Abstract
Of 41 methicillin-resistant coagulase-negative staphylococcal clinical isolates collected during a 5-month period between late 1995 and early 1996, 28 showed tube dilution teicoplanin MICs of 4 to 8 μg/ml which increased to 16 to 32 μg/ml upon prolonged incubation. Cultures of such bacteria were heterogeneous; they contained subpopulations with frequencies of 10−5 to 10−4 that could grow on up to 50 μg of teicoplanin per ml. The same cultures were also heterogeneous with respect to susceptibility to vancomycin; while the MICs for the majority of cells were 2 to 4 μg/ml, subpopulations that could grow on 6 to 12 μg of vancomycin per ml were also present at frequencies of 10−5 to 10−7. Selective enrichment of such cultures for the resistant subpopulation occurred with relative ease under laboratory conditions. Heterogeneous phenotypes for teicoplanin (but not for vancomycin) susceptibility were also identified in several Staphylococcus epidermidis isolates collected during the preantibiotic era. The addition of half the MIC of teicoplanin inhibited autolysis and caused formation of cellular aggregates which disintegrated to individual bacteria in the stationary phase when the titer of teicoplanin in the medium fell to undetectable levels, indicating removal of the antibiotic from the culture medium by the bacteria.
The appearance of resistance mechanisms against glycopeptide antibiotics among clinical isolates of enterococci (for a review, see reference 25) and the laboratory demonstration of the transfer of the vanA gene complex to Staphylococcus aureus (15) have raised concern about the occurrence of such a genetic transfer in clinical isolates of methicillin-resistant staphylococci, against which the most frequently used chemotherapy is vancomycin based. While clinical isolates of staphylococci with the enterococcal glycopeptide resistance mechanism have not been reported so far, clinical failure of teicoplanin and vancomycin treatment in coagulase-negative staphylococci (CNS) (see reference 25) and, recently, in methicillin-resistant S. aureus (MRSA) infections in Japan (10) and the United States (4, 5) have been reported. The MRSA strains involved exhibited low-level vancomycin resistance through a novel and as-yet-undefined mechanism(s) that differed from that of the enterococcal vancomycin resistance.
A different mechanism of vancomycin resistance associated with removal of the drug molecules from the growth medium during growth of the bacteria has been described recently in a laboratory mutant of MRSA (22). The vancomycin-resistant MRSA showed several unique properties: bacteria grown in the presence of vancomycin formed large cellular aggregates, produced large amounts of extracellular material resembling (by electron microscopy) cell wall material, and produced cell wall peptidoglycan with altered muropeptide composition, and the vancomycin removed from the growth medium (bound to the cell walls) could be recovered in biologically active form from the bacteria (22). An identical set of observations was made with a recently isolated highly teicoplanin-resistant laboratory mutant of S. aureus which removed teicoplanin from the medium into a cell-bound form (22a). In order to determine whether this type of glycopeptide resistance mechanism also existed among clinical isolates, we examined a number of methicillin-resistant coagulase-negative staphylococci (MR-CNS) with low-level teicoplanin resistance which were recovered from a hospital in New York City.
MATERIALS AND METHODS
Bacterial strains were grown in tryptic soy broth (TSB; Difco, Detroit, Mich.) at 37°C, with aeration. Speciation was done by the API test system (bioMerieux Vitek, Inc., Hazelwood, Mo.). Population analysis profiles (PAPs) (24) were constructed as follows. Overnight cultures of bacteria (≥109 CFU/ml) were plated at a series of dilutions on tryptic soy agar plates containing antibiotic-free medium or twofold dilutions of the test antibiotic within the drug concentration range of 0.1 to 100 μg/ml (for vancomycin and teicoplanin) and up to 800 μg/ml (for methicillin). Plates were incubated at 37°C for 48 h, and the number of bacterial colonies was counted. Plotting colony counts against drug concentrations provides a graphic display (PAP) of the composition of the bacterial culture as to the homogeneity or heterogeneity of the antibiotic susceptibility phenotype. Strains with heterogeneous expression of resistance to methicillin or glycopeptides show PAPs with one or more inflection points, since they contain, in addition to the majority of cells, one or more subpopulations of bacteria for which MICs are elevated. Prolonged incubation of such hetero cultures of MRSA at an antibiotic concentration above the MIC for the majority of the cells was shown to select for the more resistant subpopulation which can “take over” the culture, causing an upward shift in tube dilution MICs and “treatment failure” (24). The high resolution of the PAP method allows the detection of highly resistant subpopulations, even if they are present with low (10−4 to 10−8) frequencies in the culture. Methicillin and glycopeptide MICs for the majority of cells were calculated as the lowest concentration of the antibiotic causing a 99.9% loss of the inoculum. MICs of teicoplanin and vancomycin were also determined by the standard broth microdilution method, following the recommendations of the National Committee for Clinical Laboratory Standards (14). MICs were evaluated after 48 and 72 h of incubation at 37°C. The preparation of chromosomal DNA for pulsed-field gel electrophoresis (PFGE) and the separation of SmaI-restricted fragments in a CHEF apparatus (CHEF-DRII; Bio-Rad, Richmond, Calif.) were carried out as described previously (9). Autolysis was induced by suspending bacteria in buffer containing Triton X-100 (8). Titer of teicoplanin in the growth medium was determined by a bioassay (22). Aggregation of cells and the ultrastructure of bacteria were determined by phase-contrast microscopy and electron microscopy by a procedure described previously (23).
RESULTS
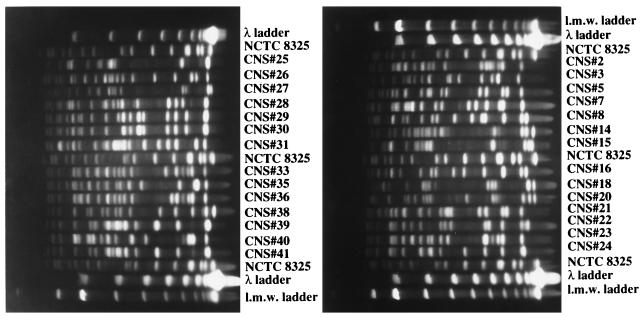
Summarized in Table 1 are the relevant properties of the 28 teicoplanin-resistant and MR-CNS isolates recovered between 25 September 1995 and 12 February 1996 in a hospital in New York City. Of the 41 methicillin-resistant strains, 28 showed low-level resistance to teicoplanin. Clinical isolates of CNS with decreased levels of susceptibility to teicoplanin from several countries have been described, and treatment failures associated with the increased MICs have been reported (see reference 25). The observations reported here point to the high frequency of such isolates; close to 70% of all MR-CNS recovered in a single hospital during a 5-month period exhibited increased teicoplanin MICs. Most of these isolates were suspected to be involved with infection. PFGE of SmaI digests of chromosomal DNA indicated a wide diversity of the strains analyzed (24 different PFGE patterns in 28 isolates) (Fig. 1), indicating that the high frequency of strains with reduced susceptibility was not due to the spread of a clone but rather, most likely involved the frequent and independent acquisition of resistance by individual bacteria.
TABLE 1.
Some relevant properties of the 28 MR-CNS clinical isolates used in this studya
| Isolate no. | Staphylococcus species | Year isolated | Isolation site | Infection or colonization | MIC (μg/ml) of:b
|
||
|---|---|---|---|---|---|---|---|
| Methicillinc | Teicoplanind | Vancomycind | |||||
| CNS 2 | S. epidermidis | 1995 | Wound | Infection | 3 | 4 (8) | 2 (2) |
| CNS 3 | S. haemolyticus | 1995 | Wound | Infection | 12 | 8 (8) | 1 (1) |
| CNS 5 | S. epidermidis | 1995 | Wound | Infection | 6 | 4 (8) | 2 (2) |
| CNS 7 | S. epidermidis | 1995 | Eye | Infection | 3 | 4 (8) | 2 (2) |
| CNS 8 | S. epidermidis | 1995 | Wound | Infection | 6 | 4 (8) | 2 (2) |
| CNS 14 | S. haemolyticus | 1995 | Wound | Infection | 100 | 8 (8) | 4 (4) |
| CNS 15 | S. haemolyticus | 1995 | Wound | Infection | 800 | 16 (32) 25 | 2 (2) 3 |
| CNS 16 | S. epidermidis | 1995 | Wound | Infection | 3 | 4 (8) | 4 (4) |
| CNS 18 | S. haemolyticus | 1995 | Wound | Infection | 50 | 8 (16) 25 | 4 (4) 3 |
| CNS 20 | S. haemolyticus | Blood | Infection | 100 | 4 (8) | 1 (1) | |
| CNS 21 | S. epidermidis | 1995 | Wound | Infection | 12 | 8 (16) 12 | 4 (4) 1.5 |
| CNS 22 | S. epidermidis | 1995 | Wound | Infection | 6 | 8 (8) | 2 (2) |
| CNS 23 | S. epidermidis | 1995 | Conjunctiva | Infection | 6 | 2 (8) | 2 (2) |
| CNS 24 | S. epidermidis | 1995 | Wound | Infection | 6 | 2 (8) | 2 (2) |
| CNS 25 | S. hominis | 1995 | Wound | >800 | 8 (16) 25 | 4 (4) 3 | |
| CNS 26 | S. epidermidis | 1996 | Wound | Infection | 3 | 8 (8) | 4 (4) |
| CNS 27 | S. haemolyticus | Wound | Colonization | 100 | 8 (8) | 2 (2) | |
| CNS 28 | S. epidermidis | 1996 | Eye | Infection | 400 | 4 (8) | 4 (4) |
| CNS 29 | S. epidermidis | 1996 | Wound | Infection | 6 | 8 (8) | 4 (4) |
| CNS 30 | S. epidermidis | 1995 | Wound | Infection | 12 | 4 (8) | 4 (4) |
| CNS 31 | S. epidermidis | Wound | Infection | 3 | 4 (8) | 4 (4) | |
| CNS 33 | S. epidermidis | 1995 | Wound | Infection | 1.5 | 8 (16) 6 | 4 (4) 1.5 |
| CNS 35 | S. epidermidis | 1995 | Wound | Infection | 6 | 4 (8) | 4 (4) |
| CNS 36 | S. epidermidis | 1995 | Wound | Infection | 3 | 4 (8) | 4 (4) |
| CNS 38 | S. epidermidis | 1995 | Wound | Infection | 3 | 4 (8) | 4 (4) |
| CNS 39 | S. epidermidis | Wound | Infection | 3 | 8 (8) | 4 (4) | |
| CNS 40 | S. epidermidis | 1995 | Infection | 3 | 8 (8) | 4 (4) | |
| CNS 41 | S. epidermidis | Infection | 12 | 8 (16) 12 | 2 (2) 1.5 | ||
Bold-faced type indicates isolates examined in more detail.
Underlined data are PAP results.
Determined from PAPs, as described in reference 24.
Determined by broth microdilution with readings after 48 and (72) h.
FIG. 1.
PFGE patterns of 28 CNS isolates for which teicoplanin MICs are elevated. For identification of strains, see Table 1. Chromosomal DNA was prepared, SmaI restricted, and separated by PFGE as described in reference 9. Low-molecular-weight (l.m.w.) standards were included.
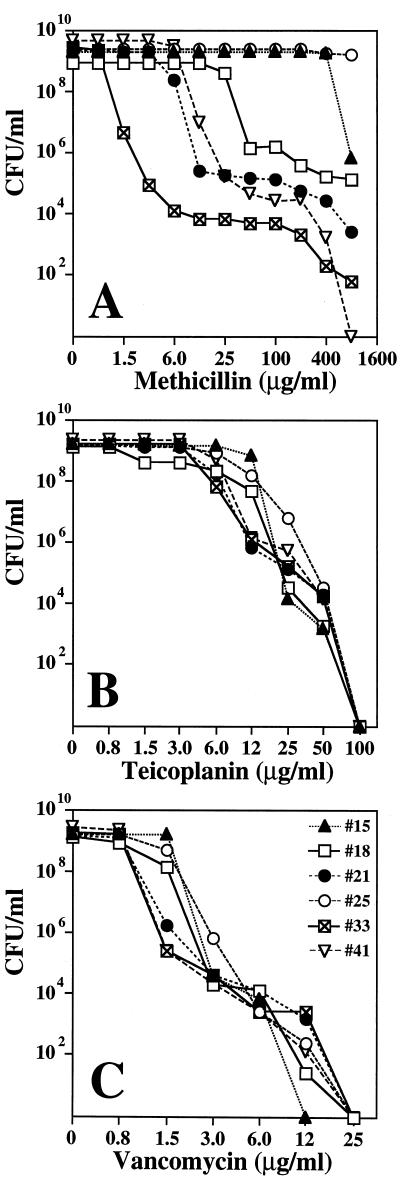
The vast majority of the 28 isolates listed in Table 1 showed heterogeneous resistance to methicillin, and the methicillin MICs (in micrograms per milliliter) listed in Table 1 refer to the resistance level of the majority of the bacteria in cultures of these strains. It should be noted that teicoplanin MICs appear to increase upon prolonged incubation of the microtiter plates (Table 1). In order to examine this phenomenon more closely, we chose six MR-CNS isolates (strains 15, 18, 21, 25, 33, and 41) for a more detailed study.
Figures 2A through C show antibiotic susceptibility profiles of these six strains examined by the quantitative method of population analysis (see Materials and Methods). Five of the six strains expressed heterogeneous methicillin resistance (Fig. 2A), and all six strains also showed heterogeneous phenotypes with respect to susceptibility to teicoplanin (Fig. 2B) and vancomycin (Fig. 2C). While the vancomycin MICs for the majority of the bacteria were within the range of susceptibility, each one of the cultures also contained cells capable of growing at elevated concentrations of vancomycin (for instance, strain 33 contained bacteria capable of forming colonies on agar containing 12 μg of vancomycin per ml, and these cells were present in the cultures at a frequency of approximately 10−6). Subpopulations of bacteria with similar increased vancomycin MICs were also present in each of the rest of the five strains examined (Fig. 2C). Such more substantially resistant bacteria may not be detectable by the routine methods used in clinical microbiology laboratories because of their low frequency. Nevertheless, such highly resistant cells may grow out and take over the bacterial population during prolonged incubation and/or treatment. The stepwise increase in teicoplanin and vancomycin MICs observed during extended incubation times may represent such a phenomenon (see MICs after 48 and 72 h incubation in Table 1). In order to test this possibility, we examined conditions favoring the selection of subpopulations with increased vancomycin MICs.
FIG. 2.
Phenotypic expression of methicillin (A), teicoplanin (B), and vancomycin (C) resistance of selected six isolates, determined by population analysis. Strain numbers and symbols are identified in panel C.
Enrichment in the absence of selective pressure.
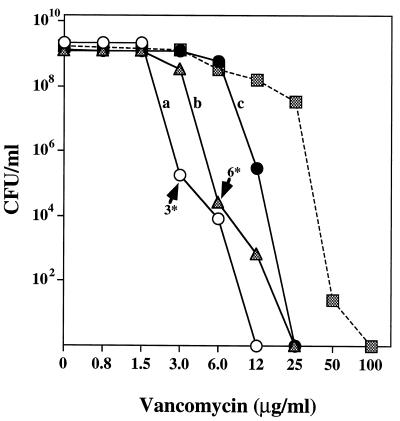
Strain CNS 15 had a vancomycin MIC of 3.0 μg/ml (for the majority of the cells) but also produced subpopulations capable of growing on agar supplemented with 3 μg (frequency, 10−4) and 6 μg (frequency, 10−5) of antibiotic per ml (Fig. 3). A colony from the 3-μg/ml-vancomycin plate was suspended in 1 ml of TSB and diluted and used to inoculate 5 ml of TSB at an initial cell concentration of about 20 to 30 CFU per ml. After overnight growth at 37°C with aeration, the turbid culture, estimated to have undergone at least 26 doublings in the nonselective medium, was plated for population analysis on agar containing different concentrations of vancomycin. The upward shift in the vancomycin MIC from 3 to 6 μg/ml for the majority of the bacteria is documented in Fig. 3. The enrichment procedure was repeated by selecting a colony from the agar containing 6 μg of vancomycin per ml. Overnight growth from small inocula in drug-free medium produced a culture in which the vancomycin MIC for the majority of the bacteria shifted upward to 12 μg/ml (Fig. 3). A culture from the second cycle of enrichment contained bacteria capable of growing on plates containing 25 μg of the antibiotic per ml at a frequency of 10−3.
FIG. 3.
Enrichment of CNS cultures for the subpopulations of bacteria for which vancomycin MICs were elevated. S. epidermidis CNS 15 was plated for population analysis on agar containing different concentrations of vancomycin, as described in Materials and Methods. The PAP of the culture is shown by open circles (a). A rare bacterial colony (frequency, about 10−4) growing on the agar containing 3 μg of vancomycin per ml was diluted in TSB and used as inoculum for an overnight culture in TSB, which was subsequently plated for population analysis (see PAP with solid triangle, b). A bacterial colony (frequency, about 10−5) capable of growing on 6 μg of vancomycin per ml was used as inoculum for a new overnight culture in drug-free medium and was plated for population analysis (solid circles, c). The gradual increase in the MICs for the majority of the cultures is illustrated in cultures a through c. A culture of CNS 15 was serially diluted into TSB containing increasing concentrations of vancomycin (eventually reaching 12 μg/ml), grown overnight to turbid cultures, and plated for population analysis, shown by the solid squares and dashed line.
Enrichment in the presence of vancomycin selection.
The culture of strain CNS 15 was diluted 1,000-fold into 5 ml of TSB containing 3 μg of vancomycin per ml and was grown overnight. Next, this culture was backdiluted into 5 ml of TSB containing 6 μg of drug per ml, and a culture of the latter was then used as inoculum for a third TSB culture containing 12 μg of antibiotic per ml. The PAP of this third serial culture is shown in Fig. 3 (dashed line). Substantial enrichment in bacteria with higher vancomycin MICs (up to 50 μg/ml) is apparent. Such bacteria, however, grew perceptibly slower than the original strain CNS 15.
The original teicoplanin MIC for CNS 15 was 25 μg/ml. After the third cycle of vancomycin selection, the teicoplanin MIC had increased to over 100 μg/ml (data not shown).
Inhibition of autolysis and aggregation of cells.
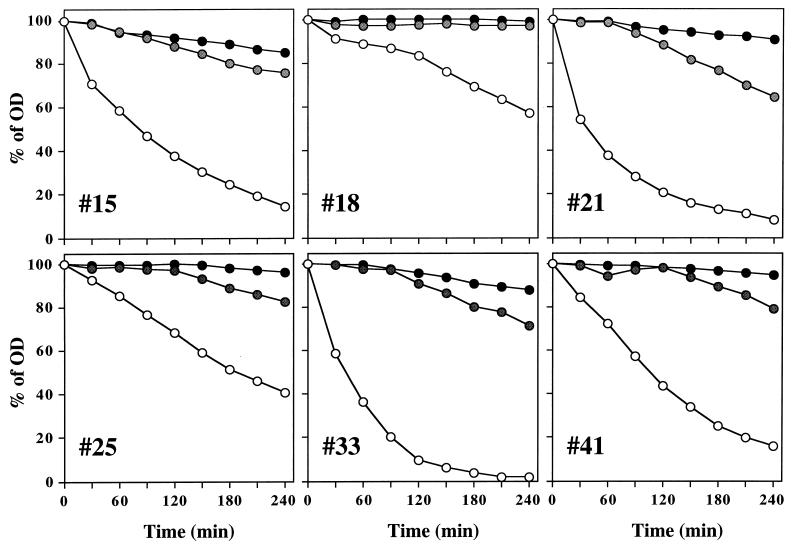
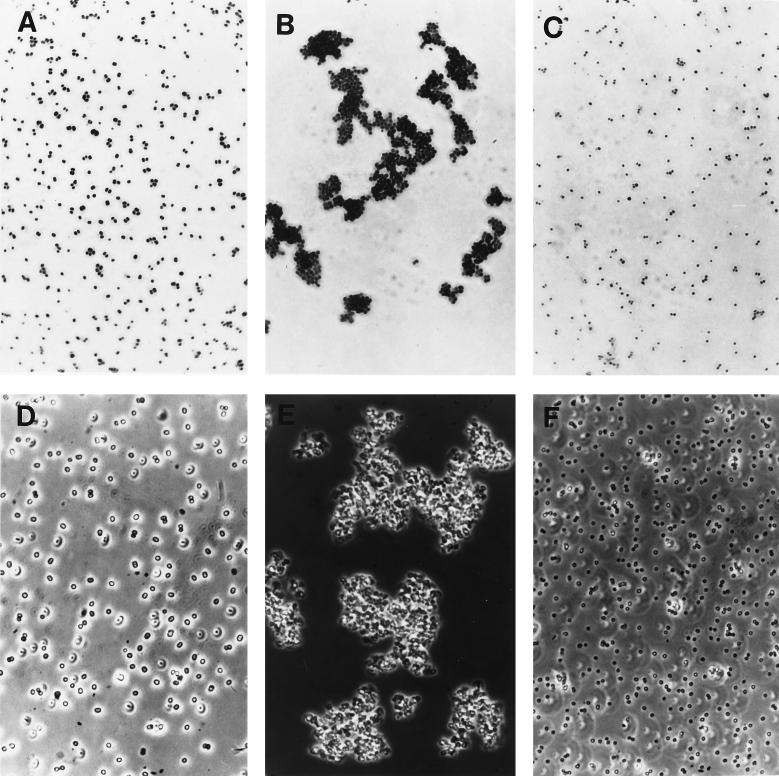
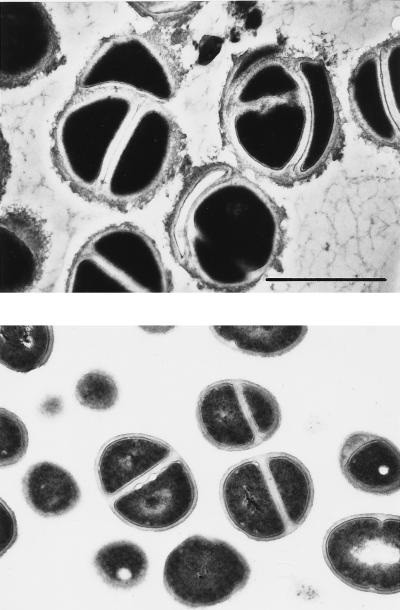
Resuspension of the six teicoplanin-resistant strains in autolysis buffer containing 0.5 times the teicoplanin MIC caused a drastic reduction in the rate of autolysis (Fig. 4). Most likely as a consequence of this, bacteria grown under the same conditions formed large aggregates of cells (Fig. 5B and E) visible even to the naked eye, and examination of thin sections of such cells by electron microscopy showed the production of large amounts of extracellular material which had staining properties similar to that of cell wall material (Fig. 6).
FIG. 4.
Effect of teicoplanin on autolysis rate. Cultures of selected CNSs were suspended in lysis buffer to an initial optical density (OD) of ≅1.0, and the rates of autolysis were monitored, as described in reference 8, in lysis buffer without teicoplanin (○) or in buffer containing teicoplanin at 8 (except for CNS 15, for which the concentration was 16) μg/ml ( ) and 50 μg/ml (•).
FIG. 5.
Appearance of teicoplanin-resistant staphylococci under optical microscope. Bacteria (CNS 15) were observed when grown in TSB (A and D) and when grown in the presence of 8 μg of teicoplanin per ml (B and E) and in the presence of teicoplanin after having reached stationary phase (C and F). (A, B, and C) Gram-stained bacteria. (D, E, and F) Cells observed by phase-contrast microscopy.
FIG. 6.
Morphology of teicoplanin-resistant staphylococci grown in the presence of teicoplanin. Bacteria (CNS 18) were grown in the presence of 8 μg of teicoplanin per ml. The culture was harvested at the mid-exponential (top panel) or stationary (bottom panel) phase of growth and then prepared for transmission electron microscopy, as described in reference 23. Magnification, ×25,000; bar, 1 μm.
Removal of teicoplanin from the medium.
The concentration of teicoplanin in the supernatant medium of cultures was determined at intervals by a bioassay. No detectable amounts of antibiotic could be found in cultures grown from small inocula to stationary phase in the presence of half the MIC equivalents of teicoplanin (8 to 16 μg/ml) (Fig. 7). About when the teicoplanin was no longer detectable in the culture medium, the aggregates of bacteria disintegrated into single cells (Fig. 5C and F) with the same appearance of cells grown in antibiotic-free medium (Fig. 5A and D).
FIG. 7.
Removal of teicoplanin from the growth medium of selected CNS isolates. Bacteria were grown in TSB containing 8 μg of teicoplanin per ml except for CNS 15, for which the concentration was 16 μg per ml. After the cultures reached stationary phase, sterile filtrates of the cultures were collected and used to determine the titers of teicoplanin in the medium supernatants by the bioassay described in reference 22. Numbers represent strains; A, initial concentration of antibiotic in the growth medium; B, final concentration found in medium supernatants collected from stationary-stage cultures.
Teicoplanin and vancomycin susceptibility of “historic” isolates of Staphylococcus epidermidis and Staphylococcus haemolyticus.
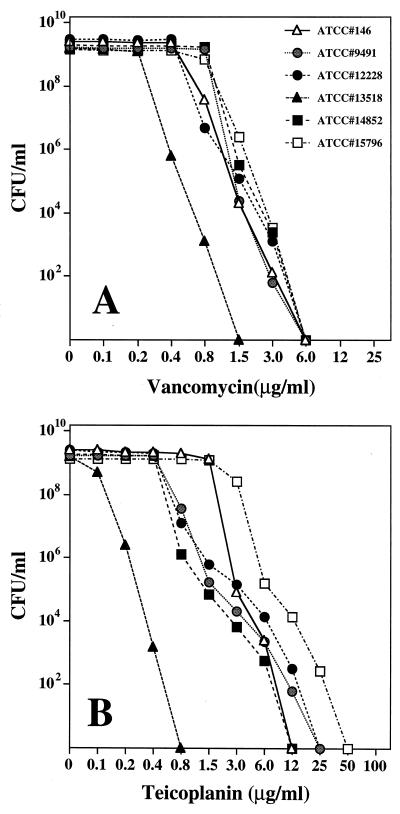
Five isolates of S. epidermidis and one isolate of S. haemolyticus were obtained from the American Type Culture Collection. The corresponding ATCC numbers (and isolation dates) are as follows: S. epidermidis 146 (1925), 9491 (1944), 12228 (1955), 13518 (1959), and 14852 (1962) and S. haemolyticus 15796 (1964). Vancomycin was approved for clinical use by the Food and Drug Administration in 1958 (6); teicoplanin is not used routinely in the United States. Population analysis of overnight cultures indicated homogeneous susceptibility of all six historic strains to vancomycin (MICs were between 0.4 and 1.5 μg/ml) (Fig. 8A), but heterogeneous susceptibility profiles for teicoplanin (MICs for the majority of strains were between 0.2 and 6.0 μg/ml) in all but one (13518) of the strains (Fig. 8B). The hetero-resistant strains included isolates 146 and 9491, collected in 1925 and 1944, respectively, i.e., long before the introduction of glycopeptide antibiotics. Cultures of most of these strains contained cells capable of growing on agar containing 12 to 25 μg of teicoplanin per ml at frequencies of 10−5 to 10−6 (Fig. 8B).
FIG. 8.
Vancomycin and teicoplanin susceptibility of “historic” isolates of CNS. CNS recovered in the preantibiotic era or close to the introduc- tion of vancomycin into clinical practice were tested for their susceptibilities to vancomycin (A) and teicoplanin (B) by population analysis (see Materials and Methods). Strain numbers and symbols are identified in panel A.
Strains 146, 9491, 12228, 14852, and 15796 were grown from small inocula in TSB containing half the respective MIC equivalents of teicoplanin. When the optical density reached 1.0, bacteria were removed by centrifugation and sterile filtration and the supernatants were tested for the concentration of teicoplanin with the highly sensitive strain 13518 (teicoplanin MIC, 0.2 μg/ml) for bioassay. No teicoplanin was detectable in any of the spent media.
DISCUSSION
The recent report on clinical failures during vancomycin therapy of MRSA disease in Japanese hospitals (10) and reports from the United States on clinical isolates of methicillin-resistant staphylococci for which vancomycin MICs are reduced (4, 5) has renewed concern about the appearance of glycopeptide resistance among methicillin-resistant and multidrug-resistant strains of S. aureus against which this generic class of antibiotics is often the only available chemotherapeutic regimen. The observations described in this study suggest that decreased susceptibility to teicoplanin and vancomycin may be frequent among current methicillin-resistant clinical isolates of CNS.
Our data with the preantibiotic era isolates suggest that heterogeneous teicoplanin phenotypes, including bacteria with reduced susceptibility to this antibiotic, are intrinsic to the species. On the other hand, data in the literature (2, 7, 19, 22) and experiments described in this report clearly show that under laboratory conditions, vancomycin can select for bacteria with increased teicoplanin MICs and that the reverse is also true (1, 11, 20). Enrichment of heterogeneous CNS cultures for the more resistant subpopulations of bacteria was surprisingly easy; when drug-free growth medium was inoculated with as few as 20 to 30 cells derived from colonies that grew on agar with 3 to 6 μg of vancomycin per ml, the overnight cultures produced bacteria in which the MICs for the vast majority of the cells were 6 and 12 μg/ml, respectively (Fig. 3), and the increased MICs for these bacteria were retained during multiple passages in antibiotic-free medium. Similar and even more rapid selection of the more resistant subpopulation was achieved by directly diluting a heterogeneous culture into antibiotic-containing growth medium (Fig. 3). These experiments clearly show that under laboratory conditions, CNS for which glycopeptide MICs are increased could originate from the subpopulations of bacteria present in heterogeneous cultures. To what degree the decreased glycopeptide susceptibility and heterogeneous phenotypes of clinical isolates described in this study emerged through a similar mechanism, i.e., selection by the clinical use of vancomycin, is not clear. However, in contrast to the relatively homogeneous vancomycin susceptibility of preantibiotic-era isolates, the cultures of clinical isolates of CNS examined were clearly heterogeneous and included subpopulations of bacteria for which vancomycin MICs were elevated.
The surprisingly high frequency of decreased teicoplanin susceptibility among MR-CNS as documented in this study, together with the extensive use of vancomycin in hospitals, may lead to further selection for the subpopulations of bacteria for which vancomycin MICs are elevated, which appear to be present already in the form of hetero resistance among MR-CNS strains. The same factors may also increase the probability that this type of glycopeptide resistance mechanism will emerge in clinical strains of MRSA. Clearly, such an increase in the MICs of glycopeptide antibiotics may then begin to jeopardize chemotherapy.
The ease with which isolates for which vancomycin MICs are increased can be selected under laboratory conditions makes one wonder why more highly glycopeptide-resistant CNS have not yet been identified among clinical isolates. The conditions under which the enrichment of cultures for such resistant subpopulations occurs in vitro suggest that the critical factors defining selective conditions in vivo are the pharmacokinetics of vancomycin and the dosing regimen used in therapy.
While several different mechanisms have been considered (3, 12, 13, 21, 22), the basis of staphylococcal glycopeptide resistance remains unknown. The addition of teicoplanin at half the respective MICs for CNS strains caused the appearance of cellular aggregates, the inhibition of autolysis, and the removal of the antibiotic from the growth medium. Regarding these features, the properties of the CNS strains resemble those of a recently described highly vancomycin-resistant laboratory mutant of MRSA (22). The formation of aggregates, morphological abnormalities, and the disappearance of glycopeptide antibiotic from the growth medium have also been described in several reports in the clinical microbiology literature (16–18). As already concluded in the studies with the highly vancomycin-resistant MRSA strain, the capacity of bacteria to sequester antibiotic molecules from the medium may contribute to their antibiotic resistance but cannot be the sole mechanistic basis for the increased MICs (22). A better understanding of the nature of the glycopeptide resistance mechanism(s) in MR-CNS and S. aureus (22) is important, since most of the current attention and drug development is limited to the glycopeptide resistance of enterococci, which seems to be completely different from the mechanism(s) presented by staphylococci.
ACKNOWLEDGMENTS
These investigations received partial support from the Lounsbery Foundation and the Bodman-Achelis Fund.
We thank E. H. W. Böhme of Hoechst Marion Roussel for providing teicoplanin.
REFERENCES
- 1.Aubert G, Passot S, Lucht F, Dorche G. Selection of vancomycin- and teicoplanin-resistant Staphylococcus haemolyticus during teicoplanin treatment of Staphylococcus epidermidisinfection. J Antimicrob Chemother. 1990;25:491–493. doi: 10.1093/jac/25.3.491. [DOI] [PubMed] [Google Scholar]
- 2.Biavasco F, Giovanetti E, Montanari M P, Lupidi R, Varaldo P E. Development of in-vitro resistance to glycopeptide antibiotics: assessment in staphylococci of different species. J Antimicrob Chemother. 1991;27:71–79. doi: 10.1093/jac/27.1.71. [DOI] [PubMed] [Google Scholar]
- 3.Billot-Klein D, Gutmann L, Bryant D, Bell D, van Heijenoort J, Grewal J, Shlaes D M. Peptidoglycan synthesis and structure in Staphylococcus haemolyticusexpressing increasing levels of resistance to glycopeptide antibiotics. J Bacteriol. 1996;178:4696–4703. doi: 10.1128/jb.178.15.4696-4703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control. Staphylococcus aureuswith reduced susceptibility to vancomycin—United States, 1997. Morbid Mortal Weekly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 5.Centers for Disease Control. Update: Staphylococcus aureuswith reduced susceptibility to vancomycin—United States, 1998. Morbid Mortal Weekly Rep. 1997;46:813–815. [PubMed] [Google Scholar]
- 6.Cooper G L, Given D B. Vancomycin. A comprehensive review of 30 years of clinical experience. Indianapolis, Ind: Lilly Research Laboratories; 1986. [Google Scholar]
- 7.Daum R S, Gupta S, Sabbagh R, Milewski W M. Characterization of Staphylococcus aureusisolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J Infect Dis. 1992;166:1066–1072. doi: 10.1093/infdis/166.5.1066. [DOI] [PubMed] [Google Scholar]
- 8.De Jonge B L M, de Lencastre H, Tomasz A. Suppression of autolysis and cell wall turnover in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureusstrain. J Bacteriol. 1991;173:1105–1110. doi: 10.1128/jb.173.3.1105-1110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin resistant Staphylococcus aureusclinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 11.Kaatz G W, Seo S M, Dorman N J, Lerner S A. Emergence of teicoplanin resistance during therapy of Staphylococcus aureusendocarditis. J Infect Dis. 1990;162:103–108. doi: 10.1093/infdis/162.1.103. [DOI] [PubMed] [Google Scholar]
- 12.Milewski W M, Boyle-Vavra S, Moreira B, Ebert C C, Daum R S. Overproduction of a 37-kilodalton cytoplasmic protein homologous to NAD+-linked d-lactate dehydrogenase associated with vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:166–172. doi: 10.1128/aac.40.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreira B, Boyle-Vavra S, de Jonge B L M, Daum R S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 15.Noble W C, Virani Z, Cree R G A. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;93:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 16.O’Hare M D, Reynolds P E. Novel membrane proteins present in teicoplanin-resistant, vancomycin-sensitive, coagulase-negative Staphylococcusspp. J Antimicrob Chemother. 1992;30:753–768. doi: 10.1093/jac/30.6.753. [DOI] [PubMed] [Google Scholar]
- 17.Sanyal D, Greenwood D. An electron microscope study of glycopeptide antibiotic-resistant strains of Staphylococcus epidermidis. J Med Microbiol. 1993;39:204–210. doi: 10.1099/00222615-39-3-204. [DOI] [PubMed] [Google Scholar]
- 18.Sanyal D, Johnson A P, George R C, Edwards R, Greenwood D. In-vitro characteristics of glycopeptide resistant strains of Staphylococcus epidermidisisolated from patients on CAPD. J Antimicrob Chemother. 1993;32:267–278. doi: 10.1093/jac/32.2.267. [DOI] [PubMed] [Google Scholar]
- 19.Schwalbe R S, Ritz W J, Verma P R, Barranco E A, Gilligan P H. Selection for vancomycin resistance in clinical isolates of Staphylococcus haemolyticus. J Infect Dis. 1990;161:45–51. doi: 10.1093/infdis/161.1.45. [DOI] [PubMed] [Google Scholar]
- 20.Shlaes D M, Shlaes J H. Teicoplanin selects for Staphylococcus aureusthat is resistant to vancomycin. Clin Infect Dis. 1995;20:1071–1073. doi: 10.1093/clinids/20.4.1071. [DOI] [PubMed] [Google Scholar]
- 21.Shlaes D M, Shlaes J H, Vincent S, Etter L, Fey P D, Goering R V. Teicoplanin-resistant Staphylococcus aureusexpresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob Agents Chemother. 1993;37:2432–2437. doi: 10.1128/aac.37.11.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieradzki K, Tomasz A. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J Bacteriol. 1997;179:2557–2566. doi: 10.1128/jb.179.8.2557-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Sieradzki, K., and A. Tomasz. Unpublished data.
- 23.Tomasz A, Jamieson J D, Ottolenghi E. The fine structure of Diplococcus pneumoniae. J Cell Biol. 1964;22:453–467. doi: 10.1083/jcb.22.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35:124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodford N, Johnson A P, Morrison D, Speller D C E. Current perspectives on glycopeptide resistance. Clin Microbiol Rev. 1995;8:585–615. doi: 10.1128/cmr.8.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]