbols: •, BN102; ○, BN122; □, CAG12072; ⧫, BN1038; ◊, BN1039; ▪, BN1042.
TABLE 2.
Phenotypic effects of folP alleles in various backgrounds
| Strain | Derivation | MIC (μg/ml) |
|---|---|---|
| BN102 | 0.25 | |
| MG1655 | 0.25 | |
| CAG12072 | MG1655, zha-203::Tn10 | 0.25 |
| BN122 | 10 | |
| BN1039 | BN122, zha-203::Tn10 | 12 |
| BN1038 | BN122, folP+ zha-203::Tn10 | 2 |
| BN1042 | MG1655, folP122 | 1 |
| BN123 | 14 | |
| BN1041 | BN123, zha-203::Tn10 | 14 |
| BN1040 | BN123, folP+ zha-203::Tn10 | 1 |
| BN1083 | MG1655, folP123 | 1 |
| BN1057 | BN123, sux+ zef-3129::Tn10 | 4 |
| BN1079 | MG1655, sux123 zef-3129::Tn10 | 1 |
Nucleotide sequences of folP mutations.
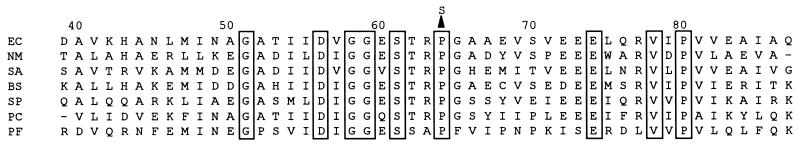
A 1,065-bp DNA fragment containing the entire coding region of folP was amplified by PCR from strains containing wild-type and mutant folP alleles. The entire folP gene was sequenced directly from the amplified DNA. The sequences of folP from E. coli MG1655 and BN102 were identical to the previously published sequence from E. coli MC4100 (9). E. coli BN122 and BN123 folP sequences were identical to one another, but they contained a single difference from the wild-type nucleotide sequence. The difference was a C-to-T transition at nucleotide 184, resulting in a Pro-to-Ser substitution at amino acid 64. The sequence of this region of folP aligned with DHPS sequences from a variety of additional sources is shown in Fig. 2. Pro64 lies very close to the active site of DHPS, adjacent to Arg63, whose side chain bonds in a hydrogen bond with an oxygen of the sulfanilamide inhibitor (1). Substitution of Pro64 by Ser is likely to alter the local structure of the peptide, in turn altering the ability of Arg63 to contact the inhibitor.
FIG. 2.
Amino acid sequence of a portion of the DHPSs from several organisms. EC, E. coli; NM, N. meningitidis; SA, S. aureus; BS, B. subtilis; SP, S. pneumoniae; PC, P. carinii; PF, P. falciparum. The numbering scheme is taken from the E. coli sequence. The arrowhead above the E. coli sequence illustrates the amino acid substitution in DHPS from E. coli BN122 and BN123. Boxes are drawn around residues conserved in all sequences.
Characterization of sur.
sur was previously identified as a sulfathiazole resistance determinant present on an amplified segment of the chromosome of E. coli BN122 (21). Deletion analysis was used to localize sur on plasmid pBNSurB, and a 2.6-kb BglII-BamHI fragment spanning the sur region was sequenced. Since pBNSurB was derived from a fourfold, 18.5-kb chromosomal DNA amplification present in E. coli BN122, an equivalent region was isolated from λEMBLsur3C (21) and sequenced. The sequence of the sur region of pBNSurB was identical to that of the wild-type allele derived from λEMBLsur3C. The sequence contained a 396-codon open reading frame preceded by a Shine-Dalgarno sequence and starting with a GTG. A search for similar sequences in the various databases by using the BLAST program (24) indicated a match with E. coli bcr, a gene isolated by its ability to confer resistance to bicyclomycin when it is overproduced (3). In the previously reported sequence, the predicted initiation site was equivalent to the Met at our position 22, and amino acid 99 was Val instead of our Asp (the previously reported nucleotide and amino acid sequences are available in database entries GenBank U00008 and Swiss-Prot P28246, respectively). The product of sur (bcr) is similar to that of the family of proton-motive force-dependent drug-H+ antiporters.
The reduction of amplification of sur in the E. coli BN122 background from four copies to one copy resulted in a decrease in the MIC from 10 to 4 μg/ml, consistent with the results reported previously (21). The presence of pBNSurB in E. coli BN102 increased the MIC from 0.25 to 1 μg/ml. The presence of pBNSurB in E. coli BN1042 (folP122) similarly altered the MIC for that strain. Thus, the increased gene dosage of sur (bcr) either by chromosomal amplification or by the presence on a multicopy plasmid contributed a 2.5- to 4-fold increase in sulfathiazole resistance.
In order to test the function of the sur (bcr) product, sur (bcr) was interrupted with a Knr cassette and crossed into the chromosome of several strains. Disruption of sur (bcr) in E. coli BN102 and BN123 had no effect on the phenotype of the strains, but disruption of sur (bcr) in BN122 proved difficult, because recipients of the Knr cassette contained both the disrupted and complete copies of sur (bcr) due to the amplification of the sur (bcr) locus. Strains with a single copy of the interrupted sur (bcr) locus were obtained after several generations of nonselective growth on rich medium containing kanamycin, and when transferred to minimal medium, these strains displayed a requirement for adenine. The adenine auxotrophy was mapped to 11 to 16 min by Hfr mating and to Kohara phage λ6E7(157) (14) by lytic complementation tests. Since this phage contained the purEK genes (30), we confirmed the complementation by subcloning a 3.0-kb BglII DNA fragment containing purEK (38) and demonstrated its ability to complement the adenine auxotrophy revealed by the sur (bcr) interruption.
Stability of pBNSurB.
Like many plasmids that overproduce integral membrane proteins similar to proton motive force-dependent antiporters, pBNSurB proved to be unstable in E. coli BN102 and its derivatives. The instability was observed as a differential plating efficiency on LB-ampicillin and minimal-ampicillin plates following transformation with pBNSurB. Supplementation of minimal medium plates revealed that either PABA (1 μg/ml) or methionine (20 μg/ml) was capable of increasing the plating efficiency of freshly transformed cells at least 100-fold. Isolation of chromosomal mutations capable of increasing E. coli BN102/pBNSurB transformation efficiency took advantage of the fact that E. coli BN102 transformed with pBNSurB and plated on LB-ampicillin produced two distinct colony sizes. When strains cured of plasmid were retransformed with pBNSurB and plated on minimal-ampicillin and minimal-ampicillin plates, strains derived from the smaller colonies produced equal numbers of colonies on both LB-ampicillin and minimal-ampicillin plates, while strains derived from the larger colonies still showed differential plating efficiencies.
Six strains selected for increased plating efficiency were tested for elevated levels of PABA or methionine synthesis. One of the strains conferred resistance to 40 mM ethionine, whereas the parental strain, E. coli BN102, was resistant to 20 mM ethionine, indicating elevated levels of synthesis of methionine (11). Two of the strains were resistant to 70 μg of sulfanilamide per ml, while the parent strain was resistant to 20 μg of sulfanilamide per ml, suggesting an increase in the level of PABA biosynthesis. The levels of PABA biosynthetic enzymes in extracts of the sulfanilamide-resistant strains were determined, and each sulfanilamide-resistant strain had a twofold increase in the relative specific activity of 4-amino-4-deoxychorismate synthase CoI (PabB) compared to that for the parental strain. Three additional strains did not display elevated levels of PABA or methionine, suggesting that pBNSurB stability could be affected by additional mutations.
An additional mutation contributing to sulfathiazole resistance in E. coli BN123.
A sulfathiazole resistance determinant distinct from sur (bcr) was mapped to 44 min in E. coli BN123 by Hfr mating and cotransduction analysis. Replacement of the new determinant, designated sux, with its wild-type allele resulted in a 2.5-fold decrease in sulfathiazole resistance. In a wild-type background, the sux-123 allele increased the level of sulfathiazole resistance fourfold (Table 2). The effect of sux appeared to be mediated through a balance of tetrahydrofolate and its one-carbon derivatives, as determined by differences in supplements required to relieve the adenine sensitivity conferred upon these strains by their purU defect (19, 20). A differential response to histidine was observed during adenine inhibition in the otherwise isogeneic sux-123 and sux+ strains (data not shown). Because the sux mutation lay close to or in his and it had affects on the histidine response to adenine sensitivity consistent with alterations in histidine regulation, we suggest that sux was likely to be a mutation that affected the regulation of histidine biosynthesis.
DISCUSSION
We have selected E. coli strains for sulfathiazole resistance and have shown that each contains a resistant DHPS. Two independently isolated mutant folP alleles each contained the same Pro64→Ser amino acid substitution. The site of the substitution lay within a highly conserved sequence block, -GGESTRPG-, which is identical among E. coli, Bacillus subtilis, Neisseria meningitidis, and Streptococcus pneumoniae (9, 26, 33). The Pro64→Ser mutation is distinct from two other mutations reported in E. coli folP, each of which was selected for sulfathiazole resistance and simultaneous temperature sensitivity. Both of the ts mutations lie at amino acid position Phe28 but have different substitutions, Leu (9) and Ile (10). While the determination of differences in the kinetic constants of the Pro64→Ser enzyme is in process, we have not noted a significant difference in the specific activity of the DHPS present in crude extracts prepared from wild-type or mutant strains (data not shown).
Pro64 is in a flexible loop of the protein that forms a portion of the binding site for PABA and sulfanilamide, with the guanidinium group of Arg63 forming a hydrogen bond with one of the oxygens of sulfanilamide (1). Replacement of the chain-bending proline with serine is likely to alter the structure of the α-carbon chain in this region and thus alter the ability of the Arg63 side chain to make stable contact with the inhibitor.
The conserved region from position 58 to 65 is also strongly conserved in the DHPS domains of multifunctional enzymes from Plasmodium falciparum (6, 35) and Pneumocystis carinii (37), and amino acid substitutions at positions 62 and 63, as well as other positions, have been found in sulfadoxine-resistant strains of P. falciparum (6, 35). Interestingly, only G58 and P64 are conserved in the highly resistant sulI- and sulII-encoded DHPS enzymes present on several R determinants. While amino acid substitutions and duplications at other sites within DHPS have been established as being responsible for sulfonamide resistance in a variety of organisms, the combined data also implicate the region between positions 58 and 65 as an important site for substitutions that alter inhibitor or substrate recognition.
Although the mutant chromosomal folP allele conferred a fourfold increase in the MIC of sulfathiazole, the low level of resistance is surprising in light of the fact that a resistant DHPS has been characterized as the major mechanism of the acquisition of resistance to sulfonamides in E. coli. High-level resistance has been observed in E. coli by introduction of low-copy-number R-determinant plasmids, indicating that resistance can be attributed solely to a resistant DHPS. It is possible that significant resistance can be achieved only if the regulation of DHPS expression is uncoupled from its chromosomal context.
The prolonged selection procedure that we have used to obtain E. coli strains resistant to high levels of sulfathiazole has resulted in several resistant strains with secondary mutations, that is, mutations outside of folP. One of the secondary mutations that we have characterized extensively is sur (bcr), originally detected as a determinant present on an amplified segment of the chromosome of E. coli BN122 (20). We have determined that the cloned copy of sur is identical to the wild-type gene and that it is identical to bcr, whose overexpression has also been correlated with bicyclomycin resistance (3). The sur (bcr) product is a member of the major facilitator family of membrane translocases (15). Several members of this family are proton motive force-dependent drug-H+ antiporters, and it may be that sur (bcr) functions in an analogous manner, effluxing sulfathiazole from the cell. While a portion of the observed instability of pBNSurB was very likely due to overexpression of an integral membrane protein, it is of interest that an end product (methionine) and an intermediate (PABA) of folate metabolism could stabilize the plasmid in strains grown on minimal medium. It is possible that PABA or another intermediate in folate biosynthesis, in addition to sulfathiazole, was also an efflux substrate for sur (bcr). It is not clear why the cloned sur (bcr), in conjunction with folP122 in a wild-type background, did not elevate the level of resistance to that of E. coli BN122. It may be that the strains were compromised due to overexpression of the membrane protein or that additional mutations that are necessary for the manifestation of high levels of resistance to sulfathiazole in conjunction with elevated levels of sur (bcr) are present in E. coli BN122.
Another mutation, designated sux, has been detected in E. coli BN123. sux is distinct from folP and sur (bcr) and has been determined to play a large role in the resistance of E. coli BN123 to sulfathiazole. As with folP and sur (bcr), sux does not confer significant levels of resistance when taken out of the context of the E. coli BN123 background, suggesting that it, too, requires the context within which it arose to exert its maximum effect on sulfathiazole resistance.
The secondary mutations sur (bcr) and sux both affected sulfathiazole resistance primarily by influencing the metabolic pathways dependent upon one-carbon metabolism, especially the purine biosynthetic pathway. This result may in turn be influenced by the presence of the purU lesion in the parental strain. Further investigation of the role of secondary mutations in sulfathiazole resistance is under way with strains not initially compromised in their one-carbon metabolism.
ACKNOWLEDGMENTS
We thank all of those who participated in discussions of this work, especially Jacalyn Green, V. K. Viswanathan, and William Merkel.
This work was supported by Public Health Service grants AI25106 and GM44199 from the National Institutes of Health.
REFERENCES
- 1.Achari A, Somers D O, Champness J N, Bryant P K, Rosemond J, Stammers D K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nature Struct Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- 2.Baumstark B R, Spremulli L L, RajBhandary U L, Brown G M. Initiation of protein synthesis without formylation in a mutant of Escherichia coli that grows in the absence of tetrahydrofolate. J Bacteriol. 1977;129:457–471. doi: 10.1128/jb.129.1.457-471.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley J, Hyatt L S, Ainley K, Parish J H, Herbert R B, White G R. Cloning and sequence analysis of an Escherichia coli gene conferrring bicyclomycin resistance. Gene. 1993;127:117–120. doi: 10.1016/0378-1119(93)90625-d. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim H, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blakely R L. Crystalline dihydropteroylglutamic acid. Nature (London) 1960;188:231–232. [Google Scholar]
- 6.Brooks D R, Wang P, Read M, Watkins W M, Sims P F, Hyde J E. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown G M. The biosynthesis of folic acid. Inhibition by sulfonamides. J Biol Chem. 1962;237:536–540. [PubMed] [Google Scholar]
- 8.Dallas W S, Dev I K, Ray P H. The dihydropteroate synthase gene, folP, is near the leucine tRNA gene, leuU, on the Escherichia coli chromosome. J Bacteriol. 1993;175:7743–7744. doi: 10.1128/jb.175.23.7743-7744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dallas W S, Gowen J E, Ray P H, Cox M J, Dev I K. Cloning, sequencing, and enhanced expression of the dihydropteroate synthase gene of Escherichia coli MC4100. J Bacteriol. 1992;174:5961–5970. doi: 10.1128/jb.174.18.5961-5970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fermér C, Kristiansen B-E, Sköld O, Swedberg G. Sulfonamide resistance in Neisseria meningitidis as defined by site-directed mutagenesis could have its origin in other species. J Bacteriol. 1995;177:4669–4675. doi: 10.1128/jb.177.16.4669-4675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafner E W, Tabor C W, Tabor H. Isolation of a metK mutant with a temperature-sensitive S-adenosylmethionine synthetase. J Bacteriol. 1977;132:832–840. doi: 10.1128/jb.132.3.832-840.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamplele I C, D’Arcy A, Dale G E, Kostrewa D, Neilsen J, Oefner C, Page M G P, Schönfeld H-J, Stüber D, Then R L. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J Mol Biol. 1997;268:21–30. doi: 10.1006/jmbi.1997.0944. [DOI] [PubMed] [Google Scholar]
- 13.Harvey R J. Growth and initiation of protein synthesis in Escherichia coli in the presence of trimethoprim. J Bacteriol. 1973;114:309–322. doi: 10.1128/jb.114.1.309-322.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohara Y, Akiyama K, Inoso K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 15.Lewis K. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 16.Lopez P, Espanosa M, Greenberg B, Lacks S A. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol. 1987;169:4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 19.Nagy P L, Marolewski A, Benkovic S J, Zalkin H. Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J Bacteriol. 1995;177:1292–1298. doi: 10.1128/jb.177.5.1292-1298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy P L, McCorkle G M, Zalkin H. purU, a source of formate for purT-dependent phosphoribosyl-N-formylglycinamide synthesis. J Bacteriol. 1993;175:7066–7073. doi: 10.1128/jb.175.21.7066-7073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichols B P, Guay G G. Gene amplification contributes to sulfonamide resistance in Escherichia coli. Antimicrob Agents Chemother. 1989;33:2242–2248. doi: 10.1128/aac.33.12.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols B P, Seibold A, Doktor S Z. para-Aminobenzoate synthesis from chorismate occurs in two steps. J Biol Chem. 1989;264:8597–8601. [PubMed] [Google Scholar]
- 23.Pato M L, Brown G M. Mechanisms of resistance of Escherichia coli to sulfonamides. Arch Biochem Biophys. 1963;103:443–448. doi: 10.1016/0003-9861(63)90435-1. [DOI] [PubMed] [Google Scholar]
- 24.Pearson W, Lipman D. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rådström P, Swedberg G. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob Agents Chemother. 1988;32:1684–1692. doi: 10.1128/aac.32.11.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rådström P, Fermér C, Kristiansen B-K, Jenkins A, Sköld O, Swedberg G. Transformational exchanges in the dihydropteroate synthase gene of Neisseria meningitidis: a novel mechanism for acquisition of sulfonamide resistance. J Bacteriol. 1992;174:6386–6393. doi: 10.1128/jb.174.20.6386-6393.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richey D P, Brown G M. The biosynthesis of folic acid. IX. Purification and properties of the enzymes required for the formation of dihydropteroic acid. J Biol Chem. 1969;244:1582–1592. [PubMed] [Google Scholar]
- 28.Roberts T M, Lauer G D. Maximizing gene expression in a plasmid using recombination in vitro. Methods Enzymol. 1979;68:473–482. doi: 10.1016/0076-6879(79)68036-9. [DOI] [PubMed] [Google Scholar]
- 29.Roland S, Ferone R, Harvey R J, Styles V L, Morrison R W. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J Biol Chem. 1979;254:10337–10345. [PubMed] [Google Scholar]
- 30.Rudd K E. Alignment of E. coli DNA sequences to a revised, integrated genomic restriction map. In: Miller J H, editor. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 31.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sköld O. R-factor-mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Chemother. 1976;9:49–54. doi: 10.1128/aac.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slock J, Stahly D P, Han C-Y, Six E W, Crawford I P. An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. J Bacteriol. 1990;172:7211–7226. doi: 10.1128/jb.172.12.7211-7226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 35.Triglia T, Cowman A F. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel H, Bonner D. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 37.Volpe F, Ballantine S P, Delves C J. The multi-functional folic acid synthesis fas gene of Pneumocystis carinii encodes dihydroneopterin aldolase, hydroxymethyl-dihydropterin pyrophosphokinase, and dihydropteroate synthase. Eur J Biochem. 1993;216:449–458. doi: 10.1111/j.1432-1033.1993.tb18163.x. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe W, Sampei G-I, Aiba A, Mizobuchi K. Identification and sequence analysis of Escherichia coli purE and purK genes encoding 5′-phosphoryl-5-amino-4-imidazole carboxylase for de novo purine biosynthesis. J Bacteriol. 1989;171:198–204. doi: 10.1128/jb.171.1.198-204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1987. pp. 2.4.1–2.4.5. [Google Scholar]